Abstract
Background
Extensive attention has been given to the role of myeloid-derived suppressor cells (MDSCs) in driving tumor progression and treatment failure. Preclinical studies have identified multiple agents that eliminate MDSCs. However, none have been authorized in the cliniccal ues due to the safety reasons. In the present study, we investigated the efficacy and mechanism of sulforaphane (SFN) to eliminate MDSCs in the tumor microenvironment (TME).
Methods
We monitored SFN effect on tumor growth and the percents or apoptosis of immune cell subsets in mice models bearing LLC or B16 cells. Flow cytometry, quantitative reverse transcription-PCR, immunohistochemistry, ELISA, immunofluorescence, imaging flow cytometry and western blot were performed to validate the role of SFN on MDSCs function in vivo and in vitro. RNA sequencing was then used to interrogate the mechanisms of how SFN regulated MDSCs function. Tumor xenograft models were established to evaluate the involvement of IL-12RB2/MMP3/FasL induced MDSCs apoptosis in vivo. We verified the effect of SFN on MDSCs and CD8+ T cells in the blood samples from a phase I clinical trial (KY-2021–0350).
Results
In this study, we elucidated that SFN liberated CD8+ T-cell antitumor ability by reducing MDSCs abundance, leading to repressed tumor growth. SFN treatment suppressed MDSCs accumulation in the peripheral blood and tumor sites of mice, but had no effect on the bone marrow. Mechanistically, SFN activates IL-12RB2, which stimulates the MMP3/FasL signaling cascade to trigger caspase 3 cleavage and induce apoptosis in MDSCs. Clinically, SFN treatment eliminates peripheral MDSCs and increases the percentage and activation of CD8+ T cells.
Conclusions
Collectively, we uncovered the role of SFN in eliminating MDSCs to emancipate CD8+ T cells through IL-12RB2/MMP3/FasL induced apoptosis, thus providing a strategy for targeting MDSCs to control tumors and improve clinical efficacy.
Keywords: Myeloid-Derived Suppressor Cells, Adaptive Immunity
WHAT IS ALREADY KNOWN ON THIS TOPIC
Myeloid-derived suppressor cells (MDSCs) have been recognized as the main contributor to tumor progression and clinical treatment failure. Multiple preclinical researches convinced that eliminating MDSCs could inhibit tumor deterioration and enhance antitumor therapies. However, no agent translated to the clinical cancer treatment for safety concern.
WHAT THIS STUDY ADDS
We found that sulforaphane (SFN) suppressed tumor growth and remodeled the tumor immune microenvironment. SFN not only activated CD8+ T cells function, but eliminated MDSCs through activating IL-12RB2/MMP3/FasL pathway.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our study identified SFN, an extracted isothiocyanate that is often used as nutraceutical, could eliminate MDSCs and enhance CD8+ T cells antitumor response, which may define a promising synergistic strategy for tumor immunotherapy.
Introduction
Cancer is a major cause of morbidity and mortality worldwide.1 Despite hundreds of years of development, traditional therapies have not improved cancer clinical efficacy. Recently, immunotherapies such as chimeric antigen receptor T cell (CAR-T) and immune checkpoint blockade have shown promising clinical efficacy in hematological cancers.2 However, the efficacy of immunotherapies in solid tumors is far from expected.3 The complex tumor microenvironment (TME) contributes to tumor progression and clinical treatment failure. Myeloid-derived suppressor cells (MDSCs), the main immune-repressive cell population, are commonly present in the TME and inhibit the functions of cytotoxic immune cells.4 MDSCs can directly or indirectly suppress T-cell activation through PD-L1 (Programmed cell death 1 ligand 1)/programmed cell death 1 (PD-1) contact and secrete immunosuppressive cytokines or exosomes. High abundance of MDSCs is associated with accelerated tumor growth, poor clinical prognosis, and treatment failure in solid tumors.5 Moreover, eliminating MDSCs could increase treatment efficacy and prolong survival time in preclinical investigations. Although numerous drugs have been confirmed to effectively eliminate MDSCs in fundamental research, none have been authorized for clinical use.6
Evidence suggests that dietary factors play a crucial role in cancer etiology and clinical treatment.7 Vegetables from the Brassicaceae family can reduce the risk of cancer development because of their rich phytochemical compounds and precursors. The anticancer ability of these plants depends on glucosinolates, which undergo enzymatic degradation to isothiocyanates (ITC). Sulforaphane (SFN) is the most commonly studied ITC.8 SFN, a naturally occurring ITC, has received increasing attention in recent years owing to its antitumor abilities.9 Numerous studies have verified the anticancer effects of SFN and its underlying mechanisms, including reactive oxygen species (ROS) elevation,10 Nrf2 activation,11 NF-κB signaling inhibition,12 and Akt/mTOR suppression.13 The ability of SFN to restrain tumors has been validated in mouse models, and no apparent side toxicity has been observed 14 15. Furthermore, the safety and bioactivity of SFN has been confirmed in clinical trails.16 17 In addition, SFN has been reported to modulate tumor immunity, including dendritic cell (DC) stimulatory ability,18 T-cell function,19 and macrophage migration.20 However, whether and how SFN regulates MDSCs reduction in the TME remain unclear.
In the present study, we determined that SFN represses tumor growth in solid tumors by enhancing CD8+T cells antitumor function and eliminating MDSCs. We observed significant changes in the numbers of CD8+ T cells, B cells, natural killer (NK) cells, and CD11b cells in the SFN-treated group. Since we have previously revealed the role of SFN in T cells,19 we focused on the modulation of CD11b cells by SFN in the present study. We determined that SFN reduced MDSCs accumulation, but had no effect on macrophages. Mechanistically, SFN induces MDSCs apoptosis through IL-12RB2 signaling, which further activates the MMP3/FasL pathway. This was confirmed in the malignant pleural effusion (MPE) samples. Furthermore, we collected the blood samples from an ongoing “Phase I-II clinical study of sulforaphane enteric-coated capsules in patients with malignant tumors” and observed marked tumor control, reduced MDSCs percentages, and elevated CD8+ T-cell function in SFN-treated patients compared with those in the control group. Collectively, our data support that SFN induces the antitumor response of CD8+ T cells through IL-12RB2/MMP3/FasL signaling-induced MDSCs apoptosis.
Materials and methods
Cell culture
B16, B16-OVA, and LLC mice cell lines were used to confirm the absence of Mycoplasma or other pathogenic infections. Cells were purchased from Shanghai Cell Bank of the Chinese Academy of Sciences and cultured at 37°C in 5% CO2. LLC cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, Cat.D6429, Sigma, USA) supplemented with 10% fetal bovine serum (FBS, Cat. S711-011, Lonseral, Suzhou, China), penicillin, and streptomycin. B16 cells were cultured in Roswell Park Memorial Institute−1640 (Cat.R8758, Sigma, USA) medium supplemented with 10% FBS, penicillin, and streptomycin.
MDSCs induction
Monocytes were obtained from the bone marrow of C57/B6 mice. After erythrocyte lysis, MDSCs were induced by the addition of granulocyte colony-stimulating factor (G-CSF, Cat.250–50, PeproTech, USA, 50 ng/mL) or granulocyte-macrophage colony-stimulating factor (GM-CSF, Cat.315–03, PeproTech, USA, 50 ng/mL) for 72 hours. The induction efficacy was analyzed using flow cytometry.
MDSCs apoptosis detection
MDSCs were induced in vitro with or without SFN treatment (10 µM, Sigma-Aldrich, St. Louis, Missouri, USA). After 72 hours, the cells were harvested and labeled with annexin V (allophycocyanin/fluorescein (APC/FITC, Cat.640920, BioLegend, USA)/propidium iodide/phycoerythrin (PI/PE, Cat.421301, BioLegend, USA) or cleaved caspase 3 (Cat.9661, Cell Signaling Technology, USA)). Finally, apoptosis was analyzed using flow cytometry.
MDSCs were induced in vitro with or without SFN treatment (10 µM). After 72 hours, the cells were harvested and labeled with PI (PE), and fluorescence microscopy was used to photograph and analyze the apoptotic cells.
Imaging flow cytometry
MDSCs were induced in vitro with or without SFN treatment (10 µM). After 72 hours, the cells were harvested and labeled for CD11b (PerCP/Cyanine5.5, Cat.101227 BioLegend, USA), Gr-1 (APC, Cat.108412, BioLegend, USA), cleaved caspase-3 (PE, Cat.9661, Cell Signaling Technology, USA), PD-L1 (PE/Cyanine7, Cat.124314 BioLegend, USA), and IL-12RB2 (FITC, Cat.ab96097, Abcam, USA). Imaging flow cytometry was used to measure protein levels.
