Abstract
Silver nanoparticles (AgNPs) were prepared using a one-step reduction of silver nitrate (AgNO3) with sodium borohydride (NaBH4) in the presence of polyvinylpyrrolidone (PVP) as a capping agent. Plant extracts from D. sissoo (DS) and A. calamus L. (AC) leaves were incorporated during the synthesis process. The crystalline nature of the AgNPs was confirmed through X-ray diffraction (XRD), confirming the face-centered cubic structure, with a lattice constant of 4.08 Å and a crystallite size of 18 nm. Field Emission Gun Transmission Electron Microscopy (FEG-TEM) revealed spherical AgNPs (10–20 nm) with evident PVP adsorption, leading to size changes and agglomeration. UV–Vis spectra showed a surface plasmon resonance (SPR) band at 417 nm for AgNPs and a redshift to 420 nm for PVP-coated AgNPs, indicating successful synthesis. Fourier Transform Infrared Spectroscopy (FTIR) identified functional groups and drug-loaded samples exhibited characteristic peaks, confirming effective drug loading. The anti-cancer potential of synthesized NPs was assessed by MTT assay in human adenocarcinoma lung cancer (A549) and lung normal cells (WI-38) cells. IC50 values for all three NPs (AgPVP NPs, DS@AgPVP NPs, and AC@AgPVP NPs) were 41.60 ± 2.35, 14.25 ± 1.85, and 21.75 ± 0.498 μg/ml on A549 cells, and 420.69 ± 2.87, 408.20 ± 3.41, and 391.80 ± 1.55 μg/ml respectively. Furthermore, the NPs generated Reactive Oxygen Species (ROS) and altered the mitochondrial membrane potential (MMP). Differential staining techniques were used to investigate the apoptosis-inducing properties of the three synthesized NPs. The colony formation assay indicated that nanoparticle therapy prevented cancer cell invasion. Finally, Real-Time PCR (RT-PCR) analysis predicted the expression pattern of many apoptosis-related genes (Caspase 3, 9, and 8).
Keywords: Silver nanoparticles, A549, Cytotoxicity, Apoptosis, D. sissoo Roxb. Ex DC., A. calamus L.
Highlights
-
•
Synthesis of plant extract capped AgPVP NPs.
-
•
Characterization of plant extract capped AgPVP NPs (XRD, FEG-TEM, UV, Zeta potential, FTIR, etc).
-
•
Apoptosis induction capability of NPs in A549 Cells (MTT, Morphological changes, differential staining, ROS measurement, etc).
-
•
Mechanism of action of plant extract capped AgPVP NPs.
1. Introduction
The second-most lethal kind of cancer in the world is lung cancer [1]. In 2022, 1, 30, 180 projected fatalities and 2, 36,740 additional cases were predicted [2]. Small-cell lung cancer, which accounts for 20 % of cases diagnosed, and non-small-cell lung cancer, which accounts for 80 % of cases diagnosed, are the two forms of lung cancer. There are several therapies for lung cancer, including radiation, chemotherapy, and surgical removal, but sadly they all have a lot of adverse effects. Drug resistance in cancer cells has become a significant factor recently. To trigger apoptosis in lung cancer with fewer side effects and at a reasonable price, a new medication with biocompatible treatment methods is desperately needed [3,4].
With the least amount of expenditure and side effects, nanotechnology provides tools and resources to diagnose and treat a range of malignancies [5,6]. Production of green nanoparticles is affordable, secure, non-toxic, and ecologically responsible [7]. Silver (Ag) is the most commercially successful nano-compound, according to the Woodrow-Wilson database on nano-products, due to its physicochemical properties, antibacterial activity and treatments, bimolecular recognition, biolabeling, catalysis, and microelectronics [8]. Various consumer goods, including electronics, cosmetics, household appliances, textiles, food processing, and medical supplies, use silver nanoparticles (AgNPs). Numerous plant extracts are thought to be effective natural reducing agents with strong antioxidant activity [9,10]. When compared to other metal nanoparticles, AgNPs are less toxic to humans [11]. Due to the development of implantable biomaterials, molecular imaging, wound healing, and drug administration, to name just a few of the growing number of biomedical applications, green production of nanoparticles with a restricted range of toxicity has become a hot study topic [[12], [13], [14], [15]]. In particular, the use of silver nanoparticles for cancer detection and treatment has increased, not only as ideal platforms for targeted therapeutic administration or as early cancer screening probes but also as a potential therapeutic molecule on its own [[16], [17], [18]]. Silver nanoparticles showed potential cytotoxicity when tested on a variety of cancer cell lines (A549, MCF-7, HT29, HeLa), as well as Dalton's lymphoma ascites tumor [[19], [20], [21], [22], [23]].
The leaves of D. sissoo and A. calamus L. have been shown to contain a variety of secondary metabolites, including phenols, tannins, alkaloids, anthraquinones, saponins, and flavonoids, as determined by Gas Chromatography-High-Resolution Mass Spectrometry (GC-HRMS) [[24], [25]]. They also exhibit strong cytotoxic action against A549 cells. Therefore, to create a new treatment strategy, we chose to synthesize DS@AgPVP NPs and AC@AgPVP NPs. Using multiple spectroscopic techniques and microscopic inspection, we have developed stable nanoparticles with a limited size distribution for the current investigation. We then investigated the anticancer properties of our green-synthesized NPs on A549 cells in vitro.
2. Experimental section
2.1. Chemicals and materials
Silver nitrate (AgNO3), sodium borohydride (NaBH4), and PVP40 (C6H9NO)n were used for the synthesis of AgNPs. Silver nitrate extra pure was purchased from SRL (94118, India), and Sodium borohydride was purchased from Merck (106371, India) and utilized without any further purification. Polyvinylpyrrolidone (PVP) with an average F.M. 40,000 ≥ 99 %, High Purity (K30), was purchased from Fisher Scientific (Amresco, 0507-500G), USA. Milli-Q water was used throughout the experimentation.
2.2. Preparation of plant extracts
Plant extracts were prepared from D. sissoo (DS) and A. calamus L. (AC) leaves, as mentioned previously [24,25]. The extracts were stored at 4 °C until further use.
2.3. Synthesis of colloidal PVP-coated silver nanoparticles
Silver nitrate (AgNO3) was reduced with sodium borohydride (NaBH4) in one step to produce the colloidal silver solution. Silver nanoparticles (AgNPs) were produced by adding a 2 mM reducing agent (NaBH4) drop-by-drop to a 1 mM silver salt (AgNO3) at room temperature. For 30 min, the mixture was regularly stirred, and during that time, the solution quickly became yellow. As a capping agent, 0.1 % PVP was added to the AgNO3 solution to create the PVP-coated silver nanoparticles. Next, NaBH4 was added drop by drop, and the mixture was vigorously stirred for 30 min. Then, add drop-by-drop plant extracts (DS and AC) to a different tube mixer and swirl for 40–45 min at 500 rpm. The PVP Ag nanoparticles that had been coated with extract were ready after 45 min. TEM, UV, particle size analysis, zeta potential analysis, and FTIR analysis characterized the NPs.
2.4. Characterization of synthesized PVP-coated silver nanoparticles
The crystalline phases of the samples were analyzed using a Rigaku Ultima diffractometer equipped with a Cu(kα) radiation source (λ = 1.546 Å) through powder X-ray diffraction technique. The morphology of AgPVP NPs was characterized using a transmission electron microscope (TEM) (Tecnai 20, Philips, and Holland). Particle size and zeta potential were determined using a particle size analyzer with a zeta potential measuring system (HORIBA, SZ100) at a scattering angle of 173° and a temperature of 25 °C using samples. The Fourier transform infrared spectra (Spectrum GX, PerkinElmer, and U.S.A.) at a resolution of 0.15 cm−1 were used to estimate the structural features of nanoparticles in the 400–4000 cm−1 range using KBr pellets.
2.5. In-vitro anticancer studies
2.5.1. Cell lines
The National Centre for Cell Sciences (NCCS) in Pune, India, has a repository of human adenocarcinoma lung cancer (A549) and Lung Normal cells (WI-38), which were obtained and preserved as per following previous instructions mentioned [26].
2.5.2. Cell viability assay
To study the cytotoxic capability of all NPs (AgPVP NPs, DS@AgPVP NPs, and AC@AgPVP NPs) on A549 and WI-38 cells, an MTT test was performed [27].
2.5.3. Detection of morphological alteration in A549 cells
An inverted fluorescent phase contrast microscope was used to examine the morphological changes within NP-treated A549 cells. 5 X 104 cells were incubated overnight in a 96-well plate at 37 °C with 5 % CO2 before being treated for 24 h with the IC50 concentrations of NPs. Morphological changes were seen and photographed using a 40× inverted fluorescence phase-contrast microscope (Carl Zeiss, Axio observer A1).
2.5.4. Estimation of intracellular reactive oxygen species (ROS) in A549 cells
ROS production was analyzed using the fluorescent marker 2,7-dichlorodihydrofluorescein diacetate in diacetate (DCFH-DA) probe, as previously described [28].
2.5.5. Estimation of mitochondrial membrane potential in A549 cells
The activity of NPs on 3 X 104 A549 cells/well was determined using mitochondrial membrane potential (MMP) by using JC-10 dye (Sigma Aldrich, MAK159). After 24 h of the incubation period, cells were stained with JC-10 dye for 30–35 min. Then, the cells were washed with 1× PBS. The fluorescence intensity of control and treated cells was analyzed using a microplate reader (Molecular Devices, USA, SpectramaxM2e) at 485 nm excitation and 530 nm emission, respectively.
2.5.6. Nuclear assessment in A549 cells by 4-6-diamidino-2-phenylindole staining
Nuclear morphology was identified by 4-6-diamidino-2-phenylindole (DAPI) staining (HiMedia, India) in (1.5 X 104) A549 cells with the capacity to create fluorescence in DS-DNA after treatment with IC50 doses of NPs respectively. After 24 h, cells were washed 2–3 times with 1× PBS, stained with 50 μl of DAPI dye, and incubated in a CO2 incubator for 30 min. Following incubation, the cells were rinsed with 1× PBS to remove excess dye. A fluorescence-inverted phase-contrast microscope with a DAPI filter was used to investigate the cells [29].
2.5.7. Live/dead cell differentiation in A549 cells by double fluorescence staining with Acridine Orange/Ethidium Bromide (AO/EB)
A549 cells (2 X 105) were seeded in 6-well plates overnight at 37 °C in a CO2 incubator. After 24 h, they were treated with the IC50 concentration of NPs and re-incubated for 24 h. Cells were fixed with ice-cold methanol for 15–20 min at room temperature, then washed 2–3 times with 1× PBS. Cells were stained with 10 μg/ml AO/EB in each well and then incubated for 15 min at 37 °C in a CO2 incubator. After incubation, cells were washed with 1× PBS and visualized by a fluorescence inverted microscope (40× magnification, Axio Observer A1, Carl Zeiss) under Fluorescein Isothiocyanate (FITC) and Tetramethylrhodamine Isothiocyanate (TRITC) filters [29].
2.5.8. Estimation of apoptosis in A549 cells by Giemsa staining
2 X 104 cells were seeded in a 6-well culture plate for 24 h. After 24 h, cells were treated with their IC50 concentrations of NPs respectively. After 24 h of incubation, the culture media was removed, and the cells were washed with 1× PBS. For the fixation of the cells, ice-cold methanol was used. Giemsa staining was used to determine the morphology of proliferative and apoptotic cells.
2.5.9. Assessment of clonogenic assay in A549 cells
To determine the Anti-invasion effects on in vitro cell proliferation, the colony formation assay was performed [30,31].
