Abstract
Recently, we demonstrated that Salmonella enterica serovar Typhimurium can form biofilm on HEp-2 cells in a type 1 fimbria-dependent manner. Previous work on Salmonella exopolysaccharide (EPS) in biofilm indicated that the EPS composition can vary based upon the substratum on which the bacterial biofilm forms. We have investigated the role of genes important in the production of colanic acid and cellulose, common components of EPS. A mutation in the colanic acid biosynthetic gene, wcaM, was introduced into S. enterica serovar Typhimurium strain BJ2710 and was found to disrupt biofilm formation on HEp-2 cells and chicken intestinal tissue, although biofilm formation on a plastic surface was unaffected. Complementation of the wcaM mutant with the functional gene restored the biofilm phenotype observed in the parent strain. A mutation in the putative cellulose biosynthetic gene, yhjN, was found to disrupt biofilm formation on HEp-2 cells and chicken intestinal epithelium, as well as on a plastic surface. Our data indicate that Salmonella attachment to, and growth on, eukaryotic cells represent complex interactions that are facilitated by species of EPS.
Historically, microbiological investigations of pathogenic bacteria have focused on the study of broth-grown (planktonic) bacteria and their ability to cause disease. More recently, the importance of a growth phase on solid surfaces, known as biofilm, has been recognized and has become the focus of research efforts for a variety of pathogenic microorganisms (6-8). Biofilms are formed in a variety of settings and are often composed of multiple species of bacteria and other organisms. In nature, bacteria are speculated to use biofilms as a prevalent mode of growth, whereas smaller fractions of the total population are believed to be free-living (8). Biofilms can also be a persistent source of infections (8). For instance, the ability of Pseudomonas aeruginosa to form biofilms is a significant virulence property when colonizing patients with cystic fibrosis because the organisms in the biofilm are more resistant to antibiotic treatment and the presence of the biofilm provides a constant source of infecting bacteria (35).
An important feature in biofilm development of many bacterial pathogens is a mucoid-like substance known as exopolysaccharide (EPS) or extracellular matrix (9). The EPS matrices from different bacteria are composed of polysaccharides, including alginate in Pseudomonas aeruginosa when forming biofilm in cystic fibrosis patients (11), cellulose in Salmonella enterica serovar Enteritidis (30), and colanic acid in Escherichia coli (9). It has been suggested that most bacteria can produce EPS in specific environmental conditions, but production is typically lost by culture on bacteriological media in a laboratory setting (19). Thus, while bacterial species have genes that determine the amount and composition of the EPS produced, environmental surfaces and conditions are also important factors in EPS production and play a role where specific bacterial species can form biofilms (4, 5).
Colanic acid is a polysaccharide comprised of repeating subunits (32) that is believed to be expressed extracellularly when E. coli cells attach to abiotic surfaces (9, 23). Danese et al. (9) found that production of colanic acid is not necessary for initial bacteria attachment but is required for subsequent three-dimensional biofilm development on abiotic surfaces. These researchers concluded that colanic acid plays a role in biofilm development after the initial bacterial attachment phase. The colanic acid biosynthetic gene cluster of E. coli has been identified and found to be composed of 19 genes (25). A similar set of genes has been identified in S. enterica serovar Typhimurium, which is likely to have similar functions (20, 31).
In a recent study (24), biofilms formed by Salmonella species were found to have different compositions depending upon the environmental conditions in which the biofilms are formed. Flagellum expression, lipopolysaccharide composition, and EPS composition varied depending upon whether a biofilm formed on glass or gallstones (24). Solano et al. (30) showed that cellulose is a primary component of EPS in Salmonella biofilms on glass surfaces, and Prouty and Gunn (24) found that neither cellulose nor colanic acid are significant components of Salmonella EPS in biofilms formed on gallstones since mutants in genes involved in the synthesis of these polysaccharides showed no difference in biofilm formation on gallstones.
Our research group has recently shown that serovar Typhimurium has the ability to adhere to and form a biofilm on eukaryotic cellular surfaces in a type 1 fimbria-dependent manner (3). S. enterica serotypes continue to be a cause of human food-borne salmonellosis throughout the world (16). The bacteria colonize the chicken intestinal tract, invade the intestinal epithelium and oviducts, and persist for long periods of time in the host (2, 18, 21, 29). Adherence and biofilm formation are likely to play significant roles in establishing long-term colonization of poultry and may play yet unknown roles in the Salmonella virulence strategy.
To characterize serovar Typhimurium biofilm formation, we first constructed Salmonella strains with mutations in genes that might affect biofilm development. The linear transformation method of mutant construction described by Datsenko and Wanner (10) made this approach efficient and time effective. Gene knockouts were created in several putative colanic acid biosynthesis genes (wcaM, wcaA, and wza), as well as the cellulose biosynthesis gene, yhjN. Biofilm formation by these strains, carrying a green fluorescent protein (GFP)-encoding plasmid, was compared to the parent strain, BJ2710. Our data confirm the findings of other groups that Salmonella colanic acid biosynthetic genes are not required for biofilm formation on abiotic surfaces (24, 30). However, in contrast to other work, we find that colanic acid biosynthetic genes do play a role in the three-dimensional architecture of the biofilm on HEp-2 cells. Interestingly, we also found that colanic acid biosynthetic genes are involved in the development of biofilm structures on the surface of mammalian tissue culture cells and chicken intestinal epithelium. We found that hns, a gene important in modulating a wide variety of E. coli genes (26), including colanic acid regulation (12, 27), influences biofilm formation. Finally, we demonstrate that the putative cellulose biosynthetic gene, yhjN, is involved in biofilm formation on eukaryotic cells, as well as biofilm formed on a plastic surface.
MATERIALS AND METHODS
Tissue culture cells.
HEp-2 tissue culture cells (ATCC CCL-23) were used for in vitro biofilm experiments and were purchased from the American Type Culture Collection. Cells were maintained in RPMI 1640 tissue culture media with 10% fetal calf serum and passaged every 48 to 72 h. For biofilm experiments, 105 HEp-2 cells from cultures passaged fewer than 10 times were seeded to sterile biofilm chambers in a 1-ml volume of RPMI 1640 growth medium and allowed to adhere to the surface overnight. The following day the monolayers were checked microscopically for appearance prior to use.
Growth conditions, bacterial strains, and plasmids.