T-cell suppression assay
CD8+ T cells were recovered from the spleens using magnetic sorting (Cat.480035, BioLegend, USA) and activated by the addition of CD3 (Cat.100238, BioLegend, USA, 10 ng/mL) and CD28 (Cat.102116, BioLegend, USA, 5 ng/mL). After labeling with the Cell Proliferation Dye eFluor 670 (Cat.65-0840-85, Thermo Fisher Scientific, USA) according to the manufacturer’s instructions (Thermo Fisher Scientific), CD8+ T cells were co-cultured with MDSCs pretreated with or without SFN (10 µM). After 48 hours, the cells were collected for flow cytometry analysis.
B16-OVA cells were co-cultured with MDSCs and CD8+ T cells from OT-1 mouse spleens. After incubation at 37°C for 24 hours, the adherent B16-OVA cells were digested and labeled with annexin V and PI. Apoptosis was detected using flow cytometry.
Phagocytic capacity detection
MDSCs were induced and pretreated with SFN for 48 hours. Then, MDSCs were added to the LLC/B16-GFP cells for another 48 hours. We collected MDSCs to examine the GFP signal by using flow cytometry. Also, we co-cultured OVA peptide (OVA 257–264, Chicken egg albumin peptide, Catalog code: vac-sin) with MDSCs after treated with or without SFN. 48 hours later, APC/Fire 750 anti-mouse H-2Kb bound to SIINFEKL Antibody (BioLegend, USA) was added. Flow cytometry was used to detect the APC signal in MDSCs.
Western blotting
Total protein was isolated from MDSCs using radioimmunoprecipitation assay (Cat.P0013B, Beyotime, China) buffer. After determining the protein concentration using a bicinchoninic acid (Cat.P0010, Beyotime, China) kit, a metal bath was used for protein thermal deformation. Equal amounts of protein were separated using 10–12% sodium dodecyl-sulfate-polyacrylamide gel electrophoresis (SDS_PAGE, Cat.8012011, Dakewe, China), transferred onto a polyvinylidene difluoride (Cat.10600001, Cytiva Amersham, USA) membrane, and blocked with 5% skimmed milk. The primary antibodies, PD-L1 (Cat.124301, BioLegend, USA), caspase 3 (Cat.14220, Cell Signaling Technology, USA), cleaved caspase 3 (Cat.9661, Cell Signaling Technology, USA), MMP3 (Cat.ab52915, Abcam, USA), IL-12-RB2 (Cat.ab96097, Abcam, USA), FasL (Cat.AF0157, Affinity Bioscience, USA), and Arg-1 (Cat.89872, Cell Signaling Technology, USA) were added and incubated overnight at 4°C. The corresponding secondary antibodies were then incubated at room temperature. Finally, an electrochemiluminescence kit (Cat.P0018S, Beyotime, China) was used for staining.
Pleural effusion from patients with lung cancer
Pleural effusions were aseptically obtained from patients with lung cancer through centrifugation and erythrocyte lysis. Human GM-CSF (10 ng/mL) was used to obtain live MDSCs treated with or without SFN (10 µM). After 72 hours, cells were harvested for flow cytometry or quantitative reverse transcription-PCR (qRT-PCR) analyses.
MDSCs messenger RNA sequencing
MDSCs from tumor tissues of LLC mice were purified using a Myeloid-Derived Suppressor Cell Isolation Kit (Cat.130-126-725, Miltenyi Biotec, Germany). The cells were then harvested and stored in TRIzol (Cat.9109, Takara, Japan). The samples were transported to Shenzhen Huada Gene for RNA sequencing (RNA-seq) (contract number F20FTSCCWLJ3077). Data were analyzed using Gene Ontology/Kyoto Encyclopedia of Genes and Genomes/Gene Set Enrichment Analysis (GO/KEGG/GSEA) based on the differentially expressed genes.
Clinical samples collection
Our college is performing a phase 1 human study, named as “phase I–II clinical trial on the application of SFN enteric-coated capsules in patients with malignant tumors (2021-KY-0350)”. The extracted SFN (Pu’er Qiyun Biotechnology, China) is used for clinical patients treatment. The dosage of oral SFN is 100 mg/person/day. We collected the blood samples from 10 patients with lung cancer, including one woman and nine men, with ages ranging from 52 to 66 years (58.1±4.72). This study is ongoing, with a planned sample size of 60 patients. The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University. We also collected the blood samples from 10 patients with lung cancer without SFN treatment, including one woman and nine men, with ages ranging from 44 to 64 years (55.7±6.88). All patients signed the informed consent form. We collected the plasma concentrations of liver and kidney profiles, including ALT (Alanine Aminotransferase), AST (Aspartate Aminotransferase), Cre (creatinine), ALP (alkaline phosphatase), total protein, albumin, direct bilirubin, indirect bilirubin, urea, uric acid, GGT (Gamma Glutamyl Transferase), and GFR (glomerular filtration rate). The clinical profile of controls and SFN patients with lung cancer is presented in table 1.
Table 1.
Clinical features of patients with lung cancer in this study
| Patients | Gender | Smoking | Alcohol | Pathologic diagnosis | Specimen origin | Stage |
| Patient 1 | Male | Yes | No | Lung cancer | Adenocarcinoma | IV |
| Patient 2 | Male | No | No | Lung cancer | Adenocarcinoma | IV |
| Patient 3 | Female | No | No | Lung cancer | Adenocarcinoma | IV |
| Patient 4 | Male | Yes | Yes | Lung cancer | Adenocarcinoma | IV |
| Patient 5 | Male | No | No | Lung cancer | Adenocarcinoma | IIIA |
| Patient 6 | Male | Yes | No | Lung cancer | Squamous | IV |
| Patient 7 | Male | Yes | Yes | Lung cancer | Squamous | IV |
| Patient 8 | Male | Yes | No | Lung cancer | Squamous | IV |
| Patient 9 | Male | Yes | No | Lung cancer | Adenocarcinoma | IV |
| Patient 10 | Male | Yes | No | Lung cancer | Adenocarcinoma | IV |
| Control 1 | Female | No | No | Lung cancer | Squamous | IV |
| Control 2 | Male | No | No | Lung cancer | Squamous | IIIB |
| Control 3 | Male | Yes | Yes | Lung cancer | Squamous | IV |
| Control 4 | Male | Yes | No | Lung cancer | Adenocarcinoma | IV |
| Control 5 | Male | No | No | Lung cancer | Squamous | IIIB |
| Control 6 | Male | Yes | No | Lung cancer | Squamous | IIIB |
| Control 7 | Male | No | No | Lung cancer | Adenocarcinoma | IV |
| Control 8 | Male | Yes | Yes | Lung cancer | Adenocarcinoma | IV |
| Control 9 | Male | Yes | Yes | Lung cancer | Adenocarcinoma | IV |
| Control 10 | Male | Yes | No | Lung cancer | Squamous | IV |
Analyzing Arg1, ROS, and nitric oxide levels within MDSCs
Analysis of intracellular Arg1 within MDSCs treated with or without SFN (10 µM) was performed using a Mouse Argininase Kit (Cat.MAK112, Sigma, USA). Cells were harvested and permeabilized according to the manufacturer’s protocol. Absorbance was measured at 430 nm.
For ROS detection, the cells were harvested and stained with an ROS probe (FITC; Cat.88-5930-74, Thermo Fisher Scientific, USA) diluted in ROS Assay Buffer. After washing to remove non-specific binding, CD11b and Gr1 flow antibodies were added. Fluorescence was analyzed using flow cytometry.
An NO Test Kit (Cat.BC1475, Solarbio, China) was used to detect nitric oxide (NO) within MDSCs. Briefly, the cells were harvested and centrifuged. Then, the supernatant was transferred to a new tube, and added to a 96-well plate with 100 µL supernatant and 100 µL chromogenic solution. After gentle shaking, absorbance was measured at 550 nm. Standard samples were used to generate standard curves. NO in the sample was calculated according to the formula for standard sample detection.
RNA extraction and real‐time PCR analysis
RNA was extracted using RNAiso Plus (Cat.9108, Takara, Japan), according to the manufacturer’s instructions. Complementary (cDNA) was generated using a high-capacity cDNA Reverse Transcription Kit (Cat.PD101-01, Vazyme, China). Real-time quantitative PCR analysis was performed on a Bio-Rad instrument (Agilent, USA), using the SYBR Green master mix (Cat.Q712-02, Vazyme, China) and 40 cycles of universal cycling conditions (95°C for 10 min, followed by 40 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 10 s). GAPDH was used as an internal reference, and the relative expression level of the target gene was calculated based on the obtained Ct value using the following formula: Folds=2–ΔΔCt. The primers are listed in table 2.
Table 2.
