Highlights
-
•
Immune infiltrate differs between HPV-associated oropharyngeal and cervical tumors.
-
•
Lymphocyte-rich infiltrate in oropharyngeal cancer.
-
•
Shift toward lymphocyte exclusion and MDSC infiltration in cervical cancer.
-
•
Oropharyngeal tumor cells express high levels of CXCL9 and CXCL10.
-
•
Cervical tumor cells express high levels of MDSC chemoattractant CXCL1.
Keywords: HNSCC, Cervical cancer, MDSC, Tumor microenvironment
Abstract
The profile of the antitumor immune response is an important factor determining patient clinical outcome. However, the influence of the tissue contexture on the composition of the tumor microenvironments of virally induced tumors is not clearly understood. Therefore, we analyzed the immune landscape of two HPV-associated malignancies: oropharyngeal squamous cell carcinoma (OPSCC) and squamous cell carcinoma of uterine cervix (CESC). We employed multiplex immunohistochemistry and immunofluorescence to evaluate the density and spatial distribution of immune cells in retrospective cohorts of OPSCC and CESC patients. This approach was complemented by transcriptomic analysis of purified primary tumor cells and in silico analysis of publicly available RNA sequencing data. Transcriptomic analysis showed similar immune profiles in OPSCC and CESC samples. Interestingly, immunostaining of OPSCC tissues revealed high densities of immune cells in both tumor stroma and tumor epithelium, whereas CESC samples were mainly characterized by the lack of immune cells in the tumor epithelium. However, in contrast to other immune cell populations, polymorphonuclear myeloid-derived suppressor cells (PMN-MDSCs) were abundant in both segments of CESC samples and CESC-derived tumor cells expressed markedly higher levels of the PMN-MDSC chemoattractants CXCL1, CXCL5, and CXCL6 than OPSCC tumor cells. Taken together, despite their having the same etiologic agent, the immune infiltration pattern significantly differs between OPSCC and CESC, with a noticeable shift toward prominent MDSC infiltration in the latter. Our data thus present a rationale for a diverse approach to targeted therapy in patients with HPV-associated tumors of different tissue origins.
Introduction
Human papillomavirus (HPV) is the etiologic agent of more than 600,000 new cancer cases per year. Despite a dramatic increase in HPV-associated head and neck squamous cell carcinoma (HNSCC) in developed countries, cervical cancer still represents 83 % of HPV-attributable malignancies worldwide. Not surprisingly, the majority of cervical cancer patients are diagnosed in less developed countries with limited access to regular screening and vaccination [1].
HPV-associated malignant tumors are caused by high-risk HPV subtypes, with HPV16 being detectable in more than 80 % of oropharyngeal squamous cell carcinomas and in 50–70 % of cervical cancers [1], [2], [3]. In general, HPV infects basal epithelial cells of the skin and mucosae. The targets of HPV in the cervix are cells in the transformation zone in the proximity of the squamocolumnar junction [4]. In the oropharynx, HPV infects and transforms the specialized reticulated epithelium of tonsillar crypts [5]. Although the process of carcinogenesis seems to be similar in these two anatomical locations, a substantial difference in the form of the HPV genome present has been described recently. In more than 80 % of cervical cancer samples, the HPV genome is stably integrated into the host DNA [6], but the majority of HPV-associated HNSCC contains episomal HPV genomes or a mixture of episomal and integrated HPV DNA [7]. There is evidence that early genes in addition to the classical oncoproteins E6 and E7 are expressed, such as E1, E2, and E5, when the HPV genome is episomally maintained [7,8]. The broader spectrum of the antigenic repertoire may thus lead to a stronger immune response in HPV-associated HNSCC than in cervical cancer [2].
It is widely accepted that the profile of the antitumor immune response is a determining factor for patient clinical outcome. Responses to standard of care protocols as well as to immunotherapy have been shown in many cases to be associated with a specific type of tumor microenvironment [9], [10], [11]. According to pancancer analysis of RNA sequencing data, both HPV+ HNSCC and cervical cancer belong to the top ten most immune-infiltrated tumors [12]. Although the abundance of CD8+ T cells [12], [13], [14], B cells [15,16], and tertiary lymphoid structures (TLSs) [16] in the TME correlates clearly with a favorable outcome in HPV-associated HNSCC, contradictory results have been reported for cervical cancer patients. Enwere et al. [17] showed no prognostic impact of CD8+ T cells on cervical cancer patient outcome, while Ohno et al. [18] reported a positive correlation between densities of CD8+ T cells and overall survival in a small cohort of cervical cancer patients treated with chemoradiotherapy. More consistently, a positive association between the density of activated memory CD4+ T cells and cervical cancer patient outcome has been reported [19,20]. The prognostic impact of B cells and TLSs has not been reported in cervical carcinomas so far. These discrepancies indicate a significant impact of tissue contexture on the immune landscape of HPV-associated tumors.
This study provides an in-depth analysis of the tumor microenvironments (TMEs) of two HPV-associated malignancies, namely oropharyngeal squamous cell carcinoma (OPSCC) and cervical squamous cell carcinoma (CESC). According to our observations, the type of tumor microenvironment and the spatial distribution of immune cells differ significantly between these two malignancies. Thus, despite their having the same etiological agent, the clinical benefit of targeted immunotherapeutic approaches, such as HPV therapeutic vaccines, should be thoroughly evaluated due to the distinct tissue contexture and TME subtype in OPSCC and CESC patients.
Materials and methods
Patients and samples
Cohort 1. Formalin-fixed paraffin-embedded (FFPE) primary HPV+ OPSCC specimens were obtained from 62 patients who underwent radical surgery at the University Hospital Hradec Kralové in the Czech Republic between 2001 and 2014.
