Abstract
Dark brown haloes of melanin around colonies are an easily visualized phenotype displayed by many Streptomyces strains harboring plasmid pIJ702 carrying the melC operon of Streptomyces antibioticus IMRU3270. Spontaneous melanin-negative mutants of pIJ702 occur with a frequency of ca. 1%, and often mutation occurs in the melC operon, which removes the BglII site as part of an inverted repeat. Other melanin-negative mutations seem to occur spontaneously in Streptomyces lividans, resulting in white colonies from which intact, melanin-producing pIJ702 can be isolated by introduction into a new host. S. lividans ZX66 was found to be such a mutant and to have a secondary mutation influencing expression of the melC operon on the chromosome. A 3.3-kb DNA fragment was isolated from its progenitor strain, JT46, and a gene able to restore melC operon expression was found to encode a member of an AraC family of transcriptional regulators, which was equivalent to AdpAc in Streptomyces coelicolor and therefore was designated AdpAl. Lack of melC operon expression was correlated with a single A-to-C transversion, which altered a single key amino acid residue from Thr to Pro. The transcription of the melC operon was found to be greatly reduced in the adpA mutant background. The counterpart gene (adpAa) in the S. antibioticus strain in which the melC operon carried on pIJ702 originated was also isolated and was found to have an identical regulatory role. Thus, we concluded that the melC operon is under general direct positive control by AdpA family proteins, perhaps at the transcriptional level and certainly at the translational level via bldA, in Streptomyces.
Tyrosinase (EC 1.14.18.1) is a ubiquitous copper-containing monooxygenase that catalyzes both the O hydroxylation of monophenols and the oxidation of O-diphenols to O-quinones (29, 32) responsible for the biosynthesis of melanin pigment from tyrosine (29) in bacteria. In Streptomyces antibioticus (23) and Streptomyces glaucescens (16), the tyrosinase gene (melC2) is preceded by the melC1 gene encoding a conserved protein essential for the expression of melanin (3, 23, 28) in a polycistronic operon. The MelC1 protein, which behaves like a molecular chaperone, has dual roles; it regulates copper incorporation, and it promotes secretion of apotyrosinase via a transient MelC1-MelC2 complex (7, 8, 28, 30).
The Streptomyces melC operon has been used extensively as a phenotypic marker for Streptomyces plasmids (23) and for the construction of promoter-probe vectors for Streptomyces (36) and Escherichia coli (9). Investigation of the control of the melC operon revealed that a cloned gene of Streptomyces lividans 66, cutR, phenotypically suppresses defective melC1 (44). The CutR protein resembles the response regulator OmpR of the osmoregulatory signal transduction system in E. coli. Next to cutR is cutS, which encodes the histidine protein kinase counterpart of OmpS. Thus, the putative cutR-cutS operon was postulated to regulate copper metabolism in Streptomyces (44). In Rhizobium leguminosarum biovar phaseoli, transcription of a gene needed for melanin synthesis was found to be activated by nifA of Rhizobium and Klebsiella pneumoniae (15), which is an activator of nifH of K. pneumoniae in E. coli cells grown with low oxygen concentrations. Analysis of Streptomyces griseus mutants defective in melanogenesis showed that there was a close regulatory correlation between melanogenesis and morphological and physiological differentiation. Also, introduction of plasmids carrying the melC operon failed to confer melanin production in the melanin-negative mutants, and disruption of melC2 barely affected melanin productivity, suggesting that another unknown enzyme is involved in melanogenesis in S. griseus (11).
Here we demonstrate unambiguously that expression of the melC operon in S. lividans and Streptomyces coelicolor and in its native host, S. antibioticus, is under strict positive control by a gene encoding a multifunctional regulatory protein (AdpAl). A counterpart protein in S. coelicolor (AdpAc) was shown to be the product of the bldH gene (47), which contains a rare leucine codon (TTA) controllable by bldA tRNA at the translational level, and to be closely associated with aerial mycelium development (41).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Streptomyces and E. coli strains used in this study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype and/or characteristicsa | Reference or source |
|---|---|---|
| S. coelicolor A3(2) strains | ||
| M145 | Wild type | 18 |
| ZD2 | M145-derived adpAc disruptant, aac(3)IV | This study |
| S. lividans 66 strains | ||
| 1326 | Wild type, Dnd+/φHAU3r | 31 |
| JT46 | Dnd+/φHAU3r | 43 |
| ZX64 | JT46-derived dndA disruptant, Dnd−/φHAU3r | Zhou, unpublished data |
| HXY1 | JT46-derived dndA disruptant, Dnd−/φHAU3r | He, unpublished data |
| ZX66 | Same as JT46 but adpA1 | This study |
| ZD1 | ZX66 derivative with an autonomous pIJ702 but integrative, pJTU1452, aac(3)IV/Mel+ | This study |
| ZD3 | JT46-derived adpA1 disruptant, aac(3)IV | This study |
| S. antibioticus strains | ||
| IMRU3720 | Wild type, Mel+ | |
| ZD4 | IMRU3720-derived adpAc disruptant, aac(3)IV | |
