Highlights
-
•
Novel techniques are encouraged to identify nontuberculosis mycobacteria (NTM).
-
•
An algorithm incorporating direct molecular identification of NTM is suggested.
-
•
Rapid NTM identification can assist in decision-making and prompt treatment initiation.
Keywords: Nontuberculous mycobacterium, Mycobacteria-other-than-tuberculosis, Molecular technique, Guideline, Pulmonary, Culture
Abstract
Nontuberculous mycobacteria (NTM) are a group of acid-fast mycobacteria other than Mycobacterium tuberculosis complex (MTBC) that cause pulmonary disease that is similar to the disease caused by MTBC. International guidelines for the diagnosis of pulmonary NTM disease are rigid and have remained unchanged for nearly 2 decades. In this opinion piece, we provide a new perspective on the traditional criteria by suggesting a diagnostic algorithm that incorporates direct molecular identification of NTM performed on raw sputum specimens (using Sanger or targeted deep sequencing approaches, among others) paired with traditional culture methods. Our approach ensures a more rapid diagnosis of pulmonary NTM disease, thus, facilitating timeous clinical diagnosis, and prompt treatment initiation, where indicated, and leverages recent advances in novel molecular techniques into routine NTM identification practice.
Introduction
Nontuberculous mycobacteria (NTM) are a collective group of more than 190 species and subspecies other than Mycobacterium leprae or Mycobacterium tuberculosis complex (MTBC) [1]. Globally, NTM infections are steadily increasing due to increased immune suppression in patients and aging populations with increased diagnosis being driven by improved diagnostic techniques and heightened awareness of NTM infection [2].
International guidelines on diagnosing pulmonary NTM infections in adults, jointly prepared by the American Thoracic Society, the European Respiratory Society, the European Society of Clinical Microbiology and Infectious Diseases, and the Infectious Diseases Society of America were updated in 2020 [1]. However, the diagnostic criteria for pulmonary infection remain largely unchanged from the previous iteration of the guidelines in 2017 and include the necessity of having two independent sputum NTM cultures (indicating the same species on culture, collected ≥1 week apart) to infer pulmonary infection. This rigid requirement aims to ensure the distinction between potential colonization or contamination of the airway from true pathogenic pulmonary NTM disease. Thus, although the guidelines have been updated regularly, the ancillary use of novel molecular diagnostics to directly identify NTM species from a sputum sample has not been incorporated.
This commentary discusses some of the advantages and obstacles of incorporating the use of molecular tools when performed directly on sputum specimens. We also propose the integration of this strategy as part of the clinical diagnostic algorithm paired with an independent culture result. In addition, a review of recent molecular advances in achieving direct sputum mycobacteria identification is also presented to provide context and future direction.
Current state-of-the-art molecular diagnostics for nontuberculous mycobacteria identification
The direct identification of NTM from clinical samples shows promising results from smear-positive samples on various platforms. The reverse blot hybridization assay (REBA) Myco-ID (YD Diagnostics, Yongin, South Korea) was able to detect 20 different NTM species from a respiratory specimen that is smear-positive with acid-fast bacilli (AFB). The test had a laboratory concordance of 98.6% with culture identification of NTM species and was able to differentiate them from MTBC [3]. More recently, the REBA Myco-ID identified 436 NTM isolates among 696 clinical patients from a cohort of individuals with mycobacterium pulmonary disease. Notably, this included instances of mixed infections, with 32 specimens demonstrating coexistence of M. intracellulare and M. abscessus, and 10 cases showing mixed infection of M. avium and M. abscessus [4]. Moreover, the positive predictive value (PPV) of REBA Myco-ID was found to be statistically significantly higher (P = 0.004) compared to Mycobacteria Growth Indicator Tube TBc identification (MGIT 960-TBc ID) [4]. Additionally, when combined with HybREAD480, an instrument for automating the post-polymerase chain reaction (PCR) step, REBA Myco-ID exhibited an accuracy rate of 94.3% (181/192; 95% confidence interval [CI]: 90.0-97.1%) across 192 clinical samples, as compared to multigene sequencing-based typing [5]. A multi-center study evaluated a commercial biochip assay (Capital Bio, Beijing, China) based