Abstract
Amyloid-β (Aβ) and tau are important biomarkers to predict the progression of cognitively unimpaired (CU) to dementia due to Alzheimer’s disease (AD), according to the diagnosis framework from the US National Institute on Aging and the Alzheimer’s Association (NIA-AA). However, it is clinically difficult to predict those subjects who were already with Aβ positive (A +) or tau positive (T +). As a typical characteristic of neurodegeneration in the diagnosis framework, the hypometabolism of the posterior cingulate cortex (PCC) has significant clinical value in the early prediction and prevention of AD. In this paper, we proposed the glucose metabolism in the PCC as a biomarker supplement to Aβ and tau biomarkers. First, we calculated the standard uptake value ratio (SUVR) of PCC based on fluorodeoxyglucose positron emission computed tomography (FDG PET) imaging. Secondly, we performed Kaplan–Meier (KM) survival analyses to explore the predictive performance of PCC SUVR, and the hazard ratio (HR) was calculated. Finally, we performed Pearson correlation analyses to explore the physiological significance of PCC SUVR. As a result, the PCC SUVR showed a consistent downward trend along the AD continuum. KM analyses showed better predictive performance when we combined PCC SUVR with cerebro-spinal fluid (CSF) Aβ42 (from HR = 2.56 to 3.00 within 5 years; from HR = 2.76 to 4.20 within 10 years) and ptau-181 (from 2.83 to 3.91 within 5 years; from HR = 2.32 to 4.17 within 10 years). There was a slight correlation between Aβ42/Aβ40 and PCC SUVR (r = 0.14, p = 0.02). In addition, several cognition scales were also correlated to PCC SUVR (from r = –0.407 to 0.383, p < 0.05). Our results showed that glucose metabolism in PCC may be a potential biomarker supplement to the Aβ and tau biomarkers to predict the progression of CU to AD.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00897-0.
Keywords: Cognitively unimpaired, FDG PET, Kaplan–Meier analysis, Neurodegeneration, Posterior cingulate cortex
Introduction
Alzheimer’s disease (AD) is the most common form of dementia. The course of AD is irreversible, which makes AD early diagnosis and prediction significant. The National Institute of Aging and Alzheimer’s Association (NIA-AA) criteria for AD suggested three important biomarkers, including amyloid-β (Aβ), pathologic tau, and neurodegeneration [AT(N)] [1]. Researches on brain imaging biomarker genomics are also in full swing [2]. However, the current research showed that the AT(N) framework had limitations, especially the low predictive accuracy of the progression from cognitively unimpaired (CU) with evidence of Aβ positive (A +) or tau positive (T +) [3]. Therefore, additional biomarkers are needed clinically.
Abnormal glucose metabolism is considered to be an important biomarker for AD [4–6]. The fluorine-18-labeled-fluorodeoxyglucose positron emission computed tomography (18F-FDG PET) has been frequently used as an imaging technique to visualize abnormal glucose metabolism in the human brain in vivo [7]. Especially, the hypometabolism in the parietotemporal association area, posterior cingulate cortex (PCC), and precuneus were typical characteristics of AD [8]. Among these, as one part of the default mode network (DMN), PCC plays a certain role in spatial memory, morphological learning, and cognitive control [9].
Recently, several researchers have used glucose metabolism to predict the progression of CU to AD [10]. A small sample trial used the regional FDG PET standard uptake value ratio (SUVR) in predicting imminent progression to memory decline [11]. Another study showed that the transition status of individuals was significantly associated with some baseline periods of glucose metabolism in PCC [12]. However, few studies focus on the combination of glucose metabolism and Aβ or tau biomarkers in predicting CU progression. Therefore, the study hypothesized that abnormal glucose metabolism was useful to predict the progression of CU to dementia due to AD. Especially, we supposed that abnormal glucose metabolism in PCC could be a supplement biomarker supplement to the Aβ and tau biomarkers to predict the progression of CU to AD.
The purposes of this paper are as follows: (1) to test whether the glucose metabolism in PCC could be a potential biomarker for the prediction of CU progression. PCC SUVR was used to represent the glucose metabolism in PCC in this study. (2) To explore whether PCC SUVR combining Aβ or tau biomarkers could improve the predictive performance for CU conversion. (3) To explore the physiological significance of glucose metabolism in PCC by correlation analyses between PCC SUVR existing biomarkers and cognition scales.
Materials and methods
Participants
A total of 849 participants were included in this study, including subjects from Alzheimer’s Disease Neuroimaging Initiative (ADNI, cohort 1) and Xuanwu Hospital, Beijing, China (cohort 2). Cohort 1 is composed of 413 CU individuals, 76 patients with mild cognitive impairment (MCI), and 197 patients with AD from ADNI. Detailed inclusion information related to participant consent in ADNI is available at https://adni.loni.usc.edu/. All ADNI participants received baseline 18F-FDG PET scans, T1-weighted structural MRI scans, and demographic information, including age, gender, apolipoprotein E (ApoE) status, Mini-Mental State Examination (MMSE) values, Functional Activities Questionnaire (FAQ), and Montreal Cognitive Assessment (MOCA). Among them, 253 subjects underwent quantitative measurements of cerebrospinal fluid (CSF) Aβ42 and Aβ40. In total, 302 subjects underwent quantitative measurement of CSF ptau-181. Standardization of measurement methods for CSF can be found at https://adni.loni.usc.edu/.
In addition, cohort 2 was enrolled as an independent test dataset, including 148 CU individuals and 15 patients with MCI. The entry criteria for CU individuals have been described previously [13]. The following exclusion criteria for CU subjects were applied: (1) a history of neurological or psychiatric illness; (2) prior exposure to neuroleptic agents or drug use; (3) an abnormal neurological examination; (4) a history of stroke, high blood pressure, brain disease, or mental illness. MCI participants were diagnosed using the Jak/Bondi actuarial neuropsychological test method [14]. Specifically, participants were considered to be MCI patients if any one of the following three criteria was met: (1) they had an impaired score, defined as > 1 SD below the age-corrected normative mean, on both measures within at least one cognitive domain (i.e., memory, language, or speed/executive function); (2) they had one impaired score, defined as > 1 SD below the age-corrected normative mean, in each of the three cognitive domains sampled; or (3) they had a score on the FAQ = 9 indicating dependence in three or more daily activities. All participants in cohort 2 received baseline 18F-FDG PET scans, T1-weighted structural MRI scans, and demographic information, including age, gender, ApoE status, MMSE, FAQ, and Montreal Cognitive Assessment Basic (MOCA-B). CSF was not collected in this cohort.
To validate PCC SUVR as a potential biomarker, we selected participants who converted from CU to cognitive impairment (CI, including MCI and AD) from the ADNI database as cohort 3. This cohort included 58 CU subjects who progressed to CI within 5 years and 84 CU subjects who progressed to CI within 10 years.
This study was approved by ADNI, Beijing Xuanwu Hospital Institutional Review Board, and written informed consent was obtained from all participants or authorized representatives.
Acquisition protocol and data preprocessing
Detailed information on the data acquisition of ADNI can be found on the website (https://adni.loni.usc.edu/). In cohort 2, all 18F-FDG PET images were acquired from an integrated simultaneous 3.0-T TOF PET/MR (SIGNA PET/MR, GE Healthcare, Milwaukee, Wisconsin, USA) at Xuanwu Hospital. Each participant was instructed to fast for at least 6 h and must have a confirmed serum glucose level below 8 mmol/L. A 35-min dynamic scan was acquired approximately 40 min after the intravenous injection of 3.7 MBq/kg of 18F-FDG. 3D T1-weighted MRI images were adopted at the sagittal plane of the gradient-echo sequence. Parameters are as follows: FOV = , matrix , slice thickness , gap , slice number , TR , TE , TI , flip angle , voxel size .
The preprocessing procedures contain format conversion, correction, spatial standardization, and smoothness. First, all DICOM images were converted into NIfTI format. Then the head movement correction of images was underway. Next, each PET image was co-registered to the T1 image and co-registered PET image was normalized into the Montreal Neurological Institute (MNI) standard space with 3 3 3 mm3 voxel size. Finally, the images were smoothed using an 8-mm full width at half maximum Gaussian kernel to increase signal-to-noise ratios. All the above steps were performed by using Statistical Parametric Mapping 12 software (SPM12, Department of Imaging Neuroscience, Institute of Neurology, London, UK) implemented in MATLAB 2018b (Mathworks Inc.).
