Abstract
Chronic lymphocytic leukemia (CLL) is an incurable indolent non-Hodgkin lymphoma characterized by tumor B cells that weakly express a B-cell receptor. The mutational status of the variable region (IGHV) within the immunoglobulin heavy chain (IGH) locus is an important prognosis indicator and raises the question of the CLL cell of origin. Mutated IGHV gene CLL are genetically imprinted by activation-induced cytidine deaminase (AID). AID is also required for IGH rearrangements: class switch recombination and recombination between switch Mu (Sm) and the 3’ regulatory region (3’RR) (Sm-3’RRrec). The great majority of CLL B cells being unswitched led us to examine IGH rearrangement blockade in CLL. Our results separated CLL into two groups on the basis of Sm-3’RRrec counts per sample: Sm-3’RRrecHigh cases (mostly unmutated CLL) and Sm-3’RR-recLow cases (mostly mutated CLL), but not based on the class switch recombination junction counts. Sm-3’RRrec appeared to be ongoing in Sm-3’RRrecHigh CLL cells and comparison of Sm-3’RRrec junction structural features pointed to different B-cell origins for both groups. In accordance with IGHV mutational status and PIM1 mutation rate, Sm-3’RRrecHigh CLL harbor a non-germinal center experienced B-cell imprint while Sm-3’RRrecLow CLL are from AID-experienced B cells from a secondary lymphoid organ. In addition to the proposals already made concerning the CLL cell of origin, our study highlights that analysis of IGH recombinatory activity can identify CLL cases from different origins. Finally, on-going Sm-3’RRrec in Sm-3’RRrecHigh cells appeared to presumably be the consequence of high c-MYC expression, as c-MYC overexpression potentiated IGH rearrangements and Sm-3’RRrec, even in the absence of AID for the latter.
Introduction
Being one of the most frequent B-cell cancers of the elderly, chronic lymphocytic leukemia (CLL) is characterized by lymphocytosis exceeding ≥5.0×109/L, and is composed of small circulating monomorphic round CD19+ CD23+ CD5+ B cells, as well as bone marrow and secondary lymphoid organ infiltration in most cases.1 CLL evolution is highly variable, with overall survival ranging from a few years to decades and is still incurable despite the development of new therapeutics such as Bruton tyrosine kinase or Bcl2 inhibitors.
Binet and Rai classifications are still the most reliable staging systems to predict CLL course and are the keystones of clinical decision for treatment.2,3 However, patients show marked karyotypic and genetic heterogeneity which also influences overall survival rate and prediction of therapeutic response. Major prognosis factors are chromosomal abnormalities such del17p, del11q, trisomy 12, isolated del13q or complex karyotypes which highlight genomic instability in CLL pathogenesis.4 Among poor prognosis genetic abnormalities are those involving the Notch pathway (Notch1 mutations), NF-kB activation (BIRC3 or MYD88 mutations), splicing (SF3B1), the DNA lesion sensor AT M or the TP53 anti-oncogene.5
Underscoring the role of the B-cell receptor (BCR) in this B-cell cancer, another important prognosis indicator is the mutational status of the variable region (IGHV) within the immunoglobulin heavy chain (IGH) locus, which separates CLL patients into two groups: those with an unmutated variable region and those with a mutated IGHV rearranged gene (umCLL and mCLL, respectively). The former bestows a poor prognosis while patients with a very long survival rate are found in the latter group.6,7 Enforcing the role of the BCR is the fact that 30% of CLL patients express a socalled “stereotyped receptor”, which suggests the role of common antigenic determinants in the promotion of B-cell transformation.8
The fact that mCLL are genetically imprinted by activation-induced cytidine deaminase (AID)-dependent IGHV somatic hypermutation (SHM) raises the question of a CLL group with a post-germinal center (GC) B-cell counterpart. Recent methylome analyses suggest proximities between mCLL and GC experienced memory B cells on the one hand and umCLL and naïve B cells on the other hand.9–11 However, CLL cells exhibit a unique CD5+, CD23+, CD27+, CD43+ with low levels of surface immunoglobuln (Ig)M and IgD immunophenotype, which is different from that of any normal B cell.12,13 Gene expression profiles revealed that both CLL groups share a characteristic gene expression signature that is close to that of antigen-experienced B cells.12,14 Reconstruction of B-cell differentiation trajectories indeed suggested that precursors of both umCLL and mCLL have reached the antigen-experienced memory B-cell stage.13 It has also been proposed that CLL may originate from an extra-follicular B-cell response since maturation of these cells is antigen driven and can be either mutated or not.12 Therefore, more than 20 years after the papers of Hamblin and Damle,6,7 this question of the CLL cell of origin (COO) has not been clearly answered and umCLL could differ from mCLL mainly by expressing BCR-related mitogenic markers.14
The fact that most CLL cells express an IgM raises the question of class switch recombination (CSR) blockade in this B-cell cancer. Low levels of CSR can be observed in a small fraction of CLL tumor cells and seem to correlate with AID expression in these intraclonal switched CLL cells.15–17 AID has been repeatedly detected in CLL B cells independently of the IGHV mutational status.15 AID likely contributes to CLL evolution and seems to generate intraclonal diversity targeting the IGH locus and non-Ig off-targets.18 Physiologically, AID induces CSR in activated proliferating B cells.19,20 CSR results from IGH intrachromosomal recombination between the switch m (Sm) region and another switch region (so-called Sx) located upstream of one of the constant genes.21 CSR requires double strand DNA breaks (DSB). Converting a cytosine into uracil, AID creates a U:G mismatch that is targeted by the base excision repair (BER) pathway to excise the uracil base using uracil-DNA-glycosylase (UNG). This results in an abasic site which is processed and generates a single strand DNA break (SSB). Several SSB on both DNA strands then produce DSB which are repaired through the ubitquitous DSB repair (DSBR) response. In B cells, during CSR, DSB repair occurs by the joint action of the non-homologous end joining (NHEJ) and alternative end-joining (Alt-EJ) pathways.22,23 With XRCC4 for the ligation step, NHEJ depends on 53BP1/ Rif124,25 which recruits the Shieldin complex. Both 53BP1/ Rif1 and Shieldin complexes are essential for protection against the resection of DNA ends.26–28 Less understood and described, Alt-EJ involves PARP1, POLQ and/or LigIII. These DSBR pathways differently shape the structure of the repair junction; each single junction is thus unique.