on 16S ribosomal ribonucleic acid (rRNA) gene sequences that can identify 17 different NTM species within 6 hours from smear-positive sputum specimens showed 98.8% sensitivity (95% CI: 96.6-99.6) and 100% specificity (95% CI: 99.7-100) when compared to sequencing the traditional microbiology culture [6]. The Anyplex Plus Mycobacterium tuberculosis (MTB)/nontuberculous mycobacteria (NTM) multi-drug resistant-TB Assay (Seegene Inc, Seoul, South Korea) is a real-time multiplex PCR platform that detected 76.7% (19/26) of NTM in sputum specimens and 100% (4/4) from extra-pulmonary sites that were subsequently found to be culture-positive [7]. Sawatpanich et al. [8] conducted a study assessing the efficacy of Anyplex MTB/NTM in the detection of NTM. The results revealed that the assay successfully identified NTM nucleic acids in 44.9% (124/276) of culture-independent specimens that tested positive for NTM culture. Remarkably, it also exhibited a positive yield in 2.3% (214/9299) of specimens deemed NTM culture-negative. For pulmonary specimens, the assay demonstrated a sensitivity of 42.6%, specificity of 96.6%, a PPV of 36.3%, a negative predictive value (NPV) of 97.4%, and an overall percent agreement of 94.3% with culture-based GenoType Mycobacterium Common mycobacteria (CM)/Additional species (AS) line probe assay. Conversely, in the case of extra-pulmonary specimens, the assay exhibited a sensitivity of 56.5%, specificity of 99%, 38.2% PPV, 99.5% NPV, and an overall percent agreement of 98.5% with GenoType Mycobacterium CM/AS line probe assay performed from cultures. These findings underscore the diagnostic potential and discriminatory capabilities of Anyplex MTB/NTM in the context of NTM detection across diverse specimen types [8]. The reverse hybridization assay GenoType Mycobacteria Direct (GTMD; Hain Lifescience GmbH, Nehren, Germany) that detects MTBC and four NTM species (M. avium, M. intracellulare, M. kansasii, and M. malmoense) was evaluated among 1570 specimens that were sputum smear-negative [9]. The GTMD method could only detect 4/10 NTM species (0/1 M. kansasii, 1/1 M. avium, 3/8 M. intracellulare) compared to culture from smear-negative pulmonary samples [9]. Another two deoxyribonucleic acid (DNA)/DNA hybridization molecular assays recently tested to identify NTM directly from sputum were the GenoType CM direct (GTCMd) (Bruker, Billerica, MA, USA) and Vision Array Myco (VAM) (ZytoVision, Bremerhaven, Germany). GTCMd and VAM assays identified 24/27 (88.9%) and 19/27 (70.3%) of NTMs accurately to species level compared to culture [10]. Importantly, GTCMd could identify Mycobacterium species among 3/3 smear-negative sputum samples for AFB, but VAM failed to identify the organism in any of the three samples.
Emerging molecular diagnostics to identify nontuberculous mycobacteria
In a recent multi-center study involving 15 laboratories across nine European countries, a comprehensive evaluation of 1330 NTM Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) identifications was conducted. The study revealed an identification accuracy of 100% at the species level in eight participating centers when MALDI-TOF MS was applied to cultures cultivated on Löwenstein-Jensen media (Becton Dickinson, Franklin Lakes). These findings highlight the efficacy and reliability of MALDI-TOF MS in providing precise NTM identifications, particularly when applied to cultures [11]. However, the same level of precision for NTM identification has not been reported when MALDI-TOF MS is directly applied to clinical samples.
Rapid identification of NTM species from respiratory specimens was explored using nucleotide MALDI-TOF MS, which detects any NTM species based on the presence of specific single nucleotide polymorphisms. In the study that involved 108 patients diagnosed with NTM pulmonary disease and 67 control patients with other respiratory diseases, the nucleotide MALDI-TOF MS exhibited the following diagnostic performance metrics for the detection of NTM: sensitivity 77.8% (95% CI: 68.6-85.0%), specificity 92.5% (95% CI: 82.8-97.2%), PPV 94.4% (95% CI: 86.8-97.9%), NPV 72.1% (95% CI: 61.2-81.0%), and accuracy 83.4% (95% CI: 76.9-88.5%), based on the reference clinical diagnostic standard for NTM pulmonary disease [12].
Although advances in protein extraction methods and database expansion to increase the number of recognized NTM patterns hold potential promise for the future [13,14], a concern remains that sputum samples have a protein composition closely related to the lungs [15] thereby potentially masking NTM specific protein peak profiles. Hence, definitive identification of NTM using MALDI-TOF MS is anticipated to require a pure culture.
