Abstract
FtsZ, the bacterial tubulin homologue, is the main player in at least two distinct processes of cell division during the development of Streptomyces coelicolor A3(2). It forms cytokinetic rings and is required for the formation of both the widely spaced hyphal cross walls in the substrate mycelium and the specialized septation that converts sporogenic aerial hyphae into spores. The latter developmentally controlled septation involves the coordinated assembly of large numbers of FtsZ rings in each sporulating hyphal cell. We used an FtsZ-enhanced green fluorescent protein (EGFP) translational fusion to visualize the progression of FtsZ ring assembly in vivo during sporulation of aerial hyphae. This revealed that the regular placement of multiple FtsZ rings and initiation of cytokinesis was preceded by a protracted phase during which spiral-shaped FtsZ intermediates were detected along the length of the aerial hyphal cell. Time course experiments indicated that they were remodeled and gradually replaced by regularly spaced FtsZ rings. Such spiral-shaped filaments could also be detected with immunofluorescence microscopy using an antiserum against FtsZ. Based on our observations, we propose a model for the progression of Z-ring assembly during sporulation of S. coelicolor. Furthermore, mutants lacking the developmental regulatory genes whiA, whiB, whiG, whiH, and whiI were investigated. They failed in up-regulation of the expression of FtsZ-EGFP in aerial hyphae, which is consistent with the known effects of these genes on ftsZ transcription.
A prokaryotic homologue of tubulin, FtsZ, is the main cell division protein in bacteria, many archaea, and eukaryotic organelles. The first recognizable step of bacterial cell division is the assembly of FtsZ into a ring-like structure, the Z ring, at the future division site (2, 11, 35, 38, 44). The Z ring is associated with the cytoplasmic membrane and drives cytokinesis. This process involves several other division proteins, which require FtsZ for their recruitment to the division site and direct the synthesis of the septal peptidoglycan. These proteins have been identified in Escherichia coli and Bacillus subtilis, but the subsets of known division proteins in different bacterial species vary considerably (11, 38). The Z ring determines the division plane in bacteria (11), and the temporal and spatial regulation of Z-ring formation is remarkably precise. The Z ring is highly dynamic, and the assembly and behavior of the ring appears to be strongly influenced by the balance between polymerization and depolymerization of FtsZ (43, 48). In E. coli and B. subtilis, the intracellular FtsZ concentration is higher than the critical concentration for polymerization. Thus, factors that inhibit ectopic polymerization are needed. They include MinC, which prevents Z-ring assembly at the cell poles (23), and EzrA, which has a negative impact on polymerization along most of the cell (19, 31). In addition, the nucleoid inhibits Z-ring assembly (50, 51, 58), and SulA is an inhibitor of FtsZ assembly that is induced as part of the SOS response to DNA damage (9). On the other hand, proteins that stabilize the FtsZ polymers or promote formation or bundling of protofilaments are also known, including ZipA in E. coli and ZapA in B. subtilis (18, 20, 42). It is not yet clear how these and other factors interplay in order to bring about the correct placement of the Z ring in space and time, and the underlying molecular mechanisms are only partially understood for the main model organisms and remain largely unknown for many bacterial lineages.
The gram-positive soil bacterium Streptomyces coelicolor A3(2) is a hyphal organism with a complex developmental cycle that imparts unique features to its cell division (8, 16). S. coelicolor A3(2) has a single ftsZ gene, which, unlike in any other bacterium in which this has been investigated, is dispensable for growth and survival (39). FtsZ and the same basic cell division machinery are involved in at least two distinct types of septum formation, and Z rings are assembled in both cases (46). Infrequent septation without detachment of the daughter cells takes place in the substrate mycelium. This leads to formation of widely spaced cross walls that separate the syncytial cells in vegetative hyphae. In contrast, during sporulation of aerial hyphae, multiple septa are laid down synchronously to convert the multigenomic aerial hyphae into chains of unigenomic spores (5). This process requires the synchronous assembly of multiple and regularly spaced Z rings along the sporogenic aerial hyphal cells (17, 46). This is an interesting case of developmental control of cell division in which none of the systems that have been implicated in selection of division sites in other bacteria appear to have a role. Thus, while nucleoid occlusion affects the formation of Z rings and division septa in E. coli and vegetative cells of B. subtilis (22, 37), the early stages of sporulation septation in Streptomyces often occur over nonsegregated chromosomes, and separation of nucleoids is not observed until septal constriction has started (13, 40, 46). The cell division inhibitor MinC, together with the ATPase MinD, prevents septation near cell poles in many bacteria, and they are given a topological specificity through interaction with either MinE, as in E. coli, or DivIVA, as in B. subtilis (11, 37). The genome sequences of S. coelicolor and Streptomyces avermitilis lack any obvious homologues of minC and minE, which argues against the presence of a bona fide Min system in Streptomyces (13, 16). Furthermore, the S. coelicolor homologue of DivIVA was shown to primarily affect polar growth and morphogenesis, and it had no overt effect on cell division (12). This implies that neither the Min system nor nucleoid occlusion is used during Streptomyces sporulation and that other factors of unknown nature control this form of cytokinesis.
The developmental control of this synchronous cell division involves a sporulation-specific up-regulation of ftsZ expression in sporogenic hyphae in both S. coelicolor and Streptomyces griseus (15, 30). This occurred at one specific promoter and depended on a set of regulators of early stages of sporulation—the whiA, whiB, whiG, whiH, whiI, and whiJ genes (15). In addition, the isolation of a missense mutation in ftsZ that preferentially affected sporulation septation indicated that there are differences in the Z-ring assembly between vegetative and sporogenic aerial hyphae (17). Additional genes have been described that have effects on sporulation septation, but their modes of action and whether they affect the cell division machinery directly or indirectly are not clear. Examples include ssgA, ssgB, crgA, and samR (10, 26, 52, 55, 59).
In this paper, we address the mechanism of the synchronous assembly of multiple Z rings in sporulating aerial hyphae of S. coelicolor. By constructing an FtsZ-EGFP translational fusion, we were able to visualize the Z-ring assembly in vivo at different stages of development. We found that FtsZ assembly is developmentally modulated during sporulation—it involves the formation of spiral-shaped FtsZ intermediates that are remodeled into regularly spaced Z rings before initiation of septation. The impact of some developmental regulatory genes on Z-ring assembly was also studied, and the possible interplay between chromosome partitioning and FtsZ polymerization is discussed.
MATERIALS AND METHODS
Strains.
The S. coelicolor A3(2) strains that were used are listed in Table 1. E. coli strain DH5α (21) was used as a host for recombinant plasmids, and strain ET12567/pUZ8002 was used to drive conjugative transfer of nonmethylated DNA from E. coli to S. coelicolor as described previously (27). Cultivation of strains and general procedures for manipulation of DNA were performed as described for E. coli (45) and Streptomyces (27). S. coelicolor strains were cultivated on mannitol soy flour agar plates (MS agar) or in tryptone soy broth (TSB) (27).
