Abstract
The Klebsiella pneumoniae fimbrial adhesin, MrkD, mediates adherence to the basolateral surfaces of renal and pulmonary epithelia and to the basement membranes of tissues. Although all isolates possessing the MrkD adhesin mediate the agglutination, in vitro, of erythrocytes treated with tannic acid, the mrkD gene is not conserved within species. The ability of a plasmid-borne mrkD gene product to mediate binding to type V collagen is associated frequently with strains of K. oxytoca and rarely with strains of K. pneumoniae. In K. pneumoniae, the MrkD adhesin is located within a chromosomally borne gene cluster and mediates binding to collagen types IV and V. The plasmid-borne determinant, mrkD1P, and the chromosomally borne gene, mrkD1C, are not genetically related. Some strains of enterobacteria possess a mrkD1C allele that is associated with hemagglutinating activity but does not bind to either type IV or type V collagen.
Type 3 fimbriae are produced by many members of the Enterobacteriaceae, including Klebsiella, Enterobacter, Proteus, Providencia, and Serratia species (5, 7, 10, 19, 22, 23, 25). This fimbrial type is detected by agglutination, in vitro, of erythrocytes treated with tannic acid, and hemagglutination can occur in the presence or absence of d-mannose (7, 21). This characteristic was originally demonstrated in Klebsiella, and the associated adherence phenotype is often referred to as the mannose-resistant Klebsiella-like hemagglutination (MR/KHA) reaction (7, 24, 26). MR/KHA activity is mediated by the MrkD adhesin polypeptide of the type 3 fimbrial gene cluster (1, 5, 16), and the adhesin facilitates binding to the basement membranes of human tissues (14, 33).
The expression of type 3 fimbriae in Klebsiella requires the presence of at least six mrk genes which have been cloned and sequenced (1, 5). The gene (mrkD) encoding the fimbrial adhesin is distinct from that which encodes the major fimbrial subunit (mrkA) (1, 5). In one strain of Klebsiella pneumoniae, the MrkD adhesin has been shown to mediate adherence to human basement membrane and basolateral surfaces of renal and pulmonary epithelia (14, 16). Specifically, this adhesin has been shown to bind to type V collagen and is an extracellular matrix binding protein (32). However, we have previously demonstrated that the mrkD gene is not conserved among all fimbriate and hemagglutinating strains of Klebsiella (28). Southern hybridization analysis indicated that in one isolate of K. pneumoniae, the mrkD gene is present on a large native plasmid (16). It has not been determined whether most isolates of K. pneumoniae possess a plasmid-borne copy of the mrk gene cluster. Also, since all mrkD genes are not identical, the ability of different mrkD gene products to bind to type V collagen has not been investigated. In the studies described below, the MrkD-mediated receptor-binding specificity of type 3 fimbria-associated adhesins encoded by mrkD alleles is reported. Differences in receptor-binding specificities can be associated with variability in the amino acid sequences of the MrkD adhesin.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The sources of the clinical and environmental isolates of Klebsiella strains used in this study have previously been described (14, 15, 28). Table 1 lists the recombinant plasmids used, and these plasmids were maintained in Escherichia coli HB101 (4), JM109 (36), or DH12S (20). Transformations were performed by electroporation with an ECM600 pulse generator (BTX Inc., San Diego, Calif.), and transformants were selected following growth in L-broth supplemented with the appropriate antibiotics.
TABLE 1.
Klebsiella strains and recombinant plasmids used in this study
| Strain, vector, or plasmid | Relevant genotype or description | Source or referencea |
|---|---|---|
| K. pneumoniae | ||
| IA565 | Chromosomal and plasmid-borne mrk gene clusters; fimbriate and hemagglutinating | 9 |
| IApc35 | IA565 lacking the plasmid-borne gene cluster; fimbriate and nonhemagglutinating | 16 |
| UIR079 | Fimbriate and hemagglutinating, with no detectable plasmid | UIHC |
| 43816 | Fimbriate and hemagglutinating, with no detectable plasmid | ATCC |
| Cloning vectors | ||
| pBluescript II | Stratagene | |
| pACYC184 | New England Biolabs | |
| Recombinant plasmids | ||
| pFK10 | The plasmid-borne mrk gene cluster of K. pneumoniae IA565 cloned into pACYC184 | 1 |
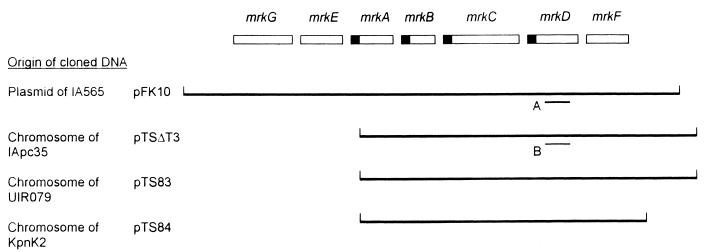
| pTSΔT3 | 6.2-kb EcoRI DNA fragment possessing mrk gene cluster from K. pneumoniae IApc35 cloned into pACYC184 | This work |
| pTS83 | 6.2-kb EcoRI DNA fragment possessing mrk gene cluster from K. pneumoniae UIR079 cloned into pBluescript II | This work |
| pTS84 | 6.0-kb EcoRI DNA fragment possessing mrk gene cluster from K. pneumoniae 43816 cloned into pBluescript II | This work |
UIHC, University of Iowa Hospital and Clinics; ATCC, American Type Culture Collection.
