Abstract
INTRODUCTION:
Magnetic Resonance Imaging (MRI) research has advanced our understanding of neurodegeneration in sporadic Early-Onset Alzheimer’s Disease (EOAD), but includes small samples, mostly amnestic EOAD, and has not focused on developing an MRI biomarker.
METHODS:
We analyzed MRI scans to define the sporadic EOAD-Signature atrophy in a small sample (n=25) of Massachusetts General Hospital (MGH) EOAD patients and investigated its reproducibility in the large LEADS sample (n=211), and investigated the relationship of the magnitude of atrophy to cognitive impairment.
RESULTS:
The EOAD-Signature atrophy replicated across the two cohorts with prominent atrophy in caudal lateral temporal cortex, inferior parietal lobule, and posterior cingulate and precuneus cortices, with relative sparing of medial temporal lobe. The magnitude of EOAD-Signature atrophy was associated with the severity of cognitive impairment.
DISCUSSION:
The EOAD-Signature atrophy is a reliable and clinically valid biomarker of AD-related neurodegeneration that could be used in clinical trials for EOAD.
Keywords: Magnetic Resonance Imaging (MRI), Disease Signature, Early-Onset Alzheimer’s Disease
Background
Sporadic early-onset Alzheimer’s disease (EOAD; symptomatic onset ≤ 65 years) is a rare and understudied but particularly devastating form of AD, affecting patients at a point in life, where they may be balancing careers, family responsibilities, and community roles. Compared to the more common typical late-onset AD (LOAD), EOAD patients often present with prominent impairments in executive function, language, visuospatial abilities, and/or somatosensory or motor functions rather than memory 1–4. These differences reflect more prominent neurodegeneration in posterior lateral temporal, lateral and medial parietal, frontal, or occipital cortex relative to the medial and ventral temporal cortical localization of typical LOAD 4–8. Convergently, neuropathological investigations have identified a “hippocampal-sparing” form of AD that is often present in younger patients 9. For these reasons, imaging biomarkers of neurodegeneration in EOAD, such as Magnetic Resonance Imaging (MRI) measures of atrophy, likely need to be different than those of neurodegeneration in typical LOAD.
Although prior neuroimaging studies have advanced our understanding of anatomical abnormalities in EOAD, these studies have included small samples, mostly comprised of patients with amnestic EOAD dementia 5,7,10,11. No studies have focused on developing an MRI biomarker specific to EOAD. Such a disease-signature MRI biomarker has been established and validated in LOAD dementia 12 and has proven to be powerful in predicting progressive decline in people with MCI 13,14 or who were cognitively unimpaired 15–17 and in predicting molecular biomarker status 14,16. An a priori-defined regional atrophy signature may also be helpful in constraining the analysis of MRI data in studies of putative disease-modifying therapies 18.
As recognition of sporadic EOAD grows, it is important to define and investigate the utility of an MRI signature of neurodegeneration in this patient population. Here we define the EOAD-Signature MRI biomarker using data from a well-characterized discovery sample from Massachusetts General Hospital (MGH) and test the hypothesis that it is reproducible in a large validation sample of participants with EOAD (the largest to date in the United States) from the Longitudinal Early-onset Alzheimer’s Disease Study (LEADS) 1. Based on prior work, we hypothesized that compared to controls and patients with early-onset non-AD (EOnonAD) cognitive impairment or dementia, the EOAD group will show cortical atrophy in posterior cingulate cortex and precuneus, inferior parietal lobule, and caudal lateral temporal cortex (and to a lesser degree lateral prefrontal cortex). We also hypothesized that the EOAD-Signature MRI biomarker will demonstrate better sensitivity and specificity in EOAD compared with LOAD-Signature and other well-accepted morphometric measures used in clinical trials for AD, such as volumes of the hippocampal formation, ventricles, or whole brain, supporting its potential utility in that setting. Finally, we hypothesized that the EOAD-Signature MRI biomarker will show clinical validity in its correlation with global cognitive impairment.
Methods
Participants
Data were obtained from two independent cohorts; for clinical and demographic characteristics of both cohorts, see Table 1. The MGH EOAD cohort included 25 patients who were participants in the ongoing MGH Frontotemporal Disorders Unit Longitudinal Cohort. All participants received a standard clinical evaluation comprising a structured history obtained from both patient and informant and patient examination to inform scoring of the global Clinical Dementia Rating score (CDR®; Morris, 1993)19, as well as neuropsychological assessment. Clinical diagnostic formulation was performed through consensus conference by our multidisciplinary team of neurologists, neuropsychologists, and speech and language pathologists. Each patient was classified based on a 3-step diagnostic formulation; mild cognitive impairment or dementia (Cognitive Functional Status), specific Cognitive-Behavioral Syndrome, and likely etiologic diagnosis 20. Age at symptomatic onset was estimated during a structured interview of the patient and informant and was less than 65 in all cases. Regarding the cognitive-behavioral syndrome, three individuals met diagnostic criteria for Posterior Cortical Atrophy (PCA) 21–23, four individuals met criteria for a dysexecutive variant of Alzheimer’s disease 24,25, and 18 individuals met criteria for an amnesic-dysexecutive multidomain syndrome 26,27. All MGH participants underwent an MRI session that was reviewed to rule out other potential neurologic contributors to cognitive impairment and to visually assess for regional atrophy. No patients included in this sample had brain lesions beyond mild white matter signal abnormalities.
Table 1.
Clinical and demographic characteristics of both cohorts.
| MGH | LEADS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EOAD (n = 25) | Control (n = 25) | Group difference (p) | EOAD (n = 211) | EOnonAD (n = 68) | Control (n = 88) | Omnibus Test (p) | EOAD vs. CN (p) | EOnonAD vs. CN (p) | EOAD vs. EOnonAD (p) | |
| Age (years) | 57.28 ± 4.18 | 67.43 ± 4.84 | .001 | 58.76 ± 3.92 | 57.97 ± 6.05 | 56.06 ± 6.01 | .001 | .001 | .05 | .75 |
| Sex (M/F) | 10/15 | 13/12 | .40 | 100/111 | 44/24 | 33/55 | .003 | .12 | .001 | .013 |
| Education (years) | 15.42 ± 2.62 | 15.69 ± 2.25 | .75 | 15.41 ± 2.35 | 15.44 ± 2.57 | 16.68 ± 2.19 | .001 | .001 | .004 | .99 |
| Race | 96% White, 4% Black | 76% White, 12% Black or African American, 12% Unknown | - | 94.31% White, 2.84% Black or African American, 0.95% Asian, 1.42% More than one race, 0.47% unknown | 91.18% White, 4.41% Black or African American, 1.47% Asian, 1.47% More than one race, 1.47% unknown | 73.86% White, 17.05% Black or African American, 5.68% Asian, 2.27% More than one race, 1.14% unknown | - | - | - | - |
| Ethnicity | 52% Non-Hispanic, 48% unknown | 80% Non-Hispanic, 20% unknown | - | 97.16% Not Hispanic or Latino, 2.84% Hispanic or Latino | 95.59% Not Hispanic or Latino, 4.41% Hispanic or Latino | 93.18% Not Hispanic or Latino, 6.82% Hispanic or Latino | .28 | .11 | .52 | .52 |
| CDR global | 0.5 (24%), 1 (64%), 2 (12%) | 0 (100%) | - | 0.5 (66.35%), 1 (33.65%) | 0.5 (82.35%), 1 (17.65%) | 0 (100%) | - | - | - | .012 |
| CDR-SOB | 5.12 ± 2.60 | 0 (100%) | - | 3.63 ± 1.72 | 2.86 ± 1.87 | 0.01 ± 0.05 | - | - | - | .002 |
| MMSE Total | 19.72 ± 5.70 | - | - | 21.92 ± 5.00 | 25.74 ± 4.15 | 29.19 ± 0.95 | .001 | .001 | .001 | .001 |
The diagnosis of EOAD was supported in six cases by amyloid PET using 11C-Pittsburgh Compound B (PiB) PET. Amyloid (Aβ) positivity was determined by visual read according to previously published procedures 28 and a summary distribution volume ratio (DVR) of frontal, lateral temporoparietal, and retrosplenial (FLR) regions greater than 1.2 29. In 10 cases, the diagnosis of EOAD was supported by cerebrospinal fluid amyloid and tau biomarkers obtained via Athena Diagnostics 30. Six other cases were followed to autopsy met neuropathological criteria for AD with A3B3C3 ratings 31. The remaining three cases had FDG-PET scans demonstrating temporoparietal hypometabolism consistent with AD 32.
We included a group of 25 amyloid-negative (Aβ-) cognitively normal (CN) individuals. CN individuals had a CDR of 0, performed within normal limits on neuropsychological testing, had normal brain structure based on MRI, and low cerebral amyloid based on quantitative analysis of PiB PET data (FLR DVR < 1.2). Individuals were excluded from our patient and control groups if they had a primary psychiatric or other neurologic disorder including major cerebrovascular infarct or stroke, seizure, brain tumor, hydrocephalus, multiple sclerosis, HIV-associated cognitive impairment, or acute encephalopathy. This work was carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. All participants and their informants/caregivers provided informed consent in accordance with the protocol approved by the MassGeneral Brigham HealthCare System Human Research Committee Institutional Review Board in Boston, Massachusetts.
