Abstract
In the model plant Arabidopsis (Arabidopsis thaliana), the absence of the essential macro-nutrient phosphate reduces primary root growth through decreased cell division and elongation, requiring alterations to the polysaccharide-rich cell wall surrounding the cells. Despite its importance, the regulation of cell wall synthesis in response to low phosphate levels is not well understood. In this study, we show that plants increase cellulose synthesis in roots under limiting phosphate conditions, which leads to changes in the thickness and structure of the cell wall. These changes contribute to the reduced growth of primary roots in low-phosphate conditions. Furthermore, we found that the cellulose synthase complex (CSC) activity at the plasma membrane increases during phosphate deficiency. Moreover, we show that this increase in the activity of the CSC is likely due to alterations in the phosphorylation status of cellulose synthases in low-phosphate conditions. Specifically, phosphorylation of CELLULOSE SYNTHASE 1 (CESA1) at the S688 site decreases in low-phosphate conditions. Phosphomimic versions of CESA1 with an S688E mutation showed significantly reduced cellulose induction and primary root length changes in low-phosphate conditions. Protein structure modeling suggests that the phosphorylation status of S688 in CESA1 could play a role in stabilizing and activating the CSC. This mechanistic understanding of root growth regulation under limiting phosphate conditions provides potential strategies for changing root responses to soil phosphate content.
Phosphate starvation changes root growth by increasing cellulose deposition through alterations in cellulose synthase complex activity and phosphorylation.
Introduction
Phosphorus, an essential nutrient in all life forms, can only be obtained by plants as inorganic phosphate (Pi). In many environments, both native and managed, the amount of Pi in the soil is below that required to support plant growth; thus, plants have developed various mechanisms to increase their survival during Pi limitation. These mechanisms include the production of carboxylic acids to release complexed Pi, expressing high-affinity transporters to maximize Pi uptake, and altering metabolism to prioritize essential processes (Chiou and Lin 2011). Moreover, the root system architecture (RSA) of Arabidopsis (Arabidopsis thaliana) undergoes substantial changes in response to low Pi levels (Abel 2017). When Pi is limited, the plant promotes the growth of secondary roots and root hairs while halting the growth of primary roots. This adaptation allows the plant to increase its root volume in the topsoil, where Pi is more readily available, in an effort to maximize the acquisition of this essential nutrient (Paz-Ares et al. 2022). Thus, RSA remodeling results in a morphological adaption to enable survival under Pi starvation conditions. The recent argument that root growth inhibition in low Pi conditions is not a biological response but rather a result of artificial root exposure to blue light that triggers photo-Fenton chemistry has been challenged by Gao et al. (2021). They showed that blue light illumination in shoots leads to reduced primary root growth under Pi-deficient conditions through long-distance shoot-to-root signaling.
The transcription factor PHOSPHATE STARVATION RESPONSE 1 (PHR1) plays a major role in coordinating most of the plant’s metabolic responses to low internal Pi (Bustos et al. 2010). In contrast, RSA remodeling is influenced by external Pi availability and is controlled by the gene LOW PI ROOT 1 (LPR1) and its homolog LPR2 (Abel 2017). LPR1, a cell wall ferroxidase in the cell wall, is involved in Fe redox cycling and the production of reactive oxygen species (ROS) and callose in response to low Pi (Naumann et al. 2022). These cell wall changes restrict cell expansion and division. It is thought that ROS and cell wall modifications work together to stiffen the cell wall (Balzergue et al. 2017).
Plant cell walls are composed of various polysaccharides, including cellulose, hemicelluloses, callose, and pectins, and play a critical role in directing cell growth and shaping the plant (Lampugnani et al. 2018). Every plant cell forms a primary cell wall, which dynamically expands during growth, but once cellular growth ceases, specific cells, such as tracheary elements and fibers, deposit thick secondary cell walls to support the cell against stress (Nicolas et al. 2022). Cellulose, consisting of β-(1→4)-D-glucan chains, forms the framework of the cell wall and is synthesized by cellulose synthase complexes (CSCs) in the plasma membrane (Polko and Kieber 2019).
In the Arabidopsis genome, there are 10 CELLULOSE SYNTHASE (CESA) genes, with CESA1, CESA3, CESA6, CESA2, CESA5, and CESA9 involved in primary cell wall formation and CESA4, CESA7, and CESA8 involved in secondary cell wall formation (Richmond 2000; Taylor et al. 2003; Persson et al. 2007). The function of CESA10 is suggested to be redundant to CESA1 in the seed coat (Griffiths et al. 2015). The Arabidopsis CESAs have a 60% sequence identity and contain 7 transmembrane helices, with the catalytic domain of the CESAs located between the 2nd and 3rd transmembrane helices (Purushotham et al. 2020). This domain contains a canonical motif and is surrounded by 2 plant-specific domains, the plant-conserved region (PCR) and class-specific region (CSR), which play important roles in CESA oligomerization and CSC assembly (Atanassov et al. 2009). Multiple oligomeric states of CESAs, including homodimers, have been reported, and it is hypothesized that their oligomerization plays a role in the early stages of cellulose CSC assembly (Atanassov et al. 2009). CSCs move at a speed of 200 to 350 nm/min on the plasma membrane and are responsible for synthesizing and depositing cellulose microfibrils (Paredez et al. 2006). The movement of CSCs is thought to be driven by the catalytic activity of the complexes and is regulated by various factors, including protein phosphorylation of the CESA proteins (Chen et al. 2010; Sánchez-Rodríguez et al. 2017). While subcellular processes that underpin the guidance of cellulose biosynthesis to establish cell shape and organs are increasingly better understood (Lampugnani et al. 2018), a mechanistic connection between the perception of low nutrient abundance, such as Pi deficiency, and a defined modulation of cell walls, required for changes in RSA, is not yet known.
Here, we show that low Pi levels lead to increased cellulose deposition in roots. These changes correspond to a decrease in phosphorylation of CESA1 at the S688 site in the CSR domain, which likely regulates CESA1 activity to mediate root growth and cellulose deposition during Pi starvation. Our findings suggest that the regulation of cellulose synthesis plays a role in regulating root growth in response to low Pi.
Results
Cellulose synthesis is induced in plant roots in response to low Pi
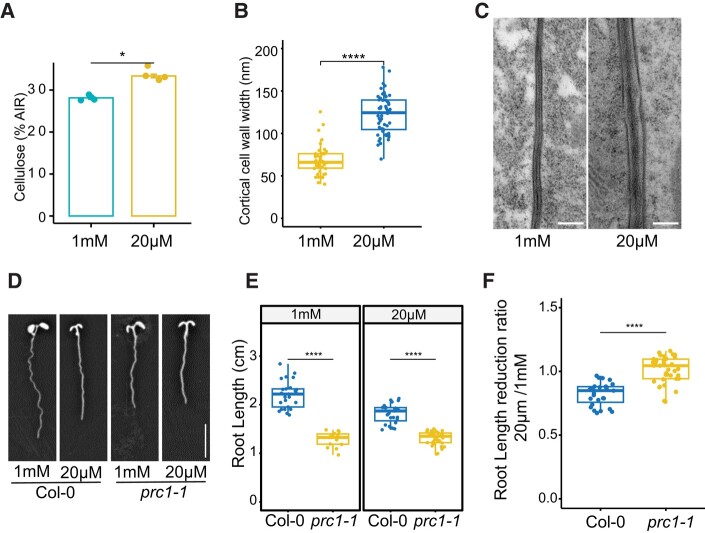
To understand cell wall changes in response to low Pi, we quantified cellulose in the roots of seedlings grown on Pi-deficient (20 μM) and Pi-sufficient (1 mM) conditions. The roots grown on low Pi accumulate significantly higher levels of cellulose compared with the ones grown on sufficient Pi, suggesting that cellulose synthesis is influenced by Pi availability (Fig. 1A). Because we observed an increase in cellulose deposition, we next assessed whether the cell wall thickness was also changed in response to Pi starvation. We, therefore, prepared root sections for transmission electron microscopy (TEM) and imaged cell walls of cortex cells in the root elongation zone. We found that the corresponding cell walls in Pi-deficient conditions were significantly thicker than those grown in sufficient Pi conditions (Fig. 1, B and C). To understand the role of cellulose changes in root growth regulation, we next measured the primary root length of the mildly cellulose-deficient mutant procuste1-1 (prc1-1) (Fagard et al. 2000) under Pi-deficient conditions. As prc1-1 mutants already have shorter roots as compared with Col-0 when grown on Pi-sufficient media, it is difficult to make a direct comparison for changes in primary root length in response to Pi deficiency. Therefore, we calculated the ratio of primary root length of seedlings grown on Pi-deficient media as compared with Pi-sufficient media for each genotype. The prc1-1 mutant roots are constitutively shorter than those of the wild type, and they do not show a substantial change in their root growth when exposed to low Pi conditions (Fig. 1, D to F). These findings suggest that controlling cellulose synthesis is likely necessary for altering root growth in response to low Pi conditions.