2.5.10. RT-PCR studies in A549 cells
Around 1 X 105 lung cancer cells were treated with the IC50 concentrations of NPs in 6-well plates for 24 h at 37 °C in a CO2 incubator, followed by total RNA extraction using the TRIzol reagent method. 2 μg of extracted RNA was further used for cDNA synthesis by using the Thermo Fisher cDNA synthesis kit as per the protocol. The primer sequences (0.5 μM each of forward and reverse primers) used in this gene expression study are listed below, as described previously [24]. Furthermore, an RT-PCR assay was performed using the Biorad SYBR Green qPCR Kit following the manufacturer's instructions.
2.6. Brine shrimp lethality assay
Brine prawn (Artemia salina) eggs were hatched in artificial seawater with 38 g/L of table salt. A lamp was placed above the tank's open side to lure freshly born prawns near the tank wall. The prawns were ready for the test after 24 h of development into nauplii (Artemia salina). The nanoparticles were subjected to the usual brine shrimp lethality bioassay [32]. To get concentrations ranging from 10 to 100 μg/ml, all nanoparticles (1 mg) were dissolved in 1 ml of 1 M NaOH (pH 8). A Petri plate containing 1 M NaOH (pH 8) in 5 ml of salt water was used as the negative control. As a positive control, potassium dichromate was dissolved in 1 M NaOH (pH 8) and serially diluted to concentrations of 5 mg/ml. A suspension of larvae (0.1 ml), containing about 10 larvae, was added to each Petri plate and incubated for 24 h. The Petri plate was then examined, and the number of dead larvae in each bottle was counted after 24 h. The death percentage was calculated as per the following equation:
| Percent of Death= (Total shrimp-Alive shrimp) / (Total Shrimp) X 100 ------ | (Equation No. 1) |
2.7. Statistical analysis
All reported data are expressed as the mean ± SEM of three individual experiments performed in triplicates. Statistical analysis among different treatment groups was determined using a one-way ANOVA followed by Tukey as a post-assay through GraphPad Prism 9.4.1. Significance: *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.001.
3. Results and discussion
3.1. Characterization of nanoparticles
3.1.1. XRD analysis
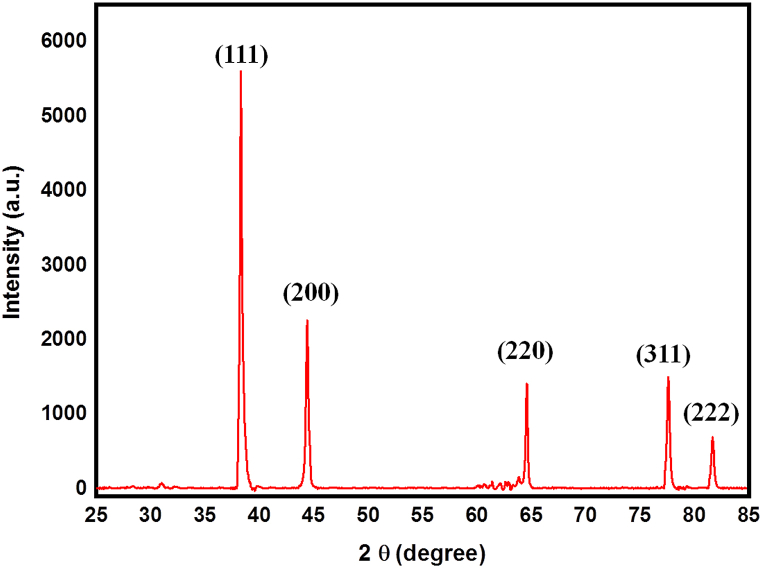
The powder X-ray diffraction (PXRD) pattern of the AgNPs is presented in Fig. 1. The obtained diffraction pattern was analyzed and indexed using Powder-X software. The XRD pattern of the AgNPs exhibited distinct and prominent peaks at 2θ angles of 38.12°, 44.28°, 64.39°, 77.49°, and 81.56°, which corresponded to the crystal planes (111), (200), (220), (311), and (222), respectively. These peaks matched well with the face-centered cubic crystal structure, confirming the crystalline nature of the sample, and agreed with the reference data from the JCPDS card (04–0783). The lattice constant (a) was determined using the formula
| (Equation No. 2) |
where represents the interplanar spacing. The calculated lattice parameter for the sample was found to be 4.08 Å. The crystallite size (D) of the samples was calculated using the Debye-Scherrer formula, as reported in Ref. [33]. The calculated value for the crystallite size was determined to be 18 nm.
Fig. 1.
XRD analysis of AgNPs.
3.1.2. Field emission Gun transmission electron microscope (FEG-TEM) analysis
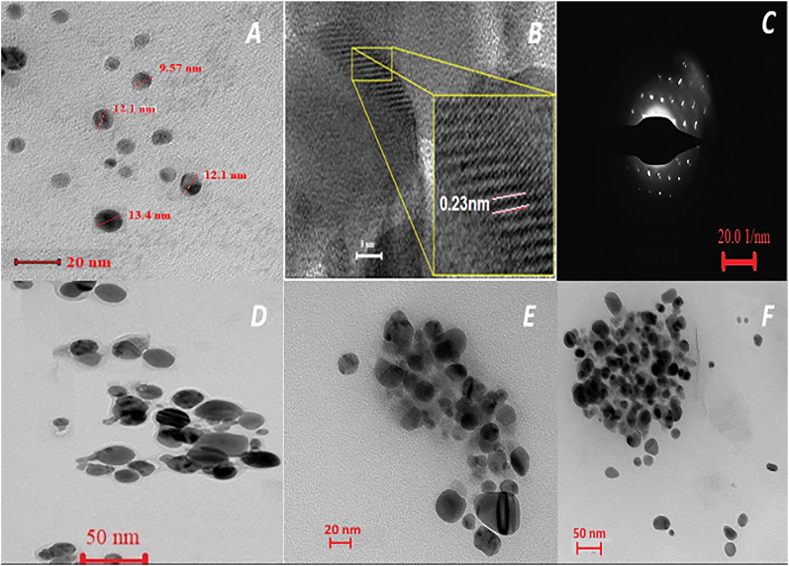
The shape and size of the particles were determined using a FEG-TEM investigation. A carbon-coated copper grid was utilized to contain dilutions of an Ag nanoparticle solution, which was then allowed to dry naturally while FEG-TEM pictures were captured. The FEG-TEM micrographs indicated that the produced AgNPs were spherical and agglomerated less (Fig. 2A). The average diameter of AgNP was determined to be between 10 and 20 nm. The clear and consistent lattice fringes visible in the inset FEG-TEM image (Fig. 2B) show the crystalline nature of the produced AgNPs. The observed distance between lattices is around 0.23 nm, which is attributable to the (111) planes of a silver crystal lattice.
Fig. 2.
FEG-TEM Analysis
(A) TEM and (B) High-resolution TEM image of AgNPs (inset shows lattice fringers) (C) selected area diffraction pattern of AgNPs. TEM images of the (D) AgPVP NPs (E) AC@AgPVP NPs (F) DS@AgPVP NPs.
Furthermore, it implies that the prominent faces of silver nanoparticles are in good agreement with the face-centered cubic structure's lattice fringe (111). Approaching the electron beam perpendicular to one of the AgNPs spheres resulted in the selected area electron diffraction (SAED) pattern (Fig. 2C). The diffraction spot pattern's strong symmetry suggests that the produced AgNPs are well crystalline. Fig. 2D shows a slight increase in particle size, significant agglomeration, and noticeable morphological alterations, indicating that PVP was successfully adsorbing onto the surface of AgNPs. Conspicuous changes in color intensity are seen in Fig. 2(E) and (F), with separate areas showing considerable blackness and brightness between two individual particles. This contrasting color disparity signifies the effective encapsulation and loading of a drug onto the PVP-coated AgNPs.
3.1.3. UV–Vis spectra
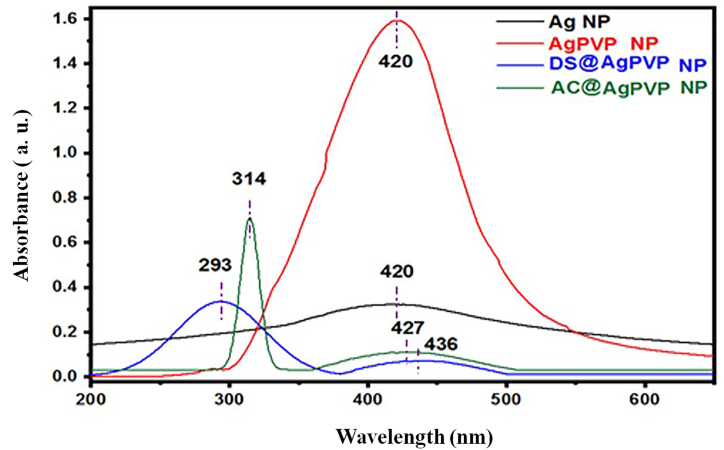
Visual examination of the solution was used to monitor the reduction of Ag ions (Ag+) to Ag nanoparticles. The color shift from colorless to yellow in the AgNO3:NaBH4 solution during the NPs synthesis was the first sign that AgNPs were effectively created. Silver nanoparticles look bright yellowish in an aqueous solution because of the surface plasmon resonance (SPR) of metal nanoparticles, according to Ref. [34]. The surface plasmon resonance (SPR) of metal nanoparticles is widely understood to be created by numerous excitations of electrons near the nanoparticle's surface in resonance with a light wave.
The UV–Vis spectra of silver nanoparticles (Fig. 3) at room temperature indicate a prominent surface plasmon resonance (SPR) band at 417 nm, confirming the synthesis of AgNPs. Whereas of the surface plasmon absorption peak for PVP-coated silver nanoparticle appears at 420 nm, which exhibits redshift. This red shift of the surface plasmon absorption in PVP-coated silver nanoparticles can be attributed to the presence of PVP as a capping agent. Experimentally, it is reported that the intensity of the surface plasmon resonance depends on the particle size, shape, capping agent, and environment of the particle [35,36]. The UV–Vis spectra can be used to determine the size distribution of nanoparticles in a colloidal solution. Sharma and his colleagues suggest that the full width at half maximum (FWHM) of the UV–Vis absorption peaks can serve as an indicator of the extent of nanoparticle aggregation [37]. In the present study, a narrower peak broadening (FWHM) in the UV–Vis absorbance spectra of silver nanoparticles is correlated with a decrease in nanoparticle polydispersity. In addition, UV–Vis absorbance spectra of AC@AgPVP NPs and DS@AgPVP NPs displayed strong absorption peaks at 314 nm and 293 nm, respectively. The existence of particular chromophores or phytochemical elements inside the leaves can explain these different peaks.
Fig. 3.
UV–Vis spectra analysis.
3.1.4. Particle size distribution and zeta potential
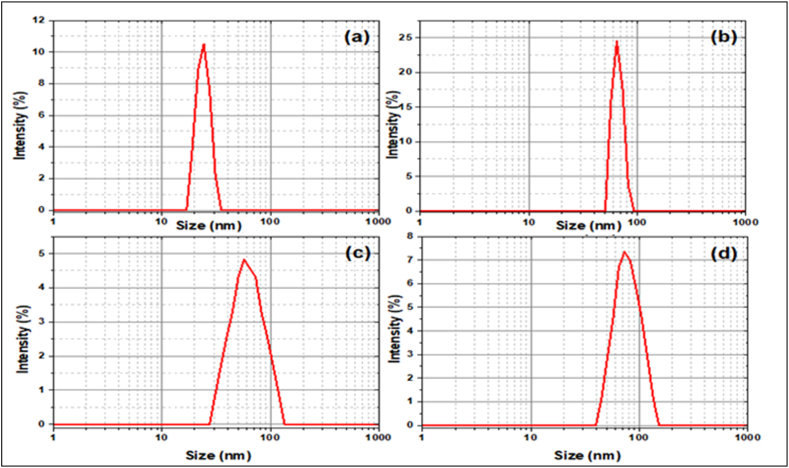
The size of the particles created had to be determined to prove their nanoscale nature. As a result, using a Zeta sizer, the Z-average size, particle size distribution, and polydispersity index (PDI) of drug-loaded Ag NPs were measured.
