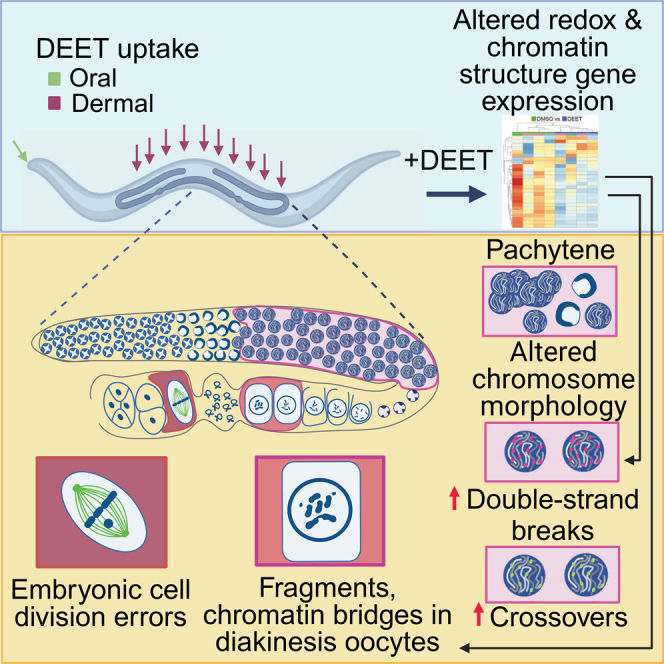
Summary
N,N-diethyl-meta-toluamide (DEET) is a commonly used synthetic insect repellent. Although the neurological effects of DEET have been widely investigated, its effects on the germline are less understood. Here, we show that exposure of the nematode Caenorhabditis elegans, which is highly predictive of mammalian reprotoxicity, resulting in internal DEET levels within the range detected in human biological samples, causes activation of p53/CEP-1-dependent germ cell apoptosis, altered meiotic recombination, chromosome abnormalities, and missegregation. RNA-sequencing analysis links DEET-induced alterations in the expression of genes related to redox processes and chromatin structure to reduced mitochondrial function, impaired DNA double-strand break repair progression, and defects during early embryogenesis. We propose that Caenorhabditis elegans exposure to DEET interferes with gene expression, leading to increased oxidative stress and altered chromatin structure, resulting in germline effects that pose a risk to reproductive health.
Subject areas: Biological sciences, Environmental toxicology, Molecular toxicology, Cell biology
Graphical abstract
Highlights
-
•
C. elegans DEET levels, within the range in many human samples, impairs meiosis
-
•
DEET compromises genomic integrity by deregulating meiotic recombination
-
•
DEET exposure causes chromosomal abnormalities and impairs early embryogenesis
-
•
Redox and chromatin genes link DEET to mitochondrial and germline dysfunction
Biological sciences; Environmental toxicology; Molecular toxicology; Cell biology
Introduction
N,N-diethyl-meta-toluamide (DEET), the major active ingredient of common topical insect repellents, was first developed by the US Department of Agriculture as protection for the military in 1946 and then registered for use by the general public in 1957.1,2,3 DEET-containing products are available in various forms including liquids, lotions, sprays, and treated materials. It is estimated that 30% of the US population uses repellents containing DEET and that approximately 1,800 tons of DEET are used annually in the United States.4,5 DEET can be absorbed topically or through the consumption of contaminated water. Trace levels of DEET have been detected in drinking water globally.6,7,8 In humans, DEET is metabolized by cytochrome P450 enzymes into oxidative metabolites including N,N-diethyl-3-(hydroxymethyl)benzamide (DHMB) and 3-(diethylcarbamoyl)benzoic acid (DCBA).9 DEET and its metabolites are detected in human urine, serum, semen, and cord serum, and an estimated 84% of the US population has detectable concentrations of DCBA in their urine.10,11,12,13,14 Concerns about the safety of DEET use began in the 1980s after reports of human encephalopathy following DEET exposure.15 Although the role of DEET in these cases was unclear, it triggered further investigations and reviews on the safety of DEET.16,17,18
Despite the widespread use of DEET in individuals of all ages, there is limited data on the effects of DEET exposure on reproductive health, and the available toxicological studies on animals have had mixed results. Early studies following the application of high doses of DEET to the skin of pregnant female rats showed an increase in embryo mortality, decreased birthweight, delayed development, and high postnatal death rate.19 A high frequency of malformations was reported when DEET was applied in a mineral oil formulation to the chorioallantoic membrane of chick embryos.20 Additionally, the sperm of male rats treated with DEET exhibited abnormal morphology and reduced motility.21 However, there was no evidence of embryotoxicity, impaired perinatal survival, or increased external, visceral, or skeletal malformations in the offspring of either rabbits or rats using different delivery methods and doses of DEET, although reduced fetal weight was observed in rats at a dose of 750 mg/kg.22,23 A study of rats exposed to a combination of the drug pyridostigmine bromide, the insecticide permethrin, and DEET for 28 days detected increased apoptosis in testicular germ cells.24 A more recent study showed that exposure of gestating female rats to a mixture of DEET and permethrin during the time of gonadal sex determination causes pubertal abnormalities, azoospermia, seminiferous tubule defects, and sloughed spermatogenic cells in the F1 generation male descendants, as well as pubertal abnormalities and ovarian disease in the F1 generation female descendants. All of these defects persisted to the F3 generation and were associated with changes in DNA methylation to the sperm genome.25 A study of DEET use during the second and third trimesters of human gestation, involving 897 pregnant women in Thailand, showed no significant differences between the control and exposed groups in birth weight, newborn anthropometrics, newborn neurologic examination, or developmental milestones in the first year of life.26 DEET use in the first trimester of human pregnancy has not been studied.
Although some of the reproductive outcomes from DEET exposure have been evaluated, there is a lack of understanding of the mechanisms underlying the observed effects, and, to our knowledge, there have not been studies assessing the effects of DEET directly on female meiosis. This is in part because meiosis is not easily recapitulated in a tissue culture setting, and female mammalian meiosis can span several months in mice and even decades in humans. The integrity of this cell division program is critical because failure to achieve accurate chromosome segregation during meiosis causes aneuploidy and can lead to infertility, stillbirths, miscarriages, and birth defects.27,28 Here, we address how DEET affects oogenesis using the nematode Caenorhabditis elegans, which is a genetically and cytologically tractable model organism that is highly predictive of mammalian reproductive toxicities and provides many advantages for the study of meiosis, including sharing a high degree of conservation of its genes and biochemical pathways with humans, carrying a well-defined and characterized germline, and exhibiting a rapid life cycle (it develops from an egg into an adult in approximately 3 days at 20°C).29,30,31,32,33 We found that DEET exposure resulting in internal levels in C. elegans within the range also found in some human biological samples causes activation of p53/CEP-1-dependent germ cell apoptosis and elevated levels of meiotic DNA double-strand breaks (DSBs) and crossover (CO) recombination. Altered expression of genes related to oxidation and reduction processes and chromatin structure, as revealed by RNA-sequencing (RNA-seq) analysis following DEET exposure, is coupled with mitochondrial dysfunction, increased levels of germline chromosome abnormalities, and impaired early embryogenesis. We propose that DEET alters gene expression, resulting in increased oxidative stress and altered chromatin structure in the germline, thereby compromising germline genomic integrity and increasing embryonic lethality.