SUVR map calculation
For each participant, the standard uptake value (SUV) of the whole brain is calculated by using the automated anatomical labeling (AAL) atlas as a template, and then the SUV of each voxel is standardized with reference to the average SUV of the whole brain. The formula is as follows:
In the above formula, is the SUV of each voxel, and is the average SUV of the whole brain. Then we obtain the SUVR map of each participant.
Definition of PCC hypometabolism in CU subjects
In this paper, the PCC brain region is defined throughout the AAL brain regions, numbered 35 and 36. The left side of PCC is with Talairach coordinates of (− 2, − 36, 37), and the right side of PCC is with Talairach coordinates of (3, − 45, 11).
First, we calculated the PCC SUVR of 413 CU participants in cohort 1; the formula is as follows:
In the above formula, is the sum of SUV in the ROI, is the number of voxels in the ROI, and is the average SUV of the whole brain.
As there is no definition of the hypometabolism of CU, the median value of PCC SUVR in cohort 1 was proposed as the threshold according to previous studies [15, 16]. In this study, the PCC SUVR of 413 CU participants was ranked from the largest to smallest, and the median value was chosen as the partition threshold. The bottom 50% of the participants below the threshold were defined as CU (PCC +), and the top 50% of the participants above the threshold were defined as CU (PCC-). To verify the effectiveness of the median value method, we have performed cross-cohort verification of the rationality of the median division between cohorts 1 and 2 (see Additional file 1).
Distribution of PCC SUVR across the AD continuum
To observe the distribution of PCC SUVR in the AD continuum, we calculated the PCC SUVR of MCI and AD in cohorts 1 and 2. For cohort 1, we obtain PCC SUVR for CU (PCC-), CU (PCC +), MCI, and AD, and then we executed one-way ANOVA analysis and Tukey’s multiple-comparisons test among different groups. The above analyses were also performed in cohort 2. For comparison, we also employed CSF Aβ42 and ptau-181 as risk factors to repeat the above analyses in cohort 1. Positive status for these biomarkers is defined by < 980 pg/mL for Aβ42, and > 21.8 pg/mL for ptau-181 [17]. Based on the dichotomized biomarkers, the participants were divided into a positive subgroup and a negative subgroup for each biomarker.
Validation of PCC SUVR as a predictor
In order to verify whether PCC SUVR can be used as an effective predictor, Kaplan–Meier (KM) analyses on CU (PCC +) and CU (PCC-) in cohort 3 were carried out. First, we collected the participants in the CU group who were converted to CI and recorded the conversion time. Then, we applied PCC SUVR as the predictor to conduct KM analysis between CU (PCC +) and CU (PCC-). The differences in survival curves between groups were analyzed by the Log-rank test, and the hazard ratio (HR) was also calculated. As a comparison, Aβ42 and ptau-181 were employed as Aβ and tau predictors, and KM analyses were performed among CU (A + /A-) and CU (T + /T-), respectively. The differences in survival curves between groups were analyzed by the Log-rank test. Finally, we combined the PCC SUVR with Aβ42 and ptau-181 respectively and performed KM analyses in CU (PCC + A + / PCC- A-) and CU (PCC + T + / PCC-T-) groups. In order to verify whether PCC SUVR is useful for prediction in different periods, the above analyses were carried out at two time points: the progression occurred within 5 years and the progression occurred within 10 years. In order to verify the effectiveness, we also calculated SUVR in cuneus, occipital, and parietal, repeated the above experiments, and compared them with PCC.
Correlation analyses
In this paper, Pearson correlation analyses of PCC SUVR and cognition scales were done in cohort 1, including MMSE, FAQ, MOCA, and Everyday Cognition scales (Ecog). The correlation analyses between PCC SUVR and CSF Aβ42, Aβ42/Aβ40, and ptau-181 were also done.
Statistical analyses
Demographic and clinical characteristics are compared by analyses of variance, Chi-square test, and two-sample t-test as appropriate. The difference of PCC SUVR is compared using one-way ANOVA test, and the differences between groups are corrected by Tukey’s multiple-comparisons tests. The level of significance is set at p < 0.05. Statistical analyses are carried out by using SPSS 23.0 software (SPSS Inc., Chicago, IL), MATLAB 2018b (Math Works Inc., Sherborn, MA), and GraphPad Prism version 9.4.1. (GraphPad Software, San Diego, California, USA).
Results
Participants
Table 1 presents the demographic information of the participants from each cohort. In cohort 1, the level of Aβ42/Aβ40 in the CU (PCC +) group is significantly lower than that in the CU (PCC-) group (p < 0.01). There is no statistical difference in other parameters between the above two groups. The age of the MCI group is significantly higher than that of the CU and AD groups, which has no significant difference between the AD group and the CU group. The scores of MMSE and MOCA in the MCI group and AD group are significantly lower than those in the CU group (p < 0.001). The scores of MOCA in the MCI group are significantly higher than AD group (p < 0.05). The FAQ scores in the MCI group and AD group are significantly higher than those in the CU group (p < 0.001). AD group is significantly higher than MCI group of FAQ (p < 0.05). The Chi-square test shows that the frequency of ApoE-4 carriers in the MCI group and the AD group is significantly higher than that in the CU group (p < 0.001). The number of ApoE-4 carriers in the AD group is significantly higher than that in MCI group (p < 0.01). The deposition levels of ptau-181 in the MCI group and AD group are significantly higher than those in the CU group (p < 0.001). The level of ptau-181 deposition in AD group is significantly higher than that in MCI group (p < 0.05).
Table 1.
Characteristics of participants
| Cohort 1 (ADNI) | Cohort 2 (Xuanwu hospital) | Cohort 3 (ADNI) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CU | MCI | AD | CU | MCI | CU converters (within 5 years) | CU converters (within 10 years) | |||
| PCC + | PCC- | PCC + | PCC- | ||||||
| Female / male | 106 / 100 | 111 / 96 | 30 / 46 b | 79 / 118 b | 46 / 28 | 54 / 20 | 5 / 10 | 24 / 34 | 38 / 46 |
| Age | 73.40 (9.20) | 72.60 (9.05) | 78.90 (8.97) d | 75.30 (11.80) | 64.00 (6.00) | 65.00 (7.00) | 71.50 (20.00) c | 78.60 (8.15) | 80.00 (7.60) |
| SMC / CN | 60 / 146 | 42 / 165 | N/A | N/A | 37 / 37 | 35 / 39 | N/A | N/A | N/A |
|
Converters / non-converters (within 5 years) |
34 / 172 | 24 / 183 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
|
Converters / non-converters (within 10 years) |
48 / 158 | 36 / 171 | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Aβ42 | 1115.00(996.25) | 1486.00(739.00) | 920.00(990.50) | 718.00(368.00) | N/A | N/A | N/A | 903.50(648.50) | 802.50(635.25) |
| Aβ42 / Aβ40 | 0.15 (0.10) | 0.19 (0.07) a | 0.12 (0.10) c, d | 0.10 (0.03) c | N/A | N/A | N/A | 0.11 (0.08) | 0.11 (0.07) |
|
ApoE-4 (noncarrier / carrier) |
141 / 64 | 153 / 54 | 40 / 36 b, e | 59 / 127b | 53 / 21 | 58 / 16 | 7 / 8b | 35 / 23 | 50 / 34 |
| MMSE | 29.00 (2.00) | 30.00 (1.00) | 25.50 (6.50) c | 23.00 (4.00) c | 29.00 (2.00) | 29.00 (2.00) | 21.50 (8.50) c | 27.00 (3.50) | 27.00 (5.00) |
| FAQ | 0.00 (0.00) | 0.00 (0.00) | 8.50 (17.25) c, d | 13.00 (11.00) c | 0.00 (0.00) | 0.00 (0.00) | 6.50 (11.00) c | 1.00 (4.00) | 1.00 (6.00) |
| MOCA | 26.00 (4.00) | 26.00 (4.00) | 22.00 (7.00) c, d | 18.00 (7.00) c | 26.00 (2.00) | 26.00 (3.00) | 14.50 (10.00) c | 25.00 (3.50) | 24.00 (2.00) |
| ptau-181 | 20.68 (10.56) | 18.57 (12.98) | 25.39 (5.12) c, d | 33.17 (20.18) c | N/A | N/A | N/A | 37.61 (22.26) | 34.48 (22.50) |
Age, Aβ42/Aβ40, MMSE, FAQ, MOCA, and ptau-181 are given as median (interquartile range). The MOCA of the Xuanwu cohort is MOCA-B
ADNI, Alzheimer Disease Neuroimaging Initiative; PCC, posterior cingulate cortex; CU, cognitively unimpaired; MCI, mild cognitive impairment; AD, Alzheimer’s disease; SMC, significant memory concern; CN, cognitive normal; MMSE, Mini-Mental State Examination; FAQ, Functional Activities Questionnaire; MOCA, Montreal Cognitive Assessment
aTwo-sample t-test, CU (PCC +) and CU (PCC-)
b test with CU
cTwo-sample t-tests with CU
dTwo-sample t-tests with AD
e test with AD
In cohort 2, the age of the MCI group is higher than that of the CU group, and the difference is statistically significant (p = 0.026). The Chi-square test shows that the frequency of ApoE-4 carriers in the MCI group is significantly higher than that in the CU group (p = 0.025), and the frequency of ApoE-4 carriers in the CU (PCC +) group is significantly higher than that in the CU (PCC-) group (p = 0.043). There is no significant difference in gender distribution between MCI and CU groups. There are no statistical differences in other parameters between the CU (PCC +) group and the CU (PCC-) group.