Direct sequencing of specific genes for NTM was identified in cystic fibrosis respiratory samples with culture-independent 16S rRNA sequencing. Targeted sequencing was used to sequence the V4 region of the 16S rRNA gene on bronchoalveolar and NTM culture-positive sputum [16]. Sequencing raw sputum detected 8/15 (53%) NTMs among 15 culture-positive (11 sputa and four bronchoalveolar lavages) samples (four M. abscessus complex and M. avium complex, respectively) [16]. Although 16S rRNA is a typical housekeeping gene used in molecular techniques to identify bacteria, it shows poor discrimination between NTM species. This is mainly due to inter-species sequence similarity and some NTM species having identical sequence homology. Therefore, it is not uncommon for certain NTMs to have 0% interspecies divergence, rendering 16S rRNA sequencing a less suitable form for NTM speciation [17]. On the other hand, sequencing of the rpoB and hsp65 genes allows for fast and reliable results that produce robust phylogenetic trees to speciate NTMs [17]. Sequencing these regions allows for an additional spectrum of less frequently isolated NTMs to be identified and not included in commercial kits. The benefit of using these genes for detecting NTMs over traditional diagnostic culture-based methods is highlighted in a study performed on respiratory specimens in which quantitative PCR of the hsp65 gene identified three NTMs (two M. avium and one M. flavescens) reported as culture-negative [18]. A recent approach employed a combination of loop-mediated isothermal amplification and lateral flow biosensors, specifically targeting the rpoB gene of M. kansassi. This innovative method enabled the detection of NTM at low concentrations, detecting as little as 1 femtogram/microliter (fg/μl) of DNA and 50 colony-forming units (CFU) of bacilli in sputum, under a constant amplification temperature [19].
There is a growing trend toward the adoption of novel multiplex assays designed to identify NTM/MTB co-infections. Sarro et al. [20] developed and optimized a platform specifically tailored for the detection of Mycobacterium avium complex (MAC). This platform targeted S1311/DT1 for MAC and IS6110/SenX3-RegX3 for MTBC. The MAC component of the assay exhibited a detection limit of five CFU in artificially spiked sputum samples. The diagnostic performance of the Multiplex MTB/NTM assay, overall, was found to be inferior to sputum culture results. However, it demonstrated comparability to the GeneXpert Mycobacterium tuberculosis (MTB)/Rifampicin (RIF) assay across various patient groups, including the TB culture-negative control group, TB/HIV positive group, individuals with treatment failure, and TB-positive individuals naive to treatment. The GeneXpert MTB/RIF assay exhibited higher sensitivity, while the Multiplex MTB/NTM assay demonstrated superior specificity across all evaluated patient cohorts [20].
Whole-genome sequencing (WGS) and next-generation sequencing (NGS) metagenomics [21,22] represent a promising frontier for characterizing new NTM species, discovering therapeutic targets, and identifying gene mutations that encode antibiotic resistance and pathogenic virulence factors [23]. In a comprehensive cohort study comprising 422 patients suspected of nontuberculous mycobacterial pulmonary disease, Wei et al. [24] demonstrated that metagenomic NGS (mNGS) exhibited a diagnostic sensitivity of 81.4%. This surpassed the 53.6% sensitivity observed with MGIT 960. Conversely, the specificity of mNGS in diagnosing nontuberculous mycobacterial pulmonary disease was 97.8%, closely aligning with the specificity of MGIT 960 at 100%. The integration of mNGS with MGIT 960 resulted in a heightened diagnostic sensitivity of 91.8%. The sensitivities for pulmonary puncture tissue fluid, bronchoalveolar lavage fluid, and sputum were determined to be 80.6%, 84.8%, and 77.5%, respectively. All of these values surpassed the corresponding sensitivities of MGIT 960 (P <0.05) [24].
While WGS has been useful in identifying novel NTM species from cultures, its application with direct sputum specimens is limited. He et al. [25] showed that MTBC and NTM targeted capture sequencing, directly on 14 sputum and 16 bronchial alveolar lavage fluid samples, displayed a sensitivity of 91.3% (CI: 70.5-98.5), specificity of 42.9% (CI: 11.8-79.8), and accuracy 83.3% (CI: 82.3-84.3), compared to BD BACTECTM MGITTM 960 (Becton Dickinson Microbiology System, Sparks, NV, USA) cultures used as the gold standard for comparison. In a separate experiment, NGS directly on sputum provided a complete drug-resistant profile faster than MGIT cultures in M. tuberculosis [26]. Interestingly, direct WGS from sputum samples revealed more within sample diversity than sequencing from the cultured M. tuberculosis isolate [27]. Recognizing genetic diversity enhances the detection of resistant variants at low frequencies [27]. Therefore, sequencing success among the MTBC directly from sputum should justify this novel diagnostic tool to identify NTMs in the same way, but further investigation is needed in this field [28]. Whether detecting these underlying populations will lead to outcome benefits or confuse clinicians further by restricting antibiotic choice still needs clarification.
