Abstract
Introduction:
Functional connectivity in the brain is often studied with blood oxygenation level dependent (BOLD) resting state functional magnetic resonance imaging (rsfMRI), but the BOLD signal is several steps removed from neuronal activity. Arterial spin labeling (ASL), particularly pulsed ASL (PASL), has also the capacity to measure the blood-flow changes in response to activity. In this paper, we investigated the feasibility of extracting major brain networks from PASL data, in contrast with rsfMRI analsyis.
Materials and methods:
In this retrospective study, we analyzed a cohort dataset that consists of 21 mild traumatic brain injury (mTBI) patients and 29 healthy controls, which was collected in a previous study. By extracting 10 major brain networks from the data of both PASL and rsfMRI, we contrasted their similarities and differences in the 10 networks extracted from both modalities.
Results:
Our data demonstrated that PASL could be used to extract all 10 major brain networks. Eight out of 10 networks demonstrated over 60 % similarity to rsfMRI data. Meanwhile, there are similar but not identical changes in networks detected between mTBI patients and healthy controls with both modalities. Notably, the PASL-extracted default mode network (DMN), other than the rsfMRI-extracted DMN, includes some regions known to be associated with the DMN in other studies. It demonstrated that PASL data can be analyzed to identify resting state networks with reasonable reliability, even without rsfMRI data.
Conclusion:
Our analysis provides an opportunity to extract functional connectivity information in heritage datasets in which ASL but not BOLD was collected.
1. Introduction
Analysis of functional brain networks or intrinsic connectivity networks (ICNs) has become a popular avenue of investigation in both normal brain research and studies of diseases.1–3 These studies rely heavily on the use of functional magnetic resonance imaging (fMRI) and the blood-oxygenation level dependent (BOLD) effect,4 which allows for visualization of changes in metabolic activity level due to a coupling between activity and blood flow.
In short, as activity increases, local blood flow increases, resulting in an increase in local oxygenation and a decrease in deoxygenated hemoglobin.4 This decrease in deoxyhemoglobin concentration leads to an increase in intensity on echo planar imaging (EPI). When fMRI data is collected in the absence of an explicit task, known as resting state fMRI (rsfMRI), fluctuations in neuronal activity cause changes in the BOLD signal, which can be analyzed to find common patterns of activity changes across brain regions.5–8 Brain regions that vary in activity together are considered to be functionally connected.
However, rsfMRI and the BOLD effect rely on a tight coupling between metabolic activity and cerebral blood flow (CBF), as well as the assumption that the hemodynamic response function (HRF) is consistent across the brain and across study subjects.5 Without these two assumptions, the existence or absence of the temporal dependency between BOLD time courses of brain regions are not directly linked to the temporal dependency between neural activity, and interpretation of the results becomes more difficult. Considering that the HRF has been demonstrated to change across the brain and is known to be affected by some disorders, including traumatic brain injury (TBI),9 rsfMRI and BOLD may not be giving us the entire story about functional connectivity (FC).3–5 As such, several groups have suggested adding additional methods of assessing FC, including arterial spin labeling (ASL), to provide additional information about these changes in FC.10–14
ASL depends on the labeling of inflowing blood, usually at the level of the carotid or internal carotid arteries, followed by detection in the brain, usually with EPI. By interleaving these labeled images with non-labeled images, we can subtract the two to see the blood that has entered the brain in the short delay between the labeling pulse and the imaging.15 Because this is a relatively low signal-to-noise ratio (SNR) method, we perform it multiple times and average the results together to obtain a less noisy result with better SNR. Like with rsfMRI and the BOLD effect, when done repeatedly, this produces some slight changes in intensity over the series of images that are associated with changes in CBF. Because of this similarity, there has been an enduring but not remarkably popular interest in analyzing ASL data with the same methods as rsfMRI, such as independent component analysis (ICA) or seed-based analyses (SBA), either alone or in combination with the rsfMRI results.10–14 By combining the ASL and rsfMRI data information, we can use the complementary information that they provide to obtain information about ICNs that is perhaps more reliable or interpretable than either method alone would offer. Additionally, rsfMRI is a relatively recent method, and while it has caught on quite quickly and enjoys widespread application leading to large datasets in a variety of studies, there are still many existing datasets that included ASL in their imaging protocol even before rsfMRI was widely used. By analyzing this ASL data alone, we can obtain information about ICNs in these datasets where rsfMRI was not collected. Finally, ASL also offers another piece of information that rsfMRI does not, which is a map of the static CBF across the brain. Since CBF is also often found to be altered in many diseases or disorders, combining the result of CBF and functional connectivity analyses could prove valuable.
Recent advances in ASL have made this an even more attractive possibility, as pseudocontinuous ASL (PCASL) offers improvements over both continuous and pulsed ASL methods (CASL and PASL), with good labeling efficiency, easy applicability on most scanners, and good SNR.16,17 This led to a recent push in ASL-based assessments of FC in healthy subjects, supporting the use of ASL to identify ICNs. However, despite the lesser SNR of PASL, it was much more widely used in the time before PCASL and rsfMRI both became standard research practices, meaning that many older datasets that would benefit from the use of ASL to assess FC would only have PASL data available to them. This augments the importance of studying the feasibility of functional connectivity analyses using PASL data. To better demonstrate the importance of evaluating brain FC in ASL data, it is of interest to examine PASL functional connectivity analysis for a clinical study.
The proposed method of ASL FC analysis and the information obtained through using it could be of particular importance in the study of mild traumatic brain injury (mTBI), which affects over 1.3 million Americans each year.18 Patients with mTBI can experience symptoms in the absence of structural brain abnormalities, which are often hypothesized to be related to changes in metabolic activity or functional connectivity between brain regions,19,20 particularly in the default mode network (DMN).21–23 Because PASL has been a standard part of mTBI imaging studies for some time but rsfMRI is a newer development, PASL in existing datasets may be able to be examined in new ways to provide information about FC changes following mTBI using appropriate analysis methods.
Here, we investigate the use of PASL and rsfMRI both separately and together in one study of healthy controls and mTBI patients. Our major aims were: 1) to investigate the reliability of intrinsic brain networks extracted from PASL in contrast with those extracted from rsfMRI data; 2) to evaluate the possibility of using a recently-developed approach, group information guided ICA (GIG-ICA) as the standard tool, to calculate brain networks of individuals using PASL data24; and 3) to evaluate the feasibility of performing connectivity analysis using PASL data for clinical studies.
2. Material and methods
2.1. Approach
First, to address the first aim, we demonstrate that ASL data can yield similar ICNs to those obtained using rsfMRI data by applying group ICA (GICA) on the PASL and rsfMRI data separately and comparing their results. This provides a foundation for future research in analyzing brain intrinsic connectivity using PASL data. Next, we assessed our proposition of considering GIG-ICA as the standard ICA approach for analyzing brain intrinsic connectivity in ASL datasets. Finally, we used GIG-ICA to calculate the subject-level ICNs for each modality and evaluated the feasibility of using ICNs obtained from PASL in clinical studies by comparing the PASL results with the rsfMRI results in an mTBI data set. We expected that the PASL results would confirm the rsfMRI findings and add new information, as well.
2.2. Participants
This study was approved by both the Human Investigation Committee of Wayne State University and the Institutional Review Board of the Detroit Medical Center, Detroit, Michigan, USA. The cohort included 50 subjects including 29 healthy subjects and 21 mTBI patients (Table 1). All participants were at least 18 years old and written informed consent was collected before enrollment. Participant were excluded for the following criteria: pregnancy, a history of brain injury, neurological disorder or psychoactive medication, and/or a history of substance or alcohol abuse.
Table 1.
Demographic characteristics of patients and healthy controls enrolled in the study.
| Patients | Controls | p-value | ||
|---|---|---|---|---|
| Age | Mean, years (SD) | 39.42 (15.10) | 29.71 (8.70) | 0.013 |
| Median, years | 43.11 | 27.54 | ||
| Range, years | 19.24–63.42 | 19.39–50.57 | ||
| Sex | Male (%) | 15 (0.71) | 20 (0.69) | 0.851 |
| Race | African American | 17 | 10 | 0.005 |
| White | 3 | 12 | ||
| Other | 1 | 7 | ||
| Delay to scan | Mean, hours (SD) | 83:57 (4.99) | NA | |
| Median, hours | 10:15 | NA | ||
| Range, hours | 4:00–446:00 | NA |
All patients with mTBI were recruited from the Detroit Receiving Hospital (DRH) Emergency Department (ED), a Level-I trauma center. Patient eligibility was based on the mTBI definition by the American Congress of Rehabilitation Medicine.25 All patients had an initial Glasgow Coma Scale (GCS) score of 13–15 in the ED, and patients with a GCS of 15 had either loss of consciousness, post-traumatic amnesia, or a change in mental status. Loss of consciousness did not exceed 30 min and post-traumatic amnesia did not exceed 24 h. All patients had a CT scan as part of their clinical evaluation. All of them were able to speak English.