The second cohort included 211 EOAD patients, 68 amyloid-negative cognitively impaired participants (EOnonAD) patients, and 88 CN participants enrolled in the 18-site multicenter LEADS (www.leads-study.org) 1. All participants are between 40–64 years old at study enrollment, fluent in English, in good general health and absent other neurological or psychiatric disorder, and had a knowledgeable informant. Because LEADS focuses on sporadic early-onset dementia, impaired individuals with genetic mutations in Amyloid Precursor Protein (APP), Presenilin-1 (PSEN1) or Presenilin-2 (PSEN2), Microtubule Associated Protein Tau (MAPT), Chromosome 9 Open Reading Frame 72 (C9ORF72), or Progranulin (GRN) were excluded (see Nudelman et al, this issue). All diagnoses within LEADS are made via consensus criteria 1. Both EOAD and EOnonAD) participants had a global CDR of 0.5 or 1.0 at the time of enrollment. CN participants were free of cognitive deficits on neuropsychological testing, and had a Mini-Mental State Examination (MMSE)33 score ≥ 24 and a CDR global score of 0. Institutional Review Board (IRB) approval was obtained through a central IRB overseen by Indiana University, and informed consent was obtained in written form from study participants or authorized representatives. All LEADS participants received a standard clinical assessment including medical and family history, concurrent medication, and medical/neurological examinations as well as a comprehensive clinical assessment (see Hammers et al., this issue).
All LEADS participants additionally underwent structural MRI and 18F-Florbetaben (FBB) PET neuroimaging at baseline. Central reads of baseline FBB PET scans were performed in cognitively impaired participants using a hybrid approach that included visual reads and global quantification of standardized uptake value ratio (SUVR) (see Cho et al. this issue). FBB PET scans were visually read as Aβ+ or Aβ- based on visual read1,34. A composite neocortical FBB SUVR90–110 was calculated using the whole cerebellum as a reference region and converted to Centiloid units 35 using the ADNI formula. A global SUVR ≥ 1.18 (corresponding to 39.2 Centiloids) was used as a quantitative threshold for amyloid PET positivity. If the visual read and quantitative assessment agreed that a scan was Aβ+, the participant was assigned to the EOAD cohort. If visual read and quantification agree that a scan was Aβ-, the participant was assigned to the EOnonAD cohort. If there was discordance between the visual read and quantification, a “tie breaker” visual read was provided by an additional reader (blinded to quantification results), and this was considered the consensus read for cohort assignment.
Structural MRI data acquisition and processing
MRI data for the MGH cohort were acquired with a Siemens Tim Trio 3.0 Tesla scanner using a T1-weighted magnetization prepared rapid acquisition sequence (MPRAGE): repetition time [TR] = 2300 ms, echo time [TE] = 2.98 ms, flip angle = 9°, field of view = 240 × 256 mm, in-plane voxel size = 1 mm isotropic, 192 sagittal slices. MRI data for the LEADS cohort were also acquired with 3.0 Tesla scanners using a sagittal 3D accelerated MPRAGE/IRSPGR T1-weighted sequence. The typical parameters for this sequence were the following, although these varied slightly by vendor and system type: TR = 2300 ms, TE = 2.98 ms, flip angle = 9°, field of view = 240 × 256 mm, in-plane voxel size = 1 mm isotropic, 208 sagittal slices, and 2x acceleration. Hardware and imaging parameters across all sites part of the LEADS cohort are summarized in Supplementary Table 1. All MRI data were visually inspected for gross artifacts (e.g., subject motion) and evaluated on image quality prior to data processing 36.
Once it passed quality control, each participant’s MPRAGE data underwent intensity normalization, skull stripping, and automated segmentation of cerebral white matter to locate the gray matter/white matter boundary via FreeSurfer v6.0, which is documented and freely available for download online (http://surfer.nmr.mgh.harvard.edu). Defects in the surface topology were corrected 37, and the gray/white boundary was deformed outward using an algorithm designed to obtain an explicit representation of the pial surface. We visually inspected each participant’s cortical surface reconstruction for technical accuracy and excluded participants similarly to our prior work, e.g.,38. Three participants from the LEADS Cohort were excluded from the sample due to failed cortical reconstruction. For the remainder of the participants, cortical thickness was calculated as the closest distance from the gray/white boundary to the gray/CSF boundary at each vertex on the tessellated surface 39. Individual maps of cortical thickness were registered to template surface space (fsaverage) and smoothed geodesically with full-width-half-maximum (FWHM) of 15 mm.
Statistical analysis
Using individual cortical thickness maps as inputs, we first created a whole-cortex vertex-wise two-class general linear model (GLM) in FreeSurfer to identify areas of the cerebral cortex where EOAD patients showed abnormal cortical thickness (i.e., atrophy) relative to Aβ- CN participants in the MGH cohort. For this analysis, statistical significance was assessed at p < .0001 corrected for false discovery rate (FDR) 40. The resulting vertex-wise significance map was converted to a binary mask. This was used as the EOAD-Signature mask, which was not constrained by any specific parcellation of the cerebral cortex. Within this EOAD-Signature mask, we examined the magnitude of mean group differences in cortical thickness at each vertex in tenths of millimeters in both cohorts. We thresholded the group difference maps at 0.2 mm in both cohorts; that is, only cortical regions which were at least 0.2 mm thinner in patients than CN controls were retained and defined as part of the EOAD signature map for each cohort. To demonstrate the topographical specificity of the effect, we additionally quantified the magnitude of cortical atrophy in the LEADS cohort without the EOAD-Signature FDR-corrected mask derived from the MGH cohort (that is, we also performed a whole-cortex analysis in the LEADS sample). Using data from the LEADS cohort, we additionally constructed a whole-cortex vertex-wise GLM to identify areas of the cerebral cortex where EOnonAD patients showed atrophy relative to CN participants.
While we tried to standardize imaging hardware and parameters across all sites in the LEADS cohort to the best of our ability, small differences do exist. To account for the potential effect of such factors, we performed harmonization of cortical thickness values using ComBat, a technique that estimates site-specific effects on features of interest using empirical Bayes, which are corrected via linear regression41. We modeled age, sex, and diagnosis (EOAD, EOnonAD, CN) of each participant as covariates to preserve the variance potentially attributable to these factors. We then created a vertex-wise GLM to identify group differences in cortical thickness, similarly to the analysis of unharmonized data.
Next, we investigated the magnitude of AD-related cortical atrophy in both cohorts within cortical regions identified in the EOAD Signature. Using the vertex-wise EOAD-Signature mask derived from the MGH cohort, we drew a set of regions of interest (ROI) labels on the fsaverage surface following the procedure similar to our prior work on LOAD 12. Nine EOAD-Signature ROIs were drawn in the left hemisphere, four of which were also identified in the homologous locations of the right hemisphere. Given this lateralization, our primary ROI-based analysis focused on the left hemisphere; results of our secondary analysis examining the right hemisphere are reported in supplementary material. In addition to the 9 EOAD-Signature ROIs, we included primary visual cortex (known in Desikan-Killiany parlance as the “pericalcarine” ROI) defined based on the standard Desikan-Killiany atlas 42 as a control ROI. Using the spherical registration of each participant to fsaverage space, the 10 cortical ROIs were mapped back to individual participants’ native surface space. For each participant, mean cortical thickness within each ROI was calculated by deriving an average of all the thickness estimates from vertices that fell within the labeled ROI. For each ROI, the effect size (Cohen’s d) was calculated as the difference in group means (e.g., between EOAD and CN) divided by the pooled standard deviation. We conducted a series of paired sample t-tests to test whether group differences were replicable in the LEADS cohort. Cohen’s d effect sizes were similarly quantified and compared between EOnonAD and CN participants in the LEADS cohort.
In addition, we further examined the relationships between the magnitude of atrophy in the EOAD Signature with severity of cognitive impairment as measured by CDR Sum of Boxes (CDR-SB) and MMSE. We created a single composite EOAD-Signature ROI label by taking the union of all 9 individual ROIs. Cortical thickness was then calculated by averaging thickness estimates of all vertices falling within this composite ROI. We computed Pearson’s product moment correlations, r, between mean cortical thickness within EOAD Signature and the CDR-SB and MMSE scores in the total sample of EOAD participants and separately within each cohort. To account for global differences in cortical thickness across cohorts, we converted each participants’ cortical thickness estimates to W-scores 38,43,44, which are analogous to Z-scores adjusted for specific covariates of no interest, which in this study were age and sex. Briefly, we first performed a multiple regression analysis using mean cortical thickness data obtained from CN participants, which resulted in beta coefficient values for age and sex as well as individual values of residuals. Using these parameters, we then computed W-scores for all EOAD patients with the following formula:
Where = the observed mean cortical thickness of patient i, = the predicted mean cortical thickness of patient i based on age and sex of this patient as well as beta coefficients obtained from CN participants, and SD = the standard deviation of the individual residuals obtained from CN participants. Because W-scores in this study were calculated using cortical thickness, more negative values indicate greater cortical atrophy relative to what would be expected solely based on age and sex.
