Abstract
Females live longer than males, and there are sex disparities in physical health and disease incidence. However, sex differences in biological aging have not been consistently reported and may differ depending on the measure used. This study aimed to determine the correlations between epigenetic age acceleration (AA), and other markers of biological aging, separately in males and females. We additionally explored the extent to which these AA measures differed according to socioeconomic characteristics, clinical markers, and diseases. Epigenetic clocks (HorvathAge, HannumAge, PhenoAge, GrimAge, GrimAge2, and DunedinPACE) were estimated in blood from 560 relatively healthy Australians aged ≥ 70 years (females, 50.7%) enrolled in the ASPREE study. A system-wide deficit accumulation frailty index (FI) composed of 67 health-related measures was generated. Brain age and subsequently brain-predicted age difference (brain-PAD) were estimated from neuroimaging. Females had significantly reduced AA than males, but higher FI, and there was no difference in brain-PAD. FI had the strongest correlation with DunedinPACE (range r: 0.21 to 0.24 in both sexes). Brain-PAD was not correlated with any biological aging measures. Significant correlations between AA and sociodemographic characteristics and health markers were more commonly found in females (e.g., for DunedinPACE and systolic blood pressure r = 0.2, p < 0.001) than in males. GrimAA and Grim2AA were significantly associated with obesity and depression in females, while in males, hypertension, diabetes, and chronic kidney disease were associated with these clocks, as well as DunedinPACE. Our findings highlight the importance of considering sex differences when investigating the link between biological age and clinical measures.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00941-z.
Keywords: Sex differences, Epigenetic aging, DNA methylation, Frailty, Brain-predicted age difference, Health markers
Introduction
In 2020, approximately 10% of the world’s population was over 65 years of age [1]; and the proportion of older adults is growing at a faster rate than all other age groups. As aging represents the most profound risk factor for many chronic diseases, the prevalence of age-related diseases such as dementia and ischemic heart disease is also proportionately increasing [2]. The geroscience hypothesis posits that by targeting aging mechanisms, we can potentially reduce the risk of several age-related diseases and extend the health span (i.e., duration of life lived in good health) [3]. As such, increasing attention has been placed on the hallmarks of aging which may better capture underlying biological differences and thus an individual’s risk of disease.
One of the most studied and promising aging biomarkers is epigenetic age. It estimates biological age based on DNA methylation (DNAm) levels at multiple cytosine-phosphate-guanine dinucleotides (CpGs) across the genome [4–9]. While epigenetic age can be estimated from any tissue, when measured in the blood, it reflects system-wide biological age [4, 10]. Over the previous decade, a number of epigenetic clock algorithms have been developed [4–9]. The first-generation epigenetic clocks, namely, HorvathAge [5] and HannumAge [6], were generated to predict chronological age using the specific CpGs sites which are strongly associated with chronological age. HorvathAge [5] was developed on 353 CpGs sites across multiple tissue samples whereas HannumAge [6] was trained on 71 CpGs sites from blood samples. Second-generation clocks such as PhenoAge [7] and GrimAge [8, 11] were trained in blood samples by incorporating additional age-related clinical markers. PhenoAge [7] used 513 CpGs sites which are related to chronological age and nine phenotypic aging biomarkers including albumin, creatinine, and C-reactive protein levels whereas GrimAge [8] considered 1030 CpGs sites associated with smoking-pack-years and a selection of plasma proteins including adrenomedullin, beta-2-microglobulin, and cystatin-C. Further, using the same set of 1030 CpGs that underline the original GrimAge, GrimAge2 was recently developed by additionally incorporating two new DNAm-based plasma proteins—high sensitivity C-reactive protein, and hemoglobin A1C [11]. The second-generation clocks have been found to outperform the first-generation clocks in the risk prediction of age-related diseases and all-cause mortality [5–8, 11, 12]. For the first- and second-generation clocks, the residual taken from regressing epigenetic age on chronological age is referred to as epigenetic age acceleration (AA) [10]. A positive value of AA indicates an older epigenetic age relative to chronological age, which has been shown to be predictive of several age-related diseases [10]. More recently, a third generation, the Pace of Aging (DunedinPACE) was developed using 173 CpGs sites in blood samples [9]. In contrast to prior generations, DunedinPACE was built by incorporating longitudinal within-person physiological change together with methylation pattern and thus specifically designed to measure the rate of aging [9].
Other measures have also been developed to estimate biological aging in recent years. Another system-wide measure of biological aging is the deficit accumulation frailty index (FI), a multidimensional tool that measures the accumulation of health deficits across a range of physical, psychological, and mental processes [13]. Unlike AA, FI can capture the clinical state of age-related physiological decline in which a person is more susceptible to developing adverse health outcomes [14, 15]. There are also aging biomarkers that target specific organs. Brain age is estimated using neuroimaging data, and an older brain relative to chronological age, positive brain-predicted age difference (brain-PAD), indicates accelerated brain aging [16]. In recent years, evidence has found that accelerated brain aging was predictive of cognitive performance, and physical disability, particularly in older people [17–19]. In literature, few studies have investigated the associations between epigenetic clocks and frailty and/or brain age, showing inconsistent relationships between these biological aging measures [20–22]. Further, all these previous studies were investigated using only first or second generations of epigenetic clocks. Thus, it is yet unclear to what extent these other biological aging indices are strongly correlated with different generations of epigenetic AA, and thus, it remains unclear to what extent these capture different aspects of the aging process.
On average, females have a longer lifespan than males [23] but may carry a higher disease and disability burden [24]. Such discrepancies may reflect differences in biological phenotypes, genetic constitutions, and physiological traits between males and females [23, 25], In terms of social behaviors, there are recognized sex differences in health-related behaviors, and in healthcare seeking, including accessing social and familial support [26]. Given these disparities, it is important to understand how the relationships between AA and health status might differ between males and females.
This study aimed to determine the correlation between epigenetic aging, with other measures of biological aging (i.e., system-wide FI, and brain-PAD), in an initially healthy population of males and females aged ≥ 70 years. We additionally explored the extent to which these AA measures were associated with sociodemographic characteristics, clinical measures, and chronic conditions, separately in males and females.
Methods
Study population
This study sub-sampled 560 Australians from the ASPREE (ASPirin in Reducing Events in the Elderly) study, who provided blood samples at baseline [27]. Full details regarding the sampling procedure, inclusion and exclusion criteria, and study design of the ASPREE study have been published previously [27, 28]. Briefly, in Australia, participants aged ≥ 70 years were recruited through a partnership with general practices, from March 2010 through December 2014. The ASPREE exclusion criteria included a history of cardiovascular disease (CVD), atrial fibrillation, anemia, dementia, a score of < 78 on the Modified Mini-Mental State examination (3MS), uncontrolled high blood pressure (i.e., ≥ 180/ ≥ 105 mm Hg), or other major life-limiting health conditions likely to cause death within 5 years.
