Abstract
The extracellular proteome of Streptomyces coelicolor grown in a liquid medium was analyzed by using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization-time of flight peptide mass fingerprint analysis. Culture supernatants became protein rich only after rapid growth had been completed, supporting the idea that protein secretion is largely a stationary phase phenomenon. Out of about 600 protein spots observed, 72 were characterized. The products of 47 genes were identified, with only 11 examples predicted to be secreted proteins. Mutation in bldA, previously known to impair the stationary phase processes of antibiotic production and morphological differentiation, also induced changes in the extracellular proteome, revealing even greater pleiotropy in the bldA phenotype than previously known. Four proteins increased in abundance in the bldA mutant, while the products of 11 genes, including four secreted proteins, were severely down-regulated. Although bldA encodes the only tRNA capable of efficiently translating the rare UUA (leucine) codon, none of the latter group of genes contains an in-frame TTA. SCO0762, a serine-protease inhibitor belonging to the Streptomyces subtilisin inhibitor family implicated in differentiation in other streptomycetes, was completely absent from the bldA mutant. This dependence was shown to be mediated via the TTA-containing regulatory gene adpA, also known as bldH, a developmental gene that is responsible for the effects of bldA on differentiation. Mutation of the SCO0762 gene abolished detectable trypsin-protease inhibitory activity but did not result in any obvious morphological defects.
Bacteria of the genus Streptomyces, known for their importance as producers of antibiotics and other secondary metabolites, are highly adapted to live in soil (3). This environment is extremely diverse in composition, and its organic component consists largely of insoluble polymers accessible only to organisms having suitable extracellular enzymes and systems for their regulation. As a broad generalization, extracellular functions of bacteria, such as production of hydrolytic exoenzymes, small signaling molecules, and antibiotics, are most prevalent after rapid growth has been completed. This is because more readily assimilated soluble nutrients are used preferentially and because a high cell density may be necessary for the concentration of extracellular molecules to reach levels high enough to have meaningful activity.
In streptomycetes, some aspects of stationary phase biology have common control elements, as revealed by the finding that mutations in some genes have drastic effects on both antibiotic production and aerial reproductive growth. Among such bld genes (named because the colonies lack aerial growth and thus are “bald”), bldA is unusual: it specifies the only tRNA in the genome capable of translating efficiently the leucine codon UUA (16, 17). Streptomyces DNA typically contains more than 70% G+C, making TTA codons in the genome quite rare; indeed, the Streptomyces coelicolor genome annotation predicts that only 145 genes contain TTA codons and so should be directly affected by bldA mutations (3; S. coelicolor database at http://streptomyces.org.uk/). This number includes 15 encoding proteins whose primary sequences indicate that they are likely to be secreted. The presence of TTA codons in an additional 14 likely regulatory genes further increases the potential for bldA mutations to alter the range of proteins exported from cells.
Mutations in bldA pleiotropically affect production of the three chromosomally specified antibiotics in S. coelicolor (actinorhodin, undecylprodigiosin, and calcium-dependent antibiotic) and of a plasmid-determined antibiotic, methylenomycin; they also abolish morphological differentiation but only on certain media (19). In this work we explore the possibility that bldA mutations might also affect aspects of stationary phase biology other than secondary metabolism and morphological differentiation. We reasoned that much of the extracellular proteome might be specific for stationary phase and could, therefore, be altered in a bldA mutant. To explore this aspect of the influence of bldA, we used two-dimensional (2D) gel electrophoresis coupled with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) peptide mass fingerprint analysis to compare proteins in culture supernatants of a wild-type S. coelicolor strain and an isogenic bldA deletion mutant. Significant differences were indeed found, all of which involved proteins not encoded by TTA-containing genes. The implication that these effects might be due to dependence of the relevant genes on the products of TTA-containing regulatory genes was explored in the case of a bldA-dependent protease inhibitor. Remarkably, the gene for this protein proved to be dependent on the TTA-containing pleiotropic regulatory gene adpA (also known as bldH), which mediates the effects of bldA on morphological differentiation (24, 35).
MATERIALS AND METHODS
Strains and growth conditions.
S. coelicolor M600 is a prototrophic, plasmid-free strain derived from S. coelicolor A3(2) (12). (Note that this strain has a duplication of 1,005 genes compared with the sequenced strain M145 [39].) In the M600 ΔbldA strain, kindly provided by M. Tao, bldA is completely replaced by an apramycin resistance cassette. The M600 ΔadpA strain similarly contains an in-frame replacement of the adpA gene (mutant construction identical to that reported for strain M145 ΔadpA [35]). For proteomic analysis and S1 nuclease mapping, strains were cultivated with vigorous agitation at 30°C in SMM (liquid minimal medium supplemented with 0.2% casamino acids) as previously described (12). Briefly, spores (about 1010 CFU ml−1) were pregerminated in 2× yeast extract-tryptone medium for 7 h at 30°C. Germlings were harvested by centrifugation (5 min at 4,000 × g), resuspended in SMM, and briefly sonicated to disperse any aggregates before inoculation into 50 ml of SMM in 250-ml siliconized flasks containing coiled stainless steel springs. Each flask received the equivalent of 5 × 107 spore CFU. Growth curves for producing secreted protein extracts to compare M600 and M600 ΔbldA strains were performed in duplicate, as were cultures for isolating RNA for comparing transcription between M600 and M600 ΔadpA strains.
Escherichia coli strains DH5α (29) and ET12567 (dam dcm hrdS) (18) were grown and transformed according to Sambrook et al. (29). Strain ET12567 was used to propagate unmethylated cosmid DNA for introduction into S. coelicolor by conjugation. E. coli BW25113/pIJ790 was the host for λ Red-mediated PCR-targeted mutagenesis (5).
Preparation of extracellular protein fractions from cultures.
Cultures were harvested by brief centrifugation (30 s at 4,000 × g) at room temperature. Harvested cell pellets were immediately frozen in liquid nitrogen and stored at −80°C for use in analysis of the membrane proteome to be reported elsewhere. The culture supernatants (50 ml) were filtered, mixed with an equal volume of 20% (wt/vol) trichloroacetic acid (TCA) in acetone, and chilled at −20°C for 1 h. Precipitated proteins were harvested by centrifugation (4°C for 10 min at 6,000 × g), and the pellets were washed with acetone (10 ml, −20°C) and repelleted by centrifugation (4°C for 10 min at 6,000 × g). Washed protein pellets were air dried (15 min at room temperature) and then resuspended in 50 to 200 μl of the denaturing isoelectric focusing buffer UTCHAPS containing 7 M urea, 2 M thiourea, 4% (wt/vol) CHAPS (3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate), 40 mM Tris (pH 9.0), 1 mM EDTA, 50 mM dithiothreitol, 4 mM Pefabloc SC protease inhibitor (Roche). Extracts were stored at −80°C until use.
2D gel electrophoresis and image analysis.
