Abstract
Broth culture supernatants from Tox+ Helicobacter pylori strains induce vacuolation of HeLa cells in vitro and contain VacA in concentrations that are higher than those found in supernatants from Tox− H. pylori strains. To investigate the basis for this phenomenon, we analyzed the transcription of the vacuolating cytotoxin gene (vacA) in eight Tox+ strains (each with a type s1/m1 vacA genotype) and nine Tox− strains (each with a type s2/m2 vacA genotype). Most of the Tox+ and Tox− strains tested used the same vacA transcriptional start point, but Tox+ strains yielded significantly stronger primer extension signal intensities than did Tox− strains (mean densitometry values of 15.8 ± 1.9 versus 8.9 ± 1.7, P = 0.0016). Correspondingly, when we introduced vacA::xylE transcriptional fusions into the chromosomes of a Tox+ strain (60190) and a Tox− strain (86-313), the level of XylE activity in 60190 vacA::xylE was about 30-fold higher than that in 86-313 vacA::xylE. Sequence analysis and promoter exchange experiments indicated that the different levels of vacA transcription in these two strains cannot be explained solely by a difference in promoter strength. These data indicate that Tox+ and Tox− H. pylori strains typically differ not only in the VacA amino acid sequence but also in the level of vacA transcription.
Helicobacter pylori organisms are curved, gram-negative bacteria found associated with the gastric epithelia of humans and other primates. Colonization of the human stomach with H. pylori consistently results in the development of gastric mucosal inflammation and is a risk factor for the development of peptic ulcer disease and gastric adenocarcinoma (7, 17, 21). One putative virulence determinant of H. pylori is a unique toxin (VacA) that induces vacuolation of epithelial cells (5, 22). VacA is initially translated as a 140-kDa protoxin, which subsequently undergoes both N-terminal and C-terminal processing to yield an ∼90-kDa mature secreted toxin (10, 23–25). Deep-etch electron microscopic analysis indicates that VacA forms large, six- or seven-sided complexes comprised of 12 or 14 subunits (9, 20).
Considerable variation exists among different H. pylori strains in the production of vacuolating cytotoxin activity. Thus, broth culture supernatants from some H. pylori strains (designated Tox+) induce vacuolation of HeLa cells in vitro, whereas other H. pylori strains (designated Tox−) lack detectable vacuolating activity in this assay (2, 8, 18). In previous studies, it has been shown that all H. pylori isolates hybridize with vacA probes (2, 10, 24, 25), but the vacA alleles in Tox+ strains are typically considerably different from those in Tox− strains (2, 10). A system for classifying vacA alleles has been developed in which specific families of vacA alleles are associated with the production of detectable vacuolating cytotoxin activity (2). Specifically, most H. pylori strains with a type s1 vacA signal sequence and a type m1 vacA midregion induce prominent cell vacuolation, whereas strains with a type s2 signal sequence and type m2 midregion consistently fail to induce cytotoxic effects (2). In addition to these vacA sequence differences, there is also evidence that concentrations of VacA are higher in broth culture supernatants from Tox+ strains than in supernatants from Tox− strains (6, 8).
In this report, we demonstrate that vacA is transcribed in both Tox+ and Tox− strains, but transcription typically occurs at higher levels in Tox+ strains than in Tox− strains. This variation is not attributable to differences in vacA transcriptional start points and is not due solely to differences in vacA promoter strength. Heterogeneity in vacA transcription levels among H. pylori strains may be a factor that contributes to different vacuolating cytotoxin phenotypes.
MATERIALS AND METHODS
Bacteria and culture conditions.
H. pylori strains were cultured at 37°C in ambient air containing 5% CO2. The wild-type H. pylori strains used in this study are listed in Table 1. The vacA genotypes of all strains were determined by a PCR-based typing method as previously described (2). Complete or partial vacA sequences from several of these strains have been reported previously (Table 1).
TABLE 1.