The bacterial strains and plasmids used in the present study are listed in Table 1. Antibiotics were added at the following concentrations: ampicillin, 100 μg ml−1; kanamycin, 25 μg ml−1; and chloramphenicol, 25 μg ml−1. For biofilm experiments, strains were grown statically in Lennox broth (0.5% NaCl) (Gibco/Invitrogen, Carlsbad, CA) at 37°C supplemented with appropriate antibiotics as necessary. RPMI 1640 agar plates were made by adding agar (Difco) to a final concentration of 1.5% to RPMI 1640 tissue culture medium. When inoculated into the biofilm chambers, the bacteria were grown in RPMI 1640 tissue culture medium (Gibco/Invitrogen). Screening of strains for cellulose production was performed by assessing the level of calcofluor (fluorescent brightener 28; Sigma) binding by colonies growing on Lennox agar plates with 200 μg of calcofluor per ml at room temperature for 24 to 48 h. The strains were exposed to a 366 nm UV light source, and the fluorescence of the yhjN mutant BJ3512 was compared to the wild-type strain BJ2710.
TABLE 1.
Bacterial strains and plasmids used in the study
| Strain or plasmid | Genotype or phenotypea | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli DH12S | mcrA Δ(mrr-hsdRMS-mcrBC) F′ lacIq ΔM15 | Invitrogen |
| Salmonella enterica serovar Typhimurium | ||
| SL1344 | Virulent wild-type strain | 36 |
| LB5010 | Serovar Typhimurium LT2 strain containing a complete fim gene cluster | 36 |
| BJ2508 | BJ2710 derivative with a fimH::kan insertion; Kanr | 3 |
| BJ2710 | SL1344 derivative containing an adhesive fimH gene | This work |
| BJ3402 | BJ2710 derivative with a deletion of the wcaM gene | This work |
| BJ3458 | BJ2710 derivative with a disruption of the hns gene | This work |
| BJ3512 | BJ2710 derivative with a disruption of the yhjN gene | This work |
| BJ3537 | BJ2710 derivative with a disruption of wcaA | This work |
| BJ3538 | BJ2710 derivative with a disruption of wza | This work |
| BJ3468 | BJ2710 derivative with a disruption of wcaM and hns | This work |
| Plasmids | ||
| pBDJ254 | pGEM-T carrying the hns gene; Ampr | This work |
| pKD3 | pANTSγ vector containing the cam template gene cloned from pSC140; Ampr | 10 |
| pKD4 | pANTSγ vector containing the kan template gene cloned from pCP15; Ampr | 10 |
| pKD46 | Temperature-sensitive red helper plasmid expressing araC-ParaB and γβexo from λ phage; Ampr | 10 |
| pMRP9-1 | GFP-expressing plasmid; Ampr | E. P. Greenberg |
| pNAL100 | pGEM-T vector carrying the wcaM gene; Ampr | This work |
| pISF204 | pBR322 vector containing fimH and fimF; Ampr | 17 |
| pBBRMCS-1 | Plasmid encoding GFP; Camr | 22 |
Ampr, ampicillin resistant; Kanr, kanamycin resistant; Camr, chloramphenicol resistant.
To create plasmid pNAL100, which carries an intact wcaM gene, genomic DNA isolated from strain SL1344 was used as a template for PCR amplification of the wcaM gene with primers WcaM5′ and WcaM3′, primer sequences will be provided upon request. The amplified PCR fragment was directly ligated into the TA overhang cloning vector pGEM-T (Promega, Madison, WI) so that expression of the wcaM gene is driven by the lac promoter. To create plasmid pBDJ254, which carries an intact Salmonella hns gene, SL1344 genomic DNA was used as template DNA in a PCR. Primers hns5′ and hns1000 were used to synthesize an 1,188-bp fragment that was cloned into the TA overhang cloning vector pGEM-T (Promega). Plasmid or genomic DNA was isolated from overnight cultures grown shaking at 37°C in Lennox broth supplemented with appropriate antibiotics. Cultures were pelleted by centrifugation, and DNA was extracted by using Qiagen kits for plasmid isolation or genomic DNA isolation according to the instructions of the manufacturer.
Construction of serovar Typhimurium strains.
S. enterica serovar Typhimurium strain BJ2710 was constructed by using the λ red recombinase system described by Datsenko and Wanner (10). First, a selectable kanamycin marker was inserted into the Salmonella chromosome downstream of the fimH gene in strain LB5010. The PCR primers fimU5′ and fimU3′ were used to construct a linear PCR fragment encoding kanamycin resistance that was inserted into the chromosome downstream of the fimH gene by recombination of the linear DNA fragment (10). Transformant colonies were demonstrated to have the kanamycin resistance cassette inserted at the proper site by PCR analysis. A P22 lysate was prepared on the LB5010 recombinant strain, and serovar Typhimurium strain SL1344 was transduced to kanamycin resistance to create strain BJ2710 that possesses the adherence phenotype of LB5010. Antibody agglutination and hemagglutination of guinea pig erythrocytes assays were performed to confirm that BJ2710 and the parent strain, SL1344, had functional type 1 fimbriae that displayed similar binding activities (17, 33). In addition, the two strains are equally virulent for mice by the oral and intraperitoneal routes of infection (data not shown). However, the BJ2710 strain expressed high-level adherence to HEp-2 cells (∼50 organisms per cell) compared to SL1344 (1 to 2 organisms per cell).
By using the linear transformation technique of Datsenko and Wanner (10), serovar Typhimurium mutants were made with mutations in wcaM using the primers WcaM5′ and WcaM3′, with mutations in wcaA using the primers WcaA5′ and WcaA3′, with mutations in wza using the primers wza5′ and wza3′, and with mutations in yhjN using the primers yhj5′ and yhjN3′, and the hns gene mutant was made using the primers HNS5 and HNS3. Primers for PCRs were purchased from IDT (Coralville, IA), and sequences will be provided upon request.
Growth curves in Lennox broth and RPMI 1640 tissue culture broth were performed for parent strains and each mutant strain. No significant difference in growth rate was observed for any strain.
Detection of type 1 fimbriae.
Bacteria were subcultured in 10 ml of LB broth and incubated without shaking for 48 h. Preparation of bacterial suspensions to detect mannose-sensitive hemagglutination of guinea pig erythrocytes was performed as previously described (33). The presence of fimbrial antigens on the surface of bacteria was detected by using monospecific fimbrial antiserum as described in Hancox et al. (17).
Biofilm formation on tissue culture plastic, HEp-2 cells and chicken intestinal epithelium.
The ability of bacterial strains to form biofilms on the surface of HEp-2 cells was investigated by using a modification of the flowthrough continuous culture system described by Parsek and Greenberg (22). Flow chambers were seeded with HEp-2 cells grown in RPMI 1640 (Gibco-Invitrogen, Carlsbad, CA) with 10% fetal calf serum (Gibco-Invitrogen), and biofilm was cultivated in a manner identical to that described by Boddicker et al. (3). All bacteria used in the assay were transformed with a plasmid (pMRP9-1) expressing the green fluorescent protein (GFP) (22). For biofilm assays, HEp-2 cells were stained with 1 μM cell tracker orange (CMTMR; Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Imaging of the biofilm was performed by using either a Bio-Rad MRC600 or a Zeiss LSM510 confocal scanning laser microscope and appropriate software to analyze the images. Biofilm development was monitored at 24 h, and all experiments were repeated on at least three separate occasions. The data presented in the figures are representative of the general appearance of each biofilm in multiple microscopic fields. Confocal images of bacterial growth on the HEp-2 cells are presented as composite images of the x-y plane.
