Abstract
Cellular senescence, a cell fate defined by irreversible cell cycle arrest, has been observed to contribute to chronic age-related conditions including non-healing wounds, such as diabetic foot ulcers. However, the role of cellular senescence in the pathogenesis of diabetic foot ulcers remains unclear. To examine the contribution of senescent phenotypes to these chronic wounds, differential gene and network analyses were performed on publicly available bulk RNA sequencing of whole skin biopsies of wound edge diabetic foot ulcers and uninvolved diabetic foot skin. Wald tests with Benjamini–Hochberg correction were used to evaluate differential gene expression. Results showed that cellular senescence markers, CDKN1A, CXCL8, IGFBP2, IL1A, MMP10, SERPINE1, and TGFA, were upregulated, while TP53 was downregulated in diabetic foot ulcers compared to uninvolved diabetic foot skin. NetDecoder was then used to identify and compare context-specific protein–protein interaction networks using known cellular senescence markers as pathway sources. The diabetic foot ulcer protein–protein interaction network demonstrated significant perturbations with decreased inhibitory interactions and increased senescence markers compared to uninvolved diabetic foot skin. Indeed, TP53 (p53) and CDKN1A (p21) appeared to be key regulators in diabetic foot ulcer formation. These findings suggest that cellular senescence is an important mediator of diabetic foot ulcer pathogenesis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00854-x.
Keywords: Wound healing, Diabetic foot ulcer, Cellular senescence, Skin aging, Network analysis
Introduction
Wound healing is a complex and dynamic phenomenon entailing tightly regulated intracellular and extracellular signals that coordinate to clear damage and regenerate tissue [1, 2]. The diabetic wound bed represents a chronic stalled wound state that affects 1 to 3.5 million persons in the USA with diabetic foot ulcers, resulting in frequent clinic visits, poor quality of life, and significant healthcare burden [3]. Cell signaling impairments in diabetic patients, including deregulated inflammation, epidermal hyperproliferation, reduced angiogenesis, and abnormal stem cell function, predispose them to non-healing vascular wounds [4–6]. As a result of these dysfunctional processes, diabetic foot ulcers are often chronic and recurrent, leading to significant morbidity, mortality, and healthcare burden [7–10]. Notably, the genetic pathways underlying diabetic foot ulcers, including those associated with cellular senescence, are poorly understood.
Cellular senescence is a cell fate characterized by essentially irreversible growth arrest, resistance to apoptosis, and a senescence-associated secretory phenotype (SASP) [11, 12]. It is triggered as a defense mechanism by intrinsic or extrinsic stresses, such as DNA damage, lipid-based signaling, mitochondrial dysfunction, aggregates of abnormal proteins, inflammation, and danger signals [13]. Senescent cells are not only byproducts of aging and disease processes, but they have been demonstrated to play active roles in mediating age-related skin dysfunction [14–16]. SASP factors, which include pro-inflammatory cytokines, matrix metalloproteinases, and growth factors, can modulate the local microenvironment and disrupt neighboring cells [17, 18]. Accumulation of senescent cells could thereby contribute to skin deterioration by disrupting physiological functions, including epidermal stem cell renewal [19] and extracellular matrix deposition [20].
Cellular senescence is implicated in both normal and impaired wound healing [1, 12, 21]. Early senescence may promote regeneration in early or acute wound healing [2, 22] and protect against cancer cell proliferation [16]. Acute SASP (i.e., CCL2, CCL5, PAI-1, PDGFα) is postulated to benefit the pro-inflammatory wound cascade [1, 2]. However, gene expression and secretory signals are observed to change when cellular senescence progresses from early to late phases [23]. Not only do their phenotypes change, but late senescent cells have been shown to be detrimental to tissue function and health [24]. Elevated or persistent senescence and a chronic SASP (i.e., CXCL1, CXCL2, IL-1Ra, IL-6, RANTES, TIMP1, TNFα) are associated with impaired and delayed healing, as well as chronic non-healed wound beds [25, 26].
Increased senescent cell burden has been observed in chronic diabetic wounds, but the contribution of senescence to the pathophysiology of diabetic foot ulcers remains unclear [12, 26]. Herein, we delineate the role of senescence-associated genes and associated protein–protein interaction networks in diabetic foot ulcers. Gene expression profiles in diabetic foot ulcers and uninvolved diabetic foot skin were compared to contrast their wound healing cascades. Protein–protein interaction networks were identified to discern pathways and key regulators differentiating wounded skin. Senescence-associated genes were specifically highlighted in expression and network analyses to elucidate the role of cellular senescence in diabetic foot ulcers.
Methods
Datasets and preprocessing
Bulk RNA sequencing data were retrieved from gene expression values (RPKM, reads per kilobase exon per million reads) generated by deep sequencing (Illumina NextSeq500) using publicly available data from the Gene Expression Omnibus database, under accession number GSE134431 (https://www.ncbi.nlm.nih.gov/geo/) [6]. These data were obtained from sequencing full-thickness skin biopsies of wound edges of diabetic foot ulcers (DFUs) from 13 patients (age 56 ± 13 years old; 13 males). For comparison, skin samples were obtained from uninvolved diabetic foot skin (DFS) without ulcers from 8 patients (ages 66 ± 13 years old; 7 males and 1 female). All samples were obtained from patients receiving care at the University of Miami Hospital Wound Care, after receiving written informed consent.