Furthermore, we tested the hypothesis that the EOAD-Signature ROIs can detect AD-related cortical atrophy with greater sensitivity compared with traditional ROIs defined by automatic parcellation of anatomical boundaries. To do this, we identified an ROI label from the Desikan-Killiany atlas that best corresponded with each of the 9 EOAD-Signature ROIs (based on the degree of spatial overlap), resulting in a separate set of 9 anatomically defined ROIs. For each set, we created a single composite EOAD-Signature ROI label by taking the union of all 9 individual ROIs. This resulted in a data-driven EOAD-Signature composite ROI and a parcellation-driven EOAD-Signature composite ROI. Cortical thickness within each of these was then calculated by averaging thickness estimates of all vertices falling within each composite ROI. In each cohort, we then compared Cohen’s d effect sizes of group differences. Moreover, we performed additional comparisons of effect sizes with mean cortical thickness derived within the 9 “LOAD-signature” ROIs 12 as well as three global volumetric measures (whole brain volume, ventricular volume, and average hippocampal volume), which were derived based on FreeSurfer’s automatic segmentation algorithms and were adjusted for the estimated total intracranial volume. In all analyses, statistical significance was assessed at p < .05, Bonferroni-corrected for multiple comparisons per analysis family. None of our analyses of cortical thickness data involved adjustments for the total intracranial volume given that these measures are typically not correlated with each other45,46.
Finally, we generated an EOAD-Signature mask from the LEADS cohort. This mask was created following the same procedure described above for the MGH cohort, except that we conducted a vertex-wise GLM analysis comparing EOAD patients and CN participants from the LEADS cohort. Statistical significance was assessed at p < 10−12 corrected for FDR.
Results
Demographic and clinical characteristics
Table 1 reports the demographic and clinical characteristics of the two cohorts. The MGH EOAD sample had slightly greater cognitive impairment with higher global CDR scores (t = 5.95, p < .001), higher CDR-SB scores (t = 3.85, p < .001), and lower MMSE scores (t = 2.05, p ≤ .041) than the LEADS cohort. The average age of the MGH EOAD CN sample was greater (t = 8.68, p ≤ .001) than the LEADS CN sample. The two cohorts did not differ in terms of sex composition (χ2 = .49, p ≤ .48) or average years of education (t = 0.01, p ≤ .99). Within the LEADS cohort, the EOAD group was slightly more impaired than the EOnonAD group.
The EOAD signature is robustly replicable across independent patient cohorts
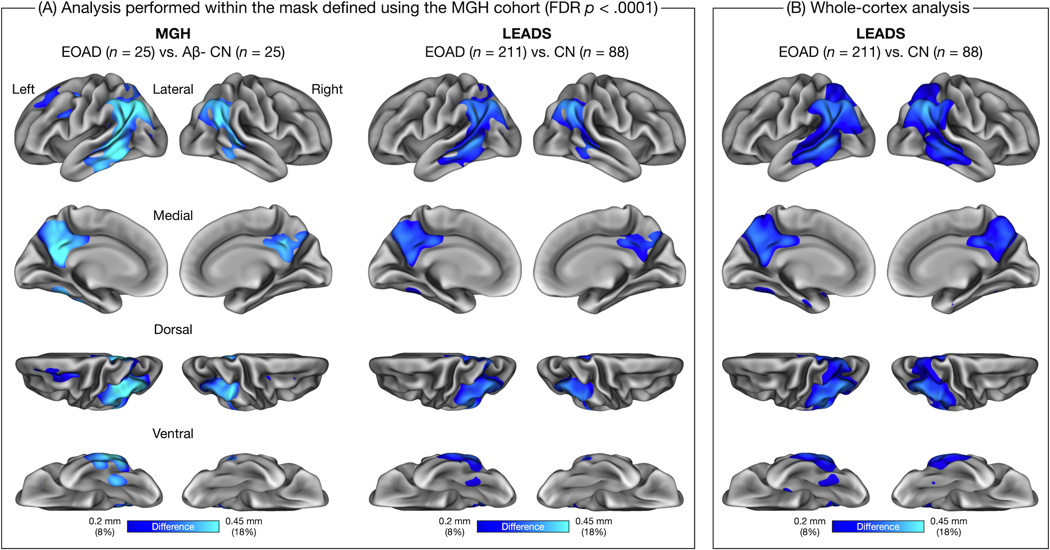
In the MGH cohort, prominent atrophy was present in the posterior cortical regions including bilateral posterior cingulate cortex (PCC) and precuneus, inferior parietal lobule, and caudal lateral temporal cortex. In the left hemisphere, atrophy was additionally identified in the superior frontal gyrus, caudal middle frontal gyrus, mid lateral temporal cortex, and fusiform gyrus. This pattern of cortical atrophy was largely replicated in the LEADS cohort, although no involvement of the left lateral frontal cortex was observed at the same threshold and the magnitude of atrophy was overall slightly weaker than the MGH cohort (Figure 1A). To examine the specificity of the topography of cortical atrophy, we generated a vertex-wise map of group differences in cortical thickness in the LEADS cohort without the EOAD-Signature mask (the binary mask of FDR-corrected statistical significance derived from the MGH cohort) (Figure 1B). This whole-cortex analysis revealed that the topography of the EOAD signature was remarkably consistent across the two cohorts.
Figure 1. Spatial topography of the early-onset Alzheimer’s disease (EOAD) signature across two independent cohorts.
(A) Colored vertices on the cortical surface maps indicate areas where patients with EOAD showed cortical atrophy compared to low-amyloid cognitively normal (CN) participants within each cohort. These group difference (“gamma”) maps were thresholded at 0.2 mm, corresponding to an ≈8% reduction in cortical thickness relative to the pooled sample of CN participants. Because the Massachusetts General Hospital (MGH) and longitudinal early-onset Alzheimer’s disease study (LEADS) cohorts differed substantially in sample size, which would influence the statistical significance of effects, we masked both group difference maps by a vertex-wise map representing areas showing statistically significant group differences in the MGH cohort (assessed at P < 0.0001, false discovery rate (FDR)-corrected). (B) Colored vertices on the cortical surface map indicate areas where LEADS EOAD patients showed cortical atrophy relative to LEADS CN participants. This map was generated similarly to the map shown in (A), but without the FDR-corrected mask derived from the MGH cohort. This map (B) demonstrates the specificity of the EOAD signature in the LEADS replication cohort compared to the MGH discovery cohort (A).
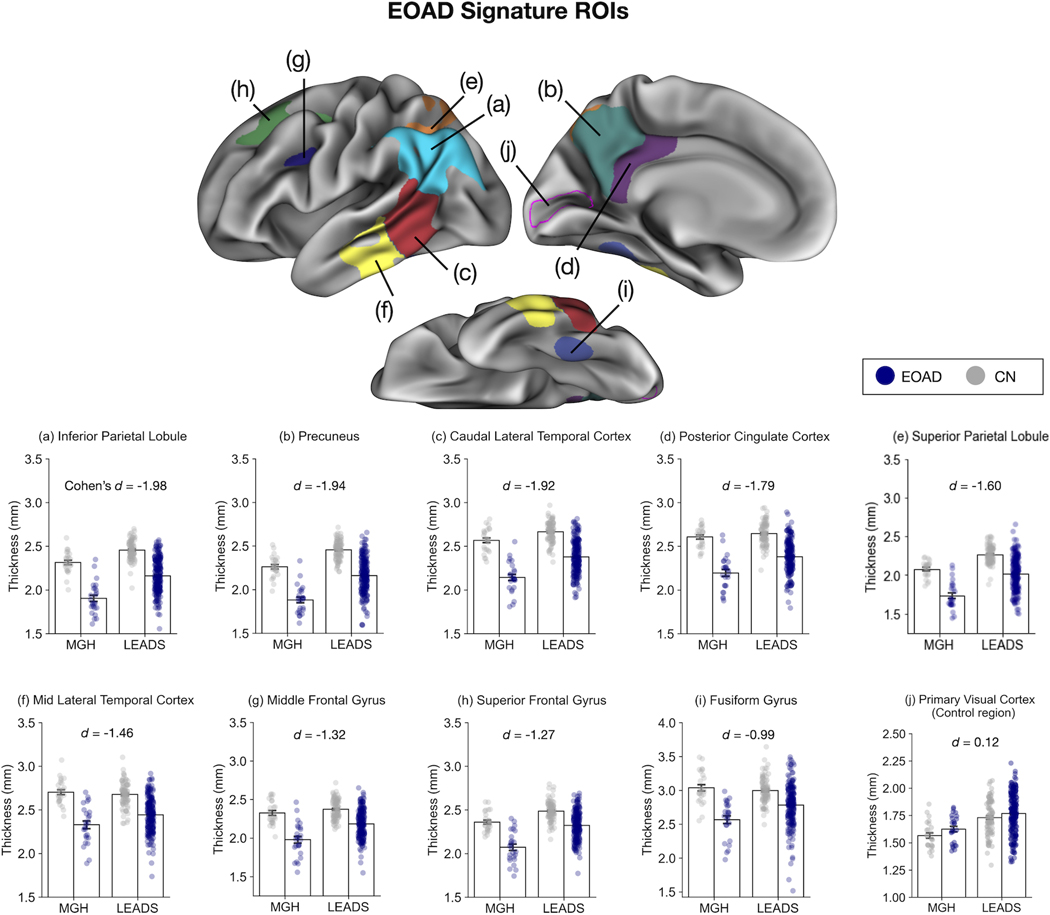
Next, we defined 9 EOAD-Signature ROIs in the left hemisphere representing areas of cortical atrophy in patients with EOAD relative to CN participants based on the results of our vertex-wise GLMs using the MGH cohort (Figure 2, top) (see Supplementary Figure 1 for ROIs in the right hemisphere). Within this set of ROIs, we investigated the consistency of the magnitude of regionally specific cortical atrophy between the MGH and LEADS cohorts (see Supplementary Figure 2 for bar plots without individual observations shown as overlaid circles). Figure 2 illustrates the mean thickness within the EOAD-Signature ROIs calculated separately for patients with EOAD and CN participants across the two cohorts; Table 2 provides details of measurements from both cohorts showing the magnitude of group differences (EOAD vs. CN) and effect size estimates. Across the two cohorts, the posterior medial (precuneus and posterior cingulate cortex) and lateral parietal (inferior and superior parietal lobule) areas tended to show larger effect sizes of cortical atrophy in EOAD, followed by the caudal and mid lateral temporal cortex, lateral prefrontal (middle and superior frontal) areas, and the fusiform gyrus. Primary visual cortex showed no effect in either cohort. Our analysis of site-harmonized cortical thickness data showed that the topography of group differences is remarkably similar with or without data harmonization via ComBat, suggesting that the EOAD signature of atrophy is robust against across-sites differences in imaging hardware and parameters (Supplementary Figure 3).