Vacuolating cytotoxin activities and vacA transcriptional activities of H. pylori strains used in this study
| Strain | vacA genotype | cagA | Vacuolating activity for HeLa cellse | vacA primer extension sig- nal intensityf |
|---|---|---|---|---|
| 60190 (ATCC 49503) | s1a/m1a | + | 160 | 25.0 |
| 84-183 (ATCC 53726) | s1b/m1b | + | 80 | 11.9 |
| 87-33 | s1b/m1 | + | 20 | 12.9 |
| 87-81 | s1b/m1b | + | 320 | 20.1 |
| 92-25 | s1b/m1 | + | 20 | 16.3 |
| 92-29 | s1b/m1 | + | 80 | 15.8 |
| 92-26 | s1b/m1 | + | 20 | 6.8 |
| 87-199 | s1a/m1 | + | 320 | 17.4 |
| 86-338 | s2/m2 | − | <10 | 19.1 |
| Tx30a (ATCC 51932) | s2/m2c | − | <10 | 8.3 |
| 86-313 | s2/m2b | − | <10 | 8.9 |
| 87-75 | s2/m2 | − | <10 | 4.8 |
| 87-203 | s2/m2d | − | <10 | 8.9 |
| 92-28 | s2/m2 | − | <10 | 8.6 |
| 87-90 | s2/m2 | − | <10 | 3.3 |
| 87-230 | s2/m2 | − | <10 | 9.6 |
| 92-20 | s2/m2 | − | <10 | 0 |
Partial vacA sequence from this strain has been reported previously (2).
Reciprocal titer of the maximum supernatant dilution that produced vacuolation of HeLa cells.
Quantified by laser densitometry (OD per square millimeter).
Analysis of VacA production.
H. pylori strains were cultured in sulfite-free brucella broth containing 5% fetal bovine serum (FBS) for approximately 24 h and harvested after reaching an optical density at 600 nm (OD600) of about 0.5. After centrifugation of the cultures, the supernatants were concentrated by ultrafiltration and tested for vacuolating cytotoxin activity by adding serial dilutions to HeLa cells in tissue culture medium containing 10 mM ammonium chloride as described previously (8). The broth culture supernatants were immunoblotted with rabbit anti-VacA serum prepared by immunizing a rabbit with purified, denatured VacA from H. pylori 60190 as described previously (6). As another approach for analyzing concentrations of VacA in culture supernatants, H. pylori 60190, 86-338, and 86-313 were grown in sulfite-free brucella broth containing 0.5% activated charcoal, and oligomeric VacA was purified from the broth culture supernatants as described previously (9). Yields of purified VacA were assessed by measuring the OD280 of VacA-containing fractions and by semiquantitative analysis of the density of VacA bands after sodium dodecyl sulfate-polyacrylamide gel electrophoresis and silver staining.
Molecular biology methods.
To prepare genomic DNA from H. pylori, cells were suspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and lysed by the addition of sodium dodecyl sulfate and proteinase K (final concentrations of 0.5% and 0.1 mg/ml, respectively) at 37°C for 45 min. Sodium chloride was then added to a final concentration of 0.7 M, and a solution of 10% hexadecyltrimethylammonium bromide–0.7 M sodium chloride was added to yield a final hexadecyltrimethylammonium bromide concentration of 1%. Cell lysates were incubated at 65°C for 10 min. Following the addition of an equal volume of chloroform, cell debris was cleared by centrifugation for 10 min at 10,000 × g. Supernatants were then extracted sequentially with equal volumes of chloroform and phenol-chloroform (1:1) and precipitated with isopropanol (3).
All PCRs were carried out in 100-μl volumes with 1.5 mM magnesium chloride and 200 μM dATP, dCTP, dGTP, and dTTP. AmpliTaq DNA polymerase (Perkin-Elmer) was added to a final concentration of 2.5 U/100 μl. Primers were used at a concentration of 1 μM. The template DNA concentration was 100 ng of chromosomal DNA per reaction or 25 to 100 ng of plasmid DNA per reaction. Denaturation was uniformly at 94°C for 1 min, and annealing temperatures were 5°C below the melting temperature of the primers. Extension at 72°C was for 1 min/kb of amplification product.
Inverse PCR was performed as described above but by using oppositely oriented primers with BglII restriction sites incorporated at their 5′ ends. After completion of thermal cycling, the template DNA was eliminated by DpnI digestion. The sample then was digested with BglII and purified by phenol-chloroform extraction and ethanol precipitation. Inverse PCR products were recircularized with T4 DNA ligase and transformed into Escherichia coli DH5α.
Primer extension analysis.
