Abstract
Breastfeeding not only reduces infection-related morbidity, but also increases growth of preterm infants. Advantages of breast milk (BM) for preterm infants are significant. They continue to be studied. However, because not all preterm infants can receive breastfeeding, bovine-based infant formula (IF) is used as an alternative, which may increase the risk of several preterm complications. Exosomes isolated from biofluids are emerging as biomarkers in research of various diseases. Here, we characterized miRNA contents of exosomes in urine and serum samples of preterm infants who were BM and IF fed and performed transcriptomic analysis of small RNA libraries. We identified significantly up-regulated 6 miRNAs and 10 miRNAs, respectively. Gene Ontology (GO) analysis revealed that target genes of these miRNAs might participate in neuronal development, immunity modulation, detoxification of reactive oxygen species, and transmembrane exchange. Our data suggest that exosome-based systemic screening for preterm infants with breastfeeding might be a screening tool for identifying target molecules involved in therapy for preterm infants in neonatal intensive care unit (NICU) and for future application as nutraceutical formulations or pharmaceuticals.
Keywords: preterm infant, exosomes, miRNA, small RNA sequencing, breastfeeding, serum, urine
1. Introduction
Preterm infants who are born extremely early (before 37 completed weeks) commonly have several complicated health problems (1, 2). According to WHO data, about 15 million infants are born prematurely every year. This trend does not tend to decrease. Because preterm infants are born at a crucial period for brain and body development, more than 1 million preterm infants die in their first year or are treated in neonatal intensive care units (NICUs) (3). Most NICUs encourage parents to keep the baby with kangaroo care because this can stimulate breast milk (BM) expression and decrease the stress level of both mother and infant (4). Advantages of BM for preterm infants are significant. They continue to be studied (5, 6). Breastfeeding is an effective way to enhance health and development, particularly for preterm infants, although this is controversial (7, 8). BM includes non-nutrient factors containing oligosaccharides, lipids, antibodies, immunoglobulins and lactoferrin (9). Premature infants are at high risk for neurodevelopmental and respiratory impairments later in life (10, 11). BM consumption is positively associated with later cognitive outcomes in full-term cohorts (7, 12, 13). However, such association in preterm infants has not been clearly identified yet (14, 15). A recent study has suggested a positive effect of BM containing linoleic acid (LA) and α-linolenic acid (ALA) on brain development of preterm infants. These precursors of long-chain polyunsaturated fatty acids (PUFAs) ω3 and ω6 are critical for brain development of preterm infants (16). In addition, BM fed premature infants have significantly lower values of oxidative stress metabolites in urine compared formula fed groups (17). Despite these beneficial effects of BM, the precise role of BM on premature infants about systemic modulations remains unclear. Furthermore, mothers’ milk is sometimes unavailable or inadequate for a few reasons, leading to preterm infants being fed with artificial formula alternative (18). Infant formula (IF) is designed as a substitute for BM to satisfy nutritional requirements of infants. Despite IF containing a higher caloric density and protein than BM, some studies reported significantly lower prevalence of necrotizing enterocolitis in BM-fed group than in IF-fed group (19), especially in preterm infants (20). Moreover, BM feeding can reduce cases of comorbidities such as death, sepsis, bronchopulmonary dysplasia, and severe retinopathy of prematurity (ROP) in infants more than IF feeding (21–26).
Mammalian cells can secrete exosomes that are found in most types of human biofluids such as serum and urine (27). Exosomes are lipid bilayer nano-sized (50–200 nm) vesicles that can protect their cargos against degradation. They can also transport regulatory cargos including miRNAs to adjacent and distant cells. However, they cannot replicate (28). Exosomes can promote a particular phenotype. They are involved in cell-to-cell communication by transferring their cargos (29, 30). Analysis of exosomes in biofluid samples collected non-invasively could detect pathological disorder or alterations (31). Thus, it can be used to evaluate conditions of premature infants in NICU. MicroRNAs (miRNAs) are endogenous small molecules with lengths of 18–25 nucleotides. They are involved in the regulation of gene expression after transcription (32, 33). They play a critical role in a wide range of biological processes and serve as useful diagnostic biomarkers in various diseases (29, 30). Recent studies have demonstrated the theoretical role of serum miRNA-495-5p levels in premature infants associated with bronchopulmonary dysplasia (34). Some plasma miRNAs have diagnostic potential in preterm infants with respiratory distress syndrome (RDS) (35). Preliminary evidence has indicated that urine extra cellular vesicles derived miRNAs can be used as biomarkers for necrotizing enterocolitis in premature infants (36). Moreover, many studies have investigated the application of exosomal miRNAs as biomarkers or therapies in the treatment of preterm infants (37–40). Multiple studies have reported changes in compositions and miRNAs within the body through diet or dietary regulation. There are also attempts to apply these changes to treatments. Some studies have reported that miRNAs themselves can be modulated by feeding, suggesting that dietary manipulation might be a promising therapeutic strategy (41–43).
Numerous studies have compared BM feeding and IF feeding in premature infants (44–46). However, few studies have measured gene expression systemically modified by BM and IF feeding using exosomal miRNAs derived from serum or urine samples of preterm infants. The aim of this research was to gain a deeper molecular understanding of the influence of BM or IF intake by preterm infants. To achieve this aim, exosomal miRNA transcriptional profiling was performed for preterm infant urine and serum samples to compare between BM-fed and IF-fed groups. Our hypothesis was that miRNAs differentially expressed in urine and serum after BM consumption would be highly relevant to lower health risk and that they could be harnessed as potential therapeutics for treating related complications in preterm infants. We therefore tried to acquire transcriptional features of exosomes derived from preterm infants after breastfeeding for future applications in neutraceutical formulas or pharmaceutical substances for the care of preterm infants.
2. Results
A total of 33 preterm infants were recruited for this study. Complete overview of distinct experiments and analyses accomplished in this study is shown in Figure 1. The figure denotes sample collection process and morphological characterization of isolated exosomes with figuration of results. The lower pink arrow represents bioinformatic pipelines applied for small RNAs sequencing data analysis.
Figure 1.
Experimental overview and bioinformatic pipeline (lower pink arrow) of this study.
2.1. Characteristics of objectives
We recruited 33 premature infants in our study. Infants were classified with two groups: the BM group (n = 12) that was fed human milk and the IF group (n = 21) that was fed exclusively infant formula. The IF group consumed one commercial brand containing 67 kcal and 2.1 g/100 mL of proteins. There were no significant differences in gestational age, birth weight, height, head circumference, maternal age, diabetes, hypertensions, or body mass index between the two groups. Only multiple birth rate was significantly different (p < 0.001) between BM and IF groups. Demographic and clinical characteristics of premature infants are shown in Table 1. Urine and serum samples were collected before infants were discharged from NICU. Average age was 57 ± 25.24 days for the BM group and 55.76 ± 39.95 days for the IF group.
Table 1.
Demographic and clinical characteristics of the study groups.
| Characteristic | Breast-fed (n = 12) | Formula-fed (n = 21) | p-value |
|---|---|---|---|
| Gestational age (wk) | 29 + 0.86 ± 3.39 | 30.65 ± 2.55 | 0.487 |
| Birth weight (g) | 1263.12 ± 433.7 | 1496.67 ± 552.62 | 0.1888 |
| Birth height (cm) | 38.25 ± 4.92 | 39.17 ± 3.89 | 0.5861 |
| Birth head circumference (cm) | 30.96 ± 3.61 | 32.24 ± 2.63 | 0.2963 |
| Male | 6 (50%) | 12 (57.14%) | 0.9736 |
| Multiple birth | 0 (0%) | 15 (71.43%) | <0.001 |
| SGA | 4 (33.33%) | 4 (19.05%) | 0.6178 |
| Apgar at 1 min | 4.92 ± 2.57 | 5 ± 1.87 | 0.9228 |
| Apgar at 5 min | 7.08 ± 1.68 | 7.71 ± 1.85 | 0.3261 |
| Maternal Age (yrs) | 34.08 ± 3.45 | 36.29 ± 3.95 | 0.1067 |
| Maternal diabetes* | 5 (41.67%) | 6 (28.57%) | 0.7011 |
| Maternal hypertension* | 6 (50%) | 13 (61.9%) | 0.7645 |
| Maternal body mass index (kg/m2) | 29.22 ± 3.3 | 28.55 ± 2.68 | 0.5687 |
| Sample collection date (d) | 57 ± 25.24 | 55.76 ± 39.95 | 0.3994 |
Value are expressed as mean ± standard deviation or number (%).
SGA, small for gestational age.