Protein extracts were separated by 2D gel electrophoresis as previously described (8). Briefly, proteins were separated in the first dimension for 100,000 V-hours by using 18-cm immobilized pH gradient strips pH 4 to 7 (Amersham Biosciences) in a Phaser isoelectric focusing unit (Genomic Solutions). Focused strips were separated in the second dimension by using in-house fabricated sodium dodecyl sulfate-12.5% polyacrylamide gels and the Investigator 5000 vertical format system from Genomic Solutions. For quantitative analysis of protein abundance profiles, gels were stained with Sypro Ruby (Bio-Rad), according to the manufacturer's instructions, and scanned by using the Perkin-Elmer ProXPRESS proteomic imaging system using excitation and emission wavelengths of 480 and 630 nm, respectively. Image analysis was performed by using PHORETIX 2D version 5.1 (NonLinear Dynamics) as follows: spot detection was optimized automatically by using the Spot Detection Wizard and then manually edited; background subtraction was performed automatically by using the Mode of Non-Spot setting; and images were then normalized to the total spot volume for each gel for quantification. Spot filtering was not used, although all spots were manually edited. Histograms of normalized spot volumes displaying changes in spot abundance during growth and between the M600 strain and the ΔbldA mutant were generated by the software. Differences between strains were considered significant if normalized volumes for a particular spot were changed twofold or more in both biological duplicate growth curves in at least one of the three samples.
Protein identification by mass spectrometry.
Protein spots of interest were excised from the Sypro Ruby-stained gels by using the Investigator ProPic robot from Genomic Solutions and were identified by tryptic digestion and MALDI-TOF mass spectrometry as previously described (8).
Construction of a mutant with a deletion of the SCO0762 gene.
A mutant allele of the SCO0762 gene, in which the entire coding region was replaced by an apramycin resistance cassette, was constructed by PCR-targeting essentially as described by Gust et al. (5). Briefly, oligonucleotide primers (5′-) with 5′ ends homologous to the 5′ and 3′ ends adjacent to the coding sequence of the SCO0762 gene and 3′ (priming) ends designed to amplify the apramycin resistance disruption cassette were used in a PCR with pIJ773 as template. The PCR product was introduced into E. coli BW25113/pIJ790 containing cosmid SCF81 (28), preinduced for λ Red functions by the addition of arabinose, to obtain a SCF81 cosmid with a disruption in the SCO0762 gene. The disrupted cosmid was isolated and transferred via E. coli ET12567/pUZ8002 to S. coelicolor M600 by conjugation. Exconjugants were selected on MS agar (mannitol-soy) (12) containing apramycin, and the products of double crossovers were identified by screening for kanamycin sensitivity. Deletions were confirmed by PCR and Southern blotting.
S1 nuclease mapping.
RNA was isolated from exponential and early stationary phase cultures as described by Strauch et al. (33). For each S1 nuclease reaction, 20 μg of RNA was hybridized in NaTCA buffer (23) to about 0.2 pmol (approximately 105 Cerenkov counts min−1) of each of the following radiolabeled probes. For the gene SCO0762, the oligonucleotide 5′-TCTCTCCGTGCCCCACGGTCAGCA-3′, which anneals within the SCO0762 gene coding region, was uniquely end labeled at its 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase. This was used in the PCR together with the unlabeled probe 5′-AGGCGATCCTAGTCGATCAAGAAACGCCCAGTTC-3′ (which anneals upstream of the SCO0762 gene) and cosmid SCF81 as template to generate a 365-bp probe. PCR was performed in the presence of 7% dimethyl sulfoxide under the following conditions: 94°C for 4 min followed by 26 cycles of 45 s at 94°C, 45 s at 58°C, and 60 s at 72°C, with a final hold at 72°C for 5 min to finish. The probe for the SCO4253 gene was made in a similar way but using 5′-GATCTGACCGATCCTCCTGACACGCCGTCACCGT-3′ as the unlabeled primer, 5′-GACCGACGAAGGCCGCCACCGA-3′ as the labeled primer, and cosmid SCD49 (28) as the template. This produced a 432-bp uniquely end-labeled probe product. Hybridizations were carried out at 45°C for 14 h after denaturation at 65°C for 15 min. S1 nuclease digestions and analyses of RNA-protected fragments were performed as described previously (11). High-resolution mapping of the transcription start site of the SCO0762 gene was achieved by generating a sequencing ladder with the same labeled primer as was used for the probe by using a Thermo Sequenase cycle sequencing kit (USB) according to the manufacturer's instructions.
Fermentation conditions for stirred jar fermentors.
Preparation of the inoculum used to seed the main jar fermentor was as follows. Spores and mycelium from cultures grown on solid MS agar plates (12) were inoculated into 50 ml of the first seed medium (GG1), consisting of 1.5% (wt/vol) glucose, 1.5% (wt/vol) glycerol, 1.5% (wt/vol) soya peptone, 0.3% (wt/vol) NaCl, and 0.1% (wt/vol) CaCO3 and incubated in submerged shaken-batch culture at 30°C for 48 h. This was then used to inoculate a second seed culture at a ratio of 10% (vol/vol) and incubation continued for 40 h. The second seed culture medium consisted of 1% (wt/vol) glucose, 1.5% (wt/vol) yeast extract. Finally, this was used to inoculate (10% [vol/vol]) 4 liters of the main culture medium (modified SMM) contained in a jar fermentor (Model KF-5 L; Korea Fermentor Co.). The culture temperature was maintained at 30°C, and the initial pH was adjusted to 7.2. Agitation and aeration conditions were 250 rpm and 1 volume per volume per min, respectively. Modified SMM was prepared according to the following recipe: 1% (wt/vol) glucose, 0.2% (wt/vol) sodium caseinate, 0.0078% (wt/vol) NaH2PO4 · 2H2O, 0.0087% (wt/vol) K2HPO4, 0.573% (wt/vol) TES (N-tris[hydroxymethyl]methyl-2-aminoethanesulfonic acid), 0.06% (wt/vol) MgSO4, and trace elements. Trace elements solution of SMM was used as previously described (12).
Measurement of growth and glucose utilization in the fermentor.
Dry cell weight measurements were taken to assess growth of the strains in the cultures. Mycelium was harvested aseptically by centrifugation at 10,000 × g for 10 min, washed twice with physiological saline solution and once with distilled water, and then collected by vacuum filtration (Whatman filter paper GF/C). Dry cell weights were measured following drying at 80°C for 24 h. The concentration of glucose in the cultures was followed by analysis using a commercially available glucose assay kit (Young Dong Co., South Korea) according to the manufacturer's instructions. The assay is based on glucose oxidase methods.
Assay of extracellular trypsin protease activity and trypsin protease inhibitory activity in culture supernatants.
Culture aliquots were harvested aseptically from the fermentors and centrifuged at 10,000 × g for 10 min, and the supernatant was retained for the assays. Trypsin-like protease activity was measured by assaying the amount of ρ-nitroanilide liberated from enzymatic hydrolysis of the synthetic substrate N-benzoyl arginine ρ-nitroanilide (BAPNA). Aliquots of culture supernatant (100 μl) were incubated with 200 μM BAPNA (ɛ405 = 9620 M−1 cm−1) at 35°C and pH 7.5 (0.1 M Tris-HCl buffer). The activity was calculated in a linear range for 5 min. One unit of hydrolytic activity was defined as the amount of enzyme needed to produce 1 μmol ρ-nitroanilide per minute at 35°C (30).
