Abstract
We have recently shown that multiple tRNA synthetase inhibitors can greatly increase lifespan in multiple models by acting through the conserved transcription factor ATF4. Here, we show that these compounds, and several others of the same class, can greatly upregulate mammalian ATF4 in cells in vitro, in a dose dependent manner. Further, RNASeq analysis of these cells pointed toward changes in protein turnover. In subsequent experiments here we show that multiple tRNA synthetase inhibitors can greatly upregulate activity of the ubiquitin proteasome system (UPS) in cells in an ATF4-dependent manner. The UPS plays an important role in the turnover of many damaged or dysfunctional proteins in an organism. Increasing UPS activity has been shown to enhance the survival of Huntington’s disease cell models, but there are few known pharmacological enhancers of the UPS. Additionally, we see separate ATF4 dependent upregulation of macroautophagy upon treatment with tRNA synthetase inhibitors. Protein degradation is an essential cellular process linked to many important human diseases of aging such as Alzheimer’s disease and Huntington’s disease. These drugs’ ability to enhance proteostasis more broadly could have wide-ranging implications in the treatment of important age-related neurodegenerative diseases.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-023-00938-8.
Keywords: Gcn4, ATF-4, ATF4, tRNA synthetase, Ubiquitin proteasome system
Introduction
The ubiquitin proteasome system (UPS) is essential for the turnover of many polypeptides in eukaryotes, but is especially responsible for the turnover of the dysfunctional, mutated, misfolded, or damaged proteins [1]. The UPS has been hypothesized as a high-impact target to treat many diseases such as Huntington’s disease, Alzheimer’s disease, and Parkinson’s disease, among others [2–7]. Increased UPS activity has also been hypothesized to improve an organism’s ability to maintain protein homeostasis, a hallmark of aging [3, 4, 8–15]. Drugs that activate or enhance proteasome activity are rare, especially in comparison to proteasome inhibitors [6].
Activating transcription factor 4 (ATF4) is a conserved transcription factor of recent interest in its wide variety of diseases such as neurodegeneration, diabetes, cancer, and skeletal muscle aging [16–27]. The Atf4 orthologs GCN4 / atf-4 have been linked to increased lifespan in both the budding yeast S. cerevisiae and the nematode C. elegans [18, 21, 28–31]. Further, long-lived mice have been shown to have elevated levels of ATF4 [32, 33].
There are four kinases in vertebrates that are known to upregulate ATF4 translation through the integrated stress response (ISR), which can be activated in response to endoplasmic reticulum stress, amino acid deprivation, the presence of double stranded RNA, and, in erythroid cells, heme deficiency [16]. These four kinases phosphorylate the alpha subunit of eukaryotic initiation factor 2 (eIF2α), an essential protein in the formation of the translation pre-initiation complex in eukaryotes. This eIF2α phosphorylation results in the delay in translational re-initiation and subsequent ATF4 translation [16, 34]. General control non-derepressible kinase 2 (GCN2) is an eIF2α kinase and the activator of the amino acid response leg of the ISR [35]. GCN2 autophosphorylates and activates upon the kinase’s binding to uncharged tRNA [20, 34, 36, 37].
There are two important modes of protein degradation in the cell. First, autophagy is a degradation process in which cellular components, such as proteins and organelles, are delivered to the degradative organelles for breakdown and re-purposing of macromolecules, like amino acids [38]. There are many different flavors of autophagy, such as, but not limited to, mitophagy, selective autophagy, and ribophagy, all of which are only starting to be understood especially in the context of aging [39–43]. Macroautophagy, hereafter ‘autophagy’, is the most widely-studied, and has been established as an important biological process in many long-lived organisms [28, 40, 44–48]. Autophagy genes and their role in aging and disease phenotypes are widely studied and have been recently reviewed [49]. ATF4 is known to aid in the induction of autophagy through its role as a transcription factor [50–57]. Second, the UPS degrades “tagged” or poly-ubiquitinated proteins [1]. In contrast, autophagy is a form of bulk component recycling, while the proteasome turns over proteins on a more individual level, resulting in the proteasome’s tight connection to the regulation of many cellular responses [1]. Increased proteasomal capacity has been found to be responsible for some extremely long-lived phenotypes in yeast, worms and flies, indicating that further research exploring this process in mammals is of great interest to the aging field [3, 4, 8–10, 18]. Altogether, the promotion of these two processes to increase protein degradation has been shown to increase health in many models, underscoring the importance of an organism’s ability to maintain protein homeostasis in healthy aging [3, 5, 40, 58–63].
Although there is great interest in the treatment potential of UPS activators, there are few known pharmacological agents that can do so [5]. Here, we show that seven different tRNA synthetase inhibitors can dramatically induce proteasome activity in an Atf4-dependent manner in mammalian cells in vitro. We also show that these same drugs can upregulate proteasomal activity at the same doses, as well as macroautophagy, suggesting these drugs’ potential to treat important diseases of aging characterized by protein aggregation in vivo. As we have recently shown that some of these same compounds dramatically increase healthy wild-type lifespan in multiple model organisms, this also leaves open the possibility that these potential treatments for known and significant diseases of aging might also act directly on aging itself [27].
Methods
Mouse embryonic fibroblast cell culturing and ATF4 KO cell line generation
Wild type and GCN2 KO mouse embryonic fibroblast (MEFs) were purchased from American Type Culture Collection global biological resource center. The ATF4 KO cell line was made in house using utilizing optimized guide RNAs optimized from the available CRISPR/Cas9 genome editing tools with Integrated DNA Technologies targeting the Atf4 gene. Transfection of the assembled Cas9 complex utilized the NEON electroporation system by ThermoFisher and fluorescence-activated cell sorting with the sy3200 Cell Sorter from Sony Biotechnology in collaborations with the UNM Comprehensive Cancer Center Support Grant NCI P30CA118100 and the UNMHSC Flow Cytometry shared resource (Supplementary Fig. 2). All cell culture procedures were done in a BSL-2 laminar flow cabinet with cells of a low progeny. All sub-culturing procedures utilized guidance from peer-published resources, without Penicillin–Streptomycin [64, 65]. All plasmid transfections utilized Viafect (Promega) following the manufacturer instructions. Halofuginone was purchased from Ambeed [CAS No. 64924–67-0]. Borrelidin was purchased from BioViotica (BVT-0098). Thapsigargin was purchased from AdipoGen (AG-CN2-0003). Bafilomycin A1 was purchased from Sigma-Aldrich (19–148). LysRS-IN-2 was purchased from GLPBio (GC65058). REP3123 and REP8839 were purchased from Axon Medchem (1704, 1705). Mupirocin was purchased from BOC Sciences (B0084-056590). MG-132 was purchased from Sigma-Aldrich (M7449). Tavaborole was purchased from Cayman Chemical (23101).
Fluorescence and luciferase assays in MEFs
ATF4 translation experiments utilized a fluorescent translational reporter for ATF4:ATF4 5:5’ATF4.GFP was a gift from David Ron (Addgene plasmid # 21852; http://n2t.net/addgene:21852; RRID:Addgene_21852) [36]. ATF4 translation over time was measured with the BioTek Synergy HTX Multi-Mode Microplate Reader shared resource in the UNMHSC Autophagy, Inflammation, and Metabolism Center for Biomedical Research Excellence (AIM) core. The AIM core is supported by NIH grant P20GM121176 from NIGMS. All luciferase assays utilized a Victor NIVO multimode plate reader. ATF4 downstream activity was measured using the pGL4[luc2P/ATF4-RE/Hygro] Vector, which was purchased from Promega utilizing a previously well-understood ATF4 binding element [66, 67]. Proteasomal assays utilized (1) Promega’s Proteasome-Glo Caspase-like Cell-Based Assay (G8660) and (2) Abcam’s Proteasomal Activity Kit (ab107921) according to the manufacturers’ procedures respectively [68, 69]. Protein aggregation assays utilized Enzo Life Sciences’ PROTEOSTAT® (51023) according to the manufacturers’ protocols.
All LIVE/DEAD, autophagy, and protein synthesis assays were quantified by the Cellinsight CX7 high-content screening platform by ThermoFisher in the UNMHSC AIM core. LIVE/DEAD assay utilized the ATT Bioquest Live or Dead Cell Viability Assay Kit (Cat. No. 22789). Autophagy assays utilized two autophagic flux reporters. pMRX-IP-GFP-LC3-RFP-LC3ΔG was a gift from Noboru Mizushima (Addgene plasmid # 84572; http://n2t.net/addgene:84572; RRID:Addgene_84572) and ptfLC3 was a gift from Tamotsu Yoshimori (Addgene plasmid # 21074; http://n2t.net/addgene:21074; RRID:Addgene_21074) [70, 71]. Widefield microscopy to supplement the findings utilized the UNMCCC Fluorescence Microscopy and Cell Imaging Shared Resource which is supported by University of New Mexico Comprehensive Cancer Center Support Grant NCI P30CA118100. Protein synthesis assays were conducted with the Click-iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit (ThermoFisher Cat. No. C10428) following the manufacturer’s protocol, where fluorescence surrounding the cell nucleus was quantified in a high-throughput manner, representing the rate of protein synthesis for each cell.
MEF RNA-seq and analysis
RNA was extracted from ~ 70% confluent MEFs after 7 h of condition exposure in accordance with the block design in 60-mm cell culture dishes. DMSO of 1% v/v was used for the controls and 600 nM borrelidin delivered in DMSO at congruently 1% v/v was used in accordance with the high downstream ATF4 transcriptional activity evidenced from the ATF4 downstream luciferase assays (Fig. 2E, 2F). RNA was extracted using Zymo’s RNA extraction kit according to the manufacturer’s protocols. A total of 48 samples were sent to GeneWiz and all except two passed RNA quality control tests, resulting in n ≥ 11 for each condition. After read quality control and fastp processing, fastq files were aligned and quantified utilizing HISAT2 and feature counts [72–75]. Data analysis and differential expression utilized R and the libraries limma, for differential expression analysis and creation of the linear model, and DESEQ2, for data visualization and differential expression analysis [76, 77].