The real case of PCC + in CU subjects
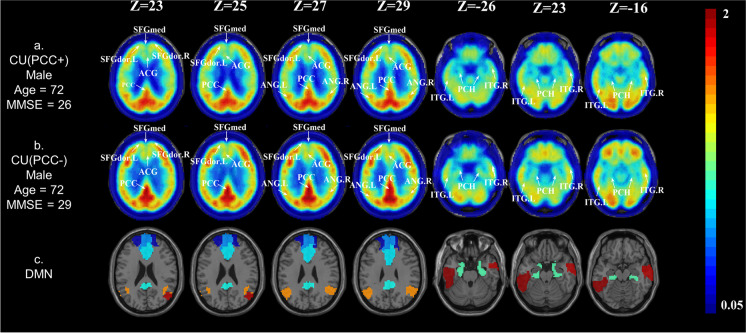
We obtained the SUVR map of each participant. Figure 1 depicts the SUVR maps of the whole brain of two sample participants. The participant in Fig. 1a has SUVR results as follows: PCC SUVR = 1.04, DMN SUVR = 0.93; while the participant in Fig. 1b has SUVR results as follows: PCC SUVR = 1.30, DMN SUVR = 0.93. For the two participants, the SUVRs in the DMN region are similar, but the participant in Fig. 1a has obviously lower metabolism in the PCC brain region than the participant in Fig. 1b.
Fig. 1.
Real cases of PCC hypometabolism in CU subjects. The age, gender, and ApoE of the two subjects matched. a A case of CU (PCC +), male, age = 72.0 years, ApoE-4 noncarrier. b A case of CU (PCC-), male, age = 72.0 years, ApoE-4 noncarrier. c The spatial location of DMN in the AAL template
Distribution of PCC SUVR across the AD continuum
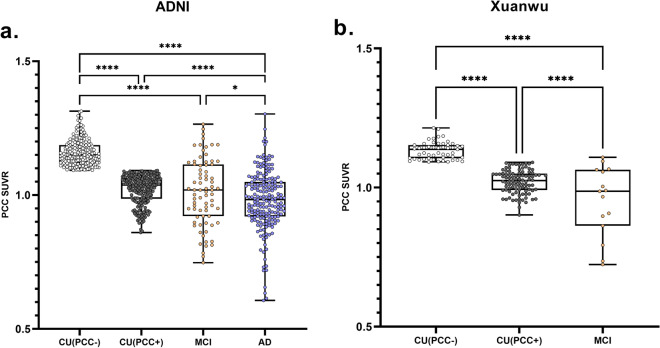
The PCC SUVR distributions of the two cohorts are shown in Fig. 2. In cohort 1, the CU group is divided into CU (PCC +) (SUVR = 1.02 ± 0.05) and CU (PCC-) (SUVR = 1.16 ± 0.05) with the median value as the segmentation threshold (SUVR threshold = 1.092). The PCC SUVRs of MCI and AD are 1.00 ± 0.13 and 0.98 ± 0.11, respectively. The results of one-way ANOVA test show that there are significant differences between the groups (p < 0.001). PCC SUVR shows a downward trend in cohort 1. The threshold (SUVR = 1.092) was applied to cohort 2 and it also indicates a downward trend. For comparison, we have applied the median PCC SUVR of cohort 2 to the two cohorts (see Additional file 1), and the results exhibit that cohort 1 does not show a stable downward trend. In addition, we pool the two cohorts to take the median, and both cohorts show a steady downward trend (see Additional file 1).
Fig. 2.
The PCC SUVR distribution of ADNI cohort (a) and Xuanwu cohort (b). *p < 0.05; ****p < 0.0001. A two-sample t-test was performed
As a comparison, the PCC SUVR differences between CU (A +) and CU (A-) in cohort 1 are compared. The results of the two-sample t-test indicate that there is a significant difference (p = 0.009) between CU (A +) (SUVR = 1.07 ± 0.84) and CU (A-) (SUVR = 1.10 ± 0.82). No significant difference (p = 0.434) between CU (T +) (SUVR = 1.08 ± 0.83) and CU (T-) (SUVR = 1.09 ± 0.85) is observed. The relevant results can be found in the supplementary material (see Additional file 2).
Validation of PCC SUVR as a predictor
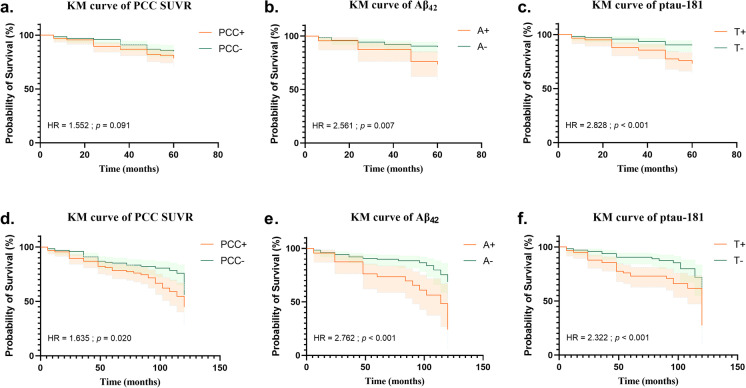
In this paper, KM survival analyses were performed on the two subgroups of CU based on whether the progression occurred within 5 or 10 years, and the survival curves were statistically analyzed by the Log-rank test. As shown in Fig. 3a to c, the PCC SUVR cannot distinguish high risk and low risk in the 5 years group, while Aβ42 and ptau-181 have a good ability to distinguish high-risk and low-risk groups, and the HRs are 2.561 and 2.828 respectively. In the 10 years group, the KM curves are shown in Fig. 3d to f. All three predictors are able to distinguish high risk and low risk, the HRs of PCC SUVR, ptau-181, and Aβ42 are 1.635, 2.322, and 2.762, and the p values are smaller than the 5 years group.
Fig. 3.
