Abstract
Mutants of Salmonella enterica lacking polyphosphate kinase (ppk) grow poorly in the presence of the weak organic acids acetate, propionate, and benzoate. This sensitivity is corrected by methionine and seems to result from destabilization of MetA (homoserine transsuccinylase), the first enzyme in methionine biosynthesis. The MetA protein is known to be sensitive to thermal inactivation, and ppk mutants are more sensitive to heat-induced methionine auxotrophy. Peroxide increases the sensitivity of ppk mutants to both heat and acid and may oxidatively damage (carbonylate) destabilized MetA. While acid appears to impair methionine biosynthesis, it leads to derepression of MetA and may inhibit growth by causing toxic accumulation of denatured protein. This is supported by the observation that the overexpression of MetA in ppk mutants causes acid sensitivity that is not corrected by methionine. We propose that polyphosphate acts as a chemical chaperone that helps refold MetA and/or may stimulate proteolysis of toxic denatured protein. The instability of MetA protein may provide a metabolic fuse that blocks growth under conditions that denature proteins; the sensitivity of this fuse is modulated by polyphosphate.
The acid sensitivity of ppk mutants was noted in the course of work on ethanolamine metabolism. The ethanolamine operon (eut) encodes five proteins that resemble components of carboxysomes, organelles thought to concentrate CO2 (30). Certain mutants lacking carboxysomes grow better with a high concentration of CO2 (5%), and this growth is prevented by ppk mutations (19). It seemed possible that the ppk mutant phenotype was due to reduced internal pH, an expected side effect of high CO2. Consistent with this possibility, ppk mutants were also sensitive to several weak organic acids, suggesting that polyphosphate protects cells against reduced internal pH.
Polyphosphate (polyP) is important for growth and survival of both bacterial and eukaryotic species, but a single precise role has been difficult to define (reviewed in references 12 and 31). PolyP is synthesized in response to high salt, nitrogen limitation, phosphate limitation, and amino acid starvation and can act as a storage reservoir for phosphate and high-energy phosphate bonds. The negative charges on this polymer buffer cellular compartments and chelate various cations (Mn, Mg, Ca, Zn, Fe, Cu, and Cd). Lack of polyP reduces expression of the transcription factor RpoS (57) and impairs induction of the SOS system of DNA repair (61). PolyP stimulates ATP-dependent proteolysis of certain ribosomal proteins after a shift from rich to minimal media and thereby provides bacterial cells with a source of amino acids during adaptation to growth in minimal medium (32, 33).
Weak organic acids, such as acetate and propionate, impose a stress and are used commercially to inhibit bacterial growth. In nature, these acids are fermentation by-products produced by many bacteria (reviewed in references 2, 34, and 50). It is not entirely clear how acids inhibit growth, but they are concentrated within cells and reduce internal pH. It is suggested here that this pH drop destabilizes proteins.
The MetA protein is sensitive to heat inactivation. At temperatures approaching the upper limit of growth (42°C), MetA unfolds reversibly and is turned over by ATP-dependent proteases (5); at higher temperatures, denatured MetA protein forms insoluble aggregates (23). Evidence is presented that MetA is also sensitive to acid and oxidizing conditions. These results suggest that MetA may be a sort of metabolic fuse, sensing conditions that destabilize proteins (heat, acid, and oxidation). Loss of MetA blocks multiple aspects of metabolism, including initiation and elongation of proteins (via formyl-methionyl- and methionyl-tRNA) and methyl transfer reactions (via S-adenosylmethionine). Sensitivity of MetA to denaturing conditions increases in ppk mutants.
MATERIALS AND METHODS
Bacterial strains and crosses.
All strains are derived from Salmonella enterica serovar Typhimurium (previously Salmonella typhimurium) LT2 (Table 1). Derivatives of transposon Tn10, TPOP1 and TPOP2, were described previously (43). Transduction crosses were mediated by phage P22 (HT105; int) (54). Growth of phage and procedures for crosses were described previously (7).
TABLE 1.
Bacterial strains and plasmidsa
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strain | ||
| TR10,000 | Wild-type S. enterica (serovar Typhimurium, strain LT2) | |
| TT23986 | Del333(eutPQTD) eutJGHABCLK337::Tn10dTc(TPOP1) | This work |
| TT25376 | ppk-1::MudJ Del333(eutPQTD) eutJGHABCLK337::Tn10dTc(TPOP1) | This work |
| TT23980 | ppk-4::Tn10d(T-POP2)[del 20,del 26] | This work |
| TT23985 | rpoS1071::Tn10d-Cm | 11 |
| TT23989 | rpoS1071::Tn10d-Cm, ppk-4::Tn10d-Tc[del 20,del 26] | This work |
| TT24553 | ppk-4::Tn10d(T-POP2)[del 20,del 26]/pSPK1 | This work |
| TT24811 | ppk-4::Tn10d(T-POP2)[del 20,del 26], resistant to heat and acid | This work |
| TT24869 | metA252::Tn10 | This work |
| TT24874 | metA252::Tn10/pMetA | This work |
| TT25269 | ppk-1::MudJ metA252::Tn10 | This work |
| TT25270 | ppk-1::MudJ metA252::Tn10/pMetA | This work |
| TT25435 | metJ572::Tn10dCm | This work |
| TT25436 | metJ572::Tn10dCm, ppk-1::MudJ | This work |
| Top10 | E. coli F−mcrA Δ(mrr hsdRMS mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 deoR | This work |
| TT24554 | E. coli Top10/pHIS-ppk | Invitrogen |
| Plasmid | ||
| pSPK1 | pMMB206 (Cmr, Ptac medium copy plasmid), ppk gene from E. coli K-12 | 62 |
| pHIS-ppk | E. coli ppk gene from pSPK1 in pTricHis Topo (Invitrogen), bla+ (Apr) | This work |
| pMetA | LT2 metA gene in pTricHis Topo (Invitrogen), bla+ (Apr) | This work |
Cmr, chloramphenicol resistant.
Chemical reagents and growth media.
Unless otherwise noted, chemicals were from Sigma, Inc. The rich medium for standard aerobic cell culture was either Difco nutrient broth supplemented with 0.1 mM NaCl (15) or Luria-Bertani (LB) broth. The minimal medium was either the E or the no-citrate E (NCE) medium of Vogel and Bonner (15) or MOPS (morpholinepropanesulfonic acid) minimal medium (38). Carbon sources were provided at the following concentrations: 11 mM for glucose, 43 mM for glycerol (Fisher), 10 mM for ethanolamine (Aldrich Chemical Co.), 25 mM for sodium acetate, and 40 mM for sodium pyruvate. Aerobic liquid cultures with ethanolamine as a carbon source were supplemented with 30 μM glycerol, which is insufficient to support growth alone. Cyanocobalamin (vitamin B12) was used at 200 nM. Unless otherwise indicated, NCE medium was supplemented with trace metals (0.3 μM CaCl2, 0.1 μM ZnSO4, 0.045 μM FeSO4, 0.2 μM Na2Se2O3, 0.2 μM Na2MoO4, 2 μM MnSO4, 0.1 μM CuSO4, 3 μM CoCl2, and 0.1 μM NiSO4), which were found to enhance growth on poor carbon sources. Amino acid concentrations were as suggested previously (15). Agar for minimal plates was molecular biology grade from U.S. Biochemicals. Antibiotics were added at the following concentrations: for kanamycin, 50 μg/ml; for tetracycline, 20 μg/ml for rich medium and 10 μg/ml for minimal medium; for chloramphenicol, 20 μg/ml for selection of chromosomal insertions and 50 μg/ml for maintenance of plasmids; and for ampicillin, 100 μg/ml for maintenance of plasmids. Hydrogen peroxide was from Mallinckrodt.
Growth conditions.
Aerobic liquid cultures were grown either in fluted 125-ml flasks with a final volume of 25 ml, or in 18-mm-diameter culture tubes with a culture volume of 5 ml and were shaken in a 37°C water bath at 175 rpm. Plates were incubated anaerobically with high (5%) CO2 using an airtight box (AnaeroPack System) containing a disposable CO2-generating packet (Pack-CO2; Mitsubishi Gas Chemical Co.). Standard anaerobic growth conditions for plates were provided by an anaerobic chamber (Forma Scientific) under an atmosphere of 84% N2, 10% H2, and 6% CO2. Anaerobic liquid cultures were grown in 18-mm-diameter tubes filled to the top, crimp capped, and incubated without shaking at 37°C.
Peroxide sensitivity was tested as described in the legend to Fig. 7. Filter paper disks (Whatman type 1) were autoclaved, moistened with 5 μl of concentrated (8.8 M) hydrogen peroxide, and air dried at room temperature for 20 min. Disks were placed on the cell lawns, and plates were incubated for 2 days at 37°C before scoring of growth (the appearance of a visible lawn) and growth inhibition (the distance from the outer edge of the disk to the beginning of visible growth outside the growth-inhibited zone).
