To the Editor,
Genetic factors and early-life exposures1 are both important determinants of childhood asthma. Among the more than 150 loci identified in genome-wide association studies (GWAS), 17q12-q21 is the most replicated childhood-onset asthma locus.2 Fine-mapping at this locus has been difficult in populations of European ancestry because of extensive linkage disequilibrium (LD) in this region. Taking advantage of the reduced LD in African ancestry populations, we3 and others4,5 have shown that single nucleotide polymorphisms (SNPs) in GSDMB are likely causal for childhood-onset asthma at this locus. However, it remains unclear whether the same variants underlie the many genotype-exposure interaction (GEI) effects. Moreover, most previous interaction studies were conducted in subjects of European ancestry,2 and it is not known whether the same GEIs are present in children of non-European ancestry. To address these questions, we asked whether genotype at the 17q12-q21 locus modifies the risk of asthma at age 7 as a function of early-life exposures. We investigated GEIs between nine 17q12-q21 SNPs and early-life exposures in 262 Black children with a family history of asthma or allergy in the Urban Environment and Childhood Asthma (URECA) cohort.
Of the 262 unrelated children in this study, 85 (32%) were diagnosed with asthma at age 7 years (Table 1). We were able to recapitulate in the Black children the previously reported relationships between environmental exposures and asthma observed in the larger cohort (442 participants),1 correcting for sex, study site, and 3 ancestry principal components. Exposure to common indoor allergens (cockroach, cat, dog, mouse), cumulative over first 3 years of life and at 3 months of age, was inversely associated with asthma risk at age 7 (cumulative exposure: odds ratio (OR) = 0.59 [95% CI 0.35-0.99], p= 0.048; exposure at 3 months: OR= 0.53 [95% CI 0.32-0.88], p= 0.015). Having more colds during the first year or the first 3 years of life was associated with an increased risk of asthma (OR= 2.03 [95% CI 1.24-3.33] p= 0.0052 and OR= 3.51 [95% CI 1.87-6.57] p= 8.68x10−5). Diversity of house dust microbiota in early life was not associated with asthma at age 7 (OR= 1.00 [95% CI 0.99-1.01], p= 0.49).
Table 1.
Characteristics of study population (Black children of the URECA cohort)
| Combined sample (n=262) | Children with asthma at age 7 (n=85) | Children without asthma by age 7 (n=177) | OR (95% CI) | p-value | |
|---|---|---|---|---|---|
| Sex (male) | 129 (49%) | 48 (56%) | 81 (46%) | 1.53 (0.91-2.59) | 0.11 |
| Mother has asthma | 117 (45%) | 47 (55%) | 70 (40%) | 1.89 (1.12-3.19) | 0.017 |
| Father has asthma | 80 (30%) | 28 (33%) | 52 (29%) | 1.18 (0.68-2.06) | 0.56 |
| Cesarean section delivery | 77 (29%) | 28 (33%) | 49 (28%) | 1.28 (0.73-2.25) | 0.38 |
| Breastfeeding | 135 (52%) | 46 (54%) | 81 (46%) | 1.40 (0.83-2.35) | 0.21 |
| Age of mother at time of birth (y) | 24±5.7 | 24.7±6.3 | 23.7±5.3 | 1.02(0.98-1.08) | 0.21 |
| No. of hours/week of day care in y1 | 13.3±18.6 | 14.5±19.4 | 12.7±18.3 | 1.01 (0.99-1.02) | 0.47 |
| No. of children in home | 1.32±1.56 | 1.38±1.79 | 1.30±1.44 | 1.03 (0.88-1.22) | 0.71 |
| Cord blood cotinine detected1 | 44 (18%) | 19 (23%) | 25 (16%) | 4.3 (0.97-19) | 0.055 |
| Atopy2 | 166 (63%) | 63 (74%) | 103 (58%) | 2.06 (1.16-3.64) | 0.013 |
| FEV1 % predicted3 | 101.3±14.9 | 100.5±15.9 | 101.7±14.5 | 0.99 (0.97-1.02) | 0.60 |
| FEV1/FVC4 | 0.82±0.08 | 0.81±0.08 | 0.83±0.07 | 0.12 (0.002-7.85) | 0.32 |
| Asthma controller medication in the past 12 months5 | 34 (13%) | 27 (31%) | 7 (4%) | 10.64 (4.39-25.75) | NA |
| Albuterol use in the past 12 months5 | 86 (34%) | 71 (84%) | 15 (9%) | 51.4 (23.5-112.2) | NA |
| Prednisone in the past 12 months5 | 54 (21%) | 35 (41%) | 19 (11%) | 5.45 (2.86-10.38) | NA |
Values are counts (percentages) or means ± SDs,
available for 242 children,
positive for at least one serum or skin test for aeroallergen-specific IgE at age 7,
measured in 175 children,
measured in 177 children,
available for 252 children.