Figure 1.
Cellulose deposition is increased in roots grown under Pi-deficient conditions. A) Cellulose quantification of 8-d-old roots grown on Pi-sufficient (1 mM) or Pi-deficient (20 µM) conditions. Data are presented as means ± Se, n = 4. AIR, alcohol insoluble residue B) The thickness of the cell walls in cortical cells within the root elongation zone was quantified using TEM. This analysis was conducted on plants that had been grown for 8 d under either Pi-sufficient or Pi-deficient conditions. Six seedlings per condition were used to quantify cell wall thickness, with more than 15 cell walls measured for each. C) Representative TEM images of cell walls between cortical cells. Scale bar = 200 nm. D) Representative images of 8-d-old Col-0 and prc1-1 roots grown on Pi-sufficient and Pi-deficient conditions. Scale bar = 1 cm. E) Root length of 8-d-old seedlings grown on Pi-sufficient and Pi-deficient media. n ≥ 15. F) Ratio of primary root length reduction of data shown in E). For B), E), and F), the boxes represent the interquartile range (IQR), with medians marked. Whiskers extend 1.5 times IQR, and dots indicate individual data points. Asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001) according to Student's t test. The graphics and statistical analysis were generated using the ggpubR package in R.
Primary wall CESA levels do not change under Pi deficiency
To gain further insights into cellulose regulation under Pi deficiency, we quantified the expression of primary wall CESAs in Arabidopsis roots grown under Pi-sufficient and Pi-deficient conditions. Primary wall CESA genes did not show any changes in their expression in response to Pi deficiency (Fig. 2A), indicating that posttranscriptional changes are likely responsible for the observed cellulose regulation. In contrast, the Pi transporter PHOSPHATE TRANSPORTER 1;3 (PHT1;3) (Nussaume et al. 2011) showed a significant expression increase (Fig. 2A), confirming that roots were indeed experiencing a Pi deficiency. To determine if changes in CESA protein levels instead could contribute to the cellulose changes, we performed shotgun proteomics analyses and performed label-free quantification of proteins. We identified 7,798 proteins from 44,795 peptide groups. We found that 232 proteins were differentially regulated in response to Pi starvation. Proteins commonly associated with Pi starvation, such as PURPLE ACID PHOSPHATASE 17 (PAP17), NON-SPECIFIC PHOSPHOLIPASE C4 (NPC4), and PHT1;3 were induced, indicating that the roots were experiencing Pi starvation. In contrast, proteins involved in RNA translation were repressed (Supplemental Table S1). The analysis of differentially abundant proteins revealed an enrichment of Gene Ontology (GO) terms related to Pi ion homeostasis, protein phosphorylation, and translation (Fig. 2C). The alteration in translation observed may be due to the reallocation of resources in low Pi conditions, which is in line with previous observations (Cheng et al. 2021). However, we found no changes in the abundance of primary wall CESA proteins in response to Pi deficiency (Fig. 2B). These results suggest that primary wall CESA protein levels do not change in response to Pi deficiency.
Figure 2.
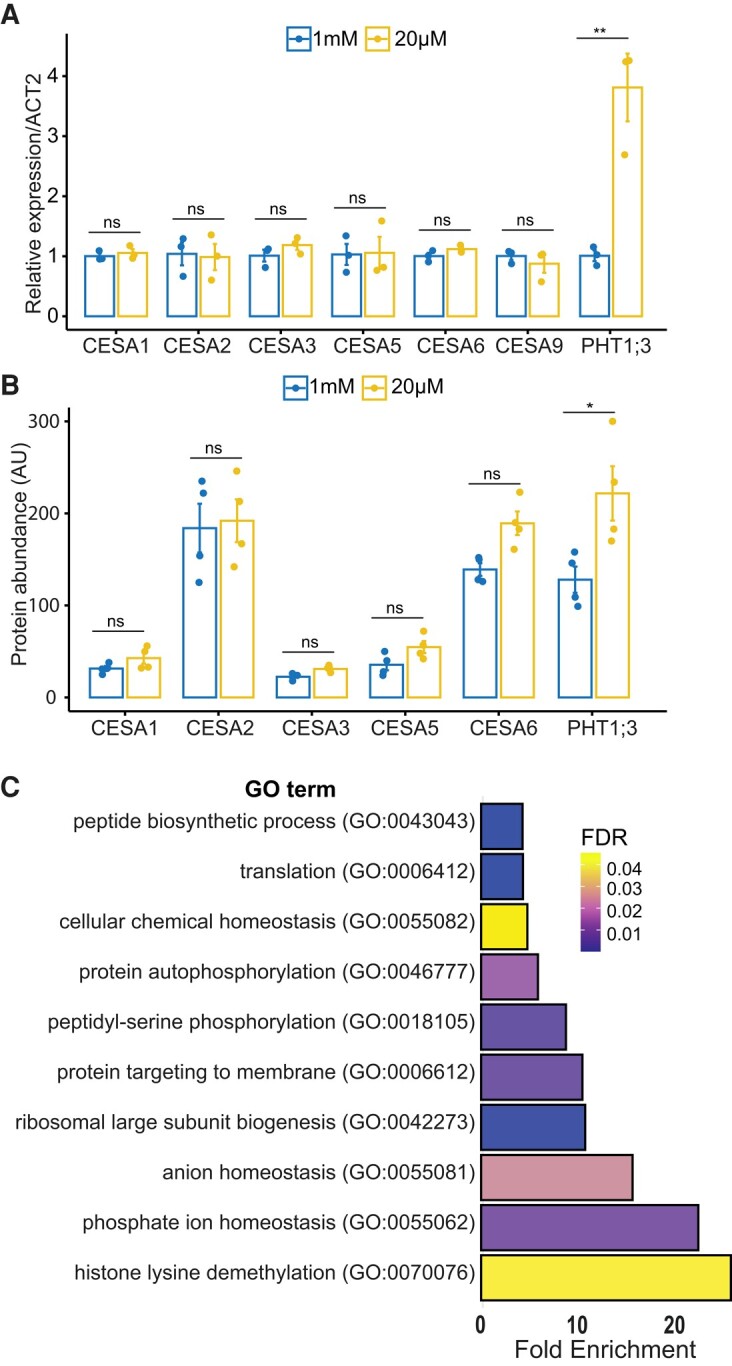
Primary wall CESA transcript and protein levels under Pi deficiency. A) Relative transcript abundance of primary wall CESAs and PHT1;3 (marker gene for Pi deficiency) in the 8-d-old roots of Arabidopsis seedlings grown under Pi-sufficient (1 mM) and Pi-deficient (20 µM) conditions. At least 12 plants were pooled in a single biological replicate for each treatment, and 3 biological replicates were applied. Data are presented as means ± Se. B) Protein abundance of primary wall CESAs and PHT1;3 in the roots of Arabidopsis plants grown in Pi-sufficient and Pi-deficient conditions. At least 30 plants were pooled in a single biological replicate for each treatment, and 4 biological replicates were applied. Data are presented as means ± Se. Asterisks denote statistical significance (ns, nonsignificant; *P < 0.05; **P < 0.01) according to Student’s t test. The graphics and statistical analysis were generated using the ggpubR package in R. C) Go term enrichment of differentially expressed proteins in Pi-deficient media compared with Pi-sufficient media. Enrichment of GO term biological process was determined by using the PANTHER database (pantherdb.org). FDR, false discovery rate.
Cellulose synthase activity is increased in response to low Pi
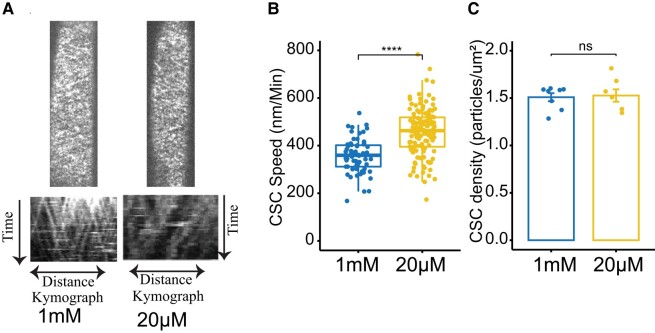
To synthesize cellulose, CSCs move in the plasma membrane, and their speed is thought to be dependent on the catalytic activity of the CESAs (Pedersen et al. 2023). To test if the cellulose changes in response to Pi deficiency were due to the modulation of CESA activity, we estimated the CESA speed at the plasma membrane. Indeed, we found that the speed of CESAs was significantly faster in the elongating root cells of plants grown on Pi deficient media compared with plants grown on Pi sufficient media (Fig. 3, A and B). This indicates that the increase in cellulose deposition in plants grown in Pi deficient is at least partially due to increased CESA activity at the plasma membrane. The density of CSCs at the plasma membrane did not differ significantly between roots grown on Pi-deficient media and those grown on sufficient Pi media (Fig. 3C). Overall, these results show that Pi availability regulates CESA activity in root cells.
Figure 3.