Figure 2. Consistency of regional cortical atrophy in early-onset Alzheimer’s disease (EOAD).
Our EOAD-signature regions of interest (ROIs) (top) were generated from a vertex-wise general linear model analysis of cortical thickness data in the Massachusetts General Hospital (MGH) cohort. The same ROIs were applied to the longitudinal early-onset Alzheimer’s disease study (LEADS) cohort to test the hypothesis that these regions are thinner in EOAD than cognitivevly normal (CN) controls. Bar plots at the bottom show mean cortical thickness within each ROI across the two cohorts, illustrating the consistency of cortical thickness. Error bars denote one standard error of the mean. Each colored circle overlaid on top of the bar plot represent one participant. Primary visual cortex (pericalcarine ROI; indicated by the magenta outline) was used as a control region to illustrate absence of effects in this region. ROIs are presented above in descending order of effect size (Cohen’s d). Cohen’s d values shown here were calculated by first computing the effect size estimate for each cohort and taking the weighted average of the two cohorts accounting for their sample sizes. See Table 2 for cohort-specific effect sizes along with other relevant statistics.
Table 2.
Quantitative metrics of cortical atrophy by region in the MGH and LEADS cohorts.
| MGH |
LEADS |
|||||||
|---|---|---|---|---|---|---|---|---|
| Mean thickness: mm (SD) |
Mean thickness: mm (SD) |
|||||||
| EOAD (n = 25) | CN (n = 25) | Group mean difference (mm) | Effect size (d) | EOAD (n = 211) | CN (n = 88) | Group mean difference (mm) | Effect size (d) | |
| Inferior parietal | 1.90 (0.18) | 2.31 (0.13) | −0.41*** | −2.63 | 2.16 (0.19) | 2.46 (0.11) | −0.3*** | −1.87 |
| Precuneus | 1.88 (0.16) | 2.26 (0.12) | −0.38*** | −2.65 | 2.16 (0.20) | 2.46 (0.11) | −0.3*** | −1.82 |
| Caudal lateral temporal | 2.14 (0.18) | 2.57 (0.14) | −0.43*** | −2.7 | 2.38 (0.19) | 2.67 (0.12) | −0.29*** | −1.79 |
| Posterior cingulate | 2.19 (0.19) | 2.61 (0.12) | −0.41*** | −2.57 | 2.38 (0.19) | 2.64 (0.13) | −0.26*** | −1.66 |
| Superior parietal | 1.73 (0.18) | 2.08 (0.10) | −0.34*** | −2.32 | 2.02 (0.21) | 2.27 (0.11) | −0.25*** | −1.48 |
| Mid lateral temporal | 2.33 (0.22) | 2.70 (0.14) | −0.37*** | −2.03 | 2.44 (0.20) | 2.68 (0.14) | −0.24*** | −1.37 |
| Middle frontal | 1.98 (0.22) | 2.33 (0.16) | −0.35*** | −1.82 | 2.18 (0.18) | 2.37 (0.13) | −0.19*** | −1.23 |
| Superior frontal | 2.07 (0.18) | 2.36 (0.11) | −0.29*** | −1.92 | 2.32 (0.16) | 2.49 (0.11) | −0.17*** | −1.17 |
| Fusiform | 2.57 (0.28) | 3.04 (0.24) | −0.47*** | −1.81 | 2.78 (0.30) | 3.00 (0.19) | −0.22*** | −0.85 |
| Primary visual | 1.63 (0.13) | 1.57 (0.12) | 0.06 | 0.47 | 1.77 (0.19) | 1.73 (0.16) | 0.04 | 0.22 |
Note: Effect size = Cohen’s d.
p < .001
The EOnonAD patient group does not show a consistent pattern of cortical atrophy
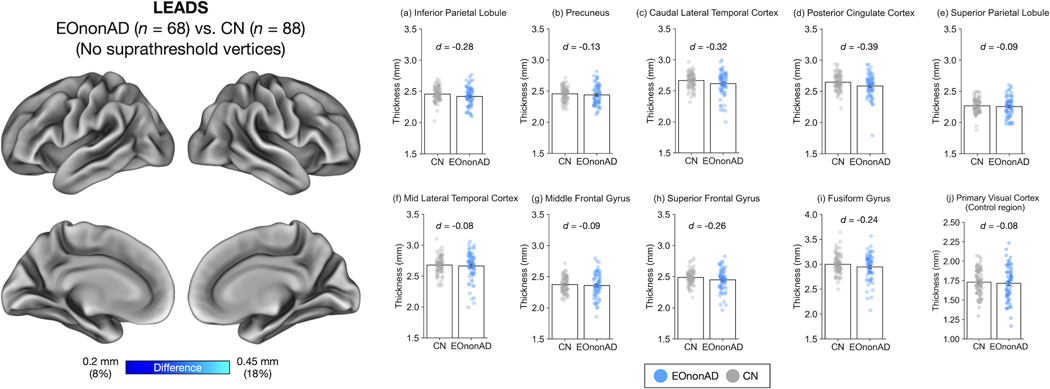
To assess whether EOAD-Signature cortical atrophy is specifically related to AD pathology, we performed vertex-wise and ROI-based analyses of cortical thickness and conducted group comparisons between EOnonAD patients and CN participants in the LEADS cohort. A vertex-wise GLM revealed no suprathreshold atrophy when examined at the same threshold as the EOAD-CN comparison above (Figure 3, left). Consistent with this result, comparisons of mean cortical thickness across 9 EOAD-Signature ROIs also revealed no effects that survived a statistical threshold corrected for multiple comparisons (Figure 3, right, and Supplementary Table 2). Although some individual patients showed patterns of atrophy suggestive of Frontotemporal Lobar Degeneration, the group as a whole did not show a consistent pattern of atrophy.
Figure 3. Absence of cortical atrophy in the longitudinal early-onset Alzheimer’s disease study (LEADS) early-onset non-Alzheimer’s Disease (EOnonAD) cohort.
Left: Results of a vertex-wise general linear model analysis comparing cortical thickness between patients with EOnonAD and cognitively normal (CN) participants. This analysis revealed no areas where EOnonAD patients showed cortical atrophy of 0.2 mm or greater compared with CN participants. Right: Consistent with these vertex-wise GLM results, a region of interest (ROI)-based analysis of cortical thickness revealed no or minimal effects of EOnonAD.