Kaplan–Meier survival curve based on PCC SUVR, Aβ42, and ptau-181 respectively. The top row is the Kaplan–Meier survival curve of subjects whose conversion occurred within 5 years. The bottom row is the Kaplan–Meier survival curve of subjects whose conversion occurred within 10 years
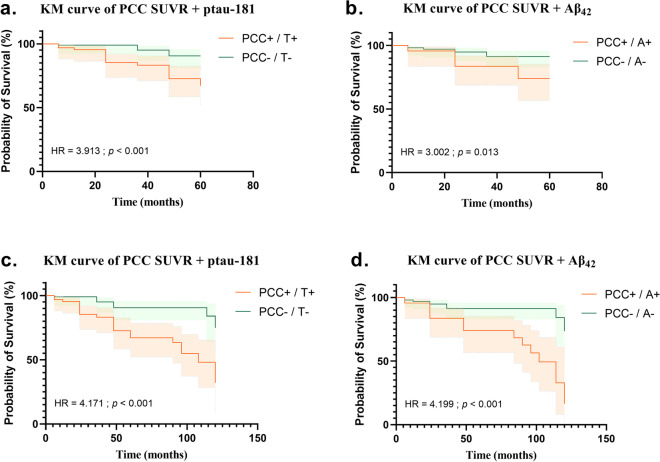
Then we superimposed PCC SUVR with CSF Aβ42 and ptau-181. KM curve results for the 5 years group are shown in Fig. 4a to b. Under the joint action of the two groups, the high- and low-risk groups are well distinguished. Among them, PCC SUVR combining ptau-181 has a stronger prediction ability (HR = 3.913) and followed by PCC SUVR combining Aβ42 (HR = 3.002). The above steps were repeated in the 10 years group, and the similar results were obtained as shown in Fig. 4c to d. The prediction ability of PCC SUVR combining ptau-181 is weaker (HR = 4.171) than PCC SUVR combining Aβ42 (HR = 4.199). The results demonstrated that Aβ42 and ptau-181 had an impact on the survival prediction performance of PCC SUVR. The above experiments were repeated with cuneus, occipital, and parietal SUVR. None of the three brain regions was able to predict CU transformation (p > 0.05), and the prediction performance was not improved after combing CSF Aβ42 and ptau-181 (see Additional file 3).
Fig. 4.
Kaplan–Meier survival curve based on PCC SUVR combing Aβ42 and ptau-181, respectively. The top line is the Kaplan–Meier survival curve of whose conversion occurred within 5 years. The bottom line is the Kaplan–Meier survival curve of whose conversion occurred within 10 years
Correlation analyses
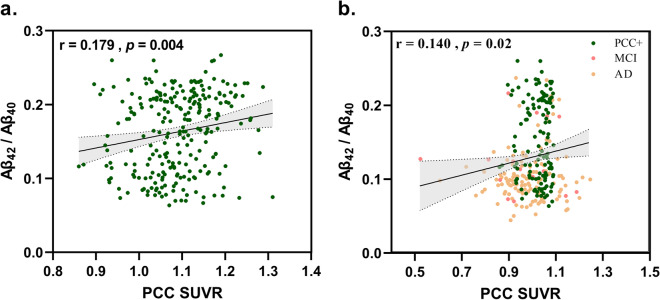
There is a slight correlation between PCC SUVR and Aβ42/Aβ40 in the CU group (r = 0.179, p = 0.004) as shown in Fig. 5a. When we counted the CU (PCC +), MCI, and AD together, the correlation still existed (r = 0.140, p = 0.02) (Fig. 5b).
Fig. 5.
Pearson correlation between PCC SUVR and Aβ42/Aβ40. a In CU group. b In CU (PCC +), MCI, and AD groups
In addition, there is a slight correlation between PCC SUVR and Ecog scales in the CU group, including EcogPtLang (r = –0.118, p = 0.036), EcogPtMem (r = –0.166, p = 0.003), EcogPtTotal (r = –0.137, p = 0.014), and ADASQ4 (r = –0.128, p = 0.009). When we combined CU, MCI, and AD groups together, the correlations became stronger. The correlation coefficients between PCC SUVR and EcogPtLang, EcogPtMem, EcogPtTotal, and ADASQ4 are 0.129, 0.214, –0.191, and –0.407 respectively (p < 0.01 for all) in cohort 1. For relevant charts, please refer to the supplementary materials (see Additional file 4).
In cohort 1, in addition to the clinical information mentioned above, PCC SUVR also has a significant correlation with EcogPtPlan (r = –0.179, p < 0.001), EcogPtDivatt (r = –0.113, p = 0.006), and EcogPtVisspat (r = –0.190, p < 0.001). Significant correlations between PCC SUVR and clinical neurological test questionnaires, including FAQ (r = –0.369, p < 0.001), MOCA (r = 0.348, p < 0.001), and MMSE (r = 0.383, p < 0.001), were observed. For relevant charts, please refer to supplementary materials (see Additional file 5).
Discussion
In this paper, we found that glucose metabolism in PCC could be used as a potential biomarker for the prediction of CU conversion. The results of survival analyses showed that the supplementation of PCC SUVR to Aβ42 and ptau-181 could improve the predictive performance of CU progression. The correlation results found a slight correlation between PCC SUVR and participants’ subjective cognition. The results suggested that glucose metabolism in PCC had potential to be used as a complement to Aβ and tau biomarkers. Notably, two cross-racial cohorts were included in this paper.
It has been well known that brain histopathological changes preceded cognitive symptoms for many years. Although the evidence obtained by FDG PET in the CU stage is limited, glucose metabolism from FDG PET scans plays an important role in both clinical and preclinical diagnoses [8]. According to previous studies, hypometabolism in PCC was observed in identifying individuals at high risk of AD [15]. In this study, the PCC SUVR showed the same trend in the AD continuum in both Chinese and Western cohorts as shown in Fig. 2, which is consistent with the literature. Cross-cohort experiments showed that almost each cohort showed a stable downward trend when the median value of PCC SUVR was used to distinguish the CU(PCC +) and CU(PCC-) (please refer to Additional file 1). Therefore, we believed that it was reasonable to divide CU(PCC +) and CU(PCC-) by the median value method. We applied the median value of PCC SUVR from cohort 2 to the two cohorts and the results showed that cohort 1 did not show a stable downward trend, which may be due to a smaller sample size in cohort 2.
The cingulate gyrus is an area of early AD-associated pathological protein deposition [18], as well as an area of early atrophy, reduced thickness, or reduced metabolism [8, 19]. Aβ, tau, and metabolic damage are common phenomena of AD, but there are differences for the three kinds of biomarkers in time distribution. Available studies suggested that the preclinical stage of AD was characterized by amyloid deposition [18], which was followed by the spread of neurofibrillary tangle tau pathology from the medial temporal lobes into the neocortex. We found that CSF Aβ42 plus PCC SUVR could improve the predictive performance of CU progression as shown in Figs. 3 and 4. We speculated that it was the ceiling effect of Aβ, which made them not sensitive enough to the progression of the disease course when they were used as predictors alone. The ceiling effect may have occurred in the CU phase. To verify our conjecture, we performed the KM survival analyses of CU under two time points of 5 and 10 years. First, in Fig. 3, the predictive performance of the 10 years group was better than that of the 5 years group when we used Aβ42 and PCC SUVR alone as the predictor, which indicated that the predictive performance of the biomarker deposition level in the baseline was different for different time points and that the time point of CU progression was closer to 10 years. However, when ptau-181 was used as an independent predictor, the prediction performance of 5 years was better than that of 10 years. We hypothesized that it was because tau was accumulated along with the course of the disease, and tau deposition level in the baseline period had less influence on tau deposition level after 10 years than that after 5 years, so the prediction performance was weaker. Secondly, when PCC SUVR was combined with Aβ42 or ptau-181 as predictors as Fig. 4 shows, the predictive performance was significantly improved compared with that used alone, indicating the effectiveness of PCC SUVR as a supplementary biomarker for Aβ or tau biomarkers. In addition, the predictive performance of the 10 years group was still better than that of the 5 years group.
In CU subjects, we found a slight correlation between Aβ42/Aβ40 and PCC SUVR as Fig. 5 shows. Considering that the ADNI cohort included in this study covers multiple countries and centers, under the influence of several complicated factors, we got these acceptable results compared to reported literatures [20–22]. Another large study in elderly cognitive normal subjects showed a significant negative correlation between local amyloid levels and metabolic signaling in several AD-typical regions, including PCC [15]. Two other studies showed correlations between local amyloid and PCC SUVR [23, 24]. These studies were consistent with the results of this paper. These results suggested that glucose metabolism in PCC and Aβ deposition already existed at the cognitive normal stage. However, we did not find a correlation between PCC SUVR and tau. Some investigators have previously found an inverse correlation between CSF tau levels and glucose metabolism in the right frontal, temporal, and parietal lobes of the brain [25, 26]. Other studies demonstrated no significant correlation between glucose metabolism and CSF tau levels [27, 28]. This may be due to pathological heterogeneity. Some studies found that CSF tau levels remained stable during dementia [29], whereas cognitive symptoms and hypometabolism deteriorated during the progression of the disease [30]. This made the mechanism of correlation between CSF tau and metabolism more complex. Therefore, the relationship between the two upstream and downstream in the CU phase needs further research.
