Abstract
To establish the role of the two putative type I leader peptidases (LepB1 and LepB2) encoded in the genome of the cyanobacterium Synechocystis sp. strain PCC 6803, we generated independent knockout mutants for both genes by introducing kanamycin resistance cassettes into the two open reading frames (sll0716 [lepB1] and slr1377 [lepB2], respectively). Although the insertion was successful in both instances, it was not possible to select homozygous mutant cells for lepB2, suggesting that the function of this gene is essential for cell viability. In contrast, LepB1 is apparently essential only for photoautotrophic growth, because homozygous lepB1::Kmr cells could be propagated under heterotrophic conditions. They were even capable to some extent of photosynthetic oxygen evolution. However, the photosynthetic activity decreased gradually with extended incubation in the light and was particularly affected by high light intensities. Both features were indicative of photooxidative damage, which was probably caused by inefficient replacement of damaged components of the photosynthetic machinery due to the lack of a leader peptidase removing the signal peptides from photosynthetic precursor proteins. Indeed, processing of the PsbO precursor polypeptide to the corresponding mature protein was significantly affected in the mutant, and reduced amounts of other proteins that are synthesized as precursors with signal peptides accumulated in the cells. These results strongly suggest that LepB1 is important for removal of the signal peptides after membrane transport of the components of the photosynthetic machinery, which in turn is a prerequisite for the biogenesis of a functional photosynthetic electron transport chain.
The biogenesis of many prokaryotic proteins involves their translocation across membranes. Such proteins are generally synthesized as precursor polypeptides with amino-terminal targeting signals, called signal peptides or leader peptides, which are removed after membrane transport. This processing is performed by specific type I leader peptidases, which are important not only for the generation of the mature proteins but also for their release from the membrane into the target compartment and thus for their correct localization (7, 24, 33). In Escherichia coli, there is a single type I leader peptidase (LepB) which has been shown to be essential for cell viability (8, 13). In contrast, Bacillus subtilis contains five type I leader peptidases that have overlapping substrate specificities and differ in importance for the cell (35). This redundancy suggests that the enzymes have different roles in the cellular processes (4).
In cyanobacteria, there are two independent membrane systems, the cytoplasmic membrane and the thylakoid system, which are targets for proteins carrying signal peptides. Both types of membranes carry leader peptidase activity, as shown, for example, for Phormidium laminosum (2). In line with this, most cyanobacterial genomes analyzed so far contain at least two genes that encode proteins with homology to type I leader peptidases. In Synechocystis sp. strain PCC 6803, the products of the open reading frames sll0716 and slr1377 (15, 21) show significant homology to LepB from E. coli, as well as to the thylakoid processing peptidase of chloroplasts, particularly in the catalytic domains (6). This suggests that these enzymes operate with similar mechanisms, and indeed, there are numerous examples which show that precursor proteins from all three sources (chloroplasts, cyanobacteria, and bacteria) are substrates for the respective heterologous leader peptidases (10, 11, 17, 37).
In order to examine the specific functions of the two putative type I leader peptidases of Synechocystis sp. strain PCC 6803, knockout mutants were generated for both genes by insertion of kanamycin resistance cassettes into the respective open reading frames. Synechocystis sp. strain PCC 6803 is particularly well suited for such experiments, because (i) it is not an obligatory photoautotrophic organism but can also grow heterotrophically if fermentable carbon sources are provided in the medium, (ii) it can be transformed easily with foreign DNA, (iii) it performs homologous recombination at high rates (30), and (iv) the complete genomic sequence is available (14). These features make Synechocystis sp. strain PCC 6803 an appropriate model to study photosynthetic processes at the molecular level.
MATERIALS AND METHODS
Strains and growth conditions.
Unless otherwise indicated, wild-type and mutant strains of Synechocystis sp. strain PCC 6803 were propagated at 30°C under photomixotrophic conditions with a light intensity of 5 microeinsteins m−2 s −1 in BG11 liquid medium (23) supplemented with 5 mM glucose. For growth on plates, agar was added to a concentration of 1.0% (wt/vol). For the growth of all mutant strains, the medium was supplemented with kanamycin (50 μg ml−1). Growth rates were monitored by measuring the optical density at 730 nm.
Mutant construction.
DNA fragments covering parts of the coding regions of lepB1 (sll0716), lepB2 (slr1377), and slr1378 were amplified by PCR by using chromosomal DNA from Synechocystis sp. strain PCC 6803 as the template and the following primers (Fig. 1A): for lepB1, F1 (5′-CCTGCTGCTGCGTTTCTTTGT-3′) and R1 (5′-GGGGTGTCGGGTATTAGGTATTG-3′); for lepB2, F2 (5′-CCCACCAGGAAGAAGAAGAGGAA-3′) and R2 (5′-GAGCAGCAGTGGCGATGGTTTTA-3′); and for slr1378, F3 (5′-CCCAGTGAAAGTGCCCGATG3-′) and R3 (5′-GTGGGCTGCTTTGGTTCCCC-3′). The resulting PCR fragments were cloned with the vector pGEMT-Easy (Promega). A kanamycin resistance gene cassette isolated from plasmid pUC4K (Amersham Biosciences) was introduced into either the HincII restriction site of lepB1, the BamHI restriction site of lepB2, or the HincII restriction site of slr1378 (Fig. 1A). The resulting plasmids were transformed into Synechocystis sp. strain PCC 6803 wild-type cells and selected for growth on kanamycin as described previously (14).
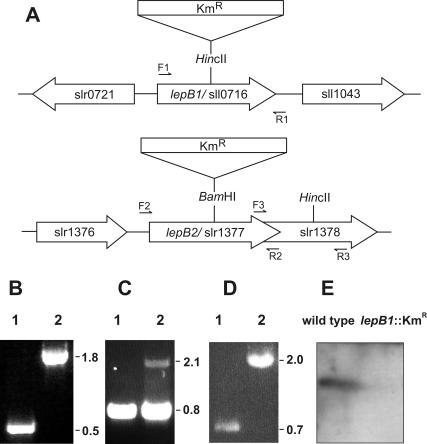
FIG. 1.
