Abstract
11q13 amplification is a frequent event in human cancer and in particular in squamous cell carcinomas (SCC). Despite almost invariably spanning 10 genes, it is unclear which genetic components of the amplicon are the key driver events in SCC. A combination of computational, in vitro, ex vivo, and in vivo models leveraging efficient primary human keratinocyte genome editing by Cas9-RNP electroporation, identified ORAOV1, CCND1, and MIR548K as the critical drivers of the amplicon in head and neck SCC. CCND1 amplification drives the cell cycle in a CDK4/6/RB1-independent fashion and may confer a novel dependency on RRM2. MIR548K contributes to epithelial–mesenchymal transition. Finally, we identify ORAOV1 as an oncogene that acts likely via its ability to modulate reactive oxygen species. Thus, the 11q13 amplicon drives SCC through at least three independent genetic elements and suggests therapeutic targets for this morbid and lethal disease.
Implications:
This work demonstrates novel mechanisms and ways to target these mechanisms underlying the most common amplification in squamous cell carcinoma, one of the most prevalent and deadly forms of human cancer.
Introduction
Cancer evolves through distinct types of mutations, including single-nucleotide variations, indels, chromosomal gains and losses, focal amplifications, and complex rearrangements. Among the most common chromosomal amplifications in cancer is the 11q13 amplification. 11q13 amplification occurs in approximately 6% of all tumors previously studied, making it the third most common amplification across cancers, following MYC and PIK3CA amplifications (ref. 1; Supplementary Fig. S1a).
Squamous cell carcinomas (SCC) are enriched for amplification of 11q13 (1–4). Amplifications of 11q13 tend to be an early event in SCC tumorigenesis (2–4) and co-occur alongside loss of function mutations in tumor suppressor genes such as TP53 and CDKN2A (5, 6). Although several studies have described a role for individual 11q13 genes in SSC tumorigenesis (5, 7), these studies have been limited by their focus on individual genes on the amplicon without consideration of the roles of other genes on the amplicon and the greater genetic context in which 11q13 amplification occurs. In particular, the role of ORAOV1 (LTO1) has been incompletely explored. Thus, despite the high frequency of 11q13 amplification, it still remains unclear which genes comprise the functional core of the 11q13 amplicon in SCC and through which mechanisms they contribute to SCC tumorigenesis.
The frequency of this CCND1 containing amplicon generated enthusiasm for the use of CDK4/6 inhibitors, such as palbociclib, as potential therapeutics in SCC. This is based upon the assumption that CDK4 and/or CDK6 are critical downstream targets of amplified CCND1 in SCC. One phase II study of palbociclib in lung SCC (LSCC) patients with known amplification of CCND1 failed to demonstrate significant disease controlling activity (8). A double-blind randomized placebo controlled trial in 125 patients of palbociclib in combination with cetuximab in patients with human papillomavirus (HPV)-negative recurrent or metastatic head and neck SCC (HNSCC) failed to demonstrate any additive benefit of Palbociclib (9). Further, a phase II study of palbociclib and cetuximab in patients who previously progressed on cetuximab yielded minimal, if any, response (10). These disappointing results raise the possibility that mechanisms other than CDK4/6 activation may be the critical events downstream of CCND1 amplification in SCC.
In this study, we comprehensively characterize the role of the 11q13 amplification during HNSCC tumorigenesis. HNSCC is a common type of SCC that originates from epithelial cells lining the upper aerodigestive tract. In contrast to other cancer types, SCCs display a remarkable genomic similarity, even between different tissues of origin (5), suggesting similar oncogenic mechanisms across lineage of origin.
Using a combination of engineered primary human keratinocytes and established HNSCC cell lines, we created novel in vitro, ex vivo, and in vivo models of HNSCC to systematically analyze the role of each component of the 11q13 amplicon during HNSCC tumorigenesis. Through this approach, we are able to model the 11q13 amplification in a way that accounts both for the timing and the genetic context in which it occurs during SCC tumorigenesis. In doing so, we identified three independent driver oncogenes present on the 11q13 amplicon and elucidated mechanisms through which the 11q13 amplification drives SCC tumorigenesis. In particular, we identify a CDK4/6 independent activity of CCND1 amplification through RRM2 upregulation that drives tumorigenesis. We also identify ORAOV1 as an oncogene in SCC, which likely exerts its effect through a thioredoxin dependent anti-oxidant activity.
Materials and Methods
Cell culture
FaDu (ATCC, catalog no. HTB-43, RRID:CVCL_1218, male) and Detroit562 (ATCC, catalog no. CCL-138, RRID:CVCL_1171, female) cells were grown in Eagle Minimum Essential Medium with l-glutamine (Fisher Scientific, catalog no. 50983283) supplemented with 10% FBS (Corning, catalog no. MT35010CV) and 1% Penicillin/Streptomycin (P/S, Corning, catalog no. MT30002CI). A-253 (ATCC, catalog no. HTB-41, RRID:CVCL_1060, male) cells were grown in McCoy's 5A (Modified) Medium (Fisher Scientific, catalog no. 16–600–082) supplemented with 5% FBS and 1% P/S. SCC-15 (ATCC, catalog no. CRL-1623, RRID:CVCL_1681) and SCC-9 (ATCC, catalog no. CRL-1629, RRID:CVCL_1685, male) cells were cultured in DMEM:F12 (Gibco, catalog no. 11039021) supplemented with 400 ng/mL hydrocortisone (EMD Millipore, catalog no. 386698), 10% FBS, and 1% P/S. HEK293T (ATCC, catalog no. CRL-3216, RRID:CVCL_0063, female) cells were grown in DMEM (Gibco, catalog no. 12491023) supplemented with 5% FBS and 1% P/S. All cell lines were purchased and authenticated at ATCC in 2016 and tested yearly for mycoplasma through PCR. Human primary fibroblasts were isolated and collected from patient derived skin samples and cultured in DMEM supplemented with 5% FBS and 1% P/S. Human primary keratinocytes were collected and isolated from patient derived mucosal (5 donors), foreskin (2 donors), or tympanic membrane (1 donor) samples and cultured in Medium 154 and Keratinocyte Serum Free Medium (1:1, Life Technologies, catalog no. M154500 and catalog no. 17005042), supplemented with 5 mL/L Human Keratinocyte Growth Supplement (Life Technologies, catalog no. S0015), 25 mg/L Bovine Pituitary Extract (Life Technologies, catalog no. 17005042), 2.5 μg/L EGF Human Recombinant (Life Technologies, catalog no. 17005042), and 1% P/S. No cells were passaged for longer than 5 weeks.
CRISPR interference constructs and cloning
Stable dCas9 cells were generated by transducing cells with pHR-SFFV-dCas9-BFP-KRAB (Addgene, catalog no. 46911) lentivirus. Stable dCas9 cells were transduced with pU6-sgRNA EF1Alpha-puro-T2A-BFP (Addgene, catalog no. 60955) lentivirus to introduce guideRNAs (gRNA; Supplementary Table S1). OR2B6-targeting gRNAs were used as negative control.
Genome engineering
To generate gene knockouts, cells were transfected via electroporation using the MaxCyte ATX electroporation platform with Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) Cas9-ribonucleoproteins (RNP) and gene-specific gRNAs (Supplementary Table S1). For electroporation, sub-confluent cells were trypsinized and washed 1x in DMEM and 1x in Opti-MEM (Life Technologies, catalog no. 31985070). Cells were resuspended to a concentration of 2.5E7 cells/mL in Opti-MEM. crRNAs and Alt-R CRISPR-Cas9 tracrRNA (Integrated DNA Technologies) were hybridized in a 1:1 ratio to a final concentration of 50 μmol/L. crRNA:tracrRNA were complexed with Cas9-RNP at a 1:1:1 ratio for 20 minutes at room temperature and subsequently mixed with the cells to a final concentrations of 2.5 μmol/L crRNA:Cas9 and 2.0E7 cells/mL. Cells were electroporated in 25 μL or 400 μL reactions with the ‘Optimization 7′ (keratinocytes, fibroblasts, Detroit562, and SCC-9) or ‘DLD-1′ (FaDu) electroporation protocols. After electroporation, cells were immediately collected from the processing assembly, plated into a 6-well plate and recovered 20 minutes at 37°C, before resuspending in 2 mL culture medium. Transfection efficiency was determined 24 hours post electroporation through flow cytometry. After 96 hours, gene disruption was confirmed through TIDE (Tracking of Indels by Decomposition) analysis (primers: Supplementary Table S1). To control for multiple edits in the amplified 11q13 region, a negative control “Safe Control” SC1 crRNA was designed to target the 11q13 region in a non-coding and non-regulating region.
LNA knockdown
Non-targeting scrambled control A and MIR548K targeting 3′ FAM miRCURY Locked Nucleic Acid (LNA) miRNA Power Inhibitors were obtained from Qiagen (Catalog no. 339160). 2 mL of cells were seeded at a density of 80.000 cells/mL in a 6-well plate and transfected 24 hours later at a final concentration of 50 nmol/L Inhibitor and 2.5 μL TransIT-LT1 Transfection Reagent (Fisher Scientific, catalog no. MIR2300). Cells were plated for the appropriate assay 48 hours after transfection. To control for target inhibition during lysis, 50 nmol/L inhibitor was added to control cells during lysis.
Exogenous 11q13 gene constructs and cloning
To clone overexpression constructs, RNA was purified from oral keratinocytes (OKC) and total cDNA was synthesized with poly-A specific primers using SuperScript III First-Strand Synthesis System (Invitrogen, catalog no. 18080051) according to the manufacturer's instructions. To create Gateway compatible PCR products of the gene of interest, coupled to an mCherry tag through a T2A sequence, primers were designed with AttB and T2A adapters (Supplementary Table S2). PCRs were performed to create AttB-gene-T2A and T2A-mCherry-AttB products from whole genome cDNA (for larger coding sequences and fluorescent tags, PCRs were performed using the following plasmids as template: CCND1K112E: Rc/CMV Cyclin D1 K112E, Addgene, catalog no. 8951, CCND2: R777-E019 Hs.CCND2, Addgene, catalog no. 70303, CTTN: pGFP Cortactin, Addgene, catalog no. 50728; FADD: pCI-hFADD-FLAG, Addgene, catalog no. 31814; EBFP: pU6-sgRNA EF1Alpha-puro-T2A-BFP, Addgene, catalog no. 60955; EGFP: pSPCas9(BB)-2A-GFP, Addgene, catalog no. 48138; mCherry: pHR_Gal4UAS_pGKmCherry, Addgene, catalog no. 79124). PCR products were purified and coupled in a subsequent PCR. Full attB products were cloned into the pDONR221 Vector (Thermofisher Scientific, catalog no. 12536017) using Gateway Technology according to the manufacturer's instruction. Coding sequences were transferred into the pLEX_307 vector (Addgene, catalog no. 41392). To create a MIR548K gBlock gene fragment for the MIR548K overexpression system, the 5′ oligo and 3′ oligo were annealed and cloned into the SGEP vector (Addgene, catalog no. 111170) using EcORI and XhOI restriction sites. As negative control, pLEX_307-EBFP was generated.
Lentiviral production and infection
HEK293T cells were transfected with 750 ng transfer plasmid, 375 ng psPAX2 (Addgene, catalog no. 12259), 750 ng pMD2.G (Addgene, catalog no. 12260), and 5.5 μL Lipofectamin 2000 Transfection reagent (Life Technologies, catalog no. 11668027) per mL culture medium. Medium was replaced 16 hours post transfection and viral supernatant was collected 72 hours later, filtered through a 0.45-μm polyvinylidene difluoride (PVDF) filter (MilliporeSigma, catalog no. SLHVM33RS), concentrated using LentiX concentrator (Takara Bio, catalog no. 631231) and stored at −80°C. Cells were transduced with viral pellets resuspended in appropriate medium with 8 μg/mL Polybrene Transfection reagent (EMD Millipore, catalog no. TR1003G) for 16 hours.