Being highly conserved across mammalian species (human, mouse, dog, rabbit etc.), the 3’ regulatory regions (3’RR) of the IGH locus are key regulatory regions for CSR. 3’RR exhibits a singular structure with three DNaseI hypersensitive (HS) sites (HS3, HS1-2, and HS4) harboring strict specific B-lineage transcriptional enhancer activity related to a “quasi-palindrome” organization where inverted repeated sequences flank the HS1-2 sequence which is the symmetry center element.29 The 3’RR can also be recombined with the Sμ region in an AID-dependent manner (Sm-3’RRrec).30 Sm-3’RRrec has been shown to occur in vitro and in vivo in activated murine and human B cells.30–32 It has been repeatedly detected in secondary lymphoid organs and peripheral blood mononuclear cells (PBMC) of both mice and humans. Similar to CSR in activated mature B cells, Sm-3’RRrec occurs between transcribed recombination donor and acceptor DNA segments. However, we observed that the structure of Sm-3’RRrec junctions in murine B cells was different from that of CSR.31 Indeed, Sm-3’RRrec junctions are reminiscent of the usage of NHEJ and/or Alt-EJ, with regard to the repair signature at Sm-3’RRrec joints and the recruitment of Alt-EJ but not NHEJ components at the 3’RR locus in mice.31 In contrast with CSR, Alt-EJ seems, therefore, to surface as the major contributor. Therefore, some arguments support the fact that Sm-3’RRrec is a different IGH recombination from that of CSR. When Sm-3’RRrec hits the IGH locus, it results in the excision of the whole cluster of constant IGH genes. This should kill BCR expression if occurring on the functional IGH allele. Since in vivo loss of BCR induces B-cell death,33 Sm-3’RRrec was initially called “locus suicide recombination”. However, to date, the search for such B cells lacking BCR due to Sm-3’RRrec has failed. Moreover, high throughput sequencing studies on human BCR-positive circulating B cells revealed that amplicons covering Sm-3’RRrec junctions certainly came from the non-functional IGH allele.32 Thus, at present, the function of Sm-3’RRrec in B-cell physiology remains unclear.
In this study, we raised the question of IGH switch blockade in CLL. For that purpose, we analyzed both CSR and Sm-3’RRrec junctions as reflections of putative switch activity. Our results revealed that CLL patients could be separated into two groups with different prognoses on the basis of Sm-3’RRrec counts but not CSR junction counts. Cases with increased Sm-3’RRrec were indeed related to umCLL. Comparison between both groups revealed that Sm-3’RRrec was likely to be ongoing in tumor cells from CLL patients with increased Sm-3’RRrec counts. Structural features of Sm-3’RRrec junctions revealed an imprint that pointed to a different B-cell origin for both groups. Moreover, Sm-3’RRrec appeared to be potentiated by MYC overexpression even in the absence of AID.
Methods
Human materials and ethics
The project was conducted according to the guidelines of the Declaration of Helsinki. CLL peripheral blood mononucelar cells (PBMC) were obtained from CRBioLim, CHU Dupuytren, Limoges Hospital (authorization no.: DC-2008-604, AC-2016-2758, and AC-2019-3418). Tonsils were obtained from children scheduled for elective tonsillectomy from CRBioLim (authorization no.: DC-2008-604, AC-2018-3157). PBMC from healthy volunteers (HV) were collected through the research project approved by CPP Sud MéditerranéeI (authorization no.: 2021-A00778-33).
Human class switch recombination and Sm-3’RRrec junction counts
Human CSR and Sm-3’RRrec junctions were amplified as described32 and used to prepare next-generation sequencing (NGS) libraries (Ion Xpress™ Plus Fragment Library Kit, Life Technologies, Thermofisher, 447269) sequenced with an Ion Proton or S5 chip (Life Technologies). FastQ were analyzed using CSReport.34 CSR and Sm-3’RRrec junction diversities were estimated through the Shannon Diversity Index (see the Online Supplementary Appendix). The Jurkat cell line and naïve B cells sorted from PB of healthy donors (n=2) served as negative controls for Sm-3’RRrec junction detection (no Sm-3’RRrec junctions were detected).
CH12F3 class switch recombination and Sm-3’RRrec junction counts
The CH12F3 cells were transfected or not by MYC expression vector (Plasmid#74164, Addgene) and cultured in RPMI1640 with Ultra Glutamine, 10% fetal calf serum (FCS) (Lonza), sodium pyruvate (Lonza), penicillin/streptomycin (Lonza), non-essential amino acids (Lonza) and β2-mercaptoethanol. Cells were stimulated for CSR toward IgA for 72 hours with murine interleukin (IL)-4 (5 ng/mL; PeproTech), human TGF-β1 (1 ng/mL; R&D Systems), and murine anti-CD40 antibody (Ab) (1 mg/mL; eBioscience). CSR and Sm-3’RRrec junctions were amplified by nested polymerase chain reaction (PCR) using specific primers (Online Supplementary Table S3) as described.34
IGHV sequence analysis
V, D, and J rearranged genes were amplified using the Biomed-2 strategy with FR1 and FR2 primers and sequence analyses were performed as described.35 The IGHV intra-clonal diversity analysis workflow is described in the Online Supplementary Appendix. Diffuse large B-cell lymphoma IGHV sequences are from Rizzo et al.36
Flow cytometry analysis
Immunophenotyping was done on a Navios-flow cytometer (Beckman Coulter) with the protocol for routine CLL diagnosis using: CD5-APC (Beckman Coulter PN a60790, clone BL1a), CD19-ECD (Beckman Coulter A07770, clone J3-119) and anti-human k light chain/anti-human λ light chains/RPE (Dako, FR481 X0935). Results were analyzed with Kaluza software version 2.1 (Beckman Coulter).
RNA extraction, cDNA synthesis and quantitative real time polymerase chain reaction
Total RNA was isolated (TRIzol™ Reagent, 15596018) and reverse-transcribed (Advantage RT-for-PCR kit Applied Biosystems™, Thermofisher 4368814/10400745). Quantitative real time PCR (qRT-PCR) were performed with the SYBR-Green PCR mix (SensiFast hi ROX Syber Green BIO820025) and primers (Online Supplementary Table S3) or with Taqman PCR mix (SensiFast Probe Hi-Rox kit BIO820025) and MYC probe (4331182 Hs00905030_m1, Thermofisher). Normal centroblasts and naïve B cells were sorted from tonsils as described.37
Relative telomere length assay
DNA (25 ng) extracted from PBMC was used in triplicate to assess relative telomere assay (RTL) by qPCR as described previously.38
Mutation analysis of PIM1
We amplified PIM1 exon 4 (Online Supplementary Table S3), containing a CLL AID-targeted nucleotide,18 using Phusion High Fidelity Taq (Thermo Scientific, F-530XL). Products were used to build NGS libraries. The analysis workflow is described in the Online Supplementary Appendix.
Statistical analysis
Graphs, histograms, curves, and standard statistical analyses were designed using GraphPad Prism 6x software. Fisher tests were done with R (version 4.3.0) using the RStudio interface (RStudio 2023.03.0 Build 386). Kaplan Meyer survival curves and Cox univariate and multivariates analyses were done using the R Survival package (URL: https://github.com/therneau/survival).
