Abstract
mRNA‐based molecular subtypes have implications for bladder cancer prognosis and clinical benefit from certain therapies. Whether small extracellular vesicles (sEVs) can reflect bladder cancer molecular subtypes is unknown. We performed whole transcriptome RNA sequencing for formalin fixed paraffin embedded (FFPE) tumour tissues and sEVs separated from matched tissue explants, urine and plasma in patients with bladder cancer. sEVs were separated using size‐exclusion chromatography, and characterized by transmission electron microscopy, nano flow cytometry and western blots, respectively. High yield of sEVs were obtained using approximately 1 g of tissue, incubated with media for 30 min. FFPE tumour tissue and tumour tissue‐derived sEVs demonstrated good concordance in molecular subtype classification. All urinary sEVs were classified as luminal subtype, while all plasma sEVs were classified as Ba/Sq subtype, regardless of the molecular subtypes indicated by their matched FFPE tumour tissue. The comparison within urine sEVs, which may exclude the sample type specific background, could pick up the different biology between NMIBC and MIBC, as well as the signature genes related to molecular subtypes. Four candidate sEV‐related bladder cancer‐specific mRNA biomarkers, FAM71E2, OR4K5, FAM138F and KRTAP26‐1, were identified by analysing matched urine sEVs, tumour tissue derived sEVs, and adjacent normal tissue derived sEVs. Compared to sEVs separated from biofluids, tissue‐derived sEVs may reflect more tissue‐ or disease‐specific biological features. Urine sEVs are promising biomarkers to be used for liquid biopsy‐based molecular subtype classification, but the current algorithm needs to be modified/adjusted. Future work is needed to validate the four new bladder cancer‐specific biomarkers in large cohorts.
Keywords: bladder cancer, extracellular vesicles, liquid biopsy, molecular subtype, tissue derived EV
1. INTRODUCTION
Bladder cancer (BCa) is the most common cancer type in the urinary system in both males and females. More than 82,000 new cases and 16,000 BCa‐caused deaths were estimated to occur in the United States in 2023 (Siegel et al., 2023). Approximately 80% of newly diagnosed BCa cases are non‐muscle invasive BCa (NMIBC) which has limited lethal potential, and is commonly treated by transurethral resection (TUR), followed by intravesical chemotherapy or Bacillus Calmette‐Guérin (BCG) instillations. Instead, muscle invasive BCa (MIBC), counting for the rest 20%, is characterized by rapid progression to metastatic disease and is associated with poor prognosis. Radical cystectomy with or without neoadjuvant platinum‐based chemotherapy is the standard treatment modalities for MIBC (Kamat et al., 2016; Knowles & Hurst, 2015). Besides the depth of invasion, in recent years, mRNA‐based molecular subtypes have been established by different research groups to describe distinct biological features of BCa (Cancer Genome Atlas Research Network, 2014; Choi et al., 2014; Damrauer et al., 2014; Robertson et al., 2017). To overcome the diversity of MIBC classifications by different research groups, a consensus classification system was developed, and an R package is now publicly available to assign single tumour to one of the six consensus subtypes: luminal‐papillary (LumP), luminal‐nonspecified (LumNS), luminal‐unstable (LumU), Stroma‐rich, basal/squamous (Ba/Sq), and neuroendocrine‐like (NE‐like) (Kamoun et al., 2020). BCa molecular subtype has implications for prognosis and clinical benefit from certain therapies. For example, the Ba/Sq tumours are associated with squamous differentiation and frequent mutations of TP53 and RB1. Patients with Ba/Sq tumours have poor survival outcomes, but seem to be good candidates for chemotherapy and immunotherapy (Necchi et al., 2020; Seiler et al., 2017). Since LumP tumours are enriched with papillary histological morphology and activation of the FGFR3 transcriptional pathway, patients with LumP tumours may be good candidates for FGFR3 targeted therapy (McConkey, 2021). The vast majority of NMIBCs can be assigned to luminal tumours (Fong et al., 2020). Currently, tissue‐based mRNA sequencing is the only way to identify the molecular subtype of BCa, which has largely limited the preoperative use of this essential prognostic biomarker. There has been an unmet need for developing non‐invasive liquid biopsy‐based molecular subtype classification for BCa management.
Extracellular vesicles (EVs) are lipid bilayer encapsulated particles that can be released by all living cells into the extracellular space (Kalluri & LeBleu, 2020). EV molecular cargos include, but are not limited to, proteins, metabolites, DNAs, and RNAs (mRNAs, miRNAs, lncRNAs, etc.) (Malloci et al., 2019; Yokoi et al., 2019). EVs are considered to play an important role in cell‐to‐cell communication by transfer of their cargo to target cells, thus, participating in a wide range of physiological and pathological processes (Dong et al., 2019). Since EVs can be found in almost all types of biofluids, the last decade has witnessed a robust increase in the utility of EVs as biomarkers in liquid biopsies. The most commonly used biofluid samples for EV separation in urological cancer research are plasma and urine (Roy et al., 2018). However, plasma EVs and urinary EVs are a mixture of EVs from different origin. It is hard to identify organ‐specific or disease‐specific EVs in these biofluids (Dong et al., 2020; Karimi et al., 2018; Webber & Clayton, 2013). The target EV “signal” can be easily lost due to the sample type‐specific background “noise”. In recent years, fresh tissue derived EVs have gained an increasing level of attention (Li et al., 2021). These EVs are considered to be released from extracellular interstitium when the tissue explants are dissociated ex vivo (Kuczler et al., 2022). The other hypothesis is that the cells can still secrete EVs in ex vivo culture conditions shortly after the dissection. Compared to plasma or urinary EVs, tissue‐derived EVs contain minimal contaminations of EVs from other origin, thus, can more directly reflect the biological features of certain organ or disease (Li et al., 2021; Webber & Clayton, 2013). Hoshino and colleagues investigated the proteomic profile of EVs in 426 human samples from tissue explants and biofluids, and identified cancer‐specific EV biomarkers for cancer detection (Hoshino et al., 2020).
In this study, we performed whole transcriptome RNA sequencing for formalin fixed paraffin embedded (FFPE) tumour tissues and small EVs (sEVs) separated from matched tissue explants, urine and plasma in patients with BCa. We investigated whether the tissue‐based molecular subtype and other cancer biological features can be identified by sEVs from different sample resources. And by comparing to adjacent normal tissue‐derived sEVs, we aimed to identify BCa‐specific sEV biomarkers in urine or plasma.
