Summary
The discovery of exercise-regulated circulatory factors has fueled interest in organ crosstalk, especially between skeletal muscle and adipose tissue, and the role in mediating beneficial effects of exercise. We studied the adipose tissue transcriptome in men and women with normal glucose tolerance or type 2 diabetes following an acute exercise bout, revealing substantial exercise- and time-dependent changes, with sustained increase in inflammatory genes in type 2 diabetes. We identify oncostatin-M as one of the most upregulated adipose-tissue-secreted factors post-exercise. In cultured human adipocytes, oncostatin-M enhances MAPK signaling and regulates lipolysis. Oncostatin-M expression arises predominantly from adipose tissue immune cell fractions, while the corresponding receptors are expressed in adipocytes. Oncostatin-M expression increases in cultured human Thp1 macrophages following exercise-like stimuli. Our results suggest that immune cells, via secreted factors such as oncostatin-M, mediate a crosstalk between skeletal muscle and adipose tissue during exercise to regulate adipocyte metabolism and adaptation.
Keywords: exercise, adipose tissue, crosstalk, oncostatin-M, immune cells, inflammation, skeletal muscle, type 2 diabetes, human
Graphical abstract
Highlights
-
•
Exercise unveils release of circulatory factors from adipose tissue
-
•
Exercise elicits a sustained increase in inflammatory genes in type 2 diabetes
-
•
Oncostatin-M is a key exercise-responsive metabolic regulator
-
•
Immune cells play a role in exercise-induced tissue crosstalk
Dollet et al. reveal intricate interplay between skeletal muscle, adipose tissue, and immune cells during exercise and identify a role for circulatory factors to regulate adipose tissue metabolism. Immune cells, via the secretion of exercise-responsive factors such as oncostatin-M, are important in crosstalk between peripheral tissues to regulate adipocyte lipolysis.
Introduction
Lifestyle intervention, including physical activity, is a recommended first-course intervention for the management of type 2 diabetes and associated cardiovascular risks. Regular exercise has beneficial effects on glucose tolerance, insulin sensitivity, and the circulating lipid profile, coincident with improvements in skeletal muscle and liver function.1 The discovery of exercise-regulated circulatory factors (“exerkines”) has fueled interest in the role of organ crosstalk, especially between skeletal muscle, white adipose tissue, and liver, in mediating the beneficial effects of exercise on insulin sensitivity and metabolism.2 In mouse models, the beneficial effects of exercise training on glucose homeostasis are conferred in part via adipose tissue browning and modifications of the lipid profile.3,4 However, in humans, the molecular mechanisms driving the adaptive response of adipose tissue to acute exercise, as well as the relevance of adipose tissue to exercise-induced improvements in metabolism, remain unclear. In healthy individuals, 6 months of exercise training increased the expression of genes involved in pathways related to oxidative phosphorylation in subcutaneous adipose tissue,5,6 which may contribute to the systemic improvements in energy homeostasis, while a 12-week training intervention in obese men did not replicate these results.7 Therefore, the extent to which exercise alters subcutaneous adipose tissue gene expression and metabolism in older individuals or people with type 2 diabetes remains to be investigated.
Adipose tissue is a major regulator of whole-body energy homeostasis and communicates with other organs through the secretion of factors (adipokines) including small molecules, lipids, and proteins.8,9 The complex cellular composition of adipose tissue, which includes adipocytes, as well as populations of progenitor cells, immune cells, endothelial cells, and nerve endings, suggests that these cells and their communications play a role in widespread whole-body effects, including energy homeostasis. Exercise may influence cell-cell communication through the release of exerkines such as the adipokine transforming growth factor β2 (TGF-β2), which is secreted in response to exercise in both rodents and humans.10 This highlights the importance of the adipose tissue secretome in the context of exercise-induced metabolic adaptations and its potential role in inter- and intraorgan crosstalk.
To gain insight into the pleotropic effects of exercise on adipocyte biology and metabolism, we mapped the transcriptomic response to acute exercise in subcutaneous adipose tissues in individuals with normal glucose tolerance or type 2 diabetes. Our analysis revealed time-dependent transcriptomic changes, with sustained markers of inflammation in adipose tissue from individuals with type 2 diabetes. We characterized the exercise-induced changes in the adipose tissue secretome profile and show that oncostatin-M is increased by exercise, irrespective of glycemic status. We showed that oncostatin-M expression originates from adipose tissue immune cells and alters lipolysis in mature human adipocytes, suggesting a role of immune cell-adipocyte crosstalk during exercise.
Results
Exercise exerts a differential transcriptomic response in adipose tissue between individuals with normal glucose tolerance and those with type 2 diabetes
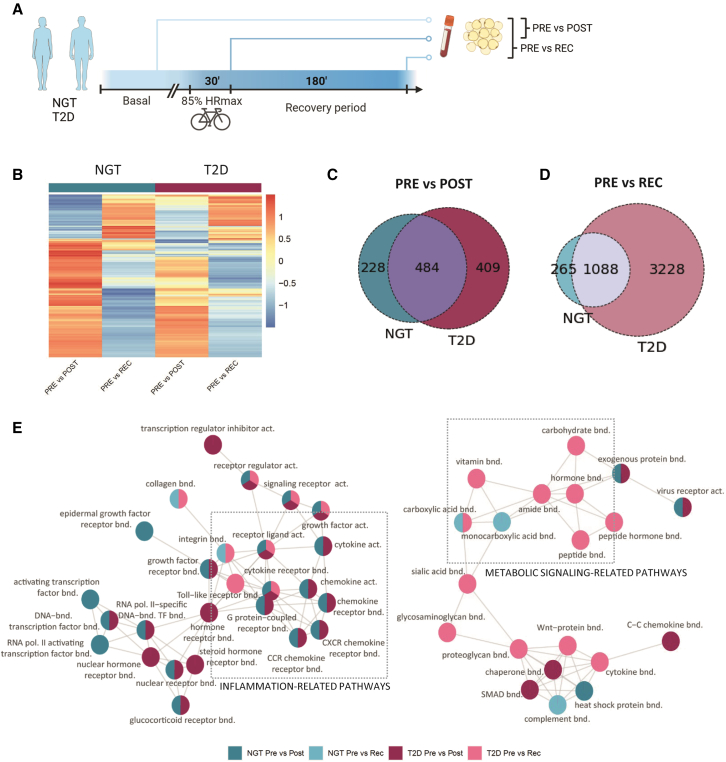
The adipose tissue transcriptomic response to an acute bout of exercise was determined in 20 individuals (11 men, 9 women) with normal glucose tolerance and 28 individuals (16 men, 12 women) with type 2 diabetes (Figure 1A ). Clinical parameters are presented in Table 1. In the basal state (pre), 589 genes were altered in adipose tissue of people with type 2 diabetes versus those with normal glucose tolerance. These genes were associated with pathways linked to oxidative phosphorylation and lipid metabolism, consistent with a type 2 diabetes-associated mitochondrial dysfunction (lists in Data S1 and S2). Exercise resulted in robust time-dependent changes in the adipose tissue transcriptome, with distinct differences between the post-exercise time point (0′ after the exercise bout) versus the recovery time point (3 h after the exercise bout) in both individuals with normal glucose tolerance and type 2 diabetes (Figure 1B). Immediately after exercise, most of the significantly differentially regulated transcripts were upregulated compared to the basal, pre-exercise condition (60% and 70%, respectively, total differentially expressed genes), and the magnitude of the response was similar between individuals with normal glucose tolerance or type 2 diabetes, with 712 and 893 genes, respectively (Figures 1B and 1C). At recovery, compared to the pre-exercise condition, the adipose tissue transcriptome displayed larger changes than immediately post-exercise. This response was more evident in adipose tissue of individuals with type 2 diabetes, with 4,316 genes differentially regulated (50% of which were downregulated) and 3,228 distinctive genes compared to the response in adipose tissue from individuals with normal glucose tolerance (Figures 1B and 1D).
Figure 1.
Adipose tissue displays a differential transcriptomic response to exercise in individuals with NGT and T2D
(A) Experimental design.
(B) Heatmap of differentially expressed genes in adipose tissue from individuals with normal glucose tolerance (NGT; n = 20) or type 2 diabetes (T2D; n = 28) analyzed between pre- and post-exercise or recovery conditions.
(C) Overlay of significantly differentially expressed genes between pre- and post-exercise in adipose tissue of participants with NGT or T2D.
(D) Overlay of significantly differentially expressed genes between pre-exercise and recovery in adipose tissue of individuals with either NGT or T2D.
(E) Visualization of the main pathways’ clusters derived from the GO hierarchy pathway analysis. Color of the circles indicates the condition in which the pathways are significantly altered.
Table 1.
Clinical parameters
| Normal glucose tolerance (NGT) | Type 2 diabetics (T2D) | t test | |
|---|---|---|---|
| n | 20 (11 men, 9 women) | 28 (18 men, 12 women) | N/A |
| Age | 60.2 ± 4.3 | 60.1 ± 4.6 | NS |
| BMI (kg/m2) | 26.9 ± 3.0 | 27.6 ± 2.7 | NS |
| W/H ratio | 0.9 ± 0.1 | 1.0 ± 0.1 | ∗∗ |
| P-Triglycerides (mmol/L) | 0.9 ± 0.3 | 1.3 ± 0.5 | ∗∗ |
| P-Cholesterol (mmol/L) | 5.3 ± 0.9 | 4.4 ± 1.1 | ∗∗ |
| HbA1c (mmol/mol) | 35.5 ± 2.4 | 48.2 ± 9.2 | ∗∗∗ |
| P-Glucose 0 OGTT (mmol/L) | 5.2 ± 0.5 | 7.4 ± 1.6 | ∗∗∗ |
| AUC glucose OGTT (mmol/L) | 877.0 ± 181.5 | 1,639.9 ± 340.1 | ∗∗∗ |
| AUC insulin OGTT | 7,850.0 ± 4,075.8 | 7,177.9 ± 4,678.3 | NS |
| AUC glucose exercise (mmol/L) | 1,101.2 ± 126.1 | 1,507.3 ± 326.7 | ∗∗∗ |
| AUC insulin exercise | 1,480.2 ± 583.9 | 3,018.0 ± 1,454.7 | ∗∗∗ |
| VO2max (mL/min) | 2,884.6 ± 759.5 | 2,431.9 ± 703.1 | ∗ |
| Max heart rate (beats/min) | 169.3 ± 11.8 | 163.7 ± 15.4 | NS |
| HOMA1-IR | 1.7 ± 0.8 | 4.0 ± 2.2 | ∗∗∗ |
NS, not significant; W/H, waist to hip; AUC, area under the curve; OCTT, oral glucose tolerance test; HOMA-IR, homeostatic model assessment for insulin resistance.
Gene Ontology enrichment analysis was performed to identify the pathways altered by exercise (Figure S1). Pathways were clustered based on the Gene Ontology (GO) network, and the two largest clusters are presented in Figure 1E. Within these clusters, pathways related to inflammation, such as chemokine binding or cytokine activity, were enriched post-exercise independent of glycemic status. Similarly, pathways related to regulation of transcription were activated post-exercise, suggesting active tissue remodeling. In recovery, most pathways represented in the network were only altered in individuals with type 2 diabetes, with a sustained enrichment of pathways related to inflammation, metabolites, and hormone binding. To maximize data availability and promote sharing of omics datasets, an online tool (https://adipomax.serve.scilifelab.se/) was generated for browsing of individual genes in response to exercise. This application allows exploration of the dataset for genes of interest and visualization of the response to exercise. For example, the peroxisome proliferator activated receptor gamma (PPARG) gene, included in the carbohydrate-binding pathway, was downregulated only in recovery state in adipose tissue from individuals with type 2 diabetes (Figure S2A). CXCL2, included in the “receptor ligand activity” pathway, remained upregulated in the recovery state only in people with type 2 diabetes (Figure S2B).