Optimal expression of type 3 fimbriae in Klebsiella strains was achieved by cultivation on glycerol-Casamino Acids agar as previously described (15, 16). All strains were incubated at 37°C for 18 to 24 h. Large, native plasmids from Klebsiella isolates were purified by a commercially available technique (Bigger Prep; 5 prime-3 prime, Inc., Boulder, Colo.). Plasmids were restricted with endonucleases from commercial sources according to the manufacturers’ instructions. Restriction fragments were analyzed by agarose gel electrophoresis as described elsewhere (27).
Southern hybridization analysis.
Total genomic DNA was prepared by standard procedures (3), and Southern hybridization analysis with both total genomic DNA and plasmid DNA preparations was performed as previously described (28). DNA probes were constructed by PCR amplification with relevant plasmids as the template (Table 1) and primers derived from sequences within appropriate mrk genes. The construction and use of the mrkA gene probe and one of the mrkD probes have been described elsewhere (15, 28). All DNA probes were nonradioactively labeled according to standard procedures (Genius System; Boehringer Mannheim, Indianapolis, Ind.), and hybridization was carried out under high-stringency conditions as previously described (28).
Isolation of mrkD1C genes.
The mrkD genes from K. pneumoniae UIR079 and 43816 were isolated with a gene probe derived from K. pneumoniae IApc35 (Table 1) (16). The probe was prepared with the primers 5′-TTCTGCACAGCGGTCCC-3′ and 5′-GATACCCGGCGTTTTCGTTAC-3′ and comprises 581 bp within the region flanked by mrkC and mrkF on the chromosome of K. pneumoniae IApc35 (Fig. 1, probe B). Following isolation of genomic DNA from the two K. pneumoniae strains (3), the DNA was partially digested with EcoRI, and DNA fragments of approximately 5 to 7 kb in size were isolated (27). Subsequently, these fragments were ligated into the EcoRI site of pBluescript KS (Stratagene, La Jolla, Calif.), and transformants in E. coli JM109 or DH12S were isolated by conventional techniques (27). Recombinants possessing Klebsiella-derived DNA were identified with the mrkD gene probe and by colony hybridization techniques described in detail elsewhere (3, 28).
FIG. 1.
Genetic organization of the mrk gene cluster. The Klebsiella-derived DNA fragments of recombinant plasmids are shown by the solid lines. The regions and sources of the mrkD1P-specific gene probe (A) and the mrkD1C-specific gene probe (B) are as indicated. The function of the mrk gene products has previously been reviewed (5).
The nucleotide sequences of the mrkD genes from K. pneumoniae UIR079 and 43816 were determined with the fmol sequencing reagents (Promega Corporation, Madison, Wis.) according to the manufacturer’s instructions. The plasmids pTS83 and pTS84 (Table 1 and Fig. 1) were used as templates for these reactions, and the DNA sequences of both strands were determined. The predicted amino acid sequences of the mrkD determinants were derived, and all amino acid sequence comparisons were performed with the basic local alignment search tool (BLAST) program (2) or with Eugene software from UNIX systems (Molecular Biology Information Resource, Baylor College of Medicine, Houston, Tex.).
Purification of type 3 fimbriae.
Fimbriae were purified from Klebsiella isolates as previously reported (11). Briefly, bacteria were grown under conditions to optimize fimbrial production, harvested, resuspended in 5 mM Tris-HCl (pH 7.5), and homogenized in a Waring blender at 4°C. After homogenization, the bacteria were removed by centrifugation, and following a second homogenization and centrifugation step, ammonium sulfate (10% [wt/vol]) was added to the supernatant. After 30 min at ambient temperature, the precipitate was collected by centrifugation and discarded. Ammonium sulfate (30% [wt/vol]) was added to the supernatant, and the suspension was allowed to stand at 4°C for 18 h. The precipitate was subsequently collected and dissolved in distilled water. Cesium chloride was added (42% [wt/vol]), and the protein solution was centrifuged in a vertical angle rotor for 6 h at 55,000 × g. The fimbriae were collected, concentrated, and resuspended in sterile distilled water.
Detection of type 3 fimbriae and fimbria-associated proteins.
MR/KHA activity was determined as previously described with tanned erythrocytes (26). The presence of type 3 fimbriae on the surface of bacteria was detected with fimbria-specific antiserum as described elsewhere (22, 26). Transmission electron microscopy was used to confirm phenotypic expression of type 3 fimbriae by bacteria (24, 26).
The purity and size of fimbrial polypeptides were determined by sodium dodecyl sulfate-polyacrylamide electrophoresis. Western blotting was performed by standard procedures with either anti-type 3 fimbrial serum or serum raised against a synthetic oligopeptide representing the first 10 amino acids of the mature MrkD adhesin (12, 28).
Binding to ECMPs.
