Highlights
-
•
Nine spirostanol-type saponins, including five novel saponins Macrostemonoside U-Y, were found in Allium macrostemon Bunge.
-
•
All saponins were tested for DPPH, ABTS radical scavenging ability and Fe3+ reducing ability.
-
•
Macrostemonoside U exhibited the strongest in vitro antioxidant activity.
-
•
Twelve batches of Allium macrostemon Bunge from different origins and processing methods were examined for the content of nine saponins.
-
•
The content of Macrostemonoside W and X is influenced by factors such as its origin and whether it undergoes heating during processing.
Keywords: Allii Macrostemonis Bulbus, Steroidal saponin, Antioxidant activity, Processing, Content determination
Abstract
Allii Macrostemonis Bulbus (AMB), a traditional Chinese edible and medicinal plant, is considered beneficial to health. In this study, we isolated and purified nine steroidal saponins (compounds 1–9) from AMB. Their structures were characterized using physicochemical properties, HR-ESI-MS, 1D and 2D NMR spectroscopy. Among these compounds, compounds 1–5 were newly discovered named macrostemonoside U-Y, respectively. We assessed the in vitro antioxidant properties of the nine steroidal saponins through free radical scavenging and reducing power assays. This provides options for developing natural antioxidants. Additionally, an HPLC-ELSD quantitative analysis method was developed for the nine saponins in 12 batches of AMB from different origins and processing methods. The results showed that the contents of the nine steroidal saponins in AMB varied greatly among different growing environments and processing methods.
1. Introduction
Allii Macrostemonis Bulbus (AMB), known as ''Xiebai'' in China, refers to the dried bulb of either Allium macrostemon Bunge or Allium chinense G. Don from the Allium genus within the Liliaceae family (Chinese Pharmacopoeia Commission, 2020). The whole plant of AMB is edible, with a unique flavor and pungent taste, which not only improves appetite, but also has an antiseptic effect, and is one of the common dishes on the table; its bulb can be used as medicine for treating coronary heart disease, epigastric and abdominal distension, and diarrhea. Remarkably, AMB has earned official recognition as both a homology of medicine and food, as stipulated by the National Health and Wellness Commission of the People's Republic of China, attesting to its elevated status in terms of both edibility and medicinal efficacy (Wu et al., 2023b). Previous investigations have revealed a diverse array of bioactive constituents within AMB. Among these, steroidal saponins have emerged as its predominant pharmacological components (Zhao, et al., 2022). Early pharmacological inquiries unveiled the capacity of AMB's saponin constituents to clinically address myocardial infarction and angina pectoris via their antiplatelet aggregation properties (Deng, et al., 2022). Studies have shown that the development of cardiovascular disease is closely related to the level of free radicals (Maxwell and Lip, 1997). As research has progressed, an expanding array of effects has been discovered, encompassing antioxidant, antitumor, antidepressant, antimicrobial, anti-atherosclerotic, hypoglycemic, hypolipidemic, and analgesic attributes (Yao, et al., 2016). Over 90 steroidal saponin-like components have been isolated from AMB. However, their quality markers remain unidentified, and few articles have been published concerning the determination of their steroidal saponin content. Hence, the primary objective of this study is to isolate additional steroidal saponin-like constituents from AMB and conduct a preliminary evaluation of their efficacy through in vitro antioxidant assays. The aim is to provide potential avenues for developing natural antioxidants. Moreover, the identified components will be evaluated for their presence in AMB products subjected to various processing techniques.
2. Materials and methods
2.1. Materials
Column chromatography (CC) analysis was conducted employing silica gel (Qingdao Ocean Chemical Factory, Qingdao, China) and C18 reverse silica gel packing (50 μm, YMC Co., Ltd., Tokyo, Japan). The analysis was performed using an Acchrom S6000 high-performance liquid chromatograph (HPLC) (Acchrom Tech Technology Co., Ltd., Beijing, China), coupled with an ELSD-UM 5800 Plus detector (Unimicro Technologies Co., Ltd., Shanghai, China). HR-ESI-MS was conducted utilizing an LTQ-Orbitrap XL spectrometer (Thermo Fisher Scientific, Boston, MA, USA). The SPECTRO star Nano full-wavelength enzyme labeler (BMG LABTECH, Offenburg, Germany) was used for absorbance measurements. Telstar* LyoQuest Vacuum Freeze Dryer (TELSTAR TECHNOLOGIES. S. L, Barcelona, Spain) is used for freeze drying of samples. DHG-9070A Electrothermal constant temperature blast drying oven (Shanghai Shenxian Constant Temperature Equipment Factory, Shanghai, China). Extraction and CC separation utilized analytically pure solvents—petroleum ether (PE), ethanol (EtOH), methanol (MeOH), n-butanol, and dichloromethane (CH2Cl2) (Beijing Chemical Factory, Beijing, China). HPLC analysis employed chromatographically pure methanol obtained (Thermo Fisher Scientific, Waltham, MA, USA). For nuclear magnetic resonance spectroscopy, deuterated solvents including deuterated pyridine and deuterated dimethyl sulfoxide (Aladdin Biochemical Technology Co., Ltd., Shanghai, China).
2.2. Nuclear magnetic resonance (NMR) analysis
1H- (600 MHz) and 13C NMR (150 MHz), heteronuclear multiple quantum correlation (HMQC), heteronuclear multiple band correlation (HMBC), correlation spectroscopy (1H–1H COSY), and nuclear overhauser effect spectroscopy (NOESY). The spectra were obtained using a Bruker Avance III 600 spectrometer (Bruker, Billerica, Germany). All compounds use the deuterated solvent signal as an internal reference and chemical shift values are expressed as δ values.
2.3. Plant material
The experimental plants were all obtained from online suppliers in 2021 and originated from Changchun, Jilin; Benxi, Liaoning; Luoyang, Henan; and Zigong, Sichuan. Identification as Allium macrostemon Bunge, belonging to the Allium genus within the Liliaceae family, was confirmed by Prof. Jing Zhang from the College of Traditional Chinese Medicine at Jilin Agricultural University. The voucher specimen (20210971) has been cataloged and deposited within the Herbal Library of the College of Traditional Chinese Medicine, Jilin Agricultural University.
2.4. Preparation of different processed products of AMB
Fresh AMB from various regions in China underwent different processing methods to produce various products: Freeze-dried AMB: The fresh AMB was pre-cooled at −20 °C and then subjected to vacuum freeze-drying in a lyophilizer. Directly dried AMB: The fresh AMB was placed in an electrically heated constant-temperature blower drying oven and dried at 50 °C to remove moisture. Steam-dried AMB: The fresh AMB was steamed, wiping off surface water, and then the remaining moisture was eliminated using an electrically heated blast dryer at 50 °C. Details regarding the distinct processed AMB products in each production area can be found in Table S1.
2.5. Extraction and separation of compounds 1–9
A 3.0 kg sample of AMB was pulverized, sieved through a 40-mesh sieve, and subsequently dried, yielding 2.7 kg of AMB powder. This powder was subjected to extraction by combining it with an 80 % EtOH solution at a ratio of 1:5 (m: V, g: mL) for a duration of 1 h. This extraction process involved the use of 100 W ultrasonic waves for 30 min, followed by filtration and repetition for a total of six times. The combined resulting filtrates were concentrated under reduced pressure until devoid of alcohol flavor. Subsequently, the concentrated solution underwent lyophilization, yielding 1.4 kg of AMB ethanol extract. Following this, 0.5 kg of the AMB ethanol extract was dissolved in water (at a ratio of mass to volume of 1:15, m: V, g: mL) and successively extracted with PE, CH2Cl2, and n-butanol in a sequential gradient. Four extractions were conducted using each solvent. The portions extracted with PE and CH2Cl2 were discarded to eliminate fat-soluble and less polar components. The n-butanol extract was concentrated under reduced pressure until it had no n-butanol odor, then lyophilized, and finally the n-butanol extract fraction of AMB was isolated, weighing 11.3 g. Subsequently, the n-butanol extract fraction (10 g) of AMB was subjected to CC analysis using 200–300 mesh silica gel. Elution was performed with a gradient of CH2Cl2-MeOH-H2O (5:2:1 ∼ 13:8:2, if the solvents were immiscible, the underlayer solvent was selected), resulting in the separation of 12 parts (P. 1 ∼ 12). P. 3 (382 mg) underwent CC on ODS silica gel, eluted with a gradient of MeOH-H2O (ranging from 10 % to 80 %), resulting in the isolation of three subparts designated as P. 3.1 to P. 3.5. Specifically, 81 mg from P. 3.2 was further subjected to CC on ODS silica gel using a MeOH-H2O gradient of 20 % to 40 %, ultimately yielding compound 6 (48 mg). P. 3.3 (72 mg) underwent CC on ODS silica gel, employing a MeOH-H2O gradient spanning 40 % to 60 %, resulting in the isolation of compound 8 (33 mg) and compound 9 (28 mg). Separately, P. 3.5 (42 mg) underwent CC on ODS silica gel using a MeOH-H2O gradient ranging from 50 % to 80 %, yielding compound 4 (13 mg). P. 4 (365 mg) underwent CC on ODS silica gel with a MeOH-H2O gradient ranging from 20 % to 90 %, resulting in the separation of three subparts labeled as P. 4.1 to P. 4.3. Specifically, P. 4.2 (59 mg) underwent recrystallization in MeOH, yielding compound 7 (34 mg). Meanwhile, P. 4.3 (42 mg) underwent CC on ODS silica gel using a MeOH-H2O gradient from 40 % to 80 %, leading to the isolation of compound 5 (19 mg). P. 11 (115 mg) underwent CC on ODS silica gel using a MeOH-H2O gradient ranging from 40 % to 90 %, yielding three subparts labeled as P. 11.1 to P. 11.5. Further separation involved subjecting P. 11.3 (32 mg) to CC on ODS silica gel with a MeOH-H2O gradient of 40 % to 60 %, resulting in the isolation of compound 3 (12 mg). Additionally, P. 11.5 (46 mg) underwent to CC on ODS silica gel employing a MeOH-H2O gradient spanning from 60 % to 90 %, leading to the isolation of compound 1 (9 mg) and compound 2 (11 mg).