| E. coli strains | ||
| DH10B | recA | GIBCO BRL |
| ET12567/pUZ8002 | recE dam dcm hsdS Cmr Strr Tetr Kmr | 35 |
| Plasmids | ||
| pIJ702 | pIJ101 derivative, tsr mel | 23 |
| pSET152 | aac(3)IV lacZ reppuc attφC31 oriT | 5 |
| pOJ260 | apr oriT reppuc lacZ | 26 |
| pMD18-T | pUC18 derivative | TaKaRa |
| pJTU1452 | ca. 3.5-kb PCR fragment (obtained by using pSET152-FP and pSET152-RP as primers and ZD1 DNA as the template) inserted into the EcoRV site of pSET152 | This studyb |
| pJTU1454 | Religation of pJTU1452 after digestion with BamHI and Bg/II, AdpA1+ | This studyb |
| pJTU1455 | Religation of pJTU1452 after partial digestion with ApaI, AdpA1− | This studyb |
| pJTU1457 | Religation of pJTU1452 after digestion with ApaI, AdpA1+ | This studyb |
| pJTU1458 | 921-bp AluI fragment after PCR amplification (obtained by using sgb118 and sgb218 as primers and M145 DNA as the template) inserted into the EcoRV site of pOJ260 | This study |
| pJTU1459 | 921-bp AluI fragment from pJTU1452 inserted into the EcoRV site of pOJ260 | This study |
| pJTU1461 | ApaI fragment (1.7 kb) of ZX66 containing complete adpA1* inserted into the ApaI site of Bluescript SK (+) | This study |
| pJTU1465 | ApaI fragment (1.5 kb) of IMRU3720 containing complete adpAa inserted into the ApaI site of Bluescript SK(+) | This study |
| pJTU1466 | 958-bp PCR fragment (obtained by using A1296 and pSET152-FP as primers and pJTU1465 as the template) internal to adpAa inserted into the EcoRV site of the pMD18-T vector | This study |
| pJTU1467 | HindIII-EcoRI fragment carrying, the 958-bp PCR fragment of pJTU1466 inserted into the corresponding sites of pOJ260 | This study |
mel, tyrosinase gene for melanin production; oriT, origin of transfer of plasmid RK2; tsr, thiostrepton resistance gene; aac(3)IV, apramycin resistance gene; Cmr, chloramphenicol resistance.
See Fig. 2.
General methods and techniques.
The general growth media and conditions used for E. coli and Streptomyces strains and the standard methods used for handling E. coli and Streptomyces in vivo and in vitro, such as preparation of plasmid and chromosome DNAs, transformation, etc., were those described previously (26, 38), unless indicated otherwise. E. coli strains were cultivated at 37°C in Luria-Bertani medium or on Luria-Bertani agar plates. Streptomyces strains were routinely grown at 30°C on SFM solid medium (17) for conjugation between E. coli and Streptomyces, on R2YE (42) for protoplast transformation, on MMT for tyrosinase expression and melanin production, or in YEME and TSBY liquid media containing 34% sucrose for mycelial growth. Plasmid and total DNAs were isolated from Streptomyces strains as described by Kieser et al. (26). Unmethylated DNA was prepared from E. coli ET12567. In vivo generation of targeted mutations in Streptomyces was achieved by conjugation between E. coli and Streptomyces strains as described by Sun et al. (40). DNA sequencing was carried out by using ABI Prism BigDye terminator cycle sequencing Ready Reaction kits from PE Applied Biosystems. DNA fragments were subcloned into pBluescriptII SK(+), and this was followed by unidirectional subcloning with the Erase-a-Base system (Promega). The double-stranded plasmid DNA used as the template was purified by polyethylene glycol 8000 precipitation before sequencing. About 200 to 400 ng of template DNA was mixed with 4 μl of a terminator Ready Reaction mixture and the appropriate primer, and the total volume was adjusted to 20 μl with deionized water. The sequencing reaction mixtures were subjected to cycle sequencing with a DNA thermal cycler (Perkin-Elmer) before the sequencing gel was examined with an ABI 377 autosequencer. The sequence data were analyzed with the Frame-Plot online program (20). DNA and deduced protein sequence homology searches were performed by using BLAST (1, 2, 39) and FASTA (37). Multiple alignments of sequences were constructed by using the BioEdit sequence alignment editor (14). The copy numbers of plasmids were estimated by densitometric scanning of band intensity after agarose gel electrophoresis as described by Kieser et al. (25). Preparations that were compared were always run on the same gel. For Streptomyces, apramycin and thiostrepton were used at concentrations of 30 and 10 μg ml−1, respectively, in SFM agar and at concentrations of 15 and 5 μg ml−1, respectively, in liquid media.
Cloning of a DNA fragment restoring the melanin phenotype to ZX66 in S. lividans.
S. lividans JT46 total DNA was partially digested with MboI to obtain a fragment size of 2 to 8 kb, treated with alkaline phosphatase to minimize formation of multiple inserts, and ligated with BamHI-digested DNA of an integrative vector, pSET152 (26). The ligation mixtures were used to transform ZX66/pIJ702 protoplasts, and colonies with restored production of black pigment were selected. The DNA insert obtained from one black colony (ZD1) was recovered by using a pair of PCR primers (pSET152-FP [5′-CCAGTCACGACGTTGTAAAACGA-3′] and pSET152-RP [5′-ACAGGAAACAGCTATGACATGAT-3′]) flanking the multiple cloning sites of pSET152 and chromosomal DNA of ZD1 as the template.
PCR primers and conditions and Southern and colony hybridizations.
