Abstract
Genome-wide analysis of temporal gene expression profiles in Escherichia coli following exposure to cadmium revealed a shift to anaerobic metabolism and induction of several stress response systems. Disruption in the transcription of genes encoding ribosomal proteins and zinc-binding proteins may partially explain the molecular mechanisms of cadmium toxicity.
Cadmium (Cd) has no known biological function and is of concern as an environmental contaminant due to its extreme toxicity, ability to cause mutations (35), and potential carcinogenicity in higher animals (2). Although Cd is not a redox-active metal, it has been speculated that Cd causes damage to cells primarily by the generation of reactive oxygen species (ROS) (53), which causes single-strand DNA damage and disrupts the synthesis of nucleic acids and proteins (37, 38). Cd is also an inhibitor of the DNA mismatch repair system (23, 35). Studies using two-dimensional gel electrophoresis also have shown that several stress response systems are expressed in response to Cd exposure, including those for heat shock, oxidative stress, stringent response, cold shock, and SOS (6, 17, 57). However, the overall response of cells to Cd toxicity at the genome level is not yet completely understood.
To examine bacterial cell responses to Cd exposure at the genome level, the present study employed a high-density oligonucleotide microarray (Affymetrix), which permits monitoring of 4,290 annotated open reading frames and intergenic regions (33, 52, 55) in Escherichia coli. Results of this study confirmed that Cd toxicity caused profound changes in gene expression in which several stress response systems were induced simultaneously. Changes in gene expression were highly dynamic as the bacterial cells responded to the ensuing toxicity which was manifested by a shift to anaerobic metabolism, up-regulation of genes related to energy conservation, and disruption in the synthesis of r-proteins and zinc-binding proteins.
Bacteria and growth conditions.
A single colony of E. coli K-12 (MG1655) was suspended in 80 ml of defined MOPS (morpholinepropanesulfonic acid) medium (http://www.genome.wisc.edu/), divided into two 250-ml Erlenmeyer flasks (replicate flasks), and grown in a shaker at 300 rpm and 37°C with constant aeration until the contents of the flasks reached an optical density at 600 nm of 0.6. The cells were exposed to Cd by adding a final concentration of 1 μg/ml of Cd (as CdCl2) to the growth medium. Cells were collected at 0 (no Cd addition, control), 5, 15, and 25 min. The cells were frozen immediately and stored at -80°C until further processing.
RNA purification and microarray procedures.
A Masterpure RNA purification kit (Epicentre Technologies) was used to purify RNA. The quality and quantity of the purified RNA were determined spectrophotometrically and by gel analysis. Affymetrix E. coli gene chips were used to run the microarray analysis. Targets were prepared by following the Affymetrix protocols. In brief, mRNAs were enriched by eliminating 16S and 23S rRNA and then fragmented and labeled with biotin. Hybridization and scanning were performed by the University of California, Irvine, Microarray Center.
Data analysis.
Data analysis was performed using the DNA-Chip Analyzer (dChip) 1.3 software program (http://www.dchip.org/) (29, 30). The data were normalized using the invariant set method (29). The array with median file cell intensity values was chosen as the baseline array (default). The model-base (perfect match/mismatch) expression value was calculated. To minimize occurrence of false positives, two steps were performed to filter the data. The first step compared samples (arrays) in a pair-wise fashion with the threshold for change at greater than twofold with a 90% confidence interval and with a difference in expression values between the genes of >100. A total of 1,525 transcripts satisfied these criteria (see supplemental Table 1). The 1,525 transcripts were subjected to secondary filtering using the filter gene function in dChip by setting the coefficient of variation at >0.4 (n = 5) and the gene present call at >20%. After the filtering process, a total of 674 transcripts (see supplemental Table 2) showed significant changes. These 674 transcripts were subjected to cluster analysis and classification analysis.
Global gene expression profile.
Genechips from Affymetrix and real-time reverse transcription-PCR (see supplemental Fig. 1 at http://www.envisci.ucr.edu/downloads/crowley/cadmiumsupplement.html) were employed to profile the genome-wide gene expression patterns in E. coli cells during mid-log-phase growth after exposure to 1 μg/ml Cd, which is nonlethal to E. coli (17).
All supplemental tables and figures are available at http://www.envisci.ucr.edu/downloads/crowley/cadmiumsupplement.html.
Results of the microarray analysis revealed that Cd caused significant changes (>2-fold) in the expression of 1,525 transcripts (see supplemental Table 1). After secondary filtering was done (29, 30), the 1,525 transcripts were reduced to a more conservative set of 674 transcripts (see supplemental Table 2), including 303 genes with known functions (see supplemental Table 3), 117 open reading frames (ORFs) with unknown functions, 223 intergenic regions, and 31 nonclassified transcripts. Gene annotations were based on published data (5) and an internet database (http://bmb.med.miami.edu/EcoGene/EcoWeb/). The 303 genes with known functions were distributed among 21 of the 22 gene annotation groups of E. coli (5) (see supplemental Table 3), the only exception being the “putative chaperone” group. Among the transcripts that were affected, 239 transcripts were up-regulated only at one or more time points (see supplemental Table 3-1). These included 111 known genes, 48 unknown ORFs, and 80 intergenic regions. In contrast, a similar number (270) of transcripts were down-regulated only at one or more time points, including 136 known genes, 35 unknown genes, and 99 intergenic regions. Another set of 134 transcripts was dynamic and underwent both significant up- and down-regulation over the tested time course. Gene groups in which transcripts primarily underwent down-regulation included those for amino acid biosynthesis (3 up-regulated and 8 down-regulated), energy metabolism (15 up-regulated and 22 down-regulated), and protein translation (12 up-regulated and 40 down-regulated). Gene groups that were primarily up-regulated included regulatory genes (3 up-regulated and 1 down-regulated), putative regulatory genes (6 up-regulated and 2 down-regulated), and genes involved in DNA replication (6 up-regulated and 4 down-regulated).