Results
Exposure to different levels of DEET results in increased embryonic lethality, germ cell apoptosis, and germline defects
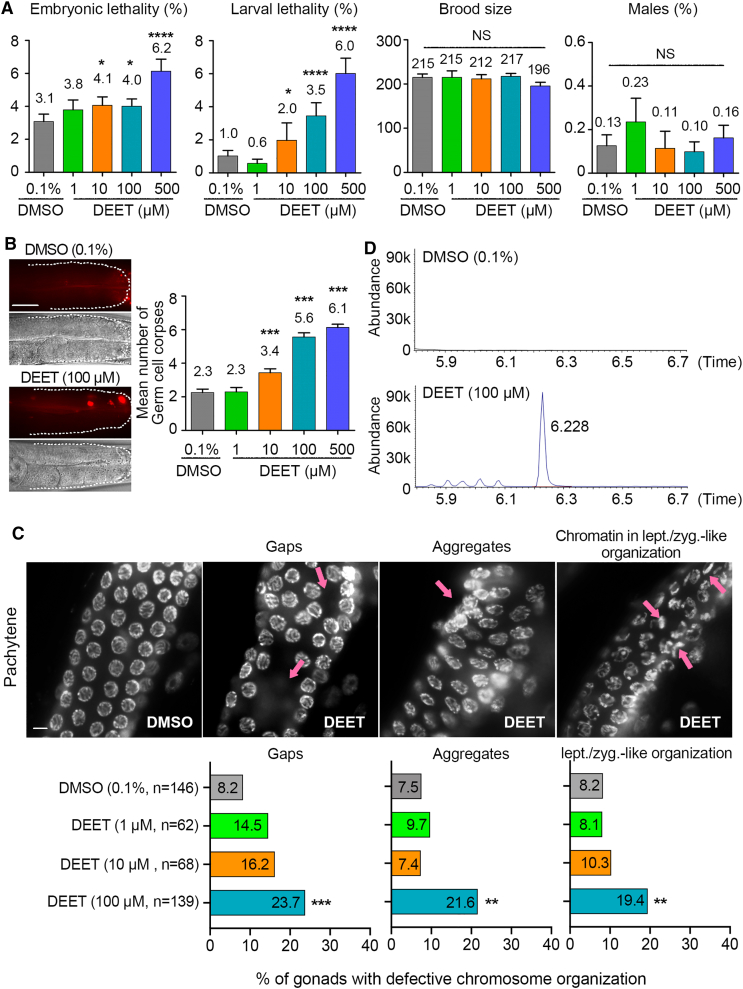
To identify levels of DEET exposure that result in meiotic defects in C. elegans, we tested various concentrations of DEET to establish dose responses. We exposed worms for 24 h to 1, 10, 100, and 500 μM DEET starting at late L4 (the last larval stage) until the completion of the first day of adulthood. DEET exposure was performed at this period to specifically capture its impact during oogenesis in the germline at the developmental stage in the worms when their gonads are fully formed and they start to actively lay eggs. We used worms carrying a collagen gene mutation (col-121(nx3)) that increases cuticle permeability without affecting the worm’s life cycle.34 This mutation enables the analysis of lower doses of chemicals (≤100 μM), which circumvent worm lethality, are frequently used in chemical screens in C. elegans, and correlate well with mammalian reproductive endpoints.30,32,33,35 Synchronized animals at the L1 larval stage were placed on regular NGM plates until the L4 stage. Animals were moved to 24-well plates where they underwent liquid exposure to either vehicle alone (0.1% DMSO) or different concentrations of DEET. This liquid exposure likely results in a combination of both dermal and oral intake of DEET. After 24 h of exposure, the total number of eggs laid, embryonic lethality, larval lethality, and incidence of male progeny were scored. col-121 worms exhibited normal motility and exterior appearance following these exposures, suggesting absence of systemic toxicity. We observed significantly increased embryonic lethality for exposures starting at 10 μM of DEET (p = 0.0463; Fisher’s exact test) and a 2-fold increase at 500 μM of DEET (p < 0.0001) (Figure 1A). We also observed significantly increased larval lethality starting at 10 μM (p = 0.0125; Fisher’s exact test). The total number of eggs laid (brood size) and male progeny, which can be indicative of sterility and X chromosome nondisjunction, respectively, were not significantly altered at the doses tested (Figure 1A and Data S1A). However, a limiting factor in this analysis is the sample size that can be compounded by subject variability, thereby obscuring the identification of effects.36 In fact, high-throughput screening of approximately 5,000 col-121 worms exposed to 100 μM DEET revealed a 2-fold increase in male progeny relative to vehicle alone.32 Therefore, to circumvent these issues, we evaluated levels of germ cell apoptosis at different doses of exposure by acridine orange staining as in.37 This is an extremely sensitive assay for germline dose-response assessment,32,35,38,39 and its use in assessing DEET exposures is further supported by the observed increase in testicular germ cell apoptosis in rats following the combined exposure to DEET, the insecticide permethrin, and the drug pyridostigmine bromide.24 In C. elegans, up to three germ cell corpses observed close to the bend of the gonad (late pachytene stage of meiosis) reflect physiological germ cell death.40,41 We observed a significant dose-dependent increase in germ cell apoptosis levels starting at 10 μM of DEET (10 μM: p = 0.0008, 100 μM and 500 μM: p < 0.0001) (Figure 1B and Data S1B), further supporting effects in the germline.
Figure 1.
Exposure to different levels of DEET results in increased embryonic lethality, germ cell apoptosis, and germline defects
(A) Embryonic lethality, larval lethality, the total number of eggs laid (brood size), and the total number of male progeny were scored at the indicated doses of exposures and compared with vehicle alone (0.1% DMSO). Error bars represent SEM. ∗p < 0.05, and ∗∗∗∗p < 0.0001 by the Fisher’s exact test, 95% CI.
(B) DEET exposure caused a significant increase in the number of germ cell corpses observed at late pachytene. (Left) Representative images of acridine-orange-stained germ cell corpses (red; top) and the same germlines visualized by Normarski optics (below). Gonads are traced to facilitate visualization. Scale bar, 50 μm. (Right) Graph shows quantification of levels of germ cell corpses for two biological repeats, with more than 30 gonads scored for each dose. Error bars represent SEM. ∗∗∗p < 0.001 by the two-tailed Mann-Whitney test, 95% CI.
(C) Images show pachytene nuclei in the germlines of whole, undissected worms fixed with Carnoy’s fixative and stained with DAPI that were exposed to vehicle alone (0.1% DMSO) and 1, 10, and 100 μM DEET. A minimum of 62 gonads from at least 4 biological repeats were scored for each indicated exposure. Arrows in the second panel from the left indicate gaps (discontinuities or spaces lacking germ cell nuclei) in the gonad. Arrow in the third panel indicates presence of DAPI-bright chromatin forming aggregates. Arrows in the fourth panel indicates nuclei with chromatin in leptotene/zygotene-like organization. n = number of gonad arms analyzed. Scale bar, 5 μm. Graphs show the percentage of gonads with defective chromosome organization corresponding to defects shown in images. ∗∗p < 0.01 and ∗∗∗p < 0.001 by the two-sided Fisher’s exact test.
(D) Representative gas chromatography-mass spectrometry (GC-MS) chromatograms. (Top) Chromatogram of worms exposed to vehicle alone (0.1% DMSO; control). (Bottom) Chromatogram of worms exposed to 100 μM of DEET. y axis represents relative abundance of signal intensity, and x axis represents retention time in minutes.
To further define the lowest dose of exposure that elicits the strongest germline effect, we assessed germline chromosome morphology defects in worms exposed to 1, 10, and 100 μM of DEET. Worms exposed to 1 and 10 μM of DEET did not show significant differences in germline chromosome organization compared with the control. However, the exposure to 100 μM DEET elicited alterations in chromosome organization in the germlines of exposed worms compared with vehicle alone, including areas with a reduced density of nuclei (gaps; 23.7% and 8.2%, respectively, p = 0.0003, Fisher’s exact test, confidence interval [CI] 95%), the presence of nuclei forming aggregates (21.6% vs. 7.5%, p = 0.0011), and the presence of nuclei with collapsed chromatin in a leptotene-/zygotene-like organization at late pachytene (19.4% vs. 8.1%, p = 0.0090) (Figure 1C and Data S1C).
To assess the uptake of DEET by C. elegans, we measured the internal concentration of DEET in whole worm extracts by isotopic dilution mass spectrometric analysis (Figure 1D). We found that an exposure to an external dose of 100 μM DEET results in an internal level of 0.067 μg/mL, which is within the range detected in various human samples such as urine, maternal serum, and cord serum.11,12,42,43 Therefore, all subsequent analysis in our study was performed at the 100 μM dose. Taken together, these results show evidence of increased embryonic lethality, larval lethality, germ cell apoptosis, and germline chromosome organization defects after exposure to DEET.
DEET exposure affects DNA double-strand break repair progression and crossover recombination levels during meiosis
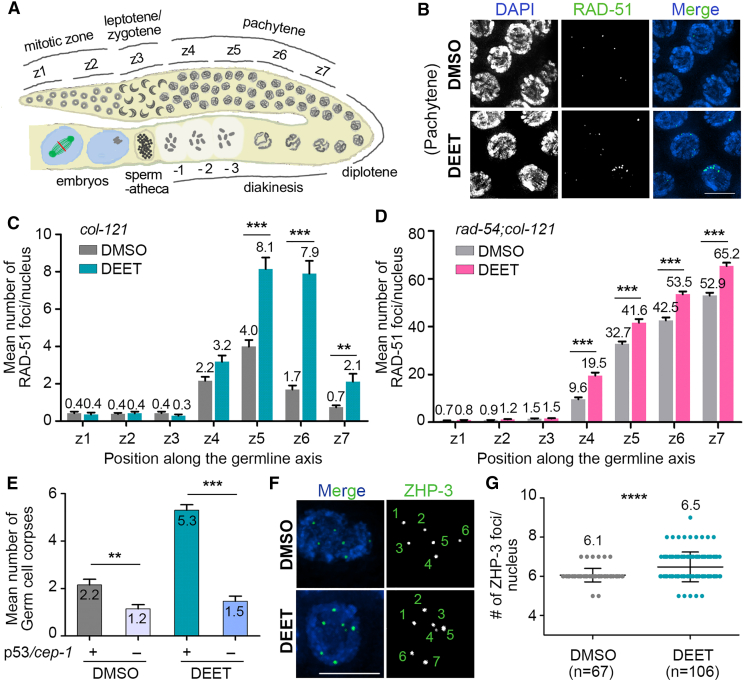
Given the elevated germ cell apoptosis and germline chromosome organization defects observed during pachytene, we examined whether DEET exposure causes defects in DNA double-strand breaks (DSBs) during prophase I. We quantified the levels of foci for the RAD-51 recombinase, an ortholog of human RAD51 that binds to 3′ ssDNA ends at DSBs to promote strand invasion and exchange for DSB repair,44 on immunostained whole-mounted gonads to observe DSB formation and repair progression as in.45 In DMSO-exposed worms, like in wild type, low levels of RAD-51 foci are observed at the premeiotic tip (zones 1–2) as well as upon entrance into meiosis at the leptotene/zygotene stage (transition zone = zone 3), which then peak during early to mid-pachytene (zone 5) and decrease significantly by late pachytene (zone 6) as repair is completed (Figures 2A–2C and Data S1D). In contrast, levels of RAD-51 foci were significantly increased in mid to late pachytene nuclei in the germlines of DEET-treated worms compared with control (zone 5: p < 0.0001, zone 6: p < 0.0001, zone 7: p = 0.0079 by the two-tailed Mann-Whitney test, 95% CI) (Figures 2B and 2C and Data S1D). To determine whether the elevated levels of RAD-51 foci were due in part to an increase in the number of DSBs, we examined RAD-51 foci in rad-54;col-121 double mutants following DEET exposure. RAD-54 is a DNA repair protein promoting RAD-51 displacement during strand exchange, and therefore, in a rad-54 mutant, although DSBs are formed and RAD-51 associates with DSB repair sites, further repair is blocked, resulting in “trapped” DSB-bound RAD-51.46 Levels of RAD-51 foci in the DEET-exposed gonads were comparable with the vehicle-treated gonads throughout the premeiotic tip (zones 1 and 2) and in leptotene/zygotene (zone 3), but in all other later stages of meiosis, DEET-exposed gonads showed a significant increase in RAD-51 foci compared with vehicle alone (Figure 2D). These data suggest that exposure to DEET results in elevated meiotic DSB levels. Unrepaired DSBs and/or aberrant recombination intermediates persisting until late pachytene can activate a DNA damage checkpoint, which results in p53/CEP-1-dependent germ cell apoptosis.41 Indeed, we observed a significant decrease in the number of germ cell corpses in cep-1;col-121 mutants, showing that DEET exposure results in p53/CEP-1-dependent germ cell apoptosis (p < 0.0001; two-tailed Mann-Whitney test, CI 95%) (Figure 2E and Data S1E). We also observed a reduction in germ cell corpses in cep-1; col-121 mutants following DMSO exposure, which indicates a basal level of toxicity for DMSO as previously described.33,47 Activation of a DNA damage checkpoint was further supported by the increased signal detected in pachytene nuclei for phosphorylated CHK-1, a checkpoint kinase involved in DNA damage sensing48 (Figure S1).