Copy number and RT-qPCR
For Reversed Transcriptase quantitative PCR (RT-qPCR), RNA was isolated with the RNeasy Plus Mini Kit (Qiagen, catalog no. 74136) and converted into cDNA using SuperScript III First-Strand Synthesis SuperMix for qRT-PCR (Thermo Fisher Scientific, catalog no. 11752250). PrimeTime qPCR Probe-based assays and Gene Expression Master Mix were purchased from Integrated DNA Technologies with 6-FAM/ZEN/IBFQ labeling (Supplementary Table S3). ACTB probes and primers for loading control were designed with JOE NHS/ZEN/3′ IBFQ labeling to allow multiplex RT-qPCR. 10 μL RT-qPCR reactions were prepared containing 500 nmol/L of each primer (gene of interest, ACTB: forward and reversed), 250 nmol/L of each probe, 5 μL Mastermix, and 10 to 50 ng cDNA. Reactions were run in triplicates on the Quantstudio 6 (Applied Biosystems). Relative gene expression levels were calculated using the ΔCT method against ACTB. For DNA Copy number qPCR, DNA was isolated using the QIAamp DNA mini kit (Qiagen, catalog no. 51306). Custom PrimeTime qPCR Probes (Integrated DNA Technologies, Supplementary Table S3) were designed such that primers and probes are spanning an intronic region of the DNA. qPCR reactions were prepared and run as described above, but with 20 to 40 ng DNA. Copy number was determined using the ΔCT method against RNAse P. For RT-qPCR of MIR548K, small RNAs were isolated using the miRNeasy Mini Kit (Qiagen, catalog no. 217004). cDNA was synthesized using the Mir-X miRNA First Strand Synthesis Kit (Takara Bio, catalog no. 638315). Each 10 μL reaction contained 100 (cell lines) – 200 (OKC) ng cDNA, 5 μL TB Green Advantage qPCR premix (Takara Bio, catalog no. 639676), 10 μmol/L forward and reversed primer for U6 and target (MIR548K specific forward primer: aaaaguecuugcggauuuugcu).
Organotypic invasion model
Frozen human dermis was thawed in DPBS (Life Technologies, catalog no. 14190250) for 48 hours at 37°C, and dermis was separated from epidermis. Dermis was treated with 0.1% Peracetic Acid (Sigma-Aldrich, catalog no. 77240) in DPBS for 2 hours and washed 3×60 minutes with DPBS. Dermis was cut into ∼1 cm2 pieces to fit stands, placed in individual wells of a 12-well plate with the dermal side up and left to airdry for 2 hours. 1 mL of fibroblasts at a concentration of 36,000 cells/mL was added to each well and plates were spun down at 1,000 rpm for 60 minutes. Dermis and fibroblasts were cocultured for 8 days before the dermis was placed on stands with the epidermal side face up. Fifty to 60 μL Matrigel (Corning, catalog no. 354234) was added to cover the dermal side. After polymerization of the Matrigel, 500 μL of 3D organotypic media (3:1 DMEM:Ham's F12 medium (Thermo Scientific, catalog no. 12–615F) with 10% FBS, 1% P/S, and 1% antibiotic-antimycotic (Gibco, catalog no. 15240062), supplemented with 24 μg /mL adenine (Sigma, catalog no. A9795–1G), 8.4 ng/mL cholera toxin (EMD Millipore, catalog no. 227036), 0.4 μg/mL hydrocortisone (EMD Millipore, catalog no. 386698), 5 μg/mL insulin (Sigma, catalog no. I1882–100MG), 10 ng/mL EGF (Life Technologies, catalog no. PHG0315), 1.4 ng/mL triiodothyronine (Sigma, catalog no. T5516), 1 μg/mL ciprofloxacin hydrochloride (Sigma, catalog no. PHR1044)) was added. 50 μL FaDu cells were added on top of the dermis at a concentration of 3.75×106 cells/mL. Media was changed every 2 days. After 10 days, dermis was removed from stand and fixed in 10% Neutral Buffered Formalin (Thermo Scientific, catalog no. 22050104) overnight, followed by an overnight wash in DPBS. Dermis was subsequently incubated in 15% sucrose in DPBS, 30% sucrose in DPBS, and 1:1 30% sucrose in DPBS mixed with Optimal Cutting Temperature compound (OCT, Fisher Scientific, catalog no. 23730571), 60 minutes each. Dermis was embedded in OCT and stored at −80°C until sectioning. OCT blocks were sectioned on a CryoStar NX70 cryostat (Thermo Scientific) at 5 μm, with 500 μm between planes. Sections were stained with Gill III Hematoxylin (Thermo Scientific, catalog no. 72611) and Eosin-Y (Fisher Scientific, catalog no. 22220104). Each organotypic resulted in approximately 10 sections. Invasion was quantified as the fraction of sections with cells that show invading cells.
Cell growth assays
For viability assays, cells were seeded into Black Greiner Cellstar 96-well plates (Sigma-Aldrich, catalog no. M9936). Starting at 72 hours after plating or drug treatment, cells were incubated with 10% Alamar Blue (Bio-Rad, catalog no. 100234–634) according to the manufacturer's instruction. Fluorescence was read out on a GloMax Explorer plate reader (Promega) at an excitation of 520 nm and emission of 580 to 640 nm. Cell viability was calculated relative to untreated or day 1 condition. For Spheroid assays, serial dilutions were made to concentrate cells at 200, 100, 50, or 20 cells/mL. 100 μL of cells were transferred to an Ultra-Low Attachment Multiwell Plate (Corning, catalog no. 3474), with 5 replicates per concentration. 72 hours after seeding, wells were qualified to be either positive or negative for sphere formation. Percentage of cells with sphere forming capacity was determined using Extreme Limiting Dilution Analysis (11). Statistical power was calculated with χ2 test. For Competition assays, TP53 and CDKN2A knockout OKC (TC-OKC) were transduced with pLEX_307 or SGEP lentivirus for the gene of interest or EBFP control. 72 hours post transduction, gene and EBFP control overexpressing OKCs were mixed 1:1 and immediately analyzed on a LSR II Flow Cytometer (BD) to determine the ratio of gene-mCherry:EBFP. Cells were kept in culture and analyzed over time to determine the change in ratios. Ratios were normalized to day 1. Experiments were performed with OKC from at least 2 different donors (1 donor per replicate).
Drug treatments
Palbociclib (MedChem Express, catalog no. HY-50767) was diluted in 0.1 mol/L HCL, triapine (3-AP, MedChemExpress, catalog no. 501871763) was diluted in DMSO, and Tert-Butyl hydroperoxide (TBHP, Life Technologies, catalog no. 180340050) was diluted in water. All drugs were added to culture media for 72 hours at indicated concentrations. For palbociclib and triapine, a three parameter nonlinear dose–response curve was fit against the cellular viability scores.
Immunoblot analysis
Cell pellets were lysed in Pierce RIPA buffer (Thermo Scientific, 62249), supplemented with phosphatase inhibitor and protease inhibitor cocktail sets (Calbiochem, catalog no. 524625 and catalog no. 539134). Protein extracts were resolved on Nu PAGE 4% to 12% Bis-Tris gradient gels (Invitrogen, catalog no. WG1401A) and transferred to PVDF membranes using the Trans-Blot Turbo system. Membranes were blocked in 5% milk in TBS-T and probed with primary antibodies overnight at 4°C, and then with horseradish peroxidase-conjugated secondary antibodies. Signals were visualized with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo scientific, catalog no. PI34577) on the Bio-Rad ChemiDox XRS+ System. The following antibodies were used: Anoctamin 1 (Cell Signaling Technology, catalog no. 14476, RRID:AB_2798491, 1:500), Beta-Tubulin (Abcam, catalog no. 6046, RRID:AB_2210370, 1:500), CDKN2A/P16INK4a (Abcam, catalog no. ab108349, RRID:AB_10858268, 1:300), Cortactin (Cell Signaling Technology, catalog no. 3503, RRID:AB_2115160, 1:1,000), Cyclin D1 (Abcam, catalog no. 134175, RRID:AB_2750906, 1:5000), Cyclin D2 (Cell Signaling Technology, catalog no. 3741, RRID:AB_2070685, 1:800), FADD (Cell Signaling Technology, catalog no. 2782, RRID:AB_2100484, 1:1000), ORAOV1 (Invitrogen, catalog no. PA5101219, RRID:AB_2850660, 1:400), P53 (Cell Signaling Technology, catalog no. 9282, RRID:AB_331476, 1:800), PPFIA1 (Abcam, catalog no. 204406, 1:200), RB1 (Abcam, catalog no. 181616, RRID:AB_2848193, 1:1000), Rb phospho S780 (Abcam, catalog no. 184702, 1:800).
Cell cycle analysis
Cells were fixed for 60 minutes in 70% ethanol, washed with DPBS and stained with FxCycle Violet Ready Flow Reagent (Fisher Scientific, catalog no. R37166) for 30 minutes at room temperature. DNA content was analyzed on a LSR II Flow Cytometer (BD).
ROS assays
Reactive oxygen species (ROS) levels were detected using CellROX Green flow cytometry assay kits (Life Technologies, catalog no. C10492). Cells were concentrated 5 × 105 cells/mL in complete medium. Negative controls were incubated with N-acetylcysteine at 1,000 μmol/L for 60 minutes at 37°C, positive controls were incubated with TBHP hydroperoxide at 400 μmol/L for 30 minutes at 37°C. CellROX Green reagent was added at 500 nmol/L for 40 minutes at 37°C. Samples were analyzed by flow cytometry.
In vivo experiments
NOD scid gamma (NSG) mice were purchased from the Jackson laboratory (Catalog no. 005557, RRID:IMSR_JAX:005557). All experimental procedures were approved by and in compliance with UCSF IACUC. TP53 and CDKN2A knockout OKCs were transduced with pLEX_307 virus to induce expression of target genes. Six days post transduction, cells were resuspended in a 1:1 ratio of Matrigel and OKC culture medium at a concentration of 1.0E7 cells/mL. NSG mice (n = 45, mixed male and female) were subcutaneously injected in the hind flank with 1.0E6 cells. Tumor growth was monitored weekly until endpoint and tumors were measured using a caliper. Tumors were dissected and origin was confirmed through RT-qPCR on target genes. For the triapine experiment, parental FaDu cells were injected into mice (mixed males and females, between 4 and 7 months old. DMSO control: n = 7, triapine: n = 9) as described above. Tumor growth was monitored at least twice per week. Once tumors were palpable, mice were randomized and drug administration started. Triapine (10 mg/kg, dissolved in SBE-β-CD in saline) or 2% DMSO were administered through an intraperitoneal injection for 5 consecutive days per week until tumors reached endpoint.
RNA sequencing and gene set enrichment analysis
Cells were transfected as indicated and RNA was collected 6 days after. RNA sequencing (RNA-seq) libraries were prepared with the QuantSeq 3′ mRNA-seq Library Prep Kit (Lexogen, catalog no. 015.24) kit according to the manufacturer's instructions. Library quality was assessed with a High Sensitivity DNA Assay on the Agilent 2100 Bioanalyzer. Samples were sequenced by the Center of Advanced Technologies (UCSF) on the HiSeq SE50/65 (Illumina). Samples of sufficient quality were analyzed for Differential Expression analysis using the DESeq2 pipeline. Gene set enrichment analysis (GSEA) on the differentially expressed genes was performed using the BiocManager, fgsea, and clusterProfiler packages for R.