Cohort 2. Formalin-fixed paraffin-embedded (FFPE) primary CESC specimens were obtained from 71 patients who underwent radical surgery at the University Hospital Hradec Kralové in the Czech Republic between 2010 and 2020. A comparison of cohorts 1 and 2 is summarized in Table 1. Data on long-term clinical outcomes were obtained retrospectively from municipality registers. The protocols were approved by the local ethics committees.
Table 1.
Clinico-pathological characteristics of the patients.
| Variable | HPV+ HNSCC | CESC | ||
|---|---|---|---|---|
| No. | % | No. | % | |
| Total No. of Patients | 62 | 71 | ||
| Age | ||||
| Median | 57 | 51 | ||
| Range | 41–77 | 26–77 | ||
| Stage | ||||
| I | 46 | 74.2 | 57 | 80.3 |
| II | 13 | 21.0 | 8 | 11.3 |
| III–IV | 3 | 4.8 | 6 | 8.6 |
| Therapy | ||||
| Radiotherapy | 62 | 100 | 15 | 21.1 |
| Chemotherapy | 21 | 33.9 | 15 | 21.1 |
| HPV subtype | ||||
| 16 | 57 | 91.9 | 50 | 70.4 |
| 18 | 1 | 1.6 | 5 | 7 |
| 33 | 1 | 1.6 | 5 | 7 |
| 35 | 3 | 4.9 | 0 | 0 |
| 45 | 0 | 0 | 4 | 5.6 |
| Other (31, 58, 59, 70, 73) | 0 | 0 | 7 | 10 |
| Co-infection | 1 | 1.6 | 9 | 12.7 |
| Outcome | ||||
| Relapse | 6 | 9.7 | 7 | 9.9 |
| Exitus | 12 | 19.4 | 7 | 9.9 |
| Smoking history | ||||
| Smoker | 15 | 24.2 | 30 | 42.6 |
| Ex-smoker | 23 | 37.1 | NA | NA |
| Non-smoker | 24 | 38.7 | 40 | 55.9 |
| NA | 0 | 0 | 1 | 1.5 |
Cohort 3. Fresh primary OPSCC specimens were obtained from 14 patients immediately after therapeutic surgery at the University Hospital Motol in Prague in the Czech Republic between 2018 and 2023. Fresh primary CESC specimens were obtained as punch biopsies from 15 patients at the University Hospital Královské Vinohrady in Prague in the Czech Republic between 2021 and 2023. Written informed consent was obtained from patients before their inclusion in the study.
None of the patients enrolled in this study had received any neoadjuvant chemo- or radiotherapy. The pathological staging of OPSCC was reviewed and classified by an experienced pathologist according to the eighth edition of the American Joint Committee on Cancer. The pathological staging of CESC was reviewed and classified by an experienced pathologist according to the 2018 FIGO staging classification. All procedures performed in this study were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Declaration of Helsinki and its later amendments.
Immunohistochemistry
Staining was carried out on FFPE sections following deparaffinization and antigen retrieval. Endogenous peroxidase was blocked with 3 % hydrogen peroxide. The sections were incubated with protein block (DAKO) and stained with primary antibodies against CD4 (RBT-CD4, LSBio), CD8 (SP16, Spring Bioscience), CD20 (L26, Dako), FoxP3 (236A/E7, Abcam), BDCA2 (polyclonal goat IgG, R&D Systems), DC-LAMP (1010E1.01, Dendritics), CD68 (KP1, Abcam), CD163 (EPR19518, Abcam), PNAd (MECA-79, Biolegend), Nitrotyrosine (A-21285, Invitrogen), and Hif-1α (EP1215Y, Abcam) followed by the manifestation of enzymatic activity and hematoxylin or Nuclear Fast Red (Vector Laboratories) counterstaining. Images were acquired using an Aperio AT2 scanner (Leica).
Immunofluorescence
Staining was carried out on 4 µm-thick FFPE sections following deparaffinization and antigen retrieval using MDSC FixVUE Panel (Ultivue) according to the manufacturer's instructions. Monocytic MDSCs (M-MDSCs) were defined as CD11b+CD14+CD15-HLA-DR- cells; polymorphonuclear MDSCs (PMN-MDSCs) were defined as CD11b+CD14-CD15+HLA-DR- cells. Images were acquired using an Aperio AT2 scanner (Leica).
Quantification of tumor-infiltrating immune cells and tertiary lymphoid structures
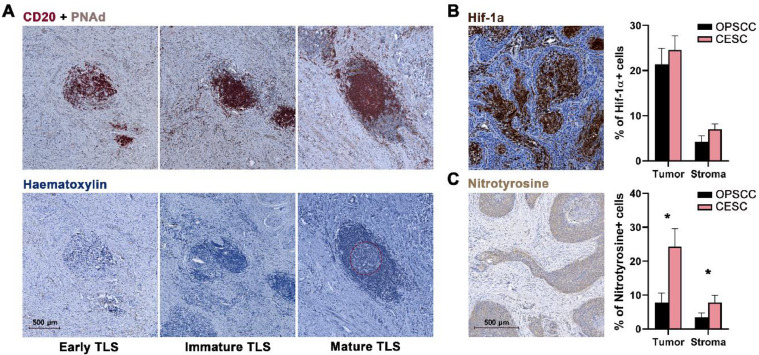
Each section was scanned and evaluated for immune cell infiltration in the tumor nest and tumor stroma using Aperio ImageScope (Leica) and Halo (IndicaLabs) software. Tertiary lymphoid structures (TLSs) were analyzed in two sequential slides based on CD20 + PNAd staining in the first section and hematoxylin staining in the subsequent section. Aggregates of > 20 CD20+ B lymphocytes accompanied by T lymphocytes were assessed as early TLSs. Structures with B-cell follicles without a visible germinal center were considered immature TLSs and structures with visible germinal centers were considered mature TLSs. Examples of these structures are shown in Fig. 4A.
Fig. 4.