Figure 2.
DEET exposure affects DNA double-strand break repair progression and crossover recombination levels during meiosis
(A) Schematic representation of a C. elegans germline indicating the position of the equally sized seven zones (z1 to z7) scored for RAD-51 foci. Nuclei in z1 and z2 are undergoing mitosis and then enter meiosis at z3, which corresponds to the leptotene/zygotene stages. Nuclei proceed through pachytene (z4 to z7), diplotene, and diakinesis on their way to the uterus. The diakinesis oocyte closest to the spermatheca is indicated as the −1 oocyte.
(B) High-resolution images of mid-pachytene nuclei (zone 5) from whole-mounted gonads stained with RAD-51 (green) and co-stained with DAPI (blue). Scale bar, 5 μm.
(C and D) Histograms show the mean number of RAD-51 foci scored per nucleus for each zone from col-121 worms (C) and rad-54;col-121 worms (D). Five gonads from three independent biological repeats (C) and four gonads from two independent biological repeats (D) were scored for each indicated exposure. Error bars represent SEM. ∗∗p < 0.01 and ∗∗∗p < 0.001 by the two-tailed Mann-Whitney test, 95% CI.
(E) Mean number of germ cell corpses scored by acridine orange staining were significantly reduced in a p53/cep-1-dependent manner. Analysis was done for three independent biological repeats, and more than 30 gonads were scored for each exposure. Error bars represent SEM. ∗∗p < 0.01, ∗∗∗p < 0.001 by the two-tailed Mann-Whitney test, 95% CI.
(F) High-resolution images of late pachytene nuclei from whole-mounted gonads stained with ZHP-3 (green) and co-stained with DAPI (blue). Numbers indicate ZHP-3 foci. DMSO shows six ZHP-3 foci representing six COs (one per homolog pair), and DEET shows seven ZHP-3 foci. Scale bar, 5 μm.
(G) Scatterplot shows the mean number of ZHP-3 foci detected in late pachytene for each indicated exposure. Analysis was done for two independent biological repeats; n = number of nuclei analyzed. Sixty-seven nuclei from 9 gonads for DMSO and 106 nuclei from 15 gonads for DEET were scored. Error bars represent SD. ∗∗∗∗p < 0.001 by the two-tailed Mann-Whitney test.
Given the observed increased levels of DSB formation and the alterations in DSB repair progression, we used ZHP-3/Zip3/RNF212 as a marker to determine whether DEET might also affect crossover (CO) designation levels. CO frequency and distribution along each pair of homologous chromosomes are tightly regulated throughout species.49 In C. elegans, only one CO occurs per homolog pair such that six ZHP-3 foci per nucleus are expected in wild type.50 While six ZHP-3 foci are clearly visible in late pachytene nuclei from germlines exposed to vehicle alone (mean of 6.1, n = 67), seven to nine ZHP-3 foci were observed in nuclei from DEET-treated germlines (mean of 6.5, n = 106, two-tailed Mann-Whitney test, CI 95%, p < 0.0001) (Figures 2F and 2G and Data S1F). This significantly increased number of ZHP-3 foci may be due to the increased levels of DSBs formed following DEET exposure, akin to the increased numbers of COs proposed to result from the elevated levels of DSBs in gamma-irradiated C. elegans germlines.51 Alternatively, DEET may be influencing the processing of DSBs into COs. Taken together, these data show that exposure to DEET results in elevated meiotic DSB levels and altered DSB repair progression, leading to increased germ cell apoptosis due to activation of a p53/CEP-1-dependent DNA damage checkpoint response and increased CO levels. Importantly, elevated CO levels have been shown to impede faithful meiotic chromosome segregation in C. elegans,52 supporting the embryonic lethality (this study) and X chromosome nondisjunction observed by high-throughput screening following DEET exposure.32
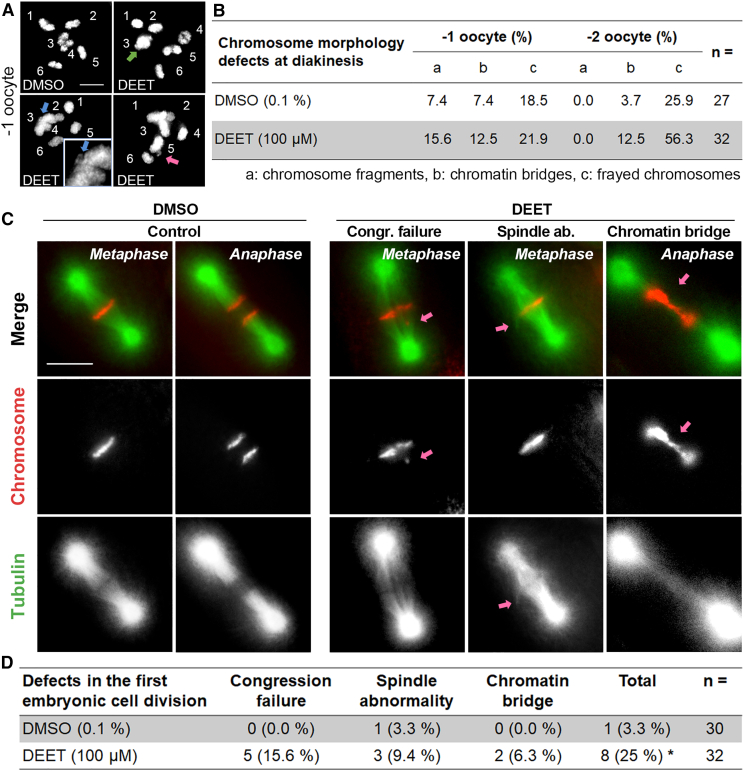
DEET exposure results in chromosomal abnormalities in late prophase I and impaired early embryogenesis
To evaluate defects at late prophase I following DEET exposure, we examined chromosome morphology in the two last oocytes proximal to the spermatheca (−1 and −2 oocytes; Figure 2A). Oocytes in DMSO-exposed worms showed mostly six clear DAPI-stained bodies representing the six pairs of attached homologs (bivalents) (Figures 3A and 3B). In contrast, we observed elevated levels of chromosome fragments (15.6%, n = 5/32), chromatin bridges (12.5%, n = 4/32), and frayed chromosomes (21.9%, n = 7/32) in oocytes from DEET-exposed worms. Although we cannot rule out the possibility that chromosomal abnormalities detected in diakinesis oocytes are the result of DEET effects directly at the diakinesis stage, they may also stem from elevated DSB levels and/or impaired DSB repair earlier in prophase I and have not been culled by germ cell apoptosis in late pachytene (Figures 3A and 3B and Data S1G).
Figure 3.
DEET exposure results in chromosomal abnormalities in late prophase I and impaired early embryogenesis
(A) High-resolution images of oocytes at diakinesis positioned right before the spermatheca (−1 oocyte). Six intact bivalents are observed in oocytes from worms exposed to vehicle control (0.1% DMSO), whereas chromosome fragments (green arrow), chromatin bridges (blue arrow), and frayed chromosomes (pink arrow) are observed in the oocytes of DEET-exposed worms. Numbers indicate each bivalent. Inset is from a partial z stack projection for magnified bivalent number 4 to facilitate visualization. Scale bar, 5 μm.
(B) Quantification of the chromosome morphology defects observed in diakinesis. n = total number of oocytes scored.
(C) Representative images from time-lapse analysis of the first embryonic division in vehicle alone (0.1% DMSO) and 100 μM DEET-exposed H2B::mCherry; ɣ-tubulin::GFP;col-121(nx3) worms. Normal configurations of metaphase and anaphase are shown for vehicle alone (control) with chromosomes (red) and tubulin (green). Arrows show examples of chromosomes that failed to align at the metaphase plate (congression failure), spindle abnormalities, and chromatin bridges at the metaphase to anaphase transition. Scale bar, 5 μm.
(D) Quantification of defects observed in the first embryonic cell division by time-lapse analysis. Eight out of 30 embryos exhibited defects in the case of DEET exposure, and among them 2 embryos exhibited more than one class of defect. n = total number of embryos scored. ∗p < 0.05 by Fisher’s exact test.