The Cancer Genome Atlas data analyses
Publicly available copy number and gene expression data from The Cancer Genome Atlas (TCGA) was accessed through diverse applications. For overall 11q13 amplification frequencies in tumors or cancer cell lines, TCGA PanCancer (12) data and the Cancer Cell Line Encyclopedia were analyzed through cBioPortal, respectively. To determine individual gene amplification frequencies in tumors, TCGA data sets [GDC TCGA esophageal (ref. 13; stratified for SCC tumors), GDC TCGA HNSCC (5), GDC TCGA LSCC (14), and GDC TCGA liver hepatocellular carcinoma (HCC); ref. 15] were explored for copy number through the Xenabrowser. Copy number amplification was called at > 3.0 copies. To determine correlations between copy number and gene expression, TCGA data sets were mined for copy number, RNA expression, and miRNA expression data. Shapiro–Wilk tests showed that data was not linearly distributed, thus pairwise Spearman coefficients between expression and copy number were computed. P values were corrected for multiple testing with the Holm-Bonferroni method. For survival analysis, TCGA HNSCC (5) data was mined for HPV status, 11q13 gene copy number, and survival in months. Kaplan–Meier curves were computed with the Survival and Survminer packages for R. For cancer dependency analysis, the Cancer Dependency Map was accessed via the depmap package for R. DepMap Release: DepMap, Broad (2020): DepMap 20Q3 Public. figshare. Dataset doi:10.6084/m9.figshare.9201770.v2 (CRISPR knockout screens from project Achilles). For HNSCC and esophageal SCC (ESCC) specific analysis, the following cell lines were analyzed: HNSCC: BHY, BICR18, BICR22, BICR31, BICR6, CAL27, DETROIT562, FADU, HSC3, HSC4, PECAPJ15, PECAPJ41CLONED2, SCC15, SCC25, SCC4, YD38, YD8. ESCC: COLO680N, KYSE140, KYSE180, KYSE410, KYSE510, OE19, OE21, TE10, TE11, TE14, TE15, TE4, TE5, TE6, TE8, TE9, TDOTT. LSCC: HCC15, LK2, HARA, NCIH2170, NCIH2882, HCC95, SW900, SQ1, EBC1, LUDLU1, CORL32, NCIH520, NCIH1869, VMRCLCP, NCIH1703, HCC2450, GT3TKB, LOUNH91, HCC1897, KNS62, EPLC272H, RERFLCAI, LC1SQ, LC1F, NCIH1385, SKMES1, RERFLCSQ1, CALU1, HCC2814, LC1SQSF, NCIH157DM. For RNA-seq analysis on HNSCC TCGA PanCancer tumors (5), expression data was accessed through cBioPortal. Tumors were selected on bearing TP53 and CDKN2A mutations and stratified on the basis of amplification of all 11q13 genes. See Supplementary Table S4 for tumor selection. Genes were considered significantly altered at q < 0.05.
Statistical analysis
All data are represented as mean ± SEM unless stated otherwise. All experiments were independently reproduced at least 3 times. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Data availability
RNA-seq data generated in this study are publicly available at the Gene Expression Omnibus under accession code GSE216849. All other raw data generated in this study are available upon request from the corresponding author.
Results
The 11q13 amplification in SCC spans from CCND1 to CTTN
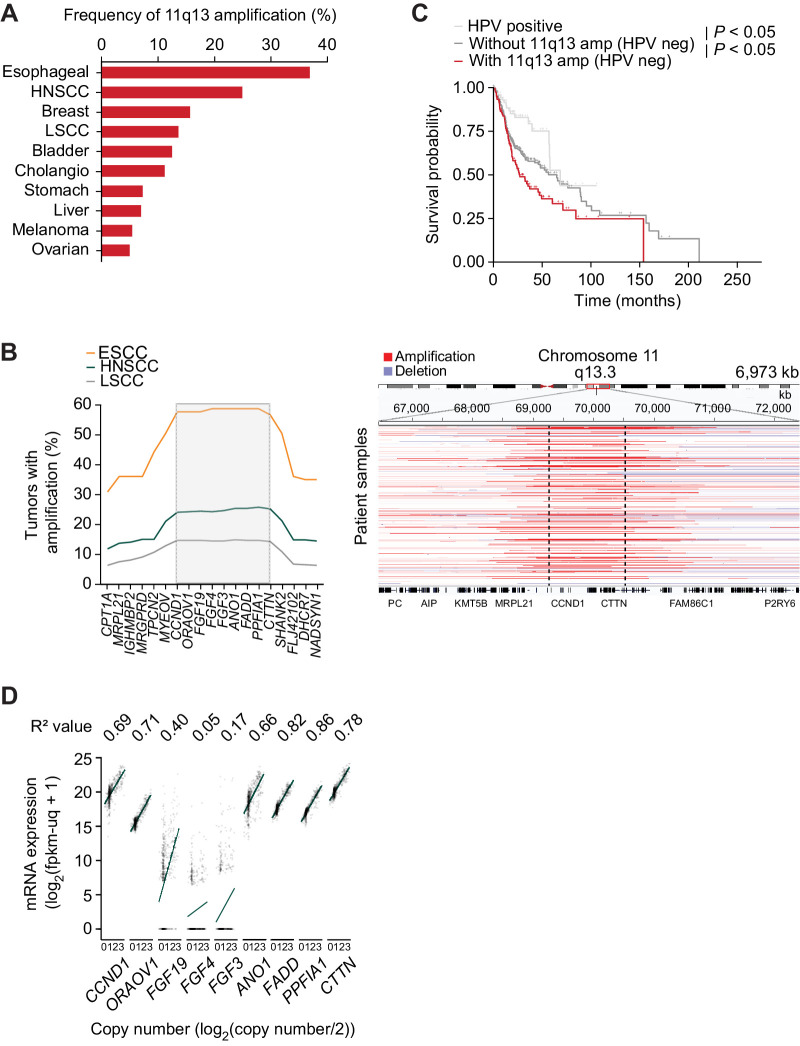
We first sought to identify the frequency of 11q13 amplification across tumor types. Among tumors analyzed in the TCGA dataset, SSCs harbored the 11q13 amplification most frequently with 37% of esophageal cancers, of which many are SCCs, and 25% of HNSCCs carrying amplifications of genes in this region, with LSCC harboring amplification slightly less frequently (1) (Fig. 1A). We next sought to define the genes that mark the edge of the minimal critical region of the amplicon in SCC. Using TCGA data for SCC (5, 13, 14), we identified 10 genes (CCND1, ORAOV1, FGF19, FGF4, FGF3, ANO1, FADD, PPFIA1, MIR548K, CTTN) as focally co-amplified across a total of 1,119 SCC tumors (Fig. 1B). From this analysis, we identified that CCND1 and CTTN form the 5′ and 3′ boundaries of the minimal critical region of the amplicon, respectively, as there is a significant drop in amplification frequency beyond these genes. Therefore, in this work, we define the minimal 11q13 amplicon as amplification of these 10 genes. The genes in this 800 kB span are nearly always amplified together, with only rare cases exhibiting a smaller amplicon not containing all 10 genes (Supplementary Fig. S1b). Amplification of these genes appears to be further selected for in either the isolation or propagation of SCC cell lines, as 60% of HNSCC and 77% of ESCC lines harbor the 11q13 amplification (Supplementary Fig. S1c), compared with 25% and 58% of primary tumors, respectively. Consistent with prior reports (5, 16, 17), we find that patients whose HNSCC tumors carry the 11q13 amplification have decreased survival (Fig. 1C). The particular core of 10 amplified genes at this locus is characteristic of this amplicon in SCC, as analysis of 378 HCC tumors identified a narrower co-amplification window, with breakpoints at CCND1 and FGF19 (Supplementary Fig. S1d). In HCC, FGF19 is aberrantly expressed and targeting the FGFR4–FGF19 axis has been demonstrated to be a therapeutic strategy for 11q13-amplified HCC (17, 18). The difference in amplification window thus implies a distinct set of genetic elements driving SCC as opposed to HCC.
Figure 1.
The 11q13 amplification in SCC spans from CCND1 to CTTN.A, Frequency of 11q13 amplification in tumors (TCGA, PanCancer Atlas). B, Left, frequency of amplification of genes in 11q13 region in ESCC (n = 97), HNSCC (n = 519), and LSCC (n = 503). Right, representative integrative genome browser view on copy number fragments of 11q13-amplified HNSCCs. C, Kaplan–Meier survival graphs for patients separated by HPV and 11q13 status (HPV negative, n = 415. HPV positive, n = 72). D, Correlation between gene expression and copy number in HNSCC (TCGA, n = 496). R2 value represents Spearman coefficient, P values calculated with Holm correction (all but FGF3 and FGF4 significant).
The driver oncogenes of the amplicon are likely overexpressed as a consequence of increased DNA copy number. Using TCGA (5) data, we determined the correlation between expression level and copy number of each protein coding gene in the amplicon in HNSCC. Six of the genes—CCND1, ORAOV1, ANO1, FADD, PPFIA1, and CTTN—were highly expressed and had a significant positive correlation between expression level and copy number (Fig. 1D). Similar trends were observed in correlation analysis for ESCC and LSCC (Supplementary Fig. S1e and S1f). One gene, FGF19, was expressed at significantly lower levels and had a weaker correlation with copy number when compared with the aforementioned six genes, an observation we confirmed with RT-qPCR gene expression analysis of the 11q13 genes in HNSCC cell lines (Supplementary Fig. S1g). The other 2 protein coding genes in the amplicon—FGF4 and FGF3—are expressed at low or undetectable levels in tumors with a weak correlation between expression and copy number, thus making them unlikely to be oncogenic drivers of 11q13 amplification.
CCND1 and ORAOV1 drive the pathogenic effect of 11q13 amplification
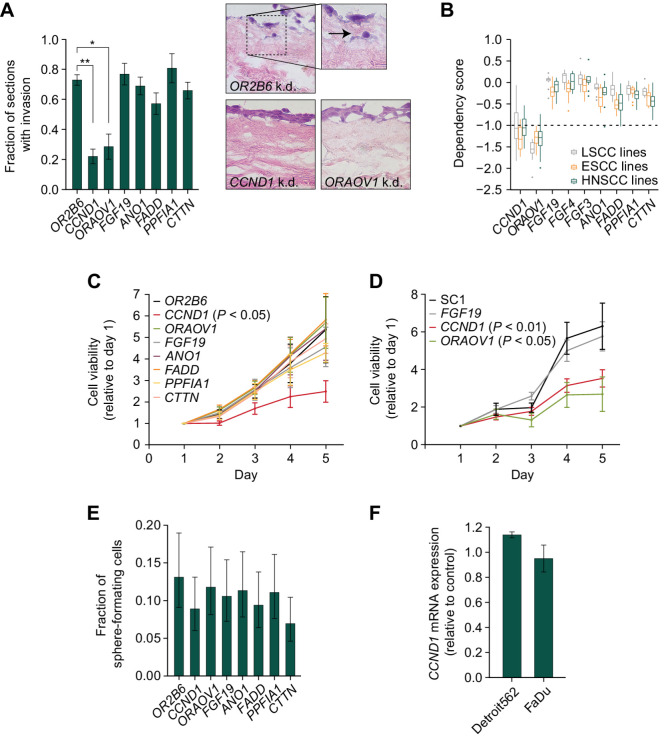
To study the necessity of the remaining 6 protein coding genes on tumor associated cellular phenotypes, we used two HNSCC cell lines—FaDu and Detroit562—which harbor the 11q13 amplification in loss of function assays (Supplementary Fig. S2a and S2b). For CRISPR interference (CRISPRi) experiments, we compared knockdown of 11q13 amplicon genes against knockdown with a guide directed against OR2B6, a non-expressed gene not on the 11q13 amplicon. Because cell viability is known to decrease in proportion to number of cut sites within the genome, for CRISPR knockout experiments, we designed a “SC1” negative control that targets a non-transcribed region of the 11q13 region to control for the multiplicity of edits introduced in the amplified 11q13 region to our target genes. We first examined the effect of 11q13 gene knockdown on cell invasion in an ex vivo organotypic model of SCC. This organotypic model enables us to model cell invasion through the basement membrane by closely recreating the tumor microenvironment with physiological layers of a dermis scaffold, fibroblasts, and keratinocytes at an air–liquid interface (19). CRISPRi-mediated knockdown of the protein coding 11q13 genes in FaDu cells seeded on the organotypic cultures revelated that only depletion of CCND1 and ORAOV1 affected invasion of cells through the basement membrane (Fig. 2A; Supplementary Fig. S2b and S2c).