The primers used for quantitative reverse transcription-PCR
| Gene name | Primer |
| Human-GAPDH | |
| Forward | GGAGCCAAAAGGGTCATCATCTC |
| Reverse | GAGGGGCCATCCACAGTCTTCT |
| Human-Arg-1 | |
| Forward | GTGGAAACTTGCATGGACAAC |
| Reverse | AATCCTGGCACATCGGGA ATC |
| Human-iNOS2 | |
| Forward | TTCAGTATCACAACCTCAGCAAG |
| Reverse | TGGACCTGCAAGTTAAAATCCC |
| Human-CYBB | |
| Forward | ACCGGGTTTATGATATTCCACCT |
| Reverse | GATTTCGACAGACTGGCAAGA |
| Human-TGF-b | |
| Forward | CCATCCCGCCCACTTTCTAC |
| Reverse | AGCTCAATCCGTTGTTCAGGC |
| Human-NCF4 | |
| Forward | CGGGCCGAGAGTGACTTTG |
| Reverse | TCTTCACCTCGATGACGAAAAC |
| Human-IL-10 | |
| Forward | TCAAGGCGCATGTGAACTCC |
| Reverse | GATGTCAAACTCACTCATGGCT |
| Human-PD-L1 | |
| Forward | TGGCATTTGCTGAACGCATTT |
| Reverse | TGCAGCCAGGTCTAATTGTTTT |
| Mouse-GAPDH | |
| Forward | TGGCCTTCCGTGTTCCTAC |
| Reverse | GAGTTGCTGTTGAAGTCGCA |
| Mouse-Arg-1 | |
| Forward | CTCCAAGCCAAAGTCCTTAGAG |
| Reverse | AGGAGCTGTCATTAGGGACATC |
| Mouse-iNOS2 | |
| Forward | GTTCTCAGCCCAACAATACAAGA |
| Reverse | GTGGACGGGTCGATGTCAC |
| Mouse-CYBB | |
| Forward | AGTGCGTGTTGCTCGACAA |
| Reverse | GCGGTGTGCAGTGCTATCAT |
| Mouse-TGF-b | |
| Forward | TCGACATGGATCAGTTTATGCG |
| Reverse | CCCTGGTACTGTTGTAGATGGA |
| Mouse-NCF4 | |
| Forward | GTCATCGAGGTCAAAACAAAAGG |
| Reverse | GCCCATGTAGACTTTGGCTG |
| Mouse-IL-10 | |
| Forward | TCAAGGCGCATGTGAACTCC |
| Reverse | GATGTCAAACTCACTCATGGCT |
| Mouse-PD-L1 | |
| Forward | GCTCCAAAGGACTTGTACGTG |
| Reverse | TGATCTGAAGGGCAGCATTTC |
| Mouse-FasL | |
| Forward | TCCGTGAGTTCACCAACCAAA |
| Reverse | GGGGGTTCCCTGTTAAATGGG |
| Mouse-Fas | |
| Forward | TATCAAGGAGGCCCATTTTGC |
| Reverse | TGTTTCCACTTCTAAACCATGCT |
Animal model and experiments
All animal experiments were approved by the Ethics Committee of the Zhengzhou University (2021–13). C57/B6 mice (female, 4–6 weeks of age) were purchased from Beijing Weitong Lihua Company and maintained under standard optimized conditions at the Experimental Animal Center of Zhengzhou University. Subcutaneous mouse models bearing B16 or LLC cells were constructed. When the tumor volume reached approximately 50 mm3, the mice were randomly grouped and treated with SFN (LKT Labs, St. Paul, Minnesota, USA, ip, 50 mg/kg) or anti-Gr1 antibody (5 mg/kg). The tumor growth was measured using a caliper. The tumor volume was calculated using the following formula: (length×width2)/2.
Tumor tissues were digested using a Tumor Dissociation Kit (Cat.130-096-730, Miltenyi Biotec, USA) and processed using gentleMACS to obtain a single-cell suspension. Single-cell suspensions were also obtained from mouse spleens for flow cytometry and labeled with CD3 (Apc/Cy7, Cat.100221, BioLegend, USA), CD8 (FITC, Cat.100705, BioLegend, USA), CD11b (PE, Cat.101208, BioLegend, USA), Gr-1 (FITC, Cat.108405, BioLegend, USA), B220 (PerCP/Cyanine5.5, Cat.103236, BioLegend, USA), CD11c (PE, Cat.117308, BioLegend, USA), and F4/80 (Apc, Cat.123116, BioLegend, USA).
Histology experiments
The tumor tissues were fixed in 4% paraformaldehyde (Cat.G1101, Servicebio, China) and embedded in paraffin. Sections were cut and processed for H&E staining and immunohistochemistry (IHC) to detect Gr-1, CD8, MMP3, FasL, IFN-γ, and IL-2. Immunofluorescence (IF) was performed to detect Gr-1 and CD8 expression. We sent all samples to the Servicebio Company for H&E, IHC and IF staining (Wuhan, China). All antibodies were provided by the company.
Statistical analysis
All data were presented as mean±SEM. The significance of the tumor growth curve and in vitro experiments was determined using the Student’s t-test for two groups. Statistical comparisons of pretreatment and post-treatment MDSC numbers in human patients were performed using a paired Student’s t-test. Correlation analysis was performed using Pearson analysis. Throughout all figures: *p<0.05, **p<0.01, and ***p<0.001, ****p<0.0001. Statistical significance was set at p<0.05.
Results
SFN suppressed tumor progression by inhibiting MDSCs-restrained CD8+ T-cell function
Several studies have demonstrated the role of SFN in activating antitumor immune responses and suppressing tumor progression.21 In this study, we observed significant tumor regression treated with SFN in mouse models bearing LLC (figure 1A) and B16 cells (online supplemental figure S1A). To determine the immune pattern in the SFN-induced tumor regression, we examined the percentages of myeloid cells, NK cells, B cells, DC cells, and CD8+ T cells. We observed elevated CD8+ T cells and decreased CD11b+ myeloid cells in the tumors and spleens (figure 1B and online supplemental figure S1B). Because we previously investigated the role of SFN in T-cell function,19 we focused on its role in myeloid cells. Notably, we did not investigate the correspondence between NK and B cells in the detection location and mouse models. Furthermore, we observed that SFN reduced the percentage of MDSCs but had no effect on macrophage accumulation in both LLC (figure 1C–D) and B16 mouse models (online supplemental figure S1B,C). IF and IHC confirmed the decreased role of SFN on MDSCs from the LLC tumor mice samples (figure 1E,F, online supplemental figure S1D). In addition, we observed a significant negative correlation between Gr1 and CD8 at the tumor sites (figure 1F), suggesting that SFN could partly liberates CD8+ T cells by eliminating MDSCs. To test the role of MDSCs in SFN-mediated tumor control, we depleted these cells using an anti-Gr1 antibody (online supplemental figure S1E,F). Eliminating MDSCs enhanced the ability of SFN to suppress tumor growth (figure 1G,H). In addition, it enhanced CD8+ T-cell infiltration and elevated function-related cytokine expression at tumor sites (figure 1I). Collectively, these results suggest that SFN liberates MDSC-induced CD8+ T-cell immune function.
Figure 1.
SFN suppressed tumor progression by inhibiting MDSCs and restraining CD8+T cells function. (A) Tumor volume of mice bearing LLC cells treated with or without SFN. (B) Flow cytometry analysis of immune cell subsets in spleens and at tumor sites from LLC mice following SFN treatment. (C) Flow cytometry analysis of macrophage and MDSCs in spleens and at tumor sites from LLC mice following SFN treatment. (D) Flow cytometry analysis of PMN-MDSCs and M-MDSCs in spleens and at tumor sites from LLC mice following SFN treatment. Immunofluorescence (E) and IHC (F) analysis of CD8 and Gr1 in tumor samples from tumor sites of LLC mice following SFN treatment. The correlation between Gr1 and CD8 was analyzed. (G) Tumor volume of mice bearing LLC cells treated with SFN, anti-Gr1, or SFN+anti-Gr1. (H) Flow cytometry analysis of MDSCs in tumor sites from LLC mice treated with SFN, anti-Gr1, or SFN+anti-Gr1. (I) IHC analysis of CD8, TNF-α, and IFN-γ in tumor samples from LLC mice treated with SFN, anti-Gr1, or SFN+anti-Gr1. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. IFN, interferon; IHC, immunohistochemistry; PMN-MDSCs, polymorphonuclear myeloid-derived suppressor cells; M-MDSCs, monocytic myeloid-derived suppressor cells; TNF, tumor necrosis factor; SFN, sulforaphane.
