Summary
Obstructive sleep apnea (OSA) induces intermittent hypoxia (IH), an independent risk factor for non-alcoholic fatty liver disease (NAFLD). While the molecular links between IH and NAFLD progression are unclear, immune cell-driven inflammation plays a crucial role in NAFLD pathogenesis. Using lean mice exposed to long-term IH and a cohort of lean OSA patients (n = 71), we conducted comprehensive hepatic transcriptomics, lipidomics, and targeted serum proteomics. Significantly, we demonstrated that long-term IH alone can induce NASH molecular signatures found in human steatohepatitis transcriptomic data. Biomarkers (PPARs, NRFs, arachidonic acid, IL16, IL20, IFNB, TNF-α) associated with early hepatic and systemic inflammation were identified. This molecular link between IH, sleep apnea, and steatohepatitis merits further exploration in clinical trials, advocating for integrating sleep apnea diagnosis in liver disease phenotyping. Our unique signatures offer potential diagnostic and treatment response markers, highlighting therapeutic targets in the comorbidity of NAFLD and OSA.
Subject areas: Natural sciences, Biological sciences, Physiology, Animal physiology, Human Physiology
Graphical abstract

Highlights
-
•
A mouse model of long-term IH captures a human steatohepatitis transcriptional signature
-
•
Long-term IH exposure boosts neutrophil and monocyte infiltration into the liver
-
•
Multi-omics identify dysregulated markers implicated in sleep apnea and steatohepatitis
-
•
In practice, our data advocate systematic sleep apnea tests for liver disease phenotyping
Natural sciences; Biological sciences; Physiology; Animal physiology; Human Physiology
Introduction
Obstructive sleep apnea (OSA) is one of the most frequent chronic diseases, affecting nearly one billion people worldwide.1 The central component of OSA is the repetitive occurrence of upper airway collapse during sleep leading to the cyclical sequences of desaturation-reoxygenation known as intermittent hypoxia (IH). The severity of IH (i.e., “the hypoxic burden”) is the major contributor to the occurrence, aggregation, and progression of other common chronic diseases such as diabetes, hypertension, heart failure, non-alcoholic fatty liver disease (NAFLD), and cerebrovascular diseases.2,3,4 It has been established in animal models of NAFLD and from clinical studies that IH and OSA trigger and accelerate the transition from steatosis to non-alcoholic steatohepatitis (NASH) and fibrosis.4,5,6,7 Inflammation plays a crucial role in the pathophysiology of NASH and OSA but to date the molecular mechanisms underlying this comorbid association are poorly characterized.8,9,10,11 In humans and in mouse models, the specific role of IH is often masked by major confounders such as obesity, diabetes, and/or reduced physical activity. To circumvent these issues, we investigated both a model of lean mice exposed to long-term IH (16 weeks) and a cohort of lean OSA patients free of comorbidities (n = 71) using high-throughput hepatic transcriptomics, lipidomics, and targeted serum proteomics.
Our hypothesis was that long-term IH is sufficient to induce the biological pathways found in human steatohepatitis transcriptomic datasets. In addition, we asked whether there is a unique set of biomarkers in mice and humans associated with hepatic and systemic inflammation. Confirmation of these hypotheses would support the notion that IH is an independent “hit” that can autonomously induce NASH in humans.
Results
A unique standard-diet mouse model exposed to long-term IH
To explore the consequences of long-term exposure to IH during sleep on hepatic physiology, mice were housed under a 12 h light-dark cycle and fed with a regular chow diet. They were randomly assigned to either IH (1-min cycles of 5%–21% FiO2) or normoxic control (NC, 1-min cycles of 21% FiO2), 8 h per day during their sleep cycle (between ZT0 and ZT8, Figure 1A) for up to 16 weeks. The body weight of mice subjected to IH decreased rapidly during the first 2 weeks and remained significantly and consistently lower throughout the experiment compared to the NC group (Figure 1B). Food intake tends to decrease during the first week of exposure and then remains similar in between groups (Figure S1A). It is interesting to note that fasted glycemia starts to increase significantly after 8 weeks of IH (Figure 1C). A major strength of our experimental design is that mice exposure to 16 weeks of IH is quite unique in the field and reflects more accurately the long-term consequences of IH in human OSA.
Figure 1.
Hepatic transcriptomic signatures of long-term intermittent hypoxia in mice
(A) Experimental design of mouse model of sleep apnea. Lean male mice fed with regular chow diet were exposed to 16 weeks of intermittent hypoxia (IH) or normoxic cycles (NC).
(B) Body weight of mice exposed to intermittent hypoxia (IH, n = 15) and normoxic control (NC, n = 15) measured once a week over the time course of the experiment. Dark bars and circles are for NC mice and gray bars and triangles for IH mice. Body weight was presented as mean + standard error of mean (SEM). Significance was calculated per each time point using a Student’s t test and ∗∗∗ indicates a p value ≤0.001.
(C) Fasted glycemia of mice exposed to intermittent hypoxia (IH, n = 15) and normoxic control (NC, n = 15) measured once a week over the time course of the experiment. Fasted glycemia was presented as mean + standard error of mean (SEM). Significance was calculated per each time point using a Student’s t test and ∗∗∗ indicates a p value ≤0.001.
(D) Pie chart representing the number of differentially expressed genes in the liver after 16 weeks of IH. Upregulated genes are shown in red and downregulated genes are shown in blue. Genes were selected using a p value ≤0.01 indicating a significant difference between IH and NC.
(E) Gene ontology (GO) analysis showing the top six biological processes enriched in upregulated genes upon IH (top) denoted in red and downregulated genes upon IH (bottom) represented in blue. The number of dysregulated genes in our transcriptome over the total number of genes for each GO category is indicated on the graph. On the left panel, genes were selected using a p value ≤0.01 indicating a significant difference between NC and IH and ranked GO were ranked per p value. On the right panel, the top 500 genes with the highest expression difference between IH and NC were selected and GO were ranked per p value.
Specific hallmarks of long-term IH on the hepatic transcriptome
We first examined whether specific hepatic transcriptional reprogramming occurred after long-term exposure to IH. Hepatic RNA sequencing (RNA-seq) revealed that IH significantly dysregulates 1203 genes (p ≤ 0.01, Figure 1D). To identify related biological functions, we performed pathway enrichment analysis using the Gene Ontology Biological Processes database, and upstream transcriptional regulator enrichment analysis using the ChIP enrichment (ChEA) database. Gene ontology analysis identified distinct enriched biological pathways differentially expressed in response to IH (Figures 1E, and S1B). Specifically, metabolic pathways involving lipids and nutrient utilization were significantly enriched in downregulated genes, while inflammatory processes were overrepresented in upregulated genes (Figure 1E, top panel). Likewise, downregulated genes were mostly targets of PPARg, a transcription factor involved in fatty acid and glucose metabolism, whereas enriched upregulated genes were targets of GATA1 and STAT3, which are transcription factors implicated respectively in the maturation of blood cells and inflammation (Figure S1). These observations were consolidated with different gene ontology resources to reinforce the robustness of the data (Figure S1B). Collectively, these data demonstrate that IH is sufficient to induce molecular pathways found in steatohepatitis in humans mostly characterized by an increase in hepatic inflammation.
Exposure to long-term IH alone in mice is enough to capture a human steatohepatitis molecular signature
To evaluate the relevance of our IH mouse model to human pathology, we compared our RNA-seq with transcriptome that has been reported in humans with steatohepatitis. We performed biological pathway enrichment and upstream transcriptional regulator enrichment analysis on the top 500 most significantly dysregulated genes in human and mice transcriptomes and compared the 20 most significantly dysregulated pathways (Figures 2A and S2; Table S2) and associated transcription factors (Figure 2B and S2; Table S2). Downregulated genes present a common signature for fatty acid oxidation and PPARg target genes. Upregulated genes are enriched for neutrophils, inflammation, and GATA1 and NRF2 target genes (Figures 2 and S2; Table S2). These results further support the notion that IH in mice is an independent “hit” sufficient to induce molecular markers implicated in human NASH. This occurred in the context of individuals under standard diet but subject to long-term IH exposure.