Insertional inactivation of the lepB1 and lepB2 genes in Synechocystis sp. strain PCC 6803. (A) Schematic maps of the chromosomal regions covering open reading frames sll0716 and slr1377. The restriction sites used for insertion of the kanamycin resistance cassette (Kmr) are indicated. The positions of the primers used for PCR analysis (forward primers F1, F2, and F3 and reverse primers R1, R2, and R3) are indicated by small arrows. (B to D) Segregation analyses of the lepB1::Kmr (B), lepB2::Kmr (C), and slr1378::Kmr (D) insertion mutants. Chromosomal DNA of wild-type Synechocystis sp. strain PCC 6803 (lanes 1) or the mutants (lanes 2) was amplified by PCR by using primers F1 and R1 (B), primers F2 and R2 (C), and primers F3 and R3 (D) and separated on 1% agarose gels. The sizes of relevant PCR products (in kilobases) are indicated on the right. (E) Western analysis of thylakoid proteins from wild-type and lepB1::Kmr mutant cells. Thylakoid proteins (100 μg of protein per lane) were separated by electrophoresis on SDS-10 to 17.5% polyacrylamide gradient gels, transferred to nylon membranes, and analyzed with antisera raised against LepB1 from Synechocystis sp. strain PCC 6803. For further details see the text.
Determination of photosynthetic electron transport.
Photosynthetic electron transport was measured by using a Clark-type electrode (Hansatech, Norfolk, United Kingdom). The temperature of the electrode chamber was maintained at 30°C throughout all experiments. Saturating red light (RG1-610) was provided by a KL-1500 light source (Schott, Mainz, Germany). The light intensity in the reaction chamber was 1,500 microeinsteins m−2 s−1 in short-term experiments (5 min) or 100 to 500 microeinsteins m−2 s−1 in long-term experiments (30 to 90 min). Electron transport was determined with intact cells resuspended in BG11 medium. The chlorophyll concentrations in the samples were adjusted to 10 and 5 μg ml−1 in short-term experiments and to 0.5 and 0.25 μg ml−1 in long-term experiments for the wild type and the lepB1::Kmr mutant, respectively. Whole-chain electron transport was measured in the presence of 10 mM sodium bicarbonate, and photosystem II-mediated electron transport was measured in the presence of 0.5 mM 2,6-dichloro-p-benzoquinone and 1 mM K3[Fe(CN)6].
Light treatment.
Cells in the late exponential growth phase were harvested by centrifugation and resuspended in fresh BG11 medium. The suspension was subjected to heat-filtered white light (1,000 microeinsteins m−2 s−1), and we were careful that the temperature did not exceed 32°C. Aliquots were collected after 30, 45, 60, and 120 min.
Two-dimensional gel electrophoresis.
Cells from a 50-ml culture in the late exponential growth phase were harvested by centrifugation and resuspended in 300 μl of chloroform supplemented with 0.07% β-mercaptoethanol. The suspension was frozen in liquid nitrogen for 5 min and thawed at room temperature with vigorous mixing. Proteins were precipitated by adding 3 ml of acetone supplemented with 10% trichloroacetic acid and 0.07% β-mercaptoethanol and were solubilized after they were washed with acetone-0.07% β-mercaptoethanol in 200 μl of buffer containing 7 M urea, 2 M thiourea, 2% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), and 2% β-mercaptoethanol. The proteins were subsequently separated on immobilized pH gradient strips (pH 4 to 7; 180 mm; Amersham Biosciences) used according to the manufacturer's instructions. In brief, the immobilized pH gradient strips were rehydrated for 12 h at 20°C in 350 μl (maximum volume) of 8 M urea-19 mM dithiothreitol-2% CHAPS-0.001% bromophenol blue-0.2% carrier ampholytes (Pharmalyte pH 4 to 7; Amersham Biosciences) supplemented with 200 μg of the solubilized protein. Isoelectric focusing was carried out with an IPGphor device (Amersham Biosciences) at 20°C at 500 V for 1 h, at 1,000 V for 1 h, and at 8,000 V for 4 h. After electrophoresis, the proteins on the strips were equilibrated for 15 min in 50 mM Tris-HCl (pH 6.8)-6 M urea-30% (vol/vol) glycerol-2% (wt/vol) sodium dodecyl sulfate (SDS)-1% (wt/vol) dithiothreitol, followed by 10 min in 50 mM Tris-HCl (pH 6.8)-6 M urea-30% (vol/vol) glycerol-2% (wt/vol) SDS-2% iodine acetamide-0.001% bromophenol blue. After this, the strips were placed on top of SDS-polyacrylamide gels, and the proteins separated in the second dimension on the basis of their molecular masses. After Coomassie brilliant blue staining, the gels were scanned and analyzed by using the ImageMaster 2D Elite software package (Amersham Biosciences).
In-gel proteolysis and mass spectrometry.
The protein spots were cut out of the gels and then washed three times with water, twice with 50 mM ammonium bicarbonate, and finally with 50 mM ammonium bicarbonate in 50% acetonitrile. The gel pieces were dried under a gentle stream of nitrogen, reswollen in 20 μl of 50 mM ammonium bicarbonate (pH 8.0), and incubated with trypsin (Promega, Madison, Wis.) overnight at 37°C.
Matrix-assisted laser desorption ionization mass spectra were recorded with a Bruker REFLEX II mass spectrometer (Bruker-Daltonik, Bremen, Germany) upgraded with a SCOUT ion source and pulsed ion extraction. Data were analyzed with the XMASS software supplied with the spectrometer. For analysis of the tryptic digests a matrix thin-layer preparation was made (31). A saturated solution of α-cyano-4-hydroxycinnamic acid in acetone was mixed at a 4:1 (vol/vol) ratio with a 10-mg ml−1 solution of nitrocellulose (Transblot transfer medium; Bio-Rad, Hercules, Calif.) in acetone. Then 0.5 μl of the matrix was deposited on the sample probe. One microliter of the sample was injected into a small drop of 1% trifluoroacetic acid previously deposited onto the matrix surface in order to prevent dissolution of the matrix layer due to the basic pH of the digest solution. The sample was allowed to dry, and the dried spot was rinsed three times with 10 μl of 0.1% trifluoroacetic acid. Mass spectra were calibrated by using trypsin autolysis products as internal standards.