Trypsin inhibitory activity was measured by analyzing the effect of culture supernatant aliquots on enzymatic digestion of N-benzoyl-l-arginine ethyl ester (BAEE) by trypsin (type I from bovine pancreas; Sigma). The activity is given by the following formula: percentage inhibition = 100 [(A − B)/A], where A is trypsin activity without the inhibitor and B is trypsin activity in the presence of the inhibitor (2). Activities are expressed as leupeptin units by comparison to values obtained in a standard inhibition curve measuring the effect of different concentrations of the serine-protease inhibitor leupeptin (Sigma) on trypsin activity. For the assay, 1 ml of 0.1 M Tris-HCl buffer (pH 7.5), 1 ml of trypsin solution at a concentration of 4 μg ml−1, and 200 μl of culture supernatant were mixed and preincubated for 5 min at 30°C. One milliliter of BAEE solution at a concentration of 0.18 mg ml−1 was then added, and the increase in N-benzoyl-l-arginine (ɛ253 = 1,150 M−1 cm−1) liberated from BAEE was monitored by measuring the absorbance at 253 nm (14).
RESULTS
Changes in the extracellular proteome of S. coelicolor during growth and as a result of mutation in bldA.
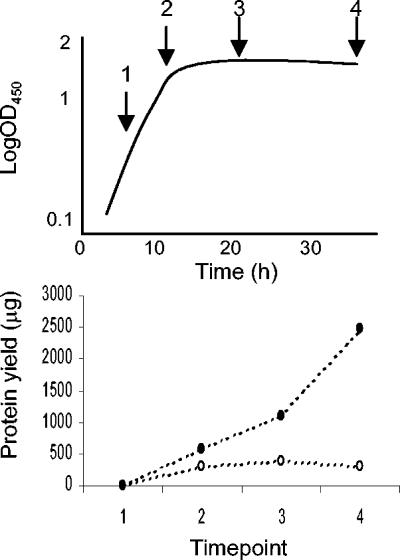
When extracellular proteins were harvested from culture supernatants of S. coelicolor M600 and M600 ΔbldA cultures by precipitation in TCA-acetone, significant levels resulted only from transition and stationary phase supernatants: no detectable protein was obtained from any exponential phase samples (Fig. 1). Typically, strain M600 yielded more protein than the ΔbldA mutant. Analysis of the precipitated proteins by 2D gel electrophoresis revealed about 600 separated protein spots, of which 21 showed differences in abundance levels between the two strains. These proteins, together with 51 other spots whose abundance profiles were of interest, were characterized by peptide mass fingerprint analysis. Certain genes often gave rise to more than one protein spot, suggesting a significant level of posttranslational protein modification (as also previously observed in analysis of the whole-cell proteome [8]).
FIG. 1.
Difference in extracellular protein yield between cultures of S. coelicolor M600 and its ΔbldA mutant. Time points correspond to the four sampling arrows in the stylized growth curve, and figures are the average of two separate experiments that produced reproducible results. Filled circles, strain M600; open circles, strain M600 ΔbldA.
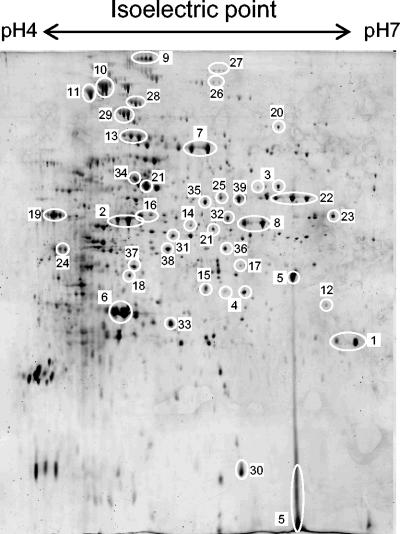
Only a minority of proteins identified in the extracellular proteome are predicted to be translocated. Table 1 groups all the identified proteins into broad functional classes, and Fig. 2 shows the positions of many of them on a typical 2D gel separation. Surprisingly, only 11 of the 47 genes in Table 1 were predicted to encode proteins with signal sequences targeting them for export from the cell. Five of these 11 gene products (namely SCO2780, 4142, 5113, 5477, and 7677) had predicted lipid attachment sites that should anchor them to the outside of the cytoplasmic membrane, and indeed all but SCO7677 were also identified in an analysis of the corresponding membrane fractions (D.-W. Kim, K. Chater, K.-J. Lee, and A. Hesketh, submitted for publication). Their appearance in culture supernatants suggests a turnover of these proteins from the membrane. Of the other six predicted secreted proteins identified, only the protease inhibitor SCO0762 had an assigned function.
TABLE 1.
Proteins identified in 2D gel separations of secreted protein extracts of S. coelicolor M600
| ORF SCO no.a | Gene productb | Tmc | Cleavage sited | Spot no. in Fig. 2e |
|---|---|---|---|---|
| Predicted extracellular proteins | ||||
| 0131 | Putative secreted protein | 1 | 25/26 | 1 |
| 0297 | Putative secreted protein | 1 | 38/39 | 2 |
| 0762 | Protease inhibitor precursor | 1 | 79/80 | 5 |
| 1968 | Putative secreted hydrolase | 1 | 27/28 | 6 |
| 2116 | Putative secreted protein | 2 | 49/50 | 23 |
| 2780 | Putative lipoprotein | 2 | 29/30 | 18 |
| 4142 | Phosphate-binding protein precursor, PstS | 1 | 21/22 | 24 |
| 5113 | Oligopeptide ABC transport lipoprotein, BldKB | 1 | 25/26 | 13 |
| 5477 | Putative oligopeptide-binding lipoprotein | 1 | 28/29 | 7 |
| 6198 | Putative secreted protein | 6 | 34/35 | 9 |
| 7677 | Putative secreted solute-binding lipoprotein | 1 | 30/31 | 25 |
| Central carbon metabolism | ||||
| 1945 | Triosephosphate isomerase | 0 | 33 | |
| 1947 | Glyceraldehyde-3-phosphate dehydrogenase | 0 | 31 | |
| 2180 | Probable dihydrolipoamide dehydrogenase | 2 | None | 22 |
| 2736 | Citrate synthase, CitA | 2 | None | 8 |
| 3096 | Enolase | 1 | None | 19 |
| 3123 | Ribose-phosphate pyrophosphokinase | 0 | ||
| 3649 | Fructose 1,6-bisphosphate aldolase | 0 | ||
| 4809 | Succinyl CoA synthase alpha chain | 0 | ||
| 4827 | Malate dehydrogenase | 1 | None | 37 |
| 5999 | Aconitase | 2 | None | 10 |
| 7000 | Isocitrate dehydrogenase | 1 | None | 29 |
| Amino acid metabolism | ||||
| 1378 | Putative glycine dehydrogenase | 4 | None | 26 |
| 2198 | Glutamine synthetase (Glnl) | 0 | 21 | |
| 4089 | Valine dehydrogenase | 1 | None | 36 |
| 4366 | Putative phosphoserine aminotransferase | 0 | ||
| 4837 | Serine hydroxymethyltransferase, GlyA1 | 1 | None | 39 |
| 4958 | Cystathionine gamma-synthase, MetB | 0 | ||
| 5212 | 3-phosphoshikimate 1-carboxyvinyltransferase | 0 | 16 | |
| 5523 | Branched-chain amino acid aminotransferase | 2 | None | 38 |
| Other primary metabolism | ||||
| 0379 | Catalase, KatA | 0 | 3 | |
| 3792 | Putative methionyl tRNA synthetase | 0 | 34 | |
| 3961 | Seryl-tRNA synthase | 1 | None | 35 |
| 4771 | Putative inosine-5′-monophosphate dehydrogenase | 2 | None | |
| 5399 | Probable acetoacetyl-CoA thiolase | 1 | None | |
| 5625 | Elongation factor Ts | 0 | ||
| 5737 | Guanosine pentaphosphate synthetase/polyribonucleotide nucleotidyltransferase | 2 | None | 28 |
| 6564 | 3-oxoacyl-[acyl-carrier-protein] synthase II, FabH2 | 2 | None | 15 |
| 7510 | Peptidyl-prolyl cis-trans isomerase | 0 | 30 | |
| Proteases | ||||
| 0805 | Putative prolyl aminopeptidase | 1 | None | 32 |
| 2549 | Putative protease | 1 | None | 27 |
| 2643 | Aminopeptidase N, PepN | 0 | 11 | |
| Others | ||||
| 0499 | Probable formyltransferase | 2 | None | 4 |
| 3790 | Conserved hypothetical protein | 1 | None | 20 |
| 5078 | Hypothetical protein, ActVA-ORF3 | 1 | None | 12 |
| 5262 | Putative dehydrogenase | 2 | None | 17 |
| 6621 | Hypothetical protein | 2 | None | 14 |
ORF SCO no., open reading frame S. coelicolor identifier.