CESC tissues show high levels of nitrotyrosine positivity. (A) Representative IHC staining and differentiation of TLSs. TLSs were characterized by the presence of CD20+ B cells and PNAd+ high endothelial venules. The dashed line marks the germinal center in the mature TLS. (B) Representative IHC staining of Hif-1α. Columns represent mean (+ SEM) proportion of Hif-1α+ cells in tumor epithelium (Tumor) and tumor stroma (Stroma). (C) Representative IHC staining of nitrotyrosine. Columns represent mean (+ SEM) proportion of nitrotyrosine+ cells in tumor epithelium (Tumor) and tumor stroma (Stroma). * p < 0.05 (Mann–Whitney U test).
TME stratification
To stratify the samples into three types of TME, namely hot, excluded, and cold, cutoffs based on the density of CD8+ T cells within the tumor stroma and tumor epithelium were calculated for four different cohorts of patients: HPV+ OPSCC (n = 55), CESC (n = 67), HPV- HNSCC (n = 63), and high-grade serous ovarian carcinoma (n = 58). The cutoff value was the median CD8+ Tcell density in the stroma (M = 171.12 CD8+ T cells/mm2) and in the tumor epithelium (M = 105.46 CD8+ T cells/mm2). Samples with densities of CD8+ T cells above the median in both compartments were considered hot, samples with densities of CD8+ T cells below the median in the tumor epithelium and above the median in the stroma were considered excluded, and samples with densities of CD8+ T cells below the median in both compartments were assessed as cold.
Processing of fresh tumor tissues
Fresh tissues were minced with scissors and digested in RPMI 1640 (Thermo Fisher Scientific) containing 1 mg/ml collagenase D (Roche) and 0.05 mg/ml DNase I (Roche) for 30 min at 37 °C under a gentle rocking motion. Subsequently, the specimens were passed through a 100-µm nylon cell strainer (BD Biosciences) and washed with PBS (Lonza). The obtained single-cell suspensions were frozen in liquid nitrogen until use.
Tumor cell sorting and RNA isolation
Tumor-derived single-cell suspensions were stained with antibodies against EpCAM (A488), CD45 (PE-Cy7), and CD90 (APC) (all BioLegend). Cells were sorted using a FACSAria IIu (BD Biosciences). EpCAM+CD45-CD90- tumor cells were collected in XB buffer, heated at 42 °C for 30 min, and centrifuged for 2 min. The cell lysates were stored at -80 °C until use.
Total RNA was isolated from the tumor cell lysates using an Arcturus Picopure RNA isolation kit (Thermo Fisher Scientific) according to the manufacturer's instructions. The concentration and purity of the samples were determined by spectrophotometry using a NanoDrop 2000c (Thermo Fisher Scientific), and the RNA integrity was assessed using a 2100 Bioanalyzer (Agilent).
Next-generation sequencing data analysis
Raw fastq files were trimmed with TrimGalore (v. 0.6.5), aligned to the human transcriptome ensemble reference (build GRCh38), quantified with salmon (v. 1.8), and imported with tximport (v. 1.24.0). Raw counts were normalized with DESeq2 (v. 1.36.0) and log-transformed in R. Selected genes were visualized with the R ComplexHeatmap package (v. 2.12.1).
The Cancer Genome Atlas (TCGA) data analysis
Estimates of immune cell populations for patients with HPV+ OPSCC (n = 33) and CESC (n = 307) were downloaded from TIMER2.0 (http://timer.cistrome.org/). CIBERSORT-ABS estimates were used for immune profile comparison between HPV+ OPSCC and CESC samples.
HPV detection
Immunohistochemical analysis
An antibody against p16INK4a (Purified Mouse Anti-Human p16, Clone G175-405, BD Pharmingen TM, dilution 1:100) or the CINtec Histology Kit (Roche) was used. The intensity of staining and the proportion of stained cells were evaluated. Samples positive for p16 expression showed more than 70 % positive cells and revealed nuclear and/or cytoplasmic staining.
PCR
HPV DNA from the paraffin-embedded tissue was extracted with a MagCore Genomic DNA FFPE One-Step Kit (RBC Bioscience) according to the manufacturer´s protocol.
HPV DNA detection and genotyping were performed by qualitative real-time PCR with an AmoyDx Human Papillomavirus Genotyping Detection Kit (Amoy Diagnostics). The test is designed for specific amplification of the L1 gene in HPV DNA to detect and genotype 19 high-risk HPVs and 2 low-risk HPVs (HPV 6 and 11). The sensitivity of the test is 100 copies of HPV DNA per reaction. An internal control is provided in the assay to test for sample quality and the presence of inhibiting factors.
Statistical analysis
Statistical analyses were performed using Statistica 10.0 software (StatSoft). Parametric assumptions of the data were verified using the Kolmogorov–Smirnov test for normality. Homogeneity of variances was tested by the Levene test. Differences between HPV+ OPSCC samples and CESC samples were analyzed using the Mann–Whitney U test. Correlations between myeloid cell and CD8+ Tcell densities were evaluated using Pearson's χ2 test. For survival analyses, patients were divided according to the median of the indicated variable. Differences between groups with high and low densities of immune cells were assessed using the log-rank test. Results were considered statistically significant at p < 0.05.