TABLE 1.
Strains of Streptomyces coelicolor A3(2) used in this work
| Strain | Genotype | Reference |
|---|---|---|
| HU133 | M145 ΔftsZ::aphI | 39 |
| J2400 | M145 whiG::hyg | 14 |
| J2401 | M145 whiA::hyg | 14 |
| J2402 | M145 whiB::hyg | 14 |
| J2408 | M145 ΔwhiH::ermE | 14 |
| J2417 | HU133 attBφC31::pKF32[ftsZ+] | 15 |
| J2450 | M145 whiI::hyg | 3 |
| K202 | M145 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K203 | HU133 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K205 | HU133 attBφC31::pKF101[Φ(ftsZ17(Spo)-egfp)Hyb] | This work |
| K206 | J2401 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K207 | J2002 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K208 | J2400 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K209 | J2408 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| K210 | J2450 attBφC31::pKF41[Φ(ftsZ-egfp)Hyb] | This work |
| M145 | Prototrophic, SCP1−SCP2−Pg1+ | 27 |
| O17 | HU133 attBφC31::pO17[ftsZ17(Spo)] | 17 |
Plasmid constructions.
To construct plasmid pKF40, which contains S. coelicolor ftsZ translationally fused at its 3′ end to egfp, the gene encoding enhanced green fluorescent protein (EGFP), the ftsZ coding region, its upstream region, and the 3′ end of ftsQ were amplified by PCR from pJR92 (39) using the primers FOR20 (CGCCAGGGTTTTCCCAGTCA) and KF46 (TCCGGATCCTTCAGGAAGTCCGGCACGT). The latter primer was designed to replace the stop codon of ftsZ with a BamHI restriction site. The PCR product was cloned as a BamHI-HindIII fragment into the pEGFP-1 vector (BD Biosciences-Clontech), which contains a promoterless egfp gene. The gene fusion, which has codons for Asp-Pro-Pro-Val-Ala-Thr linking the last codon of ftsZ with the first one of egfp, was excised from pKF40 as a BglII-NotI fragment and cloned between the BglII and NotI sites of the pIJ8600 vector (49), which is a pSET152 derivative containing the ΦC31 integration system and an aac(3)IV apramycin resistance gene. The resulting pKF41 plasmid was transformed into the methylation-deficient ET12567/pUZ8002 strain and transferred by conjugation into S. coelicolor strains, in which it integrates into the chromosomal ΦC31 attB site. The expression of the FtsZ-EGFP fusion protein in pKF41 is driven by the natural ftsZ promoters, which are included on the plasmid.
Plasmid pKF100, which contains the ftsZ17(Spo) allele (17) translationally fused at its 3′ end to egfp, was created by amplifying the ftsZ17(Spo) allele, its upstream region, and the 3′ end of ftsQ by PCR from plasmid pO17 (17) using the primers KF66 (CGGAATTCGTCAAGGTCCGCTCCTAC) and KF46 (TCCGGATCCTTCAGGAAGTCCGGCACGT), as described above. The PCR-amplified DNA was ligated into the pGEM-T vector (Promega Corp.) and subsequently cut out as a BamHI-EcoRI fragment and ligated into the pEGFP-1 vector (see above). The resulting pKF100 plasmid was digested with BglII and NotI, and the resulting fragment was cloned into pIJ8600 as described above. This yielded plasmid pKF101, which was transferred by conjugation into Streptomyces as described above.
The ftsZ genes carried on plasmids pKF41 and pKF101 were sequenced using dye termination chemistry and an ABI Prism 877 robotic workstation (Applied Biosystems). The readings from an ABI Prism 377 DNA Sequenator (Applied Biosystems) were assembled into “contigs” by the Staden Package Program (Medical Research Council, Laboratory of Molecular Biology, Cambridge, United Kingdom) and checked manually for mismatches.
Fluorescence microscopy.
For visualization of the FtsZ-EGFP hybrid protein in vegetative mycelium, liquid cultures were grown overnight in TSB medium. Ten microliters of the overnight culture were spread on microscope slides coated with 1% agarose in H2O. For visualization of the FtsZ-EGFP hybrid protein in aerial hyphae, cultures were set up by inserting a sterile coverslip into the MS agar and inoculating along the acute angle between the glass and the agar surface (6). Alternatively, cultures were grown on MS agar plates for 2 to 4 days and impression preparations were taken by lightly pressing a coverslip on the surface of the colony (6). Coverslips with attached aerial mycelium were mounted in 50% glycerol in phosphate-buffered saline on poly-l-lysine-coated slides.
Immunofluorescence microscopy of FtsZ was carried out as described previously (17, 46). All fluorescence and phase-contrast microscopy was performed using an Axioplan II imaging fluorescence microscope equipped with appropriate filter sets, an Axiocam charge-coupled device camera, and Axiovision software (Carl Zeiss Light Microscopy). Digital images were processed using Adobe Photoshop version 7.0 software.
Deconvolution microscopy was performed on FtsZ-EGFP-expressing aerial hyphae from cultures grown on MS agar for 30 to 40 h. The microscopy was carried out as described above, and Z stacks containing 15 to 35 images were collected at a spacing of 0.05 to 0.1 μm and deconvolved using the Regularized Inverse Filter or the Constrained Iterative algorithm of the Axiovision software (Carl Zeiss Light Microscopy).
Sporulation assay.
The sporulation capacities of the studied strains were estimated by inoculating an equal amount of spores from each strain on MS agar and quantifying the number of spores produced by the cultures after 6 days of incubation.
In most of the cases, 107 CFU of spores was spread on MS agar. After 6 days of incubation at 30°C, the newly formed spores from each plate were collected and prepared as described previously (27). Serial dilutions were made from each spore preparation and plated on MS agar in duplicate. The plates were scored after 4 to 5 days of incubation, and the number of spores in each preparation was calculated as CFU/ml.
RESULTS
FtsZ-EGFP fusion protein assembles into ring-like structures in both vegetative and aerial hyphae of S. coelicolor.
To monitor the behavior of FtsZ in both vegetative and aerial hyphae of S. coelicolor, we created a translational fusion between FtsZ and EGFP. The fusion was introduced on an integrating plasmid (pKF41) into the chromosomal ΦC31 attachment site of the wild-type strain M145. The resulting K202 strain showed unaffected vegetative growth and sporulation as judged by its appearance in liquid culture and on plates in comparison to its parent (for further analysis, see below). Fluorescence microscopy revealed that FtsZ-EGFP assembled, presumably as mixed polymers with the native FtsZ in this strain, into bands or ring-like structures perpendicular to the hyphal length axis in both vegetative and aerial hyphae of K202 (Fig. 1). These structures could be clearly seen to have a ring shape only in some cases, but we make the assumption that they are FtsZ rings and will refer to them here as Z rings. The appearance and distribution of these Z rings were similar to those observed previously by immunofluorescence microscopy (17, 46), indicating that this strain was useful for examining FtsZ behavior during development.