Potential benefits, limitations, and recommendations of incorporating the proposed diagnostic algorithm in routine clinical practice
The main benefit of the proposed algorithm is to shorten the time to establish the diagnosis of NTM pulmonary disease, thus, enabling prompt treatment initiation and limiting pre-treatment loss to follow-up (PTLTFU). In our setting, the rates of PTLTFU are likely substantial since a second sputum sample is only sent to the laboratory for testing in ∼30% of patients who had previously grown an NTM on sputum culture (unpublished data from the National Health Laboratory Service [NHLS], TB Laboratory, Western Cape, South Africa, 2020). It is likely that a firm diagnosis of NTM pulmonary disease (in accordance with current criteria) is achieved in even fewer patients since there is further attrition attributed to the long culture time of the second sputum sample. NHLS data from the Western Cape, South Africa, on NTM cultures between 2016-2022 indicated 1389 isolates with an average processing time of 23 days (range 3-45) from sample registration to entering the final result for clinician review. Direct molecular testing can reduce turnaround times to a few days. As is the case with TB, the reasons for PTLTFU in patients with NTM pulmonary disease are also likely complex, however, having a rapid diagnostic test (such as GeneXpert MTB/RIF Ultra) significantly reduces PTLTFU and time-to-treatment initiation [29,30]. We are expecting the same to hold true for NTM where a rapid result from molecular testing of smear-positive sputum (second sample) may reduce time-to-treatment initiation and prevent PTLTFU. However, this needs to be confirmed in future prospective studies since there is no controlled data confirming the impact of rapid molecular tests on patient-important outcomes.
We recommend the utilization of the Sanger sequencing method for the identification of targeted genes such as rpoB or hsp65 as a rapid and dependable means of discerning NTM [31]. This molecular platform exhibits a track record characterized by reliability and accuracy [32]. It boasts a minimal error rate (0.001%) [33], rendering it particularly well-suited for the sequencing of relatively lengthy fragments, typically spanning 800 base pairs. Furthermore, its cost-effectiveness, especially in the context of smaller-scale sequencing initiatives, is a noteworthy advantage [34]. It demands no extensive laboratory infrastructure or specialized expertise, as conventional PCR can be employed to obtain the requisite sequences. In addition to its practical attributes, Sanger sequencing obviates the need for advanced bioinformatics proficiency, as readily accessible alignment tools such as Basic Local Alignment Search Tool (BLAST) are freely available for the analysis of sequencing results [35]. This accessibility simplifies the data analysis process. Importantly, Sanger sequencing has demonstrated its efficacy when applied directly to sputum samples, further underscoring its practical utility in the rapid identification of NTM [36]. These collective advantages render Sanger sequencing a compelling choice for diagnosticians seeking a rapid, reliable, and accessible method for the molecular identification of NTM species, especially in low-resource settings.
A cutting-edge platform with prospective implications is the targeted, amplicon-based, deep sequencing modality known as Deeplex (GenoScreen, Lille, France). This molecular testing technique has demonstrated expeditious results and genetic characterization directly from sputum specimens for MTB, earning an endorsement from the World Health Organization [37], [38], [39]. Its utilization is particularly recognized for its efficacy in detecting drug-resistant tuberculosis and serving as a surveillance tool. This assay can identify more than 100 different mycobacterial species [40]. The current impediment to its widespread implementation lies in cost considerations, particularly in low to middle-income countries. Nevertheless, a broad-scale adoption of Deeplex holds the potential to ameliorate operational expenses. In addition, DeeplexR®Myc-TB exhibits the capability to discern co-infections between NTM and MTBC. Anticipating future developments, dedicated versions of Deeplex designed for NTM susceptibility testing hold promise in alleviating the reliance on cultures for the execution of drug susceptibility testing.
We also acknowledge that this proposed algorithm has several limitations: Firstly, given the high cost associated with some molecular assays and their poor performance in low burden (smear-negative) disease, we propose evaluating only smear-positive specimens with rapid molecular tools. This could limit its usefulness; however, it may be argued that patients that would most likely benefit from such an algorithm would be those with high burden diseases who are at increased risk for poor treatment outcomes. Secondly, the algorithm may over-diagnose NTM pulmonary disease. This would subject patients to unnecessary toxic treatment and may promote resistance. However, this limitation is mitigated by pairing the molecular diagnostic test, which has high specificity [18], to a previous culture, and confirming concordance of the results. This strategy, when applied in conjunction with the appropriate clinical context, should limit over-treatment. Thirdly, unlike culture, direct molecular testing of sputum, cannot predict sputum culture conversion to negative and does not allow for quantitative culture evaluations that can assist in assessing the severity of the disease, monitor the course of treatment, and differentiate chromogenic from nonchromogenic mycobacteria that is not possible with direct molecular testing [41], [42], [43]. Lastly, due to the variable treatment response of NTM pulmonary disease, it is possible that achieving an earlier diagnosis may not guarantee a favorable treatment response. Thus, we propose evaluating the impact of this algorithm in a larger cohort as part of operational research to validate this strategy. The algorithm proposes no changes to bronchoalveolar lavage specimen, or any other specimen obtained from usually sterile sites.