The ASPREE study complies with the Declaration of Helsinki and was approved by multiple Institutional Review Boards (www.aspree.org). All participants signed written informed consent on participation. The present sub-study was approved by the Monash University Human Research Ethics Committee (MHREC 30734).
Epigenetic age
Blood samples were processed within 4 h of the blood collection and stored at ultra-low − 80° temperatures. All samples have barcodes and are prepared for long-term storage at a purpose-built large biorepository near the ASPREE National Co-ordinating Centre in Melbourne (www.aspree.org).
DNA methylation (DNAm) from peripheral blood samples (buffy coat) collected at baseline was extracted using Qiagen DNeasy Blood and Tissue Kits (https://www.qiagen.com/au). Cytosine-phosphate-guanine (CpG) probe methylation was measured using the Illumina Infinium Methylation EPIC BeadChip (EPIC) and run at the Australian Genome Research Facility, Melbourne, Victoria (https://www.agrf.org.au/). The programming platform R version 4.1.3 with R package minfi was used for preprocessing data for use in Horvath’s online “New DNAm Age Calculator” (https://dnamage.genetics.ucla.edu/new). The preprocessNoob method was used to normalize the EPIC data [29]. According to the online age calculator’s expectation, the beta values from the EPIC data set were limited to the probes that the 450 K array could measure, and the beta values of missing probes that are not present on the 450 K array (33,124 probes) were filled with “NA” in the data set. The list of 33,124 missing probes has been described previously [30].
Next, the sample methylation data file (including beta measurements) and the sample annotation file, which included sample ID, age, sex, and tissue type, were uploaded to the online calculator. The default option (i.e., “Normalize Data”) and an additional option—“Advanced Analysis”—were selected before submitting the data. The output of the online age calculator consisted of HorvathAge [5], HannumAge [6], PhenoAge [7], GrimAge [8], and their AA measures (residuals from regressing epigenetic age on chronological age). A positive value of AA indicates an older epigenetic age relative to chronological age. Further, version-2 of GrimAge [11] and its AA were estimated through the new online software of Clock Foundation (https://dnamage.clockfoundation.org/). We also considered other DNAm estimates—telomere length [31] and DNAm-based components of original GrimAge and GrimAge2 [8, 11] (adrenomedullin (ADM), beta-2-microglobulin (B2M), cystatin-C, growth differentiation factor 15 (GDF-15), leptin, smoking-pack-years, plasminogen activation inhibitor 1 (PAI-1), tissue inhibitor of metalloproteinases 1 (TIMP-1), high sensitivity C-reactive protein (CRP), and hemoglobin A1C (A1C)). The Pace of Aging (DunedinPACE) is a measure of the ongoing rate of deterioration in system integrity. DunedinPACE was estimated using R software (version 4.1.3) and the codes described in Belsky et al. [9].
Brain age
ASPREE participants living in the Greater Melbourne region and without MRI contraindications (e.g., specific foreign bodies or electronic implants, claustrophobia) were invited to participate in the ASPREE-NEURO sub-study [32]. Brain scans were performed at the Monash Biomedical Imaging Facility, Melbourne, Australia, using a 3 Tesla Siemens Skyra scanner (Siemens, Erlangen, Germany). Raw 3D T1-weighted images were pre-processed using the MATLAB Statistical Parametric Mapping software package (University College London, London, UK) [33]. Full details regarding the pre-processing of this data have been described in detail previously [19]. Brain age was calculated using a trained, publicly accessible processing pipeline and prediction algorithm created by Cole et al. (BrainageR, version 2.1, https://github.com/james-cole/brainageR) [16].
The brain-predicted age difference (brain-PAD) refers to the difference between an individual’s estimated brain age and their chronological age. A positive brain-PAD value indicates that the individual’s brain is older than its chronological age.
Frailty index
A deficit accumulation frailty index (FI) of 67 items was constructed based on the method of Rockwood and Mitnitski [13]. The index included chronic disease diagnosis and indicators (24 items), physical deficits interfering in completing the activities of daily living (26 items), mental and psychosocial deficits (11 items), and measures of physical functional engagement and cognitive function (6 items), as described in detail previously [34]. Each item was scored from 0 (deficit absent) to 1 (deficit present). The FI was derived by summing the number of deficits noted for each person and dividing this by the total number of possible deficits. Higher scores indicate greater frailty.
Sociodemographic characteristics
Sociodemographic characteristics included in this study were chronological age, sex (male or female), years of education (< 12 years or ≥ 12 years), living situation (at home alone or with family/others), and socioeconomic status (SES) (very low, low, middle, high, very high). SES was estimated through the Socio-Economic Indices for Areas-Index of Relative Socioeconomic Advantage and Disadvantage (SEIFA-IRSAD) based on the information from the 2011 Australian Census using the residential postcodes of participants [35]. Smoking was categorized into never, former, or current. Alcohol consumption was defined as never, former, current-low risk (i.e., ≤ 10 standard drinks per week and ≤ 4 standard drinks on any 1 day), or current-high risk (i.e., > 10 standard drinks per week or > 4 standard drinks on any 1 day) [36].
Clinical measures, chronic conditions, and self-reported outcomes
Clinical measures included systolic blood pressure (SBP) and diastolic blood pressure (DBP) in mmHg, dominant hand grip strength in kilograms, and gait speed in meters/seconds. Full detailed procedures for taking measurements have been previously reported [37]. In brief, for SBP and DBP, the average scores after three measurements were used. The grip strength of both hands was assessed three times using an isometric handheld dynamometer (Jamar; Lafayette Instruments). The mean score for each participant’s self-reported dominant hand was used in the analysis. Gait speed was assessed as the time spent walking 3 m at a habitual pace on a flat surface from a standing start. Gait speed was measured twice, and the mean score was used in the analysis.
Self-reported outcomes were assessed through the SF-12 version-2 questionnaire [38]. Self-rated health ranged from 1 (excellent) to 5 (poor). Physical (PCS) and mental component scores (MCS) of health-related quality of life (HRQoL) were generated using a norm-based scoring method [38]. Higher scores indicate better health-related quality of life.