An enzyme-linked immunosorbent assay was developed to demonstrate specific binding mediated by type 3 fimbriae. The wells of flat-bottom microtiter plates were coated following incubation overnight at 4°C with optimal concentrations of extracellular matrix proteins (ECMPs) diluted in carbonate-bicarbonate buffer, pH 9.6 (34). Stock solutions of commercially available purified types I, IV, V, and X collagens, fibronectin, and laminin were prepared as recommended by the manufacturer, and the optimal coating concentrations were determined as described elsewhere (32). Prior to incubation with bacteria or purified fimbriae, nonspecific binding sites were blocked by incubation for 2 h at 22°C with a 1% (wt/vol) solution of bovine serum albumin. Subsequently, 100 μl of serial twofold dilutions of either bacterial suspensions (1010 bacteria/ml) or purified type 3 fimbriae (50 μg), prepared in phosphate-buffered saline-Tween 20 (PBS-T) (pH 7.4; 0.5 ml Tween 20 in 1 liter of PBS), was added to the wells. Following incubation for 2 h at 22°C with gentle shaking, unattached bacteria were removed by washing three times in PBS-T. For each well, the adherence assay was developed with a rabbit monospecific antifimbrial serum (100 μl) diluted in PBS-T. Then, after being washed in PBS-T, 100 μl of goat anti-rabbit immunoglobulin G conjugated to alkaline phosphatase was added to the wells and allowed to incubate for 1 h at 37°C. Finally, the plates were washed thoroughly, p-nitrophenyl phosphate (5 mg/ml in diethanolamine buffer) (Sigma Chemical Co., St. Louis, Mo.) was added, and the reaction was allowed to proceed for 40 min at 37°C. All tests were performed in triplicate, and color development was determined with an enzyme-linked immunosorbent assay plate reader set to an optical density of 405 nm.
RESULTS
Plasmid-borne and chromosomally borne mrkD genes.
Of 44 strains of Klebsiella examined, all 10 isolates of K. oxytoca and 6 of 34 K. pneumoniae strains possessed a gene related to the mrkD determinant carried on pFK10 (Fig. 1 and Table 2). All of these strains express type 3 fimbriae and demonstrated the characteristic MR/KHA activity. Seven of the K. oxytoca strains were shown to possess a plasmid-borne mrkD gene, designated mrkD1P, since plasmid preparations from these strains possessed sequences homologous to the gene probe (Fig. 1, probe A). Three K. pneumoniae strains also possessed an mrkD1P determinant on a plasmid isolated from these strains. The remaining 28 isolates of K. pneumoniae were MR/KHA positive and produce type 3 fimbriae but do not possess sequences related to the mrkD gene carried on pFK10.
TABLE 2.
Genetic relatedness of mrkD genes in bacterial strains
| Bacterium | No. of strains | No. of strains exhibiting hybridization to indicated gene probe:
|
||
|---|---|---|---|---|
| mrkD1P | mrkD1C | both mrkD | ||
| K. pneumoniae | 34 | 3 | 28 | 3 |
| K. oxytoca | 10 | 10 | 0 | 0 |
| E. cloacae | 8 | 1 | 7 | 0 |
K. pneumoniae IApc35 is a derivative of K. pneumoniae IA565 that has lost its plasmid-borne copy of the mrk gene cluster but retains a copy on the chromosome (16). Strain IApc35 was used to prepare an mrkD gene probe comprising sequences from within the chromosomally borne mrkD (Fig. 1, probe B). Plasmid pTSΔT3 carries the mrkD gene, designated mrkD1C, cloned from K. pneumoniae IApc35 (Table 1 and Fig. 1). A 581-bp DNA fragment consisting solely of mrkD-derived nucleotides was used as a gene probe to detect homologous sequences among the Klebsiella strains. With this gene probe, 31 of 34 K. pneumoniae isolates could be shown to carry homologous sequences, whereas none of the type 3 fimbria-producing strains of K. oxytoca possess related nucleotide sequences (Table 2). Three of the 34 isolates of K. pneumoniae did not possess nucleotides related to the probe derived from pTSΔT3, although these three strains do retain an mrkD1P gene homologous to that carried on pFK10 (Table 2). Also, in all three of these strains, the mrkD1P gene was detected in plasmid DNA preparations. Three strains of K. pneumoniae, including strain IA565, from which the mrk gene cluster was originally cloned (9, 12), possess sequences related to both mrkD gene probes.
A total of eight fimbriate isolates of Enterobacter cloacae were also examined for the presence of mrkD genes because this genus expresses type 3 fimbriae related to those of Klebsiella (22). Seven isolates possess a gene comprised of sequences related to those derived from mrkD1C, whereas one strain carried a gene similar to the mrkD1P DNA probe. In this latter strain, the mrkD gene was carried on DNA isolated either as total genomic DNA or from plasmid preparations.
Characterization of mrkD sequences and their gene products.
K. pneumoniae IApc35 is a nonhemagglutinating derivative of IA565 (16). Therefore, this strain and K. pneumoniae UIR079, an MR/KHA-positive isolate, were used as a source of DNA to determine the nucleotide sequences of their mrkD1C genes. The mrkD gene of K. pneumoniae 43816 was also isolated, since this gene is closely related to that of strain IApc35 as determined by Southern hybridization, and the bacteria are hemagglutinating but do not bind to collagen (see below).