2.6. In vitro antioxidant tests
The methods were referred to in the literature with some modifications (Sharma and Bhat, 2009, Sinan et al., 2021, Wang et al., 2017). The antioxidant capacity of the resulting monomeric components was evaluated by DPPH and ABTS radical scavenging ability tests and Fe3+ reduction ability tests. Precisely 100 μL of sample solutions at varying concentrations (1, 0.5, 0.25, 0.125, 0.0625 mg·mL−1) were aspirated in a 96-well plate, while 100 μL of DPPH solution (0.2 M) was added, mixed and allowed to react in a thermostat at 28 °C for 35 min under light protection. Upon completion of the reaction, absorbance values (Ax) were measured at 517 nm. Anhydrous ethanol was utilized in the control group instead of the DPPH solution, and its corresponding absorbance was recorded as (Ay). In the blank group, anhydrous ethanol was used instead of the sample solution (A0). The ABTS master batch was obtained by adding 19.2 mg of ABTS and 3.31 mg of potassium persulfate, dissolving them in water, and fixing them in a 5 mL volumetric flask. After allowing the reaction to proceed for 15 h at room temperature under light protection, the resulting solution was diluted with anhydrous ethanol to achieve an absorbance reading of 0.7 ± 0.1 at 734 nm, yielding the ABTS solution. Following the completion of the reaction, the absorbance value (Ax) was measured at 734 nm. Anhydrous ethanol was employed in the control group instead of the ABTS solution, and its corresponding absorbance was recorded as (Ay). In the blank group, anhydrous ethanol was used instead of the sample solution (A0). The scavenging rates of DPPH and ABTS radicals were calculated according to Equation (1). Carefully pipette 200 μL of the aforementioned sample solutions with varying concentrations into individual 1 mL centrifuge tubes. Subsequently, add phosphate buffer solution (pH 6.6) followed by 200 μL of 1 % potassium hexacyanoferrate solution. After thorough mixing, allow the reaction to proceed in a thermostat at 55 °C for 40 min. Upon completion, cool the mixture to room temperature, then add 200 μL of 10 % trichloroacetic acid solution to halt the reaction. Centrifuge the reaction mixture at 4000 r/m for 10 min. Aspirate 200 μL of the supernatant, followed by the addition of 200 μL of deionized water. Subsequently, introduce 40 μL of a 0.1 % ferric chloride solution, and then pipette 100 μL of the resulting mixture into a 96-well plate. Measure the absorbance (Ax) at 700 nm. Deionized water was used to replace the sample solution in the blank group (A0). The Fe3+ reduction rate was calculated according to Equation (2). Vitamin C (VC) served as the positive control within each group, following the same procedure outlined for the aforementioned groups. Each measurement was conducted with three replicate wells arranged in parallel for consistency across all analyses. The experimental data for DPPH and ABTS free radical scavenging abilities, as well as Fe3+ reduction ability, were analyzed using GraphPad Prism software. IC50 values, representing the concentration of the test sample necessary to scavenge 50 % of free radicals or reduce 50 % of Fe3+, were calculated using non-linear regression analysis through GraphPad Prism software. These values are presented as mean ± standard deviation (SD).
| (1) |
| (2) |
2.7. Preparation of standards and test material
The standards (compounds 1–9) were weighed precisely, dissolved in 80 % methanol–water solution and diluted to the corresponding concentrations, and then passed through 0.22 μm microporous filter membrane to obtain the mixed standard solutions of different concentrations. Precisely weigh 2.0 g of different batches of AMB powder (M1-12, Table S1), add 60 mL of 80 % methanol–water solution, ultrasonic extraction at 50 °C for 60 min, repeated three times, combined and recovered the solvent, add 2 mL of 80 % methanol–water to dissolve the extracts, and pass through 0.22 μm microporous filtration membrane that is to obtain the different batches of the test solution.
2.8. HPLC-ELSD analysis of spirostanol-type saponins in AMB
The chromatographic column utilized was an Agilent TC-C18 (250 mm × 4.6 mm, 5 µm) maintained at 35 °C. The injection volume was set at 20 μL, with a flow rate of 0.8 mL·min−1. Additionally, the evaporation temperature was set to 90 °C, the gas flow rate to 2.0 L·min−1, and the gain value was adjusted to 3. Mobile phase A consisted of a 0.05 % formic acid–water solution, while mobile phase B comprised acetonitrile. The following gradient system was used for elution, 0–5 min, 5–10 % B; 5–10 min, 10–15 % B; 10–13 min, 15–18 % B; 13–25 min, 18–21 % B; 25–40 min, 21–25.5 % B; 40–43 min, 25.5–26.5 % B; 43–45 min, 26.5––27 % B; 45–52 min, 27–27.5 % B; 52–55 min, 27.5–28 % B; 55–58 min, 28 % B; 58–60 min, 28–29 % B; 60–63 min, 29 % B; 63–70 min, 29–30 % B; 70–78 min, 30–75 % B; 78–85 min, 75–85 % B; 85–90 min, 85–90 % B; 90–110 min, 90 % B.
Calibration curves were plotted by injecting the corresponding concentration (x) of the standard and the peak area (y) corresponding to it (Table S4) and were used to determine the content of compounds 1–9 in different batches of AMB. The limit of detection (LOD) and limit of quantification (LOQ) were 3 and 10 times the signal-to-noise ratio, respectively. The precision test was performed by injecting the mixed standard solution into the sample six times consecutively and calculating the relative standard deviation (RSD) value of each standard. Sample M1 was injected six times consecutively and the RSD value of each standard was calculated to assess its repeatability. Sample M1 was left for 0, 2, 4, 8, 16 and 24 h before injection and the RSD value of each standard was calculated to assess its stability. The same batch of AMB powder (M1) with known content was weighed, mixed control solution with equal content of each component of the herb powder was added, and six sample solutions were prepared as described above and then injected into the sample for determination, and the spiked recoveries and RSD values of each standard were calculated.
3. Results and discussion
3.1. Five new spirostanol-type saponins (compounds 1–5)
Nine spirostanol-type saponins were isolated from post-steam-dried AMB produced in Changchun, Jilin (Fig. 1). Their structural characterization was determined by combining HR-ESI-MS, 1D, and 2D NMR spectroscopic data (Supplementary Material). The types and proportions of sugar molecules in the compounds were determined by acid hydrolysis experiments. Among the compounds, compounds 1–5 are newly discovered compounds, and the 2D NMR analysis is shown in Fig. 2. The 1H- and 13C NMR of their saponinogenin are listed in Table 1, and the sugar groups are listed in Table S2.
Fig. 1.
Chemical structures of compounds 1–9, compounds 1–5 are new compounds and 6–9 are known ones.
Fig. 2.
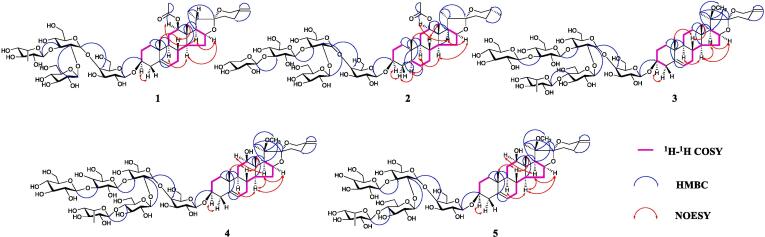
Key 1H–1H COSY (in pink bold), HMBC (blue arrows) and NOESY (red arrows) correlations of compounds 1–5. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Table 1.