The peptide mass fingerprint spectra were searched against the nonredundant protein database (NCBInr) for exact matches by using the MASCOT search program (Matrix Science Ltd., London, United Kingdom) or MS-FIT (Protein Prospector; University of California at San Francisco, San Francisco, Calif.).
To confirm the results, peptide maps were desalted with ZipTip C18 (Millipore Corp., Bedford, Mass.), and mass spectrometry-mass spectrometry spectra were recorded with an ESI-Q-TOF (electrospray ionization quadrupole time-of-flight) mass spectrometer (Micromass, Manchester, United Kingdom) equipped with a nanospray source.
Miscellaneous.
Total membranes were isolated from Synechocystis sp. strain PCC 6803 cells as described previously (34) by using thylakoid buffer (20). The chlorophyll concentration was determined as described by Lichtenthaler (19). Gel electrophoresis of proteins under denaturing conditions was carried out as described by Laemmli (18). After staining, the gels were analyzed by using the Image Quant software package (Amersham Biosciences). Western blot analysis was performed as described by Vachereau (36). For all other methods we used the protocols described by Sambrook et al. (25).
RESULTS
Insertional knockout of the two genes encoding leader peptidases in Synechocystis sp. strain PCC 6803.
Synechocystis sp. strain PCC 6803 possesses two genes that presumably encode leader peptidases, sll0716 and slr1377 (15, 21), which are referred to here as lepB1 and lepB2, respectively. In order to examine the functions of these two putative peptidases, we independently inactivated both genes by inserting a kanamycin resistance cassette into the two open reading frames via homologous recombination (Fig. 1A). After transformation, the cells were propagated under dim light on agar plates supplemented with glucose and kanamycin. In order to promote complete genetic segregation of the mutated gene, the concentration of kanamycin in the medium was increased stepwise. After several rounds of restreaking, the insertionally inactivated lepB1 gene had fully segregated, which resulted in the homozygous lepB1::Kmr mutant. PCR analysis showed that the wild-type lepB1 gene, which was represented by a 0.5-kb fragment, was completely replaced by the mutant allele, as indicated by a 1.8-kb PCR fragment (Fig. 1B). In line with this, Western analyses performed with antisera raised against the gene product of lepB1 confirmed that a protein with a molecular mass of approximately 21 kDa found in wild-type cells, whose size corresponded well with the size predicted for LepB1 (22 kDa), was not present in the mutant cells (Fig. 1E).
In contrast, it was not possible with this approach to completely eliminate the wild-type copy of lepB2. While insertional inactivation of the gene by homologous recombination was also successful in this case, the cells remained heterozygous even if the concentration of kanamycin in the growth medium was raised to 200 μg ml−1 to select for higher copy numbers of the mutant allele (Fig. 1C). This suggests that LepB2 is essential for the viability of Synechocystis sp. strain PCC 6803. However, since the open reading frame of lepB2 overlaps by 49 nucleotides the open reading frame of the adjacent gene (slr1378) (Fig. 1A), which encodes a protein with an unknown function (15, 21), we could not rule out the possibility that insertional inactivation of lepB2 had a polar effect on the transcription of slr1378, which itself might be essential for growth. In order to examine this possibility, slr1378 was inactivated by insertion of a kanamycin resistance cassette as described above. PCR analysis of cultures selected on kanamycin-containing media showed that the homozygous slr1378::Kmr mutant could be obtained easily (Fig. 1D). The cells did not exhibit a phenotype different from that of wild-type cells (data not shown), which clearly demonstrated that it must have been the function of lepB2 and not any polar effect on the expression of slr1378 which prevented the elimination of the wild-type copy of slr1377. Thus, since one of the two leader peptidases of Synechocystis sp. strain PCC 6803 could not be completely inactivated without a loss of cell viability, we concluded that the functions of the two homologous proteins are not redundant.
Homozygous lepB1::Kmr cells are sensitive to high light intensities.
Since it was impossible to fully inactivate the wild-type allele of lepB2, in all further work we focused on analysis of the homozygous lepB1::Kmr mutant. This mutant differed significantly from the wild type with respect to its growth behavior. Under photoautotrophic conditions, the mutant did not show any measurable growth (Table 1), indicating that it was not able to perform photosynthesis at significant rates. Furthermore, even if the cultures were supplemented with fermentable carbon sources like glucose (photomixotrophic conditions), the mutant cells could be propagated only if they were exposed to dim light (5 microeinsteins m−2 s−1) which was not sufficient for efficient photosynthesis (1). Under these conditions, the growth rates of the wild-type and mutant cultures were essentially identical (Table 1), demonstrating that the mutant was not affected in energy metabolism in general. However, if the lepB1::Kmr mutant was incubated in the presence of glucose but at a light intensity that allowed efficient photosynthesis (40 microeinsteins m−2 s−1), growth was again completely abolished (Table 1).
TABLE 1.
Photosynthetic properties of the wild type and the homozygous lepB1::Kmr mutant
| Property | Wild type | lepB1::Kmr mutant |
|---|---|---|
| Doubling times (h) | ||
| Photoautotrophic growth (without glucose and DCMU; 40 microeinsteins m−2 s−1) | 12.3 ± 1.3 | No growth |
| Photomixotrophic growth (with glucose and without DCMU; 5 microeinsteins m−2 s−1) | 20.9 ± 2.2 | 20.9 ± 1.0 |
| Photomixotrophic growth (with glucose and without DCMU; 40 microeinsteins m−2 s−1) | 8.0 ± 0.9 | No growth |
| Photoheterotrophic growth (with glucose and DCMU; 40 microeinsteins m−2 s−1) | 16.2 ± 1.8 | 26.6 ± 1.2 |
| Pigment characteristics | ||
| Chlorophyll content (μg ml−1A730−1)a | 3.8 ± 0.2 | 1.7 ± 0.1 |
| Phycobiliprotein/chlorophyll ratiob | 0.85 | 1.16 |
| Oxygen evolution (μmol of O2A730−1 liter−1 h−1)c | ||
| Whole-chain electron transport | 995 ± 53 | 783 ± 51 |
| Photosystem II-mediated electron transport | 1,956 ± 173 | 1,543 ± 187 |
Chlorophyll was extracted from cells grown under photomixotrophic conditions at 5 microeinsteins m−2 s−1.