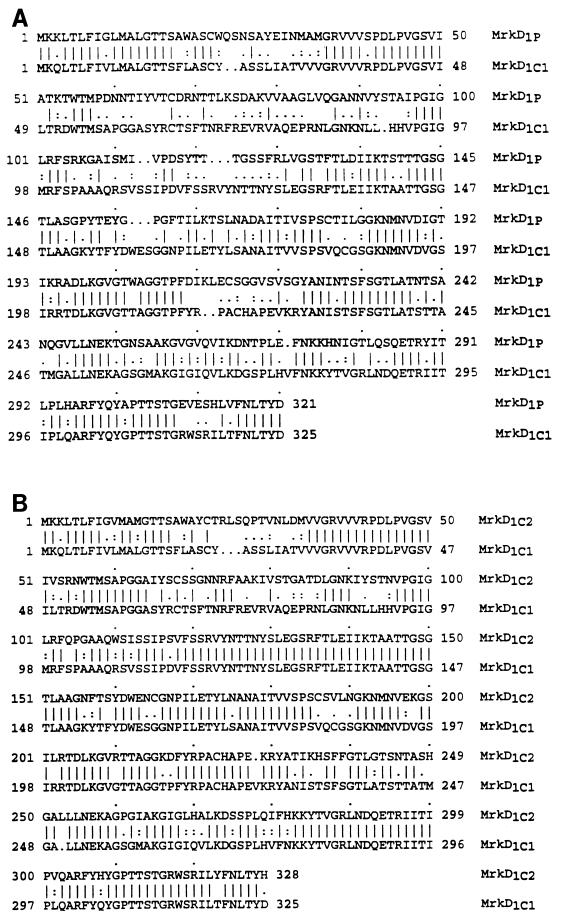
The plasmid pTS83 (Table 1 and Fig. 1) contains a 6.2-kb DNA fragment derived from K. pneumoniae UIR079 and possesses an mrkD determinant that is 978 bp in length. The predicted size of the MrkD polypeptide is 34.9 kDa, which is slightly larger than the 33.8-kDa gene product encoded by the mrkD1P gene carried on pFK10 (Fig. 1) (5). The mrkD gene of K. pneumoniae 43816 is carried on a 6.0-kb DNA fragment of plasmid pTS84 and comprises an open reading frame of 987 bp. The predicted size of the MrkD polypeptide encoded by this region is 35.4 kDa, and a comparison of the MrkD proteins of K. pneumoniae UIR079 and 43816 with that of the previously described plasmid-borne MrkD adhesin (5, 12) is shown in Fig. 2. At the amino acid level, MrkD1C of K. pneumoniae UIR079 is 55% identical to the MrkD1P polypeptide, and the two genes encoding these molecules possess 62.5% identity at the nucleotide level. A comparison of the amino acid and nucleotide sequences of the MrkD1C polypeptides and their genes from K. pneumoniae UIR079 and 43816 indicates 74 and 86% identity, respectively.
FIG. 2.
Comparison of the deduced amino acid sequences of the MrkD polypeptides. MrkD1P is the gene product encoded by a plasmid of K. pneumoniae IA565; MrkD1C1 is encoded by a chromosomally borne gene of K. pneumoniae UIR079; MrkD1C2 is the gene product of the mrkD allele located on the chromosome of K. pneumoniae 43816. Identical amino acids are indicated by solid lines, and isofunctional amino acids are represented by dots.
Bacterial binding to ECMPs.
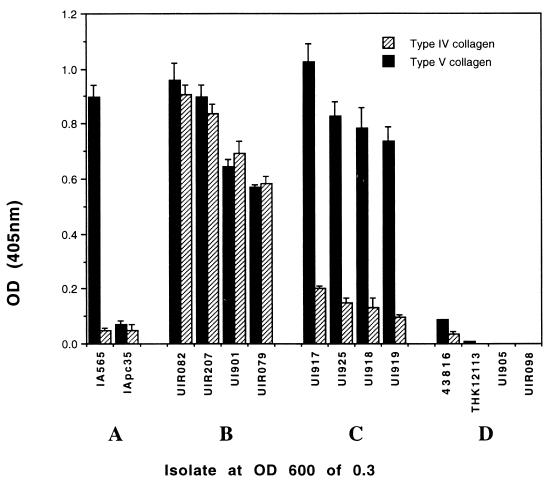
The results of the ECMP-binding assays for strains possessing the mrkD genes are summarized in Table 3 and Fig. 3. None of the Klebsiella isolates adhere, in vitro, to type I or type X collagen, fibronectin, laminin, or bovine serum albumin. All nine Klebsiella strains tested that possess the mrkD1P gene related to that carried on pFK10 bound only to type V collagen (Table 3). Nineteen of 28 K. pneumoniae strains that are type 3 fimbriate and exhibit MR/KHA but do not possess the mrkD1P gene bind to type IV and type V collagen. All of these strains possess mrkD1C sequences related to those isolated from K. pneumoniae IApc35 and UIR079. E. coli transformants possessing pTS83 also adhere to type IV and type V collagens, whereas transformants lacking the mrk gene cluster do not. Nine hemagglutinating and fimbriate isolates of K. pneumoniae, including strain 43816, possess nucleotide sequences homologous to those of strain IApc35 but did not bind to any of the ECMPs used in the assays. Six of the 10 K. oxytoca strains that were found to possess sequences homologous to the pFK10-derived mrkD gene probe were also examined for their ability to bind collagens. All six exhibited binding only to type V collagen.
TABLE 3.
Relationship between collagen binding and mrkD genotype
| Bacterium | No. of strains examined | MR/KHA | mrkD1Pa | mrkD1Ca | Binding to indicated collagen:
|
|
|---|---|---|---|---|---|---|
| Type IV | Type V | |||||
| K. pneumoniae | 19 | + | − | + | + | + |
| 9 | + | − | + | − | − | |
| 3 | + | + | − | − | + | |
| K. oxytoca | 6 | + | + | − | − | + |
| E. cloacae | 6 | + | − | + | + | + |
| 1 | + | + | − | − | + | |
Hybridization to the mrkD1P- or mrkD1C-specific gene probes.