For biofilm experiments with tissue culture plastic as the solid surface, the flow cell was assembled as described by Parsek and Greenberg (22), with the modification that a plastic tissue culture slide was used instead of glass. Chambers were inoculated with 109 CFU of bacteria and allowed to adhere to the plastic surface for 30 min prior to initiating flow of the RPMI 1640 plus 10% newborn calf serum tissue culture medium. All other parameters were the same as those for the HEp-2 biofilm experiments.
For chicken intestinal biofilm experiments, intestinal tissue was removed from euthanized 1- to 2-week-old Black Australorp chickens. The distal third of the small intestine was removed and washed with phosphate-buffered saline. Peristaltic tubing (1/16-in. diameter) was inserted into each end of a 10-cm length of excised intestine, and the tubing was secured with 6-0 silk sutures (Ethicon, Piscataway, NJ). The intestinal loops were inoculated with 105 bacteria and incubated at 37°C with 5% CO2 for 30 min to allow initial bacterial adherence. Subsequently, the loops were infused with RPMI 1640 tissue culture medium supplemented with 7% bovine calf serum and 3% chicken serum (Invitrogen, Carlsbad, CA) at a rate of 130 μl/min and incubated for 24 h in a CO2 incubator. Biofilm formation on the mucosal surfaces was examined over the entire length of the intestinal segment under low power by using a Zeiss LSM510 confocal scanning laser microscope, and representative images were generated and analyzed as described above.
Colorimetric detection of polysaccharides.
Polysaccharides were quantitated via the method described by Wehland and Bernhard (34). Bacteria were grown on LB agar overnight at 37°C, harvested by scraping, and resuspended in 5 ml of phosphate-buffered saline. The cell number was determined from the turbidity at 600 nm, as well as by plating dilutions of culture on Lennox plates. EPS was separated from the bacteria by vortexing each sample for 3 min, followed by ultracentrifugation of the bacterial suspension in a Beckman L8-70 centrifuge by using a SW40 Ti rotor at 30,000 rpm (160,000 × g) for 45 min at 10°C. The supernatant was removed and dialyzed in ddH2O for 3 h (34). Polysaccharides were quantitated by adding 60 μl of an 80% phenol solution to 2 ml of the dialyzed EPS. Then, 5 ml of concentrated sulfuric acid (Fisher Scientific, Pittsburg, PA) was added, and the tubes were incubated at room temperature for 10 min and then shaken and incubated at 30°C for 20 min longer. The absorbance at 490 nm (which is a measure of the polysaccharide content) was plotted on a standard curve of glucose (13).
RESULTS
Biofilm production by serovar Typhimurium strain BJ2710.
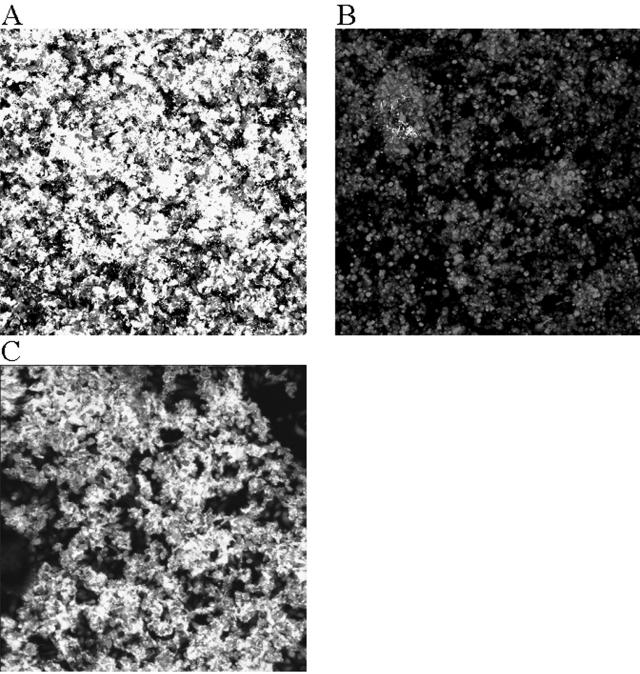
We have previously demonstrated that the serovar Typhimurium LT2 allele of the fimH adhesin gene of type 1 fimbriae mediates high-level adherence to tissue culture cells and murine intestinal tissue (3). A serovar Typhimurium SL1344 derivative, designated BJ2710, which carries this fimH allele on the chromosome, was tested for its ability to form biofilm. As shown in Fig. 1A, strain BJ2710 produced abundant, dense biofilm that covers the majority of the surface of HEp-2 tissue culture cells after 24 h of using the flowthrough system. Increasing the incubation time to 48 h did not significantly increase the amount of biofilm that was present at 24 h (data not shown). Two other serovar Typhimurium strains, LT2 and ATCC strain 14028, also produced confluent biofilm similar to that observed for BJ2710 (data not shown). As a negative control, serovar Typhimurium strain BJ2508 (BJ2710 fimH::Kan) displayed virtually no ability to attach to and form biofilm on HEp-2 cells after 24 h (Fig. 1B) or 48 h (data not shown). Previous work has also shown that the parent strain SL1344 forms no biofilm on HEp-2 cells (3). Complementation of BJ2508 with the functional LT2 fimH allele, carried on plasmid pISF204, restored the ability to form biofilm (Fig. 1C). These results demonstrate that biofilm formation by serovar Typhimurium BJ2710 is dependent upon the adherent type 1 fimbriae present in the LT2 strain (3). All subsequent experiments use strains in the BJ2710 (SL1344) genetic background.
FIG. 1.
Comparison of biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium strain BJ2508 (BJ2710 ΔfimH) on cultured HEp-2 cells. The composite image of biofilm formed by adherent serovar Typhimurium strain BJ2710 (A) or nonadherent BJ2508 (BJ2710 fimH) (B) was recorded after 24 h of incubation in the biofilm flowthrough system. (C) Biofilm formed by the fimH mutant strain BJ2508 complemented with pISF204, which carries an intact LT2 fimH gene. Panel A demonstrates that extensive biofilm is formed by strain BJ2710. Mutation of the fimH gene (B) abrogates the ability to form biofilm. The bacteria were labeled with GFP and appeared green in the confocal microscope, and the HEp-2 cells were stained with CMTMR and appeared red in the confocal microscope. The level of cell stain for each of the samples was comparable to what is observed in panel B, which is almost exclusively HEp-2 cell staining.
Putative colanic acid biosynthetic genes are required for biofilm formation on tissue culture cells in serovar Typhimurium.