*The case of both Maternal diabetes and Materanl hypertension (n = 3 in Breast-fed and n = 4 in Formula-fed).
2.2. Morphologic characterization of exosomes
To identify exosomes in urine and serum samples of BM and IF groups, we purified exosomes and performed transmission electron microscopy (TEM). Purified exosome particles were spherical with diameters of less than 200 nm and a complete membrane structure (Figure 2A). Nanoparticle tracking analysis (NTA) revealed that diameters of exosomes isolated from urine samples of preterm infants had a mean size of 111.5 nm and a mode of 81.0 nm. Exosomes isolated from serum samples of preterm infants were characterized by a mean size of 134.5 nm and a mode of 85.6 nm. Concentrations of exosomes in urine and serum samples were approximately 7.9 × 1011 and 9.94 × 1011 particles/ml, respectively (Figure 2B). Immunoblotting showed that CD9, an exosome marker, was detected in both exosomes, but not in cell lysates (Figure 2C). An endoplasmic reticulum marker CALNEXIN was used to evaluate potential contamination by non-exosomal molecules. Compared with dark bands of cell lysates, only faint bands could be detected in both exosomes separated from urine and serum samples, indicating that there were a few contaminations of non-exosomal vesicles in isolated exosomes from biofluids.
Figure 2.
Morphological characterization of isolated exosomes. (A) Transmission electron microscopy (TEM) image revealing the morphology of exosomes and presence of vesicles with sizes under 200 nm. (B) Size distribution of exosomes measured with a Nanosight NS300 (NTA). The peak of the particle size was 82 nm for urine exosomes and 86 nm for serum exosomes. (C) Expression levels of exosomal membrane marker CD9 (Cluster of Differential 9) and endoplasmic reticulum marker CALNEXIN in isolated vesicles were determined by western blotting.
2.3. Overview of small RNA sequencing
To characterize small RNA profiles of urine and serum exosomes, we prepared small RNA libraries from all premature infants in the test cohort. All reads were mapped to the human genome (hg38) and small RNA databases for miRNA (Supplementary Table S1). From 66 libraries, we obtained 2,982,090,421 raw reads from Illumina paired-end sequencing. After trimming and cleaning low-quality reads, 1,483,794,616 processed reads were generated. An average of 9,171,629 reads per library were mapped to the reference genome, with an average mapped rate of 39.6% (Supplementary Table S1). Through the alignment step, mappable cleansed reads were annotated to miRNA and a total of 2,656 miRNAs were detected in urine and serum exosomes.
2.4. Differentially expressed miRNAs in exosomes
To identify miRNAs of exosomes derived from urine and serum samples that might be associated with complications in preterm infants, differentially expressed miRNAs (DEmiRNAs) between breastmilk-fed and formula-fed groups were analyzed. A total of 2,656 mature miRNAs with zero counts across more than 51% of all samples were excluded, leaving 154 and 469 mature miRNAs for BM and IF groups, respectively. After statistical test using exactTest and Fold Change, nine and 23 miRNAs had an abundance with small p-value (p < 0.05) in urine and serum samples, respectively. Results of comparing differential expression levels of miRNAs in urine and serum of BM and IF groups are depicted in Figure 3. Table 2 presents the most predominantly up- and downregulated miRNAs in BM-fed group of urine and serum samples, respectively. The heat map of DEmiRNAs was illustrated using Pheatmap v1.0.12 in Figures 3B,D. Our sequencing data revealed that six miRNAs (hsa-miR-619-5p, hsa-miR-664a-5p, hsa-miR-484, hsa-miR-19a-3p, hsa-miR-10a-5p, hsa-miR-328-3p) were differentially expressed in urinary EVs and ten miRNAs (hsa-miR-627-5p, hsa-miR-545-5p, hsa-miR-539-5p, hsa-miR-33a-5p, hsa-miR-1296-5p, hsa-miR-4284, hsa-miR-548o-3p, hsa-miR-338-3p, hsa-miR-12136, hsa-miR-483-3p) were significantly up-regulated in serum EVs of BM-fed preterm infants (Table 2). In further analysis, we focused on these six and ten upregulated miRNAs in urine and serum EVs of BM-fed preterm infants, respectively.
Figure 3.
Volcano plots (A,C) of differentially expressed urine and serum exosomal miRNAs in the two groups using DESeq2 for data analysis. The log2 fold change (log2 FC) indicates difference of comparison, while −log10 (p-value) shows the level of significance of each miRNA between two groups. |Fold change| ≥ 2 and p-value < 0.05 were used to identify differentially expressed miRNAs between the two groups. Differences were visualized using R 3.6.1 (www.r-project.org). The heatmap (B,D) of DEmiRNAs was constructed using “pheatmap” R package (Kolde, Raivo (2019). Pheatmap: Pretty Heatmaps. R package version 1.0.12. https://CRAN.R-project.org/package=pheatmap).
Table 2.
Most differentially expressed exosomal miRNAs in urine and serum (breast-fed vs. formula-fed).
| Biofluid | miRNA | logFC | logCPM | p-value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Urine | hsa-miR-619-5p | 2.980822 | 11.204031 | 0.01015517 | |||||||
| hsa-miR-664a-5p | 2.472770 | 8.552271 | 0.03498467 | ||||||||
| hsa-miR-484 | 2.298957 | 9.091072 | 0.0073081 | ||||||||
| hsa-miR-19a-3p | 2.184316 | 8.971845 | 0.04986058 | ||||||||
| hsa-miR-10a-5p | 2.169134 | 12.274405 | 0.01795504 | ||||||||
| hsa-miR-328-3p | 2.007457 | 9.626131 | 0.02175376 | ||||||||
| hsa-miR-4286 | −5.763903 | 8.438199 | 0.006421627 | ||||||||
| hsa-miR-1273c | −3.556804 | 10.060786 | 0.022904048 | ||||||||
| hsa-miR-8485 | −2.482458 | 13.708561 | 0.012333532 | ||||||||
| Serum | hsa-miR-627-5p | 5.717146 | 6.305259 | 9.3367E-05 | |||||||
| hsa-miR-545-5p | 5.475290 | 5.706910 | 0.002119379 | ||||||||
| hsa-miR-539-3p | 3.564859 | 6.816660 | 0.001055754 | ||||||||
| hsa-miR-33a-5p | 3.487216 | 6.859864 | 0.006475274 | ||||||||
| hsa-miR-1296-5p | 3.269009 | 4.937124 | 0.008332218 | ||||||||
| hsa-miR-4284 | 3.113500 | 9.124149 | 0.016633993 | ||||||||
| hsa-miR-548o-3p | 2.480360 | 5.009451 | 0.01880367 | ||||||||
| hsa-miR-338-3p | 2.402597 | 6.500702 | 0.014093223 | ||||||||
| hsa-miR-12136 | 2.334674 | 10.901402 | 0.027091412 | ||||||||
| hsa-miR-483-3p | 2.312663 | 13.216990 | 0.010354444 | ||||||||
| hsa-miR-542-3p | −5.248159 | 5.573964 | 0.000299216 | ||||||||
| hsa-miR-1304-5p | −4.975675 | 4.801230 | 0.002427662 | ||||||||
| hsa-miR-296-3p | −4.930382 | 5.013698 | 0.010972196 | ||||||||
| hsa-miR-1290 | −3.543079 | 10.821049 | 0.024603236 | ||||||||
| hsa-miR-935 | −3.394161 | 5.265165 | 0.003353104 | ||||||||
| hsa-miR-1270 | −3.009292 | 4.379843 | 0.011267873 | ||||||||
| hsa-miR-214-5p | −2.752111 | 4.452986 | 0.033535364 | ||||||||
| hsa-miR-23a-5p | −2.556573 | 8.431524 | 0.016217351 | ||||||||
| hsa-miR-378q | −2.381636 | 5.007624 | 0.040298643 | ||||||||
| hsa-miR-139-3p | −2.361248 | 5.775051 | 0.000495777 | ||||||||
| hsa-miR-4446-3p | −2.184703 | 8.113635 | 0.024205779 | hsa-miR-340-5p | −2.132230 | 5.346058 | 0.02229045 | hsa-miR-92b-5p | −2.014595 | 4.895494 | 0.044094996 |
2.5. miRNA-target evaluation and pathway analysis
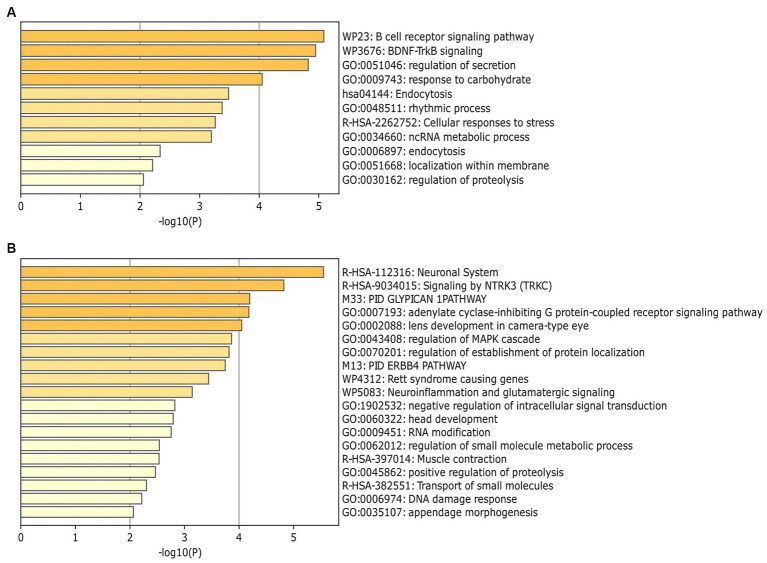