Besides, we found that there was a certain correlation between PCC SUVR and several items of the Ecog scale in the stage of CU (please refer to Additional file 4). The Ecog can be used to measure relatively mild functional changes that may predate loss of independence in major activities of daily living, and assess functional abilities that are clearly linked to specific cognitive abilities, in other words, the everyday correlates of specific neuropsychological impairments [31]. Studies have shown that subjective cognitive decline is the first clinical manifestation on the AD continuum, manifesting as a decline in the self-experience of cognitive function without evidence of objective cognitive impairment [32–34]. The results indicated that the change of metabolic damage occurred in the subject cognitive impairment period.
This study had some limitations. First, the median value threshold (to divide PCC + and PCC-) used in this paper was obtained by the data-driven method, which was reasonable, but there may be the problem of false positives or false negatives. Further study can be considered by expanding the sample size and including follow-up FDG PET data to test the threshold. Second, the value of CSF Aβ42 and Aβ40 from ADNI was not a commercially available IVD assay. It was an assay that was currently under development and for investigational use only. The measuring range of the assay is 200 (lower technical limit)–1700 pg/mL (upper technical limit). The performance of the assay beyond the upper technical limit has not been formally established. Therefore, the values above the upper technical limit were restricted to exploratory research purposes and were excluded for clinical decision-making. This may have an impact on the results of this study. And biological sex differences, as an important phenotype in the study of neurodegenerative diseases [35, 36], may lead to different degrees of hypometabolism, which may be a direction of future research. Finally, in addition to AD-like pathology, many factors may affect PCC SUVR, such as vascular stenosis, inflammation, and tumor, but such information was lacking and not investigated in this study, and we plan to further investigate the other possible contributing factors in the future.
Conclusions
Cross-cohort experiments indicated that PCC SUVR exhibited a steady downward trend across the disease continuum. The results of survival analysis showed that the supplement of Aβ42 and ptau-181 by PCC SUVR could improve the prediction performance of CU progress. In summary, we found the important role of glucose metabolism in PCC in AD prediction. The PCC SUVR can be used as a supplement to the prediction of CU progression for Aβ and tau biomarkers. The results of this study are helpful for researchers to understand the early pathology of AD and provide theoretical support for the early prevention of AD clinically.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This study was supported by the Science and Technology Innovation 2030 Major Projects (2022ZD0211606) and the Open Project Program of Nuclear Medicine and Molecular Imaging Key Laboratory of Sichuan Province (HYX21004). Data collection and dissemination for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI): the National Institutes of Health (grant number U01 AG024904), and the Department of Defense (award numberW81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
The Alzheimer’s Disease Neuroimaging Initiative
Michael W. Weiner9, Paul Aisen10, Ronald Petersen11, Clifford R. Jack11, William Jagust12, John Q. Trojanowski13, Arthur W. Toga14, Laurel Beckett15, Robert C. Green16, Andrew J. Saykin17, John Morris18, Leslie M. Shaw13, Zaven Khachaturian15, 19, Greg Sorensen20, Lew Kuller21, Marcus Raichle18, Steven Paul22, Peter Davies23, Howard Fillit24, Franz Hefti25, David Holtzman18, Marek M. Mesulam26, William Potter27, Peter Snyder28, Adam Schwartz29, Tom Montine30, Ronald G. Thomas30, Michael Donohue30, Sarah Walter30, Devon Gessert30, Tamie Sather30, Gus Jiminez30, Danielle Harvey15, Matthew Bernstein11, Paul Thompson31, Norbert Schuff9, 15, Bret Borowski11, Jeff Gunter11, Matt Senjem11, Prashanthi Vemuri11, David Jones11, Kejal Kantarci11, Chad Ward11, Robert A. Koeppe32, Norm Foster33, Eric M. Reiman34, Kewei Chen34, Chet Mathis21, Susan Landau12, Nigel J. Cairns18, Erin Householder18, Lisa Taylor-Reinwald18, Virginia Lee13, Magdalena Korecka13, Michal Figurski13, Karen Crawford14, Scott Neu14, Tatiana M. Foroud17, Steven G. Potkin35, Li Shen17, Kelley Faber17, Sungeun Kim17, Kwangsik Nho17, Leon Thal10, Neil Buckholtz36, Marylyn Albert37, Richard Frank38, John Hsiao36, Jeffrey Kaye39, Joseph Quinn39, Betty Lind39, Raina Carter39, Sara Dolen39, Lon S. Schneider14, Sonia Pawluczyk14, Mauricio Beccera14, Liberty Teodoro14, Bryan M. Spann14, James Brewer10, Helen Vanderswag10, Adam Fleisher10, 34, Judith L. Heidebrink32, Joanne L. Lord32, Sara S. Mason11, Colleen S. Albers11, David Knopman11, Kris Johnson11, Rachelle S. Doody40, Javier Villanueva-Meyer40, Munir Chowdhury40, Susan Rountree40, Mimi Dang40, Yaakov Stern40, Lawrence S. Honig40, Karen L. Bell40, Beau Ances18, Maria Carroll18, Sue Leon18, Mark A. Mintun18, Stacy Schneider18, Angela Oliver18, Daniel Marson41, Randall Griffith41, David Clark41, David Geldmacher41, John Brockington41, Erik Roberson41, Hillel Grossman42, Effie Mitsis42, Leyla de Toledo-Morrell43, Raj C. Shah43, Ranjan Duara44, Daniel Varon44, Maria T. Greig44, Peggy Roberts44, Chiadi Onyike37, Daniel D’Agostino37, Stephanie Kielb37, James E. Galvin45, Brittany Cerbone45, Christina A. Michel45, Henry Rusinek45, Mony J. de Leon45, Lidia Glodzik45, Susan De Santi45, P Murali Doraiswamy46, Jeffrey R. Petrella46, Terence Z. Wong46, Steven E. Arnold13, Jason H. Karlawish13, David Wolk13, Charles D. Smith47, Greg Jicha47, Peter Hardy47, Partha Sinha47, Elizabeth Oates47, Gary Conrad47, Oscar L. Lopez21, MaryAnn Oakley21, Donna M. Simpson37, Anton P. Porsteinsson48, Bonnie S. Goldstein48, Kim Martin48, Kelly M. Makino48, M Saleem Ismail48, Connie Brand48, Ruth A. Mulnard35, Gaby Thai35, Catherine McAdams-Ortiz35, Kyle Womack49, Dana Mathews49, Mary Quiceno49, Ramon Diaz-Arrastia49, Richard King49, Myron Weiner49, Kristen Martin-Cook49, Michael DeVous49, Allan I Levey50, James J. Lah50, Janet S. Cellar50, Jeffrey M. Burns50, Heather S. Anderson51, Russell H. Swerdlow51, Liana Apostolova31, Kathleen Tingus31, Ellen Woo31, Daniel H. S. Silverman31, Po H. Lu31, George Bartzokis31, Neill R. Graff-Radford52, Francine Parfitt52, Tracy Kendall52, Heather Johnson52, Martin R. Farlow17, Ann Marie Hake17, Brandy R. Matthews17, Scott Herring17, Cynthia Hunt17, Christopher H. van Dyck53, Richard E. Carson53, Martha G. MacAvoy53, Howard Chertkow54, Howard Bergman54, Chris Hosein54, Ging-Yuek Robin Hsiung55, Howard Feldman55, Benita Mudge55, Michele Assaly55, Charles Bernick56, Donna Munic56, Andrew Kertesz57, John Rogers57, Dick Trost57, Diana Kerwin26, Kristine Lipowski26, Chuang-Kuo Wu26, Nancy Johnson26, Carl Sadowsky58, Walter Martinez58, Teresa Villena58, Raymond Scott Turner59, Kathleen Johnson59, Brigid Reynolds59, Reisa A. Sperling16, Keith A. Johnson16, Gad