2. MATERIALS AND METHODS
2.1. Patients and sample collection
All 27 patients included in this study had histologically confirmed urothelial carcinoma of bladder. The mean age was 74 years (range: 59–88 years). Twenty‐two patients were male and five patients were female. According to the American Joint Committee on Cancer (AJCC) pathological staging (8th edition), 19 patients were diagnosed as MIBC and were treated by radical cystectomy, while eight patients were diagnosed with NMIBC and were treated by TUR of the tumour (Table S1). Preoperative whole blood from fasted patients was collected in an ethylenediaminetetraacetic acid (EDTA) coated tube (BD Biosciences, San Jose, CA, USA). Non‐first urine samples were obtained from catheter right before the surgery. Fresh tissue samples for sEV separation were collected from surgical pathology before formalin fixation. The tissue was collected in ice‐cold phosphate‐buffered saline (PBS) and rapidly transferred to the laboratory for further processing. Adjacent normal tissue was only obtained in cases undergoing radical cystectomy. The sampling of paired cancer and non‐cancer tissue was confirmed by further histology staining. Eventually, there were 22 urinary sEV samples, 26 plasma sEV samples, 20 tumour tissue derived sEV samples, 11 adjacent normal tissue derived sEV samples and 16 FFPE tumour tissue samples enrolled in this sequencing study (Figure S1 and Table S2). All sample collections were preceded by signed written informed consent and approved by the Johns Hopkins Office of Human Subjects Research Institutional Review Board.
2.2. Sample preparation
The preparation of plasma, urine and tissue explants for sEV separation was performed as previously described (Dong et al., 2020; Zieren et al., 2020). Briefly, the plasma separated from the whole blood was further pre‐cleaned by two centrifugations at 2500×g for 15 min. The supernatant was then centrifuged at 10,000×g at 4°C for 20 min. Urine was pre‐cleaned by centrifugation at 1000×g for 10 min, and the supernatant was then centrifuged at 10,000×g for 20 min at 4°C. For tissue derived sEV separation, approximately 1 g of tissue was cut into 2–3 mm pieces, followed by PBS wash. The tissue pellet was incubated with 15 mL of serum‐free RPMI 1640 (Thermo Fisher Scientific, Waltham, MA, USA) with Collagenase D (2 mg/mL) and DNase I (40 U/mL) at 37°C for 30 min. The tissue‐conditioned media was then processed by the same sequential centrifugation protocol as urine samples. All processed plasma, urine and tissue‐conditioned media was eventually filtered through a 0.45 μm hydrophilic PVDF membrane syringe filter (Thermo Fisher Scientific), then was either used fresh for sEV separation or stored at −80°C until use, limiting freeze‐thaw cycles to a maximum of one.
2.3. Size exclusion chromatography with ultrafiltration
Pre‐processed plasma (2 mL), urine (40 mL) and tissue‐conditioned media (15 mL) was first concentrated to 0.5 mL by using 10K molecular weight cut off (MWCO) Amicon Ultra‐2 Centrifugal Filters (MilliporeSigma, Burlington, MA, USA), 10K MWCO Centricon Plus‐70 Centrifugal Filters (MilliporeSigma) and 10K MWCO Amicon Ultra‐15 Centrifugal Filters (MilliporeSigma), respectively, according to the manufacturer's instructions. qEV original 70 nm columns (IZON Science, Cambridge, MA) were used for sEV separation. Briefly, up to 0.5 mL of sample was loaded to the column. EV enriched fractions (fractions 7–10) were pooled and further concentrated using 10K MWCO Amicon Ultra‐2 Centrifugal Filters (MilliporeSigma) to a final volume of 100 μL.
2.4. Transmission electron microscopy
The size and shape of separated sEVs was evaluated by transmission electron microscopy (TEM) as described previously (Dong et al., 2020). First, 10 μL of each sample was adsorbed to an ultra‐thin carbon coated 400 mesh copper grid which was glow‐discharged (EMS GloQube) by flotation for 2 min. Then, the grids were rapidly blotted on filter paper and rinsed three times in tris‐buffered saline (TBS) for 1 min. Next, the grids were negatively stained in two continuous drops of 1% uranyl acetate with methylcellulose (filtered twice through 0.22 μm filter), while the plethoric stain was rapidly aspirated and blotted. When totally dried in darkness, the grids were observed using a Philips CM‐120 TEM operating at 80 kV with an AMCT XR80 CCD sensor.
2.5. Western blotting
Western blotting was performed to assess the expression of EV protein markers and non‐EV protein markers as previously described (Dong et al., 2020). After boiling at 95°C for 5 min, equal amounts of protein from sEVs were run on 12% SDS Mini‐PROTEAN TGX Stain‐Free Protein Gel (Bio‐Rad Laboratories), under denaturing and non‐reductive conditions. The nitrocellulose membranes (Trans‐Blot Turbo Mini Nitrocellulose, Bio‐Rad Laboratories) were incubated in 4°C for one night with antibodies as follows: Flot‐1 (1:2500, rabbit monoclonal EPR604, #ab133497, Abcam); CD63 (1:250, mouse monoclonal TDS63, #10628D, Thermo Fisher Scientific); CD81 (1:100, mouse monoclonal 1.3.3.22, #sc‐7637, Santa Cruz Biotechnology) and Calnexin (1:2500, rabbit polyclonal, #ab22595, Abcam). After washing process, incubation with secondary antibodies was performed for 1 h at RT with IRDye 680RD Goat anti‐Mouse IgG (1:20,000, #92668070, LI‐COR Biosciences) and IRDye 800CW Goat anti‐Rabbit IgG (1:15,000, #92632211, LI‐COR Biosciences). The blots were imaged using the Odyssey 9120 Infrared Imaging System (LI‐COR Biosciences).
2.6. Nano‐flow cytometry
sEV samples were analysed by nano‐flow cytometry (nFCM) (NanoFCM, Inc., Xiamen, China) for particle concentration and size distribution as previously described (Dong et al., 2020). The instrument was calibrated for particle concentration using 200 nm PE and AF488 fluorophore‐conjugated polystyrene beads and for size distribution using Silica Nanosphere Cocktail (NanoFCM, Inc., S16M‐Exo). Any particles that passed by the detector during a 1 min interval were recorded in each test. The flow rate and side scattering intensity were converted into corresponding particle concentration and size distribution using the calibration curve on the NanoFCM software (NanoFCM Profession V1.0).
2.7. RNA extraction
Total RNAs were extracted from sEVs using the mirVana™ miRNA isolation kit (Thermo Fisher, Inc) according to manufacturer's instructions in RNase free conditions. RNA purity and integrity were measured by NanoDrop One (Thermo Fisher, Inc) and Agilent TapeStation 4200 (Agilent Technologies, Santa Clara, CA, USA), respectively.