FIG. 7.
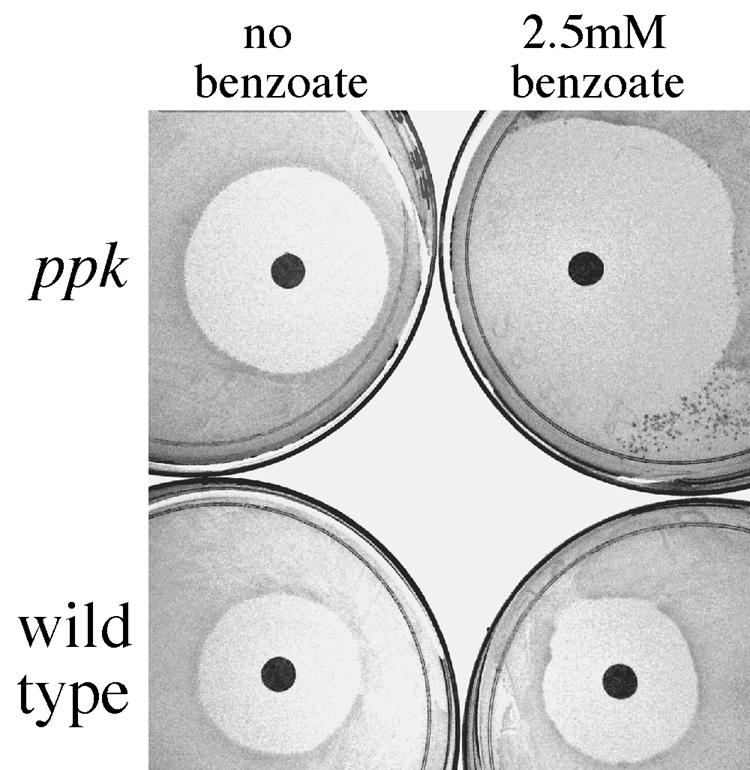
Weak acids render ppk mutants peroxide sensitive. Cells pregrown to stationary phase were spread (0.05 ml) onto NCE glucose plates containing the indicated concentration of sodium benzoate. Filter disks containing 5 μl of a 8.8 M peroxide solution were added and the plates grown for 2 days at 37°C.
To assay changes in the polyphosphate level after a shift from rich to minimal media, cells were grown to mid-log phase in LB broth, washed twice, and then incubated for various times with shaking at 37°C in MOPS minimal medium (38) containing 0.1 mM phosphate.
Construction of a strain lacking most of the eut operon.
The deletion mutant eutJGHABCLK337::T-POP1, whose eut genes are replaced by the T-POP1 element, was transduced into strain TT10648 (eutE12::MudA) selecting tetracycline resistance (Tcr). The resulting double mutant strain was used as a donor in a cross with strain TT20581, which carries the nonpolar deletion mutation eutPQTD333, selecting Tcr and screening for transductants that did not inherit the donor MudA insertion (and were more likely to retain the recipient deletion). Recombinants were shown by PCR to carry both deletion eutPQTD333 and the Tn10-associated eut-337 deletion. Strain TT20588, which carries both deletions (and therefore lacked all eut genes except eutS, M, N, E, and R) was used in the isolation of ppk mutants (below).
Isolation of ppk insertion mutants.
Mutagenesis by MudJ insertion was described previously (27). Briefly, the strain to be mutagenized (TT20588) was exposed to transducing phage grown on donor strain TT10288, which carries a chromosomal transposition-defective MudJ element in close proximity to the transposase gene of a Mud1 (ampicillin-resistant [Apr]) element. Transduced fragments that include the MudJ(Kan) element often include the nearby transposase gene. When this occurs, transposase is expressed from the transduced fragment and acts in cis to catalyze transposition of MudJ(Kan) from the fragment into the recipient chromosome. The recipient parent strain (described above) carries two eut deletions but is able to grow anaerobically on ethanolamine/B12 tetrathionate medium (41) with high CO2 (T. Fazzio, unpublished results). Transposition mutants were selected on NB-kanamycin medium and screened for those that failed to use ethanolamine anaerobically on tetrathionate medium with high CO2 but could grow on the same medium with pyruvate. These mutants had lesions in either the ppk or nuo gene; only ppk mutations are discussed here.
Insertions of Tn10dCm in ppk were isolated by transducing a pool of random Tn10dCm insertions into recipient TT20588 and screening as described above. Insertions of Tn10dTc linked to the ppk operon were obtained by transducing a pool of random Tn10dTc insertions into a ppk::MudJ insertion mutant; Tcr transductants were selected and screened to identify those that had lost the recipient MudJ(Kn) element (and the ability to grow on NB-kanamycin medium). These clones had inherited a donor fragment that carried a donor Tn10dTc element in or near ppk. The same strategy was used to obtain insertions of the T-POP2 element (43) in and near the ppk operon.
Determination of the ppk insertion sites.
Outward-directed PCR primers complementary to one end (or the other) of inserted elements were used in single-primer PCR to amplify the ppk sequence adjacent to the insertion sites (30). After several cycles of high-stringency PCR to linearly amplify adjacent sequences, stringency was reduced so that the same primer could initiate replication in the opposite direction at imperfectly matched sites. The amplified fragments were purified with Wizard or QIAGEN PCR columns and sequenced according to the method of Sanger et al. (52) at the University of Utah Health Sciences DNA Sequence Facility.
Characterizing a metA Tn10 mutation.
A Tn10 insertion (met-252) was inferred to affect either metA or metB based on nutritional tests; it could be fed methionine, homocysteine, or cystathionine. It was assigned to metA based on its 60% cotransducibility with an aceB::MudJ insertion known to lie near the metA gene. The methionine requirement of this mutation was complemented by a cloned metA gene.
Cloning the ppk and metA genes.
Primers for PCR were synthesized in the laboratory of Bob Shackmann. To produce a histidine-tagged version of polyphosphate kinase, primers corresponding to the first 24 bases and the last 24 bases of the Escherichia coli K-12 ppk gene (GenBank accession number AE000336) were used to amplify ppk from plasmid pSPK1 (62). Amplification was done with Pfu polymerase (Stratagene). An overhanging A residue, needed for cloning into pTrcHis TOPO vectors, was added to the ppk PCR fragment by incubating it with Taq polymerase for an additional 15 min at 72°C. The final PCR fragment was inserted into the pTrcHis-TOPO vector as described in the manual accompanying the kit.
A similar strategy was used to produce clones of the Salmonella metA gene. One primer used for the metA amplification (MA start, 5′ TAA AGGAGGTA TATATA ATG CCG ATT CGC GTG CTG GAC G 3′) includes a stop codon to prevent addition of the plasmid-encoded His tag to the N terminus of the metA gene product, a consensus Shine-Dalgarno sequence, a seven-base spacer, and the first 22 bases of Salmonella metA (GenBank accession number AE008895). The other primer (MA end, 5′ TTA ATC CAG GGT TGG ATT CAT GTG ACG C 3′) includes the reverse complement of the last 28 bases of the metA coding strand and its stop codon. The amplified fragment was cloned as described above.
Expression and purification of histidine-tagged Ppk.
The histidine-tagged Ppk protein was purified from the membrane fraction of lysed cells using a modified version of the protocol described by Aikiyama et al. (1). The His-tagged enzyme was purified on a nickel cartridge provided by Novagen and used to assay polyphosphate by the method of Ault-Riche et al. (4). The tagged enzyme preparation behaved like the wild-type polyphosphate kinase generously provided by J. D. Keasling but showed a twofold-higher background level of ATP production in the absence of added polyphosphate.
Polyphosphate assays.
Polyphosphate was extracted from cells and purified as described previously (4). Polyphosphate were assayed by mixing 1-μl samples with 10 μl of 5× assay mix [250 mM Tris (pH 7.4), 200 mM (NH4)2SO4, 20 mM MgCl2], 1 μl of 250 μM ADP (Sigma), and 5 μl (1 μg) of His-tagged polyphosphate kinase in a final volume of 50 μl and incubating for 45 min at 37°C to generate ATP from the polymers. Reactions were quenched by incubating at 90°C for 2 min. ATP concentrations were determined with the ATP bioluminescence assay kit from Sigma. The sample (5 μl) was mixed with 100 μl luciferase enzyme that had been diluted 1/25 in the dilution buffer supplied with the kit. The quantity of luminescence produced was measured in a 96-well plate in a Top Count NXT Microplate scintillation and luminescence counter (Packard). Levels were compared to those produced with an ATP standard supplied with the kit. Levels of cell protein were assayed with a Bio-Rad protein assay reagent using bovine serum albumin (Sigma) as the standard.
Western blot analysis.