1H NMR and 13C NMR data of the aglycones of compounds 1–5 (δ in ppm).
| Position |
1 |
2 |
3 |
4 |
5 |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| δC | δH (mult., J in Hz) | δC | δH (mult., J in Hz) | δC | δH (mult., J in Hz) | δC | δH (mult., J in Hz) | δC | δH (mult., J in Hz) | |
| 1 | 37.48 | 0.96 (1H, m)1.65 (1H, m) |
37.64 | 0.78 (2H, m) | 37.46 | 1.44 (2H, m) | 37.62 | 1.43 (2H, m) | 37.46 | 1.42 (2H, m) |
| 2 | 29.72 | 1.11 (2H, d, 6.96) | 30.36 | 1.58 (2H, m) | 29.71 | 1.49 (2H, m) | 29.73 | 1.50 (2H, m) | 29.70 | 1.10 (2H, d, 6.90) |
| 3 | 81.56 | 4.53 (1H, m) | 81.61 | 4.54 (1H, m) | 81.55 | 4.29 (1H, m) | 81.60 | 4.28 (1H, m) | 81.54 | 4.31 (1H, m) |
| 4 | 40.29 | 1.68 (2H, m) | 35.26 | 1.22 (2H, m) | 40.28 | 1.60 (2H, m) | 40.58 | 1.57 (2H, m) | 40.27 | 1.63 (2H, m) |
| 5 | 141.39 | – | 45.07 | 0.88 (1H, m) | 141.88 | – | 141.42 | – | 141.40 | – |
| 6 | 122.12 | 5.30 (1H, m) | 29.38 | 1.52 (2H, m) | 122.09 | 5.31 (1H, d, 7.92) | 122.10 | 5.31 (1H, d, 8.04) | 122.08 | 5.30 (1H, m) |
| 7 | 32.83 | 1.44 (2H, m) | 32.86 | 1.43 (2H, m) | 32.83 | 1.41 (2H, m) | 32.84 | 1.40 (2H, m) | 32.82 | 1.39 (2H, m) |
| 8 | 35.67 | 1.33 (1H, m) | 35.69 | 1.37 (1H, m) | 35.66 | 1.34 (1H, m) | 35.67 | 1.33 (1H, m) | 35.65 | 1.32 (1H, m) |
| 9 | 50.67 | 0.90 (1H, s) | 54.82 | 0.50 (1H, m) | 50.66 | 0.79 (1H, m) | 50.67 | 0.79 (1H, m) | 50.64 | 0.78 (1H, m) |
| 10 | 36.25 | – | 36.26 | – | 36.24 | – | 36.25 | – | 36.23 | – |
| 11 | 30.58 | 1.52 (2H, m) | 32.27 | 1.35 (2H, m) | 21.53 | 1.37 (2H, m) | 30.36 | 1.96 (2H, m) | 30.34 | 1.50 (1H, m)1.96 (1H, m) |
| 12 | 78.05 | 3.88 (1H, m) | 78.05 | 4.11 (1H, m) | 39.66 | 2.42 (1H, m)2.67 (1H, m) |
79.02 | 4.38 (1H, m) | 79.13 | 4.00 (1H, m) |
| 13 | 40.89 | – | 41.24 | – | 40.88 | – | 41.23 | – | 41.22 | – |
| 14 | 57.05 | 1.05 (1H, m) | 56.89 | 0.90 (1H, m) | 56.86 | 0.94 (1H, m) | 56.87 | 0.92 (1H, m) | 57.03 | 0.97 (1H, m) |
| 15 | 33.64 | 1.74 (2H, m) | 35.23 | 1.73 (2H, m) | 35.23 | 1.71 (2H, m) | 35.24 | 1.70 (2H, m) | 35.23 | 1.70 (2H, m) |
| 16 | 81.92 | 4.18 (1H, m) | 81.97 | 4.40 (1H, m) | 81.96 | 4.27 (1H, s) | 81.98 | 4.26 (1H, s) | 81.96 | 4.55 (1H, m) |
| 17 | 63.31 | 4.45 (1H, m) | 63.47 | 3.93 (1H, m) | 63.30 | 4.56 (1H, m) | 63.31 | 4.55 (1H, m) | 63.29 | 4.44 (1H, d, 8.52) |
| 18 | 16.83 | 0.83 (3H, s) | 17.11 | 0.83 (3H, s) | 16.82 | 0.83 (3H, s) | 16.83 | 0.83 (3H, s) | 16.81 | 0.82 (3H, s) |
| 19 | 17.81 | 0.70 (3H, d, 5.64) | 12.77 | 0.64 (3H, d, 4.98) | 12.74 | 0.63 (3H, s) | 12.76 | 0.63 (3H, s) | 12.73 | 0.63 (3H, d, 2.28) |
| 20 | 42.42 | 1.97 (1H, q, 6.72) | 42.45 | 1.96 (1H, t, 6.78) | 81.81 | – | 81.82 | – | 81.91 | – |
| 21 | 15.53 | 1.15 (3H, d, 6.90) | 15.55 | 1.15 (3H, d, 6.90) | 15.52 | 1.13 (3H, d, 6.90) | 15.54 | 1.15 (3H, d, 6.96) | 15.51 | 1.15 (3H, d, 6.96) |
| 22 | 109.74 | – | 109.72 | – | 109.72 | – | 109.74 | – | 109.71 | – |
| 23 | 32.65 | 1.39 (2H, m) | 32.60 | 1.40 (2H, m) | 32.25 | 1.60 (2H, m) | 32.26 | 1.60 (2H, m) | 32.24 | 1.59 (2H, m) |
| 24 | 29.44 | 1.59 (2H, m) | 29.74 | 2.04 (2H, m) | 29.35 | 1.01 (2H, m) | 29.37 | 1.01 (2H, m) | 29.34 | 1.02 (2H, m) |
| 25 | 144.88 | – | 31.08 | 1.61 (1H, d, 12.18) | 144.88 | – | 144.88 | – | 144.87 | – |
| 26 | 67.31 | 3.57 (2H, m) | 67.33 | 3.52 (2H, m) | 67.30 | 3.52 (1H, m)3.60 (1H, m) |
67.32 | 3.52 (1H, m)3.61 (1H, m) |
67.29 | 3.51 (1H, m)3.61 (1H, m) |
| 27 | 109.24 | 4.82 (2H, m) | 17.84 | 0.70 (3H, d, 5.58) | 109.69 | 4.72 (2H, q, 8.76) | 109.70 | 4.72 (2H, q, 8.82) | 109.21 | 4.77 (2H, q, 8.82) |
| OMe | – | – | – | – | 45.04 | 2.96 (3H, d, 5.58) | 45.06 | 2.96 (3H, d, 5.64) | 45.03 | 2.95 (3H, d, 5.58) |
| Ac-Me | 21.58 | 2.06 (3H, s) | 21.60 | 1.82 (3H, s) | – | – | – | – | ||
| Ac-CO | 171.09 | – | 171.13 | – | – | – | – | – | ||
Assignments were made by a combination of 1D and 2D NMR experiments. 1H NMR and 13-NMR were measured at 150 MHz and 600 MHz, respectively, using deuterated Pyridine‑d5 as the deuterated solvent.