Results
The distribution of immune cells in the TME differs markedly between CESC and HPV-associated OPSCC
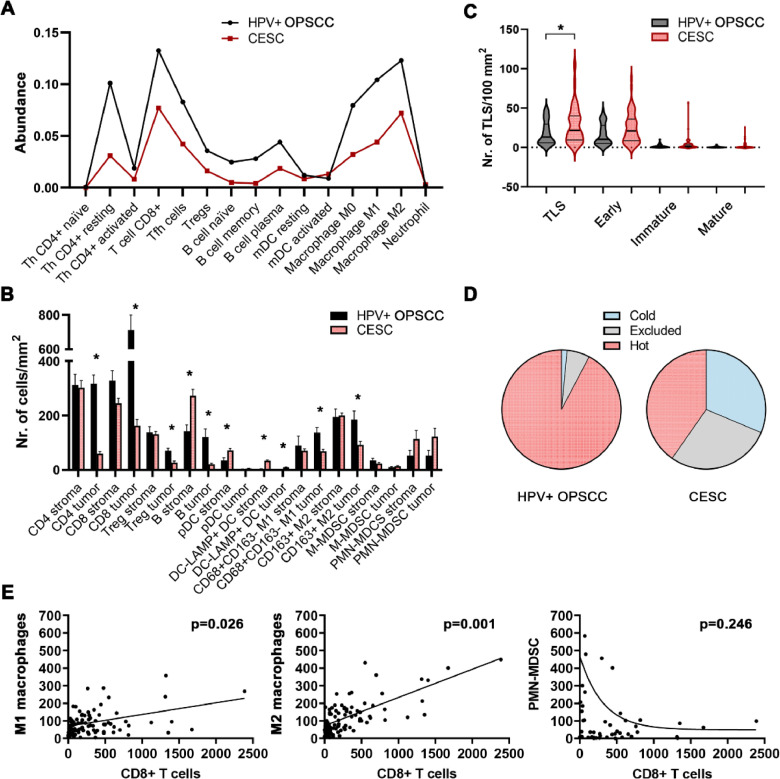
To compare the immune profiles between CESC and HPV+ OPSCC samples, we analyzed the abundance of tumor-infiltrating immune cells estimated using the CIBERSORT-ABS tool for patients selected from the publicly available TCGA database. The results show a similar profile of leukocytes in the TME, with generally lower levels of tumor-infiltrating immune cells in CESC samples than in HPV+ OPSCC samples (Fig. 1A).
Fig. 1.
Differences in frequencies of tumor-infiltrating leukocytes in patients with OPSCC and CESC. (A) The figure shows the abundance of immune cells estimated by the CIBERSORT-ABS tool from TCGA datasets. (B) Columns represent mean (+ standard error of mean, SEM) densities of immune cells in tumor stroma and tumor epithelium of immunohistochemically stained FFPE sections of HPV+ OPSCC and CESC samples. (C) Violin plot shows the distribution of TLS densities in OPSCC and CESC samples. Bold lines represent medians, and thin lines represent quartiles. (D) Pie charts show the proportions of patients with cold, excluded, and hot tumor microenvironment types. The stratification is based on CD8+ Tcell density in the tumor stroma and tumor epithelium. (E) Scatterplots show the correlation between densities of myeloid cells and CD8+ T cells in the tumor epithelium of combined cohorts of OPSCC and CESC patients. * p < 0.05 (Mann–Whitney U test).
Nevertheless, analysis of the spatial distribution of immune cells within the TME showed substantially higher levels of all cell types analyzed in the tumor epithelium of HPV+ OPSCC than in CESC. The only exception was myeloid-derived suppressor cells (MDSCs), which had migrated to the tumor epithelium in both types of HPV-associated carcinoma (Fig. 1B). Additionally, we observed markedly higher levels of CD11b+CD14-CD15+HLA-DR- PMN-MDCSs in CESC than in HPV+ OPSCC. Surprisingly, we also observed significantly higher levels of tertiary lymphoid structures (TLSs) in the TME of CESC patients than in the TME of HPV+ OPSCC patients (Fig. 1C). Examples of staining are shown in Fig. 2 and in Fig. 4A (TLS).
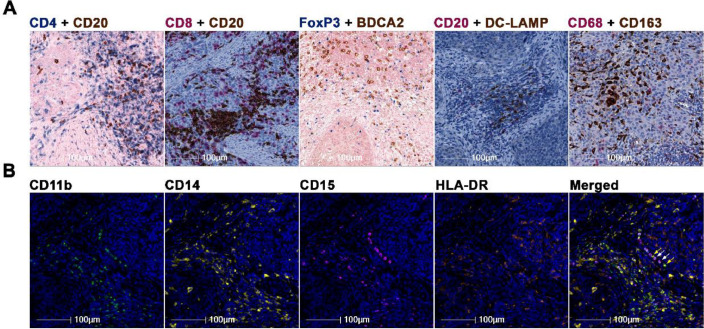
Fig. 2.
Representative IHC and high-plex immunofluorescence staining. (A) Representative IHC staining of CD4+ T cells, CD20+ B cells, FoxP3+ Tregs, BDCA2+ pDCs, DC-LAMP+ DCs, CD68+CD163− M1 macrophages, and CD68+CD163+ M2 macrophages. (B) Representative MDSC staining. Yellow arrows show CD11b+CD14+CD15−HLA-DR− M-MDSCs, and white arrows mark CD11b+CD14−CD15+HLA-DR− PMN-MDSCs.
To evaluate whether the excluded phenotype with most of the immune cells retained in the tumor stroma is a general feature of the CESC TME, we set CD8+ Tcell density-based cutoffs for hot, cold, and excluded TME. The results showed that 92.3 % of HPV+ OPSCC samples rank among hot tumors, but that only 40.3 % of CESC samples were considered hot. In contrast, the proportions of samples with excluded TME were 6.2 % and 28.4 % in HPV+ OPSCC and CESC, respectively. Similarly, the proportions of immunologically cold samples were 1.5 % and 31.3 % in HPV+ OPSCC and CESC, respectively (Fig. 1D). Thus, hot TME is markedly more common in HPV+ OPSCC, whereas the distribution of hot, excluded, and cold TME is equable in CESC.
Additionally, although the densities of macrophages correlated positively with the densities of CD8+ T cells within the tumor epithelium in both indications, mutual exclusion was detected between PMN-MDSCs and CD8+ T cells (Fig. 1E).