Cell samples (3 ml) were pelleted by centrifugation in a Sorval centrifuge at 6,000 rpm for 15 min at 4°C with an SS-34 rotor and stored at −80°C. For whole-cell samples, pellets were resuspended in 62.5 μl of 1× loading dye (4% sodium dodecyl sulfate [SDS], 10% 2-mercaptoethanol, 20% glycerol, and 0.125 M Tris-Cl [pH 6.8]), sonicated (10 cycles at 50% duty cycle, power level 1, in a Branson Sonifier), and boiled for 10 min, and 12.5 (±0.1) μg protein was loaded onto a 10% SDS-polyacrylamide gel. To prepare soluble cell fractions, the pellet from 3 ml of cell culture was vortex mixed with 10 μl of added lysis buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 20% sucrose, 1 mg/ml hen egg white lysozyme [Sigma], and 243 mg/ml phenylmethylsulfonyl fluoride [Sigma]). The suspensions were held on ice for 30 min, diluted with 40 μl of buffer B (0.5 M Tris [pH 7.4], 0.1 M EDTA, and 243 mg/ml phenylmethylsulfonyl fluoride), sonicated (eight cycles at 50% duty cycle, power level 2), and centrifuged for 20 min at 13,000 rpm in an Eppendorf centrifuge. A volume of supernatant equivalent to that used for the whole cell extract was loaded onto the sodium dodecyl sulfate-polyacrylamide gel.
After electrophoresis, proteins were transferred to nitrocellulose membrane (Life Science Products) with a Bio-Rad TransBlot SD semidry transfer cell. MetA protein was detected using anti-MetA antibodies (a generous gift from Eliora Ron) that were raised against a synthetic peptide that corresponded to the first 20 N-terminal amino acids of E. coli K-12 MetA. Nitrocellulose membranes were incubated in Tris-buffered saline (20 mM Tris [pH 8.0] and 0.5 M NaCl), with 3% dry milk, 0.5% Tween 20, and 1/7,500-diluted MetA serum for 18 h, washed, and incubated in Tris-buffered saline with peroxidase-conjugated anti-immunoglobulin G antibodies and processed with SuperSignal West Pico peroxide and luminol enhancer solutions from Pierce.
RESULTS
Isolation of ppk insertion mutants.
The ability of Salmonella to catabolize ethanolamine is encoded in the 17-gene eut operon (30, 49). Six of these genes (eutABCDER) are needed for ethanolamine use under standard growth conditions (10, 30, 37, 40, 49, 56). Five of the others encode proteins homologous to shell proteins of an organelle, the carboxysome, that is thought to concentrate CO2 in other organisms (8, 24, 58). In pursuing the role of carboxysomes, CO2 was tested for its effect on growth of various eut mutants. Surprisingly, high CO2 (5 to 6%) allowed the use of ethanolamine by a eut deletion mutant that lacked ethanolamine ammonia lyase (EutBC, the first degradative enzyme) and EutM and N (carboxysome shell proteins). High CO2 allowed this mutant to produce acetaldehyde from ethanolamine by an alternative route, as inferred by the fact that growth required the next enzyme, EutE (acetaldehyde oxidoreductase). High CO2 might improve growth by driving some CO2-fixing reaction or, alternatively, by lowering internal pH when uncharged CO2 enters cells and is converted internally to bicarbonate. In agreement with the latter idea, the effect of CO2 was mimicked by sodium benzoate (see below), which also reduces internal pH, suggesting that lowered internal pH (rather than CO2 per se) was critical to the ethanolamine phenotype.
Mutants that eliminated growth of the eut deletion strain in the presence of high CO2 were isolated. These mutants included insertions in the ppk gene. Three ppk mutants isolated this way (and their position in the ppk coding sequence) are ppk-1::MudJ (bp 1427), ppk-7::MudJ (bp 1428), and ppk-6::Tn10dCm (bp 2024). Several independently isolated ppk mutations showing the same phenotype were ppk-2::Tn10dTc (bp 1702), ppk-3::Tn10d(T-POP2) (bp 663), ppk-4::Tn10d(TPOP2) (bp 251), and ppk-5::Tn10d(TPOP2) (bp 1669).
Effect of ppk mutations on use of ethanolamine.
Figure 1 shows the effect of benzoate on aerobic growth of the parent eut double deletion strain and a derived ppk mutant. Benzoate stimulated growth of ppk+ cells on ethanolamine (compare Fig. 1A and B). Growth in the presence of benzoate was eliminated by a ppk mutation (Fig. 1A). In the absence of benzoate, growth was reduced only slightly by a ppk mutation (Fig. 1B). Growth of wild-type (eut+) cells on ethanolamine was similarly stimulated by benzoate (and CO2) and was similarly reduced by an added ppk mutation (data not shown). Thus, benzoate (and CO2) stimulated growth on ethanolamine, perhaps because they reduce internal pH. Mutations in ppk impair growth severely in the presence of these acids, suggesting that polyphosphate might help protect cells from side effects of a reduced internal pH.
FIG. 1.
A ppk insertion mutation prevents growth on ethanolamine in the presence of acid. Three independent clones of the double eut deletion strains TT23986 (ppk+) and TT25376 (ppk) were pregrown to stationary phase in NCE pyruvate, washed, and diluted 1/200 into 5 ml of NCE ethanolamine medium. Tetracycline (10 μg/ml) was included to induce EutE (from a T-POP promoter); EutE is required for growth under these conditions. Sodium benzoate (5 mM) was present in all cultures in panel A. Growth at 37°C was monitored as optical density at 650 nm (OD650) using a Spectronic 20D spectrometer. Growth conditions are described in Materials and Methods, and full genotypes are in Table 1.
Sensitivity of ppk mutants to weak organic acids under various conditions.
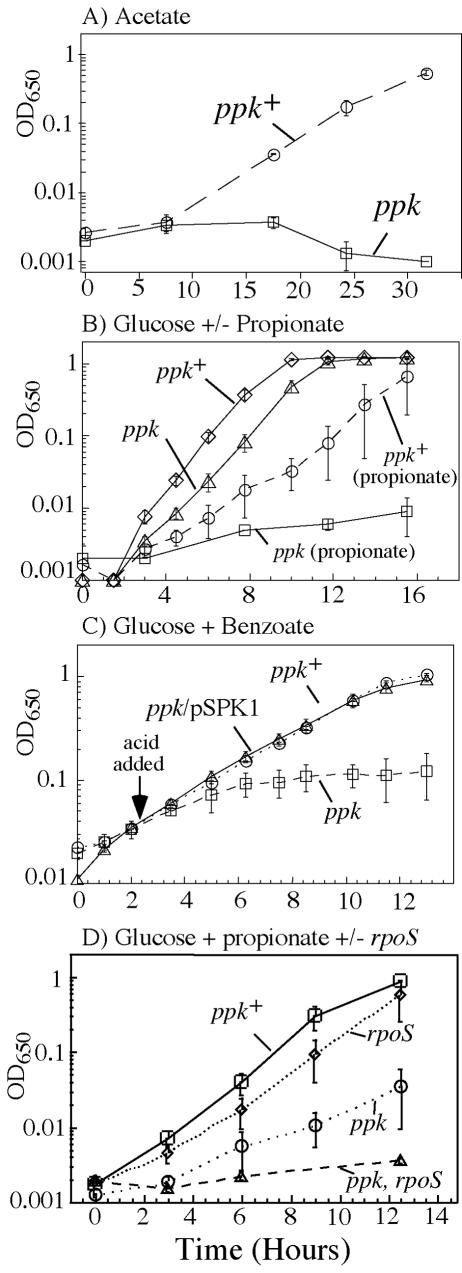
The ppk mutations identified above were moved into a wild-type strain and tested for their effects on growth using carbon sources other than ethanolamine. In initial tests on solid medium, ppk mutants failed to use the weak acids acetate or propionate as sole carbon source. This was confirmed for acetate in liquid cultures. Cells with a ppk mutation failed to grow when diluted into minimal acetate medium following pregrowth in minimal pyruvate (Fig. 2A). Pregrowth on pyruvate avoided a down-shift from rich to minimal medium, which is known to impair the growth of ppk mutants in both E. coli and Salmonella (32, 33).
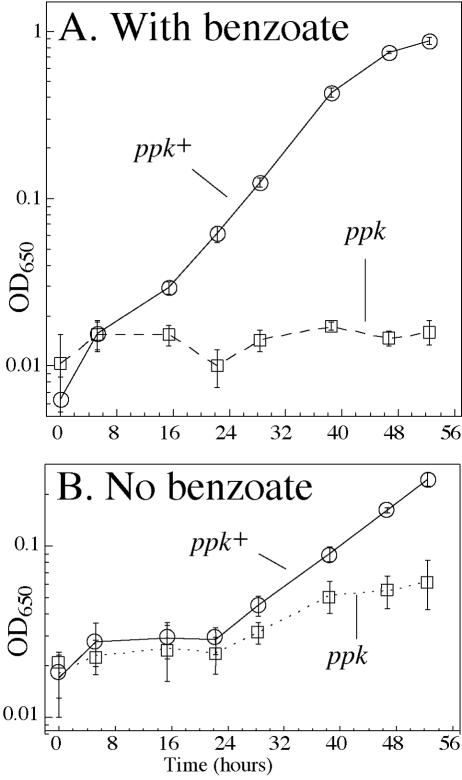
FIG.2.