2.8. Library preparation and sequencing
Whole transcriptome RNA sequencing was performed using Ion Torrent's AmpliseqRNA platform (Thermo Fisher, Inc) and an S5XL sequencer (Thermo Fisher, Inc). Twenty nanograms of purified RNA was transcribed into cDNA using SuperScript VILO kit. Then cDNA was amplified using Ion Ampliseq Transcriptome Human Gene Expression Core panel, followed by the ligation of adapters and barcodes to amplicons and purification. Purified libraries were quantified using Ion Library Quantification kit (Thermo Fisher, Inc) according to the manufacturer's instructions. Libraries were diluted to 100 pM and pooled in sets of 16. Pooled libraries were amplified on the Ion Sphere TM particles (ISP) using emulsion PCR and enriched on the IonChef (Thermo Fisher, Inc). Template‐positive ISPs were loaded into Ion550 chips and run on the S5XL instrument.
2.9. RNA‐Seq gene expression analyses
Primary analyses of RNA sequencing data were performed using AmpliSeqRNA analysis plugin in the Torrent Suite Software. This plugin aligns the raw sequence reads to a human reference genome that contains 20,802 RefSeq transcripts (hg19 Ampliseq Transcriptome_ERCC_V1.fasta) using Torrent Mapping Alignment Program (TMAP). Then, the numbers of reads mapped per gene were counted to generate raw counts that were normalized by the Bioconductor package DESeq2 (Love et al., 2014). To visualize gene expression patterns, specific gene expression values, adjusted to a median of zero, were used for clustering using Cluster 3.0 and TreeView (Eisen et al., 1998) or Bioconductor package ComplexHeatmap (Gu et al., 2016).
2.10. Pathway analyses
Functional and pathway analyses were performed using Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, CA), which contains a database for identifying networks and pathways of interest in genomic data. “Transcriptional factors as molecule type in upstream regulator” categories within IPA were used to interpret the biological properties of the bladder tumour subtypes. For upstream regulator analyses, IPA performs statistical analyses for overlap p values and an activation Z‐score. Based on the IPA knowledge database, p values and Z‐scores can be calculated based on how many targets of each transcriptional factor were overwrapped (p values) and the extent of concordance of the known effects (activation or inhibition) of the targets in the gene lists (Z‐score).
2.11. Identification of molecular subtypes
MDA Subtype classifiers were applied using normalized RNA sequencing data (Choi et al., 2014). R packages for other subtype classifications (Consensus classification, BASE47, and TCGA) was obtained from Github (Kamoun et al., 2020).
2.12. EV‐TRACK
We have submitted all relevant data of our experiments to the EV‐TRACK knowledgebase (EV‐TRACK ID: EV230963) (Van Deun et al., 2017).
3. RESULTS
3.1. EV characterization
The characterization of sEVs separated from plasma, urine and tissue samples were performed according to the Minimal Information for Studies of EVs (MISEV) guideline. TEM revealed cup‐shaped structures of EVs with a diameter of about 100 nm on negative staining in all three sample types (Figure 1a). The presence of the EV markers (CD63, CD81 and FLOT1 for urinary and tissue‐derived sEVs; CD81 and FLOT1 for plasma sEVs) and the absence of the contamination marker (Calnexin) was confirmed by western blotting (Figure 1b). nFCM demonstrated the particle size distributions and concentrations of sEV samples. The particle sizes of all three sample types ranged from 50 to 150 nm, with no significant differences between MIBC and NMIBC (Figure 2a). The particle concentrations of tissue‐derived sEVs (×1010) were higher than those of plasma (×109) and urinary sEVs (×108). The concentrations of sEVs derived from MIBC tumour tissues were significantly higher than those derived from NMIBC tumour tissues (Figure 2b).
FIGURE 1.
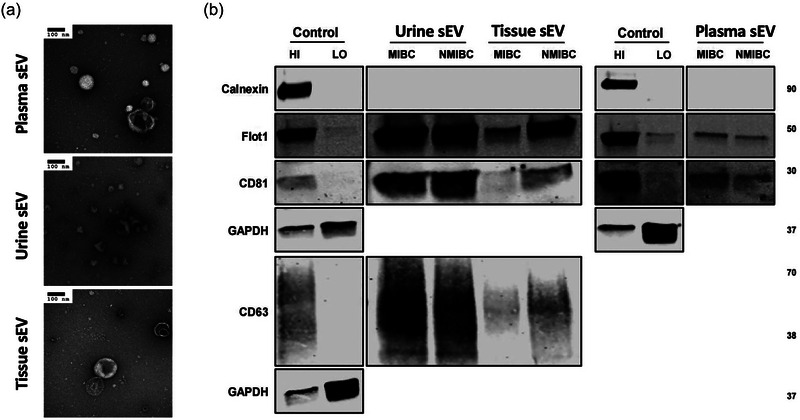
(a) TEM images confirming the presence of negative‐stained sEVs separated from plasma, urine and tissue explants, seen as cup‐shaped vesicles. Scale bars are 100 nm. (b) Western blots of flotillin‐1, CD63, CD81, and calnexin for sEVs separated from plasma, urine and tissue explants. MCF7 membrane and cytosolic protein fractions served as positive and negative controls for flotillin‐1, CD63, CD81 and calnexin.
FIGURE 2.
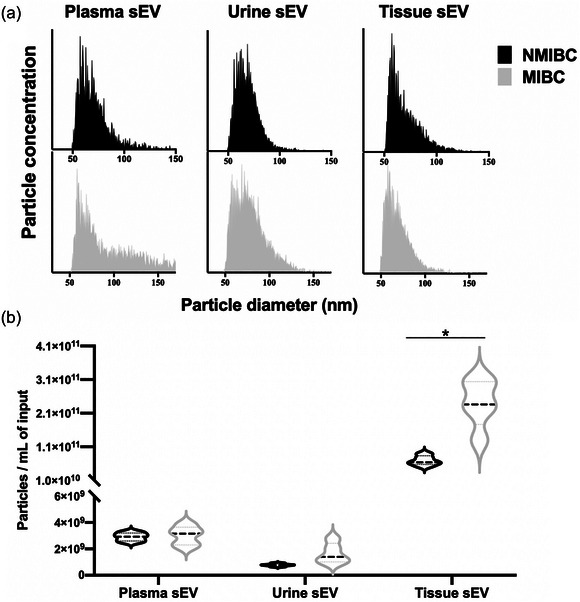
(a) Particle size distributions of sEVs separated from plasma, urine and tissue explants, measured by nFCM. The bin width was 0.5 nm. In order to make the size distribution histogram visually comparable, the Y axis was adjusted to make the concentration of particles with modal size (the peak of the curve) as 95% of maximum scale in each figure. (b) Particle concentrations of sEVs separated from plasma, urine and tissue explants, measured by nFCM. The particle concentrations have been corrected for sample input volumes. *p < 0.05.