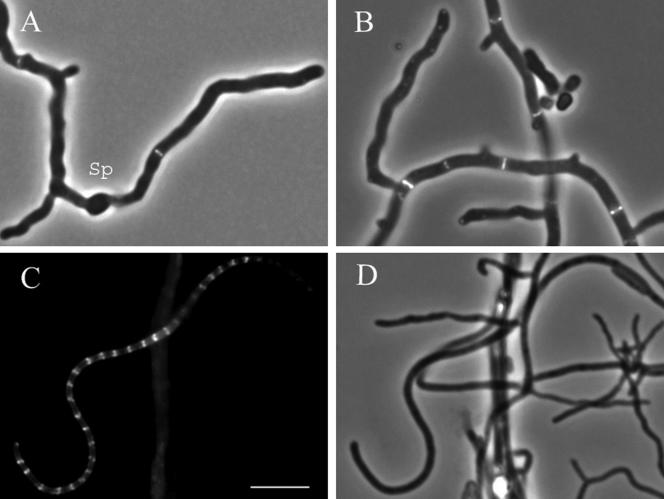
FIG. 1.
Visualization of Z rings in vegetative hyphae (A and B) and sporogenic aerial hyphae (C and D) of the strain K202, which expresses both FtsZ-EGFP and the native FtsZ. The strain carries plasmid pKF41 with an ftsZ-egfp translational fusion. Panels A and B show overlays of fluorescence (EGFP) and phase-contrast micrographs of vegetative mycelium grown from spores in liquid TSB medium for 10 h (A) or 18 h (B). A germinated spore is indicated by “Sp.” Panels C and D show fluorescence (EGFP) (C) and phase-contrast (D) micrographs of aerial hyphae after 40 h of incubation on MS agar. One example of a sporogenic hypha with clearly visible multiple Z rings is shown. Size bar, 5 μm.
FtsZ-EGFP revealed extensive spiral-shaped FtsZ polymers during sporulation.
The assembly of FtsZ in sporulating aerial hyphae was studied in detail in strain K202 [ftsZ+ Φ(ftsZ-egfp)Hyb]. The strain was grown on MS agar, and living hyphae attached to coverslips were examined microscopically after 28 to 48 h of growth. Many aerial-hyphal cells showed increased levels of fluorescence, indicating that developmentally controlled up-regulation of ftsZ expression had commenced, and in some of them, regularly spaced multiple Z rings had formed. Intriguingly, the distribution of the fluorescence signal within the individual hyphae repeatedly showed characteristic patterns different from regular ladders of Z rings, and these patterns appeared to change in relative abundance depending on the time of sampling. These observations pointed toward the possibility of the Z-ring assembly being a multistep process involving remodeling of the FtsZ polymers.
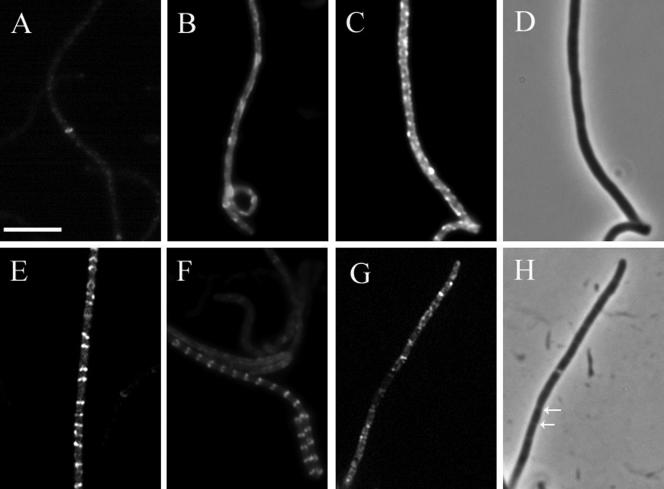
A number of distinct types of FtsZ-EGFP patterns could be distinguished, and in order to facilitate their interpretation, they were arbitrarily divided into six classes as follows (representative examples of each class are shown in Fig. 2). The hyphae from class 1 had one or two Z rings in their basal parts and contained only low levels of fluorescence (Fig. 2A). They were considered sporogenic based on their characteristic smooth shape and because they were often sticking up from the network of aerial hyphae. Class 1 aerial hyphae appeared after around 28 h of incubation with some variation between different experiments. In the hyphae of class 2, the intensity of the fluorescence was higher and the signal was mainly diffuse except for one or two Z rings, often in the basal part of the hypha, and/or occasional spiral-shaped structures (Fig. 2B). Class 3 hyphae showed extensive, often irregular, spiral-shaped structures and had a high level of diffuse FtsZ-EGFP fluorescence (Fig. 2C). In class 4 hyphae, variable numbers of short spirals, irregular or tilted Z-ring-like structures, and even some apparently completed Z rings were seen (Fig. 2E), and they were generally observed as distinct structures in hyphae where the level of diffuse fluorescence was lower than in classes 2 and 3. Class 5 represents regular ladders of multiple Z rings, as exemplified in Fig. 1C and 2F and in previous reports (17, 46). Class 6 represents hyphae with visibly invaginating septa, as detected in phase-contrast illumination (Fig. 2H). Thus, any fluorescent structures seen in these hyphae are not directly relevant to the events leading up to initiation of division and have not been further considered here.
FIG. 2.
Classification of the patterns of FtsZ-EGFP localization in sporogenic aerial hyphae. S. coelicolor strain K202 (M145 with the plasmid pKF41 [ftsZ-egfp]) was grown on MS agar, and fluorescence micrographs of FtsZ-EGFP assembly in aerial hyphae were taken after 28 to 38 h of colony development. The hyphae were subdivided into developmental classes according to their characteristic pattern of FtsZ localization (as described in the text). A representative example of a hypha from each class is shown: class 1 (A); class 2 (B); class 3 (C); class 4 (E); class 5 (F); class 6 (G). A phase-contrast micrograph of the hypha represented in panel C reveals no signs of septal constrictions (D), while the phase-contrast micrograph of the hypha represented in panel G shows constrictions at the sites of sporulation septation (H). Arrows indicate examples of visible constrictions. Size bar, 5 μm.