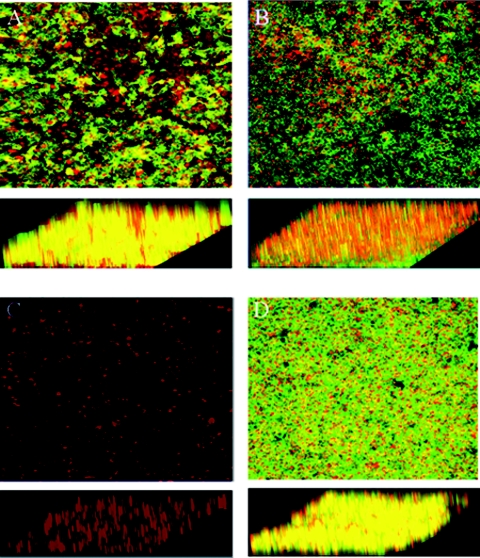
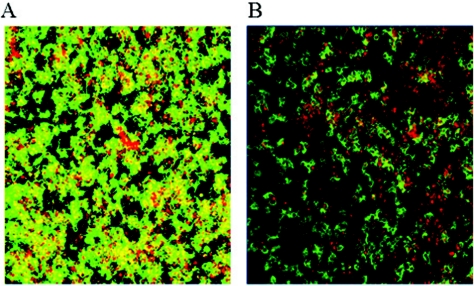
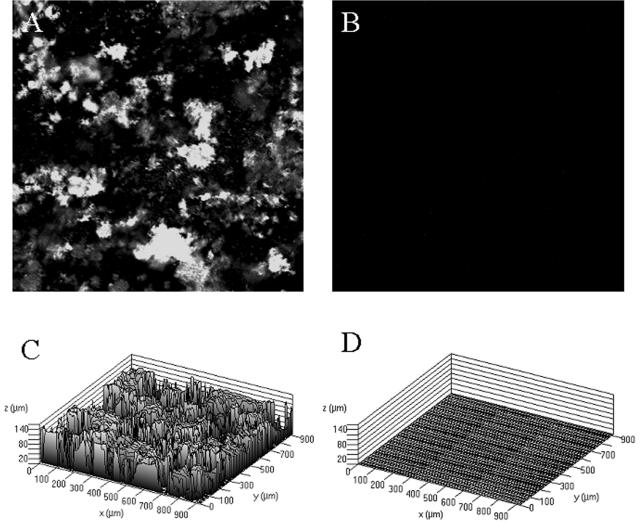
Our efforts to identify additional Salmonella genes necessary for growth and accumulation on tissue culture cells focused on genes involved in EPS synthesis. To this end, we constructed a serovar Typhimurium deletion mutation in wcaM, which is a colanic acid biosynthetic gene. Biofilm formation by the parent strain BJ2710 or strain BJ3402 (BJ2710 ΔwcaM) was compared for coverage of the apical surface of the HEp-2 cells and for height above the tissue culture cells after 24 h of growth. Representative areas of biofilm formed by each strain are shown as composite and slant images of the x-y plane (Fig. 2). As expected, serovar Typhimurium strain BJ2710 grew as an extensive biofilm (Fig. 2A), and the BJ2508 fimH mutant formed no detectable biofilm (Fig. 2C). The BJ2710 bacteria covered the underlying HEp-2 cell monolayer, and columns of biofilm structures were observed rising above the surface of the cells. Strain BJ3402 (BJ2710 ΔwcaM) retained the ability to adhere to the majority of the apical surface of the tissue culture cell monolayer (Fig. 2B), but the thickness of the bacterial community was considerably less than that observed for strain BJ2710, as seen in the slanted views. This observation suggests that the wcaM gene product contributes to the ability of the bacteria to create three-dimensional biofilm structures, which is separate from the initial colonization events. A plasmid containing the wcaM gene, pNAL100, complemented the ability to form biofilms, although the strain seemed to cover the apical surface of the tissue culture cells even more completely than BJ2710 (Fig. 2D). Two additional colanic acid biosynthetic genes, wcaA and wza, were examined for their role in biofilm accumulation. The biofilms produced by the wcaA- or wza-deficient mutants were nearly identical to that observed for the wcaM mutant (data not shown).
FIG. 2.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium BJ3402 (BJ2710 ΔwcaM) on cultured HEp-2 cells. The composite image of biofilm formed by adherent serovar Typhimurium strain BJ2710 (A) or nonadherent BJ2508 (BJ2710 fimH) (C) was captured after 24 h of incubation. Panel B is a biofilm formed by the BJ3402 (BJ2710 ΔwcaM) mutant strain, and panel D is a biofilm formed by BJ3402 (BJ2710 ΔwcaM) complemented with plasmid pNAL100 (wcaM+). Panel A demonstrates that extensive biofilm is formed by strain BJ2710. Mutation of the wcaM gene (B) results in a thin, short biofilm across the surface of the HEp-2 cells. As a negative control, disruption of the fimH gene yields virtually no bacterial accumulation at all (C). Bacteria were labeled with GFP (green) and the HEp-2 cells with CMTMR (red), and colocalization of bacteria with cells appears yellow. Slanted images provide a perspective of the depth of the biofilm.
The median height of biofilm from multiple experiments was calculated for each strain by using Zeiss confocal microscope software (Table 2). The median height of biofilm formed by parental strain BJ2710 across the entire section of the HEp-2 monolayer was 40 ± 6 μm but only ∼1 ± 1 μm for the fimH mutant, which does not bind to cells nor form biofilms. The wcaM mutant, BJ3402, produced biofilm with a median height of 14 ± 6 μm, a nearly threefold reduction compared to the parent strain. The complemented strain, BJ3402 pNAL100, produced biofilm that was slightly shorter (32 ± 2 μM) than the parent strain (40 ± 6 μM) but ∼2.5-fold thicker or taller than the wcaM mutant.
TABLE 2.