OR: odds ratio for asthma, NA: not applicable
We then tested for the main effect associations of the nine SNPs. One SNP showed nominal association with asthma (rs2517955 OR= 1.91 [95% CI 1.17-3.12], p= 0.01). However, the effect estimates for two SNPs (rs2305480 and rs8076131) that were associated with asthma in a larger cohort of Black children, which included the URECA children, were similar to the effect estimates that we reported here.3
We next asked whether genotypes at the nine SNPs modified associations between the five early-life exposures and asthma at age 7. Although no GEIs were significant after adjusting for multiple testing (adjusted p=0.05/(5x9)=0.0011), five interactions were nominally significant (p<0.05) and three interactions had p<0.01. A nominally significant interaction was observed between rs7216389, located in an intron of GSDMB, and indoor allergen concentration (pint=0.038). Stratifying by genotype, only children with the asthma-associated rs7216389-TT genotype showed the previously reported inverse relationship between early-life allergen exposure and asthma risk1 (OR= 0.46 [95% CI 0.024-0.87], p=0.017), whereas children with the rs7216389-CC or -CT genotypes had similar and non-significant risks across all exposure levels (OR=1.07 [95% CI 0.36-3.16], p=0.898). No significant interactions were observed when testing for each allergen concentration independently. These results, while only nominally significant, support previous interactions observed for this SNP with early-life exposure to animal sheds6 and cats.7
Genotypes at two intronic SNPs in GSDMA (rs8069202 and rs3859192) showed nominally significant interactions with the number of colds in the first year of life (pint =0.0082 and pint=0.0099, respectively; Figure 1) and in the first 3 years of life (pint =0.021 and pint=0.0093, respectively). These two SNPs are in LD (r2=0.64) and may not represent independent signals. Having more colds in the first year of life was associated with a higher risk of asthma only among children carrying the rs8069202-A or rs3859192-T allele (OR= 3.71 [95% CI 1.67-8.25], p=0.0012 and OR= 4.36 [95% CI 1.87-10.17], p=6.45x10−4, respectively). The risk of asthma in children with the rs8069202-GG or rs3859192-CC genotypes was not related to the number of colds (OR= 1.11 [95% CI 0.51-2.42], p=0.797 and OR= 1.12 [95% CI 0.54-2.33], p=0.757 respectively). No interactions were observed between the nine SNPs and microbiota diversity at 3 months.
Figure 1.
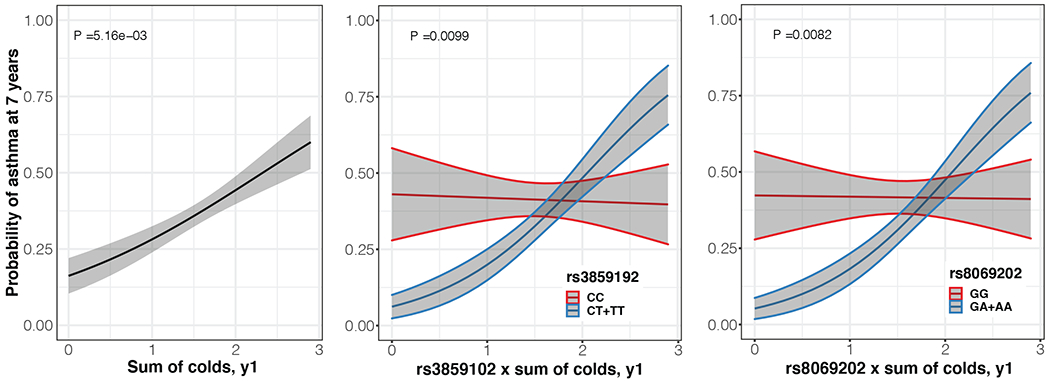
Relationship between asthma at age 7 and sum of colds in the first year of life in Black children in the URECA cohort (left panel, n=262), stratified by genotype at rs3859192 (CC vs. CT+TT) (middle panel, n=259), and stratified by genotype at rs8069292 (GG vs. GA+AA) (right panel, n=260). The fitted lines represent Least-square means and the shaded grey areas show the associated standard error and 95% confidence interval, which were calculated using the R package emmeans V1.6.3.