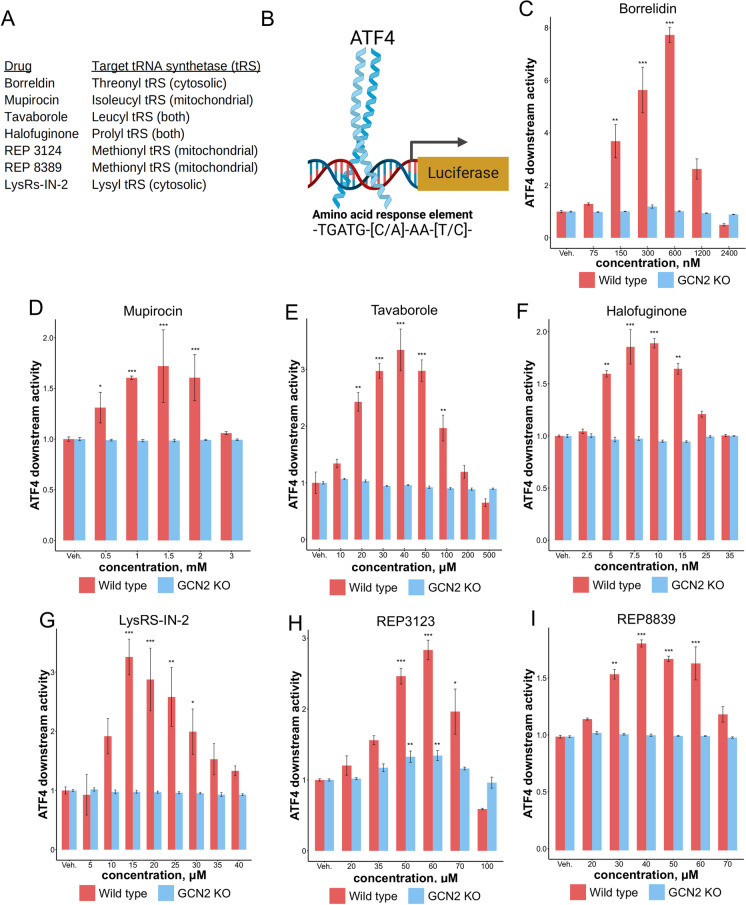
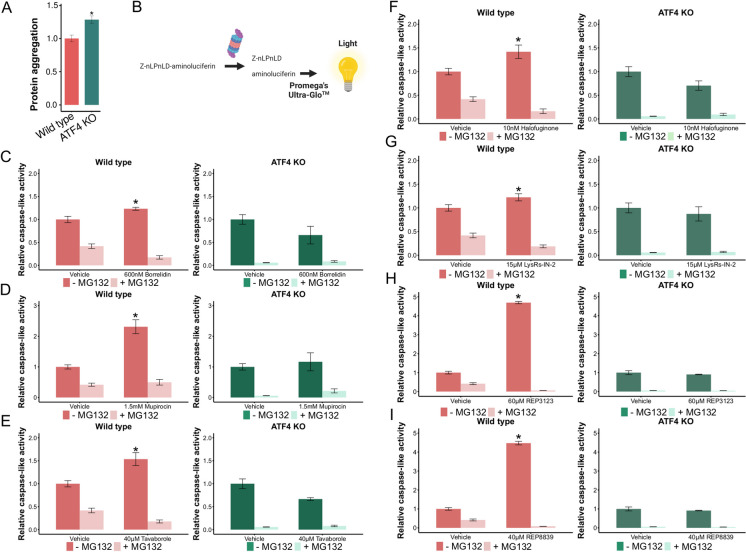
Fig. 2.
tRNA synthetase inhibitors upregulate ATF4 activity through Gcn2. A Drugs used in this study and their associated targets. B The amino acid response element measures ATF4 downstream activity. C–I Various tRNA synthetase inhibitors can increase ATF4 downstream activity (*p < 0.05, **p < 0.01, ***p < 0.001; Dunnett’s multiple testing procedure)
Western blotting analysis of MEF proteins
Cell protein extraction and Western blot analyses were done using standard procedures. Briefly, protein samples were extracted using RIPA Lysis and Extraction Buffer (Genesee Sci. Cat. No. 18–415) using manufacturer’s protocol followed by suspension in Laemmli Sample Buffer (Bio-rad Cat. No. #1610737EDU) and heating to 98 °C for 5 min to denature the proteins. Protein was quantified and loaded equally into electrophoresis gels using Pierce™ BCA Protein Assay Kit (ThermoFisher Cat. No. 23225). The protein samples were loaded into Novex™ 4 to 20% or 10% Tris–Glycine Plus, 1.0 mm, Midi Protein Gels (Invitrogen Cat. No. WXP42012BOXA) in Tri/Glycine/SDS buffer. After electrophoresis, the gel was transferred to an Immun-Blot® PVDF Membrane (Bio-rad Cat. No. #1620175) in Tris/glycine transfer buffer with 10% methanol. Membranes were blocked in 5% dried milk in TBST with tween. The membranes were immunoblotted with the following primary antibodies: LC3 (Sigma, Cat. No. L8918), ATF4 (Proteintech Cat. No. 10835–1-AP), eIF2α (ThermoFisher Cat. No. PA5-41916), Phopho-eIF2α (ThermoFisher Cat. No. 44-728G). 10 μg/well of protein was loaded for ATF4 and LC3 western blots while 20 μg/well was loaded for eIF2α and Phospho-eIF2α blots. The secondary antibody used was an appropriate horseradish peroxidase-conjugated antibody, in which the membranes were incubated in for 1 h at room temperature. The antibody-antigen complex was visualized by a Clarity™ Western ECL Substrate (Bio-rad Cat. No. 170–5061). The intensities of the bands were quantified using the Gel Imager program, by normalizing the band intensity of proteins of interest to the lane’s total protein quantified by Ponceau [78].
Statistical analysis
Statistical significance of ATF4 fluorescent and luciferase assays was calculated using the Dunnett’s multiple comparison procedure versus the vehicle. Statistical significance for all autophagy fluorescence assays were calculated using the Dunnett’s multiple comparison procedure versus the vehicle. Statistical significance of all western blots were calculated using a one-way ANOVA. All data displaying the changes in ATF4 levels, translation, or transcriptional reporter activity are normalized to the vehicle control, representing fold change (FC) with error bars representing the standard error of the normalized fold change mean. Unless specified otherwise, all error bars are standard error of the mean. Any noted statistical significance is reported in the figure legend. Significantly differentially expressed genes were found from the limma R package, using the Kenward-Roger approximation for linear models. Over-represented ontologies were calculated using both Panther and ClueGo using Benjamini–Hochberg multiple testing correction for False Discovery Rate calculation. All caspase-like and chemotrypsin-like assay p-values were calculated utilizing a one-way ANOVA. All caspase-like and chemotrypsin-like assay data presented is presented as fold change (FC) in relation to the vehicle control. All statistics and display of data was done with R programming.
Results
Multiple tRNA synthetase inhibitors can increase ATF4 in mammalian cells
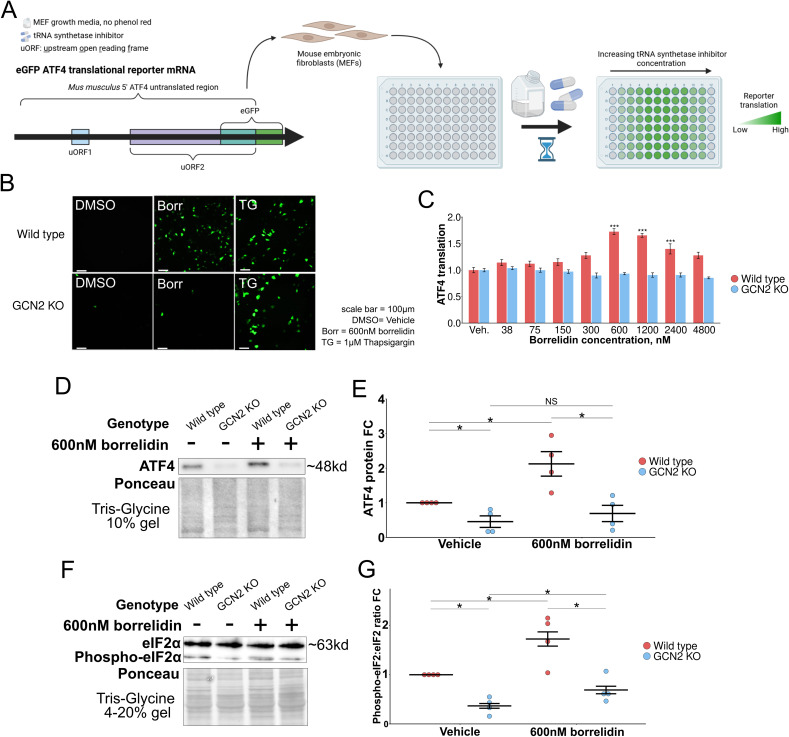
ATF4 is regulated translationally by two upstream open reading frames (uORFs) in its 5′ untranslated region, and a delay in translation re-initiation results in increased ATF4 translation [36, 37]. We first measured ATF4 translation in response to tRNA synthetase inhibitors using an eGFP-ATF4 translational reporter containing the 5′ untranslated region of ATF4 fused to eGFP in place of ATF4 in mouse embryonic fibroblasts (MEFs) (Fig. 1A) [36]. We first sought to ask if borrelidin, a threonyl tRNA synthetase inhibitor, could increase ATF4 translation through the GCN2-mediated amino acid response leg of the integrated stress response [79, 80]. To find the therapeutic window in culture, we first assessed cell viability in an increasing doses of drug and found that 24 h of 2 mM borrelidin treatment in media almost completely killed the cells (Supplemental Fig. 1A). From there, we dialed in the treatment incubation time over a dose range of 0 to 4.8 mM and found that 7 h was consistently sufficient to upregulate ATF4 translation at doses from 150 to 2400 nM (Supplemental Fig. 1B). After that, we found that this ATF4 upregulation was dependent on Gcn2, utilizing thapsigargin (TG), an inducer of the ISR through the PKR-like endoplasmic reticulum kinase, as a positive control (Fig. 1B, C, Supplemental Fig. 1B). We confirmed this ATF4 upregulation with western blots (Fig. 1D, E, Supplemental Fig. 1C, 1D). To further test if tRNA synthetase inhibitors are acting through the AAR, we assessed the ratio of phosphorylated-eIF2α to eIF2α with western blots (Fig. 1F, G, Supplemental Fig. 1E). We found both wild type and GCN2 KO MEFs had a significant rise in eIF2α phosphorylation levels in relation to its vehicle counterpart, however the GCN2 KO cells, however, they were not significantly higher than the wild type vehicle condition. Together, these data suggest that borrelidin acts through the ISR, specifically through the phosphorylation of eIF2α by the GCN2 uncharged tRNA sensor, in order to increase ATF4 translation.