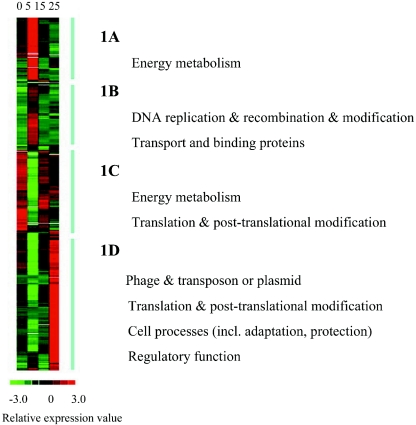
To further examine the dynamics of gene expression, the 674 transcripts were divided into four distinct clusters according to their temporal expression patterns (Fig. 1). These clusters were predominated by three functional groups (P < 0.001) that included genes for translation and posttranslational modification (clusters 1C and D), energy metabolism (clusters 1A and C), and transcripts related to phage, transposon, and plasmid DNA (cluster 1D). These data indicated that arrest of protein synthesis, alteration of energy metabolism, and cell rescue were overall features of Cd toxicity. Because the general cluster profiles may hide some significant patterns within these gene groups (29, 30), the four clusters were subjected to further analyses. Several other significant clusters were revealed (P < 0.05) and are discussed in detail below.
FIG. 1.
Temporal transcription profiles induced by Cd in E. coli drawn by cluster analysis. Each column represents one time point, i.e., 0 min (no Cd), 5 min, 15 min, and 25 min, from left to right, respectively. Each row represents one gene. Color represents change in relative expression value: red, up-regulation; black, no change; green, down-regulation; and white, missing. The profiles were divided into four clusters: 1A, dominated by energy metabolism; 1B, DNA replication and recombination and modification, transport and binding proteins; 1C, energy metabolism, translation and posttranslational modification; 1D, phage and transposon or plasmid, translation and posttranslational modification, cell processes (including adaptation and protection), regulatory function.
Translation and posttranslational modification.
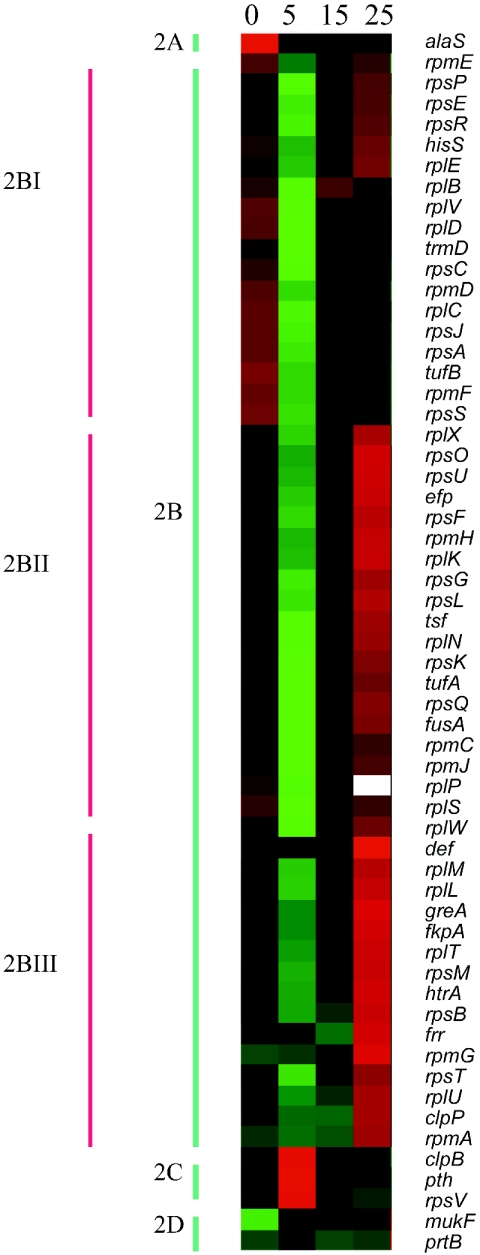
Expression patterns of genes in the translation and posttranslational modification group underwent the greatest changes in response to Cd (Fig. 2). All genes for protein translation machinery (clusters 2A and 2B) were down-regulated in the early phase (EP; at 5 min), after which some resumed expression in the late phase (LP; at 25 min) (cluster 2B), while genes for stress proteins (clusters 2C and 2D) were mostly up-regulated in the EP. This finding is consistent with the decline in the overall rate of protein synthesis that has previously been observed in response to Cd (3) and is in agreement with earlier studies showing up-regulation of heat shock proteins that are induced by Cd (3, 17). Among the stress protein genes that were upregulated in EP were clpB (ClpB) and sra (RpsV, S22) (cluster 2C). ClpB functions primarily with DnaK to prevent protein aggregation, while S22 helps with 100S ribosomal protein formation (22, 60).