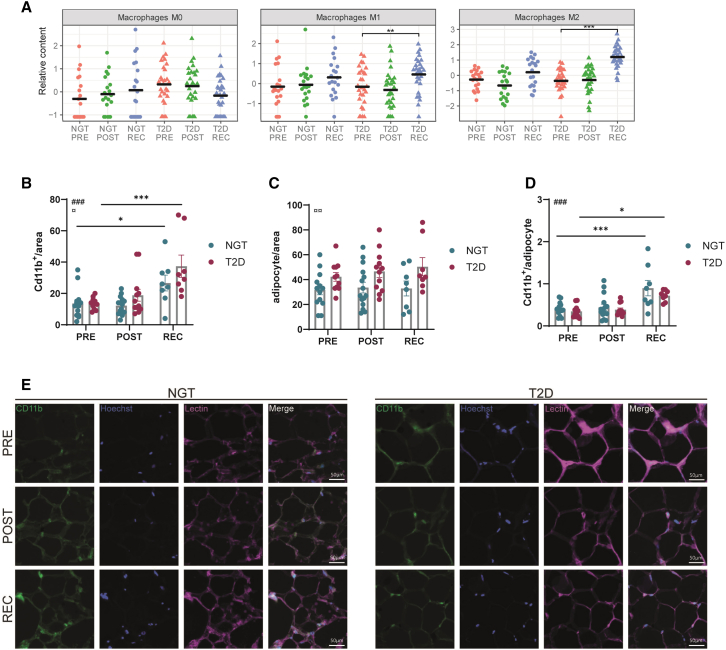
Adipose tissue is a heterogeneous tissue, containing a high proportion of non-adipocyte cells such as progenitors, immune cells, and endothelial cells.11 To further investigate the source of the inflammatory genes, a CIBERSORTx analysis was performed to estimate immune cell fractions from the bulk tissue gene expression profile.12 Uniform manifold approximation and projection analysis indicated an enhanced immune cell infiltration in the type 2 diabetes recovery samples (Figure S2C). Especially, the proportion of both M1 and M2 macrophages was increased in adipose tissue from individuals with type 2 diabetes after recovery (Figure 2A), suggesting immune cell activation and/or infiltration. To further quantify immune cell infiltration, we stained for CD11b, a marker of myeloid cells, including monocytes, in adipose tissue of a subset of subjects (Figures 2B–2E). Staining quantification showed that CD11b+ cells were enriched at recovery, and this effect was stronger in biopsies from people with type 2 diabetes, suggesting increased immune cell infiltration in response to exercise (Figure 2B). Of note, adipose tissue from people with type 2 diabetes displayed a higher number of adipocytes per area; therefore, no statistical difference between groups was visible after normalization per adipocyte (Figures 2C and 2D). Overall, our findings revealed that the adipose tissue transcriptome is robustly altered by exercise in a time-dependent manner. Moreover, this response is stronger in people with type 2 diabetes, with a sustained increase in an inflammatory signature.
Figure 2.
Recovery is associated with increased immune cell signature in adipose tissue from subjects with T2D
(A) Estimation of macrophages populations M0, M1, and M2 proportions using CIBERSORTx analysis.
(B) Quantification of Cd11b+ cells per area.
(C) Quantification of adipocyte cells per area.
(D) Ratio of Cd11b+ cells per adipocyte. Each dot represents an image, n = 3 subjects per group. Two-way ANOVA; # represents the main effect of exercise, and ¤ represents the main effect of disease. Post-tests in comparison to the pre condition: ∗∗∗∗p < 0,0001; ∗∗∗p < 0.001; ∗p < 0,05.
(E) Representative images for each condition. Hoechst staining shows nuclei. Lectin staining shows the cells structure. The scale represents 50 μm.
Exercise induces the expression of genes encoding for secreted proteins in adipose tissue
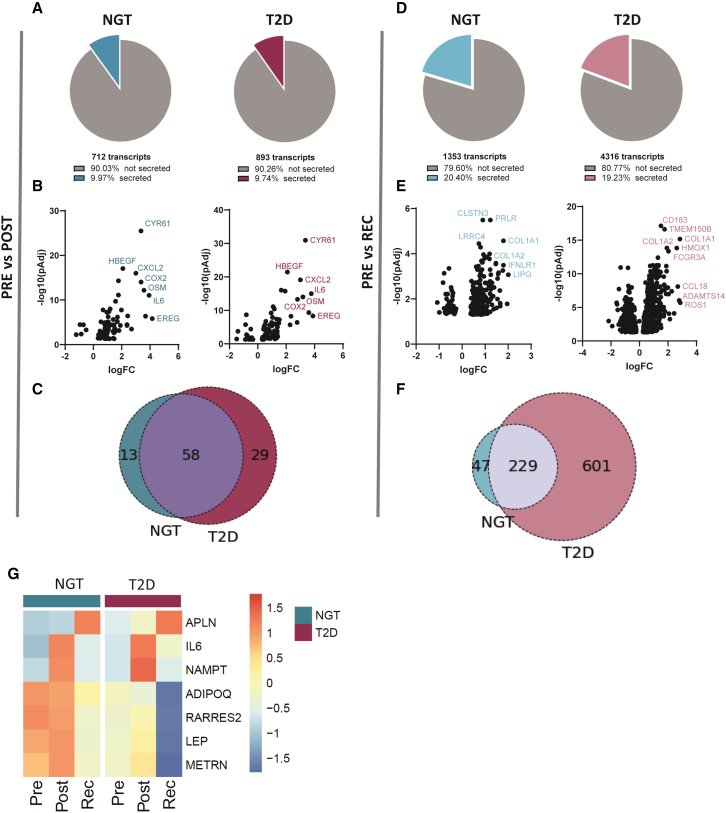
To study the effects of exercise on the adipose tissue secretome, we predicted secreted proteins encoded by differentially expressed genes in a similar manner to previous work.10,13 In the post-exercise condition, 71 genes, representing ∼10% of the total genes altered by exercise, were annotated as encoding for secreted proteins in people with normal glucose tolerance (Figure 3A). A similar profile was seen in adipose tissue from people with type 2 diabetes, with ∼10% of the altered genes encoding for secreted proteins. The most highly induced genes were associated with inflammation and tissue repair, including CYR61, CXCL2, HBEGF, IL6, and COX2 (Figure 3B), consistent with the pathway analysis (Figure 1). The majority of exercise-regulated transcripts at the post- time point were common in people with normal glucose tolerance or type 2 diabetes (Figure 3C). In the recovery state, the proportion of genes encoding for secreted proteins doubled, reaching 20% of the altered transcripts in both groups independent of glycemic status (Figure 3D). Most of these genes (normal glucose tolerance: 84%; type 2 diabetes: 66%) were upregulated, with the largest fold change noted for genes associated with extracellular matrix remodeling (Figure 3E). In contrast to the large proportion of shared genes responding in both groups immediately post-exercise (Figure 3C), 72% of the genes altered in recovery in type 2 diabetes were specific to this group (Figure 3F).
Figure 3.
Exercise induces the expression of genes encoding for secreted proteins, with a stronger effect in recovery
(A) Percentage of the genes encoding for secreted proteins from the total number of genes significantly altered in individuals with either NGT (blue) or T2D (purple) between the post- and pre-exercise conditions.
(B) Volcano plots showing log fold change (logFC) and significance of genes encoding for secreted proteins identified in (A).
(C) Overlay of genes identified in adipose tissue from individuals with NGT or T2D encoding for secreted proteins altered post-exercise.
(D) Percentage of the genes encoding for secreted proteins of the total genes significantly altered in adipose tissue from individuals with NGT (light blue) or T2D (pink) between the pre-exercise and recovery conditions.
(E) Volcano plots showing logFC and significance of genes encoding for secreted proteins identified in (D).
(F) Overlay of genes identified in adipose tissue from individuals with either NGT or T2D encoding for secreted proteins that are altered at recovery.
(G) Heatmap of gene expression profile of selected adipokines identified pre- and post-exercise and recovery in adipose tissue from individuals with either NGT or T2D. The color scale represents mean scale CPM.
Among the significantly altered genes, numerous genes encoded for adipokines, as well as secreted factors previously reported as exercise responsive in skeletal muscle (Figure 3G). The expression of leptin, adiponectin, and chemerin was reduced in individuals with type 2 diabetes, and levels were decreased after recovery in both groups. IL6, encoding an exercise-induced myokine with pro-lipolytic activity in adipose tissue,14 was highly induced in adipose tissue after exercise. NAMPT, also induced by exercise in skeletal muscle,15 followed the same pattern. METRNL (encoding Meteorin-like protein) was increased post-exercise only in individuals with normal glucose tolerance. APLN, encoding the exerkine Apelin,16 was only induced at recovery. Collectively, our results help delineate the temporal modulation of the adipose secretomic profile, highlighting a stronger response after recovery in both groups. We found that several genes encoding secreted factors previously described as exercise-responsive in skeletal muscle were also altered in adipose tissue, suggesting that this dataset represents a valuable resource for the identification of exercise-induced adipokines.
Oncostatin-M is acutely induced by exercise and regulates basal lipolysis in isolated adipocytes
In our search for exercise-induced secreted factors, we interrogated the dataset for putative secreted proteins based on (1) magnitude of the fold change in response to exercise and level of statistical significance, (2) specificity to a diagnosis group or exercise time point, and (3) validation of the secretion of the protein based on the literature. From these criteria, we selected four targets for further validation.