To determine if this effect of DEET exposure extends to early embryogenesis, we performed live imaging analysis of the first embryonic cell division in transgenic worms carrying H2B::mCherry; ɣ-tubulin::GFP and the col-121(nx3) mutation. Chromosome alignment at the metaphase plate and segregation at anaphase were normal in DMSO-treated animals except for one embryo with a spindle abnormality (3.3%, n = 1/30). In contrast, congression failure as evidenced by the presence of lagging chromosomes, spindle abnormalities, and chromatin bridges at the metaphase to anaphase transition was observed in 25% of one-cell embryos from DEET-exposed worms (n = 8/32, where 2 embryos exhibited both congression failure and spindle abnormalities; p = 0.0269 by Fisher’s exact test) (Figures 3C and 3D and Data S1G). These results suggest that DEET exposure disrupts chromosome morphology in late prophase I and leads to chromosome segregation defects and spindle abnormalities in early embryogenesis, supporting the embryonic lethality observed following this treatment.
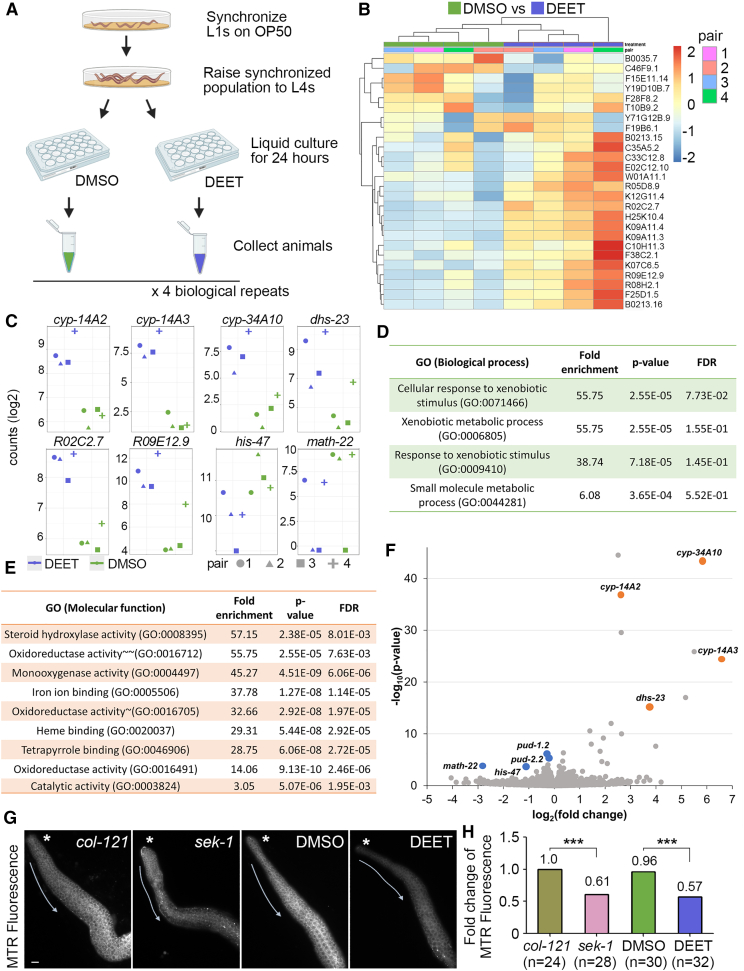
DEET exposure alters the expression of genes regulating cellular oxidation and chromatin structure
To determine whether the germline defects observed following DEET exposure might stem from alterations in gene expression, we performed RNA sequencing (RNA-seq) comparing DMSO- and DEET-exposed whole worm samples (four independent biological repeats; Figure 4A and Data S1H and S2). Twenty-six statistically significant differentially expressed (DE) genes show clear clustering between DMSO- and DEET-exposed animals as depicted in a heatmap (Figure 4B). This contrast in gene expression is further exemplified by the top 6 genes exhibiting significantly upregulated gene expression—R02C2.7, cyp-34A10, cyp-14A2, R09E12.9, dhs-23, and cyp-14A3—and the top 2 downregulated genes—his-47 and math-22—in the DEET-treated compared with DMSO-treated group (Figure 4C). Gene ontology (GO) enrichment analysis of the DE genes indicates the “cellular response to xenobiotic stimulus” (GO:0071466) and “xenobiotic metabolic process” (GO:0006805) as the top biological processes (Figure 4D). Among top ontology enrichment for molecular function, “monooxygenase activity” (GO:0004497); “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen” (GO:0016712); and “oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen” (GO:0016705) are related to oxidation-reduction (redox) reactions (Figure 4E). These processes include cyp-34A10, cyp-14A2, and cyp-14A3, and dehydrogenase dhs-23, which are among the top significantly DE genes, as well as other oxidases and reductases (Figure 4F).
Figure 4.
DEET exposure alters the expression of genes regulating cellular oxidation and chromatin structure
(A) Schematic of experimental design for RNA-seq from four independent biological repeats. Each repeat was divided in two samples (pairs), DMSO and DEET.
(B) Heatmap of DE genes (26 genes with Benjamini-Hochberg correction; BH adjusted p values <0.1).
(C) Top eight genes with significant DE. y axis shows gene counts (log2) for each pair in DMSO- and DEET-exposed samples.
(D and E) GO enrichment analysis of DE genes for biological process and molecular function, respectively. Geneontology.org was used with WormBase. Molecular function is for molecular-level activities performed by gene products, whereas biological process encompasses larger processes accomplished by multiple molecular activities53,54,55: ∼oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen (GO:0016705), ∼∼ oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen (GO:0016712).
(F) Volcano plot showing DE genes. Key genes are depicted as blue dots (downregulated in DEET) and brown dots (upregulated in DEET). They are annotated with their gene names.
(G) Mitochondrial dysfunction in the germline was measured with Mitotracker Red (MTR), and its signal is significantly decreased in the germlines of DEET-exposed worms compared with control. Shown are representative images of dissected gonads from untreated col-121 worms, sek-1 mutants, which exhibit mitochondrial dysfunction and increased oxidative stress due to lack of an MAPKK that activates p38 MAPK under oxidative stress conditions,56,57 and col-121 worms exposed to either vehicle (0.1% DMSO) or DEET. Asterisks are positioned adjacent to the premeiotic tip, and arrows indicate the direction of germline progression. Scale bar, 5 μm.
(H) Quantification of Mitotracker Red (MTR) signal intensity is shown as fold change. Analysis was done for two biological repeats; n = number of gonads scored. ∗∗∗p < 0.001 by the two-tailed Mann-Whitney test, 95% CI.
Because mitochondria are at the center of redox processes,58 we evaluated the effects of DEET exposure on mitochondrial morphology or function by using a mitochondrial-targeted dye (Mitotracker Red CMXRos, MTR hereafter) that accumulates within mitochondria according to the mitochondrial membrane potential. Similar to germlines exposed to the plasticizer bisphenol A, which exhibited reduced MTR signal corresponding to mitochondrial dysfunction and increased oxidative stress,39 gonads from DEET-exposed worms showed reduced MTR signal compared with control gonads (Figure 4G). Quantification showed an average fold change from 1.0 to 0.59 in MTR fluorescence intensity following DEET exposure (p < 0.0001 by the two-tailed Mann-Whitney test, 95% CI; Figure 4H and Data S1I). Altogether, these results show that DEET exposure induces altered gene expression of a specific set of genes including genes responsible for redox reactions, leading to increased oxidative stress further supported by mitochondrial dysfunction.
Discussion
Insights into molecular pathways impacted by DEET exposure
In the present study, we show that C. elegans exposure to 100 μM of DEET results in worm internal levels of 0.067 μg/mL. This dose of exposure leads to activation of a p53/CEP-1-dependent DNA damage checkpoint, elevated meiotic DSB and CO levels, along with germline chromosome abnormalities and chromosome missegregation. Our analysis suggests a link between DEET-induced alterations in the expression of genes regulating redox processes and chromatin structure to the observed reduction in mitochondrial function and increased frequency of germline defects. Comparison of the internal level detected in this C. elegans study with the levels detected in several human samples suggests that our findings may be relevant for mammalian reproductive health and requires further investigation.
Briefly, in a small-scale study, a median value of 1.3 μg/mL of DEET was recovered in children’s urine after 8 h of a single dermal application of 10 mL of insect repellent containing 12% DEET.12 Additionally, application of higher doses of DEET, similar to levels used by workers constantly exposed to insect bites, resulted in a median value of 1.68 μg/mL in urine.12 A study of 150 women who underwent an elective cesarean delivery in New Jersey reported 3.21 ng/g (∼0.0032 μg/mL) of DEET in maternal serum and 3.12 ng/g (∼0.0031 μg/mL) of DEET in cord serum from occasional use of pesticides during pregnancy.11 Another study of pregnant women that applied a 20% DEET solution daily during the second and third trimesters of pregnancy (median total dose, 214.2 g DEET per pregnancy) observed a range between 1 μg/mL and 2.44 μg/mL of DEET in 8% of the cord blood samples.
Intrinsic differences between humans and model systems used in the laboratory require caution in extrapolating results, in this case from worms onto humans. However, the results presented here provide insights into the mechanisms by which DEET may impact meiosis and open the door for further investigation.
Changes in gene expression may be linked to DEET-induced alterations in the C. elegans germline
Among the upregulated genes following DEET treatment, we found cyp-34A10, cyp-14A2, and cyp-14A3, which are orthologs of the human cytochrome P450 protein family. Proteins of this family include enzymes that are responsible for metabolizing external substances, such as environmental chemicals and medications, as well as endogenous intracellular substances.59,60 Therefore, the cytochrome P450 system may be upregulated to metabolize the foreign compound DEET (Figures 4C and 4D). Indeed, it has been reported that in mammals DEET is metabolized by cytochrome P450 enzymes into oxidative metabolites, including N,N-diethyl-3-hydroxymethylbenzamide (DHMB) and 3-(diethylcarbamoyl)benzoic acid (DCBA). In vitro human studies indicated the involvement of CYP2B6 and CYPA12 in DEET metabolism.61 Although we did not measure DEET metabolites in our samples, it is likely that the increased expression levels of the P450 protein-encoding cyp-34A10, cyp-14A2, and cyp-14A3 genes correlate with an increase in the proteins that potentially process DEET in C. elegans.