Patients included in the DFU group met inclusion criteria for diabetes mellitus type II, ulcer on plantar aspect of foot at least 0.5 cm2 in size, peripheral neuropathy, at least 21 years old, wound duration of at least 4 weeks, and HbA1c ≤ 13.0%. Exclusion criteria for the DFU group included active cellulitis, osteomyelitis, gangrene, vascular insufficiency, measured by ABI < 0.7 or ABI > 1.3 with revascularization in the last 6 weeks, and experimental drugs taken in the preceding 4 weeks.
Reads were aligned, and missing values were handled as previously described [6]. They were pre-processed and analyzed using R (version 4.2). Genes with reads of less than 1 RPKM were pre-filtered to increase efficiency and clarity of visualization. Regularized logarithmic transformation to log2 scale was performed on the reads to obtain log fold changes for DESeq2.
Differential gene expression analysis
The data were processed and normalized using the DESeq2 package (release 3.16) in R (version 4.2) [27]. This package allowed for assessment of similarity between samples using hierarchical clustering of sample distances and principal component analysis to determine whether DFU and uninvolved DFS groups had significantly different overall gene expression. Additionally, heat maps of the count matrix and sample-to-sample distances were obtained to evaluate the detection of differentially expressed genes across the two groups of samples. Next, senescence-associated markers, including SASP genes, were compared between DFU and uninvolved DFS groups. After reviewing literature for senescence-associated markers, senescence profiling was performed with intrinsic cellular senescence genes CDKN1A, CDKN2A, and TP53 [1]; skin-specific proliferation and differentiation-related genes KRT14, MITF, and TGFα [28–30]; and SASP genes CCL2, CXCL1, CXCL8, CXCR2, CYR61, IGFBP2, IGFBP4, IL-1α, IL-6, INHBA, MMP2, MMP3, MMP9, MMP10, MMP12, PAPPA, PLAT, PDGFα, SERPINE1, SPP1, and VCAM1 [2, 18, 31–36]. In addition, the gene set SenMayo was evaluated to corroborate the findings with results validated in human studies [35].
Statistical analysis
Statistical tests were performed on differential gene expression using the DESeq2 package (release 3.16) in R (version 4.2) [27]. Wald tests were performed on each gene to compare DFU and uninvolved DFS groups, and Benjamini–Hochberg adjusted p-values were reported to reduce false discovery rates. Differences between groups with adjusted p-values less than 0.05 were deemed significant.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) was performed with default settings, specifically, 1000 permutations with no collapse [37, 38]. DFS and DFU transcriptomes were compared in unbiased analysis of the human hallmark (H), curated (C2), ontology (C5), and SenMayo gene sets [35, 37, 39].
Network analysis
NetDecoder (https://github.com/HuLiSyspharm/NetDecoder) was used to analyze protein–protein interaction networks, comparing the DFU group with the DFS group as a control [40]. Co-expression networks were derived for each phenotype from the bulk RNA-seq (Illumina NextSeq 500) global transcriptome, and Pearson’s correlation coefficient matrices were used to produce their respective edge-weighted interactomes.
For the first set of network analyses, the source gene list was comprised of known cellular senescence markers listed above. NetDecoder default parameters were used, specifically, a size of functional neighborhood (SNF) of 0.95, threshold of flow ratios between phenotypes (ratioThreshold) of 5, and flow threshold (corThreshold) of 0.5.
For the second set of network analyses, the source gene list was comprised of the top 20 genes in human SenMayo-enriched cells [35]. To allow for adequate visualization, NetDecoder parameters were set for a size of functional neighborhood (SNF) of 0.95, threshold of flow ratios between phenotypes (ratioThreshold) of 1, and flow threshold (corThreshold) of 0.1.
Pathway enrichment analysis
Web-based gene set analysis toolkit (WebGestalt; www.webgestalt.org/) with Gene Ontology (GO) biological process functional databases was used for pathway enrichment analysis [41]. Target gene symbol lists were input and analyzed for overrepresentation against the Homo sapiens genome protein-coding reference set.
Data availability
Bulk RNA sequencing data are available under accession number GSE134431 on Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) [6]. All data presented in the current study will be made available by the investigative team upon reasonable request.
Results
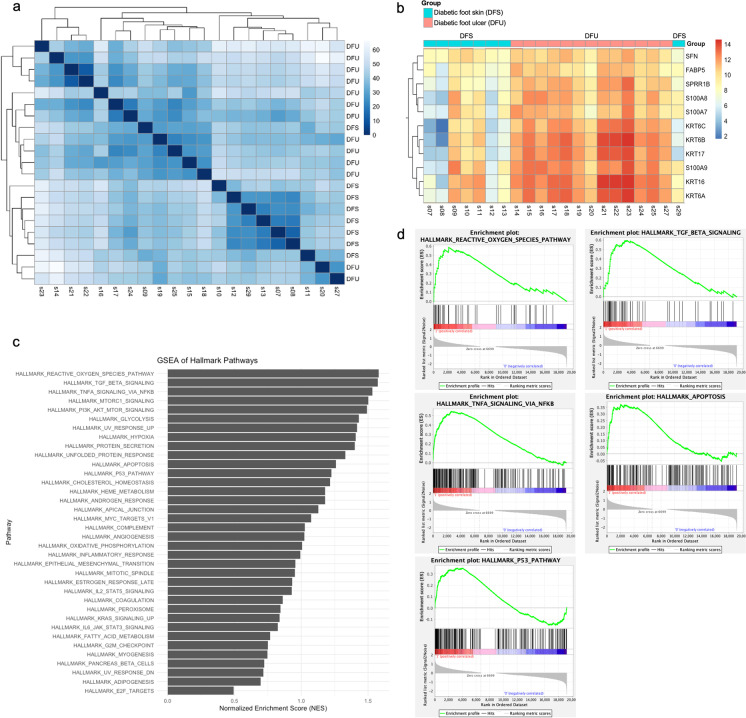
Differential gene expression analysis detected significant differences between diabetic foot ulcer (DFU) and uninvolved diabetic foot skin (DFS) groups. Principal component analysis demonstrated that DFU and DFS samples could be segregated by their gene expression profiles (Fig. S1). Moreover, hierarchical clustering of gene expression could divide the DFU and DFS patients into two distinct groups (Fig. 1a, b).