Oligonucleotide primers pSET152-FP and pSET152-RP were used to recover by PCR amplification a ca. 3.5-kb DNA fragment that included adpAl cloned and integrated into the ZD1 chromosome before it was inserted into the EcoRV site of pSET152 to obtain pJTU1452. The primers were synthesized commercially. PCRs were performed by using a HYBAID PCR machine. A typical PCR mixture (50 μl) consisted of 1.5 mM MgCl2, 5 ng of genomic DNA as the template, 5% dimethyl sulfoxide, each deoxynucleoside triphosphate at a concentration of 100 μM, and 25 pmol of the required primers in 1× PCR buffer. After addition of 5 U of Pfu polymerase (13), the DNA template was denatured at 95°C for 3 min. Amplification was carried out by 30 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 55°C, and extension for 4 min at 72°C, followed by extension for 5 min at 72°C. PCR products were then purified from 0.8% agarose gels by using a Gene Recovery kit (12). For Southern hybridization experiments, DNA was cleaved with restriction enzymes, separated in 0.8% agarose gels, and transferred onto a Hybond-N+ nylon membrane (Amersham-Pharmacia). α-[32P]dCTP-labeled radioactive probes and a Random Priming kit (Roche) were used for both Southern blotting and in situ colony hybridization.
Cloning of adpAl* from S. lividans ZX66 and adpAa from S. antibioticus IMRU3720.
Total DNAs of ZX66 and IMRU3720 were digested with ApaI (a 1.5-kb ApaI fragment in IMRU3720 DNA was found to contain adpAa by Southern hybridization), and ca. 1.5- to 1.7-kb fragments were recovered from an agarose gel for cloning into ApaI-digested and calf intestinal alkaline phosphatase-treated pBluescript SK(+). pJTU1461 was found to carry a 1.7-kb ApaI fragment of adpAl from ZX66, and pJTU1465 carried a ca. 1.5-kb ApaI fragment of adpAa from IMRU3720, as determined by colony hybridization in which a 1.7-kb ApaI fragment of adpAl was used as the probe.
Constructs for generation of an adpAl mutant of S. lividans JT46 (ZD2), an adpAc mutant of S. coelicolor M145 (ZD3), and an adpAa mutant of S. antibioticus IMRU3720 (ZD4) by targeted gene disruption.
The pJTU1458 vector mediating disruption of adpAc was constructed by PCR amplification of a ca. 3.3-kb DNA fragment covering adpAc from the S. coelicolor M145 chromosome by using oligonucleotide primers 1458-FP (5′-GACAAGCCCCGCAACCTC-3′) and 1458-RP (5′-GGCCTCGTCCTCAAACGC-3′), followed by digestion with AluI to generate a 921-bp internal fragment for insertion into the EcoRV site of E. coli plasmid pOJ260. Similarly, vector pJTU1459 mediating disruption of adpAc was constructed by recovery of a 921-bp DNA fragment after AluI digestion of pJTU1452 for insertion into the EcoRV site of pOJ260. Vector pJTU1467 mediating disruption of adpAa was constructed by cloning a 958-bp PCR fragment covering part of adpAa from pJTU1465 by using oligonucleotide primers pSET152-FP (5′-CCAGTCACGACGTTGTAAAACGA-3′) and A1296 (5′-GAGAGTTCCATACCGCTGTC-3′) into the EcoRV site of the pMD18-T vector to produce pJTU1466, from which an EcoRI-HindIII fragment was recovered for insertion into the corresponding site of pOJ260 to obtain pJTU1467.
Reverse PCR for transcript analysis.
Total RNAs from JT46/pIJ702 and ZD3/pIJ702 were isolated by using an RNeasy mini kit (QIAGEN) and were treated with DNase. Reverse transcription RCR (RT-PCR) was performed by using a OneStep RT-PCR kit and Q-Solution (QIAGEN). The cycling parameters used were 30 min at 50°C and 15 min at 95°C for the reverse transcription step, followed by 30 PCR cycles of amplification (30 s at 94°C, 30 s at 55°C, and 30 s at 72°C). Primers melF (5′-ATGCGGGGTCGTCAAC-3′) and melMR (5′-CGTAGTGGCTGACGACGCTGA-3′) were used as the oligonucleotides primers for RT-PCR experiments to amplify a 395-bp fragment internal to melC.
Expression of AdpAl in E. coli BL21(DE3), preparation of total proteins with or without AdpA, and gel mobility shift assay.
The complete adpAl sequence was amplified by PCR by using pJTU1457 DNA as the template DNA and oligonucleotide primers 1464F-NdeI (5′-GGGGGGCTTAGCCATATGAG-3′) and 1464R-XhoI (5′-GGAGCCGTCTGCTCGAGTCAC-3′). The amplified fragment was digested with NdeI plus XhoI and cloned in pET15b to generate pJTU1464. The overproduced AdpAl could be detected as both soluble and insoluble forms, but we failed to obtain a sufficient amount of the pure His-tagged AdpAl protein using nickel columns; thus, total proteins were isolated for a gel mobility shift assay essentially by the method of Vujaklija et al. (46). In brief, 3,000 cpm of 32P-labeled probe DNA was incubated with total proteins with or without AdpA (0.2 to 1 μg) at 30°C for 30 min in 40 to 50 μl (total volume) of a buffer containing 50 mM sodium phosphate buffer (pH 8.0), 10% glycerol, 1 μg of poly(dI-dC)-poly(dI-dC), and 0.01% bovine serum albumin as a protein stabilizer. After incubation, complexes and free DNA were resolved on nondenaturing 4 or 6% polyacrylamide gels (3% acrylamide-0.8% bisacrylamide) with a running buffer containing 40 mM Tris-HCl (pH 7.8), 20 mM sodium acetate, and 1 mM EDTA. The gels were dried, and the results were detected with a phosphorimager (FUJIFILM FLA-3000). A 295-bp sequence containing the promoter region of melC (from position −261 to position 34 with respect to the transcriptional start point) was prepared by PCR by using primers melupF (5′-GACCCGCAAACCCCTTGATCCGC-3′) and melupR (5′-CCTCCTGGGGTGCGTTGGGTTGA-3′), and the 5′ ends were labeled with [γ-32P]ATP and T4 polynucleotide kinase.