Chronic conditions investigated included hypertension (systolic blood pressure ≥ 140 mmHg or diastolic blood pressure ≥ 90 mmHg and/or treatment for high blood pressure), diabetes (fasting glucose ≥ 126 mg/dL or treatment or self-report), dyslipidemia (cholesterol-lowering medications or low-density lipoprotein, LDL > 160 mg/dL (> 4.1 mmol/L), or serum cholesterol ≥ 212 mg/dL (≥ 5.5 mmol/L)), obesity (body mass index ≥ 30 kg/m2) [39], chronic kidney disease (estimated glomerular filtration rate (eGFR) < 60 ml/min per 1.73 m2 or spot urinary albumin: creatinine ratio (UACR) ≥ 3 mg/mmol with eGFR ≥ 60 ml/min per 1.73 m2) [40], and depression (Center for Epidemiological Studies-Depression CES-D-10 score ≥ 8) [41].
Statistical analysis
First, the correlations between different measures of biological age (i.e., HorvathAge, HannumAge, PhenoAge, GrimAge, GrimAge2, DunedinPACE, brain age, and FI) and chronological age were examined. Next, Pearson’s correlation, with pairwise deletion, was used to investigate the correlation between the different biological aging measures. Pearson’s correlation, t-tests, or one-way ANOVA, as appropriate, were also used to investigate the correlations between the AA measures and other DNAm-based estimates and sociodemographic characteristics, clinical measures, and chronic diseases, separately in males and females. Analyses were undertaken in STATA statistical software version 17.0, and the figures and graphs were plotted using the programming platform R version 4.1.3.
Results
Participants’ characteristics
The characteristics of the 560 study participants are shown in Table 1. Approximately, half were female (50.7%), and the chronological age ranged from 70.0 to 92.0 years, with a mean of 74.5 years. The estimated brain age (mean, 72.1 ± 6.9 years), HorvathAge (mean, 70.6 ± 6.1 years), GrimAge (mean, 70.1 ± 5.0 years), and GrimAge2 (mean, 74.1 ± 5.3 years) were similar to the mean chronological age, whereas HannumAge (mean, 59.5 ± 6.1 years) and PhenoAge (mean, 58.9 ± 7.7 years) were younger than the chronological age. DunedinPACE ranged from 0.69 to 1.30, with a mean of 0.97, and the mean FI was 0.10. Compared to females, males had significantly higher epigenetic aging whereas females had higher deficit accumulation frailty scores than males. Brain age was identical between males and females (Table 1).
Table 1.
The characteristics of study participants
| All participants (n = 560) | Males (n = 276, 49.3%) | Females (n = 284, 50.7%) | |
|---|---|---|---|
| Chronological age (years); mean (SD) | 74.5 (4.2) | 74.5 (4.2) | 74.5 (4.2) |
| Epigenetic age (years) | |||
| HorvathAge; mean (SD) | 70.6 (6.1) | 71.0 (6.1) | 70.1 (6.0) |
| HannumAge; mean (SD) | 59.5 (6.1) | 60.4 (6.1) | 58.6 (6.0) |
| PhenoAge; mean (SD) | 58.9 (7.7) | 59.7 (7.7) | 58.1 (7.6) |
| GrimAge; mean (SD) | 70.1 (5.0) | 71.5 (5.1) | 68.6 (4.5) |
| GrimAge2; mean (SD) | 74.1 (5.3) | 75.4 (5.5) | 72.8 (4.8) |
| DunedinPACE; mean (SD) | 0.97 (0.11) | 0.99 (0.11) | 0.95 (0.11) |
| Epigenetic age acceleration (AA, years) | |||
| HorvathAA; mean (SD) | 0.08 (4.96) | 0.49 (4.96) | − 0.31 (4.92) |
| HannumAA; mean (SD) | 0.07 (4.84) | 0.79 (5.04) | − 0.64 (4.54) |
| PhenoAA; mean (SD) | 0.09 (6.37) | 0.76 (6.46) | − 0.55 (6.21) |
| GrimAA; mean (SD) | 0.02 (3.52) | 1.35 (3.34) | − 1.27 (3.19) |
| Grim2AA; mean (SD) | 0.02 (4.04) | 1.16 (3.99) | − 1.08 (3.78) |
| Other measures of biological aging | |||
| Mean brain agea; mean (SD) | 72.1 (6.9) | 72.1 (7.4) | 72.1 (6.4) |
| Brain-PADa; mean (SD) | − 1.50 (6.05) | − 1.70 (6.39) | − 1.27 (5.67) |
| Deficit accumulation frailty index; mean (SD) | 0.10 (0.06) | 0.09 (0.06) | 0.12 (0.06) |
| Participants’ characteristics | |||
| Years of education; n (%) | |||
| < 12 years | 234 (41.8) | 111 (40.2) | 123 (43.3) |
| ≥ 12 years | 326 (58.2) | 165 (59.8) | 161 (56.7) |
| Living situation; n (%) | |||
| At home alone | 159 (28.4) | 47 (17.0) | 112 (39.4) |
| With family or others | 401 (71.6) | 229 (83.0) | 172 (60.6) |
| Socioeconomic status (SES)b; n (%) | |||
| Very low | 48 (8.6) | 23 (8.4) | 25 (8.8) |
| Low | 44 (7.9) | 19 (6.9) | 25 (8.8) |
| Middle | 85 (15.2) | 43 (15.6) | 42 (14.8) |
| High | 121 (21.7) | 61 (22.2) | 60 (21.1) |
| Very high | 261 (46.7) | 129 (46.9) | 132 (46.5) |
| Smoking; n (%) | |||
| Never | 312 (55.7) | 129 (46.7) | 183 (64.4) |
| Former | 227 (40.5) | 135 (48.9) | 92 (32.4) |
| Current | 21 (3.8) | 12 (4.4) | 9 (3.2) |
| Alcohol consumption, n (%) | |||
| Never | 80 (14.3) | 25 (9.1) | 55 (19.4) |
| Former | 21 (3.8) | 12 (4.4) | 9 (3.2) |