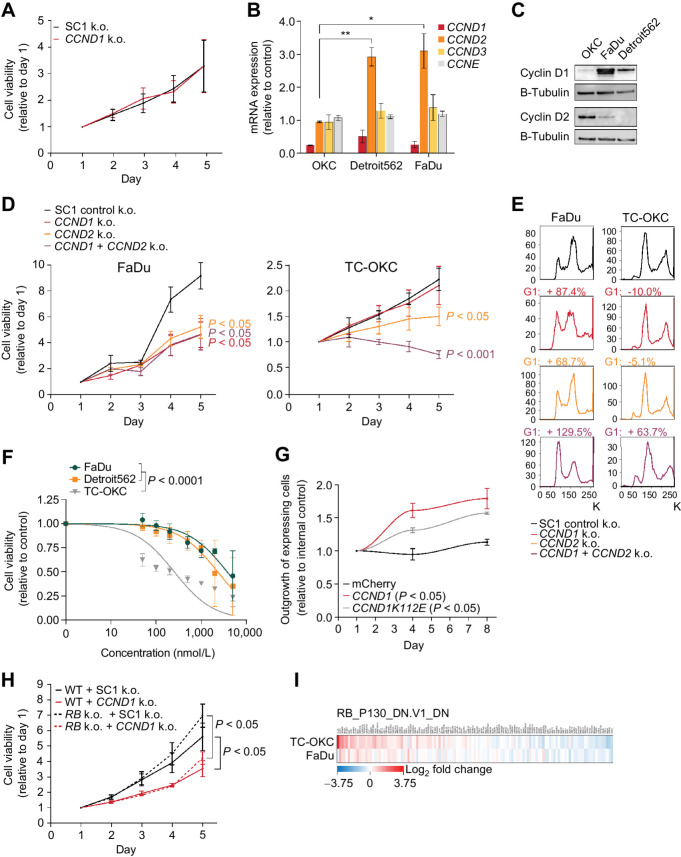
Figure 2.
CCND1 and ORAOV1 drive the pathogenic effect of 11q13 amplification. A, Left, fraction of sections with invading FaDu cells in an organotypic model upon silencing of 11q13 gene expression through CRISPR-i (*, P < 0.05; **, P < 0.01). Right, representative hematoxylin and eosin (H&E) images at 20x magnification. Arrow indicates invading cell. B, Dependency scores on 11q13 genes for LSCC, ESCC, and HNSCC cell lines, from CRISPR knockout screens from project Achilles (DepMap). Upper and lower whiskers represent the largest and smallest observed values within 1.5 times the interquartile range from the ends of the box. Scores below −1.0 show significant dependency. C, Relative cell viability values based on Alamar Blue assay upon CRISPR-i mediated knockdown of 11q13 genes in FaDu cells. P values calculated with two-way ANOVA test. Average doubling times in hours; OR2B6: 38.3, CCND1: 73.0, ORAOV1: 38.3, FGF19: 43.9, ANO1: 39.4, FADD: 37.8, PPFIA1: 45.7, CTTN: 41.6. D, Relative cell viability values based on Alamar Blue assay upon CRISPR mediated knockout of 11q13 genes in FaDu cells. Average doubling times in hours: SC1: 36.1, FGF19: 38.0, CCND1: 52.7, ORAOV1: 67.1. E, Spheroid formation frequency upon CRISPR-i mediated knockdown of 11q13 genes in FaDu cells. Error bars represent 95% confidence interval, P values calculated with χ2 test (nonsignificant). F,CCND1 expression upon ORAOV1 knockout in FaDu and Detroit562 cells.
To identify the phenotype of knockout of 11q13 genes in a larger set of cell lines, we analyzed data using the DepMap (project Achilles CRISPR knockout screens) database. We confirmed CCND1 and ORAOV1 as the only protein coding genes on the 11q13 amplicon whose knockout results in lower cancer cell viability in HNSCC, ESCC, and LSCC (Fig. 2B). In cell proliferation experiments with the FaDu and Detroit562 cell lines using both CRISPRi and CRISPR-mediated knockout, we were able to recapitulate in vitro proliferation effects upon CCND1 and ORAOV1 disruption observed in the DepMap data (Fig. 2C and D; Supplementary Fig. S2d, S2e, and S2f), whereas sphere formation was not affected upon knockdown (Fig. 2E; Supplementary Fig. S2g). Considering the genomic proximity between the genes in the 11q13 region, we also sought to verify that the observed effects were gene specific and not due to effects of perturbations on neighboring genes. We validated that silencing of the 11q13 genes does not affect expression of the neighboring genes, confirming the independence of the observed effects (Fig. 2F; Supplementary Fig. S2h and S2i).
MIR548K is a putative driver of tumorigenicity on the 3′ end of the 11q13 amplicon
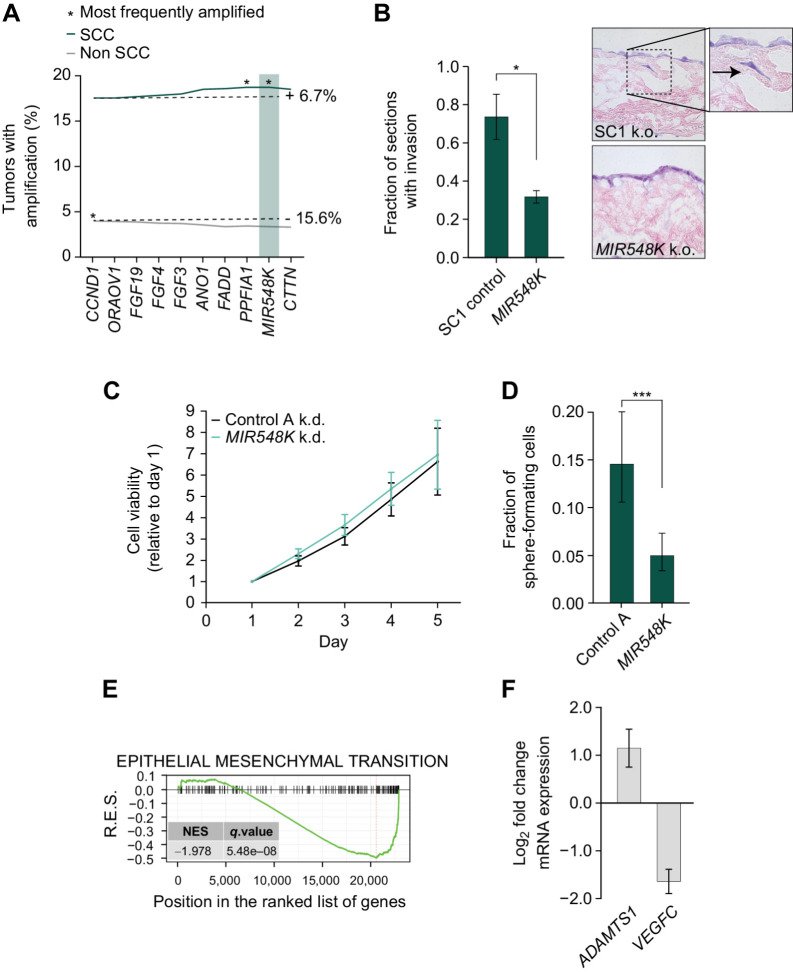
Although we identified CCND1 and ORAOV1 as potential drivers of 11q13 amplification in SCC, these 2 genes are both located on the 5′ end of the amplicon and thus are unlikely to explain the selection for the entire 800kb amplicon in SCC (Fig. 1B). Therefore, we hypothesized that there is an additional genetic element at the 3′ end of the amplicon that leads to selection of the full amplicon in SCC and drives the oncogenic effect of the amplicon in cooperation with CCND1 and ORAOV1. The 3′ end of the amplicon contains the actively transcribed miRNA MIR548K, which was previously implicated in ESCC (4, 16, 20). MIR548K is the most frequently amplified gene on the amplicon (18.73%, vs. 17.55% for CCND1) in SCC (Fig. 3A). Analysis of TCGA miRNA expression data confirmed that there is a significant positive correlation between copy number and gene expression, indicating that MIR548K expression is upregulated upon 11q13 amplification (Supplementary Fig. S3a).
Figure 3.
MIR548K is a putative driver of tumorigenicity on the 3′ end of the 11q13 amplicon. A, Amplification rates of 11q13 genes in all SCC (n = 1,356) and all non-SCC (n = 9,594) tumors in TCGA PanCancer database. SCC: CCND1 amplification rate = 17.55%, MIR548K amplification rate = 18.73%. Non-SCC: CCND1 amplification rate = 3.98%, MIR548K amplification rate = 3.36%. B, Left, Fraction of sections with invading FaDu cells in an organotypic model upon LNA-mediated knockdown of MIR548K (*, P < 0.05). Right, representative hematoxylin and eosin (H&E) images at 20x magnification. Arrow indicates invading cell. C, Relative cell viability values based on Alamar Blue assay upon LNA mediated knockdown of MIR548K in FaDu cells. Average doubling times in hours; A: 35.2, MIR548K: 34.3. D, Spheroid formation frequency upon LNA mediated knockdown of MIR548K in FaDu cells. Error bars represent 95% confidence interval, P value calculated with χ2 test (***, P = 0.001). E, Running Enrichment Scores of GSEA upon MIR548K knockout in FaDu cells for the Hallmark_Epithelial_Mesenchymal_Transition Gene Set. F, Log2FoldChange of ADAMTS1 (n = 3) and VEGFC (n = 2) expression upon MIR548K knockout in FaDu cells.
To investigate the functional role of MIR548K in SCC tumorigenic behavior, we returned to our in vitro and ex vivo models of SCC with loss of function assays. CRISPR-mediated knockout of MIR548K showed > 90% efficiency, whereas knockdown using LNAs showed a reduction of 60% to 80% in MIR548K expression as compared with non-targeting scrambled LNA “A” (Supplementary Fig. S3b and S3c). Using our organotypic model of SCC, we found that knockout of MIR548K decreases the invasive phenotype of FaDu cells to a similar extent as knockout of CCND1 or ORAOV1 (Fig. 3B). However, unlike knockdown of CCND1 and ORAOV1, knockdown of MIR548K does not affect growth of the 11q13-amplified cancer cells in 2D culture (Fig. 3C; Supplementary Fig. S3d), but does decrease the tumor sphere formation capabilities of the HNSCC lines (Fig. 3D; Supplementary Fig. S3e). These two phenotypes are specific to depletion of the MIR548K, suggesting a role for MIR548K in driving tumorigenesis that is distinct from that of CCND1 or ORAOV1. This observation is supported by previous studies reporting an essential role for MIR548K in migration and invasion in ESCC (4, 16, 20). Indeed, GSEA (21) of RNA-seq data of MIR548K knockout in FaDu cells points to involvement of MIR548K in the epithelial–mesenchymal transition, exemplified by downregulation of the E-cadherin pathway (Fig. 3E; Supplementary Fig. S3f). Moreover, consistent with previous findings (4), downregulation of MIR548K resulted in upregulation of ADAMTS1 and downregulation of VEGFC, indicating a role for MIR548K in the epithelial–mesenchymal transition through regulation of VEGFC levels (Fig. 3F). Thus, CCND1, ORAOV1, and MIR548K appear to exert effects on tumorigenesis via distinct mechanisms.