Determined from intact cells by using the equation of Richaud et al. (22).
Determined in short-term experiments (5 min) with cells grown in photomixotrophic conditions at 5 microeinsteins m−2 s−1.
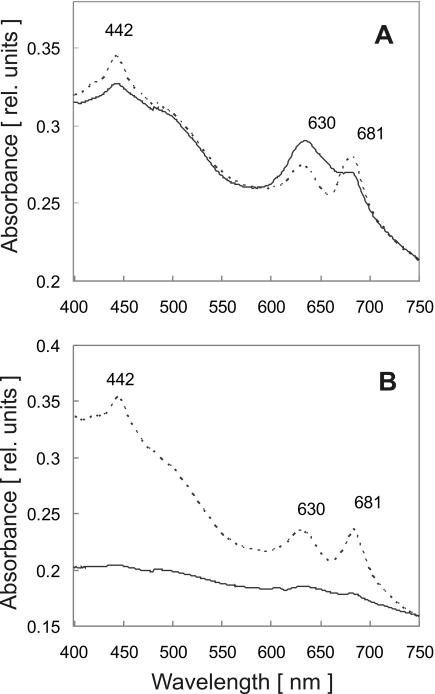
This indicated that the mutant was not only incapable of performing photosynthesis but that light, even at relatively moderate intensities, was harmful for the cells. The light sensitivity of the mutant was also reflected by the variability of its pigment content in response to different light intensities. If the mutant was propagated in dim light (5 microeinsteins m−2 s−1), all photosynthetic pigments were present, although the composition was somewhat different from that of wild-type cells (Table 1 and data not shown). In particular, the chlorophyll content was reduced (Fig. 2A), which led to an increase in the phycobiliprotein/chlorophyll ratio from 0.85 in the wild type to 1.16 in mutant cells (Table 1) and, consequently, to a bluish phenotype. After transfer of the cultures to higher light intensities (40 microeinsteins m−2 s−1 or more), significant bleaching of the mutant cells was observed, which was presumably caused by degradation of the photosynthetic pigments. This was confirmed by spectral analyses which demonstrated that there was a complete absence of the absorption peaks for chlorophyll (at 442 and 681 nm) and phycobiliproteins (at 630 nm) in the mutant samples (Fig. 2B). Remarkably, supplementation of the medium with 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), which blocked photosynthetic electron transport from photosystem II to plastoquinone, prevented this light-induced bleaching of the mutant cells and furthermore restored the ability to grow under these conditions, although the growth rate was somewhat reduced compared to that of wild-type cultures (Table 1). This suggested that the electron flow from photosystem II caused photodestruction in the mutant. It is worth noting in this context that a similar phenotype (i.e., light sensitivity which was suppressed by inhibition of photosystem II) was described for mutants which either lacked photosystem I (28) or showed an imbalance in the ratio of the two photosystems (32).
FIG. 2.
Light sensitivity of photosynthetic pigments in the lepB1::Kmr mutant. Cultures of wild-type Synechocystis sp. strain PCC 6803 (dotted line) and the lepB1::Kmr mutant (solid line) were grown under photomixotrophic conditions in dim light (5 microeinsteins m−2 s−1). The absorption spectra from identical amounts of cells were determined either directly (A) or after exposure of the cultures to a light intensity of 40 microeinsteins m−2 s−1 for 2 days (B). The absorption maxima for chlorophyll a (442 and 681 nm) and phycocyanin (630 nm) are indicated. rel. units, relative units.
Photosynthetic electron transport in the lepB1::Kmr mutant is inhibited by high light.
In order to examine the photosynthetic properties of the mutant in more detail, photosynthetic oxygen evolution was determined for wild-type and mutant cells that were grown photomixotrophically at a low light intensity (5 microeinsteins m−2 s−1). Unexpectedly, the mutant was able to perform photosynthetic electron transport rather efficiently in short-term experiments. When identical cell numbers were compared, the level of photosynthetic oxygen production of the mutant was approximately 80% of the wild-type level, irrespective of whether the entire photosynthetic electron transport chain or only photosystem II-mediated electron transport was measured for 5 min (Table 1). Thus, despite the lack of competence to grow photoautotrophically, the mutant was competent for photosynthetic electron transport, which indicated that the photosynthetic complexes of the thylakoid membrane were able to operate.
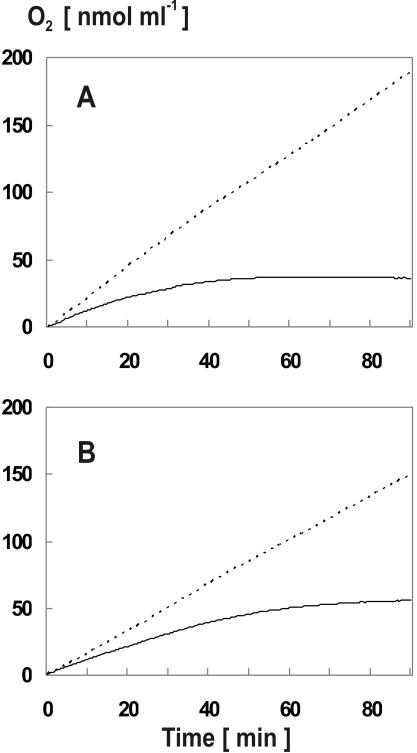
However, photosynthetic electron transport was inhibited in the mutant cells by high light intensities, as shown by two complementary approaches. In the first approach, measurement of the entire photosynthetic electron transport chain was repeated with diluted culture samples, which allowed us to determine photosynthetic oxygen evolution with the Clark electrode for extended periods. The idea behind this experiment was to potentially identify effects on electron transport which became visible only after long-term exposure of the cells to high light intensities (e.g., 500 microeinsteins m−2 s−1 in the example shown in Fig. 3A). Indeed, with this experimental design stronger differences between mutant and wild-type cells became apparent. While oxygen production in wild-type cultures remained essentially unaltered during the entire incubation time, the mutant cells showed a clear drop in oxygen evolution after approximately 20 min (Fig. 3A). When cells were incubated for more than 60 min, photosynthetic oxygen production was even terminated in the mutant culture. Similar results were obtained when the experiment was performed with more moderate light intensities (e.g., 100 microeinsteins m−2 s−1), except that the photoinhibitory effect was milder and slightly retarded (Fig. 3B). Taken together, these data strongly suggested that extended exposure to photosynthetic light inactivated the photosynthetic electron transport machinery of the mutant cells.