Compound 1, identified as an amorphous white powder, exhibited a positive reaction to Molisch and Liebermann-Burchard tests, while showing a negative response to Ehrlish reagent. Upon heating with 10 % concentrated H2SO4-EtOH on a thin-layer silica gel plate, it initially displayed a purplish-red hue, transitioning to a yellowish-green color over time—a characteristic suggestive of a spirostanol-type saponin. Positive ion HR-ESI-MS recorded its m/z as 1125.5114 [M + Na]+ (theoretical value of 1125.5094, Figure S1). Further analysis, incorporating its 1H and 13C spectra, facilitated the determination of its molecular formula as C53H82O24. The 1H NMR (600 MHz, Pyridine‑d5, δH 8.74) analysis of compound 1 (Table 1, Figure S2) revealed distinct signals representing tertiary methyl protons in the high-field region, notably at δH 0.83 (3H, s, Me-18), 0.70 (3H, d, J = 5.64 Hz, Me-19), alongside a characteristic signal of a secondary methyl proton at δH 1.15 (3H, d, J = 6.90 Hz, Me-21). Concurrently, the 13C NMR (150 MHz, Pyridine‑d5, δC 150.35) analysis of compound 1 (Table 1, Figure S3) exhibited 53 carbon signals, with 29 attributed to the saponinogenin and 24 to the four sugar molecules (Table S2). We detected characteristic double bond signals at δC 141.39, 122.12, 144.88, and 109.24 double bond characteristic signals and δC 171.09 carbonyl characteristic signal in the low-field region, and hypothesized that there might be two double bonds and one carbonyl group in the compound structure. Three methyl signals were present at δC 16.83, 17.81, and 15.53 in the high-field region, corresponding to C-18, C-19, and C-21, respectively, as identified in prior studies (Wu et al., 2023a). In addition, we found that the parent nucleus of compound 1 had two extra carbon signals at δC 171.09 and 21.58, and according to HMQC (Figure S4) and literature reports (Wang, et al., 2023), δC 21.58 is associated with δH 2.06 (3H, s), suggesting the presence of an acetyl group, which echoes our previous speculation. Simultaneously, the absence of the methyl signal corresponding to C-27 was noted within the spirostanol saponinogenin's parent nucleus. Additionally, two terminal alkenyl hydrogen proton signals were identified at 4.82 (2H, m), showing a remote correlation with δC 145.03 (C-25) (HMBC, Figure S5), thus determining the existence of a double bond between C25-C27, in line with the previous inference. The proton signal δH 1.15 (3H, d, J = 6.90 Hz) of Me-21 is remotely correlated with δC 42.42, 63.31, and 109.74, assigning it to C-20, C-17, and C-22, respectively. The proton signal at δH 0.83 (3H, s) corresponding to Me-18 displayed remote correlations with δC 40.89, 57.05, 63.31, and 78.05, assigned to C-13, C-14, C-17, and C-12, respectively. Meanwhile, we found that δH 3.88 (H-12) and 2.06 (Ac-Me) are remotely correlated with δC 171.09, respectively, due to the attachment of this acetyl group to the oxygen atom on C-12, and the chemical shift value of C-12 is shifted to 78.05 ppm toward the lower field, again proving our previous inference. The proton signal of Me-19, δH 0.70 (3H, d, J = 5.64 Hz), is remotely correlated with δC 36.25, 37.48, 50.67, and 141.39, which were assigned to C-10, C-1, C-9, and C-5, respectively. In addition, the proton signals of C-4 (δH 1.68), C-7 (δH 1.44), and the alkenyl hydrogen proton signal at position C-6 (δH 5.30) are all remotely correlated with the presence of C-5 (δC 141.39), and thus the existence of a C5-C6 double bond is determined, which corroborates our previous inference. In the 1H–1H COSY spectrum (Figure S6), there was a correlation between H2-1/H2-2/H-3, and then combined with the HMQC spectrum, therefore, δC 29.72 was assigned to C-2, and δC 81.56 was assigned to the signal at the C-3 position. At the same time, given the glycosidic shifts influencing C-3, resulting in a chemical shift towards the lower field at 81.56 ppm, it was hypothesized that the C-3 position was associated with the sugar group. In the NOESY spectrum (Figure S7), a correlation is evident between the hydrogen signal at the C-12 position (δH 3.88) and the C-17 (α-type, δH 4.45) signal, identifying the oxygen-containing group at the C-12 position as a β-type (Kim, et al., 2017).
The acid hydrolysis test exhibited a 1:1:2 ratio of galactose, rhamnose, and glucose. These sugar molecules were identified as d-galactose, l-rhamnose, and d-glucose by gas chromatography-mass spectrometry (GC–MS) analysis after derivatization. Analysis of the four anomeric proton signals at δH 4.90 (1H, dd, J = 1.86, 7.56 Hz, H-1a), 5.33 (1H, d, J = 7.62 Hz, H-1b), 5.63 (1H, t, J = 8.16 Hz, H-1c) and 5.10 (1H, dd, J = 3.06, 7.92 Hz, H-1f), alongside the HMQC correlations with the respective anomeric carbons at 103.17, 104.85, 105.02, and 105.14 ppm, confirmed the presence of four sugar molecules and supported this supposition. Observing the HMBC spectrum, the anomeric proton signal at δH 4.90 for galactose-a displayed a remote correlation with C-3 of the parent nucleus (δC 81.56). Hence, it was concluded that the sugar was linked to the C-3 position of the saponinogenin's parent nucleus, aligning with our earlier inference. Additionally, the anomeric proton δH 5.33 (H-1b) of glucose-b is remotely related to C-4a (δC 81.96) of galactose-a, presumed to be connected in a 1 → 4 manner. The anomeric proton δH 5.63 (H-1c) of glucose-c is remotely related to C-2b (δC 80.64) of glucose-b, presumed to be connected in a 1 → 2 manner. The anomeric proton of rhamnose-f δH 5.10 (H-1f) is remotely related to C-3b (δC 89.27) of glucose-b, presumably connected in a 1 → 3 manner. The structure of compound 1 was established as spirostane-5,25(27)-dien-12β-O-Ac-3-O-β-d-glucopyranosyl(1 → 2)-O-[α-l-rhamnopyranosyl(1 → 3)]-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranosid, and it has been designated as macrostemonoside U.
The physicochemical characteristics of compound 2 closely resembled those of compound 1, indicating its potential classification as a spirostanol-type saponin. The positive ion HR-ESI-MS recorded an m/z of 1277.5792 [M + Na]+ (theoretical value of 1277.5778, Figure S8). The molecular formula was determined to be C58H94O29 by combining with its 1H and 13C spectra. The 1H NMR (600 MHz, Pyridine‑d5, δH 8.74) analysis of compound 2 (Table 1, Figure S9) revealed two distinct signals of tertiary methyl protons in the high-field region at δH 0.83 (3H, s, Me-18), 0.64 (3H, d, J = 4.98 Hz, Me-19), and two signals characteristic of secondary methyl protons at δH 1.15 (3H, d, J = 6.90 Hz, Me-21), 0.70 (3H, d, J = 5.58 Hz, Me-27). Concurrently, the 13C NMR (150 MHz, Pyridine‑d5, δC 150.35) analysis of compound 2 (Table 1, Figure S10) displayed 58 carbon signals, with 29 attributed to the saponinogenin and 29 to the presence of five sugar molecules (Table S2). We identified a δC 171.13 carbonyl character signal in the low-field region and speculated that a carbonyl group might be present in the compound structure, and four methyl signals at δC 12.77, 15.55, 17.11, and 17.84 in the high-field region, which, based on previous studies (Peng, et al., 1992), we assigned to C-19, C-21, C-18, and C-27, respectively. In addition, the C-27 proton signal at δH 17.84 (Me-27) displayed remote correlations with δC 29.24, 31.08, and 67.33 (HMBC, Figure S12), corresponding to C-24, C-25, and C-26, respectively, and C-25 was determined to be the β-configuration based on the chemical shifts of the three (Cheng, 2013). Therefore, we hypothesize that compound 2 is similar to the saponinogenin structure of compound 1, with only the loss of the alkene bonds at C5-C6 and C25-C27. The proton signal at δH 0.83 (3H, s) corresponding to Me-18 exhibited remote correlations with with δC 41.24, 56.89, and 78.05, assigning it to C-13, C-14, and C-12, respectively. According to HMQC (Figure S11) and HMBC, the proton signals δH 4.11 (H-12) and δC 1.82 (Ac-Me) of C-12 are remotely correlated with δC 171.13, respectively. The chemical shift value of C-12 is shifted toward the lower field to 78.05 ppm due to the acetyl group, which is consistent with our previous speculation that the acetyl group is connected to the oxygen atom on C-12. The proton signal δH 1.15 (3H, d, J = 6.90 Hz) of Me-21 is remotely correlated with δC 42.45, 63.47, and 109.72, which were assigned to C-20, C-17, and C-22, respectively. Additionally, the proton signal at δH 0.64 (3H, d, J = 4.98 Hz) of Me-19 exhibited remote correlations with δC 36.26, 37.64, 45.07, and 54.82, which were assigned to C-10, C-1, C-5, and C-9, respectively. In addition, the proton signal δH 0.88 (H-5) of C-5 is remotely correlated with C-4 (δC 35.36) and C-6 (δC 29.38), respectively. Given that the proton signal of Me-18 (δH 0.83) appears at a lower field than that of Me-19 (δH 0.64), H-5 was determined to possess the α-configuration (Yang, 2021). In the 1H–1H COSY spectrum (Figure S13), there was a correlation between H2-1/H2-2/H-3, further supported by the HMQC spectrum. Consequently, δC 30.36 was assigned to C-2, and δC 81.61 was assigned to the signal at the C-3 position. Moreover, considering the glycosidic shifts affecting C-3 and resulting in a chemical shift towards the lower field to 81.61 ppm, it was inferred that the C-3 position is linked to the sugar group. The NOESY spectrum (Figure S14) illustrates a correlation between the hydrogen signal at the C-12 position (δH 4.11) and the H-17 (α-type, δH 3.93) signal, thereby identifying the oxygen-containing group at the C-12 position as a β-type (Kim, et al., 2017).