FIG. 3.
Binding of Klebsiella strains to collagen. Binding to type V collagen is indicated by the solid bars, and type IV collagen binding is represented by the striped bars. All bacterial suspensions were normalized to an optical density at 600 nm (OD600) of 0.3 corresponding to approximately 6 × 108 bacteria/ml. (A) K. pneumoniae IA565 and its derivative, IApc35, lacking mrkD1P. (B) Representative strains possessing mrkD1C without mrkD1P. (C) Klebsiella isolates possessing mrkD1P but no mrkD1C. (D) Strains of Klebsiella with mrkD1C and no mrkD1P but not binding to collagen. For each column, the value is expressed as the mean ± standard error of the mean; all tests were performed at least three times.
As shown in Table 3, six of the seven Enterobacter strains examined possess sequences homologous to IApc35-derived mrkD1C and bound to type IV and type V collagen. One E. cloacae isolate demonstrated binding only to type V collagen, and DNA from this strain hybridized to the pFK10-derived mrkD1P gene probe.
Binding of purified fimbriae to ECMPs.
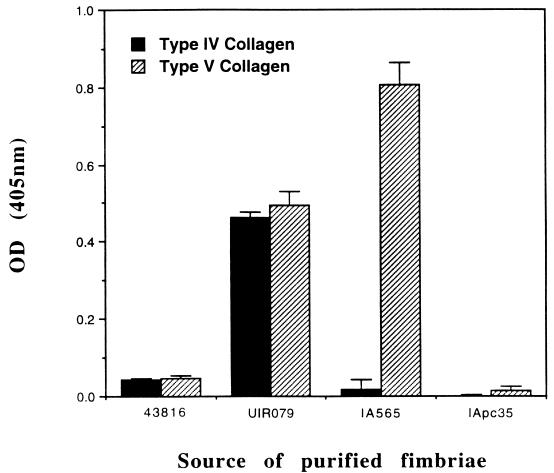
Cell-free fimbriae from four representative isolates of Klebsiella were prepared. Fimbriae from K. pneumoniae IA565 and its nonhemagglutinating derivative, IApc35, were used in the binding assay. Also, purified type 3 fimbriae from K. pneumoniae UIR079 and 43816 were prepared. The fimbriae of K. pneumoniae UIR079 binds to type IV and type V collagen, whereas fimbriae from 43816 do not bind to either collagen type, and both strains are strongly hemagglutinating. Also, both strains possess mrkD1C sequences related to those carried on the chromosome of K. pneumoniae IApc35. The pattern of collagen binding by the purified fimbriae was identical to that found for whole bacteria (Fig. 4). Fimbriae from strain IA565 bound only to type V collagen, whereas those isolated from K. pneumoniae IApc35 did not adhere to either collagen type.
FIG. 4.
Binding of purified type 3 fimbriae to collagen. Binding of type V and type IV collagen is shown by the striped and solid bars, respectively. All assays were performed with 12.5 μg of fimbrial protein. Standard errors of the means were calculated after each assay was performed at least three times.
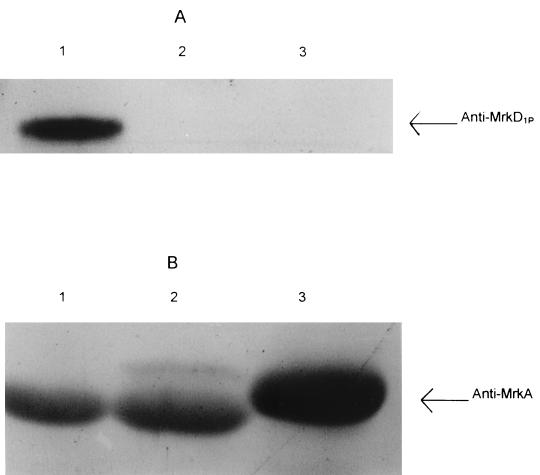
The presence of a 34-kDa polypeptide in the fimbrial preparations, encoded by the mrkD adhesin carried on pFK10, was determined with a mrkD-specific antiserum (28). Western blot analyses indicated the presence of this polypeptide in fimbrial preparations from K. pneumoniae IA565 but not in those from other strains (Fig. 5). All fimbrial preparations were recognized by serum raised against the MrkA major fimbrial subunit protein of strain IA565.
FIG. 5.
Immunoblots of purified fimbriae with anti-MrkD (A) and anti-MrkA (B) sera. Fimbriae were purified from K. pneumoniae IA565 (lane 1), K. pneumoniae IApc35 (lane 2), and K. pneumoniae UIR079 (lane 3). The anti-MrkD serum was raised against a synthetic polypeptide representing the first 10 amino acids of MrkD1P, and the anti-MrkA serum was raised against fimbriae purified from E. coli (pFK10).