Figure 2.
Comparison of hepatic signatures of human NASH and mouse long-term IH
(A) Venn diagrams representing the comparison of the top twenty GO biological processes ranked by p value enriched in the top 500 upregulated genes (red circles, left panel) or the top 500 downregulated genes (blue circles, right panel) in IH versus NC mice (dark blue and dark red circles) and NASH versus healthy humans (light blue and light red circles). The top 500 genes were selected using a p value ≤0.01.
(B) Venn diagrams representing the upstream transcriptional regulators ranked by p value associated with the top 500 upregulated genes (red circles, left panel) or the top 500 downregulated genes (blue circles, right panel) in IH versus NC mice (dark blue and dark red circles) and NASH versus healthy humans (light blue and light red circles). The top 500 genes were selected using a p value ≤0.01.
Long-term IH exposure reprograms hepatic lipid metabolism
The “multiple hit” hypothesis proposes that a continuum and potentially a combination of hepatic insults trigger and accelerate NAFLD development. Deregulation of lipid metabolism has been strongly associated with the onset and progression of NAFLD and our transcriptomic analyses confirmed that the PPARg transcription factor and the mitochondrial fatty acid oxidation pathway are central to liver injury pathogenesis in IH (Figures 3A, 3B, S2, and S3). This is exemplified by the expression profiles of target genes such as Cd36, Pparg, and Hadh (Figures 3C, S3, and S6). Therefore, this signature prompted us to analyze the hepatic lipid content through a mass spectrometry-based lipidomic approach (Figure 3D). We found no consistent differences in terms of overall hepatic content of lipids but we discovered significative variations in the abundance of specific fatty acids species potentially involved in signaling/inflammation (Figures 3D, 3E, and S3). Among them, arachidonic acid (C20:4) is significantly increased in the liver during IH while eicosanoic acid (C20:0) is decreased (Figure 3D and S3). Arachidonic acid is the precursor for the synthesis of the pro-inflammatory molecules prostaglandins and leukotrienes (Figure 3F). Interestingly, arachidonic acid is also increased in the serum of mice exposed to 16 weeks of IH (Figure 3D and S3). Hence, both lipidomic and transcriptomics analysis converge to indicate the occurrence of an inflammatory response to long-term IH.
Figure 3.
IH reprograms fatty acid metabolism and PPARg-dependent gene expression
(A) GSEA showing the enrichment of genes involved in mitochondrial fatty acid beta-oxidation and mitochondrial ATP synthesis coupled electron transport in the livers from the 16 weeks IH group.
(B) Heatmap representing significant (p value ≤0.01) differentially expressed PPARg target genes in the livers from the 16 weeks IH group.
(C) Representative expression of PPARg target and fatty acid metabolic genes determined by RT-qPCR at different length of IH exposure (4 and 16 weeks). Gene expression was normalized to beta-actin and presented as mean + standard error of mean (SEM, n = 4–5 biological replicates per group). Significance was calculated using a Student’s t test and ∗indicate a p value of 0.05.
(D) Heatmap representing fatty acid profiles in the liver and serum of mice exposed to 16 weeks of IH determined by mass spectrometry. Each fatty acid abundance was calculated and normalized according to the internal standard and presented as mean + standard error of mean (SEM, n = 5 biological replicates per group). Significance was calculated using Student’s t test and ∗ indicates a p value ≤0.05.
(E) Illustration depicting arachidonic acid metabolism and significant dysregulated fatty acid under IH and their link with inflammation.
Long-term IH exposure promotes immune cell infiltration into the liver
The distinctive inflammatory signature mediated by IH likely arises from the immune cell contribution in the bulk liver transcriptomic analysis. Remarkably, the top 500 most upregulated genes are typically found in leukocytes and are associated with inflammatory response (Figure 1E, right panel). Furthermore, GSEA corroborates the overrepresentation of the leukocytic signature upon IH (Figure 4A) and the comparison of mice and human transcriptomes pinpoints a predominant presence of neutrophils (Figures 1E and S2). These observations were further supported by immunohistochemistry experiments showing an increased number of neutrophils and monocytes in the liver of mice exposed to 16 weeks IH (Figure 4B). It is important to note that no difference in leukocyte staining was found after 4 weeks of IH exposure (Figure 4B) demonstrating again that long-term IH is required to boost hepatic immune cell infiltration and inflammation.
Figure 4.
IH fosters immune cell infiltration and activation in the liver
(A) GSEA showing the enrichment leukocytes, neutrophils, monocytes, and lymphocytes-specific genes in the livers from the 16 weeks IH group.
(B) Representative immunohistochemistry of the neutrophil marker MPO, monocytes marker CD68, and lymphocytes marker CD3 in liver biopsies at 4 and 16 weeks of IH exposure and their respective controls. A comparative observation of cell content in the liver sections from different groups was quantified and expressed as a number of positive cell staining per section area. Two slides per animal (n = 4–5) were analyzed and are presented as mean + standard error of mean (SEM). Statistical significance was calculated per each time point using a Student’s t test,∗,∗∗ and ∗∗∗ indicates respective p value ≤0.05, 0.01, and 0.001.
Long-term IH exposure in lean mice is associated with a hepatic and systemic inflammatory signature
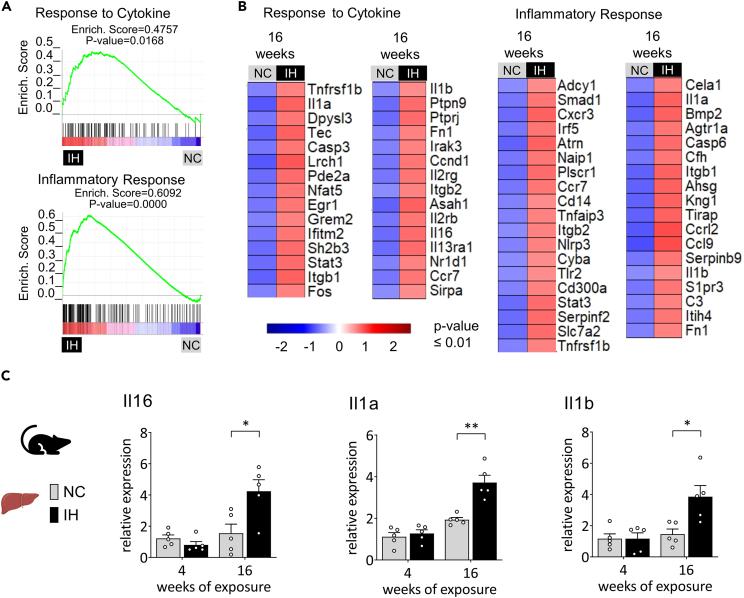
Consequently, we investigated whether long-term IH generates a peculiar inflammatory signature in the liver. GSEA confirmed the hepatic cytokine response and highlighted a specific set of inflammatory markers during IH (Figures 5A and S4). Among them, IH significantly upregulates the NLRP3 inflammasome and targets genes Il1a and Il1b, all involved in the development of chronic inflammation-related diseases (Figure 5B). We then assessed the longitudinal time course of hepatic expression by RT-qPCR of representative inflammatory markers. Strikingly, the increased expression of Il1a, Il1b, and Il16 occurs only after long-term IH exposure (Figure 5C).
Figure 5.
IH induces a set of hepatic inflammatory biomarkers
(A) GSEA showing the enrichment of genes involved in response to cytokine and inflammatory response in the livers from the 16 weeks IH group.
(B) Heatmap illustrating genes characterizing the response to cytokine and the inflammatory response significantly upregulated in the 16 weeks IH group (p-value ≤0.01).
(C) Representative inflammatory markers expression determined by RT-qPCR at different length of IH exposure (4 and 16 weeks). Gene expression was normalized to beta-actin and presented as mean + standard error of mean (SEM, n = 4–5 biological replicates per group). Significance was calculated using a Student’s t test and ∗, ∗∗ indicates respective p value of 0.05 and 0.01.