| Current-low riskc | 336 (60.0) | 158 (57.3) | 178 (62.7) |
| Current-high riskd | 123 (22.0) | 81 (29.4) | 42 (14.8) |
| Clinical measures | |||
| Systolic blood pressure (mmHg); mean (SD) | 137.8 (16.0) | 140.7 (15.3) | 134.9 (16.2) |
| Diastolic blood pressure (mmHg); mean (SD) | 75.8 (10.2) | 77.3 (10.1) | 74.2 (10.0) |
| Grip strength (kg)e; mean (SD) | 28.5 (9.9) | 35.9 (7.6) | 21.3 (5.6) |
| Gait speed (m/s); mean (SD) | 1.1 (0.2) | 1.1 (0.2) | 1.0 (0.2) |
| Self-reported health outcomes | |||
| Self-rated health; n (%) | |||
| Excellent | 89 (15.9) | 43 (15.6) | 46 (16.2) |
| Very good | 279 (49.8) | 133 (48.2) | 146 (51.4) |
| Good | 172 (30.7) | 90 (32.6) | 82 (28.9) |
| Fair | 19 (3.4) | 10 (3.6) | 9 (3.2) |
| Poor | 1 (0.2) | 0 (0.0) | 1 (0.4) |
| Health-related quality of life | |||
| Physical component score; mean (SD) | 48.5 (8.7) | 49.3 (8.1) | 47.7 (9.1) |
| Mental component score; mean (SD) | 55.5 (7.4) | 56.1 (6.4) | 54.9 (8.2) |
| Chronic conditions | |||
| Hypertension; n (%) | |||
| Yes | 381 (68.0) | 197 (71.4) | 184 (64.8) |
| No | 179 (32.0) | 79 (28.6) | 100 (35.2) |
| Diabetes; n (%) | |||
| Yes | 65 (11.6) | 43 (15.6) | 22 (7.8) |
| No | 495 (88.4) | 233 (84.4) | 262 (92.3) |
| Dyslipidemia; n (%) | |||
| Yes | 351 (62.7) | 140 (50.7) | 211 (74.3) |
| No | 209 (37.3) | 136 (49.3) | 73 (25.7) |
| Obesityf; n (%) | |||
| Yes | 151 (27.1) | 70 (25.6) | 81 (28.6) |
| No | 406 (72.9) | 204 (74.5) | 202 (71.4) |
| Chronic kidney diseaseg; n (%) | |||
| Yes | 107 (20.4) | 53 (20.4) | 54 (20.4) |
| No | 418 (79.6) | 207 (79.6) | 211 (79.6) |
| Depression; n (%) | |||
| Yes | 57 (10.2) | 26 (9.4) | 31 (10.9) |
| No | 503 (89.8) | 250 (90.6) | 253 (89.1) |
aNumbers of participants, total n = 400, males n = 209, and females n = 191
bSES, total n = 559, males n = 275, and females n = 284
cCurrent-low risk, ≤ 10 standard drinks per week and ≤ 4 standard drinks on any 1 day
dCurrent-high risk, > 10 standard drinks per week or > 4 standard drinks on any 1 day
eGrip strength, total n = 558, males n = 275, and females n = 283
fObesity, total n = 557, males n = 274, and females n = 283
gChronic kidney disease, total n = 525, males n = 260, and females n = 265
Correlation between different measures of biological age, and chronological age
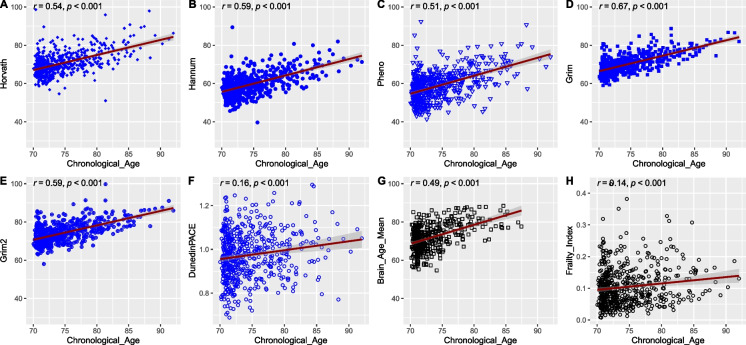
Epigenetic age, brain age, and FI were positively correlated with chronological age (Fig. 1). The strongest correlation was found for GrimAge (r = 0.67), while DunedinPACE (r = 0.16) and FI (r = 0.14) had the weakest correlations with chronological age.
Fig. 1.
Correlations between estimated biological age (y-axis) and chronological age (x-axis). A HorvathAge vs. chronological age (n = 560); B HannumAge vs. chronological age (n = 560); C PhenoAge vs. chronological age (n = 560); D GrimAge vs. chronological age (n = 560); E GrimAge2 vs. chronological age (n = 560); F DunedinPACE vs. chronological age (n = 560); G Brain age vs. chronological age (n = 400); H deficit accumulation frailty index vs. chronological age (n = 560)
Correlation between AA, brain-PAD, and FI
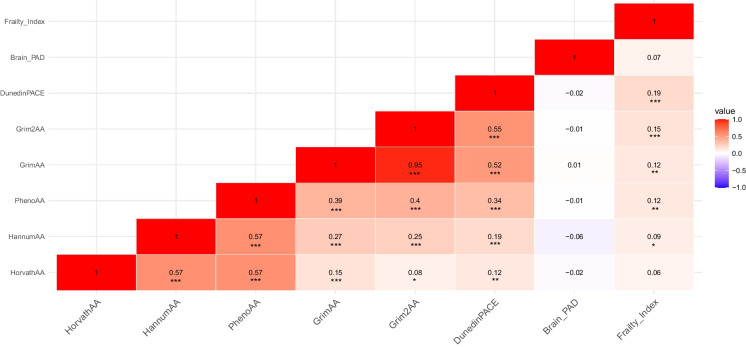
All AA measures were positively correlated with one another (r ranged from 0.08 to 0.57), but DunedinPACE showed weak correlations, particularly with 1st generation HorvathAA and HannumAA (Fig. 2). GrimAA and Grim2AA were strongly correlated with each other (r = 0.95). Brain-PAD was not correlated with any of the AA measures, or FI. FI had the strongest correlation with DunedinPACE (r = 0.19) and Grim2AA (r = 0.15), and significant but weak correlations with HannumAA, PhenoAA, and GrimAA (r ≤ 0.12). Similar correlations were also found in males and females (Supplementary Tables 1 and 2).
Fig. 2.
Pearson’s correlation matrix on epigenetic age acceleration (HorvathAA, HannumAA, PhenoAA, GrimAA, Grim2AA, DunedinPACE), brain-PAD, and deficit accumulation frailty index. *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001
AA variations according to the sociodemographic characteristics
In males (Supplementary Table 3), we observed an association between smoking and both versions of GrimAA (p-value < 0.001) and DunedinPACE only (p-value = 0.003). In females, however (Supplementary Table 4), we observed associations across all the sociodemographic characteristics. In particular, less than 12 years of education was associated with accelerated DunedinPACE; living alone was linked to an accelerated HorvathAA, low SES, current smoking, and former alcohol consumption were associated with higher in both versions of GrimAA (p-values < 0.05).