Fig. 1.
Hierarchical clustering of RNA sequencing of samples from the diabetic foot ulcer (DFU, n = 13) and uninvolved diabetic foot skin (DFS, n = 8) groups. a Heat map of sample-to-sample distances. b Heat map of count matrix. c Gene set enrichment analysis of hallmark gene set, with pathways enriched in DFU compared to DFS. d Gene set enrichment analysis of top 3 upregulated pathways in DFU (reactive oxygen species, TGF-β signaling, and TNF-α signaling via NF-κB), as well as apoptosis and p53 pathways
Next, gene set enrichment analysis was performed to compare pathways in DFU vs. DFS. None of the hallmark, curated, or ontology gene sets were significantly (p-adj < 0.05) upregulated in DFU vs. DFS, suggesting that DFU and DFS shared enrichment in common pathways. In the hallmark (H) gene set, 36 gene sets were upregulated in DFU, with greatest enrichment in genes upregulated by reactive oxygen species (normalized enrichment score (NES) = 1.58, false discovery rate (FDR) q = 0.84), genes upregulated in response to TGF-β1 (NES = 1.57, FDR q = 0.45), and genes regulated by NF-κB in response to TNF-α (NES = 1.53, FDR q = 0.42) (Fig. 1c, d). Notably, upregulation of reactive oxygen species is associated with cellular senescence [42], TGF-β1 is a canonical senescence marker [43], and TNF-α is both an inducer of senescence and SASP protein [44, 45]. Other senescence-related gene sets with NES greater than 1 in DFU included genes mediating apoptosis (NES = 1.26, FDR q = 0.57) and genes involved in p53 pathways and networks (NES = 1.23, FDR q = 0.57). After establishing that DFU and DFS samples had distinctive patterns of gene expression, the roles of specific cellular senescence markers in each group were investigated.
Upregulation of cellular senescence profile in diabetic foot ulcers
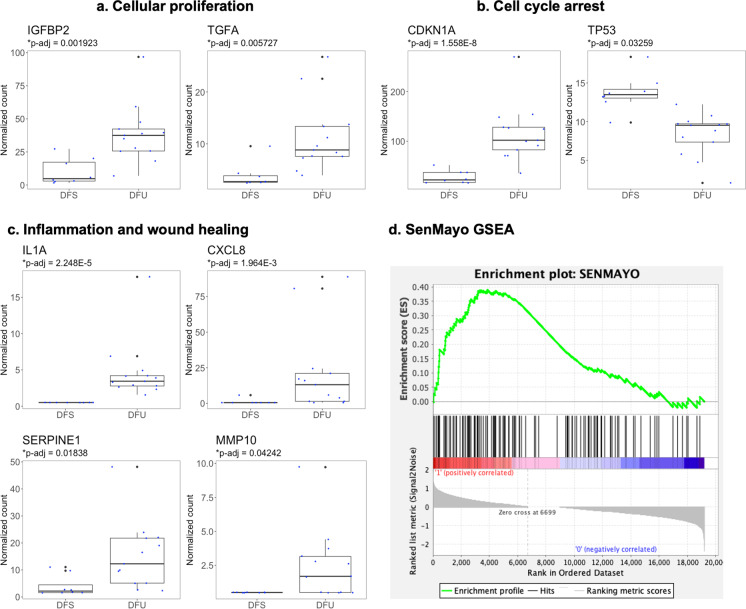
Senescence marker expression was compared between the two groups: DFU vs. DFS. Cellular proliferation markers, including IGFBP2 (Benjamini–Hochberg adjusted p = 0.0019) and TGFα (p = 0.0057), were significantly upregulated in the DFU group (Fig. 2a). Cell cycle arrest genes differed in expression between the DFS and DFU groups (Fig. 2b). CDKN1A (p = 1.6 × 10−8), an inhibitor of G1 cyclin-dependent kinases, was significantly upregulated in the DFU group. In contrast, TP53 (p = 0.033), a tumor suppressor gene with a role in DNA damage foci formation and different stress responses, was significantly downregulated [46]. Furthermore, inflammatory and wound healing markers were significantly upregulated in the DFU group, including IL-1α (p = 2.3 × 10−5), CXCL8 (p = 0.0020), SERPINE1 (p = 0.018), and MMP10 (p = 0.042) (Fig. 2c). Specific gene expression levels suggest that DFU and DFS groups have distinct phenotypes with respect to cellular senescence and wound healing.