Development of a novel primary keratinocyte cell engineering workflow
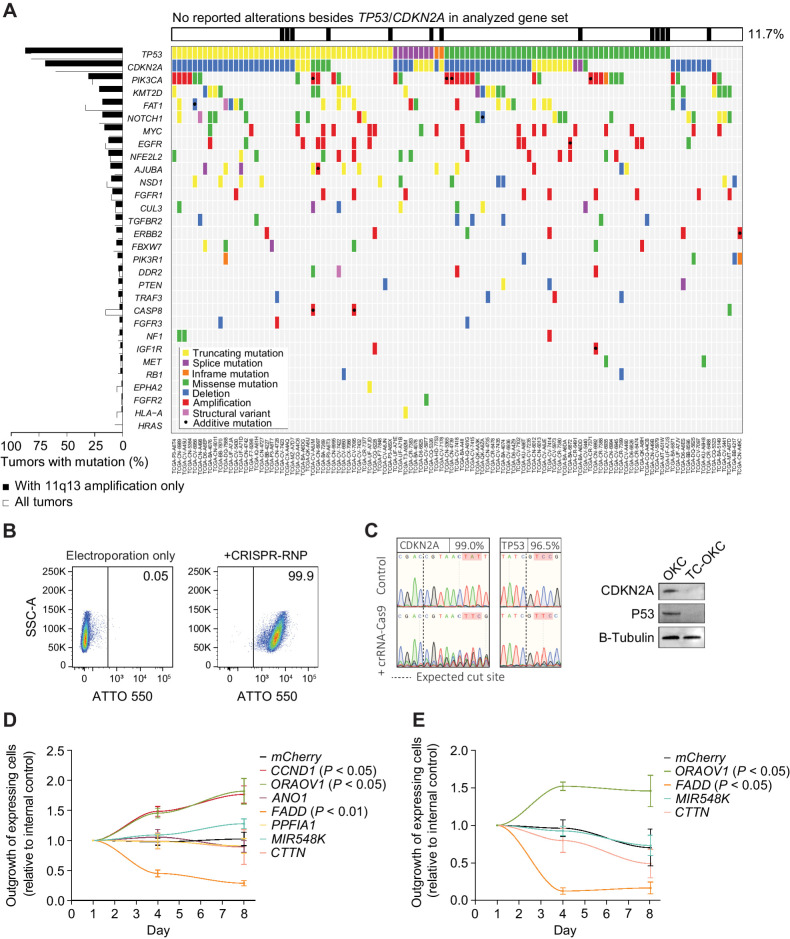
We next sought to identify whether these candidate oncogenes are sufficient to promote tumorigenic behavior in a primary cell model, using patient derived OKCs. To recreate the genetic background in which 11q13 amplification occurs in HNSCC, we engineered the OKCs to carry deleterious TP53 and CDKN2A mutations, as 88% and 70% of tumors with 11q13 amplification have these genes either mutated or deleted, respectively (Fig. 4A). To engineer the primary OKCs, we developed an electroporation-based method for efficient delivery of CRISPR-RNPs (see Methods). Because of the challenges of culturing and manipulating keratinocytes, single cell selection or sorting to improve population purity following transfection typically results in cell death, placing limitations on creating engineered keratinocyte lines.
Figure 4.
CCND1 and ORAOV1 provide a competitive growth benefit in a novel primary cell model. A, Mutational pattern of the most frequently and significantly altered genes in 11q13-amplified HNSCC tumors (TCGA, n = 111). B, Flow cytometry data on uptake of ATTO-550 labeled Cas9:crRNA:tracrRNA by primary OKC 24 hours post electroporation. C, Left, Sanger Sequencing traces of engineered OKC 96 hours post electroporation. Right, immunoblot for CDKN2A (P16) and P53 6 days post electroporation. D, Outgrowth of 11q13-mCherry/mCherry overexpressing TC-OKC versus EBFP overexpressing TC-OKC. E, Outgrowth of CCND1-EGFP + 11q13-mCherry/mCherry double overexpressing TC-OKC versus EBFP overexpressing TC-OKC.
Multiplexed electroporation of CRISPR RNPs resulted in high transfection efficiency (Fig. 4B) and efficiently created deleterious indels in TP53 and CDKN2A (Fig. 4C), without substantially affecting cell viability or cell proliferation (Supplementary Fig. S4a and S4b). We were able to repeat this process with other targets and achieve similar efficiencies (Supplementary Fig. S4c), thus allowing us to efficiently model cancer-specific mutations in primary keratinocytes with far greater efficiency than previous methods. In addition, we demonstrate that target indels are maintained in the engineered cells over several weeks of culturing (Supplementary Fig. S4d) and that the cell engineering process is gentle enough to allow for multiple rounds of genome editing without sacrificing efficiency or cell health (Supplementary Fig. S4e). Further, this method is highly scalable, enabling the electroporation of up to 8 × 10⁶ keratinocytes in a single reaction for the creation of cryopreserved engineered human keratinocyte cell banks (Supplementary Fig. S4f). Finally, we demonstrate that we can adapt this workflow to keratinocytes of multiple origins as well as fibroblasts, thereby establishing a highly versatile strategy for the engineering of primary human keratinocytes (Supplementary Fig. S4g).
CCND1 and ORAOV1 provide a competitive growth benefit in OKCs
We next determined the effects of amplification of the 11q13 genes on primary OKC cultures from at least two different donors harboring loss-of-function mutations in both TP53 and CDKN2A via lentivirus-mediated overexpression of the 11q13 genes (Supplementary Fig. S4h and S4i). Lentiviral expression of either CCND1 or ORAOV1 resulted in a competitive proliferation benefit over expression of a control lentivirus (EBFP only; Fig. 4D). Consistent with previous results, expression of MIR548K did not endow the TC-OKCs with a significant proliferation advantage in 2D culture. Furthermore, when we expressed CCND1 in combination with a second 11q13 gene, the OKCs generally grew more slowly, except when CCND1 was co-expressed with ORAOV1 (Fig. 4E). Because these effects are additive, these data suggest that amplification of ORAOV1 and CCND1 may drive SCC proliferation through distinct pathways and co-amplification of both CCND1 and ORAOV1 can cooperate in SCC tumorigenesis.
Expression of the pro-apoptotic gene FADD in the TC-OKCs significantly hindered cell proliferation when compared with control TC-OKCs, suggesting that increased expression of FADD is detrimental to SCC proliferation. Despite the negative effect that FADD amplification has on cell proliferation, virtually all SCCs with 11q13 amplification have an amplification that extends past the 3′ boundary of CCND1/ORAOV1/FGF19, thereby including the FADD gene. The inclusion of the FADD gene in the SCC-specific 11q13 amplicon therefore suggests that the oncogenic benefit of the 3′ end of the amplicon, which contains MIR548K, outweighs the suppressive effects of FADD. In addition, the benefit of the MIR548K amplification may be lineage restricted to SCCs as HCCs and other tumor types less frequently exhibit 11q13 amplification beyond focal gain of CCND1/ORAOV1/FGF19.
11q13-mediated amplification of CCND1 rewires CCND1 signaling in SCC
Having established CCND1 and ORAOV1 as drivers of SSC proliferation on the 11q13 amplicon, we next aimed to understand the mechanisms through which these two oncogenes drive cell proliferation. Because CCND1 is a well-established oncogenic driver in several tumor types, including SCC, we first decided to investigate the mechanism through which CCND1 promotes proliferation in SCC and how this relates to its normal function in nonmalignant cells.
To compare the effect of CCND1 on proliferation between noncancerous and cancerous cells, we used our keratinocyte engineering workflow to knockout CCND1 in TC-OKCs. In contrast to the necessity of CCND1 in SCC cells (Fig. 2B–D; Supplementary Fig. S2d and S2f), knockout of CCND1 in TC-OKCs had no effect on cell proliferation (Fig. 5A). We identified that loss of CCND1 expression in SCC lines results in a strong upregulation of CCND2 expression (Fig. 5B; Supplementary Fig. S5a), implying the activation of a compensatory mechanism. In contrast, TC-OKCs did not upregulate any members of the Cyclin family upon loss of CCND1. CRISPRi-mediated knockdown of CCND1 in the SCC lines recapitulated the CCND2 compensation, establishing that this effect is intrinsic to the 11q13 amplified lines and not an artifact of CRISPR-mediated gene cutting (Supplementary Fig. S5b). These data suggest that 11q13 amplification induces CCND1 oncogene dependence as CCND1-amplified lines – but not TC-OKCs - exhibit a dependence on CCND1 for cell proliferation.
Figure 5.
11q13-mediated amplification of CCND1 rewires CCND1 signaling in SCC. A, Relative cell viability values based on Alamar Blue assay upon CCND1 knockout in TC-OKC. Average doubling times in hours; SC1: 54.9, CCND1: 55.0. B, Gene expression values 96 hours post CCND1 knockout, relative to control (*, P < 0.05; **, P < 0.01). C, Immunoblot for Cyclin D1, Cyclin D2, and B-tubulin. D, Relative cell viability values based on Alamar Blue assay upon knockout of Control SC1, CCND1, CCND2, or CCND1+CCND2 in FaDu cells (left) and TC-OKC (right). Average doubling times for FaDu cells in hours; SC1: 30.0, CCND1: 43.4, CCND2: 40.1, CCND1+CCND2: 43.1. Average doubling times for OKC in hours: SC1: 83.2, CCND1: 89.3, CCND2: 164.3, CCND1+CCND2: not determined due to significant cell loss. E, Cell cycle profile through DNA staining with FxCycle Violet Ready Flow upon knockout of Control SC1, CCND1, CCND2, or CCND1+CCND2 in FaDu cells (left) and TC-OKC (right). F, Dose–response curve fitted to nonlinear regression model in TC-OKC, FaDu, and Detroit562 depicting the effect of Palbociclib treatment for 72 hours (0.5–5 μmol/L). P value calculated with extra sum of square F-test. G, Outgrowth of CCND1-mCherry or CCND1K112E-mCherry overexpressing TC-OKC versus EBFP overexpressing TC-OKC. H, Relative cell viability values based on Alamar Blue assay upon CCND1 knockout in RB1/RBL1/RBL2 knockout FaDu cells. Average doubling times in hours; WT/SC1: 39.6, WT/CCND1: 52.1, RB/SC1: 35.7, RB/CCND1: 46.3. I, RNA-seq expression data on the RB_P130 gene set upon CCND1 knockout in TC-OKC and FaDu cells.
Analysis of Basal Cyclin D1 and Cyclin D2 levels in TC-OKCs and 11q13-amplified SSC lines revealed Cyclin D2 as the primary expressed Cyclin D in OKCs, in contrast to the high Cyclin D1 levels in FaDu and Detroit562 (Fig. 5C). Both FaDu and Detroit562 cells express low levels of Cyclin D2 relative to Cyclin D1, thereby leading us to investigate whether CCND2 is necessary in the 11q13-amplified setting. Knockout of CCND1 and CCND2—either individually or in combination—in both 11q13-amplified lines resulted in significantly slowed cell proliferation with accompanying G1–S cell-cycle arrest (Fig. 5D and E; Supplementary Fig. S5a, S5c, and S5d). In contrast, TC-OKCs exhibited an intermediate proliferation defect with no cell-cycle arrest upon CCND2 knockout and only display a robust proliferation defect upon dual CCND1/2 knockout (Fig. 5D and E). Because 11q13-amplified cell lines require both CCND1 and CCND2 for proliferation, and knockout of both CCND1 and CCND2 results in additive G1–S cell-cycle arrest, CCND1 and CCND2 therefore may be acting through distinct pathways in the 11q13-amplified setting, but may be redundant in noncancerous keratinocytes. The difference in Cyclin D dependence between our noncancerous and cancerous cell models suggests that 11q13 amplification rewires cyclin-dependent proliferation pathways, leading CCND1 to have a distinct and non-redundant role from CCND2 in amplified SCC cells compared with non-amplified OKCs.
Because the 11q13 amplified cancer cells depend on high levels of Cyclin D1, we hypothesized that they would be sensitive to treatment with the CDK4/6 inhibitor palbociclib. As palbociclib inhibits both Cyclin D1 and D2 function via targeting CDK4/6, we observed a proliferation defect upon palbociclib treatment in TC-OKCs (Fig. 5F). In contrast, neither 11q13-amplified SSC cell line showed nanomolar sensitivity to palbociclib despite their sensitivity to combined CCND1/2 knockout. This disconnect between the CDK4/6-i and CCND1/2 knockout phenotypes suggest that the Cyclin D dependency in these cells may be independent of CDK4/6 activity. To validate this hypothesis, we expressed the CCND1 mutant CCND1K112E, which is unable to bind to CDK4/6 in TC-OKC (22). Overexpression of CCND1K112E still showed a competitive proliferation benefit in TC-OKCs, consistent with a CDK4/6 independent activity of CCND1 amplification (Fig. 5G).
If Cyclin D1 solely functions through interaction with CDK4/6 and subsequent inhibition of RB1, one would predict that the proliferation inhibitory effect of CCND1 knockout in 11q13 amplified cells could be rescued by loss of RB function. Therefore, we generated RB1/RBL1/RBL2 triple knockout FaDu and Detroit562 lines and assessed whether CCND1 deletion would cause inhibition of proliferation (Supplementary Fig. S5e). CCND1 knockout on this background still abrogated proliferation in these cells (Fig. 5H; Supplementary Fig. S5f). Moreover, immunoblot analysis of pRB levels shows no change in RB1 phosphorylation upon CCND1 knockout in these cells (Supplementary Fig. S5g). These findings are consistent with the interpretation that elevated levels of CCND1 in the 11q13-amplified setting rewire its function in a fashion independent of the CDK4/6–RB1 axis.