To further evaluate potential functional effects of the SNPs with pint<0.01 (rs8069202 and rs3859192) we studied gene expression in unstimulated nasal epithelial cells (NECs) collected from 189 URECA Black children at 11 years of age. We performed expression quantitative trait locus (eQTL) analysis for the two GSDMA SNPs and genes at this locus that are expressed in NECs (PGAP3, ERBB2, MEN1, IKZF3, GSDMB, ORMDL3, GSDMA and GRB7). Both SNPs were eQTLs for GSDMA (p=8.27x10−4 and p=2.67x10−4, respectively) in these cells, but were not eQTLs for any of the other genes (P>0.05 after Bonferroni correction). The rs8069202-A or rs3859192-T alleles were associated with decreased expression of GSDMA in NECs, consistent with previous studies in lung tissue.8
To our knowledge, these are the first GEI effects on asthma risk reported for SNPs that regulate the expression of GSDMA. Recent studies used the reduced LD on African American chromosomes to identify SNPs that regulate the expression of GSDMB as the main drivers of the 17q12-q21 locus-associated asthma risk.3,5 Here, we demonstrate that the number of colds in early life increases the risk of asthma in Black children with GSDMA variants (rs8069202-A or rs3859192-T) which are associated with reduced expression of GSDMA in NECs.
GSDMA is expressed in skin,9 and in the airway mucosa including airway epithelial cells.8 It belongs to the gasdermin family of proteins that mediate pyroptosis, a form of cell death that is accompanied by secretion of pro-inflammatory cytokines.9 Therefore, in the absence of infection, expression of GSDMA at low levels may help to minimize damage to airway cells and inflammation. Accordingly, our findings showed that SNPs associated with low GSDMA expression (rs8069202-A or rs3859192-T) were inversely associated with asthma in Black children (OR 0.83 and 0.77 respectively). On the other hand, during an infection, high-level GSDMA expression could promote more rapid clearance of pathogens and also initiate an antimicrobial inflammatory response at the site of infection. Under these circumstances, an insufficient GSDMA response in airway cells could inhibit clearance of respiratory pathogens, leading to prolonged infections that consequently lead to airway remodeling, airway obstruction, and ultimately asthma. This theory could explain why the number of colds in early life was positively associated with the probability of developing asthma in children with the rs8069202-A or rs3859192-T alleles (reduced GSDMA expression, Figure 1). Our findings suggest that GSDMA expression may modify asthma risk at baseline and during viral respiratory infections, perhaps through distinct mechanisms. Further studies are needed to understand the relationship between GSDMA expression on baseline airway inflammation and during illnesses.
Strengths of this study include the availability of longitudinal exposure and asthma outcomes data in Black children, which enabled us to differentiate genotype effects across the locus and identify GSDMA as a modifier of childhood asthma risk with exposure to viral respiratory infections in early-life. Study limitations include the small sample size limiting power and may be the reason for the lack of significant findings. To our knowledge there are no other studies of African American children with similar environmental exposure measurements available for replication. However, despite having low power, the nominally significant interaction detected for the GSDMB SNP shows similar trends to previously detected interactions for this same SNP with early-life allergen exposure. Furthermore, the Bonferroni correction we used is very conservative considering the high correlations between the allergen exposure indices at 3 months and cumulative over 3 years, between the sum of colds at year 1 and cumulative over 3 years, as well as the LD between selected SNPs, despite there being less LD in those with Afircan ancestry. The lack of associations with a microbiota diversity index in house dust should be re-examined in a larger cohort. However, our findings are consistent with earlier studies in the URECA cohort reporting associations between house dust microbiota with decreased risk of wheeze at 3 years but not with asthma at age 7 years.1 Finally, the eQTL studies were limited to RNA from NECs from children at routine study visits. It is possible that sampling cells during colds or acute allergen exposures might reveal additional eQTL effects that are not observed in our studies. Such investigations will be critical for fully characterizing the effects of GEIs.
In summary, our study suggests that variants in GSDMA modify the effects of early-life exposure to colds on the risk of developing asthma at age 7 among Black children. These findings, together with studies in African ancestry cohorts which focused on GSDMB,3–5 suggest that the gasdermin genes at the 17q12-q21 locus are important determinants of childhood-onset asthma risk in children with African ancestry. Understanding the mechanisms that underlie these GEI effects may lead to improved identification of children at risk and novel intervention strategies.