Oxidative stress associated with mitochondrial dysfunction is known to produce DNA DSBs.62 Therefore, our observation that DEET leads to elevated levels of DSBs in the worm germline (Figures 2C and 2D) suggests that this is due, at least in part, to altered gene expression that is responsible for redox reactions further supported by mitochondrial dysfunction (Figures 4C–4H). Moreover, DEET has been shown to induce DNA oxidative damage as determined by the presence of 8-hydroxy-2′-deoxyguanosine (8-OHdG) in rat urine following DEET exposure.63
Noticeably, DEET exposure also led to a mild downregulation of a very small set of genes (6 in total), suggesting that the response to this chemical may be very specific and tightly regulated (Figures 4B, 4C, and 4F). Among the downregulated genes are genes related to chromatin structure (his-47) and genes regulated by the daf-2/insulin-like signaling pathway (math-22, pud-1.2, pud-2.2), which is critical for regulating development, longevity, metabolism, and stress resistance.64,65,66 his-47 is an ortholog of human H2AX and is predicted to be a structural constituent of chromatin. Its homolog in S. cerevisiae, histone H2A, is a core histone protein required for chromatin assembly and chromosome function that undergoes DNA damage-dependent phosphorylation by Mec1p to facilitate DNA repair.67 Based on the function of its predicted homologs, we hypothesize that downregulation of his-47 gene expression may result in the accumulation of DNA damage, contributing to the elevated levels of RAD-51 foci observed following DEET exposure. Perturbations to chromosome structure influence the frequency of DSBs in the genome and, hence, of COs, as has been observed for meiotic condensin mutants in C. elegans.46,68 Studies in yeast suggest that histone H2A underexpression leads to location-specific disruptions in chromatin.69,70 Therefore, we propose that his-47 decreased expression levels could potentially alter chromosome structure, which may be linked to the elevated levels of DSBs and increased CO designated sites observed following DEET exposure.
Albeit with low coverage in our RNA-seq analysis, the downregulated math-22 gene is predicted to encode an understudied protein with a meprin-associated TRAF homology (MATH) domain, and its expression has been found to be regulated by the insulin receptor-like gene daf-2 and skn-1 transcription factor in other RNA-seq studies, thereby linking it to oxidative stress response.66,71 Modulation of insulin/insulin growth factor (IGF) signaling in C. elegans is the central determinant of the endocrine control of stress response, diapause, and aging. Insulin-like molecules, through the activity of the DAF-2/insulin/IGF-1-like receptor and the DAF-16/FKHRL1/FOXO transcription factor, have been proposed to regulate a set of genes that control the ability of the organism to deal with oxidative stress and interfere with metabolic programs that help to determine lifespan.72 math-22 expression decreases when daf-2 function is decreased,66 suggesting that this gene may be implicated in oxidative stress response in the worm following DEET exposure. The SKN-1 transcription factor is an evolutionarily conserved xenobiotic stress regulator and a pro-longevity factor that protects against oxidative stress by mobilizing the conserved phase 2 detoxification response.72,73,74 math-22 expression is significantly increased in skn-1(RNAi)-depleted worms, further supporting a role for math-22 in regulating the response observed in DEET exposed worms. Finally, pud-1.2 and pud-2.2 are two additional genes regulated by daf-2 and skn-1 that are among the group of DEET-downregulated genes.66,71 pud-1.2 and pud-2.2 encode proteins of unknown function but our results suggest that they may be implicated in oxidative stress response in DEET-exposed worms.
We propose that DEET exposure alters the expression of genes related to redox processes and oxidative stress, coupled with mitochondrial dysfunction, and genes regulating chromatin structure, which altogether contribute to elevated DSB levels, resulting in activation of a DNA damage checkpoint in late pachytene, chromosomal abnormalities in oocytes at diakinesis, and impaired early embryogenesis. Our study provides evidence of how exposure to DEET can lead to germline dysfunction and may interfere with reproductive health.
Limitations of the study
This study explores DEET-exposure effects during female meiosis. We used C. elegans, a well-established model for meiosis studies, and all the experiments contain internal controls, are appropriately replicated, and consist of sample sizes that are standard in the field. However, the translation of these findings into humans requires caution and further investigation. One important limitation of the study is that worms were exposed to DEET constantly for a 24-h period to evaluate the meiotic effects. Although the internal concentration of DEET was measured at the end of the treatment, this window of exposure is different from the transient DEET exposure that humans are normally subjected to. Therefore, future studies will need to evaluate the effects of DEET on meiosis using a range of shorter windows of exposure. In addition, it remains unclear how DEET is processed in the worm, and therefore, direct comparisons of DEET metabolism cannot be made between worms and humans at this time. Specific measurement of DEET metabolites by mass spectrometry could shed some light on this. Finally, preventive measures to combat transmission of insect-borne diseases such as Zika, Chikungunya, malaria, Dengue, and West Nile virus are of paramount importance, and insect repellents can reduce human risk of exposure. The findings in this study underscore the need for further research into the mechanisms of action of DEET on meiosis and the development of alternative insect repellents.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-pCHK-1 | Santa Cruz | Cat# 17922; RRID: N/A |
| rabbit anti-RAD-51 | Novus Biological (SDI) | Cat# 29480002; RRID: AB_2616441 |
| guinea pig anti-ZHP-3 | Bhalla, N.et al.75 | N/A |
| donkey anti-rabbit Cy3 | Jackson Immunoresearch | Cat# 711-165-152; RRID: AB_2307443 |
| mouse anti-goat Alexa 647 | Jackson Immunoresearch | Cat# 205-605-108; RRID: AB_2339076 |
| donkey anti-rabbit Alexa 488 | Jackson Immunoresearch | Cat# 711-545-152; RRID: AB_2313584 |
| donkey anti-guinea pig Alexa 488 | Jackson Immunoresearch | Cat# 706-545-148; RRID: AB_2340472 |
| Chemicals, peptides, and recombinant proteins | ||
| DMSO (Dimethyl sulfoxide) | MilliporeSigma | Cat# D8418 |
| DEET (N,N-Diethyl-m-toluamide) | MilliporeSigma | Cat# 36542 |
| DAPI | Thermo Fisher Scientific | Cat# D1306 |
| VectaShield | Vector Laboratories | Cat# H-1000 |
| Deposited data | ||
| RNAseq data | NCBI’s Gene Expression Omnibus76 | GEO Series accession number GEO:GSE242682 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE242682) |
| Experimental models: Organisms/strains | ||
| N2 (wild type) | https://cgc.umn.edu/strain/N2 | WB ID: WBStrain00000001 |
| col-121(nx3) IV | Watanabe, M.et al.34 | N/A |
| CV651 cep-1(lg12501)/hT2 I;col-121(nx3) IV | Shin, N.et al.32 | N/A |
| CV652 rad-54(ok615)/hT2 I;col-121(nx3) IV | Shin, N.et al.32 | N/A |
| CV639 H2B::mCherry;ɣ-tubulin::GFP;col-121(nx3) IV | Shin, N.et al.32 | N/A |
| LD1757 sek-1(km4) X | Hourihan, J.M.et al.57 | N/A |
| Oligonucleotides | ||
| See Table S1 for primers | ||
| Software and algorithms | ||
| SoftWoRx | GE Healthcare Life Sciences | N/A |
| Prism 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Monica P. Colaiácovo (mcolaiacovo@genetics.med.harvard.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus76 and are accessible through GEO Series accession number GEO:GSE242682 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE242682).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Worm strains
C. elegans strains were cultured at 20°C under standard conditions as described in.77 The following mutations and chromosome rearrangements were used in this study: LGI: cep-1(lg12501), rad-54(ok615); LGIV: col-121(nx3); LGV: yIs34[Pxol-1::GFP, rol-6]; LGX: sek-1(km4), transgenes: H2B::mCherry; γ-tubulin::GFP.
Method details
Chemical exposure and plate phenotyping
Embryos were collected from gravid adults following sodium hypochloride treatment and subjected to overnight starvation.78 Age-matched L1-stage worms were thoroughly washed with M9 and grown on regular NGM plates up to the L4 stage. Animals were then resuspended in M9 buffer with freshly cultured OP50 bacteria (OD600 = 24) together with either DMSO alone (0.1%; vehicle) or DEET (0.1 M) dissolved in DMSO for a 24-hour exposure. 300 worms in 250 μL of each condition were then dispensed into individual wells in 24-well plates. After 24 hours, the exposed worms were transferred to 1.5 mL tubes, washed five times with M9, and utilized for subsequent experiments. DMSO and DEET were purchased from MilliporeSigma (Burlington, MA). For plate phenotyping, individual worms were moved every 24 hours to new NGM plates.
Mass spectrometric analysis
Worm lysates were used for measuring the internal concentration of DEET in worm samples (both control and treated) using gas chromatography mass spectrometric (GC-MS) analysis. The sample extraction was performed as described in32 with slight modifications. Briefly, 100 μL of lysate was spiked with 40 ng of labelled internal standard (DEET-D6, 98%), allowed to equilibrate for 15 min at room temperature and extracted with 1 mL of hexane/dichloromethane mixture (1:2 v/v). The extract was centrifuged and filtered (0.2 μM nylon membrane filter) before transferring into GC vials for analysis. A Thermo trace 1310 GC with a HP-5MS capillary column (30 m×0.25 mm×0.25 μm) coupled with ISQ single quadrupole mass spectrometer (Waltham, MA, USA) was used for the analysis, with the aid of Xcalibur software. Process blank and matrix spike were included for the quality control along with the analysis of control/vehicle and treated worms. There was no detectable level of DEET found in control (blank) and the matrix spike recovery was 107%.