RESULTS
ZX66, an S. lividans ZX64 derivative incapable of expressing the melC operon.
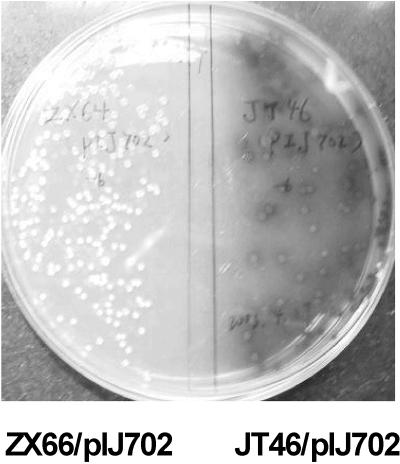
ZX64 was a mutant that was generated from S. lividans JT46 by insertion of a spectinomycin-streptomycin resistance cassette (spc-str) into dndA, one of the five open reading frames (ORFs) in the dnd gene cluster (which contained genes that rendered DNA susceptible to degradation during electrophoresis [the Dnd phenotype] [X. Zhou and Z. Deng, unpublished]). When pIJ702 carrying the melanin (melC) operon consisting of melC1 and melC2 from S. antibioticus (23) was introduced into S. lividans ZX64 by protoplast transformation, however, mostly black colonies and occasionally a rare white colony (ZX66) were obtained, while all JT46 transformants were black. The inability of ZX66 to produce melanin was found to be stable (Fig. 1), and the sporulation phenotype of this strain seemed to be poorer than that of JT46.
FIG. 1.
melC operon is expressed in JT46 (right) but not in ZX66 (left). Both strains were transformed with pIJ702 carrying melC1 and melC2 and were grown on MMT containing thiostrepton.
Lack of expression of the melC operon is not due to melC mutation and could be restored by a 3.5-kb DNA fragment from JT46, a progenitor of ZX46.
The copy number of pIJ702 seemed to be unchanged, and pIJ702 did not seem to have undergone any rearrangement because its banding patterns remained the same as those of pIJ702 present in the starting strain (JT46). The MelC1and MelC2 genes on pIJ702 did not seem to be mutated as the production of black pigment after pIJ702 was reisolated from ZX66 and reintroduced into JT46 was normal. A library with 2- to 8-kb DNA inserts generated by MboI partial digestion of JT46 total DNA was ligated with BamHI-digested pSET152, a plasmid vector able to integrate into the S. lividans chromosome. When pSET152-derived ligation mixtures were used to transform ZX66/pIJ702 protoplasts, a black transformant (ZD1) was obtained.
The DNA insert of ZD1 was recovered by using a pair of PCR primers (pSET152-FP and pSET152-RP) flanking the multiple cloning sites of pSET152 and chromosomal DNA of ZD1 as the template. A 3.5-kb DNA fragment was amplified, and reinsertion of this fragment into pSET152 (resulting in pJTU1452) and introduction by transformation into ZX66 could complement full expression of the melC operon (data not shown).
Localization of a gene complementing melC expression deficiency.
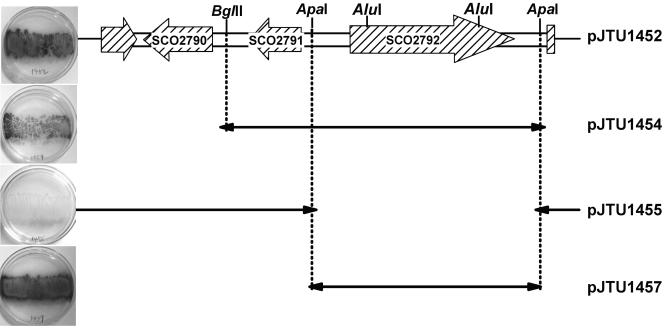
The 3.5-kb DNA insert from pJTU1452 was sequenced. BLAST searches immediately revealed strong similarity or even identity to a region (nucleotides 3045346 to 3048682) of the sequenced S. coelicolor genome which contained three complete ORFs, SCO2790, SCO2791, and SCO2792 (Fig. 2). Two incomplete ORFs (SCO2789 and SCO2793) (Fig. 2) seemed to be irrelevant to the control of melC expression and thus were not included in further analysis. One of the three complete ORFs (SCO2792; nucleotides 3047135 to 3048331) was clearly identified as an araC family transcriptional regulator, a potential candidate for controlling melC operon expression; SCO2090 showed significant homology to the USP domain of a family of universal Streptomyces proteins; and SCO2091 encoded a hypothetical protein without obvious conservation with other known proteins in the database.
FIG. 2.
Localization of a gene activating melC expression. The DNA sequence and genetic organization of the original 3,336-bp insert isolated from S. lividans in pJTU1452 are almost identical to the DNA sequence and genetic organization of the corresponding region carrying three complete ORFs (SCO2790 to SCO2792) in S. coelicolor, which is shown at the top. pJTU1452 derivatives are shown below pJTU1452, and their abilities to activate melC operon expression (black colonies on plates) are shown on the left.
To localize the responsible gene(s), religation of pJTU1452 after digestion with BamHI (flanking one side of the multiple cloning sites of the pSET152 vector) and BglII yielded pJTU1454; this removed SCO2790 (Fig. 2). In a similar way, religation of pJTU1452 after digestion with ApaI produced pJTU1455; this removed SCO2792 completely but left SCO2790 and SCO2791 intact (Fig. 2). Meanwhile, the 1.7-kb ApaI fragment carrying intact SCO2092 was inserted into the corresponding site in pSET152, resulting in pJTU1457, which carried only SCO2792 (Fig. 2).