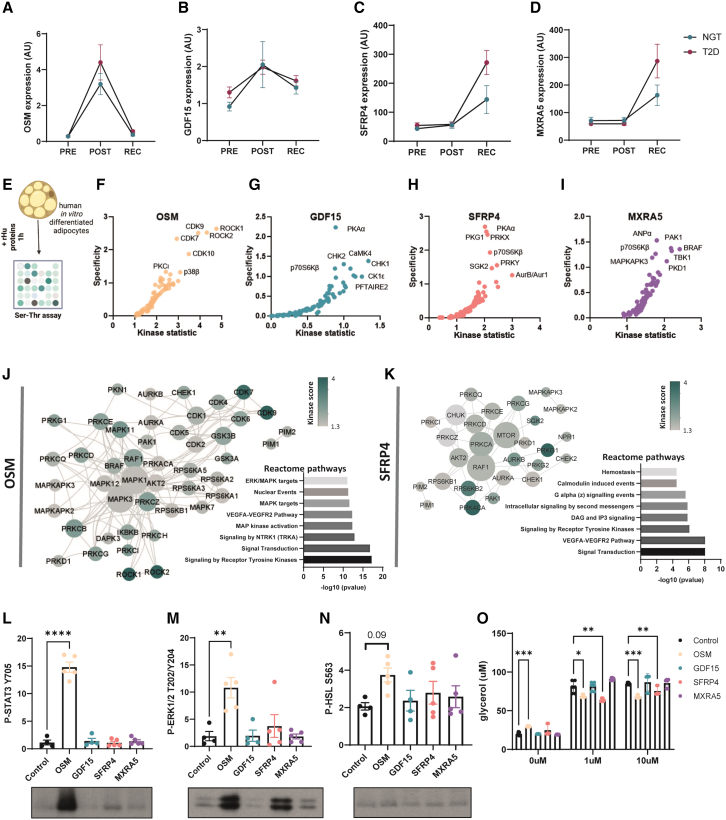
OSM, encoding for oncostatin-M, was one of the most highly induced genes identified in both groups post-exercise with a transient peak followed by restoration to starting levels at recovery (Figure 4A). GDF15, encoding an exercise-training-induced organokine in humans,17 was significantly increased in individuals with normal glucose tolerance only in the post-exercise state, while individual expression analysis revealed that GDF15 levels tended to increase in adipose tissues from both groups at post-exercise and recovery (Figure 4B). SFRP4, a modulator of Wnt signaling,18 was significantly increased at recovery only in individuals with type 2 diabetes (Figure 4C). Similarly, MXRA5, previously identified as a secreted protein in human adipose tissue,19 was significantly induced at recovery in the type 2 diabetes group (Figure 4D). To interrogate the potential molecular pathways involved in the signaling of these candidate proteins, we performed an activity-based serine-threonine kinase profiling analysis of in-vitro-differentiated human adipocytes after exposure to recombinant human oncostatin-M, GDF15, SFRP4, or MXRA5 (Figure 4E). Kinase activity was assessed through computational analysis of the differentially phosphorylated peptide signatures (Figures 4F–4I). Oncostatin-M and SFRP4 induced the strongest response, with higher specificity and statistical scores of predicted kinase activities. To better understand how the activity of these kinases integrates in a signaling pathway, we constructed a protein-protein interaction network using STRING. The oncostatin-M network centered around members of the MAPK family, with a second node of the CDK family (Figure 4J). Accordingly, reactome pathway analysis of differentially active kinases indicated an enrichment for MAPK/ERK signaling. Similar interaction network construction for SFRP4 revealed an enrichment for the PKA/PKC and mTOR signaling network (Figure 4K). To further validate these findings, we analyzed protein phosphorylation by western blot. Recombinant oncostatin-M increased STAT3 and ERK phosphorylation, consistent with an activation of MAPK/ERK signaling (Figures 4L and 4M). Phospho-HSLS563, a surrogate for PKA activation, showed a non-significant increase in response to oncostatin-M and was unchanged in response to the other recombinant proteins (Figure 4N). Interestingly, oncostatin-M increased basal lipolysis (Figure 4O), while in the presence of the beta-adrenergic agonist isoproterenol, both oncostatin-M and SFRP4 suppressed the lipolytic response (Figure 4O). Thus, our data provide evidence that exercise-induced secreted proteins can directly regulate adipocyte metabolism.
Figure 4.
Oncostatin-M is acutely induced by exercise and increases basal lipolysis in human adipocytes
(A–D) Gene expression of candidate secreted factors oncostatin-M, GDF15, SFRP4, and MXRA5 selected for further validation.
(E) Human adipocytes were treated with recombinant proteins for 1 h, and kinase activity was assessed using serine-threonine substrate array (n = 5).
(F–I) Kinase activity in response to oncostatin-M, GDF15, SFRP4, and MXRA5 was predicted by computational analysis of differentially phosphorylated peptide signatures. The kinase statistic parameter reflects the change in kinase activity, whereas the specificity parameter indicates how specific the peptide signature is to a particular kinase.
(J) Protein-protein interaction (PPI) network and reactome pathway analysis of differentially active kinases. Kinase score reflects kinase activation in response to oncostatin-M compared to control cells, and the node degree is a measure of how connected each node is within the network.
(K) PPI network and reactome pathway analysis of differentially active kinases. Kinase score reflects kinase activation in response to SFRP4 compared to control cells, and the node degree is a measure of how connected each node is within the network.
(L–N) Western-blot quantification and representative image of P-STAT3 Y705, P-ERK1/2 T202/Y204, and P-HSL S563 after 1 h incubation with recombinant proteins in isolated human adipocytes (one-way ANOVA) (n = 5).
(O) Lipolysis assay in control, 1 μM, and 10 μM isoprenaline conditions (two-way ANOVA). ∗∗∗∗p < 0,0001; ∗∗∗p < 0.001; ∗∗p < 0,01; ∗p < 0,05.
Error bars show SEM.
Oncostatin-M is released from immune cells in response to exercise-like stimuli
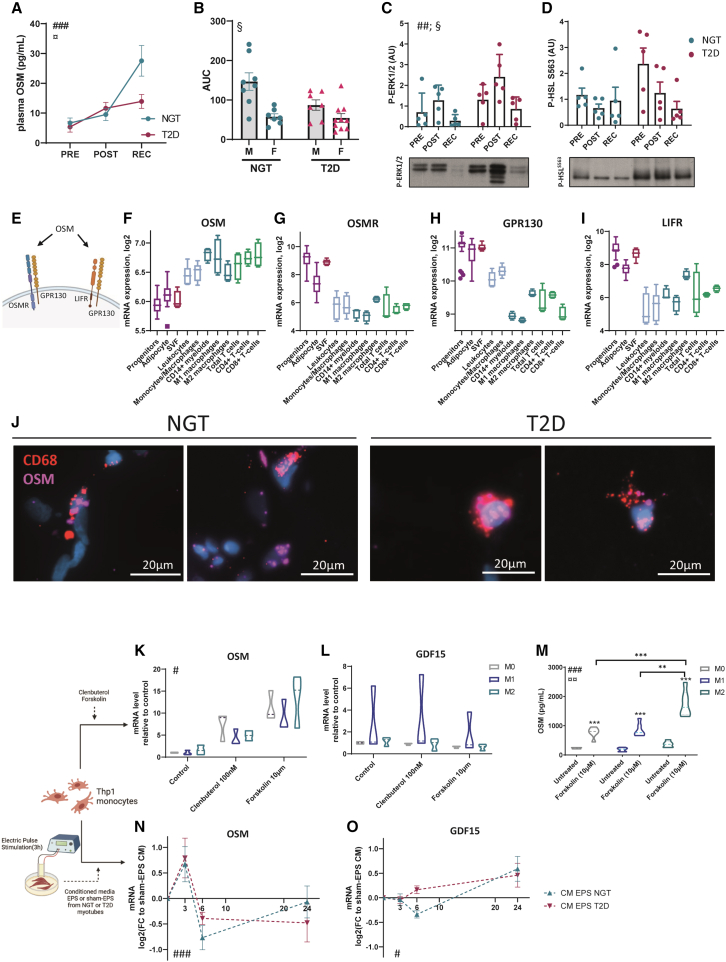
Exercise-induced secreted factors act in a paracrine/autocrine manner and may also be released into the circulation to act on distant organs. Plasma levels of oncostatin-M were increased after exercise in both groups (Figure 5A). In contrast to the decreased oncostatin-M gene expression at recovery, the plasma oncostatin-M level continued to rise, indicating an accumulation of oncostatin-M in the circulation. Plasma oncostatin-M was more potently increased by exercise in men as compared to women, suggesting a sex-specific response (Figures 5B and S3A). To assess whether the nutritional status of participants could result in increased plasma oncostatin-M or some of the changes noted in the adipose tissue transcriptome, we collected adipose tissue biopsies and plasma samples at T = 0 and T = 3 h from a subset of the normal glucose-tolerant subjects (n = 7) before and after the standardized breakfast. Plasma oncostatin-M levels did not change between fasting and 3 h after breakfast (Figure S3B). Oncostatin-M receptor gene (OSMR) expression, as well as some of the top recovery-responsive genes (MKI67, LIPG, COL1A1, and TOP2A), are unchanged 3 h after breakfast intake, indicating that the alteration in the gene expression profile is driven by exercise (Figure S3B).
Figure 5.
OSM expression arises from macrophages in response to exercise-like stimuli
(A) Oncostatin-M level in plasma from individuals with either NGT or T2D (n = 15 and 17 per group). #Time effect in both groups (two-way ANOVA test), ###p < 0.001. ¤Interaction between time and diagnosis group, ¤p < 0.05.
(B) Area under the curve of plasma oncostatin-M after separation between men and women and NGT and T2D (n = 8 per subgroup). §Sex effect (two-way ANOVA on log-transformed values), §p < 0.05.
(C and D) Western blot quantification and representative image of P-ERK1/2 and P-HSL in adipose tissue biopsies (n = 5 per group). #Time effect in both groups (two-way ANOVA test), ##p < 0.01. §Condition effect (two-way ANOVA test), §p < 0.05.
(E) Schematic of OSM binding to type I and type II receptors in humans.
(F–I) Expression of oncostatin-M and oncostatin-M receptor subunits genes OSMR, LIFR, and GPR130 in cells derived from adipose tissue after fractionation. Kruskal-Wallis testing shows significant differences between cell populations for all 4 genes (p < 0.001).
(J) Spatial detection of OSM and macrophage markers CD68 mRNAs by fluorescent in situ hybridization in frozen section of adipose tissue of subjects with NGT and T2D (n = 4 and 3).
(K and L) Thp1 macrophages were treated with 10 μM forskolin or 100 nM clenbuterol for 3 h, and expression of oncostatin-M and GDF15 was measured. #Treatment effect (two-way ANOVA, log-transformed values), #p < 0.05. n = 3 experiments.
(M) Oncostatin M measurement in the conditioned media in response to 10 μM forskolin (two-way ANOVA, log-transformed values). #Treatment effect, ###p < 0.001; ¤polarization effect, ¤¤p < 0.01; ∗Sidak’s post-tests.
(N and O) Thp1 macrophages were exposed to conditioned media from 3 h electric-pulse-stimulated (EPS) or 3 h sham EPS human myotubes derived from individuals with either NGT or T2D (n = 5 and 6) for 0, 3, 6, or 24 h, and oncostatin-M and GDF15 gene expressions were measured and presented normalized to expression in response to sham EPS. #Time effect (two-way ANOVA test), ###p < 0.001, #p < 0.05.
Error bars show SEM.
As specific adipose tissue depots could respond differently due to their direct proximity (or not) to the exercising muscle, we have investigated the regulation of OSM expression by exercise in a second cohort of fasted healthy men from a previous study.20 In this material, OSM expression increased post-exercise in subcutaneous abdominal adipose tissue, suggesting that other adipose depots can contribute to the plasma level of oncostatin-M (Figure S3C). In contrast, by analyzing mRNA data from skeletal muscle biopsies obtained from men performing a similar bout of exercise,21 it was found that OSM expression was below the detection level (with 98% of the samples <5 counts), while OSMR expression increased in response to exercise (Figure S3D). Markers of oncostatin-M signaling were assessed in adipose tissue from a subset of the participants (n = 5 per group). Phospho-ERK abundance was altered by both exercise and diagnosis, with an increase noted post-exercise. (Figure 5C). Phospho-HSL was unaffected by exercise (Figure 5D). While the plasma oncostatin-M level was increased, the exact source of oncostatin-M was not identified. Indeed, our transcriptomic analysis was performed on whole adipose tissue, raising the question of which cell type is responsible for the increased oncostatin-M expression. Bioinformatic analysis of publicly available data from microarray transcriptomes of human adipose tissue after fractionation and fluorescence activated cell sorting (FACS)22,23 revealed that oncostatin-M is primarily expressed in immune cells, such as monocyte/macrophages and T cells (Figure 5F). In contrast, oncostatin-M receptors, OSMR, IL6ST, and LIFR (Figure 5E), were specifically enriched in adipocyte and adipocyte precursors, as well as in the stromal vascular fraction (Figures 5G–5I). Furthermore, spatial detection of OSM mRNA by in situ hybridization in adipose tissue biopsies showed that OSM expression is localized in non-adipocyte cells and is partly co-expressed with CD68, a marker of monocytes and macrophages (Figure 5J), suggesting a potential expression in both monocyte and non-monocyte lineage immune cells. Combined with the increased signature of immune cells during exercise (Figure 2), collectively, our findings suggest that immune cells are the main source of oncostatin-M in response to exercise. Thus, oncostatin-M is secreted within the tissue and can act locally on adipocytes.