Results
Chronic lymphocytic leukemia patients can be separated into two groups according to Sm-3’RR recombination
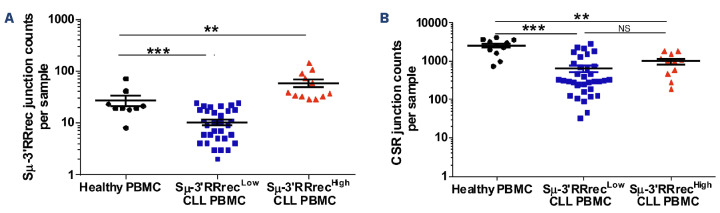
We analyzed counts of Sm-3’RR recombination (Sm-3’RR-rec) in DNA samples collected at CLL diagnosis from 47 patients. Blood tumor infiltration was over 90% circulating lymphocytes in 42 of 47 (89%) cases and over 98% in 37 of 42 cases (79%) (Online Supplementary Figure S1A). Comparison of junction counts was performed with results obtained from DNA from PBMC of nine HV. As negative controls, we used the Jurkat cell line and naïve B cells sorted from PB of healthy donors (n=2), in which no Sm-3’RRrec junctions could be detected (data not shown). Even at low levels, Sm-3’RRrec was found at comparable levels in both HV and CLL (Online Supplementary Figure S1B), and was undetectable in only three of 47 (6.3%) CLL patients. We separated CLL patients into two groups, using as a threshold value the mean of Sm-3’RRrec counts in HV (Online Supplementary Table S1), called Sm-3’RRrecHigh (12/47 patients =26%), and Sm-3’RRrecLow (35/47 patients =74%) (Figure 1A). Analysis of CSR and Sm-3’RR recombinations was based on a nested-PCR approach. Somatic hypermutation (SHM) could theoretically introduce mutations in primer binding DNA, particularly in CLL.39 But here, low levels of Sm (mutation rate average +/- standard error of the mean for Sm-3’RRrecLow, Sm-3’RRrecHigh and HV PBMC respectively are: 1.828+/-0.306, 1.475+/-0.302 and 0.870+/-0.090) and 3’RR2 (mutation rate average +/- standard error of the mean for Sm-3’RRrecLow, Sm-3’RRrecHigh and HV PBMC respectively: 0.048+/-0.018, 0.062+/-0.020 and 0.056+/-0.033) mutation frequency. In DNA segments from Sm-3’RRrec junctions, absence of significant differences between samples ruled out significant bias in amplification of the Sm-3’RRrec junctions and comparison between Sm-3’RRrecHigh and Sm-3’RRrecLow CLL. Sm-3’RRrec counts were not dependent on CLL B-cell richness as shown in the Online Supplementary Figure S1C. Moreover, the percentages of CLL B cells were similar in Sm-3’RRrecHigh and Sm-3’RRrecLow CLL samples (Online Supplementary Figure S1D).
As expected in this IgM+ B-cell cancer, CSR junction levels were much lower in CLL than in HV samples. CSR counts were similarly low in both the Sm-3’RRrecHigh and Sm-3’RRrecLow CLL groups (Figure 1B). We did not find any significant association between increased Sm-3’RRrec and CSR counts as shown in the Online Supplementary Table S2. Moreover, the correlation between CSR and Sm-3’RRrec counts was poor (correlation coefficient r=0.2; data not shown). Thus, Sm-3’RRrecHigh CLL specifically exhibited increased Sm-3’RRrec counts when compared to CSR.
Most patients with Increased Sm-3’RRrec counts had unmutated chronic lymphocytic leukemia
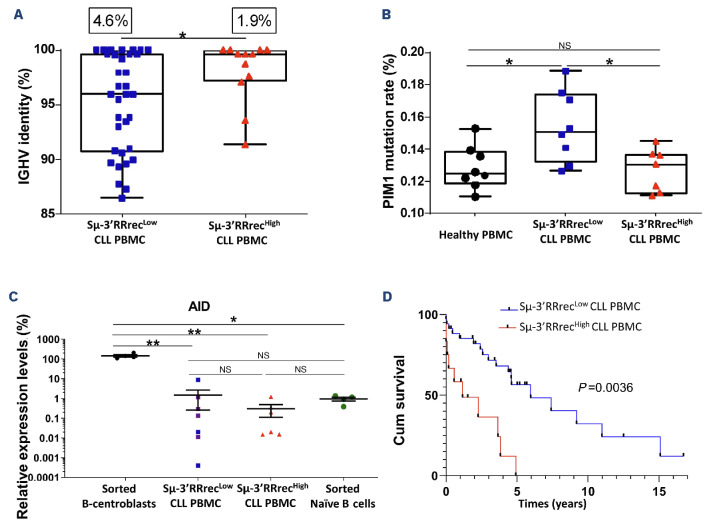
In order to further study Sm-3’RRrecHigh and Sm-3’RRrecLow CLL, we analyzed the IGHV mutational status, an important prognosis indicator of poor outcome. Even if being cautious for small numbers, no significant IGHV gene repertoire bias was found between CLL groups (Online Supplementary Figure S2). IGHV clonal rearrangements of Sm-3’RRrecHigh CLL cases exhibited stronger homology to IGHV reference sequences (Figure 2A). With the threshold of 98% homology, nine of 12 (75%) Sm-3’RRrecHigh CLL were not or only weakly mutated (mean IGHV mutation rate =98.1%). One additional Sμ-3’RRrecHigh CLL had 97% IGHV sequence homology with the reference. Sμ-3’RRrecLow cases were mCLL for which 20 of 35 (57%) cases had a mean IGHV mutation rate =4.8%; P=0.043. Consistently, we found that Sμ-3’RRrecHigh patients exhibited low rates of AID off-target PIM1 mutations (Figure 2B). As CD19 transcription and expression at the cell surface are specific for the B-cell compartment and were similar between Sμ-3’RRrecLow and Sμ-3’RRrecHigh CLL (Online Supplementary Figure S3A, B), transcript expression levels were normalized to those of CD19. When compared to centroblasts and naïve B cells sorted from benign inflammatory tonsils, AID transcript levels were comparable in both Sμ-3’RRrecHigh and Sμ-3’RRrecLow CLL, being as low as in naïve B cells, regardless of the mutated or unmutated IGHV status (Figure 2C).
Figure 1.
Sm-3’RRrec is detectable in chronic lymphocytic leukemia patients and Sm-3’RRrec counts are significantly increased in Sm-3’RRrecHigh samples. (A) Sm-3’RR recombination (Sm-3’RRrec) junction counts analyzed by next-generation sequencing and CSReport in healthy volunteer peripheral blood mononuclear cells (PBMC) (N=9, 239 Sm-3’RRrec junctions) and chronic lymphocytic leukemia (CLL) patients, divided into 2 groups based on the mean of junction counts obtained in healthy PBMC: Sm-3’RRrecLow CLL (≤27 junctions per sample, N=35, 357 junctions) and Sm-3’RRrecHigh CLL (>27 junctions per sample, N=12, 703 junctions). (B) Class switch recombination (CSR) junction counts were at comparable levels in both CLL groups (Sm-3’RRrecLow: N=35, 22,247 junctions; Sm-3’RRrecHigh: N=11, 10,739 junctions) and were lower than in HV PBMC (N=11, 27,528 junctions). Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test. 3’RR: 3’ regulatory region; NS: not significant; **P<0.01; ***P<0.001.