Taken from the S. coelicolor database (ScoDB) annotation of the S. coelicolor genome sequence at http://streptomyces.org.uk in October 2004. Products that are significantly altered in abundance in the ΔbldA mutant are underlined.
Number of transmembrane helices predicted on submission of the protein sequence to the TmPred server at www.ch.embnet.org. Scores lower than 600 were not considered significant.
Predicted N-terminal signal peptide cleavage site obtained by submission of the protein sequence to the SignalP server at www.cbs.dtu.dk or the DOLOP lipoprotein prediction tool at www.mrc-lmb.cam.ac.uk/genomes/dolop/. Only sequences already predicted to contain an N-terminal transmembrane helix were analyzed.
For clarity, not all spots are marked in Fig. 2.
FIG. 2.
2D gel separation of secreted proteins harvested from culture supernatants of S. coelicolor M600. Numbers correspond to protein spot identifications in Table 1.
Many enzymes predicted to be involved in primary metabolic processes were identified on the gels, and several of these from central carbon metabolism (Fig. 2, spots 8, 10, 22, and 19) were at least as abundant as spots from predicted secreted proteins (Fig. 2, spots 1, 6, 7, and 13). This raised the important question of whether the unexpected presence of this class of proteins in the extracellular medium is biologically significant or an artifact resulting from cell lysis as cells enter stationary phase in what were very artificial growth conditions for the organism. We take up this question in the Discussion.
The identification in the culture supernatants of several proteins of unknown function not predicted to possess signal peptides prompted a reexamination of their database sequences, and in one case a revision was indicated. The SCO3790 gene has three alternative ATG start codons in the first 180 bp of its sequence, and protein products originating from any of these give a strong prediction for the existence of a signal peptide. It is therefore probable that this protein is in fact targeted for secretion.
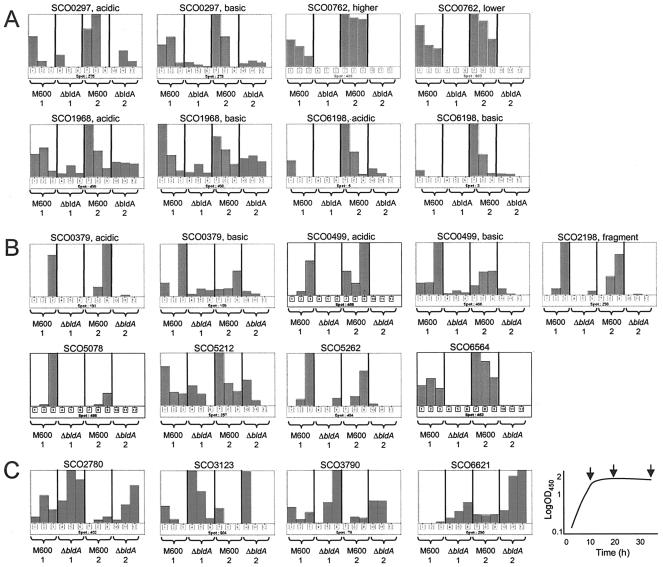
Eleven proteins were less abundant in the ΔbldA mutant, including the protease inhibitor SCO0762. Extracellular protein products from 11 genes were significantly less abundant in the ΔbldA mutant (Fig. 3A and B; note that six of these genes yielded two different spots), including four predicted to encode secreted proteins. Of these, SCO0297 has no significant database homologues, while SCO6198 resembles only another alanine-rich protein of unknown function in the S. coelicolor genome, SCO6593 (which from sequence analysis is not predicted to be secreted). SCO1968 is a putative secreted hydrolase possessing the Pfam domain 03009, characteristic of glycerophosphoryldiester phosphodiesterases involved in the metabolism of glycerol. SCO0762 is the only one of the four to possess a well-defined function and appears to be completely dependent on bldA. It is a member of the Streptomyces subtilisin inhibitor (SSI) family of serine-protease inhibitors and closely resembles the extensively characterized SSI from Streptomyces albogriseolus (Sal-SSI) (Fig. 4). Sal-SSI shows almost exclusive specificity towards subtilisin (21, 22), but site-directed modification of the P1 methionine (after the Schecter and Berger notation [31]) to lysine or arginine also confers potent inhibition of trypsin (20). The crystal structures of Sal-SSI complexed with subtilisin and of engineered Sal-SSI complexed with bovine trypsin have both been solved (9, 36, 37). SCO0762 has an arginine residue at position P1 and therefore should have trypsin inhibitory activity. It is therefore termed STI (Streptomyces trypsin inhibitor). Remarkably, the genes encoding these down-regulated proteins do not contain TTA codons that might account for their observed bldA dependence.
FIG. 3.
Time course profiles showing protein spots with significantly different abundance levels in strain M600 ΔbldA compared with strain M600. (A) Proteins predicted to be secreted that are significantly down-regulated in the ΔbldA strain. (B) Other proteins significantly down-regulated in the ΔbldA strain. (C) Proteins up-regulated in the ΔbldA strain. The numbers 1 and 2 indicate duplicate experiments in which protein samples were obtained from cultures at three time points, as illustrated in the stylized growth curve shown at the bottom right. Histogram bars are normalized spot intensities following staining with Sypro Ruby, arranged from left to right in the same order as the arrows in the growth curve indicating when the three samples were taken. For each protein, the bar extending to the top of the display represents the greatest abundance observed. The labels acidic and basic are used to distinguish between two protein spots derived from a single gene, acidic indicating the form of the protein with the most acidic pI when both forms exhibit similar molecular weights. Similarly, higher and lower are used when two protein spots derived from a single gene differ significantly in observed molecular weights. OD450, optical density at 450 nm.
FIG. 4.
Sequence alignment of the putative protease inhibitor SCO0762 with the previously characterized SSI protein from S. albogriseolus S-3253 (27). The P1 amino acid position known to influence substrate specificity (M preferentially for subtilisin, R or K for subtilisin and trypsin) is marked.