Weak organic acids inhibit growth of ppk mutants on a variety of carbon sources. Several independent cultures were grown to stationary phase in NCE pyruvate (A) or NCE glucose (B, C, and D). Precultures of cells harboring plasmid pSPK1(ppk+) were supplemented with 20 μg/ml chloramphenicol. Cells were diluted 1/200 (A and C) or 1/1,000 (B and D) into liquid test medium, and cultures were incubated with shaking at 37°C. Aerobic cultures with either 25 mM potassium acetate (A) or 5 mM sodium propionate with glucose as a carbon source (B and D) are shown. In panel C, cells were grown for 2.5 doublings before adding sodium benzoate (5 mM). Error bars represent the standard deviation from triplicate cultures (A). For B, C, and D, the averages for two independent cultures are plotted, and error bars represent the differences between the two determined values. Complete genotypes are in Table 1. ppk+ indicates TR10,000; ppk is TT23980; ppk/pSPK1 is TT24552; rpoS is TT23985,; and ppk, rpoS is TT23989. OD650, optical density at 650 nm.
Propionate inhibited the growth of ppk mutants on glucose (Fig. 2B) or on glycerol (not shown). Cells were pregrown in minimal glucose (or glycerol) and diluted into the same medium with or without added propionate. The ppk mutation had little effect on growth in the absence of propionate, demonstrating that cells experienced no shift down. On glucose and glycerol media, the strongly inhibited ppk mutant cultures reached full density after extended (24-h) incubation. This was not due to accumulation of suppressor mutants, since cells that had been through one experiment were still sensitive to acid when retested.
Cloned ppk+ eliminated benzoate inhibition.
A third weak organic acid, benzoate, which cannot be catabolized by S. enterica, also inhibited growth of a ppk mutant on glucose (Fig. 2C). In this experiment, benzoate was added to cultures that had initiated growth on glucose. Benzoate inhibited growth of ppk mutants both aerobically (Fig. 2C) and anaerobically (data not shown). Since benzoate is not catabolized, the inhibition seems unlikely to result from toxic metabolic products, as has been shown for propionate (26). Furthermore, the possibility of shift-down was eliminated because cells were growing before the addition of benzoate.
The sensitivity of a ppk mutant to benzoate was corrected by an E. coli ppk+ gene (Fig. 2C) provided on plasmid pSPK1 and expressed from a constitutive lac promoter (construction described in reference 62). Thus, acid sensitivity was due to lack of Ppk activity per se rather than to polar effects of the insertion mutation on the downstream polyphosphatase (ppx) gene or to cryptic mutations in the genetic background.
The sensitivity of ppk mutants to weak organic acid is not due to lack of RpoS.
Mutations in ppk are known to impair the expression of the starvation sigma factor RpoS (57), and weak acids are known to stimulate RpoS-mediated transcription (11, 36, 53). In order to determine whether ppk mutant phenotypes reflected lack of RpoS, an rpoS insertion mutation was moved into the wild type and ppk mutants. The rpoS mutation alone did not cause propionate sensitivity (Fig. 2D), suggesting that acid inhibition of ppk mutants is not due to lack of this sigma factor. It should be noted that the sensitivity caused by a ppk mutation was made more intense by the addition of an rpoS mutation, suggesting that in the absence of a functional polyphosphate kinase, RpoS contributes to expression of genes that defend against acid stress.
Acid-induced polyphosphate accumulation was not detected.
The assay of Ault-Riche and coworkers (4) was validated by shifting E. coli cells from rich (LB) to poor medium (see Materials and Methods) and observing polyphosphate increases comparable to published values (120 nM polyP/mg protein). Similar shift-down experiments with Salmonella showed about 20-fold lower accumulation (5.3 nM polyP/mg protein). However, in wild-type Salmonella cells growing on glucose medium, benzoate induced no increase in polyphosphate detectable above the background luminescence measured without added substrate.
It is possible that the failure to detect polyphosphate is due to its loss from Salmonella (but not from E. coli) during the cell washing prior to assays. However, no polyphosphate accumulation in E. coli was induced in response to heat, pH extremes, or oxidative conditions (4), conditions under which ppk mutations caused growth phenotypes. The polyphosphate assay used detects longer polymers of phosphate (>60), and is less accurate when low concentrations of polyP are being detected (4). We suggest that very low levels or very short chains of polyphosphate may be sufficient for the effects described here.
Methionine prevents growth inhibition by weak acids.
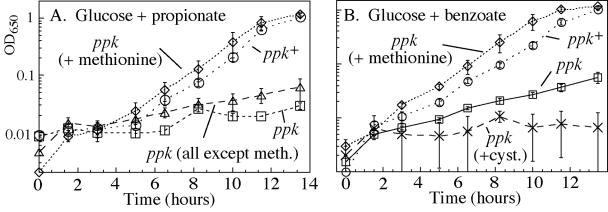
Amino acids relieve the growth lag experienced by E. coli ppk mutants following shift-down from rich to poor medium (33). Similarly, Casamino Acids (0.003%) prevented inhibition of Salmonella ppk mutants by weak acids (data not shown). Testing of individual amino acids showed that methionine (0.3 mM) completely relieved inhibition by propionate (Fig. 3A), while a mixture of all other amino acids caused no change. Growth inhibition by benzoate was also corrected by methionine alone (Fig. 3B). A slight correction of benzoate sensitivity was provided by lysine alone, serine alone, or the combination of isoleucine, leucine, and valine (not shown). Cysteine increased sensitivity to benzoate (Fig. 3B).
FIG. 3.
Methionine corrects growth inhibition of ppk mutants by organic acids. Stationary phase NCE glucose cultures were used to inoculate glucose minimal medium containing the indicated amino acids and either 5 mM sodium propionate or 2 mM sodium benzoate. The averages for two independent cultures are plotted, and error bars represent the differences between the two determined values. Relevant genotypes for strains are described in Table 1. ppk+ is TR10,000; ppk− is TT23980. OD650, optical density at 650 nm.
Organic acids affect an early step in methionine synthesis.
Correction by methionine suggested that acids imposed a block in methionine biosynthesis. To identify the position of the block, crystals of methionine biosynthetic intermediates were added to lawns of ppk cells on minimal glucose medium with benzoate (5 mM). Homocysteine (third and last intermediate) showed a ring of growth correction around a central region of no obvious growth, suggesting that homocysteine corrects the acid inhibition but is inhibitory at high concentrations. Cystathionine (second intermediate) protected well, suggesting a block in one of the first two steps, MetA or B. Added O-succinyl homoserine (first intermediate) provided poor correction of an acid-inhibited ppk mutant and a similar weak correction of ability of a metA auxotroph to grow without methionine, suggesting that O-succinyl homoserine corrected but was poorly transported. The observation of some correction by all intermediates suggested acid inhibition of the first enzyme (MetA) but did not eliminate the possibility that sensitivity was relieved by repression of the methionine biosynthetic pathway. Both possibilities were tested.
Acid-inhibited ppk mutants accumulate insoluble MetA protein.
Inhibition of methionine synthesis (at any point) would be expected to lead to derepression of methionine biosynthetic enzymes (22). We tested the MetA protein because both the nutritional tests (and additional evidence, below) suggested that it was the target and because this enzyme is known to be sensitive to heat (47, 48).
Samples were withdrawn from glucose-grown cultures 3 and 5 h after adding benzoate. Cell extracts were analyzed by Western blots using MetA antibodies generously provided by Eliora Ron. Acid increased the MetA level slightly in wild-type cells; in ppk mutant cells, the basal MetA level was higher and increased further in response to acid (Fig. 4). The addition of methionine to the cultures eliminated detectable MetA protein, presumably by repressing de novo synthesis. When soluble proteins were analyzed apart from the membrane fraction, acid appeared to cause no change in the level of soluble MetA protein in wild-type or ppk cells. The results suggested that organic acids destabilized MetA, leading to methionine limitation and derepression of metA and that excess denatured MetA protein accumulates as an insoluble aggregate, particularly in ppk mutant cells.
FIG. 4.