Tumor cells isolated from CESCs express high levels of chemokines attracting neutrophiles/PMN-MDSCs
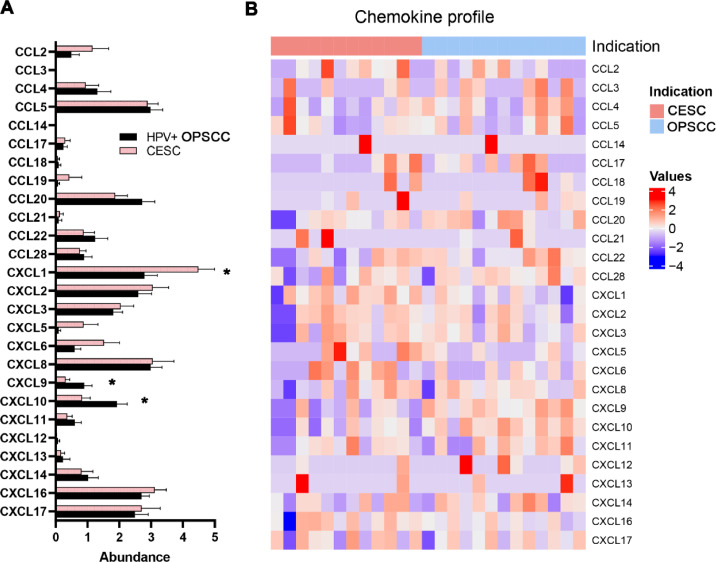
To compare expression of chemokines in HPV+ HNSCC and CESC, we purified EpCAM+CD45-CD90- tumor cells from native carcinoma samples and performed bulk RNA sequencing. The primary tumor cells derived from CESC samples produced markedly higher levels of chemokines associated with neutrophils and MDSC trafficking, namely CXCL1, CXCL5, and CXCL6. In contrast, the tumor cells derived from HPV+ HNSCC produced significantly higher levels of lymphocyte/NK cell chemoattractants CXCL9 and CXCL10 in comparison to CESC (Fig. 3).
Fig. 3.
CESC-derived tumor cells produce high levels of CXCL1. (A) Columns represent mean (+ SEM) mRNA expression of the indicated chemokines in tumor cells purified from HPV+ OPSCC and CESC primary samples. (B) The heatmap expresses z scores of relative mRNA expression of the indicated genes in tumor cells purified from HPV+ OPSCC and CESC primary samples. * p < 0.05 (Mann–Whitney U test).
HPV-associated OPSCC and CESC show comparable levels of hypoxia
To analyze the level of hypoxia in the tumor tissue, we assessed expression of hypoxia-inducible factor 1-alpha (Hif-1α) in both the tumor epithelium and the tumor stroma. There was no statistically significant difference in the proportions of Hif-1α-expressing tumor cells between HPV+ OPSCC and CESC. The distribution of hypoxic regions in the tumor epithelium was also similar, with apparent central necrotic cores surrounded by living tumor cells (Fig. 4B).
Expression of nitrotyrosine is markedly higher in CESC tissues than in HPV+ OPSCC tissues
PMN-MDSCs are known to produce large amounts of peroxynitrite (ONOO−), which can cause nitration of proteins, including chemokines. The level of nitrotyrosine in the tumor tissue may thus reflect the activity of peroxynitrite and, indirectly, the activity of MDSCs.
Indeed, we detected significantly higher levels of nitrotyrosine in both the tumor epithelium and tumor stroma of CESC than in HPV+ OPSCC samples (Fig. 4C). These results are in accordance with the observed higher densities of PMN-MDSCs in cervical carcinoma tissues.
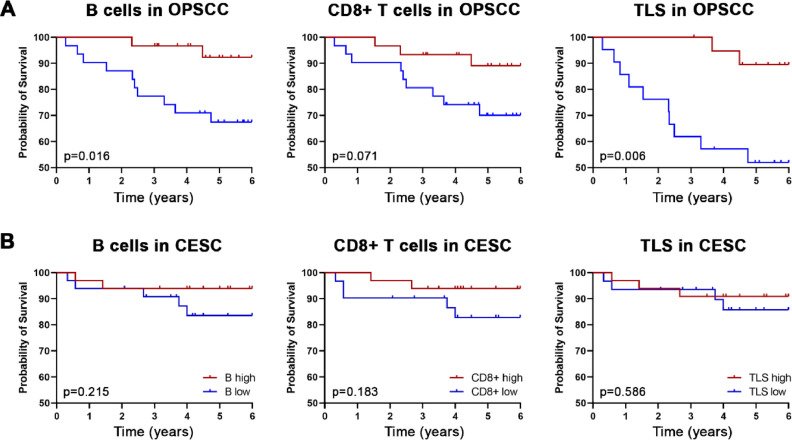
Densities of B cells, CD8+ T cells, and TLSs are positive prognostic markers in HPV+ OPSCC but not CESC
Previously we showed that high densities of CD20+ B cells and CD8+ T cells in the tumor epithelium are positive prognostic markers in HNSCC [15]. In this study, the presence of high levels of intratumoral CD20+ B cells and high densities of TLS were associated with significantly improved overall survival (OS) in HPV+ OPSCC patients (p = 0.016 and p = 0.006, respectively; Fig. 5A). In contrast, there was no correlation between the abundance of B cells, CD8+ T cells, or TLSs and the OS of CESC patients (Fig. 5B).
Fig. 5.
Prognostic value of tumor-infiltrating CD20+ B cells, CD8+ T cells, and TLSs in HPV+ OPSCC (A) and CESC (B). Kaplan–Meier curves show the overall survival of patients according to densities of the indicated cells in the tumor epithelium. p values were determined using the log-rank test.
Discussion
High-risk HPV subtypes are etiologic agents of almost all cervical carcinomas and a significant proportion of oropharyngeal squamous cell carcinomas [1]. Due to expression of viral antigens, both malignancies are considered highly immunogenic and belong to the top ten most immune-infiltrated tumors [12]. However, only a few studies have directly compared the immune profile of different HPV-associated carcinomas, suggesting a strong impact of tissue contexture on the shape of the immune response [21]. The spatial distribution of immune cells within the TME has not been compared between OPSCC and cervical carcinoma so far.