2.3. Data acquisition
All images were acquired using a 3T Siemens Verio scanner (Siemens, Munich, Germany). Structural high-resolution T1-weighted imaging was collected using an MPRAGE sequence with TR (repetition time) = 1950 ms, TE (echo time) = 2.26 ms, slice thickness = 1 mm, flip angle (FA) = 9°, field of view (FoV) = 256 × 256 mm, matrix size = 256 × 256, and voxel size = 1 mm isotropic. The rsfMRI images from this dataset have already been used for connectome-scale analyses.26 This paper focuses on the use of PASL data to extract ICNs. rsfMRI data was acquired using a gradient echo EPI sequence with in plane resolution = 3.125 × 3.125 mm, slice thickness = 3.5 mm, slice gap = 0.595 mm, matrix size = 64 × 64, TR/TE = 2000/30 ms, FA = 90°, 240 vol, number of excitations (NEX) = 1, and an acquisition time of 8 min. During rsfMRI scans, participants were instructed to relax, keep their eyes closed, avoid falling asleep, and not to think about anything specific. PASL data was collected using a gradient echo EPI sequence with 91 vol, TR/TE = 2830.2/11 ms, FA = 90°, FoV = 96 × 96 mm, and voxel size = 3.125 × 3.125 × 3.5 mm3 for the first 12 healthy controls and 10 patients, and with 101 vol, TR/TE = 2612.8/13 ms, FA = 90°, FoV = 80 × 80 mm, and voxel size = 44 mm isotropic for the other 17 healthy controls and 11 patients.
2.4. Data analysis
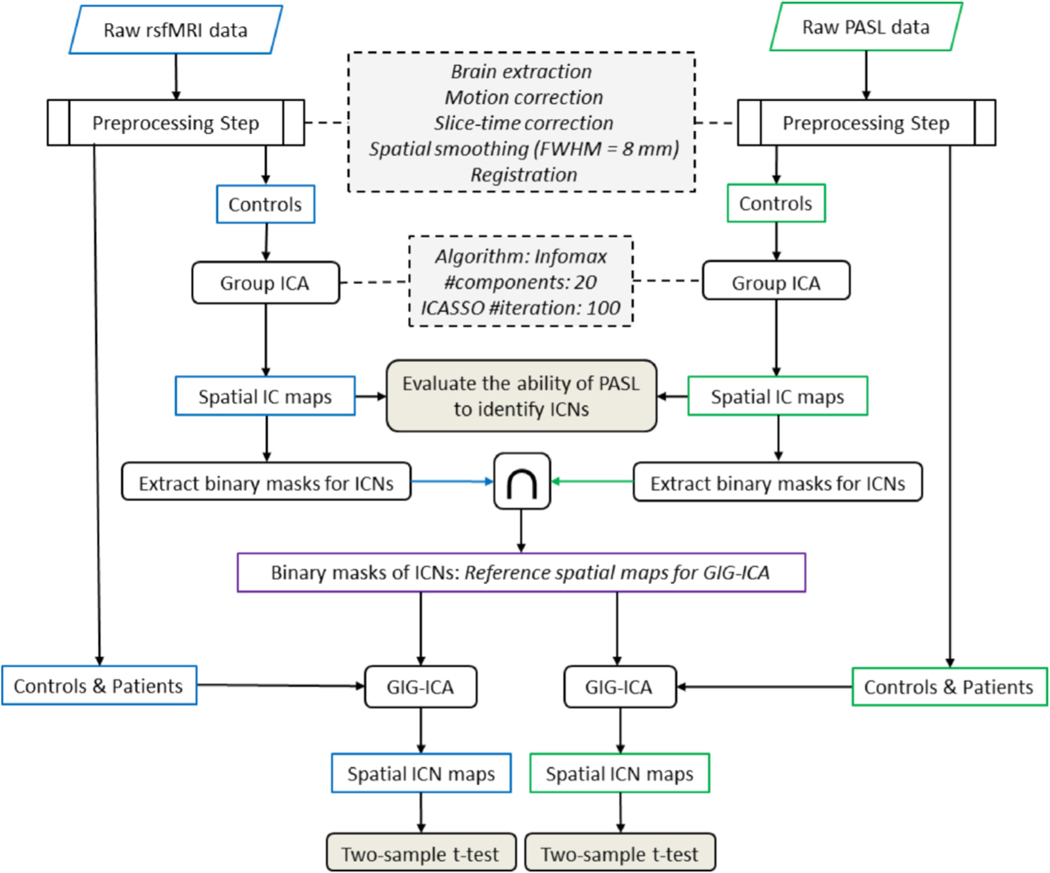
The schematic of the functional connectivity analysis pipeline is demonstrated in Fig. 1 and described below.
Fig. 1.
A schematic of the analysis pipeline. Both rsfMRI and PASL data were preprocessed and group independent component analysis (ICA) was applied using the infomax algorithm to evaluate the ability of PASL data to identify intrinsic connectivity networks (ICNs). Common regions of interest between two modalities were identified for each ICN and used as input for group information guided ICA (GIG-ICA) to obtain each individual’s ICNs. Default mode networks were compared between healthy subjects and patients using a two-sample t-test for each modality and the results were compared between modalities.
2.4.1. rsfMRI and PASL preprocessing
Data were preprocessed using the FSL software package (www.fmrib.ox.ac.uk/fsl/) including brain extraction, motion correction, slice-time correction, spatial smoothing with an 8 mm full width at half-maximum (FWHM = 8 mm). For registration, the structural T1-weighted data was registered to the Montreal Neurological Institute (MNI) standard space using nonlinear registration with 10 mm warp resolution. rsfMRI and PASL data were registered to the MNI standard space and resampled to 4 mm isotropic voxel size. This involved first registering each to the participant’s own T1-weighted image using linear registration with a 6 degrees of freedom and then performing the nonlinear registration from the T1-weighted image space to the MNI atlas.
2.4.2. ICA analysis
Group ICA analysis was performed using the GIFT software package from MIALAB (http://mialab.mrn.org/software/gift/).27,28 For group ICA, ICASSO was used to obtain reliable and stable independent components,29 and Infomax algorithm with 20 components was run 100 times with different initial values and different bootstrapped datasets and the central solution was used for further analysis.30–32 Group ICA was performed on the data of each modality separately to allow examination of the ability of PASL to extract ICNs. Independent components obtained from PASL were compared with ICNs identified using rsfMRI analysis. For comparative analyses, we avoided combining BOLD and ASL (i.e. temporal concatenation) to obtain corresponding ICNs across modalities. This prevented the information of one modality from influencing the other. A reference-based back-reconstruction approach within GIFT was utilized for individual-level ICNs. This allows the ICNs of an individual to be independent of the data of other individuals in the study which is an important concern for clinical studies, especially with a small number of subjects.33 For this purpose, we used the recently developed approach known as group information guided ICA (GIG-ICA),24,34 in which a multi-objective optimization solver is utilized to calculate independent components using a set of spatial maps as a reference. GIG-ICA has shown higher spatial and temporal accuracy than alternative back-reconstruction approaches such as spatiotemporal (dual) regression35 and has also been shown to be more robust to artifacts than single-subject ICA approaches.36
2.4.3. PASL and ICNs
While recent studies use more advanced ASL approaches such as PCASL, which offers better temporal resolution and higher SNR,16,17 this study used PASL data to extract ICNs to evaluate the feasibility of using multivariate approaches like ICA in ASL data with lower qualities. For our first aim, a blind group ICA (infomax) was applied to each modality separately to investigate the possibility of obtaining brain networks using PASL. The necessity of using blind ICA for this step is that blind ICA identifies ICNs solely based on the information which exists in the data. Thus, we prevent information of one modality (for example, BOLD) from influencing the calculation of the IC maps of another modality (for example, ASL). The rsfMRI-ICNs, which were obtained from BOLD information of rsfMRI, were identified manually by an expert (AI). The PASL-ICNs, which were obtained using both BOLD and perfusion information of PASL, were identified both manually and by identifying independent components of PASL which have maximum spatial similarity with the identified rsfMRI-ICNs. The time-courses of the ICs identified as ICNs in both modalities are dominated by low-frequency power, which was reported as the dominant frequency band for resting state neural activities.6 The results of two modalities were compared to see how well we can identify ICNs using PASL data.