3.2. Quality of RNA sequencing reads
RNA purity was evaluated based on ratios of A260/A280 optical density ratio. The purity of FFPE tumour RNA was superior to that of sEV RNA from all four sample types (adjacent normal tissue, tumour tissue, plasma and urine). Plasma sEV RNA had the lowest purity among sEV RNA isolates (Figure 3a). Sequencing libraries from FFPE tumour RNA, tumour tissue‐derived sEV RNA and adjacent normal tissue‐derived sEV RNA yielded higher valid reads (%) compared to the libraries from plasma sEV RNA and urinary sEV RNA (Figure 3b). Amplicon coverage for transcript varied among samples. Plasma sEVs had the lowest amplicon coverage. Compared to FFPE tumour RNA, RNA isolates from sEVs exhibited high heterogeneity in amplicon coverage within each sample type. Tumour tissue‐derived EVs had the highest amplicon coverage in at least “one” read, but had intermediate coverage in at least “100” reads, demonstrating high coverage on low expressed transcripts (Figure 3c). To evaluate if batch effects associated with library preparation or sequencing may have affected the sequencing results, principle component analysis (PCA) was performed. The results indicated that batch effects did not influence on PCA results, but rather, samples clustered according to sample type (Figure 3d–f). In particular, the plasma sEV‐derived RNAseq data had the lowest valid reads and amplicon coverage, resulting in distinct clustering pattern on the PCA plot.
FIGURE 3.
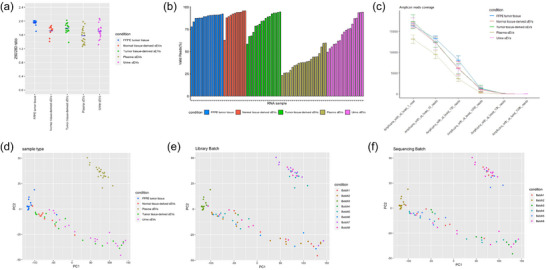
(a) The purity of RNA isolates from five sample types (FFPE tumour tissue, adjacent normal tissue‐derived sEVs, tumour tissue‐derived sEVs, plasma sEVs, urine sEVs) were measured by Nanodrop absorbance 260/280 ratio. Sequencing results were measured by valid reads (%) (b) and reads coverage (c), based on sample types. Sequencing results were also PCA‐plotted based on sample type (d), library batch (e), and sequencing batch (f).
3.3. Molecular subtype classification
Various BCa subtype classifiers were applied to RNAseq expression data, including the BASE47, MDA, TCGA, and Consensus classifications (Figure 4a) (Choi et al., 2014; Damrauer et al., 2014; Kamoun et al., 2020; Robertson et al., 2017). According to consensus classification schema of FFPE tumour tissue mRNA sequencing, three patients were assigned to Ba/Sq subtype, six patients were assigned to Stroma‐rich subtype, and seven patients were assigned to luminal subtype (six LumP and one LumU) (Figure 4b). All NMIBC tumours were assigned to LumP. Nine patients failed to be classified as any subtype, due to the suboptimal micro‐dissection, or RNA acquirement. Across different classification schema, all plasma sEVs were classified as Ba/Sq subtype except one sample in MDA subtype, while the most urinary sEVs were classified as luminal subtype (LumP or LumU), regardless of the molecular subtypes identified by their matched‐FFPE tumour tissue. Tumour tissue‐derived sEVs and FFPE tumour tissue demonstrated good concordance in molecular subtype calls (Figure 4b). One out of fourteen patients were identically classified as Ba/Sq subtype, five out of fourteen were identically classified as Stroma‐rich subtype, and six out of fourteen were identically classified as LumP subtype, based on their FFPE tissue and tumour tissue‐derived sEVs sequencing. Only one patient was assigned to Stroma‐richFFPE/LumPT‐EV, and one patient was assigned to LumUFFPE/Stroma‐richT‐EV. Although plasma sEVs were all classified as Ba/Sq subtype, the only Ba/Sq biomarker they contained was KRT6B/C. Notably, several immune markers were found in plasma sEVs (Figure 4c). Since molecular classification classifiers were developed based on solid tumour gene expression signatures, the current classifiers may not accurately predict subtypes of samples from liquid biopsy. To test this hypothesis, differentially carried transcripts were analysed between urinary sEVs from patients with different molecular subtypes based on corresponding FFPE tumour tissues. KRT6A was significantly more abundant in urinary sEVs in patients with FFPE tissue‐based Ba/Sq subtype compared to the rest subtypes. Heatmap demonstrated that differentially carried transcripts among urinary sEVs are in concordance with the signature genes of basal and luminal molecular subtypes assigned by their matched‐FFPE tumour tissue (Figure 4d). However, the urinary stroma‐rich subtype tumours expressed very few fibroblast/stroma gene signatures.
FIGURE 4.
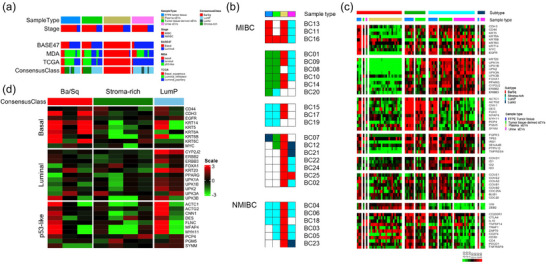
(a) Molecular subtype classification of the same sample using BASE47, MDA, TCGA, and Consensus classifiers, respectively. (b) Molecular subtype classifications based on different sample types of the same patient, using Consensus classifiers. The white box means the sample was not used for this study, either because the sample was not available, or due to the QC failure. (c) Heatmap demonstrating detailed consensus molecular subtype signature gene expressions sample. (d) Heatmap demonstrating differentially carried transcripts were analysed between urinary sEVs from patients with different molecular subtypes based on corresponding FFPE tumour tissues. The horizontal labels were patients’ molecular subtypes based on FFPE tumour tissues using Consensus classifier. The vertical labels were patients’ molecular subtypes based on urine sEVs using MDA classifier.