FIG. 2.
Temporal transcription profiles for genes involved in translation and posttranslational modification (associated with clusters 1C and 1D). The 59 genes in this cluster were further grouped into four clusters based on their dynamics over time: 2A (1 gene), featured by synthetase with down-regulation in all phases; 2B (53 genes), comprised of genes encoding r-proteins and translation factors with diverse regulation, all down-regulated in EP; 2C (3 genes), stress proteins, up-regulation in EP; 2D (2 genes), killing proteins, up-regulation. Cluster 2B was further divided into three subclusters: 2BI (18 genes), essential translation factors, down-regulation; 2BII (20 genes), elongation factors, up-regulated in LP; and 2BIII (15 genes), regulators, up-regulated in LP.
Previously, modulation of r-proteins in E. coli has been shown to occur in response to superoxide, sodium salicylate, low temperature, and H2O2 (11, 46, 48) but has not been investigated in relation to Cd toxicity. Our data revealed that 40 of the 59 transcripts that encode r-proteins were affected by Cd. To further explore the temporal shifts in r-proteins following exposure to Cd, the genes were further divided into three subclusters: 2BI, 2BII, and 2BIII (Fig. 2). Transcripts contained in 2BI were down-regulated in all phases (EP, intermediate phase [IP], and LP) after exposure to Cd and included predominantly essential translation factors, such as L2, L3, L4 (essential elements for peptidyl transferase), S5 (rpsE; a suppressor) (28), S3 (an inducer of antiapoptotic proteins) (47), and S16 (an essential element for 30S assembly) (45). Transcripts in subcluster 2BII underwent slight up-regulation in the LP and primarily included elongation factors, such as S17 (rpsQ; an antisuppressor) (32), L19, L16, and S7. Transcripts in subcluster 2BIII were significantly up-regulated in the LP and primarily encoded regulators, such as L9 (rplI; involved in negative ribosome slippage) (21) and L20 (rplT; involved in negative autogenous regulation) (44). The whole profile revealed that E. coli shuts down transcription and translation immediately after exposure to Cd (within 5 min; subclusters 2BI, 2BII, and 2BIII) but reassembles its r-proteins by the LP (within 25 min; subclusters 2BI, 2BII, and 2BIII). In comparison to the cells that were not exposed to Cd, the overexpression of regulators and elongation factors (subclusters 2BIII and 2BII) and the repression of essential translation factors (subcluster 2BI) in the LP suggest that ribosome reassembly may be imbalanced by Cd toxicity.
Energy metabolism.
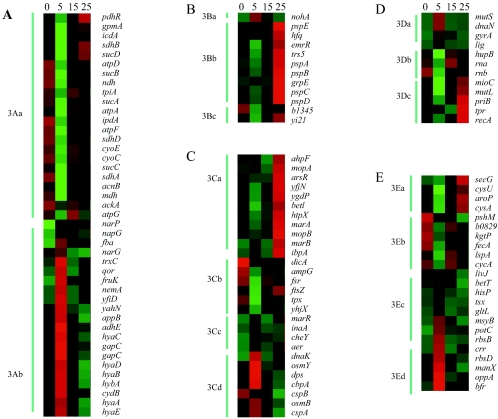
The second transcript group that underwent a significant change following exposure to Cd included the genes involved in energy metabolism, which grouped in clusters 1A and 1C in Fig. 1 and in clusters 3Aa and 3Ab in Fig. 3. Genes in cluster 3Aa, such as those in the atp operon (encoding F0F1-ATPase), sdhCDAB-sucABCD, and pdhR-lpdA, are primarily associated with aerobic metabolism, while genes in cluster 3Ab, such as the hya operon, adhE, gapC, hybA (61), and narG, are predominantly associated with anaerobic metabolism. The overexpression of genes in cluster 3Ab and depressed expression of genes in cluster 3Aa indicate that the cells switched from aerobic respiration to anaerobic metabolism following exposure to Cd. Likewise, repression of pdhR, sdhCDAB, and sucABCD (cluster 3Aa), which is believed to mark the switch from the citric acid cycle to its branched or noncyclic anaerobic form (12), and overexpression of genes encoding energy-conserving dehydrogenases, like HyaABCD (56) (cluster 3Ab), suggest that cellular metabolism was altered to promote energy conservation. Altogether, these results suggest that E. coli switches to an energy conservation mode after exposure to Cd.
FIG. 3.