To investigate the potential role for intertissue communication, we studied the regulation of oncostatin-M expression using Thp1 monocytes, treated with drugs mimicking the adrenergic activation present during exercise. These drugs included the beta-adrenergic agonist clenbuterol and forskolin, an adenylyl cyclase inhibitor that increases cellular cAMP levels. OSM expression was induced in both M1 and M2 macrophage subsets in response to clenbuterol or forskolin (Figure 5K). As a control, GDF15 expression was unchanged in response to these treatments (Figure 5L). Accordingly, forskolin increased oncostatin-M protein secretion in the media, which was induced at a larger degree in M2- as compared to M1-type macrophages. (Figure 5M). Furthermore, interleukin-6 (IL-6) and Leukemia inhibitory factor (LIF) expression, but not ciliary neurotrophic factor (CNTF), was increased by exercise and exercise -mimetics in Thp1 cells (data not shown). In contrast, forskolin treatment in human adipocytes did not induce OSM expression but did increase the expression of the receptor subunits LIFR and OSMR, suggesting a heightened sensitivity to OSM in adipocytes after exercise (Figures S4D–S4G). To further probe for a role of a skeletal muscle-to-adipose crosstalk, similar expression analysis of Thp1 cells was performed after exposure to conditioned media from in-vitro-“exercised” or control cultured skeletal muscle cells, using electric pulse stimulation to mimic exercise. This revealed a time-dependent transient increase in OSM expression in response to exercised media (as compared to control media) (Figure 5N). GDF15 expression displayed a different response to conditioned media from “exercised” cultured skeletal muscle cells, with an increase after 24 h exposure (Figure 5O). Thus, our results suggest that immune cells may be an important intermediary cell type orchestrating crosstalk between skeletal muscle and adipose tissue during exercise through the secretion of factors such as oncostatin-M.
Discussion
A role for the adipose tissue in the whole-body adaptive response to exercise has been recently brought to light by the discovery of exercise-induced adipokines.10,24,25,26 Our transcriptomic analysis revealed both temporal and type 2 diabetes-related changes in the transcriptomic profile of leg subcutaneous adipose tissue in response to acute exercise. Notably, we observed an activation of inflammatory pathways immediately after exercise and a distinct profile during recovery in individuals with type 2 diabetes, which was characterized by sustained alteration in pathways related to inflammation and metabolism. Upon a deeper investigation of putative secreted factors, we identified oncostatin-M as an exerkine within adipose tissue. This increase in oncostatin-M likely originates from immune cells in a response to exercise-induced secreted factors. Moreover, we provide evidence that oncostatin-M directly regulates lipolysis in human adipocyte cell culture, suggesting a role for immune-cell-derived factors in mediating the skeletal muscle-adipocyte crosstalk during exercise.
Exercise is a first-line therapy in the prevention and treatment of metabolic disturbances associated with type 2 diabetes due in part to the beneficial effect on glycemic control and insulin sensitivity.27 Here, we provide evidence that acute exercise provokes an inflammatory signature in adipose tissue in people with type 2 diabetes, with elevated markers of both M1- and M2-like predicted cells remaining upregulated at recovery. Exercise is a potent stimulus for the recruitment of immune cells in the circulation, with increased neutrophil and lymphocyte populations28 and lasting effects on immunity.29,30 Accordingly, we observed an infiltration of CD11b+ immune cells in adipose tissue at recovery, which was higher in biopsies from people with type 2 diabetes. The sustained inflammatory profile in adipose tissue from people with type 2 diabetes may arise from a differential priming of the immune system, associated with chronic low-grade inflammation.31 Alterations in the metabolic status, such as hypo- and hyperglycaemia, impair immune cell function and promote inflammatory responses.32 Thus, an increased glucose excursion during acute exercise in individuals with type 2 diabetes may also contribute to the sustained inflammation signature in adipose tissue. Interestingly, pathways related to substrate metabolism are also altered during recovery from exercise in individuals with type 2 diabetes. While it is difficult to decipher whether this arises from a modification of the cellular composition of adipose tissue, our results are consistent with decreased mitochondrial and fatty acid metabolism pathways in basal state33 (Data S1). Exercise training reduces adipose tissue inflammation both in humans and in mouse models.34,35,36 Whether the sustained inflammation reported here in individuals with type 2 diabetes after acute exercise is transient and/or detrimental to adipose tissue metabolism remains to be determined. Nevertheless, our results suggest that exercise duration and modalities may need to be fine-tuned37 in order to achieve an optimal exercise range to confer the greatest benefits on insulin sensitivity and metabolism for people with type 2 diabetes.
Several exercise-induced factors regulate adipose tissue metabolism and contribute to the adaptive response to training,38,39 although to date most studies have focused on skeletal muscle for the discovery of new exerkines.40 Here, we provide evidence that cytokines and chemokines, such as oncostatin-M studied herein, are among the most upregulated genes in adipose tissue after acute exercise, while during recovery, a large array of secreted factors showed altered gene expression. Of note, several classical adipokines displayed decreased expression during recovery in adipose tissue from people with type 2 diabetes, although this may be a consequence of a “dilution” of the adipocyte signature due to an increased immune cell population within the tissue, as the plasma level of adiponectin or leptin was unaffected by exercise. In contrast, oncostatin-M expression significantly correlates with HOMA-IR index and BMI at recovery, suggesting a link between the immune cell signature and oncostatin-M expression (Figure S4H). Interestingly, the candidates induced in recovery that we selected for further validation, SFRP4 and MXRA5, most likely arise from the adipocyte fraction (Figures S4A–S4C), suggesting a time- and cell-dependent modulation of the adipose tissue secretome in response to exercise. In skeletal muscle, immune cell-myotube crosstalk has been postulated to play a role in skeletal muscle remodeling and repair after strenuous exercise.41 In a recent analysis, we identified CXCL12 as an exercise-induced cytokine that regulate myotube proliferation and glucose metabolism.21 Of note, while oncostatin-M expression in exercising skeletal muscle was below transcriptomic detection level, the expression of OSMR in skeletal muscle increased at recovery, possibly enhancing the skeletal muscle tissue sensitivity to oncostatin-M.
While immune cells may regulate adipose tissue metabolism in obesity42 or with thermogenesis,43,44 the role of immune cells in adipose tissue after acute exercise has yet to be fully delineated. Targeted spatial transcriptomics in adipose tissue detected oncostatin-M expression in immune cells, both from a monocyte lineage and in non-monocytes cells, suggesting that other cells such as lymphocytes could also contribute to the increase in OSM expression. Accordingly, we found that oncostatin-M expression is induced in cultured macrophages in response to adrenergic signaling, as well as conditioned media from “exercised” myotubes, while the receptors were localized predominantly on the adipocyte fraction and increased in vitro by adrenergic stimulation. Collectively, these results suggest that oncostatin-M acutely contributes to tissue crosstalk between immune cells and adipose tissue in response to exercise. Thus, circulating factors secreted during exercise, possibly arising from the working skeletal muscle, activate immune cell populations within adipose tissue and, through modulation of an immunometabolic signature, may contribute to whole-body systemic adaptations.45 Indeed, we provide evidence that oncostatin-M expression was increased in abdominal subcutaneous adipose tissue after an acute exercise bout in a cohort of young men, indicating that the adipose tissue response is likely responding to a circulatory exercise-induced factor and that both subcutaneous depots could contribute to the plasma oncostatin-M level. Interestingly, in mice, oncostatin-M has been reported to increase after exercise in skeletal muscle and has been implicated in the recruitment of M2 macrophages,46,47 while oncostatin-M expression was not found to be regulated by exercise in adipose tissue,48 underlining the species specificity of exercise-induced adaptations. Additionally, plasma oncostatin-M levels were lower in women than men throughout the exercise intervention, highlighting a sex-specific regulation.
The increased oncostatin-M expression and secretion immediately after exercise may alter the lipolytic response of adipocytes. Indeed, the rise of plasma oncostatin-M levels during exercise and recovery is consistent with the activation of lipolysis post-exercise, where fatty acids are used as the major fuel source.49 Oncostatin-M is part of the gp130 cytokine family, involved in the regulation of a plethora of metabolic process, such as hematopoiesis, inflammation, proliferation, and neuronal survival.50 Oncostatin-M signals through binding to a receptor complex composed of GPR130 (IL6ST)-oncostatin-M receptor or -LIF receptor and activates JAK/STAT and MAPK pathways.51 Other members of the GP130 cytokine family, such as IL-6 or CNTF, regulate lipid and energy metabolism.52,53 In our study, IL6 and LIF expression was also increased by exercise, suggesting that several members of the IL-6 family may be regulated and act in a similar fashion. Several lines of evidence implicate oncostatin-M in obesity and energy homeostasis. In obesity, oncostatin-M is increased, whereas knockout for either the oncostatin-M receptor or GP130 increases inflammation and browning of white adipose tissue.54,55 Conversely, treatment of high-fat-diet-induced obese mice with oncostatin-M reduces body fat and the adipose tissue inflammatory profile.56 This apparent divergent response to oncostatin-M signaling may be determined by the receptor complex involved and/or by an acute or chronic pathological upregulation of oncostatin-M. Either of these scenarios could affect oncostatin-M action on processes involved in adipose tissue adaptation and remodeling. While we cannot decipher whether the acute exercise-induced alterations in oncostatin-M affect metabolic homeostasis over time, comparisons can be made with other exerkines. For example, a recent study provided evidence that exercise-associated loss of visceral adipose tissue mass is associated with IL-6 signaling.57 While IL-6 is a mediator of obesity-associated low-grade inflammation in adipose tissue, injection of IL-6 receptor blockers inhibits the beneficial effect of exercise on body fat composition in obese individuals.57 Our data add to the growing understanding that inflammatory mechanisms contribute to healthy tissue remodeling. Indeed, pro-inflammatory signaling in white adipose tissue is required for metabolic health and adipose tissue remodeling and expansion.58 Thus, oncostatin-M could have a similar mode of action and contribute to a beneficial regulation of lipid metabolism during exercise.
Collectively, our study highlights a role of immune cells activation/infiltration in the response to acute exercise in adipose tissue. We also unravel a potential role of immune-cell-derived cytokines in the skeletal muscle-to-adipose crosstalk during exercise. Further studies are warranted to delineate the role of inflammation in the adaptation to exercise, as well as to define the most optimal exercise modalities to achieve improvements in insulin sensitivity and metabolism, considering the metabolic status and the sex of the participant.