Seventeen different H. pylori strains were inoculated into sulfite-free brucella broth containing 5% FBS such that the initial OD600 was approximately 0.05. Cultures were harvested when the OD600 reached approximately 0.5. Total cellular RNA was extracted from the bacterial pellets by using the hot phenol method (12). Standardized (40-μg) RNA samples from each strain were heated to 90°C for 2 min in a buffer consisting of 20 mM Tris (pH 8.0), 100 mM sodium chloride, 0.1 mM EDTA, and 20 ng of a 32P-end-labeled oligonucleotide (5′ TTTTTGCACAAAGGGTGCGAC). Following primer annealing at 50°C for 3 h, extension of the labeled primer was accomplished by incubation in 50 mM Tris (pH 8.2)–6 mM MgCl2–10 mM dithiothreitol–0.2 mM deoxynucleoside triphosphates–5 U of avian myeloblastosis virus reverse transcriptase (Promega) for 1 h at 42°C. Primer extension products and sequencing reaction ladders generated by using the same primer were analyzed on 7 M urea–8% polyacrylamide gels. Signal intensities were quantified by densitometry using a GS-670 densitometer and Molecular Analyst version 1.4.1 software (Bio-Rad).
Construction of a vacA::xylE transcriptional fusion in Tox+ H. pylori 60190.
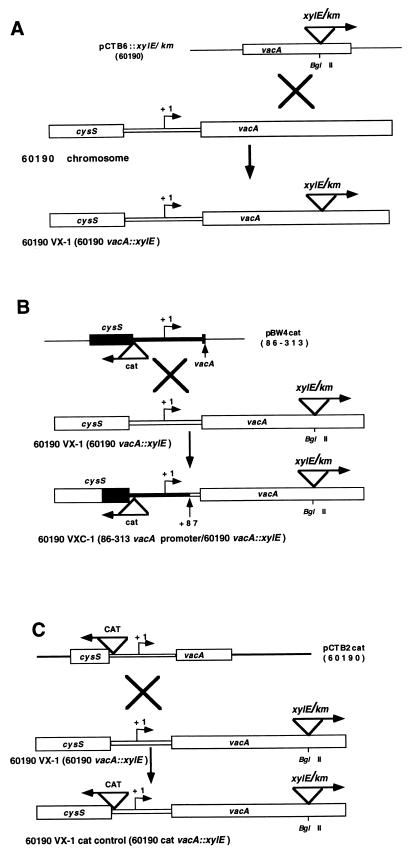
The promoterless xylE gene, encoding Pseudomonas putida catechol 2,3-dioxygenase, was fused upstream of a kanamycin resistance gene (hereafter designated km) such that these two genes were transcribed in the same direction and the kanamycin resistance gene retained its native promoter sequence (16). This xylE/km cassette was cloned into the unique BglII site within pCTB6, which contains a vacA gene fragment of H. pylori 60190 (10). The resultant plasmid construct, pCTB6::xylE/km, was used to introduce the xylE/km cassette into vacA of H. pylori 60190 by natural transformation and allelic exchange (10). Transformants were selected on brucella agar plates supplemented with 5% FBS and kanamycin (40 μg/ml). The orientation of the cassette in the transformants was determined by PCR with a xylE-specific forward primer (5′ CATGACGTCACCTCTTCATAG) and a vacA-specific reverse primer (5′ GCCTTTTTTACAACCGTGATC). The resultant Tox+ vacA reporter strain, with xylE in the same orientation as vacA transcription, is designated 60190 VX-1 (see Fig. 4A).
FIG. 4.
Construction of vacA::xylE transcriptional fusions in Tox+ H. pylori 60190. Sequences derived from Tox+ strain 60190 are represented by open boxes. Sequences derived from Tox− strain 86-313 are indicated by black boxes. Vector sequences are shown as thin, single lines. The vacA transcriptional start point (+1) is represented by a bent arrow. The directional arrow in the xylE/km cassette denotes the orientation of xylE. The kanamycin resistance gene (km) is transcribed under the control of its native promoter and in the same direction as xylE. (A) Construction of a vacA transcriptional reporter strain. The xylE/km cassette was cloned into the BglII site of pCTB6, which contains a 3.2-kb vacA fragment from Tox+ strain 60190 (yielding pCTB6::xylE/km). This construct was introduced into the chromosome of strain 60190 by natural transformation and allelic exchange, and the resultant strain (60190 vacA::xylE) was designated 60190 VX-1. (B) Introduction of Tox− vacA promoter region sequences into Tox+ strain 60190. To place the vacA::xylE fusion in 60190 VX-1 under the control of a heterologous vacA promoter from Tox− strain 86-313, the cat gene was cloned into a fragment from strain 86-313 containing the entire cysS-vacA intergenic region (to yield pBW4cat). Natural transformation and allelic exchange were used to introduce this sequence into the 60190 VX-1 chromosome. The extent of vacA sequence exchange in the chimera was experimentally determined to be up to +87 relative to the vacA TSP. The resultant strain, which now contains the vacA promoter region from strain 86-313, was designated 60190 VXC-1. (C) Construction of a chloramphenicol-resistant control strain. The construction of the chimeric reporter strain outlined in panel B required the use of a marker for chloramphenicol resistance. To determine the effect of the cat gene alone on vacA transcription, an isogenic control strain was constructed by transforming Tox+ vacA reporter strain 60190 VX-1 with plasmid pCTB2cat. The resultant Cmr Kmr strain bears the cat gene at the same location and in the same orientation as in the chimeric reporter strain, 60190 VXC-1, described above. This isogenic control strain was designated 60190 VX-1 cat control.