DLS was utilized to investigate the average nanoparticle diameter and size distribution profile of colloidally produced materials. The average diameter of nanoparticles was calculated using the Stokes-Einstein equation.
| (Equation No. 3) |
where is the hydrodynamic diameter, is Boltzmann's constant, T is the absolute temperature, is the viscosity of the medium, and D is the diffusion coefficient [38]. In terms of intensity-weighted particle size distribution, Fig. 4 (A-D) depicts the DLS size measurement for AgNPs, AgPVP NPS, DS@AgPVP NPs, and AC@AgPVP NPs. The calculated average particle size was found to be 23.06 ± 3.62 nm for AgNPs, whereas PVPAg NPs had a particle size of 66.04 ± 16.02 nm. Both sizes acquired by DLS were somewhat larger than those obtained by HRTEM. This might be because HRTEM is based on physical size, but DLS is based on hydrodynamic size. As a result, the DLS has a greater size portion. Table 1 demonstrates that with the addition of PVP and drug, the average size of AgNPS rises, confirming the adherence of PVP and medication to the surface of AgNPs. Another essential statistic for measuring particle size heterogeneity in the medium is the PDI. The PDI scale is from 0 to 1. Where 0 implies a restricted size distribution and 1 denotes a relatively broad size distribution with the possibility of big particles or aggregates. The estimated PDI value for AgNPS and AgPVP NPs confirms the nanoparticles' monodispersity. Furthermore, the significant PDI values in AgNPS samples containing pharmaceuticals show that biological molecules also ensure drug loading to the surface of the NPs, which is consistent with prior findings.
Fig. 4.
Dynamic light scattering size distribution graph of the (A) AgNPs (B) AgPVP NPs (C) DS@AgPVP NPs and (D) AC@Ag PVPNPs.
Table 1.
Average nanoparticles size.
| Sample | Average Particle size (nm) | PDI | D50 (nm) | D95 (nm) |
|---|---|---|---|---|
| Ag Nps | 23.06 3.62 | 0.157 | 24 | 32.51 |
| AgPVP NPs | 66.04 16.02 | 0.242 | 67 | 84.41 |
| DS@AgPVP NPs | 66.15 58.70 | 0.887 | 69 | 101.62 |
| AC@AgPVP NPs | 81.16 70.02 | 0.864 | 95 | 135.89 |
3.1.5. Fourier Transform Infrared Spectroscopy (FTIR)
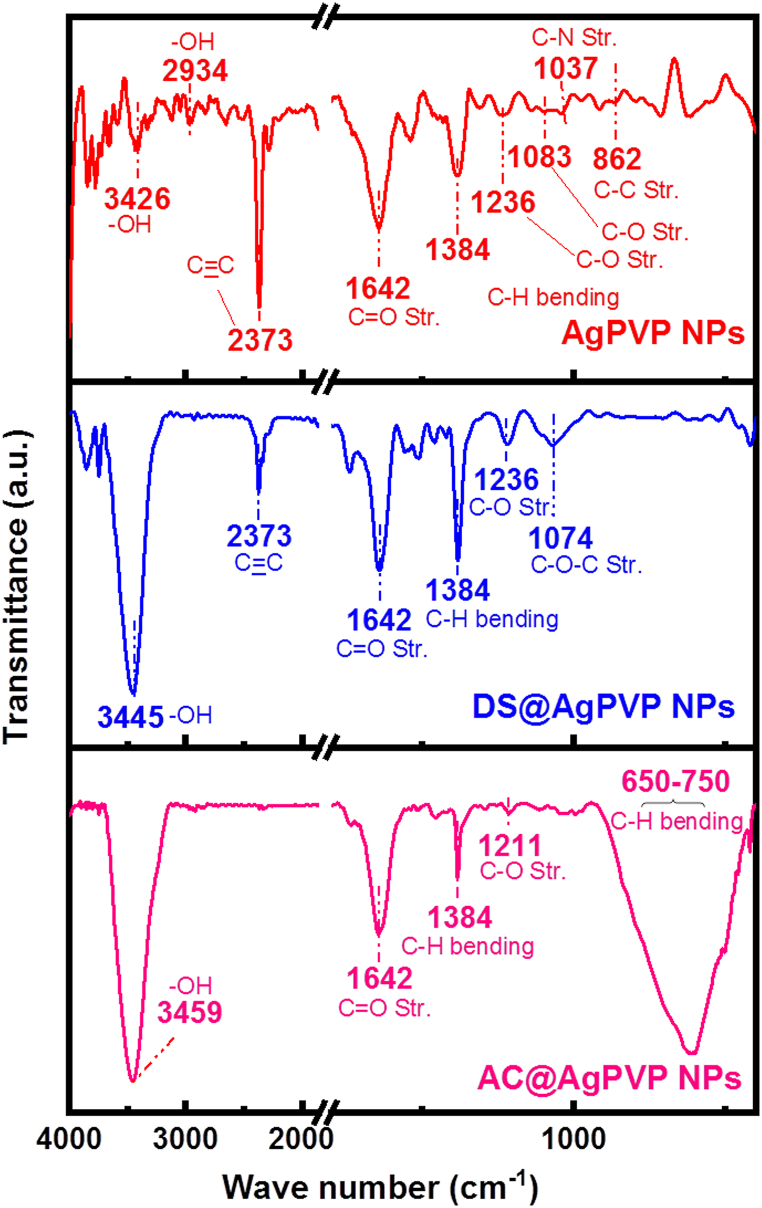
Fig. 5 presents the FTIR spectra results spanning the 500-4000 cm−1 range, utilized for the identification of functional groups on NPs. The peak observed at 862 cm−1 indicated the presence of C–C stretching vibrations, while the peak at 1037 cm−1 represented the C–N stretching vibrations originating from the PVP polymer backbone. Furthermore, the peak observed at 1083 cm−1 was associated with the C–O stretching vibrations of the PVP molecule. The peaks at 1236 cm−1 and 1384 cm−1 corresponded to the C–O and C–N stretching vibrations, respectively, arising from the amide groups within the PVP structure. Soni et al. suggest that a strong, sharp peak at 1642 cm−1 can be attributed to C O stretching vibration, which exhibits shift when compared to pure PVP [39]. This wavenumber shift in the C O bond could be caused by bond weakening induced by the partial donation of lone pair electrons from oxygen in PVP to the vacant orbital of Ag. The presence of an intense peak at 2373 cm−1 indicated a C C triple bond stretching vibration. Additionally, the peaks at 2934 cm−1 and 3426 cm−1 were attributed to the asymmetric and symmetric stretching vibrations of the O–H groups present in the PVP polymer. The peak centered at nearly 1384 cm−1 can be assigned to C–H bending from CH3 hydrocarbon groups which are likely to be present in all samples. In the spectra obtained after drug loading, a distinct and broadband was observed at approximately 3445–3459 cm−1, indicative of the stretching vibration associated with hydroxyl (-OH) groups. This characteristic band confirms the presence of hydroxyl groups, which are essential constituents of diverse phenolic phytochemical compounds, including flavonoids, phenolic acids, and polyphenols. The detection of this band further confirms the presence of these bioactive compounds in the drug-loaded samples. The weak peak at 1074 cm−1 appearing in drug-loaded spectra corresponded to the C–O–C stretching of alkyl-substituted functional units. The prominent peak (Fig. 5) region around 650-750 cm−1 belongs to C–H out of a plane from mononuclear aromatic benzene, which suggests the presence of a Flavonoid derivative with a Quercetin-like structure [40]. Moreover, the overall intensity shift and additional absorption peaks in the FTIR spectra further confirmed the presence of the drug on the PVP-Ag NPs surface.
Fig. 5.
FTIR analysis of Nanoparticles.
3.2. Cell culture studies
3.2.1. Anti-proliferative study
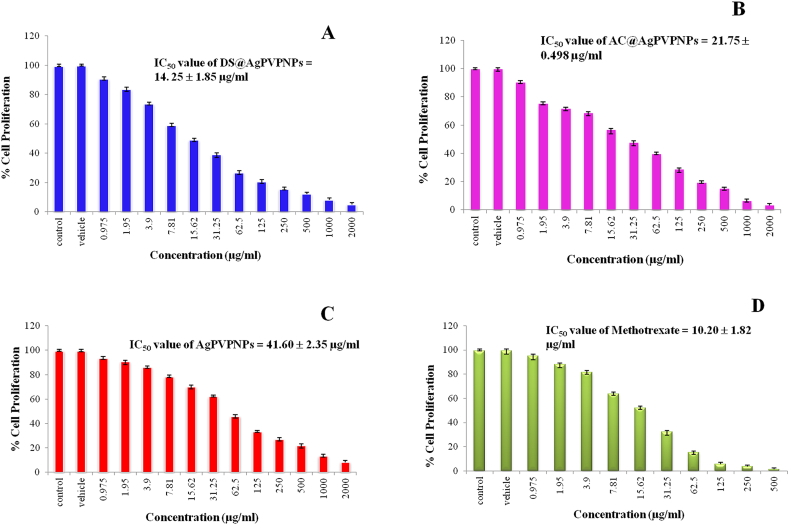
To estimate the cytotoxic or anticancer effects of NPs on A549 cells, an MTT assay was performed. A549 cells were treated with different concentrations of NPs for 24 h. The IC50 of DS@AgPVP NPs, AC@AgPVP NPs, and AgPVP NPs against A549 cells were found to be 14.25 ± 1.85, 21.75 ± 0.49, and 41.60 ± 2.35 μg/ml. The IC50 values DS@AgPVP NPs, AC@AgPVP NPs, and AgPVP NPs against WI-38 cells were 420.69 ± 2.87, 408.20 ± 3.41, and 391.80 ± 1.55 μg/ml respectively. The standard drug methotrexate (positive control) showed an IC50 value of 10.20 ± 1.82 μg/ml on A549 cells and 26.21 ± 1.14 μg/ml on WI-38 cells. NPs treatments significantly reduced the cell viability of A549 cells, as shown in Fig. 6A (A-D), compared to untreated cells. The well-known chemotherapy drug methotrexate has some serious negative side effects. Low dosages are therefore ideal for the patient's therapy. The NPs employed in this investigation demonstrated cytotoxicity exclusively against lung cancer cells, not toward healthy lung cells. It is because of the natural plant extract being embedded. Natural plant extracts have a reputation for being non-toxic to the body's natural cells. The toxicity of methotrexate against Human Embryonic Kidney Cells (HEK 293T) was also documented by Patel et al., in 2011 [41], indicating that the medicine might have negative effects on healthy human cells as well. The formation of nanoparticles increased the efficacy as evidenced by a reduction in IC50 values. DS and AC hydromethanolic crude extracts showed IC50 values of 90.56 ± 2.32 μg/ml and 92.83 ± 1.98 μg/ml respectively [24,25]. These lower IC50 values of nanoparticles of identical chemicals imply that the AgPVP NPs have a more cytotoxic impact than crude extracts. Akter et al. [42] also reported a link between particle size and toxicity, demonstrating that smaller particle sizes induce greater toxicity. It is crucial to note that the capping material can alter the bioactivity of coated AgNPs since it helps the maintenance of AgNP surface chemistry by stabilizing, giving a clear shape, and reducing Ag+ [43,44]. This section investigates the possible effects of AgNP coatings on toxicological phenomena. The kind of coating materials utilized can influence the cytotoxicity of AgNPs.
Fig. 6.
