Abstract
Purpose:
Myeloproliferative neoplasms (MPN) dysregulate JAK2 signaling. Because clinical JAK2 inhibitors have limited disease-modifying effects, type II JAK2 inhibitors such as CHZ868 stabilizing inactive JAK2 and reducing MPN clones, gain interest. We studied whether MPN cells escape from type ll inhibition.
Experimental Design:
MPN cells were continuously exposed to CHZ868. We used phosphoproteomic analyses and ATAC/RNA sequencing to characterize acquired resistance to type II JAK2 inhibition, and targeted candidate mediators in MPN cells and mice.
Results:
MPN cells showed increased IC50 and reduced apoptosis upon CHZ868 reflecting acquired resistance to JAK2 inhibition. Among >2,500 differential phospho-sites, MAPK pathway activation was most prominent, while JAK2-STAT3/5 remained suppressed. Altered histone occupancy promoting AP-1/GATA binding motif exposure associated with upregulated AXL kinase and enriched RAS target gene profiles. AXL knockdown resensitized MPN cells and combined JAK2/AXL inhibition using bemcentinib or gilteritinib reduced IC50 to levels of sensitive cells. While resistant cells induced tumor growth in NOD/SCID gamma mice despite JAK2 inhibition, JAK2/AXL inhibition largely prevented tumor progression. Because inhibitors of MAPK pathway kinases such as MEK are clinically used in other malignancies, we evaluated JAK2/MAPK inhibition with trametinib to interfere with AXL/MAPK-induced resistance. Tumor growth was halted similarly to JAK2/AXL inhibition and in a systemic cell line–derived mouse model, marrow infiltration was decreased supporting dependency on AXL/MAPK.
Conclusions:
We report on a novel mechanism of AXL/MAPK-driven escape from type II JAK2 inhibition, which is targetable at different nodes. This highlights AXL as mediator of acquired resistance warranting inhibition to enhance sustainability of JAK2 inhibition in MPN.
Translational Relevance.
Myeloproliferative neoplasms (MPN) dysregulate JAK2 signaling, however current type I JAK2 inhibitors provide limited disease-modifying potential. Type II JAK2 inhibitors such as CHZ868 attenuate MPN clonal fitness in preclinical models and are developed towards clinical application. We reveal AXL/MAPK signaling as mediator of acquired resistance to type II JAK2 inhibition. Phosphoproteomic analyses show activated MAPK signaling in MPN cells upon continuous type II JAK inhibitor exposure. Profiling of chromatin accessibility and gene expression reveals upregulated AXL kinase and MAPK target gene patterns. AXL depletion or inhibition with bemcentinib or gilteritinib resensitizes MPN cells. Combined JAK2/AXL inhibition suppresses resistant cell growth in vivo. JAK2/MAPK inhibition with trametinib, clinically used for other malignancies, shows similar efficacy. These observations uncover AXL/MAPK signaling as mediator of escape from type II JAK inhibition, which is targetable at proximal and distal kinase nodes, highlighting the potential of mechanism-based therapies to restore sensitivity to JAK2 inhibition in MPN.
Introduction
Myeloproliferative neoplasms (MPN) are hematologic malignancies, which derive from clonal hematopoietic stem/progenitor cells and show constitutive activation of JAK2 signaling (1). Patients with MPN present with excessive myeloid cell production and increased risk for thrombo-hemorrhagic complications, progression to bone marrow (BM) fibrosis, and transformation to acute myeloid leukemia (2). Somatic driver mutations in JAK2, Calreticulin (CALR), and the thrombopoietin receptor MPL are common in MPN and functionally converge on dysregulated JAK2 signaling (3, 4). Activated JAK2 signaling is also detected in triple-negative MPN without detectable JAK2, CALR, or MPL mutations, which highlights JAK2 activation as a characteristic alteration mediating MPN development (5). Consequently, JAK2 inhibitor treatment represents a clinical standard of care with important benefits for patients with MPN (6). Ruxolitinib and other clinical JAK2 inhibitors improve splenomegaly and constitutional symptoms, which means reduced MPN burden and improved quality of life along with a moderate survival benefit (7–11). However, important limitations have become evident because JAK2 inhibitors have been used. Disease-modifying effects are modest with limited impact on MPN clone size and BM fibrosis while clonal evolution progresses despite JAK inhibitor therapy (12, 13). Response to JAK2 inhibition is lost in a substantial proportion of patients after 1 to 5 years of therapy due to several mechanisms including reactivation of JAK2 signaling, which has been related to molecular adaptations of the JAK signaling network employing alternative JAK family members for activation (10, 14–16). Of note, prognosis after ruxolitinib failure and discontinuation is found to be poor and second-line treatments show moderate responses (6, 13, 14). It has been hypothesized that the limited corrective potential may in part relate to insufficient JAK2 inhibitory activity by type I inhibitor binding, the mechanism of all current clinically used JAK2 inhibitors which blocks the JAK2 kinase in the active, DFG-motif-in confirmation. This notion is in line with observations that genetic ablation of dysregulated JAK2, as evaluated in several inducible JAK2V617F mutant in vivo models, demonstrated thorough correction of MPN phenotypic features as well as resolution of the malignant clone (refs. 17, 18; bioRxiv 2022.05.18.492332). These functional in vivo validations of JAK2 as an essential driver oncogene of MPN cells have reinforced investigations into more effective targeting of activated JAK2 signaling.
Drug discovery efforts have led to the identification of type II JAK2 inhibitors, which stabilize the JAK2 kinase in an inactive, DFG-out conformation, similarly to type II BCR-ABL1 inhibitors, which have proven very effective for disease control in chronic myeloid leukemia (19). Preclinical characterization of the type II JAK2 inhibitor CHZ868 in JAK2-driven MPN and acute leukemia suggested enhanced therapeutic efficacy as compared with type I JAK2 inhibitors with improved correction of cytoses and splenomegaly, reduction of malignant clones, and maintained efficacy upon ruxolitinib failure (20, 21). On the basis of these profiles, type II JAK2 inhibitors have garnered increasing interest and clinical compounds are being developed (22). Given the advent of type II JAK2 inhibitors and based on the profound correction of MPN disease features upon inducible JAK2 ablation (refs. 17, 18; bioRxiv 2022.05.18.492332), it is imperative to further characterize gains and limitations of JAK2 inhibition and study the therapeutic profiles seen with alternative JAK2 inhibitors. Specifically, further insight into escape from JAK2 inhibition mediating acquired resistance is warranted to enhance sustainability of JAK inhibitor therapies in MPN. Here we delineated a novel mechanism of escape from type II JAK2 inhibition which is therapeutically targetable.
Materials and Methods
Inhibitors
JAK2 inhibitors CHZ868, BBT594 (type II), ruxolitinib (type I), and MEK inhibitor trametinib were from Novartis through Material Transfer Agreement; AXL inhibitors bemcentinib and gilteritinib were purchased from BioVision and Sellekchem. CHZ868 was administered by oral gavage at 15 mg/kg every day, bemcentinib at 50 mg/kg twice a day, and trametinib at 0.3 mg/kg every day. Inhibitors for in vitro use were stored at −20°C in DMSO.
Cells and proliferation assays
SET2 cells cultured in RPMI1640/20%FCS were exposed to increasing concentrations of type II JAK2 inhibitor CHZ868 to generate JAK2 inhibitor resistant SET2 cells with gradually increased IC50. JAKi-r SET2 cells were then kept at 0.3 μmol/L CHZ868 and JAKi-R SET2 cells at 0.5 μmol/L of CHZ868 as maintenance concentrations. Ba/F3 cells stably expressing Jak2V617F along with erythropoietin receptor (EPOR) cultured in RPMI1640/10%FCS (20, 23) were analogously exposed to CHZ868 and kept at 0.5 μmol/L CHZ868 maintenance concentration. AXL-deficient JAKi-R SET2 cells were generated by transduction with AXL-specific short hairpin RNAs (shRNA; Supplementary Table S1) in pLKO-Tet-On vector and puromycin selection followed by doxycycline induction. Cell proliferation was assessed upon AXL knockdown or inhibitor exposure for 48 hours using Cell Viability Luminescent Assay (Promega) in JAK inhibitor resistant (JAKi-r, JAKi-R) vs. sensitive (JAKi-S) cells. IC50 was determined with Prism 9.0 (GraphPad). Effects of inhibitor combinations were assessed in R-Studio with SynergyFinder package and Zero Interaction Potency (ZIP) model as described (24, 25). JAKi-R SET2 cells for intravenous engraftment in NOD/SCID gamma (NSG) mice stably expressed firefly luciferase upon transduction with GFP/luciferase as described before (26).
Induction of apoptosis
JAK inhibitor resistant (JAKi-r, JAKi-R) and sensitive (JAKi-S) cells at 106 cells/ml were cultured for 48 hours with increasing inhibitor concentrations, washed in ice-cold PBS, and assessed for apoptosis induction by PE Caspase-3 Apoptosis Kit (BD) on CytoFLEX Flow Cytometer (Beckman Coulter). Data analysis was with FlowJo 10 (BD).