In summary, we recommend an algorithm for diagnosing NTM pulmonary disease that incorporates direct molecular testing of smear-positive sputum samples from patients who already have a single culture-confirmed NTM result (Figure 1) instead of the mandatory requirement of having two independent cultures confirming NTM pulmonary disease as per the current international guidelines. Although the application of novel technology looks promising for direct molecular identification of NTM from sputum, before it can be widely embraced, more clinical application studies are needed to develop and integrate this algorithm into routine practice. Such an approach will ensure optimal resource allocation in systems with proven clinical benefits and favorable patient outcomes.
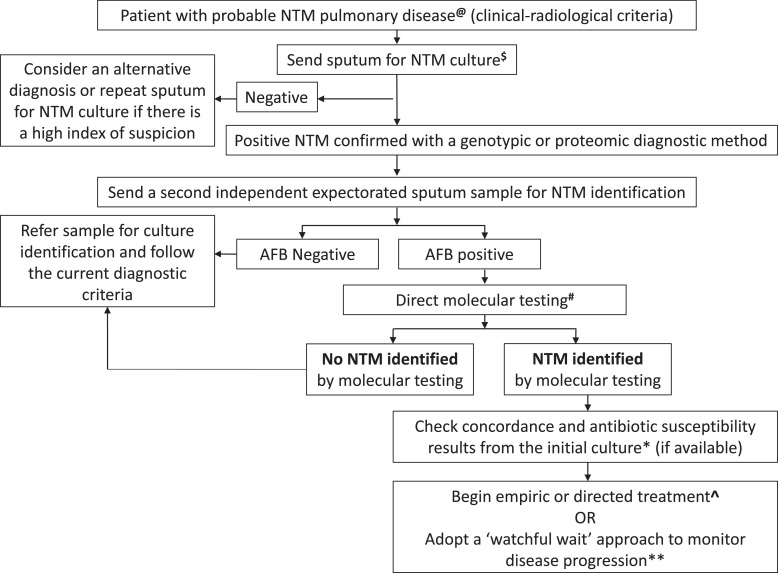
Figure 1.
The proposed algorithm for diagnosing and managing nontuberculous mycobacteria pulmonary disease. A second independent sputum sample (ideally taken ≥1 week after the initial sputum sample) is requested if the first sputum is culture-positive for NTM in a patient with a clinical and radiological picture consistent with pulmonary NTM disease. In cases of inconclusive or negative results from molecular testing and when smear microscopy is negative for AFB, sputum culture should be performed on that specimen. In addition, if the NTM identified with a direct molecular method from the sputum correlates with the NTM isolated on the initial sputum culture, susceptibilities can be performed from the culture. The clinician can consider treatment or the watchful wait management approach without needing a second culture.
AFB, acid-fast bacilli; NTM, nontuberculous mycobacteria.
Footnote:
@ Active pulmonary tuberculosis must be excluded using a standard diagnostic algorithm that includes a GeneXpert MTB/RIF Ultra negative in most settings.
$ In cases with an isolated culture that signals NTM growth, the clinician must ensure that the results correlate with the clinical and radiological profile consistent with NTM pulmonary infection.
# Direct molecular testing will only be performed depending on the availability of the appropriate assay suitable to detect the NTM that was cultured using the initial sputum sample.
* A second culture must be sent for discordance between direct molecular testing and initial culture results.
^ Microbiological treatment response can be evaluated using serial cultures/smears. These must be evaluated in conjunction with the clinical and radiological response to treatment.
** Many NTM species may not cause significant pulmonary disease (low pathogenic potential), e.g., M. gordonae, M. mucogenicum, M. nonchromogenicum, M. haemophilum, M. flavescens, M. gastri, M. terrae, or M. triviale. Thus the ‘watchful waiting’ strategy may be applied with adequate patient follow-up, incorporating clinical and radiological parameters.
Declarations of competing interest
The authors have no competing interest to declare.
Acknowledgments
Funding
RMW acknowledges the funding from the South African Medical Research Council. WJG acknowledges the funding received from the Wellcome Foundation (Grant # 222941/Z/21/Z). CO receives funding from the NHLS Research Trust Development Grant (Reference: PR2232714) and the Harry Crossley Foundation.
