Abstract
Purpose:
Pancreatic ductal adenocarcinoma (PDAC) trials have evaluated CTLA-4 and/or PD-(L)1 blockade in patients with advanced disease in which bulky tumor burden and limited time to develop antitumor T cells may have contributed to poor clinical efficacy. Here, we evaluated peripheral blood and tumor T cells from patients with PDAC receiving neoadjuvant chemoradiation plus anti–PD-1 (pembrolizumab) versus chemoradiation alone. We analyzed whether PD-1 blockade successfully reactivated T cells in the blood and/or tumor to determine whether lack of clinical benefit could be explained by lack of reactivated T cells versus other factors.
Experimental Design:
We used single-cell transcriptional profiling and TCR clonotype tracking to identify TCR clonotypes from blood that match clonotypes in the tumor.
Results:
PD-1 blockade increases the flux of TCR clonotypes entering cell cycle and induces an IFNγ signature like that seen in patients with other GI malignancies who respond to PD-1 blockade. However, these reactivated T cells have a robust signature of NF-κB signaling not seen in cases of PD-1 antibody response. Among paired samples between blood and tumor, several of the newly cycling clonotypes matched activated T-cell clonotypes observed in the tumor.
Conclusions:
Cytotoxic T cells in the blood of patients with PDAC remain sensitive to reinvigoration by PD-1 blockade, and some have tumor-recognizing potential. Although these T cells proliferate and have a signature of IFN exposure, they also upregulate NF-κB signaling, which potentially counteracts the beneficial effects of anti–PD-1 reinvigoration and marks these T cells as non-productive contributors to antitumor immunity.
Translational Relevance.
We performed single-cell transcriptomics and TCR clonotype tracking in paired blood and tumor samples from patients with resectable pancreatic ductal adenocarcinoma (PDAC) who received neoadjuvant chemoradiation with or without PD-1 blockade in the context of a phase II trial. PD-1 blockade induces CD8 T-cell expansion in the blood, and some of these T-cell clonotypes are also found in the tumor, suggesting tumor specificity. These results suggest that PDAC elicits an endogenous T-cell response that is amenable to reinvigoration by PD-1 blockade; however, the reinvigorated T cells show a signature of NF-κB signaling that is associated with poor response. In patients with PDAC, PD-1 blockade with chemoradiation, compared with chemoradiation alone, leads to improved tumor-specific T-cell responses observable in peripheral blood but does not elicit a fully protective signature in the reinvigorated T cells.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a recalcitrant tumor type with a 5-year survival of 11% (1). Approximately 80% of patients are diagnosed with advanced or locally advanced disease, making them ineligible for surgical removal. Of the patients who present with resectable or borderline resectable disease, post-surgical outcomes are still poor, with more than half of patients experiencing disease recurrence within the first two years (2–5). Recurrence can be local or distant, with distant metastases occurring most frequently in liver, peritoneum, and lung. The rapid recurrence of distal metastases suggests that although these patients presented with local tumors, they already had micrometastatic disease with tumor cells disseminated before or at the time of surgery (6). Standard practice for borderline resectable PDAC now includes neoadjuvant combination chemotherapy with or without radiotherapy as a means of potentially clearing tumor cells from undetectable metastatic sites (3). The two most commonly used neoadjuvant chemotherapy regimens are FOLFIRINOX or gemcitabine/n(ab)paclitaxel, with similar outcomes for overall survival (OS; refs. 2, 7).
Immunotherapy has transformed the practice of oncology (8). Checkpoint blockade antibodies, notably those targeting PD-1 or its ligand PD-L1, induce reinvigoration of exhausted CD8 T cells. In patients who have a pool of tumor-specific T cells, checkpoint blockade can lead to tumor control and in some cases, long-term durable remissions. Two key features of antitumor immunity are that antitumor T cells can survey distant tissues providing whole-body level surveillance, and that T cells form life-long memory. Both features would be advantageous for patients with resectable PDAC, where induction of a memory T-cell response could potentially clear disseminated tumor cells and lead to long-term survival (9, 10). In a promising study comparing short- and long-term survival after surgical removal of PDAC, long-term survival was correlated with the presence of tumor-specific memory CD8 T cells in blood that persisted for life (11).
Unfortunately, clinical trials for checkpoint blockade have proven largely unsuccessful in patients with advanced PDAC (12). In a phase II trial of durvalumab (anti–PD-L1) plus tremelimumab (anti–CTLA-4) versus durvalumab monotherapy in patients previously treated with chemotherapy, response rates were 3.1% versus 0% (13). Addition of radiotherapy to checkpoint blockade in second-line metastatic PDAC resulted in a similarly low-response rate of 5.1% (14). Even in the 1% of patients with PDAC who have MSI-high disease for which PD-1 blockade is approved, response rates are 19%, well below that of other MSI-high cancers (15). Several non-mutually exclusive factors could account for the poor responsiveness of PDAC to checkpoint blockade and other T-cell–focused strategies. These include: (i) Effects of concurrent chemotherapies; (ii) Testing immunotherapy in the metastatic setting where there is a limited time to generate a meaningful immune response; (iii) Lack of tumor-specific T cells available for reactivation by checkpoint blockade, potentially due to profound defects in naïve T-cell priming; (iv) Other factors in the tumor microenvironment that preclude effective T-cell responses. Late-stage PDAC progresses rapidly and is complicated by significant morbidity (3, 16). To determine whether PD-1 blockade would be more successful when administered at an earlier stage of disease, we conducted a phase II trial of capecitabine and radiotherapy (CRT) with or without pembrolizumab (anti–PD-1) in patients with resectable or borderline resectable PDAC (NCT02305186). Clinical results, reported separately, suggest that addition of PD-1 blockade was safe, but did not improve OS, nor did it increase the frequency of tumor-infiltrating CD8 T cells. Thus, moving anti-PD1 to localized rather than metastatic disease was inadequate to provide clinical benefit.
We therefore hypothesized that either tumor-specific T cells capable of reactivation with PD-1 blockade are not present in patients with pancreatic cancer, or that other factors impede successful antitumor immunity. To distinguish between these two possibilities and to determine whether PD-1 blockade had any measurable impact on T-cell responses in patients with pancreatic cancer, we evaluated peripheral blood and tumor samples from patients in the trial by single-cell transcriptional profiling and TCR clonotype tracking (10). These results suggest that PD-1–expressing exhausted T cells exist in patients with pancreatic cancer, and at least some of these are likely to be tumor-reactive. We confirm that PD-1 blockade can induce T-cell reactivation as defined by increased proliferation and IFN response. However, reactivated T cells from patients with PDAC show a marked signature of NF-κB activation, unlike T cells from patients with other GI malignancies who mount a productive response to PD-1 blockade. We therefore conclude that, although patients with pancreatic cancer have T cells that can expand in response to checkpoint blockade, the quality of response elicited is not sufficient to provide protective immunity.
Materials and Methods
Human samples
Human samples from this study were obtained from clinical trial NCT02305186. The study was conducted at 6 sites: University of Virginia, Dana-Farber Cancer Institute, The University of Texas MD Anderson Cancer Center, Mayo Clinic, Hartford Healthcare Cancer Center, and University of Miami. Written informed consent was provided by all study participants, and the protocol was approved by the relevant IRB at each site. The study was conducted in accordance with the Declaration of Helsinki and the International Ethical Guidelines for Biomedical Research involving Human Subjects. Inclusion criteria comprised the presence of resectable or borderline resectable PDAC as classified according to the Alliance for Clinical Trials in Oncology criteria, with measurable disease identified by the RECIST version 1.1 guidelines and an Eastern Cooperative Oncology Group performance score of 0 or 1. Exclusion criteria comprised the presence of metastatic disease; administration of immunosuppressive therapy within 7 days of the first trial treatment; and prior surgery, chemotherapy, targeted small-molecule therapy, immunotherapy, or radiotherapy for pancreatic cancer. Both sexes were enrolled.
