Abstract
Hypoplastic Left Heart Syndrome (HLHS) is a complex Congenital Heart Disease (CHD) that was almost universally fatal until the advent of the Norwood operation in 1981. Children with HLHS who largely succumbed to the disease within the first year of life, are now surviving to adulthood. However, this survival is associated with multiple comorbidities and HLHS infants have a higher mortality rate as compared to other non-HLHS single ventricle patients. In this review we (a) discuss current clinical challenges associated in the care of HLHS patients, (b) explore the use of systems biology in understanding the molecular framework of this disease, (c) evaluate induced pluripotent stem cells as a translational model to understand molecular mechanisms and manipulate them to improve outcomes, and (d) investigate cell therapy, gene therapy, and tissue engineering as a potential tool to regenerate hypoplastic cardiac structures and improve outcomes.
Keywords: Congenital heart disease, Hypoplastic left heart syndrome
1. The HLHS patient
1.1. Incidence and prevalence
Hypoplastic left heart syndrome (HLHS) is a severe form of congenital heart disease (CHD) that affects the left sided systemic blood flow. It afflicts 2 in 10,000 pregnancies and occurs in 0.016% to 0.036% of all live births [1–4]. Despite the low incidence, it is responsible for up to 23% of cardiac deaths during the first week of life and 15% of cardiac deaths within the first month of life [5]. Left heart lesions present with a spectrum of complexity, with isolated aortic valve malformations such as bicuspid aortic valve being the most common, to HLHS which has severely underdeveloped left heart structures including the mitral valve, left ventricle, aortic valve, aorta, in combination of varying degrees of severity. Seven in ten patients are male and 13.5% of siblings of HLHS patients have some form of congenital heart disease suggesting a complex interplay of multiple genetic factors. Only in the past 38 years, after the introduction of the Norwood 3-stage palliation procedure, have infants with HLHS been able to survive beyond the first decade of life. Current reported 15-year-survival is only 48% [6] with significant mortality during the first year of life [7].
1.2. Anatomy of the hypoplastic left heart
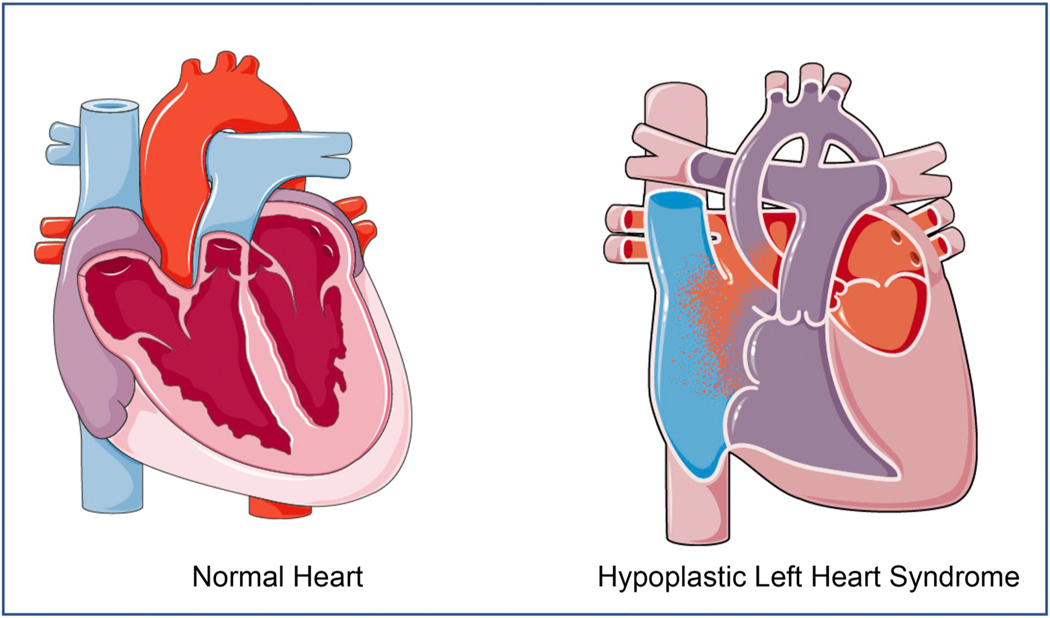
The hypoplastic left heart has underdeveloped left heart structures including the mitral valve, the left ventricle, the aortic valve as well as the aorta (Fig. 1). Representative phenotypic variants of HLHS hearts as seen in the clinic can be found in a recent review by Grossfeld et al. [8]. HLHS is distinguished from other defects with LV hypoplasia by mitral and aortic atresia or hypoplasia with an intact ventricular septum [9,10]. Towards the mild end of the HLHS spectrum, patients have valvular hypoplasia and a relatively larger ventricular cavity size, which are termed hypoplastic left heart complex (HLHC) [11]. Severe HLHS has a non-existent left ventricle with mitral and aortic atresia as well as a hypoplastic aorta and aortic arch. The complex repair needed for the aortic atresia and hypoplasia of the ascending aorta and arch distinguishes HLHS from other types of single ventricle heart defects. Severity of HLHS can determine the corrective surgical pathway – severe HLHS patients are surgically corrected into a single ventricle physiology, whereas the milder forms of HLHC can undergo a biventricular repair. In addition to having an underdeveloped left heart, HLHS patients most commonly present with coarctation of the aorta (in 67–80% cases) [12]. The aortic hypoplasia compromises systemic outflow of blood which is maintained by a patent ductus between the pulmonary artery and aorta until the Norwood operation is performed in the first few days of life. This is followed by the Fontan operation [13], which has been revolutionary in improving survival in children with complex CHD.
Fig. 1.
Anatomy of the Hypoplastic Left Heart: Structures in the left heart are underdeveloped to varying degrees. These can include an underdeveloped LV cavity size, mitral valve, aortic valve and aorta. Hypoplastic left hearts can also have intra-atrial septal abnormalities that facilitates pulmonary venous flow into the right heart circulation as shown in the figure.
1.2.1. Stage 1: The Norwood procedure
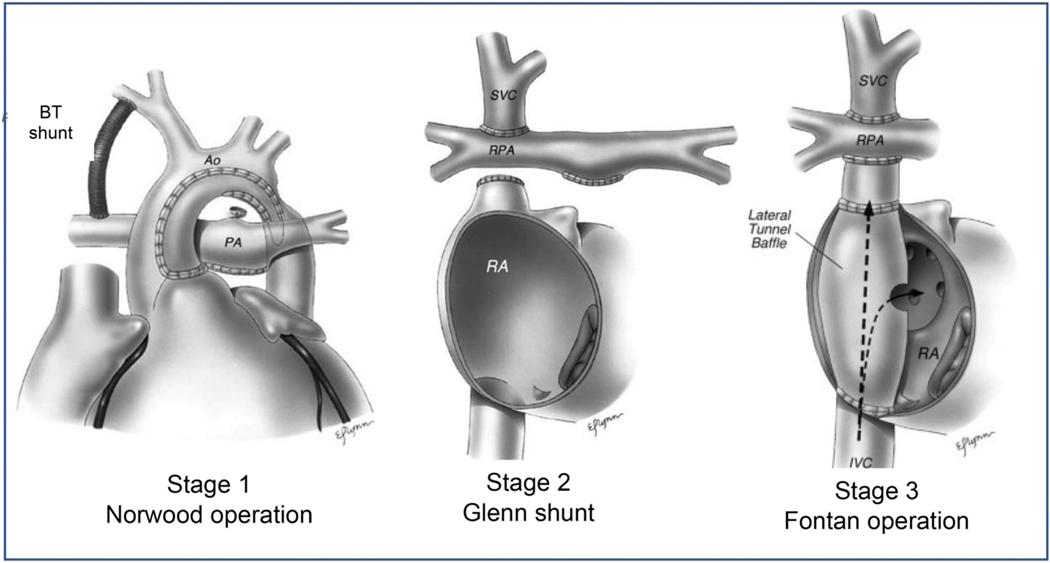
The Norwood procedure was momentous in improving the survival of HLHS patients from 0% in 1979–1984 to 42% in 1999–2005 [7]. The Norwood operation involves (a) atrial septectomy to allow pulmonary venous flow to reach the right ventricle, (b) reconstruction of the aorta using the tissue from the proximal pulmonary artery, and (c) a systemic to pulmonary artery shunt such as the Blalock-Taussig (BT) shunt that allows perfusion of the lungs (Fig. 2). The operation allows the systemic right ventricle to supply blood to the neoaorta and the body, and a portion of that blood is shunted to the lungs via the BT shunt.
Fig. 2.
Staged surgical management of HLHS infants. Stage 1: Norwood operation. Main pulmonary artery is ligated and anastomosed to the hypoplastic aorta connecting the right ventricle to the systemic circulation. Pulmonary circulation is established with a modified Blalock-Taussig (BT) shunt. The ductus arteriosus is ligated and an atrial septectomy is done. Stage 2: Glenn shunt. The superior vena cava is ligated and anastomosed end-to-side to the right pulmonary artery. The lower part of the superior vena cava is over sewn. The main pulmonary artery is usually ligated, though sometimes left open as an additional source of pulmonary blood flow. Stage 3: Total cavopulmonary connection. Intra-atrial lateral tunnel baffle directs inferior vena cava flow to the superior vena cava which is anastomosed to the right pulmonary artery. Occasionally, fenestration is placed in baffle to relieve elevated pressure in lateral tunnel in early postoperative period. Reproduced with permission from O’Brian et al. [115]
1.2.2. Stage 2: Glenn shunt
As the infant grows, stage 2 surgery is performed at around 3 months. The BT shunt is replaced with a direct connection between the superior vena cava (SVC) and the branch pulmonary arteries, also called the Glenn shunt.