Ethical approval
Ethical approval with a waiver for informed consent was obtained from the Human Research Ethics Committee of Stellenbosch University (HREC reference number: S22/10/191).
Acknowledgment
We would like to acknowledge staff at the National Health Laboratory Service, TB laboratory, Green Point, Cape Town, South Africa for their continuous NTM work.
Author contributions
CJO conceptualized the manuscript. All authors edited and approved the final version.
Data sharing
Data sharing is not applicable to this article as no new data were created in this study.
Disclaimer
The views and opinions expressed in this article are those of the authors and do not necessarily reflect official policy or position of any affiliated agency of the authors.
References
- 1.Daley CL, Iaccarino JM, Lange C, Cambau E, Wallace RJ, Jr, Andrejak C, et al. Treatment of nontuberculous mycobacterial pulmonary disease: an official ATS/ERS/ESCMID/IDSA clinical practice guideline. Eur Respir J. 2020;56 doi: 10.1183/13993003.00535-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos A, Carneiro S, Silva A, Gomes JP, Macedo R. Nontuberculous mycobacteria in Portugal: trends from the last decade. Pulmonology. 2022 doi: 10.1016/j.pulmoe.2022.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Wang HY, Bang H, Kim S, Koh WJ, Lee H. Identification of Mycobacterium species in direct respiratory specimens using reverse blot hybridisation assay. Int J Tuberc Lung Dis. 2014;18:1114–1120. doi: 10.5588/ijtld.14.0140. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Q, Xiao H, Yan L. PCR-reverse blot hybridization assay in respiratory specimens for rapid detection and differentiation of mycobacteria in HIV-negative population. BMC Infect Dis. 2021;21:264. doi: 10.1186/s12879-021-05934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim JA, Yu HJ, Hwang YY, Kang OK, Shim HJ, Jhun BW, et al. Performance evaluation of MolecuTech REBA Myco-ID using HybREAD480 for identification of nontuberculous mycobacteria. Clin Lab. 2023;69 doi: 10.7754/Clin.Lab.2022.221115. [DOI] [PubMed] [Google Scholar]
- 6.Fang H, Shangguan Y, Wang H, Ji Z, Shao J, Zhao R, et al. Multicenter evaluation of the biochip assay for rapid detection of Mycobacterial isolates in smear-positive specimens. Int J Infect Dis. 2019;81:46–51. doi: 10.1016/j.ijid.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Sali M, De Maio F, Caccuri F, Campilongo F, Sanguinetti M, Fiorentini S, et al. Multicenter evaluation of Anyplex Plus MTB/NTM MDR-TB assay for rapid detection of Mycobacterium tuberculosis complex and multidrug-resistant isolates in pulmonary and extrapulmonary specimens. J Clin Microbiol. 2016;54:59–63. doi: 10.1128/JCM.01904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sawatpanich A, Petsong S, Tumwasorn S, Rotcheewaphan S. Diagnostic performance of the Anyplex MTB/NTM real-time PCR in detection of Mycobacterium tuberculosis complex and nontuberculous mycobacteria from pulmonary and extrapulmonary specimens. Heliyon. 2022;8:e11935. doi: 10.1016/j.heliyon.2022.e11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bicmen C, Gunduz AT, Coskun M, Senol G, Cirak AK, Ozsoz A. Molecular detection and identification of Mycobacterium tuberculosis complex and four clinically important nontuberculous mycobacterial species in smear-negative clinical samples by the genotype Mycobacteria direct test. J Clin Microbiol. 2011;49:2874–2878. doi: 10.1128/JCM.00612-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schildhaus HU, Steindor M, Kölsch B, Herold T, Buer J, Kehrmann J. GenoType CM Direct® and VisionArray Myco® for the rapid identification of Mycobacteria from clinical specimens. J Clin Med. 2022;11:2404. doi: 10.3390/jcm11092404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodriguez-Temporal D, Alcaide F, Mareković I, O'Connor JA, Gorton R, van Ingen J, et al. Multicentre study on the reproducibility of MALDI-TOF MS for nontuberculous mycobacteria identification. Sci Rep. 2022;12:1237. doi: 10.1038/s41598-022-05315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao L, Gui X, Wu X, Yang J, Fang Y, Sun Q, et al. Rapid Identification of nontuberculous mycobacterium species from respiratory specimens using nucleotide MALDI-TOF MS. Microorganisms. 2023;11:1975. doi: 10.3390/microorganisms11081975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alcaide F, Amlerová J, Bou G, Ceyssens PJ, Coll P, Corcoran D, et al. How to: identify non-tuberculous mycobacterium species using MALDI-TOF mass spectrometry. Clin Microbiol Infect. 2018;24:599–603. doi: 10.1016/j.cmi.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 14.Albay A, Hoşbul T, Uçarman SN, Özcan H, Tekin K, Arslantürk A. Investigation of the efficacy of three different protein extraction protocols and matrix-assisted laser desorption ionization-time of flight mass spectrometry for the identification of nontuberculous mycobacteria. Mikrobiyol Bul. 2022;56:206–217. doi: 10.5578/mb.20229802. [DOI] [PubMed] [Google Scholar]