Height of biofilm produced by various S. enterica serovar Typhimurium strains
| Serovar Typhimurium strain BJ2710 and derivatives | Median ht of biofilm formed (μm) ± SD |
|---|---|
| BJ2710 (parent) | 40 ± 6 |
| BJ2508 (fimH) | 1 ± 1 |
| BJ3402 (wcaM) | 14 ± 6 |
| BJ3402 (wcaM) and pNAL100 (wcaM+) | 32 ± 2 |
| BJ3458 (hns) | 33 ± 4 |
| BJ3458 (hns) and pBDJ254 (hns+) | 3 ± 2 |
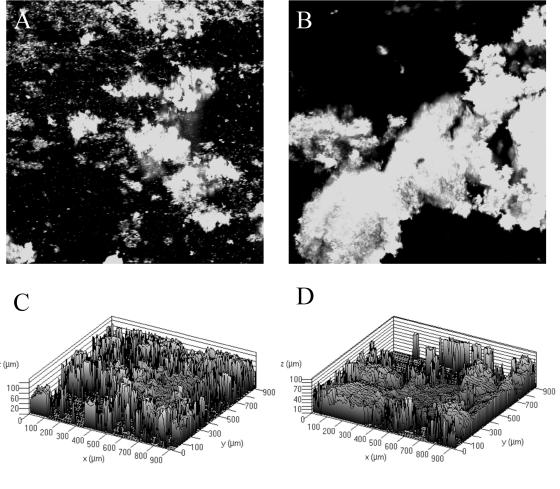
These observations led us to explore Salmonella biofilm formation on two other surfaces. First, we examined the ability of BJ2710 or BJ3402 (BJ2710 ΔwcaM) to form biofilm on plastic slides. As shown in Fig. 3A and B, strains BJ2710 and BJ3402 produced uneven, patching biofilm towers on plastic that were up to 100 μM in height (Fig. 3C and D). There were no apparent biofilm differences between the strains, indicating that the colanic acid biosynthetic gene wcaM does not contribute to the patchy attachment observed on plastic slides. These results are consistent with those reported by Prouty and Gunn (24). Next, we examined biofilm formation on chicken intestinal epithelium by using explanted chicken intestines as described in Materials and Methods. In contrast to biofilm formation on plastic, we observed a dramatic difference in the biofilm formed on chicken intestinal epithelium by BJ2710 and BJ3402 (BJ2710 ΔwcaM). Strain BJ2710 formed thick, confluent, complete biofilms (30 to 50 μM in height) after 24 h of flow through the chicken intestine (Fig. 4A and C). In contrast, diffuse, small foci of bacteria were formed on the tissue colonized by the BJ3402 (BJ2710 ΔwcaM) mutant (Fig. 4B and D). Occasionally, a few small spikes of organisms, up to 10 μM tall, were present. These data suggest that the BJ3402 strain could attach to the chicken epithelium but was unable to develop into a mature biofilm colony. The complemented strain BJ3402 pNAL100 formed biofilm on the chicken intestinal epithelium that was indistinguishable from the parent strain (data not shown).
FIG. 3.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium BJ3402 (BJ2710 ΔwcaM) on a plastic slide. (A and B) GFP-labeled BJ2710 and BJ3402, respectively, after growth for 24 h as biofilm on the plastic slides. (C and D) Three-dimensional representations of the same data shown in panels A and B. Both organisms adhere incompletely to the plastic surface but were able to form biofilm towers up to 100 μM tall after the original attachment event.
FIG. 4.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium BJ3402 (BJ2710 ΔwcaM) on chicken intestinal epithelium. (A and B) z-sections generated by confocal microscopy of BJ2710 and BJ3402, respectively, that were able to adhere to and grow as biofilm on the chicken intestinal epithelial surface. The organisms were originally labeled with GFP and appear white in the image. Panels C and D correspond to BJ2710 and BJ3402, respectively, and show a dramatic difference in the height and extent of BJ2710 biofilm which is 50 to 60 μm in height and covers the entire epithelial surface and BJ3402, which has adherent patches of organisms and occasional spikes of growth that are up to 10 μm in height.
Quantitation of extracellular polysaccharide production in S. enterica serovar Typhimurium strains.
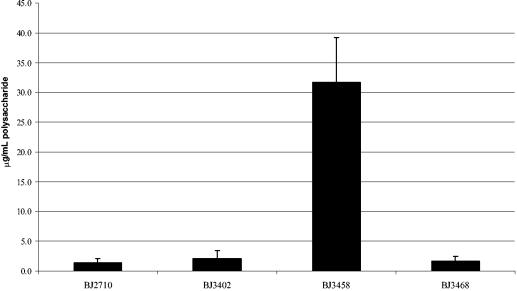
Since the serovar Typhimurium colanic acid biosynthetic genes have only been predicted from the genome sequence and have not been proven to be functional, we attempted to demonstrate that these mutations altered extracellular polysaccharide production. Extracellular polysaccharides were isolated from Salmonella strains grown on Lennox agar plates and quantitated as described in Materials and Methods. It is worth noting that the polysaccharide quantitation assay utilized detects colanic acid, as well as other polysaccharides. Little difference in EPS production was observed between strains BJ2710 (1.4 ± 0.7 μg of EPS/ml per 109 CFU) and BJ3402 (BJ2710 ΔwcaM) (2.1 ± 1.4 μg of EPS/ml per 109 CFU) after growth on Lennox agar plates (Fig. 5). Since the BJ3402 (BJ2710 ΔwcaM) mutant produced 50% more EPS than the parent strain BJ2710, it was likely, as reported by others (15, 19) that in vitro growth on bacteriological media was unfavorable for EPS production in Salmonella. Since the hns gene product represses colanic acid production in E. coli on bacteriological media (27, 28), we examined whether a serovar Typhimurium hns mutant is derepressed for EPS production on Lennox agar plates as well. Accordingly, we isolated and quantitated EPS from the hns mutant strain BJ3458, which has a mucoid colony phenotype. The strain produced 15- to 22-fold (31.7 ± 7.4 μg/ml) more EPS per 109 CFU than either BJ2710 or BJ3402 (BJ2710 ΔwcaM) in vitro. Introduction of a wcaM mutation into the BJ3458 hns strain to create an hns wcaM double mutant reduced EPS production to 1.6 ± 0.8 μg of EPS/ml (∼20-fold reduction). Since our data indicate that conditions present in developing biofilm induce expression of EPS, we hypothesize that the levels of EPS produced by an hns mutant on agar plates are more likely to be reflective of biofilm EPS production than the parent strain grown on agar plates. A similar set of experiments was performed with the strains grown on RPMI 1640 agar plates, as described in Materials and Methods, with results for EPS production similar to those observed on Lennox agar (data not shown). Thus, the wcaM gene is required for synthesis, regulation, or export of polysaccharide (i.e., colanic acid) in serovar Typhimurium. Although isolation of EPS from biofilm-grown cultures was not technically feasible, it seems likely that organisms growing in biofilm conditions are induced for EPS synthesis. Presumably, in biofilm growth conditions the production of EPS is comparable to that of the genetically derepressed hns strain.
FIG. 5.
Quantitation of EPS production by Salmonella strains. Extracellular polysaccharide from each bacterial strain was purified and quantitated by using a colorometric polysaccharide assay. Strains: BJ2710 (parent strain), BJ3402 (BJ2710 ΔwcaM), BJ3458 (BJ2710 hns::cam), BJ3468 (BJ2710 ΔwcaM hns::cam). Micrograms of polysaccharide per ml are expressed per 109 CFU.
Biofilm formation by a S. enterica serovar Typhimurium hns mutant.