1.2.3. Stage 3: The Fontan procedure
In the last stage of the procedure, which typically occurs at preschool age, the inferior vena cava (IVC) is connected to the pulmonary arteries. This last operation shunts all systemic deoxygenated blood to the lungs through a combination of changes in intrathoracic pressures and diastolic filling of the heart bypassing the need for a subpulmonic ventricle.
1.3. Mechanisms of failure in HLHS
In some studies, mortality associated with HLHS is higher than that seen with other single ventricle Fontan patients. Early stage mortality is highest between stage 1 and stage 2 operation, known as the interstage, at 8–12% [14] and has been associated with arrhythmias, decreased ventricular function at hospital discharge [15] cardiopulmonary bypass time and circulatory arrest time [16] as well as obstruction in pulmonary venous return [17]. Survival is between 63 and 74% during the first year of life and remains stable thereafter [7]. This morbidity can be attributed to impairment in coronary perfusion, obstruction of pulmonary artery blood flow, obstruction of flow through the neoaorta (noted in up to 33% of Fontan patients [18]), pulmonary hyperperfusion and right ventricular failure [19]. In addition, the Fontan circulation itself imposes long term consequences to the single ventricle HLHS patient.
In the seminal paper by Fontan et al. [20], which examined short and long term outcomes in Fontan patients 20 years after initial operation, there appeared to be an increase in mortality and decrease in functional status beyond 6 years after initial Fontan operation. Over the years, Fontan patients have been found to have multiple comorbidities including arrhythmias, myocardial and valvular dysfunction and hepatic cirrhosis caused from prolonged hepatic venous congestion. Clinically, Fontan failure is progressive with phenotypic variability across patients. These varied etiologies of Fontan failure have been classified into four distinct Fontan failure phenotypes [21]; however, the underlying cellular and molecular mechanisms contributing to their pathophysiology remain obscure. Additionally, the structure of the cavo-pulmonary connection itself- including significant flow differences between atriopulmonary (AP), lateral tunnel (LT) and extra-cardiac conduits (ECC) as well as subtle differences within each Fontan-subtype due to the location of the anastomosis—causes significant differences in flow patterns and Fontan pressures that have long term implications in outcomes. To better understand these implications, multiple studies involving computational modeling have been conducted to improve the design of the Fontan anatomy [17,22–24]. Understanding the complex interplay between Fontan hemodynamics, intrinsic cardiac architectural differences between anatomically normal versus HLHS patients, as well as serological factors that influence pathology at a molecular level could provide insights on Fontan failure and the development of new therapies.
1.4. Comorbidities associated with HLHS Fontan
Table 1 summarizes the early and late comorbidities associated with the Fontan operation. Perioperative comorbidities specific to HLHS patients include a higher incidence of stroke. Studies have reported that HLHS patients have a higher rate of late adverse events as well, including atrioventricular valve regurgitation, Fontan failure, reoperation, cardiac percutaneous interventions, pacemaker requirement, thromboembolic events and supraventricular tachycardia (SVT) [25].
Table 1.
Co-morbidities associated with Fontan circulation.
1.4.1. Heart Failure
The systemic RV in HLHS is an independent risk factor for Fontan failure with a 3.8-fold increased likelihood as compared to other Fontan anatomies [26]. One in five (19.6%) children with HLHS (versus 7.0% non-HLHS) have abnormal ventricular function and 23.4% (versus 9.2% non-HLHS) have moderate to severe atrioventricular valve (tricuspid) regurgitation. Additionally, late sequelae of the aortic reconstructions can increase afterload on the systemic right ventricle, potentially contributing to heart failure and ischemia. By adulthood, the incidence of heart failure after Fontan procedure is nearly 50% [27,28].
1.4.2. Arrythmias
Arrhythmias in Fontan patients are mostly intra-atrial and re-entrant. These arrhythmias are associated both with surgical suture lines as well as other complex genetic factors that control myocardial architecture, making catheter ablation challenging with a high incidence of recurrence. Onset of arrhythmias is an independent risk predictor of late Fontan failure and reduces post Fontan 15-year survival rates to 70% [29]. In a recent study evaluating the clinical outcomes in Fontan patients >16 years of age, 15% of patients had experienced arrhythmic events including SVT, atrial flutter and atrial fibrillation. Six percent of deaths in Fontan patients were attributed to lethal arrhythmias [18].
1.4.3. Pulmonary Hypertension
A severely restrictive atrial septal defect (ASD) in neonates with HLHS results in pulmonary venous hypertension, pulmonary edema, and intractable hypoxia [30] and has been attributed to a higher mortality without an atrial septostomy. Additionally, HLHS patients have a higher incidence of pulmonary hypertensive events post-operatively, and a significantly higher incidence of in-hospital death [31] as compared to other CHDs.
1.4.4. Plastic Bronchitis (PB)
PB is one of the rare but devastating sequalae of Fontan circulation and has a prevalence of 4–14% [32], which occurs at a median of 2.5 years after Fontan completion and is associated with an increasing rate of death, with a transplant free survival of 8.3 years [33]. Hence, it is more commonly seen in children than adults.
1.4.5. Protein Losing Enteropathy (PLE)
PLE occurs in up to 24% of Fontan patients [34] and presents with symptoms of abdominal and lower extremity swelling and diarrhea. HLHS patients have a 2.81 times higher predisposition of developing PLE [34]. PLE decreases 5-year survival from 88% [35] to 50% [36] in Fontan patients and has been attributed to elevated Fontan pressures >15 mmHg, decreased cardiac index and increased pulmonary vascular resistance.
1.4.6. Hepatic Dysfunction
After the Fontan operation, the systemic venous pressures are obligatorily elevated to allow passive filling of the systemic ventricle. This leads to systemic venous congestion where the liver acts as a reservoir between the upstream splanchnic vascular bed and the downstream pulmonary vascular bed. Almost all patients develop Fontan associated liver disease (FALD) by adulthood, which is characterized by hepatic fibrosis with normal synthetic function, until the late stages of cirrhosis. FALD is pervasive, and associated with cirrhosis, hepatocellular carcinoma, and failing Fontan physiology. In a recently published multicenter study [37], 94% of Fontan patients had at least 1 abnormal liver related finding with nearly all patients showing some abnormalities on imaging. Additionally, 99% of patients had sinusoidal dilation and 97% had sinusoidal fibrosis on histology. Multifocal hyperechoic lesions were noted in the range of 35% [38]–55% [39] of Fontan patients.
1.4.7. Thromboembolic Events
Hepatic venous congestion in Fontan circulation and endothelial dysfunction causes an increased risk of thromboembolism in Fontan patients. Thromboembolic events, either from arrhythmias or from systemic perturbations, have an incidence of 18% at 7 years [40] to 55% at 10 years post [41] Fontan surgery. The incidence of thromboembolic events decreases by a hazard ratio of 2.5 with anticoagulation [41]. Multiple studies have shown that there is no significant difference between aspirin and warfarin in decreasing the risk of thromboembolic events in the long term [40,42–44]. Amongst HLHS patients who have undergone the Norwood procedure there are multiple single case reports and a case series describing an increased risk of thrombus formation in the neoaorta months to years following the Norwood procedure [45–47]. An aortic thrombus further increases the risk of occlusion of the coronary arteries causing sudden death in some of these patients.
2. From bedside to bench
2.1. Systems biology approach to HLHS
Growing evidence supports the hypothesis that HLHS is not a monogenic disease [48] that follows a Mendelian pattern of inheritance. Together with epigenetic and environmental influences, HLHS is a multigenic disease with a multifactorial inheritance pattern which contributes to the variability in HLHS severity seen in patients. While we have yet to unravel the genetic complexity associated with HLHS, it is evident that a reductionist approach of focusing on a single culprit gene to predict HLHS severity or to determine disease prognosis for a patient is inadequate.
Systems biology, within the context of CHD, is a powerful tool to understand heart development, as it provides a framework for understanding various molecular processes in the context of each other. In the context of HLHS, it also has the potential to be used for understanding differential expression of comorbidities seen in patients. The morbidity and mortality associated with HLHS patients spans all ages and it is unclear at this time why some patients continue to survive beyond their 3rd decade of life whereas some others succumb within months after undergoing the same palliative Fontan operation. Therefore, a systems biology approach to examine the interactions between genes, gene expression, proteins and metabolites (genomics, transcriptomics, proteomics and metabolomics respectively) and the dynamic perturbations within this network that initiate pathophysiology, is paramount to the understanding of disease progression in HLHS.
2.1.1. Genomics
There is evidence from animal models and familial genetic studies that supports a genetic cause for HLHS. HLHS is associated with many syndromic diseases including Turner syndrome, Trisomy 18, DiGeorge syndrome, Jacobson syndrome, Noonan syndrome, Holt-Oram syndrome, Rubinstein-Taybi syndrome, Smith-Lemli-Opitz syndrome and heterotaxy [49]. However, the unique genetic mutations that cause HLHS versus other forms of CHD remain largely elusive and most HLHS mutations occur sporadically. Single gene mutations for HLHS are very rare and familial segregation analysis reveals alleles with low penetrance and wide expressivity. Families of patients with HLHS have an increased prevalence of CHD that ranges in severity from bicuspid aortic valve, a relatively benign CHD, to very severe forms of HLHS. Given the overall low prevalence of HLHS, early in utero mortality of fetuses with HLHS and overall high variability in the general population of genetic polymorphisms that are functionally inconsequential, identifying genetic mutations that can be attributed to HLHS alone by linkage analysis is challenging. However, studies have identified mutations in many of the core transcription factors that have been conserved across species with respect to heart development. These include NKX2.5 and NOTCH1. In addition, microdeletions in the FOX transcription family cluster have been associated with HLHS and are implicated in endothelial malformations. Mutations are also attributed to sub-cellular structures including cadherin family of adhesion proteins [50,51] as well as those attributed to dysfunction in the sarcoplasmic reticulum. Somatic mutations in transcription factors such as HAND1 [52], GATA4 and NKX2.5 [53–55] were initially implicated as a possible cause of HLHS. However, these mutations were identified in formalin-fixed tissue and subsequent independent studies in frozen cardiac samples have failed to confirm these initial findings [56,57]. Somatic mutations as a cause of HLHS, or most forms of CHD, are no longer considered to be a predominant mechanism contributing to cardiac malformations [58–60]. While additional germline mutations in genes such as GJA1 [51], PTCH1 [61], BMP2/BMPR2 [61,62], HEY2 [61], HUWEI1 [61], MLL2/KT2D [63], IRX4 [61], JAG1 [61] have been identified through whole exome sequencing (WES), their prevalence within the HLHS patients is relatively low and has not been confirmed by multiple studies. Newer and more powerful techniques associated with genetic analysis continue to be employed in understanding complex diseases such as HLHS. Emerging technologies associated with next generation sequencing are extensively described by experts in the field [64–67]. Ongoing studies through the Congenital Heart Disease Genetic Network Study seek to evaluate genetic mutations of nearly 10,000 patients with all complexities of CHD including 454 HLHS patients [68]. Specific mutations associated with HLHS are noted in Table 2. Additionally, the “Human Phenotype Ontology” (HPO) database actively collects information about phenotypic abnormalities and genes associated with diseases [69].