Fig. 2.
Differences in cellular senescence profiles, comparing samples from the diabetic foot ulcer (DFU, n = 13) and uninvolved diabetic foot skin (DFS, n = 8) groups. Boxplots include 25th (Q1) and 75th (Q2) percentiles, interquartile range (IQR), median, and potential outliers (Q1 − 1.5IQR or Q3 + 1.5IQR). Wald tests with Benjamini–Hochberg correction were performed to compare DFS and DFU for each gene. Only markers with significantly different expression levels between the two groups (p-adj < 0.05) are presented
Similarly, DFU and DFS gene expression was compared for the validated senescence gene set, SenMayo (Fig. S2a) [35]. Of the 125 genes in SenMayo, 14 were significantly upregulated in DFU, including IL-1α, CXCL8, SERPINE1, and MMP10 evaluated above, as well as EREG (Benjamini–Hochberg corrected p = 2.3 × 10−8), AREG (p = 3.6 × 10−4), IL1B (p = 1.4 × 10−3), PGF (p = 1.8 × 10−3), IGFBP2 (p = 1.9 × 10−3), JUN (p = 4.2 × 10−3), IGFBP6 (p = 0.013), PLAUR (p = 0.014), BMP2 (p = 0.029), and VEGFA (p = 0.043). Two of the SenMayo genes were downregulated: C3 (p = 3.4 × 10−4), a complement protein, and CXCL12 (p = 3.4 × 10−3), a homeostatic chemokine. Among the top 20 genes found in SenMayo-enriched human cells, 5 were significantly upregulated in DFU, and none were downregulated (Fig. S2b). The upregulated genes were CSTA (Benjamini–Hochberg corrected p = 1.0 × 10−10), CXCL8 (p = 2.0 × 10−3), S100A11 (p = 6.4 × 10−3), S100A12 (p = 0.012), and S100A8 (p = 0.046). Three of these upregulated genes are in the S100 family, which consists of calcium-binding proteins involved in inflammation, cancer, and epidermal differentiation [47]. Next, gene set enrichment analysis of the SenMayo gene set was performed. Like the other gene sets analyzed, the SenMayo gene set was not significantly upregulated, but it showed enrichment in DFU (NES = 1.04, FDR q = 0.45) (Fig. 2d).
Cellular senescence profile in diabetic foot ulcer protein–protein interaction networks
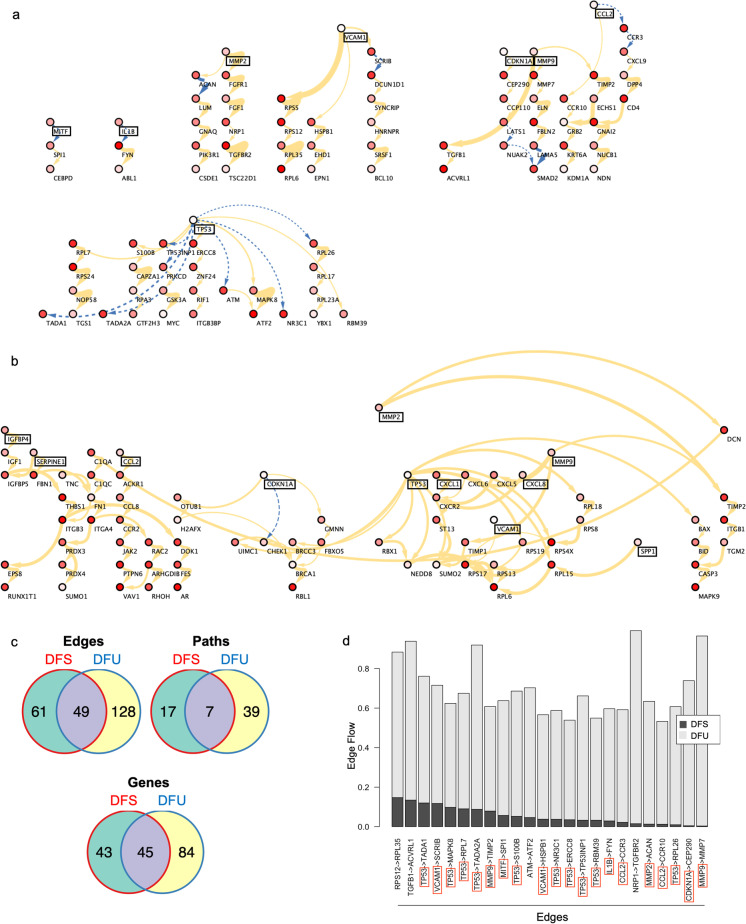
To gain more insight into protein–protein interactions, gene network analysis was performed. Prioritized DFU and edge-centered DFS protein–protein interaction networks were derived, and overall networks were significantly different between the DFU and DFS groups (Fig. 3). Paths obtained for the DFS edge-centered network were mostly isolated from each other and included eight senescence-associated markers (MITF, IL-1β, MMP2, VCAM1, CDKN1A, MMP9, CCL2, and TP53), which were all sources of their genetic paths (Fig. 3a). DFS networks had a mixture of activating and inhibiting protein–protein interactions, with half of the senescence-associated genes activating (MMP2, VCAM1, CDKN1A, and MMP9), three inhibiting (MITF, IL-1β, CCL2), and TP53 activating five and inhibiting six downstream genes. In stark contrast, DFU-prioritized and edge-centered networks had paths that were interconnected, and most genes had higher degrees, or more protein–protein interactions from each gene (Fig. 3b, S3). As senescence-associated genes were used as sources, greater interactions and paths suggest that senescence had greater impact and more downstream consequences in DFU. Eleven senescence-associated markers (IGFBP4, SERPINE1, CCL2, CDKN1A, MMP2, TP53, CXCL1, VCAM1, CXCL8, MMP9, and SPP1) were included in the prioritized network as sources (Fig. 3b). All protein–protein interactions were activating except for the one from CDKN1A. Notably, while all senescence-associated markers in the DFS network were in the DFU network except MITF and IL-1β, these markers interacted with different genes in the DFU network, leading to different genetic paths in DFS and DFU states.