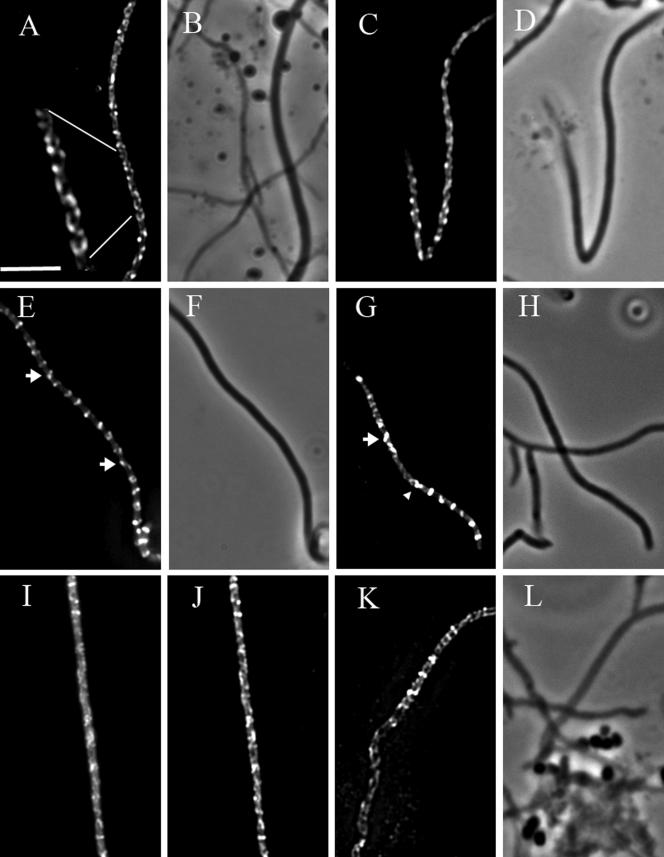
The spiral-like structures observed in these experiments were further examined by subjecting examples of hyphae from classes 3 and 4 to deconvolution microscopy in order to reduce the haze in the images caused by the out-of-focus light (Fig. 3). The spiral-shaped intermediates extended over variable distances along the sporogenic hyphae. Both the length and the pitch of the spirals were variable. In many cases, helical filaments were seen throughout long stretches of the sporogenic compartment (Fig. 3A and C), sometimes along the whole length of the sporogenic hypha, but the continuity of individual filaments was not clear and there may often have been overlapping structures. In class 4, several individual very short helical structures were observed with just one or a few turns (one example is shown in Fig. 3G), often with a much shorter pitch than the longer spirals. In several hyphae of class 4, structures reminiscent of tilted Z rings could also be seen, sometimes interrupting the main helical filament (Fig. 3E and G). These different structures are further addressed below and in the Discussion. As shown in Fig. 3I and J, the fluorescence patterns of FtsZ-EGFP localization could be discerned in the unprocessed image and are not an artifact of the digital image processing of the Z stacks. Rather, the deconvolution made them sharper and easier to interpret.
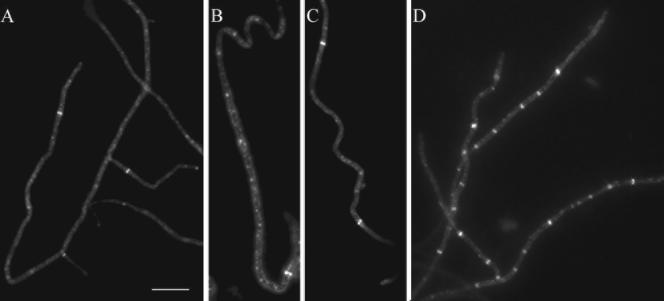
FIG.3.
Spiral-shaped FtsZ filaments in sporogenic aerial hyphae of S. coelicolor. Fluorescence micrographs showing spiral-shaped FtsZ-EGFP filaments in sporogenic aerial hyphae of K202 (A, C, E, G). The micrographs were generated by deconvolving Z stacks of images, taken after about 35 h of development on MS agar. The hyphae shown in the separate micrographs originated from independent experiments. Each micrograph is a single image from a focal plane lying roughly in the middle of the cell after the blur was removed by deconvolution. The corresponding phase-contrast micrographs of the hyphae depicted in the fluorescence images are shown (B, D, F, H). One example is shown of fluorescence micrographs showing spiral-shaped FtsZ-EGFP filaments before (I) and after (J) being subjected to deconvolution. An example of immunolocalization of native FtsZ in sporogenic aerial hyphae of M145 is shown as a fluorescence (K) and a phase-contrast (L) micrograph. The arrowhead in panel G marks an example of a short spiral, and arrows in panels E and G indicate structures reminiscent of tilted Z rings. The nonfluorescent hyphae that are visible in some panels had not entered sporulation and therefore exhibit very little ftsZ expression. Size bar, 5 μm.
Dynamics of FtsZ assembly during sporulation.
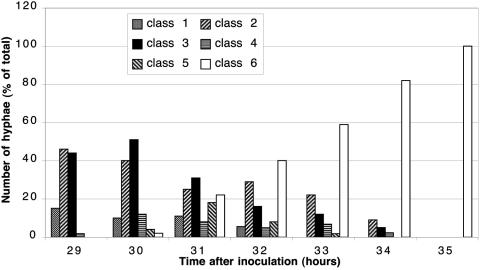
To clarify whether the different patterns of FtsZ-EGFP signals that were observed could be intermediates in the assembly of Z rings, time course experiments were carried out. We monitored the localization of FtsZ-EGFP in living aerial hyphae every 1 to 2 h between 28 and 42 h of growth on MS agar, an interval during which aerial-mycelium formation and sporulation commenced in a semisynchronous fashion in the cultures. At each time point, two coverslips with attached aerial mycelium were taken from separate plates and examined microscopically. The aerial hyphae that showed signs of increased fluorescence were classified according to the patterns of distribution of the fluorescence signal as described above. The numbers of hyphae from different classes were scored for each time point and plotted as percentages of the total number of sporogenic hyphae counted (Fig. 4). The timing of the onset of sporulation, and therefore the assembly of FtsZ in sporogenic hyphae, varied between different experiments. That may be explained by differences in the batches of MS plates, their dryness, or variable sizes of inocula. Nevertheless, the sequence and relative timing of the major intermediate steps in the FtsZ assembly was reproducible, with the whole process spanning 8 to 10 h. Figure 4 shows the results of one representative experiment. A succession was revealed in the abundances of the different classes of hyphae over time. Notably, hyphae of class 3, which had extensive spiral-shaped FtsZ filaments, were abundant only at 29 to 31 h and were then gradually replaced by apparently normal spore chains (as shown by both microscopical examinations and sporulation assays). These observations allowed us to conclude that the FtsZ assembly into regularly spaced Z rings during sporulation was a dynamic process and that it included a protracted phase during which spiral-shaped FtsZ polymers of various pitches and lengths were prevalent. They appeared to coalesce into several very short structures, often spiral shaped, and eventually into regular Z rings distributed along the entire length of the sporogenic cell.
FIG. 4.
Time course of the dynamic assembly of FtsZ-EGFP. Two coverslips with attached aerial mycelium were examined at each time point. According to the pattern of the fluorescence signal from FtsZ-EGFP, the overtly sporogenic aerial hyphae in each examined field were assigned to any of the arbitrary classes shown in Fig. 2. The histogram shows the relative number of hyphae from a particular class at a particular time point, represented as a percentage of the total number of sporogenic hyphae discerned in the microscopical preparation at that time point. Between 100 and 400 hyphae were counted for each time point.