Germ cell apoptosis
Germ cell corpses were scored using acridine orange staining, as described in,37 and a Leica DM5000B fluorescence microscope. The germlines of more than 30 worms were scored for each exposure condition from at least two independent biological repeats (see Data S1B and S1E).
Immunofluorescence microscopy
Whole mount preparation of dissected gonads and immunostainings were performed as in.45 Primary antibodies were used at the following dilutions: rabbit α-pCHK-1 (1:100; Santa Cruz), rabbit α-RAD-51 (1:10,000; Novus Biological (SDI)), and guinea pig α-ZHP-3 (1:500;75). The following secondary antibodies from Jackson ImmunoResearch Laboratories (West Grove, PA) were used at a 1:200 dilution: α-rabbit Cy3, and at a 1:500 dilution: α-rabbit Alexa 488, and α-guinea pig Alexa 488. Vectashield from Vector Laboratories (Burlingame, CA) was used as a mounting media and anti-fading agent. Immunofluorescence images were collected at 0.2 μm intervals with an IX-70 microscope (Olympus, Waltham, MA) and a cooled CCD camera (CH350; Roper Scientific) controlled by the Delta Vision system (Applied Precision, Pittsburgh, PA). Images were subjected to deconvolution by using the SoftWoRx 3.3.6 software (Applied Precision).
Live imaging
Live imaging was performed as described in32 with strain CV639 (H2B::mCherry; γ-tubulin::GFP;col-121(nx3)). The first embryonic divisions of more than 30 worms were evaluated for each exposure condition from seven independent biological repeats.
Quantification and statistical analysis
All statistical analysis were performed using GraphPad Prism Version 8.4.3. Statistical details of experiments can be found in Results, Figure legends and Data S1. Specifically, for plate phenotyping the total number of eggs laid, the total number of eggs hatched, the number of progeny that reached adulthood, and the frequency of males detected among hatched progeny were scored see Data S1A. The data was compared using the Fisher’s exact test. For apoptosis statistical comparisons between groups were performed using the two-tailed Mann-Whitney test, 95% C.I.
Time course analysis for RAD-51 foci
Quantitative analysis of RAD-51 foci for all seven zones composing the germline was performed as in.45 At least 4 gonads were examined for each condition and the average number of nuclei scored per zone was as follows, ±standard deviation: For the col-121 line: zone 1 to zone 7 (n=73±5, 88±5, 96±10, 78±7, 61±3, 53±4, 62±11). For the rad-54;col-121 line: zone 1 to zone 7 (n=67±10, 75±12, 65.5±4.5, 61.5±6.5, 56±8.0, 44.5±9.5, 34.5±1.5) See Data S1D. Statistical comparisons were performed using the two-tailed Mann-Whitney test, 95% C.I.
RNA sequencing and data analysis
Samples of 1,000 col-121 worms each following 24-hour exposure to either DMSO alone (0.1%) or DEET dissolved in DMSO (0.1M) were collected in 500 μl of Trizol (Invitrogen) and prepared using the Direct-zol RNA miniprep kit (Zymo Research) by following the manufacturer’s instructions. Four independent biological repeats per condition were used for Illumina NextSeq High 2x38 using Kapa mRNA HyperPrep kit (Roche).
Reads were processed to counts through the bcbio RNA-seq pipeline implemented in the bcbio-nextgen project (https://bcbio-nextgen.readthedocs.org/en/latest/). Raw reads were examined for quality issues using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure library generation and sequencing were suitable for further analysis. If necessary, adapter sequences, other contaminant sequences such as polyA tails, and low quality sequences with PHRED quality scores less than five were trimmed from reads using cutadapt.79 Trimmed reads were aligned to Wormbase assembly WBcel235, Annotation Version WS272, using STAR.80
Alignments were checked for evenness of coverage, rRNA content, genomic context of alignments (for example, alignments in known transcripts and introns), complexity and other quality checks using a combination of FastQC, Qualimap81 MultiQC (https://github.com/ewels/MultiQC) and custom tools.
Counts of reads aligning to known genes were generated by featureCounts.82 In parallel, Transcripts Per Million (TPM) measurements per isoform were generated by quasialignment using Salmon.83 Differential expression at the gene level was called with DESeq2,84 using counts per gene estimated from the Salmon quasialignments by tximport85 see Data S1H. Quantitating at the isoform level has been shown to produce more accurate results at the gene level. QC and differential expression analyses were done using R programming language,v.3.6.0.
RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus76 and are accessible through GEO Series accession number GEO:GSE242682 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE242682).
Quantitative RT-PCR analysis
Three samples of 30 animals each were collected in 100 μL of Trizol (Invitrogen) and RNA was extracted by following the manufacturer’s instructions. Extracted RNA was subjected to reverse transcription using iScript (Biorad) and quantitative real time PCR was performed using the SsoFast EvaGreen supermix (Biorad) according to the manufacturer’s instructions with specific primers (Table S1). Each sample was run in triplicate. Cq numbers were normalized to gpd-1 and then normalized values from DEET exposed samples were statistically compared to the normalized values from vehicle (DMSO) treated samples, see Data S1J. Statistical comparisons were performed using the unpaired two tailed t-test, 95% C.I.
Acknowledgments
We thank the members of the Colaiacovo laboratory for critical reading of this manuscript. This work was supported by the McKenzie Family Trust (M.P.C.).
Author contributions
Conceptualization, N.S. and M.P.C.; investigation, N.S., L.I.L.-L., A.L.H., M.M.-G., V.B., and R.K.; resources, V.B., S.H.S, and K.K.; writing—original draft, N.S., L.I.L.-L., and M.P.C.; writing—review & editing, N.S., L.I.L.-L., A.L.H., M.M.-G., R.K., S.H.S., K.K., and M.P.C.; supervision, S.H.S., K.K., and M.P.C.; funding acquisition, M.P.C.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 4, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108699.
Supplemental information
(A) Plate phenotyping. Total brood size counts. The total number of fertilized eggs laid, hatched, the number of progeny that reached adulthood, and the frequency of male progeny are shown for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1, 10, 100, and 500 μM of DEET. (B) Apoptotic nuclei count. Number of acridine-orange-stained nuclei detected in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1, 10, 100, and 500 μM of DEET. (C) Germline defects. The frequency of germline morphology defects observed in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1,10, and 100 μM of DEET. (D) RAD-51 foci counting analysis. The number of RAD-51 foci per nucleus quantified in each zone (Z1 to Z7) of the gonad is shown in col-121 and col-121;rad-54 worms exposed to control conditions (DMSO 0.1%) or 100 μM of DEET. Z1-Z2 = premeiotic tip, Z3 = leptotene/zygotene, Z4 = early pachytene, Z5-Z6 = mid-pachytene, and Z7 = late pachytene. (E) Levels of apoptotic nuclei scored in the cep-1 mutant background. Number of acridine-orange-stained nuclei detected in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET in a cep-1 background. (F) ZHP-3 analysis. The number of ZHP-3 foci detected per nucleus is shown in worms exposed to control conditions (DMSO 0.1%) or 100 μM of DEET. (G) Quantification of chromosome morphology defects observed for DAPI-stained bodies in oocytes at diakinesis. Shown are the different categories of chromosomal abnormalities observed in the indicated genotypes and conditions. (H) RNA sequencing. List of the 26 differentially expressed genes detected by RNA-seq analysis. Shown are the number of counts observed in the four biological repeats for worms exposed to control conditions (DMSO 0.1%) or to 100 μM of DEET. (I) Mitotracker intensity. Mitotracker Red (MTR) signal intensity values are shown for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET. (J) Quantitative RT-PCR. Cq values (specific genes indicated) observed in each technical (TecRep) and biological repeat are indicated for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET
References
- 1.Fradin M.S., Day J.F. Comparative efficacy of insect repellents against mosquito bites. N. Engl. J. Med. 2002;347:13–18. doi: 10.1056/NEJMoa011699. [DOI] [PubMed] [Google Scholar]
- 2.Kitchen L.W., Lawrence K.L., Coleman R.E. The role of the United States military in the development of vector control products, including insect repellents, insecticides, and bed nets. J. Vector Ecol. 2009;34:50–61. doi: 10.1111/j.1948-7134.2009.00007.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen-Hussey V., Behrens R., Logan J.G. Assessment of methods used to determine the safety of the topical insect repellent N,N-diethyl-m-toluamide (DEET) Parasites Vectors. 2014;7:173. doi: 10.1186/1756-3305-7-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rivera-Cancel G., Bocioaga D., Hay A.G. Bacterial degradation of N,N-diethyl-m-toluamide (DEET): cloning and heterologous expression of DEET hydrolase. Appl. Environ. Microbiol. 2007;73:3105–3108. doi: 10.1128/AEM.02765-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z.-F., Ying G.-G., Lai H.-J., Chen F., Su H.-C., Liu Y.-S., Peng F.-Q., Zhao J.-L. Determination of biocides in different environmental matrices by use of ultra-high-performance liquid chromatography–tandem mass spectrometry. Anal. Bioanal. Chem. 2012;404:3175–3188. doi: 10.1007/s00216-012-6444-2. [DOI] [PubMed] [Google Scholar]