MiRNAs listed in Table 2 were exploited as input for MiRWalk analysis, acquiring their validated targets for all binding sites (3′UTR, 5′-UTR and CDS of the target) (Supplementary Table S2). For each biofluid, a distinct target list was established and filtered based on the number of miRNA hits. A threshold of at least five of was set (Supplementary Table S2). These targets were enriched in “B cell receptor signaling pathway” and “BDNF–TrkB signaling” in urine (Figure 4A, Supplementary Table S3). They were enriched in “neuronal system,” “signaling by NTRK3,” and “lens development in camera-type eye” in serum (Figure 4B, Supplementary Table S3). In addition, acquired targets were used to create a Protein–Protein Interaction Network (PPI) to demonstrate clusters of proteins retrieved from the IMEx database. A total of 2,877 nodes with 5,814 edges were produced for urine, while serum showed 900 nodes and 1,409 edges. Major protein clusters were selected as criteria with more than 200 interaction thresholds in numerous protein clusters, resulting in five clusters for urine and two clusters for serum (Figure 5, Supplementary Figures S1, S2). These protein clusters represented central nodes, corresponding to target genes having the highest number of interactions with other proteins correlated to similar biological functions. Gene Ontology (GO) enrichment analysis with ClueGO app was performed for these major clusters (Supplementary Tables S4, S5). LRRK2 was one of the central node proteins for urine. GO terms for the LRRK2 cluster related to neuronal function and brain development such as “nervous system development,” and “axon guidance.” Regarding NTRK3-ERBB2-CRK cluster central node, “immune system” and “neurotrophin signaling pathway” were enriched terms. The most enriched GO terms for the HRAS cluster associated with recognizing toxic DNA structures and membrane networking such as “ATP-dependent DNA damage sensor activity” and “membrane trafficking.” “Nervous system development,” “cellular response to stress,” and “neuron death in response to oxidative stress” were enriched GO terms for the cluster with RRP1B central node, RRP1B. Analogous with other central nodes, “peroxidase activity” and “detoxification of reactive oxygen species” were enriched biological processes found for the cluster of SOD1 central nodes. Among the serum cluster, KRAS-NTRK3 was one of the central nodes involved in neural development, namely “axon guidance,” “neuron projection and morphogenesis,” and “neuron differentiation.” Another central node of serum KDM1A seemed to be related epigenetic modification such as “histone modification,” “chromatin organization,” and “regulation of gene expression, epigenetic.” Complete GO term data from ClueGO analysis for all PPI central clusters are listed in Supplementary Tables S4, S5.
Figure 4.
Identification of enrichment ontology (GO and KEGG) terms of miRNA-target genes. (A) Top 10 enriched terms of differentially expressed miRNAs (DEmiRNA) associated genes (colored by p-values) in urine comparison between BM and IF groups. (B) Top 20 enrichment analysis for target of DEmiRNAs in serum comparison between BM and IF groups. Heatmaps of (A,B) were produced via Metascape (https://metascape.org).
Figure 5.
Clusters of proteins generated by clusterMaker 2.0 Cytoscape application with the gLay option. They were derived from protein–protein interaction (PPI) network. Central nodes (gray) represent proteins with higher numbers of interactions. Clusters in urine and serum samples were filtered with a False Discovery Rate (FDR) < 0.05 (Benjaminin Hockberg correction).
3. Materials and methods
3.1. Ethics approval and informed consent
The present study was approved by the Institutional Review Board (IRB) of Soon Chun Hyang University Hospital Cheon-An (SCHCA 2021-01-029). For all infants participating in this study, their parents or guardians signed a consent form approved by the IRB.
3.2. Sample collection
Urine samples (5–25 mL) from 33 infants were collected and kept on ice. They were then transported to a laboratory. Protease inhibitors 1 tablet (cOmpleteTM Protease inhibitor cocktail, Basel, Switzerland) was added to samples immediately to prevent degradation of exosome-associated proteins. Specimens were stored at −80°C. Venous EDTA-stabilized blood samples (n = 33) were obtained from infants who were breastfed and infant-formula-fed infants aged 3–4 weeks. Approximately 3 mL of whole blood was collected from each subject. EDTA tubes were clotted at room temperature at least 30 min and centrifuged at 1,300 g for 10 min. Supernatants were collected and transferred to new tube to obtain fresh serum from whole blood after centrifuging at 3,000 g for 20 min at 4°C. These serum samples were kept in a −80°C deep freezer until processed.
3.3. Exosomes isolation
Exosomes were isolated from serum samples using ExoQuick (cat. no. EXOQ20A-1; System Biosciences, Palo Alto, CA, United States) to precipitate particles in serum. A high centrifugation method was not suitable to extract exosomes from serum samples because the initial volume of serum was small. In addition, proteins and lipoprotein particles are abundant in the serum Thus, exosomes were isolated from serum samples using ExoQuick according to the manufacturer’s instructions. The pellet was collected and re-suspended in 100 μL of sterilized 1X PBS and then stored at −80°C. Urine samples were centrifuged at 2,000 g for 30 min at 4°C to get rid of cells and debris. Urinary exosomes can be compromised by the presence of huge amounts of Tamm-Horsfall Protein (THP), a urinary protein. This oligomerized protein can trap and sequester exosomes during centrifugation. Remaining THP proteins were removed by further centrifugation at 17,000 g for 60 min at 4°C. The supernatant was saved and the pellet containing THP was incubated with DL-dithiothreitol (DTT, final concentration of 200 mg/mL). DTT reduced disulfide bonds linking the monomers in the THP. During the incubation of DTT and pellet at 37°C for 10 min, samples were well mixed every 2 min. Before transferring supernatant and incubated pellet to clean tubes, they were syringe filtered with 0.80 μm, 0.45 μm, and 0.20 μm filters consecutively for removal of large particles and sterilization. Filtered samples were loaded onto Amicon Ultra-15 Centrifugal Filter Units with Ultracel-30 membrane (MWCO = 30 kDa; Merck Millipore, Billerica, MA, United States) and concentrated to ≤500 μL by centrifugation at 4,000 g repeatedly. The concentrate was transferred to a clean tube for further ultracentrifugation at 110,000 g for 2 h at 4°C using a swinging bucket rotor P40ST (Hitachi, Tokyo, Japan) of himac CP100NX ultracentrifuge. The pellet was collected and re-suspended in 200–300 μL of sterilized 1X PBS and then stored at −80°C until processed
3.4. Exosomes characterization
3.4.1. Western blotting
Exosomes derived from all samples were subjected to western blot analyses. Exosome samples were first treated with a RIPA lysis buffer with protease inhibitor. Protein concentration was quantified using the bicinchoninic acid (BCA) method. Absorbance values were detected with a Multiskan GO (Thermo Fisher Scientific, Waltham, MA, United States). Lysates then centrifuged at 14,000 g for 15 min at 4°C. The supernatant was resolved by SDS-PAGE and then immunoblotted with standard techniques. Exosome protein markers against CD9 (1:1000, Abcam, Cambridge, UK) and as a negative control, Calnexin (1:1000, Cell signaling Technology, Danvers, MA, United States), were validated. 293 T cells were used to obtain a cell lysate control compared to exosome markers. Signals of membranes were captured and imaged with a ChemiDocTM Touch Imaging System (Bio-Rad Laboratories, Hercules, CA, United States).