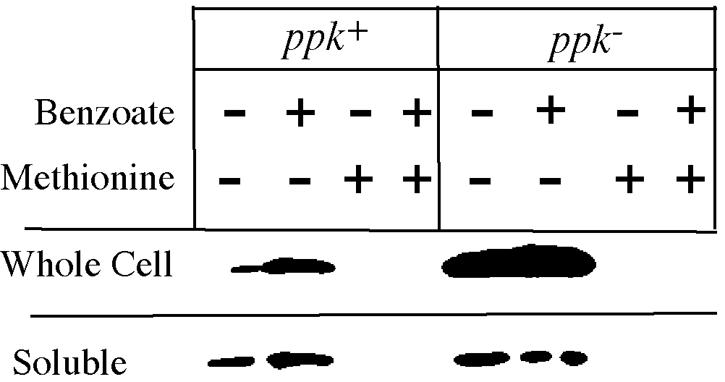
Insoluble MetA protein accumulates in acid-inhibited ppk mutants. Samples were taken (and frozen) 3 h after adding water (control) or 5 mM sodium benzoate to NCE glucose cultures. Whole and soluble cell samples were processed as described in Materials and Methods and loaded onto a 10% SDS polyacrylamide gel. After electrophoresis, the gel was Western blotted with a primary antibody to the N terminus of E. coli MetA (provided by Eliora Ron). Complete genotypes are in Table 1. WT is TR10,000; ppk− is TT23980.
This was tested directly using Western blots to observe the disappearance of MetA protein after the addition of methionine (to repress MetA synthesis) at mid-log phase. Following methionine addition, MetA disappeared within 30 min in ppk+ cells grown without acid, but persisted more than an hour longer with acid or in ppk mutant cells grown with or without acid (data not shown). Acid caused a fivefold-greater accumulation of MetA in ppk than in ppk+ cells, suggesting that polyphosphate either minimized the accumulation or accelerated the breakdown of denatured MetA. A similar accumulation of denatured MetA protein was observed previously during growth of E. coli at high temperatures (23).
Partial correction of acid sensitivity by a cloned metA gene.
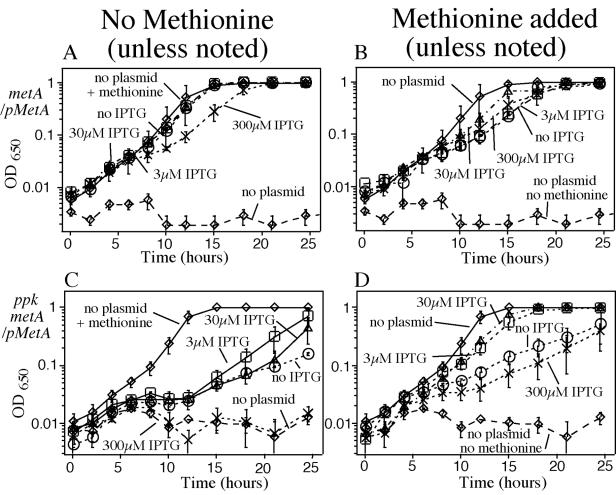
If organic acids denature the MetA protein and limit methionine production, one might expect the acid-induced methionine requirement to be corrected by increases in MetA protein. In contrast, if growth is inhibited by denatured MetA aggregate, overproduction might increase acid sensitivity. To test this, a plasmid (pmetA+) with the metA gene under control of the lac promoter was added to a metA mutant. In this strain, all MetA production is regulated by lac control elements and is not repressed by methionine.
The plasmid-encoded MetA protein is functional, since it corrected the methionine requirement of a metA ppk+ strain on standard medium with or without induction (data not shown). The plasmid also corrected in the presence of benzoate (Fig. 5A), except that the highest level of inducer impaired growth slightly. Added methionine had little effect on growth of the ppk+ metA strain carrying the plasmid (Fig. 5B).
FIG. 5.
Effects of a cloned metA gene on growth inhibition by acid. Cultures were inoculated by 1/1,000 dilution of a stationary phase minimal culture into 5 ml of NCE glucose medium containing 2.5 mM sodium benzoate and the indicated amount of IPTG. Ampicillin (0.1 mg/ml) was present for the strain carrying the pMetA plasmid. The averages for two independent cultures are plotted, and error bars represent the differences between the two determined values. OD650, optical density at 650 nm.
In the ppk metA double mutant, the uninduced plasmid provided only partial restoration of methionine-independent growth, which improved slightly at intermediate inducer levels (Fig. 5C). We suggest that in ppk mutants, MetA is more sensitive to destabilization by acid than was the case for ppk+ strains (above), making it more difficult for the plasmid to correct the methionine requirement (compare Fig. 5A and C). Two aspects of these data suggested that acid does more than reduce MetA activity. In ppk metA strains (Fig. 5C), no level of MetA expression allowed full methionine-independent growth in the presence of acid. Furthermore, the highest level of MetA induction (300 mM IPTG [isopropyl-β-d-thiogalactopyranoside]) provided no growth improvement at all.
Evidence that acid sensitivity is due to toxicity of denatured MetA protein.
Added methionine completely corrected the acid sensitivity of a ppk metA+ strain without the plasmid (Fig. 3). However, methionine did not correct growth when MetA protein was expressed from a lac promoter that is not repressed by methionine. In the latter case, methionine improved growth only at intermediate levels of inducer (3 or 30 μM IPTG). At the highest level of inducer (300 μM IPTG) or at the lowest (no IPTG), growth on acid was reduced significantly, even in the presence of methionine (Fig. 5C and D). This is consistent with the idea that acid inhibits growth because of toxic effects of aggregated denatured MetA protein. This possibility was initially suggested by Gur et al. (23) as a factor that might contribute to the MetA-mediated effects of heat.
We do not understand why methionine failed to correct acid inhibition of strains with uninduced MetA while intermediate levels of induction provided some correction. However, data like those in Fig. 5 were obtained repeatedly. It should be kept in mind that the plasmid used here is a pUC derivative which is present at very high copy number; therefore, a substantial amount of MetA protein is made without induction (enough to fully complement a simple metA defect), and the induced level may exceed that achieved in normal cells. However, this metA plasmid, even when fully induced, impaired growth (with or without added methionine) only in the presence of acid, and strong inhibition was seen only with ppk strains.
Constitutively overexpressed MetA prevents methionine correction.
To confirm the conclusion that high levels of MetA make cells sensitive to acid without relying on use of plasmids, a metJ constitutive mutant was used. The metJ mutation causes high constitutive expression of MetA that is not repressed by methionine, as demonstrated by Western hybridization (data not shown). Strains with a metJ ppk+ genotype grew normally and were not sensitive to benzoate. The metJ ppk double mutant showed standard acid sensitivity. However, unlike metJ+ ppk strains, this sensitivity was not corrected by methionine (data not shown). This test, like tests using the metA plasmid, suggests that a high level of MetA protein causes acid sensitivity in a ppk mutant and that acid inhibition is caused by toxic accumulation of denatured MetA protein rather than by reduction of methionine biosynthesis.
Effect of ppk mutations on thermal denaturation of MetA.
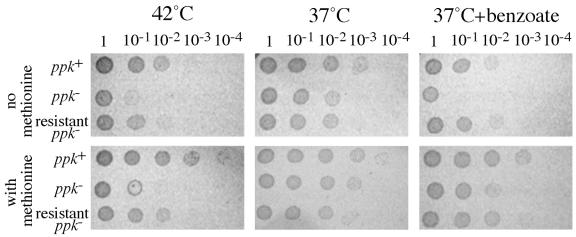
The thermosensitivity of MetA protein has been studied extensively and appears to establish the upper temperature limit to growth of wild-type E. coli on minimal medium (5, 23, 46, 48). The effects of high temperature and acid are compared in Fig. 6. The center panel shows growth of wild-type and ppk mutant cells under normal conditions (37°C; no added acid). The left panel shows the effect of elevated temperature and the right panel the effect of added benzoate. For each set of conditions, a dilution series of a cell suspension was spotted on solid medium. It is clear that a ppk mutation sensitizes cells to both acid and heat and that both effects are corrected (at least in part) by added methionine. Also included in Fig. 6 is an acid-resistant mutant strain isolated from a ppk strain for its improved growth in the presence of benzoate on glucose medium. This mutation also provided resistance to high temperature. The responsible mutation does not map in the metA gene and may resemble previously described variants of E. coli ppk mutants that are resistant to heat and peroxide (42).
FIG. 6.
Heat sensitivity of ppk mutants. Aliquots were removed from culture growing logarithmically in NCE glucose and serially diluted in NCE salts medium (no glucose) before spotting 5 μl onto E glucose plates. Where indicated, sodium benzoate was added to the medium at a final concentration of 0.5 mM. Cells were incubated overnight at the indicated temperatures.
Peroxide increases acid sensitivity of ppk mutants.
The results above suggested a model (detailed later) in which acid (or heat) destabilizes MetA protein, which can either refold or be irreversibly denatured to a toxic form. Polyphosphate may favor refolding or (more likely) may minimize accumulation of aggregated denatured protein by stimulating proteolysis of the aggregates or their unfolded precursors. This model predicts that the partially unfolded protein might be sensitive to oxidative damage (carbonylation) as described by Dukan et al. (16-18) and reviewed by Stadtman (59). Oxidative damage might lead to irreversible protein denaturation.