3.4. Muscle invasive bladder cancer versus non‐muscle invasive bladder cancer
To investigate if sEVs reflect the biology between MIBC versus NMIBC, the differentially expressed genes/carried transcripts were extracted. Volcano plots show the fold changes and p value differences for the comparison of NMIBC and MIBC in different sample types. The comparisons between tumour tissue‐derived sEVs were not performed, because the tumour tissue of MIBC was obtained from radical cystectomy specimens, while the tumour tissue of NMIBC was obtained from TUR procedures. There were 672 genes, 87 transcripts, and 18 transcripts significantly differentially expressed/carried in FFPE tumour tissue samples, urinary sEVs and plasma sEVs, respectively (Figure 5a–c). The top 10 significantly differentially expressed genes/carried transcripts in different sample types between NMIBC versus MIBC were listed, respectively (Figure 5d–f). Pathway analysis was performed using significant genes/transcripts from FFPE tumour tissue, urinary sEVs and plasma sEVs. Due to the small number of differentially‐carried transcripts in plasma sEVs, no pathways were significantly activated in plasma sEVs. FFPE tumour tissue and urinary sEV samples demonstrated shared pathway activation (Figure 5g, h). Wound healing signalling pathway, tumour microenvironment pathway, Phagosome formation, IL‐8 signalling, IL‐17 signalling, Hepatic fibrosis signalling pathway, Cardiac hypertrophy signalling (enhanced) were significantly activated in both FFPE tissue and urinary sEVs. Detailed gene lists in each pathway have been demonstrated in Table S3. To be noted, we used all significantly differentially expressed genes/carried transcripts (two folds cut‐off with adjp < 0.05) to perform the pathway analysis. The pathways with some top differentially expressed genes/carried transcripts may not be significantly enriched by IPA analysis.
FIGURE 5.
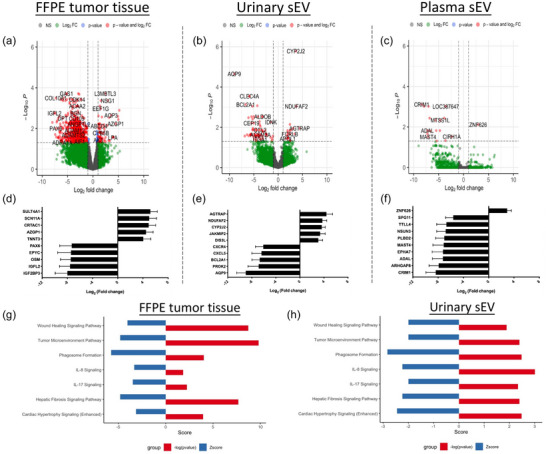
Volcano plot demonstrating significances versus means of differential fold changes for the comparison between MIBC versus NMIBC in FFPE tumour tissue (a), urine sEVs (b), and plasma sEVs (c). The top 10 differentially expressed/incorporated mRNAs with significance between MIBC versus NMIBC in FFPE tumour tissue (d), urine sEVs (e), and plasma sEVs (f). Significantly activated signalling pathways between MIBC versus NMIBC in FFPE tumour tissue (g), and urine sEVs (h).
3.5. Urinary bladder cancer‐specific sEV biomarkers
For biomarker discovery, we only enrolled patients who have adjacent normal tissue‐derived sEVs (n = 11) (Figure S1). In order to identify urinary BCa‐specific sEV mRNA cargos, the significantly differentially expressed mRNA transcripts in urinary sEVs versus adjacent normal tissue derived sEVs (adjusted p < 0.01, Log2Fold change > 9) were extracted from patients having paired samples, as well as the significantly differentially expressed mRNA transcripts in tumour tissue derived sEVs versus adjacent normal tissue derived sEVs. Ten mRNA transcripts were shared by these two gene lists, including ATP2B2, SPDYC, SLC22A7, FAM71E2, OR4K5, FAM138F, TTBK1, KRTAP26‐1, ITPRIP, CDH16 (Figure 6a). These mRNAs were thought to be highly expressed in both urinary sEVs and tumour tissue derived sEVs, but absent in adjacent normal tissue derived sEVs. The expression levels of these 10 candidate sEV mRNA biomarkers were first validated using the whole transcriptome RNA sequencing data of 16 non‐cancerous urinary sEVs samples from exoRBase (http://www.exorbase.org/exoRBaseV2/toIndex) (Figure 6b). Four mRNA biomarkers were not detected in non‐cancerous urinary sEVs, including FAM71E2, OR4K5, FAM138F and KRTAP26‐1. The expression levels of four candidate sEV mRNA biomarkers were further confirmed in all urinary sEVs (n = 21), tumour tissue derived sEVs and adjacent normal tissue derived sEVs in BCa patients enrolled in this study, regardless of matched or not. All four biomarkers had high expression levels in urinary sEVs separated from BCa patients, and their expression levels were significantly higher in urinary sEVs than in adjacent normal tissue derived sEVs (Figure 6c, d). We also tried to validate these markers in EV samples from patients with other types of cancers using publicly available database, and it demonstrated that these four mRNA markers are very likely to be BCa specific (Figure S2).
FIGURE 6.
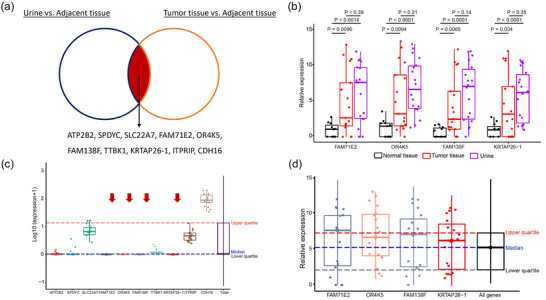
(a) Ten shared mRNA transcripts identified between paired urine sEVs versus adjacent normal tissue‐derived sEVs and tumour tissue‐derived sEVs versus adjacent normal tissue‐derived sEVs. (b) Expression levels of identified ten mRNA transcripts in an external non‐cancerous cohort (urine sEVs) from exoRBase. (c) Four mRNA transcripts which are not detected in non‐cancerous urine sEVs demonstrated significantly higher expression levels in urine sEVs and tumour tissue‐derived sEVs, comparing to normal tissue‐derived sEVs. (d) Validation of the four mRNA biomarkers in all urine sEV samples in this study (twenty‐two BCa patients), with or without paired plasma or tissue.
4. DISCUSSION
Initially, EVs were only considered to be a way cells dispose of waste (Johnstone et al., 1987). Though the detailed EV cargo sorting mechanisms still largely remained unknown and we previously demonstrated that EVs may not carry exactly the same molecules as their parental cells, it has been widely accepted that the appearance of certain molecules in EVs can reflect the real‐time biological condition of the cells (Desdín‐Micó & Mittelbrunn, 2017; Dong et al., 2021). This is why EVs are adopted as a rising star in the field of liquid biopsy. For example, Del Re and colleagues demonstrated that the level of programmed death‐ligand 1 (PD‐L1) mRNA in plasma EVs is significantly associated with response to immune checkpoint inhibitors in lung cancer patients, and the androgen receptor splicing‐variant 7 (AR‐V7) mRNA level in plasma EVs predicts resistance to hormonal therapy in patients with metastatic prostate cancer (mPCa) (Del Re et al., 2017, 2018). However, the liquid biopsy‐based BCa molecular subtype classification has long been an unmet need.