We next examined the effect of the hns gene function in Salmonella biofilm formation. The H-NS protein is known to modulate the expression of a large number of bacterial genes (1). Relevant to EPS expression, hns represses rcsA activation by binding to the rcsA promoter and an hns mutant overexpresses extracellular polysaccharide (14, 27). Biofilms produced by strains BJ2710, BJ3458 (BJ2710 hns::cam) or BJ2710 pBDJ254, which overexpresses hns, were compared after 24 h in the flowthrough system (Fig. 6). Strain BJ3458 (hns::cam) produced biofilm structures indistinguishable from those produced by the wild-type strain BJ2710, suggesting that the flowthrough chamber conditions induce EPS production to the same or nearly the same level as that observed in the hns mutant. Biofilm synthesized by this strain covered the majority of the apical surfaces of the HEp-2 cells and possessed the large “bacterial towers” characteristic of strain BJ2710 (Fig. 6A). Biofilm produced by the hns mutant had a median height of 33 ± 4 μm (Fig. 6B), as measured by confocal image analysis, which was similar to that of strain BJ2710 (40 ± 6 μm) (Table 2). In contrast to the hns mutant, strain BJ2710 containing the hns-overexpressing plasmid pBDJ254, which is believed to repress colanic acid production by overexpression of the hns gene, exhibited a biofilm that was significantly reduced in thickness (∼3 ± 2 μM) and almost completely lacked column structures (Fig. 6D).
FIG. 6.
Comparison of biofilm formation by serovar Typhimurium strain BJ2710 and serovar Typhimurium BJ3458 (BJ2710 hns::cam) on cultured HEp-2 cells. Composite images of biofilm formed by adherent serovar Typhimurium strain BJ2710 (A) or nonadherent BJ2508 (BJ2710 fimH) (C) were recorded after 24 h of incubation. Panel B is an image of biofilm formed by BJ2710 hns (BJ3458), and panel D is biofilm formed by BJ2710 hns (BJ3458) complemented with plasmid pBDJ254, which carries an intact hns gene. Absence of the hns gene has little effect on biofilm formation (B), but overexpression of the hns gene resulted in a reduced ability to form biofilm (D). The bacteria carried GFP and appeared green with the confocal microscope, and the HEp-2 cells were labeled with CMTMR and appeared red with the microscope.
Biofilm formation in S. enterica serovar Typhimurium lacking yhjN, a putative cellulose synthetase gene.
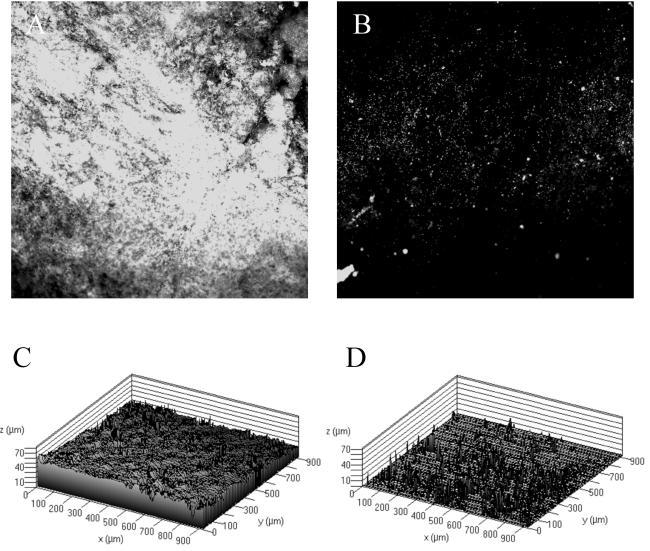
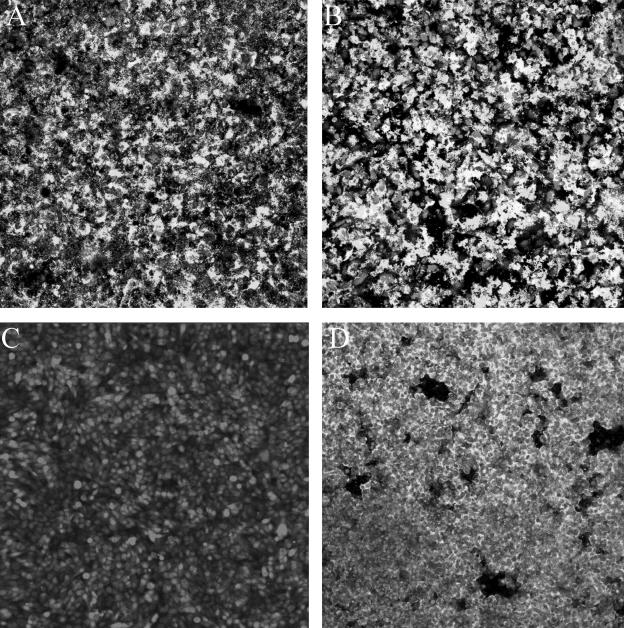
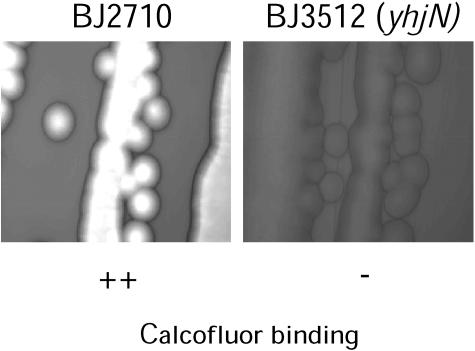
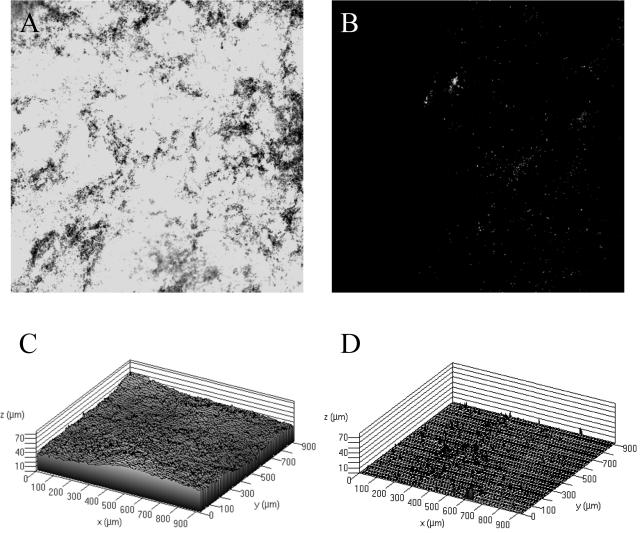
Since others have examined the role of cellulose in Salmonella biofilm production, we chose to evaluate the role of this polysaccharide in our system by constructing a strain with a mutation in the cellulose biosynthetic gene, yhjN. To determine whether the yhjN mutant produced extracellular cellulose, qualitative binding to calcofluor was used as described by Solano et al. (30). As observed in Fig. 7, the difference in fluorescence between BJ2710 grown on Lennox agar plates with calcofluor and BJ3512 (BJ2710 yhjN::cam) grown on the same plates indicates that BJ3512 has a defect in cellulose biosynthesis. Subsequently, biofilm production by BJ2710 or BJ3512 (BJ2710 yhjN::cam) was examined 24 h after infection of HEp-2 monolayers (Fig. 8). Strain BJ3512 (yhjN::cam) produced short biofilms on HEp-2 cells (Fig. 8B) which were similar to those observed for the colanic acid mutants. Consistent with the findings of Prouty and Gunn (24), the cellulose biosynthetic mutant did not form detectable biofilms on plastic slides (Fig. 9B and D), whereas the parent strain, BJ2710, formed biofilms that were uneven and patchy but also fairly tall (Fig. 9A and C). Similar to the wcaM mutant, the yhjN::cam cellulose biosynthesis mutant was profoundly defective in its ability to form a substantial biofilm on the chicken intestinal cells (Fig. 10B and D).