Fig. 1.
tRNA synthetase inhibitors can increase ATF4 levels in mouse embryonic fibroblasts. A The ATF4 eGFP translation reporter utilizes ATF4’s 5′ untranslated region upstream of the GFP start codon [37]. The study design first used the eGFP ATF4 translation reporter to find concentrations of tRNA synthetase inhibitor that upregulate ATF4 translation. B, C ATF4 translation measured in response to varying concentrations of borrelidin (*p < 0.05, **p < 0.01, ***p < 0.001; Dunnett’s multiple testing procedure). D, E ATF4 protein levels in response to borrelidin, normalized to total protein level detected by ponceau stain. F, G Phospho-eIF2 and eIF2 protein measured ratio in response to borrelidin, normalized to total protein level detected by ponceau stain (*p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA). eGFP = enhanced green fluorescent protein, Veh. = Vehicle, FC = fold change
We next tested six additional inhibitors targeting different tRNA synthetases (Fig. 2A). We found that a 7-h incubation time with these drugs was sufficient to increase ATF4 translation in wild type MEFs, similar to what we saw with borrelidin (Supplemental Fig. 1F). We next utilized an amino acid response element luciferase reporter where luciferase is only transcribed when ATF4 binds to its known consensus binding sequence (Fig. 2B) [66, 67, 81]. All of these inhibitors showed a bell-shaped response curve of ATF4 downstream activity with inhibitor dose, after 7 h of treatment (Fig. 2C–I). At lower doses, activation increases with increasing drug concentration via GCN2-mediated increased translation of ATF4. At higher doses, these drugs likely inhibit overall translation enough to offset the specific induction of ATF4. Borrelidin was the most potent ATF4 activity inducer, increasing its activity by more than sevenfold at 600 nM after 7 h of treatment (Fig. 2C). Mupirocin, despite targeting the mitochondrial isoleucyl tRNA synthetase, was similarly found to increase ATF4 downstream activity and translation through Gcn2 (Fig. 2D, Supplemental Fig. 1G) [82, 83]. The antifungal agent tavaborole, a leucyl tRNA synthetase inhibitor, was found to increase ATF4 translation and downstream activity through Gcn2 (Fig. 2E, Supplemental Fig. 1G) [82]. Previous work utilizing halofuginone, a prolyl tRNA synthetase inhibitor, has been shown to increase ATF4 levels in vitro, and our studies confirm this (Fig. 2F, Supplemental Fig. 1D) [54]. Lys-RS-IN-2, a lysyl tRNA synthetase inhibitor, was also found to increase ATF4 activity dependent on Gcn2 (Fig. 2G, Supplemental Fig. 1G) [84]. REP3123 and REP8839, two methionine tRNA synthetase inhibitors known for their potent affinity for the prokaryotic tRNA synthetase enzyme, were similarly found to increase ATF4 activity (Fig. 2H, I) [85, 86]. For all inhibitors except for REP3123, induction of ATF4 downstream activity was completely dependent on Gcn2. We reasoned that methionine tRNA synthetase inhibitors may be able to induce ATF4 independently of Gcn2, as charged methioninyl-tRNA is a component of the translation pre-initiation complex and thus translation initiation may be delayed when methionine tRNA synthetase activity is impacted [34, 87]. Taken together, these data suggest that ATF4 translation and downstream activity can be upregulated in response to multiple inhibitors of different tRNA synthetases in mammalian cells.
ATF4 upregulation causes the differential expression of genes associated with protein turnover
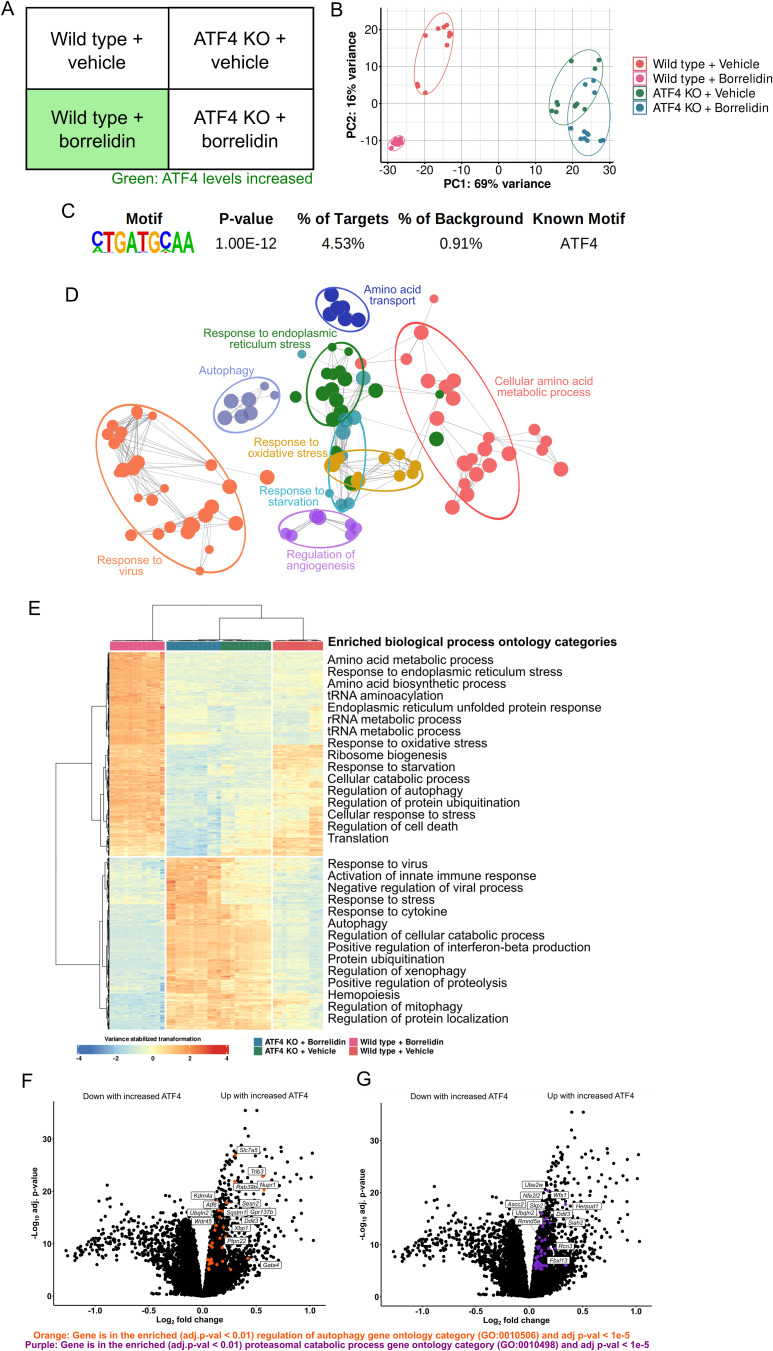
After we identified highly inducing doses of tRNA synthetase inhibitors in MEFs, we sought to analyze the ATF4-dependent changes in the cell’s transcriptome. We knocked out Atf4 using an Integrated DNA Technologies-adapted CRISPR-Cas9 protocol (Supplementary Fig. 2) and we designed an RNA-sequencing experiment to analyze the ATF4 transcriptional targets under high activity using linear model analysis of a block design with high biological replicates (n ≥ 11 per treatment) (Fig. 3A) [76, 88]. The block design features the ability to simultaneously control for the two variables: (1) tRNA synthetase inhibitor treatment and (2) ATF4 activity levels. We chose to conduct the RNASeq with borrelidin as it was the most potent inducer of the ATF4 transcriptional response. Principal component analysis of the 46 samples revealed that principal component 1 was able to separate the samples by their genotype while principal component 2 separated based on the treatment of drug (Fig. 3B). Principal component 2 separated the wild type samples based on their drug treatment much better than it did the ATF4 KO samples, suggesting that borrelidin impacted the two different cell lines differently, and in line with our expectation that many of the transcriptional changes upon borrelidin treatment might depend on increased ATF4 translation.
Fig. 3.