When the three pJTU1452 derivatives (pJTU1454, pJTU1455, and pJTU1457) were introduced by transformation into ZX66, black colonies were observed only when inserts carried SCO2792 alone (pJTU1457) or in combination with SCO2791 (Fig. 2). This experiment unambiguously demonstrated that SCO2792 is a required positive regulator of melanogenesis.
The locus that positively controls melC expression is equivalent to adpA (adpAl) in S. coelicolor, and analysis of the adpAl mutant allele in ZX66.
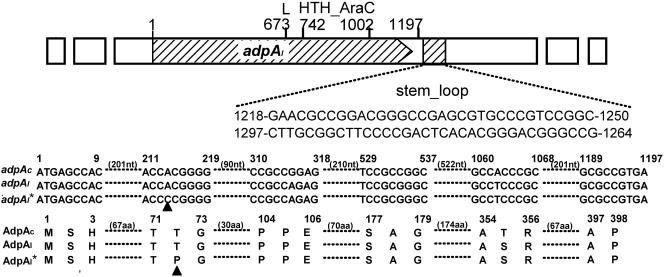
The gene that positively controls melC operon expression in S. lividans, SCO2792, is an adpAc homolog that has been reported to contain a rare leucine codon (TTA) whose translation depends on bldA tRNA and is involved in aerial mycelium development in S. coelicolor (41). We used the subscripts c and l, as used by Takano et al. (41), to distinguish between the genes of S. coelicolor and S. lividans, respectively. adpAc in S. coelicolor and adpAl in S. lividans are 99% identical at the nucleotide level, and four of the different nucleotides resulted in only one variation at the amino acid level. Similarly, the N-terminal portion of AdpAl (and AdpAc) has sequence homology with a group of proteins in the ThiJ-PfpI-DJ-1 family, which is widely distributed in bacteria, archaea, and eukaryotes, and has structural similarity to the type I glutamine amidotransferase domain (19), although the amino acid sequences of the members of this family were not dramatically conserved. This potential AraC family transcriptional regulator has a typical conserved DNA-binding domain with two helix-turn-helix motifs at the C terminus and a sequence with the potential to form a stem-loop structure 21 bp downstream of the stop codon of AdpAc or AdpAl (Fig. 3). A rare leucine codon (TTA) was found at the same position in both proteins (AdpAc and AdpAl) (Fig. 3), which must be the target for bldA regulation. When pIJ702 was introduced into an S. coelicolor M145-derived bldA mutant strain (J1700), none of the many independent thiostrepton-resistant transformants produced a black pigment, as expected.
FIG. 3.
Overall characteristics of the adpAl gene and alignment of adpAc (from S. coelicolor M145), adpAl (from S. lividans ZX64), and adpAl* (from S. lividans ZX66) and the protein sequences encoded by these genes. Nucleotides are numbered from the ATG start codon. L is the rare leucine (TTA) codon. HTH_AraC is a region with a helix-turn-helix motif for the typical AraC family of bacterial regulatory proteins, and the shaded region (with two alternative sequences) has the potential to form a stem-loop structure. Only the regions with differences at the nucleotide (nt) and amino acid (aa) levels between alleles are shown, and mutant positions are indicated by triangles.
A 1.7-kb ApaI fragment carrying a hypothetical mutant adpAl locus corresponding to SCO2792 in ZX66 was cloned into the corresponding site of pBluescript SK(+) by colony hybridization by using the 1.7-kb ApaI fragment carrying the JT46 adpAl gene as a probe and sequenced. A transversion (A to C) was found at nucleotide 214, which changed a codon from ACG (for Thr) to CCG (for Pro) (Fig. 3). Indeed, alignment of the nucleotide (and amino acid) sequences of adpAc and adpAl, as well as other sequenced adpA genes (and the corresponding proteins) in Streptomyces avermitilis (adpAa) and S. griseus (adpAg), revealed that none of the adpA genes has any variation at nucleotide 214 (A is part of the ACG codon encoding Thr).
Expression of the melC operon is under the same positive control by AdpAc in S. coelicolor M145 and by AdpAa in its native host, S. antibioticus.
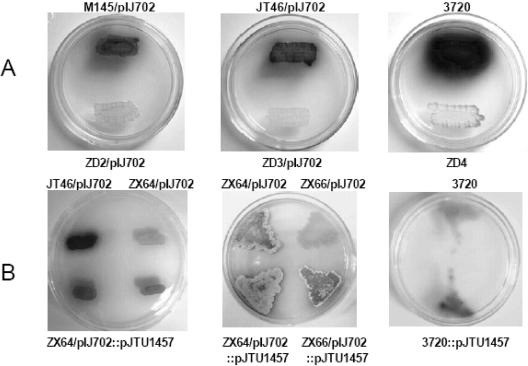
pIJ702 was introduced into ZD2 (an M145 derivative with a mutation in adpAc) and ZD3 (a JT46 derivative with a mutation in adpAl). No black pigment was secreted from ZD2 or ZD3 carrying pIJ702 or from ZD4 (an S. antibioticus IMRU3720 derivative with a mutation in adpAa), while a black pigment was clearly observed with M145 and JT46 carrying the same plasmid, as well as with the wild-type strain S. antibioticus IMRU3720 on the same plates (Fig. 4), suggesting that the melC operon is under the same positive control in all three organisms. Meanwhile, a sharp increase in the amount of black pigment was observed when an extra copy of adpAl was added by integrating pJTU1457 into the attB sites of either S. coelicolor or S. lividans strains carrying pIJ702 or S. antibioticus IMRU3720, from which the melC operon carried on pIJ702 originated (Fig. 4), by site-specific recombination through the attP site of φC31 on pSET152.