Other bldA-dependent proteins included one involved in the synthesis of the polyketide antibiotic actinorhodin (SCO5078; ActVA-ORF3). Although it was unexpected to find this protein outside the cytoplasm, it was not a surprise to find that it was bldA dependent, since actinorhodin production is switched off in bldA mutants (the pathway-specific regulator for this pathway, ActII-ORF4, is encoded by a TTA-containing gene [4]). Another protein encoded within a gene cluster for secondary metabolism, the putative formyltransferase SCO0499 involved in coelichelin biosynthesis, was also greatly reduced in abundance in the ΔbldA mutant sample, in full agreement with results from an analysis of the total cell proteome (A. Hesketh, unpublished data). In contrast, SCO6564 (FabH2), SCO0379 (catalase A), and a truncated form of SCO2198 (glutamine synthetase), all of which were severely reduced in the extracellular proteome of the ΔbldA mutant strain, showed no significant change in abundance in total cell protein extracts (Hesketh, unpublished data). There are no data for the abundance of 3-phosphoshikimate 1-carboxyvinyltransferase SCO5212 and the putative dehydrogenase SCO5262 in total protein extracts.
Four proteins are more abundant in the ΔbldA mutant.
Interestingly, four protein spots were significantly more abundant in the bldA mutant (Fig. 3C). One (SCO2780) was predicted to be a siderophore-binding lipoprotein, and its pattern of abundance was quite different in the extracellular fraction from that observed in the membrane fraction where it tended to decrease with increasing culture age (Kim et al., submitted). Because it decreased in the membrane fraction with time, simultaneously increasing in the extracellular fraction, it is tempting to think that the extracellular form originated from the membrane-bound form and that this release was more effective in the bldA mutant, either because of a physiological difference from the wild type or because of some as yet undiscovered difference in the cell wall. This could also be the case for the hypothetical membrane-associated protein SCO6621, which was more abundant in the culture supernatant of the bldA mutant than the parent strain but not significantly different in membrane fractions (data not shown). The abundance of another up-regulated spot, SCO3123, which was predicted to be ribose phosphate pyrophosphokinase (nucleic acid biosynthesis), was not significantly altered in total cellular proteins of the bldA mutant strain (Hesketh, unpublished data). SCO3790, although not annotated as being a secreted protein, could possess a convincing N-terminal signal sequence depending on which of the alternative translational start sites present in the gene is used (see above). The observed increase of this protein in the extracellular proteome of the bldA mutant might therefore have resulted from a change in the regulation of transcription or translation of this open reading frame.
Further characterization of SCO0762.
Among the four secreted proteins down-regulated in the bldA mutant, the serine-protease inhibitor SCO0762 was particularly noteworthy both because of its apparent complete dependence on bldA and because of the existence of previous evidence implicating trypsin-like proteases in the formation of aerial mycelium in several streptomycetes, including S. coelicolor (15, 34). This raised the possibility that the defects in morphological differentiation in the bldA mutant could be attributable to an inability to produce SCO0762. The function and bldA dependence of this protein were consequently investigated in more detail.
(i) Transcription of the protease-inhibitor gene SCO0762 depends on adpA, a TTA-containing regulatory gene that plays a key role in the developmental biology of S. coelicolor.
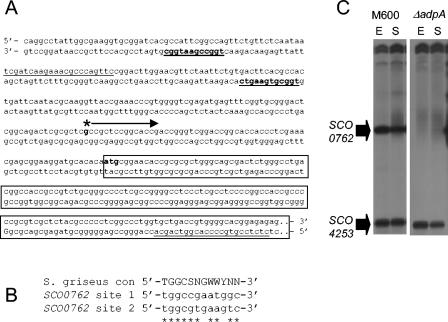
Analysis of the DNA sequence upstream of the gene SCO0762 revealed two motifs closely resembling the consensus sequence determined as being necessary for binding of the AdpA regulatory protein of Streptomyces griseus, a protein that plays a key role in differentiation and secondary metabolism (41) (Fig. 5A and B). The adpA gene in S. coelicolor contains a TTA codon, raising the possibility that the observed defect in production of SCO0762 in strain M600 ΔbldA is directly linked to an absence of AdpA. In order to investigate this further, transcription of the SCO0762 gene was analyzed in an adpA deletion mutant using S1 nuclease protection (Fig. 5C). In the M600 parent strain, transcription of the SCO0762 gene was readily detected in both exponential and stationary phase cultures, with transcript abundance decreasing slightly at the later time. In the ΔadpA mutant, the SCO0762 gene transcription was completely absent, while transcription of the SCO4253 gene, used as an internal control for RNA quality, was similar to that observed in strain M600.
FIG. 5.
Sequence and transcription analysis of the SCO0762 gene. (A) Sequence upstream of the SCO0762 gene, indicating the transcription start point (asterisk with arrow), the N terminus of SCO0762 (boxed sequence), the primers used to generate the S1 probe (sequence underlined), and putative AdpA-binding sites (in bold and underlined). (B) Comparison of the putative AdpA-binding sequences upstream of the SCO0762 gene with the consensus sequence determined for AdpA in S. griseus (41). SCO0762 site 1 is designated as the more upstream of the two putative AdpA-binding motifs, and site 2 is designated as the motif closer to the ATG start codon. (C) Transcription of the SCO0762 gene during growth of strains M600 (adpA+) and M600 ΔadpA in SMM. RNA was isolated during the mid-exponential (E) and early stationary (S) phases of growth and subjected to S1 nuclease protection analysis by using a uniquely end-labeled PCR-generated probe. Transcription of the SCO4253 gene was also analyzed as an internal control for RNA quality.
(ii) A mutant lacking the gene SCO0762 is defective in trypsin inhibitory activity but differentiates normally.
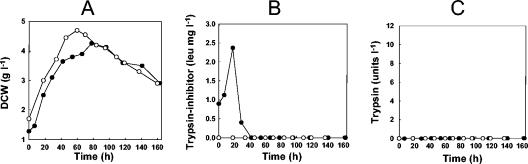
The presence of a TTA codon in the adpA gene of S. coelicolor has recently been implicated as being solely responsible for the morphological phenotype of bldA mutants (24, 35). The observation that transcription of the SCO0762 gene is completely dependent on adpA raised the possibility that the absence of this protease inhibitor in the adpA (and bldA) mutant is at least partly responsible for the defects in differentiation. To test this hypothesis, the SCO0762 gene was deleted from strain M600 by PCR-directed mutagenesis, replacing the entire open reading frame with an apramycin resistance cassette. No changes in the gross morphological phenotype during growth on MS medium or SMM were observed in the deletion mutant, which appeared to sporulate normally. The parent and mutant strains were also cultivated in liquid SMM in fermentors, and samples were assayed throughout growth for trypsin and trypsin-inhibitory activities. Dry cell weight measurements indicated that the strains grew similarly (Fig. 6A), but a significant peak in trypsin-inhibitory activity detected in the parent strain M600 was completely absent in the mutant strain lacking the SCO0762 gene(Fig. 6B), verifying that SCO0762 does indeed encode such an activity. No trypsin-like activity could be detected in either strain at any time-point (Fig. 6C).
FIG. 6.
Analysis of strain M600 and strain M600 lacking the SCO0762 gene grown in stirred jar fermentors. (A) Biomass accumulation with time, measured by dry cell weight (DCW). (B) Trypsin-protease inhibitory activity in culture supernatants measured by assaying the percent inhibition of digestion of the synthetic substrate BAEE by trypsin. Values are expressed in leupeptin units following comparison to a standard inhibition curve determined for leupeptin (see Materials and Methods). (C) Trypsin-protease activity in culture supernatants measured by assaying the liberation of ρ-nitroanilide from the synthetic substrate BAPNA. One unit of hydrolytic activity was defined as the amount of enzyme needed to produce 1 μmol of ρ-nitroanilide per minute at 35°C (30).