- 15.Dao TL, Hoang VT, Ly TDA, Lagier JC, Baron SA, Raoult D, et al. Sputum proteomic analysis for distinguishing between pulmonary tuberculosis and non-tuberculosis using matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS): preliminary results. Clin Microbiol Infect. 2021;27:1694. doi: 10.1016/j.cmi.2021.02.031. e1–6. [DOI] [PubMed] [Google Scholar]
- 16.Caverly LJ, Carmody LA, Haig SJ, Kotlarz N, Kalikin LM, Raskin L, et al. Culture-independent identification of nontuberculous mycobacteria in cystic fibrosis respiratory samples. PLoS One. 2016;11 doi: 10.1371/journal.pone.0153876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mwangi ZM, Mukiri NN, Onyambu FG, Wallace BD. Genetic diversity of nontuberculous mycobacteria among symptomatic tuberculosis negative patients in Kenya. Int J Mycobacteriol. 2022;11:60–69. doi: 10.4103/ijmy.ijmy_224_21. [DOI] [PubMed] [Google Scholar]
- 18.Scoleri GP, Choo JM, Leong LE, Goddard TR, Shephard L, Burr LD, et al. Culture-independent detection of nontuberculous mycobacteria in clinical respiratory samples. J Clin Microbiol. 2016;54:2395–2398. doi: 10.1128/JCM.01410-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C, Lu J, Long B, Rao Z, Gao Y, Wang W, et al. Detection of Mycobacterium kansasii using a combination of loop-mediated isothermal amplification (LAMP) and lateral flow biosensors. Int Microbiol. 2021;24:75–82. doi: 10.1007/s10123-020-00143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarro YDS, Butzler MA, Sanogo F, Kodio O, Tolofoudie M, Goumane MS, et al. Development and clinical evaluation of a new multiplex PCR assay for a simultaneous diagnosis of tuberculous and nontuberculous mycobacteria. EBiomedicine. 2021;70 doi: 10.1016/j.ebiom.2021.103527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Ma X, Chen J, Wang H, Yu Z. Nontuberculous mycobacteria by metagenomic next-generation sequencing: three cases reports and literature review. Front Public Health. 2022;10 doi: 10.3389/fpubh.2022.972280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marais G, Hardie D, Brink A. A case for investment in clinical metagenomics in low-income and middle-income countries. Lancet Microbe. 2023;4:e192–e199. doi: 10.1016/S2666-5247(22)00328-7. [DOI] [PubMed] [Google Scholar]
- 23.Gao CA, Huston JC, Toro PV, Gautam S, Dela Cruz CS. Precision in pulmonary, critical care, and sleep medicine. Springer Nature; London (Global): 2020. Molecular diagnostics in pulmonary infections; pp. 167–184. [DOI] [Google Scholar]
- 24.Wei W, Cao J, Wu XC, Cheng LP, Shen XN, Sha W, et al. Diagnostic performance of metagenomic next-generation sequencing in non-tuberculous mycobacterial pulmonary disease when applied to clinical practice. Infection. 2023;51:397–405. doi: 10.1007/s15010-022-01890-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Y, Gong Z, Zhao X, Zhang D, Zhang Z. Comprehensive determination of Mycobacterium tuberculosis and nontuberculous Mycobacteria from targeted capture sequencing. Front Cell Infect Microbiol. 2020;10:449. doi: 10.3389/fcimb.2020.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nimmo C, Doyle R, Burgess C, Williams R, Gorton R, McHugh TD, et al. Rapid identification of a Mycobacterium tuberculosis full genetic drug resistance profile through whole genome sequencing directly from sputum. Int J Infect Dis. 2017;62:44–46. doi: 10.1016/j.ijid.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 27.Nimmo C, Shaw LP, Doyle R, Williams R, Brien K, Burgess C, et al. Whole genome sequencing Mycobacterium tuberculosis directly from sputum identifies more genetic diversity than sequencing from culture. BMC Genomics. 2019;20:389. doi: 10.1186/s12864-019-5782-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohál M, Porvazník I, Solovič I, Mokrý J. Whole genome sequencing in the management of non-tuberculous mycobacterial infections. Microorganisms. 