3.4.2. Transmission electron microscopy and nanoparticle tracking analysis
To confirm morphology of exosome particles and determine the presence and purity of exosome, five to ten microliters of exosome from all body fluids were applied to a glow-discharged formvar film on copper 300 mesh grids (catalog no. FCF300-CU) for transmission electron microscopy. Dried grids were washed with distilled water twice and stained for contrast using 2% uranyl acetate 5 μL for 3 s. Samples were examined with a TEM (LIBRA 120, Carl Zeiss, Germany) and images were captured. To confirm concentrations and size distributions of exosome particles, random samples from body fluids were analyzed using a nanoparticle tracking analysis instrument (Nanosight NS300; Malvern Panalytical Ltd., Malvern, UK). Nanoparticle Tracking Analysis is a technique for gaging the size and concentration of nanoparticles in suspension in real time. It is established by tracking the light scattered from suspended particles sustaining Brownian motion. Approximately 10 μL exosomes samples were diluted 1:1,000 to 1:100,000 in sterilized 1X PBS. The dilution was loaded onto a 1 mL syringe and injected into a green laser module. Sizes of particles were analyzed using NTA 3.4 Build 3.4.003 version with an sCMOS camera type. The level of the camera was 15. The number of gains was 250–350 and the temperature was 25°C. The exposure time was automatically placed in the program.
3.5. RNA extraction and library preparation
Small size RNAs containing microRNAs were isolated from exosomes samples obtained from body fluids using an miRNeasy Mini kit (Qiagen, Hilden, Germany). Exosome samples were homogenized (IKA Works, Staufen, Germany) with 700 μL QIAzol lysis buffer (Qiagen). Small size RNAs were isolated according to the manufacturer’s instructions. Finally, small size RNAs were precipitated with RNase-free water. Sizes of miRNAs were confirmed using an Agilent RNA 6000 Pico Kit and a Small RNA Kit on Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, United States).
3.6. Bioinformatic analysis
3.6.1. From sequencing to dataset
RNAs (10 ng) from each sample were used to construct sequencing libraries with a SMARTer smRNA-Seq Kit (Takara Bio, Kusatsu, Japan) following the manufacturer’s protocol. Validation of libraries was performed using Agilent Technologies 2100 Bioanalyzer by checking the size, purity, and concentration. We estimated the quantity of libraries using qPCR according to qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms). These libraries were certified using TapeStation D1000 ScreenTape (Agilent Technologies, Waldbronn, Germany). Libraries were pooled with equimolar amounts and sequenced on an Illumina HiSeq 2500 (Illumina, San Diego, CA, United States) for generating 51 bp reads. Image decomposition and quality values calculation were carried out using modules of the Illumina pipeline. Sequence alignment and detection of known microRNAs were carried out using miRDeep2 software algorithm (Berlin Institute for Medical Systems Biology at the Max-Delbruck-Center for Molecular Medicine, Berlin-Buch, Germany). Prior to executing sequence alignment, Homo sapiens hg38 reference genome was searched from UCSC genome browser and indicated using Bowtie (1.1.21) to align sequencing reads to reference sequences. Sequence alignment was then executed for Homo sapiens mature and precursor miRNAs obtained from miRBase v21.2 All steps of next-generation sequencing analysis were fulfilled with Rokit Genomics (Seoul, Korea).
3.6.2. Differential gene expression analysis
Reads for each miRNA were subjected to Trimmed mean of M-values (TMM) normalization with edge R library (Genome Biology Unit, European Molecular Biology Laboratory, Heidelberg, Germany). For pre-processing, mature miRNAs with zeroed counts across more than 50% of all samples were excluded. We added 1 with normalized read count of filtered miRNAs to facilitate log2 transformation to draw a correlation plot. For each miRNA, logCPM (Counts Per Million) and log fold change were calculated between test and control. A statistical hypothesis test for comparing groups was conducted using exactTest in edgeR. |Fold change| ≥ 2 and p-value < 0.05 were used to identify differentially expressed miRNAs between groups. Hierarchical clustering analysis was performed using complete linkage and Euclidean distance to display expression patterns of differentially expressed miRNAs that satisfied the criteria, i.e., |fold change| ≥ 2 and p-value < 0.05. Differentially expressed miRNAs were analyzed and visualized using R 3.6.1.3
3.6.3. miRNA targets retrieving and functional analysis
To determine functions of the differentially expressed miRNAs in exosomes from biofluids, predicted targets were annotated on miRWalk 3.0,4 which was provided data obtained from a machine learning algorithm including experimentally verified miRNA-target interactions. Moreover, functional enrichment analysis was performed using Metascape databases (47) with p-value < 0.05 as the cut-off criterion. A protein–protein interaction (PPI) analysis was performed for predicted targets using Cytoscape software v3.9.1 suite (48) and IMEx database (49), which contained nonredundant information derived from the major public protein databases. Afterwards, the clusterMaker 2.0, a Cytoscape application (50) with the “gLay” option, was used to highlight different clusters within the network based on the number and type of connections between nodes. Clusters with several interactions greater than 30 were examined in Gene Ontology (GO) enrichment analysis carried out with ClueGO and CluePedia (51, 52). Results were filtered with an FDR < 0.05 (Benjaminin Hockberg correction).
4. Discussion
Evidence is accumulating that exosomes have a potential role in providing us knowledge of pathologic processes and in aiding biomarker discovery for commonly faced conditions in preterm infants (53). Especially, exosomes are easily found in various biological fluids (54–58). They are widely used as research sources by ensuring stability, quality, and quantity of exosomal miRNAs (59, 60). Studies have reported that BM is associated with a reduced incidence of common neonatal complications (19, 21–26). However, the role of miRNAs of exosomes in biological fluids when BM is consumed in preterm infants has not been reported yet. Our first objective was to determine differences in prototypical exosomal miRNAs between BM and IF groups as differences about metabolites in infants fed with BM and IF had been previously documented (17, 61). Our sequencing data revealed that six miRNAs were differentially expressed in urinary exosomes and that 10 miRNAs were significantly up-regulated in serum exosomes of BM-fed preterm infants than in IF-fed infants (Table 2).
A total of 16 upregulated DEmiRNAs in BM were recognized in this study, including miR-619-5p, miR-664a-5p, miR-484, miR-19a-3p, miR-627-5p, miR-33a-5p, and miR-12136. Of them, hsa-miR-619-5p has been reported to be able to bind fully complementary to mRNAs of more than 200 human genes (62), suggesting that it has a broad biological function. MiR-664a-5p has been linked to neuronal differentiation (63) and osteogenic differentiation (64). MiR-484 is located in the Meiosis arrest female1 (MARF1) promoter region on chromosome 16q13.11 in human related to nervous system (65–67). It is increased in miR-484 alleviated cerebral ischemia/reperfusion injury of a mouse model (68). Especially, it has been reported that miR-19a-3p is correlated with sepsis-induced coagulopathy (69) and that it can function as a potential therapeutic target in neonates with sepsis-induced disseminated intravascular coagulation (70). In upregulated miRNAs in the serum, hsa-miR-627-5p can restrain pulmonary artery smooth muscle cell dysfunction by targeting MAP2K4 and PI3K/AKT signaling (71) and chronic lung disease, the most common complication in preterm infants (72). MiR-33a-5p can inhibit viral infection by targeting eukaryotic translation elongation factor 1A1 (EEF1A1) (73). Moreover, it has been reported that miR-12136 is highly expressed in the brain tissue and that it plays an crucial role in the development of the nervous system (74). Therefore, these differentially expressed miRNAs in BM (Table 3) can contribute to nervous system development or alleviate neonatal complications, including coagulation, chronic lung disease, and viral infection.
Table 3.