The bidirectional interactions between local and systemic inflammation prompted us to search for a comprehensive immune response set of markers in serum from mice exposed either to short (4 weeks) or long-term IH (16 weeks). Cytokine profiling revealed a significant induction of circulating biomarkers IL16, INFB1, and TARC after 16 weeks of IH (Figures 6A, 6B, and S5). It is important to note that the circulating cytokines are not altered after a short exposure to IH, except the significant increase of IL1A. There is also a progressive evolution in hepatic inflammation and cytokine profile during exposure to NC or IH.
Figure 6.
Cytokine profiling of IH mice and OSA patients
(A) Cytokine analysis was performed using serum from NC and IH mice collected after 4 or 16 weeks exposure (n = 5 biological replicates per time point per group). Results are displayed as a heatmap and significance was calculated using Student’s t test and ∗ indicates a p value ≤0.05.
(B) Comparison of cytokines profiles from serum from IH mice and OSA patients. Data are shown as mean + standard error of mean (SEM). Significance was calculated using Student’s t test and ∗ indicates a p value ≤0.05.
Inflammatory signature in OSA patients
We then investigated the relevance of this circulating cytokine signature in the serum of lean OSA patients. These patients came from a unique cohort of lean, middle-age OSA individuals free of any cardiovascular, hepatic, or metabolic comorbidities (cohort characteristics are shown in supplementary Table S1). The goal here was to mirror the situation of lean mice exposed to long-term IH. Among the inflammatory markers found in mice, only IL16 and interferon B (IFNB) were significantly higher in OSA patients than in control subjects (Figure 6B). Nevertheless, cytokine profiling of OSA patient samples also revealed a significant induction of IL20, LIF, TNF-α, and IFNg, consistent with trends found in mouse results (Figure 6 and S5).
Discussion
OSA and NAFLD are among the most frequent chronic diseases with a high burden for patients, society, and health systems.12,13 Consistent data from clinical studies advocate that OSA contributes to the development and progression of NAFLD. However, these observational data are partly flawed by the difficulty in delineating the respective role of IH versus comorbid obesity, sedentarity, and/or diabetes. To identify specific IH-driven molecular alterations at the early stage of NAFLD development, we used a mouse model fed with a standard diet (i.e., lean mice) exposed for up to 16 weeks to IH to reflect long-term exposure to a hypoxic burden as it occurs in patients with OSA. The design of our study focused on how IH in isolation may impact hepatic molecular pathways rather than the parallel histological feature of NASH. This approach has allowed us to minimize secondary events such as steatosis and fibrosis and confounders that would be encountered using mouse models of NAFLD. The 16-week exposure to IH is a clear strenght as most of the studies available in the field do not extend beyond 4 weeks of exposure. This is of crucial importance as early acute adaptation of the animals to IH is known to have a significant impact on body weight and food intake (Figures 1B and S1) that might widely interfere with identification of mechanisms specific to IH.
To depict the molecular portrait of the hepatic consequences of IH, we employed unbiased transcriptomic analysis and revealed that long-term IH without confounders is powerful enough to drive a gene expression cascade observed also in human NASH. This is an important addition to previous observational clinical studies showing that OSA severity (assessed through the apnea-hypopnoea index) is an independent predictor of liver disease progression.4
Comparison of IH mice and human NASH hepatic transcriptomes has revealed a common set of dysregulated genes, targets of PPARg and NRF2 transcription factors. These factors are well known to be implicated in liver disease and linked to metabolism and inflammation but they have not been described previously in rodent models of sleep apnea. It is important to note that current data are not supporting or providing a rationale for a pivotal role of hypoxia-inducible factors (HIF1a and HIF2) in the context of long-term IH (Figure S6) suggesting distinct adaptive mechanisms to short- and long-term IH. In NASH patients, it has been demonstrated that NRF2 activation is correlated with liver inflammation.14 Interestingly, we have previously shown that short-term IH (2 weeks) is sufficient to alter NRFs-dependent hepatic gene expression without mediating any transcriptional changes in inflammatory genes. The current data constitute an addition to our knowledge in showing that sustained deregulation of NRFs target genes might contribute to the progression of liver disease. PPARs are key regulators of fatty acid and glucose metabolism, inflammation, and fibrosis and PPARs agonists have demonstrated promising improvement in liver disease in randomized controlled clinical trials.11,15 Our data suggest that the association between NASH and OSA might be of particular interest for this pharmacological approach and this merits to be investigated in further studies dedicated to this comorbid association. Interestingly, PPARg lipolytic target genes are downregulated after 16 weeks of IH exposure in association with a remodeling of hepatic fatty acid content. Among them, arachidonic acid has been shown to be an early indicator of inflammation during NAFLD development.16,17 Accordingly, our group has previously reported that the arachidonic acid-derived metabolite, leukotriene B4 (LTB(4)), is increased during OSA and is linked to early vascular remodeling.18,19,20
We demonstrated that long-term IH boosts leukocyte infiltration into the liver, which probably accounts for the strong inflammatory signature revealed by our bulk liver transcriptomic analyses. It would have been compelling to complement this analysis with single-cell and spatial transcriptomics and/or to characterize hepatic immune cells population by flow cytometry in order to precisely map the immune and inflammatory signature.21 Despite its limitations, this strategy allowed us to identify a set of inflammatory markers associated with long-term IH. Among them, IH significantly upregulates the NLRP3 inflammasome and the interleukins Il1a, Il1b, and Il16. Interestingly, the induction of IL16 has also been found in serum of mice exposed to long-term IH as well as in OSA patients. IL16 is a pro-inflammatory cytokine22,23 that is chemotactic for CD4+ T lymphocytes, monocytes, and eosinophils linked to various diseases such as asthma,24 rheumatoid arthritis,25 and inflammatory bowel disease.26,27 Furthermore, recent studies have shown that IL16 expression in the liver correlates with inflammation in patients with primary biliary cholangitis and progression of liver hepatocellular carcinoma,28,29 making IL16 an interesting therapeutic target. We also found a significant induction of IFNB in both IH mice and OSA patients. In addition, OSA patients present a significant increase of other interferons such as IFNg and IFNw. IFNs are essential cytokines of the immune system that serve as key effectors. Initially recognized for their crucial role in defending against viral infections, their involvement in diverse diseases has come to light in recent studies, revealing both protective and potentially harmful roles. Interestingly, several studies have shown that interferons affect the progression of non-alcoholic fatty liver disease.30
The cytokine profiling also revealed a significant induction of IL20, LIF, and TNF-α in OSA patients that parallels with the trends toward an increase in mice. It would be worthwhile to increase the number of biological replicates in mice to fully characterize the conserved signature between mice and humans. TNF-α has been described in various inflammatory diseases including NASH and OSA31,32,33 while little is known about IL20 and LIF in these pathologies.34 IL20 has been implicated in several inflammatory diseases35 and ongoing clinical trials are investigating the potential therapeutic effects of anti-IL20 in psoriasis and rheumatoid arthritis (NCT01038674, NCT01261767) making IL20 an interesting target to explore in the context of OSA and NASH.
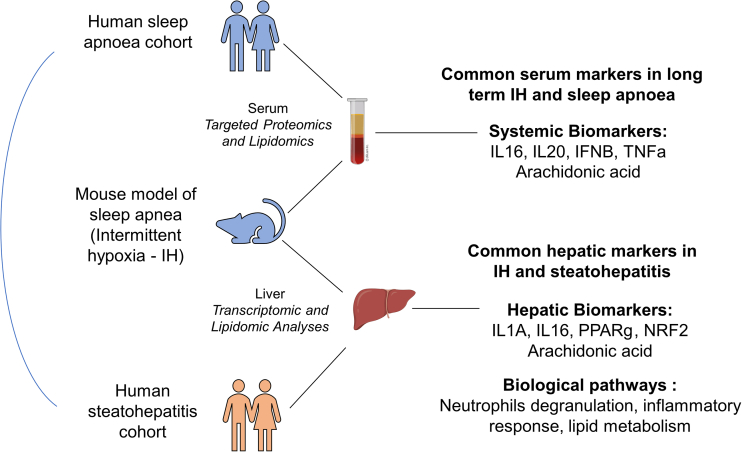
Overall, our unbiased multi-omics approach has revealed molecular signatures linking IH, NASH, and OSA. We identified group of dysregulated markers (PPARg, NRF2, arachidonic acid, IL16, IL20, IFNB, and TNF-α) that warrant further investigation in clinical trials as they might impinge on both hepatic and extrahepatic organs and participate toward the development of metabolic and cardiovascular complications associated with OSA8,36,37,38 (Figure 7).