2021;9:2237. doi: 10.3390/microorganisms9112237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas BE, Suresh C, Lavanya J, Lindsley MM, Galivanche AT, Sellappan S, et al. Understanding pretreatment loss to follow-up of tuberculosis patients: an explanatory qualitative study in Chennai, India. BMJ Glob Health. 2020;5 doi: 10.1136/bmjgh-2019-001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mwansa-Kambafwile JRM, Chasela C, Ismail N, Menezes C. Initial loss to follow up among tuberculosis patients: the role of Ward-Based Outreach Teams and short message service (SMS) technology (research proposal) BMC Res Notes. 2019;12:737. doi: 10.1186/s13104-019-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hendrix J, Epperson LE, Tong EI, Chan YL, Hasan NA, Dawrs SN, et al. Complete genome assembly of Hawai'i environmental nontuberculous mycobacteria reveals unexpected co-isolation with methylobacteria. PLoS One. 2023;18 doi: 10.1371/journal.pone.0291072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashemzadeh M, Dezfuli AA, Khosravi AD, Bandbal MM, Ghorbani A, Hamed M, et al. Molecular identification of non-tuberculous mycobacterial species isolated from extrapulmonary samples using real-time PCR and rpoB sequence analysis. AMB Express. 2023;13:43. doi: 10.1186/s13568-023-01553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng C, Xiao P. Evaluation of the correctable decoding sequencing as a new powerful strategy for DNA sequencing. Life Sci Alliance. 2022;5 doi: 10.26508/lsa.202101294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clarke C, Kerr TJ, Warren RM, Kleynhans L, Miller MA, Goosen WJ. Identification and characterisation of nontuberculous mycobacteria in African buffaloes (Syncerus caffer), South Africa. Microorganisms. 2022;10:1861. doi: 10.3390/microorganisms10091861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pascapurnama DN, Zavitri NG, Koesoemadinata RC, Cahyadi AI, Chaidir L. Identification of significant pathogenic nontuberculous mycobacteria species from presumptive TB patients using partial hsp65 gene sequencing. Infect Drug Resist. 2023;16:6923–6930. doi: 10.2147/IDR.S419956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mirlohi SH, Eftekhari K, Shirzadi R, Fateh A, Masoumi M, Modaresi M. The value of sputum polymerase chain reaction for detection of nontuberculous Mycobacteria in cystic fibrosis patients with negative nontuberculous Mycobacteria sputum culture. Med J Islam Repub Iran. 2022;36:7. doi: 10.47176/mjiri.36.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kambli P, Ajbani K, Kazi M, Sadani M, Naik S, Shetty A, et al. Targeted next generation sequencing directly from sputum for comprehensive genetic information on drug resistant Mycobacterium tuberculosis. Tuberculosis (Edinb) 2021;127 doi: 10.1016/j.tube.2021.102051. [DOI] [PubMed] [Google Scholar]
- 38.World Health Organization. Use of targeted next-generation sequencing to detect drug-resistant tuberculosis: rapid communication, https://www.who.int/publications/i/item/9789240076372; 2023 [accessed 05 December 2023].
- 39.World Health Organization. The use of next-generation sequencing for the surveillance of drug-resistant tuberculosis: an implementation manual, https://www.who.int/publications/i/item/9789240078079; 2023 [accessed 05 December 2023].
- 40.Deeplex Myc-TB user manual. From clinical sample to drug resistance. profile, https://www.illumina.com/content/dam/illumina/gcs/assembled-assets/marketing-literature/deeplex-myc-tb-user-manual/user-guide-genoscreen-deeplex-myc-tb.pdf; 2023 [accessed 05 December 2023].
- 41.Mingora CM, Garcia BA, Mange KC, Yuen DW, Ciesielska M, van Ingen J, et al. Time-to-positivity of Mycobacterium avium complex in broth culture associates with culture conversion. BMC Infect Dis. 2022;22:246. doi: 10.1186/s12879-022-07250-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Park KG, Lee G, Park J, Park YG, Park YJ. Assessment of the quantitative ability of AdvanSure TB/NTM real-time PCR in respiratory specimens by comparison with phenotypic methods. Ann Lab Med. 2014;34:51–55. doi: 10.3343/alm.2014.34.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pennington KM, Vu A, Challener D, Rivera CG, Shweta FNU, Zeuli JD, Temesgen Z. Approach to the diagnosis and treatment of non-tuberculous mycobacterial disease. J Clin Tuberc Other Mycobact Dis. 2021;24 doi: 10.1016/j.jctube.2021.100244. [DOI] [PMC free article] [PubMed] [Google Scholar]