Blood sample preparation
Peripheral blood monocytic cells (PBMC) were isolated from the blood of patients with pancreatic cancer enrolled in a multicenter clinical trial (NCT02305186) using Ficoll gradient centrifugation. They were stored at −80°C until the time of processing. On the day of preparation for sequencing, the samples were stained with the Zombie NIR live/dead stain (BioLegend #423105) and the following fluorophore-conjugated antibodies: Anti-CD3 PE-CF594 (clone UCHT1; BD Biosciences #562280; RRID:AB_11153674), anti-CD4 PacificBlue (clone OKT4; BioLegend #317429; RRID:AB_1595438), and anti-CD8 BV711 (clone SK1; BioLegend #344734; RRID:AB_2565243). In addition, a “hashtag” antibody bound to a unique DNA barcode was added to each sample separately during staining to allow for downstream multiplexing [BioLegend TotalSeq-C0251 (RRID:AB_2801031), -C0252 (RRID:AB_2801032), -C0253 (RRID:AB_2801033), and -C0254 (RRID:AB_2801034)]. Samples were then sorted on a BD FACS Aria II SORP machine, selecting for live CD3+ CD4+ and live CD3+ CD8+ cells separately. The two sorted populations were mixed at equal ratios to create one single-cell suspension for each blood sample.
Tumor sample preparation
Tumor tissue was obtained from patients enrolled in the trial described above at the time of surgical resection and mechanically disrupted into 1-mm pieces using a blade while on ice in RPMI media. They were then enzymatically digested for 30 minutes in a 37°C water bath using a tumor dissociation kit (Miltenyi Biotech #130–095–929). The samples were passed through 40-μm mesh filters, washed with PBS, and cryopreserved. Frozen tumor-infiltrating cells were thawed and stained with the Zombie NIR live/dead stain (BioLegend #423105) and the fluorophore-conjugated antibody anti-CD45 PacificBlue (clone HI30; BioLegend #304029; RRID:AB_2174123). Samples were then sorted on a BD FACS Aria II SORP machine, selecting for live CD45+ cells.
Single-cell RNA sequencing
Single-cell RNA sequencing (scRNA-seq) was performed as previously described (10). Briefly, sorted single-cell suspensions prepared from each sample were washed twice with 0.05% UltraPure BSA (Invitrogen #AM2618) in PBS. For PBMC samples, 6,000 cells were loaded into a 10x Chromium controller instrument along with Chromium Next GEM Single Cell 5′ beads (10x Genomics PN-1000263). Up to four PBMC samples were multiplexed together after being tagged with unique DNA-barcoded antibodies as described above. For tumor-infiltrating lymphocyte (TIL) samples, all sorted cells were loaded, and no multiplexing was performed. After RT-PCR, cDNA was purified, and a library was constructed from each sample using a 10x Library Construction Kit (10x Genomics PN-1000190) following the standard 10x protocol. An additional VDJ-enriched library was created for each sample using a specialized Chromium Single Cell Human TCR Amplification Kit (PN-1000252). Libraries were then sequenced on an Illumina NovaSeq system operated by Azenta/Genewiz generating paired-end 150bp reads.
Data analysis
The 10x CellRanger “multi” pipeline (v6.0.1; RRID:SCR_023221) was used to align reads to the GRCh38 reference genome and generate a single-cell feature count matrix for each library using default parameters. The count matrices were imported for downstream analysis into R using the “Seurat” package (v4.0.3; RRID:SCR_016341). Empty droplets were removed using the “DropletUtils” package (v1.8.0) initially by defining them as any droplet from which less than 100 unique RNA molecules were sequenced, then by selecting only droplets whose RNA content was significantly dissimilar to those considered definitively empty at an FDR-adjusted P value threshold of 0.01. Genes with zero expression across all cells were discarded from further analysis. For PBMC samples, cells with more than 20% mitochondrial reads were discarded. For tumor samples, a 30% cutoff value was used. Data from each sample were log-normalized and combined into one batch-corrected expression matrix by Canonical Correlation Analysis (RRID:SCR_021745). Counts were then scaled and subjected to dimensionality reduction using Principal Component Analysis (PCA; RRID:SCR_014676). Uniform Manifold Approximation and Projection (RRID:SCR_018217) embedding was generated from the top 30 dimensions of the PCA for PBMC samples and 25 dimensions for tumor samples. Clusters were identified first by constructing a Shared Nearest Neighborgraph based on each cell's k-nearest neighbors (k = 15 for PBMCs, k = 10 for TILs) and then applying the Smart Local Moving algorithm to the graph. Markers for each cluster were identified by comparing gene expression using Model-based Analysis of Single-cell Transcriptomics (MAST; RRID:SCR_016340). Potentially viral-reactive clonotypes were identified by cross-referencing CDR3β amino acid sequences against the VDJdb (version 3/30/22; RRID:SCR_014356), an annotated database of known viral TCR sequences (17).
Clonotype identification and tracking
The following steps were performed separately for each patient. First, the TCR contigs sequenced in the pre- and posttreatment sample were collapsed into one list to allow for clonotype assignment independent of time. The list was filtered to eliminate any nonproductive rearrangements and sequences obtained from cells that did not pass quality control, as described above. Then, a tentative clonotype was assigned to each unique combination of 1 to 4 TCR chains that co-occurred in one cell; a TCR chain was defined as the productive combination of a V gene, D gene (for β chains only), J gene, and complementarity determining region 3 (CDR3). Clonotypes were finalized by reassigning any cell if its TCR chain set formed a subset of another clonotype. A clonotype was considered to be shared between the blood and the tumor sample of a given patient if at least one match occurred in the α-chain or the β-chain CDR3 amino acid sequence.
Gene set enrichment analysis
The hallmark (H) gene sets were downloaded from the Molecular Signatures Database (MSigDB; RRID:SCR_016863). Gene Set Enrichment Analysis (GSEA) was performed using the R package “fgsea” (v1.14.0; RRID:SCR_020938) with default parameters, producing Benjamini–Hochberg-adjusted P values.
Comparative analysis with Griffiths and colleagues 2020
The single-cell RNA count matrix was obtained from GEO under the accession GSE130157. Metadata were also downloaded describing each cell's assigned cluster in the original work and each patient's relevant clinical background. We selected the subset of the data corresponding to the responder patient with PDAC identified as “TH8LA0” and only those cells designated as belonging to the “T_cell” supercluster.
Statistical analysis
For differential expression analysis, testing was performed using MAST as described above, and P values were adjusted for multiple hypothesis testing using the Bonferroni method. All other reported P values are the results of nonparametric Mann–Whitney tests. Statistical analysis was done in R (4.0.2).
Data availability
The sequencing data files generated in this study are not publicly available in keeping with the limitations imposed by patient consent and privacy concerns. Raw, unnormalized, and unfiltered, RNA counts for all sequenced droplets as well as raw TCR contigs are provided as online Supplementary Information. Additional data requests may be directed to the corresponding author.
Results
We performed scRNA-seq on T cells sorted from banked PBMC samples obtained from patients enrolled in a clinical trial of chemoradiation ± anti-PD1, with three patients treated on the CRT-only arm and three patients on the CRT + anti-PD1 arm for whom paired PBMCs were available (Fig. 1A and Table 1). Blood samples had been drawn immediately before the start of treatment and again at the end of neoadjuvant treatment. In addition to sequencing standard libraries that randomly sample each cell's transcriptome, we prepared libraries enriched for TCRα/β transcripts. This approach, often referred to as single-cell TCR sequencing (scTCR-seq), allowed us to construct the full TCR sequence of each cell, which then served as a barcode to track the fate of that clone over the treatment period (9, 10, 18, 19).