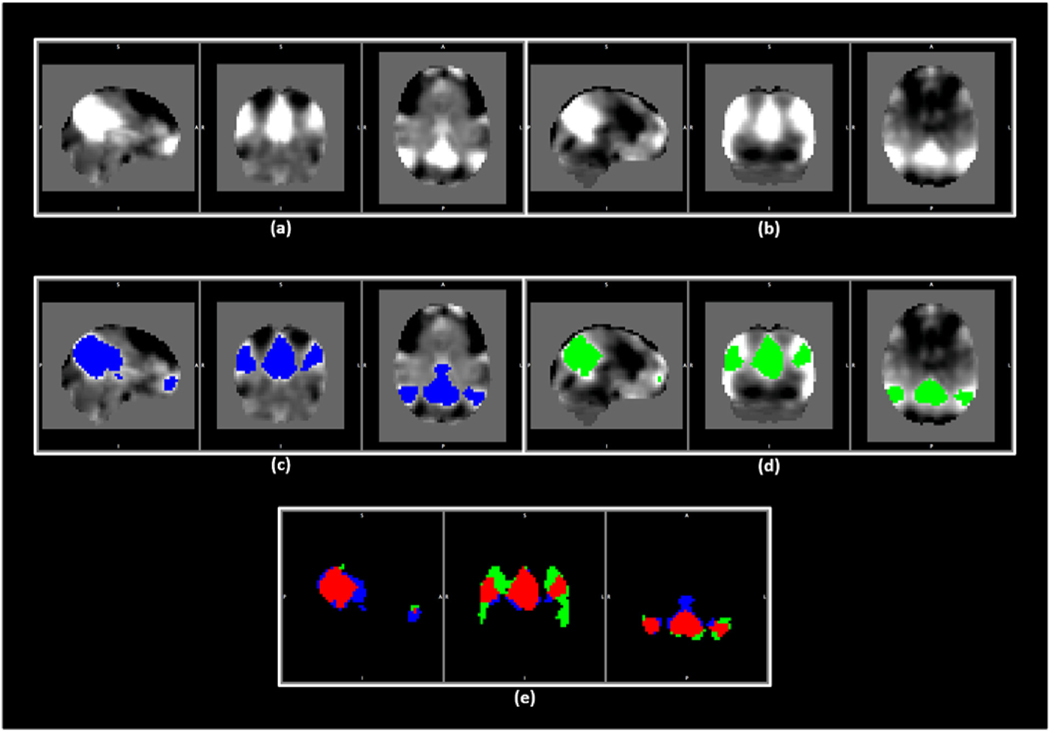
After demonstrating the ability of obtaining brain networks (i.e., ICNs) from PASL data, our next goal was to compare the results of analysis between two modalities. To accomplish this, we first identified corresponding spatial maps across modalities. This can be obtained by using GIG-ICA with the same reference for both modalities. The spatial maps chosen for use as a reference should resemble the ICNs that we expect to exist in the investigated data of both modalities but without being biased toward one of the modalities over the other. This can be achieved by selecting corresponding ICNs in both modalities, using the results of the previous ICA step, and creating regions of interest (ROIs) which are associated with each ICN in both modalities (See an example for the DMN in Fig. 2). In other words, after identifying the ICNs for each modality at the first step, we identify which voxels are significantly associated with a given ICN by thresholding spatial t-value maps and then select those voxels which are significantly associated with the ICN in both modalities, or the statistically significant overlap between the ICN maps in both modalities, as our ROIs. This produces a set of binary spatial masks that represent regions associated with each given ICN in both modalities. Next, we applied GIG-ICA using this conjoined reference to obtain corresponding ICNs in both modalities, and the obtained ICNs were used for further statistical analysis to compare the findings of two modalities. Assuming the results of the two modalities are well-aligned with each other, and that PASL produces similar findings as rsfMRI, we can move forward by evaluating the result of performing ICA analysis on PASL separately from rsfMRI, using the group IC spatial maps of each modality as the reference for that modality.
Fig. 2.
An example for the process of obtaining the same set of spatial maps to be used as the reference for GIG-ICA in both modalities. (a) and (b) show the default mode network (DMN) for rsfMRI and PASL data. (c) and (d) demonstrate the areas in blue and green associated with the DMN in rsfMRI and PASL data, respectively. (e) demonstrates these areas from (c) and (d) with the overlap indicated in red. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The next goal after demonstrating the ability of PASL to extract ICNs is to assess the performance of functional connectivity analysis using PASL data for clinical studies. The group comparison was performed between mTBI patients and healthy controls for both modalities, and their findings were compared. We first used the conjoined reference (the binary masks of 10 ICNs) and GIG-ICA to obtain the DMN and compare between two groups for each modality (see section GIG-ICA with conjoined reference findings). After demonstrating similar findings for both modalities using the same reference of GIG-ICA, we evaluated the similarity of the findings of two modalities when they were studied separately. For this purpose, the 20 IC maps obtained from each modality’s group ICA was used as reference for GIG-ICA and the result of two modalities compared with each other (see section GIG-ICA with independent references findings).
2.4.4. PASL on CBF-based perfusion images and ICNs
While our main aim in this study was to demonstrate the reliability of PASL data for FC analysis and to propose an analysis procedure for this purpose, it is also interesting to obtain ICNs from CBF-specific maps (perfusion-weighted ICNs) as well as the raw time series images. Although it can be advantageous to use the entire time series for given the lower SNR, lower temporal resolution, and fewer volumes available than the fMRI, since it provides more information, it is also important to demonstrate that we can obtain ICNS using only CBF-based (perfusion-weighted, or post-subtraction) data, too, because it is unique information which cannot be obtained using conventional fMRI modalities. Another benefit is that compared to BOLD ICNs, perfusion-weighted ICNs provide more direct measures of the physiology and metabolism of ICNs.
Therefore, even though obtaining CBF-based ICNs is not our main goal, we are going to demonstrate the feasibility of obtaining ICNs using perfusion-weighted, or subtracted, data. Using CBF-based measurements (label image minus control image), we minimize the contribution of BOLD information in identification of the ICNs, resulting in ICNs which are more closely related to neural activity. The CBF-based ICNs here were obtained by performing ICA analysis on perfusion-weighted images calculated using sinc-subtraction of label and control images. The previous study has been shown that sinc-subtraction minimizes the BOLD effect within ASL data as compared to other subtraction methods.37
2.4.5. Functional network connectivity analysis (FNC)
Finally, evaluating the temporal coherence between ICNs has raised much interest and become a popular approach to assess both static and dynamic functional connectivity of the brain. In this approach, FC is calculated at the network level and has demonstrated promising results among various brain disorders like schizophrenia and bipolar disorder. Considering the importance of FNC analysis, it is valuable to assess the suitability of PASL data for FNC analysis. We hypothesize that, similar to FNC obtained from rsfMRI data, FNC obtained from PASL data will reveal a specific pattern across healthy individuals (significantly different from correlations between uncorrelated time series). In our data, the FNC was calculated among 10 identified ICNs using a Pearson correlation and resulted in a symmetric 10 × 10 matrix which includes 45 distinct values. To evaluate our hypothesis, we created an empirical null distribution of rsfMRI-FNC. For this purpose, we used a resampling technique and created 5000 samples from time courses of rsfMRI-ICNs of 29 healthy subjects. For each sample, the time courses of ICNs were randomly assigned from different subjects. For instance, for sample i, the time course of the default mode network (DMN) was assigned from the time course of the DMN of the subject m, and the time course of the dorsal attention network (DAN) was assigned from the time course of the DAN of the subject n. In other words, the time courses of different ICNs in each sample come from different subjects. Next, we calculated the FNC for these 5000 samples and used the results to create an empirical null distribution for rsfMRI-FNC.