The acid hydrolysis test showed a ratio of 1:1:3 for galactose, xylose, and glucose. These sugar molecules were identified as d-galactose, d-xylose, and d-glucose by GC–MS analysis after derivatization. Analysis of the five anomeric proton signals at δH 4.90 (1H, dd, J = 2.28, 7.68 Hz, H-1a), 5.24 (1H, d, J = 7.80 Hz, H-1b), 5.60 (1H, t, J = 7.74 Hz, H-1c), 5.73 (1H, t, J = 9.60 Hz, H-1d) and 5.18 (1H, d, J = 7.92 Hz, H-1 g) indicated the presence of five sugar molecules. This inference was corroborated by their respective correlations in the HMQC with the five corresponding anomeric carbons at 102.85, 104.85, 105.15, 105.02, and 105.31 ppm, respectively. In the HMBC spectrum, the remote correlation observed between the anomeric proton signal at δH 4.90 of galactose-a and the parent nucleus C-3 (δC 81.61) led to the conclusion that the sugar is linked to the C-3 position of the saponinogenin's parent nucleus, aligning with our previous deduction. Additionally, the remote correlation between the anomeric proton signal δH 5.24 (H-1b) of glucose-b and C-4a (δC 80.77) of galactose-a suggests a presumed linkage in a 1 → 4 manner. The anomeric proton signal δH 5.60 (H-1c) of glucose-c is remotely correlated with C-2b (δC 80.64) of glucose-b and is presumed to be connected in a 1 → 2 manner. The anomeric proton signal δH of glucose-d 5.73 (H-1d) is remotely related to C-3b of glucose-b (δC 89.26) and is presumed to be connected in a 1 → 3 manner. The anomeric proton signal δH 5.18 (H-1 g) of xylose-g is remotely related to C-3d of glucose-d (δC 88.91) and is presumed to be connected in a 1 → 3 manner. The structure of compound 2 was established as (25R)-spirostane-12β-O-Ac-3-O-β-d-glucopyranosyl(1 → 2)-O-[β-d-xylopyranosyl(1 → 3)-O-β-d-glucopyranosyl(1 → 3)]-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranosid, and it has been designated as macrostemonoside V.
The physicochemical properties of compound 3 closely resembled those of compound 1, indicating a potential classification as a spirostanol-type saponin. The positive ion HR-ESI-MS displayed an m/z value of 1421.6221 [M + Na]+ (theoretical value of 1421.6201, Figure S15), and the molecular formula was determined to be C64H102O33 by combining with its 1H and 13C spectra. The 1H NMR (600 MHz, Pyridine‑d5, δH 8.74) analysis of compound 3 (Table 1, Figure S16) reveal the presence of four distinctive signals corresponding to methyl protons in the high-field region. Upon comparison with compound 1, these signals were identified as δH 0.83 (3H, s, Me-18), 0.63 (3H, s, Me-19), 1.15 (3H, d, J = 6.90 Hz, Me-21) and 0.87 (3H, s, Rha-6f). In addition, we found a methyl signal at δH 2.96 (3H, d, J = 5.58, OMe), which was identified as a methoxyl signal (δC 45.04) based on the HMQC (Figure S18) and literature reports (Kim, et al., 2017). The 13C NMR (150 MHz, Pyridine‑d5, δC 150.35) analysis of compound 3 (Table 1, Figure S17) revealed the presence of 64 carbon signals. Among these, 28 were attributed to the saponin element, while the remaining 36 were associated with the six sugar molecules (Table S2). We found the presence of δC 141.88, 122.09, 144.88, and 109.69 double bond characteristic signals in the low-field region, and speculated that two double bonds may exist in the compound structure, and presumed that compound 3 is similar to the saponin parent nucleus of compound 1. Therefore, we compared the carbon spectral data of compound 3 with that of compound 1 and found that the saponinogenins of the two were highly compatible. The two noteworthy points are that the acetyl group at the C-12 position disappears and we did not find the original chemical shift value of C-20, but a quaternary carbon signal of δC 81.81 was found in the low-field region, which is presumed to be the effect of methoxy resulting in the chemical shift value of the C-20 position shifted towards the low field to 81.81 ppm. According to HMBC (Figure S19), the proton signal of methoxy (δH 2.96) was remotely correlated with δC 81.81 (C-20), (C-17), and (C-22), respectively, which validates our previous speculation. The 1H–1H COSY spectrum (Figure S20) displayed a correlation between H2-1/H2-2/H-3/H2-4. This, coupled with the HMQC spectrum, led to the assignment of δC 29.71 to C-2, and δC 81.55 to the signal corresponding to the C-3 position. Similar to compound 1, the chemical shift of C-3 was influenced by glycosidic shifts, resulting in a shift towards the lower field to 81.55 ppm.
The acid hydrolysis test showed a 1:1:4 ratio of galactose, rhamnose, and glucose. These sugar molecules were identified as d-galactose, l-rhamnose, and d-glucose by GC–MS analysis after derivatization. The presence of six sugar molecules was inferred from the six anomeric proton signals δH 4.91 (1H, t, J = 7.50 Hz, H-1a), 5.53 (1H, d, J = 7.56 Hz, H-1b), 5.18 (1H, d, J = 7.74 Hz, H-1c), 5.61 (1H, d, J = 7.74 Hz, H-1d), 5.17 (1H, d, J = 7.68 Hz, H-1e) and 5.33 (1H, d, J = 7.86 Hz, H-1f) inferred the presence of six sugar molecules, which was verified by the association of HMQC with the six anomeric carbons at 102.82, 104.48, 105.42, 105.02, 105.22, and 103.16 ppm, respectively. The HMBC spectrum revealed a remote correlation between the anomeric proton signal of galactose-a at δH 4.91 and the parent nucleus, C-3 (δC 81.55). Consequently, it was concluded that the sugar was linked to the C-3 position of the parent nucleus of the saponinogenin, consistent with our earlier assessment. The anomeric proton δH 5.53 (H-1b) of glucose-b is remotely correlated with C-4a (δC 80.73) of galactose-a and is presumed to be connected in a 1 → 4 manner. The anomeric proton δH 5.18 (H-1c) of glucose-c, displayed a remote correlation with C-2b (δC 79.17) of glucose-b, suggesting a probable 1 → 2 linkage between them. The anomeric proton δH 5.61 (H-1d) of glucose-d is remotely correlated with C-3b (δC 88.90) of glucose-b and is presumed to be connected in a 1 → 3 manner. The anomeric proton δH 5.17 (H-1e) of glucose-e is remotely correlated with the C-3d (δC 87.72) of glucose-d and is presumed to be connected in a 1 → 3 manner. The anomeric proton δH 5.33 (H-1f) of rhamnose-f is remotely correlated with C-4c (δC 78.45) of glucose-c and is presumed to be connected in a 1 → 4 manner. The structure of compound 3 was established as 20β-methoxyl-spirostane-5,25(27)-dien-3-O-β-d-glucopyranosyl(1 → 3)-O-β-d-glucopyranosyl(1 → 3)-O-[α-l-rhamnopyranosyl(1 → 4)-O-β-d-glucopyranosyl(1 → 2)]-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranosid, and it has been designated as macrostemonoside W.
The similarities in physicochemical properties between compound 4 and compound 1 imply that compound 4 might also be a spirostanol-type saponin. The positive ion HR-ESI-MS exhibited an m/z value of 1437.6174 [M + Na]+ (the theoretical value is 1437.6150, Figure S22). Through correlation with its 1H and 13C spectra, the molecular formula of compound 4 was established as C64H102O34. The 1H NMR (600 MHz, Pyridine‑d5, δH 8.74) analysis of compound 4 (Table 1, Figure S23) suggested the presence of four methyl protons in the high-field region with characteristic signals δH 0.83 (3H, s, Me-18), 0.63 (3H, s, Me-19), 1.15 (3H, d, J = 6.96 Hz, Me-21) and 0.87 (3H, s, Rha-6f). In addition, a methoxy signal was suggested at δH 2.96. The 13C NMR (150 MHz, Pyridine‑d5, δC 150.35) analysis of compound 4 (Table 1, Figure S24) indicated the existence of 64 carbon signals, distributed with 28 attributions to the saponin element and 36 to the six sugar molecules (Table S2). We detected characteristic double bond signals at δC 141.42, 122.10, 144.88, and 109.70 in the low-field region. Additionally, we observed four distinct methyl signals in the high-field region at δC 16.83 (Me-18), 12.76 (Me-19), 15.54 (Me-21), and 19.85 (Rha-6f). This observation indicates the probable presence of two double bonds within the compound's structure. Upon comparison, the carbon spectral data of compound 4 is highly consistent with compound 3, except for C-12, and both have the same parent nucleus. Notably, the HMBC (Figure S26) correlation from δH 4.38 (1H, m) to C-12 (δC 79.02) suggests a hydroxyl group attached to C-12. In addition, the hydrogen signal at C-12 (δH 4.38) correlates with the H-17 (α-type, δH 4.55) signal as shown by NOESY (Figure S28). Hence, confirming the hydroxyl group at C-12 as β-type (Kim, et al., 2017). As with compound 3, combined with the 1H–1H COSY spectrum (Figure S27) and the HMQC spectrum (Figure S25), the saponinogenin has a sugar group attached at C-3 and is subjected to glycosidic shifts resulting in a shift of chemical shifts towards the lower field to 81.60 ppm.