A. Concentration-response curve of cytotoxicity bioassay in A549 cells treated with Different concentration Nanoparticles and Methotrexate drug
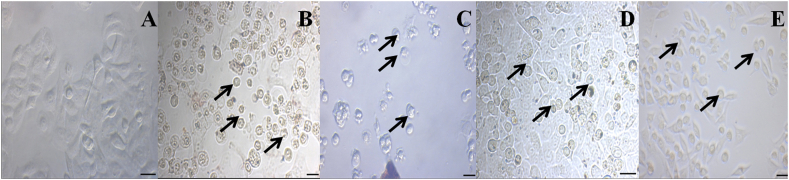
Treated with (A) DS@AgPVP NPs (B) AC@AgPVP NPs (C) AgPVP NPs (D) Methotrexate (positive control/standard drug) and Vehicle (DMSO) is used as a negative control. B. Effect of Nanoparticles on the Morphology of Lung Adenocarcinoma Cells (A549)
Morphology of (A) untreated A549 cells (B) A549 cells treated with DS@AgPVP NPs (C) A549 cells treated with AC@AgPVP NPs (D) A549 cells treated with AgPVP NPs (D) A549 cells treated with Methotrexate. The treatment period is 24 h. Arrows indicate the changes. The scale bar indicates 10 μm.
Typically, the processes that lead to the induction of toxicity include ROS generation, the depletion of antioxidant defense systems, and the loss of mitochondrial membrane potential. The surface coating of AgNPs can influence their aggregation, dissolution ratio, and shape. The types of coatings utilized and their characteristics are critical in determining the cytotoxicity of AgNPs. Fig. 6B depicts the concentration-response curve of the cytotoxicity experiment in A549 cells treated with NPs, methotrexate (positive control), and vehicle (hydromethanol). The vehicle has no discernible influence on the cytotoxic activity of A549 cells. In control and vehicle, the percentage of cell proliferation remains the same. The increased efficacy of NPs is corroborated by contemporary reports. Venugopal et al. [45] reported that AgNPs have good cytotoxicity activity against MCF-7 and A549 cells with IC50 values of 60 and 50 μg/ml. Tian and his [14] team discovered that AgNPs also have cytotoxicity with IC50 values of 50 μg/ml against A549 cells. Vivek et al. [46] reported that AgNPs have good cytotoxic activity against MCF-7, with IC50 values of 50 μg/ml for 24 h and IC50 values of 30 μg/ml for 48 h treatment. Hublikar et al. [[47], [48], [49]], reported that Green synthesis AgNPs from different extracts have good activity against A549 cells (IC50 value of 85.47 μg/ml, and 49.52 g/ml) and also have good antibacterial properties towards the bacteria E. coli.
3.2.2. Effect of nanoparticles on the morphology of A549 cells
The morphological changes were observed in NPs treated A549 cells when compared with the control cells (Fig. 6B (A)). The loss of membrane integrity, reduced cell development, and cytoplasmic condensation resulted in polygonal or bigonal-shaped lung cancer cells being transformed into round-shaped cells, which was the most notable morphological alteration of NPs-treated cells seen in this work (Fig. 6B (B-E)).
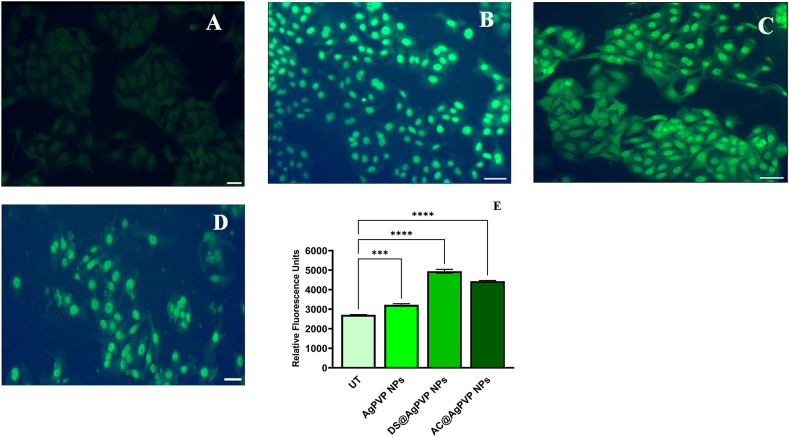
3.2.3. Detection of intracellular ROS
An important study focuses on the ROS formation in cancer cells that is caused by oxidative stress reactions that are induced by drugs [50]. When the IC50 concentrations of various NPs were used to treat A549 cancer cells in our investigation, higher ROS levels were observed in the cancer cells than in normal cells. The ability of the NPs to stimulate the creation of ROS would prevent the continued progression of the cell cycle, including cellular proliferation, attachment, and maturation, which elevate the genes for apoptosis and necrosis [51]. Because of the oxidation process, the NPs produced ROS in Lung cancer cells after the appropriate time interval, and these ROS accumulated on the DNA granules. The DNA then lost its capacity to transfer and prevented the synthesis of the polymerase enzyme [52]. Immature colonies were seen outdoors where the surrounding membrane was destroyed. Using DCFH-fluorescent DA's dye, the oxidation potential was found. Condensed immature DNA was released and bound to a specific DCFH-DA fluorescent dye when the DNA ability was destroyed, resulting in a sparse cell shape. In comparison to cells that weren't treated, treated cells displayed a severely condensed shape and necrotic structure (Fig. 7 B-D). While the clumped colonies of untreated cells revealed their smooth shape (Fig. 7A). It was undeniably proven that the AgNPs' effects on A549 cells caused the ROS to be produced intracellularly [53]. Furthermore, the activator genes were silenced, and the cells' continuous production of ROS entered a decline phase, resulting in cell death. As a result, the current findings show that NPs have a stronger ability to suppress cancer cell proliferation due to oxidative stress-mediated ROS formation. According to experimental research, AgNP-generated ROS triggers the cancer cells' intrinsic apoptotic mechanism. The chemical conversion of Ag° to Ag+, Ag–O–, and Ag–S– increases the generation of ROS in cells. Overproduction of ROS within cells causes lipid peroxidation, protein oxidation, and DNA damage, all of which can trigger the cell's intrinsic apoptotic pathway. When cellular damage is high, the intrinsic apoptotic pathway is triggered by up-regulating pro-apoptotic Bcl-2 family members and down-regulating anti-apoptotic proteins [54,55].
Fig. 7.
Qualitative and Quantitative determination of reactive oxygen species (ROS)
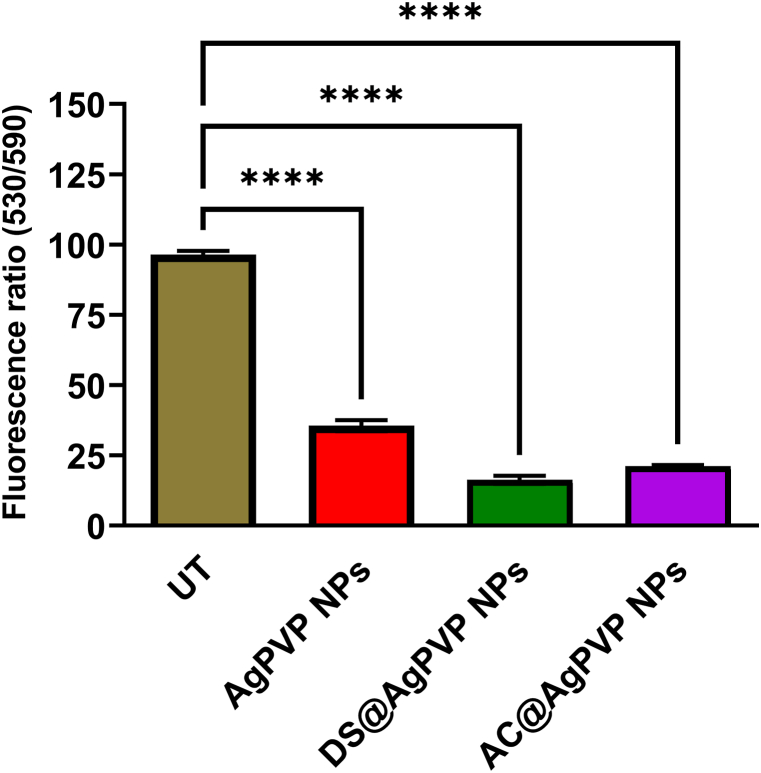
(A) The typical image indicates no generation of ROS in untreated A549 cells. A549 cells treated with (B) DS@AgPVP NPs (C) AC@AgPVP NPs (D) AgPVP NPs. The scale bar represents 10 μm. (E) Quantification of fluorescence intensity in terms of corrected total cell fluorescence (CTCF) of untreated and treated A549 cells (with DS@AgPVP NPs, AC@AgPVP NPs, and AgPVPNPs).
According to Naveen Kumar [56], ROS is a crucial target for cancer cell suppression, and AgNPs significantly boosted oxidative stress responses in A549 cells. Bhakya [52] obtained a similar result against A549 cells by using a higher concentration of AgNPs. Furthermore, the oxidative stress response blocked the activation of apoptosis-related genes and resulted in programmed cell death due to intracellular breaches in the mitochondrial membrane. Padmini et al. [57] reported that Allium sativum silver nanoparticle with an IC50 value of 22 μg/ml induced ROS-mediated apoptosis in A549 cells. The results showed conclusively that the increased levels of ROS production are directly related to the enhanced apoptotic effectiveness of nanoparticles. Recent research has linked the production of reactive oxygen species (ROS) within cells to the cytotoxic effect of AgNPs [58,59].
3.2.4. Detection of MMP
After 24 h of incubation, the fluorescent dye JC-10 was absorbed, permitting viewing of the mitochondrial membrane from cytochrome c leakage under a fluorescence microscope (Fig. 8). The dye JC-10 is quite useful for determining the form of a damaged cancer cell membrane. It interrupts the intrinsic route of the cancer cell cycle. Because they are connected to wounded cells, they can cling to torn mitochondrial membranes. When cells were harmed, apoptosis and necrotic cells were constantly created, and they appeared in a variety of hues ranging from orange to green. It is critical to keep an eye on the depolarized membrane of the mitochondria due to the dysfunction of responsive genes. When the mitochondrial membrane is disturbed, caspases are activated, and Bcl-2 suppressive genes are expressed less frequently. Following the passage of the JC-10 through the mitochondrial membrane, the internal leakage materials were lost and changed color from red to green, suggesting apoptosis [60]. Our results were consistent with the guidelines and inhibited A549 cells at IC50 NP concentrations. It triggered a cascade of apoptotic cell receptors in A549 cells, resulting in repeated life cycle arrests due to increased responsive gene activation. It is the first step in initiating apoptosis. This technique was most effective in treating cancer cells, similar to how mitochondrial membrane damage induced by instability and functional change promotes mortality. Our findings agree with Du et al., 2017 [61], who stated that mitochondria are essential for cell differentiation, death, and cell cycle growth control.
Fig. 8.
Determination of mitochondrial membrane potential (MMP) in A549 cells
Loss of mitochondrial membrane potential after treatment with DS@AgPVP NPs, AC@AgPVP NPs, and AgPVP NPs on A549 cancer cells. * Indicates the significance.
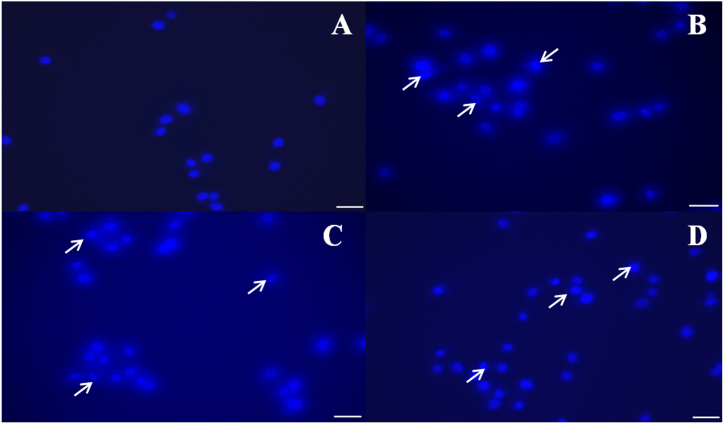
3.2.5. Effect on the integrity of nuclei by DAPI staining
Nuclear fragmentation was caused by NPs, as detected by DAPI, a prominent nuclear counter-stain. Untreated cells had normal nuclei (smooth nuclear), but A549 cells treated with NPs had apoptotic nuclei (condensed or fragmented chromatin), as illustrated in Fig. 9 (A-D). In the A549 cells, nuclear morphology studies revealed typical apoptotic alterations such as chromatin condensation, nucleus fragmentation, and the production of apoptotic bodies. Surprisingly, recent research has found that AgNPs might cause DNA damage and death in cancer cells [62]. The number of apoptotic cells rose as NP concentration increased, suggesting that nanoparticles might trigger cell death (Fig. 9 B-D). The cells' characteristics, which included spikes, shrinkage, and other signs of DNA fragmentation, were comparable to those previously reported as indicators of apoptosis in cells [[63], [64], [65], [66], [67]]. In agreement with these results, cancer cells have been shown to undergo DNA damage and apoptosis when exposed to AgNPs [21].