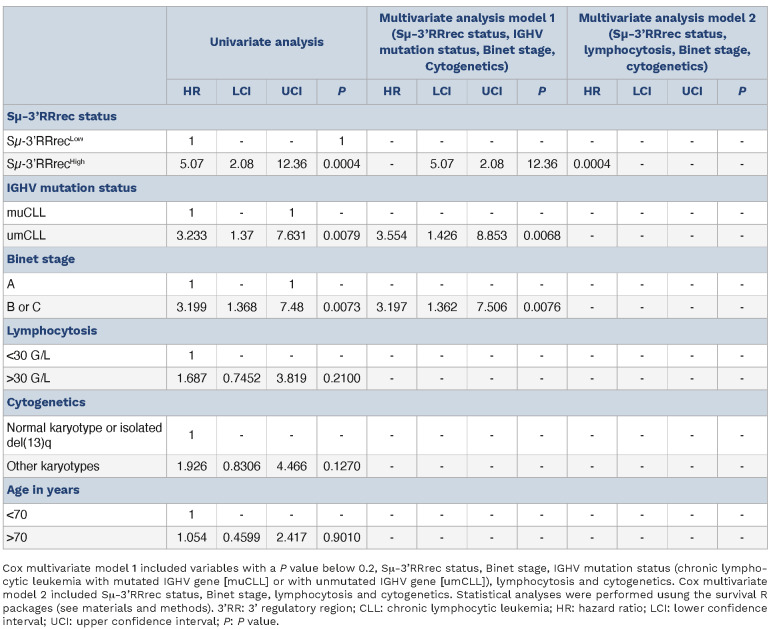
In agreement with the strong predominance of umCLL in this group, Sμ-3’RRrecHigh CLL were associated with decreased treatment-free survival (TFS) (≈14 months compared to ≈71 months; P<0.001; Figure 2D). In comparison, separating patients into CSRLow and CSRHigh groups did not result in significant differential TFS even if survival curves were separated (Online Supplementary Figure S1E). For this series, TFS also strongly depended on the Binet stage and the IGHV mutation status, and marginally depended on lymphocytosis and cytogenetics (Online Supplementary Figure S5). In order to search for independent variables, a Cox univariate analysis was first done for Sm-3’RRrec status, Binet stage, lymphocytosis, age, IGHV mutation status and cytogenetics (Table 1). A first Cox multivariate model was constructed with variables with a P value <0.2, id est Sm-3’RRrec status, Binet stage, IGHV mutation status, and cytogenetics (Table 1). In this model, IGHV mutation status and Binet stage were the two independent variables. However, a Cox model including only IGHV mutation and Sm-3’RRrec status suggested that the confounding variable was the IGHV mutation. Indeed, a second model replacing IGHV mutation status by lymphocytosis pointed on Sm-3’RRrec status as the sole independent variable (Table 1). Revealing strong overlaps between IGHV mutation and Sm-3’RRrec status in terms of TFS, these analyses reflect the enrichment Sm-3’RRrecHigh group in umCLL, which are very well known to have a poor prognosis.6,7
Figure 2.
Enrichment in unmutated chronic lymphocytic leukemia and poor prognosis of chronic lymphocytic leukemia patients with increased Sμ-3’RRrec counts. (A) Low Sμ-3’RR recombination (Sμ-3’RRrecLow) chronic lymphocytic leukemia (CLL) cases (N=34) had lower percentages of sequence identity with the reference sequence compared to the high homology of the variable region of the immunoglobulin heavy chain variable region (IGHV) segments in Sμ-3’RRrecHigh CLL (N=12). For each CLL group, the somatic hypermutation rate mean is indicated above the graph. (B) Sequence analysis of PIM1, activation induced-cytidine deaminase (AID) off-target gene. The mutation rate of PIM1 was significantly increased in Sμ-3’RRrecLow patients (N=8) compared to healthy peripheral blood mononuclear cells (PBMC) (N=8) and Sμ-3’RRrecHigh CLL (N=7). (C) AID transcripts, relative to CD19 transcripts, were lower in Sμ-3’RRrecLow CLL (N=7) and Sμ-3’RRrecHigh CLL (N=6) compared to normal B centroblasts (N=4) used as positive controls and comparable to AID transcript levels in sorted naïve B cells (N=4) used as negative controls. Purple dots correspond to mutated IGHV CLL samples. (D) Cumulative survival (Cum survival) time (years) without treatment (treatment-free survival [TFS]) for patients indicated shorter TFS in Sμ-3’RRrecHigh CLL (N=12) than Sμ-3’RRrecLow CLL (N=34). Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test (A, B, C) or Χ2 test (D). 3’RR: 3’ regulatory region; NS: not significant; *P<0.05; **P<0.01; ***P<0.001.
In contrast to poorly-diversified IGHV clonal rearrangements, Sm-3’RRrecHigh chronic lymphocytic leukemia exhibited increases in both Sm-3’RRrec and class switch recombination diversities with increased IGH locus accessibility
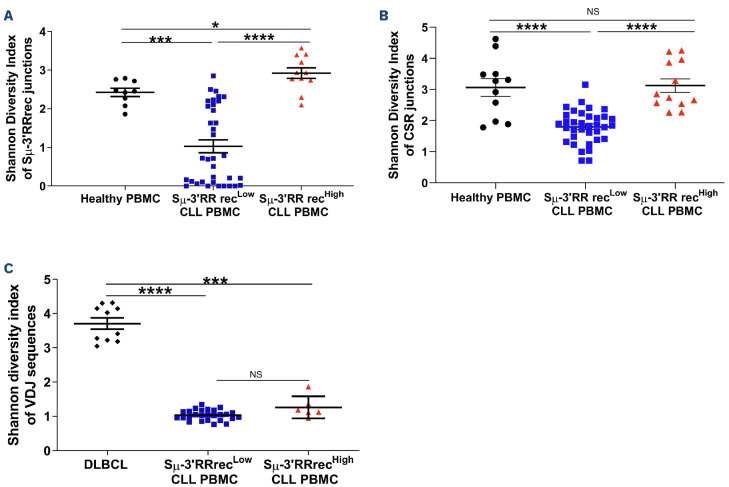
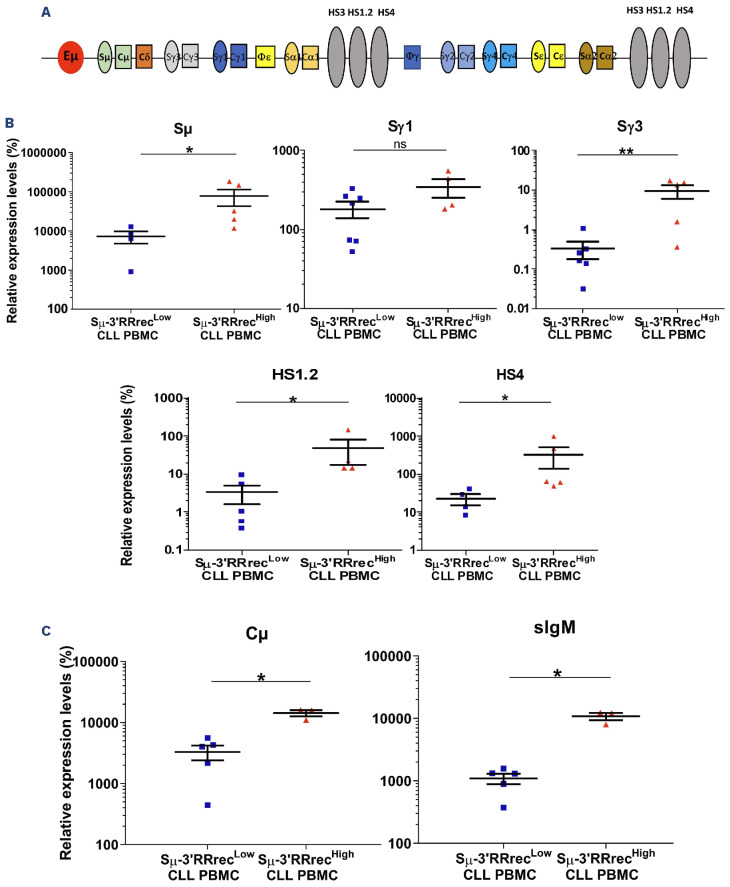
Sm-3’RRrec and CSR result from random IGH recombination involving two DNA DSB, one in the Sm donor region and one in the 3’RR or Sx acceptor region respectively. Since CLL is IgM+, Sm-3’RR rearrangements have to occur on the non-productive IGH allele; this raises the question of Sm-3’RRrec clonality. Because CSR can also take place on the non-productive allele, both Sm-3’RRrec and CSR junction diversities were evaluated using the Shannon index which measures the number of different junctions in sequencing libraries.39 HV were used here as controls of “polyclonal” junctions. Diversities of both Sm-3’RRrec and CSR junctions were strongly decreased in Sm-3’RRrecLow CLL samples, a result that should be expected in this clonal IgM+ B-cell cancer. Strikingly, the Shannon diversity index was significantly higher in Sm-3’RRrecHigh CLL samples than in Sm-3’RRrecLow CLL (Figure 3A, B). Both the absolute numbers of B cells (Online Supplementary Figure S4) and diversity indexes of CLL IGHV clonal rearrangements (Figure 3C) were similar in both the Sm-3’RRrecHigh and Sm-3’RRrecLow groups. CLL IGHV diversity was much lower than DLBCL, known to harbor intra-tumoral subclones with divergent IGHV after SHM and taken here as positive controls of intra-tumor diversity. Therefore, increased Sm-3’RRrec and CSR junction diversities were not likely to be influenced by B-cell richness but would rather reflect diversification of heavy chain rearrangements in Sm-3’RRrecHigh CLL. Because increased diversities of Sm-3’RRrec and CSR junctions in Sm-3’RRrecHigh CLL are evocative of an on-going process, we evaluated whether IGH locus DNA was accessible to recombination machinery. In order to assess locus aperture, we analyzed the expression of non-coding and coding transcripts from the constant part of the IGH locus (Figure 4A). We found higher levels of Sm, Sγ1, Sγ3, HS1.2 and HS4 sterile transcripts in Sm-3’RRrecHigh than in Sm-3’RRrecLow CLL (Figure 4B), meaning that the IGH locus was accessible to the recombination machinery in these patients. While levels of surface Ig were comparable between the two groups (data not shown), coding Cm transcripts were also increased in Sm-3’RRrecHigh CLL (Figure 4C). Altogether, our results suggest abnormal intratumoral IGH remodeling activity in the Sm-3’RRrecHigh CLL group.