In this study, we employed transcriptomic analysis of a TCGA-based dataset as well as immunohistochemical and multiple immunofluorescence staining of two retrospective cohorts of patients with HPV-associated carcinomas, including 62 OPSCC and 71 CESC patients. Although transcriptomic analysis showed similar immune profiles in OPSCC and CESC with lower frequencies of immune cells in CESC samples, spatial analysis revealed substantial differences in immune cell distribution in the CESC TME and that of OPSCC. Indeed, OPSCC samples were considered immunologically hot, with high densities of immune cells in both the tumor stroma and tumor epithelium, but CESC samples had high densities of immune cells, especially lymphocytes, only in the tumor stroma. However, higher levels of PMN-MDSCs were found in both the tumor stroma and tumor epithelium of CESC samples than in OPSCC samples.
It has been widely accepted that the density and distribution of tumor-infiltrating immune cells are significant factors that determine patient clinical outcome [22], [23], [24]. Galon et al. [25] introduced a Tcell-based classification of tumors and distinguished 4 types of TME: hot, immunosuppressed, excluded, and cold. In accordance with this approach, Someya et al. [26] described three types of TME in cervical cancer patients, namely, hot, excluded, and cold, with the cold type being associated with poor prognosis. Similarly, we were able to distinguish three types of TME based on the density and distribution of CD8+ T cells. While hot TME prevailed significantly in OPSCC samples, CESC samples mostly fell into the excluded or cold type. Interestingly, although the densities and distribution of M1 and M2 macrophages correlated positively with the densities and distribution of CD8+ T cells, near mutual exclusion between the density of PMN-MDSCs and CD8+ T cells was observed. In agreement with our findings, a strong association between the presence of MDSCs and the lack of T cells was previously described in a mouse model of pancreatic cancer [27].
Multiple chemokines, including CXCL1, CXCL2, CXCL8, CXCL12, CCL1, CCL2, CCL3, and CCL5, have been shown to mediate MDSC trafficking into the TME [28], [29], [30], [31]. Comparison of chemokine profiles of OPSCC and CESC tumor cells showed significantly higher expression of CXCL1 in tumor cells isolated from primary CESC samples and markedly higher expression of CXCL9 and CXCL10 in tumor cells isolated from OPSCC samples. As CXCL1 was shown to be associated with CXCR2+ MDSC migration [29,31,32], our data suggest that tumor cells of cervical origin directly attract MDSCs into the TME. In contrast, chemokines CXCL9 and CXCL10 produced by OPSCC-derived epithelial cells play crucial roles in the chemotaxis of lymphocytes and are associated with an immunologically hot TME [33,34]. In addition to the chemokines listed above, hypoxia-up-regulated expression of VEGF was shown to contribute to accumulation of MDSCs within the TME [35]. However, we did not observe any statistically significant differences in expression of the hypoxia marker Hif-1α between OPSCC and CESC samples.
MDSCs employ a wide range of mechanisms to impact both innate and adaptive immune responses; however, the main targets of MDSC-mediated suppression are T cells [36], [37], [38]. PMN-MDSCs produce large amounts of reactive oxygen species (ROS) and reactive nitrogen species (RNS). The reaction of superoxide with NO leads to production of peroxynitrite (PNT), which is capable of strongly inhibiting the activity of T cells via nitration/nitrosylation of Tcell receptors, MHC molecules on tumor cells, and Tcell-specific chemokines [36,39]. To assess the level of nitration in the TME, we analyzed the proportion of nitrotyrosine-positive cells in the tumor epithelium and tumor stroma of OPSCC and CESC samples. As expected, we detected significantly higher proportions of nitrotyrosine+ cells in CESC samples than in OPSCC tissues. These data are in accordance with the high abundance of MDSCs in CESC samples and may indicate high activity of PMN-MDSCs in the CESC TME. Importantly, as peroxynitrite-induced posttranslational modification of chemokines has been shown to impair Tcell trafficking into the TME [39], a high level of PNT activity might at least partly explain the lack of CD8+ T cells in the tumor epithelium of CESC samples highly infiltrated by PMN-MDSCs.
Our former study revealed a strong prognostic impact of B cells in HNSCC patients [15]. Compared with OPSCC, we did not confirm the density of B cells, CD8+ T cells, or TLSs as valid prognostic markers in our cohort of CESC patients. However, because the majority of patients were in the early stage of the disease and the number of deceased patients was low in the CESC cohort, the survival analyses should be repeated in a larger independent cohort of patients with more disease-related deaths.
In summary, our study provides an extensive comparison of the TME of two different HPV-associated squamous cell carcinomas, oropharyngeal and cervical, confirming the strong impact of tissue contexture on the profile of the antitumor immune response. Our data provide a rationale for a diverse approach to targeted therapy in patients with OPSCC and CESC. Although immunologically hot OPSCCs may benefit from a combination of HPV therapeutic vaccines and immune checkpoint blockade, MDSC-rich CESCs may benefit from a combination of HPV vaccines with MDSC-depleting chemotherapy or inhibitors of MDSC activity/recruitment.
Funding
The major sponsor of this study was Sotio a.s. JL and AR were supported by the project BBMRI-CZ LM2023033, by the European Regional Development Fund-Project BBMRI-CZ No: EF16_013/0001674, and by the Cooperatio Program, research area DIAG. The study was partly supported by the COOPERATIO program and Ministry of Health of the Czech Republic (Grants NU21-03-00273 and NU21-08-00280).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics approval
All patient studies were approved by the Ethics Committee of the University Hospital Hradec Králové, the Ethics Committee of the University Hospital Královské Vinohrady, and the Ethics Committee of the University Hospital Motol. All patients from prospective cohorts signed informed consent forms prior to inclusion in the study, which was approved by the Ethics Committee of the Motol University Hospital, Prague, Czech Republic and the Ethics Committee of the University Hospital Královské Vinohrady, Prague, Czech Republic. All procedures performed in this study were in accordance with the 1964 Declaration of Helsinki and its later amendments.