Detection of spiral-shaped FtsZ structures using immunofluorescence microscopy.
Since the possibility that the FtsZ spirals were defective polymers or artifacts caused by the EGFP fusion had to be considered, we reinvestigated the behavior of the wild-type protein in strain M145 using anti-FtsZ antibodies and immunofluorescence microscopy. Previous reports on the use of this technique have not mentioned spiral-shaped structures (17, 46). However, with the detailed knowledge of the time course of development (see above) it was possible to obtain samples from the appropriate time of development when hyphae at the desired stage of differentiation were abundant. Although immunofluorescence gives a lower resolution than observing EGFP fusion proteins in living cells, and delicate or highly dynamic structures may not survive the fixation procedure, we were able to observe spiral-shaped FtsZ structures (one example is shown in Fig. 3K and L). They were similar in appearance to the helical intermediates of the FtsZ-EGFP fusion protein and were only observed during a short time interval of colony development. These observations supported the conclusions that the assembly of multiple Z rings during sporulation in S. coelicolor involves extensive spiral-shaped intermediates and that these were not artifacts caused by the presence of the EGFP tag on FtsZ.
Functionality of the FtsZ-EGFP hybrid protein.
In order to investigate how functional the product of the ftsZ-egfp fusion carried on pKF41 was, it was introduced into the ftsZ-null mutant HU133, yielding the strain K203. This strain had lost the blue halo characteristic of HU133 and grew to an almost normal colony size on MS agar (Fig. 5A). K203 was capable of producing grey aerial mycelium, but sporulation was delayed. Microscopic examination of the aerial mycelium showed that spores were formed at a lower frequency than is typical for wild-type strains and that they were of variable shape and size, and many nonseptated aerial hyphae were observed (Fig. 5C). These observations showed that the fusion protein was partially functional, since it restored apparently normal vegetative growth to the ftsZ-null mutant HU133, but not sufficiently functional to restore normal sporulation septation to this strain. Immunoblot analysis of strain K202 showed that FtsZ-EGFP was present at a relatively low level in comparison to native FtsZ (data not shown), suggesting that it may be an unstable protein and that cleavage of the EGFP off the hybrid protein could contribute to the partial functionality of this protein.
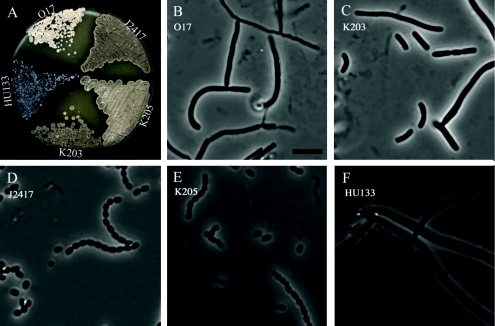
FIG. 5.
Sporulation phenotypes of strains carrying the ftsZ-egfp translational fusion. Panel A shows the colony appearance after 5 days of growth on MS agar, and panels B to F are phase-contrast micrographs of examples of spore chains and aerial hyphal fragments from the surfaces of colonies after 5 days of growth on MS agar. The S. coelicolor strains were the following: B, O17 {HU133 with plasmid pO17 [ftsZ17(Spo)]}; C, K203 (HU133 with plasmid pKF41 [ftsZ-egfp]); D, J2417 (HU133 with plasmid pKF32 [ftsZ+]); E, K205 {HU133 with plasmid pKF101 [ftsZ17(Spo)-egfp]}; F, HU133 (ΔftsZ::aphI).
Although FtsZ-EGFP was not fully functional on its own, it worked well in the heterozygous ftsZ+ Φ(ftsZ-egfp)Hyb context in strain K202, and growth and development of this strain on plates was indistinguishable from that of the parent ftsZ+ strain M145 (data not shown). The sporulation capacities of the strains bearing the ftsZ-egfp fusions were quantified, and the sizes of the spores were scored. The sporulation capacity of strain K202 was estimated as being equal to that of the parent, M145. Both strains yielded spores with similar average sizes after 6 days of growth on MS agar (average length, 1.02 ± 0.21 μm for M145 and 1.17 ± 0.43 μm for K202). The difference was due to a somewhat larger number of irregular and elongated spores and spore-like compartments in K202. In occasional vegetative or aerial hyphae, often morphologically deviating from normal hyphae, conspicuous strongly fluorescent and irregularly shaped structures were seen, presumably reflecting accumulation of nonfunctional FtsZ aggregates. Cells with such structures were rare and had no impact on quantitative analyses like those presented in Fig. 4. The production of normal amounts of regularly sized spores, and the observation of FtsZ-EGFP at expected locations (Fig. 1) in the great majority of cells, showed that, although the EGFP tag disturbed the functionality of FtsZ, there were no strong dominant-negative effects of the FtsZ-EGFP fusion. These observations support the notion that FtsZ-EGFP could form mixed polymers with the native FtsZ and be used to detect the subcellular localization of FtsZ polymers.
Interestingly, the fusion between the FtsZ17(Spo) mutant protein, which is unable to support sporulation (17), and EGFP restored sporulation to the ftsZ-null strain HU133 when expressed from the pKF101 integrating plasmid. The resulting strain was called K205, and it grew and developed as well as the congenic ftsZ+ strain J2417 on MS agar in terms of colony size and ability to produce grey spore pigment (Fig. 5A). In contrast, the O17 strain, which contains the ftsZ17(Spo) mutant allele on an integrating plasmid, was unable to sporulate, remained white during development, and gave fewer colonies than K205 upon restreaking (Fig. 5A). In accordance with these observations, sporulation assays and microscopic examination showed abundant production of spores of regular size in K205 (0.94 ± 0.20 μm), while O17 produced only aberrantly septated or nonseptated aerial hyphae (Fig. 5B and E). Thus, there was a mutual suppression of the negative effects on ftsZ function between the ftsZ17(Spo) mutation and the EGFP tag. This suppression appears to work only intramolecularly, since no suppression was seen when the ftsZ17(Spo) and Φ(ftsZ-egfp)Hyb alleles were present in trans in the same strain (17).
Strain K204, which carries ftsZ17(Spo)-egfp in addition to the native ftsZ, showed spiral-shaped FtsZ structures during sporulation similar to those of strain K202, which carries the normal ftsZ-egfp fusion (data not shown). Since ftsZ17(Spo)-egfp produces a more functional protein than ftsZ-egfp according to the assays described above, this further argues against the possibility that the spiral-shaped structures were defective polymers or artifacts caused by the disturbed functionality of the EGFP fusion protein.
Effects of developmental regulators whiA, whiB, whiG, whiH, and whiI on expression and assembly of FtsZ-EGFP.