2.4.6. Statistical analysis
Spatial similarity was measured using spatial correlation (Equation (2)) to identify ICNs of PASL data based on similarity to rsfMRI-ICNs.33,38 Corresponding maps were identified between the two analyses based on the highest spatial similarity, and this pairing was compared with the result of manually selecting paired ICNs.
| (2) |
where X and Y are spatial maps, and i is an index for corresponding voxels in the spatial maps.
A non-parametric two-sample t-test was performed to compare mTBI patients and healthy control subjects in each modality. The significant p-value was chosen to be 0.05, and the statistical maps were corrected using spatial thresholding. The size of spatial (cluster) thresholding was obtained using the MonteCarlo simulation, part of Resting-State fMRI Data Analysis Toolkit (http://www.restfmri.net/), which is equal to 2.5 cm3.33
3. Results
3.1. Participants
Demographic characteristics are presented in Table 1. The dataset includes 21 mTBI patients (6 female and 15 male) and 29 healthy control subjects (9 female and 20 male). There is no significant sex difference between two groups. However, there is a significant age difference (two sample t-test, p-value = 0.013) between the two groups. For mTBI patients, the mean age is 39.42 ± 15.10 (mean ± standard deviation) years, and for healthy control subjects it is 29.71 ± 8.70 years. Therefore, we considered the effect of age in our statistical analyses when we compared the data of the two groups.
mTBI patients and healthy controls were also statistically significantly different in race in our sample (p value = 0.005). However, previous studies reported similar patterns of ICNs, particularly in the DMN, despite different race,39 so the effect of race is usually not considered in intrinsic functional connectivity analyses, especially in studies with a small sample size.26 Moreover, to the best of our knowledge, there is no rsfMRI study that reports a significant race effect on FC.
3.2. PASL can identify similar (but not identical) networks as rsfMRI
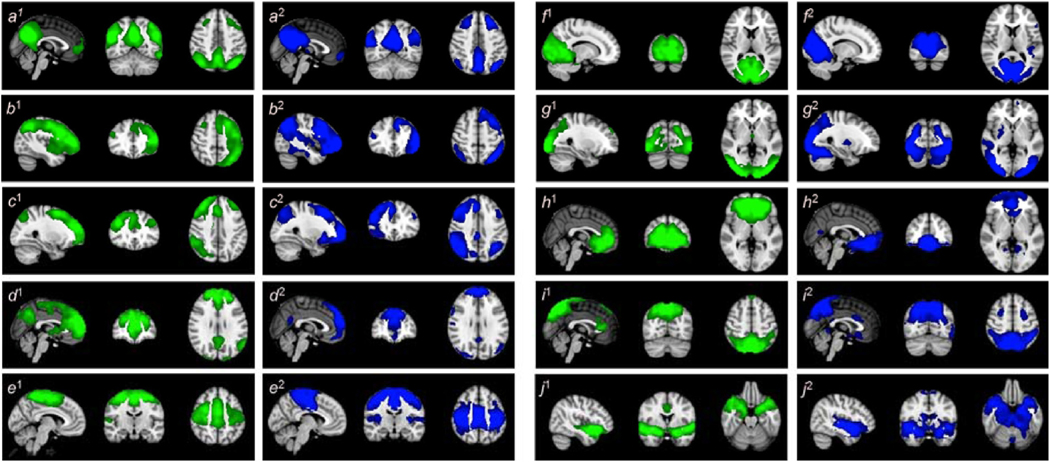
For the first step of the analysis, the data from PASL and rsfMRI sequences from the 29 healthy control subjects were analyzed using infomax separately in each modality. Ten ICNs were identified in both modalities and illustrated in Fig. 3, in which blue and green regions shows the areas associated to the ICNs in rsfMRI and PASL data, respectively.
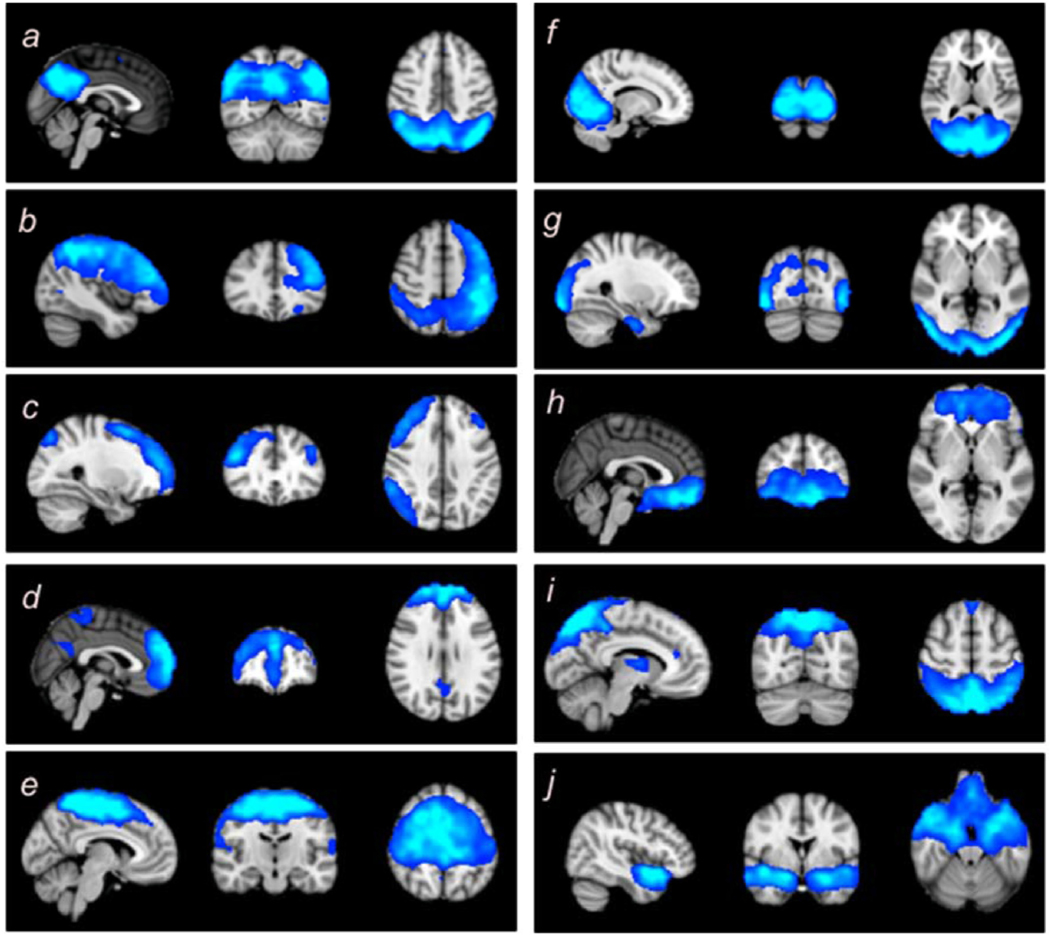
Fig. 3.
Spatial maps identified for each modality. The figures with the superscript of 1 demonstrate the brain regions associated with various networks in PASL data in a green overlay, and the figures with the superscript of 2 demonstrate the brain regions associated with various networks in rsfMRI data in a blue overlay. The identified ICNs include (a) the default mode network (DMN), (b) the left parietal–frontal network (LPFN), (c) the right parietal–frontal network (RPFN), (d) the frontal default mode network (fDMN), (e) the sensorimotor network (SMN), (f) the primary visual network (VisPri), (g) the secondary visual network (VisSec), (h) the subcallosal network (SubN), (i) the dorsal attention network (DAN), and (j) the insula functional network (IFN). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The 10 ICNs identified include (1) the default mode network (DMN),5–8,40–44 (2) and (3) the left and right frontoparietal networks (LPFN and RPFN),5–8,40–42,44 (4) the frontal default mode network (fDMN),5–7,41,42,45,46 (5) the sensorimotor network (SMN),7,8,41–44,47 (6) the primary visual network (VisPri),5–8,40–44 (7) the secondary visual network (VisSec),6–8,40,41,43,45 (8) the subcallosal network (SubN),7,39,44,48,49 (9) the dorsal attention network (DAN),5,44 and (10) the insula functional network (IFN).50,51 This demonstrates that ICNs can be obtained from PASL data, and that they are very similar to the commonly-identified rsfMRI-ICNs, except for DAN and SubN. Quantitatively, the spatial correlation between ICN maps from the two modalities demonstrates a high level of similarity between the ICNs obtained from the two modalities (Table 2).