ATF4 causes the differential expression of genes involved with protein turnover. A RNASeqblock design to control for borrelidin treatment and Atf4 knockout sequencing. B Principal component analysis of the 46 samples sequenced. C HOMER motif enrichment from the differentially expressed genes associated with ATF4 (adj. p-value < 1e-10). D) ClueGo enriched biological process ontology categories (adj. p-value < 0.01) of differentially expressed genes (adj. p-value < 1e-10) from the linear model result (Design = ~ ATF4 + Condition). E Heatmap and hierarchical clustering of linear model results (p adj < 1e-10). Biological process gene ontology enrichment utilized Panther’s ontology resource. F, G Volcano plots of genes differentially expressed from linear model. Both regulation of autophagy (in orange) and proteasomal protein catabolic process (in purple) were found to be enriched in the genes upregulated (p adj < 1e-10) with ATF4 (Panther’s ontology resource, adj. p-value < 0.01). “Up with increased ATF4” and “Down with increased ATF4” are labeled on either side of the volcano plots presented in association with the limma linear model fit to ATF4 activity given by the linear model study design (Design = ~ ATF4 + Genotype)
After alignment of reads and quantification, genes with changed mRNA levels associated with ATF4 activity were pulled out based on the linear model fit to increased ATF4 levels (Interactive volcano plot of results provided in File 1) [76]. Figure 3 C–F present the findings of this linear model design (Design = ~ ATF4 + Genotype), which upon fitting, results in the log fold change and adjusted p-values of transcripts in association with the documented ATF4 activity from Fig. 2. Using HOMER motif analysis, we confirmed that the immediately upstream presumptive promoter sequences of genes whose transcript levels changed with ATF4 activity (padj < 1e-5) showed enrichment of the known ATF4 amino acid response binding element [A/G]-TT-[G/T]-CATCA (p < 1e-12) (Fig. 3C) [66, 67, 89]. The enriched biological process ontology categories from the genes differentially expressed from the linear model fit (padj < 1e-10) were plotted on an edge-node graph utilizing ClueGo (Fig. 3D) [90].
We next used hierarchical clustering of the genes found to be significantly (padj < 1e-10) associated with linear model fit (Fig. 3E). Encouragingly, the samples (columns) clustered cleanly in alignment with their genotype and drug treatment. This clustering further revealed that genes associated with autophagy, protein catabolism, translation, and other stress responsive genes are differentially expressed downstream of ATF4. We also analyzed the enriched biological process gene ontology categories of the genes upregulated in conditions of high ATF4 activity, and found many processes involved in protein degradation were over-represented (regulation of autophagy (GO:0010506), proteasomal protein catabolic process (GO:0010498), ERAD pathway (GO:0036503)) (Fig. 3F, G, interactive volcano plot included in File 1). Further, these data present differentially expressed genes known to impact protein synthesis, by the amino acid biosynthesis ontology categories shown to be enriched here, in alignment with what is already known about Atf4 and its orthologs [16, 91]. Altogether, these data indicate that ATF4 may be a key regulator of protein turnover, changing the expression of genes involved in protein synthesis and protein degradation.
Many novel genes were found to be significantly differentially expressed with ATF4 activity, including Wipi2, a gene essential in the LC3 lipidation step of autophagy, which was upregulated with high ATF4 activity [92, 93]. Quantitative PCR of autophagy genes Atg2a, Atg7, Atp13a2, and Wipi2 further confirmed higher levels of the associated genes’ mRNA in borrelidin-treated wild type samples, in agreement with our RNASeq results for these genes (p < 0.05) (Supplemental Fig. 3A). Our RNASeq results also showed increased expression of a significant number of genes encoding tRNA synthetases when ATF4 is induced, including Tars, Gars, Cars, Nars, Yars, Sars, Lars, Wars, Mars1, Vars, and Eprs (padj < 1e-3), consistent with other studies (Supplemental Fig. 3B, 3C, 3D) [52, 94]. Conceptually, these data suggest that ATF4 increases the expression of these tRNA synthetases to correct for the scenario that the ISR was activated by the accumulation of uncharged tRNAs, potentially due to a malfunctioning tRNA synthetase.
tRNA synthetase inhibitors and ATF4 can decrease protein synthesis
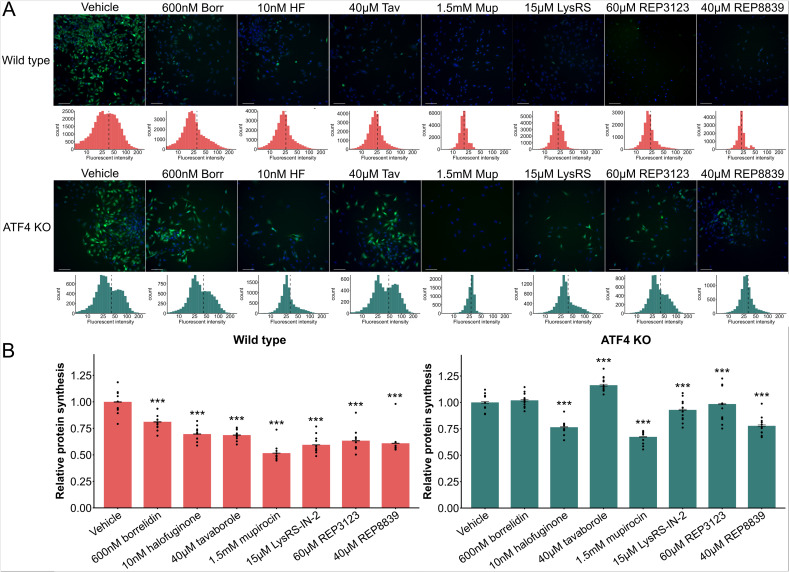
From the RNASeq results, there were many genes found to be differentially expressed with increased ATF4 activity having to do with translation, indicating that ATF4 impacts protein synthesis, aligning with other studies [16, 91, 95, 96]. As protein turnover mechanisms are tightly controlled by the balance between protein synthesis and degradation, we sought to understand the impact of our drugs and ATF4 on protein synthesis. We found that wild type cells treated with every different tRNA synthetase inhibitor at doses that increase ATF4 levels had significantly reduced translation (Fig. 4A). We next assessed the impact of tRNA synthetase inhibitors on ATF4 KO cells and found that 5/7 tRNA synthetase inhibitors had reduced translation, while borrelidin and tavaborole either had no significant change, or increased protein synthesis. Consistently, wild type cells have lower protein synthesis than ATF4 KO cells when treated with the same dosage of drug, implicating ATF4 as a reducer of protein synthesis, in agreement with other studies [16, 91, 95, 96]. However, cells treated with halofuginone, mupirocin, LysRS-IN-2, REP3123, and REP8839 still had significantly reduced protein synthesis in comparison to their vehicle counterpart in both wild type and ATF4 KO cells (Fig. 4B).
Fig. 4.
tRNA synthetase inhibitors and ATF4 can decrease protein synthesis in MEFs. A Representative images and histograms of green fluorescence, representing protein synthesis, of wild type and ATF4 KO MEFs treated with various tRNA synthetase inhibitors. Green: Alexa Fluor.™ 488. Blue: NuclearMask™ Blue Stain. B Relative protein synthesis in wild type and ATF4 KO MEFs, normalized to wild type vehicle control in all cases. Dots represent different experiments. (*p < 0.01; **p < 0.01, ***p < 0.001; Students’ t-test)
tRNA synthetase inhibitors can upregulate protein degradation in an Atf4 dependent manner
Protein turnover in an organism is balanced by both protein synthesis and protein degradation. This knowledge, along with the results from the RNASeq indicating a change in autophagy and proteasomal activity, led us to next assess peptide and protein aggregation in cell lines with and without Atf4 (Fig. 5A). Intriguingly, the untreated ATF4 KO cell line had significantly more aggregation than the untreated wild type cell line, which could be due to impaired unfolded protein response in ATF4 KO cells [97]. ATF4 is also among the individual proteins degraded by the UPS, suggesting the possible existence of a negative feedback loop [98, 99]. To further investigate ATF4’s influence on protein degradation, we assessed the proteasome’s caspase-like activity (Fig. 5B). We found that borrelidin is able to significantly increase proteasomal caspase-like activity in wild type MEFs (Fig. 5C). However, in ATF4 KO MEFs, borrelidin did not increase proteasomal caspase-like activity, suggesting that Atf4 is necessary for increased caspase-like proteasomal activity upon borrelidin treatment. Similar results were found in all of the tested tRNA synthetase inhibitors at concentrations that we found them to induce ATF4 activity (Fig. 5D–I). Interestingly, the mitochondrial methionine tRNA synthetase inhibitors, REP3124 and REP8839, induced proteasomal caspase-like activity the most, agreeing with previous work on responses to direct methionine restriction [100]. We also assessed the proteasome’s chemotrypsin-like activity, and found chymotrypsin-like proteasomal activity to be induced in an Atf4-dependent manner by all 7 inhibitors as well, mirroring our findings with caspase-like proteasomal activity (Supplemental Fig. 4A-4H). Taken together, these data show that tRNA synthetase inhibitors can increase proteasomal activity in mammalian cells in an Atf4-dependent manner.
Fig. 5.
tRNA synthetase inhibitors increase caspase-like activity through Atf4. A Peptide and protein aggregation of wild type and ATF4 KO MEFs. Protein aggregation assessed using PROTEOSTAT ® divided by the total protein in the sample, normalized to wild type. B Promega’s assay schema to measure caspase-like activity of the proteasome. C–I Caspase-like activity increases in response to different tRNA synthetase inhibitors at ATF4-inducing concentrations (*p < 0.05, **p < 0.01, ***p < 0.001; one-way ANOVA)
As genes involved in autophagy were also identified in our RNASeq results (Fig. 3D–F), we next assessed the change in autophagy in response to tRNA synthetase inhibitors, and found that autophagic flux is upregulated upon tRNA synthetase inhibitor treatment, completely dependent on Atf4. We demonstrated this utilizing three different, independent autophagy assays. The first assay used an autophagic flux GFP-LC3-RFP-(LC3ΔG) fluorescent reporter, and showed that autophagic flux increases in tRNA synthetase inhibitor treated samples, dependent on Atf4 (Supplemental Fig. 5A, 5B). We next utilized the ptfLC3 autophagic flux reporter, which again showed that autophagic flux levels rose in response to tRNA synthetase inhibitors, dependent on Atf4 (Supplemental Fig. 5C, 5D, 5E, 5F). Finally, we quantified levels of LC3II by western blot and found that they were increased upon tRNA synthetase inhibitor treatment, again dependent on Atf4 (Supplemental Fig. 5G, 5H). LC3II levels are linked to the formation of the autophagosome and increased autophagy [49]. These results further indicate that tRNA synthetase inhibitors can increase autophagy in an Atf4-dependent manner, aligning with other studies [54]. Altogether, these results show tRNA synthetase inhibitors can increase protein degradation pathways through Atf4 in mammals, outlining Atf4 as a key contributor to the regulation of protein turnover.