CSC activity increases under low Pi conditions. Roots were grown on agar plates containing Pi-sufficient (1 mM) or Pi-deficient media (20 µM). A) Plasma membrane localization and kymographs of YFP-CesA6 images taken from cells in the root elongation zone of 8-d-old Arabidopsis seedlings. B) Boxplots showing CSC speeds in the elongation zone of roots of seedlings grown on Pi-sufficient or Pi-deficient media. In box plots, the boxes represent the IQR, with medians marked. Whiskers extend 1.5 times IQR, and dots indicate individual data points, n > 5 roots. C) Bar plots showing CSC density at the plasma membrane of root cells in seedlings grown in Pi-sufficient or Pi-deficient media. Data are presented as means ± Se, n > 7. Asterisks denote statistical significance (ns, nonsignificant; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001) according to Student's t test. The graphics and statistical analysis were generated using the ggpubR package in R.
CESA1 is differentially phosphorylated under low Pi conditions
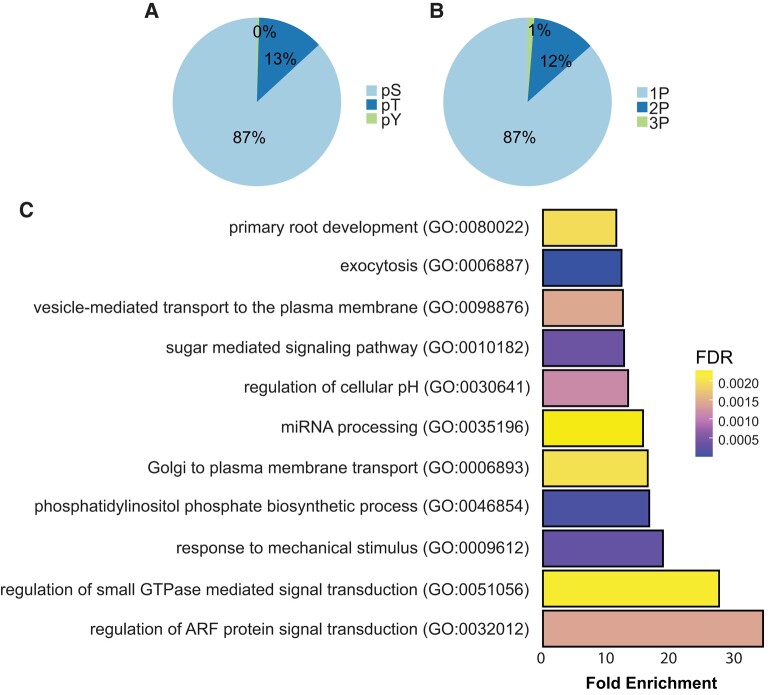
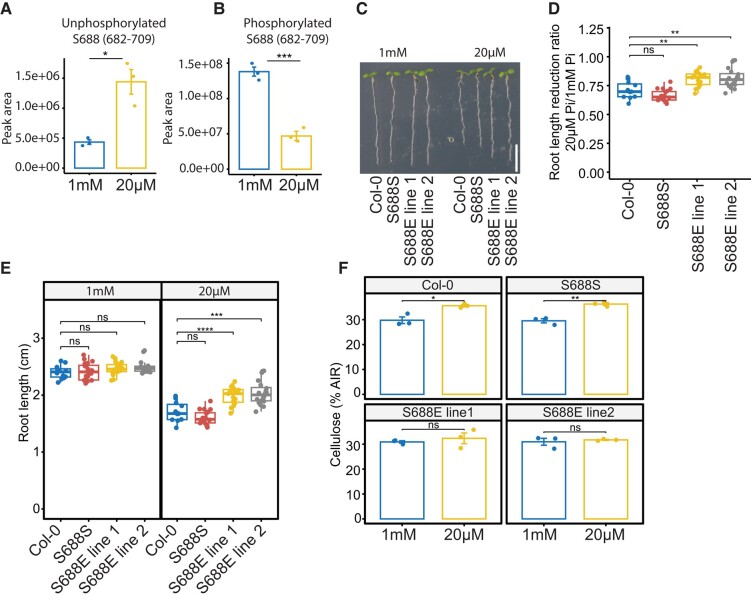
There is compelling evidence to suggest that CSC speed, and thus CESA activity, is affected by the phosphorylation status of the CESAs (Sanchez-Rodriguez et al. 2017; Speicher et al. 2018). We examined the impact of low Pi on protein phosphorylation using shotgun proteomics and phospho-enriched peptide quantification. Our results revealed 7,982 phosphorylated peptides out of 44,795 total peptides, with 6,932 being serine phosphorylations, 1,016 being threonine phosphorylations, and 34 being tyrosine phosphorylations. Of these, 6,908 were single phosphorylation, 971 were double phosphorylation, and 103 were triple phosphorylation (Fig. 4, A and B; Supplemental Table S2). A total of 552 peptides belonging to 422 proteins were differentially phosphorylated (adj. P-value ≤ 0.05) in response to Pi starvation. The data showed an overrepresentation of GO terms associated with the biosynthesis of phosphatidylinositol phosphate, miRNA processing, the detection of abiotic stimuli, and root development among differentially phosphorylated peptides in response to Pi starvation (Fig. 4C). Quantification of CESA phosphorylation showed significant changes in response to low Pi treatments as compared with sufficient Pi. Specifically, we observed a reduction in phosphorylation at the S11 and S688 sites in CESA1, as well as a decrease in the abundance of a triply phosphorylated peptide from CESA3 under these conditions (Supplemental Table S2). We focused on the role of phosphorylation at the S688 site in CESA1, as it likely plays a role in CSC complex activity and stability (Fig. 5). Additionally, we were able to identify the corresponding unphosphorylated site, which showed an increase in abundance, further supporting the changes in phosphorylation at this site in response to Pi deficiency (Fig. 5, A and B). These results indicate that phosphorylation at S688 of CESA1 is regulated in response to Pi deficiency, indicating that CESA activity might be regulated through changes in phosphorylation status in response to low Pi.
Figure 4.
Changes in protein phosphorylation in response to Pi starvation. Plants were grown in a hydroponics system for 14 d with sufficient Pi (1 mM) and then transferred to either Pi-sufficient (1 mM) or Pi-deficient (20 µM) media for 48 h before experiments. A) Pie chart displaying the percentage of serine, threonine, and tyrosine phosphorylation in our data. pS, phosphorylated serine; pT, phosphorylated threonine; pY, phosphorylated tyrosine; 1P, single phosphorylated; 2P, double phosphorylated; 3P, triply phosphorylated. B) Pie chart showing the percentage of single, double, and triply phosphorylated peptides in our data. C) GO term enrichment of differentially phosphorylated proteins in Pi-deficient media in comparison with Pi-sufficient media. Enrichment of GO terms biological process was determined by using the PANTHER database (pantherdb.org). FDR, false discovery rate.
Figure 5.
CESA1 phosphorylation is modified to regulate root growth under Pi-deficient conditions. For A) and B), roots were grown in a hydroponics system for 14 d in sufficient Pi (1 mM) and transferred to Pi-sufficient (1 mM) or Pi-deficient (20 µM) media for 48 h before experiments. A) and B) Summed peak area of unphosphorylated A) and phosphorylated B) peptide containing S688 phosphosite. At least 30 plants were pooled in a single biological replicate for each treatment, and 3 biological replicates were applied. Data are presented as means ± Se. C) Representative images of 8-d-old seedlings grown on Pi-sufficient and Pi-deficient media. Scale bar = 1 cm. D) Primary root length reduction ratio of plants grown under Pi-deficient conditions as compared with plants grown under Pi-sufficient conditions. E) Primary root length of 8-d-old plants grown under Pi-sufficient and Pi-deficient conditions. For D) and E), the boxes represent the IQR, with medians marked. Whiskers extend 1.5 times IQR, and dots indicate individual data points, n > 15. F) Cellulose quantification in the 8-d-old roots of plants grown under Pi-sufficient and Pi-deficient conditions. S688S and S688E lines are in the cesa1 (−/−) background. Data are presented as means ± Se, n = 3. Asterisks denote statistical significance (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001) according to Student's t test. The graphics and statistical analysis were generated using the ggpubR package in R.