The EOAD Signature is a clinically valid measure of AD-related neurodegeneration
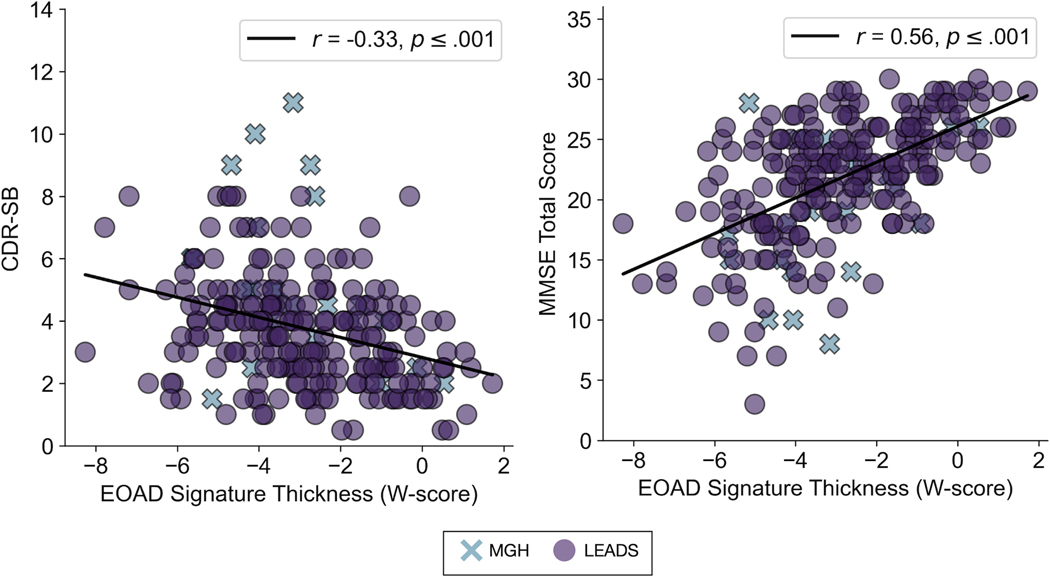
We analyzed the relationship of the EOAD Signature atrophy measure to severity of cognitive impairment within all the EOAD participants from the two independent samples. Within the entire sample, cortical thickness with the EOAD Signature expressed as W-scores correlated with CDR-SB (r = −.33, p < .001) and with MMSE scores (r = .56, p < .001) (Figure 4). Correlations between EOAD-Signature atrophy and these measures in each cohort separately are reported in supplementary materials (Supplementary Figure 4). These findings indicate that greater atrophy within the EOAD Signature was associated with greater cognitive impairment in EOAD.
Figure 4. The early-onset Alzheimer’s disease (EOAD) signature is a clinically valid biomarker of neurodegeneration in symptomatic patients with MCI or mild dementia due to EOAD.
The magnitude of atrophy in the EOAD signature correlates with severity of cognitive impairment as measured by Clinical Dementia Rating Scale-Sum of Boxes (CDR-SB) (r = −0.33, P < 0.001) and Mini-Mental State Examination (MMSE) (r = −0.56, P < 0.001) within the entire sample (N = 236). More negative W-scores indicate more severe atrophy. Higher CDR-SB and lower MMSE scores indicate greater cognitive impairment. See Figure S4 for the results of this analysis in each cohort separately.
The EOAD signature affords greater sensitivity in detecting cortical atrophy than anatomically-defined ROIs
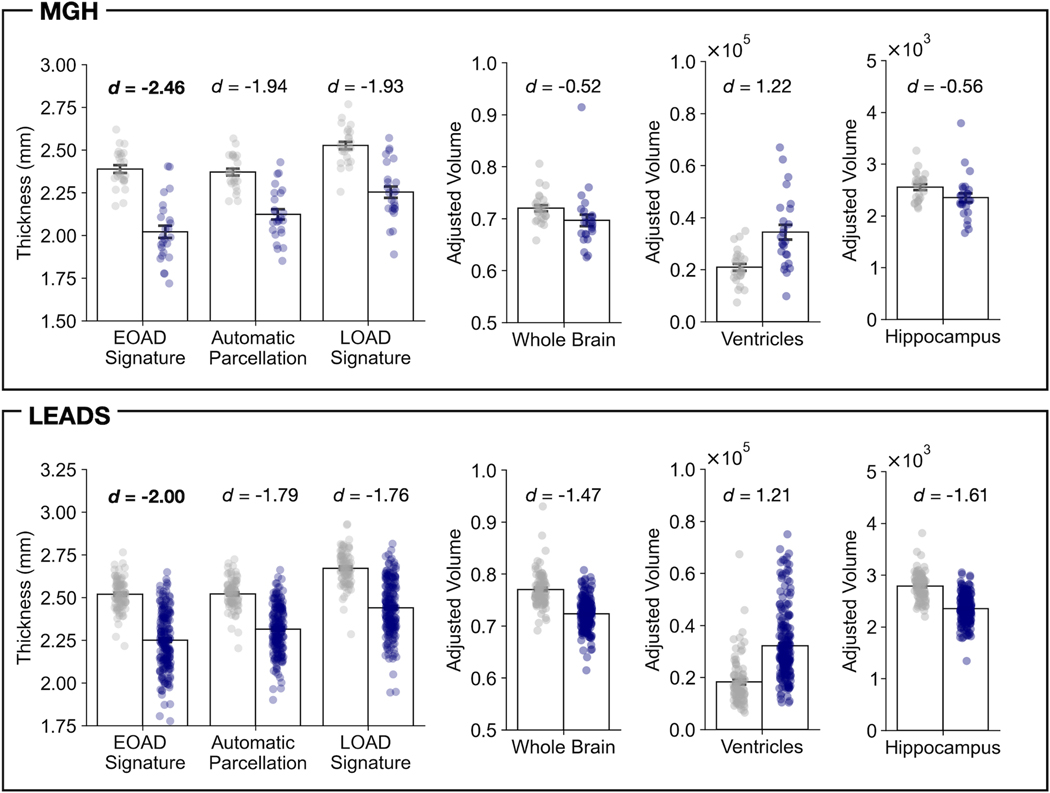
We compared the effect size of EOAD-related cortical atrophy (EOAD vs. CN) between the composite EOAD-Signature ROI and the corresponding ROI defined by gyral or sulcal anatomical boundaries using the standard Desikan-Killiany atlas. This analysis allowed us to test the hypothesis that the EOAD-Signature ROIs can detect cortical atrophy in this population with greater sensitivity compared with traditional anatomically-derived ROIs based on automatic parcellation of the cerebral cortex covering similar but larger areas. In both MGH and LEADS cohorts, we found that the magnitude of the difference in cortical thickness between patients with EOAD and CN participants was numerically larger with the composite EOAD-Signature ROI than with the composite anatomically-defined ROI. As expected, the effect size identified using the composite EOAD Signature ROI was also larger than that obtained with the LOAD signature ROIs and the volumetric measures commonly used in AD research, suggesting the superior sensitivity of our EOAD Signature cortical surface-based approach for detecting AD-related atrophy in spatially distributed areas of the cerebral cortex (Figure 5).
Figure 5. The magnitude of group differences between early-onset Alzheimer’s Disease (EOAD) patients and cognitively normal (CN) participants comparing different cortical signature methods and standard morphometric measures used in clinical trials.
Bar plots show mean cortical thickness (left) or volumetric measures (right) separately for patients with EOAD and CN participants in the Massachusetts General Hospital (MGH) and longitudinal early-onset Alzheimer’s disease study (LEADS) cohorts. Cortical thickness estimates were derived by first merging the individual region of interest (ROI) labels for each signature (EOAD signature, the analogous automatic parcellation ROIs based on the Desikan–Killiany atlas, and LOAD signature), then averaging cortical thickness estimates of all vertices falling within each label to create a composite measure. Whole brain, ventricular, and hippocampal (average of two hemispheres) volume were based on FreeSurfer’s automatic segmentation of each participant’s structural MRI data, adjusted for their estimated total intracranial volume.
The LEADS EOAD Signature: A tool for the scientific community
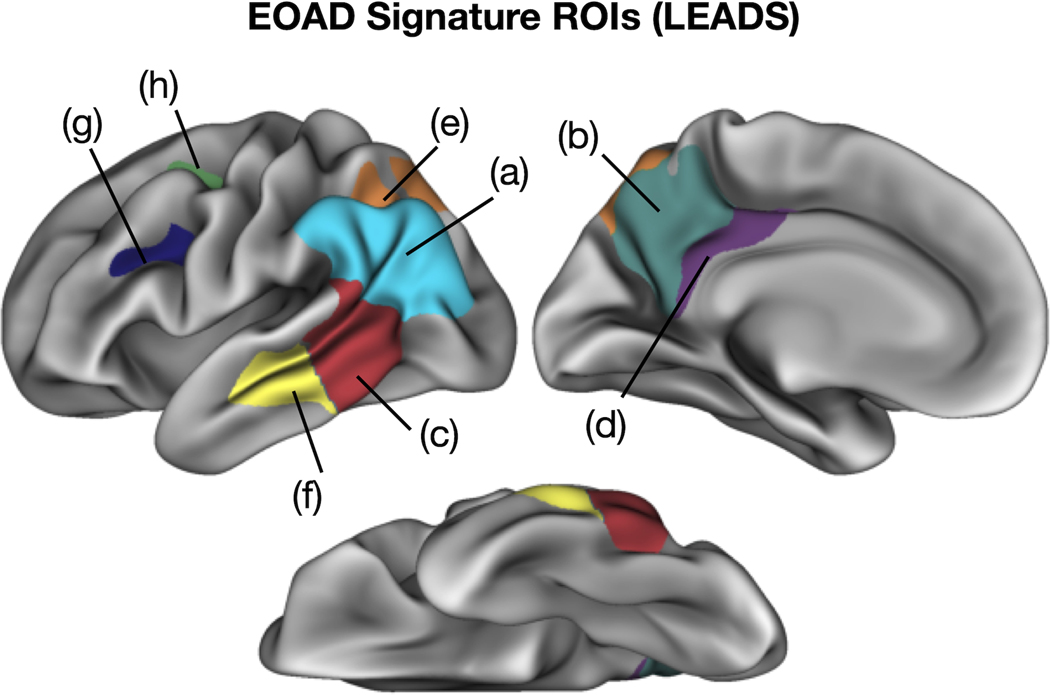
We generated a preliminary LEADS EOAD Signature from the present sample, with plans to revise this when the full sample is collected. We defined 9 EOAD-Signature ROIs representing areas of cortical atrophy in patients with EOAD relative to CN participants based on the results of our vertex-wise GLMs using the LEADS cohort (Figure 6). Similar to the MGH-generated EOAD Signature, the LEADS-generated EOAD Signature ROIs included inferior parietal, precuneus, caudal lateral temporal, posterior cingulate, superior parietal, mid lateral temporal, middle frontal gyrus, superior frontal gyrus and the fusiform gyrus.