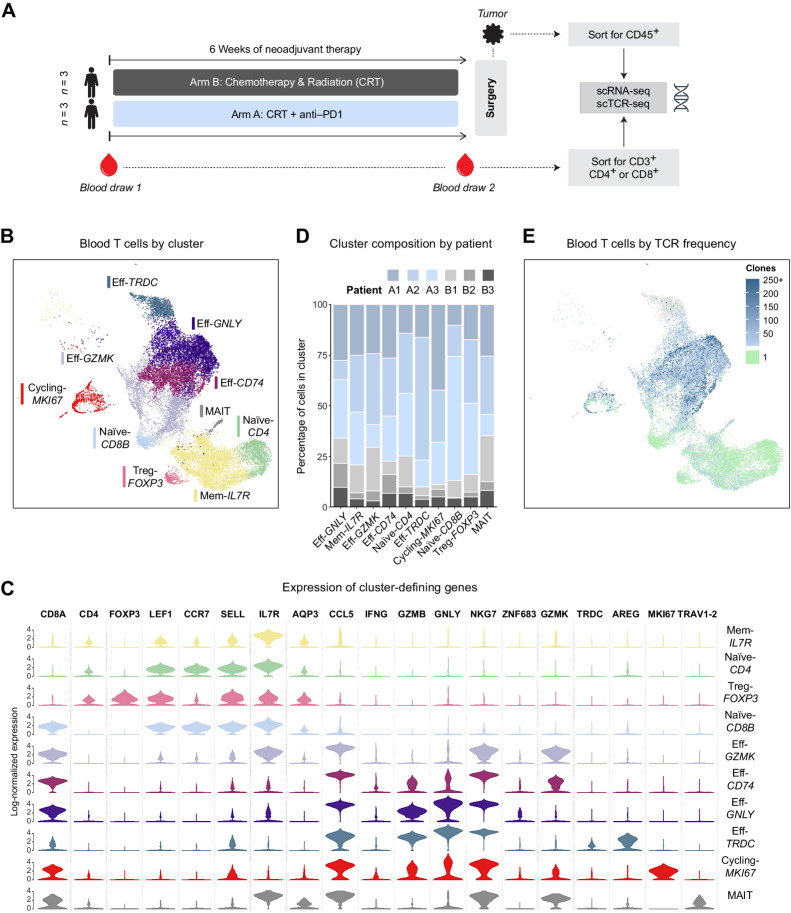
Figure 1.
Circulating T cells were analyzed by scRNA-seq before and after neoadjuvant PD-1 blockade combined with chemoradiotherapy in patients with pancreatic adenocarcinoma. A, Schema of clinical trial design and sample analysis. B, Uniform Manifold Approximation and Projection (UMAP) plot of all sequenced blood T cells that passed quality controls colored by an unsupervised clustering algorithm. C, Violin plot showing the log-normalized expression of key cluster-defining genes for each T-cell cluster. D, Bar plot showing the breakdown of each cluster's cells by patient of origin. E, UMAP of blood T cells where each cell is colored by the observed frequency of its TCR in the sample. Singleton clones are colored green; gray indicates no TCR was detected.
Table 1.
Patient sample information.
| Identifier | Tumor resectability | Treatment | Tumor analyzed | Blood analyzed | Tumor gradea | Pathologic response |
|---|---|---|---|---|---|---|
| A1 | Borderline resectable | Anti-PD1 + CRT | Yes | Yes | G3 | None |
| A2 | Borderline resectable | Anti-PD1 + CRT | Yes | Yes | G3 | Partial |
| A3 | Borderline resectable | Anti-PD1 + CRT | Yes | Yes | Other | Positive |
| B1 | Borderline resectable | CRT only | No | Yes | G3 | None |
| B2 | Resectable | CRT only | No | Yes | G2 | Partial |
| B3 | Resectable | CRT only | No | Yes | Other | None |
| B4 | Borderline resectable | CRT only | Yes | No | G2 | Partial |
| B5 | Resectable | CRT only | Yes | No | G3 | None |
| B6 | Resectable | CRT only | Yes | No | G2 | Partial |
Note: Clinical characteristics of analyzed patient cohort.
aTumors are graded histologically as G2, moderately differentiated, and G3, poorly differentiated.
In total, after applying quality control filters, our PBMC sequencing yielded approximately 20,000 cells across the six analyzed patients. The cells were roughly evenly distributed between the two time points (Supplementary Fig. S1A and S1B). We successfully identified key phenotypes of circulating T cells, including naïve, memory, early activated, several populations of effector, and cycling cells (Fig. 1B and C; Supplementary Fig. S3A). All clusters contained cells from each of the analyzed patients, and no patient-specific clusters were found, reflecting appropriate integration of the dataset to minimize patient-specific variation (Fig. 1D). We also succeeded in assigning a TCR clonotype to 85% of sequenced cells (Fig. 1E), spanning a total of about 9,000 unique clonotypes. Of these, most (88%) were singleton clonotypes composed of only one cell and generally localized to the Naive-CD4 and Naive-CD8B clusters, as expected. The highest degree of clonal expansion was observed in the effector clusters Eff-GNLY and Eff-CD74 and predominantly represented CD8 clonality, with CD8 T cells accounting for 86% of highly expanded (10+) clonotypes.
In addition to PBMCs, we obtained tumor samples from six patients at the time of the surgical resection, performed shortly after the end of the treatment period. Three samples came from the anti–PD1-treated patients whose PBMCs were also analyzed, allowing for matched analysis between circulating and tumor-infiltrating cells. The remaining three samples came from a different subset of CRT-only patients for whom blood could not be analyzed. We performed the same scRNA-seq and scTCR-seq analyses described above on the CD45+ tumor-infiltrating cells sorted from those samples.
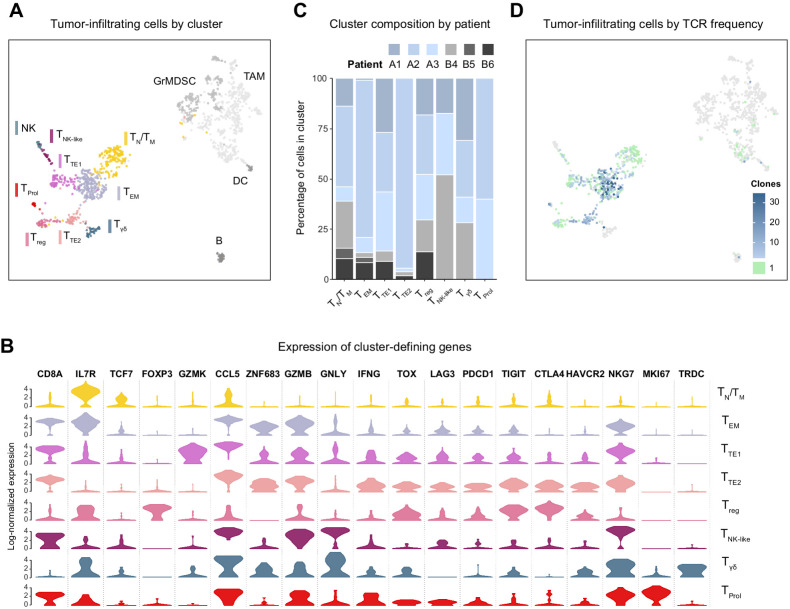
Owing to the relative difficulty of recovering viable cells from fibrotic pancreatic tumors, our total cell yield from the tumor samples was 1,368 cells (Supplementary Fig. S2A). We were able to capture the diversity of the intratumoral immune environment, finding several clusters of granulocytic myeloid-derived suppressor cells and tumor-associated macrophages in addition to the T-cell clusters on which we focused our analysis. Consistent with previous reports on tumor-infiltrating T cells, we found clusters corresponding to TN naïve, TM memory, TEM effector-memory, TTE terminally exhausted, and TNK-like cytotoxic CD8 cells (Fig. 2A and B, Supplementary Fig. S3B; ref. 20). We verified the lack of patient-specific clusters and the presence of cells from both treatment arm in all but the TProl cluster of cycling cells, which only had cells from patients who had received PD-1 blockade (Fig. 2C). The TCR clonotype of nearly all infiltrating αβ T cells was identified (Fig. 2D). As in the circulation, most clonotypes (81%) were singletons primarily found in the TN/TM cluster, and the vast majority (85%) of expanded clonotypes were comprised of CD8 cells. However, unlike in blood, expanded clonotypes belonged to the effector-memory and terminally exhausted clusters, not the active effector pool.