Table 1.
Univariate and multivariate analysis of treatment-free survival including biological parameters in the chronic lymphocyt-ic leukemia cases from the study.
In contrast with those of class switch recombination, structural features of Sm-3’RRrec junctions harbor an activated B-cell imprint and discriminate both Sm-3’RRrecLow and Sm-3’RRrecHigh chronic lymphocytic leukemia
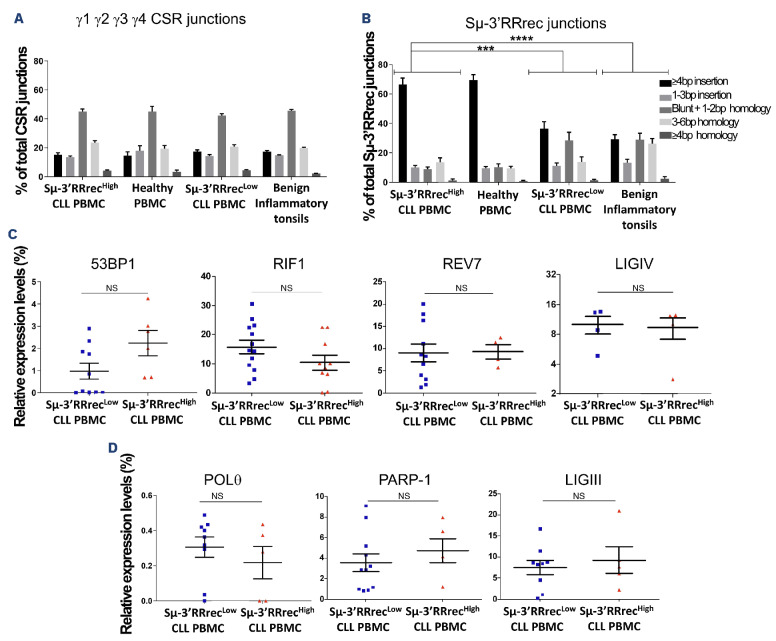
In addition to being unique, joint structure is differently shaped according to the DSBR machinery. In B cells, DS-BR occurs mainly through NHEJ and, to a lesser extent, through Alt-EJ pathways for IGH recombination.22,23 The joint structure of each single Sm-3’RRrec and CSR junction can be determined by alignment to reference sequences. We performed structural analyses of Sm-3’RRrec and CSR junctions in CLL samples, HV PBMC and benign inflammatory tonsil cells. Here, while circulating B lymphocytes from HV, included because they were exempt of any known disease, were predominantly resting, tonsils were analyzed since they are very well known benign inflammatory lymphoid tissues with highly active B-cell responses and numerous GC, which are the main site of post-medullary Ig gene recombination. CSR joint structures were comparable between HV PBMC and tonsils and were similar to those of CLL regardless of Sμ-3’RRrec status (Figure 5A). In contrast, the structure of Sμ-3’RRrec joints differed between HV PBMC and tonsils. The latter exhibited more Sn-3’RRrec junctions with small microhomologies (1-2 bp) and blunt junctions while long insertions (≥4 bp) were predominant in the former (Figure 5B). Therefore, Sμ-3’RRrec joint structures seem to be differently imprinted according to tissue origin and/or B-cell activation. This was not the case for CSR, a strong indication that these two IGH recombination events, even if mechanistically close, are not linked. Strikingly, the structures of Sμ-3’RRrec junctions differed between Sμ-3’RRrecHigh and Sμ-3’RRrecLow CLL, the former being close to those of HV PMBC and the latter more similar to benign inflammatory tonsils.
Figure 3.
Intratumoral IGH remodeling activity in the Sm-3’RRrecHigh chronic lymphocytic leukemia group. The Shannon diversity index was used to estimate class switch recombination (CSR), Sm-3’RR recombination (Sm-3’RRrec) and intra-clonal immunoglobulin heavy chain variable region (IGHV) diversities. (A) Higher Sm-3’RRrec junction diversity was observed in Sm-3’RRrecHigh chronic lymphocytic leukemia (CLL) samples (N=11) compared to Sm-3’RRrecLow (N=35) CLL and healthy peripheral blood mononuclear cells (PBMC) (N=9). (B) CSR junction diversity was increased in Sm-3’RRrecHigh CLL samples (N=12) compared to Sm-3’RRrecLow (N=35) CLL and comparable to those of healthy PBMC (N=11). (C) CLL IGHV diversities were lower than those observed in diffuse large B-cell lymphomas (DL-BCL) harboring intra-tumoral subclones with divergent IGHV after somatic hypermutation (SHM) used as positive controls (N=10). Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test. IGH: immunoglobulin heavy chain; 3’RR: 3’ regulatory region; NS: not significant; *P<0.05; **P<0.01; ***P<0.001.
Figure 4.
Increased IGH locus accessibility in Sm-3’RRrecHigh chronic lymphocytic leukemia. Quantification of immunoglobulin heavy chain (IGH) locus non-coding transcripts (Sm, Sγ1, Sγ3, HS1.2 and HS4) relative to those of CD19 (A) and coding transcripts (Cm and surface immunglobulin M [sIgM]) (B) in Sm-3’RRrecLow (N=4-7) and Sm-3’RRrecHigh (N=3-5) chronic lymphocytic leukemia (CLL). Sm-3’RRrecHigh exhibited high levels of IGH locus transcription in both productive and non-productive transcripts. Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test. 3’RR: 3’ regulatory region; NS: not significant; PBMC: peripheral blood mononuclear cells; *P<0.05; **<0.01.