Correlation between AA, clinical measures, and self-reported health outcomes
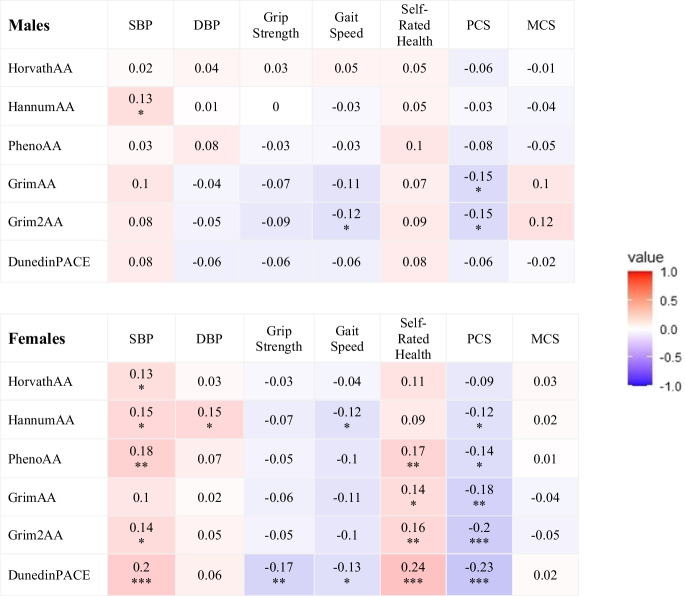
Correlations between AA measures and clinical measures/self-reported health outcomes were also more commonly found in females than males (Fig. 3). In males, correlations were observed between SBP and HannumAA (r = 0.13), between gait speed and Grim2AA (r = − 0.12), and between PCS and both versions of GrimAA (r = − 0.15). In females, all clinical measures and self-reported health status, except for MCS, were correlated with AA measures (r ranged from − 0.23 to 0.24) with DunedinPACE having the greatest number of significant correlations.
Fig. 3.
Correlation matrix between epigenetic age acceleration measures and systolic blood pressure (SBP), diastolic blood pressure (DBP), grip strength, gait speed, self-rated health (higher scores indicating worse health), and physical component score (PCS) and mental component score (MCS) of the SF-12, separately in males and females. *p-value < 0.05; **p-value < 0.01; ***p-value < 0.001
AA variations according to the chronic condition status
Hypertension, diabetes, and chronic kidney disease were associated with accelerated GrimAA (both versions) and DunedinPACE in males (Supplementary Table 5). In contrast, only obesity and depression were associated with accelerated GrimAA (both versions) in females, and obesity was also associated with accelerated DunedinPACE.
Other DNAm-based estimates and their associations with sociodemographic characteristics, clinical measures, self-reported health outcomes, and the chronic condition status
There were also sex differences in the associations between other DNA-based estimates (telomere length, plasma proteins, and smoking-pack-years) and sociodemographic factors and health markers (Supplementary Tables 6 to 15). The correlations between telomere length and clinical measures except for DBP and MCS were found only in females (Supplementary Table 8). Regarding the chronic conditions, only chronic kidney disease was associated with shorter telomere length in females, and no associations were found in males (Supplementary Table 9). Correlations between either plasma proteins or smoking-pack-years and clinical measures were commonly found in both males and females (Supplementary Tables 12 and 13). At least one plasma protein was associated with diabetes, dyslipidemia, obesity, and chronic kidney disease in both males and females, but associations were more commonly found in males than females (Supplementary Tables 14 and 15).
Discussion
In this study, we examined the extent to which different measures of epigenetic aging were correlated with a system-wide measure of biological aging (FI) and an organ-specific biological aging marker (brain-PAD), separately in males and females. We observed that FI had weak but significant positive correlations with AA in both sexes, whereas brain-PAD was not correlated with any of the other biological aging measures. This suggests that these system-wide measures of biological aging are capturing somewhat overlapping, yet distinct aspects of the aging process. We also found that epigenetic aging was correlated with participants’ sociodemographic characteristics and health status, but this varied between males and females. Together, these findings indicate that markers of biological aging may be useful clinical indicators of overall physical, cognitive, and psychosocial deficits even in relatively healthy older people who have reached 70 years without major illness, including CVD and dementia. Further, we also found that there were differences between males and females in the relationships between other DNAm estimates (i.e., telomere length, plasma proteins, and smoking-pack-years) and health status.
Not surprisingly, in our present study, the first-generation AA measures were most strongly correlated with one another, and the most recent of these measures, DunedinPACE, was most significantly associated with the second-generation AA measures where the correlation with Grim2AA was the highest. Our finding is supported by previous research [5, 6, 8, 9] and aligns with the general purpose for which these epigenetic clock algorithms were developed. The first-generation epigenetic clocks were generated to predict chronological age [5, 6] whereas the second-generation clocks, like GrimAge [8, 11], go beyond just estimating chronological age and additionally aim to predict the risk of developing health outcomes. To reflect that purpose, GrimAge algorithms were constructed by incorporating additional age-related clinical and laboratory measures as well as lifestyle factors such as smoking [8, 11]. Thus, clear evidence has been found that second-generation clocks outperform first-generation clocks in the risk prediction of age-related diseases and death [8]. The latest generation clock—DunedinPACE—was specifically designed to capture the gradual, progressive deterioration simultaneously affecting different organ systems with aging [9]. This noticeably contrasts with first-generation clocks which were designed and trained to predict chronological age (HorvathAge [5] and HannumAge [6]). Moreover, prior studies have reported significant but weak correlations between AA and system-wide FI [21, 42]. However, as ours is the first to investigate correlations with GrimAge2 and DunedinPACE, we added new evidence to the field that DunedinPACE and GrimAge2 are more strongly correlated with FI rather than other AAs. This is not surprising given both DunedinPACE and FI have a multidimensional nature. Like DunedinPACE, the FI defines biological aging through the accumulation of age-related health deficits across a range of functions (physical, psychological, and social). Noticeably, our study also demonstrated that the correlations of GrimAge2 with DunedinPACE and FI were slightly stronger than those of the original GrimAge. This finding could be due to the additional inclusion of two commonly used biomarkers (i.e., DNAm-based high-sensitivity CRP, and hemoglobin A1C) in the GrimAge2 development [11]. Further, our finding of GrimAge2 also somewhat reflects the fact that GrimAge2 outperforms the original GrimAge in terms of mortality and morbidity risk prediction [11].