Table 2.
Spatial similarities (spatial correlation %) between similar ICNs of the two modalities.
| ICNs | DMN | LPFN | RPFN | fDMN | SMN | VisPri | VisSec | SubN | DAN | IFN |
|---|---|---|---|---|---|---|---|---|---|---|
| Spatial Similarity (%) | 75.19 | 63.84 | 59.16 | 61.41 | 82.17 | 85.31 | 63.50 | 52.13 | 46.81 | 59.06 |
3.3. Comparing the results of analysis between two modalities
In the previous step, we illustrated the feasibility of extracting similar ICNs from PASL as from rsfMRI. The next step was to compare and evaluate the result of a between-groups analysis between these two modalities. We expected to observe similar findings that are confirmatory and complementary to each other from the two modalities. For instance, if we investigate the effect of a specific brain disorder using both modalities, we would expect that both modalities would show a similar trend of alterations across regions (the findings of each modality would confirm the findings of the other). Moreover, we expected to see some extra findings in each modality that are unnoticeable or invisible in the other but are not opposed to the findings of the other. For this purpose, we compared the DMN between healthy control subjects and mTBI patients and compared these findings between the two modalities.
To achieve this goal and to perform fair comparisons, we first used the conjoined reference which is the binary masks of 10 ICNs and GIG-ICA to identify corresponding spatial maps across modalities. Using the conjoined reference, insures that the references for the GIG-ICA would have equal contribution from each modality, and the obtained ICNs (including the DMN that we investigated here) would not be biased towards one or the other.
3.3.1. GIG-ICA with conjoined reference findings
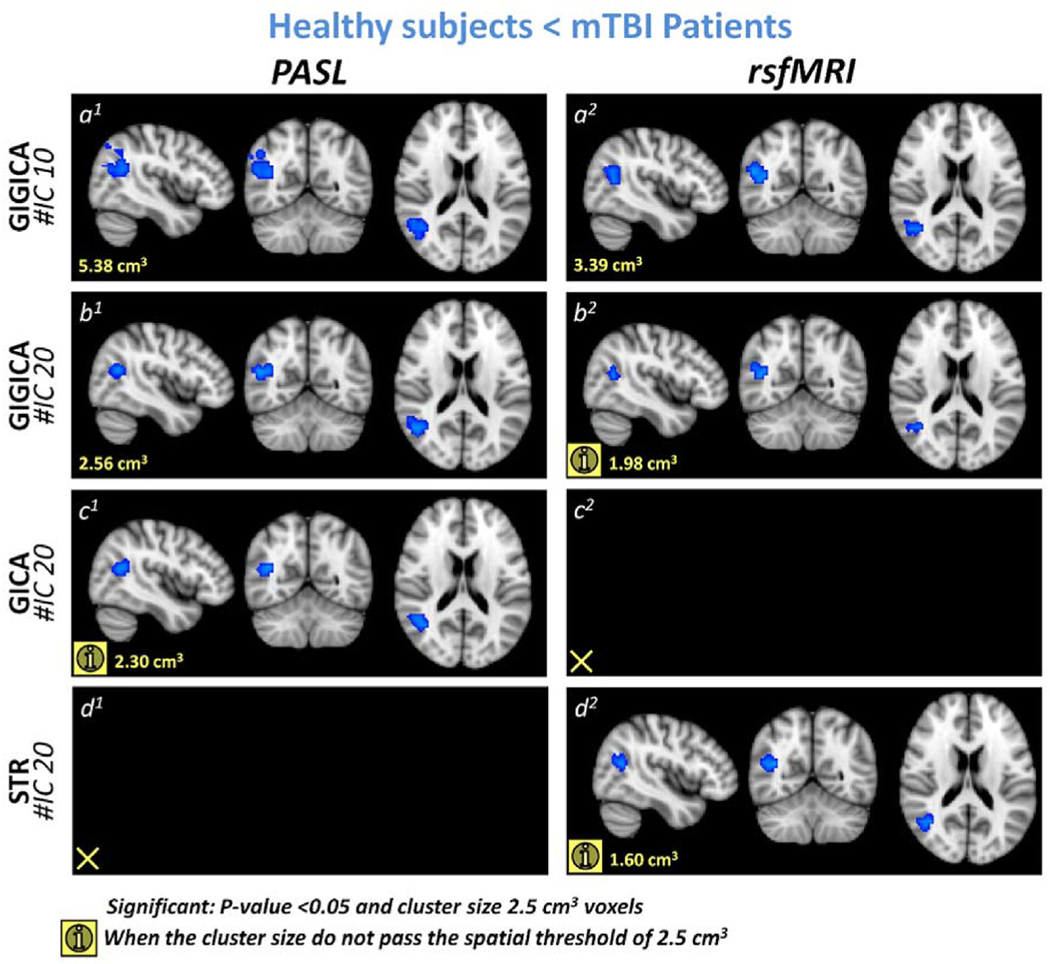
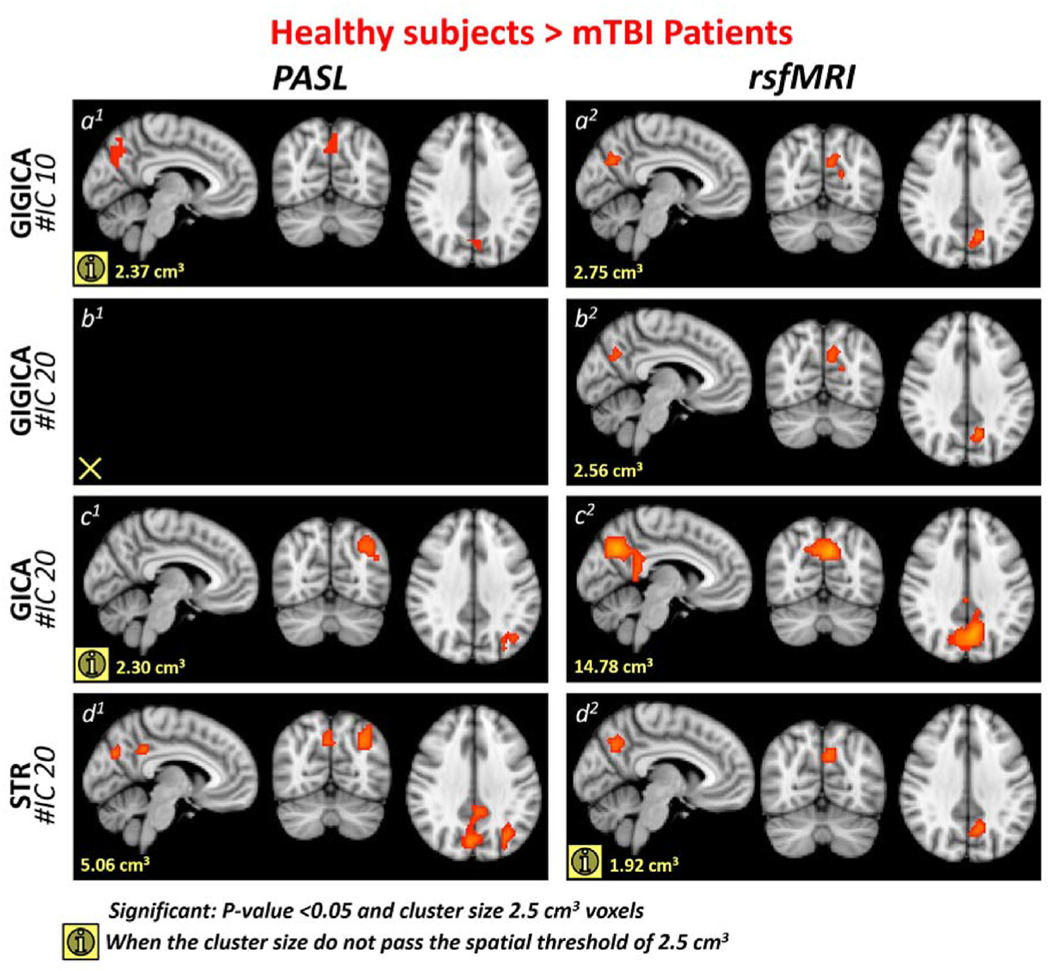
For each individual subject, ICNs were extracted using GIG-ICA and the conjoined reference. The DMN was compared between the two groups using a non-parametric two sample t-test while considering age as a confounding factor. The analysis revealed clusters of 5.38 cm3 and 3.39 cm3 that showed statistically significant increases in association of the angular gyrus with the DMN in both PASL and rsfMRI data, respectively, in mTBI patients as compared to healthy control subjects (Fig. 4(a)1 and a2). This demonstrates that the two modalities can identify similar findings. Moreover, rsfMRI data manifested a reduced association of the precuneus with the DMN for a 2.75 cm3 cluster (Fig. 5(a)2). We observed a reduced association of the precuneus with the DMN in PASL data in a 2.37 cm3 cluster (Fig. 5a1), which did not survive the spatial threshold of 2.50 cm3. In other words, while we observed a similar trend of reduced association of the precuneus with the DMN, it was not statistically significant in the PASL data. In summary, for GIG-ICA with the same reference, our analysis demonstrated similar findings for PASL and rsfMRI data in the DMN.