Figure 5.
Sμ-3’RRrec junction structural features are related to different lymphoid tissue imprints and discriminate between Sμ-3’RRrecLow and Sμ-3’RRrecHigh chronic lymphocytic leukemia. Structures at the Sμ-3’RR recombination (Sμ-3’RRrec) and class switch recombination (CSR) junctions were determined using CSReport by alignment to reference sequences for chronic lymphocytic leukemia (CLL) samples, healthy volunteer (HV) peripheral blood mononuclear cells (PBMC) and benign inflammatory tonsil cells. Structural features account for length in base pairs (bp) of nucleotide insertions at the joint, of short homology (microhomology) between acceptor and donor sequences and absence of insertions and homology (blunt) (A) Sμ-Sy1, SΜ-SY2, SΜ-SY3, SΜ-SY4 CSR joint structures were comparable between HV PBMC, benign inflammatory tonsils and CLL. (B) The Sμ-3’RRrec junction structure differed between CLL: Sμ-3’RRrecHigh samples were comparable to HV PBMC and predominantly exhibited junctions with long insertions (≥4 bp) while Sμ-3’RRrecLow CLL and benign inflammatory tonsils were enriched in Sμ-3’RRrec junctions with small microhomologies (1-2 bp) and blunt junctions. Quantification of transcripts coding for actors implicated in double strand break (DSB) repair by quantitative real-time polymerase chain reaction relative to CD19 transcripts (C) by non-homologous end joining (NHEJ) (53BP1, RIF1, Rev7 and LIGIV) and (D) by alternative end-joining (Alt-EJ) (PARP-1, POL9 and LIGIII). Statistical analyses were performed using the X2 test (A, B) or unpaired t test (C, D). 3’RR: 3’ regulatory region; NS: not significant; ***P<0.001; ****P<0.0001.
Decreased small microhomologies (1-2 bp) and blunt junctions argue against NHEJ involvement in DSBR of Sm-3’RRrec junctions from both HV PBMC and Sm-3’RRrecHigh CLL.22,23,40 We, therefore, quantified transcripts coding for actors implicated in the protection of DSB DNA ends and favoring NHEJ (53BP1, Rif1 and Rev7, a Shieldin complex component), NHEJ actor (LIGIV) and Alt-EJ components (PARP-1, POL8 and LIGIII). We did not detect any significant difference in the tested transcripts of NHEJ proteins (Figure 5C) or Alt-EJ actors (Figure 5D) between Sm-3’RRrecHigh and Sm-3’RRrecLow. As already suggested by the fact that CSR junction structures were similar between Sm-3’RRrecHigh and Sm-3’RRrecLow CLL, this suggests that Sm-3’RRrec junction structural differences observed for both Sm-3’RRrec CLL groups were not due to imbalances in NHEJ/Alt-EJ actors. Altogether, these results indicate that Sm-3’RRrecHigh and Sm-3’RRrecLow CLL have different Sm-3’RRrec imprints, the former being close to recirculating resting B cells and the latter reflecting activated B cells in a benign inflammatory secondary lymphoid organ.
Overexpression of c-MYC potentiated Sm-3’RR recombination even in the absence of AID
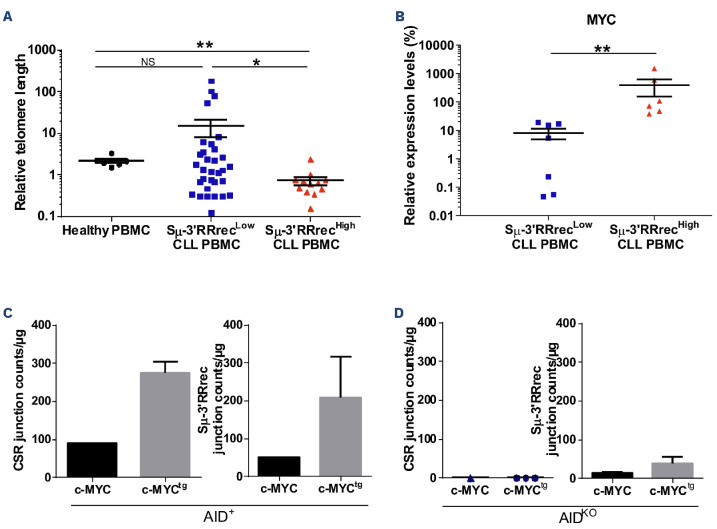
Because IGH locus accessibility is also linked to B-cell activation and proliferation,21 we evaluated the past history of CLL B-cell proliferation by measuring relative telomere length. While homogeneous in HV (mean of 2.14), telomere lengths were very heterogeneous in Sm-3’RRrecLow patients; 13 of 32 (40%) had long or very long telomeres, indicating that cells underwent few proliferation cycles. In contrast, all but one Sm-3’RRrecHigh patient homogeneously exhibited telomeres that were shorter than HV, reflecting increased proliferation cycle numbers (Figure 6A). Shorter telomeres in Sm-3’RRHigh CLL were associated with increased MYC expression (Figure 6B).
We thus raised the question of the impact of c-MYC on IGH recombination. For this purpose, we used the murine B-cell lymphoma CH12F3 cell line and its AID knockout (KO) counterpart stably transfected or not with a MYC overexpression vector and stimulated in vitro to undergo CSR and Sm-3’RRrec. As shown in Figure 6C, the levels of both CSR and Sm-3’RRrec were increased when MYC was overexpressed in the AID context (AID+MYCtg). In the absence of AID (AIDKO), CSR was undetectable either in absence or presence of MYC overexpression. Some Sm-3’RRrec junctions were detectable in the absence of both AID and the MYC overexpressing vector. Induction of MYC overexpression resulted in increased numbers of Sm-3’Rrrec events (Figure 6D). Contamination could be ruled out because sequences of these Sm-3’RRrec junctions were unique. Some of these junctions contained sequence fragments of Sε and Sy2 regions between Sm and 3’RR, which suggests that these Sm-3’RRrec events were sequentially preceded by a CSR event. These results show that MYC potentiated both CSR and Sμ-3’RRrec when AID was expressed. Even at low frequency, Sμ-3’RRrec was possible in the absence of AID but in the presence of MYC overexpression.
Discussion
In this study, we observed that Sm-3’RRrec was detectable in CLL patients. Moreover, we showed that the Sm-3’RRrec rate was increased at levels even higher than in polyclonal HV in one group of CLL cases, the Sm-3’RRrecHigh group. In the Sm-3’RRrecHigh group, Sm-3’RRrec appeared to be on-going, as supported by increased diversity of the Sm-3’RRrec junctions.