Phosphoproteomics
Cells were lysed in 2 mol/L guanidium hydrochloride, 100 mmol/L ammonium bicarbonate, 5 mmol/L TCEP, and phosphatase inhibitors (Sigma) by sonication (Bioruptor, Diagenode). Proteins were reduced by 10 min incubation at 95°C and alkylated with 10 mmol/L chloroacetamide at 37°C. Samples were diluted with 100 mmol/L ammonium bicarbonate to final guanidium hydrochloride concentration of 0.5 mol/L. Proteins were digested with sequencing-grade modified trypsin (1/50, w/w; Promega) for 12 hours at 37°C. Samples were acidified with 5% TFA and peptides purified using C18 reverse-phase spin columns (Macrospin, Harvard Apparatus) according to the manufacturer's instructions, dried under vacuum, and stored at −20°C until use. Peptide samples were enriched for phosphorylated peptides using Fe(III)-IMAC cartridges on an AssayMAP Bravo platform as described (23). Phospho-enriched peptides were resuspended in 0.1% aqueous formic acid and subjected to LC/MS-MS analysis using an Orbitrap Fusion Lumos Mass Spectrometer fitted with an EASY-nLC 1200 (Thermo Fisher Scientific) and a custom-made column heater set to 60°C. Peptides were resolved using RP-HPLC column (75 μm × 36 cm) packed in-house with C18 resin (ReproSil-Pur C18–AQ, 1.9 μm resin; Dr. Maisch GmbH) at a flow rate of 0.2 μL/min. The gradient used for peptide separation was 5% B to 8% B over 5 minutes to 20% B over 45 minutes to 25% B over 15 minutes to 30% B over 10 minutes to 35% B over 7 minutes to 42% B over 5 minutes to 50% B over 3 minutes to 95% B over 2 minutes, followed by 18 minutes at 95% B. Buffer A was 0.1% formic acid in water and buffer B 80% acetonitrile, 0.1% formic acid in water. The mass spectrometer was operated in DDA mode with cycles of 3 seconds between master scans. Each master scan was acquired in Orbitrap at resolution of 120,000 FWHM (at 200 m/z) and scan range 375 to 1,600 m/z followed by MS2 scans of the most intense precursors in the Orbitrap at resolution of 30,000 FWHM (at 200 m/z) with isolation width of the quadrupole set to 1.4 m/z. Maximum ion injection time was set to 50 milliseconds (MS1) and 54 milliseconds (MS2) with an AGC target set to 250% and “Standard”, respectively. Peptides with charge state 2–5 were included for analysis. Monoisotopic precursor selection was set to Peptide and Intensity Threshold set to 2.5e4. Peptides were fragmented by higher-energy collisional dissociation with collision energy set to 30%. One microscan was acquired for each spectrum. The dynamic exclusion duration was 30 seconds. Acquired raw files were aligned with Progenesis QI (v2.0, Nonlinear Dynamics Limited) and searched with MASCOT. Statistical analysis was performed with SafeQuant R package v.2.3.2 (24).
Immunoblotting
Cells were exposed to inhibitors or DMSO for 4 hours, washed in ice-cold PBS and lysed in presence of Protease Arrest (EMD) and Phosphatase Inhibitor (Calbiochem). Total protein was quantified by Bradford assay (Bio-Rad), separated on 4% to 12% Bis-Tris gels (Invitrogen) and blots probed for pJAK2, JAK2, STAT3, pSTAT3, pSTAT5, pMEK1/2, MEK, pERK1/2, ERK, Actin (Cell Signaling), STAT5 (Santa Cruz), and AXL (R&D). Densitometry of band intensities was with Image-Studio (LI-COR).
RNA sequencing and qRT-PCR
RNA was extracted using NucleoSpin RNA Plus kit (Macherey-Nagel) from JAK2 inhibitor sensitive (JAKi-S) and resistant cells (JAKi-r, JAKi-R), reverse transcribed with high-capacity cDNA reverse transcription (AppliedBiosystems), and RNA quality confirmed by Bioanalyzer RNA6000 Nano kit (Agilent). Sequencing libraries were prepared with TruSeq Stranded mRNA Library Prep kit (Illumina) and sequenced on NovaSeq 6000 (Illumina) on the same run (50 bp, paired end). A detailed RNA sequencing (RNA-seq) protocol and data analysis are provided in the Supplementary Methods. Expression levels were validated by qRT-PCR using gene specific oligos (Supplementary Table S2) and SyBR Green Mix (Applied Biosystems) on ABI7500 Real-Time thermocycler.
Assay for Transposase-Accessible Chromatin using sequencing
For analysis of chromatin accessibility, nuclei from JAK2 inhibitor sensitive (JAKi-S) and resistant cells (JAKi-r, JAKi-R) were isolated in triplicates and nuclear pellets processed with Nextera DNA Library prep kit FC-121–1030 (Illumina) containing Tn5 transposase in TD buffer supplemented with 0.1% Tween-20 and 0.01% digitonin. After 30 minutes at 37°C and 1,000 rpm, DNA was purified with MinElute Clean-up kit (Qiagene) and libraries prepared by PCR amplification for 12 cycles with barcoded primers. Libraries were purified with PCR purification kit (Qiagen) and sequenced on NovaSeq 6000 on the same run (50 bp, paired end). Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) protocol details and analysis steps are provided in the supplement.
Cell line–derived (CDX) mouse models of JAK2 inhibitor resistance
For studies of acquired resistance to JAK2 inhibition in vivo, JAKi-R SET2 cells were engrafted into NSG mice, because duration of treatment to induce acquired resistance in transgenic MPN mouse models is too extensive from the animal welfare perspective. For subcutaneous xenografting, 15×106 JAK2i-R SET2 cells were injected subcutaneously in 200 μL RPMI/0.5 μmol/L CHZ868 at the right flank of NSG mice aged 8 to 9 weeks. Tumor size was monitored by caliper and volume estimated as described [ref. 27; V = 0.5 × (larger diameter) × (smaller diameter)2]. At mean tumor volume of 100 mm3, mice were randomized to treatment groups according to tumor volume and inhibitors administered by oral gavage including CHZ868 15 mg/kg every day, bemcentinib 50 mg/kg twice a day, trametinib 0.3 mg/kg every day, or combinations as compared with vehicle. Mice were treated until the upper limit of tumor volume at 1,500 mm3 was reached in vehicle-treated mice, and sacrificed 2 hours after last dosing. Tumor weights were determined, tumors fixed in 4% buffered formalin and stained for hematoxylin and eosin (H&E) and IHC for human CD45 (hCD45) to confirm tumor composition by JAK2i-R SET2 cells. Alternatively, tumors were dissected, filtered through 100 μm strainers, red cell lysed, and subjected to RNA extraction for expression analyses as described above. To evaluate acquired resistance to JAK2 inhibition in a systemic model, 5×106 luciferase-tagged JAK2i-R SET2 cells were intravenously injected into NSG mice as described (28, 29) and engraftment confirmed on day 7 using bioluminescence imaging with Newton Imager 7.0 (Vilber; Fig. 4) or IVIS Lumina imager (Perkin-Elmer; Fig. 6) upon intraperitoneal injection with D-Luciferin at 150 mg/kg intraperitoneally (Goldbio). At mean bioluminescent signal of 1.75×105 p/s/cm2/sr, mice were randomized into treatment groups according to JAK2i-resistant cell burden. Mice were treated by oral gavage with CHZ868 15 mg/kg every day, trametinib 0.3 mg/kg every day, or combination as compared with vehicle and re-assessed by BLI at 7 days of treatment. Mice were sacrificed 2 hours after last dosing, bones fixed in 4% buffered formalin and stained for H&E and IHC for hCD45. BM infiltration by JAK2i-R SET2 cells was quantified by a specialized hematopathologist blinded to treatment. Images were taken on Olympus BX43 (cellSense 1.6). In addition, a competitive BM transplantation model of Jak2V617F CD45.2 cells transplanted 1:1 with Jak2 WT CD45.1 cells into CD45.1 recipient mice was used as reported before (1, 20, 23, 30). Mice were randomized according to blood counts and treated for 2 weeks with bemcentinib 50 mg/kg twice a day and reduced dose type II JAK2 inhibitor CHZ868 at 15 mg/kg every day. Myelo-erythroid progenitors were assessed by flow cytometry using lineage markers, Sca-1, c-Kit, CD41, CD71, and Ter119 (Biolegend) on LSRFortessa (BD). Allele burden was determined as fraction of CD45.2 cells. Animal care, all animal procedures, and experiments were in strict adherence to Swiss laws for animal welfare and approved by the Swiss Cantonal Veterinary Office of Basel-Stadt.
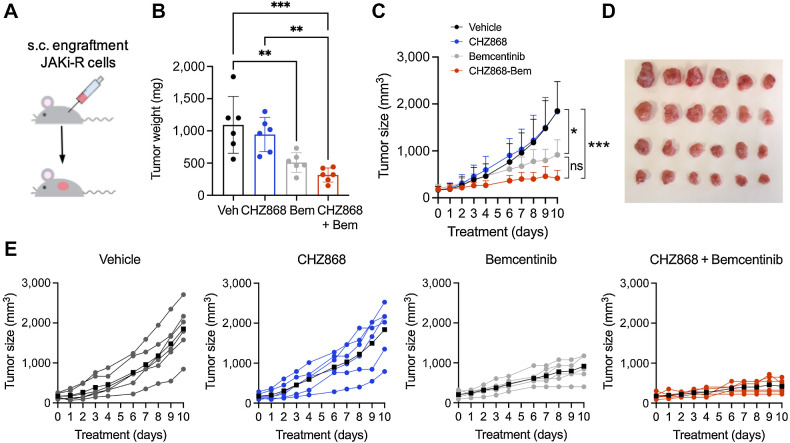
Figure 4.