Figure 2.
Tumor-infiltrating immune cells were analyzed by scRNA-seq after neoadjuvant PD-1 blockade combined with chemoradiotherapy in patients with pancreatic adenocarcinoma. A, UMAP plot of all sequenced tumor-infiltrating cells that passed quality controls colored by an unsupervised clustering algorithm. B, Violin plot showing the log-normalized expression of key cluster-defining genes for each T-cell cluster. C, Bar plot showing the breakdown of each cluster's cells by patient of origin. D, UMAP of tumor-infiltrating cells where each cell is colored by the observed frequency of its TCR in the sample.
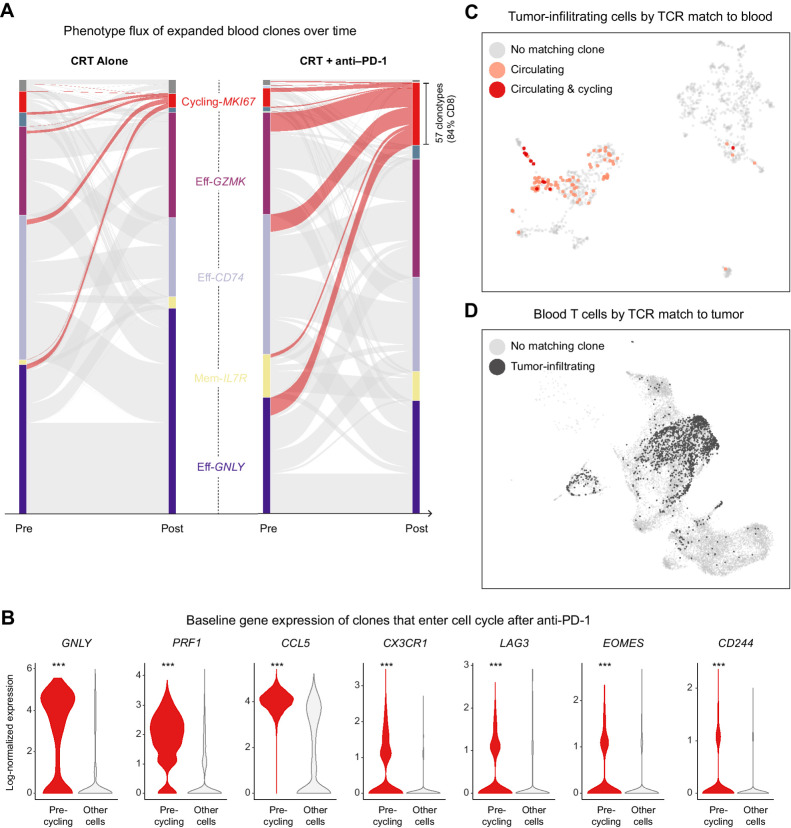
To address the fundamental question of whether a globally immunosuppressive state in patients with PDAC hampers T-cell activation in response to PD-1 blockade, we tracked the transcriptional phenotypes of expanded T-cell clones, which are naturally barcoded by their TCR sequence, before and after treatment. We found that in both arms of the trial, some subset of the tracked clonotypes in circulation entered into cell cycle, as evidenced by clones belonging to the clonotypes appearing in the TProl cluster after treatment. This phenomenon was strikingly enhanced in patients who received PD-1 blockade (Fig. 3A). The newly cycling clonotypes were mostly CD8 cells that, before treatment, belonged to the effector-memory (Eff-GZMK), early activated (Eff-CD74), and terminal effector (Eff-GNLY) clusters. Their baseline gene expression profile was marked by a cytotoxic program with markedly high expression of GNLY (coding for granulysin) and PRF1 (perforin) as well as chemokine receptors CCR5 and CX3CR1, and exhaustion markers as such as CD244 and LAG3 (Fig. 3B). Within the cohort of anti–PD1-treated patients, for whom we were able to perform matched analyses of blood and tumor, we were pleasantly surprised to find many cross-matching T-cell clones, despite relatively limited tumor cell yield. The overwhelming majority of those cells in the circulation were activated CD8s, though their clones in the tumor took on a variety of phenotypes, as evidenced by their broad cluster distribution (Fig. 3C and D); those clones in the tumor TN/TM cluster more likely represented bystanders, whereas those localizing to effector clusters were more likely to be truly tumor-reactive. Importantly, we confirmed that several of the newly cycling clonotypes were observed in the tumor as activated clones in the TTE1 and TNK-like clusters, consistent with tumor-reactive clones, rather than bystander cells (20). Cluster TTE1, which expressed more GZMK and NKG7 but lower levels of HAVRC2, CTLA4, and TIGIT than cluster TTE2, likely represents a less terminally exhausted CD8 T-cell state (21–23). We therefore suggest that cytotoxic T cells in the blood of PDAC patients remain sensitive to reinvigoration by PD-1 blockade, and that at least some of the affected cells have tumor-recognizing potential. The fact that potentially tumor-reactive clonotypes constituted approximately 5% of the treatment-expanded repertoire at baseline suggests that endogenous T-cell priming does occur in PDAC (Supplementary Fig. S2B). However, this rate is much lower than what was observed in a study by Luoma and colleagues (18) of similarly treated head and neck squamous cell carcinoma (HNSCC), which has a substantially higher response rate to immune checkpoint blockade.
Figure 3.
Neoadjuvant PD-1 blockade stimulates polyclonal expansion in circulating cytotoxic T cells with matching tumor-infiltrating clones. A, Alluvial plot showing the flux of clonotypes from pretreatment to posttreatment clusters. The thickness of a band connecting a pretreatment cluster to a posttreatment cluster is proportional to the number of clonotypes with member cells appearing in both clusters; the curves highlighted in red correspond to clonotypes with posttreatment cells in the cycling cluster. B, Violin plot showing the expression distribution of key genes among pretreatment cells belonging to clonotypes that entered cell cycle after treatment in comparison with all other pretreatment cells; ***, denotes an adjusted P < 1 × 10−14 after Bonferroni multiple-hypothesis correction. C, UMAP of tumor-infiltrating cells highlighting the subset of T cells whose TCRs were also expressed by circulating and cycling T cells. D, UMAP of blood T cells, highlighting the subset of cells whose TCRs were also expressed by tumor-infiltrating T cells.