FIG. 7.
Detection of extracellular cellulose produced by BJ2710 and BJ3512 (BJ2710 yhjN::cam) by calcofluor binding. Each strain was grown on Lennox agar containing 200 μg per ml of calcofluor. Exposure to UV light at 366 nm reveals fluorescence that is the result of calcofluor binding to cellulose polymers.
FIG. 8.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium strain BJ3512 (BJ2710 yhjN::cam) on cultured HEp-2 cells. The composite image of biofilm formed by adherent serovar Typhimurium strain BJ2710 and the cellulose biosynthesis mutant BJ3512 was recorded after 24 h of incubation. Panel A is the positive control BJ2710, and panel B is an image of biofilm formed by the BJ3512 (BJ2710 yhjN::cam) mutant strain. A mutation in the yhjN gene (panel B) results in a patchy, shorter biofilm. Bacteria are labeled with GFP (green) and the HEp-2 cells with CMTMR (red), and colocalization of bacteria with cells appears as yellow.
FIG. 9.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium 3512 (BJ2710 yhjN::cam) on a plastic slide. Images shown in panels A and B depict GFP-labeled BJ2710 or BJ3512, respectively, after growth for 24 h as biofilm on the plastic slides. Panels C and D are three-dimensional representations of the same data shown in panels A and B. BJ2710 organisms adhered incompletely to the plastic surface but were able to form biofilm towers up to 100 μM tall after the original attachment events, whereas no organisms of strain BJ3512 were detectable on the plastic after 24 h of incubation.
FIG. 10.
Biofilm formation by serovar Typhimurium strain BJ2710 or serovar Typhimurium BJ3512 (BJ2710 yhjN::cam) on a chicken intestinal epithelium. The images shown in panels A and B depict GFP-labeled BJ2710 or BJ3512, respectively, after growth for 24 h as biofilm on the epithelial tissue. Panels C and D correspond to BJ2710 and BJ3512, respectively, and show a dramatic difference in the height and extent of the BJ2710 biofilm, which is 50 to 60 μm in height and covers the entire epithelial surface, and the BJ3512 biofilm, which has almost no visible organisms.
DISCUSSION
In the present study we have taken advantage of a recently developed system in which serovar Typhimurium forms biofilms on the surface of mammalian tissue cultures (3). We have extended this model to study biofilm formation on chicken intestinal epithelium. The biofilm cultures on HEp-2 cells can be maintained for up to 48 h, and the chicken intestinal biofilms can be maintained for up to 24 h. The ability to grow biofilms on these cells is likely to be a useful and relevant tool for studying biofilm formation by Salmonella since the organism is an important member of the intestinal flora in both chickens and turkeys. In addition, Salmonella species cause invasive intestinal disease in a wide variety of other animal hosts, including humans. Due to these properties and interactions, an understanding of the genetics of Salmonella biofilm may lead to methods to modify and/or control Salmonella colonization and carriage of poultry and initiation of disease in a variety of hosts.
The colanic acid genes were of particular interest in our studies because of the putative role for colanic acid in EPS of E. coli biofilms. To investigate the role of colanic acid production in S. enterica serovar Typhimurium biofilm formation, a nonpolar mutation in the wcaM gene was created in the BJ2710 strain background. Based on homology to the E. coli colonic acid biosynthetic genes, this mutant was predicted to have a significantly reduced ability to produce colanic acid. Comparison of EPS production on agar from strains BJ2710 and BJ3402 (BJ2710 ΔwcaM) revealed no differences between the strains, but EPS levels were very low in both strains. We believe that this is due to very low production of EPS when grown on agar (19). A ΔwcaM hns::cam strain had significantly decreased levels of extracellular polysaccharide production (∼2 μg/ml) compared to an hns parent strain (∼31.5 μg/ml), which demonstrates that the wcaM gene is involved in extracellular polysaccharide production. These results also indicate that the hns mutation derepresses EPS production during agar growth. Subsequent biofilm studies with the wcaM mutant carrying a GFP plasmid showed that BJ2710 and the wcaM mutant produced biofilms on a plastic surface that were similar but that the wcaM mutant had an attenuated ability to form fully developed biofilm on tissue culture cells and a profound defect in forming biofilms on chicken intestinal epithelium compared to serovar Typhimurium BJ2710.
The regulation of colanic acid biosynthesis in Salmonella has not been studied in any detail. However, the work on colanic acid regulation in E. coli provides a starting point in understanding Salmonella colanic acid biosynthetic gene regulation. Both E. coli and S. enterica serovar Typhimurium have clusters of genes that are required for colanic acid biosynthesis (32). After the initial adherence events, it is likely that EPS production is induced by an unknown signal in the developing biofilm environment, where the EPS then functions by stabilizing the growing biofilm structure. The biofilm phenotype of the wcaM mutant also suggests that colanic acid production does occur at 37°C in Salmonella if the bacteria are exposed to eukaryotic cells in tissue culture media.
Since cellulose has also been implicated in Salmonella biofilm formation, we investigated the role of this polysaccharide in biofilm formation on mammalian cell surfaces. Two separate cellulose biosynthetic operons have been described in Salmonella strains that are adjacent to one another on the chromosome. One set of four genes is designated yhjONML (30), and the second set of three genes is designated yhjSTU (30). Construction of mutations in the individual genes, by other laboratories, has revealed that each is required for cellulose biosynthesis, and cellulose mutants were unable to form biofilm on glass or form a pellicle in LB (30). Based on these results and on the hypothesis that composition of EPS varies based on environmental conditions, we investigated the role of cellulose in S. enterica serovar Typhimurium biofilms formed on eukaryotic cell surfaces. A calcofluor binding assay confirmed that the mutant did not synthesize extracellular cellulose. Biofilm studies with the yhjN serovar Typhimurium mutant, carrying a GFP plasmid, showed that the mutant had a defect in the ability to form fully developed biofilm on tissue culture cells and a profound defect in the ability to form biofilm on chicken epithelium. The biofilm produced by the yhjN mutant resembled the wcaM mutant in that the depth was thinner than the parent strain and that it lacked the towers characteristic of the biofilm produced by the parent strain BJ2710.