Figure 7.
Summary of findings
Long-term IH in mice induces inflammatory pathways entangled in human sleep apnea and steatohepatitis.
Both OSA and NAFLD are chronic diseases with different clusters of associated co-morbidities that impact individual prognosis.39 To implement precision medicine in both conditions, biomarkers reflecting this complexity of these pathologies are needed to guide the personalized management of the patients. The progression of liver disease is controlled by a variety of factors, including inflammatory cells and cytokines.9,10 Our original data pave new avenues for investigating therapeutic targets related to the specific role of cytokines in the comorbid association between OSA and NAFLD. These observations also provide a strong rationale for studies conducted specifically on this co-morbid association. In clinical practice, our data support the systematic implementation of sleep apnea diagnostic tests in liver disease phenotyping.
Limitations of the study
Our study demonstrated that long-term IH significantly enhances leukocyte infiltration into the liver, likely contributing to the inflammatory signature identified through our bulk liver transcriptomic analyses. While our findings provide valuable insights, a more nuanced understanding could be achieved by complementing this analysis with advanced techniques such as single-cell and spatial transcriptomics. Additionally, characterizing the specific hepatic immune cell population through flow cytometry would offer a more precise mapping of the immune and inflammatory signatures, enhancing the depth of our investigation.
In our exploration, we pinpointed a distinctive cluster of biomarkers, including pivotal transcription factors such as PPARg and NRF, as well as inflammatory cytokines like IL16 and TNF-α. However, our study refrains from definitively concluding the specific roles of each element. It is crucial to acknowledge that these mechanisms may interconnect, and our current investigation cannot firmly establish the individual contributions of PPARg, NRF, IL16, and TNF-α to IH-induced alterations in cellular metabolism and inflammation. Subsequent studies are imperative to unravel the intricate relationships among these factors and ascertain their respective impacts on the observed physiological changes.
Our study uncovered a subtle, time-dependent signature, revealing distinct mechanisms of adaptation or maladaptation to short-term and long-term IH exposure, emphasizing the importance of characterizing the adaptive and maladaptive responses associated with varying durations of IH exposure.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-CD68 | Abcam | ab12512 |
| Rabbit Anti- MPO | Abcam | ab208670 |
| Rabbit Anti-CD3 | Abcam | ab5690 |
| Rabbit anti-HIF1a | Abcam | ab2185 |
| Mouse anti-HIF1a | Santa Cruz Biotechnology | sc13515 |
| Mouse anti- Tubuline-a | Sigma Aldrich | T5168 |
| HRP Goat anti-Rabbit | Jackson ImmunoResearch | 111-035-003 |
| HRP Goat anti-Mouse | Jackson ImmunoResearch | 115-035-003 |
| Biotinylated Goat Anti-rabbit | Vector Laboratories | BA-1000 |
| Biological samples | ||
| Human Serum | ClinicalTrials.gov | NCT00764218 |
| Mouse Serum | This paper | See STAR Methods |
| Mouse Liver | This paper | See STAR Methods |
| Chemicals, peptides, and recombinant proteins | ||
| TRIzol reagent | Invitrogen | 15596026 |
| DEPC treated water | Invitrogen | 4622224 |
| Actinomycin D | Sigma Aldrich | A1410 |
| Tridecanoic acid | Sigma Aldrich | 91988 |
| Phosphatidylcholine | Avanti | 840009 |
| Chloroform/methanol (1:2)-TMSH solution | Macherey-Nagel | 701520 |
| Citrate buffer pH6 | Sigma Aldrich | C999 |
| Hydrogen peroxide | Sigma Aldrich | H1009 |
| Goat Serum Blocking Solution | Vector Laboratories | S-1000 |
| HistoGreen | Novus Biological | E109 |
| Mayer hematoxylin | Sigma Aldrich | 51275 |
| Vectamount permanent medium | Vectorlab | H-5000 |
| Bovine serum albumin | Euromedex | 04100812 |
| Trichostatin A (TSA) | Santa Cruz Biotechnology | sc-3511 |
| Nicotinamide | Sigma Aldrich | N0636 |
| Dihydroethidium | Sigma Aldrich | 309800 |
| Complete protease inhibitor | Roche | 04693132001 |
| Critical commercial assays | ||
| iScript complementary DNA (cDNA) synthesis kit | BioRad Laboratories | 1708840 |
| SsoAdvanced SYBR Green Supermix kit | BioRad Laboratories | 1725270 |
| SuperScript II reverse transcriptase | Thermo Fischer Scientific | 18064014 |
| Vectastain Elite ABC kit peroxidase | Vector Laboratories | PK-6101 |
| Avidin/biotin blocking kit | Vector Laboratories | SP-2001 |
| 4–20% Mini-PROTEAN TGX Precast Protein Gels | BioRad Laboratories | 4561096 |
| Deposited data | ||
| Mouse Liver Gene Expression Dataset | GEO | GSE242668 |
| Human Liver Gene Expression Dataset | GEO | GSE33814 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6JRj | Janvier Labs | C57BL/6JRj |
| Oligonucleotides | ||
| Primers for RT-qPCR | This paper | See STAR Methods |
| Software and algorithms | ||
| R software | R project | N/A |
| Illumina software bcl2fastq v2.20.0.422 | Illumina | N/A |
| SARTools | GitHub.com | N/A |
| DESeq2 | bioconductor.org | N/A |
| GSEA | gsea-msigdb.org | N/A |
| MetaScape | metascape.org | N/A |
| AmiGO | geneontology.org | N/A |
| EnrichR | maayanlab.cloud | N/A |
| DAVID | david.ncifcrf.gov | N/A |
| Microsoft Office | Microsoft | N/A |
| GraphPad Prism | GraphPad | N/A |
| Affinity Designer | Affinity | N/A |
| BioRender | Biorender.com | N/A |
| Zen software | Zeiss | N/A |
| Other | ||
| LASQC diet | Génobios | Rod 16-R |
| Precellys 24 tissue homogenizer | Bertin Technologies | N/A |
| CK14 ceramic beads | Bertin Technologies | P000912-LYSK0-A |
| Agilent Bioanalyzer Nano RNA | Agilent | N/A |
| Nanodrop | Thermofischer | N/A |
| NovaSeq 6000 | Illumina | N/A |
| Gas chromatography-mass spectrometry | Agilent | 5977A-7890B |
| Axio Scan microscope | Zeiss | N/A |
| LSM510 microscope | Zeiss | N/A |
| Luminex™ 200 system | Eve Technologies Corp. | N/A |
| Roche Accu-Chek Guide glucometer | Roche | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jean-Louis Pépin (jpepin@chu-grenoble.fr).
Materials availability
The study did not generate new unique reagents.
Data and code availability
-
•
Data reported in this paper will be shared by the lead contact upon request. RNA-seq data have been deposited at the Biotechnology Information Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Hepatic mouse gene expression data reported in this paper is available under the accession number GSE242668. This dataset contains 10 samples, 5 from IH mice and 5 from NC mice. The hepatic human gene expression dataset used in this study was recovered from the GEO repository under the accession number GSE33814. It consisted of 25 samples, 12 of which were steatohepatitis and 13 were controls.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study details
Animal handling and ethics approval
Sixteen-week-old male C57BL/6JRj mice (Janvier Labs, France) were housed with ad libitum food and water access on a 12 h light/12 h dark cycle (light ON at 8 am = ZT0 and light OFF at 8 pm = ZT12). Mice were maintained with regular chow diet (LASQC diet, Rod 16, Altromin international) during all the experiment. Mice were randomly assigned to either intermittent hypoxia (IH) or normoxic control (NC) and directly exposed in their housing cages to 16 weeks of NC or IH, 8h per day during their sleeping period (IH from 8 am = ZT0 to 4 pm = ZT8 and NC the rest of the day) (Figure 1A).