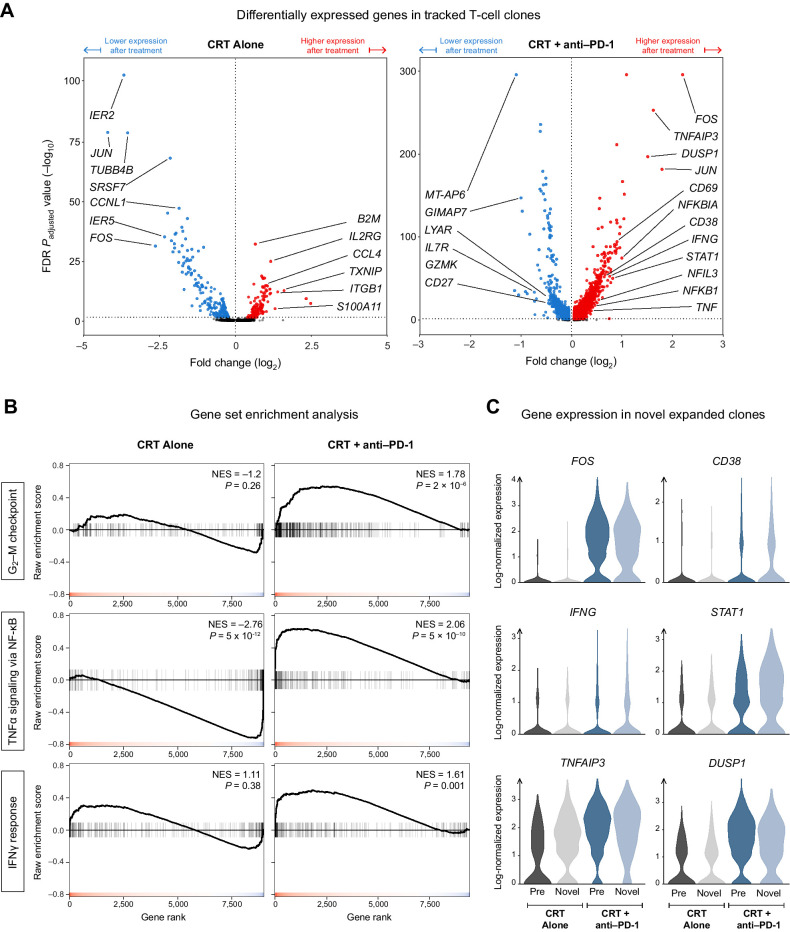
Beyond augmented cell-cycle entry, we sought to characterize the full scope of the transcriptional changes that occurred in tracked T-cell clones over the treatment period. We discovered that the cells of patients treated with PD-1 blockade were remarkably different after therapy, with over 3,849 significantly upregulated genes, compared with only 307 genes with CRT alone (Fig. 4A). Top among those uniquely upregulated genes were several members of the AP-1 (FOS/JUN) transcription factor family, IFN-program genes such as IFNG and its canonical transcription factor STAT1, as well as genes involved in the NF-κB signaling pathway such as NFKB1 (coding for p50), NFIL3, and TNFAIP3 (coding for the A20 deubiquitinase that is expressed upon NF-κB activation and serves as a negative feedback regulator).
Figure 4.
PD-1 blockade induces the AP-1, NF-κB, and IFN transcriptional programs in the T cells of patients with PDAC. A, Volcano plot showing the differentially expressed genes (DEG) over the treatment period among T-cell clones that were present in both pre- and posttreatment samples. B, Gene Set Enrichment Analysis (GSEA) of three hallmark gene sets for the DEGs obtained in (A). C, The posttreatment expression of key genes from the hallmark pathways in (B) among novel expanded cells in comparison with pre-existing expanded cells.
To confirm that these gene-level observations reflected pathway-level enrichment, we performed GSEA on the list of differentially expressed genes (DEG) from each treatment arm. Using GSEA, we are able to compute an enrichment score for any previously established gene set within our own dataset's DEGs. A high score signifies that many genes from the gene set are unusually highly upregulated in our DEG list, whereas a low score would indicate a surprisingly strong downregulation of many of those genes. We focused on the following three “hallmark gene sets” that were curated and published within the Molecular Signatures Database (MSigDB): “G2–M Checkpoint,” “TNFα Signaling via NF-κB,” and “IFNγ Response.” Each set consists of 200 genes established as members of the given pathway. Supporting our findings of enhanced cell-cycle entry, the “G2–M Checkpoint” gene set was highly enriched in the anti-PD1 arm alone (Fig. 4B). We also found significant enrichment of the “TNFα Signaling via NF-κB” and “IFNγ Response” gene sets, consistent with previous work from our group characterizing enhanced antitumor T-cell responses in mice (24). Notably, this enrichment was not seen in the CRT-only DEGs, confirming that these pathways are upregulated specifically due to the effect of PD-1 blockade on the treated patients’ T cells.
Our pre-post differential gene expression analysis leaves out an important subset of the T-cell response: The novel expanded (NE) clonotypes, which are, by definition, not detected in pretreatment samples but are encountered as expanded clones in posttreatment samples. Although some NE clonotypes are liable to be falsely labeled as such due to inevitable down-sampling of the T-cell repertoire by current sequencing technology, PD-1 blockade has been shown to boost the rate of emergence of NE clones, and this phenomenon is thought to play an important role in mounting effective antitumor responses (25). Acknowledging the potential importance of NE clones, we examined the expression levels of key genes upregulated by PD-1 blockade, such as FOS and STAT1, in those cells in the circulation. We found that NE clones in the anti-PD1 arm had similarly high expression levels of those genes when compared with pre-existing expanded cells (Fig. 4C), suggesting that the transcriptional effects of PD-1 blockade (namely, the upregulation of AP-1, NF-κB, and IFN programs) applied to NE clones as well.
The remarkable degree of transcriptional change in the circulating T cells of patients treated with PD-1 blockade stood at odds with the lack of clinical effectiveness in treated patients. To reconcile these discordant observations, we turned to a scRNA-seq dataset of PBMCs collected from a phase I clinical trial (NCT02268825) of patients with metastatic GI cancers who received the same anti-PD1 mAb used in our study (pembrolizumab) in combination with standard-of-care chemotherapy. This dataset, reported by Griffiths and colleagues (26) in 2020, was analyzed on the same scRNA-seq platform as ours, and, importantly, it included one patient with PDAC who responded well to therapy with only minimal disease burden at 5 months. Such a response to immunotherapy is rarely observed outside of microsatellite-unstable PDAC and provided an informative comparison with our non-responder patients.
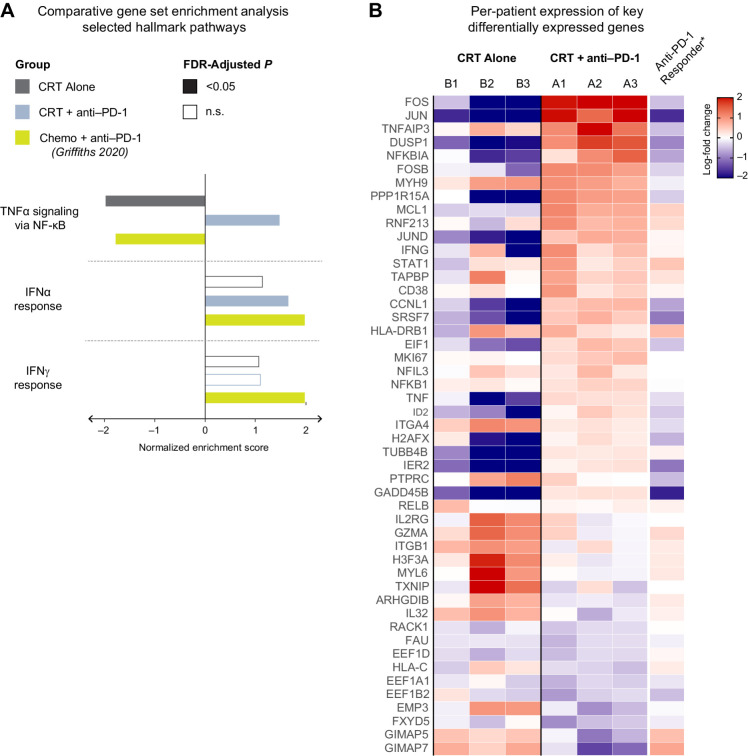
We started our comparison by compiling a list of genes that were differentially expressed in the T cells of the PDAC responder patient after one cycle of treatment relative to baseline expression before the start of treatment. We then used an extended version of our previously described GSEA to quantify the degree of enrichment of all 50 of the hallmark pathways in MSigDB (Fig. 5A). The only significantly enriched pathways found in the responder patient but not in CRT-only patients were those corresponding to IFNα/γ responses, recapitulating the same findings reported by Griffiths and colleagues (26). Our non-responder patients, on the other hand, displayed no enrichment in the IFNα pathway and a weaker enrichment of the IFNγ gene set. Intriguingly, we found that the NF-κB pathway, which was the most enriched in non-responder patients, was the most downregulated pathway in the responder patient (Fig. 5A; Supplementary Fig. S4A).