The localization of FtsZ-EGFP in living aerial hyphae of the whiA, whiB, whiG, whiH, and whiI disruption mutants containing the pKF41 plasmid was examined, since these genes are required for full sporulation septation. Aerial hyphae were investigated after 4 to 6 days of incubation on MS agar (Fig. 6), when the aerial hyphae of the mutants had developed their characteristic phenotypes.
FIG. 6.
Visualization of FtsZ-EGFP assembly into Z rings in aerial hyphae of the whiG mutant strain J2400 (A), whiB mutant J2402 (B), whiI mutant J2450 (C), and whiH mutant J2408 (D) after 96 h of development on MS agar plates. Size bar, 5 μm.
Occasional Z rings were visible in the straight aerial hyphae of the whiG mutant (Fig. 6A), presumably corresponding to the septa of vegetative type that can be seen in whiG aerial hyphae (14). Mutants lacking whiA (data not shown) or whiB (Fig. 6B) formed long, coiled aerial hyphae, and occasional Z rings could be observed in these, but they were fewer than in the whiG mutant. As observed previously for sporulation septation (14), the Z rings were seldom in the coiled segments. Also in the aerial hyphae of whiI, infrequent and widely spaced Z rings were observed (Fig. 6C). Also, when samples were taken at an earlier time of development (48 h), these mutants showed low fluorescent signal and spiral-shaped structures were not detected.
The whiH mutant K209 differed somewhat from the other mutants. Several (4 to 12) Z rings were observed in many aerial hyphae of this strain (Fig. 6D). They were irregularly spaced and of variable shape and fluorescent intensity, even with occasional short helical structures. Although much lower than in the wild-type strains, this frequency of Z rings was higher than in any of the other whi mutants that were examined and higher than can be explained by the vegetative type of septation. A slight increase in the overall fluorescence signal was seen in the whiH aerial hyphae that contained these Z rings, but this increase was small compared to the intensity in the sporogenic hyphae of the wild-type counterpart, strain K202. Thus, the absence of large numbers of Z rings can be accounted for by the previously reported failure to up-regulate ftsZ expression in the whiH mutant also, and it is not possible to determine whether there are any additional specific effects of the tested whi genes on the assembly of FtsZ.
DISCUSSION
From the results presented here, we conclude that the assembly of FtsZ into a ladder of regularly spaced Z rings during sporulation in S. coelicolor involves the formation and remodeling of spiral-shaped FtsZ intermediates. These were clearly observed in living cells by using an FtsZ-EGFP fusion protein and could also be seen after immunofluorescence detection of the native FtsZ protein. Since sporulation in this organism occurs within the aerial mycelia of colonies, the sporulating hyphae had to be removed from this environment and immersed in liquid in order to be studied microscopically. For this reason, we have not followed the behavior of these FtsZ polymers with time lapse microscopy in individual hyphae. Instead, the patterns of localization of FtsZ-EGFP in living sporogenic hyphae were monitored in samples taken out at different times of colony development. This revealed a succession of stages of FtsZ assembly in which the FtsZ spirals were abundant in sporogenic hyphae for a limited amount of time and were then replaced by hyphae with Z rings and sporulation septa (Fig. 4). This strongly supports the notion that the spiral-shaped structures are transient intermediates in a dynamic process of FtsZ assembly and remodeling.
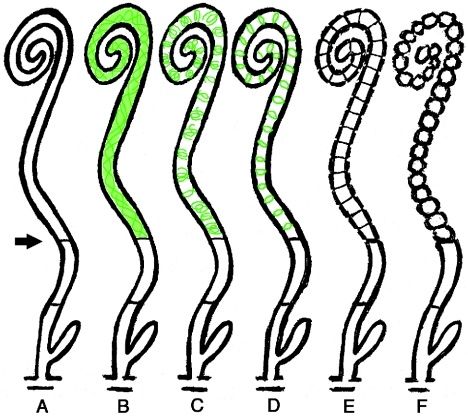
Based on these results, we propose a model for the assembly of FtsZ prior to sporulation septation in S. coelicolor (Fig. 7). The process may start with the placement of a Z ring and a basal septum in the distal part of the sporogenic hypha (stage A). For S. griseus, it has been suggested that the sporogenic cells are delimited at the base by a specialized septum, different from both vegetative cross walls and sporulation septa (30). No direct equivalent has been observed in S. coelicolor. However, we frequently see one or two hyphal cross walls, similar to vegetative septa, in the aerial branches that sporulate, but in the section below what will become the actual spore chain (unpublished observations). We suggest that such a septum demarcates the compartment in which sporulation septation later commences with a significantly increased expression of FtsZ, as judged by the increase in the fluorescent signal from FtsZ-EGFP. The latter observation is in agreement with the developmentally controlled induction of ftsZ transcription in sporogenic hyphae (15, 30). The rise in the cytoplasmic concentration of FtsZ leads to the initiation of assembly into spiral-shaped filaments (Fig. 7, stage B). The exact shape, helical pitch, and length of these polymers were for technical reasons difficult to visualize and appeared to be variable, but they formed throughout the length of the sporogenic cell, and no particular site of initiation or nucleation of the structures could be discerned. We propose that the spiral-shaped filaments are continually remodeled and eventually transformed into short helical or ring-like structures (Fig. 7, stage C) that subsequently give rise to an array of regularly spaced Z rings (Fig. 7, stage D). As judged from the low abundance of hyphae with pronounced regular ladders of Z rings in the time course experiments (class 5 in Fig. 4), we speculate that this is a short-lived stage and that septation rapidly commences once the FtsZ polymers have coalesced into stable rings. Thus, the hyphae with ladders of Z rings rapidly trigger sporulation septation and develop visible constrictions (Fig. 7, stage E). This leads to the formation of chains of unigenomic spores (Fig. 7, stage F).
FIG. 7.
Schematic representation of the proposed progression of dynamic localization of FtsZ during sporulation of S. coelicolor M145. The different stages discussed in the text are indicated by letters. FtsZ localization is shown in green. The arrow points to the putative basal septum.
Developmentally induced remodeling of FtsZ polymers has previously been reported in B. subtilis (4). In this organism, the formation of an endospore starts with an asymmetric cell division, while vegetative-cell division always occurs at the exact middle of the rod-shaped cell. Work by Ben-Yehuda and Losick showed that the reorganization from medial to polar cytokinetic rings during sporulation involved helical FtsZ intermediates and depended both on an increase in ftsZ expression and on the FtsZ-interacting sporulation protein SpoIIE (4). In S. coelicolor, the assembly of FtsZ into spiral-shaped intermediates is associated with a marked increase in ftsZ transcription in sporogenic hyphae (15). The up-regulation is required for sporulation septation, since a mutant of S. coelicolor that lacks the developmentally induced ftsZ2p promoter was unable to form spores (15). On the other hand, ectopic increase of ftsZ expression to different levels was not sufficient to trigger multiple cell divisions in vegetative hyphae (56). Thus, it is possible that some sporulation-specific protein may also be involved in the reorganization of the pattern of Z-ring formation in Streptomyces. In this context, it is interesting that ectopic overexpression of ssgA can trigger multiple sporulation-like divisions in growing mycelium (25, 55, 56). However, it is not clear whether this effect involves any alterations of ftsZ expression. The mode of action of ssgA or its family of paralogues is not yet known, but both ssgA and ssgB are developmentally regulated, and mutants have sporulation phenotypes (26, 29, 54, 59).