Gene transcript profiles. (A) Energy metabolism group, which includes 43 genes grouped into two clusters: 3Aa (23 genes), proteins for aerobic metabolism, down-regulated in EP and slightly resumed after IP; and 3Ab (20 genes), proteins for anaerobic metabolism, up-regulated only in EP. (B) Phage, transposon, or plasmid group (cluster 1D), which includes 12 genes grouped into three clusters: 3Ba (1 gene), nohA (defense against oxygen radicals), up-regulated in EP; 3Bb (9 genes), phage shock proteins, greatly increased in LP; 3Bc (2 genes), unclear function, down-regulated in EP and resuming after IP. (C) Cell processes group (cluster 1D), which includes 28 genes grouped into four clusters: 3Ca (11 genes), stress proteins, up-regulated in LP; 3Cb (6 genes), cell division, down-regulated; 3Cc (4 genes), taxis, up-regulated; 3Cd (7 genes), broader stress proteins, high overexpression in EP. (D) DNA replication, recombination, modification, and repair group (cluster 1B), which includes 12 genes in 3 clusters: 3Da (4 genes), beta-sliding-clamp-interacting proteins, up-regulated in EP; 3Db (3 genes), cleavage proteins, down-regulated; 3Dc (5 genes), restart and repair of replication proteins, overexpressed in LP. (E) Transport and binding proteins (cluster 1B), which includes 23 genes in 4 clusters: 3Ea (4 genes), cysteine-containing proteins, up-regulated in LP; 3Eb (6 genes), transport systems with high-energy-dependent down-regulation; 3Ec (8 genes), components of uptake systems of high binding affinity and low energy cost, up-regulated; 3Ed (5 genes), carbohydrate phosphotransferase system (PTS), up-regulated in EP.
The repression of genes for aerobic metabolism and stimulation of qor following Cd exposure is similar to the cell responses that are induced by other stresses, such as H2O2 and superoxide (11, 46). However, this study revealed more fundamental changes in gene transcription that are caused by Cd than have been reported for oxidative stress. Here, the promoters for pdhR, lpdA, sdhCDAB, and sucABCD were strongly corepressed in the EP (cluster 3Aa), and the transcriptional profiles of these operons (lpdA, sdhCDAB, and sucABCD) were correlated with those of their corresponding intergenic regions. The microarray probe sets IG_69127588_127911_fwd_f_st, IG_483_757629_757686_fwd_f_st (see supplemental Table 3), and IG_484_761963_762236_fwd_st (see supplemental Table 1) are designed to monitor the intergenic regions between aceF-lpdA, sdhB-sucA, and sucB-sucC, respectively. The alteration of their transcriptions in correspondence with their operons indicated that read-through occurs between the above-mentioned operons (ace-lpd, sdh-suc, sucAB-sucCD) following exposure to Cd. In contrast, the probe set for the intergenic region of glt-sdh, IG_482_753692_764375_rev_f_st, which is transcribed in the opposite direction of the sdh operon, did not show any significant change in transcription, indicating that Psdh and Ppdh coregulate sdhCDAB-sucABCD and pdhR-lpdA, respectively, during Cd toxicity. The above-described results are in agreement with prior studies of these genes using Northern blot analysis under similar experimental conditions (13).
Phage, transposon, or plasmid.
Nearly all of the genes in the phage, transposon, and plasmid category (Fig. 3B) were up-regulated following exposure to Cd. Genes in the phage shock protein operon (pspABCE), which is driven by σ54 instead of σ32, in particular, also were expressed (cluster 3Bb), along with σ54 (see supplemental Table 3). Prior studies have shown that the psp operon, which may be related to translation (9, 24), is strongly induced by phage infection, ethanol treatment, osmotic shock, heat shock, and prolonged incubation in stationary phase. In addition to these genes, grpE and hfq were also up-regulated (cluster 3Bb). GrpE, along with DnaK/DnaJ and ClpB, participates in a multichaperone system (39). Hfq (cluster 3Bb) facilitates access of the replicase for phage Qβ replication (51) and positively regulates the rpoS gene (40). Efflux systems in E. coli play a key role in stress tolerance by pumping out both drugs and heavy metals from cells (31). In general, the efficacy of these transport systems varies with different bacterial species (41-43). One of the best-studied systems in E. coli is the zinc export system (ZntA), which is a P-type ATPase that is regulated by ZntR. Although the primary role of ZntA is for Zn efflux, induction of zntA has been reported to be 13-fold greater for Cd than for Zn (4, 10). It has therefore been speculated that ZntA is a major Cd detoxification system in E. coli. However, this hypothesis was not strongly supported in the experiment performed here, as the emrAB system for multidrugs, which is repressed by EmrR (cluster 3Bb) (62), was not significantly expressed in comparison to the control. Likewise, the zinc efflux system (ZntA) was up-regulated only twofold (see supplemental Table 3).
Cell processes.
Genes encoding stress proteins were strongly up-regulated in response to Cd (Fig. 3C). Among the most important and abundant genes in this group are those that encode heat shock proteins (Hsp). In the EP, dnaK or clpB (cluster 3Cd or 2C, respectively), which encodes a bichaperone system (49), was up-regulated. In the LP, after the cells resumed cell division, marked by expression of ftsZ and dicA (cluster 3Cb), various chaperone systems were significantly activated, including the following: ibpA (hslT; cluster 3Ca); mopA and mopB (GroEL/GroES; cluster 3Ca); clpP (ClpP; cluster 2BIII); grpE (GrpE; cluster 3Bb), which couples with dnaK (cluster 3Cd); htpX (HtpX; cluster 3Ca); and rpoH (see supplemental Table 3). In addition to hsp, genes for other stress response systems were also expressed. In the EP, these included osmB and osmY (OsmB/OsmY in cluster 3Cd; hyperosmotic response), dps (Dps in cluster 3Cd; oxidative or nutritional stress), cspABCE (cspAB in cluster 3Cd and cspCE [see ST 3]; cold shock system), inaA (in cluster 3Cc; acid resistance), cheY (in cluster 3Cc; chemotaxis), and aer (in cluster 3Cc; aerotaxis), while in the LP, marAB (in cluster 3Ca; multiple antibiotic resistance), which is repressed by MarR (marR in cluster 3Cc); ahpF (in cluster 3Ca; major H2O2 degradation); and nudH (ygdP in cluster 3Ca; nudix hydrolases) were expressed.