Consent to participate
All patients and healthy donors enrolled in this study signed an informed consent, which was approved by the Ethics Committee of the Motol University Hospital, Prague, Czech Republic (OPSCC patients) and the Ethics Committee of the University Hospital Královské Vinohrady, Prague, Czech Republic (CESC patients).
CRediT authorship contribution statement
Lucie Pavelková: Data curation, Investigation, Formal analysis. Eliška Táborská: Investigation, Methodology. Linn A. Syding: Investigation, Methodology. Klára Plačková: Investigation. Ekaterina Simonova: Software, Visualization, Formal analysis. Kamila Hladíková: Investigation, Methodology. Michal Hensler: Software, Data curation, Visualization, Formal analysis. Jan Laco: Methodology, Resources, Validation. Vladimír Koucký: Data curation, Investigation, Methodology. Michal Zábrodský: Data curation, Supervision. Jan Bouček: Resources, Writing – review & editing, Supervision. Marek Grega: Methodology, Validation, Resources. Kateřina Rozkošová: Investigation, Methodology. Hana Vošmiková: Investigation, Methodology. Michael J. Halaška: Data curation, Resources, Supervision. Lukáš Rob: Resources, Data curation, Supervision. Ivan Práznovec: Data curation. Miroslav Hodek: Data curation, Resources. Milan Vošmik: Supervision, Data curation. Petr Čelakovský: Data curation. Viktor Chrobok: Data curation, Supervision. Aleš Ryška: Supervision, Resources, Validation. Lenka Palová-Jelínková: Supervision, Validation. Radek Špíšek: Conceptualization, Resources, Supervision, Writing – original draft. Anna Fialová: Conceptualization, Data curation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank the nursing and medical staff at the Department of Otorhinolaryngology and Head and Neck Surgery, Motol University Hospital, and at the Department of Obstetrics and Gynecology, University Hospital Kralovske Vinohrady, for their indispensable cooperation in the study.
References
- 1.de Martel C., Plummer M., Vignat J., Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer. 2017;141(4):664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conarty J.P., Wieland A. The tumor-specific immune landscape in HPV+ head and neck cancer. Viruses. 2023;15(6) doi: 10.3390/v15061296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hildesheim A., Schiffman M., Bromley C., Wacholder S., Herrero R., Rodriguez A., et al. Human papillomavirus type 16 variants and risk of cervical cancer. J. Natl. Cancer Inst. 2001;93(4):315–318. doi: 10.1093/jnci/93.4.315. [DOI] [PubMed] [Google Scholar]
- 4.Doorbar J., Quint W., Banks L., Bravo I.G., Stoler M., Broker T.R., et al. The biology and life-cycle of human papillomaviruses. Vaccine. 2012;30(5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. Suppl. [DOI] [PubMed] [Google Scholar]
- 5.Faraji F., Zaidi M., Fakhry C., Gaykalova D.A. Molecular mechanisms of human papillomavirus-related carcinogenesis in head and neck cancer. Microbes Infect. 2017;19(9–10):464–475. doi: 10.1016/j.micinf.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research N, Albert Einstein College of M, Analytical Biological S, Barretos Cancer H, Baylor College of M, Beckman Research Institute of City of H Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nulton T.J., Olex A.L., Dozmorov M., Morgan I.M., Windle B. Analysis of The Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget. 2017;8(11):17684–17699. doi: 10.18632/oncotarget.15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramqvist T., Mints M., Tertipis N., Nasman A., Romanitan M., Dalianis T. Studies on human papillomavirus (HPV) 16 E2, E5 and E7 mRNA in HPV-positive tonsillar and base of tongue cancer in relation to clinical outcome and immunological parameters. Oral Oncol. 2015;51(12):1126–1131. doi: 10.1016/j.oraloncology.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Cabrita R., Lauss M., Sanna A., Donia M., Skaarup Larsen M., Mitra S., et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature. 2020;577(7791):561–565. doi: 10.1038/s41586-019-1914-8. [DOI] [PubMed] [Google Scholar]
- 10.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577(7791):549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petitprez F., de Reynies A., Keung E.Z., Chen T.W., Sun C.M., Calderaro J., et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 12.Mandal R., Senbabaoglu Y., Desrichard A., Havel J.J., Dalin M.G., Riaz N., et al. The head and neck cancer immune landscape and its immunotherapeutic implications. JCI Insight. 2016;1(17):e89829. doi: 10.1172/jci.insight.89829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nasman A., Romanitan M., Nordfors C., Grun N., Johansson H., Hammarstedt L., et al. Tumor infiltrating CD8+ and Foxp3+ lymphocytes correlate to clinical outcome and human papillomavirus (HPV) status in tonsillar cancer. PLoS One. 2012;7(6):e38711. doi: 10.1371/journal.pone.0038711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordfors C., Grun N., Tertipis N., Ahrlund-Richter A., Haeggblom L., Sivars L., et al. CD8+ and CD4+ tumour infiltrating lymphocytes in relation to human papillomavirus status and clinical outcome in tonsillar and base of tongue squamous cell carcinoma. Eur. J. Cancer. 2013;49(11):2522–2530. doi: 10.1016/j.ejca.2013.03.019. [DOI] [PubMed] [Google Scholar]
- 15.Hladikova K., Koucky V., Boucek J., Laco J., Grega M., Hodek M., et al. Tumor-infiltrating B cells affect the progression of oropharyngeal squamous cell carcinoma via cell-to-cell interactions with CD8(+) T cells. J. Immunother Cancer. 2019;7(1):261. doi: 10.1186/s40425-019-0726-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruffin A.T., Cillo A.R., Tabib T., Liu A., Onkar S., Kunning S.R., et al. B cell signatures and tertiary lymphoid structures contribute to outcome in head and neck squamous cell carcinoma. Nat. Commun. 2021;12(1):3349. doi: 10.1038/s41467-021-23355-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enwere E.K., Kornaga E.N., Dean M., Koulis T.A., Phan T., Kalantarian M., et al. Expression of PD-L1 and presence of CD8-positive T cells in pre-treatment specimens of locally advanced cervical cancer. Mod. Pathol. 2017;30(4):577–586. doi: 10.1038/modpathol.2016.221. [DOI] [PubMed] [Google Scholar]