DISCUSSION
Unexpected aspects of the extracellular proteome of strain M600.
The accumulation of extracellular proteins in liquid SMM was strikingly growth phase dependent, with significant quantities of precipitated protein being obtained only from transition phase and stationary phase cultures (Fig. 1). This possibly reflects a requirement for cultures to reach a high cell density before translocated proteins begin to accumulate significantly, but it could also result from cell lysis occurring as exponential growth ceases. The occurrence of some cell lysis was indicated by the observation that the majority of the proteins identified in the extracellular samples were predicted to be cytosolic (see Table 1).
Enzymes of primary metabolism found in the extracellular proteome.
Orthologues of a number of the predicted cytosolic proteins have previously been identified in the extracellular proteomes of other organisms, perhaps suggesting an as yet uncharacterized additional extracellular function for at least some of them. For example, glutamine synthetase was detected extracellularly in Corynebacterium glutamicum (6); aconitase, malate dehydrogenase, dihydrolipoamide dehydrogenase, and peptidyl-prolyl cis-trans isomerase were detected in the plant pathogen Xylella fastidiosa (32); catalase, aminopeptidase, and inosine-5′-monophosphate dehydrogenase were detected in Pseudomonas aeruginosa (26); and catalase and glyceraldehyde-3-phosphate dehydrogenase were detected in Bacillus subtilis (10). While many of the proteins listed in Table 1 are abundant in the cytosol, and thus could be expected to appear in culture supernatants as a result of partial cell lysis, others are not. The tRNA synthetases SCO3792 and SCO3961 appear as fairly faint spots on analysis of total cellular protein extracts, as do the putative 3-phosphoshikimate 1-carboxyvinyltransferase SCO5212 predicted to be involved in amino acid metabolism, the prolyl aminopeptidase SCO0805, and FabH2 (SCO6564) from fatty acid biosynthesis (S. coelicolor proteome database at http://dbk.ch.umist.ac.uk/StreptoBASE/s_coeli/referencegel/). Furthermore, the putative glycine dehydrogenase SCO1378, protease SCO2549, putative dehydrogenase SCO5262, and the hypothetical proteins SCO3790 and SCO6621 were not observed at all in the cellular proteome. Conversely, many proteins known to be abundant intracellularly, such as the ribosomal proteins, elongation factor Tu-1, and the chaperonins DNAK and GroEL 1 and 2, were not detected in the extracellular proteome. Together, these data suggest that cell lysis alone cannot account for the observed presence in culture supernatants of all the predicted intracellular proteins listed in Table 1. Indeed, surface localization of glyceraldehyde-3-phosphate dehydrogenase in Mycoplasma genitalium is proposed to play a role in host colonization as an adhesin to mucin (1). This was taken as an example of an organism with a limited genome size maximizing its resources but sets a precedent that could equally well be true of genomically more well-endowed microorganisms.
The extracellular subproteome includes some enzymes of secondary metabolism.
The unexpected extracellular location of SCO5078, a tailoring enzyme from the actinorhodin biosynthetic cluster, could be the result of cell lysis, as discussed earlier, or could indicate that the late tailoring steps in the pathway take place extracellularly. If the latter were the case, other equally abundant tailoring enzymes in the pathway would also have been expected to be identified in this study. Thus, the products of SCO5071-3, SCO5075, SCO5079-80, and SCO5090 were readily detected in whole-cell extracts (7, 8) but were not found in the extracellular proteome. This makes it unlikely that all the late tailoring steps in the synthesis of actinorhodin are extracellular, though it is difficult to entirely exclude differential stability of the relevant proteins in the extracellular environment. SCO5074 from the actinorhodin cluster has previously been shown to be translocated into the cell wall in S. coelicolor (8) and was also abundant in culture supernatants analyzed for proteins in the pI 6 to 11 range (data not shown). It therefore appears that biosynthesis of actinorhodin does involve at least one extracellular protein, and it may be that SCO5078 also acts in part in this way. Like SCO5074, SCO5078 is predicted to be responsible for a tailoring enzymatic reaction occurring late in the biosynthesis of actinorhodin. Evidence for the existence of a C-terminal truncated form of SCO5078 in strain M600 was previously reported (7), but MALDI-TOF analysis revealed tryptic peptides representing amino acids 25 to 279 (out of 281) in the spot detected in this study, indicating that it was not modified in this way. Another example of a “secondary metabolism” protein occurring unexpectedly in the extracellular fraction was SCO0499, which belongs to the secondary metabolism cluster responsible for producing the siderophore coelichelin. This protein, a predicted formyltransferase, has also been detected in the membrane proteome (Kim et al., submitted) and in total cell protein extracts (Hesketh, unpublished data).
Comparatively few predicted secreted proteins were identified in these experiments.
Eleven of the 47 genes whose products were identified in the extracellular proteome had been predicted to encode secreted proteins. This is a very small fraction of the 819 secreted proteins predicted in the annotation of the complete genome (3). Among a host of possible explanations for this low detection rate, we point out three that might affect many genes under our experimental conditions: repression by glucose; the absence of specific inducers; and growth in liquid medium, rather than on a solid matrix like soil, to which streptomycetes are highly adapted.
Changes in the extracellular proteome are a previously unobserved consequence of bldA mutation.
A major purpose of this work was to explore the possibility that a bldA mutation might affect aspects of stationary phase biology other than secondary metabolism and morphological differentiation. Our intuition was that much of the extracellular proteome might be specific for stationary phase, and based on the results presented here, this appears to be true. Extracellular protein products from 11 genes were significantly less abundant in the bldA mutant (Fig. 3A and B), including 4 of the 11 proteins detected that were predicted to encode secreted proteins. The bldA-dependent, predicted secreted proteins tended to decrease in abundance as stationary phase progressed (Fig. 3A). In contrast, the other bldA-dependent proteins (except SCO5212, an enzyme involved in amino acid metabolism) accumulated with culture age (Fig. 3B). Two of these secreted proteins are of completely unknown function, while the third is predicted to be involved in glycerol metabolism, potentially being involved in recovery of carbon from phospholipids. Although all were more than twofold reduced in the ΔbldA mutant, none corresponded to genes containing a TTA codon. The simplest explanation for this apparent paradox is that the genes are subject to the influence of one or more TTA-containing regulatory genes. An example of this is discussed below.
We were surprised to find no products of TTA-containing genes in the extracellular fraction, especially in view of the prediction that 15 such genes encode secreted proteins. The most likely of several possible explanations is that these genes are not expressed under the conditions used, perhaps requiring specific induction by molecules absent from the minimal medium employed.
The bldA-dependent extracellular proteome includes at least one protein that depends on the pleiotropic developmental regulatory gene adpA.