- 6.Sandstrom M.W., Kolpin D.W., Thurman E.M., Zaugg S.D. Widespread detection of N,N-diethyl-m-toluamide in U.S. streams: comparison with concentrations of pesticides, personal care products, and other organic wastewater compounds. Environ. Toxicol. Chem. 2005;24:1029–1034. doi: 10.1897/04-297r.1. [DOI] [PubMed] [Google Scholar]
- 7.Costanzo S.D., Watkinson A.J., Murby E.J., Kolpin D.W., Sandstrom M.W. Is there a risk associated with the insect repellent DEET (N,N-diethyl-m-toluamide) commonly found in aquatic environments? Sci. Total Environ. 2007;384:214–220. doi: 10.1016/j.scitotenv.2007.05.036. [DOI] [PubMed] [Google Scholar]
- 8.Merel S., Snyder S.A. Critical assessment of the ubiquitous occurrence and fate of the insect repellent N,N-diethyl-m-toluamide in water. Environ. Int. 2016;96:98–117. doi: 10.1016/j.envint.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Selim S., Hartnagel R.E., Jr., Osimitz T.G., Gabriel K.L., Schoenig G.P. Absorption, Metabolism, and Excretion of N,N-Diethyl-m-toluamide Following Dermal Application to Human Volunteers. Fund. Appl. Toxicol. 1995;25:95–100. doi: 10.1006/faat.1995.1043. [DOI] [PubMed] [Google Scholar]
- 10.Smallwood A.W., DeBord K.E., Lowry L.K. N,N’-diethyl-m-toluamide (m-DET): analysis of an insect repellent in human urine and serum by high-performance liquid chromatography. J. Anal. Toxicol. 1992;16:10–13. doi: 10.1093/jat/16.1.10. [DOI] [PubMed] [Google Scholar]
- 11.Barr D.B., Ananth C.V., Yan X., Lashley S., Smulian J.C., Ledoux T.A., Hore P., Robson M.G. Pesticide concentrations in maternal and umbilical cord sera and their relation to birth outcomes in a population of pregnant women and newborns in New Jersey. Sci. Total Environ. 2010;408:790–795. doi: 10.1016/j.scitotenv.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Tian J.-N., Yiin L.-M. Urinary metabolites of DEET after dermal application on child and adult subjects. J. Environ. Health. 2014;76:162–169. [PubMed] [Google Scholar]
- 13.Calafat A.M., Baker S.E., Wong L.-Y., Bishop A.M., Morales-A P., Valentin-Blasini L. Novel exposure biomarkers of N,N-diethyl-m-toluamide (DEET): Data from the 2007-2010 National Health and Nutrition Examination Survey. Environ. Int. 2016;92–93:398–404. doi: 10.1016/j.envint.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal T.R., Mínguez-Alarcón L., Chiu Y.-H., Williams P.L., Nassan F.L., Dadd R., Ospina M., Calafat A.M., Hauser R., Earth Study Team Urinary concentrations of 3-(diethylcarbamoyl)benzoic acid (DCBA), a major metabolite of N,N-diethyl-m-toluamide (DEET) and semen parameters among men attending a fertility center. Hum. Reprod. 2017;32:2532–2539. doi: 10.1093/humrep/dex327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robbins P.J., Cherniack M.G. Review of the biodistribution and toxicity of the insect repellent N,N-diethyl-m-toluamide (DEET) J. Toxicol. Environ. Health. 1986;18:503–525. doi: 10.1080/15287398609530891. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Rahman A., Dechkovskaia A.M., Goldstein L.B., Bullman S.H., Khan W., El-Masry E.M., Abou-Donia M.B. Neurological deficits induced by malathion, DEET, and permethrin, alone or in combination in adult rats. J. Toxicol. Environ. Health. 2004;67:331–356. doi: 10.1080/15287390490273569. [DOI] [PubMed] [Google Scholar]
- 17.Osimitz T.G., Murphy J.V., Fell L.A., Page B. Adverse events associated with the use of insect repellents containing N,N-diethyl-m-toluamide (DEET) Regul. Toxicol. Pharmacol. 2010;56:93–99. doi: 10.1016/j.yrtph.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Legeay S., Clere N., Apaire-Marchais V., Faure S., Lapied B. Unusual modes of action of the repellent DEET in insects highlight some human side effects. Eur. J. Pharmacol. 2018;825:92–98. doi: 10.1016/j.ejphar.2018.02.033. [DOI] [PubMed] [Google Scholar]
- 19.Gleĭberman S.E., Volkova A.P., Nikolaev G.M., Zhukova E.V. [Study of the embrotoxic properties of the repellent diethyltoluamide] Farmakol. Toksikol. (Mosc.) 1975;38:202–205. [PubMed] [Google Scholar]
- 20.Kuhlmann R.S., Caneron R.H., Kolesari G.L., Wu A. N,N-diethyl-meta-toluamide: Embryonic sensitivity. Teratology. 1981;23 [Google Scholar]
- 21.Gleĭberman S.E., Volkova A.P., Nikolaev G.M., Zhukova E.V. [Study of the remote results of the use of repellants. Report 1. Experimental study of the long-term effects of the repellant diethyltoluamide (DETA)] Med. Parazitol. 1976;45:65–69. [PubMed] [Google Scholar]
- 22.Wright D.M., Hardin B.D., Goad P.W., Chrislip D.W. Reproductive and developmental toxicity of N,N-diethyl-m-toluamide in rats. Fund. Appl. Toxicol. 1992;19:33–42. doi: 10.1016/0272-0590(92)90025-d. [DOI] [PubMed] [Google Scholar]
- 23.Schoenig G.P., Neeper-Bradley T.L., Fisher L.C., Hartnagel R.E. Teratologic evaluations of N,N-diethyl-m-toluamide (DEET) in rats and rabbits. Fund. Appl. Toxicol. 1994;23:63–69. doi: 10.1006/faat.1994.1079. [DOI] [PubMed] [Google Scholar]
- 24.Abou-Donia M.B., Suliman H.B., Khan W.A., Abdel-Rahman A.A. Testicular germ-cell apoptosis in stressed rats following combined exposure to pyridostigmine bromide, N,N-diethyl m-toluamide (DEET), and permethrin. J. Toxicol. Environ. Health. 2003;66:57–73. doi: 10.1080/15287390306463. [DOI] [PubMed] [Google Scholar]
- 25.Manikkam M., Tracey R., Guerrero-Bosagna C., Skinner M.K. Pesticide and insect repellent mixture (permethrin and DEET) induces epigenetic transgenerational inheritance of disease and sperm epimutations. Reprod. Toxicol. 2012;34:708–719. doi: 10.1016/j.reprotox.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGready R., Hamilton K.A., Simpson J.A., Cho T., Luxemburger C., Edwards R., Looareesuwan S., White N.J., Nosten F., Lindsay S.W. Safety of the insect repellent N,N-diethyl-M-toluamide (DEET) in pregnancy. Am. J. Trop. Med. Hyg. 2001;65:285–289. doi: 10.4269/ajtmh.2001.65.285. [DOI] [PubMed] [Google Scholar]
- 27.Hunt P.A., Hassold T.J. Human female meiosis: what makes a good egg go bad? Trends Genet. 2008;24:86–93. doi: 10.1016/j.tig.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Nagaoka S.I., Hassold T.J., Hunt P.A. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 2012;13:493–504. doi: 10.1038/nrg3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lui D.Y., Colaiácovo M.P. In: Germ Cell Development in C. elegans. Schedl T., editor. Springer New York; 2013. Meiotic development in Caenorhabditis elegans; pp. 133–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allard P., Kleinstreuer N.C., Knudsen T.B., Colaiácovo M.P. A C. elegans screening platform for the rapid assessment of chemical disruption of germline function. Environ. Health Perspect. 2013;121:717–724. doi: 10.1289/ehp.1206301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hillers K.J. Meiosis. WormBook. 2017:1–43. doi: 10.1895/wormbook.1.178.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shin N., Cuenca L., Karthikraj R., Kannan K., Colaiácovo M.P. Assessing effects of germline exposure to environmental toxicants by high-throughput screening in C. elegans. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1007975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyd W.A., McBride S.J., Rice J.R., Snyder D.W., Freedman J.H. A high-throughput method for assessing chemical toxicity using a Caenorhabditis elegans reproduction assay. Toxicol. Appl. Pharmacol. 2010;245:153–159. doi: 10.1016/j.taap.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Watanabe M., Mitani N., Ishii N., Miki K. A mutation in a cuticle collagen causes hypersensitivity to the endocrine disrupting chemical, bisphenol A, in Caenorhabditis elegans. Mutat. Res. 2005;570:71–80. doi: 10.1016/j.mrfmmm.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Cuenca L., Shin N., Lascarez-Lagunas L.I., Martinez-Garcia M., Nadarajan S., Karthikraj R., Kannan K., Colaiácovo M.P. Environmentally-relevant exposure to diethylhexyl phthalate (DEHP) alters regulation of double-strand break formation and crossover designation leading to germline dysfunction in Caenorhabditis elegans. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1008529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson A.L., Colaiácovo M.P. Exposure to phthalates: germline dysfunction and aneuploidy. Prenat. Diagn. 2021;41:610–619. doi: 10.1002/pd.5921. [DOI] [PubMed] [Google Scholar]
- 37.Kelly K.O., Dernburg A.F., Stanfield G.M., Villeneuve A.M. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics. 2000;156:617–630. doi: 10.1093/genetics/156.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Allard P., Colaiácovo M.P. Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl. Acad. Sci. USA. 2010;107:20405–20410. doi: 10.1073/pnas.1010386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hornos Carneiro M.F., Shin N., Karthikraj R., Barbosa F., Kannan K., Colaiácovo M.P. Antioxidant CoQ10 restores fertility by rescuing bisphenol A-induced oxidative DNA damage in the Caenorhabditis elegans Germline. Genetics. 2020;214:381–395. doi: 10.1534/genetics.119.302939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gumienny T.L., Lambie E., Hartwieg E., Horvitz H.R., Hengartner M.O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 41.Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M.O. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- 42.Lovekamp-Swan T., Davis B.J. Mechanisms of phthalate ester toxicity in the female reproductive system. Environ. Health Perspect. 2003;111:139–145. doi: 10.1289/ehp.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis R.C., Cantonwine D.E., Anzalota Del Toro L.V., Calafat A.M., Valentin-Blasini L., Davis M.D., Baker S.E., Alshawabkeh A.N., Cordero J.F., Meeker J.D. Urinary biomarkers of exposure to insecticides, herbicides, and one insect repellent among pregnant women in Puerto Rico. Environ. Health. 2014;13:97. doi: 10.1186/1476-069X-13-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 45.Colaiácovo M.P., MacQueen A.J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A.M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 46.Mets D.G., Meyer B.J. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139:73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldstein P., Magnano L., Rojo J. Effects of dimethyl sulfone (DMSO2) on early gametogenesis in Caenorhabditis elegans: ultrastructural aberrations and loss of synaptonemal complexes from pachytene nuclei. Reprod. Toxicol. 1992;6:149–159. doi: 10.1016/0890-6238(92)90117-c. [DOI] [PubMed] [Google Scholar]