Resistance to type II JAK inhibition is overcome in vivo by combined JAK2/AXL inhibition. A, JAK2 inhibitor resistant SET2 cells (JAKi-R) were injected subcutaneously into the flank of NSG mice. Animals were treated orally with type II JAK2 inhibitor CHZ868 15 mg/kg every day, AXL inhibitor bemcentinib (bem) 50 mg/kg twice a day, combined CHZ868 15 mg/kg every day/bemcentinib 50 mg/kg twice a day or vehicle control. Treatment was initiated at 100 mm3 tumor size and continued until maximal tumor size was reached in vehicle treated mice. B, Analysis of tumor weight at end of treatment confirmed significant reduction of tumor growth in vivo by AXL inhibition with bemcentinib or by combined JAK2/AXL inhibition with CHZ868/bemcentinib (n = 6/group). C, Tumor size over time showed tumor growth in vehicle-treated mice and analogously in CHZ868-treated mice suggesting resistance to type II JAK2 inhibition in vivo. AXL inhibition by bemcentinib significantly reduced tumor growth, while combined JAK2/AXL inhibition almost suppressed tumor growth (n = 6/group). D, Photographic image of isolated tumors at end of treatment. E, Tumor growth in individual mice is shown for each treatment group. Black lines indicate the average tumor size per group. Combined CHZ868/bemcentinib almost suppressed tumor growth in all the treated mice. Data are presented as mean ± SD and analyzed by one-way ANOVA. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
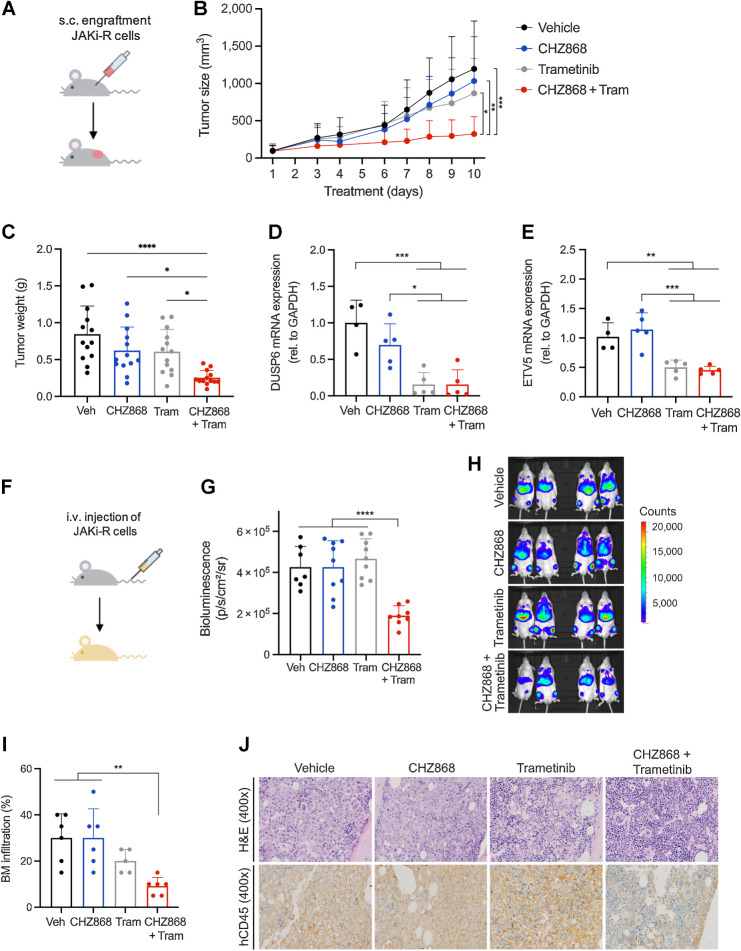
Figure 6.
Targeting MAPK pathway abrogates acquired resistance to type II JAK2 inhibition in vivo. A, Schema of in vivo model with subcutaneous engraftment. JAK2 inhibitor resistant SET2 cells (JAK2i-R) were injected into the flank of NSG mice. Animals were treated orally with type II JAK2 inhibitor CHZ868 15 mg/kg every day, MAPK inhibitor trametinib (tram) 0.3 mg/kg every day, combined CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day, or vehicle control. Treatment was initiated at 100 mm3 tumor size and continued until maximal tumor size was reached in vehicle-treated mice. B, Tumor size over time showed tumor growth in vehicle-treated mice similarly to CHZ868-treated mice. Combined JAK2/MAPK inhibition effectively reduced tumor size (n = 13/group). C, Analysis of tumor weight at end of treatment confirmed significant reduction of tumor growth in vivo by combined JAK2/MAPK inhibition with CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day (n = 13/group). D, Expression of MAPK pathway target DUSP6 was significantly reduced by combined JAK2/MAPK inhibition with CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day (n = 4–5/group). E, Expression of MAPK pathway target ETV5 was significantly reduced by combined JAK2/MAPK inhibition with CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day (n = 4–5/group). F, Schema of in vivo model with intravenous engraftment. JAK2 inhibitor resistant SET2 cells (JAK2i-R) stably expressing luciferase were injected intravenously into NSG mice and engraftment documented by bioluminescent imaging. Animals were treated orally with type II JAK2 inhibitor CHZ868 15 mg/kg every day, MAPK inhibitor trametinib 0.3 mg/kg every day, combined CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day, or vehicle control. G, Bioluminescent signal was significantly reduced in mice treated with combined CHZ868/trametinib at 7 days of treatment (n = 7–9/group). H, Representative bioluminescence images are shown. I, BM infiltration of JAK2i-R cells was determined as human CD45 positivity by IHC on BM sections. A significant reduction of BM infiltration was observed with combined CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day (n = 5–6/group). J, Representative images of BM sections stained with H&E (top) and IHC for hCD45 (lower panel) showed reduced infiltration with combined CHZ868 15 mg/kg every day/MAPK inhibitor trametinib 0.3 mg/kg every day. Original magnification x400. Data are presented as mean ± SD and analyzed by one-way ANOVA. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Patient samples
Collection of blood samples and clinical data from patients with MPN was approved by Ethik Kommission Beider Basel. Written informed consent was obtained from all patients in accordance with the Declaration of Helsinki. MPN was established according to WHO/ICC 2022. For colony formation assays, CD34+ PBMCs were plated at 3,000 cells/well into MethoCult (STEMCELL,#04435) with 0.1 to 0.25 μmol/L CHZ868 and/or 1 to 3 μmol/L bemcentinib or 0.1 to 0.5 μmol/L trametinib. Colony number and subtypes including erythroid (CFU-E, BFU-E), granulocyte-macrophage (CFU-GM), and granulocyte-erythroid-macrophage-megakaryocyte (CFU-GEMM) were scored after 14 days.
Statistical analysis
Unless otherwise specified, data are presented as mean ± SD. Comparisons between 2 groups were performed using unpaired Student t test and multiple comparisons by one-way ANOVA test with Tukey correction. P value ≤ 0.05 was considered significant (ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001). Statistical analyses were performed using GraphPad Prism, v8.0.
Data availability
Sequencing data generated in this study are publicly available in Sequence Read Archive database under the accession number PRJNA922569 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA922569). Mass spectrometry proteomics data have been deposited to ProteomeXchange Consortium via PRIDE (https://www.ebi.ac.uk/pride/archive/projects/PXD039419) partner repository with dataset identifier PXD039419. Other raw data in this study are available upon request from the corresponding author.
Results
MPN cells acquire resistance to type II JAK2 inhibition via MAPK pathway activation
Because loss of response to tyrosine kinase inhibitor therapies has become evident in several malignant settings upon extended exposures including MPN, we subjected JAK2V617F mutant MPN cells to continuous type II JAK2 inhibition with CHZ868 (Supplementary Fig. S1A). When exposed to increasing concentrations of type II JAK inhibitor, SET2 cells showed significantly higher IC50 values for CHZ868 along with reduced susceptibility for apoptosis induction as compared with JAK inhibitor naive cells (JAKi-S) suggesting escape from type II JAK2 inhibition via acquired resistance mechanisms (Fig. 1A and B). Two JAK inhibitor-resistant lines were obtained for molecular characterization and maintained at intermediate concentration of 0.3 μmol/L CHZ868 (JAKi-r) or at a higher concentration of 0.5 μmol/L CHZ868 (JAKi-R), respectively, to investigate the underlying mechanisms of acquired resistance (Supplementary Fig. S1A). Significantly increased IC50 emerged also in Ba/F3 cells stably expressing Jak2V617F and EPOR and in JAK2V617F UKE-1 cells suggesting escape from type II JAK inhibition may occur more generally in JAK2V617F MPN settings (Supplementary Fig. S1B; Supplementary Fig. S2A and S2B). Notably, JAKi-r and JAKi-R SET2 cells were similarly unresponsive to BBT594, a type II JAK2 inhibitor, which has previously been reported (19), and showed significantly and gradually increased IC50 also to type I inhibitor ruxolitinib in line with the notion that response to JAK2 inhibition was lost (Supplementary Fig. S1C and S1D).