DNA replication, recombination, modification, and repair.
There were 12 genes associated with clusters 1B and 3D. In general, the genes coding for DNA repair and recombination systems were up-regulated (clusters 3Da and 3Dc), whereas genes encoding DNA replication and translation systems (cluster 3Db), including hupB, rna, and rnb, were down-regulated. Interestingly, dnaN (β clamp) was up-regulated with mutS (MutS), lig (ligase), and gyrA (gyrase) in the same cluster (cluster 3Da). One possible function of dnaN is to negatively regulate dnaA (DnaA) (26, 58), which initiates oriC replication. After ATP became available in the LP (see cluster 2B), mioC (cluster 3Dc) was up-regulated, and the initiation of replication was possibly resumed. Another possible function of dnaN may be to bind other gene products in the same cluster (cluster 3Da), such as MutS (cluster 3Da), to perform broader repair (34). The dnaN product may also interact with another member of this cluster, DNA ligase (34), for Okazaki fragment maturation. This is in agreement with the prior observation that Cd toxicity is associated with the mismatch repair system (23).
Another interesting up-regulation occurred with the recA gene (cluster 3Dc). Expression of recA (cluster 3Dc) and dnaN indicated that both the repair and recombination systems were triggered to cope with Cd toxicity. In contrast to up-regulation of recA and dnaN, our results showed that the well-characterized systems that are induced by ROS, such as GO (36) and base excision DNA repair (15), were not significantly induced by acute exposure to Cd. In addition, OxyR was not induced. Altogether, this suggests that the primary DNA damage caused by Cd is not directly associated with ROS, which has previously been proposed as one of the mechanisms by which Cd causes cell damage (53).
Transport and binding proteins.
Cluster 3Ea was associated with cysteine-related proteins (CysU, CysA, and AroP [cysteine aromatic]). One likely function of these proteins is to bind Cd. The overexpression of genes encoding ATP-dependent systems like Cys (ATP dependent) suggests that detoxification had priority over energy conservation when energy metabolism resumed in the LP (Fig. 2B). The transcript secG (SecG), whose function is ambiguous (18), was also up-regulated (cluster 3Ea). Based on the assumption that protein translocation systems can bidirectionally transport target proteins (50), SecG may work along with other Sec proteins to transport amino acids to detoxify Cd.
Biosynthesis of amino acids is a process that requires high-energy consumption, whereas uptake of amino acids via periplasmic binding proteins with high affinity uses much less energy. Under the energy crisis experienced following exposure to Cd in the EP (cluster 2B), E. coli appeared to shift to low-energy-requiring transport systems for importing amino acids and carbon sources by up-regulating transport-related genes (clusters 3Ec and 3Ed) while shutting down the production of transport systems that required energy (cluster 3Eb).
Another function of the genes in clusters 3Ec and 3Ed may be related to detoxification of Cd, such as that performed by livJ, cycA or sbp, hisJQMP, betT (osmoregulatory), and bfr (8). Superoxide stress and sodium also activate various genes for periplasmic binding proteins and proteins in the PTS family, but the individual genes are quite different from those activated by Cd in this study (46). However, some genes induced by acetate are very similar to those that were induced by Cd, such as oppA (27). This is consistent with the idea that ROS are not a primary result of Cd toxicity and that Cd detoxification is more complex than induction of the ROS system.
Regulatory function and network.
The regulatory function and network group contained six regulator genes, osmE, lexA, ompR, rpoE, fur, and soxR (see supplemental Fig. 2). lexA was the only gene in this group that was down-regulated in the EP; osmE was up-regulated in the EP and the rest were up-regulated in the LP. Although there were only six genes in this group, they could play a pivotal role in gene regulation because regulators often work together to cope with specific stimuli. It is therefore of interest to analyze other genes that have similar regulatory functions with the above-mentioned six genes together to unravel the regulatory network that occurs during Cd toxicity in E. coli. These genes were selected from supplemental Table 3 and are listed in supplemental Table 4 and plotted in supplemental Fig. 3.
Stress responses activated here included those for osmotic stress (OsmE and OmpR), heat shock (σE), and superoxide (Fur and SoxR). osmE encodes an envelope protein of unknown function that is regulated by σ70 and σS at elevated osmotic pressure and decelerating growth, respectively (7). Similarly, the OmpR (regulator)/EnvZ (sensor) system regulates an osmosensory pathway. rpoE (σE) regulates 43 genes (14). Both σE and σ32, the latter of which is also regulated by σE (16), are activated by stress to respond to misfolded/unfolded proteins in the periplasm and cytoplasm, respectively (63). In agreement with prior research, our data suggest that σE may also mediate the expression of genes for the biosynthesis/transport of lipopolysaccharides (14), such as lpxD, nlpD, fkpA, and htrA (protease to allow E. coli to remain viable at >42°C), along with rpoD (σ70) and rpoH (σ30) (see supplemental Table 4).