Construction of a vacA::xylE transcriptional fusion in Tox− H. pylori 86-313.
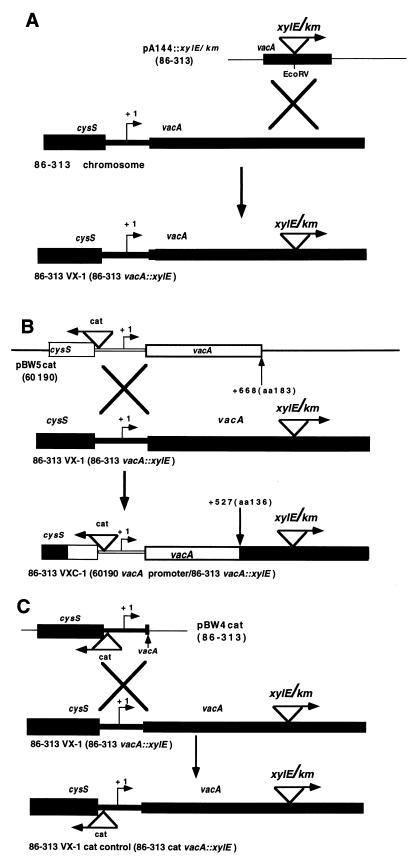
A 1.3-kb internal fragment of vacA from Tox− strain 86-313 was PCR amplified by using primers 5′ CCCACGCAAGTCATTGATGG 3′ and 5′ GGTATTATTTTTTCGCACCAC 3′ (2) and cloned into pT7 Blue (Novagen), resulting in pA144. The xylE/km cassette described above was cloned into the unique EcoRV site within this vacA sequence. The resulting plasmid, pA144::xylE/km, was introduced into strain 86-313 via natural transformation, and kanamycin-resistant colonies were selected. The resulting 86-313 vacA::xylE reporter strain, with xylE in the same orientation as vacA transcription, is designated 86-313 VX-1 (see Fig. 3A).
FIG. 3.
Construction of vacA::xylE transcriptional fusions in Tox− H. pylori 86-313. Sequences derived from Tox+ strain 60190 are represented by open boxes. Sequences derived from Tox− strain 86-313 are indicated by black boxes. Vector sequences are shown as thin, single lines. The vacA TSP (+1) is represented by a bent arrow. The directional arrow in the xylE/km cassette denotes the orientation of xylE. The kanamycin resistance gene (km) is transcribed under the control of its native promoter and in the same direction as xylE. (A) Construction of a vacA transcriptional reporter strain. The xylE/km cassette was cloned into the EcoRV site of pA144, which contains a 1.3-kb vacA fragment from Tox− strain 86-313 (yielding pA144::xylE/km). This construct was introduced into the chromosome of strain 86-313 by natural transformation and allelic exchange, and the resultant strain (86-313 vacA::xylE) was designated 86-313 VX-1. (B) Introduction of Tox+ vacA promoter region sequences into Tox− strain 86-313. To place the vacA::xylE fusion in 86-313 VX-1 under the control of a heterologous vacA promoter from Tox+ strain 60190, the cat gene was cloned into a fragment from strain 60190 containing the entire cysS-vacA intergenic region (to yield pBW5cat). Natural transformation and allelic exchange were used to introduce this sequence into the 86-313 VX-1 chromosome. The extent of vacA sequence exchange in the chimera was experimentally determined to be up to +527, relative to the TSP. The resultant strain, which now contains the vacA promoter region from 60190, was designated 86-313 VXC-1. (C) Construction of a chloramphenicol-resistant control strain. The construction of the chimeric reporter outlined in panel B required the use of a marker for chloramphenicol resistance. To determine the effect of the cat gene alone on vacA transcription, an isogenic control strain was constructed by transforming Tox− vacA reporter strain 86-313 VX-1 with plasmid pBW4cat. The resultant Cmr Kmr strain bears the cat gene at the same location and in the same orientation as in the chimeric reporter strain, 86-313 VXC-1, described above. This isogenic control strain was designated 86-313 VX-1 cat control.