The IH stimulus consisted of 60 sec cycles alternating 30 sec of hypoxia (hypoxic plateau at 5% FiO2) and 30 sec of NC (normoxic plateau at 21% FiO2). NC mice were exposed to similar air-air cycles in order to avoid bias from noise and turbulence related to gas flow. After 16 weeks of exposure, mice were sacrificed by decapitation, serums were collected and livers were harvested and immediately frozen in liquid nitrogen until further analysis.
All experimental procedures were carried out in accordance with European Directive 2010/63/UE. They were reviewed by the Institutional Ethics Committee for Animal Care and Use (Cometh 12) and authorized by the French Ministry of Research (APAFIS# 15156-2018051615245109).
Human biological samples: study population and ethics approval
Biological samples from a case-controlled study (InfraSAS) of patients without any known cardiovascular comorbidity comprised 71 patients: 19 with severe OSA (AHI>30), 27 with moderate OSA (15<AHI<30) and 25 control subjects (AHI<15) (ClinicalTrials. gov: NCT00764218). Serum samples were taken in a fasted state in the morning after an overnight polysomnography and were stored at −80°C until use. For both studies, ethical approval was obtained from our Institutional Review Board (Comité de Protection des Personnes Sud-Est V, Grenoble, France) and all subjects or patients gave written informed consent. Protocols conformed to the principles of the Declaration of Helsinki.
Method details
Food intake and glycemia
Food intake was systematically measured each day at 8 am (=ZT0), just before the onset of the hypoxia exposure period. To achieve this, the feeders in the cages, each accommodating five mice, were weighed, and the difference from the previous day was calculated to determine the food intake over the past 24 hours for a group of five mice. This approach, involving an average calculation across five mice, allowed for precise tracking of daily variations in dietary consumption.
Glycemic measurements were conducted at 2 pm (ZT6) after a 6-hour fasting period within the NC or IH exposure between ZT0 and ZT8. Blood collection was facilitated through a minor incision at the tip of the tail, and the obtained samples were promptly analyzed using the Roche Accu-Chek Guide glucometer.
RNA extraction
For RNA extraction, 50mg of frozen liver tissues were homogenized in TRIzol Reagent (Invitrogen, 15596026) using the Precellys 24 tissue homogenizer and CK14 ceramic beads (Bertin Technologies) with the following program: 6000 rpm, 3 × 10 sec operation (5 sec break). RNA was extracted after precipitation with isopropanol and ethanol washes according to the manufacturer’s protocol. RNA pellets were resuspended in DEPC treated water (Invitrogen, 4622224), flash-frozen in liquid nitrogen and stored at −80°C.
Reverse transcription and quantitative real-time PCR analysis
One μg of RNA was reverse transcribed to cDNA using iScript complementary DNA (cDNA) synthesis kit (Bio-Rad Laboratories, 1708840), according to the manufacturer’s protocol.
cDNA was used for quantitative real-time PCR (RT-qPCR) using a SsoAdvanced SYBR Green Supermix kit (Bio-Rad Laboratories, 1725270) according to the manufacturer’s protocol. Gene expression was normalized to beta actin.
Primer sequences used for analysis are listed below:
| mRNA | Forward Primer (5′-3′) | Reverse Primer (5′-3′) |
|---|---|---|
| Il1a | TTGGTTAAATGACCTGCAACA | GAGCGCTCACGAACAGTTG |
| Il1b | AGTTGACGGACCCCAAAAG | AGCTGGATGCTCTCATCAGG |
| Il16 | AAGGTCACAGACCCTTCTGG | TGGCAGCAGCTCTCTGGT |
| Ccl9 | TGGGCCCAGATCACACAT | CCCATGTGAAACATTTCAATTTC |
| Cd36 | TTGAAAAGTCTCGGACATTGAG | TCAGATCCGAACACAGCGTA |
| Cpt1a | GACTCCGCTCGCTCATTC | TCTGCCATCTTGAGTGGTGA |
| Hadh | TGGATACTACAAAGTTCATCTTGGA | AAGGACTGGGCTGAAATAAGG |
| Pparg | GAAAGACAACGGACAAATCACC | GGGGGTGATATGTTTGAACTTG |
| Vegfa | AAAAACGAAAGCGCAAGAAA | TTTCTCCGCTCTGAACAAGG |
| Slc2a1 | GACCCTGCACCTCATTGG | GATGCTCAGATAGGACATCCAAG |
RNA sequencing analysis
mRNA sequencing
RNA quality was monitored for quality control using Agilent Bioanalyzer Nano RNA chip and Nanodrop absorbance ratios of 260/280 nm and 260/230 nm. Library construction was performed according to the Illumina kit protocol (TruSeq stranded mRNA sample preparation). mRNA was enriched using oligo dT magnetic beads and chemically fragmented. First-strand synthesis of cDNA was performed in the presence of actinomycin D with random primers and SuperScript II reverse transcriptase. Second-strand synthesis of cDNA was performed by replacing dTTP by dUTP and then the 3′ ends were adenylated. Illumina barcoded adapters were ligated on the ends and the adapter ligated fragments were enriched by PCR cycles. The resulting libraries were validated by qPCR and sized by Agilent Bioanalyzer DNA high sensitivity chip. The concentrations for the libraries were normalized and then multiplexed together. The concentration used for clustering the flowcell SP was 300 pM. The multiplexed libraries were sequenced on one lane using single-end 100 cycle chemistry for the NovaSeq 6000 (Illumina).
Bioinformatic analysis
Sequencing data for 10 samples in FastQ format was produced using post-processing from Illumina software bcl2fastq v2.20.0.422 by the Montpellier GenomiX Facility at the Biocampus, Montpellier (France). Reads failing Illumina’s standard quality tests were not included in these FastQ files. The sequencing reads from each sample in the experiment were separately aligned to the reference genome mm10 and corresponding known splice junctions extracted from the UCSC Genome Browser40,41 using the ELAND v2e short-read aligner (Illumina). Reads aligned with a non-unique best match or with two or more mismatches with the reference sequences were discarded from the analyses. The remaining uniquely aligned reads were used to estimate relative transcript abundance for further analysis. Reads per Kilobase of transcript per Million mapped reads (RPKM) were used to quantify the relative abundance of each transcript in a sample and to perform gene expression analyses. Normalization, differential and statistical analyses were carried out using the R software, Bioconductor,42 DESeq2,43,44 and SARTools45 packages.
Lipidomics
Total lipid was extracted from the liver using chloroform/methanol, 1:2 (v/v) in the presence of 20 nmol of tridecanoic acid (Sigma) and 20 nmol of phosphatidylcholine (PC, C21:0/C21:0, Avanti) as internal standard. The liver samples were dissected into small pieces and ground in methanol prior to the addition of chloroform. After vigorous sonication and vortex, the organic phase was extracted by biphasic separation generated by the addition of chloroform and 0.5% acetic acid/0.2% KCl to give a chloroform/methanol/aqueous phase 2:1:0.8 (v/v/v) ratio. The resulting bottom organic phase was dried and on-line derivatized to fatty acid methyl ester by a chloroform/methanol (1:2)-TMSH solution (Macherey-Nagel) and analyzed by gas chromatography-mass spectrometry (Agilent 5977A-7890B). Each fatty acid abundance was calculated and normalized according to the internal standard and liver weight.