Table 2.
Genetic mutations associated with HLHS.
| Gene | Mutations identified | Mechanism of action |
|---|---|---|
|
| ||
| NKX2.5 | • Missense | Cardiac transcription factor: mutation causes diminished ability to bind to DNA [121] |
| NOTCH 1 | • Substitution | Cardiac transcription factor: mutation causes dysregulation of cardiac chamber development |
| • Frame shift | and valvulogenesis [62] | |
| • Splice site | ||
| RbFOX2 | • Splice site | RNA binding protein: Mutation affects epithelial to mesenchymal transition, stem cell |
| • Nonsense | differentiation, RNA splicing [65,122] | |
| • Frame shift | ||
| ETS 1 | • Copy number variant | Involved in neural crest development and development of endocardium |
| • Deletion | Implicated in Jacobsen syndrome [123] | |
2.1.2. Transcriptomics
Given the complex genetics of HLHS, it has become evident that there is great diversity within the HLHS genotype, and as yet these mutations cannot be correlated to clinical characteristics. Hence, there is great utility in using dynamic components within the cell to monitor deviations from normal phenotype, both during development and during pathologic worsening of the adult patient. The transcriptome represents all RNA transcribed by a cell. Unlike the genome, it is dynamic and changes during stages of development and progression of disease and may be specific to individual cells. Li et al. [70] utilized transcriptomics to determine gene expression profiles during embryogenesis and further used CHD associated genes to probe disrupted developmental pathways. This interaction between genome and transcriptome provided valuable information on various signaling pathways and their relative regulation over time in the context of CHD. In addition, transcriptomics allows for characterization of non-coding RNAs such as miRNA and ribosomal RNA that have structural and functional roles and can influence disease states and cell-cell signaling. Sucharov et al. [71] evaluated miRNA profile in HLHS patients and determined that the miRNA profile was different in adults and children and reflected the disease state of the patient. Furthermore, miRNA levels responded to therapies such as volume unloading of the ventricle suggesting that miRNA profiles could be useful for monitoring efficacy of drugs and therapies.
2.1.3. Proteomics
Proteomics is the study of proteins, including cytokines within a tissue or bodily fluid. Unlike the genome which is static, proteomes are dynamic and can modulate their function in response to environmental cues to maintain homeostasis without being too ephemeral. Consequently, they are excellent targets for evaluating disease states and their response to drugs and therapies. Furthermore, proteins as molecules are modular and are targets of post-translational modifications - including phosphorylation, glycosylation, alkylation and acetylation, which can change the function of the protein greatly. “Expression proteomics” comparing global protein levels in biological fluids such as serum and urine in normal and disease states is valuable to identify biomarkers for clinical monitoring [72]. However, proteins influence cell behavior at microscopic levels as well, and these differences may not be appreciated on a global scale. Hence, “Cell-map proteomics” that evaluate proteins at a more cellular level is useful in identifying protein dysregulation at a cellular level. While molecules such as brain natriuretic peptide (BNP), troponin and inflammatory cytokines (e.g., tumor necrosis factor alpha and interleukin-6) have been studied clinically as biomarkers of severity and disease progression in cardiac conditions, studies in HLHS patients alone have not been performed. Given the small population size of HLHS patients in any given center and variations in surgical interventions, co-morbid conditions and drug therapies, it is challenging to eliminate confounding factors in determining protein-based biomarkers of significance in HLHS patients. Additionally, acquiring cardiac tissue samples for “cellmap proteomics”, which could be clinically more informative for HLHS patients on a routine basis, is clinically impossible and too risky for the patients. Methods used for proteomic analysis are described in detail in multiple comprehensive reviews [72–82].
2.1.4. Metabolomics
Metabolomics analyzes biological byproducts of metabolic reactions within the body, which by far are the most dynamic molecules in the body, using tools such as nuclear magnetic resonance (NMR) and mass spectroscopy (MS). Within CHD, metabolomics has been used to detect biomarkers in pregnant mothers in the first trimester which indicated an abnormal pattern of lipid metabolism in fetuses with CHD [83]. In a separate study involving pediatric patients, a combination of betaine, taurine, glutamine, and phenylalanine were associated with CHD with high accuracy [84]. There are no known studies to date that evaluate metabolomic profile in HLHS patients. Given that metabolites are dynamic, can change within seconds, and are influenced by day to day changes in diet and activity level, identifying metabolic biomarkers needs to undergo extensive validation prior to use in mainstream medicine. However, studies combining various systems biology approaches allow for evaluation of interdependent cellular processes that contribute to disease mechanisms and can therefore be a powerful tool in CHD.
2.2. Lessons from HLHS heart and tissue explants
Explanted hearts and tissues from patients with HLHS provide valuable insights into cardiomyocyte structure and function. Atrial septal tissue explants from neonates showed an overall transcriptional repression and altered expression of key cell cycle regulators in HLHS patients [85]. Neonatal hearts also showed an uneven distribution of blood vessels with some areas having inadequate perfusion [86].
Studies by Perryman et al. [87] and Miyamoto et al. [71,88] showed that tissue from HLHS single ventricles (SV) have distinct gene expression profiles that are indicative of cardiac remodeling recapitulated in failing ventricles of adult-acquired heart failure. The hallmark of the “fetal gene pattern” (FGP) is characterized by a change in the α versus β-myosin heavy chain (MHC) ratio [87], upregulation of B-type natriuretic peptide (BNP) and atrial natriuretic peptide (ANP) [87] as well as a decrease in sarcoplasmic reticulum calcium-adenosine triphosphate 2a (SERCA2a) [87,88], a Ca+ ATPase that transports calcium into the sarcoplasmic reticulum from the cytosol, thereby modulating myocardial contraction. A similar gene expression pattern is observed in HLHS SV tissues. Similarly, β-adrenergic receptor (β-AR) density is decreased and β-AR function is altered in HLHS SV patients, which has implications in the utility of medications such as β-blockers. To date, βblockers have shown mixed results in the SV population in small studies with inadequate power to evaluate HLHS patients alone [89–92].
In adults with acquired heart failure, progression of disease is driven by cardiac fibrosis [93]. However, tissue explants from HLHS SV, both with and without heart failure, did not show histological or gene expression profiles suggestive of on-going fibrosis. While these patients were predominantly infants, where intrinsic myocardial dysfunction may dominate the pathophysiology of heart failure, this may not reflect the pathophysiology of heart failure in HLHS adults, where circulating fibrotic markers have been found to be elevated [94,95]. Similarly, studies that compared RV from biventricular hearts versus HLHS hearts showed dysregulation in gene expression of calcium handling, β-adrenergic activity and cytoskeletal structure, amongst others as noted in Table 3.
Table 3.
Histopathological and molecular findings in HLHS.
| Downregulated genes/proteins | Adrenergic receptors: β1-AR [87,88], Structural proteins: α-MHC, β-MHC [87], Calcium handling: SERCA 2a [87], Angiogenesis: VEGF [96] |
| Upregulated genes/proteins | Structural proteins: Collagen-1α, Collagen-3 (in non-failing Fontan) [124], β-MHC (in failing Fontan) [88], Transcription Factors: HIF-1α (marker of hypoxia) [96], p53 (marker of DNA damage) [96] Histones: ƔH2AX (marker of DNA damage) [96] Receptors: TGFβ1 (in fetal cardiomyocytes) [96] Adhesion molecules: PECAM-1 (inappropriately expressed in cardiomyocytes) [87] |
| Unchanged gene/protein expression | Adrenergic Receptors: β2-AR [87] Receptors: TGFβR1, TGFβR2 (markers of fibrosis) [124] Structural enzymes: MMP-2, MMP-9 [124] |
Explanted fetal HLHS hearts have provided insight into developmental differences in HLHS SV. Gaber et al. [96] demonstrated that fetal LVs had an increase in HIF-1α, a transcription factor involved in transcriptional response to hypoxia, and decreases in vascular endothelial growth factor (VEGF) and Thymosin β4, which recruit cardiac and vascular progenitor cells. Additionally, markers of DNA damage and cell senescence were upregulated in HLHS fetal hearts. These studies provide insight into possible mechanisms causing LV hypoplasia during embryogenesis. Furthermore, these studies suggest that there are structural, functional and phenotypic differences in HLHS cardiomyocytes that drive cardiac dysfunction which may have implications beyond hemodynamic limitations imposed by the single ventricle alone. Fully categorizing these differences as hallmarks of HLHS versus interpatient variability in HLHS patients is essential in guiding medical therapy.
3. The bench
3.1. Animal models of HLHS
Liu et al. described the first animal model of HLHS, generated through random mutagenesis, which supported the hypothesis of a “two hit” genetic model for the disease [57]. However, given that some of the mice had a VSD and a double outlet right ventricle (DORV) phenotype, the study was unable to show an exclusive correlation of the implicated gene mutations with the HLHS phenotype.