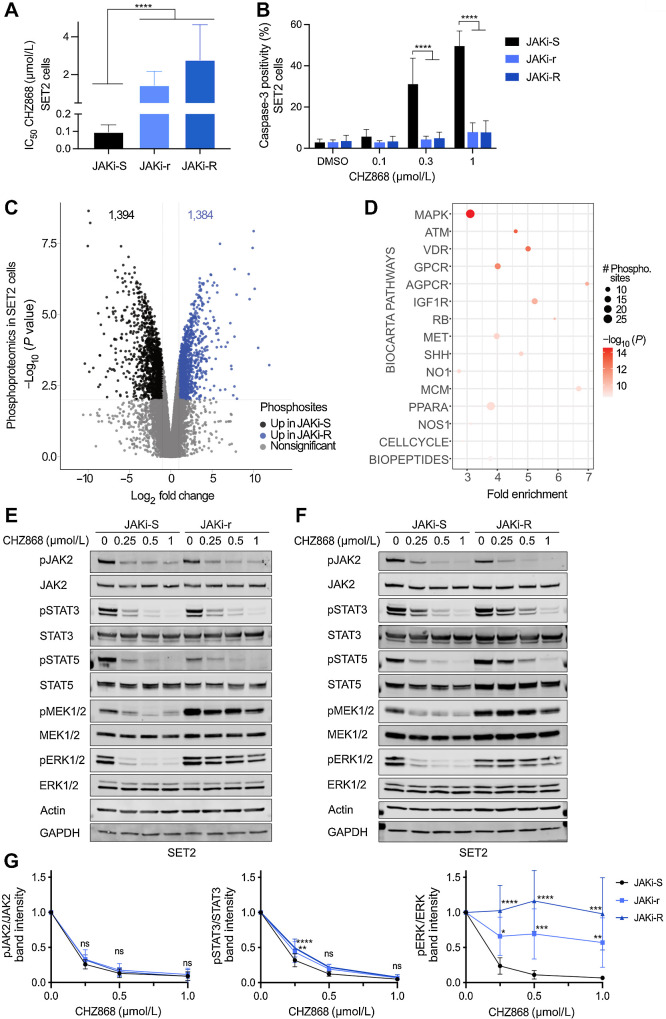
Figure 1.
Acquired resistance to type II JAK2 inhibition occurs via MAPK pathway activation in MPN cells. A, JAK2 V617F mutant SET2 cells upon continuous exposure to the type II JAK2 inhibitor CHZ868 at 0.3 μmol/L (JAKi-r) or at 0.5 μmol/L (JAKi-R) developed significantly increased IC50 for CHZ868 as compared with JAK2 inhibitor sensitive SET2 cells (JAKi-S) reflecting acquired resistance (n = 3–5). B, Susceptibility for apoptosis induction in JAKi-r and JAKi-R cells was significantly reduced as compared with JAK2 inhibitor sensitive SET2 cells (JAKi-S) as reflected by caspase 3 activation upon exposure to CHZ868 for 48 hours (n = 3–4). C, Phosphoproteomic mass spectrometry analysis revealed differentially phosphorylated residues in JAKi-R cells as compared with JAKi-S cells as indicated by Volcano plot (blue: significant upregulation in JAKi-R cells, black: significant upregulation in JAKi-S cells, grey: nonsignificant changes). D, Pathway analysis by PathfindR of differentially phosphorylated sites in JAKi-R as compared with JAKi-S cells highlighted the MAPK pathway as most differentially phosphorylated pathway based on Biocarta gene sets. E and F, Immunoblotting confirmed activated MAPK pathway signaling in JAKi-r (E) and JAKi-R cells (F) in comparison with JAKi-S cells as reflected by pERK1/2 and pMEK1/2 despite exposure to increasing concentrations of type II JAK2 inhibitor CHZ868. JAK2, STAT3, and STAT5 remained inhibited in presence of CHZ868 in JAKi-r and JAKi-R cells similarly to JAKi-S cells. G, Densitometry of phosphoproteins shown in immunoblots confirmed increased ERK1/2 phosphorylation in JAKi-r and JAKi-R cells exposed to type II JAK2 inhibition with CHZ868 whereas JAKi-S cells showed dose-dependent inhibition of pERK (right panel). Densitometry also confirmed dose-dependent suppression of JAK2 and STAT3 phosphorylation upon type II JAK2 inhibition with CHZ868 in JAKi-r and JAKi-R cells similarly to JAKi-S cells (left and middle, n = 7–9). Data are presented as mean ± SD. Comparisons between 2 groups were performed by unpaired Student t test; multiple comparisons were performed by one-way ANOVA test with Tukey correction. P value ≤ 0.05 was considered significant (ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
To explore signaling networks, which underlie MPN cell proliferation escaping JAK2 inhibition, we assessed the phosphoproteome of type II JAK inhibitor resistant cells using mass spectrometry. Differential phosphorylation was seen at >2,500 sites as compared with JAK inhibitor sensitive cells including 1,384 upregulated phospho-residues (Fig. 1C). Pathway analysis revealed most prominent activation in the MAPK signaling pathway with enhanced phosphorylation of ERK, MEK, and BRAF, while negative MAPK regulators as SOS1 showed decreased phosphorylation compared with JAK inhibitor sensitive cells (Fig. 1D; Supplementary Fig. S1E). Phospho-specific immunoblotting confirmed activation of MAPK pathway components MEK and ERK in cells with acquired resistance to JAK2 inhibition, which was maintained upon exposure to increasing concentrations of type II JAK2 inhibitor (Fig. 1E–G; Supplementary Fig. S1F). JAK2-STAT3/5 phosphorylation remained responsive to JAK inhibition in JAKi-r and JAKi-R cells showing concentration-dependent inhibition analogous to sensitive cells. This was concordant with the absence of second site mutations in JAK2 suggesting acquired resistance to type II inhibition is JAK2 independent (Fig. 1E–G; Supplementary Fig. S1F). Jak2V617F EPOR Ba/F3 and UKE-1 cells showed analogous patterns (Supplementary Fig. S1G; Supplementary Fig. S2C) highlighting MAPK pathway activation as potential mediator of escape from JAK inhibition.
MAPK pathway signatures in JAK2 inhibitor resistance relate to upregulated AXL kinase and AP-1/GATA motif exposure
We next sought to investigate the transcriptional profiles of JAK inhibitor resistant cells to characterize the alterations which instruct changes of the phospho-proteome leading to acquired resistance. Analysis of the top 500 differentially expressed genes in JAK inhibitor resistant vs. sensitive cells as assessed by RNA-seq showed JAKi-r and JAKi-R cells clustering together with gradually enhanced effects in JAKi-R cells (Fig. 2A). Principal component analysis confirmed gradual separation of JAKi-r and JAKi-R from JAKi-S cells in PC1, while JAKi-r and JAKi-R cells additionally differed in PC2 (Supplementary Fig. S3A). Concordant with enhanced MAPK pathway activation on proteomic level, transcriptional profiling revealed upregulated RAS isoform expression and upregulation of RAS targets by gene set enrichment analysis (GSEA) in resistant cells, while enhanced STAT signaling was confirmed in JAK inhibitor sensitive cells (Fig. 2A and B). Of note, we observed increased expression of AXL, a tyrosine kinase known to signal via MAPK pathway (31, 32) as one of the top upregulated genes in type II JAK inhibitor resistant cells (Fig. 2A).
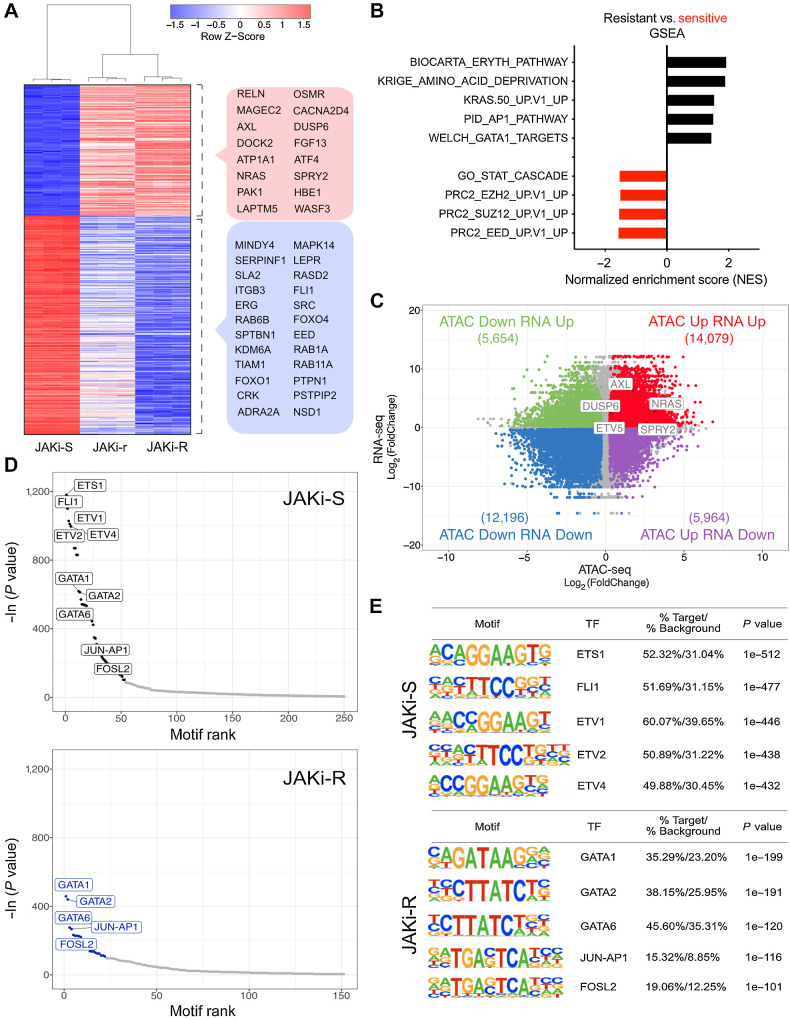
Figure 2.