Immunohistochemistry
Immunohistochemistry was adapted from the manufacturer’s protocol (PK-6101, Vectastain Elite ABC kit peroxidase, Vector Laboratories) and46 with the following modifications: 5 μm slices of paraffin embedded liver were rehydrated and processed for antigen retrieval with citrate buffer pH6 (C999, Sigma Aldrich) or tris-EDTA buffer pH9. Then, endogenous peroxidase activity was quenched with 3% hydrogen peroxide (H1009, Sigma Aldrich) and non-specific binding was prevented with an avidin/biotin blocking kit (SP-2001, Vector Laboratories) and 5% goat serum (Vector Laboratories). Afterwards, primary rabbit antibodies (CD68 ab12512; myeloperoxidase MPO ab208670; CD3 ab5690, Abcam) and secondary biotinylated goat anti rabbit antibodies (Vector Laboratories) were used. After incubations and washes with PBS, colorimetric substrate was added (E109, HistoGreen, Novus Biological) and slices were counterstained with Mayer hematoxylin (51275, Sigma Aldrich). Vectamount permanent medium (Vectorlab) was used to glue the coverslips on the slides. Images were acquired using an Axio Scan microscope (Zeiss) and Zen® software (Zeiss).
Dihydroethidium (DHE) staining
DHE staining was adapted from (15) with the following modifications: Frozen livers in OCT mounting media were cryosectioned at 10 μm thick and allowed to air dry for 15 min at room temperature onto Superfrost plus slides (Dutscher France). DHE 10 μM (Sigma-Aldrich) was added onto liver sections for 30 min at 37°C (PBS was used as control) in a dark moist chamber. After washing, cover slips were added and the fluorescent signal was recorded using confocal microscopy (Zeiss, LSM510 Meta confocal microscope) and analyzed with ImageJ software. DHE-positive cells were expressed as the percentage of liver area.
Protein extraction and Western blots analysis
The protocols were adapted from (36) with the following modifications:
For whole cell protein extracts, 50 mg of liver was homogenized in RIPA Buffer (10 mM Tris-HCl pH8, 150 mM NaCl, 1% NP40, 0.5%, DOC, 5 mM MgCl2, 0.22 μM TSA, 10 mM Nicotinamide, 20 mM NaF, 0.5 mM PMSF and Complete protease inhibitor (Roche, 04693132001)).
For Western blots analysis, 20 μg of whole cell protein extract was loaded on 4%–20% precast polyacrylamide gels (Biorad, 4561096). Antibodies used for Western blots are as follows: anti-HIF1a (Abcam, ab2185), anti-HIF1a (Santa Cruz Biotechnologies, sc13515), anti-α-TUBULIN (Sigma, T5168), anti-Rabbit HRP (Jackson laboratories, 111-035-003), anti-Mouse HRP (Jackson laboratories, 115-035-003).
Cytokine profiling
This study used Luminex xMAP technology for multiplexed quantification of 96 Human and 44 Mice cytokines, chemokines, and growth factors. The multiplexing analysis was performed using the Luminex 200 system (Luminex, Austin, TX, USA) by Eve Technologies Corp. (Calgary, Alberta). For patients, 96 markers were simultaneously measured in the samples using Eve Technologies' Human Cytokine 96-Plex Discovery Assay®. For mice, 44 markers were simultaneously measured in the samples using Eve Technologies' Mouse Cytokine 44-Plex Discovery Assay®. The 96-plex assay consists of two separate kits; the Panel A 48-plex and the Panel B 48-plex (MilliporeSigma, Burlington, Massachusetts, USA). The assay was run according to the manufacturer’s protocol. The Panel A 48-plex consisted of sCD40L, EGF, Eotaxin, FGF-2, FLT-3 Ligand, Fractalkine, G-CSF, GM-CSF, GROα, IFN-α2, IFN-γ, IL-1α, IL-1β, IL-1RA, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12(p40), IL-12(p70), IL-13, IL-15, IL-17A, IL-17E/IL-25, IL-17F, IL-18, IL-22, IL-27, IP-10, MCP-1, MCP-3, M-CSF, MDC, MIG/CXCL9, MIP-1α, MIP-1β, PDGF-AA, PDGF-AB/BB, RANTES, TGFα, TNF-α, TNF-β, and VEGF-A. The Panel B 48-plex consisted of 6CKine, APRIL, BAFF, BCA-1, CCL28, CTACK, CXCL16, ENA-78, Eotaxin-2, Eotaxin-3, GCP-2, Granzyme A, Granzyme B, HMGB1, I-309, I-TAC, IFNβ, IFNω, IL-11, IL-16, IL-20, IL-21, IL-23, IL-24, IL-28A, IL-29, IL-31, IL-33, IL-34, IL-35, LIF, Lymphotactin, MCP-2, MCP-4, MIP-1δ, MIP-3α, MIP-3β, MPIF-1, Perforin, sCD137, SCF, SDF-1, sFAS, sFASL, TARC, TPO, TRAIL, and TSLP. Assay sensitivities of these markers range from 0.05–100 pg/mL for the 96-plex. Individual analyte sensitivity values are available in the MILLIPLEX MAP protocol. Data were extracted based on cytokine-specific standards generated by Eve Technologies (Calgary, Canada).
Quantification and statistical analysis
Functional analysis of the hepatic transcriptome
Functional analysis of the RNA sequencing followed a protocol similar to the ones previously described.47,48 Normalization, differential and statistical analyses were carried out using the R software, Bioconductor,42 DESeq2,43,44 and SARTools45 packages. Then, differentially expressed genes were selected using a p value of 0.01 and assessed using multiple pathway analysis tools GSEA 49 MetaScape 50 AmiGO 51,52 DAVID 53,54 EnrichR 55,56 to identify Gene Ontology (GO) terms (biological process, molecular functions and cellular compartments) and transcription factor binding site (TFBS) enrichment.
Graphical representations and statistical analysis
Statistical analyses are detailed for each experiment in the figure legend. Heat maps were generated by R package ‘pheatmap’. Statistical analyses and graphical representations were processed using Microsoft Excel and GraphPad Prism. Graphic design and illustrations were created with Microsoft Powerpoint, Affinity Designer and BioRender.
Acknowledgments
We thank all members of the HP2 laboratory for stimulating discussions and technical assistance, especially to Antoine Boutin Paradis for his help with histology. Computations presented in this paper were performed using the GRICAD infrastructure (https://gricad.univ-grenoble-alpes.fr), which is supported by Grenoble research communities. We acknowledge the contribution of SFR Santé Lyon-Est facility (CiQLE, UAR3453 CNRS, US7 Inserm, UCBL), especially Annabelle Bouchardon and Smatti Batoule, for their help with histology. We also thank the Grenoble-Alpes animal facility (UMS-hTAG), especially Candice Hackney for her support. Finally, we thank Alison Foote (an independent medical writer based in Grenoble, France) for editing the manuscript.
The HP2 laboratory is supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the University of Grenoble-Alpes (UGA), the Fondation Agir Pour les Maladies Chroniques (APMC, Project Temporise Liver), the Agence Nationale de la Recherche (ANR, Project Temporise, ANR-19-CE14-0037-01), and the ANR Initiatives d’Excellence de l’UGA (ANR-IDEX-UGA, Project LiFE, 15-IDEX-0002). This work has been partially supported by UGA e-health chair and MIAI @ university Grenoble Alpes, (ANR-19-P3IA-0003). Fond de dotation Bastide Médical. J.G. was the recipient of the postdoctoral fellowship from the ANR-IDEX-UGA Initiative de Recherche Stratégique (ANR-IDEX-UGA-IRS, Project HypoClock). G.V. received a grant from Société Francophone du Diabète. E.M.’s work was supported by grants from the Fondation ARC and ERICAN. M.G. was financed by the Medical Research grant from Montpellier-Nimes Medical School, University of Montpellier. Apicolipid, C.Y.B. and Y.Y.B., and the Lipidomic GEMELI platform were supported by Agence Nationale de la Recherche (ApicoLipidAdapt grant ANR-21-CE44-0010; Laboratoire d’Excellence Parafrap France ANR-11-LABX-0024), Fondation pour la Recherche Médicale (FRM Equation 202103012700), Région Auvergne-Rhône-Alpes for the lipidomics analyses platform (Grant IRICE Project GEMELI), CNRS (LIA_IRP Apicolipidadapt), and CEFIPRA (Project 6003-1).