Limitations of the study
It is important to acknowledge a few limitations of the present study. These include the fact that the way in which the RNA sequencing was performed precludes the determination of transcriptomic sex differences. Furthermore, while food intake does not appear to account for the exercise-induced inflammatory responses in adipose tissue or the increased plasma OSM level, we cannot exclude the possibility that the response is not different in subjects with type 2 diabetes. Our analysis focused on leg adipose tissue, and while the exercise-induced OSM response seems shared to both abdominal and leg fat, further studies would be needed to decipher a depot-dependent response to exercise. Finally, based on our sequencing analysis, our in vitro assays focused on the role of macrophages in OSM secretion; nevertheless, the role of other immune cell types such as lymphocytes in OSM secretion and crosstalk during exercise would require further investigation.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) 1:1000 | Cell Signaling Technology | #9101; RRID:AB_331646 |
| Phospho-HSL (Ser563) 1:1000 | Cell Signaling Technology | #4139; RRID:AB_2135495 |
| Phospho-Stat3 (Tyr705) (D3A7) XP® 1:1000 | Cell Signaling Technology | #9145; RRID:AB_2491009 |
| Itgam 1:500 | Atlas Antibodies | AMAb90911; RRID:AB_2665722 |
| Biological samples | ||
| Primary human myoblasts | Previous works from the group | N/A |
| Human in vitro differentiated adipocytes | Gao et al., 201759 | N/A |
| Human subcutaneous leg fat biopsies | This paper | N/A |
| Human plasma samples | This paper | N/A |
| Human subcutaneous abdominal fat biopsies | Fabre et al.20 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Oncostatin-M | Bio-Techne LTD | 295-OM-050 |
| GDF15 | Bio-Techne LTD | 9279-GD-050 |
| SFRP4 | Bio-Techne LTD | 1827-SF-025/CF |
| MRXA5 | Nordic Biosite | LS-G26239-10 |
| Isoproterenol | Merck | I5627 |
| Clenbuterol | Merck | C5423-10MG |
| Forskolin | Merck | F6886-10MG |
| IBMX | Merck | I5879-250MG |
| Insulin | Novo Nordisk | Actrapid |
| Dexamethasone | Merck | D4902-1G |
| Rosiglitazone | GlaxoSmithKline | BRL49653-C |
| apo-transferrin | BBI solution | T100-5 |
| Recombinant Human IL4 | PeProTech | 200–04 |
| Recombinant Human IL13 | PeProTech | 200–13 |
| Recombinant Human IFNg | PeProTech | 300–02 |
| Phorbol 12-myristate 13-acetate (PMA) | Merck/Sigma-Aldrich | P1585 |
| Lipopolysaccharides | Merck/Sigma-Aldrich | L9641 |
| Hoechst 33342 | Abcam | Ab228551 |
| Lectin | Vector Laboratories | DL-1048-1 |
| TRIzol | Thermo Fisher Scientific | 15596018 |
| Triton X-100 | Sigma-Aldrich | X100-100ML |
| Tween 20 | Sigma-Aldrich | P1379-1L |
| DAKO Fluorescence mounting media | Agilent Technologies | S302380-2 |
| Protein Block | Agilent Technologies | X090930-2 |
| Protease Inhibitor Cocktail | CALBIOCHEM | 539131–10 |
| ECL reagent | GE Healthcare | RPN2106 |
| Critical commercial assays | ||
| TruSeq Stranded Total RNA with Ribo-Zero Gold | Illumina | 20020598 |
| Rneasy lipid tissue kit | Qiagen | Cat#74804 |
| Agilent RNA 6000 nano kit | Agilent | 5067–1511 |
| Qubit dsDNA HS assay kit | Invitrogen | Q32851 |
| Agilent High Sensitivity DNA chip | Agilent | 5067–4626 |
| Human Oncostatin M/OSM ELISA Kit | Abcam | ab215543 |
| EZNA total RNA kit I | Omega Bio-tek | R6834-01 |
| Pierce BCA Protein Assay Kit | ThermoFisher | Cat#23227 |
| Multiplex Fluorescent detection reagents v2 | ACDBio | 323110 |
| Free glycerol Reagent | Sigma-Aldrich | #F6428 |
| Amplex Ultrared Reagent | Invitrogen | #A36006 |
| Deposited data | ||
| Human adipose tissue in response to exercise (RNAseq) | This paper | GEO: GSE198922 |
| Experimental models: Cell lines | ||
| Thp1 monocytes cell line | ATCC | TIB-202 |
| Oligonucleotides | ||
| TaqMan probe human OSM | Applied Biosystems | Hs00171165_m1 |
| TaqMan probe human GDF15 | Applied Biosystems | Hs00171132_m1 |
| TaqMan probe human LIF | Applied Biosystems | Hs01055668_m1 |
| TaqMan probe human OSMR | Applied Biosystems | Hs00384276_m1 |
| TaqMan probe human LIFR | Applied Biosystems | Hs01123581_m1 |
| TaqMan probe human IL6ST | Applied Biosystems | Hs00174360_m1 |
| TaqMan probe human IL6 | Applied Biosystems | Hs00174131_m1 |
| TaqMan probe human CXCL2 | Applied Biosystems | Hs00601975_m1 |
| RNAscope™ Probe- Hs-OSM | ACDBio | 456381 |
| RNAscope™ Probe- Hs-CD68-C2 | ACDBio | 560591-C2 |
| Software and algorithms | ||
| GraphPad Prism 9.0 | GraphPad Software | N/A |
| R v4.3.0 | R | N/A |
| Zen Pro | Zeiss | N/A |
| Quantity One 1-D Analysis Software | Bio-Rad | N/A |
| Python | Python | https://www.python.org/ |
| Zeiss Zen Pro Software | Zeiss | N/A |
| Standard inverted multichannel Carl Zeiss Axio Obeserver Micropscope (Leica camera 20x objective) | Zeiss | N/A |
| Fiji | https://fiji.sc | |
| QuPath Image Processing Software | QuPath | https://qupath.github.io/ |
| QuantStudio 7 Real-Time PCR | Thermo Fisher Scientific | N/A |
| BioNavigator® | PamGene | N/A |
| Cytoscape | Cytoscape Team | https://cytoscape.org/ |
| NIS Elements Software | Nikon | N/A |
| Nikon inverted TiE microscope | Nikon | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Anna Krook (anna.krook@ki.se).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
(1)
The RNAsequencing source data have been deposited to the Gene Expression Omnibus repository and is publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
(2)
This paper does not report original code.
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participants details
Human intervention
The study was performed according to the Declaration of Helsinki and all participants provided informed consent. The Stockholm regional ethical committee approved the protocols. The study was performed in Stockholm and included a total of 11 men and nine women with normal glucose tolerance (NGT) and 18 men and nine women with type 2 diabetes (T2D). Exclusion criteria were blood pressure >160/95 mmHg, physical impairment, cardiovascular disease, smoking status and insulin treatment. Metformin treatment was not an exclusion criterion. Clinical characteristics of the study participants are included in Table 1. For the basal measurement, participants reported to the laboratory after an overnight fast and blood sampling and subcutaneous leg adipose biopsies from lateral thigh (“pre”) were obtained. An oral glucose tolerance test was performed, and blood glucose measurements were obtained over 2 h. Five to ten days after the initial study visit, the participants consumed a standardised breakfast consisting of a glass of milk and a sandwich with cheese and subsequently performed an acute bout of exercise on a cycle ergometer (Rodby). The workload was set to 85% maximal heart rate and the exercise bout was maintained for 30 min. Immediately after the exercise bout, additional blood samples and adipose tissue biopsies (“post”) were taken. The participants were then advised to rest for 3 h, with no food intake, before additional blood samples and a fat biopsy was taken (“recovery”). The participants were instructed to refrain from medications 24 h prior to the visit. For the breakfast study, a subset of participants with normal glucose tolerance (n = 9) were invited back to the laboratory on a separate occasion, and plasma (n = 9) and an adipose tissue biopsy (n = 7) was taken in fasting state, then the participants ate a similar breakfast as in the exercise study. A second biopsy was collected 3h30 after the breakfast. RNA was subsequently extracted using RNeasy kit (Qiagen), and gene expression was analyzed using the ΔΔCt method using an average of two housekeeping genes. RNA from abdominal adipose tissue of men after exercise (n = 15) were obtained from the study of Fabre et al.,20 and gene expression performed as described above. Skeletal muscle (vastus lateralis) expression level of OSM and OSMR from men with either NGT (n = 17) or T2D (n = 20) were extracted from the dataset GEO: GSE202295.21
In vitro studies
Human adipocytes
Cultures of in vitro differentiated human adipocytes were established and subjected to differentiation protocols as described.60 Cells were isolated from abdominal subcutaneous white adipose tissue of one male donor. The adipose tissue was incubated in type II collagenase (Sigma-Aldrich) followed by isolation of the stroma vascular fraction (SVF), which includes the adipocyte progenitors. The cells were washed and subsequently expanded for 6–8 passages in proliferation medium (DMEM, 10 mmol/L HEPES, 10% FBS, 50 μg/mL Penicillin-Streptomycin) supplemented with 2.5 ng/mL Fibroblast growth factor 2 (FGF2) (Sigma-Aldrich, St. Louis, MO). Adipogenesis was induced two days post confluence using DMEM/F12 or DMEM without glucose/FGF2 containing 0–20 mM of glutamine and 1–50 mM glucose, as well as the adipogenic cocktail; 5 mg/mL insulin, 0.25 mmol/L dexamethasone, 0.5 mmol/L 3-isobutyl-1-methylxanthine (IBMX) and 10 mmol/L rosiglitazone for two days after which the dexamethasone and IBMX were removed and the cells were allowed to undergo full adipogenic differentiation. The day of the experiments, cells were kept in DMEM/F12 medium without additives for 2 h, then incubated in presence of commercial recombinant human proteins (30–100 nM): Oncostatin-M (295-OM-050, Bio-Techne LTD), GDF15 (9279-GD-050, Bio-Techne LTD), SFRP4 (1827-SF-025/CF, Bio-Techne LTD) and MRXA5 (LS-G26239-10, Nordic Biosite). One hour after incubation, the cells were rinsed once in ice-cold PBS and thereafter directly were frozen in liquid nitrogen and stored at −80°C before protein extraction. For the lipolysis assay, the cells were incubated in phenol red free medium containing 2% Bovine Serum Albumin, supplemented with the recombinant proteins, in the absence or presence of isoproterenol (1 μM or 10 μM). The cells were incubated for 3 h, and the glycerol content in the medium was assessed using Free glycerol Reagent (#F6428, Sigma-Aldrich) and Amplex Ultrared Reagent (#A36006, Invitrogen). After 15 min incubation fluorescence was measured at Ex/Em 530/590 nm. For the RNA analysis, the cells were exposed for 3h to either 10μM forskolin or 1μM isoproterenol. RNA was extracted using the RNeasy Lipid Tissue Mini Kit (Qiagen), and cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit (ThermoFisher). Quantitative Polymerase Chain Reaction was performed using TaqMan Fast Universal PCR Master Mix (ThermoFisher).
Human primary muscle cells
Primary muscle cells were isolated from vastus lateralis biopsies derived from male donors (Age: 50 ± 10 years; BMI: 22.9 ± 2.4 kg/m2) with or without type 2 diabetes (n = 5; 6). Myoblasts were proliferated in growth medium (F12/DMEM, 25 mM glucose, 20% FBS, 1% penicillin-streptomycin [Anti-anti, Thermo Fisher Scientific, Stockholm, Sweden]). Differentiation was induced in DMEM/M199 media containing HEPES (0.02 M; Invitrogen), zinc sulfate (0.03 μg/mL), vitamin B12 (1.4 μg/mL; Sigma-Aldrich), insulin (10 μg/mL; Actrapid; Novo Nordisk), apo-transferrin (100 μg/mL; BBI Solutions), and 0.5% FBS and 1% penicillin-streptomycin. Myotubes were exposed to EPS, using the C-Pace EP Culture Pacer (IonOptix, MA). Cells were pulsed at 40 V, 1 Hz, 2 ms pulse duration for 3 h.61 Conditioned medium was collected immediately after EPS or sham-EPS, subjected to centrifugation (10,000 g, 15 min), filtered (3 μm) and frozen at −80°C before use.
THP1 monocytic cell line
THP1 human monocytes (ATCC Manassas, VA) were grown in RPMI 1640 containing 5% FBS and supplemented with 100 U/ml penicillin, 100 μg/mL streptomycin, and 250 ng/mL amphotericin B. Differentiation was induced by diluting cells to 2.106 cells/mL in medium containing 100 ng/mL phorbol 12-myristate 13-acetate. After 24h, cells were washed with PBS and allowed to stabilise for an additional 24h in regular growth medium before experiments. For the conditioned medium (CM) experiment, CM from EPS-stimulated or sham-EPS myotubes was added to differentiated macrophages for a time-course (0, 3, 6 or 24h) and RNA was extracted for analysis of gene expression. Macrophages were exposed for 24h to 20 ng/mL IL-4/IL-13 to induce M2 polarization or 20 ng/mL IFN-g and 10 pg/mL LPS to promote M1 polarization. Cells were then incubated in the presence of either Clenbuterol (100 nM) or Forskolin (10 μM) for 3h, and RNA was subsequently extracted using a commercial kit (EZNA total RNA kit I, Omega Bio-tek, GA). Gene expression was analyzed using the ΔΔCt method using an average of two housekeeping genes. Conditioned-media oncostatin-M levels were assessed using a commercial kit (ab215543) (Abcam, Cambridge, UK).