jitc-2023-007983supp001.pdf (2MB, pdf)
SFN inhibited MDSC immunosuppression and enhanced CD8+ T cells antitumor response
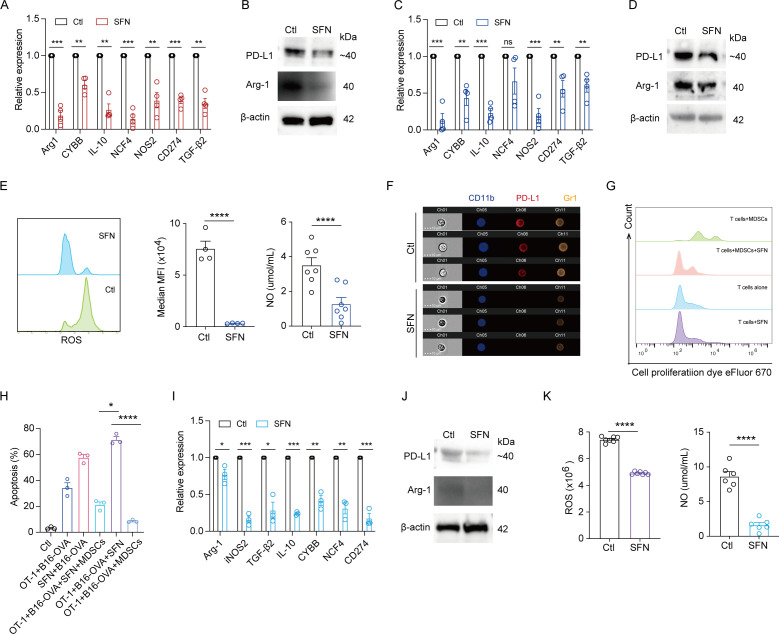
We investigated the effects of SFN on MDSCs. First, we purified MDSCs from the spleen of SFN-treated LLC mice and examined their functional genes. SFN significantly reduced the expression of MDSCs immunosuppression-related genes, including Arg1, CYBB, IL-10, NCF4, NOS2, TGF-β2 and CD274 (figure 2A). We also performed western blotting to examine the levels of PD-L1 and Arg-1, and observed a marked reduction in SFN-treated MDSCs (figure 2B). We then obtained myeloid cells from wild type C57 mice bone marrow and induced MDSCs by adding G-CSF and GM-CSF. SFN treatment significantly reduced the mRNA levels of immunosuppressive cytokines, including Arg-1, NOS2, TGF-β2, IL-10, CYBB, and CD274 (figure 2C). The reductions in Arg-1, PD-L1 (figure 2D,F), ROS, and NO (figure 2E) were further verified at the protein level. Additionally, SFN reversed the inhibitory effect of MDSCs on CD8+ T-cell proliferation (figure 2G) and restored the ability of CD8+ T cells to kill B16-OVA cells (figure 2H), suggesting that SFN can emancipate CD8+ T cells from MDSCs-induced immunosuppression. Finally, we obtained MDSCs from MPE samples of patients with cancer and treated them with SFN to examine immunosuppressive cytokines. Similar reductions in immunosuppressive cytokines, including Arg-1, NOS2, TGF-β2, IL-10, CYBB, NCF4, and CD274, were observed at the mRNA level in the SFN-treated group (figure 2I). Additionally, we observed that SFN reduced PD-L1, Arg-1 (figure 2J), ROS, and NO (figure 2K) at the protein level. Collectively, these results indicate that SFN restores the MDSCs suppressed killing ability of CD8+ T cells by reducing MDSC immunosuppression.
Figure 2.
SFN inhibited MDSC immunosuppression and enhanced CD8+T cells antitumor response. (A) qRT-PCR was performed to analyze the immunosuppression-related genes in MDSCs purified from the spleens of LLC mice treated with or without SFN. (B) Western blotting was used to detect the expressions of PD-L1 and Arg-1 in MDSCs purified from the spleens of LLC mice treated with or without SFN. (C) qRT-PCR was performed to examine the immunosuppression-related genes in induced MDSCs from bone marrow treated with or without SFN. (D) Western blotting was used to determine the expression of PD-L1 and Arg-1 in induced MDSCs from bone marrow treated with or without SFN. (E) Flow cytometry analysis of ROS in induced MDSCs from bone marrow treated with or without SFN. ELISA was conducted to examine the NO levels in MDSCs treated with or without SFN. (F) Imaging flow cytometry was used to detect the PD-L1 and Gr1 in induced MDSCs from bone marrow treated with or without SFN. Flow cytometry analysis of CD8+ T-cell proliferation (G) and killing ability (H) co-cultured induced with MDSCs from bone marrow with or without SFN treatment. (I) qRT-PCR was performed to examine the immunosuppression-related genes in MDSCs purified from patients’ MPE followed treatment with or without SFN. (J) Western blotting was used to determine the expressions of PD-L1 and Arg-1 in MDSCs purified from patients’ MPE followed treatment with or without SFN. (K) Flow cytometry analysis of ROS in MDSCs purified from patients’ MPE treated with or without SFN. ELISA was conducted to analyze the NO levels in MDSCs purified from patients’ MPE treated with or without SFN. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. MDSCs, myeloid-derived suppressor cells; MPE, malignant pleural effusion; NO, nitric oxide; qRT-PCR, quantitative reverse transcription-PCR; ROS, reactive oxygen species; SFN, sulforaphane; PD-L1, Programmed cell death 1 ligand 1; MFI, Mean Fluorescence Intensity.
SFN reduced MDSCs immunosuppression by inducing the apoptosis
Herein, we observed that SFN treatment restores the MDSCs suppressed killing ability of CD8+ T cells. To further investigate the potential mechanism of SFN-induced suppression of immunosuppression by MDSCs, we performed RNA-seq of MDSCs treated with or without SFN. Overall, 16,623 genes intersected, with 623 and 582 unique genes in the control and SFN groups, respectively (figure 3A). MDSCs are a heterogeneous population of immature suppressive innate immune cells that expand in the TME. One of the identified strategies to eliminate MDSCs is promoting its maturation and differentiation.22 We analyzed the DCs and macrophages associated genes and observed no differences in control and SFN treated MDSCs (online supplemental figure S2A). Consistently, flow analysis showed no differences of macrophages and DCs in control and SFN treated MDSCs (online supplemental figure S2B). Also, we evaluated the phagocytic capacity to examine whether SFN treatment enables the licensed recognition of cross-presented antigen of MDSCs. Flow analysis showed that SFN treatment did not promote MDSCs ability to phagocyte OVA or LLC/H322 cells (online supplemental figure S2C), suggesting that SFN treatment did not promote MDSCs maturation or elevate the licensed recognition of cross-presented antigen.
Figure 3.
SFN reduced MDSC immunosuppression by inducing apoptosis. (A) Venn analysis showing differences in gene expression between MDSCs with or without SFN. (B) Gene Set Enrichment Analysis analysis. (C) Heatmap of apoptosis-related gene expression. (D) Flow cytometry analysis of bone marrow induced MDSCs populations and apoptotic rates following treatment with SFN. (E) PI was used to stain bone marrow induced MDSCs treated with or without SFN. PI+ MDSCs were calculated and analyzed. (F) Immunofluorescence was performed to examine the apoptosis of MDSCs in tumor tissues from LLC mice using terminal deoxynucleotidyl transferase dUTP nick end labeling and Gr1. (G) Flow cytometry analysis of MDSCs in blood, bone marrow, and spleens from mice bearing LLC and B16 cells. (H) Flow cytometry analysis of PI+ MDSCs in blood, bone marrow, and spleens from mice bearing LLC and B16 cells. (I) Flow cytometry analysis of MDSCs from MPE of patients with cancer percentage and apoptotic rate in patients using MPE. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. KEGG, Kyoto Encyclopedia of Genes and Genomes; MDSCs, myeloid-derived suppressor cells; MPE, malignant pleural effusion; PI, Propidium Iodide; SFN, sulforaphane.
jitc-2023-007983supp002.pdf (585.2KB, pdf)
We went on to explore the mechanism underlying SFN reduced MDSCs immunosuppression. GSEA showed that the apoptotic pathway was enriched (figure 3B), and the heat map showed a significant elevation of apoptosis-related genes in the SFN-treated group (figure 3C). This indicates that SFN may induce MDSCs apoptosis to inhibit immunosuppression. We examined the apoptosis after SFN treatment in MDSCs and observed a marked reduced MDSCs percentage and an increased apoptotic rate (figure 3D). PI staining further confirmed the SFN-induced MDSCs apoptosis (figure 3E). Consistent with the in vitro data, we observed elevated MDSCs apoptosis in tumor tissues from mouse models bearing LLC cells using for terminal deoxynucleotidyl transferase dUTP nick end labeling staining (figure 3F). Notably, SFN treatment significantly decreased MDSCs abundance (figure 3G) and elevated PI+MDSCs (figure 3H) in the blood and spleens, but had no effect on bone marrow MDSCs, indicating that SFN may promote MDSCs elimination peripherally. Finally, we obtained MDSCs from MPE of patients with cancer and treated them with SFN to examine apoptotic markers. SFN treatment significantly reduced the percentage of MDSCs and increased apoptosis (figure 3I). These observations indicated that SFN affects peripheral MDSCs abundance by enhancing apoptosis. All cells need the normal physiological state of life to sustain the function, so as MDSCs. We examined the Janus kinase (JAK)/signal transducer and activator of transcription (STAT)5, which is a main factor of MDSCs expansion and function,23 after SFN treatment and observed that SFN significantly reduced the total-JAK/STAT5 and p-JAK/STAT5 (online supplemental figure S2D). Together, these results indicated that SFN induced MDSCs apoptosis to inhibit the immunosuppression.