The first suggestion that oxidation might contribute to acid sensitivity was the observation that cells growing on pyruvate were not sensitive to acid inhibition as were cells growing on either glucose (Fig. 2B) or glycerol (data not shown). Pyruvate is known to destroy peroxides by a nonenzymatic mechanism (21, 63), and peroxides are known to accumulate in aerobic growth media (55). We tested the general idea that acid might make proteins sensitive to oxidative damage, by assaying carbonylation levels in wild-type cells using an OxyBlot protein oxidation detection kit from Chemicon International. We found that carbonylation levels increased markedly during growth in the presence of benzoate (data not shown).
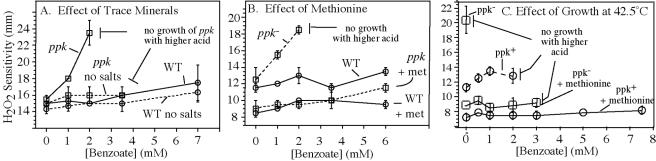
The role of oxidation in ppk mutant phenotypes was tested directly by adding hydrogen peroxide during tests of acid-induced methionine limitation. Cells (ppk or ppk+) were pregrown in minimal glucose medium and spread on plates of the same medium containing increasing concentrations either propionate or benzoate. Filter paper disks with peroxide were added to the plates and zones of growth inhibition measured. This assay is shown in Fig. 7, and results are quantified in Fig. 8. At benzoate concentrations greater than 2.5 mM, the lawn of ppk cells failed to grow. As acid concentrations approached this point, the zone of inhibition by H2O2 increased in size (Fig. 8A). Similar results were obtained on plates containing propionate in place of benzoate (data not shown). The size of the peroxide inhibition zone is not dictated by the growth rate of cells in the lawn; a his auxotroph growing on either histidine or various concentrations of the growth-limiting precursor histidinol showed essentially the same-sized zone of peroxide inhibition regardless of growth rate (data not shown).
FIG. 8.
Effect of mineral salts, methionine, and temperature on the acid-mediated peroxide sensitivity of ppk mutants. Cells were grown and plated as in Fig. 7. Peroxide sensitivity was scored as the distance from the filter disk to the point of visible cell lawn. Growth was scored as the ability to detect a visible lawn outside the peroxide inhibition zone. Cultures designated “no salts” lacked the trace minerals otherwise added to supplement growth in minimal medium. Complete genotypes are in Table 1. WT is TR10,000; ppk is TR23980. Error bars represent the standard deviations of triplicate independent cultures (TR10,000) or the differences between two values whose average is plotted (TT23980).
The sensitivity to peroxide in the presence of acid was not apparent in the absence of added trace minerals (Fig. 8A), although ppk cells still failed to grow at benzoate concentrations above 3.5 mM. Adding iron alone also increased peroxide sensitivity and full sensitivity was generated by the addition of iron plus copper, both of which are known to catalyze the formation of hydroxyl radicals from hydrogen peroxide via the Fenton reaction (28, 39). Methionine (0.3 mM) corrected the H2O2 sensitivity and allowed cells to tolerate higher acid concentrations (Fig. 8B), suggesting that MetA protein was more sensitive to acid in the presence of peroxide. While peroxide stimulated acid sensitivity, it is not essential to the basal acid sensitivity shown by ppk mutants, since some acid sensitivity is seen anaerobically (see above). We conclude that MetA is sensitive to reduced pH and this sensitivity is exacerbated when destabilized protein is subject to oxidative damage.
Increasing temperature from 37°C (Fig. 8B) to 42.5°C (Fig. 8C) increased the peroxide sensitivity of ppk mutants but not wild-type cells. With no added acid, the zone of peroxide inhibition for a ppk mutant increased from 12 mm (at 37°C) to 20 mm (at 42.5°C). Heat did not increase the peroxide sensitivity of a ppk+ strain. The effect of high temperature on peroxide sensitivity of a ppk mutant was corrected by methionine.
The combined effects of acid and temperature are quantified in Fig. 8C. High temperature increased the acid sensitivity of both ppk+ and ppk mutant cells (compare Fig. 8B and C). At 37°C, ppk+ cells could tolerate benzoate concentrations as high as 6 mM, but they failed to grow at 42.5°C on benzoate concentrations above 2 mM. Similarly, a ppk mutant tolerated 2 mM benzoate at 37°C, but at 42.5°C could not tolerate the lowest concentration tested (0.5 mM). Again, the increased sensitivity to acid at high temperature was corrected in part by methionine.
DISCUSSION
Growth of mutants lacking polyphosphate (ppk) was impaired by any of several weak organic acids that reduce internal pH (44) and by elevated temperature. These conditions are known to destabilize some proteins (13), suggesting that polyphosphate may stabilize proteins or stimulate degradation of inhibitory denatured protein aggregates (see model below). The MetA protein appears to be the target protein responsible for these effects.
A model for polyphosphate effects on protein stability.
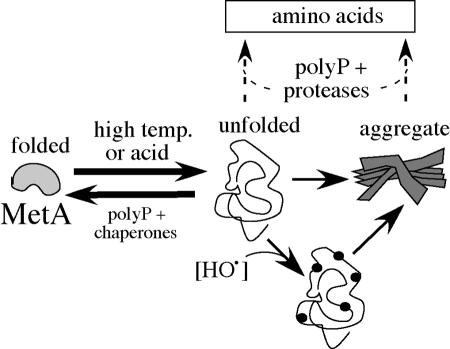
Two seemingly opposite effects are suggested by the results presented here. On the one hand, polyphosphate appeared to stabilize MetA, because it prevents the methionine requirement caused by denaturing conditions. On the other hand, polyphosphate appears to stimulate degradation of MetA, because it prevented the toxic effect of denaturing conditions in strains that overproduced MetA. We propose that impairment of MetA by acid (or heat) limits methionine production and leads to MetA derepression. The impairment could result from destabilization of the MetA protein or from inhibition of its activity by protonation of its active-site thiol at low pH (9). Consequent derepression of MetA may compensate for limitation of the methionine supply, but generates a high level of MetA protein, which can denature and form inhibitory aggregates. Added methionine satisfies any methionine shortage and prevents aggregate accumulation by repressing MetA production. Polyphosphate may protect against the affects of acid by stabilizing MetA protein and/or may stimulate proteolysis of toxic aggregates. If polyP served both to stabilize MetA and to stimulate proteolysis of aggregates, it would play a role like that of chaperonins, which also contribute to both refolding and proteolysis of proteins (64).
In the model shown in Fig. 9, weak organic acids or high temperatures destabilize protein. The destabilized form can refold to regain activity or can be irreversibly denatured, especially after oxidative damage. Refolding is stimulated by chaperonins or by interaction with polyphosphate, while aggregated denatured protein is made more accessible to proteases by the same interactions. This model is based on experiments in which denaturing conditions seem to affect a particular protein, MetA. However, the effects noted here are likely to be general and we suggest that MetA is adapted to be particularly sensitive so it can serve as a sort of metabolic fuse, by which denaturing conditions inhibit cell growth. The background for various aspects of the model are discussed below.
FIG. 9.
A model for the role of polyP in resistance of MetA to acid or heat. MetA exists in cells in an equilibrium between a native and a reversibly unfolded conformation (48). The unfolded protein can also form an insoluble aggregate (23). The more surface-exposed unfolded form is proposed to be susceptible to oxidation (16) and to proteolysis or aggregation. The unfolded form is favored at high temperature or low pH. Chaperones may favor refolding. PolyP is proposed to favor refolding and/or proteolysis of denatured protein or its aggregates.
Effects of weak acids.
Resting E. coli cells concentrate added weak acids. Acetate added externally at 8 mM (pH 6) reaches an internal concentration of 240 mM; benzoate added at 2 mM reaches an internal level of 35 mM (44). Weak acids enter cells in their uncharged protonated form and deprotonate internally, lowering the intracellular pH (20, 51). The toxic effects of organic acids have been attributed in part to this drop in internal pH and consequent decreases in proton motive force (50). The possibility that these acids (or the lowered pH) denature proteins or sensitize them to oxidative damage (carbonylation) is supported by the observation that weak organic acids induce synthesis of chaperones, peroxidases, and catalases (2, 3, 6, 16, 35, 53).
Ferguson and coworkers (20) showed that the addition of 25 mM acetate reduces the internal pH by about 0.5 pH unit in cells buffered at physiological pH. This is a small change relative to that required to denature most proteins. However, some proteins are super-sensitive to denaturation by lower pH because their structures include buried nonionized residues (particularly histidines) that can ionize after partial unfolding (13). The MetA protein, inferred here to be a sensitive target, has a total of 10 His residues, 4 of which (shown in boldface type) are located in a single hydrophobic region, HHILHPHALL (residues 170 to 176). Since MetA is subject to reversible inactivation at elevated temperature (22, 23), small perturbations, i.e., adding charges to one or more normally buried His residues, may be sufficient to favor the unfolded form. This would be favored at reduced pHs. It may be relevant to protein destabilization that the weak organic acids used here are amphipathic anions, which could denature by interactions preferentially with exposed internal regions of a partially unfolded protein (13).