Transcriptional programs and histone occupancy in acquired resistance to type II JAK2 inhibition. A, Differential expression analysis of JAK inhibitor resistant (JAKi-r and JAKi-R) vs. sensitive (JAKi-S) SET2 cells showed JAKi-r and JAKi-R cells clustered together as opposed to JAKi-S cells. The top 500 differentially expressed genes are displayed and genes with relation to MAPK signaling or epigenetic and transcriptional changes are highlighted (red box: upregulation, blue box: downregulation) including RAS and AXL tyrosine kinase. B, Differentially expressed genes between JAK2 inhibition resistant and sensitive cells were assessed by GSEA. C, RNA-seq and ATAC-seq analysis of paired samples revealed a high number of differentially expressed genes and differentially accessible chromatin regions, respectively, in JAK2 inhibition resistant as compared with sensitive cells (n = 3). Log2(FoldChange) of expression (y-axis) and accessibility (x-axis) is given for JAKi-R versus JAKi-S cells. Number of genes in each quadrant is indicated in brackets. A subset of genes relating to MAPK pathway signaling are highlighted (FDR-adjusted P value < 0.01, Fold Change > 1.2). D and E, Accessible chromatin regions were assessed for the presence of TF binding motifs by HOMER analysis in JAK2 inhibition sensitive (JAKi-S; upper panels) and resistant (JAKi-R; lower panels) cells, highlighting GATA and AP1 complex motifs in JAKi-R cells.
Because GSEA suggested negative enrichment of PRC2 complex components including SUZ12, EZH2, and EED in resistant cells (Fig. 2B), we assessed potential implications of chromatin remodeling on transcriptional profiles. ATAC-seq was performed to characterize differential histone occupancy. When both epigenetic and expression profiles were correlated, we observed a significant shift towards decreased chromatin accessibility along with reduced gene expression at respective loci (Fig. 2C, lower left quadrant; Supplementary Fig. S3B–S3D). Conversely, a similar number of loci revealed increased chromatin accessibility concordant with upregulated gene expression suggesting overall altered histone topology in type II JAK inhibitor resistant settings (Fig. 2C, upper right quadrant; Supplementary Fig. S3B–S3D). To further characterize those regions of increased accessibility, genomic sequences were analyzed for the presence of transcription factor (TF) binding motifs. Accessible chromatin sites of JAK inhibitor sensitive cells were most prominently enriched for motifs recognized by members of the ETS family including ETS1, FLI1, and ETV1, which have been associated with MPN previously (refs. 33, 34; Fig. 2D and E, top). In contrast, these motifs were not as evident in JAK inhibitor resistant cells, which showed most prominently maintained enrichment of AP-1 complex and GATA family motifs consistent with cooperativity of AP-1 and GATA in transcriptional regulation (ref. 35; Fig. 2D and E, bottom). This was also reflected by GSEA with enriched AP-1 and GATA target gene sets in JAK inhibitor resistant cells (Fig. 2B). Notably, chromatin accessibility in the proximal region of the AXL gene was increased and presence of GATA and JUN-AP1 TF binding motifs was observed (Supplementary Fig. S3E–S3F). Collectively, these findings demonstrated substantial histone remodeling upon acquired JAK2 inhibitor resistance with differential compaction of chromatin involving modified PRC2 target regions. While these changes associated with reduced ETS family TF binding motif exposure, AP-1/GATA motifs and respective transcriptional programs were maintained. Given AP-1/GATA family components have previously been implicated in AXL expression (refs. 32, 35, 36; bioRxiv 415752), we hypothesized that enriched binding motif exposure in the accessible chromatin regions related to enhanced AXL and RAS target expression.
Escape from type II JAK2 inhibition is dependent on increased AXL kinase expression and is targetable in MPN cells and cell line–derived mouse models
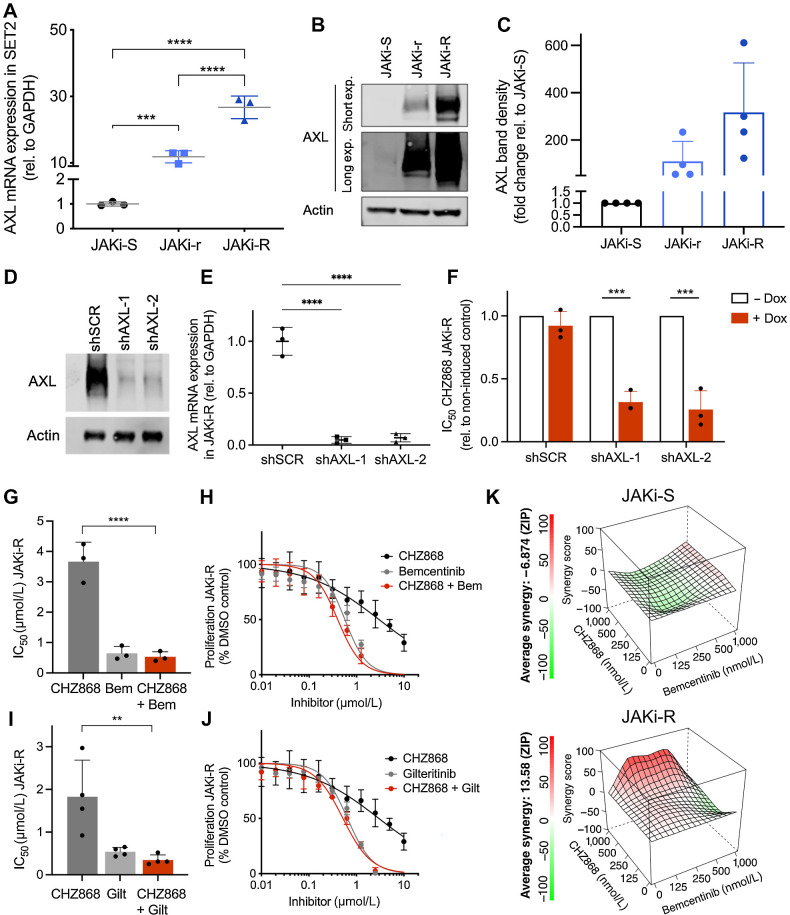
To assess implications of AXL tyrosine kinase for JAK inhibitor resistance, we first determined AXL expression in JAKi-r and JAKi-R cells. We found that AXL levels were significantly increased both when assessed at RNA and protein levels (Fig. 3A–C; Supplementary Fig. S2). We depleted AXL in JAK inhibitor resistant cells using two different, doxycycline-inducible shRNAs (shAXL-1, shAXL-2) to assess for dependencies of cell proliferation and survival on AXL overexpression (Fig. 3D and E). Of note, shRNA-induced AXL deficiency was able to sensitize JAK inhibitor resistant cells to CHZ868 with significantly reduced IC50 suggesting AXL as vulnerability, which should be addressed to overcome acquired resistance to type II JAK inhibition (Fig. 3F; Supplementary Fig. S4A). AXL inhibition with bemcentinib, an AXL specific inhibitor, or AXL/FLT3 inhibitor gilteritinib substantially reduced MPN cell proliferation in JAKi-R cells (Fig. 3G–J). Consistent with the findings upon shRNA-mediated AXL depletion, both bemcentinib and gilteritinib were effective in type II JAK2 inhibitor resistant cells as single agents and in combination with CHZ868 including a certain synergistic effect, which was not seen to the same extent in JAK inhibitor sensitive cells (Fig. 3K). Similar effects of AXL inhibitors and combined JAK2/AXL inhibition were evident in JAKi-r SET2 and UKE-1 cells (Supplementary Fig. S2; Supplementary Fig. S4B–S4E) suggesting a targetable dependency on AXL kinase in MPN models resistant to type II JAK inhibition. AXL expression was not increased in type I JAK2 inhibitor resistant SET2 cells in line with previous studies showing that reactivation of JAK2 signaling underlies type I JAK2 inhibitor resistance (ref. 15; Supplementary Fig. S5).
Figure 3.
Acquired resistance to type II JAK2 inhibition is dependent on AXL tyrosine kinase. A, AXL mRNA expression was significantly increased in JAK inhibitor resistant (JAKi-r and JAKi-R) vs. sensitive cells (JAKi-S) relative to GAPDH (n = 3) consistent with RNA-seq (Fig. 2A). B and C, Increased AXL expression in JAKi resistant cells (JAKi-r and JAKi-R) was confirmed on protein level as indicated by immunoblotting (B) and respective densitometry (C; n = 4). D, Reduced AXL protein expression shown by immunoblotting upon shRNA-induced AXL knock-down with shAXL-1 and shAXL-2 hairpins vs. shSCR control in JAKi-R SET2 cells 48 hours after doxycycline induction. E, Significantly reduced AXL mRNA expression shown by qRT-PCR upon shRNA-induced AXL knock-down with shAXL-1 and shAXL-2 hairpins vs. shSCR control in JAKi-R SET2 cells 48 hours after doxycycline induction (n = 3). F, Significantly increased susceptibility of JAKi-R SET2 cells to type II JAK2 inhibition with CHZ868 as shown by reduced IC50 upon AXL depletion by two different shRNAs (shAXL-1, shAXL-2) upon doxycycline induction (+dox) as compared with shSCR control. Non-induced control (-dox), n = 3. G, Significantly reduced IC50 values in JAKi-R SET2 cells were achieved upon combined exposure with the AXL inhibitor bemcentinib and type II JAK2 inhibition with CHZ868 (n = 3). H, Representative graph showing reduced proliferation of JAKi-R SET2 cells exposed to the AXL inhibitor bemcentinib or combined JAK2 / AXL inhibition with CHZ868 / bemcentinib as compared with CHZ868 as a single agent. I, Significantly reduced IC50 values in JAKi-R SET2 cells were achieved upon combined exposure with the AXL/FLT3 inhibitor gilteritinib and type II JAK2 inhibition with CHZ868 (n = 4). J, Representative graph showing reduced proliferation of JAKi-R SET2 cells exposed to the AXL/FLT3 inhibitor gilteritinib or combined JAK2 / AXL inhibition with CHZ868 / gilteritinib as compared with CHZ868 as a single agent. K, Synergy analysis of type II JAK2 inhibitor CHZ868 with the AXL inhibitor bemcentinib in JAKi-S (upper panel) and JAKi-R (lower panel) SET2 cells showed positive synergy in JAKi-R cells, but not in JAKi-S cells (n = 3, mean values are shown for synergy scores along with representative graphs). Data are presented as mean ± SD and analyzed by one-way ANOVA (panels A, G, I) or two-tailed Student t test (F). ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
To evaluate whether AXL expression would maintain type II JAK inhibitor resistance in MPN in vivo, we engrafted JAKi-R cells subcutaneously into NSG mice (Fig. 4A). Resistance to type II JAK inhibition was confirmed by similar growth of subcutaneous tumors composed of hCD45+ SET2 cells upon treatment with CHZ868 (15 mg/kg every day) or vehicle (Fig. 4B–E; Supplementary Fig. S6). AXL inhibition with bemcentinib (50 mg/kg twice a day) slowed down tumor growth with significantly reduced tumor weights, while dual targeting of JAK2/AXL with CHZ868/bemcentinib prevented relevant tumor growth after initiation of treatment and was tolerable (Fig. 4B–E). In addition, a competitive BM transplantation model of Jak2V617F CD45.2 mouse BM cells transplanted 1:1 with Jak2 WT CD45.1 cells into CD45.1 recipient mice was treated with bemcentinib 50 mg/kg twice a day, which mitigated splenomegaly and reticulocytosis, although not as effective as type II JAK2 inhibition with CHZ868 at a reduced dose of 15 mg/kg every day in this non-resistant model. Notably, combined JAK2/AXL inhibition was most effective in correcting the MPN phenotype and reduced the proportion of Jak2V617F CD45.2 cells in myelo-erythroid progenitor compartments, suggesting AXL targeting may increase sensitivity to type II JAK2 inhibition and enhance a disease-modifying potential (Supplementary Figs. S7–S8).