Despite low CSR junction counts, CSR diversity was also significantly higher in Sm-3’RRrecHigh CLL samples than in Sm-3’RRrecLow CLL meaning that CSR was on-going in these cases. On-going IGH CSR can be observed in a restricted subpopulation of tumoral CLL cells.15,17,41,42 In our study, increased Sm-3’RRrec and CSR diversities in Sm-3’RRrecHigh CLLs reflected IGH remodeling in the tumoral B-cell clone. Since CLL cases of this series were all IgM+, Sm-3’RRrec had occurred on the non-productive IGH allele while CSR could occur on both alleles. CSR mainly occurs on the functional allele.43,44 Here, the numbers of CSR junctions were strongly decreased in CLL without any differences between Sm-3’RRrecHigh and Sm-3’RRrecLow CLL. This indicates that despite increased IGH remodeling, Sm-3’RRrecHigh class-switched CLL cells could have been counterselected. Indeed, as reviewed recently, IgM BCR is a key component of CLL pathogenesis development and evolution, not only for its antigenic recognition properties but also likely through its structure and its signaling capacities.45 Even if the CSR junction structure appeared identical among samples, Sm-3’RRrecLow and Sm-3’RRrecHigh CLL samples exhibited different Sm-3’RRrec structural profiles. In Sm-3’RRrecHigh CLL, Sm-3’RRrec junctions were close to those of circulating B cells, while Sm-3’RRrec junctions from Sm-3’RRrecLow CLL were similar to those of tonsils B cells. According to Sm-3’RRrec junction imprints, our results are full agreement with the hypothesis that CLL can be subdivided according to two different COO. Since physiologically most circulating B cells are IgM+IgD+ pre-GC cells and since tonsils contain numerous active GC, we suggest that the imprint of Sm-3’RRrec junctions would be from circulating non-GC experienced B cells for Sm-3’RRrecHigh cases (mostly umCLL) while Sm-3’RRrecLow cases (mostly mCLL) would have Sm-3’RRrec junction imprints of AID-experienced B cells issued from secondary lymphoid organs. This correlates with IGHV mutational status and PIM1 mutation rate that were decreased in Sm-3’RRrecHigh CLL and increased in Sm-3’RRrecLow CLL.
Consistent with increased IGH recombination activity, the IGH locus was strongly transcribed in Sm-3’RRrecHigh CLL and was thus targetable by the IGH recombination machinery. Characteristic of activated and proliferating cells, shorter telomeres in Sm-3’RRrechigh CLL indicate increased numbers of past mitoses. Upon B-cell stimulation, the MYC gene frequently relocates to the transcription factory occupied by the IGH locus.46 Consistently, we observed increased c-MYC expression levels in Sm-3’RRrecHigh CLL. This result led us to evaluate the impact of c-MYC on CSR and Sm-3’RRrec. We found that c-MYC overexpression potentiated both CSR and Sm-3’RRrec recombination in the presence of AID. Moreover, c-MYC induced increases in Sm-3’RRrec counts in the absence of AID. It has been recently shown that some residual CSR can occur in absence of the AID.47 Due to specific constitutively occuring IGH loop conformation between 3’RR, Em and Sm in mature B cells,48 DNA segments from both Sm and 3’RR are likely to be in close proximity and to be recombined together when DSB-targeted even in the absence of AID. Regardless of the AID status of CLL cells, this c-MYC effect contributes to genetic instability. Indeed, CLL is known to harbor DNA repair alterations and to accumulate DSB across the genome.49,50 Moreover, both NHEJ and Alt-EJ are good candidate processes for chromosomal material exchange due to their capacity to ligate DNA ends from independent molecular origins.
Figure 6.
Increased IGH remodeling in Sm-3’RRrecHigh chronic lymphocytic leukemia cells is potentiated by high levels of c-MYC expression. (A) Relative telomere length (RTL) measured by specific quantitative real time polymerase chain reaction relative to the human β globin gene (Sm-3’RRrecLow, N=33; Sm-3’RRrecHigh, N=12; healthy volunteer [HV] peripheral blood mononuclear cells [PBMC], N=6). Telomere length was significantly shorter in the Sm-3’RRrecHigh group compared to the Sm-3’RRrecLow group and HV PBMC. (B) c-MYC expression was higher in Sm-3’RRrecHigh (N=6) compared to Sm-3’RRrecLow chronic lymphocytic leukemia (CLL) samples (N=7). (C) Detection of class switch recombination (CSR) and Sm-3’RR recombination (Sm-3’RRrec) junctions in activated CH12F3 clones overexpressing or not MYC in the presence of activation induced-cytidine deaminase (AID) (CSR: AID+, N=1 and AID+MYCtg, N=3; Sm-3’RRrec: AID+, N=1 and AID+ MYCtg, N=3) suggested that c-MYC tends to increase CSR and Sm-3’RRrec counts. This was also observed for Sm-3’RRrec in the absence of AID (D), as Sm-3’RRrec junctions, even if rare, were detectable in CH12F3 AIDKO clones (N=2) and appeared to increase with MYC overexpression (AIDKO MYCtg, N=2). No CSR junctions were detected in the absence of AID (AIDKO, N=1), even with MYC overexpression (AIDKOMYCtg, N=3). Graphs represent the mean ± standard error of the mean. Statistical analyses were performed using unpaired t test. KO: knockout; tg: transgenic; 3’RR: 3’ regulatory region; NS: not significant; *P<0.05; **P<0.01; ***P<0.001.
IGHV mutation and Sm-3’RRrec status exhibited strong overlaps in terms of TFS, which reflects that fact that Sμ-3’RRrecHigh group was enriched in umCLL. Here, TFS very likely reflects the different natural histories of the disease, which should be underpinned by biological differences. Separating CLL patients in the CLL Sμ-3’RRrecLow group that would originate from GC-experienced B cells or in the CLL Sμ-3’RRrecHigh group with probable non-GC experienced B cells and close to umCLL, our study characterizes for the first time active IGH recombination potentiated by c-Myc overexpression in Sμ-3’RRrecHigh CLL, even in the absence of AID.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Villéger at the CRBioLim and Dr. Vallejo and the INSERM CIC 1435, Dupuytren Hospital, Limoges, France, for providing human samples. We thank Dr. Jeanne Cook Moreau (UMR CNRS 7276 INSERM 1276) for careful English editing.
Funding Statement
Funding: This work was supported by grants to SP from la Ligue Contre le Cancer (Comité de la Haute-Vienne, CD87), the Fondation ARC pour la Recherche sur le Cancer (ARCP-JA2022060005138), the INCa-Cancéropôle GSO (Emergence program no. 2019-E11) and grants to CRIBL laboratory from the Institut CARNOT CALYM. IAJ is supported by Fondation pour la Recherche Medicale and Lebanese associations (AZM and Saade, LASeR). MP is supported by Région Nouvelle Aquitaine and Université de Limoges. KG is supported by Région Nouvelle Aquitaine and Délégation INSERM Nouvelle Aquitaine. JP is supported by the Institut CARNOT CALYM. SP is a National Institute of Health and Medical Research (INSERM) investigator.