Effects of oxidative stress on protein stability.
Weak acids induce peroxidases, such as KatE (2) and AhpC (6, 35), suggesting that they increase vulnerability to oxidative damage. Rao and Kornberg (42) showed previously that ppk mutants grown to mid-log phase in minimal media are more sensitive to peroxide (42 mM) than wild-type cells.
Dukan and coworkers (16) found that proteins with multiple missense substitutions (generated by error-prone mutant ribosomes or streptomycin-induced miscoding) become sensitive to oxidative damage (carbonylation). The level of mistranslation correlated with the amount of chaperone GroEL detected in cells, indicating that mistranslation generated unfolded proteins, which served to induce this chaperone. Conditions that increased miscoding had no effect on the levels of oxidative stress proteins, the level of superoxide, or the level of cysteine oxidation, suggesting that miscoding affects the sensitivity of the oxidation target molecules rather than making the cell interior more oxidizing. We propose that weak organic acid or high temperature partially unfolds proteins and (like miscoding) makes them more vulnerable to oxidative damage (reviewed in reference 59). Consistent with this, the removal of trace metals (including iron and copper) in the experiments described here prevented the peroxide sensitivity, but not the acid sensitivity; both iron and copper are known to promote hydroxyl radical formation (28, 39). It is possible that part of the polyphosphate effect is due to chelating these metals.
Aggregation of denatured protein.
Protein aggregation is associated with many diseases, including Alzheimer's disease, prion-related disorders, and type II diabetes (reviewed in reference 64). Such aggregates may also inhibit bacterial growth. Tomoyasu and coworkers (60) found that E. coli rpoH mutants, which lack the chaperones and proteases controlled by this sigma factor, accumulate aggregates and are growth deficient at 37°C. Thermotolerance is restored and aggregation prevented by high-level expression of the chaperone DnaK, suggesting that the growth defect resulted from aggregated denatured proteins. Protein aggregation is usually prevented by the action of chaperones and proteases (reviewed in reference 64).
Previously, Biran and coworkers (5) showed that MetA protein experiences rapid turnover at destabilizing temperatures. This turnover is diminished in a strain deficient in the three ATP-dependent proteases Lon, ClpP, and HslVU, suggesting that these proteases degrade MetA. At higher temperatures, insoluble MetA protein aggregates accumulate (23); this accumulation was reduced (and growth was improved) by the molecular chaperone trimethylamine oxide. Results presented here suggest that at least part of the effect of ppk mutations might be impairment of ability to proteolyse denatured MetA.
Kuroda et al. demonstrated that polyphosphate stimulates proteolysis (32, 33). They found that under shift-down conditions, the concentration of long (greater than 60) polymers of polyphosphate increases to 30 nmol/mg protein and stimulates Lon-mediated degradation of ribosomal proteins. Polyphosphate was required by two ATP-dependent proteases (Lon and ClpP), the same proteases inferred by others to degrade MetA (23).
An effect of external pH on ppk growth phenotypes was reported previously by Kim and coworkers (29), who showed that Salmonella ppk mutants grow to slightly lower densities in rich medium when the pH is reduced (to 5.8) and that stationary phase cultures of ppk mutants are more sensitive to killing by heat and acid than wild-type cells. Stationary phase cultures of E. coli ppk mutants are less able to survive lower external pH, heat, and peroxide stress (14, 42).
The above findings fit with those presented here if MetA protein is particularly unstable and derepresses in response to inactivation or inhibition by acid. While derepression might correct the methionine limitation, the increased production of an unstable protein could promote formation of a toxic aggregate of denatured MetA protein (as seen in Fig. 4). By activating proteolysis, polyphosphate could prevent the accumulation of these aggregates and added methionine could prevent their accumulation by repressing the rate of MetA production. Since MetA is feedback inhibited by methionine and S-adenosylmethionine, it is possible that binding of these effectors affects MetA stability.
Other effects of acid and oxidation on methionine biosynthesis.
Inhibition of growth by acid was attributed here to effects on the first methionine biosynthetic enzyme MetA based on correction by methionine and all of its biosynthetic precursors and on changes in sensitivity caused by varying the level of MetA protein.
Roe and coworkers (45) also showed an acid-induced methionine requirement for E. coli. However, they concluded that the effect was due to accumulation of the MetE substrate, homocysteine. We suspect that these two data sets are congruent. In the E. coli studies, strains were inhibited by weak acids at an external pH of 6, which allows more extreme internal acidification because acid enters more freely. Their conditions were sufficiently stringent to inhibit ppk+ cells and may reveal effects of acid on MetE activity. MetE protein has recently been shown to be sensitive to oxidative damage (25) and may become more vulnerable when destabilized by acid. We suggest that at a neutral pH (used here), acids destabilize MetA slightly in ppk+ cells and more severely in ppk mutants and that a pH of 6 allows more severe destabilization (even in ppk+) cells and extends the effect to MetE. This may explain the cysteine sensitivity seen with Salmonella in the presence of benzoate (Fig. 3). We suggest that added cysteine is converted to homocysteine by the action of MetB and C (cysteine + O-succinyl homoserine→→homocysteine); accumulation of this toxic intermediate may be favored when the methionine pathway is derepressed and MetE activity is inhibited.
A suggested role for polyphosphate.
We suggest that one function of polyphosphate may be to optimize transition through short-term environmental stresses that necessitate the mobilization of internal resources. This purpose may be served by its acting as a reservoir for the storage of phosphate and energy. The functions discussed here would contribute to the same ultimate goal. Many stressful environments destabilize internal proteins and lead to protein aggregates that must be degraded to avoid toxic effects and to provide amino acids for adaptation to a new set of conditions. Particularly sensitive enzymes, such as MetA, may block growth temporarily so that more cellular proteins are rendered dispensable during the period of physiological adaptation.
Acknowledgments
This work was supported by NIH grant GM34804 (J.R.R.).
We thank J. D. Keasling and his laboratory staff at U. C. Berkeley for helping one of us (M.P.-C.) with the polyphosphate assays and supplying strains. Eliora Ron (University of Tel Aviv, Israel) generously supplied antibody to MetA protein. This project benefited from suggestions and discussions with Peter Anderson, Renee Dawson, Julianne Grose, Christopher Mace, Joseph Penrod, and David Sheppard. Renee Dawson and Joseph Penrod helped with the manuscript. We thank David Blair at the University of Utah, in whose lab some of these experiments were conducted; David Wolstenholme for providing research space; and Janet Shaw and Joe Dickinson for supplies and access to equipment.
REFERENCES
- 1.Aikiyama, M., E. Crook, and A. Kornberg. 1992. The polyphosphate gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane localization of the protein. J. Biol. Chem. 267:22556-22561. [PubMed] [Google Scholar]
- 2.Arnold, C. N., J. McElhanon, A. Lee, R. Leonhart, and D. A. Siegele. 2001. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J. Bacteriol. 183:2178-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Audia, J. P., C. C. Webb, and J. W. Foster. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int. J. Med. Microbiol. 291:97-106. [DOI] [PubMed] [Google Scholar]
- 4.Ault-Riche, D., C. D. Fraley, C. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biran, D., E. Gur, L. Gollan, and E. Z. Ron. 2000. Control of methionine biosynthesis in Escherichia coli by proteolysis. Mol. Microbiol. 37:1436-1443. [DOI] [PubMed] [Google Scholar]
- 6.Blankenhorn, D., J. Phillips, and J. L. Slonczewski. 1999. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J. Bacteriol. 181: 2209-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik, T. A., M. Ailion, and J. R. Roth. 1992. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 174:2253-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobik, T. A., G. D. Havemann, R. J. Busch, D. S. Williams, and H. C. Aldrich. 1999. The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 includes genes necessary for formation of polyhedral organelles involved in coenzyme B12-dependent 1,2-propanediol degradation. J. Bacteriol. 181:5967-5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Born, T. L., and J. S. Blanchard. 1999. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 38:14416-14423. [DOI] [PubMed] [Google Scholar]
- 10.Brinsmade, S. R., and J. C. Escalante-Semerena. 2004. The eutD gene of Salmonella enterica encodes a protein with phosphotransacetylase enzyme activity. J. Bacteriol. 186:1890-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, L., and T. Elliott. 1996. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J. Bacteriol. 178:3763-3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, M., and A. Kornberg. 2004. Inorganic phosphate in the origin and survival of species. Proc. Natl. Acad. Sci. USA 101:16085-16087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Creighton, T. E. 1993. Proteins: structure and molecular properties, 2nd ed. W.H. Freeman and Co., New York, N.Y.