Our finding that males had accelerated epigenetic age in comparison to females is consistent with prior evidence demonstrating that, for individuals with the same chronological age, males tend to be biologically older than females [43]. This also aligns with the established longer life expectancy of females compared to males [23] which could be driven by a complex interaction of biological factors, including genetic influences and sex hormones, as well as behavioral factors [44]. Males may be more likely to engage in risky behaviors and unhealthy lifestyles such as smoking and heavy drinking [45, 46] and have an age-related decline in testosterone concentrations, which could lead to a greater risk of fatal illnesses including heart disease and cancer [47, 48]. There is also evidence that females are more willing to engage with health-related information and are more likely to seek medical treatment for their illness [49] which together could help initiate early detection and intervention. In this manner, females may have less fatal outcomes. In contrast, females in our study had higher deficit accumulation FI scores than males, which captures the overall disease burden. This finding also supports the conclusion of a prior meta-analysis among 37,426 participants, showing that females of all ages have a poorer health status in terms of FI, compared to males [50]. Thus, our finding from this relatively healthy older cohort might also reflect the male–female health-survival paradox, in that females live longer and are more likely to experience disabilities and disease burdens [46, 51].
Several studies have investigated whether AA measures are associated with participants’ sociodemographic characteristics and markers of their health status [52–54]. These studies have generally, but not consistently, shown that lower SES status and/or risk factors for chronic diseases, such as high blood pressure and smoking, are associated with accelerated aging. However, very few studies have considered whether these associations may vary between males and females. Our study provides an important new contribution to this field, demonstrating clear evidence of sex differences. In females, several significant correlations were observed between AA and health risk factors such as low SES and living alone, as well as markers of physical functioning (e.g., grip strength and gait speed), SBP, DBP, and self-rated overall and physical health whereas only associations between AA, especially Grim2AA, and SBP, gait speed, and self-rated physical health were found in males. Thus, our cross-sectional findings highlight that accelerated epigenetic aging could be linked with physical health deterioration in both males and females. Hence, a further longitudinal study is suggested to enhance this novel relationship and to examine whether accelerated epigenetic aging could be predictive of physical health deterioration in later life. Moreover, significant associations were found between newer accelerated AA measures and hypertension, diabetes, and chronic kidney disease in males only whereas the associations of AA measures with obesity and depression were observed only in females. Although the exact reasons for these differences are still unclear, biological disparities and previously noted sex differences in the prevalence of health conditions [55, 56] may have contributed to our results. Overall, it highlights the importance of considering separately males and females when investigating causal inference of our observed relationships in future longitudinal studies.
Our study had both strengths and limitations. The strength of this study includes investigating several epigenetic clocks including the most recent ones—GrimAge2 and DunedinPACE and their correlations with the system-wide FI, and brain-PAD, among community-based individuals. Our sex-balanced sample (females 50.7%) allowed us to investigate the relationships between AA and markers of health in males and females separately. We acknowledged that our community-based sample comprises relatively healthy older Australians without dementia or CVD, and about two-thirds had high SES. Thus, this could limit the generalizability of our findings. However, the sample had a good distribution in terms of education level (e.g., 41.8% had < 12 years of education) and ranged in age from 70 to 92 years. The findings add new evidence about the utility of biological aging measures even in relatively healthy older individuals.
In conclusion, our study showed that, in relatively healthy older individuals, epigenetic age was correlated with the system-wide deficit accumulation FI, but not with the neuroimaging-based brain aging biomarker (brain-PAD). We also showed that despite having similar chronological ages, females in our study had lower epigenetic aging, but higher FI, than males. Further, this study provides novel evidence showing sex differences in the relationships between AA measures and markers of health status. Thus, our results highlight the importance of considering sex differences when investigating biological aging and its utility as a marker of health or disease risk.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the participants of the Australian ASPREE and ASPREE-NEURO studies for volunteering their time and the general practitioners for supporting the participants throughout the trial. The authors also acknowledge the hard work of all ASPREE field staff and those at the Monash Biomedical Imaging Facility who attained magnetic resonance images.
Author contribution
Robyn Woods played a major role in the design, funding, and completion of the ASPREE study, including the acquisition of data. Joanne Ryan conceived this study. Aung Zaw Zaw Phyo had full access to all the data in the study and analyzed the data. Aung Zaw Zaw Phyo interpreted the data, with input from Joanne Ryan, Peter D. Fransquet, Jo Wrigglesworth, and Sara Espinoza. Aung Zaw Zaw Phyo wrote the initial manuscript draft and undertook revisions with assistance from Joanne Ryan. All authors provided intellectual input and approved the final manuscript.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions The ASPREE trial was mainly supported by grants from the National Institute on Aging and the National Cancer Institute at the U.S. National Institutes of Health (grant numbers U01AG029824 and U19AG062682), the National Health and Medical Research Council of Australia (grant numbers 334047 and 1127060), and the Monash University (Australia) and the Victorian Cancer Agency (Australia). ASPREE-NEURO has funding from the NHMRC (grant number 1086188) and support from Monash Biomedical Imaging. Other funding resources and collaborating organizations of the ASPREE study are listed on http://www.aspree.org. Joanne Ryan is funded by a National Health and Medical Research Council Leadership 1 Investigator Grant (2016438). Funders played no role in the design, methods, data collection, analysis, or preparation of the paper.
Data availability
The data that support the findings of this study are available from authors upon reasonable request and with permission of the ASPREE principal investigators through the website (www.ASPREE.org). Data will be shared to approved researchers through a secure web-based data portal Safe Haven.
Declarations
Ethical approval
The data of the present secondary data-analysis study was from a five-year ASPREE clinical trial (Trial Registration: International Standard Randomized Controlled Trial Number Register (ISRCTN 83772183) and clinicaltrials.gov (NCT 01038583)). The ASPREE trial was approved by multiple Institutional Review Boards in Australia and the USA (www.aspree.org). The current study was approved by the Monash University Human Research Ethics Committee (MHREC 30734).
Consent for publication
All authors gave their approval for submission.
Informed consent
All individual participants of the ASPREE study including the ASPREE Biobank and ASPREE-NEURO sub-studies signed informed consent on participation.
Human and animal rights
No animals were used for this research. The study on humans was conducted in accordance with the Declaration of Helsinki 2008 revision, NHMRC Guidelines on Human Experimentation, the federal patient privacy (HIPAA) law, the International Conference of Harmonisation Guidelines for Good Clinical Practice, and the Code of Federal Regulations. ASPREE also follows the Code of Federal Regulations as it relates to areas of clinical research. The overall management and conduct of the ASPREE clinical trial are the responsibility of the ASPREE Steering Committee.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.United Nations, Department of Economic and Social Affairs, Population Division. World population ageing 2020: highlights - living arrangements of older persons (ST/ESA/SER.A/451). New York: United Nations; 2020. Available from: https://www.un.org/development/desa/pd/sites/www.un.org.development.desa.pd/files/undesa_pd-2020_world_population_ageing_highlights.pdf. Accessed 07 Nov 2022.