We have made several observations in the present study that we believe are worth pursuing in more detail. The construction of a wcaM reporter is under way; this will be used to identify and characterize signals that lead to biofilm formation, as well as to identify various stages of biofilm assembly. Work is continuing to identify and characterize the components of the extracellular matrix of S. enterica serovar Typhimurium biofilms formed on eukaryotic cell surfaces. In the future, it would be interesting to test the in vivo relevance of our findings by examining, in competitive colonization studies, how strains that are defective in some aspect of biofilm formation can compete with Salmonella strains that are known to be efficient colonizers of chickens. Ultimately, the present study may be useful in reducing or eliminating the colonization of poultry with pathogenic Salmonella strains.
Acknowledgments
We thank Aaron Baxter, Jenny Boddicker, and Jenny Jagnow for helpful discussions. We also thank Tom Moninger and Randy Nessler of the University of Iowa Central Microscopy facility for their expert technical assistance with the confocal microscopy.
This research was funded by NIH grants AI007511 (N.A.L.), AI38268 (B.D.J.), and AI057192 (B.D.J.).
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Baba, E., H. Wakeshima, K. Fukui, T. Fukata, and A. Arakawa. 1992. Adhesion of bacteria to the cecal mucosal surface of conventional and germ-free chickens infected with Eimeria tenella. Am. J. Vet. Res. 53:194-197. [PubMed] [Google Scholar]
- 3.Boddicker, J. D., N. A. Ledeboer, J. Jagnow, B. D. Jones, and S. Clegg. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 45:1255-1265. [DOI] [PubMed] [Google Scholar]
- 4.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. Garcia-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation, and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 191:31-36. [DOI] [PubMed] [Google Scholar]
- 5.Bryan, B. A., R. J. Linhardt, and L. Daniels. 1986. Variation in composition and yield of exopolysaccharides produced by Klebsiella sp. strain K32 and Acinetobacter calcoaceticus BD4. Appl. Environ. Microbiol. 51:1304-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton, J. W., K. J. Cheng, G. G. Geesey, T. I. Ladd, J. C. Nickel, M. Dasgupta, and T. J. Marrie. 1987. Bacterial biofilms in nature and disease. Annu. Rev. Microbiol. 41:435-464. [DOI] [PubMed] [Google Scholar]
- 7.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 8.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 9.Danese, P. N., L. A. Pratt, and R. Kolter. 2000. Exopolysaccharide production is required for development of Escherichia coli K-12 biofilm architecture. J. Bacteriol. 182:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, D. G., A. M. Chakrabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dierksen, K. P., and J. E. Trempy. 1996. Identification of a second RcsA protein, a positive regulator of colanic acid capsular polysaccharide genes, in Escherichia coli. J. Bacteriol. 178:5053-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Rebers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 14.Ebel, W., and J. E. Trempy. 1999. Escherichia coli RcsA, a positive activator of colanic acid capsular polysaccharide synthesis, functions to activate its own expression. J. Bacteriol. 181:577-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant, W. D., I. W. Sutherland, and J. F. Wilkinson. 1969. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. J. Bacteriol. 100:1187-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guard-Petter, J. 2001. The chicken, the egg, and Salmonella enteritidis. Environ. Microbiol. 3:421-430. [DOI] [PubMed] [Google Scholar]
- 17.Hancox, L. S., K. S. Yeh, and S. Clegg. 1997. Construction and characterization of type 1 non-fimbriate and non-adhesive mutants of Salmonella typhimurium. FEMS Immunol. Med. Microbiol. 19:289-296. [DOI] [PubMed] [Google Scholar]
- 18.Holt, P. S. 2003. Molting and Salmonella enterica serovar Enteritidis infection: the problem and some solutions. Poultry Sci. 82:1008-1010. [DOI] [PubMed] [Google Scholar]
- 19.Junkins, A. D., and M. P. Doyle. 1992. Demonstration of exopolysaccharide production by enterohemorrhagic Escherichia coli. Curr. Microbiol. 25:9-17. [DOI] [PubMed] [Google Scholar]
- 20.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 21.Oyofo, B. A., R. E. Droleskey, J. O. Norman, H. H. Mollenhauer, R. L. Ziprin, D. E. Corrier, and J. R. DeLoach. 1989. Inhibition by mannose of in vitro colonization of chicken small intestine by Salmonella typhimurium. Poultry Sci. 68:1351-1356. [DOI] [PubMed] [Google Scholar]
- 22.Parsek, M. R., and E. P. Greenberg. 1999. Quorum sensing signals in development of Pseudomonas aeruginosa biofilms. Methods Enzymol. 310:43-55. [DOI] [PubMed] [Google Scholar]
- 23.Prigent-Combaret, C., and P. Lejeune. 1999. Monitoring gene expression in biofilms. Methods Enzymol. 310:56-79. [DOI] [PubMed] [Google Scholar]
- 24.Prouty, A. M., and J. S. Gunn. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rahn, A., J. Drummelsmith, and C. Whitfield. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181:2307-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroder, O., and R. Wagner. 2002. The bacterial regulatory protein H-NS-a versatile modulator of nucleic acid structures. Biol. Chem. 383:945-960. [DOI] [PubMed] [Google Scholar]
- 27.Sledjeski, D., and S. Gottesman. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:2003-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soerjadi, A. S., R. Rufner, G. H. Snoeyenbos, and O. M. Weinack. 1982. Adherence of salmonellae and native gut microflora to the gastrointestinal mucosa of chicks. Avian Dis. 26:576-584. [PubMed] [Google Scholar]
- 30.Solano, C., B. Garcia, J. Valle, C. Berasain, J. M. Ghigo, C. Gamazo, and I. Lasa. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793-808. [DOI] [PubMed] [Google Scholar]
- 31.Stevenson, G., K. Andrianopoulos, M. Hobbs, and P. R. Reeves. 1996. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J. Bacteriol. 178:4885-4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevenson, G., R. Lan, and P. R. Reeves. 2000. The colanic acid gene cluster of Salmonella enterica has a complex history. FEMS Microbiol. Lett. 191:11-16. [DOI] [PubMed] [Google Scholar]
- 33.Tinker, J. K., L. S. Hancox, and S. Clegg. 2001. FimW is a negative regulator affecting type 1 fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wehland, M., and F. Bernhard. 2000. The RcsAB box. Characterization of a new operator essential for the regulation of exopolysaccharide biosynthesis in enteric bacteria. J. Biol. Chem. 275:7013-7020. [DOI] [PubMed] [Google Scholar]
- 35.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 36.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25:139-143. [PubMed] [Google Scholar]