Finally, we used RNA-seq to compare the transcriptomes of TC-OKCs with CCND1 knockout versus 11q13-amplified FaDu cells with CCND1 knockout. GSEA revealed that, although loss of CCND1 in TC-OKCs significantly affects RB-related genes, these gene-sets are not affected in the FaDu cells (Fig. 5I; Supplementary Fig. S5h). Thus, the amplification of 11q13 in SCC induces CCND1-dependent oncogene addiction and rewires CCND1 to act primarily through a CDK-independent pathway that is distinct from its role in noncancerous OKCs.
RRM2 is a downstream target in 11q13-amplified SCC
To identify downstream mechanisms through which CCND1 drives tumorigenesis in 11q13-amplified SCC, we compared RNA-seq data generated from CCND1 knockout FaDu cells with expression data of 208 HNSCC tumors (139 without amplification, 69 with amplification, all TP53 and CDKN2A mutant) (5). This comparative analysis identified RRM2 as a gene whose expression is significantly upregulated in cells harboring CCND1 amplification in both data sets (Fig. 6A). The ribonucleotide reductase regulatory subunit M2 (RRM2) is the rate-limiting subunit of the ribonucleotide reductase (RNR) enzyme that catalyzes the generation of deoxyribonucleotides and has previously been shown to be a potential target for treatment of several cancers (23–26). Analysis of RRM2 expression in 1,146 SCC tumors shows a modestly but significantly higher expression of RRM2 in CCND1 amplified tumors amongst all 3 analyzed SCCs (Fig. 6B). Furthermore, RRM2 levels are significantly increased in 11q13-amplified cell lines (Supplementary Fig. S6a). Lentiviral expression of CCND1 in TC-OKC was sufficient to increase RRM2 expression to similar levels as found in the amplified cell lines, confirming RRM2 as a downstream target of CCND1 (Fig. 6C). To assess the role of RRM2 in tumor-proliferation, we knocked out RRM2 in SCC9, Detroit562, and FaDu cells using CRISPR-Cas9 gene editing. Whereas RRM2 knockout in SCC9 cells, which do not harbor the 11q13 amplicon, does not affect cell viability, RRM2 knockout in Detroit562 and FaDu cells significantly abrogated cell viability when compared with a control edited at the CCR5 locus (Fig. 6D).
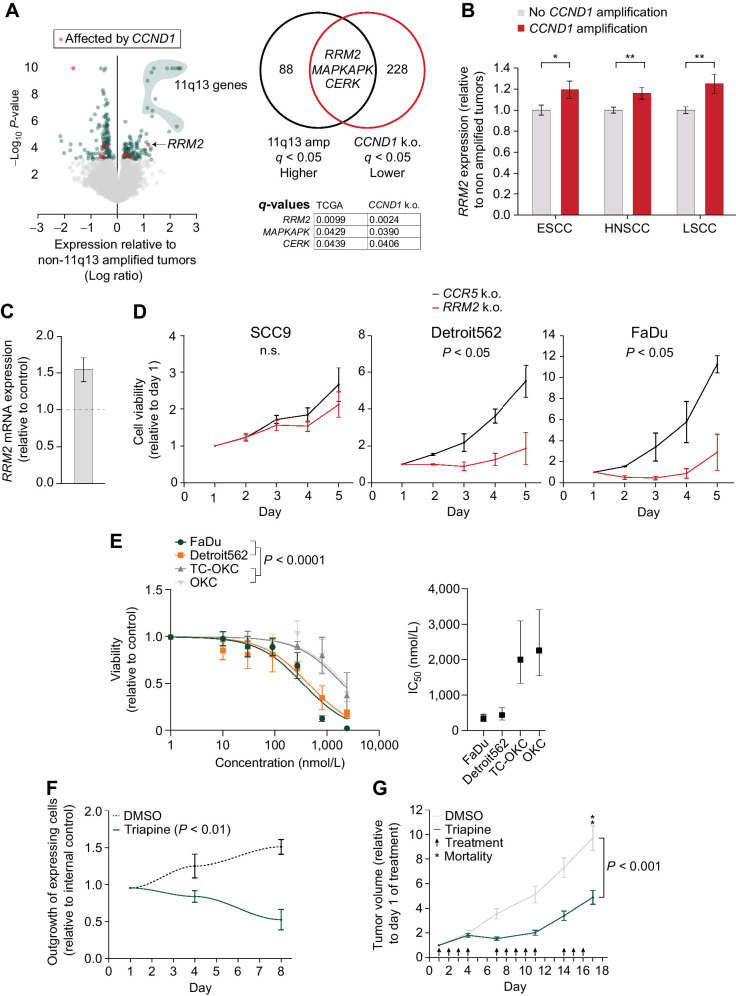
Figure 6.
RRM2 is a potential target in 11q13-amplified SCC. A, Left, Volcano plot of TCGA expression data of TP53/CDKN2A mutated (n = 139) vs. TP53/CDKN2A mutated + 11q13 amplified tumors (n = 69). Red dots indicate genes that are also affected upon CCND1 knockout in FaDu cells. Right, Venn diagram depicting overlapping differentially expressed genes between TCGA 11q13 amplified genes and CCND1 knockout RNA-seq and their q-values. B,RRM2 gene expression levels in SCC tumors with CCND1 amplification relative to tumors without CCND1 amplification (TCGA, ESCC: n = 184, HNSCC (HPV negative): n = 412, LSCC: n = 550). *, P < 0.05; **, P < 0.01. C,RRM2 expression values in TC-OKC 96 hours post transduction with CCND1 overexpression lentivirus, relative to control. D, Relative cell viability values based on Alamar Blue assay upon RRM2 knockout in SCC9, Detroit562, and FaDu cells. Average doubling times for SCC9 cells in hours; CCR5: 67.9, RRM2: 88.6. Average doubling times for Detroit562 cells in hours; CCR5: 39.0, RRM2: 106.8. Average doubling times for FaDu cells in hours; CCR5: 27.5, RRM2: 62.5. E, Left, Dose–response curve fitted to a nonlinear regression model in OKC, TC-OKC, FaDu, and Detroit562 cells depicting the effect of triapine treatment for 72 hours (1–7.3 μmol/L). P value calculated with extra sum of square F-test. Right, IC50 concentration range of the 4 cell types. F, Outgrowth of CCND1 overexpressing TC-OKC versus EBFP overexpressing TC-OKC in the presence or absence of 500 nmol/L triapine, P value calculated by one-sample t-test. G, Tumor growth relative to first day of treatment of either triapine (10 mg/kg, n = 9) or control (2% DMSO, n = 7) treated FaDu tumor xenograft bearing NSG mice.
To assess the potential of targeting RRM2 in 11q13-driven HNSCC, we measured the effect of triapine on proliferation of OKC, TC-OKC, FaDu, and Detroit562 cells. Triapine is a potent inhibitor of the RRM2 subunit of the RNR enzyme (27) and is currently being explored as a cancer treatment option in several phase I, II, and III clinical trials (28, 29). Whereas both OKC and TC-OKC show moderate sensitivity to triapine only at high doses, both 11q13-amplified SCC lines are significantly more sensitive to treatment with average IC50 values 5 to 7 times lower compared with the non-amplified OKCs (Fig. 6E). To validate that targeting RRM2 inhibits proliferation in a CCND1-dependent manner, we analyzed the competitive proliferation benefit of CCND1 overexpression in the absence or presence of triapine. Triapine treatment decreases proliferation of CCND1 overexpressing TC-OKCs, but not control TC-OKCs (Fig. 6F). These data support our hypothesis that the pro-tumorigenic effects of CCND1 in 11q13-amplified cells are dependent on RRM2.
To validate these findings in vivo, we treated FaDu-xenograft tumor bearing mice with either triapine or control vehicle (2% DMSO). Triapine administration significantly slowed down tumor growth (Fig. 6G; Supplementary Fig. S6b and S6c). Whereas in the control group at day 17, 2 of 7 mice died and 4 of 5 remaining mice reached ethical endpoint due to tumor burden, none of the mice out of the triapine group reached endpoint. Treatment was well tolerated and did not affect weight of the mice (Supplementary Fig. S6d). Thus, inhibition of the CCND1–RRM2 axis using triapine or other RNR inhibitors may be a viable strategy for targeting 11q13-amplified tumors.
Upregulation of ORAOV1 is sufficient to drive SCC tumorigenesis in vivo
While CCND1 is a well characterized oncogene across multiple tumor types, the other driver of the pro-proliferation effects of the 11q13 amplification, ORAOV1, has been less well characterized. Nearly all tumors, both SCC and non-SCC, that have CCND1 amplification also have amplification of ORAOV1 (Fig. 7A). Because ORAOV1 is able to drive proliferation of SCC cells, and ORAOV1 and CCND1 are nearly always co-amplified, we hypothesized that ORAOV1 plays a role in tumorigenesis that is distinct from CCND1. Analysis of DepMap cell viability data across all tumor types identified that expression of ORAOV1 is essential for cancer cell proliferation across all tumor types (Fig. 7B). In addition, CRISPR knockout of ORAOV1 in TC-OKCs revealed that ORAOV1 is dispensable for non-tumor keratinocyte cell proliferation, suggesting that ORAOV1 expression may be selectively necessary for proliferation of tumor cells (Fig. 7C).
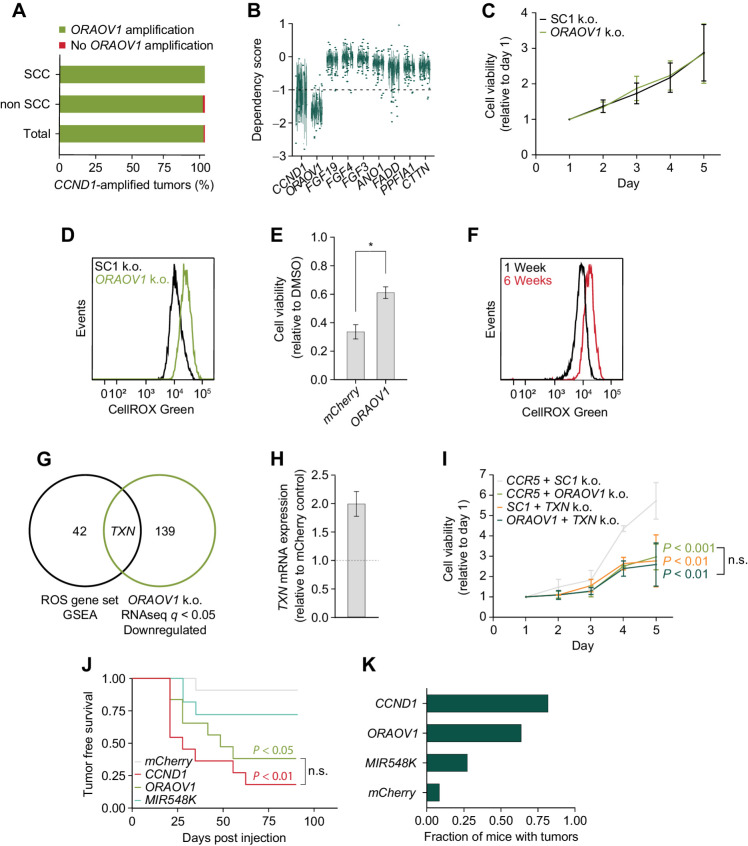
Figure 7.