Marshall16, Meghan Frey16, Barton Lane16, Allyson Rosen16, Jared Tinklenberg16, Marwan N. Sabbagh60, Christine M. Belden60, Sandra A. Jacobson60, Sherye A. Sirrel60, Neil Kowall60, Ronald Killiany61, Andrew E. Budson61, Alexander Norbash61, Patricia Lynn Johnson61, Joanne Allard61, Alan Lerner63, Paula Ogrocki63, Leon Hudson63, Evan Fletcher15, Owen Carmichae25, John Olichney15, Charles DeCarli15, Smita Kittur64, Michael Borrie65, T-Y. Lee65, Rob Bartha65, Sterling Johnson66, Sanjay Asthana66, Cynthia M. Carlsson66, Adrian Preda31, Dana Nguyen31, Pierre Tariot33, Stephanie Reeder33, Vernice Bates67, Horacio Capote67, Michelle Rainka67, Douglas W. Scharre68, Maria Kataki68, Anahita Adeli68, Earl A. Zimmerman69, Dzintra Celmins69, Alice D. Brown69, Godfrey D. Pearlson70, Karen Blank70, Karen Anderson70, Robert B. Santulli71, Tamar J. Kitzmiller71, Eben S. Schwartz71, Kaycee M. Sink72, Jeff D. Williamson72, Pradeep Garg72, Franklin Watkins72, Brian R. Ott73, Henry Querfurth73, Geoffrey Tremont73, Stephen Salloway74, Paul Malloy74, Stephen Correia74, Howard J. Rosen9, Bruce L. Miller9, Jacobo Mintzer75, Kenneth Spicer75, David Bachman75, Stephen Pasternak57, Irina Rachinsky57, Dick Drost57, Nunzio Pomara76, Raymundo Hernando76, Antero Sarrael76, Susan K. Schultz77, Laura L. Boles Ponto77, Hyungsub Shim77, Karen Elizabeth Smith77, Norman Relkin22, Gloria Chaing22, Lisa Raudin19, 22, Amanda Smith78, Kristin Fargher78, Balebail Ashok Raj78, Thomas Neylan9, Jordan Grafman26, Melissa Davis10, Rosemary Morrison10, Jacqueline Hayes9, Shannon Finley9, Karl Friedl79, Debra Fleischman43, Konstantinos Arfanakis43, Olga James46, Dino Massoglia75, J Jay Fruehling66, Sandra Harding66, Elaine R. Peskind30, Eric C. Petrie68, Gail Li68, Jerome A. Yesavage80, Joy L. Taylor80 & Ansgar J. Furst80
9 UC San Francisco, San Francisco, CA 94143, USA. 10 UC San Diego, San Diego, CA 92093, USA. 11 Mayo Clinic, Rochester, NY 14603, USA. 12 UC Berkeley, Berkeley, CA 94720, USA. 13 UPenn, Philadelphia, PA 9104, USA. 14 USC, Los Angeles, CA 90089, USA. 15 UC Davis, Davis, CA 95616, USA. 16 Brigham and Women’s Hospital/Harvard Medical School, Boston, MA 02115, USA. 17 Indiana University, Bloomington, IN 47405, USA. 18 Washington University in St Louis, St Louis, MI 63130, USA. 19 Prevent Alzheimer’s Disease 2020, Rockville, MD 20850, USA. 20 Siemens, Munich 80333, Germany. 21 University of Pittsburgh, Pittsburgh, PA 15260, USA. 22 Weill Cornell Medical College, Cornell University, New York City, NY 10065, USA. 23 Albert Einstein College of Medicine of Yeshiva University, Bronx, NY 10461, USA. 24 AD Drug Discovery Foundation, New York City, NY 10019, USA. 25 Acumen Pharmaceuticals, Livermore, CA 94551, USA. 26 Northwestern University, Evanston and Chicago, IL 60208, USA. 27 National Institute of Mental Health, Rockville, MD 20852, USA. 28 Brown University, Providence, RI 02912, USA. 29 Eli Lilly, Indianapolis, IN 46225, USA. 30 University of Washington, Seattle, W A 98195, USA. 31 UCLA, Los Angeles, CA 90095, USA. 32 University of Michigan, Ann Arbor, MI 48109, USA. 33 University of Utah, Salt Lake City, UT 84112, USA. 34 Banner Alzheimer’s Institute, Phoenix, AZ 85006, USA. 35 UC Irvine, Irvine, CA 92697, USA. 36 National Institute on Aging, Bethesda, MD 20892, USA. 37 Johns Hopkins University, Baltimore, MD 21218, USA. 38 Richard Frank Consulting, Washington, DC 20001, USA. 39 Oregon Health and Science University, Portland, OR 97239, USA. 40 Baylor College of Medicine, Houston, TX 77030, USA. 41 University of Alabama, Birmingham, AL 35233, USA. 42 Mount Sinai School of Medicine, New York City, NY 10029, USA. 43 Rush University Medical Center, Chicago, IL 60612, USA. 44 Wien Center, Miami, FL 33140, USA. 45 New York University, New York City, NY 10003, USA. 46 Duke University Medical Center, Durham, NC 27710, USA. 47 University of Kentucky, Lexington, KY 0506, USA. 48 University of Rochester Medical Center, Rochester, NY 14642, USA. 49 University of Texas Southwestern Medical School, Dallas, TX 75390, USA. 50 Emory University, Atlanta, GA 30322, USA. 51 Medical Center, University of Kansas, Kansas City, KS 66103, USA. 52 Mayo Clinic, Jacksonville, FL 32224, USA. 53 Yale University School of Medicine, New Haven, CT 06510, USA. 54 McGill University/Montreal-Jewish General Hospital, Montreal, QC H3T 1E2, Canada. 55 University of British Columbia Clinic for AD & Related Disorders, Vancouver, BC V6T1Z3, Canada. 56 Cleveland Clinic Lou Ruvo Center for Brain Health, Las Vegas, NV 89106, USA. 57 St Joseph’s Health Care, London, ON N6A 4V2, Canada. 58 Premiere Research Institute, Palm Beach Neurology, Miami, FL 33407, USA. 59 Georgetown University Medical Center, Washington, DC 20007, USA. 60 Banner Sun Health Research Institute, Sun City, AZ 85351, USA. 61 Boston University, Boston, MA 02215, USA. 62 Howard University, Washington, DC 20059, USA. 63 Case Western Reserve University, Cleveland, OH 20002, USA. 64 Neurological Care of CNY, Liverpool, NY 13088, USA. 65 Parkwood Hospital, London, ON N6C0A7, Canada. 66 University of Wisconsin, Madison, WI 53706, USA. 67 Dent Neurologic Institute, Amherst, NY 14226, USA. 68 Ohio State University, Columbus, OH 43210, USA. 69 Albany Medical College, Albany, NY 12208, USA. 70 Hartford Hospital, Olin Neuropsychiatry Research Center, Hartford, CT 06114, USA. 71 Dartmouth- Hitchcock Medical Center, Lebanon, NH 03766, USA. 72 Wake Forest University Health Sciences, Winston-Salem, NC 27157, USA. 73 Rhode Island Hospital, Providence, RI 02903, USA. 74 Butler Hospital, Providence, RI 02906, USA. 75 Medical University South Carolina, Charleston, SC 29425, USA. 76 Nathan Kline Institute, Orangeburg, NY 10962, USA. 77 University of Iowa College of Medicine, Iowa City, IA 52242, USA. 78 University of South Florida: USF Health Byrd Alzheimer’s Institute, Tampa, FL 33613, USA. 79 Department of Defense, Arlington, V A 22350, USA. 80 Stanford University, Stanford, CA 94305, USA.
Author contribution
QZ took part in data collection, performed all analyses, and generated the first and final drafts. CF took part in generating the final drafts, and critically reviewed the manuscript. LW critically reviewed the manuscript. TL took part in designing the study and data collection and critically reviewed the manuscript. YH took part in designing the study and data collection, and critically reviewed the manuscript. JJ took part in the study designed, provided funding, and critically reviewed the manuscript.
Funding
This study was supported by the Science and Technology Innovation 2030 Major Projects (2022ZD0211606) and the Open Project Program of Nuclear Medicine and Molecular Imaging Key Laboratory of Sichuan Province (HYX21004). Data collection and dissemination for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI): the National Institutes of Health (grant number U01 AG024904), and the Department of Defense (award numberW81XWH-12–2-0012).
Data availability
All data are available upon request from the authors.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qi Zhang, Chunqiu Fan, and Luyao Wang contributed equally to the work.
Contributor Information
Ying Han, Email: hanying@xwh.ccmu.edu.cn.