Animal models are integral to understanding genotype-phenotype correlation in diseases when the implicated genes are functionally preserved across species. Despite the limitations of the study by Liu et al., the study demonstrated that severe CHD involves dysregulation of the cell cycle and cell proliferation, abnormal subcellular structures including mitochondria and centrosomes and abnormalities in cardiac development and signaling molecules such as Notch, BMP, TGF and PPAR. These observations further confirm intrinsic cellular abnormalities within the myocardium that contribute to their multiple comorbidities beyond their single ventricle physiology alone.
3.2. Cell culture and potential of disease-in-a-dish model
While frozen sections of HLHS cardiac tissues and explanted HLHS hearts have provided great insights into gene expression profiles and structural variations, tissue samples from these sources cannot provide insight into the dynamic perturbations associated with HLHS nor the efficacy of therapeutic drugs in addressing pathology.
Various groups have derived iPSCs from HLHS patients and differentiated them into cardiomyocytes. Studies involving a single cell line derived from a single HLHS patients have independently shown deficiencies in HLHS derived myocyte structure and function [97]. However, given the intra- and inter-culture variability associated with iPSC derived cells, it was unclear if these differences were associated with HLHS pathology. Other studies involving multiple cell lines from various patients have been simultaneously conducted by various groups and these studies have confirmed that iPSC derived HLHS cardiomyocytes have reduced beating rate, a disorganized sarcomeric structure, reduced response to β1 agonists and dysfunction of the sarcoplasmic reticulum [87,97,98]. Furthermore, these differences correlate well to differences in gene expression analysis performed by rt-PCR as well as tissue RNA sequencing.
Given the lack of viable animal models to study HLHS in molecular detail beyond its genomic abnormalities, iPSCs are a promising alternative to evaluate cellular physiology and study mechanistic basis of the pathology driving the disease and its comorbidities. However, iPSCs generate immature cardiomyocytes and can have variable levels of expression of receptors and proteins that may not fully represent the cellular physiology of HLHS cardiomyocytes. Hence, it is essential to characterize limitations in using iPSC to model HLHS. While channelopathies [99] and metabolic diseases have been successfully translated to iPSCs, specific cellular mechanisms including calcium handling, electrophysiology, β-adrenergic stimulation and conduction velocity may have limitations that will need to be differentiated from its phenotypic disease [100].
4. From bench to bedside
4.1. Gene therapy
The ability to make molecular modification to genes with newer technologies such as “Clustered Regularly Interspaced Short Palindromic Repeats” (CRISPR) [57,101,102] carries promise towards eliminating HLHS in-utero altogether. However, the genetic factors contributing to HLHS are poorly understood and studies point to a multifactorial etiology involving genetic, epigenetic as well as environmental factors, making gene therapy a limited option with our current understanding of this disease.
4.2. Regenerative medicine and stem cell therapies
In response to pressure overload, as is experienced by the HLHS SV, the normal myocardium responds by undergoing myocyte hypertrophy, angiogenesis and increased antioxidant activity to counter the increased oxidative stress [103–105]. In HLHS patients, the systemic right ventricle does not have the same adaptive response as the left ventricle and consequently, myocyte hypertrophy is not followed by a proportional increase in capillary density [86,106,107]. This perfusion mismatch leads to ischemia and decompensated heart failure. Hence, regenerative therapies have focused on providing pro-angiogenic factors that facilitate formation of new blood vessels to support the increasing mass of the RV. Various cell sources that provide angiogenic factors in a paracrine manner have emerged as promising frontiers to facilitate angiogenesis. A comprehensive review of these various cell sources and their impact has been described recently by Bittle et al. [108] and others [109].
Mesenchymal stem cells (multipotent cells derived from the bone marrow), cardiospheres (self-assembling multicellular clusters of cardiac explants), umbilical cord blood cells (UCB), and c-kit cardiac progenitor cells (stem cells derived from the myocardial tissue) have been tested in animal models of cardiac dysfunction and clinical trials involving humans. c-kit cells have been extensively studied in the pediatric population and have been found to improve LV function and fibrosis [110]. Neonatal c-kit cells are more potent than adult cells, and also more abundant within the myocardium. Similarly, UCB cells were found to provide clinical benefit to myocardial function and Phase 1 studies have shown safety and feasibility with intra-myocardial injections [111].
While mechanistically, stem cells have been shown to improve myocardial function through paracrine signaling, current challenges associated with translating this therapy into humans are associated with retention of cells after engraftment. Studies have shown that intracoronary injection of stem cells in patients allows for only 0–0.97% of myocardial retention after 24 h from initial delivery [112]. Similarly, following intramyocardial injection of cells, studies have reported a survival and retention rate of around 11.3% [113]. Engineering delivery solutions that aid cell retention and viability will make cell therapy a potent intervention for patients across the board.
4.3. Tissue engineering and regeneration
Curing HLHS by growing the underdeveloped left heart structures would be an adventurous long-term goal for any scientist in regenerative medicine. Efforts towards engineering cardiac tissues, blood vessels and valves in a laboratory, that can then be implanted into patients such as those with HLHS is a non-trivial task and involves solving immense limitations within the tissue engineering field. Amongst these many challenges include generating complex tissue structures that have a functional vasculature, and constructs with mature cardiomyocytes that have the functional capacity to meet the cardiac demands of the patient. A detailed description of these limitations has been described by Chery et al. [109].
Given the inherent structural and functional differences of HLHS cardiomyocytes, it is likely that using endogenous multipotent cells from patients may propagate these limitations through any endogenously stimulated or engineered tissue. Hence, genetically modifying the cell source to correct these abnormalities adds an additional level of complexity in tissue engineering constructs. Allogeneic cells are considered to be more feasible for development of constructs, albeit with concerns of cellular rejection due to immunogenicity. iPSC banks that can match donor and recipient HLA types have been proposed to address these concerns of immune rejection and are still in the exploratory stage in many countries [114]. Given these challenges, tissue engineering of cardiac structures is not an immediately attainable goal for HLHS patients.
5. Conclusion
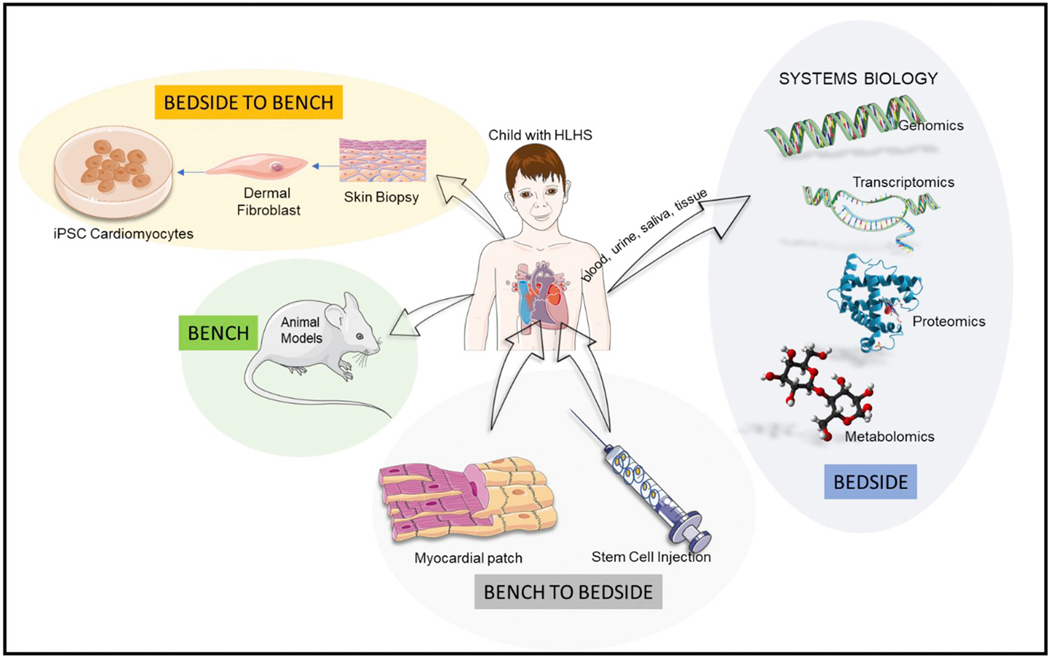
HLHS is a complex cardiac disease with an atypical and diverse molecular pathophysiology. Given the absence of viable animal models, understanding this molecular pathophysiology is particularly challenging. However, hiPSC based cardiac tissue constructs can help bridge the gap in providing a translational model for understanding this disease. While cell therapy and tissue engineering have immense potential for providing lasting cure for these patients, continued efforts in optimizing these technologies are necessary to bring these therapies to the bedside. To advance this field so that it truly benefits patients, an integrative approach that involves strategies from bedside to bench and then back to the bedside are essential as depicted in Fig. 3.
Fig. 3.
Global overview of integrative research involving HLHS.
Acknowledgements
Images in Figs. 1 and 3 were obtained from Servier Medical Art. Graphic for “metabolomics” in Fig. 2 was obtained from public domain, after being released by its author Benjah-bmm27, and graphic for “proteomics” was obtained from public domain.
Funding
AS is supported in part by the Warshaw Fellow Research Award from the Department of Pediatrics at Emory University.
Footnotes
Declaration of Competing Interest
None.