SFN induced MDSCs apoptosis by activating IL-12RB2
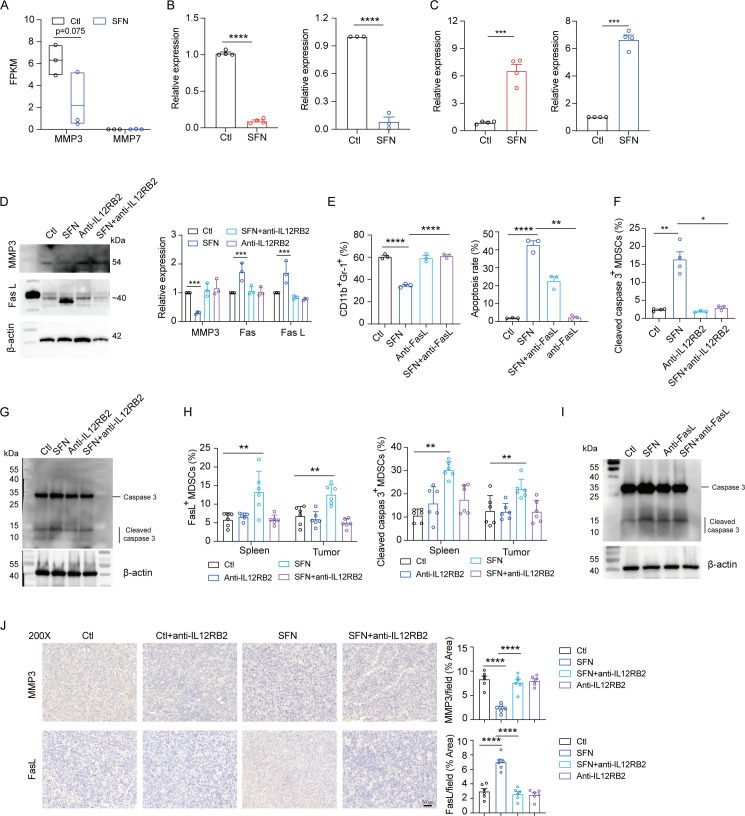
Next, we intended to explore the mechanism underlying SFN-induced MDSCs apoptosis. Based on the RNA-seq data, SFN treatment resulted in the expression of different genes (online supplemental figure S3A). GO and KEGG analyses showed enriched pathways in SFN-treated MDSCs, including regulation of the innate immune response, negative regulation of immune system processes, cytokine receptor activity, and cytokine receptor binding (figure 4A and online supplemental figure S3B). GSEA consistently revealed that cytokine–cytokine receptor interactions were enriched in SFN-treated MDSCs (figure 4B). We assayed pathway-related genes and observed elevated gene expression in the SFN-treated group, with IL-12RB2 exhibiting the highest fold change (figure 4C). Elevated IL-12RB2 expression after SFN treatment of MDSCs was validated at the protein level (figure 4D). Since interleukin (IL)-12 and IL-35 are the ligands for IL-12RB2,24 so we examined whether SFN activated IIL-12RB2 signaling pathway through binding to the ligands. While no differences were observed of EBl3, IL-12a, IL-12b, and IL-12Rb1 according to the RNA-seq data (figure 4E). Additionally, interferon-γ has been reported to induce IL-12RB2 expression via STAT5a/STAT5b pathway in T cells.25 However, we observed that SFN treatment reduced the STAT5 pathway, suggesting that SFN may not through its ligands but directly activated IL-12RB2. To confirm this, we used IL-12RB2 neutralizing antibody and determined that SFN exhibited less inhibition of MDSCs percentage and apoptotic rate (figure 4F). Plus, IL-12RB2 neutralizing antibody reversed SFN-induced MDSCs and reduced the expression of immunosuppressive genes, including Arg1, CD274, and IL-10 (figure 4G). Imaging flow cytometry confirmed that blocking IL-12RB2 reversed SFN-induced PD-L1 reduction in MDSCs (online supplemental figure S3C). Finally, we used the LLC mice model to examine the role of IL-12RB2 in the SFN-induced apoptosis of MDSCs. Combining IL-12RB2 neutralizing antibody weakened SFN-induced tumor control and IL-12RB2 neutralizing antibody alone had no effect (figure 4H). IL-12RB2 neutralizing antibody application reversed the SFN reduction in MDSCs aggregation (figure 4I). Correspondingly, CD8+ T-cell accumulation disappeared, and function-related cytokines were reduced in IL-12RB2 neutralizing antibody combing with SFN group (figure 4I). The pathological experiments showed similar results (figure 4J and online supplemental figure S3D). These observations indicate that SFN depletes MDSCs to amplify CD8+ T cells, which depends on IL-12RB2 activation-induced MDSCs apoptosis.
Figure 4.
SFN induced MDSCs apoptosis by activating IL-12RB2. Bone marrow induced MDSCs cultured with or without SFN were used for RNA-seq analysis. (A) Gene Ontology analysis. (B) Gene Set Enrichment Analysis. (C) Heat map of cytokine_cytokine receptor_interaction-related gene expression. (D) The relative expressions of IL-12a, IL-12b, IL-12rb21, Ebi3 and IL-12rb2 in MDSCs treated with or without SFN, based on RNA-seq data. (E) Western blotting was used to examine the IL-12RB2 in MDSCs treated with or without SFN. (F) Flow cytometry analysis of MDSC populations and apoptotic rates following treatment with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (G) PCR analysis of MDSC immunosuppression-related genes following treatment with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (H) Tumor volume of mice model bearing LLC cells treated with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (I) Flow cytometry analysis of MDSCs and CD8+T-cell percentages and function in tumor samples from LLC mice treated with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (J) Immunohistochemistry analysis of Gr1, CD8, TNF-α, and IFN-γ in tumor samples from LLC mice treated with SFN, anti-IL-12RB2, and SFN+anti-IL-12RB2. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. IFN, interferon; KEGG, Kyoto Encyclopedia of Genes and Genomes; MDSCs, myeloid-derived suppressor cells; RNA-seq, RNA sequencing; SFN, sulforaphane; TNF, tumor necrosis factor; FPKM, Fragments Per Kilobase of transcript per Million mapped reads.
jitc-2023-007983supp003.pdf (2MB, pdf)
MMP3/FasL was downstream of SFN-activated IL-12RB2 in MDSCs
IL12RB2 activation induces ovarian cancer cell apoptosis by promoting membrane FasL expression and reducing the expression of MMP3 and MMP7.26 To determine whether MMP3 and MMP7 respond to SFN induced IL-12RB2 activation in MDSCs, we analyzed RNA-seq data and determined that only MMP3 expression was reduced in SFN treated MDSCs (figure 5A), which was further validated in vivo and in vitro (figure 5B). MMP3 is an FasL protease.27 Next, we examined FasL expression and observed a significant elevation in FasL expression in SFN treated MDSCs in vivo and in vitro (figure 5C). Further, we verified MMP3 reduction and FasL elevation induced by SFN treatment, while IL-12RB2-neutralizing antibody reversed the phenomenon in protein level (figure 5D). In addition, when using the FasL-neutralizing antibody, SFN did not reduce the percentage of MDSCs or induce apoptosis of MDSCs (figure 5E), indicating that MMP3/FasL responds to IL-12RB2 activation in SFN treated MDSCs. FasL induces downstream caspase-3 activation to induce cell apoptosis.28 29 Herein, we observed elevated levels of cleaved caspase-3 (online supplemental figure S4A) in MDSCs with SFN treatment. And SFN induced higher cleaved caspase-3+MDSCs percentages in samples from mouse bone marrow and human MPE (online supplemental figure S4B), as well as in the spleens and tumor samples from LLC and B16 mouse models (online supplemental figure S4C). Whereas IL-12RB2-neutralizing antibody reversed this phenomenon (figure 5F). Also, we observed higher cleaved caspase 3 in SFN treated MDSCs, while blocking IL-12RB2 reduced its level by using western blot (figure 5G). Moreover, blocking IL-12RB2 reversed the SFN treatment-induced FasL+MDSCs and cleaved caspase-3+MDSCs increase in the spleens and tumor sites of the LLC mouse model (figure 5H). Additionally, blocking FasL reduced SFN induced caspase-3 cleavage, indicating that cleaved caspase-3 responds to SFN induced FasL elevation (figure 5I). Finally, we observed reduced MMP3 and elevated FasL levels in tumor samples after SFN treatment, whereas the IL-12RB2 neutralizing antibody reversed this phenomenon (figure 5J and online supplemental figure S4D). Collectively, these results suggest that SFN activates the IL-12RB2/MMP3/FasL pathway and induces caspase-3 cleavage, leading to MDSC apoptosis.