Fig. 3.
Arrow color represents activation (solid yellow) or inhibition (dashed blue) of target protein. Arrow thicknesses represents weight of interactions. Node colors represent node flows. Senescence-associated markers are boxed in black. a Uninvolved diabetic foot skin (DFS) edge-centered protein–protein interaction network. b Diabetic foot ulcer (DFU) prioritized protein–protein interaction network. c Significant edges, paths, and genes found in the prioritized protein–protein interaction networks of diabetic foot ulcers (DFU, n = 13), uninvolved diabetic foot skin (DFS, n = 8), or both. d Key edges, or protein–protein interactions, in the diabetic foot ulcer (DFU) network compared to the uninvolved diabetic foot skin (DFS) network. Senescence-associated markers are boxed in red
There were more highly expressed genes (129 DFU > 88 DFS), protein–protein interactions, or edges (177 DFU > 110 DFS), and genetic paths (46 DFU > 24 DFS) in diabetic wounding with little overlap (49 common edges, 45 common genes, and 7 common paths) between the DFU and DFS groups (Fig. 3c). Moreover, DFU samples had higher edge flows between genes, suggesting greater protein–protein interaction activity (Fig. 3d). Because cellular senescence markers were used as pathway sources, this indicates greater activity and contribution of cellular senescence in DFU compared to DFS. Key edges, which are protein–protein interactions with greatest flow differences, differed between the DFU and DFS groups. Notably, many key edges involved senescence-associated markers as sources, including TP53, VCAM1, MMP9, MITF, IL-1β, CCL2, MMP2, and CDKN1A. In fact, all senescence-associated genes were sources of protein–protein interactions or signaling pathways, in the DFS and DFU protein–protein interaction networks. In fact, senescence-associated genes comprised a large proportion of sources in both networks. Their positions indicated that senescence-associated genes were key regulators in the DFS and DFU protein–protein interaction networks.
Although senescence-associated genes were key components of both DFU and DFS protein–protein interaction networks, they differed in expression and interaction partners. In the DFU network, senescence-associated genes had greater expression and more flow to downstream genes, suggesting that these signaling pathways are critical for disease phenotype. Half of the senescence-associated genes in the DFS network had inhibitory interactions, but CDKN1A was the only gene that had an inhibitory interaction in the DFU network. Furthermore, all senescence-associated genes that were present in both DFU and DFS networks interacted with different genes in either network. In addition to having more senescence-associated genes in the DFU network, the same senescence-associated genes shifted from inhibiting a set of target genes in DFS to activating a completely different set of genes in DFU. Overrepresentation analysis of the target genes revealed different biological processes associated with DFU and DFS networks (Fig. S4a-c). While the DFS network was enriched in RNA catabolism and apoptosis, the DFU networks were enriched in inflammatory and immune responses, extracellular matrix organization, and protein metabolism.
Network analysis was also performed on protein–protein interaction networks with the top 20 genes in SenMayo-enriched human cells as the gene source list. Similarly, the networks reflected a larger impact of SenMayo genes on DFU than DFS networks (Fig. S5). Again, the SenMayo genes differed in type of interaction, i.e., activating or inhibitory, and interaction partners in DFU and DFS edge-centered subnetworks. Overrepresentation analysis was also performed on targets of the top 20 SenMayo genes in the prioritized DFU protein–protein interaction network (Fig. S4d). Like the DFU-prioritized network with senescence markers as the source list, the targets were enriched in immune responses and inflammation, specifically, upregulating migration and chemotaxis, especially of leukocytes, and cytokine signaling.
Discussion
To prevent infections and amputations, DFUs require frequent and aggressive management, including wound debridement and dressings, offloading pressure, and infection control [4, 10]. Even with current treatments, patients with DFUs are at risk for significant morbidity and mortality; they are associated with a 2.5 times higher risk of death at 5 years compared to those without DFUs [10]. Understanding the gene pathways in DFUs could reveal new and more effective targets for DFU management. In this work, we elucidated the role of senescence-associated genes in DFU compared to DFS. By leveraging a computational system biology approach, we further described senescence-associated gene activity by contrasting their interactions in DFS and DFU protein–protein interaction networks. To our knowledge, this is the first report characterizing cellular senescence pathways in human diabetic foot ulcers.