- 48.Moser S.C., von Elsner S., Büssing I., Alpi A., Schnabel R., Gartner A. Functional dissection of Caenorhabditis elegans CLK-2/TEL2 cell cycle defects during embryogenesis and germline development. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baudat F., Imai Y., de Massy B. Meiotic recombination in mammals: localization and regulation. Nat. Rev. Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 50.Meneely P.M., Farago A.F., Kauffman T.M. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics. 2002;162:1169–1177. doi: 10.1093/genetics/162.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dernburg A.F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A.M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 52.Hollis J.A., Glover M.L., Schlientz A.J., Cahoon C.K., Bowerman B., Wignall S.M., Libuda D.E. Excess crossovers impede faithful meiotic chromosome segregation in C. elegans. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi H., Muruganujan A., Ebert D., Huang X., Thomas P.D. PANTHER version 14: more genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47:D419–D426. doi: 10.1093/nar/gky1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Inoue H., Hisamoto N., An J.H., Oliveira R.P., Nishida E., Blackwell T.K., Matsumoto K. The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev. 2005;19:2278–2283. doi: 10.1101/gad.1324805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hourihan J.M., Moronetti Mazzeo L.E., Fernández-Cárdenas L.P., Blackwell T.K. Cysteine sulfenylation directs IRE-1 to activate the SKN-1/Nrf2 antioxidant response. Mol. Cell. 2016;63:553–566. doi: 10.1016/j.molcel.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Handy D.E., Loscalzo J. Redox regulation of mitochondrial function. Antioxidants Redox Signal. 2012;16:1323–1367. doi: 10.1089/ars.2011.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riddick D.S., Ding X., Wolf C.R., Porter T.D., Pandey A.V., Zhang Q.-Y., Gu J., Finn R.D., Ronseaux S., McLaughlin L.A., et al. NADPH-cytochrome P450 oxidoreductase: roles in physiology, pharmacology, and toxicology. Drug Metab. Dispos. 2013;41:12–23. doi: 10.1124/dmd.112.048991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Manikandan P., Nagini S. Cytochrome P450 Structure, Function and Clinical Significance: A Review. Curr. Drug Targets. 2018;19:38–54. doi: 10.2174/1389450118666170125144557. [DOI] [PubMed] [Google Scholar]
- 61.Das P.C., Cao Y., Rose R.L., Cherrington N., Hodgson E. Enzyme induction and cytotoxicity in human hepatocytes by chlorpyrifos and N,N-diethyl-m-toluamide (DEET) Drug Metabol. Drug Interact. 2008;23:237–260. doi: 10.1515/dmdi.2008.23.3-4.237. [DOI] [PubMed] [Google Scholar]
- 62.O’Driscoll M., Jeggo P.A. The role of double-strand break repair - insights from human genetics. Nat. Rev. Genet. 2006;7:45–54. doi: 10.1038/nrg1746. [DOI] [PubMed] [Google Scholar]
- 63.Abu-Qare A., Abou-Donia M. Increased 8-hydroxy-2’-deoxyguanosine, a biomarker of oxidative DNA damage in rat urine following a single dermal dose of DEET (N, N-diethyl-m-toluamide), and permethrin, alone and in combination. Toxicol. Lett. 2000;117:151–160. doi: 10.1016/s0378-4274(00)00257-5. [DOI] [PubMed] [Google Scholar]
- 64.Dong M.-Q., Venable J.D., Au N., Xu T., Park S.K., Cociorva D., Johnson J.R., Dillin A., Yates J.R. Quantitative mass spectrometry identifies insulin signaling targets in C. elegans. Science. 2007;317:660–663. doi: 10.1126/science.1139952. [DOI] [PubMed] [Google Scholar]
- 65.Murphy C.T., Hu P.J. Insulin/insulin-like growth factor signaling in C. elegans. WormBook. 2013;1–43:1–43. doi: 10.1895/wormbook.1.164.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kaushik N., Rastogi S., Verma S., Pandey D., Halder A., Mukhopadhyay A., Kumar N. Transcriptome Analysis of Insulin Signaling-Associated Transcription Factors in C. elegans Reveal Their Genome-Wide Target Genes Specificity and Complexity. Indian J. Manag. Sci. 2021;22 doi: 10.3390/ijms222212462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Downs J.A., Lowndes N.F., Jackson S.P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature. 2000;408:1001–1004. doi: 10.1038/35050000. [DOI] [PubMed] [Google Scholar]
- 68.Lascarez-Lagunas L.I., Martinez-Garcia M., Nadarajan S., Diaz-Pacheco B.N., Berson E., Colaiácovo M.P. Chromatin landscape, DSB levels, and cKU-70/80 contribute to patterning of meiotic DSB processing along chromosomes in C. elegans. PLoS Genet. 2023;19 doi: 10.1371/journal.pgen.1010627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Norris D., Osley M.A. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell Biol. 1987;7:3473–3481. doi: 10.1128/mcb.7.10.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Norris D., Dunn B., Osley M.A. The effect of histone gene deletions on chromatin structure in Saccharomyces cerevisiae. Science. 1988;242:759–761. doi: 10.1126/science.2847314. [DOI] [PubMed] [Google Scholar]
- 71.Rizki G., Picard C.L., Pereyra C., Lee S.S. Host cell factor 1 inhibits SKN-1 to modulate oxidative stress responses in Caenorhabditis elegans: HCF-1 regulates SKN-1 in oxidative stress response. Aging Cell. 2012;11:717–721. doi: 10.1111/j.1474-9726.2012.00831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baumeister R., Schaffitzel E., Hertweck M. Endocrine signaling in Caenorhabditis elegans controls stress response and longevity. J. Endocrinol. 2006;190:191–202. doi: 10.1677/joe.1.06856. [DOI] [PubMed] [Google Scholar]
- 73.Bowerman B., Eaton B.A., Priess J.R. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68:1061–1075. doi: 10.1016/0092-8674(92)90078-q. [DOI] [PubMed] [Google Scholar]
- 74.Blackwell T.K., Steinbaugh M.J., Hourihan J.M., Ewald C.Y., Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic. Biol. Med. 2015;88:290–301. doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bhalla N., Wynne D.J., Jantsch V., Dernburg A.F. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. j. 2011;17:10. [Google Scholar]
- 80.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.García-Alcalde F., Okonechnikov K., Carbonell J., Cruz L.M., Götz S., Tarazona S., Dopazo J., Meyer T.F., Conesa A. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics. 2012;28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 82.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 83.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Plate phenotyping. Total brood size counts. The total number of fertilized eggs laid, hatched, the number of progeny that reached adulthood, and the frequency of male progeny are shown for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1, 10, 100, and 500 μM of DEET. (B) Apoptotic nuclei count. Number of acridine-orange-stained nuclei detected in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1, 10, 100, and 500 μM of DEET. (C) Germline defects. The frequency of germline morphology defects observed in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 1,10, and 100 μM of DEET. (D) RAD-51 foci counting analysis. The number of RAD-51 foci per nucleus quantified in each zone (Z1 to Z7) of the gonad is shown in col-121 and col-121;rad-54 worms exposed to control conditions (DMSO 0.1%) or 100 μM of DEET. Z1-Z2 = premeiotic tip, Z3 = leptotene/zygotene, Z4 = early pachytene, Z5-Z6 = mid-pachytene, and Z7 = late pachytene. (E) Levels of apoptotic nuclei scored in the cep-1 mutant background. Number of acridine-orange-stained nuclei detected in worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET in a cep-1 background. (F) ZHP-3 analysis. The number of ZHP-3 foci detected per nucleus is shown in worms exposed to control conditions (DMSO 0.1%) or 100 μM of DEET. (G) Quantification of chromosome morphology defects observed for DAPI-stained bodies in oocytes at diakinesis. Shown are the different categories of chromosomal abnormalities observed in the indicated genotypes and conditions. (H) RNA sequencing. List of the 26 differentially expressed genes detected by RNA-seq analysis. Shown are the number of counts observed in the four biological repeats for worms exposed to control conditions (DMSO 0.1%) or to 100 μM of DEET. (I) Mitotracker intensity. Mitotracker Red (MTR) signal intensity values are shown for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET. (J) Quantitative RT-PCR. Cq values (specific genes indicated) observed in each technical (TecRep) and biological repeat are indicated for worms exposed to control conditions (DMSO 0.1%) and worms exposed to 100 μM of DEET
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus76 and are accessible through GEO Series accession number GEO:GSE242682 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE242682).
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.