FIG. 3.
High light intensities lead to inactivation of photosynthetic oxygen evolution in the lepB1::Kmr mutant. The photosynthetic oxygen evolution by wild-type (dotted line) and lepB1::Kmr cultures (solid line) which were exposed to continuous light at an intensity of either 500 microeinsteins m−2 s−1 (A) or 100 microeinsteins m−2 s−1 (B) was determined for identical amounts of cells by using a Clark electrode.
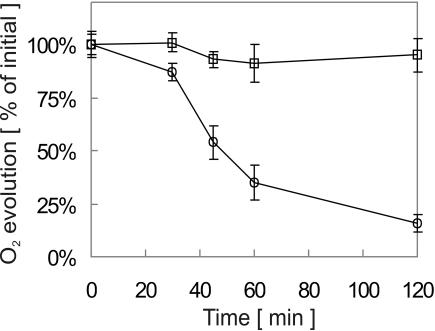
This was confirmed by an independent approach in which the oxygen evolution was determined for cultures that were exposed to light stress (30 to 120 min at 1,000 microeinsteins m−2 s−1) prior to the actual measurement. While the wild type was only mildly affected by this treatment, exhibiting more than 90% of the initial photosynthetic activity even after 120 min of light stress, the activity of the mutant declined gradually with increasing incubation time (Fig. 4). After 120 min, the residual photosynthetic activity was reduced to approximately 15% of that obtained with untreated samples. From these data we concluded that the mutant cells were able to assemble a functional photosynthetic electron transport chain when they were grown in dim light but that they were apparently not able to preserve its activity when they were exposed to high light intensities.
FIG. 4.
Photosynthetic oxygen evolution by lepB1::Kmr mutant cells is inhibited by light stress. Cultures of wild-type Synechocystis sp. strain PCC 6803 (□) and the lepB1::Kmr mutant (○) propagated under photomixotrophic conditions in dim light (5 microeinsteins m−2 s−1) were exposed to light stress (1,000 microeinsteins m−2 s−1). Samples containing identical amounts of cells were taken at different times, and photosynthetic oxygen evolution was immediately determined for 5 min by using a Clark electrode. The value for each time is the average of at least five independent measurements. The error bars indicate the standard errors.
Processing of PsbO is affected in the lepB1 mutant.
The gene analyzed here, lepB1, encodes a peptidase required for the removal of signal peptides from precursor proteins that need to cross a membrane to reach their final destination in the cell. Considering the phenotype of the mutant described so far, it appears likely that LepB1 plays an important role in the processing of precursor proteins from the photosynthetic machinery of the thylakoid membrane. This function can be assumed to be essential for the biogenesis of the photosynthetic electron transport chain and thus to be particularly important under conditions in which the photosynthetic machinery is damaged (for example, after photooxidation of the membrane complexes by light stress). Under these circumstances, the number of defective protein subunits which need to be replaced is significantly increased.
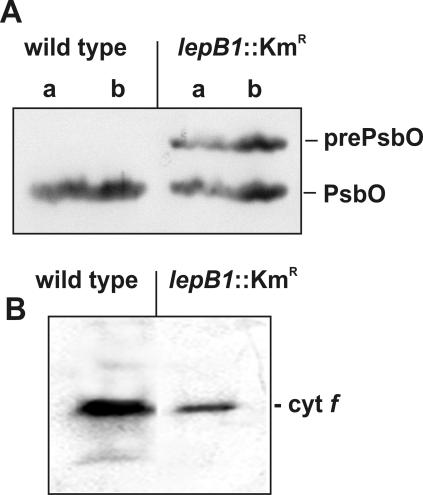
As an initial step to examine whether LepB1 is indeed involved in the processing of thylakoid proteins, Western analyses were performed for the PsbO subunit of the oxygen-evolving system associated with photosystem II. In Synechocystis, this protein is synthesized as a precursor polypeptide carrying a signal peptide for translocation across the thylakoid membrane (9). In wild-type cells, a single protein was recognized by the antisera which were raised against PsbO from spinach (Fig. 5A). Based on its mobility during SDS-polyacrylamide gel electrophoresis (PAGE), the size of this protein was estimated to be approximately 29 kDa, which is in line with the molecular mass predicted for mature PsbO (26.5 kDa). This protein was also present in the lepB1 mutant. However, in these samples a second polypeptide was detected by the antibodies, and the molecular mass of this polypeptide was approximately 32 kDa. This polypeptide probably represented the unprocessed precursor of PsbO which still carried the PsbO signal peptide with a predicted molecular mass of 3 kDa, indicating that LepB1 plays an important, although not essential, role in the maturation of PsbO.
FIG. 5.
Western analysis of thylakoid proteins in wild-type and lepB1::Kmr cells. Thylakoid proteins isolated from cultures propagated under photomixotrophic conditions in dim light were separated by electrophoresis on SDS—10 to 17.5% polyacrylamide gradient gels, transferred to nylon membranes, and analyzed with antisera raised against PsbO (A) or cytochrome f (B) from spinach. The positions of the presumed precursor and mature proteins are indicated on the right. Lanes a, 50 μg of protein; lanes b, 100 μg of protein.
Remarkably, not all thylakoid proteins carrying cleavable signal peptides were affected in maturation in cells lacking LepB1. Cytochrome f, for example, which also is synthesized as a precursor polypeptide with a thylakoid-targeting signal peptide, did not show any accumulation of precursor in the mutant cells when it was analyzed in the same manner as PsbO (Fig. 5B), although it was shown previously to be stable also in its precursor form (34). Thus, we concluded that two leader peptidases with overlapping but not identical substrate specificities are present in the thylakoid membrane system of Synechocystis sp. strain PCC 6803.