Target gene consistencies of each biofluids detailed with respect to the binding sites.
| Biofluid | miRNA | Target 3′-UTR | Target 5′-UTR | Target CDS | Filtered target |
|---|---|---|---|---|---|
| Urine | hsa-miR-619-5p | 10,023 | 1,254 | 7,650 | 40 |
| hsa-miR-664a-5p | 4,790 | 680 | 4,836 | ||
| hsa-miR-484 | 6,435 | 1,447 | 5,498 | ||
| hsa-miR-19a-3p | 488 | 37 | 294 | ||
| hsa-miR-10a-5p | 2,476 | 581 | 3,011 | ||
| hsa-miR-328-3p | 6,970 | 2,026 | 6,198 | ||
| Serum | hsa-miR-627-5p | 1,850 | 244 | 1,473 | 87 |
| hsa-miR-545-5p | 64 | 5 | 15 | ||
| hsa-miR-539-3p | 186 | 12 | 102 | ||
| hsa-miR-33a-5p | 2,774 | 319 | 2,941 | ||
| hsa-miR-1296-5p | 7,310 | 2,069 | 9,152 | ||
| hsa-miR-4284 | 5,038 | 636 | 3,811 | ||
| hsa-miR-548o-3p | 667 | 85 | 634 | ||
| hsa-miR-338-3p | 2,271 | 380 | 4,504 | ||
| hsa-miR-12136 | 904 | 73 | 803 | ||
| hsa-miR-483-3p | 7,917 | 1,782 | 5,573 |
According to gene ontology analysis of DEmiRNAs target genes, ‘B cell receptor signaling pathway’ and ‘BDNF–TrkB signaling’ were the most enriched pathways in urine. These two biological processes mainly contribute to immune responses (75) and protect neurons from cell death caused by reactive oxygen species (76), respectively. Oxygen therapy is frequently used in the treatment of neonatal diseases. However, it can cause neuronal glial cell death and result in neonatal brain injury (77). Consumption of BM is involved in the reduction of metabolites associated with reactive oxygen species in urine of preterm infants than full-term infants (17). In this regard, enrichment analysis results discovered that BM consumption in preterm infants might reduce side effects of oxygen treatment in the NICU. In addition, ‘neuronal system’, ‘signaling by NTRK3’, and ‘lens development in camera-type eye’ were enriched GO terms in the serum. A large cohort’s study of preterm infants with 30 weeks of gestation has shown that BM consumption has statistically significant effects on visual acuity (p = 0.003) and teller acuity scores rather than IF feeding (p < 0.0001) (15). Moreover, a recent study has reported that BM feeding is associated with a reduced odds of stage 2 or 3 ROP (OR = 0.25, 95% CI: 0.091–0.705; p = 0.009) in preterm infants (24). That study also claimed that early BM feeding before achieving complete enteral feeding of bovine milk-containing products might alleviate the development of ROP. Taken together, these results suggest that consumption of breast milk may reduce the incidence and severity of neonatal complications.
A total of seven central node genes were recognized by protein network analysis, including LRRK2 (Leucine-Rich Repeat Kinase 2), ERBB2-NTRK3-CRK, HRAS (Harvey rat sarcoma virus), RRP1B (Ribosomal RNA Processing 1B), and SOD1 (superoxide dismutase 1) in urine and KRAS-NTRK3 and KDM1A (Lysine Demethylase 1A) in serum. NTRK3 (Neurotrophic tyrosine kinase, receptor, type 3) was a central node protein for both biofluids. It belongs to the neurotrophic tyrosine kinase family containing c-terminal protein tyrosine kinase domain. It is essential for development and growth of the nervous system (78). Interestingly, NTRK3 is a receptor for neurotrophins. A recent study has reported significant different levels of neurotrophins in preterm infants compared to term infants (79). In line with that study, our study also showed significant differences in levels of miRNAs related to NTRK3 in both urine and serum samples of preterm infants between BM-fed and formula-fed groups. Among central nodes were ERBB2 (v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2), CRK (Cysteine-rich receptor-like-kinase), HRAS, and KRAS (Kirsten rat sarcoma virus), all of which were proto-oncogenes involved in the development of various cancer types. Their overexpression is also closely related to various diseases (80–82). Data of this study informed that miR-627-5p, miR-4284, miR-33a-5p, miR-1296-5p, and miR-12136 were involved in KRAS protein regulation (Supplementary Table S2). KRAS protein was implicated in congenital pulmonary disease in a recent study (83), suggesting that these DEmiRNAs might be related to neonatal pulmonary disease. Among them, miR-1296 has been suggested as potential biomarker in chronic lung disease in adults (84). Other miRNAs have not been reported in relation to pulmonary disease. These findings suggest that miRNA-627-5p, miR-4284, miR-33a-5p, miR-1296-5p, and miR-12136 require further investigation in cases of neonatal pulmonary disease before they can be used as biomarkers or therapeutic targets. Another function derived from the PPI cluster analysis was immune involvement of RRP1B, also known as nucleolar protein. When RRP1B is overexpressed, it can promote RNA-dependent RNA polymerase binding to capped mRNA and enhance viral transcription of influenza A viruses (85). In addition, it is contagious (86). Moreover, LRRK2 protein, another central cluster protein supported by the network analysis (Figure 5), is concerned with lysosomal activity in neurons with a role in the autophagy pathway (87). Interestingly, a recent study has shown that autophagy is dependent on oxidative stress and that inhibiting autophagy can significantly reduce ischemic damage in a neonatal rat brain model (88). SOD1 is an enzyme constituting important antioxidant defense against oxidative stress in human body that has therapeutic effects on various disease (89). Some studies on prematurity have suggested that haplotypes of SOD1 genes might be independent protective or risk indicators for complications of prematurity (90). Poggi et al. have reported that SOD1 can reduce the risk of RDS, IVH, and ROP in a study of 152 preterm infants (91). Regarding the involvement of other antioxidant genes, KDM1A that can regulate homeostasis of histone methylation (92) has been reported to play a role in regulating autophagy. It is involved in the transcriptional control of various downstream effectors (93). Especially, LSD1 participates in antioxidant gene expression with NRF2 (nuclear factor erythroid 2-related factor 2) (94). It may reduce oxidative stress. Accumulating evidence of various effects of BM on preterm infants related to immune response, nervous system, eye development, and brain development could improve neonatal conditions in NICUs. Despite advances in NICUs, preterm infants often suffer from neurological disorders in later life. Premature infants are especially vulnerable to injury caused by ROS due to their imperfect endogenous radical scavenging systems (77). In this regard, exploiting involvement in antioxidant processes through analysis of DEmiRNAs and their target genes in BM could lead to useful therapies for preterm infants with specific prevention strategies against several complications.
In summary, we found that intake of BM and IF in NICU caused practical differences in exosomal miRNA contents in urine and serum samples of preterm infants. Increased miRNAs and their targets were relevant for neural development, ROS regulation, and detoxification processes. It would be interesting to improve neonatal status and further develop neuronal system containing brain and eye. While this is intriguing, the impact of certain amounts of transcription on gene expression needs to be evaluated. The amount of efficacy on cells or tissues of preterm infants needs to be assessed in all aspects to elucidate their roles through in vitro and in vivo experiments.
Data availability statement
The original contributions presented in the study are publicly available. The raw sequence datasets generated for this study can be found in SRA. BioProject ID collecting all samples is the following: PRJNA1035999.
Ethics statement
The studies involving humans were approved by Investigational Review Board (IRB) of Soon Chun Hyang University Hospital Cheon-An (SCHCA 2021-01-029). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
E-BK: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft. JS: Investigation, Validation, Writing – review & editing. LL: Investigation, Data curation, Writing-review & editing. HK: Data curation, Methodology, Software, Writing – original draft. JK: Investigation, Methodology, Software, Writing – original draft. YS: Methodology, Software, Validation, Writing – original draft. HJ: Data curation, Software, Visualization, Writing – original draft. H-TK: Project administration, Supervision, Writing – review & editing. SR: Conceptualization, Project administration, Supervision, Validation, Writing – review & editing.
Acknowledgments
This work is part of the E-BK’s Ph.D. thesis at the Soonchunhyang University, Asan, Korea.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the Bio & Medical Technology Development Program (2019M3E5D3073092) and the Basic Science Research Program (MSIT, NRF-2021R1A2C3012633, 2020R1A2C1100479, and RS-2023-00219563), through of the National Research Foundation of Korea (NRF) funded by the Ministry of Science & ICT, the Ministry of Education (2023RS00274601), and the Soonchunhyang University Research Fund.
Footnotes
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnut.2024.1339919/full#supplementary-material
Clusters of proteins generated by clusterMaker 2.0 Cytoscape application using differential expressed miRNAs targets in urine. miRNA Targets were filtered based on established criteria.
Clusters of proteins generated by clusterMaker 2.0 Cytoscape application using differential expressed miRNAs targets in serum. miRNA Targets were filtered based on established criteria.
Mapping statistics by smallRNA library sequencing.
List of genes selected by targeting 5 or more miRNAs in each sample, and miRNA’s target gene consistency for binding sites.
Functional enrichment analysis results from Metascape with the target gene list selected in S2.
Gene Ontology enrichment results of clstuer analysis in Urine.
Gene Ontology enrichment results of clstuer analysis in Serum.