Discussion
Pharmacological inducers of the UPS are scarce but have the potential to treat many diseases such as Huntington’s disease, Alzheimer’s disease, and Parkinson’s disease [2, 3, 5, 58, 101, 102]. Pharmacological autophagy inducers also have the potential to play roles in protection against neurodegenerative diseases and aging [103–105]. Notably, we found multiple distinct tRNA synthetase inhibitors can dramatically upregulate the UPS in mammalian cells through Atf4 (Fig. 6) [98, 99]. The drugs presented here, including two tRNA synthetase inhibitors that have been shown to increase lifespan in yeast and worms, have major implications for the biological activity of ATF4 and its potential impact on the aging process [27]. Further, the unfolded protein response is a pathway ending with ATF4 that is known to increase lifespan in many model organisms [97, 106–109]. This study, along with the growing literature surrounding ATF4, underscores the highly important role ATF4 potentially plays in many longlived models that may converge on improved proteostasis, itself a hallmark of aging [11–13, 110, 111].
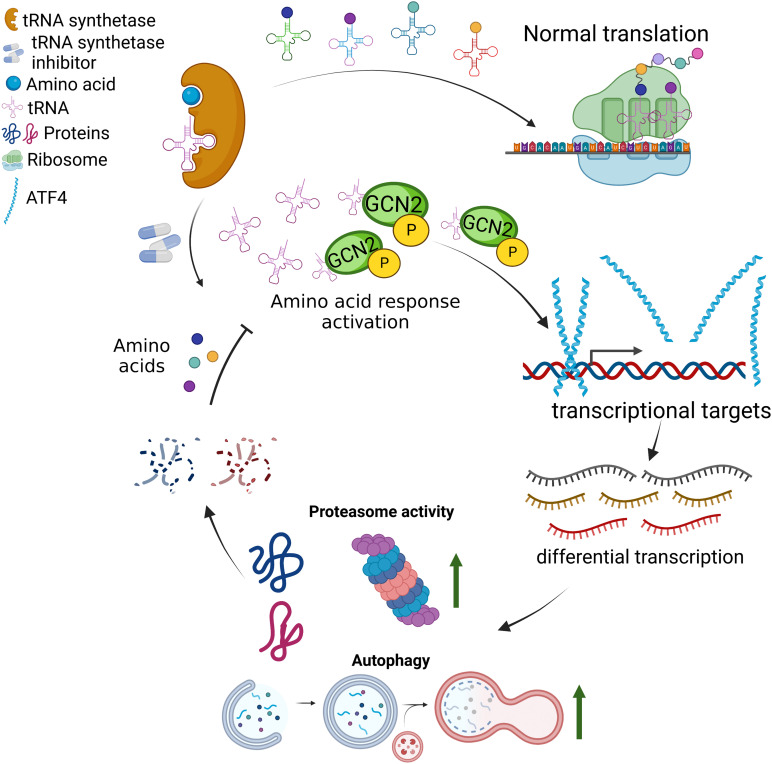
Fig. 6.
tRNA synthetase inhibitors cause the activation of the integrated stress response. Proteasome activity and autophagy play a role in the negative feedback loop of amino acid response pathway activation
Cells are likely to interpret uncharged tRNAs as a sign of amino acid starvation, regardless of whether the uncharged tRNA stems from the cytoplasm or mitochondria [112]. Given that GCN2 contains a histadyl tRNA synthetase-like domain that is thought to have the ability to recognize many types of uncharged tRNA, it logically follows that GCN2 may be able to sense both cytosolic and mitochondrial uncharged tRNA [16, 19, 112–115]. Its activation leads to the translation of ATF4 and likely similar downstream responses no matter the type of uncharged tRNA that initiated this cascade, supporting what we found from testing various tRNA synthetase inhibitors that inhibit both cytosolic and mitochondrial tRNA synthetases. To that end, we found that protein synthesis is decreased and proteasomal activity is increased in wild type cells in comparison to their ATF4 KO counterparts. While our current model is that mupirocin, REP3132, and REP8839 act only as inhibitors of their mitochondrial tRNA synthetases respectively, we have not ruled out the possibility that these compounds act on the cytosolic enzymes as well, or even on other previously undiscovered targets.
In order to further assess these drugs’ potential for therapeutic uses, researchers must ensure that the dosing of their organisms leads to the high upregulation of ATF4 or its orthologs. Of note, only certain ranges of concentrations of these drugs greatly increase lifespan in yeast and worms, dependent on their ATF4 orthologs [27]. Under-dosing may have little to no effect, and over-dosing may have a detrimental effect, aligning with other studies (and further evidenced here in Supplemental Fig. 1A) [116].
ATF4 and its orthologs are also known to impact protein synthesis [91, 95, 96, 117]. The results presented here show ATF4 can decrease protein synthesis in response to doses of some inhibitors, but it alone is not responsible for the reduction of protein synthesis in mupirocin, halofuginone, LysRS-IN-2, REP3123, and REP 8839 treated cells. Previous work outlined that borrelidin decreases protein synthesis in both wild type and gcn4Δ yeast; however, replicative lifespan is only increased in the wild type strains [27]. This, along with these data here, indicate that lowered translation is not the sole contributor to lifespan increase and that other protein degradation processes regulated by Gcn4/ATF4, such as autophagy or the UPS, may play a necessary role, warranting further investigation. However, these data uncouple elevated proteasome activity and reduced protein synthesis.
Although this manuscript outlines the argument that ATF4 is beneficial in many contexts of aging, there are contexts in which ATF4 plays a negative role [67, 118, 119]. For instance, recent work by Miller et al. has shown that ATF4 KO mice have maintained muscle mass with age [119]. This may be due to the fact that those mice developed without Atf4, suggesting that development without Atf4 can result in alternative pathways regulating the same phenomenon. Another perspective may be that since the data presented here associates Atf4 with protein degradation mechanisms, perhaps the loss of Atf4 results in impaired protein degradation capacity and thus decreased atrophy. This could explain why the maintenance of muscle mass is improved in the Miller et al. ATF4 KO mouse model. Further, ATF4 is considered a cancer target as it has been found to be pro-oncogenic; when upregulated, ATF4 can give cancerous cells the ability to live with limited nutrient delivery, potentially due to its positive relationship with protein degradation mechanisms such as autophagy [52, 53, 120–124].
Conceptually, many of the processes found to be enriched in our RNASeq results may have evolved to be influenced by ATF4 in the event the ISR was activated by amino acid deprivation (i.e. the accumulation of uncharged tRNA) to increase amino acid availability and decrease amino acid usage [16, 20, 113, 125]. Similarly, since the integrated stress response is also activated by endoplasmic reticulum stress and the presence of double-stranded RNA, it follows that ATF4 would logically impact genes that would have the ability to respond these activators of the ISR as well [16]. The RNASeq data here illustrates that even though we increased ATF4 translation through GCN2, we found many differentially expressed genes tied to the biological process categories unfolded protein response, viral presence response, and endoplasmic reticulum stress (Fig. 3). These data further outline the roles ATF4 plays in response to ISR activation, consistent with previous findings, as a multifaceted transcription factor [50–52].
Inducing processes that activate pathways known to promote protein turnover has great interest in the development of therapeutics for diseases characterized by protein aggregation, including important diseases of aging such as Huntington’s and Alzheimer’s diseases [105]. The results presented here showing that tRNA synthetase inhibitors greatly upregulate proteasomal degradation as well as autophagy open the door for further exploration of tRNA synthetase inhibitors as potential treatments for these very important diseases of aging.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Figures created with BioRender.com. We thank Jack Peterson and members of the McCormick lab and Hines lab at the University of New Mexico for the technical assistance and helpful discussions.
Author contribution
B.L.M. and M.A.M. conceived the experiments and wrote the manuscript. B.L.M. generated the mouse embryonic fibroblast ATF4 KO line, conducted the experiments with A.S.R and O.C.H., and executed the data analysis. M.A.M. provided reagents and support for the studies. All authors discussed the results and commented on the manuscript.
Funding
This study was supported by NIH 5P20GM121176, NIH R01AG07077601, a Longevity Impetus Grant, a Glenn Foundation for Medical Research / American Federation for Aging Research Junior Investigator Grant, an American Federation for Aging Research Reboot Award, a UNM SOM RAC New Investigator Award, and a Japan Agency for Medical Research and Development / New York Academy of Sciences Longevity Interstellar Initiative Award, to MM. We thank Matthew Campen, Michael A. Mandell and Curt Hines for their helpful discussions in pursuing this research.