Changes in CESA1 phosphorylation regulate root growth under Pi deficiency
To understand the role of the S688 phosphosite in Pi deficiency-mediated root growth regulation, we performed site-directed mutagenesis of S688 into phospho-null (S688A) or phosphomimic (S688E) and transformed these versions, including wild-type version (S688S) into cesa1 knockout mutant. Plants with the wild-type version of CESA1 fully rescued the lethal phenotype of cesa1 mutants (SAIL_278_E08) (Fig. 5C). We recovered 3 lines of heterozygous cesa1 mutants containing the S688 phospho-null version of CESA1. These lines were allowed to self-pollinate, and 30 descendent plants from each of these lines were genotyped. However, we were unable to recover any homozygous cesa1 mutant lines. These results suggest that S688 phospho-null version of CESA1 is unable to rescue the gametophytic lethal phenotype of cesa1 mutants. In contrast, the phosphomimetic version was able to fully complement the cesa1 mutant, indicating that some degree of phosphorylation at site S688 is critical for CESA1 function. The measurement of primary root length in plants segregating for both the cesa1 mutation and phosphomimic T-DNA revealed no significant differences between Pi-sufficient and Pi-deficient growth conditions (Supplemental Fig. S1). However, under Pi-deficient conditions, we did observe some degree of segregation in terms of primary root length (Supplemental Fig. S1). It is important to note that CESA1 forms functional cellulose synthase complexes in conjunction with other CESA proteins. In the presence of multiple forms of CESA proteins, determining which protein forms are incorporated into a functional cellulose synthase complex becomes challenging. To establish a more controlled experimental setting, we specifically isolated plants that carried the phosphomimic version in a homozygous cesa1 (−/−) background. Primary root length measurement of phosphomimetic plants in the homozygous cesa1 (−/−) background grown under Pi-deficient conditions showed significantly reduced changes in primary root length compared with the wild type (Fig. 5, C to E). Moreover, S688E plants did not show an increase in cellulose in their roots when grown on Pi-deficient conditions (Fig. 5F). These results indicate that phosphorylation at phosphosite S688 likely regulates CESA1 activity in response to Pi deficiency. To understand the impact of S688 phosphorylation on CSC activity, we crossed the phosphomimetic plants with YFP-CESA6 to enable CSC imaging in this background. However, these plants experienced silencing, likely due to the presence of 3 T-DNAs (SAIL_278_E08, S688E, and YFP-CESA6), and we were unable to recover phosphomimetic plants with a bright enough YFP-CESA6 marker suitable for CSC imaging.
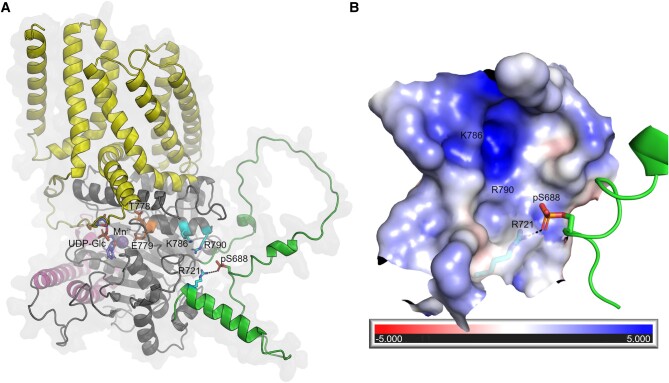
Phosphorylation of S688 in CESA1 might regulate the activity of the cellulose synthase complex via intramolecular conformational changes
To gain insights into a possible effect of the S688 phosphorylation of CESA1 function, we modeled the phosphoserine (pS688) and computed surface parameters. CESA1 S688 is located in the disordered part of the CSR (Q715-T649) modeled with low confidence which must be interpreted with caution. However, S688 is found immediately upstream in sequence from a reliably modeled α-helical segment (R716-W222), and modeling the pS688 indicates that its phosphate group could exist close to R721 (<3 Å) in the vicinity of a cavity (∼12 Å) with positive electrostatic potential (formed largely by charged residues, e.g. R721, K786 and R790) (Fig. 6, A and B). In addition, the flexibility of the partly disordered CSR region surrounding pS688 might allow it to interact with other charged residues in the nearby cavity, e.g. K786 and R790, located on the same α-helix as the strictly conserved TED catalytic motif (Cruz et al. 2019; Pedersen et al. 2023). Thus, it is possible that the phosphorylation status of S688 may influence the catalytic activity of CESA1 via intramolecular conformational changes and/or affect the multimeric state of the CSC. Furthermore, we searched the Functional Analysis Tools for Post-Translational Modifications (FAT-PTM) database (available at https://bioinformatics.cse.unr.edu/fat-ptm/), where we previously summarized phosphorylation sites in CESA proteins (Pedersen et al. 2023). Interestingly, we identified an equivalent documented phosphorylation site, pS671, in CESA3 (Wang et al. 2013). Additionally, equivalent charged residues R706, K771, and R775 were found to be present in CESA3, perhaps suggesting that this phosphorylation site might regulate the activity of CESA3 in a similar manner to CESA1.
Figure 6.
Structural basis for the effects of CESA phosphorylation. A)AtCESA1 model showing the transmembrane domain (yellow), PCR (magenta), and CSR (green). Residues R721, K786, and R790 that contribute to the positive electrostatic potential in the pocket, the modeled phosphoserine (pS688), and TED motif (T778 and E779) are shown in cyan, green, and orange, respectively. Black dashes indicate the possible interaction between pS688 and R721. The UDP-Glc (purple) and Mn2+ (sphere) ligands from the crystal structure of the AtCESA3 catalytic domain (PDB 7CK2) are superimposed onto the AtCESA1 model for comparison. B) The largely positive electrostatic potential of the pocket near the pS688 site is mapped onto the surface. The positions of residues R721, K786, and R790 are indicated. The potentials are on a (−5, 5) red–white–blue color map in units of kBTec-1.
Discussion
Our study uncovered that Arabidopsis roots had higher cellulose content under Pi limited conditions (Fig. 1A). Usually, an increase in cellulose synthesis is associated with increased plant growth and cell expansion because new cell walls are constantly needed for cells to divide and expand. However, low Pi levels lead to a decrease in both cell division and cell elongation in primary roots (Abel 2017). As a result, the increased cellulose deposition during Pi starvation likely results in stronger cell walls with reduced flexibility, negatively impacting cell expansion and growth. The cell wall thickness also increased at the cell elongation zone, which is consistent with previous research showing that cell wall thickness increases in response to Pi deprivation (Muller et al. 2015). It is important to note that normalization of cellulose quantification with alcohol insoluble residue (AIR) may introduce artifacts, as reduced accumulation of another substance under Pi-starved conditions can falsely appear as increased cellulose content. However, our conclusions are supported by 2 other lines of evidence, including an increase in CSC speed under Pi starvation and a lack of cellulose increase in CESA1 phosphomimic lines.
An increase in cellulose content under Pi-deficient conditions is likely due to an increase in CSC activity at the plasma membrane. An increase in CSC activity has been documented to alter cellulose content (Chen et al. 2010; Sánchez-Rodríguez et al. 2017). One regulatory aspect of CSC speed involves changes in the phosphorylation of CESA proteins (Speicher et al. 2018). Under low Pi conditions, the phosphorylation status of CESA1 proteins at the S688 site is reduced (Fig. 5, A and B). This is consistent with previous research showing that reducing the phosphorylation at the T157 site of CESA1 increases CSC speed and cellulose content (Sanchez-Rodriguez et al. 2017). It is, therefore, probable that the S688 site plays a similar role in CESA1 activity. In our study, the phospho-null S688A mutation was unable to rescue the cesa1 knockout mutants. A previous study reported that the S688A mutation of CESA1 partially complements a conditional cesa1 mutant, rsw1-1, leading to a decrease in CESA1 activity and cellulose content (Chen et al. 2010). However, the rsw1-1 mutant is not a cesa1 null mutant, making it difficult to interpret the outcome of the S688A version of CESA1 when another conditionally functional version of CESA1 is also present. The same study also found that the S688E version of CESA1 fully complements the rsw1-1 (Chen et al. 2010). Similarly, we were able to effectively complement the cesa1 knockout mutants with a phosphomimetic version of S688, suggesting that some level of phosphorylation is necessary for the CESA1 function. In addition, we discovered that the S688E version of CESA1 is less responsive to low Pi in terms of primary root growth reduction and cellulose synthesis (Fig. 5, C to E). These findings imply that phosphorylation at the S688 site likely plays a role in regulating CESA1 activity, which in turn might control primary root growth, under Pi deficiency. It is possible that this is the result of altered CESA complex integrity, given that S688 is found in the CSR, which has a putative role in the oligomerization of the CESA complex. In addition, although pS688 is located in a disordered region suboptimal for structural interpretation, it is bordered by a structurally ordered region (high model confidence) and the intermolecular CSR–CSR interface, which could retain pS688 approximately near the positively electrostatic cavity. Therefore, it is also possible that pS688 could interact with K786 and/or R790 located on the same α-helix as the conserved TED catalytic motif. We speculate that such interactions could lead to intramolecular conformational changes near the CESA1 active site, altering its activity.