References
- [1].Talner CN, Report of the New England Regional Infant Cardiac Program, by Donald C. Fyler, MD, Pediatrics 65 (suppl) (1980) 375–461 (Pediatrics 102(1 Pt 2) (1998) 258–9). [PubMed] [Google Scholar]
- [2].Samanek M, Slavik Z, Zborilova B, Hrobonova V, Voriskova M, Skovranek J, Prevalence, treatment, and outcome of heart disease in live-born children: a prospective analysis of 91,823 live-born children, Pediatr. Cardiol 10 (4) (1989) 205–211. [DOI] [PubMed] [Google Scholar]
- [3].Morris CD, Outcalt J, Menashe VD, Hypoplastic left heart syndrome: natural history in a geographically defined population, Pediatrics 85 (6) (1990) 977–983. [PubMed] [Google Scholar]
- [4].Ferencz C, Rubin JD, McCarter RJ, Brenner JI, Neill CA, Perry LW, Hepner SI, Downing JW, Congenital heart disease: prevalence at livebirth. The Baltimore-Washington Infant Study, Am. J. Epidemiol 121 (1) (1985) 31–36. [DOI] [PubMed] [Google Scholar]
- [5].Khairy P, Fernandes SM, Mayer JE Jr., Triedman JK, Walsh EP, Lock JE, Landzberg MJ, Long-term survival, modes of death, and predictors of mortality in patients with Fontan surgery, Circulation 117 (1) (2008) 85–92. [DOI] [PubMed] [Google Scholar]
- [6].Pundi KN, Johnson JN, Dearani JA, Pundi KN, Li Z, Hinck CA, Dahl SH, Cannon BC, O’Leary PW, Driscoll DJ, Cetta F, 40-year follow-up after the Fontan operation: long-term outcomes of 1,052 patients, J. Am. Coll. Cardiol 66 (15) (2015) 1700–1710. [DOI] [PubMed] [Google Scholar]
- [7].Siffel C, Riehle-Colarusso T, Oster ME, Correa A, Survival of children with hypoplastic left heart syndrome, Pediatrics 136 (4) (2015) e864–e870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grossfeld P, Nie S, Lin L, Wang L, Anderson RH, Hypoplastic left heart syndrome: a new paradigm for an old disease? J. Cardiovasc. Dev. Dis 6 (1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Noonan JA, Nadas AS, The hypoplastic left heart syndrome; an analysis of 101 cases, Pediatr. Clin. N. Am 5 (4) (1958) 1029–1056. [DOI] [PubMed] [Google Scholar]
- [10].Lev M, Pathologic anatomy and interrelationship of hypoplasia of the aortic tract complexes, Lab. Investig 1 (1) (1952) 61–70. [PubMed] [Google Scholar]
- [11].Tchervenkov CI, Jacobs ML, Tahta SA, Congenital heart surgery nomenclature and database project: hypoplastic left heart syndrome, Ann. Thorac. Surg 69 (4 Suppl) (2000) S170–S179. [DOI] [PubMed] [Google Scholar]
- [12].Elzenga NJ, Gittenberger-de Groot AC, Coarctation and related aortic arch anomalies in hypoplastic left heart syndrome, Int. J. Cardiol 8 (4) (1985) 379–393. [DOI] [PubMed] [Google Scholar]
- [13].Fontan F, Baudet E, Surgical repair of tricuspid atresia, Thorax 26 (3) (1971) 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ghanayem NS, Allen KR, Tabbutt S, Atz AM, Clabby ML, Cooper DS, Eghtesady P, Frommelt PC, Gruber PJ, Hill KD, Kaltman JR, Laussen PC, Lewis AB, Lurito KJ, Minich LL, Ohye RG, Schonbeck JV, Schwartz SM, Singh RK, Goldberg CS, Pediatric Heart Network I, Interstage mortality after the Norwood procedure: Results of the multicenter Single Ventricle Reconstruction trial, J. Thorac. Cardiovasc. Surg 144 (4) (2012) 896–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Simsic JM, Bradley SM, Stroud MR, Atz AM, Risk factors for interstage death after the Norwood procedure, Pediatr. Cardiol 26 (4) (2005) 400–403. [DOI] [PubMed] [Google Scholar]
- [16].Kern JH, Hayes CJ, Michler RE, Gersony WM, Quaegebeur JM, Survival and risk factor analysis for the Norwood procedure for hypoplastic left heart syndrome, Am. J. Cardiol 80 (2) (1997) 170–174. [DOI] [PubMed] [Google Scholar]
- [17].Bove EL, Lloyd TR, Staged reconstruction for hypoplastic left heart syndrome, Contemp. Results Ann. Surg 224 (3) (1996) 387–394 (discussion 394–5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Feinstein JA, Benson DW, Dubin AM, Cohen MS, Maxey DM, Mahle WT, Pahl E, Villafane J, Bhatt AB, Peng LF, Johnson BA, Marsden AL, Daniels CJ, Rudd NA, Caldarone CA, Mussatto KA, Morales DL, Ivy DD, Gaynor JW, Tweddell JS, Deal BJ, Furck AK, Rosenthal GL, Ohye RG, Ghanayem NS, Cheatham JP, Tworetzky W, Martin GR, Hypoplastic left heart syndrome: current considerations and expectations, J. Am. Coll. Cardiol 59 (1 Suppl) (2012) S1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bartram U, Grunenfelder J, Van Praagh R, Causes of death after the modified Norwood procedure: a study of 122 postmortem cases, Ann. Thorac. Surg 64 (6) (1997) 1795–1802. [DOI] [PubMed] [Google Scholar]
- [20].Fontan F, Kirklin JW, Fernandez G, Costa F, Naftel DC, Tritto F, Blackstone EH, Outcome after a “perfect” Fontan operation, Circulation 81 (5) (1990) 1520–1536. [DOI] [PubMed] [Google Scholar]
- [21].Book WM, Gerardin J, Saraf A, Marie Valente A, Rodriguez F 3rd, Clinical phenotypes of Fontan failure: implications for management, Congenit. Heart Dis 11 (4) (2016) 296–308. [DOI] [PubMed] [Google Scholar]
- [22].Bove EL, de Leval MR, Migliavacca F, Balossino R, Dubini G, Toward optimal hemodynamics: computer modeling of the Fontan circuit, Pediatr. Cardiol 28 (6) (2007) 477–481. [DOI] [PubMed] [Google Scholar]
- [23].Slesnick TC, Yoganathan AP, Computational modeling of Fontan physiology: at the crossroads of pediatric cardiology and biomedical engineering, Int. J. Card. Imaging 30 (6) (2014) 1073–1084. [DOI] [PubMed] [Google Scholar]
- [24].Trusty PM, Wei ZA, Slesnick TC, Kanter KR, Spray TL, Fogel MA, Yoganathan AP, The first cohort of prospective Fontan surgical planning patients with follow-up data: how accurate is surgical planning? J. Thorac. Cardiovasc. Surg 157 (3) (2019) 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Iyengar AJ, Winlaw DS, Galati JC, Wheaton GR, Gentles TL, Grigg LE, Justo RN, Radford DJ, Weintraub RG, Bullock A, Celermajer DS, d’Udekem Y, Australia, R. New Zealand Fontan, The extracardiac conduit Fontan procedure in Australia and New Zealand: hypoplastic left heart syndrome predicts worse early and late outcomes, Eur. J. Cardiothorac. Surg 46 (3) (2014) 465–473 (discussion 473). [DOI] [PubMed] [Google Scholar]
- [26].d’Udekem Y, Iyengar AJ, Galati JC, Forsdick V, Weintraub RG, Wheaton GR, Bullock A, Justo RN, Grigg LE, Sholler GF, Hope S, Radford DJ, Gentles TL, Celermajer DS, Winlaw DS, Redefining expectations of long-term survival after the Fontan procedure: twenty-five years of follow-up from the entire population of Australia and New Zealand, Circulation 130 (11 Suppl 1) (2014) S32–S38. [DOI] [PubMed] [Google Scholar]
- [27].Piran S, Veldtman G, Siu S, Webb GD, Liu PP, Heart failure and ventricular dysfunction in patients with single or systemic right ventricles, Circulation 105 (10) (2002) 1189–1194. [DOI] [PubMed] [Google Scholar]
- [28].Stout KK, Broberg CS, Book WM, Cecchin F, Chen JM, Dimopoulos K, Everitt MD, Gatzoulis M, Harris L, Hsu DT, Kuvin JT, Law Y, Martin CM, Murphy AM, Ross HJ, Singh G, Spray TL, Chronic heart failure in congenital heart disease: a scientific statement from the American Heart Association, Circulation 133 (8) (2016) 770–801. [DOI] [PubMed] [Google Scholar]
- [29].Carins TA, Shi WY, Iyengar AJ, Nisbet A, Forsdick V, Zannino D, Gentles T, Radford DJ, Justo R, Celermajer DS, Bullock A, Winlaw D, Wheaton G, Grigg L, d’Udekem Y, Long-term outcomes after first-onset arrhythmia in Fontan physiology, J. Thorac. Cardiovasc. Surg 152 (5) (2016) (1355–1363 e1). [DOI] [PubMed] [Google Scholar]
- [30].Atz AM, Feinstein JA, Jonas RA, Perry SB, Wessel DL, Preoperative management of pulmonary venous hypertension in hypoplastic left heart syndrome with restrictive atrial septal defect, Am. J. Cardiol 83 (8) (1999) 1224–1228. [DOI] [PubMed] [Google Scholar]
- [31].Bando K, Turrentine MW, Sharp TG, Sekine Y, Aufiero TX, Sun K, Sekine E, Brown JW, Pulmonary hypertension after operations for congenital heart disease: analysis of risk factors and management, J. Thorac. Cardiovasc. Surg 112 (6) (1996) 1600–1607 (discussion 1607–9). [DOI] [PubMed] [Google Scholar]
- [32].Caruthers RL, Kempa M, Loo A, Gulbransen E, Kelly E, Erickson SR, Hirsch JC, Schumacher KR, Stringer KA, Demographic characteristics and estimated prevalence of Fontan-associated plastic bronchitis, Pediatr. Cardiol 34 (2) (2013) 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Schumacher KR, Gossett J, Guleserian K, Naftel DC, Pruitt E, Dodd D, Carboni M, Lamour J, Pophal S, Zamberlan M, Gajarski RJ, Fontan-associated protein-losing enteropathy and heart transplant: a Pediatric heart transplant study analysis, J. Heart Lung Transplant 34 (9) (2015) 1169–1176. [DOI] [PubMed] [Google Scholar]