Cellular senescence has been postulated to be a key driver of diabetic foot ulcer pathogenesis [12, 26, 48]. Consistent with previous studies, we found that DFUs had elevated expression of senescence-associated genes and pathways compared to DFS [12, 26]. Importantly, in our studies of human chronic wounds, we found that elevated expression of CDKN1A, as obtained from QRT-PCR, predicted wound chronicity (data not shown). Moreover, we compared p16INK4a + cells in diabetic and non-diabetic chronic wounds, as p16INK4a is a canonical marker of senescence, and demonstrated that diabetic wounds had increased senescence burden. However, some senescence-related genes that were upregulated in other wound models, such as CDKN2A, CYR61, PDGFα, and CXCR2, were not significantly upregulated in DFU that we studied [2, 26, 31]. These discrepancies could be due to differences in depth of skin sampled or wound types, chronicity, and models.
Our analysis is consistent with previous reports showing that TP53 is a crucial part of the senescence response in wound healing [31, 49, 50]. We observed TP53 downregulation as opposed to upregulation found in a previous study, which could be explained by the chronicity of diabetic foot ulcers, likely resulting in a late senescence phenotype [31]. However, other studies have shown that absence of p53, the protein encoded by TP53, was associated with a markedly increased SASP, which was consistent with our findings [51]. Moreover, TP53 was shown to have different gene interactions in the context of different diseases [40]. Other analyses of senescence gene networks also describe TP53 as having some of the highest degree scores [52] and being a crucial regulator of cellular senescence [53].
The sole gene with an inhibitory interaction in the prioritized DFU network was CDKN1A, which, notably, had an activating interaction in the DFS network. p21, the protein encoded by CDKN1A, is known to protect against genotoxic stress but also mediate cellular senescence [54, 55]. According to our DFS network, these contrasting roles of CDKN1A could be mediated by interactions with different downstream partners. In fact, the inhibitory interaction of p21 in the DFU network could be consistent with actions of p21 to inhibit cell cycle and DNA repair pathways, particularly in cellular senescence [54, 55]. In addition, p21 was shown to mediate senescent phenotypes in p53-dependent and -independent pathways [55, 56]. In the DFU network, CDKN1A and TP53 had separate pathways as well as a common pathway that converged on BRCA1, supporting the idea of having both TP53-dependent and -independent gene activity.
This work shows that senescence-associated genes interact with different genes in DFS vs. DFU and that these interactions are integral to each phenotype, implicating senescence in DFU pathophysiology. Analysis of single-cell transcriptomic or proteomic data could uncover the signaling and cellular mechanisms underlying the pathogenesis of DFU and roles of senescent cells in the dynamic wound healing process. Understanding cellular mechanisms could also lead to more precise targeting of cell types or signals in the development of therapeutics for wound care management.
Our findings also support further study of the use of senolytics, agents that selectively eliminate senescent cells, and senomorphics, agents that inhibit SASP factors, to treat or prevent DFUs. The identification of key senescence-associated genes and gene interactions could aid in the design and choice of senolytics and senomorphics for DFU therapeutics and potential monitoring of the progression or improvement of DFUs.
In summary, many senescence-associated genes were found to be differentially expressed and interacted with different genes in full-thickness skin biopsies of DFU wound edges compared to uninvolved DFS. The difference in the roles of senescence-associated genes in DFS and DFU protein–protein interaction networks likely suggests that they contribute to DFU formation or progression.
Limitations of the study
Because of the lack of a rigorous, robust transcriptomic definition of cellular senescence profile in skin, some senescence markers could have been excluded from this analysis. Multiple senescence gene sets have been proposed, and some differ from the gene set used in this work [35, 57, 58]. However, multiple sources were reviewed to obtain a list of pertinent senescence-associated genes, and the targeted differential gene analysis was balanced with the use of unbiased network analysis.
The use of bulk RNA sequencing data prevented the study of distinct cellular gene expression and cell types. However, senescent cells are rare in vivo, and bulk RNA sequencing provides more depth than most single-cell transcriptomic techniques [59]. Moreover, by studying protein–protein interaction networks in addition to differential gene expression, we obtained substantially more insight into how transcriptomic activity differed in DFS vs. DFU. Future studies could validate these findings by quantifying target or downstream proteins of the senescence-associated genes in animal models or patient samples.
Furthermore, although we did not obtain the topology of the entire protein–protein interaction network, using NetDecoder, we captured the key information flows that differentiated protein–protein interaction networks in the DFS vs. DFU phenotypes [40]. This enabled us to study context-dependent roles of senescence-associated genes in the etiology of DFU. Future studies could aim to evaluate DFU transcriptomics, proteomics, or epigenomics and study signaling at a single-cell resolution. They could also contribute to unveiling sex differences in wound pathology [60].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We gratefully acknowledge Dr. Hu Li for his helpful discussion.
Funding
This work was supported by Robert and Arlene Kogod Center on Aging (S.P.W.). This work was also supported by National Institute of Health grants AG013925 (J.L.K.) and AG062413 (J.L.K.), the Translational Geroscience Network (AG061456: J.L.K.), the Connor Group (J.L.K.), Robert J. and Theresa W. Ryan (J.L.K.), and the Noaber Foundation (J.L.K.). In addition, this work was supported by the Mayo Clinic Medical Scientist Training Program institutional training grant (T32 GM065841, G.T.Y. and D.D.M) and the National Cancer Institute Ruth L. Kirschstein National Research Service Award (F30 CA250122, D.D.M.).