DISCUSSION
We have previously demonstrated that K. pneumoniae IA565 possesses two mrk gene clusters that are located on a plasmid and the chromosome (16). Also, K. pneumoniae IApc35, a derivative of strain IA565, has lost the plasmid-borne mrk sequences but retained the chromosomal copy of the mrk gene cluster (16). Using an mrkD gene probe derived from the plasmid-borne gene cluster, we demonstrated that this determinant was not highly conserved among isolates of K. pneumoniae, even though most of these strains express type 3 fimbriae. However, this plasmid-borne mrkD gene was found with a high frequency among fimbriate strains of K. oxytoca and more rarely among K. pneumoniae isolates (28). Therefore, even though most strains of K. pneumoniae express the type 3 fimbrial MR/KHA, they must possess an MrkD adhesin that is not closely related to that encoded by the plasmid of K. pneumoniae IA565. In order to determine whether strains of K. pneumoniae possess a highly conserved mrkD gene, this determinant was isolated and characterized.
Using an mrkD gene probe derived from K. pneumoniae IApc35, a strain lacking the plasmid-borne mrk gene cluster (16), we demonstrated that most isolates of K. pneumoniae possess sequences related to those of the gene probe and that these sequences were not associated with plasmids. The results of Southern hybridization analysis indicate that fimbriate K. pneumoniae isolates do retain a highly conserved mrkD determinant that is different from that associated with the plasmid-borne adhesin gene. The plasmid-borne adhesin gene is carried more frequently by strains of K. oxytoca, often on large native plasmids. Similarly, in the small number of K. pneumoniae isolates that do possess an mrkD gene related to that carried on the recombinant plasmid pFK10, the gene is commonly carried on a plasmid. Therefore, the plasmid-borne gene found in most isolates of K. oxytoca has been designated mrkD1P, and that associated with the chromosome of K. pneumoniae is termed mrkD1C. The presence of the mrkD1P gene in a small number of K. pneumoniae strains may be due to horizontal transfer from K. oxytoca. We have previously demonstrated that the remaining mrk genes of the fimbrial gene cluster are highly conserved regardless of whether they are present on the chromosome or a plasmid in Klebsiella (15, 16). Therefore, the two genes encoding the MR/KHA of the type 3 fimbriae have undergone evolutionary divergence. Similarly, in fimbriate isolates of E. cloacae, the two distinct mrkD genes can be found, and in the strain possessing mrkD1p, this determinant is carried on a plasmid.
Although it could be demonstrated by PCR analysis that K. pneumoniae IApc35 retains a region of DNA between its chromosomally borne mrkC and mrkF genes that is approximately 1,200 nucleotides in length, this strain expresses nonadhesive fimbriae (16). If, in fact, the mrkD adhesin gene is present in this location, K. pneumoniae IApc35 should be hemagglutinating. However, nucleotide sequencing demonstrated that a translation termination codon was present in the middle of this region of DNA, and therefore this strain will synthesize a truncated MrkD polypeptide. Consequently, a hemagglutinating isolate of K. pneumoniae, strain UIR079, that possessed DNA sequences related to that of the mrkD1C gene probe was used to clone and subsequently to determine the nucleotide sequence of a functional MrkD1C adhesin. The mrkD1C and mrkD1P genes are not closely related, and this lack of similarity explains the inability of each gene probe to detect sequences from the heterologous gene. A comparison of the MrkD1C and MrkD1P polypeptides indicates significant differences in the amino acid sequences of these two molecules (Fig. 2), particularly in the N-terminal regions. A greater degree of amino acid sequence conservation is found within the C termini of the two polypeptides. The C-terminal domain of various fimbrial adhesins has been reported to possess four conserved sequence motifs that are thought to be necessary for folding and assembly (13). The MrkD adhesins share these motifs in hydrophobic clusters, which display a typical configuration strongly associated with amphipathic β-strands. As discussed below, K. pneumoniae 43816 demonstrates a distinct binding specificity associated with its type 3 fimbriae, and therefore the mrkD determinant from this isolate was also cloned and characterized. This specific gene is closely related to that of K. pneumoniae UIR079 and IApc35, and a comparison of the amino sequences of the gene products indicates a close similarity (Fig. 2). In fact, the mrkD1C genes are allelic variants, and their gene products differ at only two small regions within the N terminus of the polypeptides. This close relatedness explains why the K. pneumoniae 43816 mrkD gene is recognized by the gene probe derived from strain IApc35. The lack of reactivity of the K. pneumoniae UIR079 and 43816 MrkD proteins with serum raised against the N terminus of MrkD from strain IA565 is consistent with the observed differences in the amino acid sequences at this region. Serum raised against the major fimbrial subunit, MrkA, reacts with the MrkA polypeptides of all fimbriate strains examined (Fig. 5), confirming the serologic relatedness of the type 3 fimbriae reported by Old and Adegbola (22).
We have previously demonstrated that type V collagen-binding specificity is a function of the MrkD1P polypeptide and that all strains of Klebsiella possessing this adhesin exhibit identical binding properties (16, 32). We have now demonstrated that the possession of the MrkD1C fimbria-associated protein is frequently correlated with the ability of bacteria to bind to both type IV and type V collagen. Both fimbriate bacteria and cell-free, purified fimbriae exhibited identical binding specificities with those appendages possessing the MrkD1c polypeptide adhering to collagen type IV and type V. Interestingly, the fimbriae isolated from K. pneumoniae 43816 could not be shown to bind to any of the extracellular matrix proteins used in our assays. However, the bacteria and purified fimbriae are strongly hemagglutinating, suggesting that these appendages do function in vitro as adhesins. In fact, using the mrkD1P gene, we have previously shown that the MrkD polypeptide is responsible for MR/KHA activity and binding to type V collagen (12). Therefore, hemagglutination by K. pneumoniae 43816 is most likely a function of MrkD1C. Since the only major differences in the MrkD1C polypeptides of K. pneumoniae UIR079 and 43816 are found at two sites within the N termini of these molecules, it is possible that the observed receptor-binding specificity is a function of these regions. Currently, we are investigating this hypothesis, using site-specific mutagenesis of the two mrkD genes.