Jiehui Jiang, Email: jiangjiehui@shu.edu.cn.
and for the Alzheimer’s Disease Neuroimaging Initiative:
Michael W. Weiner, Paul Aisen, Ronald Petersen, Clifford R. Jack, William Jagust, John Q. Trojanowski, Arthur W. Toga, Laurel Beckett, Robert C. Green, Andrew J. Saykin, John Morris, Leslie M. Shaw, Zaven Khachaturian, Greg Sorensen, Lew Kuller, Marcus Raichle, Steven Paul, Peter Davies, Howard Fillit, Franz Hefti, David Holtzman, Marek M. Mesulam, William Potter, Peter Snyder, Adam Schwartz, Tom Montine, Ronald G. Thomas, Michael Donohue, Sarah Walter, Devon Gessert, Tamie Sather, Gus Jiminez, Danielle Harvey, Matthew Bernstein, Paul Thompson, Norbert Schuff, Bret Borowski, Jeff Gunter, Matt Senjem, Prashanthi Vemuri, David Jones, Kejal Kantarci, Chad Ward, Robert A. Koeppe, Norm Foster, Eric M. Reiman, Kewei Chen, Chet Mathis, Susan Landau, Nigel J. Cairns, Erin Householder, Lisa Taylor-Reinwald, Virginia Lee, Magdalena Korecka, Michal Figurski, Karen Crawford, Scott Neu, Tatiana M. Foroud, Steven G. Potkin, Li Shen, Kelley Faber, Sungeun Kim, Kwangsik Nho, Leon Thal, Neil Buckholtz, Marylyn Albert, Richard Frank, John Hsiao, Jeffrey Kaye, Joseph Quinn, Betty Lind, Raina Carter, Sara Dolen, Lon S. Schneider, Sonia Pawluczyk, Mauricio Beccera, Liberty Teodoro, Bryan M. Spann, James Brewer, Helen Vanderswag, Adam Fleisher, Judith L. Heidebrink, Joanne L. Lord, Sara S. Mason, Colleen S. Albers, David Knopman, Kris Johnson, Rachelle S. Doody, Javier Villanueva-Meyer, Munir Chowdhury, Susan Rountree, Mimi Dang, Yaakov Stern, Lawrence S. Honig, Karen L. Bell, Beau Ances, Maria Carroll, Sue Leon, Mark A. Mintun, Stacy Schneider, Angela Oliver, Daniel Marson, Randall Griffith, David Clark, David Geldmacher, John Brockington, Erik Roberson, Hillel Grossman, Effie Mitsis, Leyla de Toledo-Morrell, Raj C. Shah, Ranjan Duara, Daniel Varon, Maria T. Greig, Peggy Roberts, Chiadi Onyike, Daniel D’Agostino, Stephanie Kielb, James E. Galvin, Brittany Cerbone, Christina A. Michel, Henry Rusinek, Mony J. de Leon, Lidia Glodzik, Susan De Santi, PMurali Doraiswamy, Jeffrey R. Petrella, Terence Z. Wong, Steven E. Arnold, Jason H. Karlawish, David Wolk, Charles D. Smith, Greg Jicha, Peter Hardy, Partha Sinha, Elizabeth Oates, Gary Conrad, Oscar L. Lopez, MaryAnn Oakley, Donna M. Simpson, Anton P. Porsteinsson, Bonnie S. Goldstein, Kim Martin, Kelly M. Makino, MSaleem Ismail, Connie Brand, Ruth A. Mulnard, Gaby Thai, Catherine McAdams-Ortiz, Kyle Womack, Dana Mathews, Mary Quiceno, Ramon Diaz-Arrastia, Richard King, Myron Weiner, Kristen Martin-Cook, Michael DeVous, Allan I. Levey, James J. Lah, Janet S. Cellar, Jeffrey M. Burns, Heather S. Anderson, Russell H. Swerdlow, Liana Apostolova, Kathleen Tingus, Ellen Woo, Daniel H. S. Silverman, Po H. Lu, George Bartzokis, Neill R. Graff-Radford, Francine Parfitt, Tracy Kendall, Heather Johnson, Martin R. Farlow, Ann Marie Hake, Brandy R. Matthews, Scott Herring, Cynthia Hunt, Christopher H. van Dyck, Richard E. Carson, Martha G. MacAvoy, Howard Chertkow, Howard Bergman, Chris Hosein, Ging-Yuek Robin Hsiung, Howard Feldman, Benita Mudge, Michele Assaly, Charles Bernick, Donna Munic, Andrew Kertesz, John Rogers, Dick Trost, Diana Kerwin, Kristine Lipowski, Chuang-Kuo Wu, Nancy Johnson, Carl Sadowsky, Walter Martinez, Teresa Villena, Raymond Scott Turner, Kathleen Johnson, Brigid Reynolds, Reisa A. Sperling, Keith A. Johnson, Gad Marshall, Meghan Frey, Barton Lane, Allyson Rosen, Jared Tinklenberg, Marwan N. Sabbagh, Christine M. Belden, Sandra A. Jacobson, Sherye A. Sirrel, Neil Kowall, Ronald Killiany, Andrew E. Budson, Alexander Norbash, Patricia Lynn Johnson, Joanne Allard, Alan Lerner, Paula Ogrocki, Leon Hudson, Evan Fletcher, Owen Carmichae, John Olichney, Charles DeCarli, Smita Kittur, Michael Borrie, T.-Y. Lee, Rob Bartha, Sterling Johnson, Sanjay Asthana, Cynthia M. Carlsson, Adrian Preda, Dana Nguyen, Pierre Tariot, Stephanie Reeder, Vernice Bates, Horacio Capote, Michelle Rainka, Douglas W. Scharre, Maria Kataki, Anahita Adeli, Earl A. Zimmerman, Dzintra Celmins, Alice D. Brown, Godfrey D. Pearlson, Karen Blank, Karen Anderson, Robert B. Santulli, Tamar J. Kitzmiller, Eben S. Schwartz, Kaycee M. Sink, Jeff D. Williamson, Pradeep Garg, Franklin Watkins, Brian R. Ott, Henry Querfurth, Geoffrey Tremont, Stephen Salloway, Paul Malloy, Stephen Correia, Howard J. Rosen, Bruce L. Miller, Jacobo Mintzer, Kenneth Spicer, David Bachman, Stephen Pasternak, Irina Rachinsky, Dick Drost, Nunzio Pomara, Raymundo Hernando, Antero Sarrael, Susan K. Schultz, Laura L. Boles Ponto, Hyungsub Shim, Karen Elizabeth Smith, Norman Relkin, Gloria Chaing, Lisa Raudin, Amanda Smith, Kristin Fargher, Balebail Ashok Raj, Thomas Neylan, Jordan Grafman, Melissa Davis, Rosemary Morrison, Jacqueline Hayes, Shannon Finley, Karl Friedl, Debra Fleischman, Konstantinos Arfanakis, Olga James, Dino Massoglia, JJay Fruehling, Sandra Harding, Elaine R. Peskind, Eric C. Petrie, Gail Li, Jerome A. Yesavage, Joy L. Taylor, and Ansgar J. Furst
References
- 1.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA Research Framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li L, Yu X, Sheng C, Jiang X, Zhang Q, Han Y, et al. A review of brain imaging biomarker genomics in Alzheimer’s disease: implementation and perspectives. Transl Neurodegener. 2022;11:42. doi: 10.1186/s40035-022-00315-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois B, Villain N, Frisoni GB, Rabinovici GD, Sabbagh M, Cappa S, et al. Clinical diagnosis of Alzheimer's disease: recommendations of the International Working Group. Lancet Neurol. 2021;20:484–496. doi: 10.1016/S1474-4422(21)00066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kantarci K, Boeve BF, Przybelski SA, Lesnick TG, Chen Q, Fields J, et al. FDG PET metabolic signatures distinguishing prodromal DLB and prodromal AD. Neuroimage Clin. 2021;31:102754. doi: 10.1016/j.nicl.2021.102754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murray J, Tsui WH, Li Y, McHugh P, Williams S, Cummings M, et al. FDG and amyloid PET in cognitively normal individuals at risk for late-onset Alzheimer's disease. Adv J Mol Imaging. 2014;4:15–26. doi: 10.4236/ami.2014.42003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strom A, Iaccarino L, Edwards L, Lesman‐Segev OH, Soleimani‐Meigooni DN, Jagust WJ, et al. Glucose metabolism mainly reflects local atrophy and tau pathology at symptomatic stages of Alzheimer’s disease. Alzheimer's Dementia. 2020;16:e043968. [DOI] [PMC free article] [PubMed]