Another important regulator protein that was up-regulated by Cd was Fur (fur), which controls 90 genes (20). Fur negatively regulates the uptake systems for iron and zinc and is also a positive regulator for genes that are involved in the detoxification of ROS (20), such as fecA and acnB (see supplemental Table 4). The transcription of fur is activated by SoxR (soxR), a regulatory protein of the soxRS regulon for superoxide (64). SoxR also regulates marA (see supplemental Table 4), which transcribes over 60 genes (1), and CspA (cold shock protein) (see supplemental Table 3). Another regulator that was depressed by Cd was lexA, which regulates as many as 30 proteins (54). While lexA was depressed, genes coding for the functional components of the SOS response, recA (cluster 3Dc), dnaN (cluster 3Da), dinJ (see ST 3), and uvrB (see supplemental Table 1), were up-regulated, suggesting that the main repair pathway activated by Cd is the nucleotide excision repair system, which typically responds to UV and is characterized by involvement of the Uvr system (uvrB [see supplemental Table 1]). This nucleotide excision repair system is in lieu of the system for base excision repair, which responds to oxidative damage and is characterized by glycosylase activity (59).
To appreciate the general regulation of transcription in response to Cd stress, all transcripts were sorted with respect to their degree of up- or down-regulation (see supplemental Table 5). The genes with greatest up-regulation were those which function for repair and transport with low-energy cost (EP) and for transport systems with intermediate-energy requirements (IP), as well as for stress proteins with high-energy cost (LP), while genes for biosynthesis were down-regulated in all phases. Interestingly, the intergenic region IG_2542_4117930_4117995_rev_st was the most highly expressed sequence. This region is between menA and hslU, with counterclockwise transcription. MenA (menaquinone, VK12) (see supplemental Table 1) is important in the anaerobic electron transport system and detoxification. It is therefore speculated that overexpression of the intergenic region may be related to energy conservation and Cd detoxification (Fig. 3A) at EP. In addition, some less-characterized ORFs were highly induced by Cd, such as probe set b2383 and ydbH (see supplemental Table 5). Identification of these highly expressed transcripts may be of value in designing reporter gene assays for detecting bioavailable Cd in the environment.
Replacement of Zn by Cd leads to dysfunction and structural alternation of related proteins (19). After examination of the database, 14 genes out of the 30 known genes coding for zinc-binding proteins (25) showed significant changes following exposure to Cd (see supplemental Table 6 and supplemental Fig. 4). Of these, the dominant genes (eight genes) belonged to the translation and posttranslational modification group (Fig. 2), most of which (six genes) were r-protein subunits that were up-regulated in the LP. This may be explained as a response to compensate for loss of functional zinc-binding proteins due to substitution of zinc by Cd. In higher organisms, the replacement of zinc by Cd might potentiate carcinogenesis and other disease processes (19).
Conclusions.
The present study used a microarray approach to investigate dynamic genome-wide transcription responses that are induced by Cd in a model organism, E. coli. The results showed that exposure to Cd caused an arrest in protein biosynthesis, a shift to anaerobic metabolism, and lowered energy production. Mistaken assembly of r-proteins and Cd replacement in zinc-binding and cysteine-rich proteins may account for some of the damage caused by Cd. Induction of a complex network of regulatory systems was observed, including those that function for DNA repair, heat shock, oxidative stress, cold shock, osmotic stress, taxis, acid, antibiotic, and nudix hydrolases, as well as efflux systems for heavy metals that have previously been shown to facilitate resistance to Cd. Results from this study may be valuable for guiding future research on mechanisms of Cd toxicity and for stimulating comparative studies on gene expression in other organisms in which Cd causes mutations and cancer.
Acknowledgments
This research was supported by grant DE-FG07-96ER20255 from the U.S. Department of Energy.
REFERENCES
- 1.Barbosa, T. M., and S. B. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyersmann, D. 2002. Effects of carcinogenic metals on gene expression. Toxicol. Lett. 127:63-68. [DOI] [PubMed] [Google Scholar]
- 3.Beyersmann, D., and S. Hechtenberg. 1997. Cadmium, gene regulation, and cellular signalling in mammalian cells. Toxicol. Appl. Pharmacol. 144:247-261. [DOI] [PubMed] [Google Scholar]
- 4.Binet, M. R., and R. K. Poole. 2000. Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase ZntA in Escherichia coli. FEBS Lett. 473:67-70. [DOI] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Blom, A., W. Harder, and A. Matin. 1992. Unique and overlapping pollutant stress proteins of Escherichia coli. Appl. Environ. Microbiol. 58:331-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bordes, P., F. Repoila, A. Kolb, and C. Gutierrez. 2000. Involvement of differential efficiency of transcription by esigmas and esigma70 RNA polymerase holoenzymes in growth phase regulation of the Escherichia coli osmE promoter. Mol. Microbiol. 35:845-853. [DOI] [PubMed] [Google Scholar]