A small number of the Klebsiella isolates examined in this study, as exemplified by K. pneumoniae 43816, did not mediate binding to any of the target proteins used in our assays. It is not possible to conclude that these strains represent one group exhibiting an identical binding pattern. However, all of these strains clearly possess an mrkD gene that is closely related to mrkD1C, since these strains hybridize to this gene probe. Differences in binding specificity have been associated with allelic variation in the adhesin gene of the type 1 fimbrial gene cluster of E. coli (29–31). Also, the PapG adhesin of P fimbriae demonstrates variability in binding activity dependent upon the papG gene present in the gene cluster (17, 18). The mrkD1C genes of the type 3 fimbrial gene cluster are also comprised of at least two allelic variants. However, the plasmid-borne mrkD1p gene does not demonstrate a relatedness to the mrkD1C alleles, and the nucleotide sequences of these two genes have diverged such that the gene probes do not cross-hybridize.
In summary, we have demonstrated that most strains of K. pneumoniae produce type 3 fimbriae possessing an MrkD polypeptide different from that associated with the plasmid-encoded, type-V-collagen-binding molecule (16). This latter MrkD adhesin is found primarily in fimbriate strains of K. oxytoca and less frequently among K. pneumoniae isolates. Most strains of K. pneumoniae produce an MrkD molecule that is not encoded by a plasmid. In these strains, fimbriae carrying one type of MrkD adhesin can be shown to mediate binding to type IV and type V collagen. However, in some Klebsiella strains, another MrkD variant present in fimbriae exhibits the characteristic MR/KHA activity but cannot be shown to bind in vitro to a range of ECMPs. Investigations into the molecular biology of these naturally occurring MrkD polypeptides could provide information on the nature of the receptors recognized by type 3 fimbriae. Since type 3 fimbriate enterobacteria are frequent opportunistic pathogens of immunocompromised individuals (6, 8, 15, 35), the role of MrkD molecules in binding to damaged epithelial surfaces is currently being investigated.
REFERENCES
- 1.Allen B L, Gerlach G-F, Clegg S. Nucleotide sequence and functions of mrk determinants necessary for expression of type 3 fimbriae in Klebsiella pneumoniae. J Bacteriol. 1991;173:916–920. doi: 10.1128/jb.173.2.916-920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 4.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 5.Clegg S, Korhonen K T, Hornick B D, Tarkkanen A-M. Type 3 fimbriae of the Enterobacteriaceae. In: Klem P, editor. Fimbriae: adhesion, genetics, biogenesis, and vaccines. Boca Raton, Fla: CRC Press; 1994. pp. 97–104. [Google Scholar]
- 6.Craven D E, Barber T W, Steger K A, Montecalvo M A. Nosocomial pneumonia in the 1990s: update of epidemiology and risk factors. Semin Respir Infect. 1990;5:157–172. [PubMed] [Google Scholar]
- 7.Duguid J P. Fimbriae and adhesive properties in Klebsiella strains. J Gen Microbiol. 1959;21:271–286. doi: 10.1099/00221287-21-1-271. [DOI] [PubMed] [Google Scholar]
- 8.Duma R J. Gram-negative bacillary infections. Pathogenic and pathophysiologic correlates. Am J Med. 1985;78:154–164. doi: 10.1016/0002-9343(85)90119-6. [DOI] [PubMed] [Google Scholar]
- 9.Gerlach G-F, Allen B L, Clegg S. Molecular characterization of the type 3 (MR/K) fimbriae of Klebsiella pneumoniae. J Bacteriol. 1988;170:3547–3553. doi: 10.1128/jb.170.8.3547-3553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlach G-F, Allen B L, Clegg S. Type 3 fimbriae among enterobacteria and the ability of spermidine to inhibit MR/K hemagglutination. Infect Immun. 1989;57:219–224. doi: 10.1128/iai.57.1.219-224.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gerlach G F, Clegg S. Characterization of two genes encoding antigenically distinct type-1 fimbriae of Klebsiella pneumoniae. Gene. 1988;64:231–240. doi: 10.1016/0378-1119(88)90338-1. [DOI] [PubMed] [Google Scholar]
- 12.Gerlach G F, Clegg S, Allen B L. Identification and characterization of the genes encoding the type 3 and type 1 fimbrial adhesins of Klebsiella pneumoniae. J Bacteriol. 1989;171:1262–1270. doi: 10.1128/jb.171.3.1262-1270.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girardeau J P, Bertin Y. Pilins of fimbrial adhesins of different member species of Enterobacteriaceae are structurally similar to the C-terminal half of adhesin proteins. FEBS Lett. 1995;357:103–108. doi: 10.1016/0014-5793(94)01340-7. [DOI] [PubMed] [Google Scholar]