Construction of chimeric strains with alternate vacA promoters.
To place the vacA::xylE transcriptional fusion in strain 86-313 VX-1 under the control of a Tox+ promoter, a 1.3-kb fragment was amplified from Tox+ 60190 genomic DNA by using primers 5′ AATTACTTGCTAGGGGTGCATTAT 3′ and 5′ ATCAGCACTATCCTTATAGCTTG 3′. This fragment contains 519 bp from the 3′ end of cysS, the cysS-vacA intergenic region, and 548 bp from the 5′ end of vacA and was cloned into pT7Blue (Novagen) to yield pBW5. The chloramphenicol acetyltransferase (cat) gene of Campylobacter coli (26) was then cloned into the HindIII site at the 3′ terminus of cysS, in the orientation opposite to that of vacA to yield pBW5cat (see Fig. 3B). This plasmid was used to introduce the Tox+ vacA promoter and adjacent sequences into the Tox− reporter strain 86-313 VX-1 described above. Transformants were screened on brucella agar containing 5% FBS and chloramphenicol (10 μg/ml). The extent of replacement of 86-313 sequences with 60190 sequences in the resulting Kmr Cmr 86-313 VX-1 transformants was determined by PCR amplification and sequencing of the cysS-vacA intergenic region. The chimeric strain shown in Fig. 3B is designated 86-313 VXC-1.
To place the vacA::xylE transcriptional fusion in strain 60190 VX-1 under the control of a Tox− promoter, a DNA fragment was amplified from Tox− strain 86-313 by using primers 5′ GAAGAACTGCTTGGCATCGGG 3′ and 5′ ATTCCATTTTCTTCCTTTC 3′. This fragment contains 221 bp of cysS, the entire cysS-vacA intergenic region, and the first 7 bp of the vacA structural gene. The resulting PCR product was cloned into pBluescript SK+ to yield pBW3. A unique BglII site was introduced 3 bp downstream of the stop codon of cysS in pBW3 by inverse PCR mutagenesis by using primers with BglII sites incorporated at the 5′ ends (5′ GAAGATCTAGCTTAAAAAAGCTTCTCCCAAATCGTGCC and 5′ GAAGATCTTCTTTAAATTTTACCTATTTACGCACTC) to yield pBW4. The cat gene from C. coli (26) was cloned into the BglII site after ends were made blunt by treatment with Klenow fragment (3). A construct, designated pBW4cat, was selected in which cat and vacA are divergently transcribed (see Fig. 4B). To replace the native vacA promoter in Tox+ reporter strain 60190 VX-1 with the vacA promoter from Tox− strain 86-313, strain 60190 VX-1 was transformed with pBW4cat. Cmr Kmr transformants were selected, and the extent of sequence replacement was determined by PCR amplifying and sequencing the entire cysS-vacA intergenic region of the transformants. The resulting chimeric strain shown in Fig. 4B is designated 60190 VXC-1.
The introduction of heterologous promoter sequences into either of the chimeric reporter strains required the presence of two different selectable markers (described above). To determine whether introduction of the cat gene alone altered levels of vacA transcription, this gene was introduced into the chromosomes of strains 60190 VX-1 and 86-313 VX-1 in the same orientation and at the same sites as described previously. This was accomplished by transformation of strain 86-313 VX-1 with pBW4cat to generate an isogenic Kmr Cmr Tox− reporter strain (86-313 VX-1 cat control) with a cat marker at the 3′ terminus of cysS (see Fig. 3C). A similar control for the Tox+ reporter, 60190 VX-1, was generated by transformation with pCTB2cat to yield the isogenic Kmr Cmr reporter strain 60190 VX-1 cat control (see Fig. 4C).
Assay for XylE activity.