- 8.Bou-Abdallah, F., A. C. Lewin, N. E. Le Brun, G. R. Moore, and N. D. Chasteen. 2002. Iron detoxification properties of Escherichia coli bacterioferritin. Attenuation of oxyradical chemistry. J. Biol. Chem. 277:37064-37069. [DOI] [PubMed] [Google Scholar]
- 9.Brissette, J. L., L. Weiner, T. L. Ripmaster, and P. Model. 1991. Characterization and sequence of the Escherichia coli stress-induced psp operon. J. Mol. Biol. 220:35-48. [DOI] [PubMed] [Google Scholar]
- 10.Brocklehurst, K. R., J. L. Hobman, B. Lawley, L. Blank, S. J. Marshall, N. L. Brown, and A. P. Morby. 1999. ZntR is a Zn(II)-responsive MerR-like transcriptional regulator of zntA in Escherichia coli. Mol. Microbiol. 31:893-902. [DOI] [PubMed] [Google Scholar]
- 11.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 12.Cronan, J. E., Jr., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxylate bypass, p. 206-216. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 13.Cunningham, L., and J. R. Guest. 1998. Transcription and transcript processing in the sdhCDAB-sucABCD operon of Escherichia coli. Microbiology 144:2113-2123. [DOI] [PubMed] [Google Scholar]
- 14.Dartigalongue, C., D. Missiakas, and S. Raina. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866-20875. [DOI] [PubMed] [Google Scholar]
- 15.Demple, B., and M. S. DeMott. 2002. Dynamics and diversions in base excision DNA repair of oxidized abasic lesions. Oncogene 21:8926-8934. [DOI] [PubMed] [Google Scholar]
- 16.Erickson, J. W., and C. A. Gross. 1989. Identification of the sigma E subunit of Escherichia coli RNA polymerase: a second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 17.Ferianc, P., A. Farewell, and T. Nystrom. 1998. The cadmium-stress stimulon of Escherichia coli K-12. Microbiology 144:1045-1050. [DOI] [PubMed] [Google Scholar]
- 18.Flower, A. M., L. L. Hines, and P. L. Pfennig. 2000. SecG is an auxiliary component of the protein export apparatus of Escherichia coli. Mo.l Gen. Genet. 263:131-136. [DOI] [PubMed] [Google Scholar]
- 19.Hanas, J. S., and C. G. Gunn. 1996. Inhibition of transcription factor IIIA-DNA interactions by xenobiotic metal ions. Nucleic Acids Res. 24:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 21.Herr, A. J., C. C. Nelson, N. M. Wills, R. F. Gesteland, and J. F. Atkins. 2001. Analysis of the roles of tRNA structure, ribosomal protein L9, and the bacteriophage T4 gene 60 bypassing signals during ribosome slippage on mRNA. J. Mol. Biol. 309:1029-1048. [DOI] [PubMed] [Google Scholar]
- 22.Houry, W. A. 2001. Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Peptide Sci. 2:227-244. [DOI] [PubMed] [Google Scholar]
- 23.Jin, Y. H., A. B. Clark, R. J. Slebos, H. Al-Refai, J. A. Taylor, T. A. Kunkel, M. A. Resnick, and D. A. Gordenin. 2003. Cadmium is a mutagen that acts by inhibiting mismatch repair. Nat. Genet. 34:326-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jovanovic, G., L. Weiner, and P. Model. 1996. Identification, nucleotide sequence, and characterization of PspF, the transcriptional activator of the Escherichia coli stress-induced psp operon. J. Bacteriol. 178:1936-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katayama, A., A. Tsujii, A. Wada, T. Nishino, and A. Ishihama. 2002. Systematic search for zinc-binding proteins in Escherichia coli. Eur. J. Biochem. 269:2403-2413. [DOI] [PubMed] [Google Scholar]
- 26.Katayama, T., T. Kubota, K. Kurokawa, E. Crooke, and K. Sekimizu. 1998. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the E. coli chromosomal replicase. Cell 94:61-71. [DOI] [PubMed] [Google Scholar]
- 27.Kirkpatrick, C., L. M. Maurer, N. E. Oyelakin, Y. N. Yoncheva, R. Maurer, and J. L. Slonczewski. 2001. Acetate and formate stress: opposite responses in the proteome of Escherichia coli. J. Bacteriol. 183:6466-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurland, C. G., D. Hughes, and M. Ehrenberg. 1996. Limitations of translational accuracy, p. 979-1004. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C. [Google Scholar]
- 29.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032. [Online.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, L., Z. He, G. K. Pandey, T. Tsuchiya, and S. Luan. 2002. Functional cloning and characterization of a plant efflux carrier for multidrug and heavy metal detoxification. J. Biol. Chem. 277:5360-5368. [DOI] [PubMed] [Google Scholar]
- 32.Liebman, S. W., Y. O. Chernoff, and R. Liu. 1995. The accuracy center of a eukaryotic ribosome. Biochem. Cell. Biol. 73:1141-1149. [DOI] [PubMed] [Google Scholar]
- 33.Lipshutz, R. J., S. P. Fodor, T. R. Gingeras, and D. J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20-24. [DOI] [PubMed] [Google Scholar]
- 34.Lopez de Saro, F. J., and M. O'Donnell. 2001. Interaction of the beta sliding clamp with MutS, ligase, and DNA polymerase I. Proc. Natl. Acad. Sci. USA 98:8376-8380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McMurray, C. T., and J. A. Tainer. 2003. Cancer, cadmium and genome integrity. Nat. Genet. 34:239-241. [DOI] [PubMed] [Google Scholar]
