Abstract
The nucleotide sequence of the Treponema pallidum mcp2 gene was determined. mcp2 encodes a 45.8-kDa protein whose deduced amino acid sequence has significant homology with the C-terminal region of bacterial methyl-accepting chemotaxis proteins (MCPs). The Mcp2 N terminus lacks the hydrophobic transmembrane regions present in most MCPs. An Mcp2 fusion protein was strongly reactive with antibody (HC23) to the highly conserved domain of MCPs and with rabbit syphilitic serum. Antibody HC23 reacted with six T. pallidum proteins, including a 45-kDa protein that may correspond to Mcp2. This protein was present in the aqueous phase from T. pallidum cells that were solubilized with Triton X-114 and phase partitioned.
Treponema pallidum subsp. pallidum, a motile, spiral-shaped bacterium, is the causitive agent of syphilis, a chronic, sexually transmitted disease that continues to be a public health problem worldwide. Due to the inability of researchers to continuously cultivate T. pallidum in vitro, the mechanisms that this spirochete uses to survive in the host and cause disease are poorly understood. The small genome size (∼1,000 kb) and limited biosynthetic capabilities of T. pallidum suggest that it must obtain most of its nutrients from the host (21, 22, 26). Thus, motility and chemotaxis are likely to be important for treponemal growth and dissemination.
Chemotaxis has been extensively studied for Escherichia coli, Salmonella typhimurium (29), and Bacillus subtilis (4, 12). The chemotaxis systems of these bacteria are composed of transmembrane methyl-accepting chemotaxis proteins (MCPs) and cytoplasmic chemotaxis (Che) proteins. MCPs act as receptors for environmental inputs by binding to attractants and repellents. These proteins also act as transducers by interacting with ligand-bound periplasmic proteins. Studies in our laboratory have focused on elucidation of the chemotaxis system of T. pallidum. We recently reported the identification and characterization of an operon containing four che genes from this spirochete (10). Additionally, Hagman et al. (11) identified a T. pallidum gene (mcp1) encoding a putative MCP. Since bacterial species usually synthesize several MCPs (12, 19, 25, 29, 31), the goal of this study was to identify and characterize additional treponemal MCPs.
An oligonucleotide corresponding to an amino acid sequence (GFAVVA) in the C-terminal highly conserved domain (HCD) of bacterial MCPs (16, 20) was labeled with fluorescein-11-dUTP (Amersham Life Sciences, Arlington Heights, Ill.) and used to probe a λ ZAP II T. pallidum genomic DNA library. Treponemal DNA inserts from positive recombinant λ phages were excised and recircularized in pBluescript SK−. The nucleotide sequences of the T. pallidum DNA inserts were determined with the Taq DyeDeoxy Terminator cycle sequencing kit (Applied Biosystems Inc., Foster City, Calif.) at the University of North Carolina at Chapel Hill Automated DNA Sequencing Facility on models 373A and 377 DNA sequencers (Applied Biosystems Inc.). Analysis of the nucleotide sequences revealed two open reading frames (ORFs) whose deduced C-terminal amino acid sequences showed homology to the C-terminal region of several bacterial MCPs. The first ORF was identical to the previously reported mcp1 gene (11). The second ORF, designated mcp2, contains 1,284 nucleotides with a G+C content (53.2%) similar to that reported for T. pallidum (52.0 to 53.7%) (27) (Fig. 1). Mcp2 encodes a 427-amino-acid protein with a predicted molecular mass of 45.8 kDa and a pI of 4.54. Two putative ribosome binding sites (GGA and AGGA) are located 5 and 9 nucleotides 5′ of the predicted GTG start codon, respectively. A putative ς70-like promoter is located 78 nucleotides 5′ of the GTG start codon. The −35 (TTGAGA) and −10 (TCTACT) sequences are separated by 17 nucleotides. Two potential stem-loop structures, characteristic of a rho-independent transcriptional terminator, are located 7 and 11 nucleotides 3′ of the predicted TAG stop codon. Analysis of the nucleotide sequences 5′ and 3′ of mcp2 revealed the presence of an ORF whose deduced amino acid sequence showed significant homology with a Treponema denticola chymotrypsinlike protease (prtB) (3) and an ORF that has no homology with proteins in the database, respectively.