The acid hydrolysis test displayed a ratio of 1:1:4 for galactose, rhamnose, and glucose. These sugar molecules were identified as d-galactose, l-rhamnose, and d-glucose by GC–MS analysis after derivatization. The presence of six sugar molecules was inferred from the six anomeric proton signals δH 4.90 (1H, t, J = 7.50 Hz, H-1a), 5.52 (1H, d, J = 7.56 Hz, H-1b), 5.18 (1H, d, J = 7.98 Hz, H-1c), 5.60 (1H, d, J = 7.68 Hz, H-1d), 5.16 (1H, d, J = 7.86 Hz, H-1e) and 5.33 (1H, d, J = 8.04 Hz, H-1f) inferred the presence of six sugar molecules, which was verified by the association of HMQC with the six anomeric carbons at 102.83, 104.47, 105.43, 105.02, 105.22, and 103.17 ppm, respectively. The HMBC spectrum revealed a correlation between the anomeric proton signal at δH 4.90 of galactose-a and the C-3 position (δC 81.60) of the parent nucleus. This confirms the attachment of sugar to the C-3 position of the saponinogenin, aligning with our earlier conclusion. Moreover, the anomeric proton at δH 5.52 (H-1b) of glucose-b showed correlation with C-4a (δC 80.74) of galactose-a, suggesting a 1 → 4 manner. The anomeric proton δH 5.18 (H-1c) of glucose-c is remotely correlated with C-2b of glucose-b (δC 79.16), presumably connected in a 1 → 2 manner. The anomeric proton δH 5.60 (H-1d) of glucose-d is remotely correlated with C-3b (δC 88.90) of glucose-b, with a presumed 1 → 3 connection. The anomeric proton δH 5.16 (H-1e) of glucose-e is remotely correlated with the C-3d (δC 87.75) of glucose-d, which is presumed to be connected in a 1 → 3 manner. The anomeric proton δH 5.33 (H-1f) of rhamnose-f is remotely correlated with C-4c (δC 78.44) of glucose-c, which is presumed to be connected in a 1 → 4 manner. The structure of compound 4 was established as 20β-methoxyl-spirostane-5,25(27)-dien-3β,12β-diol-3-O-β-d-glucopyranosyl(1 → 3)-O-β-d-glucopyranosyl(1 → 3)-O-[α-l-rhamnopyranosyl(1 → 4)-O-β-d-glucopyranosyl(1 → 2)]-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranosid, and it has been designated as macrostemonoside X.
The comparable physicochemical properties between compound 5 and compound 1 imply a possible spirostanol-type saponin structure for compound 5. The positive ion HR-ESI-MS displayed an m/z of 1275.5643 [M + Na]+ (the theoretical value was 1275.5622, Figure S29). Upon analysis of its 1H and 13C spectra, the molecular formula was determined to be C58H92O29. The 1H NMR (600 MHz, Pyridine‑d5, δH 8.74) analysis of compound 5 (Table 1, Figure S30) revealed four methyl protons in the high field region, presenting characteristic signals at δH 0.82 (3H, s, Me-18), 0.63 (3H, d, J = 2.28, Me-19), 1.15 (3H, q, J = 6.96 Hz, Me-21) and 0.87 (3H, s, Rha-6f). In addition, a methoxy signal was suggested at δH 2.95. The 13C NMR (150 MHz, Pyridine‑d5, δC 150.35) analysis of compound 5 (Table 1, Figure S31) indicated the existence of 58 carbon signals, comprising 28 assigned to the saponinogenin and 30 to the five sugar molecules (Table S2). We found the presence of δC 141.40, 122.08, 144.87, and 109.21 double bond characteristic signals in the low-field region, speculating that two double bonds may be present in the structure of the compounds, and suggesting the presence of four methyl signals in the high-field region δC 16.81 (Me-18), 12.73 (Me-19), 15.51 (Me-21), and 19.82 (Rha-6f). Interestingly, the carbon spectral data of compounds 5 and 4 were highly overlapping, differing only in the number of glycosyl groups attached, with the saponinogenin being identical. A correlation between the hydrogen signal at the C-12 position (δH 4.00) and the H-17 (α-type, δH 4.44) signal is evident from NOESY (Figure S35), which establishes that the hydroxyl group at the C-12 position is also of β-type (Kim, et al., 2017).
The acid hydrolysis test displayed a ratio of 1:1:3 for galactose, rhamnose, and glucose. These sugar molecules were identified as d-galactose, l-rhamnose, and d-glucose by GC–MS analysis after derivatization. Based on the detection of five distinct anomeric proton signals with chemical shifts δH 4.91 (1H, t, J = 7.50 Hz, H-1a), 5.62 (1H, d, J = 7.80 Hz, H-1b), 5.19 (1H, dd, J = 3.78, 7.86 Hz, H-1c), 5.12 (1H, d, J = 11.10 Hz, H-1d) and 5.34 (1H, d, J = 7.80 Hz, H-1f), it can be inferred that five distinct sugar molecules are present. This inference was corroborated by the HMQC (Figure S32) correlation with the respective five anomeric carbons at 102.82, 105.03, 105.60, 105.41, and 103.16 ppm. Based on the HMBC analysis (Figure S33), the remote correlation observed between the anomeric proton signal, δH 4.91 of galactose-a, and the parent nucleus, C-3 (δC 81.54), indicated the attachment of the sugar to the C-3 position of the saponinogenin. This shift in the chemical shift value of C-3 towards a lower field was attributed to the glycosidic shift phenomenon. The anomeric proton δH 5.62 (H-1b) of glucose-b is remotely correlated with C-4a (δC 80.72) of galactose-a and is presumed to be connected in a 1 → 4 manner. The anomeric proton δH 5.19 (H-1c) of glucose-c, displayed a remote correlation with C-2b (δC 79.17) of glucose-b, suggesting a probable 1 → 2 linkage between them. The anomeric proton δH 5.12 (H-1d) of glucose-d is remotely correlated with C-3b (δC 88.97) of glucose-b and is presumed to be connected in a 1 → 3 manner. The anomeric proton δH 5.33 (H-1f) of rhamnose-f is remotely correlated with C-4c (δC 78.54) of glucose-c and is presumed to be connected in a 1 → 4 manner. The structure of compound 5 was established as 20β-methoxyl-spirostane-5,25(27)-dien-3β,12β-diol-3-O-β-d-glucopyranosyl(1 → 3)-O-[α-l-rhamnopyranosyl(1 → 4)-O-β-d-glucopyranosyl(1 → 2)]-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranosid, and it has been designated as macrostemonoside Y.
Moreover, the structural elucidation of macrostemonoside A (6) (Peng, et al., 1992), dongnoside E (7) (Ding, et al., 1989), tigogenin-3-O-β-d-glucopyranosyl(1 → 4)-O-β-d-galactopyranoside (8) (Zhang, et al., 2006), and odospiroside (9) (Janeczko, et al., 1987) (the carbon spectral data are presented in Table S3) was conducted by cross-referencing their spectroscopic data with previously reported literature. Notably, compound 7, identified for the first time in this plant.
3.2. Antioxidant activities
The in vitro antioxidant activities of compounds 1–9 were assessed using VC as a positive control. These evaluations included assays for scavenging capacity against DPPH and ABTS radicals, as well as reducing capacity against Fe3+. As indicated in Table 2, all compounds displayed robust scavenging capabilities against ABTS radicals, suggesting a potentially associated scavenging mechanism. Particularly noteworthy was compound 1, demonstrating the strongest activity in scavenging both radicals and reducing Fe3+, possibly attributed to the presence of alkene bonds and acetyl groups within its structure. Additionally, compounds 2–6 also exhibited relatively strong in vitro antioxidant activity.