Fig. 9.
Identification of Apoptosis by DAPI staining
(A) Untreated A549 cells (B) A549 cells treated with DS@AgPVP NPs (C) A549 cells treated with AC@AgPVP NPs (D) A549 cells treated with AgPVP NPs. Arrows show the chromatin changes. The scale bar indicates 10 μm.
3.2.6. Detection of apoptosis by AO/EB staining
The induction of apoptosis after treatment with IC50 concentrations of NPs was assessed by fluorescence microscopy after staining with AO/EB. AO penetrates the cell membrane, and the normal cells have green fluorescence, while in apoptotic cells, apoptotic bodies are formed because of nuclear shrinkage, blebbing that is observed as orange-colored bodies. Necrotic cells were observed as red fluorescence due to their loss of membrane integrity when viewed under the TRITC filter in an inverted fluorescence microscope (Fig. 10 (A-H)) [68]. Venugopal et al. [45] reported that AgNPs synthesized from Syzygium aromaticum produced live and dead cells in MCF-7 cells confirmed by the AO/EB staining method.
Fig. 10.
Live and Dead cells differentiation by Acridine Orange/Ethidium Bromide (AO/EB) staining
Under the TRITC filter, a bright orange color nucleus with a contrasting red color cytoplasm was seen in the picture due to the loss of cell integrity in the dead cells. Live cells exhibit green fluorescence when seen through a FITC filter because their cell membranes are intact. (A) Untreated A549 cells observed in FITC filter (B) Untreated A549 cells observed in TRITC filter (C) DS@AgPVP NPs treated A549 cell observed in FITC filter (D) DS@AgPVP NPs treated A549 cell observed in TRITC filter (E) AC@AgPVP NPs treated A549 cell observed in FITC filter (F) AC@AgPVP NPs treated A549 cell observed in TRITC filter (G) AgPVP NPs treated A549 cell observed in FITC filter (H) AgPVP NPs treated A549 cell observed in TRITC filter. Arrows indicate the changes due to the treatment of Nanoparticles compared to the untreated cells. The scale bar indicates 10 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
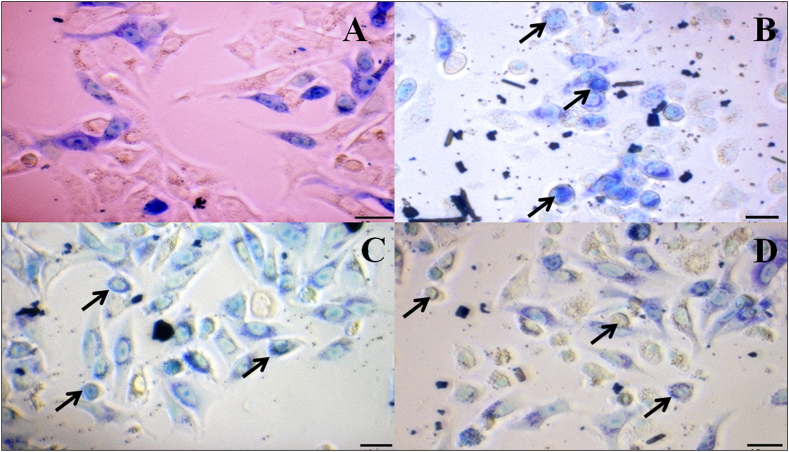
3.2.7. Detection of morphological changes in cells by Giemsa staining
The early morphological markers of apoptosis were identified using Giemsa staining. Cell shrinkage, loss of membrane asymmetry and attachment, and plasma membrane blebbing are the key features associated with morphological alterations. Cells treated with NPs had the most morphologically altered cells when compared to untreated cells (Fig. 11 A). Under a phase-contrast microscope, Giemsa staining allows normal and treated cells to be distinguished, indicating that nanoparticles were more detrimental to A549 cells than to normal cells (Fig. 11 A-D).
Fig. 11.
Assessment of Apoptosis by Giemsa staining
(A) Untreated A549 cells
(B) DS@AgPVP NPs treated A549 cells
(C) AC@AgPVP NPs treated A549 cells
(D) AgPVP NPs treated A549 cells. Arrows represent the changes. The scale bar indicates 10 μm.
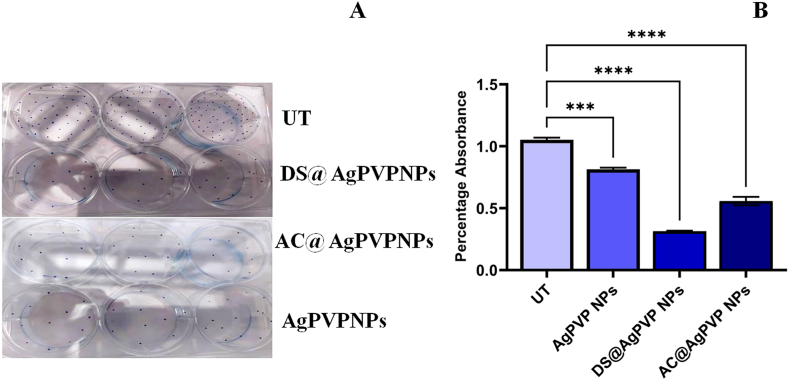
3.2.8. Colony formation assay
A cell splits from the original tumor site and spreads to different locations of the body during the metastasizing process. To measure cell adhesion, a colony formation assay is utilized. Colony formation was significantly reduced in cells treated with NPs. Within the NPs-treated and untreated groups, the number of colonies varied greatly. However, their size has not changed (Fig. 12A). A reduced colony number in the treatment group demonstrated the extract's anti-proliferative efficacy. The proportion of dye uptake in cells treated with the extract was significantly lower, according to dye quantification (Fig. 12B). NPs significantly reduced colony numbers when compared to untreated cells (p < 0.0001). Bendale et al. [30] reported that platinum nanoparticles with an IC50 value of 200 μg/ml reduced colony formation in A549 cells.
Fig. 12.
Colony formation assay
Nanoparticles treatment significantly decreased colony formation in A549 cells
(A) Colony formation in DS@AgPVP NPs, AC@AgPVP NPs, and AgPVP NPs treated and untreated A549 cells.
(B) Quantification of the crystal violet from DS@AgPVP NPs, AC@AgPVP NPs, and AgPVP NPs treated and untreated A549 cells. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
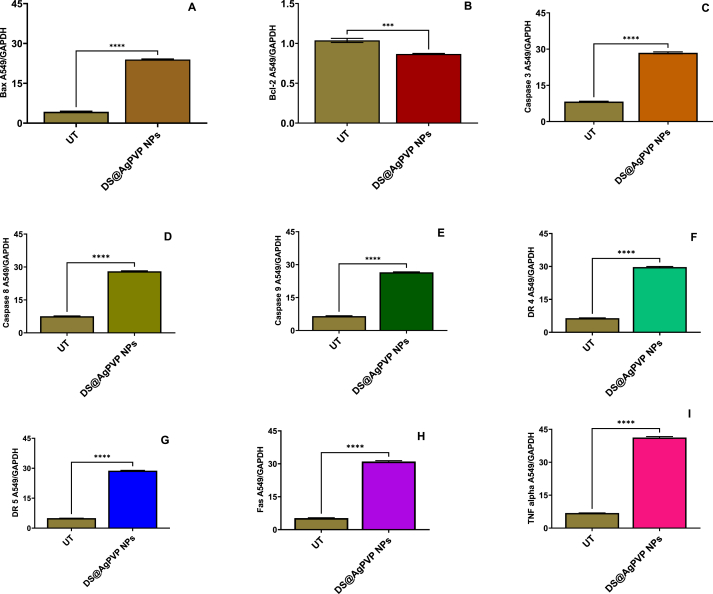
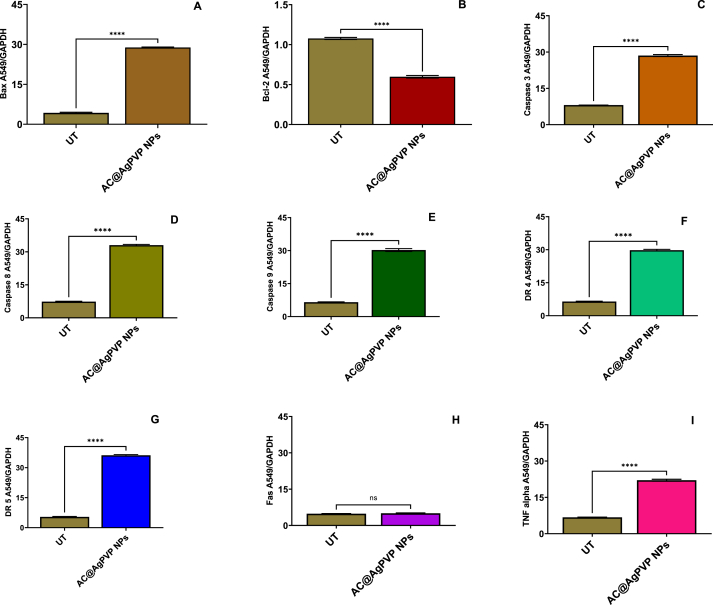
3.2.9. Investigation of the mechanism of apoptosis by gene expression studies
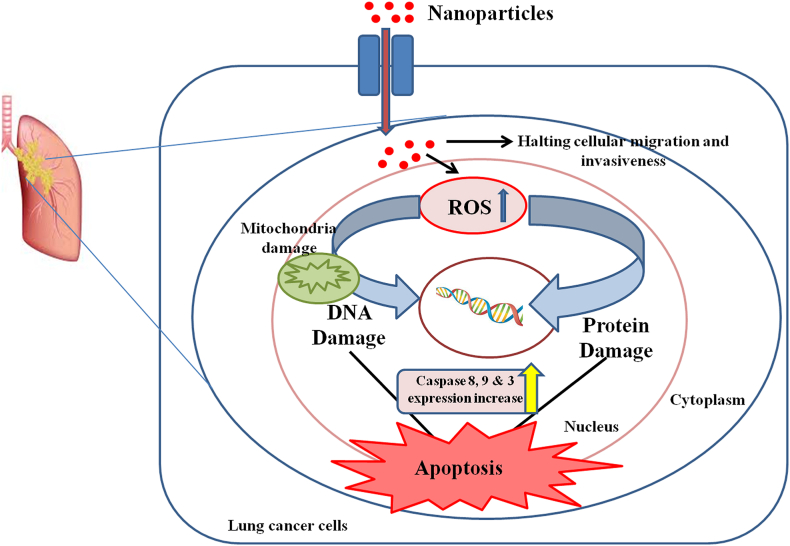
To investigate the mechanism of apoptosis induction during 24 h incubation of A549 cells treated with NPs, qRT-PCR was used to evaluate the expression of several apoptosis genes, including Bcl-2, Bax, Cas-3, Cas-9, Cas-8, Fas, TNF-α, DR4, and DR5. Treatment of the A549 cells with IC50 concentrations of NPs significantly decreased the mRNA level of Bcl-2 (Fig. 13A (A-I), 13B (A-I), 13C (A-I)). In addition, the mRNA levels of Bax, Cas-3, Cas-9, Cas-8, Fas, TNF-α, DR4, and DR5 were significantly increased after 24 h (p < 0.05). Additionally, after 24 h, the NPs were found to significantly reduce the levels of Cas-3, Cas-9, and Cas-8 in A549 cells by around 2.5-fold in comparison to the control (Fig. 13). This finding demonstrates that NPs cause apoptosis by disrupting mitochondrial membrane potential. This decrease in mitochondrial membrane potential may have triggered the apoptotic cascade in A549 cells exposed to NPs. Activation of the DR4, DR5, Fas, and TNF-α receptors is lethal to cancer cells [69,70]. According to various researchers, AgNPs interact with cell membrane proteins, activate signaling pathways, and produce ROS, which damage proteins and nucleic acids and ultimately lead to apoptosis and cell proliferation inhibition. The action of AgNPs on mitochondrial membrane permeability causes the release of ROS, resulting in oxidative stress, interruption of ATP production, and DNA damage, which may affect JNK-mediated caspase-dependent apoptosis in human cell lines. JNK belongs to the MAPK family and contributes to apoptosis by phosphorylating Bcl-2, resulting in Bcl-2 inactivation. When cytochrome c enters the cytosol, it initiates a cascade that includes caspases 3 through apaf-1 and caspase 9. The whole picture suggests that AgNPs induce apoptosis in cancer cells via ROS-mediated activation of the intrinsic pathway [[71], [72], [73], [74], [75]]. The activation of TRAIL receptors activates the caspase 8 channels, the extrinsic route, and the mitochondrial cascade, which signals the activation of caspase 9 (intrinsic pathway). Both caspases activate the common executioner (Caspase 3) in A549 cells, causing apoptosis [[76], [77], [78]]. Throughout this process, the anti-apoptotic gene Bcl-2 is downregulated, which greatly raises the Bax/Bcl-2 ratio and initiates apoptosis in A549 cells [79,80]. As a result, in this study, we proposed a mechanism of action of NPs on A549 cells (Fig. 14). In their study, Bethu et al. (2018) [81] observed that the administration of RS-AgNPs resulted in the induction of apoptosis in cancer cells. This phenomenon was attributed to the upregulation of many proapoptotic proteins, including caspase-3, caspase-8, caspase-9, p53, and Bax. Additionally, the downregulation of Bcl-2 was seen, suggesting the activation of both intrinsic and extrinsic pathways of apoptosis. According to earlier research, AgNPs induce apoptosis in A549 cells via the intrinsic pathway, whereas DS and AC, both crude extracts, induce apoptosis in A549 cells via the intrinsic and extrinsic pathways. Only AgNPs, DS, and AC crude extract nanoparticles activate apoptotic pathways internally or extrinsically in human lung adenocarcinoma cells (A549), according to gene expression analysis.