Declarations
Patents on senolytic drugs are held by Mayo Clinic. The authors have no financial interest to declare in relation to the content of this article. This research was conducted in compliance with Mayo Clinic conflict of interest policies.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rhinn M, Ritschka B, Keyes WM. Cellular senescence in development, regeneration and disease. Development. 2019;146(20). 10.1242/dev.151837. [DOI] [PubMed]
- 2.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31(6):722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baum CL, Arpey CJ. Normal cutaneous wound healing: clinical correlation with cellular and molecular events. Dermatol Surg. 2005;31(6):674–86. doi: 10.1097/00042728-200506000-00011. [DOI] [PubMed] [Google Scholar]
- 4.Singer AJ, Tassiopoulos A, Kirsner RS. Evaluation and management of lower-extremity ulcers. N Engl J Med. 2017;377(16):1559–1567. doi: 10.1056/NEJMra1615243. [DOI] [PubMed] [Google Scholar]
- 5.Eming SA, Martin P, Tomic-Canic M. Wound repair and regeneration: mechanisms, signaling, and translation. Sci Transl Med. 2014;6(265):265sr6–265sr6. doi: 10.1126/scitranslmed.3009337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawaya AP, et al. Deregulated immune cell recruitment orchestrated by FOXM1 impairs human diabetic wound healing. Nat Commun. 2020;11(1):4678. doi: 10.1038/s41467-020-18276-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice JB, et al. Burden of diabetic foot ulcers for medicare and private insurers. Diabetes Care. 2014;37(3):651–658. doi: 10.2337/dc13-2176. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P, et al. Global epidemiology of diabetic foot ulceration: a systematic review and meta-analysis. Ann Med. 2017;49(2):106–116. doi: 10.1080/07853890.2016.1231932. [DOI] [PubMed] [Google Scholar]
- 9.Singh N, Armstrong DG, Lipsky BA. Preventing foot ulcers in patients with diabetes. JAMA. 2005;293(2):217–228. doi: 10.1001/jama.293.2.217. [DOI] [PubMed] [Google Scholar]
- 10.Armstrong DG, Boulton AJM, Bus SA. Diabetic foot ulcers and their recurrence. N Engl J Med. 2017;376(24):2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 11.Tchkonia T, et al. Cellular senescence and the senescent secretory phenotype: therapeutic opportunities. J Clin Invest. 2013;123(3):966–972. doi: 10.1172/JCI64098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkinson HN, Hardman MJ. Senescence in wound repair: emerging strategies to target chronic healing wounds. Front Cell Dev Biol. 2020;8:773. doi: 10.3389/fcell.2020.00773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wyles SP, Tchkonia T, Kirkland JL. Targeting cellular senescence for age-related diseases: path to clinical translation. Plast Reconstr Surg. 2022;150:20s–26s. doi: 10.1097/PRS.0000000000009669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López-Otín C, et al. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McHugh D, Gil J. Senescence and aging: causes, consequences, and therapeutic avenues. J Cell Biol. 2018;217(1):65–77. doi: 10.1083/jcb.201708092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker DJ, et al. Naturally occurring p16Ink4a-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Demaria M, et al. Cell Autonomous and non-autonomous effects of senescent cells in the skin. J Investig Dermatol. 2015;135(7):1722–1726. doi: 10.1038/jid.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basisty N, et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18(1):e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Arcangelo D, Tinaburri L, Dellambra E. The role of p16(INK4a) pathway in human epidermal stem cell self-renewal, aging and cancer. Int J Mol Sci. 2017;18(7): 1591. [DOI] [PMC free article] [PubMed]
- 20.Wlaschek M, et al. Connective tissue and fibroblast senescence in skin aging. J Invest Dermatol. 2021;141(4s):985–992. doi: 10.1016/j.jid.2020.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Blair MJ, et al. Skin structure-function relationships and the wound healing response to intrinsic aging. Adv Wound Care (New Rochelle) 2020;9(3):127–143. doi: 10.1089/wound.2019.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiebert P, et al. Nrf2-mediated fibroblast reprogramming drives cellular senescence by targeting the matrisome. Dev Cell. 2018;46(2):145–161.e10. doi: 10.1016/j.devcel.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 23.van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herranz N, Gil J. Mechanisms and functions of cellular senescence. J Clin Invest. 2018;128(4):1238–1246. doi: 10.1172/JCI95148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanley A, Osler T. Senescence and the healing rates of venous ulcers. J Vasc Surg. 2001;33(6):1206–1211. doi: 10.1067/mva.2001.115379. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson HN, et al. Elevated local senescence in diabetic wound healing is linked to pathological repair via CXCR2. J Investig Dermatol. 2019;139(5):1171–1181.e6. doi: 10.1016/j.jid.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alam H, et al. Novel function of keratins 5 and 14 in proliferation and differentiation of stratified epithelial cells. Mol Biol Cell. 2011;22(21):4068–4078. doi: 10.1091/mbc.e10-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leclerc J, Ballotti R, Bertolotto C. Pathways from senescence to melanoma: focus on MITF sumoylation. Oncogene. 2017;36(48):6659–6667. doi: 10.1038/onc.2017.292. [DOI] [PubMed] [Google Scholar]
- 30.Brown RL, Breeden MP, Greenhalgh DG. PDGF and TGF-α act synergistically to improve wound healing in the genetically diabetic mouse. J Surg Res. 1994;56(6):562–570. doi: 10.1006/jsre.1994.1090. [DOI] [PubMed] [Google Scholar]