XylE activity was assessed qualitatively at the colony level by spraying colonies grown on brucella agar–5% FBS–kanamycin (40 μg/ml) with 20 mM catechol in distilled water and visually examining colonies for the yellow reaction product 2-hydroxymuconic semialdehyde. For quantitative assays, cells were harvested from broth cultures by centrifugation and resuspended in 50 mM potassium phosphate buffer (pH 7.5), and the cell density was quantified and standardized by measuring OD600. Catechol was added to a final concentration of 3 mM, and enzyme specific activities were determined spectrophotometrically in a Beckman DU 7400 spectrophotometer at 375 nm (11, 27). One unit of XylE activity corresponds to the formation at 22°C of 1 mmol of 2-hydroxymuconic semialdehyde/min (molar extinction coefficient, 4.4 × 104).
RESULTS AND DISCUSSION
Characterization of VacA production by a panel of H. pylori strains.
In this study, we analyzed eight H. pylori strains with a type s1/m1 vacA genotype and nine strains with a type s2/m2 vacA genotype (Table 1). Each strain was grown in broth culture until a standardized OD600 was reached, and the broth culture supernatants then were concentrated by ultrafiltration and tested in a HeLa cell vacuolation assay. Broth culture supernatants from each of the type s1/m1 strains (Tox+) induced vacuolation of HeLa cells, whereas supernatants from each of the type s2/m2 strains (Tox−) did not (Table 1). Immunoblotting studies with anti-VacA serum indicated that an immunoreactive ∼90-kDa band was present in broth culture supernatants from all 17 strains (data not shown). However, when standardized amounts of supernatant protein from different strains were immunoblotted and compared, the VacA bands in Tox+ supernatants tended to be darker than those in Tox− supernatants. To compare the concentrations of VacA in Tox+ and Tox− supernatants by another approach, VacA was purified from standardized volumes of culture supernatant from three strains (Tox+ strain 60190, Tox− strain 86-338, and Tox− strain 86-313) and the yields of purified VacA were analyzed as described in Materials and Methods. The broth culture supernatant from H. pylori 60190 yielded about 10-fold higher quantities of purified oligomeric VacA than did the supernatant from strain 86-338 and >50-fold higher quantities than the supernatant from strain 86-313 (data not shown). These data indicate that all of the H. pylori strains tested produce a VacA product, but supernatants from Tox+ strains contain higher concentrations of VacA than do supernatants from Tox− strains.
Primer extension analysis of vacA transcription.
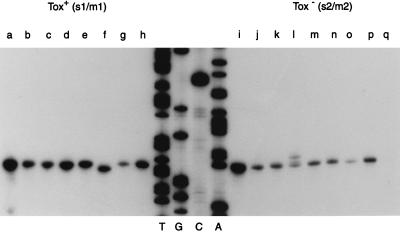
To investigate a possible basis for the different concentrations of VacA in supernatants from Tox+ and Tox− strains, we analyzed vacA transcription in the panel of 17 H. pylori strains by quantitative primer extension analysis (Fig. 1). The primer for these experiments (Fig. 2) was chosen based on the fact that its sequence was 100% complementary to the corresponding vacA sequences of the seven different Tox+ and three Tox− strains sequenced to date (including strains 60190, 84-183, 87-199, Tx30a, 86-313, and 87-203 from the current study), thereby reducing the possibility that varying signal strengths could be due to inefficient primer annealing. As shown in Fig. 1, vacA transcription was detected in all 17 strains, and 15 strains (7 Tox+ and 8 Tox−) used the same conserved single transcriptional start point (TSP). This site was located 1 nucleotide downstream from the vacA transcriptional start site identified in a previous study (24). The use of a second primer (5′AGAGGGCGATTGATTTTGCGGTGTG), which anneals farther downstream within the vacA coding region, confirmed the use of this TSP and failed to demonstrate any alternate start sites (data not shown). A variant Tox+ strain (92-29) appeared to use a TSP located 1 bp closer to the translational start codon (Fig. 1), which could potentially be due to a 1-bp deletion within the 5′ untranslated region of this strain. A variant Tox− strain (87-75) simultaneously used three different, adjacent nucleotides as TSPs (Fig. 1). The conservation of adenosine as the +1 site for vacA transcription in most strains may be important, because it has been demonstrated that the identity of the +1 site can affect transcriptional efficiency (19). Although most strains used the same vacA TSP, there was considerable variation in the intensity of primer extension signals (Fig. 1). Primer extension signals from eight Tox+ strains were significantly more intense than signals from nine Tox− strains (relative densitometry OD values [mean ± the standard error of the mean] of 15.8 ± 1.9 versus 8.9 ± 1.7, P = 0.0016) (Fig. 1), although there were outliers in both groups. These data indicate that Tox+ and Tox− H. pylori strains differ in the level of vacA transcription.