FIG. 4.
(A) Lack of mel operon expression in adpA mutants of S. coelicolor ZD2, S. lividans ZD3, and S. antibioticus ZD4 compared with their progenitors, M145 (left), JT46 (middle), and IMRU3720 (right), respectively. (B) Dosage effect of adpA. The top half of each plate contained wild-type S. antibioticus IMRU3720, S. lividans JT46 and ZX64 carrying pIJ702, or S. lividans ZX64 and ZX66 carrying pIJ702, while the bottom half of each plate contained strains corresponding to the strains in the top half but with an additionally integrated copy of adpAl (carried on pJTU1457) except that the two patches for the same strain (ZX64/pIJ702::pJTU1457) are shown in the bottom half of the left plate. For the strains at the bottom there was both increased production of black pigment and enhanced sporulation compared with the strains at the top.
AdpAl affects colony phenotypes and transcriptionally interacts with the promoter region of the melC operon.
Constructed gene disruptants with disruptions internal to adpAc in M145 (ZD2), adpAl in JT46 (ZD3), and adpAa in S. antibioticus IMRU3720 (ZD4) were compared in order to examine their colony phenotypes along with those of the corresponding parental strains. All of the mutants appeared to be obviously bald, as reported previously for the bldA mutant of S. coelicolor M145 (41), while all of the parental strains sporulated well.
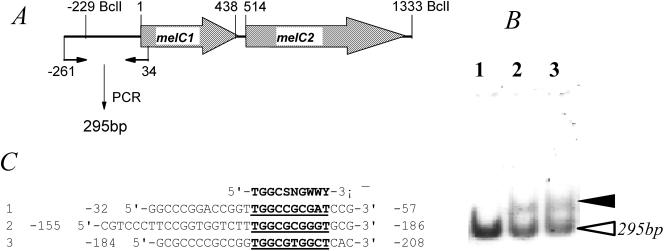
An attempt was also made to detect the possible interaction of AdpAl with the promoter region of the melC operon by a gel mobility shift assay. As shown in Fig. 5, the total proteins isolated with AdpAl from E. coli strain BL21(DE3) carrying pJTU1464 (a pET15b derivative carrying the adpA gene) obviously bound to the 295-bp DNA fragment carrying the promoter region of the melC operon radioactively labeled with [γ-32P]ATP and T4 polynucleotide kinase, while the total proteins without AdpAl from E. coli strain BL21(DE3) carrying pET15b lacked specific binding. Additionally, the band in lane 3, in which 0.02 μg of total proteins was added, was obviously more intense than the band in lane 2, in which only 0.01 μg of total proteins was added. The same experiments were repeated by using various concentrations of the same protein preparations with and without AdpAl but with an unrelated 318-bp labeled probe fragment as an additional control, and no shifted band, like that in lanes 2 and 3, was detected on the gel (data not shown).
FIG. 5.
(A and B) Gel mobility shift assay (B) for detection of the binding of AdpAl to a γ-32P-labeled 295-bp DNA fragment (A) covering the promoter region of the melC operon. The solid arrowhead in panel B indicates the position when 0.01 μg (lane 2) or 0.02 μg (lane 3) of total proteins isolated from E. coli BL21(DE3) carrying pJTU1464 was added to the radioactively labeled probe fragment (open arrowhead). Addition of 0.02 μg of total proteins isolated from E. coli BL21(DE3) carrying pET15b to the same probe was used as a negative control (lane 1). (C) Alignment of the three regions upstream of the melC operon (A) with the consensus AdpA-binding sequence (51) shown at the top (5′-TGGCSNGWWY-3′, where S is G or C, W is A or T, Y is T or C, and N is any nucleotide).
DISCUSSION
The tyrosinase (mel) gene, which is responsible for melanin synthesis, has been sequenced from at least nine Streptomyces strains (S. coelicolor, S. avermitilis, S. lincolnensis, S. galbus, S. castaneoglobisporus, S. tanashiensis, S. griseus, S. venezuelae, S. antibioticus, and S. glaucescens). Both the gene sequences and the protein sequences of all of these organisms are similar, with similar sizes and with the genes organized as a polycistronic operon (melC) consisting of two separate ORFs (melC1 and melC2), but little was known about the regulation of tyrosinase production at the molecular level in Streptomyces.
The initial isolation of many melanin-negative mutants of S. griseus revealed a close association among melanogenesis, the ability to produce streptomycin and A-factor, and the ability to form aerial mycelia (11). A-factor (2-isocapryloyl-3R-hydroxymethyl-γ-butyrolactone) (24) acts as a chemical signaling molecule that triggers morphological differentiation and secondary metabolism (6) at an extremely low concentration. A-factor exerts its signaling effects by binding to a cytoplasmic receptor protein (ArpA), which binds to the promoter region of a transcriptional regulator (AdpAg), in order to dissociate it from the DNA and permit transcription of free AdpAg to activate a number of genes required for both morphological differentiation and secondary metabolism (34). Several direct targets activated by AdpA have been identified, and these targets include strR, a pathway-specific transcriptional activator for streptomycin biosynthetic pathway genes in S. griseus (34); sgmA, a gene encoding a metalloendopeptidase that appears to affect the rate of development in S. griseus (22); adsA (bldN in S. coelicolor [4]), a gene encoding an RNA polymerase extracytoplasmic-function sigma factor needed for normal mycelial development in S. griseus (48); ssgA, which encodes a small acidic protein important for aerial hyphal septation in S. coelicolor (45) and S. griseus (21, 49); and amfR, a gene encoding a transcriptional activator of the amf operon needed for aerial mycelium formation in S. griseus (50). A direct link among melanogenesis, its absolutely required activation by the AdpA homolog, and its clear dosage effect either on melanin production or sporulation in a number of Streptomyces strains is obviously direct evidence that AdpA is a key and widespread transcription factor involved in multiple biological processes. A brief look at the promoter sequences upstream of the melC operon, whose DNA was used for the gel shift assay (Fig. 5), immediately revealed three regions with variable sequence homology to the identified consensus AdpA-binding sequence (5′-TGGCSNGWWY-3′) (51) (Fig. 5), which might be the specific target(s). The exact binding site, however, has not been determined by DNase I footprinting analysis.