Figure 6. The longitudinal early-onset Alzheimer’s disease study (LEADS) early-onset Alzheimer’s disease (EOAD) signature.
Colored vertices on the cortical surface maps indicate areas where EOAD patients showed cortical atrophy compared to low-amyloid cognitively normal (CN) participants within the LEADS cohort. See Figure 2 for the anatomic labels corresponding to each colored region shown here.
Discussion
The objective of this study was to define the utility of an MRI biomarker of neurodegeneration in sporadic EOAD—the cortical EOAD Signature. Using quantitative analysis of cortical thickness from T1-weighted MRI scans, we identified a signature pattern of distributed cortical atrophy in EOAD that replicated across two well-characterized independent samples: a discovery sample of 25 participants with EOAD from MGH and a large validation sample of 211 participants with EOAD from the multicenter LEADS consortium1. In addition to reliability across the two samples, the EOAD Signature demonstrated clinical validity against widely used measures of symptom severity and cognitive function, with greater atrophy associated with greater cognitive impairment (on both the CDR-SB and the MMSE). The EOAD Signature accurately differentiated EOAD patients from healthy controls as well as patients with EOnonAD cognitive impairment. The EOAD Signature included areas primarily in medial and lateral parietal cortex, posterior lateral temporal cortex, with lesser involvement of the lateral prefrontal cortex and fusiform gyrus. These findings are consistent with prior studies of single small samples showing atrophy largely in these same regions in sporadic EOAD in contrast with the prominent effects seen in anterior medial and ventral temporal cortex observed in typical cases of LOAD 4–8. PET studies of EOAD have also shown high tau pathology 47–50 and prominent hypometabolism 28,51 within EOAD-Signature regions.
The localization and magnitude of regional cortical atrophy in EOAD was remarkably consistent across our two independent samples despite differences in the severity of cognitive impairment across cohorts, and differences in analytic factors, including MRI hardware and software inherent in multicenter studies (although most LEADS sites employ Siemens Prisma scanners). One difference, however, was observed in the lateral prefrontal cortex with the LEADS cohort showing less atrophy compared to the MGH cohort (not meeting the stringent a priori threshold for visualization in Figure 1B but nevertheless showing a large effect size using the MGH-derived ROI in Table 2). A possible explanation is differences in the proportional phenotypic mix between the MGH group and LEADS cohorts. Additionally, longitudinal studies show that posterior cortical regions are typically impacted first in AD followed by the prefrontal cortex as the disease progresses 52,53. Thus, this difference is also likely a reflection of the lesser magnitude of cognitive impairment in LEADS relative to the MGH sample, which also likely explains the overall smaller magnitude of atrophy in the LEADS sample. With that point about relative magnitude in mind, all of the regional cortical atrophy effects predicted from the MGH discovery sample to be found in the LEADS validation sample were large (Cohen’s d values in Table 2). The relatively more prominent parietal and posterior temporal neurodegeneration in EOAD than in LOAD may explain the higher frequency of non-memory impairments 4,54, including dysfunction in attention, language, visuospatial, and executive abilities 55–59.
The observation that the magnitude of EOAD-Signature atrophy correlated with relative severity of impairment (as measured by CDR-SB and MMSE) supports the clinical validity of this summary biomarker of neurodegeneration in this patient population. We demonstrated this relationship previously in LOAD 12, which supported the use of this summary biomarker of neurodegeneration in multiple other types of studies as an a priori biomarker of AD-related neurodegeneration 12–17. As we consider outcomes in clinical trials in EOAD patients aiming to slow neurodegeneration, the EOAD-Signature should be included.
Furthermore, the spatial overlap between our observed cortical neurodegeneration in EOAD and the topography of large-scale brain networks suggests the involvement of multiple brain networks in EOAD. The EOAD Signature closely corresponds to the localization of core regions within at least five different large-scale brain networks, including (1) the default mode network (DMN) involved in memory and known to be targeted in AD with hub regions in PCC/precuneus, inferior parietal lobe, and lateral temporal cortex 55, (2) the left hemisphere language network with core regions in the perisylvian cortex 57,60, (3) the Frontoparietal network (FPN) involved in executive function with hub regions in the caudal middle frontal gyrus, superior frontal gyrus, superior parietal cortex and inferior parietal cortex/supramarginal gyrus 56,59, (4) the Dorsal Attention network including lateral intraparietal area and frontal eye field 59, and (5) the Visual Association network 61. Evidence from functional neuroimaging studies in AD further demonstrates disruption of functional connectivity between core regions within these brain networks 62–64. It remains to be seen whether atrophy in EOAD may originate within a hub region such as the precuneus 65,66 and spread through connections within multiple distributed neural networks 67–70. Longitudinal analyses of the LEADS sample will be conducted as the sample accrues to test the hypothesized spread of neurodegeneration over time.
Our findings further demonstrate the superiority of the EOAD-Signature MRI biomarker compared to other MRI biomarkers of AD in detecting EOAD-related neurodegeneration. Atrophy in the EOAD signature showed better sensitivity compared with traditional anatomically derived ROIs based on automatic parcellation or volumetric measures of the hippocampal formation, ventricles, or whole brain, supporting its potential utility in clinical trials for AD. The data-driven LOAD-Signature MRI biomarker also did not capture atrophy in EOAD as well as the EOAD Signature, providing further support for the potential utility of a measure tailored to EOAD rather than AD more generally; this is especially important if the goal is to detect small effect sizes in longitudinal analyses, such as might be expected with disease-modifying therapies. We plan to test the EOAD Signature as an outcome measure in MRI datasets from clinical trials when the opportunity arises, as we have done with the LOAD Signature in the past 18.
We will share the EOAD Signature generated from the present LEADS sample (N = 211) as a tool for the scientific community. Once the full sample (N = 400) is collected, we plan to release an updated version of this signature. It is possible that an analysis of a larger sample may change the topography or extent of atrophy in some brain regions identified here. However, the replication of the EOAD Signature in two independent samples and the general convergence of our results with previous studies suggest that, if revisions are necessary, they will likely be minor. If a user is technically proficient with FreeSurfer data analysis, the procedure for using the EOAD Signature to quantify atrophy within the relevant regions is fully automated and user-friendly with minimal additional skill required for its use.
The current study has several limitations. First, we present findings here using the LEADS cohort near the midpoint of data collection. Due to the clinical heterogeneity of EOAD, future analyses with the full LEADS sample will increase the precision with which we can estimate regional atrophy in EOAD and will allow us to do so with a large sample of very mildly affected (i.e., CDR 0.5) individuals. Furthermore, the current sample is predominantly White and highly educated. As the LEADS sample increases, we will be able to include more heterogenous samples of the population, to enrich ethnic, racial, and educational diversity. Second, these results reflect only baseline data from LEADS. Future work will focus on using the EOAD Signature of regional atrophy to examine longitudinal change. Longitudinal studies will further allow comparisons of the EOAD-Signature MRI to other MRI biomarkers of AD based on effect sizes from longitudinal measures. Furthermore, examination of longitudinal MRI data in relation to tau PET and fluid biomarker data will enable us to better understand the molecular drivers of propagation of neurodegeneration within and between large-scale brain networks. Follow-up studies will also explore the relationship between atrophy within the EOAD-Signature and impairments in different cognitive domains. Finally, as the LEADS sample size increases, we will be able to compare atrophy measures in sporadic EOAD to those in sporadic LOAD and in autosomal dominant EOAD.
Supplementary Material
Acknowledgements
We would like to thank Anne Fagan to her contribution to this work. The authors would like to express special thanks to LEADS participants and their family members and friends and to study staff and administrative personnel, without whose effort and time this research would not have been possible.