Upregulation of ORAOV1 is sufficient to drive SCC tumorigenesis in vivo.A, Fraction of CCND1 amplified tumors that carry ORAOV1 amplification in SCC (n = 238) and non SCC (n = 382; TCGA, PanCancer Atlas). B, Dependency scores on 11q13 genes for all cancer lines in database, from CRISPR knockout screens from project Achilles (DepMap). Scores below −1.0 show significant dependency. Upper and lower whiskers represent the largest and smallest observed values within 1.5 times the interquartile range from the ends of the box. C, Relative cell viability values based on Alamar Blue assay upon ORAOV1 knockout in TC-OKC. Average doubling time in hours; SC1: 54.9, ORAOV1: 55.3. D, ROS levels as measured by CellROX Green in FaDu cells upon ORAOV1 knockout. E, Cell viability values based on Alamar Blue assay of mCherry or ORAOV1 overexpressing TC-OKC after 72 hours of treatment with 150 nmol/L TBHP, relative to DMSO. F, ROS levels as measured by CellROX Green in TC-OKC 1 week or 6 weeks post engineering. G, Venn diagram showing overlap between genes in the ROS Pathways GSEA Hallmark gene set and genes downregulated (q < 0.05) in the RNA-seq dataset upon ORAOV1 knockout in FaDu cells. H, TXN expression values in OKC upon ORAOV1 overexpression. I, Relative cell viability values based on Alamar Blue assay upon knockout of Controls SC1+CCR5, CCR5+ORAOV1, SC1+TXN, or ORAOV1+TXN in FaDu cells. P values calculated with two-way ANOVA test. Average doubling times in hours; CCR5/SC1: 38.2, CCR5/ORAOV1: 61.2, SC1/TXN: 65.3, ORAOV1/TXN: 69.9. J, Tumor-free survival of NSG mice injected with 1E6 human TC-OKC cells overexpressing indicated gene (mCherry: n = 12, CCND1, ORAOV1, MIR548K: n = 11 mice). P values calculated with a Log-Rank Mantel–Cox test with post-hox P value adjustment for multiple comparisons. K, Fraction of mice with tumors at endpoint (90 days post injection).
Although not much is known about the function of ORAOV1 in mammalian cells, one study suggests its involvement in ROS metabolism (30), and several studies report that the ORAOV1 ortholog LTO1 in Arabidopsis is essential for redox regulation (31–33). To explore the possibility that ORAOV1 has a similar role in HNSCC cells, we measured ROS levels in FaDu cells with and without knockout of ORAOV1. ORAOV1 knockout markedly increased ROS levels in cancer cells, supporting a role for ORAOV1 in redox regulation in cancer cells (Fig. 7D; Supplementary Fig. S7a). An increase in ROS levels during the early stages of tumorigenesis can have significant pro-tumorigenic effects (34), but persistently high ROS levels can induce apoptosis and senescence in tumor cells (35). Therefore, balanced redox regulation is an important feature of tumors (5, 34), and upregulation of ORAOV1 via 11q13 amplification may be one mechanism cancer cells use to regulate ROS levels. This is supported by our finding that expression of ORAOV1 protects TC-OKCs from a loss in cell viability upon treatment with the oxidant TBHP (Fig. 7E).
Because 11q13 amplification most often occurs after loss of TP53 and CDKN2A in SCC, we were interested to see how these mutations affect ROS levels. We engineered OKCs with dual TP53 and CDKN2A mutations and measured ROS levels 1 week and 6 weeks after gene editing. TC-OKC accumulate higher ROS levels over time (Fig. 7F), consistent with literature that shows that TP53 mutations increase ROS levels (36). Thus, for SCC, 11q13 amplification-mediated ORAOV1 overexpression may help regulate ROS levels upon TP53 and CDKN2A mutation during early tumorigenesis. To identify possible mechanisms through which ORAOV1 counters oxidative stress, we overlaid the ROS Pathways GSEA Hallmark gene set with RNA-seq data of ORAOV1 knockout FaDu cells. We found that loss of ORAOV1 significantly decreases the level of thioredoxin (TXN; Fig. 7G, Supplementary Fig. S7b). TXN is a small reductase that plays a key role in countering oxidative stress and is upregulated in many cancers (34, 37).
To verify the RNA-seq data, we performed RT-qPCR on ORAOV1 knockout FaDu and Detroit562 cells and found similar downregulation of TXN levels (Supplementary Fig. S7c). Furthermore, we found that for HNSCC, compared with TC-OKCs, specifically the ORAOV1-amplified lines have increased TXN levels (Supplementary Fig. S7d). Finally, overexpression of ORAOV1 in TC-OKCs confirmed that ORAOV1 overexpression increases TXN levels (Fig. 7H).
To assess whether the growth effect of ORAOV1 on cancer cells is through its effect on TXN expression, we engineered FaDu cells to carry either control, ORAOV1, TXN, or combined ORAOV1 and TXN deleterious indels. Knockout of either ORAOV1 or TXN individually resulted in decreased cell growth, but TXN knockout did not have an additive effect on the growth decrease upon ORAOV1 knockout (Fig. 7I), suggesting these genes may function through a shared pathway. Thus, the ORAOV1–TXN signaling axis activated via 11q13 amplification may be a key regulator of oxidative stress in SCC.
Finally, we assessed the oncogenic driver potential of CCND1, ORAOV1, and MIR548K in vivo. We injected NSG mice with the engineered TC-OKC overexpressing either CCND1, ORAOV1, MIR548K or mCherry (control). Whereas only 1/12 of the control mice developed a tumor, overexpression of any of these 3 genes increased tumor formation in multiple mice (Fig. 7J; Supplementary Fig. S7e). CCND1 was the most potent driver of tumorigenesis, with 82% of mice developing tumors within 9 weeks of injection (Fig. 7J and K). The level of CCND1 expression was directly correlated to tumor growth, emphasizing the effect of 11q13 amplification and thus overexpression on the tumors (Supplementary Fig. S7f). We found however, that ORAOV1 is an independent contributor to tumorigenesis, as 64% of mice injected with ORAOV1-overexpressing TC-OKC developed tumors. RT-qPCR analysis showed that these tumors have a 5-fold upregulation of TXN compared with control cells, corroborating our previous results (Supplementary Fig. S7g). These data demonstrate that activation of the ORAOV1–TXN signaling axis via 11q13 amplification is a potent oncogenic event for SCC tumorigenesis and may represent a critical pathway to target for anti-cancer therapy.
Discussion
In this study, we identified the three major genomic features of the 11q13 amplicon – CCND1, ORAOV1, and MIR548K – that contribute to SCC tumorigenesis. Our data reveal that these 3 genes contribute to SCC tumorigenesis in three distinct ways, likely leading to powerful cooperation in tumorigenesis. 11q13 amplification typically occurs as an early genomic event during SCC tumorigenesis, following loss of TP53 and CDKN2A (2–4). As these previous two events are insufficient to robustly induce tumors (Fig. 7J), we believe that the amplification of the 11q13 locus—and associated amplification of CCND1, ORAOV1, and MIR548K—is a critical oncogenic event during SCC tumorigenesis of 11q13-amplified tumors. Previous studies have identified loss of TP53 or CDKN2A occurring at some level in normal skin or premalignant lesions (3, 38, 39), yet 11q13 amplification has not yet been detected in premalignant HNSCC lesions (3). Therefore, amplification of 11q13 may represent a critical event that drives neoplastic growth during the early stages of SCC tumorigenesis.
We observed that the amplification in the 11q13 region in SCC differs from the length of the amplicon in other tumor types, such as HCC. While 11q13 amplification in SSC represents the larger region that we analyzed in this study, 11q13 amplification in HCC is typically restricted to ORAOV1, CCND1, and FGF19, with FGF19 being a critical component of the amplicon in HCC in comparison to SCC (17, 18). We identify that in SCC, the 3′ end of the amplicon has a functional role that is likely mediated by amplification of the MIR548K locus. Because miRNAs are known to have significant pleiotropy dependent on cellular context (40, 41), it is feasible that MIR548K exhibits its tumorigenic effect selectively in squamous epithelia, and therefore the miRNA-containing 3′ end of the 11q13 amplicon is only selected for in the context of SCC tumorigenesis. Indeed, previous studies reporting a role for MIR548K in tumorigenesis and metastasis, predominantly do so in the context of SCC (4, 16, 20). However, future studies directly evaluating the role of MIR548K across different cellular contexts and how MIR548K elicits its effects in these scenarios are needed to clearly establish this relationship.
The primary focus of previous studies investigating the function of 11q13 amplification has largely been on CCND1 due to its established role as a canonical oncogene across multiple tumor types. Our study further supports the role of CCND1 as a key oncogene in 11q13-amplified SCC, with 11q13 amplified cancer cells exhibiting CCND1 oncogene addiction similar to what has been described in other CCND1-dependent tumors (42, 43). However, we also identify key differences between the function of CCND1 in the 11q13-amplified context versus non-amplified settings, supporting the hypothesis that CCND1 signaling is rewired depending on its level of amplification, expression, or both. Notably, the RRM2-mediated growth effect of CCND1 amplification appears to be unique to the 11q13-amplified context. Furthermore, amplification status of CCND1 in SCC also affects the role of CCND2, as CCND1 and CCND2 appear to have significant redundancy in their function in the non-amplified setting and more specialized roles in the amplified setting.
In contrast to, for example, breast cancers with CCND1 amplification, in HNSCCs CCND1 amplification occurs in a background of CDKN2A mutations or deletions (5, 44). The combination of these CCND1 and CDKN2A events gives a worse prognosis than the events by themselves (45). Loss of p16 results in activation of the CDK-pathway, yet CCND1 amplification provides an additional benefit to the cancer cells. This can be explained by an extra selective pressure as each event also contributes to other oncogenic processes/pathways besides their shared pathway. This may further support CDK-independent effects of CCND1 amplification in these cells.
This difference in CCND1 signaling may have a direct impact on how we target the CCND1 signaling axis for targeted cancer therapy, as 11q13-amplified SCC lines are significantly more resistant to CDK4/6 inhibition relative to non-amplified cells. Indeed, clinical trials of CDK4/6 inhibitors in HNSCC have met with limited success (8–10). However, this rewiring of CCND1 function in the 11q13-amplified setting may open up additional avenues for targeting the CCND1 signaling axis in SSC that have not been explored previously. Two examples of this are direct inhibition of CCND1 and inhibition of the downstream RRM2 pathway that seems to largely mediate the pro-growth effects of CCND1 in SCC. We find that these two strategies for inhibiting the CCND1 axis in 11q13-amplified SCC are significantly more effective to elicit an antitumor growth effect compared with a standard CDK4/6 inhibition strategy while avoiding significant toxicity in normal keratinocytes. Therefore, these and other CDK4/6 independent targeting strategies should be investigated further.
Despite both CCND1 and ORAOV1 being co-amplified in > 98% of tumors, previous studies have largely overlooked the role of ORAOV1 and the relationship between these two genes. We have now identified that ORAOV1 and CCND1 drive cancer growth through two distinct pathways with an additive effect, creating a potent oncogenic combination that occurs in over 6% of all tumors, regardless of tissue of origin. Although previous studies in yeast and plants have positioned ORAOV1 as a putative oncogene, our study provides a direct line of evidence that gain of ORAOV1 is sufficient to drive SCC tumor growth. Indeed, in each of our in vitro, ex vivo, and in vivo models of SCC tumorigenesis, ORAOV1 had a similar magnitude of oncogenic effect as CCND1.
Amplification of ORAOV1 and downstream activation of TXN may represent a generalizable mechanism through which cancer cells deal with oxidative stress during tumorigenesis. In addition, the ORAOV1–TXN signaling axis that is activated in the 11q13-amplified setting represents another potential target for targeted therapy, as deletion of ORAOV1 exhibits potent antitumor growth effects across multiple cancer types. Targeting the thioredoxin system has recently been explored as a strategy for treatment of several cancer types, thus providing us with a direction of targeting ORAOV1-mediated signaling (37, 46, 47).
We identified RRM2 and TXN activation as two of the major altered downstream effects of the 11q13 amplification. Remarkably, TXN was originally discovered as activator of the ribonucleotide reducing function of the RNR enzyme (48). Moreover, TXN and RRM2 have been found to be upregulated together, correlating to poor prognosis, and through their physical interaction contribute to cancer malignancy (49, 50). This points towards potential cooperation between amplicon effectors.
In conclusion, our study identifies CCND1, ORAOV1, and MIR548K as putative oncogenes that drive the pathogenic effect of 11q13 amplification through three independent oncogenic events. Continuing to investigate the role of these three oncogenes will provide critical information about potential anti-cancer therapeutic strategies for 11q13-amplified SCC and may help guide current treatment paradigms.