References
- 1.WHO . WHO: recommended definitions, terminology and format for statistical tables related to the perinatal period and use of a new certificate for cause of perinatal deaths. Modifications recommended by FIGO as amended October 14, 1976. Acta Obstet Gynecol Scand. (1977) 56:247–53. [PubMed] [Google Scholar]
- 2.Rubens CE, Sadovsky Y, Muglia L, Gravett MG, Lackritz E, Gravett C. Prevention of preterm birth: harnessing science to address the global epidemic. Sci Transl Med. (2014) 6:262sr5. doi: 10.1126/scitranslmed.3009871 [DOI] [PubMed] [Google Scholar]
- 3.Avila-Alvarez A, Boga AS, Bermúdez-Hormigo C, Carballal JF. Extrauterine growth restriction among neonates with a birthweight less than 1,500 grams. Anales De Pediatria. (2018) 89:325–32. doi: 10.1016/j.anpedi.2018.02.004 [DOI] [PubMed] [Google Scholar]
- 4.Coskun D, Gunay U. The effects of kangaroo care applied by Turkish mothers who have premature babies and cannot breastfeed on their stress levels and amount of Milk production. J Pediatr Nurs. (2020) 50:e26–32. doi: 10.1016/j.pedn.2019.09.028 [DOI] [PubMed] [Google Scholar]
- 5.Cristofalo EA, Schanler RJ, Blanco CL, Sullivan S, Trawoeger R, Kiechl-Kohlendorfer U, et al. Randomized trial of exclusive human milk versus preterm formula diets in extremely premature infants. J Pediatr. (2013) 163:1592–1595.e1. doi: 10.1016/j.jpeds.2013.07.011, PMID: [DOI] [PubMed] [Google Scholar]
- 6.Morales Y, Schanler RJ. Human milk and clinical outcomes in VLBW infants: how compelling is the evidence of benefit? Semin Perinatol. (2007) 31:83–8. doi: 10.1053/j.semperi.2007.02.002, PMID: [DOI] [PubMed] [Google Scholar]
- 7.Anderson JW, Johnstone BM, Remley DT. Breast-feeding and cognitive development: a meta-analysis. Am J Clin Nutr. (1999) 70:525–35. doi: 10.1093/ajcn/70.4.525 [DOI] [PubMed] [Google Scholar]
- 8.Roze JC, Darmaun D, Boquien CY, Flamant C, Picaud JC, Savagner C, et al. The apparent breastfeeding paradox in very preterm infants: relationship between breast feeding, early weight gain and neurodevelopment based on results from two cohorts, EPIPAGE and LIFT. BMJ Open. (2012) 2:e000834. doi: 10.1136/bmjopen-2012-000834, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr LE, Virmani MD, Rosa F, Munblit D, Matazel KS, Elolimy AA, et al. Role of human Milk bioactives on Infants' gut and immune health. Front Immunol. (2021) 12:604080. doi: 10.3389/fimmu.2021.604080, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhutta AT, Cleves MA, Casey PH, Cradock MM, Anand KJ. Cognitive and behavioral outcomes of school-aged children who were born preterm: a meta-analysis. JAMA. (2002) 288:728–37. doi: 10.1001/jama.288.6.728, PMID: [DOI] [PubMed] [Google Scholar]
- 11.Khetan R, Hurley M, Spencer S, Bhatt JM. Bronchopulmonary dysplasia within and beyond the neonatal unit. Adv Neonatal Care. (2016) 16:17–25. doi: 10.1097/ANC.0000000000000251 [DOI] [PubMed] [Google Scholar]
- 12.Drane DL, Logemann JA. A critical evaluation of the evidence on the association between type of infant feeding and cognitive development. Paediatr Perinat Epidemiol. (2000) 14:349–56. doi: 10.1046/j.1365-3016.2000.00301.x, PMID: [DOI] [PubMed] [Google Scholar]
- 13.Jain A, Concato J, Leventhal JM. How good is the evidence linking breastfeeding and intelligence? Pediatrics. (2002) 109:1044–53. doi: 10.1542/peds.109.6.1044, PMID: [DOI] [PubMed] [Google Scholar]
- 14.Furman L, Wilson-Costello D, Friedman H, Taylor HG, Minich N, Hack M. The effect of neonatal maternal milk feeding on the neurodevelopmental outcome of very low birth weight infants. J Dev Behav Pediatr. (2004) 25:247–53. doi: 10.1097/00004703-200408000-00004 [DOI] [PubMed] [Google Scholar]
- 15.O'Connor DL, Jacobs F, Hall R, Adamkin D, Auestad T, Castillo I, et al. Growth and development of premature infants fed predominantly human milk, predominantly premature infant formula, or a combination of human milk and premature formula. J Pediatr Gastroenterol Nutr. (2003) 37:437–46. doi: 10.1097/00005176-200310000-00008, PMID: [DOI] [PubMed] [Google Scholar]
- 16.Boquien CY. Human Milk: An ideal food for nutrition of preterm newborn. Front Pediatr. (2018) 6:295. doi: 10.3389/fped.2018.00295, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledo A, Arduini A, Asensi MA, Sastre J, Escrig R, Brugada M, et al. Human milk enhances antioxidant defenses against hydroxyl radical aggression in preterm infants. Am J Clin Nutr. (2009) 89:210–5. doi: 10.3945/ajcn.2008.26845, PMID: [DOI] [PubMed] [Google Scholar]
- 18.Jang HL, Cho JY, Kim MJ, Kim EJ, Park EY, Park SA, et al. The experience of human Milk banking for 8 years: Korean perspective. J Korean Med Sci. (2016) 31:1775–83. doi: 10.3346/jkms.2016.31.11.1775, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adhisivam B, Vishnu Bhat B, Banupriya N, Poorna R, Plakkal N, Palanivel C. Impact of human milk banking on neonatal mortality, necrotizing enterocolitis, and exclusive breastfeeding - experience from a tertiary care teaching hospital, South India. J Matern Fetal Neonatal Med. (2019) 32:902–5. doi: 10.1080/14767058.2017.1395012 [DOI] [PubMed] [Google Scholar]
- 20.Vazquez DC, Garcia SS, Renau MI, Iglesias-Platas I. Availability of donor Milk for very preterm infants decreased the risk of necrotizing enterocolitis without adversely impacting growth or rates of breastfeeding. Nutrients. (2019) 11:1895. doi: 10.3390/nu11081895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bharwani SK, Green BF, Pezzullo JC, Bharwani SS, Bharwani SS, Dhanireddy R. Systematic review and meta-analysis of human milk intake and retinopathy of prematurity: a significant update. J Perinatol. (2016) 36:913–20. doi: 10.1038/jp.2016.98, PMID: [DOI] [PubMed] [Google Scholar]
- 22.Cacho NT, Parker LA, Neu J. Necrotizing enterocolitis and human Milk feeding: a systematic review. Clin Perinatol. (2017) 44:49–67. doi: 10.1016/j.clp.2016.11.009 [DOI] [PubMed] [Google Scholar]
- 23.El-Mohandes AE, Picard MB, Simmens SJ, Keiser JF. Use of human milk in the intensive care nursery decreases the incidence of nosocomial sepsis. J Perinatol. (1997) 17:130–4. [PubMed] [Google Scholar]
- 24.Ginovart G, Gich I, Verd S. Human milk feeding protects very low-birth-weight infants from retinopathy of prematurity: a pre-post cohort analysis. J Matern Fetal Neonatal Med. (2016) 29:3790–5. doi: 10.3109/14767058.2016.1145648, PMID: [DOI] [PubMed] [Google Scholar]
- 25.Hock A, Miyake H, Li B, Lee C, Ermini L, Koike Y, et al. Breast milk-derived exosomes promote intestinal epithelial cell growth. J Pediatr Surg. (2017) 52:755–9. doi: 10.1016/j.jpedsurg.2017.01.032, PMID: [DOI] [PubMed] [Google Scholar]
- 26.McNelis K, Fu TT, Poindexter B. Nutrition for the extremely preterm infant. Clin Perinatol. (2017) 44:395–406. doi: 10.1016/j.clp.2017.01.012 [DOI] [PubMed] [Google Scholar]
- 27.Godoy PM, Bhakta NR, Barczak AJ, Cakmak H, Fisher S, MacKenzie TC, et al. Large differences in small RNA composition between human biofluids. Cell Rep. (2018) 25:1346–58. doi: 10.1016/j.celrep.2018.10.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thery C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. (2018) 7:1535750. doi: 10.1080/20013078.2018.1535750, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiedt S, Prestel M, Malik R, Schieferdecker N, Duering M, Kautzky V, et al. RNA-Seq identifies circulating miR-125a-5p, miR-125b-5p, and miR-143-3p as potential biomarkers for acute ischemic stroke. Circ Res. (2017) 121:970–80. doi: 10.1161/CIRCRESAHA.117.311572, PMID: [DOI] [PubMed] [Google Scholar]
- 30.Hu G, Drescher KM, Chen XM. Exosomal miRNAs: biological properties and therapeutic potential. Front Genet. (2012) 3:56. doi: 10.3389/fgene.2012.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Q, Su H, Li J, Lyon C, Tang W, Wan M, et al. Clinical applications of exosome membrane proteins. Precis Clin Med. (2020) 3:54–66. doi: 10.1093/pcmedi/pbaa007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambros V. The functions of animal microRNAs. Nature. (2004) 431:350–5. doi: 10.1038/nature02871 [DOI] [PubMed] [Google Scholar]