Our phosphoproteomics analysis revealed significant changes in phosphorylation status of proteins involved in protein trafficking, specifically the ADP-ribosylation factor (ARF) GTPase protein family (Fig. 4C). These proteins play a role in orchestrating intracellular protein trafficking along the secretory pathway, particularly from the endoplasmic reticulum (ER) and Golgi apparatus to the plasma membrane (Donaldson and Jackson 2011). The intricate process of intracellular trafficking is essential in facilitating Pi transport signaling. Notably, PHOSPHATE FACILITATOR1 (PHF1) is a key regulator governing the secretion of phosphate transporters (PHTs) to the plasma membrane (González et al. 2005). Mutations in the PHF1 gene are associated with the retention of PHT1;1 in the ER, leading to reduced accumulation of this transporter in the plasma membrane (González et al. 2005). Consequently, these molecular perturbations result in diminished levels of Pi in plants and a constitutively active Pi starvation response. Furthermore, protein trafficking may be essential for adjusting the abundance of Pi transporters at various organelles, thus regulating the distribution of phosphate across cell compartments. Additionally, PHOSPHATE1 (PHO1), a key phosphate exporter involved in xylem loading, predominantly localizes to the Golgi apparatus (Arpat et al. 2012). It is hypothesized that PHO1 is only transiently present at the plasma membrane due to its potential toxicity to plants (Arpat et al. 2012). Hence, precise regulation of protein trafficking is necessary to maintain an appropriate quantity of Pi transporters at the plasma membrane, ensuring Pi homeostasis. The identification of posttranslational regulations in protein trafficking processes within our data set presents a valuable opportunity to gain further insights into the mechanisms governing the maintenance of phosphate homeostasis.
Kinases regulate changes in root growth in response to Pi deficiency; e.g. mutants of CBL-interacting protein kinases (CIPK) are more sensitive to Pi deficiency in terms of root elongation (Lu et al. 2020). While CESA proteins can be phosphorylated at multiple sites (Speicher et al. 2018), the specific kinases that mediate this phosphorylation have not yet been identified. It is likely that protein kinases involved in Pi starvation signaling also regulate CESA phosphorylation to control primary root growth under Pi-deficient conditions. CESA1 phosphorylation is regulated by the brassinosteroid (BR) pathway (Sanchez-Rodriguez et al. 2017). Here, the BIN2 protein, which is a key inhibitor of the BR pathway, impacts crystalline cellulose levels by directly phosphorylating CESA1 (Sanchez-Rodriguez et al. 2017). In contrast, the BES1 and BZR1 proteins, which are important transcription factors in the BR pathway, increase cellulose synthesis by binding to CESA promoters (Xie et al. 2011). In addition to directly regulating cellulose synthesis, BR also affects the rearrangement of microtubules, which are necessary for the proper direction of CSCs during cellulose synthesis (Catterou et al. 2001). The CELLULOSE SYNTHASE-INTERACTIVE PROTEIN 1, CELLULOSE SYNTHASE MICROTUBULE UNCOUPLING, and COMPANION OF CELLULOSE SYNTHASE proteins are involved in the connection of CSCs to microtubules (Bringmann et al. 2012; Endler et al. 2015; Liu et al. 2016). Interestingly, the BR signaling pathway also regulates primary root growth under Pi deficiency (Singh et al. 2018). The constitutive mutants of BRASSINAZOLE RESISTANT 1 (BZR1), a key regulator of BR-mediated regulation of gene expression (Yin et al. 2002), does not show root growth reduction under low Pi (Singh et al. 2018). This is likely due to the BZR1-mediated regulation of LPR1 but may also involve cellulose synthesis regulation under low Pi (Singh et al. 2018). Another study found that CELLULOSE SYNTHASE-LIKE B5 (CSLB5) is differently regulated in seedlings experiencing Pi deficiency. Further investigation revealed that plants with a mutation in the CSLB5 gene had shorter root hairs than wild-type plants when grown in low Pi conditions, suggesting that CSLB5 plays a role in the plant’s response to low Pi levels (Lin et al. 2011). While the CSLB5 protein is part of the cellulose synthase superfamily, the CSLBs are currently not associated with any specific functions.
Apart from cellulose, other cell wall–related processes may be regulated in response to Pi deficiency. The transcriptomic comparison of 3 Pi response mutants (lpr1, lpr2, and pdr2) revealed significant changes in the expression of genes related to the cell wall in response to low Pi levels (Hoehenwarter et al. 2016). These changes occurred in 4 main areas: pectin modification, cell wall relaxation, hemicellulose/cellulose modification, and carbohydrate hydrolytic enzymes (Hoehenwarter et al. 2016). Data from several transcriptomic experiments show that low Pi levels lead to changes in the transcript abundances of numerous cell wall–related genes in Arabidopsis plants (Wu et al. 2003; Misson et al. 2005; Lin et al. 2011; Wege et al. 2016). Furthermore, low levels of Pi in plants result in an accumulation of lignin and callose in the cell walls of roots in an LPR1-dependent manner. Lignin is a structural component of the cell wall that helps to strengthen it by attaching to cellulose, but an excess of lignin can cause the cell wall to become less flexible. Pi deficiency can also cause an increase in the concentration of unesterified pectins in the cell walls of roots, particularly in the elongation zone and the root apical meristem (Hoehenwarter et al. 2016). These pectins can bind to Fe3+, Al3+, and Ca2+ and can form complex structures called “egg-boxes” through Ca2+-pectate crosslinking (Grant et al. 1973). The formation of egg-boxes can lead to the stiffening of the cell wall and reduced growth (Chebli and Geitmann 2017).
Moreover, the cell wall serves as the primary interface between the cell and its external environment, making it the initial recipient of any alterations in nutrient availability. Local Pi sensing, which is primarily regulated by LPR proteins, prominently takes place within the plant cell walls (Muller et al. 2015). Remarkably, the efficacy of local Pi sensing relies on the presence of external iron (Fe) (Muller et al. 2015). LPR proteins utilize Pi-dependent cues to monitor subtle disparities in Fe availability in the apoplast (Naumann et al. 2022).
Furthermore, plants employ modifications in the cell wall structure to release P that is bound to the cell wall under conditions of Pi deficiency (Zhu et al. 2015). It is probable that the presence of carboxylate groups (-COO-) in homogalacturonans (a type of pectin) enables the sequestration of Fe ions through the formation of -COO-Fe linkages. Under low Pi conditions, the increased abundance of pectins provides an elevated number of carboxylate groups, leading to enhanced binding of Fe and subsequent liberation of trapped phosphate from FePO4 complexes. The release of Pi from the cell wall is expected to exert a notable influence on the overall signaling response to Pi starvation. For example, in rice (Oryza sativa), mutants with cell wall synthesis defects show a constitutive Pi starvation response, and increased Pi transport was observed even when grown in high Pi conditions (Jin et al. 2015).
In conclusion, our findings, combined with previous observations, emphasize the importance of cell wall dynamics in orchestrating the plant’s response to limited Pi availability.
Materials and methods
Plant material and growth conditions
The prc1-1 mutant used in these experiments was previously described (Fagard et al. 2000). The cesa1 T-DNA insertion mutant line (SAIL_278_E08) was obtained from the Arabidopsis Biological Resource Centre (ABRC, https://abrc.osu.edu/). The homozygosity of the cesa1 mutation was confirmed by PCR using the following forward and reverse primers: 5′- CAGAAGTGACTCCAATGCTCC-3′ and 5′- TGGTTGGTGGAATGAGAAGAG-3′.
A vertical agar plate method was used to conduct experiments with varied Pi conditions. The standard media contained half-strength MS (Murashige and Skoog) Basal Salt Mixture (M407, Phytotech), 0.5% (w/v) MES (Sigma-Aldrich), 0.5% (w/v) Suc (Sigma-Aldrich), 10.3 mM NH4NO3, 9.4 mM KNO3, 0.05% (w/v), and 0.8% (w/v) Difco granulated agar (lot 6173985). For Pi-sufficient conditions, standard media was supplemented with 1 mM KH2PO4; for Pi-deficient conditions, the standard media was supplemented with 20 µM KH2PO4 and 980 µM KCl. Arabidopsis (A. thaliana) seeds were surface sterilized in chlorine gas for 3 h, resuspended in 0.1% (w/v) agarose, stratified at 4°C for 2 d, and then seeded in a single row on 12-cm-square plates that were sealed with 3 m Micropore tape. Plates were arranged vertically into racks in the growth chamber, which had a 16 h light/8 h dark cycle, 120 E m2 s1 of light intensity, 23°C (day)/19°C (night), and 60% humidity.
Cellulose quantification
Arabidopsis seedlings were grown on Pi-sufficient (1 mM) and Pi-deficient (20 µM) conditions for 10 d. The roots were separated from the shoots, and crystalline cellulose was measured as previously described (Sanchez-Rodriguez et al. 2012). At least 12 plants were employed in a single biological replicate for each treatment, and a minimum of 3 biological replicates were conducted for each experiment.