S. coelicolor and S. griseus represent quite distant poles of Streptomyces phylogeny in terms of the 16S rRNA sequence, and their last common ancestor probably existed about 200 million years ago (10). However, homologs of the S. griseus adsA, sgmA, amfR, and ssgA genes and more than eight adpA-like genes are also found in the genome of S. coelicolor, and they may be involved in a number of independent or different aspects of the morphological and physiological development processes. The adpAc gene, corresponding to adpAg in S. griseus (41), was also found to have a rare TTA codon, like adpAl. In S. coelicolor, mutation of bldA, which encodes the tRNA for the rare leucine codon UUA, causes pleiotropic deficiencies in both the development of reproductive aerial hyphae and the production of antibiotics on most media (27, 33). As adpAl also has a rare TTA codon like that found in adpAc and adpAg, it was not surprising to find that the mel gene was not expressed in a bldA mutant and hence was likely to be controlled at the translational level via bldA tRNA.
Comparative analysis of a single mutation resulted in identification of a critical nucleotide (A at position 214), whose change (to C) led to a transversion in the amino acid sequence (from Thr to Pro at position 27). Conceivably, this amino acid must be at the active site of the AdpA protein to activate the transcription of the melC operon and to affect sporulation, but whether it has a similar role in activating other biological process, such as antibiotic production, still needs to be examined closely.
Acknowledgments
We thank D. A. Hopwood for critical reading of the manuscript and many valuable comments.
This work was supported by the 973 and 863 programs of the Ministry of Science and Technology, by the National Science Foundation of China, by the PhD Training Fund of the Ministry of Education, and by the Shanghai Municipal Council of Science and Technology.
REFERENCES
- 1.Altschul, S. F., and E. V. Koonin. 1998. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci. 23:444-447. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernan, V., D. Filpula, W. Herber, M. Bibb, and E. Katz. 1985. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene 37:101-110. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., V. Molle, and M. J. Buttner. 2000. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J. Bacteriol. 182:4606-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O′Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Chater, K. F., and S. Horinouchi. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9-15. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L. Y., M. Y. Chen, W. M. Leu, T. Y. Tsai, and Y. H. Lee. 1993. Mutational study of Streptomyces tyrosinase trans-activator MelC1. MelC1 is likely a chaperone for apotyrosinase. J. Biol. Chem. 268:18710-18716. [PubMed] [Google Scholar]
- 8.Chen, L. Y., W. M. Leu, K. T. Wang, and Y. H. Lee. 1992. Copper transfer and activation of the Streptomyces apotyrosinase are mediated through a complex formation between apotyrosinase and its trans-activator MelC1. J. Biol. Chem. 267:20100-20107. [PubMed] [Google Scholar]
- 9.della-Cioppa, G., S. J. Garger, G. G. Sverlow, T. H. Turpen, and L. K. Grill. 1990. Melanin production in Escherichia coli from a cloned tyrosinase gene. Bio/Technology 8:634-638. [DOI] [PubMed] [Google Scholar]
- 10.Embley, T. M., and E. Stackebrandt. 1994. The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 48:257-289. [DOI] [PubMed] [Google Scholar]
- 11.Endo, K., K. Kamo, K. Hosono, T. Beppu, and K. Ueda. 2001. Characterization of mutants defective in melanogenesis and a gene for tyrosinase of Streptomyces griseus. J. Antibiot. (Tokyo) 54:789-796. [DOI] [PubMed] [Google Scholar]
- 12.Friedman, D. I., and M. Gottesman. 1983. Lytic mode of lambda development, p. 21-51. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 13.Gottesman, S., E. Roche, Y. Zhou, and R. T. Sauer. 1998. The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12:1338-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 15.Hawkins, F. K., and A. W. Johnston. 1988. Transcription of a Rhizobium leguminosarum biovar phaseoli gene needed for melanin synthesis is activated by nifA of Rhizobium and Klebsiella pneumoniae. Mol. Microbiol. 2:331-337. [DOI] [PubMed] [Google Scholar]
- 16.Hintermann, G., M. Zatchej, and R. Hutter. 1985. Cloning and expression of the genetically unstable tyrosinase structural gene from Streptomyces glaucescens. Mol. Gen. Genet. 200:422-432. [DOI] [PubMed] [Google Scholar]
- 17.Hobbs, G., C. M. Frazer, D. C. J. Gardner, J. A. Cullum, and S. G. Oliver. 1989. Dispersed growth of Streptomyces in liquid culture. Appl. Microbiol. Biotechnol. 31:272-277. [Google Scholar]
- 18.Hopwood, D. A. 1967. Genetic analysis and genome structure in Streptomyces coelicolor. Bacteriol. Rev. 31:373-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horvath, M. M., and N. V. Grishin. 2001. The C-terminal domain of HPII catalase is a member of the type I glutamine amidotransferase superfamily. Proteins 42:230-236. [PubMed] [Google Scholar]
- 20.Ishikawa, J., and K. Hotta. 1999. FramePlot: a new implementation of the frame analysis for predicting protein-coding regions in bacterial DNA with a high G+C content. FEMS Microbiol. Lett. 174:251-253. [DOI] [PubMed] [Google Scholar]
- 21.Jiang, H., and K. E. Kendrick. 2000. Characterization of ssfR and ssgA, two genes involved in sporulation of Streptomyces griseus. J. Bacteriol. 182:5521-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, J. Y., A. Suzuki, H. Yamazaki, Y. Ohnishi, and S. Horinouchi. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katz, E., C. J. Thompson, and D. A. Hopwood. 1983. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J. Gen. Microbiol. 129:2703-2714. [DOI] [PubMed] [Google Scholar]
- 24.Khokhlov, A. S., I. I. Tovarova, L. N. Borisova, S. A. Pliner, L. N. Shevchenko, E. Kornitskaia, N. S. Ivkina, and I. A. Rapoport. 1967. The A-factor, responsible for streptomycin biosynthesis by mutant strains of Actinomyces streptomycini. Dokl. Akad. Nauk SSSR 177:232-235. [PubMed] [Google Scholar]
- 25.Kieser, T., D. A. Hopwood, H. M. Wright, and C. J. Thompson. 1982. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol. Gen. Genet. 185:223-228. [DOI] [PubMed] [Google Scholar]
- 26.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom.