FIG. 1.
Primer extension analysis of vacA mRNA. vacA transcription was analyzed in 17 H. pylori strains (8 Tox+ and 9 Tox−) by primer extension analysis using standardized (40-μg) RNA samples from each strain and the primer 5′TTTTTGCACAAAGGGTGCGAC 3′. The sequencing ladder was generated by using the same primer and pCTB2, which contains a partial vacA sequence from H. pylori 60190, as the template (10). Tox+ strains are shown to the left of the sequencing ladder, and Tox− strains are shown to the right. Strain designations are as follows: lane a, 60190; lane b, 84-183; lane c, 87-33; lane d, 87-81; lane e, 92-25; lane f, 92-29; lane g, 92-26; lane h, 87-199; lane i, 86-338; lane j, Tx30a; lane k, 86-313; lane l, 87-75; lane m, 87-203; lane n, 92-28; lane o, 87-90; lane p, 87-230; lane q, 92-20. The ninth Tox− strain (92-20) yielded a weak primer extension product that was detectable with prolonged exposure (data not shown). The signals from Tox+ strains were significantly more intense than signals from Tox− strains (mean densitometry values of 15.8 ± 1.9 versus 8.9 ± 1.7, P = 0.0016).
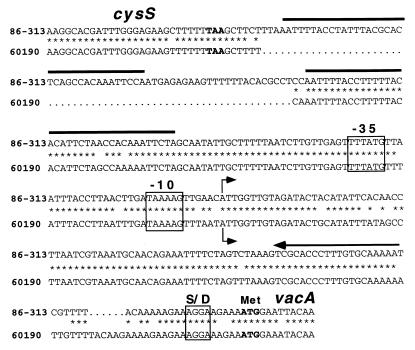
FIG. 2.
Comparison of cysS-vacA intergenic regions in H. pylori 86-313 (Tox−) and 60190 (Tox+). The cysS-vacA intergenic region from H. pylori 86-313 was PCR amplified and sequenced as described previously (14), and the sequence of the corresponding region from H. pylori 60190 has been reported previously (10). Analysis of the aligned sequences demonstrated a 63-bp insertion in the cysS-vacA intergenic region of Tox− strain 86-313. The corresponding absence of this sequence in Tox+ H. pylori 60190 is denoted by dots. Positions of nucleotide identity are denoted by asterisks. A 36-bp sequence and its direct repeat are indicated by solid bars. The vacA transcriptional start points (determined by primer extension analysis [Fig. 1]) are indicated by the bent arrows. The putative Shine-Dalgarno (S/D) sequence and putative −10 and −35 hexamers are boxed. The stop codon (TAA) of cysS and the start codon (ATG) of vacA are in boldface. The primer used to determine vacA transcriptional start points (Fig. 1) is indicated by an arrow over the complementary sequences.
Introduction of vacA::xylE transcriptional fusions into H. pylori 60190 and 86-313.
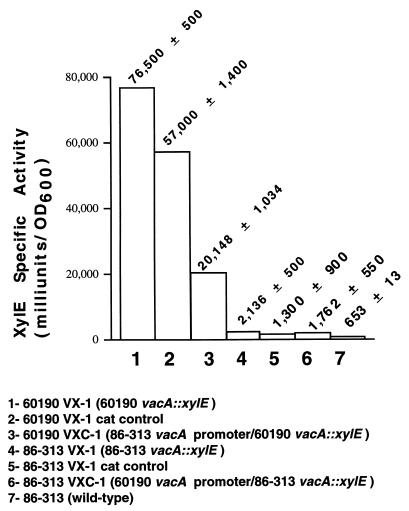
To investigate further the apparent differences among strains in the level of vacA transcription, we introduced vacA::xylE transcriptional fusions into the chromosomes of two H. pylori strains, Tox+ strain 60190 and Tox− strain 86-313. These two strains were chosen based on the primer extension data, which indicated a difference in the level of vacA transcription (Fig. 1), and because both strains were known to be naturally competent for transformation (unpublished data). The introduction of vacA::xylE transcriptional fusions into strains 60190 and 86-313 yielded strains 60190 VX-1 and 86-313 VX-1, respectively (Fig. 3A and 4A). The XylE activity was more than 30-fold higher in Tox+ reporter strain 60190 VX-1 than in Tox− reporter strain 86-313 VX-1 (76,500 ± 500 versus 2,136 ± 500 mU/OD600, P < 0.001). Thus, both primer extension analysis and vacA::xylE transcriptional fusion data indicated that these two strains differ in the level of vacA transcription.