Suppression of AXL/MAPK activation by MEK inhibition sensitizes resistant MPN cells and cell line–derived mouse models to JAK2 inhibition
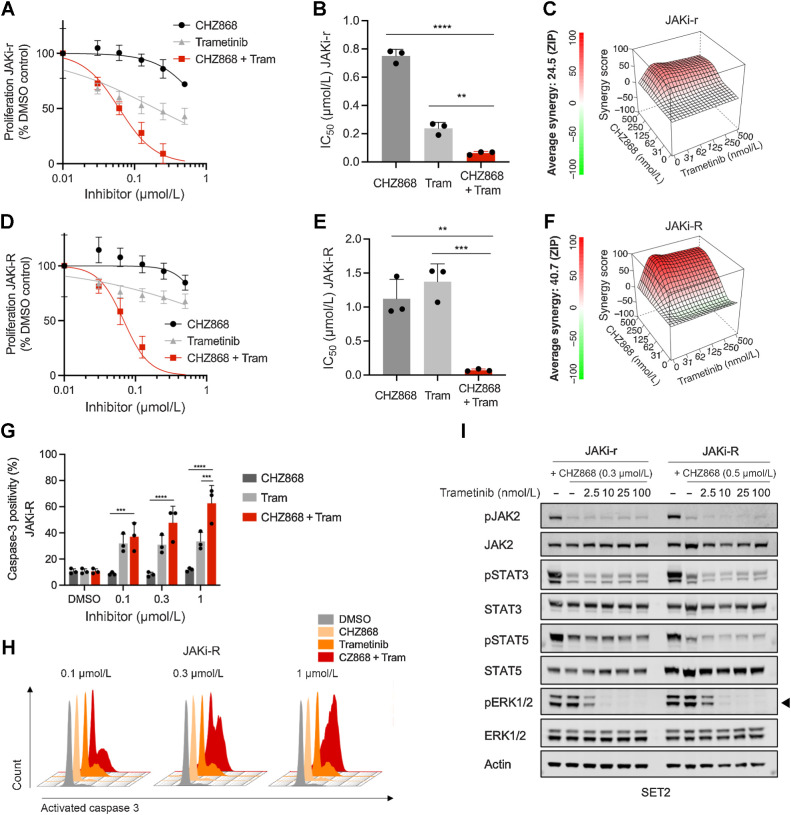
Given the dependency of type II JAK inhibitor resistance on AXL known to signal through the MAPK pathway (31, 32), and evident MAPK activation in resistant MPN cells on RNA and protein levels, we investigated targeting of the MAPK pathway as translationally relevant approach. Inhibitors of MEK1/2, a distal kinase of the MAPK pathway, are approved and in clinical use for several malignancies, which is so far not the case for AXL inhibitors. Thus, MEK inhibition may facilitate clinical application in settings of acquired resistance. Of note, JAKi-r and JAKi-R cells were sensitized to CHZ868 by combined CHZ868/trametinib showing decreased IC50 values (Fig. 5A–F). ZIP scores of 24.5 and 40.7 suggested synergistic effects of JAK2/MAPK inhibition with CHZ868/trametinib with gradual enhancement in JAKi-r and JAKi-R SET2 cells, respectively, highlighting a dependency on MAPK pathway activation similar to the effects seen with JAK2/AXL inhibition. Synergistic effects of CHZ868/trametinib with reduced proliferation was confirmed in JAK2 inhibitor resistant Jak2V617F EPOR Ba/F3 and UKE-1 cells (Supplementary Fig. S2; Supplementary Fig. S9). Also, induction of apoptosis as reflected by caspase-3 positivity was significantly enhanced by combined CHZ868/trametinib (Fig. 5G–H; Supplementary Fig. S9). Immunoblotting of phosphorylated MAPK pathway components showed on-target efficacy of trametinib with concentration-dependent suppression of pERK1/2 in JAK inhibitor resistant cells (Fig. 5I). Thus, interference with AXL as well as with the associated MAPK pathway activation by AXL or MEK inhibition, respectively, was similarly effective and instrumental to sensitize resistant cells to JAK2 inhibition.
Figure 5.
Targeting MAPK pathway abrogates acquired resistance to type II JAK2 inhibition in MPN cells. A, Proliferation of JAKi-r SET2 cells was effectively inhibited by combined JAK2/MAPK pathway inhibition with CHZ868/trametinib as shown by a representative graph of proliferation capacity upon 48 hours exposure of inhibitor exposure. B, IC50 was significantly reduced by combined CHZ868/trametinib in JAKi-r cells (n = 3). C, Synergy analysis of type II JAK2 inhibition with CHZ868 and MAPK pathway inhibition with trametinib showed positive synergy in JAKi-r cells. Mean of n = 3 experiments is shown for synergy score along with a representative graph D. Proliferation of JAKi-R SET2 cells was effectively inhibited by combined JAK2/MAPK pathway inhibition with CHZ868/trametinib as shown by a representative graph of proliferation capacity upon 48 hours of inhibitor exposure. E, IC50 was significantly reduced by combined CHZ868/trametinib in JAKi-R cells (n = 3). F, Synergy analysis of type II JAK2 inhibition with CHZ868 and MAPK pathway inhibition with trametinib showed positive synergy in JAKi-R cells. Mean of n = 3 experiments is shown for synergy score along with a representative graph G. Apoptotic cell death reflected by positivity for caspase-3 was induced by combined JAK2 / MAPK pathway inhibition by CHZ868/trametinib in JAKi-R cells upon 48 hours exposure to inhibitors (n = 3–4). H, Representative histograms of caspase-3 positivity in JAKi-R cells are shown. I, Immunoblotting showed dose-dependent suppression of MAPK pathway signaling reflected by pERK1/2 (arrowhead) in JAKi-r and JAKi-R SET2 cells upon exposure to increasing concentrations of trametinib combined with CHZ868. Data are presented as mean ± SD and analyzed by one-way ANOVA. ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
As MAPK pathway inhibition using MEK inhibitors represents an established clinical approach in several malignancies, we further characterized combined JAK2/MAPK inhibition in vivo in MPN CDX mouse models. Analogously to the studies with JAK2/AXL inhibition, JAK2/MAPK inhibitor treatment with CHZ868 15 mg/kg and trametinib 0.3 mg/kg suppressed tumor progression as reflected by tumor size and weight in NSG mice engrafted with JAK inhibitor resistant cells subcutaneously and was tolerable (Fig. 6A–C; Supplementary Fig. S10A–S10C). In contrast, tumor progression was not significantly mitigated by treatment with CHZ868 as a single agent confirming resistance. Expression of MAPK pathway downstream targets including DUSP6 and ETV6 was significantly decreased upon combined targeting of JAK2/MAPK highlighting sensitization of JAK inhibitor resistant tumor growth (Fig. 6D and E). When AXL and MAPK inhibitors were employed at lower doses of 35 mg/kg bemcentinib and 0.2 mg/kg trametinib along with CHZ868, more subtle effects on tumor growth were seen, while combined inhibitor treatment enhanced corrective effects with comparable tumor reduction as seen upon AXL or MAPK inhibition at full dosage (Supplementary Fig. S11).
For further validation in a systemic CDX mouse model, JAK inhibitor resistant (JAKi-R) cells stably expressing firefly luciferase were injected intravenously into NSG mice and BM engraftment was confirmed by bioluminescent imaging (BLI) and hCD45 IHC (Fig. 6F–J). Whereas CHZ868 as a single agent treatment was unable to reduce BM infiltration, combined JAK2/MAPK inhibition with CHZ868/trametinib significantly reduced MPN cell expansion in vivo as reflected by reduced bioluminescent signal intensity and a notable lack of signal intensity increase over time (Fig. 6H; Supplementary Fig. S10D). Significantly reduced hCD45+ cell infiltration of NSG mouse BM was noted (Fig. 6I and J). Thus, a translational relevance of targeting AXL/MAPK activation was highlighted both in subcutaneous and systemic CDX mouse models of JAK inhibitor resistant MPN cell engraftment.