FIG. 1.
Nucleotide and deduced amino acid sequences of T. pallidum mcp2 (GenBank accession no. AF016689). The −35 and −10 regions of the putative promoter are italicized and underlined. The putative ribosome binding sites (RBS) and start and stop codons are in boldface type. The downstream stem-loop structures (putative rho-independent transcriptional terminators) are indicated by arrows. The putative signal peptide is underlined, and the K1, HCD, and R1 regions are boxed.
Amino acid sequence analysis revealed that T. pallidum Mcp2 has overall identities of 20% to T. pallidum Mcp1 (11), 25.5 to 26.7% to E. coli MCPs, 26.7 to 29.5% to B. subtilis MCPs, 22.1 to 30.6% to Borrelia burgdorferi MCPs (8), and 41.4%/37.1% and 63.1% to T. denticola McpA (9)/DmcA (14) and DmcB (GenBank accession no. U84257), respectively. When the amino acid sequence of the HCD of T. pallidum Mcp2 was compared to HCDs of T. pallidum Mcp1, E. coli MCPs, B. burgdorferi MCPs, B. subtilis MCPs, and T. denticola McpA/DmcA and DmcB, the identities were 64.6%, 62.5%, 47.7 to 75.0%, 62.5 to 73.0%, and 92.0 and 98.0%, respectively. Figure 2 shows a multiple sequence alignment of the T. pallidum Mcp2 HCD with HCDs of selected bacterial MCPs.
FIG. 2.
Multiple sequence alignment of the HCD of T. pallidum (Tp) Mcp2 with T. pallidum Mcp1 (U56999 [11]), T. denticola (Td) McpA (AF012922 [9]), T. denticola DmcB (U84257), B. burgdorferi (Bb) Mcp2 (AE001161 [8]), E. coli (Ec) Tap (P07018 [16]), and B. subtilis (Bs) TlpC (P39209 [13]). Bars (|) denote identity to the T. pallidum Mcp2 amino acid sequence; colons (:) and dots (.) denote similarity to the T. pallidum Mcp2 amino acid sequence (based on the Dayhoff PAM-250 similarity matrix).
MCPs typically contain several structural features related to their functions (1, 29). The N-terminal periplasmic chemoreceptor (ligand-binding) domain, whose amino acid sequence is not conserved, is flanked by two transmembrane regions (TM1 and TM2). The highly conserved C-terminal region which includes the cytoplasmic signaling domain (HCD) has been suggested to interact with chemotaxis proteins CheA and CheW to control bacterial swimming behavior (18). The HCD region is flanked by two methylation sequences (K1 and R1). Interestingly, the structural organization of T. pallidum Mcp2 diverges from that of typical bacterial MCPs. ALOM analysis (15) indicated that Mcp2 lacks N-terminal transmembrane regions. Kyte-Doolittle (17) hydropathy analysis of Mcp2 (data not shown) revealed a single N-terminal hydrophobic stretch of 39 amino acids which has characteristics of a signal peptide (hydrophobic core and an Ala-X-Ala signal peptidase I processing site) (Fig. 1). As previously indicated, Mcp2 contains a well conserved C-terminal HCD (Fig. 1 and 2). Based on the consensus methylation sequence of E. coli MCPs ([A/S]-X-X-E-[E/Q]-X-[A/T/S]-A-[A/T/S]), in which the underlined Glu/Gln residue serves as the methylation site (5, 23), T. pallidum Mcp2 has potential K1 (SSVEQMVAN) and R1 (AMKEQSDGS) sequences flanking the HCD (Fig. 1). The assignment of these sites is tentative until further biological information is obtained.