Fig. 13.
Gene expression Studies
(A) After the DS@AgPVP NPs (IC50) treatment in A549 cells affects the gene's expression in the apoptosis process (A) Bax (B) Bcl-2 (C) Caspase 3 (D) Caspase 8 (E) Caspase 9 (F) DR4 (G) DR5 (H) Fas (I) TNF-α
(B) After the AC@AgPVP NPs (IC50) treatment in A549 cells affects the gene's expression in the apoptosis process (A) Bax (B) Bcl-2 (C) Caspase 3 (D) Caspase 8 (E) Caspase 9 (F) DR4 (G) DR5 (H) Fas (I) TNF-α
(C) After the AgPVP NPs (IC50) treatment in A549 cells affects the gene's expression in the apoptosis process (A) Bax (B) Bcl-2 (C) Caspase 3 (D) Caspase 8 (E) Caspase 9 (F) DR4 (G) DR5 (H) Fas (I) TNF-α. Images represent the value of mean ± std. error of three independent experiments with n = 3. * Indicates the significance.
Fig. 14.
Probable Molecular Mechanism of Action of Nanoparticles on Non-small cell lung cancer (A549) Cells.
3.3. Brine shrimp lethality assay
Brine shrimp cytotoxic activities of all AgNPs determine their different pharmacological properties [82]. In this study, ten different concentrations; 10–100 μg/ml of synthesized nanoparticles were used to determine their cytotoxicity using a brine shrimp lethality assay. The LD50 value of DS@AgPVP NPs was found to be 40 μg/ml which is less than AC@AgPVP NPs (70 μg/ml) and AgPVP NPs (80 μg/ml) (Table 2). The lowered LD50 value suggested the cytotoxic nature of DS@AgPVP NPs and AC@AgPVP NPs compared to AgPVP NPs due to the presence of crude extracts capping the surface of AgPVP NPs. This enhanced cytotoxicity of the nanoparticles, having an LD50 of 40 μg/ml to brine shrimp revealed the presence of toxic constituents. The Cytotoxic effects of nanoparticles on shrimp larvae can be linked with anticancer activity, and nanoparticles could be an alternative source of anticancer drugs [82]. These days, much attention is being given to metallic nanoparticles and their anticancer activity. The toxicity of these AgNPs and the mechanism of action cannot be explained in detailed [82].
Table 2.
Evaluated % Death of Shrimp and LC50 values for the synthesized NPs.
| Particles | Concentration of NPs (μg/ml) | % Death | LD50 value (μg/ml) |
|---|---|---|---|
| DS@ Ag PVP NPs | 10 | 10 | 40 μg/ml |
| 20 | 20 | ||
| 30 | 40 | ||
| 40 | 50 | ||
| 50 | 60 | ||
| 60 | 70 | ||
| 70 | 70 | ||
| 80 | 80 | ||
| 90 | 90 | ||
| 100 | 100 | ||
| AC@ Ag PVP NPs | 10 | 0 | 70 μg/ml |
| 20 | 5 | ||
| 30 | 10 | ||
| 40 | 10 | ||
| 50 | 20 | ||
| 60 | 30 | ||
| 70 | 50 | ||
| 80 | 60 | ||
| 90 | 60 | ||
| 100 | 70 | ||
| Ag PVP NPs | 10 | 0 | 80 μg/ml |
| 20 | 10 | ||
| 30 | 20 | ||
| 40 | 30 | ||
| 50 | 30 | ||
| 60 | 40 | ||
| 70 | 40 | ||
| 80 | 50 | ||
| 90 | 60 | ||
| 100 | 70 |
4. Conclusion
The use of plant extracts from D. sissoo (DS) and A. calamus L. (AC) leaves in the synthesis of silver nanoparticles (AgNPs) resulted in the formation of well-characterized face-centered cubic structured nanoparticles. The crystalline nature, shape, and effective inclusion of polyvinylpyrrolidone (PVP) as a capping agent were validated by a thorough various techniques including X-ray diffraction (XRD), Field Emission Gun Transmission Electron Microscopy (FEG-TEM), UV–Vis spectra, and Fourier Transform Infrared Spectroscopy (FTIR). MTT assay findings revealed considerably lower IC50 values on human adenocarcinoma lung cancer (A549) cells compared to lung normal cells (WI-38), indicating the anti-cancer potential of the nanoparticles (AgPVP NPs, DS@AgPVP NPs, and AC@AgPVP NPs). Nanoparticles induced the production of reactive oxygen species (ROS), changed mitochondrial membrane potential (MMP), and displayed apoptosis-inducing capabilities along with the prevention of cancer cell invasion. Notably, This study provides a comprehensive understanding of the synthesis process, characterization, and therapeutic potential of silver nanoparticles functionalized with plant extracts, opening avenues for further research and development in nano-medicine and cancer treatment.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Anjali B. Thakkar: Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. R.B. Subramanian: Writing – review & editing, Supervision. Vasudev R. Thakkar: Writing – review & editing, Supervision. Sandip V. Bhatt: Writing – review & editing, Validation, Supervision, Methodology, Conceptualization. Sunil Chaki: Writing – review & editing, Supervision. Yati H. Vaidya: Formal analysis. Vikas Patel: Formal analysis. Parth Thakor: Writing – review & editing, Writing – original draft, Validation, Supervision, Methodology, Formal analysis, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the P. G. Department of Biosciences and P. G. Department of Applied and Interdisciplinary Sciences (IICISST), DST-PURSE Programme, Sardar Patel University for the Instrumentation and infrastructure facilities.
Contributor Information
Sandip V. Bhatt, Email: sandip.bhatt08@yahoo.com.
Parth Thakor, Email: parth7218@gmail.com.
References
- 1.American Cancer Society . American Cancer Society; Atlanta, Ga: 2022. Facts & Figures 2022. [Google Scholar]
- 2.https://seer.cancer.gov/statfacts/html/lungb.html.