Method details
Library preparation and RNA sequencing
Approximately 30 mg of adipose tissue was used for RNA extraction. Tissues were homogenized using TissueLyser (Qiagen) in 1 mL TRIzol (Invitrogen). Chloroform was added, samples were mixed, subjected to centrifugation, and the aqueous phase was transferred to a new tube. Thereafter, 1:1 70% ethanol was added, and the total volume was loaded on silica-membrane columns. RNA extraction was performed using a commercially available kit (Rneasy lipid tissue kit, Qiagen). RNA concentration and purity were assessed by absorbance at 260 and 280 nm using NanoDrop One (Thermo Fisher Scientific, Waltham, MA). RNA was checked for quality using the Agilent RNA 600 nano kit and Bioanalyser instrument (Agilent Technologies, Santa Clara, CA). Aliquots of RNA (300 ng) were analyzed using the Illumina TruSeq Stranded Total RNA with Ribo-Zero Gold protocol (Illumina) as described.62 Samples were cleaned and validated for DNA concentration using the Qubit dsDNA HS assay kit (Invitrogen) and for base pair size and purity using the Agilent High Sensitivity DNA chip and Bioanalyzer instrument. The libraries were subjected to 38-bp paired-end sequencing on a NextSeq500 (Illumina).
Bioinformatic analysis
Reads were mapped to ENSEMBL hg38 release 98 using STAR (2.7.2b),63 and transcripts counted with FeatureCounts (2.0.2) using gencode release 28.64 Sequencing depth ranged from 61.1 to 13.4 million with a mean of 31.3 million reads. Transcriptomic data is deposited under accession number GEO: GSE198922. Reads were filtered using filterbyExpression, with a minimum count of 15 and design matrix modeling sex and group. Differential gene expression analysis was performed using ∼0 + group + sex, quality weights and duplicate correlation blocking for participants, an FDR cut-off of 0.05 and a fold change cut-off of 0.1. Enrichment analysis was performed using clusterProfiler,65 using the hypergeometric enrichment method as implemented in enrichGO, and molecular function GO terms, with all detected genes serving as background. GO was considered significant if FDR<0.05 (Figure S1). Graphs presented in Figure 1E visualise the largest graph clusters from the GO hierarchy. Cell deconvolution analysis was performed using CIBERSORTx12 in batch B, 1000 permutations and absolute mode with the L1 signature. Differences in immune cell infiltration was performed only on Macrophage M0, M1, and M2 cell types using log10 transformed data and hierarchical mixed model analysis66 using the formula counts ∼ diagnosis + exercise + (1 | participant) with no assumption violations as detected by DHARMa.67
The Perseus bioinformatics platform68 was used for predicting secreted proteins. Genes were first matched against the Homo Sapiens UniProt FASTA database, containing 20,591 gene entries (version 2019, downloaded October 16, 2020). Gene names were matched to their corresponding UniProt identifiers, and proteins that either contained a signal peptide (UniProt keyword: signal) or were annotated to the extracellular compartment (Gene Ontology Cellular Compartment: extracellular matrix, region, space) were predicted as high-confidence secreted proteins.
Plasma analysis
Plasma oncostatin-M levels were assessed in a subset of men and women matched for BMI and age (n = 15 NGT, age = 60.3 ± 4.7, BMI = 27.1 ± 3.0; 17 T2D, age = 60.9 ± 4.1, BMI = 27.8 ± 2.9) using a commercial ELISA kit (ab215543, Abcam, Cambridge, UK).
mRNA expression in cells derived from human adipose tissue
To interrogate cell types present in human adipose tissue GEO: GSE8065423 and GEO: GSE10079522 were downloaded from the GEO repository and raw files processed with robust multiarray normalization using the oligo package. Markers used for sorting and identification of the different cell types was created from the GEO metadata: Progenitor (CD45-/CD34+/CD31-), Monocyte/Macrophage (CD45+/CD14+), Leukocyte (CD45+/CD14-), CD14+ Myeloid (CD45+/CD14+/CD206+), Total T-cells (CD45+/CD3+), CD4+ T-cells (CD45+/CD3+/CD4+/CD8-), CD8+ T-cells (CD45+/CD3+/CD4-/CD8+), M1 Macrophage (CD45+/CD14+/CD206+/CD11c+), M2 Macrophage (CD45+/CD14+/CD206+/CD11c-), stroma vascular fraction (pellet obtained after collagenase), Adipocyte (floating cells after collagenase).
Human adipose tissue sections
Analysis was performed on a subset of the full cohort. Participants were invited back to the laboratory on a separate occasion, and an adipose tissue biopsy was taken for immunofluorescence and in situ hybridization staining. Biopsies were stored overnight in 4% formaldehyde in 0.1 M phosphate buffer solution. The following morning, they were placed in 10% sucrose in 0.1 M phosphate buffer solution for 2 to 10 days. After collecting biopsies from all time points, samples from an individual participant were placed in the same optimal cutting solution block and frozen on dry ice. Sections were cut with a thickness of 10 μm.
In situ hybridization in adipose tissue biopsies
All ISH was carried out using Advanced Cell Diagnostics RNAscope Fluorescent Multiplex Kit (Advance Cell Diagnostics, 323110) following the manufacturer’s instructions for fixed frozen tissue. After mounting tissue, slides underwent an ethanol gradient to dehydrate the tissue (5 min in 50% EtOH, 70% EtOH, 99% EtOH, 99%EtOH). Slides were allowed 5 min to dry, and a hydrophobic barrier was drawn using a Dako pen (DAKO, S2002) before proceeding with hydrogen peroxide (provided in the RNAscope kit) for a 10 min at room temperature to block endogenous peroxidases. Then adding Protease Plus (Advance Cell Diagnostics, 322381) at 40°C for 30 min secured in a HybEZ Humidity Control Tray (Advanced Cell Diagnostics, 310012) in a HybEZ II Oven (Advanced Cell Diagnostics, 321720). Slides were then hybridized with two targeted mRNA oligonucleotides probes per slide (OSM and CD68, 56381 and 560591-C2, ACDbio) at 40°C for 2 h. Then probes were sequentially hybridized with Amplification reagents 1–3 (30 min, 30 min, 15 min), incubated with horseradish peroxidase (HRP) which specifically binds to channel 1 (C1) probes at 40˚c for 15 min, and C1-signal was developed by adding Opal 520 fluorophore (1:1000, FL: TSA buffer) at 40˚c for 30 min (PerkinElmer Opal520, Cat. No. FP1487A). All unbound HRP was quenched by HRP-blocker at 40˚c for 15 min. Then the same methods were applied for the C2 probe with Opal 570 fluorophore for C2-signal (1:1000, FL: TSA buffer) (Akoya Biosciences Opal570, Cat. No. OP-001003). Slides were mounted using Antifade Prolong Gold with Dapi and imaged using a standard inverted multichannel Carl Zeiss Axio observer microscope at 20x objective with Leica camera.
Immunohistochemistry staining of adipose tissue biopsies
The slides were washed with 0.3% Triton X-100, 0.05% Tween 20 in PBS by two steps for 5 min at room temperature, then washed with PBS. The slides were then blocked with Protein Block (Agilent Technologies, X090930-2) for 30 min at room temperature. Anti-ITGAM (1:500, Atlas Antibodies, AMAb90911) was diluted in Protein Block and incubated on the sections overnight at 4°C. The sections were then washed three times in PBS containing 0.3% Triton X-100, 0.05% Tween 20 for 10 min each at room temperature. The slides were then incubated with the secondary antibody (donkey α-mouse conjugated with Alexa Flour 488 (1:200, Thermofisher, A-21202) diluted in Protein Block for 1 h at room temperature. After three washes, 100 μL Sudan Black B in 70% ethanol was added to each section and the samples were incubated for 2 min at room temperature. The slides were thereafter rinsed in PBS and incubated with Hoechst 33342 (1:5000, Abcam, Ab228551) and LCA-Dylight647 (1:500, Vector Laboratories, DL-1048-1) diluted in PBS for 20 min to stain nuclei and cell membrane respectively. Prior to mounting in DAKO Fluorescence mounting media (S302380-2, Agilent Technologies), the samples were washed with PBS and swirled in distilled water. Images were acquired using the NIS Elements software, an inverted TiE microscope (Nikon) equipped with a ×1.2 magnification lens and an Andor Zyla 4.2+ camera (pixel size 6.45 mm). A Nikon ×20/0.75 air objective was used to acquire z stack images. Quantification of adipocytes and immune cells was done using the “analyze> cell counter” function in Fiji.
Western blot
Cells or frozen adipose tissue biopsies were homogenized in ice-cold homogenization buffer [20 mM Tris, pH 7.8, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 0.5 mM Na3VO4, 1% Triton X-100, 10% glycerol, 10 mM NaF, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 5 mM Na4P2O7, and 1% (v/v) Protease Inhibitor Cocktail (Calbiochem, Darmstadt, Germany)]. Lysates were subsequently rotated for 1 h at 4°C and subjected to centrifugation at 12,000 g for 10 min at 4°C. The supernatant was then collected, and protein concentration was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Proteins were resolved with SDS-PAGE (4–12% polyacrylamide gels) and transferred to the PVDF membrane with wet electrotransfer. After the transfer, membranes were stained with Ponceau S [0.1% (w/v) in 5% (v/v) acetic acid] to evaluate uniformity of sample loading and transfer. Membranes were incubated with a primary antibody in the primary antibody buffer [20 mM Tris, 150 mM NaCl, pH 7.5, 0.1% (w/v) BSA and 0.1% (w/v) sodium azide] overnight at 4°C and subsequently secondary antibody-horseradish peroxidase conjugate in TBST with 5% (w/v) dry milk for 1 h at room temperature. Membranes were incubated with ECL reagent and then immunolabeled proteins were visualized on the X-ray films. Films were scanned with GS-800 Densitometer (Bio-Rad) and analyzed with Quantity One 1-D Analysis Software (Bio-Rad). Intensities of individual bands were expressed in arbitrary units relative to the total intensity of all the bands.
Kinase activity assay with PamChip
Cell lysates were obtained using buffer containing phosphatase and protease inhibitors. Total lysate (5 μg) was applied per well on the PamChip. For the serine-threonine peptide kinase PamChip (Pamgene), the PamChip was blocked with 2% bovine serum albumin using the standard setting of a PamStation 12 System (Pamgene). The composition of the kinase buffer was as follows: 1× PK buffer, 1× STK antibody mix, 0.5 mg/mL bovine serum albumin, and 400 μM ATP. The kinase buffer was prepared and chilled on ice, and the solution is added to 5 μg ice-cold lysates and immediately applied to the serine-threonine peptide kinase PamChip. The assay conducted on a PamStation 12 System according to standard procedures. After incubation, detection mix (1 × antibody buffer, 5 μg/mL STK antibody FITC-labeled) was prepared and added to the wells. Detection was run on a PamStation 12 System according to standard procedures. The data (spot images) were further quantified and linked to the peptide identities using BioNavigator software. Quantified data were then fitted to a curve using CurveFitHT software using the Vinip2 curve fitting algorithm; the initial velocity was derived at the first kinetic time point read. Each peptide was assigned a score based on difference of the mean signal of the peptide, divided by the estimation of the variation of the peptide, between control and experimental condition. PhosphoNET database from Kinexus was queried to retrieve top kinases for each peptide. Kinase score was calculated as an average of the scores for each associated peptide in each set. Significance scores were calculated by permutation of peptide labels and is determined by the formula Q = −10log[max(m/M,1/M)], where m is the number of times of M permutations where calculated permuted score was higher than the original calculated value without permutation. Protein-protein interaction networks were generated using STRING (string-db.org) and visualized and edited in Cytoscape. Only upstream kinases with a median final score (calculated based on specificity and significance score) higher than 1.3 were used for network generation. The distance of nodes situated on the edges of the network was manually adjusted for space optimization. Pathway analysis was performed using STRING. Overrepresentation scores were plotted as -log10(p value).