Author contributions
J.G. and J.-L.P. conceived and designed the study and drafted the manuscript. F.C., S.R., E.M., and J.G. performed bioinformatic analysis and graphical representations. J.G., M.G., and E.L. performed RNA extractions and analysis. S.B., M.G., and A.D. performed the immunohistochemical analysis. J.G. and G.V. performed mice experimentation. Y.Y.B. and C.B. performed lipidomics analysis. R.B.M. and M.J.F. administered biobanking, clinical data collection, and extraction. D.G.R., S.K., and S.R. provided technical support for project design and data analysis and participated in interpretation. All authors critically revised the manuscript for important intellectual content and approved the version for publication.
Declaration of interests
The authors declare no competing interests.
Published: January 9, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.108837.
Contributor Information
Jonathan Gaucher, Email: jonathan.gaucher@univ-grenoble-alpes.fr.
Jean-Louis Pépin, Email: jpepin@chu-grenoble.fr.
Supplemental information
References
- 1.Benjafield A.V., Ayas N.T., Eastwood P.R., Heinzer R., Ip M.S.M., Morrell M.J., Nunez C.M., Patel S.R., Penzel T., Pépin J.L., et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir. Med. 2019;7:687–698. doi: 10.1016/S2213-2600(19)30198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lévy P., Kohler M., McNicholas W.T., Barbé F., McEvoy R.D., Somers V.K., Lavie L., Pépin J.L. Obstructive sleep apnoea syndrome. Nat. Rev. Dis. Primers. 2015;1 doi: 10.1038/nrdp.2015.15. [DOI] [PubMed] [Google Scholar]
- 3.Ryan S., Arnaud C., Fitzpatrick S.F., Gaucher J., Tamisier R., Pépin J.L. Adipose tissue as a key player in obstructive sleep apnoea. Eur. Respir. Rev. 2019;28 doi: 10.1183/16000617.0006-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jullian-Desayes I., Trzepizur W., Boursier J., Joyeux-Faure M., Bailly S., Benmerad M., Le Vaillant M., Jaffre S., Pigeanne T., Bizieux-Thaminy A., et al. Obstructive sleep apnea, chronic obstructive pulmonary disease and NAFLD: an individual participant data meta-analysis. Sleep Med. 2021;77:357–364. doi: 10.1016/j.sleep.2020.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Aron-Wisnewsky J., Minville C., Tordjman J., Lévy P., Bouillot J.-L., Basdevant A., Bedossa P., Clément K., Pépin J.L. Chronic intermittent hypoxia is a major trigger for non-alcoholic fatty liver disease in morbid obese. J. Hepatol. 2012;56:225–233. doi: 10.1016/j.jhep.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Aron-Wisnewsky J., Clement K., Pépin J.L. Nonalcoholic fatty liver disease and obstructive sleep apnea. Metabolism. 2016;65:1124–1135. doi: 10.1016/j.metabol.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 7.Mesarwi O.A., Loomba R., Malhotra A. Obstructive Sleep Apnea, Hypoxia, and Nonalcoholic Fatty Liver Disease. Am. J. Respir. Crit. Care Med. 2019;199:830–841. doi: 10.1164/rccm.201806-1109TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell E.E., Wong V.W.-S., Rinella M. Non-alcoholic fatty liver disease. Lancet. 2021;397:2212–2224. doi: 10.1016/S0140-6736(20)32511-3. [DOI] [PubMed] [Google Scholar]
- 9.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nat. Rev. Gastroenterol. Hepatol. 2018;15:349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 10.Peiseler M., Schwabe R., Hampe J., Kubes P., Heikenwälder M., Tacke F. Immune mechanisms linking metabolic injury to inflammation and fibrosis in fatty liver disease - novel insights into cellular communication circuits. J. Hepatol. 2022;77:1136–1160. doi: 10.1016/j.jhep.2022.06.012. [DOI] [PubMed] [Google Scholar]
- 11.Pawlak M., Lefebvre P., Staels B. Molecular mechanism of PPARα action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015;62:720–733. doi: 10.1016/j.jhep.2014.10.039. [DOI] [PubMed] [Google Scholar]
- 12.Lyons M.M., Bhatt N.Y., Pack A.I., Magalang U.J. Global burden of sleep-disordered breathing and its implications. Respirology. 2020;25:690–702. doi: 10.1111/resp.13838. [DOI] [PubMed] [Google Scholar]
- 13.Lazarus J.V., Mark H.E., Anstee Q.M., Arab J.P., Batterham R.L., Castera L., Cortez-Pinto H., Crespo J., Cusi K., Dirac M.A., et al. Advancing the global public health agenda for NAFLD: a consensus statement. Nat. Rev. Gastroenterol. Hepatol. 2022;19:60–78. doi: 10.1038/s41575-021-00523-4. [DOI] [PubMed] [Google Scholar]
- 14.Mohs A., Otto T., Schneider K.M., Peltzer M., Boekschoten M., Holland C.H., Hudert C.A., Kalveram L., Wiegand S., Saez-Rodriguez J., et al. Hepatocyte-specific NRF2 activation controls fibrogenesis and carcinogenesis in steatohepatitis. J. Hepatol. 2021;74:638–648. doi: 10.1016/j.jhep.2020.09.037. [DOI] [PubMed] [Google Scholar]
- 15.Francque S.M., Bedossa P., Ratziu V., Anstee Q.M., Bugianesi E., Sanyal A.J., Loomba R., Harrison S.A., Balabanska R., Mateva L., et al. A Randomized, Controlled Trial of the Pan-PPAR Agonist Lanifibranor in NASH. N. Engl. J. Med. 2021;385:1547–1558. doi: 10.1056/NEJMoa2036205. [DOI] [PubMed] [Google Scholar]
- 16.Sztolsztener K., Chabowski A., Harasim-Symbor E., Bielawiec P., Konstantynowicz-Nowicka K. Arachidonic Acid as an Early Indicator of Inflammation during Non-Alcoholic Fatty Liver Disease Development. Biomolecules. 2020;10:1133. doi: 10.3390/biom10081133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnweber T., Pizzini A., Nairz M., Weiss G., Tancevski I. Arachidonic Acid Metabolites in Cardiovascular and Metabolic Diseases. Int. J. Mol. Sci. 2018;19:3285. doi: 10.3390/ijms19113285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier-Veyret E., Bäck M., Arnaud C., Belaïdi E., Tamisier R., Lévy P., Arnol N., Perrin M., Pépin J.L., Stanke-Labesque F. Cysteinyl-leukotriene pathway as a new therapeutic target for the treatment of atherosclerosis related to obstructive sleep apnea syndrome. Pharmacol. Res. 2018;134:311–319. doi: 10.1016/j.phrs.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Stanke-Labesque F., Pépin J.L., Gautier-Veyret E., Lévy P., Bäck M. Leukotrienes as a molecular link between obstructive sleep apnoea and atherosclerosis. Cardiovasc. Res. 2014;101:187–193. doi: 10.1093/cvr/cvt247. [DOI] [PubMed] [Google Scholar]
- 20.Stanke-Labesque F., Bäck M., Lefebvre B., Tamisier R., Baguet J.-P., Arnol N., Lévy P., Pépin J.L. Increased urinary leukotriene E4 excretion in obstructive sleep apnea: effects of obesity and hypoxia. J. Allergy Clin. Immunol. 2009;124:364–370.e3702. doi: 10.1016/j.jaci.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 21.Haas J.T., Vonghia L., Mogilenko D.A., Verrijken A., Molendi-Coste O., Fleury S., Deprince A., Nikitin A., Woitrain E., Ducrocq-Geoffroy L., et al. Transcriptional Network Analysis Implicates Altered Hepatic Immune Function in NASH development and resolution. Nat. Metab. 2019;1:604–614. doi: 10.1038/s42255-019-0076-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cruikshank W., Little F. lnterleukin-16: the ins and outs of regulating T-cell activation. Crit. Rev. Immunol. 2008;28:467–483. doi: 10.1615/critrevimmunol.v28.i6.10. [DOI] [PubMed] [Google Scholar]
- 23.Glass W.G., Sarisky R.T., Vecchio A.M.D. Not-so-sweet sixteen: the role of IL-16 in infectious and immune-mediated inflammatory diseases. J. Interferon Cytokine Res. 2006;26:511–520. doi: 10.1089/jir.2006.26.511. [DOI] [PubMed] [Google Scholar]