Electron microscopy
Roots were grown on Pi-sufficient (1 mM) and Pi-deficient (20 µM) for 8 d, and the imaging was performed in the root elongation zones as described below. Six independent seedlings were imaged for each treatment. The roots were first segmented into 2 mm sections and then placed onto a flat specimen carrier (Leica Microsystems, Vienna, Austria). These sections were frozen using a high-pressure freezer (EM PACT 2; Leica Microsystems). The samples were freeze substituted according to the previously described method (McDONALD and WEBB 2011) using acetone containing 1% w/v osmium tetroxide (Proscitech, Queensland, Australia) and 0.1% w/v uranyl acetate (Proscitech, Queensland, Australia). Once at room temperature, samples were rinsed in acetone and embedded in Spurr's resin (Proscitech, Queensland, Australia). The resulting blocks were then sectioned using a 45°, 2 mm ultradiamond knife (Diatome Diamond Knives, Nidau, Switzerland). Ultrathin serial sections were carefully placed onto formvar-coated 100 mesh thin bar grids. These grids underwent staining with a 2% w/v uranyl acetate solution for a duration of 10 min, followed by lead citrate solution for 1 min. Images were collected on a Jeol JEM-2100 Transmission Electron Microscope (Jeol Australasia Pty. Ltd. Australia) at 80 kV on an AMT Nanosprint II CMOS detector (Newspec, Australia). The cell wall thickness was measured using FIJI (Schindelin et al. 2012).
Live-cell imaging and data processing
Transgenic lines expressing pCESA6::YFP-CESA6 have been previously described (Paredez et al. 2006). Seedlings for image analysis were grown for 8 d in Pi-sufficient or Pi-deficient conditions. Roots were mounted in water under a pad of 0.8% w/v agarose (Bioline). Imaging was performed using a CSU-W1 spinning disk head (Yokogawa) mounted to an inverted Nikon Ti-E microscope equipped with a 100× TIRF oil-immersion objective (NA = 1.49) and a deep-cooled iXon Ultra 888 EM-CCD camera (Andor Technology). A timelapse consisting of 800 ms exposure every 10 s for 5 min was recorded. Fiji (NIH) plug-ins “stackreg” and “bleach correction” with default settings were used for drift and bleaching corrections, respectively. “kymograph evaluation” plug-in of the free tracking software FIESTA was used for CSC speed analysis (Ruhnow et al. 2011). More than 100 kymographs were analyzed, and the speed of CSCs was determined by estimating the slopes of their trajectories with a straight line.
To determine the density of CSCs at the plasma membrane, areas of interest without substantial Golgi signals were selected using the free-hand selection tool. CSCs were automatically identified on 8 bit images using the Find Maxima tool in Fiji (Schindelin et al. 2012), using the same noise threshold for all images. The density of CSCs in each area of interest was calculated by dividing the number of particles by the area of the region.
Mass spectrometry analysis
Plants were grown in sufficient Pi media (1 mM) for high root biomass production as described previously (Hetu et al. 2005). After 14 d, plants were treated with Pi-sufficient or Pi-deficient media for 48 h, and roots were harvested and snap-frozen in liquid nitrogen for subsequent protein extraction. Roots were ground in a mortal pestle, and powder was resuspended in protein extraction buffer (50 mM MOPS, 2 mM EDTA, and 2 mM EGTA) containing 1 tablet of cOmplete EDTA-free protease inhibitor cocktail (Sigma) per 50 mL of buffer and 1 tablet of phosSTOP (Sigma) per 20 mL of buffer. After 10 min centrifugation at 10,000 × g to remove the debris, the supernatant was centrifuged at 80,000 × g for 1 h to retrieve the membrane fraction in the pellet. The pellet was resuspended in the protein denaturation buffer (6 M urea and 100 mM ammonium bicarbonate), and protein quantification was performed with bicinchoninic acid (BCA, Thermo Fisher) assay kit according to the manufacturer's instructions.
A total of 250 µg of proteins were reduced with 10 mM DTT at 60°C for 1 h and acylated with 15 mM IAA for 45 min at room temperature. Urea concentrations were diluted to 1 M with 100 mM ammonium bicarbonate, and protein digestion was performed with 10 µg of Trypsin per mg of protein samples at 37°C overnight. Samples were acidified with formic acid and centrifuged at 10,000 × g for 10 min at 4°C. The supernatant was desalted with SepPak C18 cartridges (Waters) according to manufacturer instructions. The desalted peptides underwent shotgun proteomics for label-free protein quantification and were also further processed using a titanium dioxide–based phosphopeptide enrichment method as described in Sanes (2019). LC-MS/MS was used to analyze the peptide samples. The LC system, Ultimate 3000 RSLC (Thermo Fisher Scientific, San Jose, CA, USA), was set up with an Acclaim PepMap RSLC analytical column (C18, 100 Å, 75 μm × 50 cm, Thermo Fisher Scientific, San Jose, CA, USA) and Acclaim PepMap nano-trap column (75 μm × 2 cm, C18, 100 Å) and controlled at 50°C. Solvent A was 0.1% v/v formic acid and 5% v/v DMSO in water, and solvent B was 0.1% v/v formic acid and 5% DMSO in ACN. The trap column was loaded with tryptic peptide at an isocratic flow 3% v/v ACN containing 0.05% v/v TFA (Trifluoroacetic acid) at 6 µL/min for 6 min, followed by the switch of the trap column as parallel to the analytical column. The gradient settings for the LC runs, at a flow rate 300 nL/min, were as follows: solvent B 3% to 23% in 89 min, 23% to 40% in 10 min, 40% to 80% in 5 min, maintained at 80% for 5 min before dropping to 3% in 2 min, and equilibration at 3% solvent B for 7 min. A Fusion Lumos Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA) was employed at positive mode to execute the MS experiments with settings of spray voltages, S-lens RF, and capillary temperature level of 1.9 kV, 30%, and 275°C, respectively. The mass spectrometry data were acquired with a 3 s cycle time for 1 full scan MS spectra and as many data-dependent higher-energy C-trap dissociation (HCD)-MS/MS spectra as possible. Full scan MS spectra features ions at m/z of 400 to 1500, a maximum ion trapping time of 50 ms, an auto gain control target value of 4e5, and a resolution of 120,000 at m/z 200. An m/z isolation window of 1.6, an auto gain control target value of 5e4, a 35% normalized collision energy, a first mass at m/z of 110, a maximum ion trapping time of 60 ms, and a resolution of 15,000 at m/z 200 were used to perform data-dependent HCD-MS/MS of precursor ions (charge states from 2 to 5). Dynamic exclusion for 30 s was enabled. The results were analyzed using the Arabidopsis database TAIR10 and Protein Discoverer (version 2.4, Thermo Fisher Scientific) with Sequest HT. The ptmRS node was used in the analysis process. The mass tolerances were set at 10 ppm for precursor and 0.05 Da for fragment mass tolerance. The analysis allowed for up to 2 missed cleavages. Cysteine was fixed as a carbamidomethylation modification, while N-terminal protein acetylation and methionine oxidation were variable modifications. For phospho-peptides, additional parameters were set for variable modifications of phosphorylation on serine, threonine, and tyrosine. Label-free quantitation of the S688 containing peptide was performed using the Skyline software package (MacLean et al. 2010). Manual integration of the data was performed for peak integration (MacLean et al. 2010; Schilling et al. 2012). Peak areas were summed for 3 isotopic peaks (M, M + 1, M + 2) for each peptide to serve as the peptide’s quantitative measure. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the data set identifier PXD045341.
Site-specific mutagenesis and complementation
The wild-type sequence of CESA1 was obtained from Arabidopsis cDNA through PCR amplification and cloned into the D-TOPO entry vector from Invitrogen. Site-directed mutagenesis at the CESA1 sequence was performed using the infusion cloning kit from Takara Bio, following the manufacturer’s instructions. Inverse PCR was used to create the S688A mutation with the following forward and reverse primers: 5′- AATGCTCCACTTTTCAATATGGAGGACATCGATGAGGG-3′ and 5′- TATTGAAAAGTGGAGCATTGGCGTCACTTCTGTTGATG-3′. The S688E mutation was created using the following forward and reverse primers: 5′- AATGCTCCACTTTTCAATATGGAGGACATCGATGAGGG-3′ and 5′- TATTGAAAAGTGGAGCATTTTCGTCACTTCTGTTGATG-3′. The entry vector was then transferred into a modified pMDC32 destination vector that contained a CESA1 promoter. The 35S promoter in the pMDC32 vector was replaced with the CESA1 promoter using the following method: the pMDC32 vector was linearized using the HindIII and KpnI restriction enzymes at the sites surrounding the 35S promoter. The CESA1 promoter was amplified using the forward primer 5′-GGCCAGTGCCAAGCTGATTCTCATACGCCCGTCGA-3′ and reverse primer 5′-TCGAGGGGGGGCCCGCGCAGCCACCGACACACA-3′, which included a 15 bp overlap at the extremities of the linearized pMDC32 vector. The amplified promoter and linearized pMDC32 vector were ligated using the infusion cloning enzyme from Takara Bio, following the manufacturer’s instructions. The ligation reaction was transformed into Stellar competent cells, and colonies were screened for correct pCesA1 insertion.