Data Availability
All genomic data described in this study are available from the Gene Expression Omnibus (GEO) with accession number #GSE217634.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coux O, Tanaka K, Goldberg AL. structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 2.Seo H, Sonntag K-C, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of Huntington’s disease neuronal model cells. PLoS One. 2007;2(2):e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chondrogianni N, Sakellari M, Lefaki M, Papaevgeniou N, Gonos ES. Proteasome activation delays aging in vitro and in vivo. Free Radic Biol Med. 2014;71:303–320. doi: 10.1016/j.freeradbiomed.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen NN, Rana A, Goldman C, Moore R, Tai J, Hong Y, Shen J, Walker DW, Hur JH. Proteasome Β5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila Melanogaster. Sci Rep. 2019;9(1):3170. doi: 10.1038/s41598-019-39508-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yazgili AS, Ebstein F, Meiners S. The proteasome activator PA200/PSME4: an emerging new player in health and disease. Biomolecules. 2022;12(8):1150. doi: 10.3390/biom12081150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahlmann B, Becher B, Sobek A, Ehlers C, Kopp F, Kuehn L. in vitro activation of the 20s proteasome. Enzyme Protein. 1993;47(4–6):274–284. doi: 10.1159/000468685. [DOI] [PubMed] [Google Scholar]
- 7.Cascio P. PA28γ: new insights on an ancient proteasome activator. Biomolecules. 2021;11(2):228. doi: 10.3390/biom11020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, Sutphin G, Carr D, Tar K, Dittmar G, Kaeberlein M, Kennedy BK, Schmidt M. Elevated proteasome capacity extends replicative lifespan in Saccharomyces Cerevisiae. PLoS Genet. 2011;7(9):e1002253. doi: 10.1371/journal.pgen.1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghazi A, Henis-Korenblit S, Kenyon C. Regulation of Caenorhabditis Elegans lifespan by a proteasomal E3 ligase complex. Proc Natl Acad Sci. 2007;104(14):5947–5952. doi: 10.1073/pnas.0700638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jana NR. Protein homeostasis and aging: role of ubiquitin protein ligases. Neurochem Int. 2012;60(5):443–447. doi: 10.1016/j.neuint.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 11.López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Otín, C, Blasco, M, Partridge, L, Serrano, M, Kroemer, G. 2023 Hallmarks of aging: an expanding universe. Cell 186(2) 10.1016/j.cell.2022.11.001. [DOI] [PubMed]
- 13.Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20(2):214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Fernandez IA, Qi Y, Jasper H. Loss of a proteostatic checkpoint in intestinal stem cells contributes to age-related epithelial dysfunction. Nat Commun. 2019;10(1):1050. doi: 10.1038/s41467-019-08982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Davis SS, Borch Jensen M, Rodriguez-Fernandez IA, Apaydin C, Juhasz G, Gibson BW, Schilling B, Ramanathan A, Ghaemmaghami S, Jasper H. JNK modifies neuronal metabolism to promote proteostasis and longevity. Aging Cell. 2019;18(3):e12849. doi: 10.1111/acel.12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ameri K, Harris AL. Activating transcription factor 4. Int J Biochem Cell Biol. 2008;40(1):14–21. doi: 10.1016/j.biocel.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Hinnebusch AG, Natarajan K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot Cell. 2002;1(1):22–32. doi: 10.1128/EC.01.1.22-32.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick MA, Delaney JR, Tsuchiya M, Tsuchiyama S, Shemorry A, Sim S, Chou AC-Z, Ahmed U, Carr D, Murakami CJ, Schleit J, Sutphin GL, Wasko BM, Bennett CF, Wang AM, Olsen B, Beyer RP, Bammler TK, Prunkard D, Johnson SC, Pennypacker JK, An E, Anies A, Castanza AS, Choi E, Dang N, Enerio S, Fletcher M, Fox L, Goswami S, Higgins SA, Holmberg MA, Hu D, Hui J, Jelic M, Jeong K-S, Johnston E, Kerr EO, Kim J, Kim D, Kirkland K, Klum S, Kotireddy S, Liao E, Lim M, Lin MS, Lo WC, Lockshon D, Miller HA, Moller RM, Muller B, Oakes J, Pak DN, Peng ZJ, Pham KM, Pollard TG, Pradeep P, Pruett D, Rai D, Robison B, Rodriguez AA, Ros B, Sage M, Singh MK, Smith ED, Snead K, Solanky A, Spector BL, Steffen KK, Tchao BN, Ting MK, Vander Wende H, Wang D, Welton KL, Westman EA, Brem RB, Liu X, Suh Y, Zhou Z, Kaeberlein M, Kennedy BK. A Comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab. 2015;22(5):895–906. doi: 10.1016/j.cmet.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramirez M, Wek RC, Vazquez de Aldana CR, Jackson BM, Freeman B, Hinnebusch AG. Mutations activating the yeast EIF-2 alpha kinase GCN2: Isolation of alleles altering the domain related to Histidyl-TRNA synthetases. Mol Cell Biol. 1992;12(12):5801–5815. doi: 10.1128/mcb.12.12.5801-5815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 21.Molenaars M, Janssens GE, Williams EG, Jongejan A, Lan J, Rabot S, Joly F, Moerland PD, Schomakers BV, Lezzerini M, Liu YJ, McCormick MA, Kennedy BK, van Weeghel M, van Kampen AHC, Aebersold R, MacInnes AW, Houtkooper RH. A Conserved mito-cytosolic translational balance links two longevity pathways. Cell Metab. 2020;31(3):549–563.e7. doi: 10.1016/j.cmet.2020.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei N, Zhu L-Q, Liu D. ATF4: a novel potential therapeutic target for Alzheimer’s disease. Mol Neurobiol. 2015;52(3):1765–1770. doi: 10.1007/s12035-014-8970-8. [DOI] [PubMed] [Google Scholar]
- 23.Kitakaze K, Oyadomari M, Zhang J, Hamada Y, Takenouchi Y, Tsuboi K, Inagaki M, Tachikawa M, Fujitani Y, Okamoto Y, Oyadomari S. ATF4-mediated transcriptional regulation protects against β-cell loss during endoplasmic reticulum stress in a mouse model. Mol. Metab. 2021;54:101338. doi: 10.1016/j.molmet.2021.101338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. an integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11(3):619–633. doi: 10.1016/S1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 25.Tian X, Zhang S, Zhou L, Seyhan AA, Hernandez Borrero L, Zhang Y, El-Deiry WS. Targeting the integrated stress response in cancer therapy. Front Pharmacol. 2021;12:747837. doi: 10.3389/fphar.2021.747837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ebert SM, Dyle MC, Bullard SA, Dierdorff JM, Murry DJ, Fox DK, Bongers KS, Lira VA, Meyerholz DK, Talley JJ, Adams CM. Identification and small molecule inhibition of an activating transcription factor 4 (ATF4)-dependent pathway to age-related skeletal muscle weakness and atrophy. J Biol Chem. 2015;290(42):25497–25511. doi: 10.1074/jbc.M115.681445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robbins CE, Patel B, Sawyer DL, Wilkinson B, Kennedy BK, McCormick MA. Cytosolic and mitochondrial TRNA synthetase inhibitors increase lifespan in a GCN4/Atf-4-dependent manner. iScience. 2022;25(11):105410. doi: 10.1016/j.isci.2022.105410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Z, Xia B, Postnikoff SD, Shen Z-J, Tomoiaga AS, Harkness TA, Seol JH, Li W, Chen K, Tyler JK. Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged yeast while their activation extends lifespan. eLife. 2018;7:e35551. doi: 10.7554/eLife.35551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Statzer C, Meng J, Venz R, Bland M, Robida-Stubbs S, Patel K, Petrovic D, Emsley R, Liu P, Morantte I, Haynes C, Mair WB, Longchamp A, Filipovic MR, Blackwell TK, Ewald CY. ATF-4 and hydrogen sulfide signalling mediate longevity in response to inhibition of translation or MTORC1. Nat Commun. 2022;13(1):967. doi: 10.1038/s41467-022-28599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, Pak DN, Fields S, Kennedy BK, Kaeberlein M. Yeast life span extension by depletion of 60S ribosomal subunits is mediated by Gcn4. Cell. 2008;133(2):292–302. doi: 10.1016/j.cell.2008.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang M-J, Vasudevan D, Kang K, Kim K, Park J-E, Zhang N, Zeng X, Neubert TA, Marr MT, Ryoo HD. 4E-BP is a target of the GCN2-ATF4 pathway during Drosophila development and aging. J Cell Biol. 2017;216(1):115–129. doi: 10.1083/jcb.201511073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W, Li X, Miller RA. ATF4 Activity: a common feature shared by many kinds of slow-aging mice. Aging Cell. 2014;13(6):1012–1018. doi: 10.1111/acel.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Miller RA. Elevated ATF4 function in fibroblasts and liver of slow-aging mutant mice. J Gerontol Ser A. 2015;70(3):263–272. doi: 10.1093/gerona/glu040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem. 2014;83(1):779–812. doi: 10.1146/annurev-biochem-060713-035802. [DOI] [PubMed] [Google Scholar]