The identity of the putative developmental regulator(s) that modulates the cell division machinery to give rise to sporulation septation remains to be elucidated. We have examined the effects of the developmental regulatory genes whiA, whiB, whiG, whiH, and whiI on the expression and assembly of FtsZ during sporulation. The failure of whiA, whiB, whiG, and whiI mutants to make more Z rings than during vegetative growth (Fig. 6) is consistent with their deficiency in sporulation-specific induction of the ftsZ2p promoter in aerial hyphae (15). This was reflected here by the low expression of the FtsZ-EGFP translational fusion in aerial hyphae of these strains. Similar abundances of Z rings have previously been observed using immunolocalization of FtsZ in aerial hyphae of whiB- and whiG-deficient strains (46). The higher frequency of Z rings in the whiH mutant is consistent with the fact that this mutant, in contrast to the others, is capable of laying down a few sporulation septa (14, 46). Although the exact significance of these Z rings, and whether they are all used for septation, is unclear, it distinguishes whiH from the other early whi genes in relation to cell division control. Finally, although the comparatively low expression may be sufficient to explain the failure of these mutants to make sporulation septa, the possibility cannot be excluded that any of the whi genes may affect cell division by other mechanisms in addition to transcriptional control of ftsZ.
The dynamic behavior of the FtsZ polymers raises questions regarding the factors that guide the regular placement of series of Z rings in sporogenic hyphae and ensure the production of uniformly sized spores containing one copy of the genome each. The actions of these factors are likely to be different from that of the Min system and an overt “nucleoid veto.” Whereas such mechanisms inhibit FtsZ polymerization at other sites than midcell in many bacteria, an important implication of the results reported here is that in Streptomyces FtsZ can start assembling into filaments along most of the sporogenic hyphal cell without any apparent zones of inhibition. An interesting possibility, then, is that the remodeling and congregation of these polymers into evenly distributed Z rings could be influenced by the positioning of the chromosomes. There may be a stage in the process of sporulation when individual chromosomes or particular chromosomal loci are being aligned in a certain way along the lengths of sporogenic hyphae, thus influencing the distribution of the Z rings. Also, either the arrays of Z rings or the growing sporulation septa may direct the regular partitioning of the individual nucleoids. These speculations are supported by reports of irregularly placed sporulation septa in mutants defective in chromosome partitioning (28, 57) and the observed segregation defects in mutants specifically defective in making sporulation septa, such as whiH, ftsZΔ2p, and ftsZ17(Spo) strains (7, 15, 17).
A number of studies have described the ability of FtsZ to produce spiral-like filaments in vivo. Such structures have been observed both in certain mutants in E. coli and B. subtilis and upon overexpression of FtsZ in E. coli (1, 24, 36, 41, 47). Most recently, it was reported that highly mobile helix-like structures are part of the normal cycle of FtsZ assembly in E. coli (53). In the case of Streptomyces sporulation, the challenge is now to determine how such FtsZ spirals can be reshaped into a large number of Z rings. Although the structure of FtsZ protofilaments has been elucidated (32, 33), the assembly and nature of the polymers in the Z ring in vivo remains unclear, and it is not known how FtsZ protofilaments interact with each other and with other proteins (34). Such interactions should be crucial for the in vivo dynamics of FtsZ polymerization and are likely to be critical for the remodeling of the extensive spiral-shaped structures in S. coelicolor. The efficiency of sporulation septation seems to be a sensitive indicator of disturbances in the behavior of the FtsZ polymers, and this can even be detected as decreases in the amount of spore pigment being produced (15, 17). Thus, S. coelicolor may offer a powerful system for genetic analysis of the in vivo dynamics of FtsZ.
Acknowledgments
This work was supported by a grant from the Swedish Research Council.
We thank Nora Ausmees and Santanu Dasgupta for discussions and constructive criticism of the manuscript.
REFERENCES
- 1.Addinall, S. G., and J. Lutkenhaus. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22:231-237. [DOI] [PubMed] [Google Scholar]
- 2.Addinall, S. G., and B. Holland. 2002. The tubulin ancestor, FtsZ, draughtsman, designer, and driving force for bacterial cytokinesis. J. Mol. Biol. 318:219-236. [DOI] [PubMed] [Google Scholar]
- 3.Aínsa, J. A., H. D. Parry, and K. F. Chater. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2). Mol. Microbiol. 34:607-619. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Yehuda, S., and R. Losick. 2002. Asymmetric cell division in B. subtilis involves a spiral-like intermediate of the cytokinetic protein FtsZ. Cell 109:257-266. [DOI] [PubMed] [Google Scholar]
- 5.Chater, K. F. 1999. Developmental decisions during sporulation in the aerial mycelium in Streptomyces, p. 33-48. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. ASM Press, Washington, D.C.
- 6.Chater, K. F. 1972. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 72:9-28. [DOI] [PubMed] [Google Scholar]
- 7.Chater, K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667-673. [DOI] [PubMed] [Google Scholar]
- 8.Chater, K. F., and R. Losick. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p. 149-182. In J. A. Shapiro and M. Dworkin (ed.), Bacteria as multicellular organisms. Oxford University Press, New York, N.Y.