- 3.Anna R.F., Małgorzata K.L., Victor S., Silvia I., Mnuel A., Agnieszka K., Grazyna S. Development of noncytotoxic silver–chitosan nanocomposites for efficient control of biofilm forming microbes. RSC Adv. 2017;7 doi: 10.1039/C7RA08359A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vijayakumar S., Malaikozhundan B., Saravanakumar S., Duran-Lara E.F., Wang M.H., Vaseeharan B. Garlic clove extract assisted silver nanoparticle- Antibacterial, antibiofilm, antihelminthic, anti-inflammatory, anticancer and ecotoxicity assessment. J. Photochem. Photobiol. B Biol. 2019;198 doi: 10.1016/j.jphotobiol.2019.111558. 111558 1-11155838. [DOI] [PubMed] [Google Scholar]
- 5.Bano N., Siddiqui S., Amir M. Roohi, House of industrially important bioactive metabolites: a review on actinobacteria. Indian J. Biotechnol. 2019;18:293–304. [Google Scholar]
- 6.Iram S., Zahera M., Wahid I., Baker A., Raish M., Khan A., Ali N., Ahmad S., Khan M.S. Cisplatin bioconjugated enzymatic GNPs amplify the effect of cisplatin with acquiescence. Sci. Rep. 2019;9:1–16. doi: 10.1038/s41598-019-50215-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Sheddi E.S., Farshori N.N., Al-Oqail M.M., Al-Massarani S.M., Saquib Q., Wahab R., Musarrat J., Al-Khedhairy A.A., Siddiqui M.A. Anticancer potential of green synthesized silver nanoparticles using extract of nepeta deflersiana against human cervical cancer cells (HeLA) Bioinorgan. Chem. Appl. 2018:1–13. doi: 10.1155/2018/9390784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abd-Elnaby H.M., Abo-Elala G.M., Abdel-Raouf U.M., Hamed M.M. Antibacterial and anticancer activity of extracellular synthesized silver nanoparticles from marine Streptomyces rochei MHM13. Egyptian Journal of Aquatic Research. 2016;42:301–312. doi: 10.1016/j.ejar.2016.05.004. [DOI] [Google Scholar]
- 9.He Y., Du Z., Lv H., Jia Q., Tang Z., Zheng X., Zhang K., Zhao F. Green synthesis of silver nanoparticles by Chrysanthemum morifolium Ramat. extract and their application in clinical ultrasound gel. Int. J. Nanomed. 2013;8:1809–1815. doi: 10.2147/IJN.S43289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harshiny M., Matheswaran M., Arthanareeswaran G., Kumaran S., Rajasree S. Enhancement of antibacterial properties of silver nanoparticles-ceftriaxone conjugate through Mukia maderaspatana leaf extract mediated synthesis. Ecotoxicol. Environ. Saf. 2015;121:135–141. doi: 10.1016/j.ecoenv.2015.04.041. [DOI] [PubMed] [Google Scholar]
- 11.Stensberg M.C., Wei Q., McLamore E.S., Porterfield D.M., Wei A., Sepúlveda M.S. Toxicological studies on silver nanoparticles: challenges and opportunities in assessment, monitoring and imaging. Nanomedicine. 2011;6(5):879–898. doi: 10.2217/nnm.11.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaloupka K., Malam Y., Seifalian A.M. Nanosilver as a new generation of nanoproduct in biomedical applications. Trends Biotechnol. 2010;28(11):580–588. doi: 10.1016/j.tibtech.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Kohl Y., Kaiser C., Bost W., et al. Preparation and biological evaluation of multifunctional PLGA nanoparticles designed for photoacoustic imaging. Nanomedicine. 2011;7(2):228–237. doi: 10.1016/j.nano.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Tian J., Wong K.K., Ho C.M., Lok C.N., Yu W.Y., Che C.M., Chiu J.F., Tam P.K.H. Topical delivery of silver nanoparticles promotes wound healing. ChemMedChem. 2007;2(1):129–136. doi: 10.1002/cmdc.200600171. [DOI] [PubMed] [Google Scholar]
- 15.dos Santos C.A., Seckler M.M., Ingle A.P., et al. Silver nanoparticles: therapeutical uses, toxicity, and safety issues. J. Pharmaceut. Sci. 2014;103(7):1931–1944. doi: 10.1002/jps.24001. [DOI] [PubMed] [Google Scholar]
- 16.Ong C., Lim J.Z., Ng C.T., Li J.J., Yung L.Y., Bay B.H. Silver nanoparticles in cancer: therapeutic efficacy and toxicity. Curr. Med. Chem. 2013;20(6):772–781. [PubMed] [Google Scholar]
- 17.Locatelli E., Naddaka M., Uboldi C., Loudos G., Fragogeorgi E., Molinari V., Pucci A., Tsotakos T., Psimadas D., Ponti J., Franchini M.C. Targeted delivery of silver nanoparticles and alisertib: in vitro and in vivo synergistic effect against glioblastoma. Nanomedicine. 2014;9(6):839–849. doi: 10.2217/nnm.14.1. [DOI] [PubMed] [Google Scholar]
- 18.Wei X., Li H., Li Z., Vuki M., Fan Y., Zhong W., Xu D. Metal-enhanced fluorescent probes based on silver nanoparticles and its application in IgE detection. Anal. Bioanal. Chem. 2012;402(3):1057–1063. doi: 10.1007/s00216-011-5591-1. [DOI] [PubMed] [Google Scholar]
- 19.Gengan R.M., Anand K., Phulukdaree A., Chuturgoon A. A549 lung cell line activity of biosynthesized silver nanoparticles using Albizia adianthifolia leaf. Colloids and Surfaces B Biointerfaces. 2013;105(4):87–91. doi: 10.1016/j.colsurfb.2012.12.044. [DOI] [PubMed] [Google Scholar]
- 20.Jeyaraj M., Sathishkumar G., Sivanandhan G., Ali M., Rajesh M., Arun R., Kapildev G., Manickavasagam M., Thajuddin N., Premkumar K., Ganapathi A. Biogenic silver nanoparticles for cancer treatment: an experimental report. Colloids and Surfaces B Biointerfaces. 2013;106(2):86–92. doi: 10.1016/j.colsurfb.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Sanpui P., Chattopadhyay A., Ghosh S.S. Induction of apoptosis in cancer cells at low silver nanoparticle concentrations using chitosan nanocarrier. ACS Appl. Mater. Interfaces. 2011;3(2):218–228. doi: 10.1021/am100840c. [DOI] [PubMed] [Google Scholar]
- 22.Jeyaraj M., Rajesh M., Arun R., Mubarak Ali D., Sathishkumar G., Sivanandhan G., Dev G.K., Manickavasagam M., Premkumar K., Thajuddin N., Ganapathi A. An investigation on the cytotoxicity and caspase-3-mediated apoptotic effect of biologically synthesized silver nanoparticles using Podophyllum hexandrum on human cervical carcinoma cells. Colloids and Surfaces B Biointerfaces. 2013;102(1):708–717. doi: 10.1016/j.colsurfb.2012.09.042. [DOI] [PubMed] [Google Scholar]
- 23.Sriram M.I., Kanth S.B., Kalishwaralal K., Gurunathan S. Antitumor activity of silver nanoparticles in Dalton's lymphoma ascites tumor model. Int. J. Nanomed. 2010;5(1):753–762. doi: 10.2147/IJN.S11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thakkar A.B., Sargara P., Subramanian R.B., Thakkar V.R., Thakor P. Induction of apoptosis in lung carcinoma cells (A549) by hydromethanolic extract of Acorus calamus L. Process Biochem. 2022;123:1–10. doi: 10.1016/j.procbio.2022.10.028. [DOI] [Google Scholar]
- 25.Thakkar A.B., Subramanian R.B., Thakkar V.R., Thakor P. Hydromethanolic leaves extract of Dalbergia sissoo Roxb. Ex DC. Induces apoptosis in lung adenocarcinoma cells. Process Biochem. 2023;134(1):250–261. doi: 10.1016/j.procbio.2023.10.006. [DOI] [Google Scholar]
- 26.Thakor P., Subramanian R.B., Thakkar S.S., Ray A., Thakkar V.R. Phytol induces ROS mediated apoptosis by induction of caspase 9 and 3 through activation of TRAIL, FAS and TNF receptors and inhibits tumor progression factor Glucose 6 phosphate dehydrogenase in lung carcinoma cell line (A549) Biomed. Pharmacother. 2017;92:491–500. doi: 10.1016/j.biopha.2017.05.066. [DOI] [PubMed] [Google Scholar]
- 27.Gajera S.B., Mehta J.V., Thakor P., Thakkar V.R., Chudasama P.C., Patel J.S., Patel M.N. Half-sandwich iridium III complexes with pyrazole-substituted heterocyclic frameworks and their biological applications. New J. Chem. 2016;40(12):9969–9980. doi: 10.1039/C6NJ02153K. [DOI] [Google Scholar]
- 28.Degli E.M. In: Pon L.A., Schon E.A., editors. vol. 65. Academic; California: 2001. Assessing functional integrity of mitochondria in vitro in vivo; pp. 75–96. (Methods in Cell Biology). [DOI] [PubMed] [Google Scholar]
- 29.Agabeigi R., Rasta S.H., Rahmati-Yamchi M., Salehi R., Alizadeh E. Novel Chemo-Photothermal therapy in breast cancer using methotrexate-loaded folic acid Conjugated Au@SiO2 nanoparticles. Nanoscale Res. Lett. 2020;15(62):1–14. doi: 10.1186/s11671-020-3295-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendale Y., Bendale V., Paul S. Evaluation of cytotoxic activity of platinum nanoparticles against normal and cancer cells and its anticancer potential through induction of apoptosis. Integrative Medicine Research. 2017;6(2):141–148. doi: 10.1016/j.imr.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morais C., Gobe G., Johnson D.W., Healy H. Anti-angiogenic actions of pyrrolidine dithiocarbamate, a nuclear factor kappa B inhibitor. Angiogenesis. 2009;12(4):365–379. doi: 10.1007/s10456-009-9158-0. [DOI] [PubMed] [Google Scholar]
- 32.Giri R. Kr, Chaki S., Khimani A.J., Vaidya Y.H., Thakor P., Thakkar A.B., Pandya S.J., Deshpande M.P. Biocompatible CuInS2 nanoparticles as potential antimicrobial, antioxidant, and cytotoxic agents. ACS Omega. 2021;6(40):26533–26544. doi: 10.1021/acsomega.1c03795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajput P., Deshpande M.P., Bhoi H.R., Suchak N.M., Desai P.H., Chaki S.H., Pandya S.J., Mishra M., Bhatt S.V., K Tiwari D., Sathe V. Photocatalytic and antibacterial activity of Yttrium doped TiO2 nanostructure. Chemical Physics Impact. 2022;5 doi: 10.1016/j.chphi.2022.100101. [DOI] [Google Scholar]
- 34.Anandalakshmi K., Venugopal J., Ramasamy V. Characterization of silver nanoparticles by green synthesis method using Pedalium murex leaf extract and their antibacterial activity. Appl. Nanosci. 2016;6:399–408. doi: 10.1007/s13204-015-0449-z. [DOI] [Google Scholar]
- 35.Kaur A., Kumar R. Enhanced bactericidal efficacy of polymer stabilized silver nanoparticles in conjugation with different classes of antibiotics. RSC Adv. 2019;9:1095. doi: 10.1039/c8ra07980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silva E., Saraiva S.M., Miguel S.P., Correia I.J. PVP-coated silver nanoparticles showing antifungal improved activity against dermatophytes. J. Nanoparticle Res. 2014;16:726. doi: 10.1007/s11051-014-2726-2. [DOI] [Google Scholar]
- 37.Sharma R., Dhillon A., Kumar D. Mentha-Stabilized silver nanoparticles for high-Performance Colorimetric detection of Al(III) in aqueous systems. Sci. Rep. 2018;8:5189. doi: 10.1038/s41598-018-23469-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehta K., Bharatiya B., Parekh A., Bhatt S.V., Shah V., Shah D.O., Mukherjee T. Submicellar aggregates in aqueous sodium dodecyl sulphase solutions: Investigations by dynamic light scattering and water penetration through porous media. Colloids and Surface A: Physicochemical and Engineering Aspects. 2016;506:331–337. doi: 10.1016/j.colsurfa.2016.06.045. [DOI] [Google Scholar]
- 39.Soni B.H., Deshpande M.P., Bhatt S.V., Garg N., Pandya N.N., Chaki S.H. Influence of Mn doping on optical properties of ZnO nanoparticles synthesized by microwave irradiation. J. Opt. 2013;42:328–334. doi: 10.1007/s12596-013-0136-y. [DOI] [Google Scholar]
- 40.Bijauliya R.K., Kannojia P., Mishra P., Pathak G.K. Isolation and structure elucidation of Quercetin like structure from Dalbergia sissoo (Fabaceae) J. Drug Deliv. Therapeut. 2020;10(3-s):6–11. doi: 10.22270/jddt.v10i3-s.4131. [DOI] [Google Scholar]
- 41.Patel R.M., Patel Sahil K. Cytotoxic activity of methanolic extract of Artocarpus heterophyllus against A549, Hela and MCF-7 cell lines. J. Appl. Pharmaceut. Sci. 2011;1(7):167–171. [Google Scholar]