- 14.Crook, E., M. Aikiyama, N. N. Rao, and A. Kornberg. 1994. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 269:6290-6295. [PubMed] [Google Scholar]
- 15.Davis, R. W. 1980. A manual for genetic engineering: advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Dukan, S., A. Farewell, M. Ballestros, F. Taddei, M. Radman, and T. Nystrom. 2000. Protein oxidation in response to increased transcriptional or translational errors. Proc. Natl. Acad. Sci. USA 97:5746-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dukan, S., and T. Nystrom. 1998. Bacterial senescence: stasis results in increased and differential oxidation of cytoplasmic proteins leading to developmental induction of the heat shock regulon. Genes Dev. 12:3431-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dukan, S., and T. Nystrom. 1999. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem. 274:26027-26032. [DOI] [PubMed] [Google Scholar]
- 19.Fazzio, T., and J. Roth. Unpublished data.
- 20.Ferguson, G. P., D. McLaggan, and I. R. Booth. 1995. Potassium channel activation by glutathione-S-conjugates in Escherichia coli: protection against methylglyoxal is mediated by cytoplasmic acidification. Mol. Microbiol. 17:1025-1033. [DOI] [PubMed] [Google Scholar]
- 21.Giandomenico, A. R., G. E. Cerniglia, J. E. Biaglow, C. W. Stevens, and C. J. Koch. 1997. The importance of sodium pyruvate in assessing the damage produced by hydrogen peroxide. Free Radical Biol. Med. 23:426-434. [DOI] [PubMed] [Google Scholar]
- 22.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 23.Gur, E., D. Biran, E. Gazit, and E. Z. Ron. 2002. In vivo aggregation of a single enzyme limits growth of Escherichia coli at elevated temperatures. Mol. Microbiol. 46:1391-1397. [DOI] [PubMed] [Google Scholar]
- 24.Havemann, G. D., E. M. Sampson, and T. A. Bobik. 2002. PduA is a shell protein of polyhedral organelles involved in coenzyme B12-dependent degradation of 1,2-propanediol in Salmonella enterica serovar Typhimurium LT2. J. Bacteriol. 184:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hondorp, E., and R. Matthews. 2004. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2:1738-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horswill, A. R., A. R. Dudding, and J. C. Escalante-Semerena. 2001. Studies of propionate toxicity in Salmonella enterica identify 2-methylcitrate as a potent inhibitor of cell growth. J. Biol. Chem. 276:19094-19101. [DOI] [PubMed] [Google Scholar]
- 27.Hughes, K. T., and J. R. Roth. 1988. Transitory cis complementation: a method for providing transposition functions to defective transposons. Genetics 119:9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 29.Kim, K., N. N. Rao, C. D. Fraley, and A. Kornberg. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. USA 99:7675-7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kofoid, E. C., C. Rappleye, I. Stojiljkovic, and J. R. Roth. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologs of carboxysome shell proteins. J. Bacteriol. 181:5317-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 32.Kuroda, A., K. Nomura, R. Ohtomo, J. Kato, T. Ikeda, N. Takiguchi, H. Ohtake, and A. Kornberg. 2001. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science 293:705-708. [DOI] [PubMed] [Google Scholar]
- 33.Kuroda, A., S. Tanaka, T. Ikeda, J. Kato, N. Takiguchi, and H. Ohtake. 1999. Inorganic polyphosphate kinase is required to stimulate protein degradation and for adaptation to amino acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. USA 96:14264-14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwon, Y. M., and S. C. Ricke. 1998. Induction of acid resistance of Salmonella typhimurium by exposure to short-chain fatty acids. Appl. Environ. Microbiol. 64:3458-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert, L. A., K. Abshire, D. Blankenhorn, and J. L. Slonczewski. 1997. Proteins induced in Escherichia coli by benzoic acid. J. Bacteriol. 179:7595-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loewen, P. C., B. Hu, J. Strutinsky, and R. Sparling. 1998. Regulation in the rpoS regulon of Escherichia coli. Can. J. Microbiol. 44:707-717. [DOI] [PubMed] [Google Scholar]
- 37.Mori, K., R. Bando, N. Hieda, and T. Toraya. 2004. Identification of a reactivating factor for adenosylcobalamin-dependent ethanolamine ammonia lyase. 186:6845-6854. [DOI] [PMC free article] [PubMed]
- 38.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oikawa, S., and S. Kawanishi. 1996. Site-specific damage induced by NADH in the presence of copper(II): role of active oxygen species. Biochemistry 35:4584-4590. [DOI] [PubMed] [Google Scholar]
- 40.Penrod, J., T. Fazzio, C. Mace, E. C. Kofoid, and J. R. Roth. Unpublished data.
- 41.Price-Carter, M., J. Tingey, T. A. Bobik, and J. R. Roth. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463-2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rappleye, C., and J. R. Roth. 1997. A Tn10 derivative (TPOP) for isolation of insertions with conditional (tetracycline-dependent) phenotypes. J. Bacteriol. 179:5827-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roe, A. J., D. McLaggan, I. Davidson, C. O'Byrne, and I. R. Booth. 1998. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J. Bacteriol. 180:767-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roe, A. J., C. O'Byrne, D. McLaggan, and I. R. Booth. 2002. Inhibition of Escherichia coli growth by acetic acid: a problem with methionine biosynthesis and homocysteine toxicity. Microbiology 148:2215-2222. [DOI] [PubMed] [Google Scholar]
- 46.Ron, E. Z. 1975. Growth rate of Enterobacteriaceae at elevated temperatures: limitation by methionine. J. Bacteriol. 124:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ron, E. Z., and B. D. Davis. 1971. Growth rate of Escherichia coli at elevated temperatures: limitation by methionine. J. Bacteriol. 107:391-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ron, E. Z., and M. Shani. 1971. Growth rate of Escherichia coli at elevated temperatures: reversible inhibition of homoserine trans-succinylase. J. Bacteriol. 107:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roof, D. M., and J. R. Roth. 1988. Ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 170:3855-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell, J. B., and F. Diez-Gonzalez. 1998. The effects of fermentation acids on bacterial growth. Adv. Microb. Physiol. 39:205-234. [DOI] [PubMed] [Google Scholar]
- 51.Salmond, C. V., R. Kroll, and I. R. Booth. 1984. The effect of food preservatives on pH homeostasis in Escherichia coli. J. Gen. Microbiol. 130:2845-2850. [DOI] [PubMed] [Google Scholar]
- 52.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schellhorn, H. E., and V. L. Stones. 1992. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J. Bacteriol. 174:4769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmieger, H. 1971. A method of detection of phage mutants with altered transducing ability. Mol. Gen. Genet. 110:378-381. [DOI] [PubMed] [Google Scholar]
- 55.Seaver, L. C., and J. A. Imlay. 2001. Hydrogen peroxide fluxes and compartmentalization inside growing Escherichia coli. J. Bacteriol. 183:7182-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheppard, D. E., J. T. Penrod, T. Bobik, E. Kofoid, and J. R. Roth. 2004. Evidence that a B12-adenosyl transferase is encoded within the ethanolamine operon of Salmonella enterica. J. Bacteriol. 186:7635-7644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shiba, T., K. Tsutsumi, H. Yano, Y. Ihara, A. Kameda, S. Tanaka, H. Takahashi, M. Munekata, N. N. Rao, and A. Kornberg. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. USA 94:11210-11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shively, J., and R. English. 1991. The carboxysome, a prokaryotic organelle: a mini review. Can. J. Bot. 69:957-963. [Google Scholar]
- 59.Stadtman, E. R. 1993. Oxidation of free amino acids and amino acid residues in proteins by radiolysis and by metal-catalyzed reactions. Annu. Rev. Biochem. 62:797-821. [DOI] [PubMed] [Google Scholar]
- 60.Tomoyasu, T., A. Mogk, H. Langen, P. Goloubinoff, and B. Bukau. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397-413. [DOI] [PubMed] [Google Scholar]
- 61.Tsutsumi, K., M. Munekata, and T. Shiba. 2000. Involvement of inorganic polyphosphate in expression of SOS genes. Biochim. Biophys. Acta 1493:73-81. [DOI] [PubMed] [Google Scholar]
- 62.Van Dien, S. J., S. Keyhani, C. Yang, and J. D. Keasling. 1997. Manipulation of independent synthesis and degradation of polyphosphate in Escherichia coli for investigation of phosphate secretion from the cell. Appl. Environ. Microbiol. 63:1689-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Van Niel, E. W. J., K. Hofvendahl, and B. Hahn-Hägerdal. 2002. Formation and conversion of oxygen metabolites by Lactococcus lactis subsp. lactis ATCC 19435 under different growth conditions. Appl. Environ. Microbiol. 68:4350-4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wickner, S., M. R. Maurizi, and S. Gottesman. 1999. Posttranslational quality control: folding, refolding, and degrading proteins. Science 286:1888-1893. [DOI] [PubMed] [Google Scholar]