- 9.Cordell, S. C., E. J. Robinson, and J. Löwe. 2003. Crystal structure of the SOS cell division inhibitor SulA and in complex with FtsZ. Proc. Natl. Acad. Sci. USA 100:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del Sol, R., A. Pitman, P. Herron, and P. Dyson. 2003. The product of a developmental gene, crgA, that coordinates reproductive growth in Streptomyces belongs to a novel family of small actinomycete-specific proteins. J. Bacteriol. 185:6678-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Errington, J., R. A. Daniel, and D.-J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flärdh, K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523-1536. [DOI] [PubMed] [Google Scholar]
- 13.Flärdh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564-571. [DOI] [PubMed] [Google Scholar]
- 14.Flärdh, K., K. C. Findlay, and K. F. Chater. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2). Microbiology 145:2229-2243. [DOI] [PubMed] [Google Scholar]
- 15.Flärdh, K., E. Leibovitz, M. J. Buttner, and K. F. Chater. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38:737-749. [DOI] [PubMed] [Google Scholar]
- 16.Flärdh, K., and G. P. van Wezel. 2003. Cell division during growth and development of Streptomyces. Recent Res. Dev. Bacteriol. 1:71-90. [Google Scholar]
- 17.Grantcharova, N., W. Ubhayasekera, S. L. Mowbray, J. R. McCormick, and K. Flärdh. 2003. A missense mutation in ftsZ differentially affects vegetative and developmentally controlled cell division in Streptomyces coelicolor A3(2). Mol. Microbiol. 47:645-656. [DOI] [PubMed] [Google Scholar]
- 18.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haeusser, D. P., R. L. Schwartz, A. M. Smith, M. E. Oates, and P. A. Levin. 2004. EzrA prevents aberrant cell division by modulating assembly of the cytoskeletal protein FtsZ. Mol. Microbiol. 52:801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hale, C. A., A. C. Rhee, and P. A. de Boer. 2000. ZipA-induced bundling of FtsZ polymers mediated by an interaction between C-terminal domains. J. Bacteriol. 182:5153-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 22.Harry, E. J. 2001. Bacterial cell division: regulating Z-ring formation. Mol. Microbiol. 40:795-803. [DOI] [PubMed] [Google Scholar]
- 23.Hu, Z., A. Mukherjee, S. Pichoff, and J. Lutkenhaus. 1999. The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization. Proc. Natl. Acad. Sci. USA 96:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 25.Kawamoto, S., H. Watanabe, A. Hesketh, J. C. Ensign, and K. Ochi. 1997. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology 143:1077-1086. [DOI] [PubMed] [Google Scholar]
- 26.Keijser, B. J. F., E. E. E. Noens, B. Kraal, H. K. Koerten, and G. P. van Wezel. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225:59-67. [DOI] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 28.Kim, H. J., M. J. Calcutt, F. J. Schmidt, and K. F. Chater. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J. Bacteriol. 182:1313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kormanec, J., and B. Sevcikova. 2002. The stress-response sigma factor σH controls the expression of ssgB, a homologue of the sporulation-specific cell division gene ssgA, in Streptomyces coelicolor A3(2). Mol. Genet. Genom. 267:536-543. [DOI] [PubMed] [Google Scholar]
- 30.Kwak, J., A. J. Dharmatilake, H. Jiang, and K. E. Kendrick. 2001. Differential regulation of ftsZ transcription during septation of Streptomyces griseus. J. Bacteriol. 183:5092-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levin, P. A., I. G. Kurtser, and A. D. Grossman. 1999. Identification and characterization of a negative regulator of FtsZ ring formation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 96:9642-9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Löwe, J., and L. A. Amos. 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203-206. [DOI] [PubMed] [Google Scholar]
- 33.Löwe, J., and L. A. Amos. 1999. Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J. 18:2364-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Löwe, J., F. van den Ent, and L. A. Amos. 2004. Molecules of the bacterial cytoskeleton. Annu. Rev. Biophys. Biomol. Struct. 33:177-198. [DOI] [PubMed] [Google Scholar]
- 35.Lutkenhaus, J., and S. G. Addinall. 1997. Bacterial cell division and the Z ring. Annu. Rev. Biochem. 66:93-116. [DOI] [PubMed] [Google Scholar]
- 36.Ma, X., D. W. Ehrhardt, and W. Margolin. 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cell by using green fluorescent protein. Proc. Natl. Acad. Sci. USA 93:12998-13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Margolin, W. 2001. Spatial regulation of cytokinesis in bacteria. Curr. Opin. Microbiol. 4:647-652. [DOI] [PubMed] [Google Scholar]
- 38.Margolin, W. 2000. Themes and variations in prokaryotic cell division. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 39.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene, ftsZ. Mol. Microbiol. 14:243-254. [DOI] [PubMed] [Google Scholar]
- 40.Miguélez, E. M., B. Rueda, C. Hardisson, and M. B. Manzanal. 1998. Nucleoid partitioning and the later stages of sporulation septum synthesis are closely associated events in the sporulating hyphae of Streptomyces carpinensis. FEMS Microbiol. Lett. 159:59-62. [Google Scholar]
- 41.Mileykovskaya, E., Q. Sun, W. Margolin, and W. Dowhan. 1998. Localization and function of early cell division proteins in filamentous Escherichia coli cells lacking phosphatidylethanolamine. J. Bacteriol. 180:4252-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.RayChaudhuri, D. 1999. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 18:2372-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romberg, L., and P. A. Levin. 2003. Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu. Rev. Microbiol. 57:125-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rothfield, L., S. Justice, and J. Garcia-Lara. 1999. Bacterial cell division. Annu. Rev. Genet. 33:423-448. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Schwedock, J., J. R. McCormick, E. A. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25:847-858. [DOI] [PubMed] [Google Scholar]
- 47.Stricker, J., and H. P. Erickson. 2003. In vivo characterization of Escherichia coli ftsZ mutants: effects on Z-ring structure and function. J. Bacteriol. 185:4796-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stricker, J., P. Maddox, E. D. Salmon, and H. P. Erickson. 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc. Natl. Acad. Sci. USA 99:3171-3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun, J., G. H. Kelemen, J. M. Fernandez-Abalos, and M. J. Bibb. 1999. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2). Microbiology 145:2221-2227. [DOI] [PubMed] [Google Scholar]
- 50.Sun, Q., and W. Margolin. 2004. Effects of perturbing nucleoid structure on nucleoid occlusion-mediated toporegulation of FtsZ ring assembly. J. Bacteriol. 186:3951-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, Q., and W. Margolin. 2001. Influence of the nucleoid on placement of FtsZ and MinE rings in Escherichia coli. J. Bacteriol. 183:1413-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tan, H., Y. Tian, H. Yang, G. Liu, and L. Nie. 2002. A novel Streptomyces gene, samR, with different effects on differentiation of Streptomyces ansochromogenes and Streptomyces coelicolor. Arch. Microbiol. 177:274-278. [DOI] [PubMed] [Google Scholar]
- 53.Thanedar, S., and W. Margolin. 2004. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in Escherichia coli. Curr. Biol. 14:1167-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Traag, B. A., G. H. Kelemen, and G. P. van Wezel. 2004. Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2). Mol. Microbiol. 53:985-1000. [DOI] [PubMed] [Google Scholar]
- 55.van Wezel, G. P., J. van der Meulen, S. Kawamoto, R. G. Luiten, H. K. Koerten, and B. Kraal. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182:5653-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Wezel, G. P., J. van der Meulen, E. Taal, H. Koerten, and B. Kraal. 2000. Effects of increased and deregulated expression of cell division genes on the morphology and on antibiotic production of streptomycetes. Antonie Leeuwenhoek 78:269-276. [DOI] [PubMed] [Google Scholar]
- 57.Wenner, T., V. Roth, G. Fischer, C. Fourrier, B. Aigle, B. Decaris, and P. Leblond. 2003. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 50:411-425. [DOI] [PubMed] [Google Scholar]
- 58.Wu, L. J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915-925. [DOI] [PubMed] [Google Scholar]
- 59.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]