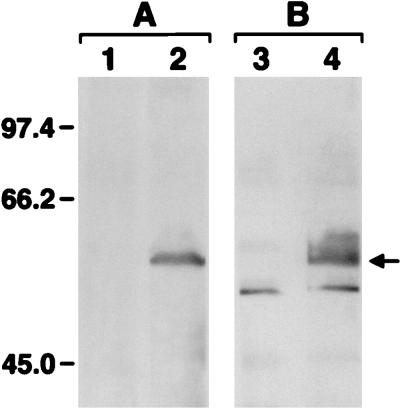
To facilitate further studies, mcp2 was PCR amplified from T. pallidum genomic DNA (Expand Long Template PCR System kit; Boehringer Mannheim, Indianapolis, Ind.) and cloned into the Pinpoint Xa-1 T-vector downstream of a sequence encoding a peptide that is biotinylated when the fusion protein is synthesized in E. coli (Technical Bulletin 234; Promega Corp., Madison, Wis.). The resulting plasmid, pmcp2, was transformed into E. coli JM109 cells. Conditions for the synthesis and detection of the Mcp2 fusion protein were those suggested by the manufacturer. Following induction with IPTG (isopropyl-β-d-thiogalactopyranoside), JM109 cells containing the PinPoint Xa-1 T-vector and pmcp2 were washed and solubilized in sample buffer. The proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS)-polyacrylamide gels (28) and electroblotted to a nitrocellulose membrane. A biotinylated Mcp2 fusion protein of 59.5 kDa (predicted molecular mass of 58.8 kDa) was identified following probing of cell extracts of JM109 (pmcp2) with streptavidin alkaline phosphatase (data not shown). A protein of similar size was not detected in cell extracts of JM109 (PinPoint Xa-1 T-vector). To further characterize the Mcp2 fusion protein, immunoblotting was performed according to Alam and Hazelbauer (1) with a site-specific multiple antigenic peptide antibody (HC23) (31) that was raised against 23 amino acids representing the highest-homology region within the HCD of MCPs. Reactivity was determined with the ECL Western Blotting Analysis System (Amersham Life Sciences). The HC23 antibody was strongly reactive with the Mcp2 fusion protein (Fig. 3A, lane 2), confirming the observed C-terminal amino acid sequence homology of Mcp2. The HC23 antibody was not reactive with cell extracts of JM109 (PinPoint Xa-1 T-vector) (Fig. 3A, lane 1). To determine if mcp2 is expressed during syphilitic infection in the rabbit model, immunoblotting was performed with rabbit syphilitic serum that was preadsorbed with cell extracts of JM109 to remove antibodies to E. coli proteins. The Mcp2 fusion protein present in cell extracts of JM109 (pmcp2) was strongly reactive with syphilitic serum, indicating that Mcp2 is synthesized during infection and elicits a humoral (immunoglobulin G antibody) response (Fig. 3B, lane 4). The Mcp2 fusion protein was not reactive with normal (preinfection) serum obtained from the same rabbit (data not shown). Additionally, an immunoreactive 59.5-kDa protein was not present in cell extracts of JM109 (PinPoint Xa-1 T-vector) (Fig. 3B, lane 3).
FIG. 3.
Immunoblot analysis of the Mcp2 fusion protein. Lanes 1 and 3 contain solubilized cell extracts of JM109 (PinPoint Xa-1 T-vector), and lanes 2 and 4 contain solubilized cell extracts of JM109 (pmcp2). Set A was blotted with the HC23 antibody (1:2,000 dilution), and set B was blotted with rabbit syphilitic serum (1:1,000 dilution). The position of the Mcp2 fusion protein is indicated by the arrow. Molecular mass standards are indicated (in kilodaltons).
The availability of the HC23 antibody prompted investigation of T. pallidum for the presence of MCPs. T. pallidum cells were extracted and purified from rabbit testes and solubilized in sample buffer (28). Treponemal proteins were separated on SDS-polyacrylamide gels and electroblotted to nitrocellulose. With the exception of a 47.5-kDa protein that reacted with secondary antibody in the absence of primary antibody, none of the treponemal proteins reacted with preimmune rabbit serum (data not shown). In contrast, immunoblotting with antibody HC23 revealed specific reactivity with six treponemal proteins ranging in size from 76.8 to 32 kDa (Fig. 4, lane A). Proteins of approximately 66 and 45 kDa likely correspond to Mcp1 and Mcp2, respectively. These results suggest that T. pallidum synthesizes several MCPs. BLAST (2) analysis of provisional data from the T. pallidum whole genome sequencing project indicates that this spirochete has a least four genes encoding MCP-like proteins.
FIG. 4.