AXL or MAPK inhibition enhance suppression of myeloid colony outgrowth from primary JAK2V617F patient cells by type II JAK2 inhibition
To further evaluate the translational potential of targeting AXL/MAPK in the setting of type II JAK2 inhibition with CHZ868, we assessed effects of combined CHZ868/bemcentinib or CHZ868/trametinib on myeloid colony outgrowth from primary JAK2V617F MPN patient CD34+ cells (Supplementary Fig. S12). Because type II JAK2 inhibitors have not been approved for clinical application yet, resistant MPN patients’ cells are unavailable and effects evaluated in JAK inhibitor sensitive settings. Bemcentinib and trametinib both enhanced effects of type II JAK2 inhibitor CHZ868 on myelo-erythroid colony outgrowth consistent with the notion of moderate AXL expression and MAPK activation in JAK inhibitor sensitive settings. More pronounced effects may potentially become evident in type II JAK inhibitor resistant conditions given enhanced dependency on more upregulated AXL/MAPK.
Discussion
Given the limited efficacy of current JAK inhibitors, additional approaches to target JAK2 are being explored including type II JAK2 inhibitors (20, 21) and mutant-selective approaches. This relates to the limited disease-modifying capacity of clinical JAK2 inhibitors restricted to modest effects on BM fibrosis, MPN clone size, and clonal evolution (13, 12). An essential role of activated JAK2 as driver oncogene in MPN has been reinforced by several studies demonstrating thorough disease modification and resolution of the MPN clone when activated JAK2 is genetically ablated (refs. 17, 18; bioRxiv 2022.05.18.492332). These reports are consistent with the notion that more effective JAK2 inhibition is required to achieve disease modification in MPN. In contrast to clinical JAK2 inhibitors acting via type I binding mode and blocking JAK2 in active, DFG-loop-in conformation, type II JAK2 inhibitors stabilize JAK2 in an inactive, DFG-loop-out conformation. Studies so far suggested enhanced corrective efficacy in JAK2-driven hematologic neoplasms including MPN (20, 21). Subsequent type II JAK2 inhibitors with improved tolerability are in development for clinical evaluation (22). Thus, it is imperative to further characterize gains and limitations of JAK2 inhibition and to study characteristics of alternative JAK2 inhibitors. Mechanisms of escape mediating acquired resistance to JAK2 inhibition are of particular importance given sustainable effects of JAK inhibitor treatment in MPN represent a significant aim.
Acquired resistance to clinical JAK2 inhibitors, which mediates JAK2 inhibitor failure, has been described in substantial proportions of patients as, e.g., in up to ≈50% of patients with initial response to ruxolitinib over a 5-year period (10, 14). Underlying mechanisms of escape have been characterized including reactivation of JAK2-STAT signaling via JAK family heterodimer formation enabling transactivation of JAK2 through JAK1 and TYK2 (15, 20). In contrast, acquired resistance to type II JAK2 inhibition does not rely on reactivation of JAK2, which remained suppressed. However, alterations of histone occupancy and transcriptional programs associated with increased accessibility of binding sites for AP-1 TFs known to promote AXL expression (refs. 32, 36; bioRxiv 415752), and indeed with AXL overexpression. Our observation that escape from type II JAK2 inhibition is dependent on AXL in MPN cells and cell line–derived mouse models and can be corrected by combined JAK2/AXL inhibition underscores a central role for upregulated AXL as mediator of acquired resistance, which will require validation in humans including trials and clinical practice.
Overexpression of AXL has been implicated in development of resistance in several malignancies and is able to mediate escape from targeted therapies and chemotherapy. As such, upregulated AXL expression has been found in patients with acute myeloid leukemia (AML) progressing on chemotherapy (37) or in acquired resistance to targeted inhibitor therapies in breast and lung cancer as well as head and neck squamous cell carcinoma (32, 38–41). Combination therapy approaches involving AXL inhibition showed a high potential to resolve drug-resistance phenotypes similar to our findings concerning combined JAK2/AXL inhibition in JAK inhibitor resistant MPN. Thus, AXL inhibitors as, e.g., bemcentinib are currently evaluated in clinical studies as single agents or in combinations for several malignancies including lung and pancreatic cancer, chronic lymphocytic leukemia, and others (ref. 42; e.g., NCT03572634, NCT02729298). Given our findings, AXL inhibition also represents a candidate for clinical validation in resistant MPN settings in the future.
AXL has also been detected in treatment-naive MPN cells, and a slightly higher level than in healthy individuals was observed (43). AXL inhibition reduced colony formation from treatment-naive MPN cells, but showed relatively high IC50 values. Substantial concentrations of bemcentinib at, e.g., 2 to 3 μmol/L were required to induce apoptosis in proportions of 20% to 25% of cells in this setting (44). AXL levels were not increased upon short term exposure to type I or type II JAK2 inhibitors in treatment-naive MPN cells (44) in line with the notion that upregulation of AXL represents a mediator of acquired resistance emerging upon extended type II JAK2 inhibitor exposure. This is further highlighted by the observation that in resistance settings, genetic AXL depletion or AXL inhibition by bemcentinib or gilteritinib was effective at low concentrations. Dual JAK2/AXL inhibition was able to resolve resistance phenotypes indicating that upregulated AXL is indeed a targetable vulnerability in type II JAK inhibitor resistance, which is not seen to the same extent in treatment-naive MPN and is warranting clinical validation, first in trials and later in clinical practice.
Because MAPK pathway inhibitors are clinically used for the therapy of other malignancies such as MEK inhibitors, we also evaluated the potential of targeting MAPK pathway activation associated with AXL overexpression in acquired resistance settings. With the MEK inhibitor trametinib we observed similar efficacy of JAK2/MAPK inhibition as seen with JAK2/AXL inhibition in MPN cells and cell line–derived mouse models. This further supports AXL/MAPK upregulation as a novel mechanism of acquired resistance to JAK2 inhibition, which is of translational relevance and should be validated in patients with MPN in the future. Given AXL upregulation is amenable to pharmacologic inhibition, its significance should be evaluated more broadly in MPN as well as in other settings of acquired resistance in hematologic neoplasms.
Supplementary Material
Codilupi et al, Supplementary figures and methods
Acknowledgments
We thank Christian Beisel from Genomics Facility Basel, Department of Biosystems Science and Engineering, ETH Zürich, for technical support, Markus Ackerknecht and Baptiste Hamelin from Mohamed Bentires-Alj laboratory and Nicole Kirchhammer from Albert Zippelius laboratory at the Department of Biomedicine, University Hospital Basel and University of Basel, for technical support.
This work was generously supported by research grants from the Swiss National Science Foundation (PZ00P3_161145, PCEFP3_181357), the Cancer League Basel and the “Stiftung für krebskranke Kinder Regio Basiliensis” (KLbB-4784–02–2019), and the Foundation for the Fight Against Cancer to S.C. Meyer, and by the MSK Cancer Center Support Grant/Core Grant P30 CA008748 to R.L. Levine.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
S. Jungius reports grants from Swiss National Science Foundation (SNSF) during the conduct of the study. A.C. Dunbar reports personal fees from Incyte outside the submitted work. A. Rovo reports grants and personal fees from Novartis AG and Alexion; grants from CSL Behring; and personal fees from Pfizer, OrPhaSwiss GmbH, Blueprint Medicines, Sanofi Aventis AG, and Swedish Orphan Biovitrum AG outside the submitted work. R.L. Levine is a supervisory board member of Qiagen; a member of the board of directors at Ajax Therapeutics (receiving compensation and equity support); is/has been a scientific advisor to Imago, Mission Bio, Syndax, Zentalis, Ajax, Bakx, Auron, Prelude, C4 Therapeutics, and Isoplexis (receiving equity support); has research support from Ajax and AbbVie; has consulted for Janssen; and received honoraria from AstraZeneca and Kura for invited lectures. S.C. Meyer reports grants from Swiss National Science Foundation, Cancer League Basel, Foundation “Stiftung für krebskranke Kinder Regio Basiliensis”, and Foundation for the Fight Against Cancer during the conduct of the study. S.C. Meyer also reports other support from Novartis, Celgene/BMS, GSK, Ajax Therapeutics Inc., OrphaSwiss GmbH, AbbVie AG, and Amgen outside the submitted work; in addition, S.C. Meyer has a patent for PAT058952-US-PSP pending and a patent for PAT058953-US-PSP pending. No disclosures were reported by the other authors.
Authors' Contributions
T. Codilupi: Formal analysis, investigation, visualization, writing–original draft. J. Szybinski: Formal analysis, investigation, visualization, writing–original draft. S. Arunasalam: Formal analysis, investigation, visualization. S. Jungius: Formal analysis, investigation. A.C. Dunbar: Formal analysis, investigation, visualization. S. Stivala: Investigation. S. Brkic: Investigation. C. Albrecht: Investigation. L. Vokalova: Investigation. J.L. Yang: Data curation, Formal analysis. K. Buczak: Investigation. N. Ghosh: Investigation. J.R. Passweg: Resources. A. Rovo: Resources. A. Angelillo-Scherrer: Resources. D. Pankov: Resources. S. Dirnhofer: Investigation. R.L. Levine: Conceptualization, methodology, writing–review and editing. R. Koche: Conceptualization, data curation, formal analysis, methodology. S.C. Meyer: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–review and editing.