Compared to EVs separated from biofluids, tissue‐derived EVs may reflect more tissue‐ or disease‐specific biological features. There is an increasing number of studies focusing on tissue‐derived EVs in recent years, covering a wide range of cancerous and non‐cancerous diseases (Li et al., 2021). The tissue types include but are not limited to melanoma tissue, kidney tissue, brain tissue, adipose tissue, liver tissue, etc. The processing protocols vary in details, but all have a similar principle, which is dissecting the tissue into small pieces, shortly after it is removed from the patient, followed by dissociating the tissue using collagenase and DNase (Qin et al., 2021). Tissue‐conditioned media/solution is then used for EV separation. In this study, we reported the separation, characterization and analyses of BCa tissue‐derived sEVs for the first time. Relatively high yield of sEVs were obtained using approximately 1 g of tissue, incubated with media for 30 min (Zieren et al., 2020). FFPE tumour tissue and tumour tissue‐derived sEVs demonstrated good concordance in molecular subtype classification. Though tissue‐derived EVs cannot be used for non‐invasive liquid biopsy, it proved the concept that the biological features associated with molecular subtype were carried and well reflected by sEVs secreted by cells from the tumour tissue. Since the tumour tissues were collected from two different procedures, TUR versus radical cystectomy, tissue‐derived sEVs were not used for the analyses between NMIBC versus MIBC in this study.
Plasma and urine are the most commonly used biofluid samples for EV research (Roy et al., 2018). Our previous study demonstrated that plasma EVs are usually co‐separated with other non‐EV contaminants, regardless of separation method used (Dong et al., 2020). Plasma sEVs‐derived sequencing data had the lowest valid reads and amplicon coverage, resulting in distinct gene expression patterns on PCA plot. All plasma sEVs were classified as Ba/Sq subtype. One hypothesis is that background sEVs from the blood cells, instead of BCa‐specific sEVs, in the circulation, dominated the molecular subtype given by the classifier. Similarly, plasma sEVs could not reveal the different biology between NMIBC and MIBC. On the other hand, all urinary sEVs were classified as luminal subtype (LumP or LumU), but the comparison within urinary sEVs could pick up the different biology between NMIBC and MIBC, as well as the signature genes related to molecular subtypes. In the comparison between NMIBC and MIBC, although there were less significantly carried mRNA transcripts in urinary sEVs, compared to matched FFPE tissue, they have the same significantly enriched pathways. This indicated that even though the biology associated with sample type might override the biology associated with the disease on molecular subtype classifier, urinary sEVs in BCa patients did carry disease‐specific biomarkers. These results implied that the current BCa molecular subtype classifier should not be applied to urinary or plasma sEV samples directly, since it is more likely to pick up the sample type features, instead of BCa biology. Urinary sEVs are promising biomarkers to be used for liquid biopsy‐based molecular subtype classification, if the current algorithm can be modified/adjusted using urinary sEVs from healthy donors to subtract the background information in urine. It will largely benefit the clinical decision making if we can acquire the molecular subtype by testing the urine before surgery.
It is essential to identify disease‐specific biomarkers in liquid biopsy. In BCa, urine‐based assays, like nuclear matrix protein 22 (NMP22), bladder tumour antigen (BTA), and UroVysion, have been approved by FDA for years (Ng et al., 2021). However, several drawbacks, including the high false positive rates, have limited the clinical application of these biomarkers (Vlachostergios & Faltas, 2019). Thus, it has been an unmet need to develop BCa‐specific biomarkers with high sensitivity and specificity. In this study, we analyzed mRNA cargos in sEVs separated from matched urine, tumour tissue and adjacent normal tissue. Theoretically, BCa‐specific sEV mRNA biomarkers are supposed to be abundant in both urinary sEVs and tumour tissue derived sEVs, but absent in adjacent normal tissue derived sEVs. Armstrong and colleagues analysed microRNA of matched FFPE‐tumour tissue, plasma, urinary sEVs and white blood cells from patients with BCa, and found miR‐720/3007a, miR‐205, miR‐200c‐3p and miR‐29b‐3p were common to both tumour and urine (Armstrong et al., 2015). In this study, ten candidate mRNAs were found using matched samples, and after validated in an external non‐cancerous urinary sEV dataset, four mRNA biomarkers were eventually identified. The abundancy of each was high in both urinary and tumour tissue derived sEVs, but low in adjacent normal tissue derived sEVs in our BCa cohort. We noticed these four urinary sEV mRNA biomarkers (FAM71E2, OR4K5, FAM138F and KRTAP26‐1) have not been widely reported before. Liu and colleagues developed an immune‐related prognostic signature in lung squamous cell carcinoma patients, in which they mentioned the high expression of FAM71E2 is associated with a worse overall survival (Liu et al., 2022). Further validation in large cohorts is needed.
This study had several limitations. First, given the relatively small number of cases, all of the findings in this study need to be further confirmed in larger cohorts with patients at different stages of BCa. Second, the four BCa‐specific biomarkers identified in this study need to be validated prospectively. Third, the pretreatment of RNase for sEVs was not applied in this study. So, we have to be aware of the potential contamination of RNAs on the sEV surface. Last but not least, all of the sEV samples were separated by SEC. Though several studies have demonstrated that the sample purity of SEC ranks high among the most commonly used EV separation methods, the current findings, especially the biomarker candidates, need to be validated by density‐gradient ultracentrifugation (Dong et al., 2020; Mol et al., 2017).
In conclusion, we performed whole transcriptome RNA sequencing for FFPE tumour tissues and sEVs separated from matched tissue explants, urine and plasma in patients with BCa. FFPE tumour tissue and tumour tissue‐derived sEVs demonstrated good concordance in molecular subtype classification. Urinary sEV and FFPE tumour tissue demonstrated shared pathway activation when comparing NMIBC versus MIBC. By comparing urinary sEVs and tumour tissue‐derived sEVs to matched adjacent normal tissue‐derived sEVs, four candidate sEV mRNA biomarkers were identified, including FAM71E2, OR4K5, FAM138F and KRTAP26‐1. Future work is needed to validate these new BCa‐specific biomarkers in large cohorts.