Supplementary Material
CRISPR sequences.
Lentiviral overexpression cloning sequences.
qPCR sequences
TCGA HNSCC tumor selection
11q13 genes are frequently co-amplified in SCC
Growth analyses upon efficient and gene specific knockdown in 11q13 amplified cells
Growth analyses upon abrogation of MIR548K
Development of a novel primary keratinocyte cell engineering workflow
Expression and growth analyses upon CCND1 or RB loss
Expression and inhibition of RRM2
Expression analysis of TXN and in vivo tumors
Acknowledgments
This work was supported, in part, by the NIH grants R01-DE029890 and R01-DC018076 (to A.D. Tward). The authors thank René Bernards, Andrei Goga, Eliah Shamir, Sonia Scaria, and Bogdan Popescu for helpful discussions, Licia Selleri for providing access to critical instrumentation, Lesley Eschinger and James Brady for enabling the collaboration with MaxCyte, and Maurizio Risolino and Andrea Yeung for technical assistance.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
A.G. Mancini reports personal fees from MaxCyte, Inc. during the conduct of the study; personal fees from MaxCyte, Inc. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
C.I. Mahieu: Conceptualization, data curation, formal analysis, investigation, methodology, writing–original draft, writing–review and editing. A.G. Mancini: Conceptualization, methodology. E.P. Vikram: Investigation. V. Planells-Palop: Resources, methodology. N.M. Joseph: Investigation. A.D. Tward: Conceptualization, resources, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gerstung M, Jolly C, Leshchiner I, Dentro SC, Gonzalez S, Rosebrock D, et al. The evolutionary history of 2,658 cancers. Nature 2020;578:122–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Veeramachaneni R, Walker T, Revil T, De Weck A, Badescu D, O'Sullivan J, et al. Analysis of head and neck carcinoma progression reveals novel and relevant stage-specific changes associated with immortalization and malignancy. Sci Rep 2019;9:11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhang W, Hong R, Li L, Wang Y, Du P, Ou Y, et al. The chromosome 11q13.3 amplification associated lymph node metastasis is driven by miR-548k through modulating tumor microenvironment. Mol Cancer 2018;17:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015;517:576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011;333:1157–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ramos-García P, Ruiz-Ávila I, Gil-Montoya JA, Ayén Á, González-Ruiz L, Navarro-Triviño FJ, et al. Relevance of chromosomal band 11q13 in oral carcinogenesis: an update of current knowledge. Oral Oncol 2017;72:7–16. [DOI] [PubMed] [Google Scholar]
- 8. Edelman MJ, Redman MW, Albain KS, McGary EC, Rafique NM, Petro D, et al. SWOG S1400C (NCT02154490)—a phase II study of palbociclib for previously treated cell cycle gene alteration–positive patients with stage IV squamous cell lung cancer (Lung-MAP Substudy). J Thorac Oncol 2019;14:1853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Adkins DR, Lin JC, Sacco A, Ley J, Oppelt P, Vanchenko V, et al. Palbociclib and cetuximab compared with placebo and cetuximab in platinum-resistant, cetuximab-naïve, human papillomavirus–unrelated recurrent or metastatic head and neck squamous cell carcinoma: a double-blind, randomized, phase II trial. Oral Oncol 2021;115:105192. [DOI] [PubMed] [Google Scholar]
- 10. Oppelt P, Ley JC, Worden F, Palka K, Maggiore R, Liu J, et al. Palbociclib and cetuximab in cetuximab-resistant human papillomavirus–related oropharynx squamous-cell carcinoma : a multicenter phase II trial. Oral Oncol 2021;114:105164. [DOI] [PubMed] [Google Scholar]
- 11. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 12. Campbell PJ, Getz G, Korbel JO, Stuart JM, Jennings JL, Stein LD, et al. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, et al. Integrated genomic characterization of esophageal carcinoma. Nature 2017;541:169–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammerman PS, Voet D, Lawrence MS, Voet D, Jing R, Cibulskis K, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ally A, Balasundaram M, Carlsen R, Chuah E, Clarke A, Dhalla N, et al. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell 2017;169:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Song Y, Li L, Ou Y, Gao Z, Li E, Li X, et al. Identification of genomic alterations in esophageal squamous cell cancer. Nature 2014;508:91–5. [DOI] [PubMed] [Google Scholar]
- 17. Sawey ET, Chanrion M, Cai C, Wu G, Zhang J, Zender L, et al. Identification of a therapeutic strategy targeting amplified FGF19 in liver cancer by oncogenomic screening. Cancer Cell 2011;19:347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim RD, Sarker D, Meyer T, Yau T, Macarulla T, Park JW, et al. First-in-human phase I study of fisogatinib (BLU-554) validates aberrant FGF19 signaling as a driver event in hepatocellular carcinoma. Cancer Discov 2019;9:1696–707. [DOI] [PubMed] [Google Scholar]
- 19. Vörsmann H, Groeber F, Walles H, Busch S, Beissert S, Walczak H, et al. Development of a human three-dimensional organotypic skin-melanoma spheroid model for in vitro drug testing. Cell Death Dis 2013;4:e719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen Z, Lin J, Wu S, Xu C, Chen F, Huang Z. Up-regulated miR-548k promotes esophageal squamous cell carcinoma progression via targeting long noncoding RNA-LET. Exp Cell Res 2018;362:90–101. [DOI] [PubMed] [Google Scholar]
- 21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hinds PW, Dowdy SF, Eaton EN, Arnold A, Weinberg RA. Function of a human cyclin gene as an oncogene. Proc Natl Acad Sci USA 1994;91:709–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mazzu YZ, Armenia J, Chakraborty G, Yoshikawa Y, Coggins SA, Nandakumar S, et al. A novel mechanism driving poor-prognosis prostate cancer: overexpression of the DNA repair gene, ribonucleotide reductase small subunit M2 (RRM2). Clin Cancer Res 2019;25:4480–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson EA, Sultana N, Shah KN, Elford HL, Faridi JS. Molecular targeting of RRM2, NF-kB, and mutant TP53 for the treatment of triple-negative breast cancer. Mol Cancer Ther 2021;20:655–64. [DOI] [PubMed] [Google Scholar]
- 25. Rasmussen RD, Gajjar MK, Tuckova L, Jensen KE, Maya-Mendoza A, Holst CB, et al. BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun 2016;7:13398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xiong W, Zhang B, Yu H, Zhu L, Yi L, Jin X. RRM2 regulates sensitivity to sunitinib and PD-1 blockade in renal cancer by stabilizing ANXA1 and activating the AKT pathway. Adv Sci 2021;8:e2100881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cory J, Cory A, Rappa G, Lorico A, Liu MC, Lin TS, et al. Structure-function relationships for a new series of pyridine-2-carboxaldehyde thiosemicarbazones on ribonucleotide reductase activity and tumor cell growth in culture and in vivo. Advan Enzym Regul 1995;35:55–68. [DOI] [PubMed] [Google Scholar]
- 28. Kunos CA, Andrews SJ, Moore KN, Chon HS, Ivy SP. Randomized phase II trial of triapine-cisplatin-radiotherapy for locally advanced stage uterine cervix or vaginal cancers. Front Oncol 2019;9:1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mannargudi MB, Deb S. Clinical pharmacology and clinical trials of ribonucleotide reductase inhibitors: is it a viable cancer therapy? J Cancer Res Clin Oncol 2017;143:1499–529. [DOI] [PubMed] [Google Scholar]
- 30. Togashi Y, Arao T, Kato H, Matsumoto K, Terashima M, Hayashi H, et al. Frequent amplification of oraov1 gene in esophageal squamous cell cancer promotes an aggressive phenotype via proline metabolism and ros production. Oncotarget 2014;5:2962–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Du JJ, Zhan CY, Lu Y, Cui HR, Wang XY. The conservative cysteines in transmembrane domain of AtVKOR/LTO1 are critical for photosynthetic growth and photosystem II activity in Arabidopsis. Front Plant Sci 2015;6:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karamoko M, Cline S, Redding K, Ruiz N, Hamel PP. Lumen thiol oxidoreductase1, a disulfide bond-forming catalyst, is required for the assembly of photosystem II in arabidopsis. Plant Cell 2011;23:4462–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu Y, Wang HR, Li H, Cui HR, Feng YG, Wang XY. A chloroplast membrane protein LTO1/AtVKOR involving in redox regulation and ROS homeostasis. Plant Cell Rep 2013;32:1427–40. [DOI] [PubMed] [Google Scholar]
- 34. Hayes JD, Dinkova-Kostova AT, Tew KD. Oxidative stress in cancer. Cancer Cell 2020;38:167–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reczek CR, Birsoy K, Kong H, Martínez-Reyes I, Wang T, Gao P, et al. A CRISPR screen identifies a pathway required for paraquat-induced cell death. Nat Chem Biol 2017;13:1274–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maillet A, Pervaiz S. Redox regulation of p53, redox effectors regulated by p53: a subtle balance. Antioxid Redox Signal 2012;16:1285–94. [DOI] [PubMed] [Google Scholar]
- 37. Jia JJ, Geng WS, Wang ZQ, Chen L, Zeng XS. The role of thioredoxin system in cancer: strategy for cancer therapy. Cancer Chemother Pharmacol 2019;84:453–70. [DOI] [PubMed] [Google Scholar]
- 38. Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei L, Christensen SR, Fitzgerald ME, Graham J, Hutson ND, Zhang C, et al. Ultradeep sequencing differentiates patterns of skin clonal mutations associated with sun-exposure status and skin cancer burden. Sci Adv 2021;7:eabd7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sood P, Krek A, Zavolan M, Macino G, Rajewsky N. Cell-type-specific signatures of microRNAs on target mRNA expression. Proc Natl Acad Sci USA 2006;103:2746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Farh KKH, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 2005;310:1817–21. [DOI] [PubMed] [Google Scholar]
- 42. Weinstein IB. Addiction to oncogenes-the Achilles heal of cancer. Science 2002;297:63–4. [DOI] [PubMed] [Google Scholar]
- 43. Shan J, Zhao W, Gu W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol Cell 2009;36:469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 2018;33:690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dale B, Cheng M, Park KS, Kaniskan HÜ, Xiong Y, Jin J. Advancing targeted protein degradation for cancer therapy. Nat Rev Cancer 2021;21:638–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stafford WC, Peng X, Olofsson MH, Zhang X, Luci DK, Lu L, et al. Irreversible inhibition of cytosolic thioredoxin reductase 1 as a mechanistic basis for anticancer therapy. Sci Transl Med 2018;10:eaaf7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang J, Li X, Han X, Liu R, Fang J. Targeting the thioredoxin system for cancer therapy. Trends Pharmacol Sci 2017;38:794–808. [DOI] [PubMed] [Google Scholar]
- 48. Laurent TC, Moore EC, Reichard P. Enzymatic synthesis of deoxyribonucleotides. Joural Biol Chem 1964;239:3436–44. [PubMed] [Google Scholar]
- 49. Cadenas C, Franckenstein D, Schmidt M, Gehrmann M, Hermes M, Geppert B, et al. Role of thioredoxin reductase 1 and thioredoxin interacting protein in prognosis of breast cancer. Breast Cancer Res 2010;12:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lou M, Liu Q, Ren G, Zeng J, Xiang X, Ding Y, et al. Physical interaction between human ribonucleotide reductase large subunit and thioredoxin increases colorectal cancer malignancy. J Biol Chem 2017;292:9136–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CRISPR sequences.
Lentiviral overexpression cloning sequences.
qPCR sequences
TCGA HNSCC tumor selection
11q13 genes are frequently co-amplified in SCC
Growth analyses upon efficient and gene specific knockdown in 11q13 amplified cells
Growth analyses upon abrogation of MIR548K
Development of a novel primary keratinocyte cell engineering workflow
Expression and growth analyses upon CCND1 or RB loss
Expression and inhibition of RRM2
Expression analysis of TXN and in vivo tumors
Data Availability Statement
RNA-seq data generated in this study are publicly available at the Gene Expression Omnibus under accession code GSE216849. All other raw data generated in this study are available upon request from the corresponding author.