Table 2.
The scavenging ability of compounds 1–9 for DPPH and ABTS radicals and their reduction of Fe3+.
| Compounds | IC50 |
||
|---|---|---|---|
| DPPH | ABTS | Fe3+ | |
| 1 (mg/mL) | 0.71 ± 0.09 | 0.03 ± 0.02 | 1.17 ± 0.23 |
| 2 (mg/mL) | 0.88 ± 0.11 | 0.05 ± 0.02 | 1.27 ± 0.27 |
| 3 (mg/mL) | 0.85 ± 0.12 | 0.04 ± 0.02 | 1.27 ± 0.14 |
| 4 (mg/mL) | 0.78 ± 0.08 | 0.04 ± 0.01 | 1.21 ± 0.17 |
| 5 (mg/mL) | 0.81 ± 0.09 | 0.04 ± 0.01 | 1.25 ± 0.21 |
| 6 (mg/mL) | 0.74 ± 0.21 | 0.04 ± 0.02 | 1.28 ± 0.22 |
| 7 (mg/mL) | 1.17 ± 0.12 | 0.26 ± 0.07 | 1.77 ± 0.37 |
| 8 (mg/mL) | 1.55 ± 0.21 | 0.21 ± 0.08 | 1.87 ± 0.24 |
| 9 (mg/mL) | 1.20 ± 0.13 | 0.17 ± 0.04 | 1.78 ± 0.21 |
| VC (μg/mL) | 1.94 ± 0.11 | 1.23 ± 0.14 | 85.45 ± 4.65 |
IC50 values of compounds 1–9 are in mg/mL, and IC50 values of VC are in μg/mL. IC50 values are expressed as means ± SD (n = 3).
3.3. Establishment of HPLC-ELSD profiles of chemical constituents in AMB and quantitative analysis of nine steroidal saponins
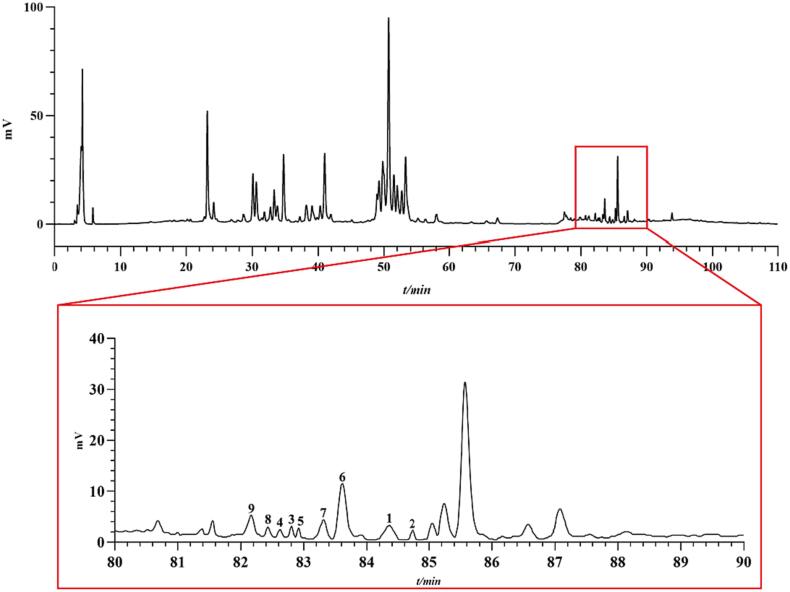
The HPLC-ELSD spectra of the chemical components in AMB are shown in Fig. 3, with good separation between each component. Mixed standard solutions were also used in the analysis (Fig. 4), which can help us to quantify the nine steroidal saponins more smoothly. The key parameters of the analytical method, such as standard curve equation, quantitative concentration range, LOD, LOQ, precision, reproducibility, stability, spiked recoveries, and retention time (tR) were listed in Table S4. Among them, the RSD values of precision, reproducibility, stability and spiked recoveries were less than 2.0 %, and the recoveries for compounds 1–9 were 99.94 %, 103.21 %, 101.69 %, 100.56 %, 97.80 %, 97.19 %, 101.63 %, 98.45 % and 98.21 %, respectively, indicating that the method is accurate and reliable and has the conditions for quantitative analysis. The contents of the nine steroidal saponins in AMB of different origins and processing methods are listed in Table 3. In all samples, the total amount of the nine steroidal saponins ranged from 352.80 to 1759.80 μg/g. It is noteworthy that, among the nine saponins, the contents of compounds 1, 6, 8, and 9 were generally higher in all batches of AMB, whereas the contents of compounds 3 and 4 were rarely or even undetectable. Among the different origins, the total amount of the nine saponins was in the order of Zigong, Sichuan > Changchun, Jilin > Benxi, Liaoning > Luoyang, Henan. Interestingly, compounds 3 and 4 were not detected in the AMB from Zigong, Sichuan, and it was hypothesized that they might be related to the geographical environment of growth. Meanwhile, among the different processing methods, the total amount of nine saponins was higher in the directly dried and lyophilized AMB, but compounds 3 and 4 were not detected in the lyophilized AMB, and it was hypothesized that the production of the two was related to whether or not they were heated during processing. To the best of our knowledge, this is the first time that saponins have been quantitatively analyzed in AMB of different origins and processing methods.
Fig. 3.
The HPLC-ELSD spectra of AMB and its local amplification.
Fig. 4.
The HPLC-ELSD spectra of mixed standards of 9 steroidal saponins.
Table 3.
Content of 9 steroidal saponins in commercial AMB of different origins and processing methods (μg/g of herbs).
| AMB | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| M1 | 77.98 ± 1.37 | 61.1 ± 0.64 | 56.92 ± 0.81 | 64.62 ± 1.50 | 56.83 ± 0.79 | 66.33 ± 1.07 | 50.80 ± 0.64 | 65.68 ± 0.99 | 63.29 ± 1.06 | 563.53 ± 5.69 |
| M2 | 98.59 ± 0.49 | 79.67 ± 0.51 | 56.06 ± 1.39 | 65.66 ± 1.57 | 57.78 ± 1.10 | 168.46 ± 1.48 | 71.55 ± 0.73 | 65.87 ± 2.58 | 216.92 ± 1.09 | 880.56 ± 6.46 |
| M3 | 198.86 ± 1.21 | 172.14 ± 1.47 | – | – | 151.25 ± 1.11 | 95.77 ± 0.61 | 72.12 ± 1.35 | 101.86 ± 1.73 | 122.36 ± 1.61 | 914.36 ± 3.11 |
| M4 | 84.27 ± 1.05 | 61.16 ± 0.48 | 56.88 ± 1.32 | 65.79 ± 2.65 | 58.25 ± 1.35 | 89.38 ± 1.03 | 58.10 ± 1.74 | 63.19 ± 1.70 | 73.56 ± 1.05 | 610.58 ± 7.04 |
| M5 | 98.09 ± 1.20 | 80.11 ± 1.49 | 64.01 ± 1.20 | 70.86 ± 0.71 | 64.43 ± 1.08 | 180.09 ± 1.50 | 71.33 ± 0.92 | 69.09 ± 0.70 | 221.35 ± 1.76 | 919.36 ± 3.34 |
| M6 | 191.63 ± 1.18 | 181.64 ± 1.12 | – | – | 109.68 ± 0.54 | 96.92 ± 0.31 | 64.00 ± 1.29 | 97.71 ± 1.08 | 79.34 ± 1.85 | 820.92 ± 3.12 |
| M7 | 92.24 ± 1.16 | 66.73 ± 1.03 | – | – | – | 75.26 ± 1.39 | 52.18 ± 1.56 | – | 66.41 ± 1.61 | 352.80 ± 4.29 |
| M8 | 97.51 ± 1.13 | 82.48 ± 1.37 | 57.88 ± 0.63 | 66.49 ± 0.89 | 59.09 ± 1.57 | 162.42 ± 2.02 | 67.59 ± 1.19 | 62.22 ± 1.35 | 210.14 ± 1.32 | 865.81 ± 6.35 |
| M9 | 79.35 ± 0.56 | 64.13 ± 1.23 | – | – | 83.58 ± 1.09 | 76.65 ± 1.89 | 71.45 ± 0.89 | 88.00 ± 1.84 | 65.69 ± 1.88 | 528.84 ± 2.21 |
| M10 | 135.57 ± 1.31 | – | – | – | – | 72.07 ± 1.62 | 76.34 ± 1.44 | 163.48 ± 2.59 | 60.99 ± 2.73 | 508.46 ± 7.84 |
| M11 | 102.24 ± 1.05 | 155.86 ± 1.62 | – | – | 212.46 ± 1.59 | 154.23 ± 1.68 | 201.62 ± 1.00 | 781.27 ± 1.82 | 152.12 ± 0.68 | 1759.80 ± 3.58 |
| M12 | 116.00 ± 1.77 | 65.31 ± 1.20 | – | – | 108.31 ± 1.75 | 63.67 ± 0.99 | 50.54 ± 0.47 | 178.41 ± 0.95 | 110.39 ± 2.45 | 692.62 ± 3.23 |
Values expressed as mean ± SD (n = 3).