AUTHOR CONTRIBUTIONS
Liang Dong: Conceptualization; data curation; methodology; writing—original draft. Mingxiao Feng: Data curation; formal analysis; methodology; visualization. Morgan D. Kuczler: Data curation; methodology. Kengo Horie: Data curation. Chi‐Ju Kim: Formal analysis; methodology. Zehua Ma: Formal analysis; visualization. Kara Lombardo: Project administration; resources. Heather Lyons: Project administration; resources. Max Kates: Resources; writing—review and editing. Trinity J. Bivalacqua: Resources; writing—review and editing. David J. McConkey: Supervision; writing—review and editing. Wei Xue: Supervision; writing—review and editing. Woonyoung Choi: Data curation; formal analysis; visualization. Kenneth J. Pienta: Conceptualization; supervision; writing—review and editing.
CONFLICT OF INTEREST STATEMENT
No conflict of interests.
Supporting information
Supporting Information
Supporting Information
Supporting Information
ACKNOWLEDGEMENTS
The authors thank Barbara Smith, microscopy specialist of the Microscope Facility at Johns Hopkins University School of Medicine, for acquiring TEM images. Liang Dong was supported by the National Natural Science Foundation of China (82103485) and Innovative Research Team of High‐level Local Universities in Shanghai. Kengo Horie was supported by Japan Society for the Promotion of Science (KAKENHI: 19K18555). Chi‐Ju Kim was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education (2021R1A6A3A14043146).
Dong, L. , Feng, M. , Kuczler, M. D. , Horie, K. , Kim, C.‐J. , Ma, Z. , Lombardo, K. , Lyons, H. , Amend, S. R. , Kates, M. , Bivalacqua, T. J. , McConkey, D. , Xue, W. , Choi, W. , & Pienta, K. J. (2024). Tumour tissue‐derived small extracellular vesicles reflect molecular subtypes of bladder cancer. Journal of Extracellular Vesicles, 13, e12402. 10.1002/jev2.12402
Liang Dong and Mingxiao Feng contributed equally.
Contributor Information
Liang Dong, Email: drdongliang@126.com.
Wei Xue, Email: uroxuewei@163.com.
Woonyoung Choi, Email: wchoi16@jhmi.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Armstrong, D. A. , Green, B. B. , Seigne, J. D. , Schned, A. R. , & Marsit, C. J. (2015). MicroRNA molecular profiling from matched tumor and bio‐fluids in bladder cancer. Molecular Cancer, 14(1), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, W. , Porten, S. , Kim, S. , Willis, D. , Plimack, E. R. , Hoffman‐Censits, J. , Roth, B. , Cheng, T. , Tran, M. , Lee, I. L. , & Melquist, J. (2014). Identification of distinct basal and luminal subtypes of muscle‐invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell, 25(2), 152–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damrauer, J. S. , Hoadley, K. A. , Chism, D. D. , Fan, C. , Tiganelli, C. J. , Wobker, S. E. , Yeh, J. J. , Milowsky, M. I. , Iyer, G. , Parker, J. S. , & Kim, W. Y. (2014). Intrinsic subtypes of high‐grade bladder cancer reflect the hallmarks of breast cancer biology. Proceedings of the National Academy of Sciences, 111(8), 3110–3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Re, M. , Biasco, E. , Crucitta, S. , Derosa, L. , Rofi, E. , Orlandini, C. , Miccoli, M. , Galli, L. , Falcone, A. , Jenster, G. W. , & van Schaik, R. H. (2017). The detection of androgen receptor splice variant 7 in plasma‐derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. European Urology, 71(4), 680–687. [DOI] [PubMed] [Google Scholar]
- Del Re, M. , Marconcini, R. , Pasquini, G. , Rofi, E. , Vivaldi, C. , Bloise, F. , Restante, G. , Arrigoni, E. , Caparello, C. , Bianco, M. G. , & Crucitta, S. (2018). PD‐L1 mRNA expression in plasma‐derived exosomes is associated with response to anti‐PD‐1 antibodies in melanoma and NSCLC. British Journal of Cancer, 118(6), 820–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdín‐Micó, G. , & Mittelbrunn, M. (2017). Role of exosomes in the protection of cellular homeostasis. Cell Adhesion & Migration, 11(2), 127–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. , Huang, C. Y. , Johnson, E. J. , Yang, L. , Zieren, R. C. , Horie, K. , Kim, C. J. , Warren, S. , Amend, S. R. , Xue, W. , & Pienta, K. J. (2021). High‐throughput simultaneous mRNA profiling using nCounter technology demonstrates that extracellular vesicles contain different mRNA transcripts than their parental prostate cancer cells. Analytical Chemistry, 93(8), 3717–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. , Zieren, R. C. , Horie, K. , Kim, C. J. , Mallick, E. , Jing, Y. , Feng, M. , Kuczler, M. D. , Green, J. , Amend, S. R. , & Witwer, K. W. (2020). Comprehensive evaluation of methods for small extracellular vesicles separation from human plasma, urine and cell culture medium. Journal of Extracellular Vesicles, 10(2), e12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, L. , Zieren, R. C. , Wang, Y. , de Reijke, T. M. , Xue, W. , & Pienta, K. J. (2019). Recent advances in extracellular vesicle research for urological cancers: From technology to application. Biochimica et Biophysica Acta (BBA)‐Reviews on Cancer, 1871(2), 342–360. [DOI] [PubMed] [Google Scholar]
- Eisen, M. B. , Spellman, P. T. , Brown, P. , & Botstein, D. (1998). Cluster analysis and display of genome‐wide expression patterns. Proceedings of the National Academy of Sciences, 95(25), 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong, M. H. , Feng, M. , McConkey, D. J. , & Choi, W. (2020). Update on bladder cancer molecular subtypes. Translational Andrology and Urology, 9(6), 2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z. , Eils, R. , & Schlesner, M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics, 32(18), 2847–2849. [DOI] [PubMed] [Google Scholar]
- Hoshino, A. , Kim, H. S. , Bojmar, L. , Gyan, K. E. , Cioffi, M. , Hernandez, J. , Zambirinis, C. P. , Rodrigues, G. , Molina, H. , Heissel, S. , & Mark, M. T. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell, 182(4), 1044–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone, R. M. , Adam, M. , Hammond, J. R. , Orr, L. , & Turbide, C. (1987). Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). Journal of Biological Chemistry, 262(19), 9412–9420. [PubMed] [Google Scholar]