- 2.Le Couteur DG, Thillainadesan J. What is an aging-related disease? An epidemiological perspective. J Gerontol A Biol Sci Med Sci. 2022;77(11):2168–74. [DOI] [PMC free article] [PubMed]
- 3.Le Couteur DG, Barzilai N. New horizons in life extension, healthspan extension and exceptional longevity. Age Ageing. 2022;51(8):afac156. doi: 10.1093/ageing/afac156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryan CP. “Epigenetic clocks”: theory and applications in human biology. Am J Hum Biol. 2021;33(3):e23488. doi: 10.1002/ajhb.23488. [DOI] [PubMed] [Google Scholar]
- 5.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife. 2022;11:e73420. doi: 10.7554/eLife.73420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen BH, Marioni RE, Colicino E, Peters M, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu AT, Binder AM, Zhang J, Yan Q, Reiner AP, Cox SR, et al. DNA methylation GrimAge version 2. Aging (Albany NY) 2022;14(23):9484–9549. doi: 10.18632/aging.204434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCrory C, Fiorito G, Hernandez B, Polidoro S, O'Halloran AM, Hever A, et al. GrimAge outperforms other epigenetic clocks in the prediction of age-related clinical phenotypes and all-cause mortality. J Gerontol A Biol Sci Med Sci. 2021;76(5):741–749. doi: 10.1093/gerona/glaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rockwood K, Mitnitski A. Frailty in relation to the accumulation of deficits. J Gerontol A Biol Sci Med Sci. 2007;62(7):722–727. doi: 10.1093/gerona/62.7.722. [DOI] [PubMed] [Google Scholar]
- 14.Ekram ARMS, Woods RL, Britt C, Espinoza S, Ernst ME, Ryan J. The association between frailty and all-cause mortality in community-dwelling older individuals: an umbrella review. J Frailty Aging. 2021;10(4):320–326. doi: 10.14283/jfa.2021.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drubbel I, de Wit NJ, Bleijenberg N, Eijkemans RJC, Schuurmans MJ, Numans ME. Prediction of adverse health outcomes in older people using a frailty index based on routine primary care data. J Gerontol A Biol Sci Med Sci. 2013;68(3):301–308. doi: 10.1093/gerona/gls161. [DOI] [PubMed] [Google Scholar]
- 16.Cole JH, Ritchie SJ, Bastin ME, Valdés Hernández MC, Muñoz Maniega S, Royle N, et al. Brain age predicts mortality. Mol Psychiatry. 2018;23(5):1385–1392. doi: 10.1038/mp.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christman S, Bermudez C, Hao L, Landman BA, Boyd B, Albert K, et al. Accelerated brain aging predicts impaired cognitive performance and greater disability in geriatric but not midlife adult depression. Transl Psychiatry. 2020;10(1):317. doi: 10.1038/s41398-020-01004-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunlop K, Victoria LW, Downar J, Gunning FM, Liston C. Accelerated brain aging predicts impulsivity and symptom severity in depression. Neuropsychopharmacology. 2021;46(5):911–919. doi: 10.1038/s41386-021-00967-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wrigglesworth J, Yaacob N, Ward P, Woods RL, McNeil J, Storey E, et al. Brain-predicted age difference is associated with cognitive processing in later-life. Neurobiol Aging. 2022;109:195–203. doi: 10.1016/j.neurobiolaging.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Teeuw J, Ori APS, Brouwer RM, de Zwarte SMC, Schnack HG, Hulshoff Pol HE, et al. Accelerated aging in the brain, epigenetic aging in blood, and polygenic risk for schizophrenia. Schizophr Res. 2021;231:189–197. doi: 10.1016/j.schres.2021.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Seligman B, Berry SD, Lipsitz LA, Travison TG, Kiel DP. Epigenetic age acceleration and change in frailty in MOBILIZE Boston. J Gerontol A Biol Sci Med Sci. 2022;77(9):1760–1765. doi: 10.1093/gerona/glac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vetter VM, Spira D, Banszerus VL, Demuth I. Epigenetic clock and leukocyte telomere length are associated with vitamin D status but not with functional assessments and frailty in the Berlin Aging Study II. J Gerontol A Biol Sci Med Sci. 2020;75(11):2056–2063. doi: 10.1093/gerona/glaa101. [DOI] [PubMed] [Google Scholar]
- 23.Austad SN. Why women live longer than men: sex differences in longevity. Gend Med. 2006;3(2):79–92. doi: 10.1016/S1550-8579(06)80198-1. [DOI] [PubMed] [Google Scholar]
- 24.Whitson HE, Landerman LR, Newman AB, Fried LP, Pieper CF, Cohen HJ. Chronic medical conditions and the sex-based disparity in disability: the Cardiovascular Health Study. J Gerontol A Biol Sci Med Sci. 2010;65A(12):1325–1331. doi: 10.1093/gerona/glq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Institute of Medicine (US) Committee on understanding the biology of sex and gender differences. Exploring the biological contributions to human health: does sex matter? Washington, D.C: National Academies Press; 2001. [PubMed]
- 26.Vlassoff C. Gender differences in determinants and consequences of health and illness. J Health Popul Nutr. 2007;25(1):47–61. [PMC free article] [PubMed] [Google Scholar]
- 27.McNeil JJ, Woods RL, Nelson MR, Murray AM, Reid CM, Kirpach B, et al. Baseline characteristics of participants in the ASPREE (ASPirin in Reducing Events in the Elderly) study. J Gerontol A Biol Sci Med Sci. 2017;72(11):1586–1593. doi: 10.1093/gerona/glw342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.ASPREE investigator group Study design of ASPirin in Reducing Events in the Elderly (ASPREE): a randomized, controlled trial. Contemp Clin Trials. 2013;36(2):555–564. doi: 10.1016/j.cct.2013.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Triche TJ, Jr, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA methylation bead arrays. Nucleic Acids Res. 2013;41(7):e90. doi: 10.1093/nar/gkt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fransquet PD, Lacaze P, Saffery R, Shah RC, Vryer R, Murray A, et al. Accelerated epigenetic aging in peripheral blood does not predict dementia risk. Curr Alzheimer Res. 2021;18(5):443–451. doi: 10.2174/1567205018666210823100721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu AT, Seeboth A, Tsai P-C, Sun D, Quach A, Reiner AP, et al. DNA methylation-based estimator of telomere length. Aging (Albany NY) 2019;11(16):5895–5923. doi: 10.18632/aging.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward SA, Raniga P, Ferris NJ, Woods RL, Storey E, Bailey MJ, et al. ASPREE-NEURO study protocol: a randomized controlled trial to determine the effect of low-dose aspirin on cerebral microbleeds, white matter hyperintensities, cognition, and stroke in the healthy elderly. Int J Stroke. 2017;12(1):108–113. doi: 10.1177/1747493016669848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cole JH, Leech R, Sharp DJ, Alzheimer’s disease neuroimaging initiative Prediction of brain age suggests accelerated atrophy after traumatic brain injury. Ann Neurol. 2015;77(4):571–81. doi: 10.1002/ana.24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan J, Espinoza S, Ernst ME, Ekram ARMS, Wolfe R, Murray AM, et al. Validation of a deficit-accumulation frailty index in the ASPirin in Reducing Events in the Elderly Study and its predictive capacity for disability-free survival. J Gerontol A Biol Sci Med Sci. 2022;77(1):19–26. doi: 10.1093/gerona/glab225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Australian Bureau of Statistics. 2033.0.55.001 - Census of Population and Housing: Socio-Economic Indexes for Areas (SEIFA), Australia. Canberra: Australian Bureau of Statistics; 2011. Available from: https://www.abs.gov.au/ausstats/abs@.nsf/DetailsPage/2033.0.55.0012011. Accessed 7 Nov 2022
- 36.Australian guidelines to reduce health risks from drinking alcohol. Commonwealth of Australia, Canberra: National Health and Medical Research Council; 2020. Available from: https://www.nhmrc.gov.au/health-advice/alcohol. Accessed 7 Nov 2022
- 37.Orchard SG, Polekhina G, Ryan J, Shah RC, Storey E, Trevor TJC, et al. Combination of gait speed and grip strength to predict cognitive decline and dementia. Alzheimers Dement (Amst) 2022;14(1):e12353. doi: 10.1002/dad2.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ware Jr. JE, Kosinski M, Turner-Bowker DM, Gandek B. User’s manual for the SF-12v2® health survey (with a supplement documenting SF-12® health survey). Lincoln, RI: QualityMetric Incorporated; 2002.