References
- 1. Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 2010;17:584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Spivak JL. Myeloproliferative neoplasms. N Engl J Med 2017;376:2168–81. [DOI] [PubMed] [Google Scholar]
- 3. Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014;123:e123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Szybinski J, Meyer SC. Genetics of myeloproliferative neoplasms. Hematol Oncol Clin North Am 2021;35:217–36. [DOI] [PubMed] [Google Scholar]
- 5. Meyer SC, Levine RL. Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res 2014;20:2051–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brkic S, Meyer SC. Challenges and perspectives for therapeutic targeting of myeloproliferative neoplasms. HemaSphere 2021;5:e516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, et al. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med 2012;366:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harrison C, Kiladjian J-J, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, et al. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med 2012;366:787–98. [DOI] [PubMed] [Google Scholar]
- 9. Vannucchi AM, Kiladjian JJ, Griesshammer M, Masszi T, Durrant S, Passamonti F, et al. Ruxolitinib versus standard therapy for the treatment of polycythemia vera. N Engl J Med 2015;372:426–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Verstovsek S, Mesa RA, Gotlib J, Gupta V, DiPersio JF, Catalano JV, et al. Long-term treatment with ruxolitinib for patients with myelofibrosis: 5-year update from the randomized, double-blind, placebo-controlled, phase 3 {COMFORT}-{I} trial. J Hematol Oncol 2017;10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Verstovsek S, Gotlib J, Mesa RA, Vannucchi AM, Kiladjian J-J, Cervantes F, et al. Long-term survival in patients treated with ruxolitinib for myelofibrosis: COMFORT-I and -II pooled analyses. J Hematol Oncol 2017;10:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deininger M, Radich J, Burn TC, Huber R, Paranagama D, Verstovsek S. The effect of long-term ruxolitinib treatment on JAK2p.V617F allele burden in patients with myelofibrosis. Blood 2015;126:1551–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Newberry KJ, Patel K, Masarova L, Luthra R, Manshouri T, Jabbour E, et al. Clonal evolution and outcomes in myelofibrosis after ruxolitinib discontinuation. Blood 2017;130:1125–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Palandri F, Breccia M, Bonifacio M, Polverelli N, Elli EM, Benevolo G, et al. Life after ruxolitinib: reasons for discontinuation, impact of disease phase, and outcomes in 218 patients with myelofibrosis. Cancer 2020;126:1243–52. [DOI] [PubMed] [Google Scholar]
- 15. Koppikar P, Bhagwat N, Kilpivaara O, Manshouri T, Adli M, Hricik T, et al. Heterodimeric JAK–STAT activation as a mechanism of persistence to JAK2 inhibitor therapy. Nature 2012;489:155–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meyer SC. Mechanisms of resistance to JAK2 inhibitors in myeloproliferative neoplasms. Hematol Oncol Clin North Am 2017;31:627–42. [DOI] [PubMed] [Google Scholar]
- 17. Bhagwat N, Koppikar P, Keller M, Marubayashi S, Shank K, Rampal R, et al. Improved targeting of JAK2 leads to increased therapeutic efficacy in myeloproliferative neoplasms. Blood 2014;123:2075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chapeau EA, Mandon E, Gill J, Romanet V, Ebel N, Powajbo V, et al. A conditional inducible JAK2V617F transgenic mouse model reveals myeloproliferative disease that is reversible upon switching off transgene expression. PLoS One 2019;14:e0221635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andraos R, Qian Z, Bonenfant D, Rubert J, Vangrevelinghe E, Scheufler C, et al. Modulation of activation-loop phosphorylation by JAK inhibitors is binding mode dependent. Cancer Discov 2012;2:512–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyer SC, Keller MD, Chiu S, Koppikar P, Guryanova OA, Rapaport F, et al. CHZ868, a type II JAK2 inhibitor, reverses Type I JAK inhibitor persistence and demonstrates efficacy in myeloproliferative neoplasms. Cancer Cell 2015;28:15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu S-C, Li LS, Kopp N, Montero J, Chapuy B, Yoda A, et al. Activity of the type II JAK2 inhibitor CHZ868 in B cell acute lymphoblastic leukemia. Cancer Cell 2015;28:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rai S, Stetka J, Usart M, Hao-Shen H, Park Y, Houston R, et al. The second generation type II JAK2 inhibitor, AJ1–10502, demonstrates enhanced selectivity, improved therapeutic efficacy and reduced mutant cell fraction compared to type I JAK2 inhibitors in models of myeloproliferative neoplasms (MPNs). Blood 2022;140:6722–3. [Google Scholar]
- 23. Brkic S, Stivala S, Santopolo A, Szybinski J, Jungius S, Passweg JR, et al. Dual targeting of JAK2 and ERK interferes with the myeloproliferative neoplasm clone and enhances therapeutic efficacy. Leukemia 2021;35:2875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He L, Kulesskiy E, Saarela J, Turunen L, Wennerberg K, Aittokallio T, et al. Methods for high-throughput drug combination screening and synergy scoring. Methods Mol Biol 2018;1711:351–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-response landscapes using an interaction potency model. Comput Struct Biotechnol J 2015;13:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dao T, Pankov D, Scott A, Korontsvit T, Zakhaleva V, Xu Y, et al. Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat Biotechnol 2015;33:1079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vulin M, Jehanno C, Sethi A, Correia AL, Obradović MMS, Couto JP, et al. A high-throughput drug screen reveals means to differentiate triple-negative breast cancer. Oncogene 2022;41:4459–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dubrovsky L, Brea EJ, Pankov D, Casey E, Dao T, Liu C, et al. Mechanisms of leukemia resistance to antibody dependent cellular cytotoxicity. Oncoimmunology 2016;5:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dubrovsky L, Pankov D, Brea EJ, Dao T, Scott A, Yan S, et al. A TCR-mimic antibody to WT1 bypasses tyrosine kinase inhibitor resistance in human BCR-ABL+ leukemias. Blood 2014;123:3296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stivala S, Codilupi T, Brkic S, Baerenwaldt A, Ghosh N, Hao-Shen H, et al. Targeting compensatory MEK/ERK activation increases JAK inhibitor efficacy in myeloproliferative neoplasms. J Clin Invest 2019;129:1596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dagamajalu S, Rex DAB, Palollathil A, Shetty R, Bhat G, Cheung LWT, et al. A pathway map of AXL receptor-mediated signaling network. J Cell Commun Signal 2021;15:143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scaltriti M, Elkabets M, Baselga J. Molecular pathways: AXL, a membrane receptor mediator of resistance to therapy. Clin Cancer Res 2016;22:1313–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bock O, Hussein K, Neusch M, Schlué J, Wiese B, Kreipe H. Transcription factor Fli-1 expression by bone marrow cells in chronic myeloproliferative disorders is independent of an underlying JAK2 (V617F) mutation. Eur J Haematol 2006;77:463–70. [DOI] [PubMed] [Google Scholar]
- 34. Hodge DR, Li D, Farrar WL, Qi SM. IL-6 induces expression of the Fli-1 proto-oncogene via STAT3. Biochem Biophys Res Commun 2002;292:287–91. [DOI] [PubMed] [Google Scholar]
- 35. Kawana M, Lee ME, Quertermous EE, Quertermous T. Cooperative interaction of GATA-2 and AP1 regulates transcription of the endothelin-1 gene. Mol Cell Biol 1995;15:4225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mudduluru G, Leupold JH, Stroebel P, Allgayer H. PMA up-regulates the transcription of Axl by AP-1 transcription factor binding to TRE sequences via the MAPK cascade in leukaemia cells. Biol Cell 2011;103:21–33. [DOI] [PubMed] [Google Scholar]
- 37. Hong C-C, Lay J-D, Huang J-S, Cheng A-L, Tang J-L, Lin M-T, et al. Receptor tyrosine kinase AXL is induced by chemotherapy drugs and overexpression of AXL confers drug resistance in acute myeloid leukemia. Cancer Lett 2008;268:314–24. [DOI] [PubMed] [Google Scholar]
- 38. Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res 2009;69:6871–8. [DOI] [PubMed] [Google Scholar]
- 39. Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Elkabets M, Pazarentzos E, Juric D, Sheng Q, Pelossof RA, Brook S, et al. AXL mediates resistance to PI3Kα inhibition by activating the EGFR/PKC/mTOR axis in head and neck and esophageal squamous cell carcinomas. Cancer Cell 2015;27:533–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Luthar N, et al. AXL mediates resistance to cetuximab therapy. Cancer Res 2014;74:5152–64. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 42. Ludwig KF, Du W, Sorrelle NB, Wnuk-Lipinska K, Topalovski M, Toombs JE, et al. Small-molecule inhibition of Axl targets tumor immune suppression and enhances chemotherapy in pancreatic cancer. Cancer Res 2018;78:246–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pearson S, Blance R, Somervaille TCP, Whetton AD, Pierce A. AXL inhibition extinguishes primitive JAK2 mutated myeloproliferative neoplasm progenitor cells. HemaSphere 2019;3:e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Beitzen-Heineke A, Berenbrok N, Waizenegger J, Paesler S, Gensch V, Udonta F, et al. AXL inhibition represents a novel therapeutic approach in BCR-ABL negative myeloproliferative neoplasms. HemaSphere 2021;5:e630. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Codilupi et al, Supplementary figures and methods
Data Availability Statement
Sequencing data generated in this study are publicly available in Sequence Read Archive database under the accession number PRJNA922569 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA922569). Mass spectrometry proteomics data have been deposited to ProteomeXchange Consortium via PRIDE (https://www.ebi.ac.uk/pride/archive/projects/PXD039419) partner repository with dataset identifier PXD039419. Other raw data in this study are available upon request from the corresponding author.