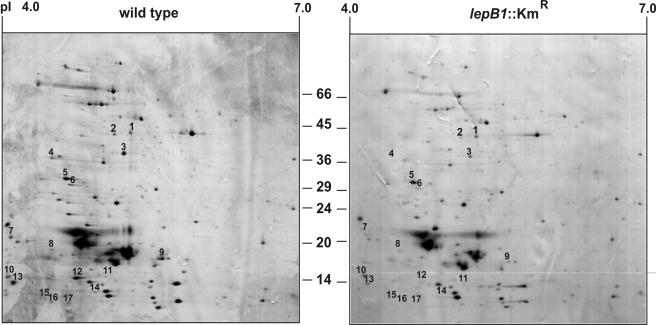
Since other heterologous antisera raised against thylakoid proteins that are synthesized with cleavable signal peptides (plastocyanin, PsaF, and CFoII) did not specifically recognize their cyanobacterial homologs when they were tested in such experiments (data not shown), two-dimensional analyses in which isoelectric focusing and SDS-PAGE were combined were used as an alternative approach to examine the effect of the mutation on the protein complement in the cells. With such assays, it was possible to simultaneously analyze approximately 200 protein species from total cells (Fig. 6). A comparison of the protein patterns obtained for the wild type and mutant in five independent experiments revealed that in this group the accumulation of 16 polypeptides was always significantly reduced in the mutant cells (Table 2). Eight of these proteins were subsequently identified by mass spectrometry, and four of them turned out to be synthesized with signal peptides (Table 2). Since only about 10% of the proteins encoded by Synechocystis sp. strain PCC 6803 are predicted to carry signal peptides (Zhbanko and Klösgen, unpublished data), four of eight proteins appeared to be a remarkably high number, although it is obvious that the limited size of the sample did not allow any serious statistical evaluation.
FIG. 6.
Two-dimensional analysis of total proteins from Synechocystis sp. strain PCC 6803. Total proteins were isolated from wild-type and lepB1::Kmr cells grown under photomixotrophic conditions in dim light and were separated in two dimensions by isoelectric focusing (pI 4.0 to 7.0) and SDS-polyacrylamide gel electrophoresis (10 to 17.5% polyacrylamide gradient gel). In each case, 200 μg of protein was loaded. Examples of gels obtained with proteins isolated from either wild-type cells (left panel) or lepB1::Kmr cells (right panel) are shown after staining with Coomassie brilliant blue. The numbers indicate protein spots that in five independent experiments always showed significant differences. These spots were recovered and analyzed by matrix-assisted laser desorption ionization—time of flight mass spectrometry (Table 2).
TABLE 2.
Proteins with altered accumulation in homozygous lepB1::Kmr cells
| Spot no. | pI | Mol mass (kDa) | Open reading frame | Protein | Localization | Size of signal peptide (amino acids) |
|---|---|---|---|---|---|---|
| Reduced amt in the mutant | ||||||
| 1 | 5.29 | 43.2 | NIa | |||
| 2 | 5.13 | 42.8 | slr0394 | Phosphoglycerate kinase | Cytoplasm | |
| 3 | 5.23 | 36.8 | slr0513 | FutA2 (iron-binding transporter) | Periplasm | 31 |
| 4 | 4.49 | 35.2 | NI | |||
| 5 | 4.65 | 30.1 | sll0427b | PsbO (33-kDa protein of photosystemII) | Thylakoid lumen | 28 |
| 6 | 4.69 | 28.8 | sll1784 | Unknown protein | Periplasm | 33c |
| 7 | 4.08 | 19.5 | NI | |||
| 8 | 4.49 | 17.5 | NI | |||
| 9 | 5.6 | 16.3 | sll1578 | CpcA (phycocyanin α chain) | Phycobilisome | |
| 11 | 4.04 | 14.3 | NI | |||
| 12 | 4.74 | 14.0 | sll1746 | Rpl12 (ribosomal protein L12) | Cytoplasm | |
| 13 | 4.1 | 13.7 | sll1194 | PsbU (12-kDa protein of photosystem II) | Thylakoid lumen | 36 |
| 14 | 4.93 | 12.6 | NI | |||
| 15 | 4.41 | 12.0 | NI | |||
| 16 | 4.48 | 11.7 | ssl2501 | Unknown protein | ||
| 17 | 4.66 | 11.6 | NI | |||
| Increased amt in the mutant | ||||||
| 10 | 5.04 | 14.3 | NI |
NI, not identified.
Confirmed by Western blotting.
Determined by using SignalP (http://www.cbs.dtu.dk/services/SignalP-2.0).
Still, these results allow several interesting conclusions. First, processing of neither of the precursor proteins identified is completely abolished in the mutant, which is consistent with the results obtained by Western analysis for PsbO (Fig. 5A). Instead, accumulation of only the terminal processing products is reduced, suggesting that an independent processing activity, presumably provided by LepB2, can replace the function of LepB1 to some extent. Second, not all of the proteins affected are components of the thylakoid system. FutA2, for example, is an iron-binding protein that is located in the plasma membrane of Synechocystis sp. strain PCC 6803 (12). This indicates that LepB1 is probably involved in the processing of proteins from both membrane systems present in cyanobacteria, which in turn suggests that the peptidase is localized in both the thylakoid and plasma membranes. Finally, among the polypeptides that accumulated to a lesser extent in the mutant are cytosolic proteins, like phosphoglycerate kinase (Table 2), whose function is not directly related to functions associated with the thylakoid system or the periplasmic space of the cells. So far, it is not known why the accumulation of the proteins is affected in the absence of LepB1, and whether this is a direct consequence of the mutation or a secondary effect remains to be determined.
DISCUSSION
In the present work, we examined the role of the two putative type I leader peptidases that are encoded in Synechocystis sp. strain PCC 6803 by the lepB1 (sll0716) and lepB2 (slr1377) genes (15, 21). By using site-specific insertion mutagenesis, both genes were independently knocked out, and the resulting cells were selected for complete loss of the wild-type alleles. Interestingly, homozygous null mutants could be isolated only for lepB1, whereas it was impossible with this approach to eliminate all wild-type copies of lepB2. This indicates that LepB2 is essential for the viability of Synechocystis sp. strain PCC 6803 and cannot functionally be replaced by LepB1. Likewise, full functional complementation was also not obtained in the reciprocal situation, since homozygous lepB1::Kmr cells lost the ability to grow photoautotrophically. Thus, despite the fact that both enzymes belong to the same class of leader peptidases (6), they are apparently not redundant but instead have functions that are at least partially specialized in the cell.