- 39.WHO Consultation on Obesity, (1999: Geneva, Switzerland), & World Health Organization. Obesity : Preventing and managing the global epidemic : Report of a WHO consultation. Geneva: World Health Organization; 2000. Available from: https://apps.who.int/iris/handle/10665/42330. Accessed 7 Nov 2022 [PubMed]
- 40.Kidney disease: improving global outcomes (KDIGO) CKD work group. KDIGO 2012 clinical practice guideline for the evaluation and management of Chronic Kidney Disease. Kidney Int Suppl. 2013;3(1):1–150.
- 41.Radloff LS. The CES-D scale: a self-report depression scale for research in the general population. Appl Psychol Meas. 1977;1(3):385–401. doi: 10.1177/014662167700100306. [DOI] [Google Scholar]
- 42.Ryan J, Wrigglesworth J, Loong J, Fransquet PD, Woods RL. A systematic review and meta-analysis of environmental, lifestyle, and health factors associated with DNA Methylation Age. J Gerontol A Biol Sci Med Sci. 2020;75(3):481–494. doi: 10.1093/gerona/glz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauvais-Jarvis F, Bairey Merz N, Barnes PJ, Brinton RD, Carrero J-J, DeMeo DL, et al. Sex and gender: modifiers of health, disease, and medicine. Lancet. 2020;396(10250):565–582. doi: 10.1016/S0140-6736(20)31561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verbrugge LM. Gender and health: an update on hypotheses and evidence. J Health Soc Behav. 1985;26(3):156–182. doi: 10.2307/2136750. [DOI] [PubMed] [Google Scholar]
- 46.Zhao E, Crimmins EM. Mortality and morbidity in ageing men: biology, lifestyle and environment. Rev Endocr Metab Disord. 2022;23(6):1285–1304. doi: 10.1007/s11154-022-09737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khaw K-T, Dowsett M, Folkerd E, Bingham S, Wareham N, Luben R, et al. Endogenous testosterone and mortality due to all causes, cardiovascular disease, and cancer in men: European Prospective Investigation Into Cancer in Norfolk (EPIC-Norfolk) Prospective Population Study. Circulation. 2007;116(23):2694–2701. doi: 10.1161/CIRCULATIONAHA.107.719005. [DOI] [PubMed] [Google Scholar]
- 48.Maggio M, Basaria S. Welcoming low testosterone as a cardiovascular risk factor. Int J Impot Res. 2009;21(4):261–264. doi: 10.1038/ijir.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ek S. Gender differences in health information behaviour: a Finnish population-based survey. Health Promot Int. 2015;30(3):736–745. doi: 10.1093/heapro/dat063. [DOI] [PubMed] [Google Scholar]
- 50.Gordon EH, Peel NM, Samanta M, Theou O, Howlett SE, Hubbard RE. Sex differences in frailty: a systematic review and meta-analysis. Exp Gerontol. 2017;89:30–40. doi: 10.1016/j.exger.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 51.Nusselder WJ, Wapperom D, Looman CWN, Yokota RTC, van Oyen H, Jagger C, et al. Contribution of chronic conditions to disability in men and women in France. Eur J Public Health. 2019;29(1):99–104. doi: 10.1093/eurpub/cky138. [DOI] [PubMed] [Google Scholar]
- 52.Hughes A, Smart M, Gorrie-Stone T, Hannon E, Mill J, Bao Y, et al. Socioeconomic position and DNA methylation age acceleration across the life course. Am J Epidemiol. 2018;187(11):2346–2354. doi: 10.1093/aje/kwy155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galkin F, Kochetov K, Koldasbayeva D, Faria M, Fung HH, Chen AX, et al. Psychological factors substantially contribute to biological aging: evidence from the aging rate in Chinese older adults. Aging (Albany, NY) 2022;14(18):7206–7222. doi: 10.18632/aging.204264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao L, Zan G, Liu C, Xu X, Li L, Chen X, et al. Associations between blood pressure and accelerated DNA methylation aging. J Am Heart Assoc. 2022;11(3):e022257. doi: 10.1161/JAHA.121.022257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Australian Bureau of Statistics. Health conditions prevalence. Canberra: Australian Bureau of Statistics; 2020–21 [cited 2023 March 29]. Available from: https://www.abs.gov.au/statistics/health/health-conditions-and-risks/health-conditions-prevalence/latest-release. Accessed 29 Mar 2023
- 56.Australian Institute of Health and Welfare. Heart, stroke and vascular disease: Australian facts. 2023 [cited 2023 March 29]. Available from: https://www.aihw.gov.au/reports/heart-stroke-vascular-diseases/hsvd-facts/contents/risk-factors/high-blood-pressure. Accessed 29 Mar 2023
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from authors upon reasonable request and with permission of the ASPREE principal investigators through the website (www.ASPREE.org). Data will be shared to approved researchers through a secure web-based data portal Safe Haven.