Immunoblot analysis of SDS-solubilized T. pallidum cell extracts and T. pallidum cell fractions obtained following Triton X-114 solubilization and phase partitioning. A 1:500 dilution of antibody HC23 was used. Lane A, SDS-solubilized cell extracts of T. pallidum; lane B, Triton X-114 insoluble pellet; lane C, aqueous phase; lane D, detergent phase. The position of the 45-kDa MCP is indicated by the arrow. Molecular mass standards are indicated (in kilodaltons).
To investigate the hydrophobic character and potential cellular location of the treponemal MCPs, T. pallidum cells were solubilized with 2% Triton X-114 and phase partitioned as previously described (7, 24). The insoluble pellet (protoplasmic cylinder) and the aqueous (hydrophilic) and detergent (hydrophobic) phases were analyzed by immunoblotting with the HC23 antibody. The 45-kDa MCP was present in the aqueous phase (Fig. 4, lane C), whereas the remaining MCPs were associated with the insoluble pellet (Fig. 4, lane B). The latter result is characteristic of integral membrane proteins that are not readily solubilized from the protoplasmic cylinder by Triton X-114 (7). Localization of the 45-kDa protein to the aqueous phase is consistent with the results of Alom and Kyte-Doolittle analyses, suggesting that Mcp2 has a cleavable N-terminal signal peptide. Soluble MCPs that lack hydrophobic transmembrane domains have been reported for Halobacterium salinarium (6, 25, 31) and Rhodobacter sphaeroides (30). The results of gene inactivation studies performed with these bacteria showed that the soluble MCPs are necessary for chemotaxis to certain substrates. Brooun et al. (6) proposed that HtrXI, a soluble H. salinarium MCP, plays an important role in the adaptation of chemotactic responses via interaction with a putative membrane-bound transducer, HtrVII. T. pallidum Mcp2 may function in a similar manner by interacting with Mcp1 or other membrane-bound MCPs.
Although Hagman et al. (11) suggested that T. pallidum synthesizes only a single MCP (Mcp1), the genetic and immunological data presented here indicate that this organism synthesizes additional MCPs, including a putative soluble MCP (Mcp2). The ability to synthesize MCPs with different specificities is consistent with the nutritional needs of T. pallidum and may be an important factor in the dissemination of treponemes during early infection. Studies to determine the effect of inactivation of mcp genes on treponemal chemotaxis are not possible due to the inability to cultivate and genetically manipulate T. pallidum. Development of a chemotaxis assay employing treponemes freshly extracted from rabbits should facilitate identification of chemoattractants, thus providing some clues as to the functional specificities of the treponemal MCPs.
Nucleotide sequence accession number.
The GenBank accession number of mcp2 is AF016689.
Acknowledgments
We thank S. Norris for the λ Zap II T. pallidum genomic DNA library and M. Alam for the HC23 antibody. We also thank N. Barnes and H. Bergen for technical assistance.
This research was supported by National Institutes of Health grant U19-AI31496 and by the University of North Carolina Minority Postdoctoral Scholars Program.
REFERENCES
- 1.Alam M, Hazelbauer G L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991;173:5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arakawa S, Kuramitsu H K. Cloning and sequence analysis of a chymotrypsinlike protease from Treponema denticola. Infect Immun. 1994;62:3424–3433. doi: 10.1128/iai.62.8.3424-3433.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff D S, Ordal G W. Bacillus subtilis chemotaxis: a deviation from the Escherichia coli paradigm. Mol Microbiol. 1992;6:23–28. doi: 10.1111/j.1365-2958.1992.tb00833.x. [DOI] [PubMed] [Google Scholar]
- 5.Boyd A, Kendall K, Simon M I. Structure of the serine chemoreceptor in Escherichia coli. Nature (London) 1983;301:623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- 6.Brooun A, Zhang W, Alam M. Primary structure and functional analysis of the soluble transducer protein HtrXI in the archaeon Halobacterium salinarium. J Bacteriol. 1997;179:2963–2968. doi: 10.1128/jb.179.9.2963-2968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham T M, Walker E M, Miller J N, Lovett M A. Selective release of the Treponema pallidum outer membrane and associated polypeptides with Triton X-114. J Bacteriol. 1988;170:5789–5796. doi: 10.1128/jb.170.12.5789-5796.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, Gwinn M, Dougherty B, Tomb J-F, Fleischmann R D, Richardson D, Peterson J, Kerlavage A R, Quackenbush J, Salzberg S, Hanson M, Vugt R V, Palmer N, Adams M D, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fujii C, Cotton M D, Horst K, Roberts K, Hatch B, Smith H O, Venter J C. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 9.Greene S R, Stamm L V. Identification, sequence, and expression of Treponema denticola mcpA, a putative chemoreceptor gene. FEMS Microbiol Lett. 1997;157:245–249. doi: 10.1111/j.1574-6968.1997.tb12780.x. [DOI] [PubMed] [Google Scholar]