- Kalluri, R. , & LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science, 367(6478), eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamat, A. M. , Hahn, N. M. , Efstathiou, J. A. , Lerner, S. P. , Malmström, P. U. , Choi, W. , Guo, C. C. , Lotan, Y. , & Kassouf, W. (2016). Bladder cancer. The Lancet, 388(10061), 2796–2810. [DOI] [PubMed] [Google Scholar]
- Kamoun, A. , de Reyniès, A. , Allory, Y. , Sjödahl, G. , Robertson, A. G. , Seiler, R. , Hoadley, K. A. , Groeneveld, C. S. , Al‐Ahmadie, H. , Choi, W. , & Castro, M. A. (2020). A consensus molecular classification of muscle‐invasive bladder cancer. European Urology, 77(4), 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, N. , Cvjetkovic, A. , Jang, S. C. , Crescitelli, R. , Hosseinpour Feizi, M. A. , Nieuwland, R. , Lötvall, J. , & Lässer, C. (2018). Detailed analysis of the plasma extracellular vesicle proteome after separation from lipoproteins. Cellular and Molecular Life Sciences, 75(15), 2873–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, M. A. , & Hurst, C. D. (2015). Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nature Reviews Cancer, 15(1), 25–41. [DOI] [PubMed] [Google Scholar]
- Kuczler, M. D. , Zieren, R. C. , Dong, L. , de Reijke, T. M. , Pienta, K. J. , & Amend, S. R. (2022). Advancements in the identification of EV derived mRNA biomarkers for liquid biopsy of clear cell renal cell carcinomas. Urology, 160, 87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S. R. , Man, Q. W. , Gao, X. , Lin, H. , Wang, J. , Su, F. C. , Wang, H. Q. , Bu, L. L. , Liu, B. , & Chen, G. (2021). Tissue‐derived extracellular vesicles in cancers and non‐cancer diseases: Present and future. Journal of Extracellular Vesicles, 10(14), e12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhao, D. , Shan, Y. , Cui, W. , Xie, Q. , Jiang, J. , Peng, W. , Zhang, C. , & Duan, C. (2022). Development and validation of a novel immune‐related prognostic signature in lung squamous cell carcinoma patients. Scientific Reports, 12(1), 20737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malloci, M. , Perdomo, L. , Veerasamy, M. , Andriantsitohaina, R. , Simard, G. , & Martínez, M. C. (2019). Extracellular vesicles: Mechanisms in human health and disease. Antioxidants & Redox Signaling, 30(6), 813–856. [DOI] [PubMed] [Google Scholar]
- McConkey, D. J. (2021). Molecular biology of bladder cancer: Potential implications for therapy. Hematology/Oncology Clinics, 35(3), 457–468. [DOI] [PubMed] [Google Scholar]
- Mol, E. A. , Goumans, M. J. , Doevendans, P. A. , Sluijter, J. P. , & Vader, P. (2017). Higher functionality of extracellular vesicles isolated using size‐exclusion chromatography compared to ultracentrifugation. Nanomedicine: Nanotechnology, Biology and Medicine., 13(6), 2061–2065. [DOI] [PubMed] [Google Scholar]
- Necchi, A. , Raggi, D. , Gallina, A. , Ross, J. S. , Farè, E. , Giannatempo, P. , Marandino, L. , Colecchia, M. , Lucianò, R. , Bianchi, M. , & Colombo, R. (2020). Impact of molecular subtyping and immune infiltration on pathological response and outcome following neoadjuvant pembrolizumab in muscle‐invasive bladder cancer. European Urology, 77(6), 701–710. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network . (2014). Comprehensive molecular characterization of urothelial bladder carcinoma. Nature, 507(7492), 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, K. , Stenzl, A. , Sharma, A. , & Vasdev, N. (2021). Urinary biomarkers in bladder cancer: A review of the current landscape and future directions. Urologic Oncology: Seminars and Original Investigations, 39(1), 41–51. [DOI] [PubMed] [Google Scholar]
- Qin, B. , Hu, X. M. , Su, Z. H. , Zeng, X. B. , Ma, H. Y. , & Xiong, K. (2021). Tissue‐derived extracellular vesicles: Research progress from isolation to application. Pathology‐Research and Practice, 226, 153604. [DOI] [PubMed] [Google Scholar]
- Robertson, A. G. , Kim, J. , Al‐Ahmadie, H. , Bellmunt, J. , Guo, G. , Cherniack, A. D. , Hinoue, T. , Laird, P. W. , Hoadley, K. A. , Akbani, R. , & Castro, M. A (2017). Comprehensive molecular characterization of muscle‐invasive bladder cancer. Cell, 171(3), 540–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S. , Hochberg, F. H. , & Jones, P. S. (2018). Extracellular vesicles: The growth as diagnostics and therapeutics; a survey. Journal of Extracellular Vesicles, 7(1), 1438720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, R. , Ashab, H. A. , Erho, N. , van Rhijn, B. W. , Winters, B. , Douglas, J. , Van Kessel, K. E. , van de Putte, E. E. , Sommerlad, M. , Wang, N. Q. , & Choeurng, V. (2017). Impact of molecular subtypes in muscle‐invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. European Urology, 72(4), 544–554. [DOI] [PubMed] [Google Scholar]
- Siegel, R. L. , Miller, K. D. , Wagle, N. S. , & Jemal, A. (2023). Cancer statistics, 2023. CA: A Cancer Journal for Clinicians, 73(1), 17–48. [DOI] [PubMed] [Google Scholar]
- Van Deun, J. , Mestdagh, P. , Agostinis, P. , Akay, Ö. , Anand, S. , Anckaert, J. , Martinez, Z. A. , Baetens, T. , Beghein, E. , Bertier, L. , & Berx, G. (2017). EV‐TRACK: Transparent reporting and centralizing knowledge in extracellular vesicle research. Nature Methods, 14(3), 228–232. [DOI] [PubMed] [Google Scholar]
- Vlachostergios, P. J. , & Faltas, B. M. (2019). The molecular limitations of biomarker research in bladder cancer. World Journal of Urology, 37(5), 837–848. [DOI] [PubMed] [Google Scholar]
- Webber, J. , & Clayton, A. (2013). How pure are your vesicles? Journal of Extracellular Vesicles, 2(1), 19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi, A. , Villar‐Prados, A. , Oliphint, P. A. , Zhang, J. , Song, X. , De Hoff, P. , Morey, R. , Liu, J. , Roszik, J. , Clise‐Dwyer, K. , & Burks, J. K. (2019). Mechanisms of nuclear content loading to exosomes. Science Advances, 5(11), eaax8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieren, R. C. , Dong, L. , Pierorazio, P. M. , Pienta, K. J. , de Reijke, T. M. , & Amend, S. R. (2020). Extracellular vesicle isolation from human renal cancer tissue. Medical Oncology, 37(4), 1–1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