- 42.Akter M., Sikder M.T., Rahman M.M., Atique Ullah A.K.M., Binte Hossain K.F., Banik S., Hosokawa T., Saito T., Kurasaki M. A systematic review on silver nanoparticles-induced cytotoxicity: physicochemical properties and perspectives. J. Adv. Res. 2017;9:1–16. doi: 10.1016/j.jare.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Retout M., Jabin I., Bruylants G. Synthesis of Ultrastable and Bioconjugable Ag, Au, and bimetallic Ag_Au nanoparticles coated with calix[4]arenes. ACS Omega. 2021;6:19675–19684. doi: 10.1021/acsomega.1c02327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fahmy H.M., Mosleh A.M., Elghany A.A., Mohamed Shams-Eldin E.M., Abu Serea E.S., Ali S.A., Shalan A.E. Coated silver nanoparticles: synthesis, cytotoxicity, and optical properties. RSC Adv. 2019;9:20118–20136. doi: 10.1039/c9ra02907a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Venugopal K., Rather H.A., Rajagopal K., Shanthi M.P., Sheriff K., Illiyas M., Rather R.A., Manikandan E., Uvarajan S., Bhaskar M., Maaza M. Synthesis of silver nanoparticles (Ag NPs) for anticancer activities (MCF 7 breast and A549 lung cell lines) of the crude extract of Syzygium aromaticum. J. Photochem. Photobiol. B Biol. 2017;167:282–289. doi: 10.1016/j.jphotobiol.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 46.Vivek R., Thangam R., Muthuchelian K., Gunasekaran P., Kaveri K., Kannan S. Green biosynthesis of silver nanoparticles from Annona squamosa leaf extract and its in vitro cytotoxic effect on MCF-7 cells. Process Biochem. 2012;47(12):2405–2410. doi: 10.1016/j.procbio.2012.09.025. [DOI] [Google Scholar]
- 47.Hublikar L.V., Ganachari S.V., Raghavendra N., Banapurmath N.R., Patil V.B., Yunus Khan T.M., Badruddin I.A. Biogenesis of silver nanoparticles and its multifunctional anti-corrosion and anticancer studies. Coatings. 2021;11:1–19. doi: 10.3390/coatings11101215. [DOI] [Google Scholar]
- 48.Hublikar L.V., Ganachari S.V., Patil V.B., Nandi S., Honnad A. Anticancer potential of biologically synthesized silver nanoparticles using Lantana camara leaf extract. Progress in Biomaterials. 2023;12:155–169. doi: 10.1007/s40204-023-00219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hublikar L.V., Ganachari S.V., Raghavendra N., Patil V.B., Banapurmath N.R. Green synthesis silver nanoparticles via Eichhornia Crassipes leaves extract and their applications. Current Research in Green and Sustainable Chemistry. 2021;4 [Google Scholar]
- 50.Hu G., Cai Y., Tu Z., Luo J., Qiao X., Chen Q., Zhang W. Reducing the cytotoxicity while improving the anticancer activity of silver nanoparticles through a tocopherol succinate modification. RSC Adv. 2015;5 doi: 10.1039/C5RA12911G. [DOI] [Google Scholar]
- 51.Selvi B.C.G., Madhavan J., Santhanam A. Cytotoxic effect of silver nanoparticles synthesized from Padina tetrastromatica on breast cancer cell line. Adv. Nat. Sci. Nanosci. Nanotechnol. 2016;7 doi: 10.1088/2043-6262/7/3/035015. [DOI] [Google Scholar]
- 52.Bhakya S., Muthukrishnan S., Sukumaran M., Grijalv M., Cumbal L., Benjamin Franklin J.H., Senthil Kumar T., Rao M.V. Antimicrobial, antioxidant and anticancer activity of biogenic silver nanoparticles - an experimental report. RSC Adv. 2016;6:81436–81446. doi: 10.1039/C6RA17569D. [DOI] [Google Scholar]
- 53.Rajivgandhi G., Maruthupandy M., Quero F., Li W.J. Graphene/nickel oxide nanocomposites against isolated ESBL producing bacteria and A549 cancer cells. Mater. Sci. Eng. C. 2019;102:829–843. doi: 10.1016/j.msec.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 54.Tosun N.G., Kaplan O., Imamoglu R., Turkekul I., Gokce I., Ozgur A. Green synthesized silver nanoparticles with mushroom extracts of Paxina leucomelas and Rhizopogon luteolus induce ROS-Induced intrinsic apoptotic pathway in cancer cells. Inorganic and nano-metal chemistry. 2022:1–10. doi: 10.1080/24701556.2022.2081200. [DOI] [Google Scholar]
- 55.Kaplan O., Tosun N.G., Imamoglu R., Turkekul İ., Gokçe I., Ozgur A. Biosynthesis and characterization of silver nanoparticles from Tricholoma ustale and Agaricus arvensis extracts and investigation of their antimicrobial, cytotoxic, and apoptotic potentials. J. Drug Deliv. Sci. Technol. 2022;69 doi: 10.1016/j.jddst.2022.103178. [DOI] [Google Scholar]
- 56.Naveen Kumar S., Rajivgandhi G., Ramachandran G., Manoharan N. A marine sponge Fascaplysinopsis sp. derived alkaloid fascaplysin inhibits the HepG2 hepatocellular carcinoma cell. Frontiers in Laboratory Medicine. 2018;2:41–48. doi: 10.1016/j.flm.2018.06.001. [DOI] [Google Scholar]
- 57.Padmini R., Uma Maheshwari Nallal V., Razia M., Sivaramakrishnan S., Alodaini Hissah Abdulrahman, Hatamleh Ashraf Atef, Al-Dosary Munirah Abdullah, Ranganathan Venkatalakshmi, Chung Woo Jin. Cytotoxic effect of silver nanoparticles synthesized from ethanolic extract of Allium sativum on A549 lung cancer cell line. J. King Saud Univ. Sci. 2022;34 [Google Scholar]
- 58.Franco-Molina M., Mendoza-Gamboa E., Sierra-Rivera C., et al. Antitumor activity of colloidal silver on MCF-7 human breast cancer cells. J. Exp. Clin. Cancer Res. 2010;29:148. doi: 10.1186/1756-9966-29-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kovacs D., Igaz N., Keskeny C., et al. Silver nanoparticles defeat p53-positive and p53-negative osteosarcoma cells by triggering mitochondrial stress and apoptosis. Sci. Rep. 2016;6 doi: 10.1038/srep27902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Momtazi-borojeni A.A., Behbahani M., Sadeghi-aliabadi H. Antiproliferative activity and apoptosis induction of crude extract and fractions of Avicennia Marina. Iranian Journal of Basic Medical Sciences. 2013;16:1–10. [PMC free article] [PubMed] [Google Scholar]
- 61.Du C., Guo Y., Cheng Y., Han M., Zhang W., Qian H. Anti-cancer effects of torulene, isolated from Sporidiobolus pararoseus, on human prostate cancer LNCaP and PC-3 cells via a mitochondrial signal pathway and the downregulation of AR expression. RSC Adv. 2017;2466:1–9. doi: 10.1039/C6RA24721K. [DOI] [Google Scholar]
- 62.May B.J., Monera AL-Abdan, Gadah A., Saud A. Effects of green silver nanoparticles on apoptosis and oxidative stress in normal and cancerous human hepatic cells in vitro. Int. J. Nanomed. 2020;15:1537–1548. doi: 10.2147/IJN.S239861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monks N.R., Bordignon S.A.L., Ferraz A., Machado K.R., Faria D.H., Lopes R.M., Souza C.A., Mondin I.C.C., Lima Martha F.S., B da Rocha A., Schwartsmann G. Anti-tumor screening of Brazilian plants. Pharmaceut. Biol. 2002;40(8):603–616. doi: 10.1076/phbi.40.8.603.14658. [DOI] [Google Scholar]
- 64.Vijayarathna S., Sasidharan S. Cytotoxicity of methanol extracts of Elaeis guineensis on MCF-7 and Vero cell lines. Asian Pac. J. Trop. Biomed. 2012;2(10):826–829. doi: 10.1016/S2221-1691(12)60237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Grbović F., et al. In vitro cytotoxic activity of Origanum vulgare L. on HCT-116 and MDA-MB-231 cell lines. Plants. 2013;2:371–378. doi: 10.3390/plants2030371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Graidist P., Martla M., Sukpondma P. Cytotoxic activity of Piper cubeba extract in breast cancer cell lines. Nutrients. 2015;7:2707–2718. doi: 10.3390/nu7042707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lukhele S.T., Motadi L.R. Cannabidiol rather than Cannabis sativa extracts inhibit cell growth and induce apoptosis in cervical cancer cells. BMC Complementary Medicine and Therapies. 2016;16:335. doi: 10.1186/s12906-016-1280-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kumar N., Biswas S., Mathew A.E., Varghese S., Mathew J.E., Nandakumar K. Pro-apoptotic and cytotoxic effects of enriched fraction of Elytrantheparasitica (L.) Danser against HepG2 Hepatocellular carcinoma. BMC Complementary Medicine and Therapies. 2016;16(420):1–11. doi: 10.1186/s12906-016-1395-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ricci M.S., Zong W.X. Chemotherapeutic approaches for targeting cell death Pathways. Oncol. 2006;11(4):342–357. doi: 10.1634/theoncologist.11-4-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin D., Kwon H.Y., Sohn E.J., Nam M.S., Kim J.H., Lee J.C., Ryu S.Y., Park B., Kim S.H. Upregulation of death receptor 5 and production of reactive oxygen species mediate sensitization of PC-3 prostate cancer cells to TRAIL induced apoptosis by vitisin a. Cell. Physiol. Biochem. 2015;36:1151–1162. doi: 10.1159/000430286. [DOI] [PubMed] [Google Scholar]
- 71.Kanipandian N., Li D., Kannan S. Induction of intrinsic apoptotic signaling pathway in A549 lung cancer cells using silver nanoparticles from Gossypium hirsutum and evaluation of in vivo toxicity. Biotechnology Reports. 2019;23:1–11. doi: 10.1016/j.btre.2019.e00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ullah I., Khalil A.T., Ali M., Iqbal J., Ali W., Alarifi S., Shinwari Z.K. Green-synthesized silver nanoparticles induced apoptotic cell death in MCF-7 breast cancer cells by generating reactive oxygen species and activating caspase 3 and 9 enzyme activities. Oxid. Med. Cell. Longev. 2020 doi: 10.1155/2020/1215395. Article ID 1215395 1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hemlata, Gupta S., Tejavath K.K. ROS-mediated apoptosis induced by BSA nanospheres encapsulated with fruit extract of cucumis prophetarum in various human cancer cell lines. ACS Omega. 2021;6:10383–10395. doi: 10.1021/acsomega.1c00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhen X., Qi F., Min W., Huange Z., Yingying L., Songlin Z. Green biosynthesized silver nanoparticles with aqueous extracts of ginkgo biloba induce apoptosis via mitochondrial pathway in cervical cancer cells. Front. Oncol. 2020;10:1–10. doi: 10.3389/fonc.2020.575415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Daei S., Ziamajidi N., Abbasalipourkabir R., Aminzadeh Z., Vahabirad M. Silver nanoparticles exert apoptotic activity in bladder cancer 5637 cells through alteration of bax/bcl-2 genes expression. Chonnam Medical Journal. 2022;58:102–109. doi: 10.4068/cmj.2022.58.3.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khosravi-Far R., Esposti M.D. Death receptor signals to mitochondria. Cancer Biol. Ther. 2004;3:1051–1057. doi: 10.4161/cbt.3.11.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ndebele K., Gona P., Jin T.G., Benhaga N., Chalah A., Degli-Esposti M., Khosravi-Far R. Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) induced mitochondrial pathway to apoptosis and caspase activation is potentiated by phospholipid scramblase-3. Apoptosis. 2008;13(7):845–856. doi: 10.1007/s10495-008-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cretney E., Shanker A., Yagita H., Smyth M.J., Sayers T.J. NF-related apoptosis inducing ligand as a therapeutic agent in autoimmunity and cancer. Immunol. Cell Biol. 2006;84:87–98. doi: 10.1111/j.1440-1711.2005.01413.x. [DOI] [PubMed] [Google Scholar]
- 79.Tsujimoto Y. Role of Bcl-2 family proteins in apoptosis: apoptosomes or mitochondria? Gene Cell. 1998;3:697–707. doi: 10.1046/j.1365-2443.1998.00223.x. [DOI] [PubMed] [Google Scholar]
- 80.Borner C. The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol. Immunol. 2003;39:615–647. doi: 10.1016/s0161-5890(02)00252-3. [DOI] [PubMed] [Google Scholar]
- 81.Bethu M.S., Netala V.R., Domdi L., Tartte V., Janapala V.R. Potential anticancer activity of biogenic silver nanoparticles using leaf extract of Rhynchosia suaveolens: an insight into the mechanism, Artificial Cells. Nanomedicine, and Biotechnology. 2018;46:S104–S114. doi: 10.1080/21691401.2017.1414824. [DOI] [PubMed] [Google Scholar]
- 82.Phull A.R., Abbas Q., Ali A., Raza H., kim S.J., Zia M., Haq I. Antioxidant, cytotoxic and antimicrobial activities of green synthesized silver nanoparticles from crude extract of Bergenia ciliate. Future Journal of Pharmaceutical Sciences. 2016;2:31–36. doi: 10.1016/j.fjps.2016.03.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.