- [34].Schumacher KR, Stringer KA, Donohue JE, Yu S, Shaver A, Caruthers RL, Zikmund-Fisher BJ, Fifer C, Goldberg C, Russell MW, Fontan-associated protein-losing enteropathy and plastic bronchitis, J. Pediatr 166 (4) (2015) 970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Veldtman GR, Webb GD, Improved survival in Fontan-associated protein-losing enteropathy, J. Am. Coll. Cardiol 64 (1) (2014) 63–65. [DOI] [PubMed] [Google Scholar]
- [36].Gobergs R, Salputra E, Lubaua I, Hypoplastic left heart syndrome: a review, Acta. Med. Litu 23 (2) (2016) 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu FM, Kogon B, Earing MG, Aboulhosn JA, Broberg CS, John AS, Harmon A, Sainani NI, Hill AJ, Odze RD, Johncilla ME, Ukomadu C, Gauvreau K, Valente AM, Landzberg MJ, Alliance I. for Adult Research in Congenital Cardiology, Liver health in adults with Fontan circulation: a multicenter cross-sectional study, J. Thorac. Cardiovasc. Surg 153 (3) (2017) 656–664. [DOI] [PubMed] [Google Scholar]
- [38].Bae JM, Jeon TY, Kim JS, Kim S, Hwang SM, Yoo SY, Kim JH, Fontan-associated liver disease: Spectrum of US findings, Eur. J. Radiol 85 (4) (2016) 850–856. [DOI] [PubMed] [Google Scholar]
- [39].Lindsay I, Johnson J, Everitt MD, Hoffman J, Yetman AT, Impact of liver disease after the fontan operation, Am. J. Cardiol 115 (2) (2015) 249–252. [DOI] [PubMed] [Google Scholar]
- [40].Alsaied T, Alsidawi S, Allen CC, Faircloth J, Palumbo JS, Veldtman GR, Strategies for thromboprophylaxis in Fontan circulation: a meta-analysis, Heart 101 (21) (2015) 1731–1737. [DOI] [PubMed] [Google Scholar]
- [41].Egbe AC, Connolly HM, McLeod CJ, Ammash NM, Niaz T, Yogeswaran V, Poterucha JT, Qureshi MY, Driscoll DJ, Thrombotic and embolic complications associated with atrial arrhythmia after Fontan operation: role of prophylactic therapy, J. Am. Coll. Cardiol 68 (12) (2016) 1312–1319. [DOI] [PubMed] [Google Scholar]
- [42].Potter BJ, Leong-Sit P, Fernandes SM, Feifer A, Mayer JE Jr., Triedman JK, Walsh EP, Landzberg MJ, Khairy P, Effect of aspirin and warfarin therapy on thromboembolic events in patients with univentricular hearts and Fontan palliation, Int. J. Cardiol 168 (4) (2013) 3940–3943. [DOI] [PubMed] [Google Scholar]
- [43].McCrindle BW, Manlhiot C, Cochrane A, Roberts R, Hughes M, Szechtman B, Weintraub R, Andrew M, Monagle P, Fontan Anticoagulation Study G, Factors associated with thrombotic complications after the Fontan procedure: a secondary analysis of a multicenter, randomized trial of primary thromboprophylaxis for 2 years after the Fontan procedure, J. Am. Coll. Cardiol 61 (3) (2013) 346–353. [DOI] [PubMed] [Google Scholar]
- [44].Monagle P, Cochrane A, Roberts R, Manlhiot C, Weintraub R, Szechtman B, Hughes M, Andrew M, McCrindle BW, A Multicenter, randomized trial comparing heparin/warfarin and acetylsalicylic acid as primary Thromboprophylaxis for 2 years after the Fontan procedure in children, J. Am. Coll. Cardiol 58 (6) (2011) 645–651. [DOI] [PubMed] [Google Scholar]
- [45].Owens ST, Gomez-Fifer C, Ensing GJ, Thrombus formation in the native aortic root in patients with hypoplastic left heart syndrome, Pediatr. Cardiol 27 (3) (2006) 385–387. [DOI] [PubMed] [Google Scholar]
- [46].Abdullah MH, Van Arsdell GS, Hornberger LK, Adatia I, Precoronary stenosis after stage I palliation for hypoplastic left heart syndrome, Ann. Thorac. Surg 70 (6) (2000) 2147–2149. [DOI] [PubMed] [Google Scholar]
- [47].Brennan TV, Rodefeld MD, Tacy TA, Reddy VM, Hanley FL, Late thrombosis of the native aortic root after Norwood reconstruction for hypoplastic left heart syndrome, J. Thorac. Cardiovasc. Surg 121 (3) (2001) 580–582. [DOI] [PubMed] [Google Scholar]
- [48].McBride KL, Pignatelli R, Lewin M, Ho T, Fernbach S, Menesses A, Lam W, Leal SM, Kaplan N, Schliekelman P, Towbin JA, Belmont JW, Inheritance analysis of congenital left ventricular outflow tract obstruction malformations: segregation, multiplex relative risk, and heritability, Am. J. Med. Genet. A 134A (2) (2005) 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Benson DW, Martin LJ, Lo CW, Genetics of hypoplastic left heart syndrome, J. Pediatr 173 (2016) 25–31. [DOI] [PubMed] [Google Scholar]
- [50].Mahtab EA, Gittenberger-de Groot AC, Vicente-Steijn R, Lie-Venema H, Rijlaarsdam ME, Hazekamp MG, Bartelings MM, Disturbed myocardial connexin 43 and N-cadherin expressions in hypoplastic left heart syndrome and borderline left ventricle, J. Thorac. Cardiovasc. Surg 144 (6) (2012) 1315–1322. [DOI] [PubMed] [Google Scholar]
- [51].Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH, Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE), Mutat. Res 479 (1–2) (2001) 173–186. [DOI] [PubMed] [Google Scholar]
- [52].Reamon-Buettner SM, Ciribilli Y, Inga A, Borlak J, A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts, Hum. Mol. Genet 17 (10) (2008) 1397–1405. [DOI] [PubMed] [Google Scholar]
- [53].Reamon-Buettner SM, Borlak J, Somatic NKX2–5 mutations as a novel mechanism of disease in complex congenital heart disease, J. Med. Genet 41 (9) (2004) 684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Reamon-Buettner SM, Borlak J, NKX2–5: an update on this hypermutable homeodomain protein and its role in human congenital heart disease (CHD), Hum. Mutat 31 (11) (2010) 1185–1194. [DOI] [PubMed] [Google Scholar]
- [55].Reamon-Buettner SM, Borlak J, Somatic mutations in cardiac malformations, J. Med. Genet 43 (8) (2006) e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Firulli BA, Toolan KP, Harkin J, Millar H, Pineda S, Firulli AB, The HAND1 frameshift A126FS mutation does not cause hypoplastic left heart syndrome in mice, Cardiovasc. Res 113 (14) (2017) 1732–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Liu X, Yagi H, Saeed S, Bais AS, Gabriel GC, Chen Z, Peterson KA, Li Y, Schwartz MC, Reynolds WT, Saydmohammed M, Gibbs B, Wu Y, Devine W, Chatterjee B, Klena NT, Kostka D, de Mesy Bentley KL, Ganapathiraju MK, Dexheimer P, Leatherbury L, Khalifa O, Bhagat A, Zahid M, Pu W, Watkins S, Grossfeld P, Murray SA, Porter GA Jr., Tsang M, Martin LJ, Benson DW, Aronow BJ, Lo CW, The complex genetics of hypoplastic left heart syndrome, Nat. Genet 49 (7) (2017) 1152–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Esposito G, Butler TL, Blue GM, Cole AD, Sholler GF, Kirk EP, Grossfeld P, Perryman BM, Harvey RP, Winlaw DS, Somatic mutations in NKX2–5, GATA4, and HAND1 are not a common cause of tetralogy of Fallot or hypoplastic left heart, Am. J. Med. Genet. A 155A (10) (2011) 2416–2421. [DOI] [PubMed] [Google Scholar]
- [59].Salazar M, Consoli F, Villegas V, Caicedo V, Maddaloni V, Daniele P, Caianiello G, Pachon S, Nunez F, Limongelli G, Pacileo G, Marino B, Bernal JE, De Luca A, Dallapiccola B, Search of somatic GATA4 and NKX2.5 gene mutations in sporadic septal heart defects, Eur. J. Med. Genet 54 (3) (2011) 306–309. [DOI] [PubMed] [Google Scholar]
- [60].Draus JM Jr., Hauck MA, Goetsch M, Austin EH 3rd, Tomita-Mitchell A, Mitchell ME, Investigation of somatic NKX2–5 mutations in congenital heart disease, J. Med. Genet 46 (2) (2009) 115–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Durbin MD, Cadar AG, Williams CH, Guo Y, Bichell DP, Su YR, Hong CC, Hypoplastic left heart syndrome sequencing reveals a novel NOTCH1 mutation in a family with single ventricle defects, Pediatr. Cardiol 38 (6) (2017) 1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Iascone M, Ciccone R, Galletti L, Marchetti D, Seddio F, Lincesso AR, Pezzoli L, Vetro A, Barachetti D, Boni L, Federici D, Soto AM, Comas JV, Ferrazzi P, Zuffardi O, Identification of de novo mutations and rare variants in hypoplastic left heart syndrome, Clin. Genet 81 (6) (2012) 542–554. [DOI] [PubMed] [Google Scholar]
- [63].Yoon JK, Ahn KJ, Kwon BS, Kim GB, Bae EJ, Noh CI, Ko JM, The strong association of left-side heart anomalies with Kabuki syndrome, Korean J. Pediatr 58 (7) (2015) 256–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Blue GM, Kirk EP, Giannoulatou E, Sholler GF, Dunwoodie SL, Harvey RP, Winlaw DS, Advances in the genetics of congenital heart disease: a clinician’s guide, J. Am. Coll. Cardiol 69 (7) (2017) 859–870. [DOI] [PubMed] [Google Scholar]