Supplementary Material
Key Messages:
We investigated genotype-exposure interactions between 17q12-q21 SNPs and early-life exposures in Black children.
Early-life colds are positively associated with asthma risk in children with intronic GSDMA SNPs.
Understanding GSDMA SNP interactions with early-life exposures could lead to new asthma prevention strategies.
Funding Statement:
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Contract numbers 1UM1AI114271-01, UM2AI117870. Additional support was provided by grants from the NIH Office of Director UH3 OD023282, the National Center for Research Resources, National Institutes of Health, under grant NCRR: UL1TR001079, 1UL1TR001430, UL1TR001873, UL1TR002345. JDG is supported by the National Institutes of Health through a Ruth L. Kirschstein National Research Service Award 5T32HL007035 and AM is supported by the Bourse de formation postdoctorale – Fonds de recherche du Québec - Santé (FRQS).
Disclosure Statement:
MCA reports consulting fees from Regeneron outside the submitted work. JEG has received grants from the NIH; is a paid consultant for AstraZeneca, Meissa Vaccines Inc., and Gossamer Bio; and has stock options in Meissa Vaccines Inc.
Abbreviations:
- SNP
single nucleotide polymorphism
- GWAS
genome-wide association study
- LD
linkage disequilibrium
- GEI
genotype-exposure interaction
- OR
Odds ratio
- eQTL
expression quantitative trait locus
- NEC
nasal epithelial cell
- URECA
Urban Environment and Childhood Asthma cohort
Footnotes
Ethical statement: Written informed consent, or assent, was provided by all participants or their legal guardians, where applicable. The Urban Environmental Factors and Childhood Asthma (URECA) study was approved by the Institutional Review Boards (IRB) of Boston Medical Center (H-33833), Columbia University Medical Center (IRB-AAAC5139), Johns Hopkins University (NA_00070888/CIR00007139), and Saint Louis Children’s Hospital (201012988), as well as the Western Copernicus Group IRB (20142570).
Supplementary material can be found at: https://doi.org/10.5281/zenodo.5748575
References:
- 1.O’Connor GT, Lynch SV, Bloomberg GR, et al. Early-life home environment and risk of asthma among inner-city children. J Allergy Clin Immunol. 2018;141(4):1468–1475. doi: 10.1016/j.jaci.2017.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stein MM, Thompson EE, Schoettler N, et al. A decade of research on the 17q12-21 asthma locus: Piecing together the puzzle. J Allergy Clin Immunol. 2018;142(3):749–764.e3. doi: 10.1016/j.jaci.2017.12.974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ober C, McKennan CG, Magnaye KM, et al. Expression quantitative trait locus fine mapping of the 17q12–21 asthma locus in African American children: a genetic association and gene expression study. Lancet Respir Med. 2020;8(5):482–492. doi: 10.1016/S2213-2600(20)30011-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Christenson SA, Modena B, et al. Genetic analyses identify GSDMB associated with asthma severity, exacerbations, and antiviral pathways. J Allergy Clin Immunol. 2020;0(0). doi: 10.1016/j.jaci.2020.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gui H, Levin AM, Hu D, et al. Mapping the 17q12–21.1 Locus for Variants Associated with Early-Onset Asthma in African Americans. Am J Respir Crit Care Med. 2021;203(4):424–436. doi: 10.1164/rccm.202006-2623oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loss GJ, Depner M, Hose AJ, et al. The Early Development of Wheeze. Environmental Determinants and Genetic Susceptibility at 17q21. Am J Respir Crit Care Med. 2016;193(8):889–897. doi: 10.1164/rccm.201507-1493OC [DOI] [PubMed] [Google Scholar]
- 7.Stokholm J, Chawes BL, Vissing N, Bønnelykke K, Bisgaard H. Cat exposure in early life decreases asthma risk from the 17q21 high-risk variant. J Allergy Clin Immunol. 2018;141(5):1598–1606. doi: 10.1016/j.jaci.2017.07.044 [DOI] [PubMed] [Google Scholar]
- 8.Hao K, Bossé Y, Nickle DC, et al. Lung eQTLs to Help Reveal the Molecular Underpinnings of Asthma. Williams SM, ed. PLoS Genet. 2012;8(11):e1003029. doi: 10.1371/journal.pgen.1003029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Xia S, Zhang Z, Wu H, Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat Rev Drug Discov 2021 205. 2021;20(5):384–405. doi: 10.1038/s41573-021-00154-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