- 35.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab. 2009;20(9):436–443. doi: 10.1016/j.tem.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167(1):27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6(5):1099–1108. doi: 10.1016/S1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 38.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 39.Wong SQ, Kumar AV, Mills J, Lapierre LR. Autophagy in aging and longevity. Hum Genet. 2020;139(3):277–290. doi: 10.1007/s00439-019-02031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubinsztein DC, Mariño G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 41.Cebollero E, Reggiori F, Kraft C. Reticulophagy and ribophagy: regulated degradation of protein production factories. Int J Cell Biol. 2012;2012:e182834. doi: 10.1155/2012/182834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tolkovsky AM. Mitophagy. Biochim Biophys Acta BBA - Mol Cell Res. 2009;1793(9):1508–1515. doi: 10.1016/j.bbamcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 43.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J Mol Biol. 2016;428(9 Part A):1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tóth ML, Sigmond T, Borsos É, Barna J, Erdélyi P, Takács-Vellai K, Orosz L, Kovács AL, Csikós G, Sass M, Vellai T. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis Elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 45.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila Melanogaster. Cell Metab. 2010;11(1):35–46. doi: 10.1016/j.cmet.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. Elegans. PLOS Genet. 2008;4(2):e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Z-J, Postnikoff S, Tyler JK. Is Gcn4-induced autophagy the ultimate downstream mechanism by which hormesis extends yeast replicative lifespan? Curr Genet. 2019;65(3):717–720. doi: 10.1007/s00294-019-00936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alvers AL, Fishwick LK, Wood MS, Hu D, Chung HS, Dunn WA, Aris JP. Autophagy and amino acid homeostasis are required for chronological longevity in Saccharomyces Cerevisiae. Aging Cell. 2009;8(4):353–369. doi: 10.1111/j.1474-9726.2009.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamamoto H, Zhang S, Mizushima N. Autophagy genes in biology and disease. Nat Rev Genet. 2023;24(6):382–400. doi: 10.1038/s41576-022-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.B’chir W, Maurin A-C, Carraro V, Averous J, Jousse C, Muranishi Y, Parry L, Stepien G, Fafournoux P, Bruhat A. The EIF2α/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013;41(16):7683–7699. doi: 10.1093/nar/gkt563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallejo M, Ron D, Miller CP, Habener JF. C/ATF, a member of the activating transcription factor family of dna-binding proteins, dimerizes with CAAT/enhancer-binding proteins and directs their binding to camp response elements. Proc Natl Acad Sci U S A. 1993;90(10):4679–4683. doi: 10.1073/pnas.90.10.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene. 2010;29(31):4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 53.Verginadis II, Avgousti H, Monslow J, Skoufos G, Chinga F, Kim K, Leli NM, Karagounis IV, Bell BI, Velalopoulou A, Salinas CS, Wu VS, Li Y, Ye J, Scott DA, Osterman AL, Sengupta A, Weljie A, Huang M, Zhang D, Fan Y, Radaelli E, Tobias JW, Rambow F, Karras P, Marine J-C, Xu X, Hatzigeorgiou AG, Ryeom S, Diehl JA, Fuchs SY, Puré E, Koumenis C. A stromal integrated stress response activates perivascular cancer-associated fibroblasts to drive angiogenesis and tumour progression. Nat Cell Biol. 2022;24(6):940–953. doi: 10.1038/s41556-022-00918-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Aniello C, Fico A, Casalino L, Guardiola O, Di Napoli G, Cermola F, De Cesare D, Tatè R, Cobellis G, Patriarca EJ, Minchiotti G. A novel autoregulatory loop between the Gcn2-Atf4 pathway and L-proline metabolism controls stem cell identity. Cell Death Differ. 2015;22(7):1094–1105. doi: 10.1038/cdd.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rolfes RJ, Hinnebusch AG. Translation of the yeast transcriptional activator GCN4 is stimulated by purine limitation: implications for activation of the protein kinase GCN2. Mol Cell Biol. 1993;13(8):5099–5111. doi: 10.1128/mcb.13.8.5099-5111.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40(2):280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deegan S, Koryga I, Glynn SA, Gupta S, Gorman AM, Samali A. A close connection between the PERK and IRE arms of the UPR and the transcriptional regulation of autophagy. Biochem Biophys Res Commun. 2015;456(1):305–311. doi: 10.1016/j.bbrc.2014.11.076. [DOI] [PubMed] [Google Scholar]
- 58.Milano A, Iaffaioli RV, Caponigro F. 2007 The proteasome: a worthwhile target for the treatment of solid tumours? Eur J Cancer Oxf Engl. 1990;43(7):1125–1133. doi: 10.1016/j.ejca.2007.01.038. [DOI] [PubMed] [Google Scholar]
- 59.Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8(8):739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- 60.Frake RA, Ricketts T, Menzies FM, Rubinsztein DC. Autophagy and neurodegeneration. J Clin Invest. 2015;125(1):65–74. doi: 10.1172/JCI73944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia K, Levine B. Autophagy is required for dietary restriction-mediated life span extension in C. Elegans Autophagy. 2007;3(6):597–599. doi: 10.4161/auto.4989. [DOI] [PubMed] [Google Scholar]
- 62.David, D. Aging and the Aggregating Proteome. Front Genet 2012; 3. [DOI] [PMC free article] [PubMed]
- 63.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C Elegans. PLOS Biol. 2010;8(8):e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu, J. Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol Chapter 2005; 28(Unit 28.1) 10.1002/0471142727.mb2801s70. [DOI] [PubMed]
- 65.Qiu, L., Lai, W., Stumpo, D., Blackshear, P. Mouse embryonic fibroblast cell culture and stimulation. BIO-Protoc. 2016; 6(13) 10.21769/BioProtoc.1859. [DOI] [PMC free article] [PubMed]
- 66.Sieber J, Hauer C, Bhuvanagiri M, Leicht S, Krijgsveld J, Neu-Yilik G, Hentze MW, Kulozik AE. Proteomic Analysis reveals branch-specific regulation of the unfolded protein response by nonsense-mediated MRNA decay. Mol Cell Proteomics. 2016;15(5):1584–1597. doi: 10.1074/mcp.M115.054056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miyake M, Nomura A, Ogura A, Takehana K, Kitahara Y, Takahara K, Tsugawa K, Miyamoto C, Miura N, Sato R, Kurahashi K, Harding HP, Oyadomari M, Ron D, Oyadomari S. Skeletal muscle-specific eukaryotic translation initiation factor 2α Phosphorylation controls amino acid metabolism and fibroblast growth factor 21-mediated non-cell-autonomous energy metabolism. FASEB J Off Publ Fed Am Soc Exp Biol. 2016;30(2):798–812. doi: 10.1096/fj.15-275990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schipper-Krom, S.; Sanz, A. S.; van Bodegraven, E. J.; Speijer, D.; Florea, B. I.; Ovaa, H.; Reits, E. A. Visualizing Proteasome Activity and Intracellular Localization Using Fluorescent Proteins and Activity-Based Probes. Front Mol Biosci 2019; 6. [DOI] [PMC free article] [PubMed]
- 69.Kisselev AF, Garcia-Calvo M, Overkleeft HS, Peterson E, Pennington MW, Thornberry NA, Goldberg AL, Ploegh HL. the caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J Biol Chem. 2003;278(38):35869–35877. doi: 10.1074/jbc.M303725200. [DOI] [PubMed] [Google Scholar]
- 70.Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, Kodama A, Ishihara T, Mizushima T, Mizushima N. An autophagic flux probe that releases an internal control. Mol Cell. 2016;64(4):835–849. doi: 10.1016/j.molcel.2016.09.037. [DOI] [PubMed] [Google Scholar]
- 71.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 72.Chung M, Bruno VM, Rasko DA, Cuomo CA, Muñoz JF, Livny J, Shetty AC, Mahurkar A, Dunning Hotopp JC. Best practices on the differential expression analysis of multi-species RNA-seq. Genome Biol. 2021;22(1):121. doi: 10.1186/s13059-021-02337-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao Y, Smyth GK, Shi W. FeatureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 74.Chen S, Zhou Y, Chen Y, Gu J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Law CW, Chen Y, Shi W, Smyth GK. Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smyth, G. K. Limma: linear models for microarray data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V. J., Huber, W., Irizarry, R. A., Dudoit, S., Eds.; Statistics for Biology and Health; Springer: New York, NY, 2005; 397–420. 10.1007/0-387-29362-0_23.
- 78.Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ. The use of total protein stains as loading controls: an alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neurosci Methods. 2008;172(2):250–254. doi: 10.1016/j.jneumeth.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Berger J, Jampolsky LM, Goldberg MW. Borrelidin, a new antibiotic with antiborrelia activity and penicillin enhancement properties. Arch Biochem. 1949;22(3):476–478. [PubMed] [Google Scholar]
- 80.Habibi D, Ogloff N, Jalili RB, Yost A, Weng AP, Ghahary A, Ong CJ. Borrelidin, a small molecule nitrile-containing macrolide inhibitor of threonyl-TRNA synthetase, is a potent inducer of apoptosis in acute lymphoblastic leukemia. Invest New Drugs. 2012;30(4):1361–1370. doi: 10.1007/s10637-011-9700-y. [DOI] [PubMed] [Google Scholar]
- 81.Kitajima S, Sun W, Lee KL, Ho JC, Oyadomari S, Okamoto T, Masai H, Poellinger L, Kato H. A KDM6 inhibitor potently induces ATF4 and its target gene expression through HRI activation and by UTX inhibition. Sci Rep. 2021;11(1):4538. doi: 10.1038/s41598-021-83857-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bouz G, Zitko J. Inhibitors of aminoacyl-TRNA synthetases as antimycobacterial compounds: an up-to-date review. Bioorganic Chem. 2021;110:104806. doi: 10.1016/j.bioorg.2021.104806. [DOI] [PubMed] [Google Scholar]
- 83.Capobianco JO, Doran CC, Goldman RC. Mechanism of mupirocin transport into sensitive and resistant bacteria. Antimicrob Agents Chemother. 1989;33(2):156–163. doi: 10.1128/AAC.33.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baragaña B, Forte B, Choi R, Nakazawa Hewitt S, Bueren-Calabuig JA, Pisco JP, Peet C, Dranow DM, Robinson DA, Jansen C, Norcross NR, Vinayak S, Anderson M, Brooks CF, Cooper CA, Damerow S, Delves M, Dowers K, Duffy J, Edwards TE, Hallyburton I, Horst BG, Hulverson MA, Ferguson L, Jiménez-Díaz MB, Jumani RS, Lorimer DD, Love MS, Maher S, Matthews H, McNamara CW, Miller P, O’Neill S, Ojo KK, Osuna-Cabello M, Pinto E, Post J, Riley J, Rottmann M, Sanz LM, Scullion P, Sharma A, Shepherd SM, Shishikura Y, Simeons FRC, Stebbins EE, Stojanovski L, Straschil U, Tamaki FK, Tamjar J, Torrie LS, Vantaux A, Witkowski B, Wittlin S, Yogavel M, Zuccotto F, Angulo-Barturen I, Sinden R, Baum J, Gamo F-J, Mäser P, Kyle DE, Winzeler EA, Myler PJ, Wyatt PG, Floyd D, Matthews D, Sharma A, Striepen B, Huston CD, Gray DW, Fairlamb AH, Pisliakov AV, Walpole C, Read KD, Van Voorhis WC, Gilbert IH. Lysyl-TRNA synthetase as a drug target in malaria and Cryptosporidiosis. Proc Natl Acad Sci U S A. 2019;116(14):7015–7020. doi: 10.1073/pnas.1814685116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Critchley IA, Young CL, Stone KC, Ochsner UA, Guiles J, Tarasow T, Janjic N. Antibacterial Activity of REP8839, a new antibiotic for topical use. Antimicrob Agents Chemother. 2005;49(10):4247–4252. doi: 10.1128/AAC.49.10.4247-4252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Critchley IA, Green LS, Young CL, Bullard JM, Evans RJ, Price M, Jarvis TC, Guiles JW, Janjic N, Ochsner UA. Spectrum of activity and mode of action of REP3123, a new antibiotic to treat Clostridium Difficile Infections. J Antimicrob Chemother. 2009;63(5):954–963. doi: 10.1093/jac/dkp041. [DOI] [PubMed] [Google Scholar]