Fig. 4.
Blue overlay represents the regions that show higher association with the default mode network (DMN) in mTBI patients compared to healthy subjects. (a) shows the results of using the conjoined reference (the binary masks of 10 ICNs) and GIG-ICA (conjoined reference). (b) demonstrates the result of using the 20 IC maps obtained from each modality’s group ICA as the reference to perform GIG-ICA (independent references). (c) and (d) illustrate the results of using two back reconstruction approaches, GICA and spatial-temporal regression (STR), respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 5.
Red overlay represents the regions that show lower association with the default mode network (DMN) in mTBI patients compared to healthy subjects. (a) (and (b) present the results of GIG-ICA using two different references. (a) GIG-ICA with conjoined reference, in which the reference is the binary masks of 10 ICNs. (b)GIG-ICA with independent references, in which the reference was obtained from performing group ICA (infomax) with 20 components on each modality. (c) and (d) illustrate the results of GICA and spatial-temporal regression (STR), respectively, the two most used back reconstruction approaches,. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3.2. GIG-ICA with independent references findings (confirmatory)
After demonstrating similar findings for both modalities in a corresponding network, the next question is how similar the findings of the two modalities are when they are investigated separately. For this step, we again used GIG-ICA, which allows comparison of the findings here with the findings of the previous step; however, we used the 20 IC maps obtained from each modality’s group ICA as our reference rather than the ROIs used as the reference in the previous step, so that each modality’s result is independent from the data of the other modality. Similar findings were observed in the angular gyrus as in the previous GIG-ICA analysis using the overlapping ROI reference. For the PASL data, we observed a significant increase in association of the angular gyrus with the DMN in a 2.56 cm3 cluster (Fig. 4b1). For the rsfMRI, the cluster in the angular gyrus that showed increased association with the DMN was only 1.98 cm3, which did not pass the spatial threshold (2.50 cm3) but clearly shows that a similar pattern exists (Fig. 4b2). We also observed a similar trend in increased association of the angular gyrus with the DMN in PASL data when we used GICA and in rsfMRI when we used spatial-temporal regression approach (STR).52 This demonstrates that PASL can identify changes in brain FC either along with or in the absence of rsfMRI. We also observed a similar pattern of decreased functional association of the precuneus with the DMN in TBI patients as we observed in the previous GIG-ICA step for rsfMRI data with a 2.56 cm3 cluster (Fig. 5b2), but this decrease in association between the DMN and the precuneus was not observed in the PASL data even as a cluster that was too small to meet the statistically significant threshold.
The results of these two steps reveal that rsfMRI and PASL complement each other and could be used together. Moreover, these results demonstrate that the increase in association of the angular gyrus with the DMN could be considered as a more significant effect of injury and more robustly associated with the effect of injury than the decrease in association between the precuneus and the DMN.
3.3.3. Back reconstruction approaches (GICA and spatial-temporal regression) did not show consistent results between PASL and rsfMRI in the DMN
We further evaluated the findings of our statistical comparisons of the DMN using two common back reconstruction approaches: GICA and spatial-temporal regression (STR).51 We did not observe a statistically significant increase in the association of the angular gyrus with the DMN using either of these modalities. However, if we reduce the spatial threshold value to less than 2.50 cm3, both of these approaches captured a similar pattern in one of the two modalities, which demonstrates GICA’s and STR’s lower sensitivity to detect this change as compared to GIG--ICA.24,35 GICA was able to identify a similar trend of increased functional association of the angular gyrus with the DMN only in PASL data with a cluster size of 2.30 cm3 (Fig. 4c1). By using STR, we observed a similar trend in only rsfMRI data for a cluster with the size of 1.60 cm3 (Fig. 4d2). We also observed decreased associations between the precuneus, PCC, and lateral occipital cortex with the DMN across approaches and modalities. However, the most consistent finding in the decreased association with the DMN is the precuneus, which was observed using GIG-ICA. GIG-ICA reveals decreased association of the precuneus with the DMN, which was a stronger effect in the rsfMRI data than in the PASL data. GICA demonstrated a similar decrease in association of the precuneus and PCC with the DMN in the rsfMRI data in a much larger cluster than in the GIG-ICA (cluster size 14.78 cm3) (Fig. 5c2), but it did not demonstrate the same effect in the PASL data; instead it showed a non-significant trend of an decrease in functional association between the lateral occipital cortex and the DMN which did not pass the statistical comparison spatial criteria (cluster size 2.30 cm3) (Fig. 5c1). For the STR approach, we did not observe the decrease in association of the precuneus with the DMN for the statistical criteria in the rsfMRI data, but we did observe the similar pattern for a cluster with the size of 1.92 cm3 (Fig. 5d2). Unlike the GICA in the PASL data, the STR showed a decrease in the association of the precuneus, PCC, and lateral occipital cortex in two clusters with a total size of 5.06 cm3 (Fig. 5d1).
3.4. CBF-based (perfusion-based) ICNs
The CBF-based ICNs were also obtained by performing ICA analysis, using infomax, on perfusion-weighted images calculated using sinc-subtraction of label and control images. The previous study has been shown that sinc-subtraction minimizes the BOLD effect within ASL data as compared to other subtraction methods.37 Fig. 6 demonstrates the ICA results generated using the CBF-based images. We were able to identify the same 10 ICNs from perfusion-weighted images that we already observed in the rsfMRI and PASL data. ICNs obtained from BOLD-contaminated images and CBF-based images, which should be less susceptible to the BOLD effect, were not compared here.
Fig. 6.
The 10 ICNs obtained from the perfusion-weighted images.
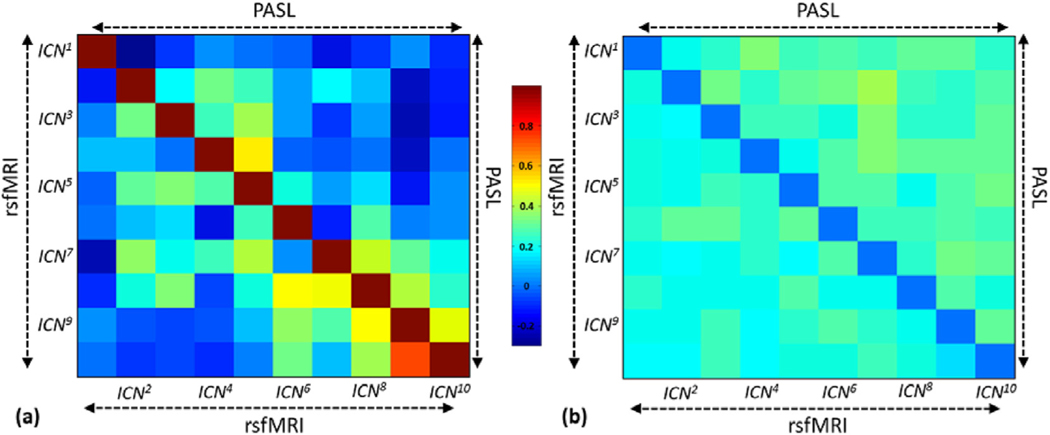
3.5. Functional network connectivity (FNC) comparison between PASLand rsfMRI-derived networks
Fig. 7 (a) and (b), respectively, show the averages and standard deviations of FNC calculated for the PASL data (upper triangle) and rsfMRI data (lower triangle). This presentation allows a better comparison between two modalities. Fig. 7(a) shows a similar FNC pattern between the two modalities. However, there are some differences which can be due to the different sources of ICNs in each modality and also different SNR and number of volumes between modalities. The effect of a lower number of volumes and lower SNR in the PASL data were also observed as increases in standard deviations in Fig. 7(b), which shows higher variations in FNC in the PASL data than in the rsfMRI data.