- 24.Laberge S., Ernst P., Ghaffar O., Cruikshank W.W., Kornfeld H., Center D.M., Hamid Q. Increased expression of interleukin-16 in bronchial mucosa of subjects with atopic asthma. Am. J. Respir. Cell Mol. Biol. 1997;17:193–202. doi: 10.1165/ajrcmb.17.2.2750. [DOI] [PubMed] [Google Scholar]
- 25.Franz J.K., Kolb S.A., Hummel K.M., Lahrtz F., Neidhart M., Aicher W.K., Pap T., Gay R.E., Fontana A., Gay S. Interleukin-16, produced by synovial fibroblasts, mediates chemoattraction for CD4+ T lymphocytes in rheumatoid arthritis. Eur. J. Immunol. 1998;28:2661–2671. doi: 10.1002/(SICI)1521-4141(199809)28:09<2661::AID-IMMU2661>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Fang G., Kong F., Zhang H., Huang B., Zhang J., Zhang X. Association between inflammatory bowel disease and interleukins, chemokines: a two-sample bidirectional mendelian randomization study. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1168188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z.-W., Sun B., Gong T., Guo S., Zhang J., Wang J., Sugawara A., Jiang M., Yan J., Gurary A., et al. GNAI1 and GNAI3 Reduce Colitis-Associated Tumorigenesis in Mice by Blocking IL6 Signaling and Down-regulating Expression of GNAI2. Gastroenterology. 2019;156:2297–2312. doi: 10.1053/j.gastro.2019.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirshahi F., Aqbi H.F., Isbell M., Manjili S.H., Guo C., Saneshaw M., Bandyopadhyay D., Dozmorov M., Khosla A., Wack K., et al. Distinct hepatic immunological patterns are associated with the progression or inhibition of hepatocellular carcinoma. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeba Y., Ohta Y., Ootaki M., Kobayashi T., Kida K., Watanabe M., Koizumi S., Otsubo T., Iiri T., Matsumoto N. Identification of interleukin-16 production on tumor aggravation in hepatocellular carcinoma by a proteomics approach. Tumour Biol. 2021;43:309–325. doi: 10.3233/TUB-211507. [DOI] [PubMed] [Google Scholar]
- 30.Møhlenberg M., Terczynska-Dyla E., Thomsen K.L., George J., Eslam M., Grønbæk H., Hartmann R. The role of IFN in the development of NAFLD and NASH. Cytokine. 2019;124 doi: 10.1016/j.cyto.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Vachliotis I.D., Polyzos S.A. The Role of Tumor Necrosis Factor-Alpha in the Pathogenesis and Treatment of Nonalcoholic Fatty Liver Disease. Curr. Obes. Rep. 2023;12:191–206. doi: 10.1007/s13679-023-00519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeo B.S.Y., Koh J.H., Tan B.K.J., Ding Y., Teo Y.H., Alkan U., See A., Loh S., Toh S.T. Improved Inflammatory and Cardiometabolic Profile After Soft-Tissue Sleep Surgery for Obstructive Sleep Apnea: A Systematic Review and Meta-analysis. JAMA Otolaryngol. Head Neck Surg. 2022;148:862–869. doi: 10.1001/jamaoto.2022.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao Y., Song Y., Ning P., Zhang L., Wu S., Quan J., Li Q. Association between tumor necrosis factor alpha and obstructive sleep apnea in adults: a meta-analysis update. BMC Pulm. Med. 2020;20:215. doi: 10.1186/s12890-020-01253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caparrós E., Francés R. The Interleukin-20 Cytokine Family in Liver Disease. Front. Immunol. 2018;9:1155. doi: 10.3389/fimmu.2018.01155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin T.-Y., Hsu Y.-H. IL-20 in Acute Kidney Injury: Role in Pathogenesis and Potential as a Therapeutic Target. Int. J. Mol. Sci. 2020;21:1009. doi: 10.3390/ijms21031009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minville C., Hilleret M.-N., Tamisier R., Aron-Wisnewsky J., Clement K., Trocme C., Borel J.-C., Lévy P., Zarski J.-P., Pépin J.L. Nonalcoholic fatty liver disease, nocturnal hypoxia, and endothelial function in patients with sleep apnea. Chest. 2014;145:525–533. doi: 10.1378/chest.13-0938. [DOI] [PubMed] [Google Scholar]
- 37.Targher G., Tilg H., Byrne C.D. Non-alcoholic fatty liver disease: a multisystem disease requiring a multidisciplinary and holistic approach. Lancet. Gastroenterol. Hepatol. 2021;6:578–588. doi: 10.1016/S2468-1253(21)00020-0. [DOI] [PubMed] [Google Scholar]
- 38.Adams L.A., Anstee Q.M., Tilg H., Targher G. Non-alcoholic fatty liver disease and its relationship with cardiovascular disease and other extrahepatic diseases. Gut. 2017;66:1138–1153. doi: 10.1136/gutjnl-2017-313884. [DOI] [PubMed] [Google Scholar]
- 39.Kendzerska T., Gershon A.S., Hawker G., Leung R.S., Tomlinson G. Obstructive sleep apnea and risk of cardiovascular events and all-cause mortality: a decade-long historical cohort study. PLoS Med. 2014;11 doi: 10.1371/journal.pmed.1001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karolchik D., Baertsch R., Diekhans M., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.ENCODE Project Consortium The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 42.Gentleman R.C., Carey V.J., Bates D.M., Bolstad B., Dettling M., Dudoit S., Ellis B., Gautier L., Ge Y., Gentry J., et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varet H., Brillet-Guéguen L., Coppée J.Y., Dillies M.-A. SARTools: A DESeq2- and EdgeR-Based R Pipeline for Comprehensive Differential Analysis of RNA-Seq Data. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaucher J., Boussouar F., Montellier E., Curtet S., Buchou T., Bertrand S., Hery P., Jounier S., Depaux A., Vitte A.-L., et al. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J. 2012;31:3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaucher J., Vial G., Montellier E., Guellerin M., Bouyon S., Lemarie E., Pelloux V., Bertrand A., Pernet-Gallay K., Lamarche F., et al. Intermittent Hypoxia Rewires the Liver Transcriptome and Fires up Fatty Acids Usage for Mitochondrial Respiration. Front. Med. 2022;9 doi: 10.3389/fmed.2022.829979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gaucher J., Kinouchi K., Ceglia N., Montellier E., Peleg S., Greco C.M., Schmidt A., Forne I., Masri S., Baldi P., et al. Distinct metabolic adaptation of liver circadian pathways to acute and chronic patterns of alcohol intake. Proc. Natl. Acad. Sci. USA. 2019;116:25250–25259. doi: 10.1073/pnas.1911189116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 54.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xie Z., Bailey A., Kuleshov M.V., Clarke D.J.B., Evangelista J.E., Jenkins S.L., Lachmann A., Wojciechowicz M.L., Kropiwnicki E., Jagodnik K.M., et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021;1:e90. doi: 10.1002/cpz1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data reported in this paper will be shared by the lead contact upon request. RNA-seq data have been deposited at the Biotechnology Information Gene Expression Omnibus (GEO) and are publicly available as of the date of publication. Hepatic mouse gene expression data reported in this paper is available under the accession number GSE242668. This dataset contains 10 samples, 5 from IH mice and 5 from NC mice. The hepatic human gene expression dataset used in this study was recovered from the GEO repository under the accession number GSE33814. It consisted of 25 samples, 12 of which were steatohepatitis and 13 were controls.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.