Funding Sources
This study is generously supported by R56 AG057195, U01AG6057195, U24AG021886, Alzheimer’s Association LEADS GENETICS-19-639372, Alzheimer’s Association LDRFP-21-818464, Alzheimer’s Association LDRFP-21-824473 and Alzheimer’s Association LDRFP-21-828356. NACC is funded by the NIA (U24 AG072122). NACC data are contributed by the following NIA-funded ADRCs: P30 AG010133, P30 AG062422, P30 AG066462, P30AG066507, P30 AG062421, P30 AG066506, P30AG072977, P30 AG066444, P30 AG066515, P30 AG062677, P30 AG072980, P30 AG072979, P30 AG066511 as well as R01 DC014296, R21 DC019567, R21 AG073744, K23 DC016912, P50 AG005134, and P30 AG062421 and by the Tommy Rickles Chair in Primary Progressive Aphasia Research. This research was carried out in part at the Athinoula A. Martinos Center for Biomedical Imaging at the MGH, using resources provided by the Center for Functional Neuroimaging Technologies, P41EB015896, a P41 Biotechnology Resource Grant supported by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health. This work also involved the use of instrumentation supported by the NIH Shared Instrumentation Grant Program and/or High-End Instrumentation Grant Program; specifically, grant number(s) S10RR021110, S10RR023043, S10RR023401. We would like to express special thanks to LEADS participants and their family members and friends and to study staff and administrative personnel, without whose effort and time this research would not have been possible.
Footnotes
Conflicts of Interest
No authors associated with this project have reported conflicts of interest that would impact these results.
References
- 1.Apostolova LG, Aisen P, Eloyan A, et al. The Longitudinal Early-onset Alzheimer’s Disease Study (LEADS): Framework and methodology. Alzheimers Dement. 2021;n/a(n/a). doi: 10.1002/alz.12350 [DOI] [PMC free article] [PubMed]
- 2.Barclay LL, Zemcov A, Blass JP, McDowell FH. Factors associated with duration of survival in Alzheimer’s disease. Biol Psychiatry. 1985;20(1):86–93. doi: 10.1016/0006-3223(85)90139-8 [DOI] [PubMed] [Google Scholar]
- 3.Heyman A, Wilkinson WE, Hurwitz BJ, et al. Early-onset Alzheimer’s disease: Clinical predictors of institutionalization and death. Neurology. 1987;37(6):980–984. [DOI] [PubMed] [Google Scholar]
- 4.Mendez MF. Early-onset Alzheimer Disease and Its Variants: Contin Lifelong Learn Neurol. 2019;25(1):34–51. doi: 10.1212/CON.0000000000000687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aziz AL, Giusiano B, Joubert S, et al. Difference in imaging biomarkers of neurodegeneration between early and late-onset amnestic Alzheimer’s disease. Neurobiol Aging. 2017;54:22–30. doi: 10.1016/j.neurobiolaging.2017.02.010 [DOI] [PubMed] [Google Scholar]
- 6.Dickerson BC, Brickhouse M, McGinnis S, Wolk DA. Alzheimer’s disease: The influence of age on clinical heterogeneity through the human brain connectome. Alzheimers Dement Diagn Assess Dis Monit. 2017;6:122–135. doi: 10.1016/j.dadm.2016.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frisoni GB, Pievani M, Testa C, et al. The topography of grey matter involvement in early and late onset Alzheimer’s disease. Brain. 2007;130(3):720–730. doi: 10.1093/brain/awl377 [DOI] [PubMed] [Google Scholar]
- 8.Migliaccio R, Agosta F, Possin KL, et al. Mapping the Progression of Atrophy in Early- and Late-Onset Alzheimer’s Disease. Galimberti D, ed. J Alzheimers Dis. 2015;46(2):351–364. doi: 10.3233/JAD-142292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. Neuropathologically defined subtypes of Alzheimer’s disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 2011;10(9):785–796. doi: 10.1016/S1474-4422(11)70156-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Contador J, Pérez-Millán A, Tort-Merino A, et al. Longitudinal brain atrophy and CSF biomarkers in early-onset Alzheimer’s disease. NeuroImage Clin. 2021;32:102804. doi: 10.1016/j.nicl.2021.102804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falgàs N, Ruiz-Peris M, Pérez-Millan A, et al. Contribution of CSF biomarkers to early-onset Alzheimer’s disease and frontotemporal dementia neuroimaging signatures. Hum Brain Mapp. 2020;41(8):2004–2013. doi: 10.1002/hbm.24925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dickerson BC, Bakkour A, Salat DH, et al. The Cortical Signature of Alzheimer’s Disease: Regionally Specific Cortical Thinning Relates to Symptom Severity in Very Mild to Mild AD Dementia and is Detectable in Asymptomatic Amyloid-Positive Individuals. Cereb Cortex. 2009;19(3):497–510. doi: 10.1093/cercor/bhn113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickerson BC, Wolk D. Biomarker-based prediction of progression in MCI: Comparison of AD-signature and hippocampal volume with spinal fluid amyloid-β and tau. Front Aging Neurosci. 2013;5:55. doi: 10.3389/fnagi.2013.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Racine AM, Brickhouse M, Wolk DA, Dickerson BC. The personalized Alzheimer’s disease cortical thickness index predicts likely pathology and clinical progression in mild cognitive impairment. Alzheimers Dement Diagn Assess Dis Monit. 2018;10:301–310. doi: 10.1016/j.dadm.2018.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickerson BC, Stoub TR, Shah RC, et al. Alzheimer-signature MRI biomarker predicts AD dementia in cognitively normal adults. Neurology. 2011;76(16):1395–1402. doi: 10.1212/WNL.0b013e3182166e96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dickerson BC, Wolk DA, On behalf of the Alzheimer’s Disease Neuroimaging Initiative. MRI cortical thickness biomarker predicts AD-like CSF and cognitive decline in normal adults. Neurology. 2012;78(2):84–90. doi: 10.1212/WNL.0b013e31823efc6c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Racine AM, Fong TG, Travison TG, et al. Alzheimer’s-related cortical atrophy is associated with postoperative delirium severity in persons without dementia. Neurobiol Aging. 2017;59:55–63. doi: 10.1016/j.neurobiolaging.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz AJ, Sundell KL, Charil A, et al. Magnetic resonance imaging measures of brain atrophy from the EXPEDITION3 trial in mild Alzheimer’s disease. Alzheimers Dement Transl Res Clin Interv. 2019;5(1):328–337. doi: 10.1016/j.trci.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris JC. The Clinical Dementia Rating (CDR): Current version and scoring rules. Neurology. 1993;43(11):2412–2414. [DOI] [PubMed] [Google Scholar]
- 20.Dickerson BC, McGinnis SM, Xia C, et al. Approach to atypical Alzheimer’s disease and case studies of the major subtypes. CNS Spectr. 2017;22(6):439–449. doi: 10.1017/S109285291600047X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crutch SJ, Schott JM, Rabinovici GD, et al. Consensus classification of posterior cortical atrophy. Alzheimers Dement. 2017;13(8):870–884. doi: 10.1016/j.jalz.2017.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendez MF, Ghajarania N, Perryman KM. Posterior cortical atrophy: Clinical characteristics and differences compared to Alzheimer’s disease. Dement Geriatr Cogn Disord. 2002;14(1):33–40. [DOI] [PubMed] [Google Scholar]
- 23.Tang-Wai DF, Graff-Radford NR, Boeve BF, et al. Clinical, genetic, and neuropathologic characteristics of posterior cortical atrophy. Neurology. Published online 2004. [DOI] [PubMed]
- 24.Ossenkoppele R, Pijnenburg YAL, Perry DC, et al. The behavioural/dysexecutive variant of Alzheimer’s disease: clinical, neuroimaging and pathological features. Brain. 2015;138(9):2732–2749. doi: 10.1093/brain/awv191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Townley RA, Graff-Radford J, Mantyh WG, et al. Progressive dysexecutive syndrome due to Alzheimer’s disease: a description of 55 cases and comparison to other phenotypes. Brain Commun. 2020;2(1):fcaa068. doi: 10.1093/braincomms/fcaa068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabinovici GD, Furst AJ, Alkalay A, et al. Increased metabolic vulnerability in early-onset Alzheimer’s disease is not related to amyloid burden. Brain. 2010;133(2):512–528. doi: 10.1093/brain/awp326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villeneuve S, Rabinovici GD, Cohn-Sheehy BI, et al. Existing Pittsburgh Compound-B positron emission tomography thresholds are too high: statistical and pathological evaluation. Brain. 2015;138(7):2020–2033. doi: 10.1093/brain/awv112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blennow K, Dubois B, Fagan AM, Lewczuk P, Leon MJ, Hampel H. Clinical utility of cerebrospinal fluid biomarkers in the diagnosis of early Alzheimer’s disease. Alzheimers Dement. 2015;11(1):58–69. doi: 10.1016/j.jalz.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montine TJ, Phelps CH, Beach TG, et al. National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathol (Berl). 2012;123(1):1–11. doi: 10.1007/s00401-011-0910-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nobili F, Arbizu J, Bouwman F, et al. European Association of Nuclear Medicine and European Academy of Neurology recommendations for the use of brain 18F-fluorodeoxyglucose positron emission tomography in neurodegenerative cognitive impairment and dementia: Delphi consensus. Eur J Neurol. 2018;25(10):1201–1217. doi: 10.1111/ene.13728 [DOI] [PubMed] [Google Scholar]