Quantification and statistical analysis
Statistical analysis of the RNA sequencing data was performed using R and is detailed in the methods above. Pearson (for parametric values) or Spearman (for non-parametric values) correlation tests were performed using the Pandas Python package. Other statistical tests have been performed using Graphpad Prism software and are detailed in the legends of each graph, as well as the n for each experiment. Normality assumption of the datasets were tested, and appropriate tests were chosen. For low n datasets or non-normally distributed values, non-parametric alternatives of the tests were chosen when available, or for two-way ANOVA, data were log-transformed to improve normality before analysis.
Acknowledgments
L.D. was supported by a Novo Nordisk postdoctoral fellowship run in partnership with Karolinska Institutet and by a Novo Nordisk Foundation postdoctoral fellowship for research abroad (NNF20OC0060969). We acknowledge The Single-Cell Omics platform at the Novo Nordisk Foundation Center for Basic Metabolic Research (CBMR) for the technical expertise and support. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen, partially funded by an unrestricted donation from the Novo Nordisk Foundation (NNF18CC0034900). This work was supported by grants from Knut and Alice Wallenberg Foundation (M.R., J.R.Z., and A.K.); the Swedish Research Council (M.R., J.R.Z., and A.K.); ERC-SyG SPHERES (856404 to M.R.); the Novo Nordisk Foundation (including the MSAM consortium NNF15SA0018346 and the MeRIAD consortium grant number 0064142 to both M.R. and A.K.); CIMED (M.R.); the Swedish Diabetes Foundation (M.R. and J.R.Z.); the Swedish Heart-Lung Foundation (K.C.); the European Foundation for the Study of Diabetes (L.D., N.J.P., and A.K.); the Region Stockholm (ALF project) (M.R., J.R.Z., K.C., and A.K.); and the Strategic Research Program in Diabetes at Karolinska Institutet (M.R., J.R.Z., and A.K.). Figures were created using BioRender.
Author contributions
L.D., N.J.P., J.R.Z., and A.K. conceived and designed experiments. L.D., A.V.C., L.A.P., N.J.P., E.L.L., O.D., M.E., S.F.-C., J.J., and T.d.C.B. performed experiments. L.S.L. performed the bioinformatic analysis. L.D., L.S.L., A.V.C., L.A.P., E.L.L., D.T.C., A.D., M.E., and N.J.P. analyzed data. K.C. managed the human study and samples collection. M.R. and R.B. provided biological materials. M.R., R.B., and H.W.-H. contributed to the scientific discussion. L.D., J.R.Z., and A.K. prepared the figures and wrote the manuscript. All authors read and approved the final version of the manuscript.
Declaration of interests
J.R.Z. serves on the advisory boards for Cell and Cell Metabolism.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: December 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101348.
Contributor Information
Lucile Dollet, Email: lucile.dollet@sund.ku.dk.
Anna Krook, Email: anna.krook@ki.se.
Supplemental information
References
- 1.Egan B., Zierath J.R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Wallberg-Henriksson H., Zierath J.R. Metabolism. Exercise remodels subcutaneous fat tissue and improves metabolism. Nat. Rev. Endocrinol. 2015;11:198–200. doi: 10.1038/nrendo.2015.24. [DOI] [PubMed] [Google Scholar]
- 3.Stanford K.I., Middelbeek R.J.W., Townsend K.L., Lee M.Y., Takahashi H., So K., Hitchcox K.M., Markan K.R., Hellbach K., Hirshman M.F., et al. A novel role for subcutaneous adipose tissue in exercise-induced improvements in glucose homeostasis. Diabetes. 2015;64:2002–2014. doi: 10.2337/db14-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.May F.J., Baer L.A., Lehnig A.C., So K., Chen E.Y., Gao F., Narain N.R., Gushchina L., Rose A., Doseff A.I., et al. Lipidomic Adaptations in White and Brown Adipose Tissue in Response to Exercise Demonstrate Molecular Species-Specific Remodeling. Cell Rep. 2017;18:1558–1572. doi: 10.1016/j.celrep.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arner P., Kriegholm E., Engfeldt P., Bolinder J. Adrenergic regulation of lipolysis in situ at rest and during exercise. J. Clin. Invest. 1990;85:893–898. doi: 10.1172/JCI114516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rönn T., Volkov P., Tornberg A., Elgzyri T., Hansson O., Eriksson K.F., Groop L., Ling C. Extensive changes in the transcriptional profile of human adipose tissue including genes involved in oxidative phosphorylation after a 6-month exercise intervention. Acta Physiol. 2014;211:188–200. doi: 10.1111/apha.12247. [DOI] [PubMed] [Google Scholar]
- 7.Stinkens R., Brouwers B., Jocken J.W., Blaak E.E., Teunissen-Beekman K.F., Hesselink M.K., van Baak M.A., Schrauwen P., Goossens G.H. Exercise training-induced effects on the abdominal subcutaneous adipose tissue phenotype in humans with obesity. J. Appl. Physiol. 2018;125:1585–1593. doi: 10.1152/japplphysiol.00496.2018. [DOI] [PubMed] [Google Scholar]
- 8.Thyfault J.P., Bergouignan A. Exercise and metabolic health: beyond skeletal muscle. Diabetologia. 2020;63:1464–1474. doi: 10.1007/s00125-020-05177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon H., Pessin J.E. Adipokines mediate inflammation and insulin resistance. Front. Endocrinol. 2013;4:71. doi: 10.3389/fendo.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takahashi H., Alves C.R.R., Stanford K.I., Middelbeek R.J.W., Nigro P., Ryan R.E., Xue R., Sakaguchi M., Lynes M.D., So K., et al. TGF-β2 is an exercise-induced adipokine that regulates glucose and fatty acid metabolism. Nat. Metab. 2019;1:291–303. doi: 10.1038/s42255-018-0030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijay J., Gauthier M.-F., Biswell R.L., Louiselle D.A., Johnston J.J., Cheung W.A., Belden B., Pramatarova A., Biertho L., Gibson M., et al. Single-cell analysis of human adipose tissue identifies depot- and disease-specific cell types. Nat. Metab. 2020;2:97–109. doi: 10.1038/s42255-019-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman A.M., Steen C.B., Liu C.L., Gentles A.J., Chaudhuri A.A., Scherer F., Khodadoust M.S., Esfahani M.S., Luca B.A., Steiner D., et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez-Franquesa A., Stocks B., Borg M.L., Kuefner M., Dalbram E., Nielsen T.S., Agrawal A., Pankratova S., Chibalin A.V., Karlsson H.K.R., et al. Discovery of thymosin β4 as a human exerkine and growth factor. Am. J. Physiol. Cell Physiol. 2021;321 doi: 10.1152/ajpcell.00263.2021. C770-c778. [DOI] [PubMed] [Google Scholar]
- 14.van Hall G., Steensberg A., Sacchetti M., Fischer C., Keller C., Schjerling P., Hiscock N., Møller K., Saltin B., Febbraio M.A., Pedersen B.K. Interleukin-6 stimulates lipolysis and fat oxidation in humans. J. Clin. Endocrinol. Metab. 2003;88:3005–3010. doi: 10.1210/jc.2002-021687. [DOI] [PubMed] [Google Scholar]
- 15.Costford S.R., Bajpeyi S., Pasarica M., Albarado D.C., Thomas S.C., Xie H., Church T.S., Jubrias S.A., Conley K.E., Smith S.R. Skeletal muscle NAMPT is induced by exercise in humans. Am J Physiol-Endoc M. 2010;298:E117–E126. doi: 10.1152/ajpendo.00318.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besse-Patin A., Montastier E., Vinel C., Castan-Laurell I., Louche K., Dray C., Daviaud D., Mir L., Marques M.A., Thalamas C., et al. Effect of endurance training on skeletal muscle myokine expression in obese men: identification of apelin as a novel myokine. Int. J. Obes. 2014;38:707–713. doi: 10.1038/ijo.2013.158. [DOI] [PubMed] [Google Scholar]
- 17.Kleinert M., Clemmensen C., Sjøberg K.A., Carl C.S., Jeppesen J.F., Wojtaszewski J.F.P., Kiens B., Richter E.A. Exercise increases circulating GDF15 in humans. Mol. Metabol. 2018;9:187–191. doi: 10.1016/j.molmet.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ehrlund A., Mejhert N., Lorente-Cebrián S., Aström G., Dahlman I., Laurencikiene J., Rydén M. Characterization of the Wnt inhibitors secreted frizzled-related proteins (SFRPs) in human adipose tissue. J. Clin. Endocrinol. Metab. 2013;98:E503–E508. doi: 10.1210/jc.2012-3416. [DOI] [PubMed] [Google Scholar]
- 19.Arner P., Petrus P., Esteve D., Boulomié A., Näslund E., Thorell A., Gao H., Dahlman I., Rydén M. Screening of potential adipokines identifies S100A4 as a marker of pernicious adipose tissue and insulin resistance. Int. J. Obes. 2018;42:2047–2056. doi: 10.1038/s41366-018-0018-0. [DOI] [PubMed] [Google Scholar]
- 20.Fabre O., Ingerslev L.R., Garde C., Donkin I., Simar D., Barrès R. Exercise training alters the genomic response to acute exercise in human adipose tissue. Epigenomics. 2018;10:1033–1050. doi: 10.2217/epi-2018-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pillon N.J., Smith J.A.B., Alm P.S., Chibalin A.V., Alhusen J., Arner E., Carninci P., Fritz T., Otten J., Olsson T., et al. Distinctive exercise-induced inflammatory response and exerkine induction in skeletal muscle of people with type 2 diabetes. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acosta J.R., Joost S., Karlsson K., Ehrlund A., Li X., Aouadi M., Kasper M., Arner P., Rydén M., Laurencikiene J. Single cell transcriptomics suggest that human adipocyte progenitor cells constitute a homogeneous cell population. Stem Cell Res. Ther. 2017;8:250. doi: 10.1186/s13287-017-0701-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrlund A., Acosta J.R., Björk C., Hedén P., Douagi I., Arner P., Laurencikiene J. The cell-type specific transcriptome in human adipose tissue and influence of obesity on adipocyte progenitors. Sci. Data. 2017;4 doi: 10.1038/sdata.2017.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanford K.I., Middelbeek R.J.W., Goodyear L.J. Exercise Effects on White Adipose Tissue: Beiging and Metabolic Adaptations. Diabetes. 2015;64:2361–2368. doi: 10.2337/db15-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao R.R., Long J.Z., White J.P., Svensson K.J., Lou J., Lokurkar I., Jedrychowski M.P., Ruas J.L., Wrann C.D., Lo J.C., et al. Meteorin-like is a hormone that regulates immune-adipose interactions to increase beige fat thermogenesis. Cell. 2014;157:1279–1291. doi: 10.1016/j.cell.2014.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nigro P., Middelbeek R.J.W., Alves C.R.R., Rovira-Llopis S., Ramachandran K., Rowland L.A., Møller A.B., Takahashi H., Alves-Wagner A.B., Vamvini M., et al. Exercise Training Promotes Sex-Specific Adaptations in Mouse Inguinal White Adipose Tissue. Diabetes. 2021;70:1250–1264. doi: 10.2337/db20-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tuomilehto J., Lindström J., Eriksson J.G., Valle T.T., Hämäläinen H., Ilanne-Parikka P., Keinänen-Kiukaanniemi S., Laakso M., Louheranta A., Rastas M., et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N. Engl. J. Med. 2001;344:1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 28.Gonçalves C.A.M., Dantas P.M.S., Dos Santos I.K., Dantas M., da Silva D.C.P., Cabral B.G.d.A.T., Guerra R.O., Júnior G.B.C. Effect of Acute and Chronic Aerobic Exercise on Immunological Markers: A Systematic Review. Front. Physiol. 2019;10:1602. doi: 10.3389/fphys.2019.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peake J.M., Neubauer O., Walsh N.P., Simpson R.J. Recovery of the immune system after exercise. J. Appl. Physiol. 2017;122:1077–1087. doi: 10.1152/japplphysiol.00622.2016. [DOI] [PubMed] [Google Scholar]