Figure 5.
Lack of response to neoadjuvant PD-1 correlates with NF-κB activation and a relatively restrained upregulation of IFN-response genes. A, Gene Set Enrichment Analysis (GSEA) of hallmark gene sets performed on the list of treatment-induced differentially expressed genes in each group of patients. B, Heat map showing the treatment-induced log-fold change of key genes, broken down by individual patient.
To validate that GSEA-based enrichment scores were not the result of one or two outlier genes, we inspected the per-patient log-fold change in the expression of 50 genes from the hallmark NF-κB and IFN gene sets (Fig. 5B). We confirmed that the enrichment scores accurately summarized concordant changes at the level of individual genes and individual patients. Genes correlated with negative regulatory pathways such as CD38 were increased in our non-responders (27). Therefore, the transcriptional profile of T cells after PD-1 blockade in our non-responder patients with PDAC reflected dual and possibly conflicting reprogramming: A likely beneficial upregulation of the type II IFN pathway and a potentially detrimental activation of NF-κB.
Discussion
Pancreatic cancer has shown a frustrating lack of response to immunotherapy, including most recently a lack of clinical benefit of neoadjuvant PD-1 blockade plus chemoradiation compared with chemoradiation alone in patients with resectable PDAC. Here, we evaluated peripheral blood and tumor from patients with PDAC receiving PD-1 blockade in the context of a phase II trial to interrogate whether PD-1 blockade induces CD8 T-cell expansion of likely tumor-reactive clonotypes. We found that PD-1 blockade does induce proliferation of T cells in the blood derived from pre-existing effector CD8 populations. These treatment-expanded clonotypes also match TCR clonotypes found in the tumor, suggesting that some of them are likely to recognize tumor antigens. Although our numbers of cells were limited, we also observed proliferating T cells in tumors of patients treated with PD-1 blockade. In peripheral blood, treatment-expanded clonotypes expressed a transcriptional signature of NF-κB signaling, which is divergent from reported signatures of expanded clonotypes from patients responding to PD-1 blockade that show a signature of IFN exposure.
Our study has several key findings. First, moving to an earlier stage of disease did not reveal a responsiveness of patients with PDAC to PD-1 blockade. The large tumor burden in patients with advanced PDAC can induce immune suppression due to systemic effects on myeloid cells complicated by concurrent therapies administered for metastatic disease (12, 28). However, patients with resectable PDAC in our study also showed no added benefit of PD-1 blockade. Administration of capecitabine with radiotherapy may have affected the T-cell response, although the dosing scheme used in this trial was intended to induce immunogenic cell death to potentiate T-cell priming and efficacy of PD-1 blockade. Comparison of pretreatment and posttreatment peripheral blood from patients receiving PD-1 blockade indicates that clonal expansion of effector CD8 T cells occurred, consistent with findings in melanoma, non–small cell lung cancer, and head and neck cancer (18, 29, 30). The phenotype of the expanded clones is that of effector-memory CD8 T cells, which have been previously linked to tumor-reactivity (20), and indeed several expanded clonotypes matched proliferating T-cell clonotypes in the resected tumor specimens. Exhausted T cells in the tumors appeared in two clusters, potentially reflecting progenitor exhausted (TTE1) and terminally exhausted (TTE2) pools (21–23). Interestingly, circulating reinvigorated clonotypes were located in the progenitor exhausted pool, consistent with the idea that these stem-like cells are able to proliferate in response to PD-1 blockade and replenish the effector pool (21, 22). We found no matches between circulating reinvigorated clones and the most highly exhausted tumor-infiltrating T cells (cluster TTE2), suggesting that those two clonotype pools are distinct, at least at the time point profiled. Our results argue against a complete lack of T-cell reactivation being the major driver of PD-1 blockade unresponsiveness in PDAC.
Many groups, including ours have suggested that T-cell priming is impaired in PDAC, resulting in a dearth of antitumor T cells for PD-1 blockade to reinvigorate (31–34). PDAC has an average tumor mutational burden that should yield an adequate number of neoantigens for T-cell priming (35). Dendritic cells, notably the Batf3+ cross-presenting cDC1 subtype, are low in pancreatic cancer mouse models, and their absence is a major cause of inadequate T-cell priming in mice (36, 37). In humans, T-cell priming has been more difficult to evaluate. Hypoxia, regulatory T cells, immunosuppressive macrophages, and factors in the tumor microenvironment can prevent T cells from infiltrating and accumulating in the tumor bed; therefore, analysis of T-cell expansion in the blood is a better measure of T-cell priming than analysis of T-cell activity in tumors that is complicated by multiple variables (12). T-cell priming has been reported for PDAC self and neoantigens, as evidenced by detection of peripheral T cells specific for the self-antigen mesothelin and the presence of neoantigen-specific T cells, particularly in patients surviving for 5 years after surgery (11, 38). Endogenous T-cell priming could be detected in our study, as pre-existing T-cell clones showed PD-1 blockade-induced expansion in the blood. This phenomenon was much more restrained than in similar analyses of paired peripheral blood samples in patients with metastatic breast cancer (10) or resectable HNSCC (18) treated with PD-1 blockade, although tumor sequencing was more extensive in the latter study, making it difficult to conclusively ascribe the observed differences to a defect in priming as opposed to better cross-matching of circulating T cells to their tumor-infiltrating counterparts. Collectively, these findings suggest that T-cell priming in PDAC does occur at a level consistent with other tumor types. Further quantitative efforts to evaluate endogenous T-cell priming across cancer types in humans are warranted.
Although patients with PDAC treated with PD-1 blockade showed evidence of treatment-induced CD8 T-cell expansion, these T cells had a strong treatment-induced signature of NF-κB signaling, as did NE clonotypes. The reason for this NF-κB signature is unclear. TNFα is a plausible candidate, and TNFα blockade has been shown in mouse models to improve responses to PD-1 blockade and other immunotherapies (39, 40). Use of TNFα-blocking antibodies in humans has been shown to be effective at reducing colitis in patients experiencing immune-related adverse events associated with checkpoint blockade (41, 42). Whether TNFα blockade also positively impacts the antitumor response remains to be evaluated. Augmenting non-canonical NF-κB signaling in T cells in pancreatic cancer mouse models can be beneficial, indicating that the context of NF-κB signaling and which NF-κB subunits are activated matters (24, 43). Other factors may also contribute to expanded T cells showing a high NF-κB signature as compared with IFN signature. IFNγ is associated with both acute responses to immunotherapy and establishment of long-term durable remissions (44, 45). Here, we find that circulating CD8 T-cell clonotypes with likely tumor reactivity are expanded in patients with PDAC upon treatment with PD-1 blockade, but that acquisition of an NF-κB signature is correlated with a nonproductive response.