- 33.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state.” J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6 [DOI] [PubMed] [Google Scholar]
- 34.Sabri O, Sabbagh MN, Seibyl J, et al. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer’s disease: Phase 3 study. Alzheimers Dement. 2015;11(8):964–974. doi: 10.1016/j.jalz.2015.02.004 [DOI] [PubMed] [Google Scholar]
- 35.Klunk WE, Koeppe RA, Price JC, et al. The Centiloid Project: Standardizing quantitative amyloid plaque estimation by PET. Alzheimers Dement. 2015;11(1):1–15.e4. doi: 10.1016/j.jalz.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jack CR, Barnes J, Bernstein MA, et al. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement. 2015;11(7):740–756. doi: 10.1016/j.jalz.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001;20(1):70–80. doi: 10.1109/42.906426 [DOI] [PubMed] [Google Scholar]
- 38.Putcha D, Katsumi Y, Brickhouse M, et al. Gray to white matter signal ratio as a novel biomarker of neurodegeneration in Alzheimer’s disease. NeuroImage Clin. 2023;37:103303. doi: 10.1016/j.nicl.2022.103303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci. 2000;97(20):11050–11055. doi: 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Genovese CR, Lazar NA, Nichols T. Thresholding of Statistical Maps in Functional Neuroimaging Using the False Discovery Rate. NeuroImage. 2002;15(4):870–878. doi: 10.1006/nimg.2001.1037 [DOI] [PubMed] [Google Scholar]
- 41.Fortin JP, Cullen N, Sheline YI, et al. Harmonization of cortical thickness measurements across scanners and sites. NeuroImage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desikan RS, Ségonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- 43.Katsumi Y, Putcha D, Eckbo R, et al. Anterior dorsal attention network tau drives visual attention deficits in posterior cortical atrophy. Brain. Published online October 14, 2022:awac245. doi: 10.1093/brain/awac245 [DOI] [PMC free article] [PubMed]
- 44.Katsumi Y, Quimby M, Hochberg D, et al. Association of Regional Cortical Network Atrophy With Progression to Dementia in Patients With Primary Progressive Aphasia. Neurology. Published online October 3, 2022. doi: 10.1212/WNL.0000000000201403 [DOI] [PMC free article] [PubMed]
- 45.Barnes J, Ridgway GR, Bartlett J, et al. Head size, age and gender adjustment in MRI studies: a necessary nuisance? NeuroImage. 2010;53(4):1244–1255. doi: 10.1016/j.neuroimage.2010.06.025 [DOI] [PubMed] [Google Scholar]
- 46.Schwarz CG, Gunter JL, Wiste HJ, et al. A large-scale comparison of cortical thickness and volume methods for measuring Alzheimer’s disease severity. NeuroImage Clin. 2016;11:802–812. doi: 10.1016/j.nicl.2016.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.La Joie R, Visani AV, Lesman-Segev OH, et al. Association of APOE4 and Clinical Variability in Alzheimer Disease With the Pattern of Tau- and Amyloid-PET. Neurology. 2021;96(5):e650–e661. doi: 10.1212/WNL.0000000000011270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nasrallah IM, Chen YJ, Hsieh MK, et al. 18F-Flortaucipir PET/MRI Correlations in Nonamnestic and Amnestic Variants of Alzheimer Disease. J Nucl Med. 2018;59(2):299–306. doi: 10.2967/jnumed.117.194282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sintini I, Schwarz CG, Martin PR, et al. Regional multimodal relationships between tau, hypometabolism, atrophy, and fractional anisotropy in atypical Alzheimer’s disease. Hum Brain Mapp. 2019;40(5):1618–1631. doi: 10.1002/hbm.24473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia C, Makaretz SJ, Caso C, et al. Association of In Vivo [18F]AV-1451 Tau PET Imaging Results With Cortical Atrophy and Symptoms in Typical and Atypical Alzheimer Disease. JAMA Neurol. 2017;74(4):427. doi: 10.1001/jamaneurol.2016.5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanner JA, Iaccarino L, Edwards L, et al. Amyloid, tau and metabolic PET correlates of cognition in early and late-onset Alzheimer’s disease. Brain. 2022;145(12):4489–4505. doi: 10.1093/brain/awac229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison TM, Joie RL, Maass A, et al. Longitudinal tau accumulation and atrophy in aging and alzheimer disease. Ann Neurol. 2019;85(2):229–240. doi: 10.1002/ana.25406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sintini I, Martin PR, Graff-Radford J, et al. Longitudinal tau-PET uptake and atrophy in atypical Alzheimer’s disease. NeuroImage Clin. 2019;23:101823. doi: 10.1016/j.nicl.2019.101823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sirkis DW, Bonham LW, Johnson TP, La Joie R, Yokoyama JS. Dissecting the clinical heterogeneity of early-onset Alzheimer’s disease. Mol Psychiatry. Published online April 7, 2022:1–15. doi: 10.1038/s41380-022-01531-9 [DOI] [PMC free article] [PubMed]
- 55.Buckner RL, Andrews-Hanna JR, Schacter DL. The Brain’s Default Network. Ann N Y Acad Sci. 2008;1124(1):1–38. doi: 10.1196/annals.1440.011 [DOI] [PubMed] [Google Scholar]
- 56.Dosenbach NUF, Fair DA, Cohen AL, Schlaggar BL, Petersen SE. A dual-networks architecture of top-down control. Trends Cogn Sci. 2008;12(3):99–105. doi: 10.1016/j.tics.2008.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hickok G, Poeppel D. The cortical organization of speech processing. Nat Rev Neurosci. 2007;8(5):393–402. doi: 10.1038/nrn2113 [DOI] [PubMed] [Google Scholar]
- 58.Touroutoglou A, Hollenbeck M, Dickerson BC, Barrett LF. Dissociable large-scale networks anchored in the right anterior insula subserve affective experience and attention. NeuroImage. 2012;60(4):1947–1958. doi: 10.1016/j.neuroimage.2012.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vincent JL, Kahn I, Snyder AZ, Raichle ME, Buckner RL. Evidence for a Frontoparietal Control System Revealed by Intrinsic Functional Connectivity. J Neurophysiol. 2008;100(6):3328–3342. doi: 10.1152/jn.90355.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomasi D, Volkow ND. Resting functional connectivity of language networks: characterization and reproducibility. Mol Psychiatry. 2012;17(8):841–854. doi:https://doi-org.ezproxy.neu.edu/10.1038/mp.2011.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yeo BTT, Krienen FM, Sepulcre J, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adriaanse SM, Binnewijzend MAA, Ossenkoppele R, et al. Widespread Disruption of Functional Brain Organization in Early-Onset Alzheimer’s Disease. PLOS ONE. 2014;9(7):e102995. doi: 10.1371/journal.pone.0102995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Filippi M, Basaia S, Canu E, et al. Brain network connectivity differs in early-onset neurodegenerative dementia. Neurology. 2017;89(17):1764–1772. doi: 10.1212/WNL.0000000000004577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gour N, Felician O, Didic M, et al. Functional connectivity changes differ in early and late-onset alzheimer’s disease: Network Reorganization Relative to AD Onset. Hum Brain Mapp. 2014;35(7):2978–2994. doi: 10.1002/hbm.22379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Buckner RL, Sepulcre J, Talukdar T, et al. Cortical Hubs Revealed by Intrinsic Functional Connectivity: Mapping, Assessment of Stability, and Relation to Alzheimer’s Disease. J Neurosci. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Margulies DS, Vincent JL, Kelly C, et al. Precuneus shares intrinsic functional architecture in humans and monkeys. Proc Natl Acad Sci. 2009;106(47):20069–20074. doi: 10.1073/pnas.0905314106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gili T, Cercignani M, Serra L, et al. Regional brain atrophy and functional disconnection across Alzheimer’s disease evolution. J Neurol Neurosurg Psychiatry. 2011;82(1):58–66. doi: 10.1136/jnnp.2009.199935 [DOI] [PubMed] [Google Scholar]
- 68.Mesulam MM. Neuroplasticity Failure in Alzheimer’s Disease: Bridging the Gap between Plaques and Tangles. Neuron. 1999;24(3):521–529. doi: 10.1016/S0896-6273(00)81109-5 [DOI] [PubMed] [Google Scholar]
- 69.Seeley WW, Crawford RK, Zhou J, Miller BL, Greicius MD. Neurodegenerative Diseases Target Large-Scale Human Brain Networks. Neuron. 2009;62(1):42–52. doi: 10.1016/j.neuron.2009.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogel JW, Iturria-Medina Y, Strandberg OT, et al. Spread of pathological tau proteins through communicating neurons in human Alzheimer’s disease. Nat Commun. 2020;11(1):2612. doi: 10.1038/s41467-020-15701-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.