The secreted protease inhibitor SCO0762 was completely absent in the ΔbldA mutant. Sequence comparison with the well-characterized inhibitor Sal-SSI strongly suggested that SCO0762 is an inhibitor of trypsin-like proteases, an inference that received very strong experimental support from the finding that such inhibitory activity was detected readily in the wild type but was undetectable in the mutant lacking the SCO0762 gene. The possible consequent release of a protease from inhibition in the bldA mutant might explain why the yield of extracellular proteins from this strain was significantly reduced compared to the parent (Fig. 1). A similar comparison of the yield of protein from membrane preparations showed no significant difference between the two strains (data not shown). Increased effective protease activity might also explain why some membrane-associated proteins seemed to be more readily released into the extracellular medium in the bldA mutant. Production of a trypsin-like protease immediately prior to the onset of aerial mycelium formation has been observed in several Streptomyces strains, including S. coelicolor, and it has been proposed that this activity is essential for morphological differentiation (13). The protease activity is believed to participate in the degradation of substrate mycelium, providing nutrients for aerial mycelial growth and sporulation. In Streptomyces exfoliatus SMF13 the trypsin-like protease activity is regulated by an inhibitor (leupeptin) that is in turn subject to specific inactivation by a proteolytic enzyme (13, 15). In Streptomyces antibioticus a trypsin-like protease processes an exocellular nuclease into a form that is active for DNA degradation in the substrate mycelium (25). This protease activity can also be inhibited by leupeptin. The possibility that SCO0762 forms part of a similar system important for differentiation in S. coelicolor was therefore investigated by construction and analysis of a deletion mutant. The mutant was clearly defective in the production of protease-inhibitory activity (Fig. 6B), but it sporulated normally on agar-grown cultures. However, no trypsin-like protease activity could be detected in the fermentation studies (Fig. 6C), and it is therefore unclear what SCO0762 acts upon and whether its role is connected to morphological differentiation or to another sphere of influence of bldA on stationary phase physiology. This will be the subject of future studies, in which surface-grown colonies will be analyzed.
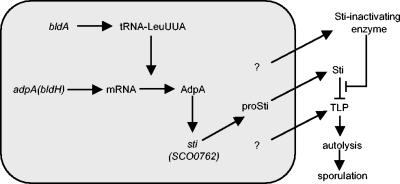
In common with the three other genes for secreted proteins down-regulated in strain M600 ΔbldA, the SCO0762 gene does not contain a TTA codon that could account for its observed bldA dependence. Examination of the DNA sequence immediately upstream of the SCO0762 gene, however, revealed two potential binding sites for the AdpA regulatory protein that is encoded by a TTA-containing gene. An M600 ΔadpA mutant was completely defective in transcription of the SCO0762 gene (Fig. 5), linking the observed absence of SCO0762 in strain M600 ΔbldA directly to the TTA-containing regulator AdpA. It is interesting that the Sal-SSI gene in S. albogriseolus S-3253 possesses a 5′-TGGCGTGAAGTG-3′ sequence just upstream of its transcription start point (27) that is in very good agreement with the AdpA-binding consensus motif. The SSI gene in Streptomyces venezuelae also has a potential (although less well conserved) AdpA-binding motif sequence of 5′-TGGCGCGCTGCC-3′ approximately 50 bp upstream of its transcription start site (38). It therefore seems possible that the regulation of this protease-inhibitory activity by AdpA is conserved between at least some Streptomyces strains. Figure 7 illustrates a cascade of events based on these observations and reports from the literature that could link bldA to autolysis of substrate mycelia in S. coelicolor via AdpA and STI. Furthermore, Yamakazi et al. (41) identified a role for AdpA in activating transcription of at least five protease genes in S. griseus, suggesting that control of proteolytic activity is a widespread function for AdpA. We are not aware of any published data concerning the presence or absence of STI-like protease inhibitors in S. griseus, though we anticipate that such inhibitors may be widespread among streptomycetes, probably including S. griseus. Finally, we note that the possibility of a developmental role for a bldA- and bldH-dependent extracellular protease cascade could prove relevant to their participation in a cascade of (still largely undefined) extracellular interactions between various classes of bld mutants (40).
FIG. 7.
Model illustrating a proposed cascade of events linking bldA to mycelial autolysis in S. coelicolor that subsequently supplies nutrients to fuel efficient aerial mycelium formation and sporulation. The model is based on the results of the present study and of other studies (13, 15, 25). The shaded box represents the inside of a cell.
Acknowledgments
We are grateful to Mike Naldrett and the staff of the JIC Proteomics Facility for their help and assistance and to Govind Chandra for help with bioinformatics analysis of the S. coelicolor genome. We thank Meifing Tao and Eriko Takano for generously providing the M600 ΔbldA and M600 ΔadpA strains, respectively, and David Hopwood for thoughtful comments on the manuscript.
This work was funded by BBSRC grant EGH16080, by a grant-in-aid to the John Innes Centre from the BBSRC, and by research grant (KISTEP)-M60301000019-04A0200-01510 from the Korea Institute of Science and Technology Evaluation and Planning. Dae-Wi Kim is also supported by a BK21 Research fellowship from the Korea Ministry of Education and Human Resource Development.
REFERENCES
- 1.Alvarez, R. A., M. W. Blaylock, and J. B. Baseman. 2003. Surface localized glyceraldehyde-3-phosphate dehydrogenase of Mycoplasma genitalia binds mucin. Mol. Microbiol. 48:1417-1425. [DOI] [PubMed] [Google Scholar]
- 2.Aoyagi, T. 1989. Protease inhibitor and biological control, p. 403-418. In M. E. Bushell, and U. Grafe (ed.), Bioactive metabolites from microorganisms: progress in industrial microbiology, vol. 27. Elsevier, New York, N.Y. [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A.-M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C.-H. Huang, T. Kieser, L. Larke, L. Murphey, K. Oliver, S. O'Niel, E. Rabbinowitsch, M.-A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 4.Fernandez-Moreno, M. A., J. L. Caballero, D. A. Hopwood, and F. Malpartida. 1991. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell 66:769-780. [DOI] [PubMed] [Google Scholar]
- 5.Gust, B., G. L. Challis, K. Fowler, T. Kieser, and K. F. Chater. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. USA 100:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hermann, T., W. Pfefferle, C. Baumann, E. Busker, S. Schaffer, M. Bott, H. Sahm, N. Dusch, J. Kalinowski, A. Puhler, A. K. Bendt, R. Kramer, and A. Burkovski. 2001. Proteome analysis of Corynebacterium glutamicum. Electrophoresis 22:1712-1723. [DOI] [PubMed] [Google Scholar]
- 7.Hesketh, A., and K. F. Chater. 2003. Evidence from proteomics that some of the enzymes of actinorhodin biosynthesis have more than one form and may occupy distinctive cellular locations. J. Ind. Microbiol. Biotechnol. 30:523-529. [DOI] [PubMed] [Google Scholar]
- 8.Hesketh, A. R., G. Chandra, A. D. Shaw, J. J. Rowland, D. B. Kell, M. J. Bibb, and K. F. Chater. 2002. Primary and secondary metabolism, and post-translational protein modifications, as portrayed by proteomic analysis of Streptomyces coelicolor. Mol. Microbiol. 46:917-932. [DOI] [PubMed] [Google Scholar]
- 9.Hirono, S., H. Akagawa, T. Mitsui, and Y. Iitaka. 1984. Crystal structure at 2.6 A resolution of the complex of subtilisin BPN′ with Streptomyces subtilisin inhibitor. J. Mol. Biol. 178:389-413. [DOI] [PubMed] [Google Scholar]
- 10.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146:65-75. [DOI] [PubMed] [Google Scholar]
- 11.Janssen, G. R., J. M. Ward, and M. J. Bibb. 1989. Unusual transcriptional and translational features of the aminoglycoside phosphotransferase gene (aph) from Streptomyces fradiae. Genes Dev. 3:415-429. [DOI] [PubMed] [Google Scholar]
- 12.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. John Innes Centre, Norwich, United Kingdom.