Supplementary Material
supplemental figures and legends
processed sequencing data
TCR contigs
Arm A count matrices
Arm B count matrices
Acknowledgments
S.K. Dougan was funded by NIH 1R01AI158488–01, U01 CA224146–01, the Hale Center for Pancreatic Cancer Research, the Ludwig Center at Harvard, and a Technology Impact Award from the Cancer Research Institute and is a Pew-Stewart Scholar in Cancer Research. C.L. Slingluff Jr was funded by research support to the University of Virginia from Celldex, GlaxoSmithKline, Merck, and Polynoma. O.E. Rahma was funded by Merck. V. Cardot-Ruffino was funded by a fellowship from the Pancreatic Cancer Action Network and the Francois Wallace Monahan Fund in loving memory of Michael Insel. J.A. Nowak was funded by the Hale Center for Pancreatic Cancer Research. B.M. Wolpin is supported by the Dana-Farber Cancer Institute Hale Family Center for Pancreatic Cancer Research, Lustgarten Foundation dedicated laboratory program, NIH grant U01 CA210171, NIH grant P50 CA127003, Stand Up to Cancer, Pancreatic Cancer Action Network, Noble Effort Fund, and Wexler Family Fund. We thank Michael Manos and the DFCI Center for Immune-Oncology for sample processing. We thank Jason Pyrdol and the DFCI Center for Cancer Immunology Research for assistance with the 10x Genomics platform.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This article is featured in Highlights of This Issue, p. 469
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
B.M. Wolpin reports grants from Celgene, Novartis, Eli Lilly, and Revolution Medicine and personal fees from Mirati, GRAIL, and Ipsen outside the submitted work. T.A. Abrams reports personal fees from AstraZeneca, Eisai, Exelixis, and Bristol-Myers Squibb outside the submitted work. J.D. Mancias reports grants from Novartis outside the submitted work. T.S. Bekaii-Saab reports research funding (to institution) from Agios, Arys, Arcus, Atreca, Boston Biomedical, Bayer, Eisai, Celgene, Lilly, Ipsen, Clovis, Seattle Genetics, Genentech, Novartis, Mirati, Merus, Abgenomics, Incyte, Pfizer, and BMS; consulting fees (to institution) from Servier, Ipsen, Arcus, Pfizer, Seattle Genetics, Bayer, Genentech, Incyte, Eisai, Merus, and Merck KGaA and Merck; consulting (to self) from Stemline, AbbVie, Blueprint Medicines, Boehringer Ingelheim, Janssen, Daiichi Sankyo, Natera, TreosBio, Celularity, Caladrius Biosciences, Exact Science, Sobi, BeiGene, Kanaph, AstraZeneca, Deciphera, Zai Labs, Exelixis, MJH Life Sciences, Aptitude Health, Illumina, Foundation Medicine, Sanofi, and GlaxoSmithKline; IDMC/DSMB participation with The Valley Hospital, Fibrogen, Suzhou Kintor, AstraZeneca, Exelixis, Merck/Eisai, PanCan, and 1Globe; scientific advisory board participation with Imugene, Immuneering, Xilis, Replimune, Artiva, and Sun Biopharma; royalties from Up-to-Date; and inventions/patents WO/2018/183488: Human PD1 peptide vaccines and uses thereof licensed to Imugene and WO/2019/055687: Methods and compositions for the treatment of cancer cachexia licensed to Recursion. C.L. Slingluff Jr reports grants from Merck during the conduct of the study as well as other support from Curevac and Polynoma and grants and other support from Merck and Celldex outside the submitted work; in addition, C.L. Slingluff Jr reports a patent for peptides used in cancer vaccines issued and licensed to GSK (but now out of patent life), a patent application for new peptides in cancer vaccines pending, and a patent for use of intratumoral agents for cancer immunotherapy issued. O.E. Rahma reports grants and personal fees from Merck during the conduct of the study as well as other support from AstraZeneca outside the submitted work; in addition, O.E. Rahma reports a patent for Methods of using pembrolizumab and trebananib pending. S.K. Dougan reports grants from Novartis, BMS, Eli Lilly, and Genocea and personal fees and other support from Kojin outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
L.R. Ali: Conceptualization, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. P.J. Lenehan: Investigation, writing–review and editing. V. Cardot-Ruffino: Investigation, writing–review and editing. A. Dias Costa: Investigation, writing–review and editing. M.H.G. Katz: Resources, writing–review and editing. T.W. Bauer: Resources, writing–review and editing. J.A. Nowak: Resources, writing–review and editing. B.M. Wolpin: Resources, writing–review and editing. T.A. Abrams: Resources, writing–review and editing. A. Patel: Resources, writing–review and editing. T.E. Clancy: Resources, writing–review and editing. J. Wang: Resources, writing–review and editing. J.D. Mancias: Resources, writing–review and editing. M.J. Reilley: Resources, writing–review and editing. C.-C.H. Stucky: Resources, writing–review and editing. T.S. Bekaii-Saab: Resources, writing–review and editing. R. Elias: Resources, writing–review and editing. N. Merchant: Resources, writing–review and editing. C.L. Slingluff Jr: Resources, writing–review and editing. O.E. Rahma: Resources, writing–review and editing. S.K. Dougan: Conceptualization, supervision, funding acquisition, writing–original draft, writing–review and editing.