The cesa1 homozygous knockout mutants are male gametophytic lethal; hence, heterozygous plants were transformed with the chosen expression vectors with the floral dip method. For the selection of T1 plants, we utilized hygromycin as a selection marker for the transformed vector and performed PCR genotyping to identify heterozygous cesa1 mutants. The selected plants were then self-fertilized to produce the next generation of seeds. T2 plants were selected for both homozygosity of the cesa1 mutation by PCR genotyping and the presence of the transformed T-DNA by hygromycin selection in the next generation.
RT-qPCR
The plant material was immediately frozen in liquid nitrogen and ground using the Invitrogen tissue lyser. Approximately 100 to 150 mg of powder was used to extract total RNA using the RNeasy Plant Mini Kit from QIAGEN. On-column DNase treatment was performed using the RNase-free DNase kit from QIAGEN. A total of 500 ng of RNA per sample and SuperScript III reverse transcriptase from Invitrogen were used for reverse transcription. RT-qPCR was performed using SYBR select master mix from Invitrogen and primers described in Supplemental Table S3.
Protein modeling
The CESA1 model was retrieved from the AlphaFold (AF) Protein Structure Database (https://alphafold.ebi.ac.uk/files/AF-W8PUJ0-F1-model_v4.pdb). Parts of the CSR are modeled with low-confidence scores (predicted local distance difference test, pLDDT < 70), indicating disorder. The position of the phosphoserine site S688 (albeit modeled with low confidence) is close in sequence to an α-helical segment (R716-E735, proximal to the catalytic domain) modeled with confident accuracy (pLDDT > 70). Phosphorylation was modeled in COOT v.0.9.8.1 (Emsley et al. 2010), choosing a rotamer with favorable geometry. Surface electrostatic potential was calculated with PDB2PQR (for partial charges and atomic radii) and Adaptive Poisson-Boltzmann Solver (APBS) (Jurrus et al. 2018) using 0.58 grid spacing.
Accession numbers
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2022) partner repository with the data set identifier PXD045341. Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: CESA1 (AT4G32410), CESA3 (AT5G05170), CESA4 (AT5G44030), CESA5 (AT5G09870), CESA6 (AT5G64740), CESA7 (AT5G17420), CESA8 (AT4G18780), CESA9 (AT2G21770), CESA10 (AT2G25540), CSLB5 (AT4G15290), PHT1;1 (AT5G43350), PHT1;3 (AT5G43360), PAP17 (AT3G17790), NPC4 (AT3G03530), PHF1 (AT3G52190), PHO1 (AT3G23430), LPR1 (AT1G23010), BZR1 (AT1G75080), BIN2 (AT4G18710), and BES1 (AT1G19350).
Supplementary Material
Acknowledgments
We would like to extend our gratitude to Joshua Heazlewood for his invaluable advice on proteomics. Furthermore, we acknowledge the Mass Spectrometry and Proteomics facility at Bio21 Institute for their support in the processing of proteomics data. We also extend our appreciation to Julian Ratcliffe and the Bioimaging Platform of La Trobe University for their contributions to TEM imaging.
Contributor Information
Ghazanfar Abbas Khan, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia.
Arka Dutta, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia.
Allison van de Meene, School of Biosciences, University of Melbourne, Parkville, VIC 3010, Australia.
Kristian E H Frandsen, Copenhagen Plant Science Center, Department of Plant & Environmental Sciences, University of Copenhagen, Frederiksberg C 1871, Denmark.
Michael Ogden, Copenhagen Plant Science Center, Department of Plant & Environmental Sciences, University of Copenhagen, Frederiksberg C 1871, Denmark.
James Whelan, Department of Animal, Plant and Soil Sciences, School of Agriculture, Biomedicine and Environment, La Trobe University, Bundoora, VIC 3086, Australia; College of Life Science, Zhejiang University, Hangzhou 310058, China.
Staffan Persson, School of Biosciences, University of Melbourne, Parkville, VIC 3010, Australia; Copenhagen Plant Science Center, Department of Plant & Environmental Sciences, University of Copenhagen, Frederiksberg C 1871, Denmark; Joint International Research Laboratory of Metabolic and Developmental Sciences, State Key Laboratory of Hybrid Rice, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai 20040, China.
Author contributions
G.A.K. and S.P. conceived and designed the research. G.A.K. conducted the experiments. A.D. and M.O. provided technical assistance to G.A.K. in the transgenic experiments. A.v.d.M. performed the TEM. K.E.H.F. performed the protein modeling. G.A.K. wrote the paper. S.P. and J.W. revised the manuscript. All authors read and approved the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Primary root length measurement of segregating phosphomimic CESA1 (S688E) plants.
Supplemental Table S1. List of proteins identified in the shotgun proteomics experiment of roots grown in phosphate-sufficient and phosphate-deficient conditions.
Supplemental Table S2. List of peptides identified in the shotgun proteomics experiment of roots grown in phosphate-sufficient and phosphate-deficient conditions.
Supplemental Table S3. Primers used in this study.
Funding
G.A.K. was funded by the Swiss National Science Foundation Fellowship (P2LAP3-168408) and the DECRA Fellowship from the Australian Research Council (DE210101200). K.E.H.F. was funded by a Novo Nordisk Foundation Industrial Biotechnology and Environmental Biotechnology Postdoctoral grant (NNF21OC0071799) and a Villum Foundation Experiment grant (VIL50427). S.P. was funded by a Villum Foundation, 2 Novo Nordisk, and Danish National Research Foundation grants (25915, 19OC0056076, 20OC0060564, and DNRF155, respectively).
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
References
- Abel S. Phosphate scouting by root tips. Curr Opin Plant Biol. 2017:39:168–177. 10.1016/j.pbi.2017.04.016 [DOI] [PubMed] [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012:71(3):479–491. 10.1111/j.1365-313X.2012.05004.x [DOI] [PubMed] [Google Scholar]
- Atanassov II, Pittman JK, Turner SR. Elucidating the mechanisms of assembly and subunit interaction of the cellulose synthase complex of Arabidopsis secondary cell walls. J Biol Chem. 2009:284(6):3833–3841. 10.1074/jbc.M807456200 [DOI] [PubMed] [Google Scholar]
- Balzergue C, Dartevelle T, Godon C, Laugier E, Meisrimler C, Teulon J-M, Creff A, Bissler M, Brouchoud C, Hagège A, et al. Low phosphate activates STOP1-ALMT1 to rapidly inhibit root cell elongation. Nat Commun. 2017:8(1):15300. 10.1038/ncomms15300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann M, Landrein B, Schudoma C, Hamant O, Hauser MT, Persson S. Cracking the elusive alignment hypothesis: the microtubule-cellulose synthase nexus unraveled. Trends Plant Sci. 2012:17(11):666–674. 10.1016/j.tplants.2012.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Perez-Perez J, Solano R, Leyva A, Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010:6(9):e1001102. 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS. Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. II. Effects of brassinosteroids on microtubules and cell elongation in the bul1 mutant. Planta 2001:212(5–6):673–683. 10.1007/s004250000467 [DOI] [PubMed] [Google Scholar]
- Chebli Y, Geitmann A. Cellular growth in plants requires regulation of cell wall biochemistry. Curr Opin Cell Biol. 2017:44:28–35. 10.1016/j.ceb.2017.01.002 [DOI] [PubMed] [Google Scholar]
- Chen S, Ehrhardt DW, Somerville CR. Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc Natl Acad Sci U S A. 2010:107(40):17188–17193. 10.1073/pnas.1012348107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Min W, Li M, Zhou L, Hsu CC, Yang X, Jiang X, Ruan Z, Zhong Y, Wang ZY, et al. Quantitative proteomics reveals that GmENO2 proteins are involved in response to phosphate starvation in the leaves of glycine max L. Int J Mol Sci. 2021:22(2):920. 10.3390/ijms22020920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol. 2011:62(1):185–206. 10.1146/annurev-arplant-042110-103849 [DOI] [PubMed] [Google Scholar]
- Cruz ER, Nguyen H, Nguyen T, Wallace IS. Functional analysis tools for post-translational modification: a post-translational modification database for analysis of proteins and metabolic pathways. Plant J. 2019:99(5):1003–1013. 10.1111/tpj.14372 [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol. 2011:12(6):362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr D Biol Crystallogr. 2010:66(4):486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler A, Kesten C, Schneider R, Zhang Y, Ivakov A, Froehlich A, Funke N, Persson S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015:162(6):1353–1364. 10.1016/j.cell.2015.08.028 [DOI] [PubMed] [Google Scholar]