- 10.Greene S R, Stamm L V, Hardham J H, Young N R, Frye J G. Identification, sequences, and expression of Treponema pallidum chemotaxis genes. DNA Sequence. 1997;7:267–284. doi: 10.3109/10425179709034046. [DOI] [PubMed] [Google Scholar]
- 11.Hagman K E, Porcella S F, Popova T G, Norgard M V. Evidence for a methyl-accepting chemotaxis protein gene (mcp1) that encodes a putative sensory transducer in virulent Treponema pallidum. Infect Immun. 1997;65:1701–1709. doi: 10.1128/iai.65.5.1701-1709.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanlon D W, Ordal G W. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem. 1994;269:14038–14046. [PubMed] [Google Scholar]
- 13.Hanlon D W, Rosario M M L, Ordal G W, Venema G, Sinderen D V. Identification of TlpC, a novel 62 kDa MCP-like protein from Bacillus subtilis. Microbiology. 1994;140:1847–1854. doi: 10.1099/13500872-140-8-1847. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka M, Li H, Arakawa S, Kuramitsu H. Characterization of a methyl-accepting chemotaxis protein gene, dmcA, from the oral spirochete Treponema denticola. Infect Immun. 1997;65:4011–4016. doi: 10.1128/iai.65.10.4011-4016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning protein. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 16.Krikos A, Mutoh N, Boyd A, Simons M I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 17.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Parkinson J S. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan D G, Baumgartner J W, Hazelbauer G L. Proteins antigenically related to methyl-accepting chemotaxis proteins of Escherichia coli detected in a wide range of bacterial species. J Bacteriol. 1993;175:133–140. doi: 10.1128/jb.175.1.133-140.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moual H L, Koshland J D E. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 21.Nichols J C, Baseman J B. Carbon sources utilized by virulent Treponema pallidum. Infect Immun. 1975;12:1044–1050. doi: 10.1128/iai.12.5.1044-1050.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Norris S J the Treponema pallidum Polypeptide Research Group. Polypeptides of Treponema pallidum: progress toward understanding their structural, functional, and immunological roles. Microbiol Rev. 1993;57:750–779. doi: 10.1128/mr.57.3.750-779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park C, Dutton D P, Hazelbauer G L. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J Bacteriol. 1990;172:7179–7189. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radolf J D, Chaberlain N R, Clausell A, Norgard M V. Identification and localization of integral membrane proteins of Treponema pallidum subsp. pallidum by phase partitioning with the nonionic detergent Triton X-114. Infect Immun. 1988;56:490–498. doi: 10.1128/iai.56.2.490-498.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudolph J, Nordmann B, Storch K-F, Gruenberg H, Rodewald K, Oesterhelt D. A family of halobacterial transducer proteins. FEMS Microbiol Lett. 1997;139:161–168. doi: 10.1111/j.1574-6968.1996.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 26.Schiller N L, Cox C D. Catabolism of glucose and fatty acids by virulent Treponema pallidum. Infect Immun. 1977;16:60–68. doi: 10.1128/iai.16.1.60-68.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smibert R M. Genus III. Treponema Schaudinn 1905, 1728AL. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams and Wilkins; 1984. pp. 49–57. [Google Scholar]
- 28.Stamm L V, Kerner T C, Jr, Bankaitis V A, Bassford P J., Jr Identification and preliminary characterization of Treponema pallidum protein antigens expressed in Escherichia coli. Infect Immun. 1983;41:709–721. doi: 10.1128/iai.41.2.709-721.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 30.Ward M J, Harrison D M, Ebner M J, Armitage J P. Identification of a methyl-accepting chemotaxis protein in Rhodobacter sphaeroides. Mol Microbiol. 1995;18:115–121. doi: 10.1111/j.1365-2958.1995.mmi_18010115.x. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]