References
- 1. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 2. Kunzmann V, Siveke JT, Algul H, Goekkurt E, Siegler G, Martens U, et al. Nab-paclitaxel plus gemcitabine versus nab-paclitaxel plus gemcitabine followed by FOLFIRINOX induction chemotherapy in locally advanced pancreatic cancer (NEOLAP-AIO-PAK-0113): a multicentre, randomised, phase 2 trial. Lancet Gastroenterol Hepatol 2021;6:128–38. [DOI] [PubMed] [Google Scholar]
- 3. Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat Cancer 2022;3:272–86. [DOI] [PubMed] [Google Scholar]
- 4. Katz MHG, Shi Q, Meyers J, Herman JM, Chuong M, Wolpin BM, et al. Efficacy of preoperative mFOLFIRINOX vs. mFOLFIRINOX plus hypofractionated radiotherapy for borderline resectable adenocarcinoma of the pancreas: the A021501 phase 2 randomized clinical trial. JAMA Oncol 2022;8:1263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Conroy T, Castan F, Lopez A, Turpin A, Ben Abdelghani M, Wei AC, et al. Five-year outcomes of FOLFIRINOX vs. gemcitabine as adjuvant therapy for pancreatic cancer: a randomized clinical trial. JAMA Oncol 2022;8:1571–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haeno H, Gonen M, Davis MB, Herman JM, Iacobuzio-Donahue CA, Michor F. Computational modeling of pancreatic cancer reveals kinetics of metastasis suggesting optimum treatment strategies. Cell 2012;148:362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sohal DPS, Duong M, Ahmad SA, Gandhi NS, Beg MS, Wang-Gillam A, et al. Efficacy of perioperative chemotherapy for resectable pancreatic adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol 2021;7:421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Middleton G. Beyond Ipilimumab: a review of immunotherapeutic approaches in clinical trials in melanoma. Immunother Adv 2021;1:ltaa010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Heckler M, Ali LR, Clancy-Thompson E, Qiang L, Ventre KS, Lenehan P, et al. Inhibition of CDK4/6 promotes CD8 T-cell memory formation. Cancer Discov 2021;11:2564–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ali LR, Garrido-Castro AC, Lenehan PJ, Bollenrucher N, Stump CT, Dougan M, et al. PD-1 blockade and CDK4/6 inhibition augment nonoverlapping features of T-cell activation in cancer. J Exp Med 2023;220:e20220729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 2017;551:512–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Balachandran VP, Beatty GL, SK D. Broadening the impact of immunotherapy to pancreatic cancer: challenges and opportunities. Gastroenterology 2019;156:2056–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Reilly EM, Oh DY, Dhani N, Renouf DJ, Lee MA, Sun W, et al. Durvalumab with or without tremelimumab for patients with metastatic pancreatic ductal adenocarcinoma: a phase 2 randomized clinical trial. JAMA Oncol 2019;5:1431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xie C, Duffy AG, Brar G, Fioravanti S, Mabry-Hrones D, Walker M, et al. Immune checkpoint blockade in combination with stereotactic body radiotherapy in patients with metastatic pancreatic ductal adenocarcinoma. Clin Cancer Res 2020;26:2318–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, et al. Efficacy of pembrolizumab in patients with noncolorectal high microsatellite instability/mismatch repair-deficient cancer: results from the phase II KEYNOTE-158 study. J Clin Oncol 2020;38:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lenehan PJ, Cirella A, Uchida AM, Crowley SJ, Sharova T, Boland G, et al. Type 2 immunity is maintained during cancer-associated adipose tissue wasting. Immunother Adv 2021;1:ltab011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goncharov M, Bagaev D, Shcherbinin D, Zvyagin I, Bolotin D, Thomas PG, et al. VDJdb in the pandemic era: a compendium of T-cell receptors specific for SARS-CoV-2. Nat Methods 2022;19:1017–9. [DOI] [PubMed] [Google Scholar]
- 18. Luoma AM, Suo S, Wang Y, Gunasti L, Porter CBM, Nabilsi N, et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022;185:2918–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luoma AM, Suo S, Williams HL, Sharova T, Sullivan K, Manos M, et al. Molecular pathways of colon inflammation induced by cancer immunotherapy. Cell 2020;182:655–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, et al. Phenotype, specificity and avidity of antitumour CD8(+) T cells in melanoma. Nature 2021;596:119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20:326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 2016;537:417–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beltra JC, Manne S, MS A-H, Kurachi M, Giles JR, Chen Z, et al. Developmental relationships of four exhausted CD8(+) T-cell subsets reveals underlying transcriptional and epigenetic landscape control mechanisms. Immunity 2020;52:825–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ventre KS, Roehle K, Bello E, Bhuiyan AM, Biary T, Crowley SJ, et al. cIAP1/2 antagonism induces antigen-specific T-cell–dependent immunity. J Immunol 2023;210:991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 2019;25:1251–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Griffiths JI, Wallet P, Pflieger LT, Stenehjem D, Liu X, Cosgrove PA, et al. Circulating immune cell phenotype dynamics reflect the strength of tumor-immune cell interactions in patients during immunotherapy. Proc Natl Acad Sci USA 2020;117:16072–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Verma V, Shrimali RK, Ahmad S, Dai W, Wang H, Lu S, et al. PD-1 blockade in subprimed CD8 cells induces dysfunctional PD-1(+)CD38(hi) cells and anti–PD-1 resistance. Nat Immunol 2019;20:1231–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cardot-Ruffino V, Bollenrucher N, Delius L, Wang SJ, Brais LK, Remland J, et al. G-CSF rescue of FOLFIRINOX-induced neutropenia leads to systemic immune suppression in mice and humans. J Immunother Cancer 2023;11:e006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kamphorst AO, Pillai RN, Yang S, Nasti TH, Akondy RS, Wieland A, et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1–targeted therapy in lung cancer patients. Proc Natl Acad Sci USA 2017;114:4993–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edner NM, Ntavli E, Petersone L, Wang CJ, Fabri A, Kogimtzis A, et al. Stratification of PD-1 blockade response in melanoma using pre- and post-treatment immunophenotyping of peripheral blood. Immunother Adv 2023;3:ltad001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morrison AH, Diamond MS, Hay CA, Byrne KT, Vonderheide RH. Sufficiency of CD40 activation and immune checkpoint blockade for T-cell priming and tumor immunity. Proc Natl Acad Sci USA 2020;117:8022–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stump CT, Roehle K, Manjarrez Orduno N, Dougan SK. Radiation combines with immune checkpoint blockade to enhance T-cell priming in a murine model of poorly immunogenic pancreatic cancer. Open Biol 2021;11:210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vonderheide RH. The immune revolution: a case for priming, not checkpoint. Cancer Cell 2018;33:563–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yasmin-Karim S, Bruck PT, Moreau M, Kunjachan S, Chen GZ, Kumar R, et al. Radiation and local anti-CD40 generate an effective in situ vaccine in preclinical models of pancreatic cancer. Front Immunol 2018;9:2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spranger S, Luke JJ, Bao R, Zha Y, Hernandez KM, Li Y, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell–inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci USA 2016;113:E7759–E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lin JH, Huffman AP, Wattenberg MM, Walter DM, Carpenter EL, Feldser DM, et al. Type 1 conventional dendritic cells are systemically dysregulated early in pancreatic carcinogenesis. J Exp Med 2020;217:e20190673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Meyer MA, Baer JM, Knolhoff BL, Nywening TM, Panni RZ, Su X, et al. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat Commun 2018;9:1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Thomas AM, Santarsiero LM, Lutz ER, Armstrong TD, Chen YC, Huang LQ, et al. Mesothelin-specific CD8(+) T-cell responses provide evidence of in vivo cross-priming by antigen-presenting cells in vaccinated pancreatic cancer patients. J Exp Med 2004;200:297–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bertrand F, Montfort A, Marcheteau E, Imbert C, Gilhodes J, Filleron T, et al. TNFalpha blockade overcomes resistance to anti–PD-1 in experimental melanoma. Nat Commun 2017;8:2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh MJ, Ali LR, Lenehan P, Kureshi CT, Kureshi R, Dougan M, et al. Blockade of innate inflammatory cytokines TNFalpha, IL-1beta, or IL6 overcomes virotherapy-induced cancer equilibrium to promote tumor regression. Immunother Adv 2023;3:ltad011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Badran YR, Cohen JV, Brastianos PK, Parikh AR, Hong TS, Dougan M. Concurrent therapy with immune checkpoint inhibitors and TNFalpha blockade in patients with gastrointestinal immune-related adverse events. J Immunother Cancer 2019;7:226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dougan M, Blidner AG, Choi J, Cooksley T, Glezerman I, Ginex P, et al. Multinational Association of Supportive Care in Cancer (MASCC) 2020 clinical practice recommendations for the management of severe gastrointestinal and hepatic toxicities from checkpoint inhibitors. Support Care Cancer 2020;28:6129–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roehle K, Qiang L, Ventre KS, Heid D, Ali LR, Lenehan P, et al. cIAP1/2 antagonism eliminates MHC class I-negative tumors through T-cell–dependent reprogramming of mononuclear phagocytes. Sci Transl Med 2021;13:eabf5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Walsh MJ, Stump CT, Kureshi R, Lenehan P, Ali LR, Dougan M, et al. IFNgamma is a central node of cancer immune equilibrium. Cell Rep 2023;42:112219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma pathway genes in tumor cells as a mechanism of resistance to anti–CTLA-4 therapy. Cell 2016;167:397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplemental figures and legends
processed sequencing data
TCR contigs
Arm A count matrices
Arm B count matrices
Data Availability Statement
The sequencing data files generated in this study are not publicly available in keeping with the limitations imposed by patient consent and privacy concerns. Raw, unnormalized, and unfiltered, RNA counts for all sequenced droplets as well as raw TCR contigs are provided as online Supplementary Information. Additional data requests may be directed to the corresponding author.