There could be various reasons for this lack of complementation. The lack of complementation might, for example, reflect different substrate specificities of the two enzymes. Since under all culture conditions analyzed LepB2 was essential for growth, the protein must be assumed to have important housekeeping functions, presumably in the processing of essential components of the cyanobacterial membrane systems. In contrast, LepB1 is dispensable for growth under heterotrophic conditions but is required both for photoautotrophic growth and for protection against light stress. This suggests that the enzyme is preferentially involved in the maturation of the components of the photosynthetic machinery, and, indeed, the PsbO subunit of the oxygen-evolving system associated with photosystem II could be identified as a putative substrate (Fig. 5). Substrate specificity might, however, also be mimicked by different localizations of the two leader peptidases in the cyanobacterial membranes. In line with this, LepB2 was identified in cell fractions that were enriched for the inner plasma membrane of Synechocystis sp. strain PCC 6803, whereas LepB1 was not found in these samples (12), suggesting that it is located in the thylakoid membranes instead. However, this apparent difference in the subcellular topology of the two leader peptidases cannot be totally stringent, because (i) one of the presumed substrates of LepB1 is a periplasmic protein, FutA (Table 2), and (ii) processing of thylakoid proteins like PsbO or cytochrome f is not completely abolished in homozygous lepB1::Kmr cells (Fig. 5).
A striking feature of the homozygous lepB1::Kmr mutant is its inability to grow photoautotrophically, despite the fact that it is able to perform photosynthetic electron transport at significant rates (Table 1). This electron transport takes place, however, only for short periods of time. During extended exposure of the mutant cells to photosynthetic light, oxygen evolution gradually declines and finally ends (Fig. 3). This suggests that the mutant is highly sensitive to light, which is indicative of photooxidative stress. In line with this, the mutant is able to grow even in the presence of carbohydrates either if it is kept in dim light or if photosynthetic electron transport is inhibited by DCMU (Table 1), which blocks the QB binding site in photosystem II. Interestingly, a similar phenotype was described for mutants affected in photosystem I. Like the lepB1::Kmr mutant, these strains showed enhanced light sensitivity which was suppressed if the activity of photosystem II was decreased either by addition of DCMU or by a reduction in the size of the light-harvesting antenna system associated with photosystem II (28). A similar effect of DCMU, notably the restoration of growth in high-light conditions, was observed for a pmgA mutant (32). This strain was not able to properly adapt the stoichiometry of the two photosystems in response to different light conditions, which presumably led to an imbalance in the photosynthetic electron flow. The lower cytochrome f accumulation in the lepB1 mutant (Fig. 5B) likewise indicates that there is an imbalance in the photosynthetic complexes. However, since blue native PAGE, which allows workers to separate membrane complexes in essentially intact forms (3, 26, 27), did not reveal significant differences in the accumulation of the thylakoid protein complexes of the wild type and mutant (Zhbanko and Klösgen, unpublished data), major alterations of the two photosystems appear to be unlikely. Thus, the cause of the photoinhibition observed with lepB1::Kmr cells is still not known.
Therefore, further work is required to determine the primary cause of the lack of photoautotrophic growth of the lepB1 mutant. PsbO, PsbU, and FutA2, all of which accumulated in reduced amounts in the mutant (Table 2), were shown to be not essential for photoautotrophy of Synechocystis sp. strain PCC 6803 (5, 16, 29) and thus cannot be the cause of the defects observed. The functions of other proteins that are affected (for example, the gene product of sll1784) are not known. Furthermore, only a very limited number of proteins have been examined so far. Considering the complexity of the processes involved in the biogenesis of the photosynthetic apparatus, it will therefore not be trivial to finally answer these questions.
Acknowledgments
We gratefully acknowledge the support of Sergey Shestakov, who was one of the initiators of this project. We thank Richard Malkin for providing anti-cytochrome f antibodies and Gerd Hause and Mario Jakob for their help with various methods, as well as for stimulating discussions.
This work was supported by grants from the Russian Foundation for Basic Research (grant 04-04-48200) and the Russian Program of Leading Scientific Institutions to V.Z., as well as by a grant in the framework of the Sonderforschungsbereich 363 provided by the German Research Foundation (DFG) to R.B.K.