- 13.Kim, I. S., S.-G. Kang, and K. J. Lee. 1995. Physiological importance of trypsin-like protease during morphological differentiation of Streptomyces spp. J. Microbiol. 33:315-321. [Google Scholar]
- 14.Kim, I. S., and K. J. Lee. 1995. Physiological roles of leupeptin and extracellular protease in mycelium development of Streptomyces exfoliatus SMF13. Microbiology 141:1017-1301. [DOI] [PubMed] [Google Scholar]
- 15.Kim, I. S., and K. J. Lee. 1996. Trypsin-like protease of Streptomyces exfoliatus SMF13, a potential agent in mycelial differentiation. Microbiology 142:1797-1806. [DOI] [PubMed] [Google Scholar]
- 16.Lawlor, E. J., H. A. Bayliss, and K. F. Chater,. 1987. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1:1305-1310. [DOI] [PubMed] [Google Scholar]
- 17.Leskiw, B. K., E. J. Lawlor, J. M. Fernandez-Abalos, and K. F. Chater. 1991. TTA codons in some genes prevent their expression in a class of developmental, antibiotic-negative, Streptomyces mutants. Proc. Natl. Acad. Sci. USA 88:2461-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacNeil, D. J., J. L. Occi, K. M. Gewain, T. MacNeil, P. H. Gibbons, C. L. Ruby, and S. J. Danid. 1992. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene 155:119-125. [DOI] [PubMed] [Google Scholar]
- 19.Merrick, M. J. 1976. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J. Gen. Microbiol. 96:299-315. [DOI] [PubMed] [Google Scholar]
- 20.Miura, K., I. Kumagai, S. Obata, S. Kojima, and S. Taguchi. 1988. Partial alteration of a protein Streptomyces subtilisin inhibitor by site-directed mutagenesis. Proc. Jpn. Acad. 64B:147-149. [Google Scholar]
- 21.Murao, S., S. Sato, and N. Muto. 1972. Studies on microbial alkaline protease inhibitor (SS-I) from Streptomyces albogriseolus S3253. 1. Isolation of alkaline protease inhibitor producing microorganisms. Agric. Biol. Chem. 36:1737-1744. [Google Scholar]
- 22.Murao, S., and S. Sato. 1973. SSI, a new alkaline protease inhibitor from Streptomyces albogriseolus S3253. Agric. Biol. Chem. 37:160-163. [Google Scholar]
- 23.Murray, M. G. 1986. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal. Biochem. 158:165-170. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, K. T., J. Tenor, H. Stettler, L. T. Nguyen, L. D. Nguyen, and C. J. Thompson. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicieza, R. G., J. Huergo, B. A. Connolly, and J. Sanchez. 1999. Purification, characterisation, and role of nucleases and serine proteases in Streptomyces differentiation. J. Biol. Chem. 274:20366-20375. [DOI] [PubMed] [Google Scholar]
- 26.Nouwens, A. S., M. D. P. Wilcox, B. J. Walsh, and S. J. Cordwell. 2002. Proteomic comparison of membrane and extracellular proteins from invasive (PA01) and cytotoxic (6206) strains of Pseudomonas aeruginosa. Proteomics 2:1325-1346. [DOI] [PubMed] [Google Scholar]
- 27.Obata, S., S. Taguchi, I. Kumugai, and K.-I. Miura. 1989. Molecular cloning and nucleotide sequence determination of gene encoding Streptomyces subtilisin inhibitor (SSI). J. Biochem. 105:367-371. [DOI] [PubMed] [Google Scholar]
- 28.Redenbach, M., H. M. Kieser, D. Danapiete, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sarath, G., R. S. De la Motte, and F. W. Wagner. 1989. Protease assay methods, p. 25-55. In R. J. Beynone and J. S. Bond (ed.). Proteolytic enzymes: a practical approach. IRL Press, Oxford, United Kingdom.
- 31.Schecter, I., and M. Berger. 1967. On the size of the active site in proteases. I. Papain. Biochim. Biophys. Res. Commun. 27:157-162. [DOI] [PubMed] [Google Scholar]
- 32.Smolka, M. B., D. Martins, F. V. Winck, C. E. Santoro, R. R. Castellari, F. Ferrari, I. J. Brum, E. Galembeck, H. Della Coletta Filho, M. A. Machado, S. Marangoni, and J. C. Novello. 2003. Proteome analysis of the plant pathogen Xylella fastidiosa reveals major cellular and extracellular proteins and a peculiar codon bias distribution. Proteomics 3:224-237. [DOI] [PubMed] [Google Scholar]
- 33.Strauch, E., E. Takano, H. A. Bayliss, and M. J. Bibb. 1991. The stringent response in Streptomyces coelicolor A3(2). Mol. Microbiol. 5:289-298. [DOI] [PubMed] [Google Scholar]
- 34.Taguchi, S., H. Kikuchi, M. Suzuki, S. Kojima, M. Terabe, K. Miura, T. Nakase, and H. Momose. 1993. Streptomyces subtilisin inhibitor-like proteins are distributed widely in streptomycetes. Appl. Environ. Microbiol. 59:4338-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takano, E., M. Tao, F. Long, M. J. Bibb, L. Wang, W. Li, M. J. Buttner, M. J. Bibb, Z. X. Deng, and K. F. Chater. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Mol. Microbiol. 50:475-486. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi, Y., Y. Satow, K. T. Nakamura, and Y. Mitsui. 1991. Refined crystal structure of the complex of subtilisin BPN′ and Streptomyces subtilisin inhibitor at 1.8 Å resolution. J. Mol. Biol. 221:309-325. [PubMed] [Google Scholar]
- 37.Takeuchi, Y., T. Nonaka, K. T. Nakamura, S. Kojima, and K.-I. Miura. 1992. Crystal structure of an engineered subtilisin inhibitor complexed with bovine trypsin. Proc. Natl. Acad. Sci. USA 89:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Mellaert, L., E. Lammertyn, S. Schacht, P. Proost, J. Van Damme, B. Wroblowski, J. Anne, T. Scarcez, E. Sablon, J. Raeymaeckers, and A. Van Broekhoven. 1998. Molecular characterization of a novel subtilisin inhibitor protein produced by Streptomyces venezuelae CBS762.70. DNA Seq. 9:19-30. [DOI] [PubMed] [Google Scholar]
- 39.Weaver, D., N. Karoonuthaisiri, H. H. Tsai, C. H. Huang, M. L. Ho, S. Gai, K. G. Patel, J. Huang, S. N. Cohen, D. A. Hopwood, C. W. Chen, and C. M. Kao. 2004. Genome plasticity in Streptomyces: identification of 1Mb TIRs in the S. coelicolor A3(2) chromosome. Mol. Microbiol. 51:1535-1550. [DOI] [PubMed] [Google Scholar]
- 40.Willey, J., J. Schwedock, and R. Losick. 1993. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes Dev. 7:895-903. [DOI] [PubMed] [Google Scholar]
- 41.Yamazaki, H., A. Tomono, Y. Ohnishi, and S. Horinouchi. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555-572. [DOI] [PubMed] [Google Scholar]