Fig. 7.
Average and standard deviation of FNC calculated in rsfMRI (lower triangle) and PASL (higher triangle). Similar FNC patterns were observed in both modalities, demonstrating that PASL can be used to track whole-brain functional network connectivity patterns.
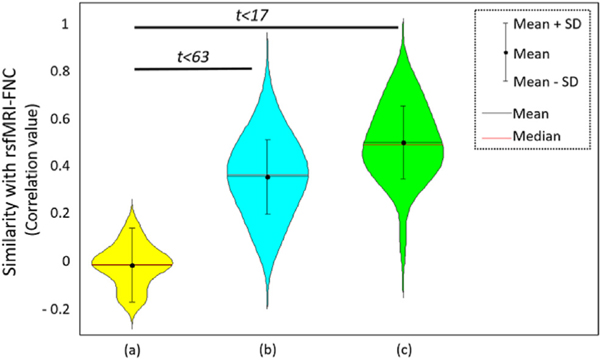
In order to quantify our FNC analysis and assess our hypothesis, we evaluated the similarity between the FNC derived from PASL and rsfMRI data. Our hypothesis was that the similarity FNC obtained from PASL (PASL-FNC) and rsfMRI (rsfMRI-FNC) is statistically significant. The results show significant similarity between FNC of PASL and rsfMRI both within and between subjects (Fig. 8). Performing two sample t-test between within subject similarity (similarity between FNC-PASL of subject i and FNC-rsfMRI of subject i) and the similarity between rsfMRI-FNC and the rsfMRI-FNC of the null distribution shows the statistically significant difference with t-value = 17.16 and confidence interval (CI) = [0.43 0.54]. Using the same analysis and comparing between subjects (similarity between FNC-PASL of subject i and FNC-rsfMRI of subject j) with the similarity between rsfMRI-FNC and the rsfMRI-FNC of the null distribution shows the statistically significant difference with t-value = 64.19 and CI = [0.33 0.35]. It should be noted that the larger t-value, despite lower confidence interval for between subjects analysis, is due to a higher degree of freedom in the analysis.
Fig. 8.
Similarity of FNC obtained from rsfMRI (FNC-rsfMRI) with FNC obtained from samples of empirical null distribution (null), PASL-FNC of different subjects (middle), and PASL-FNC of the same subjects (right). Similarity between PASL-FNC and rsfMRI-FNC were statistically significant for both within and between individuals.
4. Discussion
In this study, we were able to extract brain intrinsic connectivity networks from our PASL data. Our findings are consistent with other previously published works that used PCASL methods to identify functional connectivity, despite PASL’s lower SNR and CNR as compared to PCASL. In this dataset, we extracted 10 major brain networks from the data of both modalities, demonstrated similarities in the networks extracted from PASL and rsfMRI, and detected similar changes in networks between mTBI patients and healthy controls with both modalities. Previous works have demonstrated 5 networks and used PCASL data, which is generally considered to be of higher quality in terms of SNR. This demonstration using PASL opens new doors for new analyses of old datasets that preceded newer ASL methods and rsfMRI.
Notably, the networks extracted from the PASL data slightly differ from those extracted from the rsfMRI data, but with a priori reasons to trust these differences. For example, there are some differences in regions associated with the DMN. However, the regions that show significant association with the DMN in the PASL data are known to be involved in the DMN in others’ work, so it is not likely to be a random false positive. This offers a potential advantage of using both modalities, since the PASL picked up some FC that the rsfMRI missed, and vice versa. Thus, jointly analyzing both modalities may provide a better picture of changes in brain FC.
Other studies using different ASL methods, like PCASL, may be expected to show better results due to better SNR, as might other studies with greater subject numbers or greater consistency in data collection. Our dataset also had some variation in the TR, TE, and number of volumes collected, but was still sufficient for obtaining brain networks and demonstrating changes in their connectivity patterns in an mTBI group as compared to a healthy control group. This ability of studying FC using PASL data provides an opportunity to reprocess massive previously-collected PASL datasets for new research purposes and potentially helping to identify biomarkers for different brain disorders.
In this work, we also tested two separate ways of generating independent components from the PASL data, looking at ICA on both the whole time series and on the subtraction images generated from the labeled and unlabeled images. Each has some potential advantages and disadvantages: using the entire dataset will reduce loss of information in the subtraction step but may also include some BOLD variation that the ICA step may be using, while doing the subtraction should reduce the BOLD contamination but also lowers the temporal resolution. Both succeeded in extracting networks, suggesting that the temporal resolution is acceptable even after subtraction and that the BOLD contamination is not the sole driver of the ICA success.
5. Limitations and future directions
One limitation is that while we did demonstrate a difference between the two groups, the mTBI patients and healthy controls, they do differ in a few ways besides the presence or absence of an injury, reducing the interpretability of this work in the study of mTBI. However, the main purpose of this study was to demonstrate that we can identify networks and detect differences, so this does not hinder our main purpose despite reducing the interpretability of this work for consequences of mTBI. Our successful application of this method in mTBI indicates its potential utility in other disorders, as well. Our suggestions for future work with this method include comparing FNC and doing dynamic analyses with ASL data; performing joint analyses like joint ICA rather than analyzing each modality alone; and studying FC using ASL in existing datasets.
6. Conclusions
This study demonstrated that PASL can be analyzed to identify resting state networks with reasonable reliability. Based on our analyses, it suggests that existing datasets from studies that included PASL but not rsfMRI may contain new information that is yet uninvestigated, giving us a greater opportunity to test newer ideas about functional connectivity without requiring new patients and funding to collect new data. Additionally, constructing networks from CBF-based data rather than BOLD-based data provides a more direct measure of functional connectivity.
Acknowledgements
Data collection was funded by DoD W81XWH-11–1-0493 and a Seed Grant from the International Society for Magnetic Resonance in Medicine. Analysis was supported under NIH R21NS090153, NIH P20GM103472, NIH R01REB020407 and NIH F30 HD084144.
Footnotes
Authorship statement
Natalie Wiseman: Data analysis and manuscript drafting Armin Iraji: Data analysis, manuscript drafting and review E Mark Haacke: study design and manuscript review Vince Calhoun: study design and manuscript review Zhifeng Kou: Study design, data interpretation and manuscript review.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. The author Zhifeng Kou is the Editorial Board Member of the journal, but was not involved in the peer review procedure. This paper was handled by another Editor Board member.