- [65].Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, Jin SC, Deanfield J, Giardini A, Porter GA Jr., Kim R, Bilguvar K, Lopez-Giraldez F, Tikhonova I, Mane S, Romano-Adesman A, Qi H, Vardarajan B, Ma L, Daly M, Roberts AE, Russell MW, Mital S, Newburger JW, Gaynor JW, Breitbart RE, Iossifov I, Ronemus M, Sanders SJ, Kaltman JR, Seidman JG, Brueckner M, Gelb BD, Goldmuntz E, Lifton RP, Seidman CE, Chung WK, De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies, Science 350 (6265) (2015) 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, RomanoAdesman A, Bjornson RD, Breitbart RE, Brown KK, Carriero NJ, Cheung YH, Deanfield J, DePalma S, Fakhro KA, Glessner J, Hakonarson H, Italia MJ, Kaltman JR, Kaski J, Kim R, Kline JK, Lee T, Leipzig J, Lopez A, Mane SM, Mitchell LE, Newburger JW, Parfenov M, Pe’er I, Porter G, Roberts AE, Sachidanandam R, Sanders SJ, Seiden HS, State MW, Subramanian S, Tikhonova IR, Wang W, Warburton D, White PS, Williams IA, Zhao H, Seidman JG, Brueckner M, Chung WK, Gelb BD, Goldmuntz E, Seidman CE, Lifton RP, De novo mutations in histone-modifying genes in congenital heart disease, Nature 498 (7453) (2013) 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Genomics CPC, Gelb B, Brueckner M, Chung W, Goldmuntz E, Kaltman J, Kaski JP, Kim R, Kline J, Mercer-Rosa L, Porter G, Roberts A, Rosenberg E, Seiden H, Seidman C, Sleeper L, Tennstedt S, Kaltman J, Schramm C, Burns K, Pearson G, Rosenberg E, The congenital heart disease genetic network study: rationale, design, and early results, Circ. Res 112 (4) (2013) 698–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hoang TT, Goldmuntz E, Roberts AE, Chung WK, Kline JK, Deanfield JE, Giardini A, Aleman A, Gelb BD, Mac Neal M, Porter GA Jr., Kim R, Brueckner M, Lifton RP, Edman S, Woyciechowski S, Mitchell LE, Agopian AJ, The congenital heart disease genetic network study: cohort description, PLoS One 13 (1) (2018) e0191319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kohler S, Carmody L, Vasilevsky N, Jacobsen JOB, Danis D, Gourdine JP, Gargano M, Harris NL, Matentzoglu N, McMurry JA, Osumi-Sutherland D, Cipriani V, Balhoff JP, Conlin T, Blau H, Baynam G, Palmer R, Gratian D, Dawkins H, Segal M, Jansen AC, Muaz A, Chang WH, Bergerson J, Laulederkind SJF, Yuksel Z, Beltran S, Freeman AF, Sergouniotis PI, Durkin D, Storm AL, Hanauer M, Brudno M, Bello SM, Sincan M, Rageth K, Wheeler MT, Oegema R, Lourghi H, Della Rocca MG, Thompson R, Castellanos F, Priest J, Cunningham-Rundles C, Hegde A, Lovering RC, Hajek C, Olry A, Notarangelo L, Similuk M, Zhang XA, Gomez-Andres D, Lochmuller H, Dollfus H, Rosenzweig S, Marwaha S, Rath A, Sullivan K, Smith C, Milner JD, Leroux D, Boerkoel CF, Klion A, Carter MC, Groza T, Smedley D, Haendel MA, Mungall C, Robinson PN, Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources, Nucleic Acids Res. 47 (D1) (2019) D1018–D1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Li X, Martinez-Fernandez A, Hartjes KA, Kocher JP, Olson TM, Terzic A, Nelson TJ, Transcriptional atlas of cardiogenesis maps congenital heart disease interactome, Physiol. Genomics 46 (13) (2014) 482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Sucharov CC, Sucharov J, Karimpour-Fard A, Nunley K, Stauffer BL, Miyamoto SD, Micro-RNA expression in hypoplastic left heart syndrome, J. Card. Fail 21 (1) (2015) 83–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Young J, Stone WL, Pediatric proteomics: an introduction, Front Biosci. (Schol Ed) 4 (2012) 1078–1087. [DOI] [PubMed] [Google Scholar]
- [73].Wright I, Van Eyk JE, A roadmap to successful clinical proteomics, Clin. Chem 63 (1) (2017) 245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wiktorowicz JE, Brasier AR, Introduction to clinical proteomics, Adv. Exp. Med. Biol 919 (2016) 435–441. [DOI] [PubMed] [Google Scholar]
- [75].Savino R, Paduano S, Preiano M, Terracciano R, The proteomics big challenge for biomarkers and new drug-targets discovery, Int. J. Mol. Sci 13 (11) (2012) 13926–13948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Paik YK, Kim H, Lee EY, Kwon MS, Cho SY, Overview and introduction to clinical proteomics, Methods Mol. Biol 428 (2008) 1–31. [DOI] [PubMed] [Google Scholar]
- [77].Lam MP, Ping P, Murphy E, Proteomics research in cardiovascular medicine and biomarker discovery, J. Am. Coll. Cardiol 68 (25) (2016) 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ioannidis JP, A roadmap for successful applications of clinical proteomics, Proteomics Clin. Appl 5 (5–6) (2011) 241–247. [DOI] [PubMed] [Google Scholar]
- [79].Geyer PE, Holdt LM, Teupser D, Mann M, Revisiting biomarker discovery by plasma proteomics, Mol. Syst. Biol 13 (9) (2017) 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M, Plasma proteome profiling to assess human health and disease, Cell Syst. 2 (3) (2016) 185–195. [DOI] [PubMed] [Google Scholar]
- [81].Clough T, Thaminy S, Ragg S, Aebersold R, Vitek O, Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs, BMC Bioinforma. 13 (Suppl. 16) (2012) S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ahram M, An introduction into proteomics and its clinical applications, Saudi Med. J 28 (4) (2007) 499–507. [PubMed] [Google Scholar]
- [83].Bahado-Singh RO, Ertl R, Mandal R, Bjorndahl TC, Syngelaki A, Han B, Dong E, Liu PB, Alpay-Savasan Z, Wishart DS, Nicolaides KH, Metabolomic prediction of fetal congenital heart defect in the first trimester, Am. J. Obstet. Gynecol 211 (3) (2014) 240 e1–240 e14. [DOI] [PubMed] [Google Scholar]
- [84].Yu M, Sun S, Yu J, Du F, Zhang S, Yang W, Xiao J, Xie B, Discovery and validation of potential serum biomarkers for Pediatric patients with congenital heart diseases by metabolomics, J. Proteome Res 17 (10) (2018) 3517–3525. [DOI] [PubMed] [Google Scholar]
- [85].Gambetta K, Al-Ahdab MK, Ilbawi MN, Hassaniya N, Gupta M, Transcription repression and blocks in cell cycle progression in hypoplastic left heart syndrome, Am. J. Physiol. Heart Circ. Physiol 294 (5) (2008) H2268–H2275. [DOI] [PubMed] [Google Scholar]
- [86].Salih C, Sheppard MN, Ho SY, Morphometry of coronary capillaries in hypoplastic left heart syndrome, Ann. Thorac. Surg 77 (3) (2004) 903–907 (discussion 907). [DOI] [PubMed] [Google Scholar]
- [87].Bohlmeyer TJ, Helmke S, Ge S, Lynch J, Brodsky G, Sederberg JH, Robertson AD, Minobe W, Bristow MR, Perryman MB, Hypoplastic left heart syndrome myocytes are differentiated but possess a unique phenotype, Cardiovasc. Pathol 12 (1) (2003) 23–31. [DOI] [PubMed] [Google Scholar]
- [88].Miyamoto SD, Stauffer BL, Polk J, Medway A, Friedrich M, Haubold K, Peterson V, Nunley K, Nelson P, Sobus R, Stenmark KR, Sucharov CC, Gene expression and beta-adrenergic signaling are altered in hypoplastic left heart syndrome, J. Heart Lung Transplant 33 (8) (2014) 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Shaddy RE, Boucek MM, Hsu DT, Boucek RJ, Canter CE, Mahony L, Ross RD, Pahl E, Blume ED, Dodd DA, Rosenthal DN, Burr J, LaSalle B, Holubkov R, Lukas MA, Tani LY, G. Pediatric Carvedilol Study, Carvedilol for children and adolescents with heart failure: a randomized controlled trial, JAMA 298 (10) (2007) 1171–1179. [DOI] [PubMed] [Google Scholar]
- [90].Book WM, Shaddy RE, Medical therapy in adults with congenital heart disease, Heart Fail. Clin 10 (1) (2014) 167–178. [DOI] [PubMed] [Google Scholar]
- [91].Bradley EA, Saraf A, Book W, Heart failure in women with congenital heart disease, Heart Fail. Clin 15 (1) (2019) 87–96. [DOI] [PubMed] [Google Scholar]
- [92].Sabanayagam A, Cavus O, Williams J, Bradley E, Management of Heart Failure in adult congenital heart disease, Heart Fail. Clin 14 (4) (2018) 569–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC, Cardiac fibrosis: the fibroblast awakens, Circ. Res 118 (6) (2016) 1021–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Opotowsky AR, Baraona F, Owumi J, Loukas B, Singh MN, Valente AM, Wu F, Cheng S, Veldtman G, Rimm EB, Landzberg MJ, Galectin-3 is elevated and associated with adverse outcomes in patients with single-ventricle Fontan circulation, J. Am. Heart Assoc 5 (1) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Saraf A, De Staercke C, Rodriguez F, Kalogeropoulos A, Knezevic A, Gerardin J, Ephrem G, Hayek S, Jennings S, Katabarwa A, Biomarker profile in adults with Fontan circulation, J. Am. Coll. Cardiol 69 (11 Supplement) (2017) 650. [Google Scholar]