- 87.Mazor KM, Stipanuk MH. GCN2- and EIF2α-phosphorylation-independent, but ATF4-dependent, induction of CARE-containing genes in methionine-deficient cells. Amino Acids. 2016;48(12):2831–2842. doi: 10.1007/s00726-016-2318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.McCormick M, Chen K, Ramaswamy P, Kenyon C. New genes that extend Caenorhabditis Elegans’ lifespan in response to reproductive signals. Aging Cell. 2012;11(2):192–202. doi: 10.1111/j.1474-9726.2011.00768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime Cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. ClueGO: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinforma Oxf Engl. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mittal N, Guimaraes JC, Gross T, Schmidt A, Vina-Vilaseca A, Nedialkova DD, Aeschimann F, Leidel SA, Spang A, Zavolan M. The Gcn4 transcription factor reduces protein synthesis capacity and extends yeast lifespan. Nat Commun. 2017;8(1):457. doi: 10.1038/s41467-017-00539-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dooley HC, Razi M, Polson HEJ, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55(2):238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilson MI, Dooley HC, Tooze SA. WIPI2b and Atg16L1: setting the stage for autophagosome formation. Biochem Soc Trans. 2014;42(5):1327–1334. doi: 10.1042/BST20140177. [DOI] [PubMed] [Google Scholar]
- 94.Torrence ME, MacArthur MR, Hosios AM, Valvezan AJ, Asara JM, Mitchell JR, Manning BD. The MTORC1-mediated activation of ATF4 promotes protein and glutathione synthesis downstream of growth signals. eLife. 2021;10:e63326. doi: 10.7554/eLife.63326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5(5):897–904. doi: 10.1016/S1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 96.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, Salas Salinas C, Chinga F, Monroy AM, Fu W, Wang P, Kossenkov A, Ye J, Amaravadi RK, Ignatova Z, Fuchs SY, Diehl JA, Ruggero D, Koumenis C. ATF4 Couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol. 2019;21(7):889–899. doi: 10.1038/s41556-019-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jiang D, Cui H, Xie N, Banerjee S, Liu R-M, Dai H, Thannickal VJ, Liu G. ATF4 Mediates mitochondrial unfolded protein response in alveolar epithelial cells. Am J Respir Cell Mol Biol. 2020;63(4):478–489. doi: 10.1165/rcmb.2020-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu J, Dang N, Menu E, De Bruyne E, Xu D, Van Camp B, Van Valckenborgh E, Vanderkerken K. Activation of ATF4 mediates unwanted Mcl-1 accumulation by proteasome inhibition. Blood. 2012;119(3):826–837. doi: 10.1182/blood-2011-07-366492. [DOI] [PubMed] [Google Scholar]
- 99.Lassot I, Ségéral E, Berlioz-Torrent C, Durand H, Groussin L, Hai T, Benarous R, Margottin-Goguet F. ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(BetaTrCP) ubiquitin ligase. Mol Cell Biol. 2001;21(6):2192–2202. doi: 10.1128/MCB.21.6.2192-2202.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zou K, Rouskin S, Dervishi K, McCormick MA, Sasikumar A, Deng C, Chen Z, Kaeberlein M, Brem RB, Polymenis M, Kennedy BK, Weissman JS, Zheng J, Ouyang Q, Li H. Life span extension by glucose restriction is abrogated by methionine supplementation: cross-talk between glucose and methionine and implication of methionine as a key regulator of life span. Sci Adv. 2020;6(32):eaba1306. doi: 10.1126/sciadv.aba1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Adams J. The development of proteasome inhibitors as anticancer drugs. Cancer Cell. 2004;5(5):417–421. doi: 10.1016/s1535-6108(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 102.Kane RC, Bross PF, Farrell AT, Pazdur R. Velcade: U.S. FDA approval for the treatment of multiple myeloma progressing on prior therapy. Oncologist. 2003;8(6):508–513. doi: 10.1634/theoncologist.8-6-508. [DOI] [PubMed] [Google Scholar]
- 103.Nieto-Torres JL, Hansen M. Macroautophagy and aging: the impact of cellular recycling on health and longevity. Mol Aspects Med. 2021;82:101020. doi: 10.1016/j.mam.2021.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Levine B, Packer M, Codogno P. Development of autophagy inducers in clinical medicine. J Clin Invest. 2015;125(1):14–24. doi: 10.1172/JCI73938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang ZD, Milman S, Lin J-R, Wierbowski S, Yu H, Barzilai N, Gorbunova V, Ladiges WC, Niedernhofer LJ, Suh Y, Robbins PD, Vijg J. Genetics of extreme human longevity to guide drug discovery for healthy ageing. Nat Metab. 2020;2(8):663–672. doi: 10.1038/s42255-020-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ron D, Walter P. signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 107.Patil CK, Li H, Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2(8):e246. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woo CW, Cui D, Arellano J, Dorweiler B, Harding H, Fitzgerald KA, Ron D, Tabas I. Adaptive suppression of the ATF4–CHOP branch of the unfolded protein response by Toll-like receptor signalling. Nat Cell Biol. 2009;11(12):1473–1480. doi: 10.1038/ncb1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fusakio ME, Willy JA, Wang Y, Mirek ET, Al Baghdadi RJT, Adams CM, Anthony TG, Wek RC. Transcription factor ATF4 directs basal and stress-induced gene expression in the unfolded protein response and cholesterol metabolism in the liver. Mol Biol Cell. 2016;27(9):1536–1551. doi: 10.1091/mbc.E16-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Solis GM, Kardakaris R, Valentine ER, Bar-Peled L, Chen AL, Blewett MM, McCormick MA, Williamson JR, Kennedy B, Cravatt BF, Petrascheck M. Translation attenuation by minocycline enhances longevity and proteostasis in old post-stress-responsive organisms. eLife. 2018;7:e40314. doi: 10.7554/eLife.40314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miller BF, Drake JC, Naylor B, Price JC, Hamilton KL. The measurement of protein synthesis for assessing proteostasis in studies of slowed aging. Ageing Res Rev. 2014;18:106–111. doi: 10.1016/j.arr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhu S, Sobolev AY, Wek RC. Histidyl-TRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of EIF-2. J Biol Chem. 1996;271(40):24989–24994. doi: 10.1074/jbc.271.40.24989. [DOI] [PubMed] [Google Scholar]
- 113.Dong J, Qiu H, Garcia-Barrio M, Anderson J, Hinnebusch AG. Uncharged TRNA activates GCN2 by displacing the protein kinase moiety from a bipartite TRNA-binding domain. Mol Cell. 2000;6(2):269–279. doi: 10.1016/S1097-2765(00)00028-9. [DOI] [PubMed] [Google Scholar]
- 114.Wek RC, Jackson BM, Hinnebusch AG. Juxtaposition of domains homologous to protein kinases and Histidyl-TRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc Natl Acad Sci. 1989;86(12):4579–4583. doi: 10.1073/pnas.86.12.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wek SA, Zhu S, Wek RC. The Histidyl-TRNA synthetase-related sequence in the EIF-2 alpha protein kinase GCN2 interacts with TRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol. 1995;15(8):4497–4506. doi: 10.1128/MCB.15.8.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi: 10.15252/embr.201642195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Park Y, Reyna-Neyra A, Philippe L, Thoreen CC. MTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 2017;19(6):1083–1090. doi: 10.1016/j.celrep.2017.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Adams CM, Ebert SM, Dyle MC. Role of ATF4 in skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2017;20(3):164–168. doi: 10.1097/MCO.0000000000000362. [DOI] [PubMed] [Google Scholar]
- 119.Miller MJ, Marcotte GR, Basisty N, Wehrfritz C, Ryan ZC, Strub MD, McKeen AT, Stern JI, Nath KA, Rasmussen BB, Judge AR, Schilling B, Ebert SM, Adams CM. The Transcription regulator ATF4 is a mediator of skeletal muscle aging. GeroScience. 2023 doi: 10.1007/s11357-023-00772-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pike LRG, Singleton DC, Buffa F, Abramczyk O, Phadwal K, Li J-L, Simon AK, Murray JT, Harris AL. Transcriptional up-regulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449(2):389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 121.Rozpedek W, Pytel D, Mucha B, Leszczynska H, Diehl JA, Majsterek I. The role of the PERK/EIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr Mol Med. 2016;16(6):533–544. doi: 10.2174/1566524016666160523143937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Singleton DC, Harris AL. Targeting the ATF4 pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(12):1189–1202. doi: 10.1517/14728222.2012.728207. [DOI] [PubMed] [Google Scholar]
- 123.Rzymski T, Milani M, Singleton DC, Harris AL. Role of ATF4 in regulation of autophagy and resistance to drugs and hypoxia. Cell Cycle Georget Tex. 2009;8(23):3838–3847. doi: 10.4161/cc.8.23.10086. [DOI] [PubMed] [Google Scholar]
- 124.Yerlikaya A. Heme-regulated inhibitor: an overlooked EIF2α kinase in cancer investigations. Med Oncol Northwood Lond Engl. 2022;39(5):73. doi: 10.1007/s12032-022-01668-1. [DOI] [PubMed] [Google Scholar]
- 125.Hinnebusch AG. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces Cerevisiae. Microbiol Rev. 1988;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data described in this study are available from the Gene Expression Omnibus (GEO) with accession number #GSE217634.