- 27.Lawlor, E. J., H. A. Baylis, and K. F. Chater. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 28.Lee, Y. H., B. F. Chen, S. Y. Wu, W. M. Leu, J. J. Lin, C. W. Chen, and S. C. Lo. 1988. A trans-acting gene is required for the phenotypic expression of a tyrosinase gene in Streptomyces. Gene 65:71-81. [DOI] [PubMed] [Google Scholar]
- 29.Lerch, K. 1981. Metal ions in biological systems, vol. 13. Marcel Dekker Inc., New York, N.Y.
- 30.Leu, W. M., L. Y. Chen, L. L. Liaw, and Y. H. Lee. 1992. Secretion of the Streptomyces tyrosinase is mediated through its trans-activator protein, MelC1. J. Biol. Chem. 267:20108-20113. [PubMed] [Google Scholar]
- 31.Lomovskaya, N. D., N. M. Mkrtumian, N. L. Gostimskaya, and V. N. Danilenko. 1972. Characterization of temperate actinophage φC31 isolated from Streptomyces coelicolor A3(2). J. Virol. 9:258-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mason, H. S. 1965. Oxidases. Annu. Rev. Biochem. 34:595-634. [DOI] [PubMed] [Google Scholar]
- 33.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 34.Ohnishi, Y., S. Kameyama, H. Onaka, and S. Horinouchi. 1999. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Mol. Microbiol. 34:102-111. [DOI] [PubMed] [Google Scholar]
- 35.Paget, M. S., L. Chamberlin, A. Atrih, S. J. Foster, and M. J. Buttner. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paget, M. S., G. Hintermann, and C. P. Smith. 1994. Construction and application of streptomycete promoter probe vectors which employ the Streptomyces glaucescens tyrosinase-encoding gene as reporter. Gene 146:105-110. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Schaffer, A. A., L. Aravind, T. L. Madden, S. Shavirin, J. L. Spouge, Y. I. Wolf, E. V. Koonin, and S. F. Altschul. 2001. Improving the accuracy of PSI-BLAST protein database searches with composition-based statistics and other refinements. Nucleic Acids Res. 29:2994-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun, Y., X. Zhou, J. Liu, K. Bao, G. Zhang, G. Tu, T. Kieser, and Z. Deng. 2002. Streptomyces nanchangensis, a producer of the insecticidal polyether antibiotic nanchangmycin and the antiparasitic macrolide meilingmycin, contains multiple polyketide gene clusters. Microbiology 148:361-371. [DOI] [PubMed] [Google Scholar]
- 41.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475-486. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, C. J., J. M. Ward, and D. A. Hopwood. 1980. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature 286:525-527. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, J. F., and C. W. Chen. 1987. Isolation and characterization of Streptomyces lividans mutants deficient in intraplasmid recombination. Mol. Gen. Genet. 208:211-218. [DOI] [PubMed] [Google Scholar]
- 44.Tseng, H. C., and C. W. Chen. 1991. A cloned ompR-like gene of Streptomyces lividans 66 suppresses defective melC1, a putative copper-transfer gene. Mol. Microbiol. 5:1187-1196. [DOI] [PubMed] [Google Scholar]
- 45.van Wezel, G. P., J. van der Meulen, S. Kawamoto, R. G. Luiten, H. K. Koerten, and B. Kraal. 2000. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. J. Bacteriol. 182:5653-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vujaklija, D., S. Horinouchi, and T. Beppu. 1993. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 175:2652-2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 48.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2000. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. J. Bacteriol. 182:4596-4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamazaki, H., Y. Ohnishi, and S. Horinouchi. 2003. Transcriptional switch on of ssgA by A-factor, which is essential for spore septum formation in Streptomyces griseus. J. Bacteriol. 185:1273-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamazaki, H., Y. Takano, Y. Ohnishi, and S. Horinouchi. 2003. amfR, an essential gene for aerial mycelium formation, is a member of the AdpA regulon in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 50:1173-1187. [DOI] [PubMed] [Google Scholar]
- 51.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]