References
- 1.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98(2):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. [DOI] [PubMed] [Google Scholar]
- 3.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn Reson Med. 1995;34(4): 537–541. [DOI] [PubMed] [Google Scholar]
- 4.Ogawa S, Lee TM, Kay AR, Tank DW. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc Natl Acad Sci USA. 1990;87(24): 9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damoisaux JS, Rombous SARB, Barkhof F, Scheltens P, Stam CJ, Smith SM, et al. Consistent resting–state networks across healthy subjects. Proc Natl Acad Sci U S A. 2006;103(37):13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen Elena A, Erhardt Erik B, Damaraju Eswar, Gruner William, Segall Judith M, Silva Rogers F, et al. A baseline for the multivariate comparison of resting–state networks. Front Syst Neurosci. 2011;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armin Iraji, Calhoun Vince D, Wiseman Natalie M, Davoodi–Bojd Esmaeil, Avanaki Mohammad RN, Haacke E Mark, et al. The connectivity domain: analyzing resting state fMRI data using feature–based data–driven and model–based methods. Neuroimage. 2016;134:494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beckmann Christian F, DeLuca Marilena, Devlin Joseph T, Smith Stephen M. Investigations into resting–state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayer AR, Toulouse T, Klimaj S, Ling JM, Pena A, Bellgowan PSF. Investigating the properties of the hemodynamic response function after mild traumatic brain injury. J Neurotrauma. 2014;31(2):189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jann K, Gee DG, Kilroy E, Schwab S, Smith RX, Cannon TD, et al. Functional connectivity in BOLD and CBF data: similarity and reliability of resting brain networks. Neuroimage. 2015;106:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jann K, Orosz A, Dierks T, Wang DJJ, Wiest R, Federspiel A. Quantification of network perfusion in ASL cerebral blood flow data with seed based and ICA approaches. Brain Topogr. 2013;26(4):569–580. [DOI] [PubMed] [Google Scholar]
- 12.Zou Q, Yuan B–K, Gu H, Liu D, Wang DJJ, Gao J–H, et al. Detecting static and dynamic differences between eyes–closed and eyes–open resting states using ASL and BOLD fMRI. PLoS One. 2015;10(3):e0121757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buxton RB. Beyond BOLD correlations: a more quantitative approach for investigating brain networks. J Cerebr Blood Flow Metabol. 2016. Mar;36(3):461–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuang K–H, van Gelderen P, Merkle H, Bodurka J, Ikonomidou VN, Koretsky AP, et al. Mapping resting–state functional connectivity using perfusion MRI. Neuroimage. 2008;40(4):1595–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23(1):37–45. [DOI] [PubMed] [Google Scholar]
- 16.Wong EC. An introduction to ASL labeling techniques. J Magn Reson Imag. 2014; 40(1):1–10. [DOI] [PubMed] [Google Scholar]
- 17.Alsop DC, Detre JA, Golay X, Günther M, Hendrikse J, Hernandez–Garcia L, et al. Recommended implementation of arterial spin–labeled perfusion MRI for clinical applications: a consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn Reson Med. 2015;73(1):102–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faul M. Traumatic Brain Injury in the United States: Emergency Department Visits, Hospitalizations, and Deaths, 2002–2006. Atlanta (GA): Centers for Disease Control and Prevention, National Center for Injury Prevention and Control; 2010. [Google Scholar]
- 19.Chen CJ, Wu CH, Liao YP, Hsu HL, Tseng YC, Liu HL, et al. Working memory in patients with mild traumatic brain injury: functional MR imaging analysis. Radiology. 2012;264(3):844–851. [DOI] [PubMed] [Google Scholar]
- 20.Chen H. Longitudinal analysis of brain recovery after mild traumatic brain injury based on groupwise consistent brain network clusters. In: Navab N, et al. , eds. Medical Image Computing and Computer–Assisted Intervention – MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part II. Cham: Springer International Publishing; 2015:194–201. [Google Scholar]
- 21.Palacios EM, Yuh EL, Chang YS, Yue JK, Schnyer DM, Okonkwo DO, et al. Resting-state functional connectivity alterations associated with six–month outcomes in mild traumatic brain injury. J Neurotrauma. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sours C, Rosenberg J, Kane R, Roys S, Zhuo J, Shanmuganathan K, et al. Associations between interhemispheric functional connectivity and the Automated Neuropsychological Assessment Metrics (ANAM) in civilian mild TBI. Brain Imaging Behav. 2015;9(2):190–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Milham MP, Lui YW, Miles L, Reaume J, Sodickson DK, et al. Default–mode network disruption in mild traumatic brain injury. Radiology. 2012;265(3):882–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du Y, Fan Y. Group information guided ICA for fMRI data analysis. Neuroimage. 2013; 69:157–197. [DOI] [PubMed] [Google Scholar]
- 25.Kay T, Harrington DE, Adams R, Anderson T, Berrol S, Cicerone K, et al. Definition of mild traumatic brain injury. J Head Trauma Rehabil. 1993;8(3):86–87. [Google Scholar]
- 26.Iraji A, Chen H, Wiseman N, Zhang T, Welch R, O’Neil B, et al. Connectome–scale assessment of structural and functional connectivity in mild traumatic brain injury at the acute stage. Neuroimage Clin. 2016;12:100–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calhoun VD, Adali T, Pearlson GD, Pekar JJ. A method for making group inferences from functional MRI data using independent component analysis. Hum Brain Mapp. 2001;14(3):140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calhoun VD, Adali T. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE Rev Biomed Eng. 2012;5:60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Himberg J, Hyvarinen A, Esposito F. Validating the independent components of neuroimaging time series via clustering and visualization. Neuroimage. 2004;22(3): 1214–1222. [DOI] [PubMed] [Google Scholar]
- 30.Calhoun VD, Liu J, Adali T. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage. 2009;45(1 Suppl): S163–S172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Correa N, Adali T, Calhoun VD. Performance of blind source separation algorithms for fMRI analysis using a group ICA method. Magn Reson Imaging. 2007;25(5): 684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calhoun V, Adali T. Complex infomax: convergence and approximation of infomax with complex nonlinearities. In: Neural Networks for Signal Processing, 2002. Proceedings of the 2002 12th IEEE Workshop on. IEEE; 2002. [Google Scholar]
- 33.Iraji A, Benson RR, Welch RD, O’Neil BJ, Woodard JL, Ayaz SI, et al. Resting state functional connectivity in mild traumatic brain injury at the acute stage: independent component and seed–based analyses. J Neurotrauma. 2015;32(14):1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du Y. A group ICA based framework for evaluating resting fMRI markers when disease categories are unclear: application to schizophrenia, bipolar, and schizoaffective disorders. Neuroimage. 2015;122:272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salman MS. Group information guided ICA shows more sensitivity to group differences than dual–regression. In: Biomedical Imaging (ISBI 2017), 2017 IEEE 14th International Symposium on. IEEE; 2017. [Google Scholar]
- 36.Du Y, Allen EA, He H, Sui J, Wu L, Calhoun VD. Artifact removal in the context of group ICA: a comparison of single–subject and group approaches. Hum Brain Mapp. 2016;37(3):1005–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jann K, Gee DG, Kilroy E, Schwab S, Smith RX, Cannon TD, et al. Functional connectivity in BOLD and CBF data: similarity and reliability of resting brain networks. Neuroimage. 2015;106:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calhoun VD, Allen E. Extracting intrinsic functional networks with feature–based group independent component analysis. Psychometrika. 2013;78(2):243–259. [DOI] [PubMed] [Google Scholar]
- 39.Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, et al. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 2010;107(10):4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. fMRI resting state networks define distinct modes of long–distance interactions in the human brain. Neuroimage. 2006;29(4):1359–1367. [DOI] [PubMed] [Google Scholar]
- 41.van den Heuvel MP, Hulshoff Pol HE. Exploring the brain network: a review on resting-state fMRI functional connectivity. Eur Neuropsychopharmacol. 2010;20(8):519–534. [DOI] [PubMed] [Google Scholar]
- 42.Damoiseaux JS. Reduced resting–state brain activity in the “default network” in normal aging. Cerebr Cortex. 2008;18(8):1856–1864. [DOI] [PubMed] [Google Scholar]
- 43.Smith SM, Fox PT, Miller KL, Glahn DC, Mickle Fox P, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106(31):13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuo X–N, Kelly C, Adelstein JS, Klein DF, Xavier Castellanos F, Milham MP. Reliable intrinsic connectivity networks: test–retest evaluation using ICA and dual regression approach. Neuroimage. 2010;49(3):2163–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiviniemi V, Starck T, Remes J, Long X, Nikkinen J, Haapea M, et al. Functional segmentation of the brain cortex using high model order group PICA. Hum Brain Mapp. 2009;30(12):3865–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Bie HM, Boersma M, Adriaanse S, Veltman DJ, Wink AM, Roosendaal SD, et al. Resting–state networks in awake five– to eight–year old children. Hum Brain Mapp. 2012;33(5):1189–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo–planar mri. Magn Reson Med. 1995; 34(4):537–541. [DOI] [PubMed] [Google Scholar]
- 48.Leaver AM, Espinoza R, Joshi SH, Vasavada M, Njau S, Woods RP, et al. Desynchronization and Plasticity of Striato–Frontal Connectivity in Major Depressive Disorder. Cereb Cortex. 2015;26(11):4337–4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laird AR, Fox PM, Eickhoff SB, Turner JA, Ray KL, McKay DR, et al. Behavioral interpretations of intrinsic connectivity networks. J Cognit Neurosci. 2011;23(12): 4022–4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xie C, Bai F, Yu H, Shi Y, Yuan Y, Chen G, et al. Abnormal insula functional network is associated with episodic memory decline in amnestic mild cognitive impairment. Neuroimage. 2012;63(1):320–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor KS, Seminowicz DA, Davis KD. Two systems of resting state connectivity between the insula and cingulate cortex. Hum Brain Mapp. 2009;30(9):2731–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD. Comparison of multi–subject ICA methods for analysis of fMRI data. Hum Brain Mapp. 2011;32(12): 2075–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]