- 14.Hornick D B, Allen B L, Horn M A, Clegg S. Adherence to respiratory epithelia by recombinant Escherichia coli expressing Klebsiella pneumoniae type 3 fimbrial gene products. Infect Immun. 1992;60:1577–1588. doi: 10.1128/iai.60.4.1577-1588.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hornick D B, Allen B L, Horn M A, Clegg S. Fimbrial types among respiratory isolates belonging to the family Enterobacteriaceae. J Clin Microbiol. 1991;29:1795–1800. doi: 10.1128/jcm.29.9.1795-1800.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornick D B, Thommandru J, Smits W, Clegg S. Adherence properties of an mrkD-negative mutant of Klebsiella pneumoniae. Infect Immun. 1995;63:2026–2032. doi: 10.1128/iai.63.5.2026-2032.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson J R, Brown J J. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the Gal(alpha 1–4)Gal-binding PapG adhesins of Escherichia coli. J Infect Dis. 1996;173:920–926. doi: 10.1093/infdis/173.4.920. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J R, Swanson J L, Barela T J, Brown J J. Receptor specificities of variant Gal(alpha 1–4)Gal-binding PapG adhesins of uropathogenic Escherichia coli as assessed by hemagglutination phenotypes. J Infect Dis. 1997;175:373–381. doi: 10.1093/infdis/175.2.373. [DOI] [PubMed] [Google Scholar]
- 19.Korhonen T K, Tarkka E, Ranta H, Haahtela K. Type 3 fimbriae of Klebsiella sp.: molecular characterization and role in bacterial adhesion to plant roots. J Bacteriol. 1983;155:860–865. doi: 10.1128/jb.155.2.860-865.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J J, Smith M, Jessee J, Bloom F. DH11S: an Escherichia coli strain for preparation of single-stranded DNA from phagemid vectors. BioTechniques. 1992;12:718–721. [PubMed] [Google Scholar]
- 21.Ofek I, Beachey E H. Bacterial adherence. Adv Intern Med. 1980;25:503–532. [PubMed] [Google Scholar]
- 22.Old D C, Adegbola R A. Antigenic relationships among type-3 fimbriae of Enterobacteriaceae revealed by immunoelectronmicroscopy. J Med Microbiol. 1985;20:113–121. doi: 10.1099/00222615-20-1-113. [DOI] [PubMed] [Google Scholar]
- 23.Old D C, Adegbola R A. Haemagglutinins and fimbriae of Morganella, Proteus and Providencia. J Med Microbiol. 1982;15:551–564. doi: 10.1099/00222615-15-4-551. [DOI] [PubMed] [Google Scholar]
- 24.Old D C, Adegbola R A. A new mannose-resistant haemagglutinin in Klebsiella. J Appl Bacteriol. 1983;55:165–172. doi: 10.1111/j.1365-2672.1983.tb02661.x. [DOI] [PubMed] [Google Scholar]
- 25.Old D C, Scott S S. Hemagglutinins and fimbriae of Providencia spp. J Bacteriol. 1981;146:404–408. doi: 10.1128/jb.146.1.404-408.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Old D C, Tavendale A, Senior B W. A comparative study of the type-3 fimbriae of Klebsiella species. J Med Microbiol. 1985;20:203–214. doi: 10.1099/00222615-20-2-203. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 28.Schurtz T A, Hornick D B, Korhonen T K, Clegg S. The type 3 fimbrial adhesin gene (mrkD) of Klebsiella species is not conserved among all fimbriate strains. Infect Immun. 1994;62:4186–4191. doi: 10.1128/iai.62.10.4186-4191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokurenko E V, Chesnokova V, Doyle R J, Hasty D L. Diversity of the Escherichia coli type 1 fimbrial lectin. Differential binding to mannosides and uroepithelial cells. J Biol Chem. 1997;272:17880–17886. doi: 10.1074/jbc.272.28.17880. [DOI] [PubMed] [Google Scholar]
- 30.Sokurenko E V, Courtney H S, Maslow J, Siitonen A, Hasty D L. Quantitative differences in adhesiveness of type 1 fimbriated Escherichia coli due to structural differences in fimH genes. J Bacteriol. 1995;177:3680–3686. doi: 10.1128/jb.177.13.3680-3686.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sokurenko E V, Courtney H S, Ohman D E, Klemm P, Hasty D L. FimH family of type 1 fimbrial adhesins: functional heterogeneity due to minor sequence variations among fimH genes. J Bacteriol. 1994;176:748–755. doi: 10.1128/jb.176.3.748-755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarkkanen A M, Allen B L, Westerlund B, Holthofer H, Kuusela P, Risteli L, Clegg S, Korhonen T K. Type V collagen as the target for type-3 fimbriae, enterobacterial adherence organelles. Mol Microbiol. 1990;4:1353–1361. doi: 10.1111/j.1365-2958.1990.tb00714.x. [DOI] [PubMed] [Google Scholar]
- 33.Tarkkanen A-M, Virkola R, Clegg S, Korhonen T K. Binding of the type 3 fimbriae of Klebsiella pneumoniae to human endothelial and urinary bladder cells. Infect Immun. 1997;65:1546–1549. doi: 10.1128/iai.65.4.1546-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voller A, Bidwell D, Bartlett A. Enzyme-linked immunosorbent assay. In: Rose N R, Friedman H, editors. Manual of clinical immunology. 2nd ed. Washington, D.C: ASM Press; 1980. pp. 359–371. [Google Scholar]
- 35.Williams P, Tomas J M. The pathogenicity of Klebsiella pneumoniae. Rev Med Microbiol. 1990;1:196–204. [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]