Figure 5.
MMP3/FasL was the downstream of SFN-activated IL-12RB2 in MDSCs. (A) The relative expressions of MMP3 and MMP7 in bone marrow induced MDSCs with or without SFN, based on RNA sequencing data. (B) qRT-PCR was performed to examine the expression of MMP3 from MDSCs with or without SFN treatment in vivo and in vitro. (C) qRT-PCR was performed to examine the expression of FasL from MDSCs with or without SFN treatment in vivo and in vitro. (D) Western blotting and qRT-PCR were performed to examine MMP3 and FasL in MDSCs treated with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (E) Flow analysis of MDSCs percentages and apoptotic rates following SFN, anti-IL-12RB2, and SFN+anti-IL-12RB2 treatment. (F) Flow analysis of cleaved caspase-3+MDSCs percentages following SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2 treatment. (G) Western blotting was performed to analyze caspase-3 expression in MDSCs treated with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. (H) Flow analysis of FasL+ and cleaved caspase-3+MDSCs following SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2 treatment. (I) Western blotting was performed to examine caspase-3 expression in MDSCs treated with SFN, anti-FasL, or SFN+anti FasL. (J) Immunohistochemistry analysis of MMP3 and FasL in tumor samples from LLC mice treated with SFN, anti-IL-12RB2, or SFN+anti-IL-12RB2. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. FPKM, Fragments Per Kilobase of transcript per Million mapped reads; MDSCs, myeloid-derived suppressor cells; qRT-PCR, quantitative reverse transcription-PCR; SFN, sulforaphane.
jitc-2023-007983supp004.pdf (1.7MB, pdf)
SFN depleted MDSCs and improved CD8+ T cells activation in clinic
Finally, we collected samples from a clinical trial, named as “Phase I-II clinical study on the application of SFN enteric-coated capsules in patients with malignant tumors (2021-KY-0350)” to evaluate the function of SFN in MDSCs depletion and CD8+ T-cell activation (figure 6A). Peripheral blood samples were obtained from patients before the initiation of dosing and after SFN treatment. The samples were processed for flow cytometry. As expected, the gating strategy used for quantifying human MDSCs (CD33+CD11b+HLA-DRlow cells)30 revealed a significant reduction in the percentage of MDSCs and a marked increase in IL-12RB2-positive and FasL-positive MDSCs after SFN treatment (figure 6B). Additionally, the treatment of patients with SFN resulted in a significant increase in CTL (cytotoxic T lymphocyte) activation, as revealed by significant increases in the fraction of CD8+ T cells expressing glucocorticoid-induced TNFR(tumor necrosis factor receptor)-related31 and a reduction in CD8+ T cells expressing PD-1 (figure 6C). However, no significant trends were observed in the control group (figure 6D,E). These observations demonstrate that the antitumor function of SFN in patients with cancer depends on MDSCs depletion and CD8+ T-cell activation. Finally, we collected and analyzed the plasma concentrations of liver and kidney profiles. We observed no significant differences between the control and SFN treated groups in ALT, AST, Cre, ALP, total protein, albumin, direct bilirubin, indirect bilirubin, urea, uric acid, GGT, and GFR (figure 6F), indicating the safety of SFN treatment in clinic.
Figure 6.
SFN depleted MDSCs and improved CD8+T cells activation in the clinic. (A) The clinical features of patients in control and SFN treated lung cancer group. (B) Flow cytometry analysis of MDSCs, IL-12RB2+MDSCs, and FasL+MDSCs in blood samples from patients before and after SFN treatment. (C) Flow cytometry analysis of CD8+, GITR+CD8+, and PD-1+CD8+ T cells in blood samples from patients before and after SFN treatment. (D) Flow cytometry analysis of MDSCs, IL-12RB2+MDSCs, and FasL+MDSCs in blood samples from control group. (E) Flow cytometry analysis of CD8+, GITR+CD8+, and PD-1+CD8+ T cells in blood samples from control group. (F) Evaluation of plasma concentrations of liver and kidney profiles between control and SFN treated groups. Data were presented as the mean±SEM. ns, not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. GITR, glucocorticoid-induced TNFR-related; MDSCs, myeloid-derived suppressor cells; PD-1, programmed cell death 1; SFN, sulforaphane; AST, glutamic oxaloacetic transaminase; GGT, glutamyl transpeptidase; Cre, creatinine; ALT, glutamic-pyruvic transaminase; ALP, alkaline phosphatase; TBIL, total bilirubin; DB, direct bilirubin; IBIL, indirect bilirubin; GFR, glomerular filtration rate; CR, Complete Remission; PR, Partial Remission; PD, progressive disease; NA, Not Assessed; TNFR, tumor necrosis factor receptor.
Discussion
Malignant cells develop multiple mechanisms to escape immune surveillance and attack during cancer progression. Emerging evidence has demonstrated that MDSCs contribute to immunosuppression, cancer progression, and poor clinical outcomes.22 These observations have encouraged researchers to develop therapies to eliminate MDSCs. Preclinical investigations have demonstrated the importance of MDSCs depletion in controlling tumor progression and enhancing antitumor therapy efficiency.32 33 However, none have been applied in clinic because of the induced side effects. As a naturally occurring agent, SFN is the main constituent of health products without any safety hazards. Recent research has shown the antitumor ability of SFN by directly eliminating cancer cells34 or indirectly regulating the antitumor response.19 20 In the present study, our data showed that SFN repressed tumor growth and MDSCs abundance in vitro and in vivo, thereby driving the antitumor response of CD8+ T cells and supporting this approach as a means of augmenting antitumor immunity by eliminating MDSCs.
The close association between MDSCs and poor clinical outcomes has prompted researchers to consider MDSCs potential therapeutic targets. Four strategies have been explored to reduce MDSCs immunosuppression: (1) Prevention of expansion. MDSCs expansion is controlled by tumor-derived factors, including VEGF/GM-CSF/G-CSF/IL-6/C5a/MMP9/SCF, which subsequently trigger JAK protein family members and STAT3.35 Therefore, neutralizing antibodies against these factors or pathway inhibitors have been hypothesized to suppress MDSCs expansion. Such strategies include amino biophosphonates that target MMP9,36 the blockade of the SCF receptor.37 (2) Promotion of differentiation. Several approaches have been proposed to promote the differentiation of MDSCs into mature myeloid cells, including the use of vitamin A38 and all-trans retinoic acid.39 (3) Repression of function. Several studies have shown that MDSCs immunosuppression depends on Arg1, inducible nitric oxide synthase (iNOS), PD-L1, COX2, or exosomes. Thus, blocking the signaling pathways that regulate the production of suppressive factors has been developed to suppress MDSCs immunosuppression, such as COX2 inhibitor,40 sildenafil to suppress iNOS,41 and nitroaspirin to inhibit Arg1 and iNOS.42 (4) Direct elimination. Some therapeutic drugs containing gemcitabine,43 docetaxel,44 and 5-fluorouracil45 have been reported to remove MDSCs. Our data showed that SFN repressed MDSCs immunosuppression by directly inducing apoptosis and constitutes a basis for testing SFN as a strategy to eliminate MDSCs in patients with cancer. MDSCs need to maintain the physiological state of life to play as the immunosuppressive contributor in the TME. Moreover, SFN reduced STAT5 levels and phosphorylation to inhibit MDSCs immunosuppressioin, which reflected as reduced Arg-1, PD-L1, TGF-b and IL-10 after SFN treatment.
Several mechanisms have been reported that explain the ability of SFN to inhibit tumor development. In breast cancer models, SFN inhibited the taxanes that induced NF-kB mediated IL-6/IL-8 increase, which further contributed to stemness, to suppress tumor growth.46 SFN perturbs the pancreatic cancer cell cycle by regulating G2/M arrest and reducing Ku70/Ku80/XRCC4 expression to improve chemotherapeutic efficacy.47 SFN represses the biological functions of mesothelioma cells by inhibiting arginine methyltransferase 5 and methylosome protein 50.48 Previously, our lab reported that SFN suppressed PD-1 expression in CAR-T cells to enhance the potentiated antitumor response. Additionally, SFN reduced the PD-L1 level on tumor cells through β-TrCP activated ubiquitination-mediated proteolysis process.19 In this study, we observed that SFN activates IL-12RB2 to induce apoptosis in MDSCs, which is consistent with reports that SFN participates in the cytokine–cytokine receptor pathway in cancer.49 Both researches convinced the modulating role of SFN on liberating CD8+ T cells function through different mechanisms, which supplement the role of SFN to reprogram tumor immune microenvironment.