- 33.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. (2019) 20:5–20. doi: 10.1038/s41580-018-0059-1, PMID: [DOI] [PubMed] [Google Scholar]
- 34.Sun YF, Ma L, Gong XH, Hong WC, Cai C. Expression of microRNA-495-5p in preterm infants with bronchopulmonary dysplasia: a bioinformatics analysis. Zhongguo Dang Dai Er Ke Za Zhi. (2020) 22:24–30. doi: 10.7499/j.issn.1008-8830.2020.01.006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kan Q, Ding S, Yang Y, Zhou X. Expression profile of plasma microRNAs in premature infants with respiratory distress syndrome. Mol Med Rep. (2015) 12:2858–64. doi: 10.3892/mmr.2015.3699, PMID: [DOI] [PubMed] [Google Scholar]
- 36.Mar PK, Galley J, Rajab A, Besner GE. Urine extracellular vesicle-derived miRNA patterns in infants with necrotizing enterocolitis. Pediatrics. (2021) 147:923. doi: 10.1542/peds.147.3MA9.923a [DOI] [Google Scholar]
- 37.Lal CV, Olave N, Travers C, Rezonzew G, Dolma K, Simpson A, et al. Exosomal microRNA predicts and protects against severe bronchopulmonary dysplasia in extremely premature infants. JCI. Insight. (2018) 3:e93994. doi: 10.1172/jci.insight.93994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma M, Luo Q, Fan L, Li W, Li Q, Meng Y, et al. The urinary exosomes derived from premature infants attenuate cisplatin-induced acute kidney injury in mice via microRNA-30a-5p/ mitogen-activated protein kinase 8 (MAPK8). Bioengineered. (2022) 13:1650–65. doi: 10.1080/21655979.2021.2021686, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Umair Z, Baek MO, Song J, An S, Chon SJ, Yoon MS. MicroRNA-4516 in urinary exosomes as a biomarker of premature ovarian insufficiency. Cell. (2022) 11:2797. doi: 10.3390/cells11182797, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong X, Weng B, Zhang X, Yan C, Cai C. The molecular basis of brain injury in preterm infants with sepsis - associated encephalopathy. BMC Pediatr. (2022) 22:336. doi: 10.1186/s12887-022-03372-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quintanilha BJ, Reis BZ, Duarte GBS, Cozzolino SMF, Rogero MM. Nutrimiromics: role of microRNAs and nutrition in modulating inflammation and chronic diseases. Nutrients. (2017) 9:1168. doi: 10.3390/nu9111168, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortega FJ, Cardona-Alvarado MI, Mercader JM, Moreno-Navarrete JM, Moreno M, Sabater M, et al. Circulating profiling reveals the effect of a polyunsaturated fatty acid-enriched diet on common microRNAs. J Nutr Biochem. (2015) 26:1095–101. doi: 10.1016/j.jnutbio.2015.05.001, PMID: [DOI] [PubMed] [Google Scholar]
- 43.Tian L, Song Z, Shao W, Du WW, Zhao LR, Zeng K, et al. Curcumin represses mouse 3T3-L1 cell adipogenic differentiation via inhibiting miR-17-5p and stimulating the Wnt signalling pathway effector Tcf7l2. Cell Death Dis. (2017) 8:e2559. doi: 10.1038/cddis.2016.455, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim EJ, Lee NM, Chung SH. A retrospective study on the effects of exclusive donor human milk feeding in a short period after birth on morbidity and growth of preterm infants during hospitalization. Medicine (Baltimore). (2017) 96:e7970. doi: 10.1097/MD.0000000000007970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costa S, Maggio L, Alighieri G, Barone G, Cota F, Vento G. Tolerance of preterm formula versus pasteurized donor human milk in very preterm infants: a randomized non-inferiority trial. Ital J Pediatr. (2018) 44:96. doi: 10.1186/s13052-018-0532-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corpeleijn WE, de Waard M, Christmann V, van Goudoever JB, Jansen-van der Weide MC, Kooi EM, et al. Effect of donor Milk on severe infections and mortality in very low-birth-weight infants: the early nutrition study randomized clinical trial. JAMA Pediatr. (2016) 170:654–61. doi: 10.1001/jamapediatrics.2016.0183 [DOI] [PubMed] [Google Scholar]
- 47.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. (2019) 10:1523. doi: 10.1038/s41467-019-09234-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. (2003) 13:2498–504. doi: 10.1101/gr.1239303, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. (2014) 42:D358–63. doi: 10.1093/nar/gkt1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris JH, Apeltsin L, Newman AM, Baumbach J, Wittkop T, Su G, et al. clusterMaker: a multi-algorithm clustering plugin for Cytoscape. BMC Bioinformatics. (2011) 12:436. doi: 10.1186/1471-2105-12-436, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. (2013) 29:661–3. doi: 10.1093/bioinformatics/btt019, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. (2009) 25:1091–3. doi: 10.1093/bioinformatics/btp101, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schiller EA, Cohen K, Lin X, El-Khawam R, Hanna N. Extracellular vesicle-microRNAs as diagnostic biomarkers in preterm neonates. Int J Mol Sci. (2023) 24:2622. doi: 10.3390/ijms24032622, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. (2020) 367:eaau6977. doi: 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Admyre C, Johansson SM, Qazi KR, Filen JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. (2007) 179:1969–78. doi: 10.4049/jimmunol.179.3.1969, PMID: [DOI] [PubMed] [Google Scholar]
- 56.Liu J, Sun W, Liu C, Na Q. Umbilical cord blood-derived exosomes in maternal-fetal disease: a review. Reprod Sci. (2023) 30:54–61. doi: 10.1007/s43032-022-00879-1, PMID: [DOI] [PubMed] [Google Scholar]
- 57.Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. (2010) 16:34–8. doi: 10.1111/j.1601-0825.2009.01604.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, et al. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. (2007) 72:1095–102. doi: 10.1038/sj.ki.5002486 [DOI] [PubMed] [Google Scholar]
- 59.Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, et al. The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer. (2020) 147:2934–47. doi: 10.1002/ijc.33111 [DOI] [PubMed] [Google Scholar]
- 60.Nik Mohamed Kamal N, Shahidan WNS. Non-Exosomal and Exosomal circulatory MicroRNAs: which are more valid as biomarkers? Front Pharmacol. (2019) 10:1500. doi: 10.3389/fphar.2019.01500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aguilar-Lozano A, Baier S, Grove R, Shu J, Giraud D, Leiferman A, et al. Concentrations of purine metabolites are elevated in fluids from adults and infants and in livers from mice fed diets depleted of bovine Milk exosomes and their RNA cargos. J Nutr. (2018) 148:1886–94. doi: 10.1093/jn/nxy223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atambayeva S, Niyazova R, Ivashchenko A, Pyrkova A, Pinsky I, Akimniyazova A, et al. The binding sites of miR-619-5p in the mRNAs of human and orthologous genes. BMC Genomics. (2017) 18:428. doi: 10.1186/s12864-017-3811-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watanabe K, Yamaji R, Ohtsuki T. MicroRNA-664a-5p promotes neuronal differentiation of SH-SY5Y cells. Genes Cells. (2018) 23:225–33. doi: 10.1111/gtc.12559, PMID: [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Liu Y, Wu M, Wang H, Wu L, Xu B, et al. MicroRNA-664a-5p promotes osteogenic differentiation of human bone marrow-derived mesenchymal stem cells by directly downregulating HMGA2. Biochem Biophys Res Commun. (2020) 521:9–14. doi: 10.1016/j.bbrc.2019.09.122 [DOI] [PubMed] [Google Scholar]
- 65.Jia YZ, Liu J, Wang GQ, Song ZF. miR-484: a potential biomarker in health and disease. Front Oncol. (2022) 12:830420. doi: 10.3389/fonc.2022.830420, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Allach El Khattabi L, Heide S, Caberg JH, Andrieux J, Doco Fenzy M, Vincent-Delorme C, et al. 16p13.11 microduplication in 45 new patients: refined clinical significance and genotype-phenotype correlations. J Med Genet. (2020) 57:301–7. doi: 10.1136/jmedgenet-2018-105389, PMID: [DOI] [PubMed] [Google Scholar]