- 36.Michaels, M. L., and J. H. Miller. 1992. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine). J. Bacteriol. 174:6321-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra, R. S. 1984. Protein synthesis in Escherichia coli during recovery from exposure to low levels of Cd2+. Appl. Environ. Microbiol. 47:1012-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitra, R. S., and I. A. Bernstein. 1978. Single-strand breakage in DNA of Escherichia coli exposed to Cd2+. J. Bacteriol. 133:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Motohashi, K., Y. Watanabe, M. Yohda, and M. Yoshida. 1999. Heat-inactivated proteins are rescued by the DnaK. J-GrpE set and ClpB chaperones. Proc. Natl. Acad. Sci. USA 96:7184-7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muffler, A., D. Fischer, and R. Hengge-Aronis. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 10:1143-1151. [DOI] [PubMed] [Google Scholar]
- 41.Nies, D. H. 1992. CzcR and CzcD, gene products affecting regulation of resistance to cobalt, zinc, and cadmium (czc system) in Alcaligenes eutrophus. J. Bacteriol. 174:8102-8110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 43.Noll, M., and S. Lutsenko. 2000. Expression of ZntA, a zinc-transporting P1-type ATPase, is specifically regulated by zinc and cadmium. IUBMB Life 49:297-302. [DOI] [PubMed] [Google Scholar]
- 44.Olsson, C. L., M. Graffe, M. Springer, and J. W. Hershey. 1996. Physiological effects of translation initiation factor IF3 and ribosomal protein L20 limitation in Escherichia coli. Mol. Gen. Genet. 250:705-714. [DOI] [PubMed] [Google Scholar]
- 45.Persson, B., G. Bylund, D. Berg, and P. Wikstrom. 1995. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 177:5554-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pomposiello, P. J., M. H. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3:179-192. [DOI] [PubMed] [Google Scholar]
- 48.Sahara, T., T. Goda, and S. Ohgiya. 2002. Comprehensive expression analysis of time-dependent genetic responses in yeast cells to low temperature. J. Biol. Chem. 277:50015-50021. [DOI] [PubMed] [Google Scholar]
- 49.Schlieker, C., B. Bukau, and A. Mogk. 2002. Prevention and reversion of protein aggregation by molecular chaperones in the E. coli cytosol: implications for their applicability in biotechnology. J. Biotechnol. 96:13-21. [DOI] [PubMed] [Google Scholar]
- 50.Schnell, D. J., and D. N. Hebert. 2003. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell 112:491-505. [DOI] [PubMed] [Google Scholar]
- 51.Schuppli, D., G. Miranda, H. C. Tsui, M. E. Winkler, J. M. Sogo, and H. Weber. 1997. Altered 3′-terminal RNA structure in phage Qbeta adapted to host factor-less Escherichia coli. Proc. Natl. Acad. Sci. USA 94:10239-10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Selinger, D. W., K. J. Cheung, R. Mei, E. M. Johansson, C. S. Richmond, F. R. Blattner, D. J. Lockhart, and G. M. Church. 2000. RNA expression analysis using a 30 base pair resolution Escherichia coli genome array. Nat. Biotechnol. 18:1262-1268. [DOI] [PubMed] [Google Scholar]
- 53.Stohs, S. J., and D. Bagchi. 1995. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 18:321-336. [DOI] [PubMed] [Google Scholar]
- 54.Sutton, M. D., B. T. Smith, V. G. Godoy, and G. C. Walker. 2000. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu. Rev. Genet. 34:479-497. [DOI] [PubMed] [Google Scholar]
- 55.Tjaden, B., R. M. Saxena, S. Stolyar, D. R. Haynor, E. Kolker, and C. Rosenow. 2002. Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays. Nucleic Acids Res. 30:3732-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Unden, G., and J. Bongaerts. 1997. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim. Biophys. Acta 1320:217-234. [DOI] [PubMed] [Google Scholar]
- 57.VanBogelen, R. A., P. M. Kelley, and F. C. Neidhardt. 1987. Differential induction of heat shock, SOS, and oxidation stress regulons and accumulation of nucleotides in Escherichia coli. J. Bacteriol. 169:26-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Villarroya, M., I. Perez-Roger, F. Macian, and M. E. Armengod. 1998. Stationary phase induction of dnaN and recF, two genes of Escherichia coli involved in DNA replication and repair. EMBO J. 17:1829-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Volkert, M. R., and P. Landini. 2001. Transcriptional responses to DNA damage. Curr. Opin. Microbiol. 4:178-185. [DOI] [PubMed] [Google Scholar]
- 60.Wada, A. 1998. Growth phase coupled modulation of Escherichia coli ribosomes. Genes Cells 3:203-208. [DOI] [PubMed] [Google Scholar]
- 61.Wu, L. F., M. A. Mandrand-Berthelot, R. Waugh, C. J. Edmonds, S. E. Holt, and D. H. Boxer. 1989. Nickel deficiency gives rise to the defective hydrogenase phenotype of hydC and fnr mutants in Escherichia coli. Mol. Microbiol. 3:1709-1718. [DOI] [PubMed] [Google Scholar]
- 62.Xiong, A., A. Gottman, C. Park, M. Baetens, S. Pandza, and A. Matin. 2000. The EmrR protein represses the Escherichia coli emrRAB multidrug resistance operon by directly binding to its promoter region. Antimicrob. Agents Chemother. 44:2905-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young, J. C., and F. U. Hartl. 2003. A stress sensor for the bacterial periplasm. Cell 113:1-2. [DOI] [PubMed] [Google Scholar]
- 64.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]