Three classic pro-apoptotic networks have been reported to induce caspase-dependent cellular apoptosis, including TNF-α (tumor necrosis factor)/TNF receptor 1, Fas/FasL, and TNF-related apoptosis-inducing ligand (TRAIL)/death receptor 4 (or TRAIL-R1)/death receptor 5 (or TRAIL-R2).50 IL-12RB2 has previously been associated with the upregulation of FasL by reducing MMP-3/7 expression in ovarian cancer cells, which induces lymphocyte apoptosis.26 FasL induces apoptosis by binding to its receptor Fas (CD95), which activates the downstream signaling cascade and causes cell death.51 Some MMPs have been reported to participate in the generation of soluble CD95L. Both MMP352 and MMP729 can cleave m-CD95L to release s-CD95L. In this study, we observed elevated levels of caspase 3-positive MDSCs after SFN treatment. Consistent with previous studies, SFN stimulated IL-12RB2, which reduced MMP3 levels, elevated FasL expression to induce the caspase-3 cleavage in MDSCs, and finally induced apoptosis.53 The role of FasL in MDSCs apoptosis was also reported by Peyvandi et al.54 We verified the role of SFN on MDSCs apoptosis and CD8+ T cells activation in clinical samples. Regarding the safety of SFN, the analysis of the plasma concentrations of liver and kidney profiles showed no differences after SFN treatment.
Conclusions
Our study elucidated the role of SFN in modulating the tumor immune response in cancer cells by inducing MDSCs apoptosis. Mechanistically, SFN activated IL-12RB2, which induced the MMP3/FasL signaling pathway to cause caspase-3 cleavage-related cell death in MDSCs. We confirmed these phenomena in clinical samples in a Phase I clinical trial. This study provides evidence supporting the potential application of SFN in clinical cancer treatment as a Chinese medicinal herb that eliminates MDSCs without safety hazards.
Acknowledgments
We thank the Animal Center of Zhengzhou University for the animal feeding and related experiments. We thank Translational Medicine Center of The First Affiliated Hospital of Zhengzhou University. We thank Henan Province International Joint Laboratory of Tumor Immunology and Biotherapy.
Footnotes
JL, HC and CG contributed equally.
Contributors: JLiu conceptualized the study; HC and CG developed the methodology; CG, MZ, and ZF curated and analyzed the data; HC and ML performed the statistical analyses; ZZ, FL, and XZ provided the technical and material support; JLiu and LW provided the clinical samples; YZ and QL supervised the study; and JLiu drafted the manuscript. YZ is responsible for the overall content as the guarantor. All the authors revised and approved the final manuscript.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available upon reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Our college is performing a phase 1 human study, named as “phase I–II clinical trial on the application of SFN enteric-coated capsules in patients with malignant tumors (KY-2021-0350)”. Participants gave informed consent to participate in the study before taking part.
References
- 1. Glasner A, Rose SA, Sharma R, et al. Conserved transcriptional connectivity of regulatory T cells in the tumor microenvironment informs new combination cancer therapy strategies. Nat Immunol 2023;24:1020–35. 10.1038/s41590-023-01504-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Myers RM, Shah NN, Pulsipher MA. How I use risk factors for success or failure of CD19 CAR T cells to guide management of children and AYA with B-cell ALL. Blood 2023;141:1251–64. 10.1182/blood.2022016937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang S-W, Pan C-M, Lin Y-C, et al. Bite-secreting CAR-γδT as a dual targeting strategy for the treatment of solid tumors. Adv Sci (Weinh) 2023;10:2206856. 10.1002/advs.202206856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Qin G, Liu S, Liu J, et al. Overcoming resistance to immunotherapy by targeting GPR84 in myeloid-derived suppressor cells. Signal Transduct Target Ther 2023;8:164. 10.1038/s41392-023-01388-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chaib M, Tanveer UA, Makowski L. Myeloid cells in the era of cancer Immunotherapy: top 3 unanswered questions. Pharmacol Ther 2023;244:108370. 10.1016/j.pharmthera.2023.108370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joshi S, Sharabi A. Targeting myeloid-derived suppressor cells to enhance natural killer cell-based immunotherapy. Pharmacol Ther 2022;235:108114. 10.1016/j.pharmthera.2022.108114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martínez-Garay C, Djouder N. Dietary interventions and precision nutrition in cancer therapy. Trends Mol Med 2023;29:489–511. 10.1016/j.molmed.2023.04.004 [DOI] [PubMed] [Google Scholar]
- 8. Xu Y, Han X, Li Y, et al. Sulforaphane mediates glutathione depletion via polymeric nanoparticles to restore cisplatin chemosensitivity. ACS Nano 2019;13:13445–55. 10.1021/acsnano.9b07032 [DOI] [PubMed] [Google Scholar]
- 9. Kim KH, Moon E, Kim SY, et al. 4-Methylthio-Butanyl derivatives from the seeds of Raphanus sativus and their biological evaluation on anti-inflammatory and antitumor activities. J Ethnopharmacol 2014;151:503–8. 10.1016/j.jep.2013.11.003 [DOI] [PubMed] [Google Scholar]
- 10. Park HS, Han MH, Kim G-Y, et al. Sulforaphane induces reactive oxygen species-mediated mitotic arrest and subsequent apoptosis in human bladder cancer 5637 cells. Food Chem Toxicol 2014;64:157–65. 10.1016/j.fct.2013.11.034 [DOI] [PubMed] [Google Scholar]
- 11. Jo GH, Kim GY, Kim WJ, et al. Sulforaphane induces apoptosis in T24 human urinary bladder cancer cells through a reactive oxygen species-mediated mitochondrial pathway: the involvement of endoplasmic reticulum stress and the NRF2 signaling pathway. Int J Oncol 2014;45:1497–506. 10.3892/ijo.2014.2536 [DOI] [PubMed] [Google Scholar]
- 12. Shan Y, Wu K, Wang W, et al. Sulforaphane down-regulates COX-2 expression by activating P38 and inhibiting NF-kappaB-DNA-binding activity in human bladder T24 cells. Int J Oncol 2009;34:1129–34. 10.3892/ijo_00000240 [DOI] [PubMed] [Google Scholar]
- 13. Justin S, Rutz J, Maxeiner S, et al. Chronic sulforaphane administration inhibits resistance to the mTOR-inhibitor everolimus in bladder cancer cells. Int J Mol Sci 2020;21:4026. 10.3390/ijms21114026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abbaoui B, Riedl KM, Ralston RA, et al. Inhibition of bladder cancer by broccoli Isothiocyanates sulforaphane and erucin: characterization, metabolism, and Interconversion. Mol Nutr Food Res 2012;56:1675–87. 10.1002/mnfr.201200276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang F, Shan Y. Sulforaphane retards the growth of UM-UC-3 Xenographs, induces apoptosis, and reduces survivin in athymic mice. Nutr Res 2012;32:374–80. 10.1016/j.nutres.2012.03.014 [DOI] [PubMed] [Google Scholar]
- 16. Baenas N, Suárez-Martínez C, García-Viguera C, et al. Bioavailability and new biomarkers of cruciferous sprouts consumption. Food Res Int 2017;100:497–503. 10.1016/j.foodres.2017.07.049 [DOI] [PubMed] [Google Scholar]
- 17. Fahey JW, Wade KL, Stephenson KK, et al. Bioavailability of sulforaphane following ingestion of glucoraphanin-rich broccoli sprout and seed extracts with active myrosinase: a pilot study of the effects of proton pump inhibitor administration. Nutrients 2019;11:1489. 10.3390/nu11071489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y, Petrikova E, Gross W, et al. Sulforaphane promotes dendritic cell stimulatory capacity through modulation of regulatory molecules, JAK/STAT3- and microrna-signaling. Front Immunol 2020;11:589818. 10.3389/fimmu.2020.589818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen C, Zhang Z, Tian Y, et al. Sulforaphane enhances the antitumor response of chimeric antigen receptor T cells by regulating PD-1/PD-L1 pathway. BMC Med 2021;19:283. 10.1186/s12916-021-02161-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johler SM, Fuchs J, Seitz G, et al. Macrophage migration inhibitory factor (MIF) is induced by cytotoxic drugs and is involved in immune escape and migration in childhood rhabdomyosarcoma. Cancer Immunol Immunother 2016;65:1465–76. 10.1007/s00262-016-1896-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sundaram MK, R P, Haque S, et al. Dietary Isothiocyanates inhibit cancer progression by modulation of epigenome. Semin Cancer Biol 2022;83:353–76. 10.1016/j.semcancer.2020.12.021 [DOI] [PubMed] [Google Scholar]