REFERENCES
- 1.Anderson, S. L., and L. McIntosh. 1991. Light-activated heterotrophic growth of the cyanobacterium Synechocystis sp. strain PCC 6803: a blue-light-requiring process. J. Bacteriol. 173:2761-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbrook, A. C., J. C. L. Packer, and C. J. Howe. 1993. Components of the protein translocation machinery in the thermophilic cyanobacterium Phormidium laminosum. Biochem. Biophys. Res. Commun. 197:874-877. [DOI] [PubMed] [Google Scholar]
- 3.Berghöfer, J., and R. B. Klösgen. 1999. Two distinct translocation intermediates can be distinguished during protein transport by the TAT (delta pH) pathway across the thylakoid membrane. FEBS Lett. 460:328-332. [DOI] [PubMed] [Google Scholar]
- 4.Bonnemain, C., C. Raynaud, H. Reglier-Poupet, I. Dubail, C. Frehel, M. A. Lety, P. Berche, and A. Charbit. 2004. Differential roles of multiple signal peptidases in the virulence of Listeria monocytogenes. Mol. Microbiol. 51:1251-1266. [DOI] [PubMed] [Google Scholar]
- 5.Burnap, R. L., and L. A. Sherman. 1991. Deletion mutagenesis in Synechocystis sp. PCC6803 indicates that the Mn-stabilizing protein of photosystem II is not essential for O2 evolution. Biochemistry 30:440-446. [DOI] [PubMed] [Google Scholar]
- 6.Chaal, B. K., R. M. Mould, A. C. Barbrook, J. C. Gray, and C. J. Howe. 1998. Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana. Implications for the origin and catalytic mechanism of the enzyme. J. Biol. Chem. 273:689-692. [DOI] [PubMed] [Google Scholar]
- 7.Dalbey, R. E., and W. Wickner. 1985. Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J. Biol. Chem. 260:15925-15931. [PubMed] [Google Scholar]
- 8.Date, T. 1983. Demonstration by a novel genetic technique that leader peptidase is an essential enzyme of Escherichia coli. J. Bacteriol. 154:76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson, J. M., and J. D. Rochaix. 1992. The molecular biology of photosystem II, p. 101-177. In The photosystems: structure, function and molecular biology, vol. 11. Elsevier Press, Amsterdam, The Netherlands. [Google Scholar]
- 10.Halbig, D., B. Hou, R. Freudl, G. A. Sprenger, and R. B. Klösgen. 1999. Bacterial proteins carrying twin-R signal peptides are specifically targeted by the ΔpH-dependent transport machinery of the thylakoid membrane system. FEBS Lett. 447:95-98. [DOI] [PubMed] [Google Scholar]
- 11.Halpin, C., P. D. Elderfield, H. E. James, R. Zimmermann, B. Dunbar, and C. Robinson. 1989. The reaction specificities of the thylakoidal processing peptidase and Escherichia coli leader peptidase are identical. EMBO J. 8:3917-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang, F., I. Parmryd, F. Nilsson, A. L. Persson, H. B. Pakrasi, B. Andersson, and B. Norling. 2002. Proteomics of Synechocystis sp. strain PCC 6803. Mol. Cell. Proteomics 1:956-966. [DOI] [PubMed] [Google Scholar]
- 13.Inada, T., D. L. Court, K. Ito, and Y. Nakamura. 1989. Conditionally lethal amber mutations in the leader peptidase gene of Escherichia coli. J. Bacteriol. 171:585-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivleva, N. B., S. V. Shestakov, and H. B. Pakrasi. 2000. The carboxyl-terminal extension of the precursor D1 protein of photosystem II is required for optimal photosynthetic performance of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 124:1403-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 16.Katoh, H., N. Hagino, A. R. Grossman, and T. Ogawa,. 2001. Genes essential to iron transport in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 183:2779-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuwabara, T., R. Nagata, and K. Shinohara. 1989. Expression and processing of cyanobacterial Mn-stabilizing protein in Escherichia coli. Eur. J. Biochem. 186:227-232. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lichtenthaler, H. K. 1987. Chlorophylls and carotenoids, the pigments of photosynthetic biomembranes. Methods Enzymol. 148:350-382. [Google Scholar]
- 20.Mi, D., S. Lin, and R. E. Blankenship. 1999. Picosecond transient absorption spectroscopy in the blue spectral region of photosystem I. Biochemistry 38:15231-15237. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, Y., T. Kaneko, M. Hirosawa, N. Miyajima, and S. Tabata. 1998. CyanoBase, a www database containing the complete nucleotide sequence of the genome of Synechocystis sp. strain PCC6803. Nucleic Acids Res. 26:63-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Richaud, C., G. Zabulon, A. Joder, and J. C. Thomas. 2001. Nitrogen or sulfur starvation differentially affects phycobilisome degradation and expression of the nblA gene in Synechocystis strain PCC 6803. J. Bacteriol. 183:2989-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 24.Sambasivarao, D., H. A. Dawson, G. Zhang, G. Shaw, J. Hu, and J. H. Weiner. 2001. Investigation of Escherichia coli dimethyl sulfoxide reductase assembly and processing in strains defective for the sec-independent protein translocation system membrane targeting and translocation. J. Biol. Chem. 276:20167-20174. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schägger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 27.Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223-231. [DOI] [PubMed] [Google Scholar]
- 28.Shen, G., S. Boussiba, and W. F. J. Vermaas. 1993. Synechocystis sp. PCC 6803 strains lacking photosystem I and phycobilisome function. Plant Cell 5:1853-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen, J. R., M. Ikeuchi, and Y. Inoue. 1997. Analysis of the psbU gene encoding the 12-kDa extrinsic protein of photosystem II and studies on its role by deletion mutagenesis in Synechocystis sp. PCC 6803. J. Biol. Chem. 272:17821-17826. [DOI] [PubMed] [Google Scholar]
- 30.Shestakov, S. V., and J. Reaston. 1987. Gene-transfer and host vector systems of cyanobacteria. Oxford Surv. Plant Mol. Cell Biol. 4:137-166. [Google Scholar]
- 31.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 32.Sonoike, K., Y. Hihara, and M. Ikeuchi. 2003. Physiological significance of the regulation of photosystem stoichiometry upon high light acclimation of Synechocystis sp. PCC 6803. Plant Cell Physiol. 42:379-384. [DOI] [PubMed] [Google Scholar]
- 33.Thony-Meyer, L., and P. Künzler. 1997. Translocation to the periplasm and signal sequence cleavage of preapocytochrome c depend on sec and lep, but not on the ccm gene products. Eur. J. Biochem. 246:794-799. [DOI] [PubMed] [Google Scholar]
- 34.Tichy, M., and W. Vermaas. 1999. Accumulation of pre-cytochrome f in a Synechocystis sp. PCC 6803 mutant impaired in cytocherome c maturation. J. Biol. Chem. 274:32396-32401. [DOI] [PubMed] [Google Scholar]
- 35.Tjalsma, H., A. Bolhuis, M. L. van Roosmalen, T. Wiegert, W. Schumann, C. P. Broekhuizen, W. J. Quax, G. Venema, S. Bron, and J. M. van Dijl. 1998. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 12:2318-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vachereau, A. 1989. Luminescent immunodetection of Western-blotted proteins from Coomassie-stained polyacrylamide gel. Anal. Biochem. 179:206-208. [DOI] [PubMed] [Google Scholar]
- 37.Wexler, M., E. G. Bogsch, R. B. Klösgen, T. Palmer, C. Robinson, and B. C. Berks. 1998. Targeting signals for a bacterial Sec-independent export system direct plant thylakoid import by the delta pH pathway. FEBS Lett. 431:339-342. [DOI] [PubMed] [Google Scholar]