- [96].Gaber N, Gagliardi M, Patel P, Kinnear C, Zhang C, Chitayat D, Shannon P, Jaeggi E, Tabori U, Keller G, Mital S, Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation, Am. J. Pathol 183 (3) (2013) 720–734. [DOI] [PubMed] [Google Scholar]
- [97].Jiang Y, Habibollah S, Tilgner K, Collin J, Barta T, Al-Aama JY, Tesarov L, Hussain R, Trafford AW, Kirkwood G, Sernagor E, Eleftheriou CG, Przyborski S, Stojkovic M, Lako M, Keavney B, Armstrong L, An induced pluripotent stem cell model of hypoplastic left heart syndrome (HLHS) reveals multiple expression and functional differences in HLHS-derived cardiac myocytes, Stem Cells Transl. Med 3 (4) (2014) 416–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Yang C, Xu Y, Yu M, Lee D, Alharti S, Hellen N, Ahmad Shaik N, Banaganapalli B, Sheikh Ali Mohamoud H, Elango R, Przyborski S, Tenin G, Williams S, O’Sullivan J, Al-Radi OO, Atta J, Harding SE, Keavney B, Lako M, Armstrong L, Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis, Hum. Mol. Genet 26 (16) (2017) 3031–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Preininger MK, Jha R, Maxwell JT, Wu Q, Singh M, Wang B, Dalal A, McEachin ZT, Rossoll W, Hales CM, Fischbach PS, Wagner MB, Xu C, A human pluripotent stem cell model of catecholaminergic polymorphic ventricular tachycardia recapitulates patient-specific drug responses, Dis. Model. Mech 9 (9) (2016) 927–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Giacomelli E, Mummery CL, Bellin M, Human heart disease: lessons from human pluripotent stem cell-derived cardiomyocytes, Cell. Mol. Life Sci 74 (20) (2017) 3711–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].van Kampen SJ, van Rooij E, CRISPR craze to transform cardiac biology, Trends Mol. Med (2019), 10.1016/j.molmed.2019.06.008 (Jul 17, pii: S1471–4914(19)30170–4, [Epub ahead of print]). [DOI] [PubMed] [Google Scholar]
- [102].Nelson CE, Wu Y, Gemberling MP, Oliver ML, Waller MA, Bohning JD, Robinson-Hamm JN, Bulaklak K, Castellanos Rivera RM, Collier JH, Asokan A, Gersbach CA, Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy, Nat. Med 25 (3) (2019) 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Tomanek RJ, Torry RJ, Growth of the coronary vasculature in hypertrophy: mechanisms and model dependence, Cell. Mol. Biol. Res 40 (2) (1994) 129–136. [PubMed] [Google Scholar]
- [104].Sag CM, Santos CX, Shah AM, Redox regulation of cardiac hypertrophy, J. Mol. Cell. Cardiol 73 (2014) 103–111. [DOI] [PubMed] [Google Scholar]
- [105].Souders CA, Borg TK, Banerjee I, Baudino TA, Pressure overload induces early morphological changes in the heart, Am. J. Pathol 181 (4) (2012) 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Padalino MA, Castellani C, Toffoli S, Della Barbera M, Milanesi O, Thiene G, Stellin G, Angelini A, Pathological changes and myocardial remodelling related to the mode of shunting following surgical palliation for hypoplastic left heart syndrome, Cardiol. Young 18 (4) (2008) 415–422. [DOI] [PubMed] [Google Scholar]
- [107].Drake JI, Bogaard HJ, Mizuno S, Clifton B, Xie B, Gao Y, Dumur CI, Fawcett P, Voelkel NF, Natarajan R, Molecular signature of a right heart failure program in chronic severe pulmonary hypertension, Am. J. Respir. Cell Mol. Biol 45 (6) (2011) 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bittle GJ, Morales D, Deatrick KB, Parchment N, Saha P, Mishra R, Sharma S, Pietris N, Vasilenko A, Bor C, Ambastha C, Gunasekaran M, Li D, Kaushal S, Stem cell therapy for hypoplastic left heart syndrome: mechanism, clinical application, and future directions, Circ. Res 123 (2) (2018) 288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chery J, Wong J, Huang S, Wang S, Si MS, Regenerative medicine strategies for hypoplastic left heart syndrome, Tissue Eng. Part B Rev 22 (6) (2016) 459–469. [DOI] [PubMed] [Google Scholar]
- [110].Kazakov A, Meier T, Werner C, Hall R, Klemmer B, Korbel C, Lammert F, Maack C, Bohm M, Laufs U, C-kit(+) resident cardiac stem cells improve left ventricular fibrosis in pressure overload, Stem Cell Res. 15 (3) (2015) 700–711. [DOI] [PubMed] [Google Scholar]
- [111].Burkhart HM, Qureshi MY, Rossano JW, Peral SC, O’Leary PW, Hathcock M, Kremers W, Nelson TJ, Phase I clinical results of autologous stem cell therapy for hypoplastic left heart syndrome: safety and feasibility of intraoperative intramyocardial injections, J. Thorac. Cardiovasc. Surg (2019), 10.1016/j.jtcvs.2019.06.001 (Jun 7, pii: S0022–5223(19)31154–7, [Epub ahead of print]). [DOI] [PubMed] [Google Scholar]
- [112].Lezaic L, Socan A, Peitl PK, Poglajen G, Sever M, Cukjati M, Cernelc P, Vrtovec B, Imaging and 1-day kinetics of intracoronary stem cell transplantation in patients with idiopathic dilated cardiomyopathy, Nucl. Med. Biol 43 (7) (2016) 410–414. [DOI] [PubMed] [Google Scholar]
- [113].Hou D, Youssef EA, Brinton TJ, Zhang P, Rogers P, Price ET, Yeung AC, Johnstone BH, Yock PG, March KL, Radiolabeled cell distribution after intramyocardial, intracoronary, and interstitial retrograde coronary venous delivery: implications for current clinical trials, Circulation 112 (9 Suppl) (2005) I150–I156. [DOI] [PubMed] [Google Scholar]
- [114].Taylor CJ, Peacock S, Chaudhry AN, Bradley JA, Bolton EM, Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types, Cell Stem Cell 11 (2) (2012) 147–152. [DOI] [PubMed] [Google Scholar]
- [115].O’Brien P, Boisvert JT, Current management of infants and children with single ventricle anatomy, J. Pediatr. Nurs 16 (5) (2001) 338–350. [DOI] [PubMed] [Google Scholar]
- [116].Mahle WT, Hu C, Trachtenberg F, Menteer J, Kindel SJ, Dipchand AI, Richmond ME, Daly KP, Henderson HT, Lin KY, McCulloch M, Lal AK, Schumacher KR, Jacobs JP, Atz AM, Villa CR, Burns KM, Newburger JW, Pediatric Heart Network I, Heart failure after the Norwood procedure: An analysis of the Single Ventricle Reconstruction Trial, J. Heart Lung Transplant 37 (7) (2018) 879–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Schwartz I, McCracken CE, Petit CJ, Sachdeva R, Late outcomes after the Fontan procedure in patients with single ventricle: a meta-analysis, Heart 104 (18) (2018) 1508–1514. [DOI] [PubMed] [Google Scholar]
- [118].Mertens L, Hagler DJ, Sauer U, Somerville J, Gewillig M, Protein-losing enteropathy after the Fontan operation: an international multicenter study. PLE study group, J. Thorac. Cardiovasc. Surg 115 (5) (1998) 1063–1073. [DOI] [PubMed] [Google Scholar]
- [119].Wilson TG, Shi WY, Iyengar AJ, Winlaw DS, Cordina RL, Wheaton GR, Bullock A, Gentles TL, Weintraub RG, Justo RN, Grigg LE, Radford DJ, d’Udekem Y, Australia, R. New Zealand Fontan, Twenty-Five Year Outcomes of the Lateral Tunnel Fontan Procedure, Semin. Thorac. Cardiovasc. Surg 29 (3) (2017) 347–353. [DOI] [PubMed] [Google Scholar]
- [120].Schumacher KR, Singh TP, Kuebler J, Aprile K, O’Brien M, Blume ED, Risk factors and outcome of Fontan-associated plastic bronchitis: a case-control study, J. Am. Heart Assoc 3 (2) (2014) e000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, Grossfeld P, Fatkin D, Jones O, Hayes P, Feneley M, Harvey RP, Cardiac homeobox gene NKX2–5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome, J. Am. Coll. Cardiol 41 (11) (2003) 2072–2076. [DOI] [PubMed] [Google Scholar]
- [122].Verma SK, Deshmukh V, Nutter CA, Jaworski E, Jin W, Wadhwa L, Abata J, Ricci M, Lincoln J, Martin JF, Yeo GW, Kuyumcu-Martinez MN, Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts, Sci. Rep 6 (2016) 30896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, Mazaika E, Vardarajan B, Italia M, Leipzig J, DePalma SR, Golhar R, Sanders SJ, Yamrom B, Ronemus M, Iossifov I, Willsey AJ, State MW, Kaltman JR, White PS, Shen Y, Warburton D, Brueckner M, Seidman C, Goldmuntz E, Gelb BD, Lifton R, Seidman J, Hakonarson H, Chung WK, Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data, Circ. Res 115 (10) (2014) 884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Nakano SJ, Siomos AK, Garcia AM, Nguyen H, SooHoo M, Galambos C, Nunley K, Stauffer BL, Sucharov CC, Miyamoto SD, Fibrosis-related gene expression in single ventricle heart disease, J. Pediatr 191 (2017) 82–90 (e2). [DOI] [PMC free article] [PubMed] [Google Scholar]