- 18.Ohno A., Iwata T., Katoh Y., Taniguchi S., Tanaka K., Nishio H., et al. Tumor-infiltrating lymphocytes predict survival outcomes in patients with cervical cancer treated with concurrent chemoradiotherapy. Gynecol. Oncol. 2020;159(2):329–334. doi: 10.1016/j.ygyno.2020.07.106. [DOI] [PubMed] [Google Scholar]
- 19.Wang J., Li Z., Gao A., Wen Q., Sun Y. The prognostic landscape of tumor-infiltrating immune cells in cervical cancer. Biomed. Pharmacother. 2019;120 doi: 10.1016/j.biopha.2019.109444. [DOI] [PubMed] [Google Scholar]
- 20.Yang S., Wu Y., Deng Y., Zhou L., Yang P., Zheng Y., et al. Identification of a prognostic immune signature for cervical cancer to predict survival and response to immune checkpoint inhibitors. Oncoimmunology. 2019;8(12) doi: 10.1080/2162402X.2019.1659094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santegoets S.J., van Ham V.J., Ehsan I., Charoentong P., Duurland C.L., van Unen V., et al. The anatomical location shapes the immune infiltrate in tumors of same etiology and affects survival. Clin. Cancer Res. 2019;25(1):240–252. doi: 10.1158/1078-0432.CCR-18-1749. [DOI] [PubMed] [Google Scholar]
- 22.Fialova A., Koucky V., Hajduskova M., Hladikova K., Spisek R. Immunological network in head and neck squamous cell carcinoma-A prognostic tool beyond HPV status. Front. Oncol. 2020;10:1701. doi: 10.3389/fonc.2020.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C., et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 24.Chen H., Xia B., Zheng T., Lou G. Immunoscore system combining CD8 and PD-1/PD-L1: a novel approach that predicts the clinical outcomes for cervical cancer. Int. J. Biol. Markers. 2020;35(1):65–73. doi: 10.1177/1724600819888771. [DOI] [PubMed] [Google Scholar]
- 25.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18(3):197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 26.Someya M., Tsuchiya T., Fukushima Y., Hasegawa T., Hori M., Kitagawa M., et al. Prediction of treatment response from the microenvironment of tumor immunity in cervical cancer patients treated with chemoradiotherapy. Med. Mol. Morphol. 2021;54(3):245–252. doi: 10.1007/s00795-021-00290-w. [DOI] [PubMed] [Google Scholar]
- 27.Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., Vonderheide R.H. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 2007;67(19):9518–9527. doi: 10.1158/0008-5472.CAN-07-0175. [DOI] [PubMed] [Google Scholar]
- 28.Bronte V., Brandau S., Chen S.H., Colombo M.P., Frey A.B., Greten T.F., et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 2016;7:12150. doi: 10.1038/ncomms12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Highfill S.L., Cui Y., Giles A.J., Smith J.P., Zhang H., Morse E., et al. Disruption of CXCR2-mediated MDSC tumor trafficking enhances anti-PD1 efficacy. Sci. Transl. Med. 2014;6(237):237. doi: 10.1126/scitranslmed.3007974. ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Law A.M.K., Valdes-Mora F., Gallego-Ortega D. Myeloid-derived suppressor cells as a therapeutic target for cancer. Cells. 2020;9(3) doi: 10.3390/cells9030561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Umansky V., Blattner C., Gebhardt C., Utikal J. The Role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines. 2016;4(4) doi: 10.3390/vaccines4040036. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kapanadze T., Gamrekelashvili J., Ma C., Chan C., Zhao F., Hewitt S., et al. Regulation of accumulation and function of myeloid derived suppressor cells in different murine models of hepatocellular carcinoma. J. Hepatol. 2013;59(5):1007–1013. doi: 10.1016/j.jhep.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Guillebon E., Dardenne A., Saldmann A., Seguier S., Tran T., Paolini L., et al. Beyond the concept of cold and hot tumors for the development of novel predictive biomarkers and the rational design of immunotherapy combination. Int. J. Cancer. 2020;147(6):1509–1518. doi: 10.1002/ijc.32889. [DOI] [PubMed] [Google Scholar]
- 34.Harlin H., Meng Y., Peterson A.C., Zha Y., Tretiakova M., Slingluff C., et al. Chemokine expression in melanoma metastases associated with CD8+ T-cell recruitment. Cancer Res. 2009;69(7):3077–3085. doi: 10.1158/0008-5472.CAN-08-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ostrand-Rosenberg S., Fenselau C. Myeloid-derived suppressor cells: immune-suppressive cells that impair antitumor immunity and are sculpted by their environment. J. Immunol. 2018;200(2):422–431. doi: 10.4049/jimmunol.1701019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gabrilovich D.I. Myeloid-derived suppressor cells. Cancer Immunol. Res. 2017;5(1):3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gabrilovich D.I., Ostrand-Rosenberg S., Bronte V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012;12(4):253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mabuchi S., Sasano T. Myeloid-derived suppressor cells as therapeutic targets in uterine cervical and endometrial cancers. Cells. 2021;10(5) doi: 10.3390/cells10051073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Molon B., Ugel S., Del Pozzo F., Soldani C., Zilio S., Avella D., et al. Chemokine nitration prevents intratumoral infiltration of antigen-specific T cells. J. Exp. Med. 2011;208(10):1949–1962. doi: 10.1084/jem.20101956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.