- 31.Jun J-I, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol. 2010;12(7):676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eggert T, et al. Distinct functions of senescence-associated immune responses in liver tumor surveillance and tumor progression. Cancer Cell. 2016;30(4):533–547. doi: 10.1016/j.ccell.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang AS, Dreesen O. Biomarkers of cellular senescence and skin aging. Front Genet. 2018;9:247. doi: 10.3389/fgene.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, et al. INHBA is a novel mediator regulating cellular senescence and immune evasion in colorectal cancer. J Cancer. 2021;12(19):5938–5949. doi: 10.7150/jca.61556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saul D, et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun. 2022;13(1):4827. doi: 10.1038/s41467-022-32552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Childs BG, Li H, van Deursen JM. Senescent cells: a therapeutic target for cardiovascular disease. J Clin Investig. 2018;128(4):1217–1228. doi: 10.1172/JCI95146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha VK, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 39.Liberzon A, et al. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.da Rocha EL, et al. NetDecoder: a network biology platform that decodes context-specific biological networks and gene activities. Nucleic Acids Res. 2016;44(10):e100–e100. doi: 10.1093/nar/gkw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao Y, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–w205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Victorelli S, Passos JF. Reactive oxygen species detection in senescent cells. Methods Mol Biol. 2019;1896:21–29. doi: 10.1007/978-1-4939-8931-7_3. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Alexander PB, Wang XF. TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol. 2017;9(4). 10.1101/cshperspect.a022145. [DOI] [PMC free article] [PubMed]
- 44.Beyne-Rauzy O, et al. Tumor necrosis factor alpha induces senescence and chromosomal instability in human leukemic cells. Oncogene. 2004;23(45):7507–7516. doi: 10.1038/sj.onc.1208024. [DOI] [PubMed] [Google Scholar]
- 45.Freund A, et al. Inflammatory networks during cellular senescence: causes and consequences. Trends Mol Med. 2010;16(5):238–246. doi: 10.1016/j.molmed.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pawge G, Khatik GL. p53 regulated senescence mechanism and role of its modulators in age-related disorders. Biochem Pharmacol. 2021;190:114651. doi: 10.1016/j.bcp.2021.114651. [DOI] [PubMed] [Google Scholar]
- 47.Sedaghat F, Notopoulos A. S100 protein family and its application in clinical practice. Hippokratia. 2008;12(4):198–204. [PMC free article] [PubMed] [Google Scholar]
- 48.Berlanga-Acosta JA, et al. Cellular senescence as the pathogenic hub of diabetes-related wound chronicity. Front Endocrinol (Lausanne) 2020;11:573032. doi: 10.3389/fendo.2020.573032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Helman A, et al. p16Ink4a-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosteiro L, et al. Senescence promotes in vivo reprogramming through p16(INK)(4a) and IL-6. Aging Cell. 2018;17(2). 10.1111/acel.12711. [DOI] [PMC free article] [PubMed]
- 51.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6(12):2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Avelar RA, et al. A multidimensional systems biology analysis of cellular senescence in aging and disease. Genome Biol. 2020;21(1):91. doi: 10.1186/s13059-020-01990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Zhao L, Wang J. Searching for the mechanisms of mammalian cellular aging through underlying gene regulatory networks. Front Genet. 2020;11:593. 10.3389/fgene.2020.00593. [DOI] [PMC free article] [PubMed]
- 54.Ju Z, Choudhury AR, Rudolph KL. A dual role of p21 in stem cell aging. Ann NY Acad Sci. 2007;1100(1):333–344. doi: 10.1196/annals.1395.036. [DOI] [PubMed] [Google Scholar]
- 55.Georgakilas AG, Martin OA, Bonner WM. p21: a two-faced genome guardian. Trends Mol Med. 2017;23(4):310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Chen J, et al. Contribution of p16INK4a and p21CIP1 pathways to induction of premature senescence of human endothelial cells: permissive role of p53. Am J Physiol Heart Circulatory Physiol. 2006;290(4):H1575–H1586. doi: 10.1152/ajpheart.00364.2005. [DOI] [PubMed] [Google Scholar]
- 57.Hernandez-Segura A, et al. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol. 2017;27(17):2652–2660.e4. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jochems F, et al. The cancer SENESCopedia: adelineation of cancer cell senescence. Cell Rep. 2021;36(4):109441. doi: 10.1016/j.celrep.2021.109441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar A, Bano D, Ehninger D. Cellular senescence in vivo: from cells to tissues to pathologies. Mech Ageing Dev. 2020;190:111308. doi: 10.1016/j.mad.2020.111308. [DOI] [PubMed] [Google Scholar]
- 60.Navarro-Peternella FM, et al. Differences between genders in relation to factors associated with risk of diabetic foot in elderly persons: a cross-sectional trial. J Clin Transl Endocrinol. 2016;6:30–36. doi: 10.1016/j.jcte.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Bulk RNA sequencing data are available under accession number GSE134431 on Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) [6]. All data presented in the current study will be made available by the investigative team upon reasonable request.