- Fagard M, Desnos T, Desprez T, Goubet F, Refregier G, Mouille G, McCann M, Rayon C, Vernhettes S, Hofte H. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 2000:12(12):2409–2424. 10.1105/tpc.12.12.2409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YQ, Bu LH, Han ML, Wang YL, Li ZY, Liu HT, Chao DY. Long-distance blue light signalling regulates phosphate deficiency-induced primary root growth inhibition. Mol Plant. 2021:14(9):1539–1553. 10.1016/j.molp.2021.06.002 [DOI] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell. 2005:17(12):3500–3512. 10.1105/tpc.105.036640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GT, Morris ER, Rees DA, Smith PJC, Thom D. Biological interactions between polysaccharides and divalent cations: the egg-box model. FEBS Lett. 1973:32(1):195–198. 10.1016/0014-5793(73)80770-7 [DOI] [Google Scholar]
- Griffiths JS, Ki Š, Kushwaha R, Lam P, Tateno M, Young R, Voiniciuc C, Dean G, Mansfield SD, DeBolt S, et al. Unidirectional movement of cellulose synthase complexes in Arabidopsis seed coat epidermal cells deposit cellulose involved in mucilage extrusion, adherence, and ray formation. Plant Physiol. 2015:168:502–520. 10.1104/pp.15.00478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetu MF, Tremblay LJ, Lefebvre DD. High root biomass production in anchored Arabidopsis plants grown in axenic sucrose supplemented liquid culture. Biotechniques 2005:39(3):345–349. 10.2144/05393ST02 [DOI] [PubMed] [Google Scholar]
- Hoehenwarter W, Mönchgesang S, Neumann S, Majovsky P, Abel S, Müller J. Comparative expression profiling reveals a role of the root apoplast in local phosphate response. BMC Plant Biol. 2016:16(1):106. 10.1186/s12870-016-0790-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, Fang C, Yuan H, Wang S, Wu Y, Liu X, Zhang Y, Luo J. Interaction between carbon metabolism and phosphate accumulation is revealed by a mutation of a cellulose synthase-like protein, CSLF6. J Exp Bot. 2015:66(9):2557–2567. 10.1093/jxb/erv050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurrus E, Engel D, Star K, Monson K, Brandi J, Felberg LE, Brookes DH, Wilson L, Chen J, Liles K, et al. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 2018:27(1):112–128. 10.1002/pro.3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani ER, Khan GA, Somssich M, Persson S. Building a plant cell wall at a glance. J Cell Sci. 2018:131(2):jcs207373. 10.1242/jcs.207373 [DOI] [PubMed] [Google Scholar]
- Lin WD, Liao YY, Yang TJ, Pan CY, Buckhout TJ, Schmidt W. Coexpression-based clustering of Arabidopsis root genes predicts functional modules in early phosphate deficiency signaling. Plant Physiol. 2011:155(3):1383–1402. 10.1104/pp.110.166520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Schneider R, Kesten C, Zhang Y, Somssich M, Zhang Y, Fernie AR, Persson S. Cellulose-microtubule uncoupling proteins prevent lateral displacement of microtubules during cellulose synthesis in Arabidopsis. Dev Cell. 2016:38(3):305–315. 10.1016/j.devcel.2016.06.032 [DOI] [PubMed] [Google Scholar]
- Lu X, Li X, Xie D, Jiang C, Wang C, Li L, Zhang Y, Tian H, Gao H, Wang C. The Ca(2+) -regulated protein kinase CIPK1 integrates plant responses to phosphate deficiency in Arabidopsis thaliana. Plant Biol (Stuttg). 2020:22(5):753–760. 10.1111/plb.13137 [DOI] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010:26(7):966–968. 10.1093/bioinformatics/btq054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald KL, Webb RI. Freeze substitution in 3 h or less. J Microsc. 2011:243(3):227–233. 10.1111/j.1365-2818.2011.03526.x [DOI] [PubMed] [Google Scholar]
- Misson J, Raghothama KG, Jain A, Jouhet J, Block MA, Bligny R, Ortet P, Creff A, Somerville S, Rolland N, et al. A genome-wide transcriptional analysis using Arabidopsis thaliana affymetrix gene chips determined plant responses to phosphate deprivation. Proc Natl Acad Sci U S A. 2005:102(33):11934–11939. 10.1073/pnas.0505266102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Burstenbinder K, Abel S. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell. 2015:33(2):216–230. 10.1016/j.devcel.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Naumann C, Heisters M, Brandt W, Janitza P, Alfs C, Tang N, Toto Nienguesso A, Ziegler J, Imre R, Mechtler K, et al. Bacterial-type ferroxidase tunes iron-dependent phosphate sensing during Arabidopsis root development. Curr Biol. 2022:32(10):2189–2205.e6. 10.1016/j.cub.2022.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas WJ, Fäßler F, Dutka P, Schur FKM, Jensen G, Meyerowitz E. Cryo-electron tomography of the onion cell wall shows bimodally oriented cellulose fibers and reticulated homogalacturonan networks. Curr Biol. 2022:32(11):2375–2389.e6. 10.1016/j.cub.2022.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci. 2011:2:83. 10.3389/fpls.2011.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006:312(5779):1491–1495. 10.1126/science.1126551 [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Puga MI, Rojas-Triana M, Martinez-Hevia I, Diaz S, Poza-Carrion C, Minambres M, Leyva A. Plant adaptation to low phosphorus availability: core signaling, crosstalks, and applied implications. Mol Plant. 2022:15(1):104–124. 10.1016/j.molp.2021.12.005 [DOI] [PubMed] [Google Scholar]
- Pedersen GB, Blaschek L, Frandsen KEH, Noack LC, Persson S. Cellulose synthesis in land plants. Mol Plant. 2023:16(1):206–231. 10.1016/j.molp.2022.12.015 [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Bai J, Bandla C, García-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu Deepti J, Prakash A, Frericks-Zipper A, Eisenacher M, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022:50(D1):D543–D552. 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S, Paredez A, Carroll A, Palsdottir H, Doblin M, Poindexter P, Khitrov N, Auer M, Somerville CR. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc Natl Acad Sci U S A. 2007:104(39):15566–15571. 10.1073/pnas.0706592104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polko JK, Kieber JJ. The regulation of cellulose biosynthesis in plants. Plant Cell. 2019:31(2):282–296. 10.1105/tpc.18.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushotham P, Ho R, Zimmer J. Architecture of a catalytically active homotrimeric plant cellulose synthase complex. Science 2020:369(6507):1089–1094. 10.1126/science.abb2978 [DOI] [PubMed] [Google Scholar]
- Richmond T. Higher plant cellulose synthases. Genome Biol. 2000:1(4):REVIEWS3001. 10.1186/gb-2000-1-4-reviews3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophys J. 2011:100(11):2820–2828. 10.1016/j.bpj.2011.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rodriguez C, Bauer S, Hematy K, Saxe F, Ibanez AB, Vodermaier V, Konlechner C, Sampathkumar A, Ruggeberg M, Aichinger E, et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell. 2012:24(2):589–607. 10.1105/tpc.111.094672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Rodriguez C, Ketelaar K, Schneider R, Villalobos JA, Somerville CR, Persson S, Wallace IS. BRASSINOSTEROID INSENSITIVE2 negatively regulates cellulose synthesis in Arabidopsis by phosphorylating cellulose synthase 1. Proc Natl Acad Sci U S A. 2017:114(13):3533–3538. 10.1073/pnas.1615005114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR. Tell me a story. eLife 2019:8:e50527. 10.7554/eLife.50527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012:11(5):202–214. 10.1074/mcp.M112.017707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012:9(7):676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AP, Fridman Y, Holland N, Ackerman-Lavert M, Zananiri R, Jaillais Y, Henn A, Savaldi-Goldstein S. Interdependent nutrient availability and steroid hormone signals facilitate root growth plasticity. Dev Cell. 2018:46(1):59–72.e4. 10.1016/j.devcel.2018.06.002 [DOI] [PubMed] [Google Scholar]
- Speicher TL, Li PZ, Wallace IS. Phosphoregulation of the plant cellulose synthase complex and cellulose synthase-like proteins. Plants (Basel). 2018:7(3):52. 10.3390/plants7030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969:26(1–2):31–43. 10.1016/S0022-5320(69)90033-1 [DOI] [PubMed] [Google Scholar]
- Taylor NG, Howells RM, Huttly AK, Vickers K, Turner SR. Interactions among three distinct CesA proteins essential for cellulose synthesis. Proc Natl Acad Sci U S A. 2003:100(3):1450–1455. 10.1073/pnas.0337628100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Gu LF, Ye M, Zou H, Sun SS, He JX. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J Proteomics. 2013:78:486–498. 10.1016/j.jprot.2012.10.018 [DOI] [PubMed] [Google Scholar]
- Wege S, Khan GA, Jung JY, Vogiatzaki E, Pradervand S, Aller I, Meyer AJ, Poirier Y. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016:170(1):385–400. 10.1104/pp.15.00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 2003:132(3):1260–1271. 10.1104/pp.103.021022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Yang C, Wang X. Brassinosteroids can regulate cellulose biosynthesis by controlling the expression of CESA genes in Arabidopsis. J Exp Bot. 2011:62(13):4495–4506. 10.1093/jxb/err164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002:109(2):181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Zhu XF, Wang ZW, Wan JX, Sun Y, Wu YR, Li GX, Shen RF, Zheng SJ. Pectin enhances rice (Oryza sativa) root phosphorus remobilization. J Exp Bot. 2015:66(3):1017–1024. 10.1093/jxb/eru461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.