4. Conclusion
In conclusion, nine steroidal saponins (compounds 1–9) were isolated from AMB. Among these, compounds 1–5 marked the initial discovery, while compound 7 was newly isolated from AMB. These saponins underwent screening through in vitro antioxidant assays and served as standards to establish HPLC-ELSD profiles of AMB. The results show that compounds 1, 4, and 6 exhibited potent in vitro antioxidant activity with IC50 values for scavenging DPPH and ABTS radicals at 0.71 ± 0.09 mg/mL, 0.03 ± 0.02 mg/mL; 0.78 ± 0.08 mg/mL, 0.04 ± 0.01 mg/mL; and 0.74 ± 0.21 mg/mL, 0.04 ± 0.02 mg/mL, respectively. For Fe3+ reducing activity, their respective IC50 values were 1.17 ± 0.23 mg/mL, 1.21 ± 0.17 mg/mL, and 1.28 ± 0.22 mg/mL. These outcomes emphasize their potential as promising candidates for natural antioxidant development. In addition, the content determination results showed that compounds 1, 6, 8 and 9 were higher in AMB, whereas the content of compounds 3 and 4 may be affected by geography and processing methods. AMB, a traditional and cost-effective food and medicinal plant, warrants further exploration of these saponins' antioxidant potential for enhanced contributions to human health.
CRediT authorship contribution statement
Jianfa Wu: Writing – original draft, Methodology, Investigation. Ying Cui: Supervision, Resources, Funding acquisition, Conceptualization. Chang Liu: Investigation. Weixing Ding: Investigation. Shen Ren: Investigation. Jing Zhang: Supervision, Resources, Funding acquisition, Conceptualization. Lulu Wang: Validation, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was funded by the National Key Research and Development Program of China (2019YFE0116800), the Jilin Science and Technology Development Program Project (20240404007YY), and the Jilin Agricultural University Graduate Teaching Case Library Construction Project (2021).
I would also like to thank my love, Ms. Ying Cui, for her companionship and help in the experiment.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101144.
Contributor Information
Jing Zhang, Email: zhangjing4693@jlau.edu.cn.
Lulu Wang, Email: relulu@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Allium macrostemon Bunge from different origins and methods of processing (Table S1); 1H-NMR Data of sugar moieties of compounds 1-5 (δ in ppm) (Table S2); 13C-NMR Data of compounds 6-9 (δ in ppm) (Table S3); Methodological investigations and retention times for compounds 1-9 (Table S4); The HR-ESI-MS, 1H-NMR, 13C-NMR and 2D-NMR spectrum for compounds 1-5 (Figure S1-S35).
Data availability
The data that has been used is confidential.
References
- Cheng S.B. Zhejiang University; Zhejiang, China: 2013. Study on steroidal from the bulbs of Allium macrostemon. Masters Thesis. [Google Scholar]
- Chinese Pharmacopoeia Commission . China Medical Science Press; 2020. Pharmacopoeia of People’s Republic of China (Part 1) pp. 392–393. [Google Scholar]
- Deng K., Feng H., Wang Z.P., Wang C.S. Study on the effect and mechanism of Allium saponins on platelet aggregation rate in patients with coronary heart disease with cold phlegm blockade syndrome. Journal of Basic Chinese Medicine. 2022;25(6):783–786. doi: 10.19945/j.cnki.issn.1006-3250.2019.06.023. [DOI] [Google Scholar]
- Ding Y., Chen Y.Y., Wang D.Z., Yang C.R. Steroidal saponins from a cultivated form of Agave sisalana. Phytochemistry. 1989;28(10):2787–2791. doi: 10.1016/S0031-9422(00)98089-0. [DOI] [Google Scholar]
- Janeczko Z., Jansson P.E., Sendra J. A new stereoidal saponin from Polygonatum officinale. Planta Medica. 1987;53(1):52–54. doi: 10.1055/s-2006-962618. [DOI] [PubMed] [Google Scholar]
- Kim Y.S., Suh W.S., Park K.J., Choi S.U., Lee K.R. Allimacrosides A-E, new steroidal glycosides from Allium macrostemon Bunge. Steroids. 2017;118:41–46. doi: 10.1016/j.steroids.2016.12.002. [DOI] [PubMed] [Google Scholar]
- Maxwell S.R., Lip G.Y. Free radicals and antioxidants in cardiovascular disease. British Journal of Clinical Pharmacology. 1997;44(4):307–317. doi: 10.1046/j.1365-2125.1997.t01-1-00594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J.P., Wu Y., Yao X.S., Okuyama T., Narui T. Two new steroidal saponins from Allium macrostemon. Acta Pharmaceutica Sinica. 1992;27(12):918–922. doi: 10.16438/j.0513-4870.1992.12.008. [DOI] [PubMed] [Google Scholar]
- Sharma O.P., Bhat T.K. DPPH antioxidant assay revisited. Food Chemistry. 2009;113(4):1202–1205. doi: 10.1016/j.foodchem.2008.08.008. [DOI] [Google Scholar]
- Sinan K.I., Mahomoodally M.F., Eyupoglu O.E., Etienne O.K., Sadeer N.B., Ak G., Zengin G. HPLC-FRAP methodology and biological activities of different stem bark extracts of Cajanus cajan (L.) Millsp. Journal of Pharmaceutical and Biomedical Analysis. 2021;192 doi: 10.1016/j.jpba.2020.113678. [DOI] [PubMed] [Google Scholar]
- Wang J.L., Liu K., Li X.X., Bi K.L., Zhang Y.M., Huang J.J., Zhang R.R. Variation of active constituents and antioxidant activity in Scabiosa tschiliensis Grunning from different stages. Journal of Food Science and Technology. 2017;54(8):2288–2295. doi: 10.1007/s13197-017-2666-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Wang L.L., Zhang M.L., Guo Y.D., Zhang J., Ma G.X. Five new spirosterol saponins from Allii Macrostemonis Bulbus. Chinese Journal of Natural Medicines. 2023;21(3):226–232. doi: 10.1016/S1875-5364(23)60423-6. [DOI] [PubMed] [Google Scholar]
- Wu J.F., Li L., Liu C., Li C.Y., Cui Y., Ding W.X., Shi L.L. Two new compounds from Allii Macrostemonis Bulbus and their in vitro antioxidant activities. Molecules. 2023;28(17) doi: 10.3390/molecules28176176. Article 6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.F., Wang L.L., Cui Y., Liu F., Zhang J. Allii Macrostemonis Bulbus: A comprehensive review of ethnopharmacology, phytochemistry and pharmacology. Molecules. 2023;28 doi: 10.3390/molecules28062485. Article 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.R. Jilin Unversity; Changchun, China: 2021. Study on chemical constituents and biological activity of Allium chinense G. Don. Masters Thesis. [Google Scholar]
- Yao Z.H., Qin Z.F., Dai Y., Yao X.S. Phytochemistry and pharmacology of Allii Macrostemonis Bulbus, a traditional Chinese medicine. Chinese Journal of Natural Medicines. 2016;14(7):481–498. doi: 10.1016/S1875-5364(16)30058-9. [DOI] [PubMed] [Google Scholar]
- Zhang J.D., Xu Z., Cao Y.B., Chen H.F., Yan L., An M.M., Jiang Y.Y. Antifungal activities and action mechanisms of compounds from Tribulus terrestris L. Journal of Ethnopharmacology. 2006;103(1):76–84. doi: 10.1016/j.jep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Zhao S.T., Liu L., Qin M.M., Xie Y.F., Yu J. Progress in the study of saponin species in Allii Macrostemonis Bulbus and their pharmacological effects. Chinese Traditional Patent Medicine. 2022;44(11):3596–3603. doi: 10.3369/j.issn.1001-1528.2022.11.033. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Allium macrostemon Bunge from different origins and methods of processing (Table S1); 1H-NMR Data of sugar moieties of compounds 1-5 (δ in ppm) (Table S2); 13C-NMR Data of compounds 6-9 (δ in ppm) (Table S3); Methodological investigations and retention times for compounds 1-9 (Table S4); The HR-ESI-MS, 1H-NMR, 13C-NMR and 2D-NMR spectrum for compounds 1-5 (Figure S1-S35).
Data Availability Statement
The data that has been used is confidential.