- 7.Ishibashi K, Onishi A, Fujiwara Y, Oda K, Ishiwata K, Ishii K. Longitudinal effects of aging on (18)F-FDG distribution in cognitively normal elderly individuals. Sci Rep. 2018;8:11557. doi: 10.1038/s41598-018-29937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato T, Inui Y, Nakamura A, Ito K. Brain fluorodeoxyglucose (FDG) PET in dementia. Ageing Res Rev. 2016;30:73–84. doi: 10.1016/j.arr.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Leech R, Sharp DJ. The role of the posterior cingulate cortex in cognition and disease. Brain. 2014;137:12–32. doi: 10.1093/brain/awt162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huda A, Kartamihardja AHS, Darmawan B, Budiawan H, Wiwie M. Metabolic activity value in the posterior cingulate cortex using F-18 fluorodeoxyglucose positron emission tomography brain to predict the severity of Alzheimer's. World J Nucl Med. 2017;16:108–113. doi: 10.4103/1450-1147.203075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stonnington CM, Chen Y, Savage CR, Lee W, Bauer RJ, III, Sharieff S, et al. Predicting imminent progression to clinically significant memory decline using volumetric MRI and FDG PET. J Alzheimers Dis. 2018;63:603–615. doi: 10.3233/JAD-170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizk-Jackson A, Insel P, Petersen R, Aisen P, Jack C, Weiner M. Early indications of future cognitive decline: stable versus declining controls. PLoS One. 2013;8(9):e74062. [DOI] [PMC free article] [PubMed]
- 13.Li X, Wang X, Su L, Hu X, Han Y. Sino Longitudinal Study on Cognitive Decline (SILCODE): protocol for a Chinese longitudinal observational study to develop risk prediction models of conversion to mild cognitive impairment in individuals with subjective cognitive decline. BMJ Open. 2019;9:e028188. doi: 10.1136/bmjopen-2018-028188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bondi MW, Edmonds EC, Jak AJ, Clark LR, Delano-Wood L, McDonald CR, et al. Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J Alzheimers Dis. 2014;42:275–289. doi: 10.3233/JAD-140276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe VJ, Weigand SD, Senjem ML, Vemuri P, Jordan L, Kantarci K, et al. Association of hypometabolism and amyloid levels in aging, normal subjects. Neurology. 2014;82:1959–1967. doi: 10.1212/WNL.0000000000000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosconi L, Rinne JO, Tsui WH, Murray J, Li Y, Glodzik L, et al. Amyloid and metabolic positron emission tomography imaging of cognitively normal adults with Alzheimer's parents. Neurobiol Aging. 2013;34:22–34. doi: 10.1016/j.neurobiolaging.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansson O, Seibyl J, Stomrud E, Zetterberg H, Trojanowski JQ, Bittner T, et al. CSF biomarkers of Alzheimer's disease concord with amyloid-beta PET and predict clinical progression: a study of fully automated immunoassays in BioFINDER and ADNI cohorts. Alzheimers Dement. 2018;14:1470–1481. doi: 10.1016/j.jalz.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long JM, Holtzman DM. Alzheimer disease: an update on pathobiology and treatment strategies. Cell. 2019;179:312–339. doi: 10.1016/j.cell.2019.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pini L, Pievani M, Bocchetta M, Altomare D, Bosco P, Cavedo E, et al. Brain atrophy in Alzheimer's disease and aging. Ageing Res Rev. 2016;30:25–48. doi: 10.1016/j.arr.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Moscoso A, Grothe MJ, Ashton NJ, Karikari TK, Rodríguez JL, Snellman A, et al. Longitudinal associations of blood phosphorylated Tau181 and neurofilament light chain with neurodegeneration in Alzheimer disease. JAMA Neurol. 2021;78:396–406. doi: 10.1001/jamaneurol.2020.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terracciano A, Bilgel M, Aschwanden D, Luchetti M, Stephan Y, Moghekar AR, et al. Personality associations with amyloid and tau: results from the Baltimore Longitudinal Study of Aging and meta-analysis. Biol Psychiat. 2022;91:359–369. doi: 10.1016/j.biopsych.2021.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakker L, Köhler S, Eussen SJ, Choe K, van den Hove DL, Rutten BP, et al. Correlations between kynurenines in Plasma and CSF, and their relation to markers of Alzheimer’s disease pathology. Brain Behav Immun. 2023;111:312–19. [DOI] [PubMed]
- 23.Drzezga A, Becker JA, Van Dijk KR, Sreenivasan A, Talukdar T, Sullivan C, et al. Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 2011;134:1635–1646. doi: 10.1093/brain/awr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Rinne JO, Mosconi L, Pirraglia E, Rusinek H, DeSanti S, et al. Regional analysis of FDG and PIB-PET images in normal aging, mild cognitive impairment, and Alzheimer's disease. Eur J Nucl Med Mol Imaging. 2008;35:2169–2181. doi: 10.1007/s00259-008-0833-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiaravalloti A, Barbagallo G, Ricci M, Martorana A, Ursini F, Sannino P, et al. Brain metabolic correlates of CSF Tau protein in a large cohort of Alzheimer’s disease patients: a CSF and FDG PET study. Brain Res. 2018;1678:122. [DOI] [PubMed]
- 26.Jaillard A, Vanhoutte M, Maureille A, Schraen S, Skrobala E, Delbeuck X, et al. The relationship between CSF biomarkers and cerebral metabolism in early-onset Alzheimer’s disease. Eur J Nucl Med Mol Imaging. 2019;46:333. [DOI] [PubMed]
- 27.Okamura N, Arai H, Higuchi M, Tashiro M, Matsui T, Itoh M, et al. Cerebrospinal fluid levels of amyloid β-peptide1–42, but not tau have positive correlation with brain glucose metabolism in humans. Neurosci Lett. 1999;273:203–207. doi: 10.1016/S0304-3940(99)00644-8. [DOI] [PubMed] [Google Scholar]
- 28.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blennow K, Zetterberg H, Minthon L, Lannfelt L, Strid S, Annas P, et al. Longitudinal stability of CSF biomarkers in Alzheimer's disease. Neurosci Lett. 2007;419:18–22. doi: 10.1016/j.neulet.2007.03.064. [DOI] [PubMed] [Google Scholar]
- 30.Brier MR, Gordon B, Friedrichsen K, McCarthy J, Stern A, Christensen J, et al. Tau and Aβ imaging, CSF measures, and cognition in Alzheimer’s disease. Sci Transl Med. 2016;8:338ra366–338ra366. doi: 10.1126/scitranslmed.aaf2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomaszewski Farias S, Mungas D, Harvey DJ, Simmons A, Reed BR, Decarli C. The measurement of everyday cognition: development and validation of a short form of the Everyday Cognition scales. Alzheimers Dement. 2011;7:593–601. doi: 10.1016/j.jalz.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabin LA, Smart CM, Amariglio RE. Subjective cognitive decline in preclinical Alzheimer's disease. Annu Rev Clin Psychol. 2017;13:369–396. doi: 10.1146/annurev-clinpsy-032816-045136. [DOI] [PubMed] [Google Scholar]
- 33.Jessen F, Amariglio RE, van Boxtel M, Breteler M, Ceccaldi M, Chetelat G, et al. A conceptual framework for research on subjective cognitive decline in preclinical Alzheimer's disease. Alzheimers Dement. 2014;10:844–852. doi: 10.1016/j.jalz.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jessen F, Amariglio RE, Buckley RF, van der Flier WM, Han Y, Molinuevo JL, et al. The characterisation of subjective cognitive decline. Lancet Neurol. 2020;19:271–278. doi: 10.1016/S1474-4422(19)30368-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Lu J, Wang M, Zuo C, Jiang J. Influence of gender on tau precipitation in Alzheimer’s Disease according to ATN Research Framework. Phenomics. 2022;1–11. [DOI] [PMC free article] [PubMed]
- 36.Lu J, Wang M, Wu P, Yakushev I, Zhang H, Ziegler S, et al. Adjustment for the age-and gender-related metabolic changes improves the differential diagnosis of parkinsonism. Phenomics. 2023;3:50–63. doi: 10.1007/s43657-022-00079-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available upon request from the authors.