References
- 1.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46(2):219-234. [DOI] [PubMed] [Google Scholar]
- 3.Binet JL, Auquier A, Dighiero G, et al. A new prognostic classification of chronic lymphocytic leukemia derived from a multivariate survival analysis. Cancer. 1981;48(1):198-206. [DOI] [PubMed] [Google Scholar]
- 4.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133(11):1205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rossi D, Gaidano G. The clinical implications of gene mutations in chronic lymphocytic leukaemia. Br J Cancer. 2016;114(8):849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840-1847. [PubMed] [Google Scholar]
- 7.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 8.Stamatopoulos K, Agathangelos A, Rosenquist R, Ghia P. Antigen receptor stereotypy in chronic lymphocytic leukemia. Leukemia. 2017;31(2):282-291. [DOI] [PubMed] [Google Scholar]
- 9.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236-1242. [DOI] [PubMed] [Google Scholar]
- 10.Kulis M, Merkel A, Heath S, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wierzbinska JA, Toth R, Ishaque N, et al. Methylome-based cell-of-origin modeling (Methyl-COOM) identifies aberrant expression of immune regulatory molecules in CLL. Genome Med. 2020;12(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein U, Tu Y, Stolovitzky GA, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J Exp Med. 2001;194(11):1625-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng A, Chiorazzi N. Potential relevance of B-cell maturation pathways in defining the cell(s) of origin for chronic lymphocytic leukemia. Hematol Oncol Clin North Am. 2021;35(4):665-685. [DOI] [PubMed] [Google Scholar]
- 14.Rosenwald A, Alizadeh AA, Widhopf G, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194(11):1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palacios F, Moreno P, Morande P, et al. High expression of AID and active class switch recombination might account for a more aggressive disease in unmutated CLL patients: link with an activated microenvironment in CLL disease. Blood. 2010;115(22):4488-4496. [DOI] [PubMed] [Google Scholar]
- 16.Patten PEM, Chu CC, Albesiano E, et al. IGHV-unmutated and IGHV-mutated chronic lymphocytic leukemia cells produce activation-induced deaminase protein with a full range of biologic functions. Blood. 2012;120(24):4802-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oppezzo P, Vuillier F, Vasconcelos Y, et al. Chronic lymphocytic leukemia B cells expressing AID display dissociation between class switch recombination and somatic hypermutation. Blood. 2003;101(10):4029-4032. [DOI] [PubMed] [Google Scholar]
- 18.Morande PE, Yan X-J, Sepulveda J, et al. AID overexpression leads to aggressive murine CLL and nonimmunoglobulin mutations that mirror human neoplasms. Blood. 2021;138(3):246-258. [DOI] [PubMed] [Google Scholar]
- 19.Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102(5):565-575. [DOI] [PubMed] [Google Scholar]
- 20.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553-563. [DOI] [PubMed] [Google Scholar]
- 21.Stavnezer J, Guikema JEJ, Schrader CE. Mechanism and regulation of class switch recombination. Annu Rev Immunol. 2008;26261-26292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soulas-Sprauel P, Le Guyader G, Rivera-Munoz P, et al. Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J Exp Med. 2007;204(7):1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan CT, Boboila C, Souza EK, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449(7161):478-482. [DOI] [PubMed] [Google Scholar]
- 24.Manis JP, Morales JC, Xia Z, Kutok JL, Alt FW, Carpenter PB. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5(5):481-487. [DOI] [PubMed] [Google Scholar]
- 25.Ward IM, Reina-San-Martin B, Olaru A, et al. 53BP1 is required for class switch recombination. J Cell Biol. 2004;165(4):459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman JR, Barral P, Vannier J-B, et al. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol Cell. 2013;49(5):858-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Virgilio M, Callen E, Yamane A, et al. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 2013;339(6120):711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noordermeer SM, Adam S, Setiaputra D, et al. The shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560(7716):117-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pinaud E, Marquet M, Fiancette R, et al. The IgH locus 3’ regulatory region: pulling the strings from behind. Adv Immunol. 2011;11027-11070. [DOI] [PubMed] [Google Scholar]
- 30.Péron S, Laffleur B, Denis-Lagache N, et al. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science. 2012;336(6083):931-934. [DOI] [PubMed] [Google Scholar]
- 31.Boutouil H, Boyer F, Cook-Moreau J, Cogné M, Péron S. IgH locus suicide recombination does not depend on NHEJ in contrast to CSR in B cells. Cell Mol Immunol. 2019;16(2):201-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalloul I, Boyer F, Dalloul Z, et al. Locus suicide recombination actively occurs on the functionally rearranged IgH allele in B-cells from inflamed human lymphoid tissues. PLoS Genet. 2019;15(6):e1007721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lam KP, Kühn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90(6):1073-1083. [DOI] [PubMed] [Google Scholar]
- 34.Boyer F, Boutouil H, Dalloul I, et al. CSReport: a new computational tool designed for automatic analysis of class switch recombination junctions sequenced by high-throughput sequencing. J Immunol. 2017;198(10):4148-4155. [DOI] [PubMed] [Google Scholar]
- 35.Gachard N, Parrens M, Soubeyran I, et al. IGHV gene features and MYD88 L265P mutation separate the three marginal zone lymphoma entities and Waldenström macroglobulinemia/ lymphoplasmacytic lymphomas. Leukemia. 2013;27(1):183-189. [DOI] [PubMed] [Google Scholar]
- 36.Rizzo D, Viailly P-J, Mareschal S, et al. Oncogenic events rather than antigen selection pressure may be the main driving forces for relapse in diffuse large B-cell lymphomas. Am J Hematol. 2017;92(1):68-76. [DOI] [PubMed] [Google Scholar]
- 37.Wohlford EM, Baresel PC, Wilmore JR, Mortelliti AJ, Coleman CB, Rochford R. Changes in tonsil B cell phenotypes and EBV receptor expression in children under 5-years-old. Cytometry B Clin Cytom. 2018;94(2):291-301. [DOI] [PubMed] [Google Scholar]
- 38.Joglekar MV, Satoor SN, Wong WKM, Cheng F, Ma RCW, Hardikar AA. An optimised step-by-step protocol for measuring relative telomere length. Methods Protoc. 2020;3(2):E27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A. Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol. 2017;17(1):61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boboila C, Yan C, Wesemann DR, et al. Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4. J Exp Med. 2010;207(2):417-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oppezzo P, Magnac C, Bianchi S, et al. Do CLL B cells correspond to naive or memory B-lymphocytes? Evidence for an active Ig switch unrelated to phenotype expression and Ig mutational pattern in B-CLL cells. Leukemia. 2002;16(12):2438-2446. [DOI] [PubMed] [Google Scholar]
- 42.Oppezzo P, Navarrete M, Chiorazzi N. AID in chronic lymphocytic leukemia: induction and action during disease progression. Front Oncol. 2021;11:634383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci U S A. 2005;102(40):14362-14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakai E, Bottaro A, Alt FW. The Ig heavy chain intronic enhancer core region is necessary and sufficient to promote efficient class switch recombination. Int Immunol. 1999;11(10):1709-1713. [DOI] [PubMed] [Google Scholar]
- 45.Young RM, Phelan JD, Wilson WH, Staudt LM. Pathogenic B cell receptor signaling in lymphoid malignancies: new insights to improve treatment. Immunol Rev. 2019;291(1):190-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Osborne CS, Chakalova L, Mitchell JA, et al. Myc dynamically and preferentially relocates to a transcription factory occupied by Igh. PLoS Biol. 2007;5(8):e192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalloul I, Laffleur B, Dalloul Z, et al. UnAIDed Class switching in activated B-cells reveals intrinsic features of a self-cleaving IgH locus. Front Immunol. 2021;12:7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wuerffel R, Wang L, Grigera F, et al. S-S synapsis during class switch recombination Is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27(5):711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deriano L, Guipaud O, Merle-Béral H, et al. Human chronic lymphocytic leukemia B cells can escape DNA damage-induced apoptosis through the nonhomologous end-joining DNA repair pathway. Blood. 2005;105(12):4776-4783. [DOI] [PubMed] [Google Scholar]
- 50.Popp HD, Flach J, Brendel S, et al. Accumulation of DNA damage and alteration of the DNA damage response in monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. Leuk Lymphoma. 2019;60(3):795-804. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.