- 30.Czepluch F.S., Barrès R., Caidahl K., Olieslagers S., Krook A., Rickenlund A., Zierath J.R., Waltenberger J. Strenuous physical exercise adversely affects monocyte chemotaxis. Thromb. Haemostasis. 2011;105:122–130. doi: 10.1160/TH10-06-0363. [DOI] [PubMed] [Google Scholar]
- 31.Zatterale F., Longo M., Naderi J., Raciti G.A., Desiderio A., Miele C., Beguinot F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2019;10:1607. doi: 10.3389/fphys.2019.01607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calder P.C., Dimitriadis G., Newsholme P. Glucose metabolism in lymphoid and inflammatory cells and tissues. Curr Opin Clin Nutr. 2007;10:531–540. doi: 10.1097/MCO.0b013e3281e72ad4. [DOI] [PubMed] [Google Scholar]
- 33.Gonzalez-Franquesa A., Patti M.E. Insulin Resistance and Mitochondrial Dysfunction. Adv. Exp. Med. Biol. 2017;982:465–520. doi: 10.1007/978-3-319-55330-6_25. [DOI] [PubMed] [Google Scholar]
- 34.Bradley R.L., Jeon J.Y., Liu F.F., Maratos-Flier E. Voluntary exercise improves insulin sensitivity and adipose tissue inflammation in diet-induced obese mice. Am J Physiol-Endoc M. 2008;295:E586–E594. doi: 10.1152/ajpendo.00309.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruun J.M., Helge J.W., Richelsen B., Stallknecht B. Diet and exercise reduce low-grade inflammation and macrophage infiltration in adipose tissue but not in skeletal muscle in severely obese subjects. Am J Physiol-Endoc M. 2006;290:E961–E967. doi: 10.1152/ajpendo.00506.2005. [DOI] [PubMed] [Google Scholar]
- 36.Čížková T., Štěpán M., Daďová K., Ondrůjová B., Sontáková L., Krauzová E., Matouš M., Koc M., Gojda J., Kračmerová J., et al. Exercise Training Reduces Inflammation of Adipose Tissue in the Elderly: Cross-Sectional and Randomized Interventional Trial. J. Clin. Endocrinol. Metab. 2020;105:e4510–e4526. doi: 10.1210/clinem/dgaa630. [DOI] [PubMed] [Google Scholar]
- 37.Savikj M., Zierath J.R. Train like an athlete: applying exercise interventions to manage type 2 diabetes. Diabetologia. 2020;63:1491–1499. doi: 10.1007/s00125-020-05166-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brandão B.B., Madsen S., Rabiee A., Oliverio M., Ruiz G.P., Ferrucci D.L., Branquinho J.L., Razolli D., Pinto S., Nielsen T.S., et al. Dynamic changes in DICER levels in adipose tissue control metabolic adaptations to exercise. Proc. Natl. Acad. Sci. USA. 2020;117:23932–23941. doi: 10.1073/pnas.2011243117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts L.D., Boström P., O'Sullivan J.F., Schinzel R.T., Lewis G.D., Dejam A., Lee Y.K., Palma M.J., Calhoun S., Georgiadi A., et al. β-Aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab. 2014;19:96–108. doi: 10.1016/j.cmet.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pedersen B.K., Febbraio M.A. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 2012;8:457–465. doi: 10.1038/nrendo.2012.49. [DOI] [PubMed] [Google Scholar]
- 41.Oishi Y., Manabe I. Macrophages in inflammation, repair and regeneration. Int. Immunol. 2018;30:511–528. doi: 10.1093/intimm/dxy054. [DOI] [PubMed] [Google Scholar]
- 42.Chawla A., Nguyen K.D., Goh Y.P.S. Macrophage-mediated inflammation in metabolic disease. Nat. Rev. Immunol. 2011;11:738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajbhandari P., Arneson D., Hart S.K., Ahn I.S., Diamante G., Santos L.C., Zaghari N., Feng A.C., Thomas B.J., Vergnes L., et al. Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. Elife. 2019;8 doi: 10.7554/eLife.49501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villarroya F., Cereijo R., Villarroya J., Gavaldà-Navarro A., Giralt M. Toward an Understanding of How Immune Cells Control Brown and Beige Adipobiology. Cell Metab. 2018;27:954–961. doi: 10.1016/j.cmet.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 45.Man K., Kutyavin V.I., Chawla A. Tissue Immunometabolism: Development, Physiology, and Pathobiology. Cell Metab. 2017;25:11–26. doi: 10.1016/j.cmet.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Komori T., Morikawa Y. Essential roles of the cytokine oncostatin M in crosstalk between muscle fibers and immune cells in skeletal muscle after aerobic exercise. J. Biol. Chem. 2022;298 doi: 10.1016/j.jbc.2022.102686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hojman P., Dethlefsen C., Brandt C., Hansen J., Pedersen L., Pedersen B.K. Exercise-induced muscle-derived cytokines inhibit mammary cancer cell growth. Am J Physiol-Endoc M. 2011;301:E504–E510. doi: 10.1152/ajpendo.00520.2010. [DOI] [PubMed] [Google Scholar]
- 48.Pendergrast L.A., Lundell L.S., Ehrlich A.M., Ashcroft S.P., Schönke M., Basse A.L., Krook A., Treebak J.T., Dollet L., Zierath J.R. Time of day determines postexercise metabolism in mouse adipose tissue. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2218510120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laurens C., de Glisezinski I., Larrouy D., Harant I., Moro C. Influence of Acute and Chronic Exercise on Abdominal Fat Lipolysis: An Update. Front. Physiol. 2020;11 doi: 10.3389/fphys.2020.575363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jones S.A., Jenkins B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018;18:773–789. doi: 10.1038/s41577-018-0066-7. [DOI] [PubMed] [Google Scholar]
- 51.Björk C., Wilhelm U., Mandrup S., Larsen B.D., Bordoni A., Hedén P., Rydén M., Arner P., Laurencikiene J. Effects of selected bioactive food compounds on human white adipocyte function. Nutr. Metab. 2016;13:4. doi: 10.1186/s12986-016-0064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crowe S., Turpin S.M., Ke F., Kemp B.E., Watt M.J. Metabolic remodeling in adipocytes promotes ciliary neurotrophic factor-mediated fat loss in obesity. Endocrinology. 2008;149:2546–2556. doi: 10.1210/en.2007-1447. [DOI] [PubMed] [Google Scholar]
- 53.Petersen E.W., Carey A.L., Sacchetti M., Steinberg G.R., Macaulay S.L., Febbraio M.A., Pedersen B.K. Acute IL-6 treatment increases fatty acid turnover in elderly humans in vivo and in tissue culture in vitro. Am J Physiol-Endoc M. 2005;288:E155–E162. doi: 10.1152/ajpendo.00257.2004. [DOI] [PubMed] [Google Scholar]
- 54.van Krieken P.P., Odermatt T.S., Borsigova M., Blüher M., Wueest S., Konrad D. Oncostatin M suppresses browning of white adipocytes via gp130-STAT3 signaling. Mol. Metabol. 2021;54 doi: 10.1016/j.molmet.2021.101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piquer-Garcia I., Campderros L., Taxerås S.D., Gavaldà-Navarro A., Pardo R., Vila M., Pellitero S., Martínez E., Tarascó J., Moreno P., et al. A Role for Oncostatin M in the Impairment of Glucose Homeostasis in Obesity. J. Clin. Endocrinol. Metab. 2020;105:E337–E348. doi: 10.1210/clinem/dgz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komori T., Tanaka M., Furuta H., Akamizu T., Miyajima A., Morikawa Y. Oncostatin M is a potential agent for the treatment of obesity and related metabolic disorders: a study in mice. Diabetologia. 2015;58:1868–1876. doi: 10.1007/s00125-015-3613-9. [DOI] [PubMed] [Google Scholar]
- 57.As W.N., Lehrskov L.L., Rh C., Ge L., Dorph E., Mk L., Launbo N., Sr F., Sk S., Nymand S., et al. Exercise-Induced Changes in Visceral Adipose Tissue Mass Are Regulated by IL-6 Signaling: A Randomized Controlled Trial. Cell Metabol. 2019;29:844–855.e843. doi: 10.1016/j.cmet.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Wernstedt Asterholm I., Tao C., Morley T.S., Wang Q.A., Delgado-Lopez F., Wang Z.V., Scherer P.E. Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab. 2014;20:103–118. doi: 10.1016/j.cmet.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao H., Volat F., Sandhow L., Galitzky J., Nguyen T., Esteve D., Åström G., Mejhert N., Ledoux S., Thalamas C., et al. CD36 Is a marker of human adipocyte progenitors with pronounced adipogenic and triglyceride accumulation potential. Stem Cells. 2017;35:1799–1814. doi: 10.1002/stem.2635. [DOI] [PubMed] [Google Scholar]
- 60.Petrus P., Lecoutre S., Dollet L., Wiel C., Sulen A., Gao H., Tavira B., Laurencikiene J., Rooyackers O., Checa A., et al. Glutamine Links Obesity to Inflammation in Human White Adipose Tissue. Cell Metab. 2020;31:375–390.e11. doi: 10.1016/j.cmet.2019.11.019. [DOI] [PubMed] [Google Scholar]
- 61.Pillon N.J., Gabriel B.M., Dollet L., Smith J.A.B., Sardón Puig L., Botella J., Bishop D.J., Krook A., Zierath J.R. Transcriptomic profiling of skeletal muscle adaptations to exercise and inactivity. Nat. Commun. 2020;11:470. doi: 10.1038/s41467-019-13869-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gabriel B.M., Altıntaş A., Smith J.A.B., Sardon-Puig L., Zhang X., Basse A.L., Laker R.C., Gao H., Liu Z., Dollet L., et al. Disrupted circadian oscillations in type 2 diabetes are linked to altered rhythmic mitochondrial metabolism in skeletal muscle. Sci. Adv. 2021;7:eabi9654. doi: 10.1126/sciadv.abi9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 65.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bates D., Mächler M., Bolker B., Walker S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015;67:1–48. [Google Scholar]
- 67.Hartig F. 2022. Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.4.5. [Google Scholar]
- 68.Tyanova S., Cox J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018;1711:133–148. doi: 10.1007/978-1-4939-7493-1_7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
(1)
The RNAsequencing source data have been deposited to the Gene Expression Omnibus repository and is publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
(2)
This paper does not report original code.
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.