- 67.Fujitani M, Zhang S, Fujiki R, Fujihara Y, Yamashita T. A chromosome 16p13.11 microduplication causes hyperactivity through dysregulation of miR-484/protocadherin-19 signaling. Mol Psychiatry. (2017) 22:364–74. doi: 10.1038/mp.2016.106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu X, Wang X, Zhang L, Zhou Y, Yang L, Yang M. By targeting apoptosis facilitator BCL2L13, microRNA miR-484 alleviates cerebral ischemia/reperfusion injury-induced neuronal apoptosis in mice. Bioengineered. (2021) 12:948–59. doi: 10.1080/21655979.2021.1898134, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Y, Wang H, Zhang C, Zhang C, Yang H, Gao R, et al. Plasma Hsa-miR-92a-3p in correlation with lipocalin-2 is associated with sepsis-induced coagulopathy. BMC Infect Dis. (2020) 20:155. doi: 10.1186/s12879-020-4853-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang R, Lu S, Yang X, Li M, Jia H, Liao J, et al. miR-19a-3p downregulates tissue factor and functions as a potential therapeutic target for sepsis-induced disseminated intravascular coagulation. Biochem Pharmacol. (2021) 192:114671. doi: 10.1016/j.bcp.2021.114671, PMID: [DOI] [PubMed] [Google Scholar]
- 71.Li T, Tan X, Huang Y, Cui J, Chen F, Xiong Y. MicroRNA miR-627-5p restrains pulmonary artery smooth muscle cell dysfunction by targeting MAP 2 K4 and PI3K/AKT signaling. Genes Environ. (2022) 44:23. doi: 10.1186/s41021-022-00251-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hilgendorff A, Reiss I, Ehrhardt H, Eickelberg O, Alvira CM. Chronic lung disease in the preterm infant. Lessons learned from animal models. Am J Respir Cell Mol Biol. (2014) 50:233–45. doi: 10.1165/rcmb.2013-0014TR, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Z, Ye J, Ashraf U, Li Y, Wei S, Wan S, et al. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol. (2016) 90:3722–34. doi: 10.1128/JVI.03242-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garg P, Jamal F, Srivastava P. Deciphering the role of precursor miR-12136 and miR-8485 in the progression of intellectual disability (ID). IBRO Neurosci Rep. (2022) 13:393–401. doi: 10.1016/j.ibneur.2022.10.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tanaka S, Baba Y. B cell receptor signaling. Adv Exp Med Biol. (2020) 1254:23–36. doi: 10.1007/978-981-15-3532-1_2 [DOI] [PubMed] [Google Scholar]
- 76.Jin W. Regulation of BDNF-TrkB signaling and potential therapeutic strategies for Parkinson's disease. J Clin Med. (2020) 9:257. doi: 10.3390/jcm9010257, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reich B, Hoeber D, Bendix I, Felderhoff-Mueser U. Hyperoxia and the immature brain. Dev Neurosci. (2016) 38:311–30. doi: 10.1159/000454917, PMID: [DOI] [PubMed] [Google Scholar]
- 78.Kaplan DR, Miller FD. Neurotrophin signal transduction in the nervous system. Curr Opin Neurobiol. (2000) 10:381–91. doi: 10.1016/S0959-4388(00)00092-1 [DOI] [PubMed] [Google Scholar]
- 79.Krey FC, Stocchero BA, Creutzberg KC, Heberle BA, Tractenberg SG, Xiang L, et al. Neurotrophic factor levels in preterm infants: a systematic review and Meta-analysis. Front Neurol. (2021) 12:643576. doi: 10.3389/fneur.2021.643576, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Iqbal N, Iqbal N. Human epidermal growth factor receptor 2 (HER2) in cancers: overexpression and therapeutic implications. Mol Biol Int. (2014) 2014:852748: 1–9. doi: 10.1155/2014/852748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Birge RB, Kalodimos C, Inagaki F, Tanaka S. Crk and CrkL adaptor proteins: networks for physiological and pathological signaling. Cell Commun Signal. (2009) 7:13. doi: 10.1186/1478-811X-7-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pazik M, Michalska K, Zebrowska-Nawrocka M, Zawadzka I, Lochowski M, Balcerczak E. Clinical significance of HRAS and KRAS genes expression in patients with non-small-cell lung cancer - preliminary findings. BMC Cancer. (2021) 21:130. doi: 10.1186/s12885-021-07858-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Muntean A, Banias LE, Ade-Ajayi N, Patel SB, McKinney O, Davenport M. Neonatal congenital pulmonary airway malformation associated with mucinous adenocarcinoma and KRAS mutations. J Pediatr Surg. (2022) 57:520–6. doi: 10.1016/j.jpedsurg.2021.12.018, PMID: [DOI] [PubMed] [Google Scholar]
- 84.Deng H, Xie C, Ye Y, Du Z. MicroRNA-1296 expression is associated with prognosis and inhibits cell proliferation and invasion by Wnt signaling in non-small cell lung cancer. Oncol Lett. (2020) 19:623–30. doi: 10.3892/ol.2019.11154, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Su WC, Hsu SF, Lee YY, Jeng KS, Lai MM. A nucleolar protein, ribosomal RNA processing 1 homolog B (RRP1B), enhances the recruitment of cellular mRNA in influenza virus transcription. J Virol. (2015) 89:11245–55. doi: 10.1128/JVI.01487-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barak S, Kushnir A, Chulski E, Miron D. Influenza a/H1N1 virus in very low-birth-weight premature infant: case report. Am J Perinatol. (2010) 27:513–5. doi: 10.1055/s-0030-1247606, PMID: [DOI] [PubMed] [Google Scholar]
- 87.Wallings R, Connor-Robson N, Wade-Martins R. LRRK2 interacts with the vacuolar-type H+-ATPase pump a1 subunit to regulate lysosomal function. Hum Mol Genet. (2019) 28:2696–710. doi: 10.1093/hmg/ddz088, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu Q, Harris VA, Kumar S, Mansour HM, Black SM. Autophagy in neonatal hypoxia ischemic brain is associated with oxidative stress. Redox Biol. (2015) 6:516–23. doi: 10.1016/j.redox.2015.06.016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Younus H. Therapeutic potentials of superoxide dismutase. Int J Health Sci (Qassim). (2018) 12:88–93. PMID: [PMC free article] [PubMed] [Google Scholar]
- 90.Giusti B, Vestrini A, Poggi C, Magi A, Pasquini E, Abbate R, et al. Genetic polymorphisms of antioxidant enzymes as risk factors for oxidative stress-associated complications in preterm infants. Free Radic Res. (2012) 46:1130–9. doi: 10.3109/10715762.2012.692787 [DOI] [PubMed] [Google Scholar]
- 91.Poggi C, Giusti B, Vestri A, Pasquini E, Abbate R, Dani C. Genetic polymorphisms of antioxidant enzymes in preterm infants. J Matern Fetal Neonatal Med. (2012) 25:123–6. doi: 10.3109/14767058.2012.714976 [DOI] [PubMed] [Google Scholar]
- 92.Li Y, Zhao Y, Li X, Zhai L, Zheng H, Yan Y, et al. Biological and therapeutic role of LSD1 in Alzheimer's diseases. Front Pharmacol. (2022) 13:1020556. doi: 10.3389/fphar.2022.1020556, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fullgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. (2014) 15:65–74. doi: 10.1038/nrm3716, PMID: [DOI] [PubMed] [Google Scholar]
- 94.Lin CY, Chang CB, Wu RC, Chao A, Lee YS, Tsai CN, et al. Glucose activates lysine-specific demethylase 1 through the KEAP1/p62 pathway. Antioxidants (Basel). (2021) 10:1898. doi: 10.3390/antiox10121898 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clusters of proteins generated by clusterMaker 2.0 Cytoscape application using differential expressed miRNAs targets in urine. miRNA Targets were filtered based on established criteria.
Clusters of proteins generated by clusterMaker 2.0 Cytoscape application using differential expressed miRNAs targets in serum. miRNA Targets were filtered based on established criteria.
Mapping statistics by smallRNA library sequencing.
List of genes selected by targeting 5 or more miRNAs in each sample, and miRNA’s target gene consistency for binding sites.
Functional enrichment analysis results from Metascape with the target gene list selected in S2.
Gene Ontology enrichment results of clstuer analysis in Urine.
Gene Ontology enrichment results of clstuer analysis in Serum.
Data Availability Statement
The original contributions presented in the study are publicly available. The raw sequence datasets generated for this study can be found in SRA. BioProject ID collecting all samples is the following: PRJNA1035999.