- 22. Grover A, Sanseviero E, Timosenko E, et al. Myeloid-derived suppressor cells: a propitious road to clinic. Cancer Discovery 2021;11:2693–706. 10.1158/2159-8290.CD-21-0764 [DOI] [PubMed] [Google Scholar]
- 23. Waight JD, Netherby C, Hensen ML, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest 2013;123:4464–78. 10.1172/JCI68189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee C-C, Lin J-C, Hwang W-L, et al. Macrophage-secreted Interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun 2018;9:3763. 10.1038/s41467-018-06268-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gocher-Demske AM, Cui J, Szymczak-Workman AL, et al. IFNγ-induction of T(H)1-like regulatory T cells controls antiviral responses. Nat Immunol 2023;24:841–54. 10.1038/s41590-023-01453-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gorelik E, Edwards RP, Feng X, et al. IL-12 receptor-mediated upregulation of FasL in human ovarian carcinoma cells. Int J Cancer 2004;112:620–7. 10.1002/ijc.20482 [DOI] [PubMed] [Google Scholar]
- 27. Garcia AJ, Tom C, Guemes M, et al. ERα signaling regulates MMP3 expression to induce FasL cleavage and osteoclast apoptosis. J Bone Miner Res 2013;28:283–90. 10.1002/jbmr.1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu W, Vetreno RP, Crews FT. Hippocampal TNF-death receptors, caspase cell death cascades, and IL-8 in alcohol use disorder. Mol Psychiatry 2021;26:2254–62. 10.1038/s41380-020-0698-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Powell WC, Fingleton B, Wilson CL, et al. The metalloproteinase matrilysin proteolytically generates active soluble Fas ligand and potentiates epithelial cell apoptosis. Curr Biol 1999;9:1441–7. 10.1016/s0960-9822(00)80113-x [DOI] [PubMed] [Google Scholar]
- 30. Zhang H, Lian M, Zhang J, et al. A functional characteristic of cysteine-rich protein 61: modulation of myeloid-derived suppressor cells in liver inflammation. Hepatology 2018;67:232–46. 10.1002/hep.29418 [DOI] [PubMed] [Google Scholar]
- 31. Durham NM, Holoweckyj N, MacGill RS, et al. GITR ligand fusion protein agonist enhances the tumor antigen-specific CD8 T-cell response and leads to long-lasting memory. J Immunother Cancer 2017;5:47. 10.1186/s40425-017-0247-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lai X, Liu X-L, Pan H, et al. Light-triggered efficient sequential drug delivery of biomimetic nanosystem for multimodal chemo-, antiangiogenic, and anti-MDSC therapy in Melanoma. Adv Mater 2022;34:e2106682. 10.1002/adma.202106682 [DOI] [PubMed] [Google Scholar]
- 33. Tang KH, Li S, Khodadadi-Jamayran A, et al. Combined inhibition of SHP2 and CXCR1/2 promotes antitumor T-cell response in NSCLC. Cancer Discov 2022;12:47–61. 10.1158/2159-8290.CD-21-0369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen X, Adhikary G, Ma E, et al. Sulforaphane inhibits CD44V6/Yap1/TEAD signaling to suppress the cancer phenotype. Mol Carcinog 2023;62:236–48. 10.1002/mc.23479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conche C, Finkelmeier F, Pešić M, et al. Combining ferroptosis induction with MDSC blockade renders primary tumours and metastases in liver sensitive to immune checkpoint blockade. Gut 2023;72:1774–82. 10.1136/gutjnl-2022-327909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Melani C, Sangaletti S, Barazzetta FM, et al. Amino-Biphosphonate-mediated MMP-9 inhibition breaks the tumor-bone marrow axis responsible for myeloid-derived suppressor cell expansion and macrophage infiltration in tumor stroma. Cancer Res 2007;67:11438–46. 10.1158/0008-5472.CAN-07-1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pan P-Y, Wang GX, Yin B, et al. Reversion of immune tolerance in advanced malignancy: modulation of myeloid-derived suppressor cell development by blockade of stem-cell factor function. Blood 2008;111:219–28. 10.1182/blood-2007-04-086835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hengesbach LM, Hoag KA. Physiological concentrations of retinoic acid favor myeloid dendritic cell development over granulocyte development in cultures of bone marrow cells from mice. J Nutr 2004;134:2653–9. 10.1093/jn/134.10.2653 [DOI] [PubMed] [Google Scholar]
- 39. Kusmartsev S, Cheng F, Yu B, et al. All-trans-retinoic acid eliminates immature myeloid cells from tumor-bearing mice and improves the effect of vaccination. Cancer Res 2003;63:4441–9. [PubMed] [Google Scholar]
- 40. Zea AH, Rodriguez PC, Atkins MB, et al. Arginase-producing myeloid suppressor cells in renal cell carcinoma patients: a mechanism of tumor evasion. Cancer Res 2005;65:3044–8. 10.1158/0008-5472.CAN-04-4505 [DOI] [PubMed] [Google Scholar]
- 41. Serafini P, Meckel K, Kelso M, et al. Phosphodiesterase-5 inhibition augments endogenous antitumor immunity by reducing myeloid-derived suppressor cell function. J Exp Med 2006;203:2691–702. 10.1084/jem.20061104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. De Santo C, Serafini P, Marigo I, et al. Nitroaspirin corrects immune dysfunction in tumor-bearing hosts and promotes tumor eradication by cancer vaccination. Proc Natl Acad Sci U S A 2005;102:4185–90. 10.1073/pnas.0409783102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki E, Kapoor V, Jassar AS, et al. Gemcitabine selectively eliminates splenic Gr-1+/CD11B+ myeloid suppressor cells in tumor-bearing animals and enhances antitumor immune activity. Clin Cancer Res 2005;11:6713–21. 10.1158/1078-0432.CCR-05-0883 [DOI] [PubMed] [Google Scholar]
- 44. Zhang X, Sun S, Miao Y, et al. Docetaxel enhances the therapeutic efficacy of PSMA-specific CAR-T cells against prostate cancer models by suppressing MDSCs. J Cancer Res Clin Oncol 2022;148:3511–20. 10.1007/s00432-022-04248-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yang Y, Zhang M, Zhang Y, et al. 5-fluorouracil suppresses colon tumor through activating the P53-Fas pathway to sensitize myeloid-derived suppressor cells to Fasl(+) cytotoxic T lymphocyte cytotoxicity. Cancers (Basel) 2023;15:1563. 10.3390/cancers15051563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burnett JP, Lim G, Li Y, et al. Sulforaphane enhances the anticancer activity of Taxanes against triple negative breast cancer by killing cancer stem cells. Cancer Lett 2017;394:52–64. 10.1016/j.canlet.2017.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Naumann P, Liermann J, Fortunato F, et al. Sulforaphane enhances irradiation effects in terms of perturbed cell cycle progression and increased DNA damage in pancreatic cancer cells. PLoS One 2017;12:e0180940. 10.1371/journal.pone.0180940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ezeka G, Adhikary G, Kandasamy S, et al. Sulforaphane inhibits PRMT5 and MEP50 function to suppress the mesothelioma cancer cell phenotype. Mol Carcinog 2021;60:429–39. 10.1002/mc.23301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. AlQathama A, Prieto JM. Natural products with therapeutic potential in Melanoma metastasis. Nat Prod Rep 2015;32:1170–82. 10.1039/c4np00130c [DOI] [PubMed] [Google Scholar]
- 50. Zhang X, Tang J, Kou X, et al. Proteomic analysis of MSC-derived apoptotic vesicles identifies Fas inheritance to ameliorate haemophilia a via activating platelet functions. J Extracell Vesicles 2022;11:e12240. 10.1002/jev2.12240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang J, Cao Z, Wang P, et al. Apoptotic extracellular vesicles ameliorate multiple myeloma by restoring Fas-mediated apoptosis. ACS Nano 2021;15:14360–72. 10.1021/acsnano.1c03517 [DOI] [PubMed] [Google Scholar]
- 52. Matsuno H, Yudoh K, Watanabe Y, et al. Stromelysin-1 (MMP-3) in Synovial fluid of patients with rheumatoid arthritis has potential to cleave membrane bound Fas ligand. J Rheumatol 2001;28:22–8. [PubMed] [Google Scholar]
- 53. Leca J, Lemonnier F, Meydan C, et al. IDH2 and TET2 mutations synergize to modulate T follicular helper cell functional interaction with the AITL microenvironment. Cancer Cell 2023;41:323–39. 10.1016/j.ccell.2023.01.003 [DOI] [PubMed] [Google Scholar]
- 54. Peyvandi S, Buart S, Samah B, et al. Fas ligand deficiency impairs tumor immunity by promoting an accumulation of Monocytic myeloid-derived Suppressor cells. Cancer Res 2015;75:4292–301. 10.1158/0008-5472.CAN-14-1848 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007983supp001.pdf (2MB, pdf)
jitc-2023-007983supp002.pdf (585.2KB, pdf)
jitc-2023-007983supp003.pdf (2MB, pdf)
jitc-2023-007983supp004.pdf (1.7MB, pdf)
Data Availability Statement
Data are available upon reasonable request.