Comparison of vacA promoter strengths in H. pylori 60190 and 86-313.
One possible explanation for differential vacA transcription among strains is the occurrence of variations in vacA promoters. Such a phenomenon accounts for the active transcription of the pertussis toxin operon in Bordetella pertussis and the presence of a silent toxin operon in B. parapertussis and B. bronchiseptica (1). Although the precise locations of vacA promoter sequences in H. pylori have not been determined, putative −10 and −35 hexamers can be inferred based on spacing relative to the vacA transcriptional start point and comparison with E. coli consensus sequences. A comparison of the putative vacA −10 and −35 sequences in H. pylori 60190 and 86-313 reveals no obvious differences that would account for the different levels of vacA transcription in these two strains (Fig. 2). A second potential explanation for the demonstrated difference in vacA transcription might be varying numbers of binding sites for a trans-acting factor. This possibility is relevant because unlike that of Tox+ strain 60190, the cysS-vacA intergenic region of Tox− strain 86-313 contains a 63-bp insertion (14). This 63-bp insertion contains a 36-bp segment that is duplicated a few base pairs farther downstream (Fig. 2).
To determine experimentally whether sequence differences in the cysS-vacA intergenic region might account for different levels of vacA transcription, the vacA::xylE fusion in Tox− reporter strain 86-313 VX-1 was placed under the control of the vacA promoter region from Tox+ strain 60190. Sequence analysis confirmed that in this chimeric reporter strain (86-313 VXC-1), an exchange of promoter and signal sequences had taken place and that the 63-bp insertion had been eliminated (Fig. 3B). Nevertheless, there was no increase in XylE activity in response to the heterologous promoter sequences (Fig. 5). These data indicate that the constraint on transcription in strain 86-313 is not the consequence of either a weak promoter or cis-acting sequences in the promoter region.
FIG. 5.
XylE activity of H. pylori vacA::xylE transcriptional reporter strains. Specific XylE activities (milliunits per OD600) were determined by using bacteria that had been grown in brucella broth–5% FBS for 18 h (late-log phase to early stationary phase). In all assays, the densities of bacterial suspensions were standardized by OD600. XylE activity was quantified as described in Materials and Methods. Results represent the mean ± the standard deviation of three assays from a representative experiment. Absolute values varied slightly from trial to trial, but the overall pattern shown here is representative of three independent experiments. Results from H. pylori 86-313 (parental strain, no xylE fusion) are consistent with background levels of 2-hydroxymuconic semialdehyde production. Levels of XylE activity were significantly higher in strain 60190 VX-1 (lane 1) than in strain 86-313 VX-1 (lane 4), P < 0.001. Placement of the 86-313 vacA promoter upstream from vacA in strain 60190 VX-1 resulted in a significant decrease in XylE activity (compare lanes 2 and 3; P < 0.001) but did not reduce activity to the same level as in strain 86-313 VX-1 (lanes 4 and 5).
In a converse experiment, the vacA::xylE transcriptional fusion in Tox+ reporter strain 60190 VX-1 was placed under the control of the vacA promoter from Tox− strain 86-313. Sequence analysis of the DNA from the resulting chimeric reporter, 60190 VXC-1, confirmed that all 73 bp upstream from the promoter, the putative −35 and −10 sequences, and the 5′ untranslated region through +87 in this chimeric strain had been replaced with sequences from strain 86-313 (Fig. 4B). The level of XylE activity in this chimera was about 65% less than that of the control strain, 60190 VX-1 cat control (Fig. 5; P < 0.001). However, the level of XylE activity in the chimeric strain was still about 10-fold higher than that in Tox− reporter strain 86-313 VX-1 (Fig. 5).
The results of these promoter exchange experiments suggest that strains 60190 and 86-313 differ in vacA promoter strength. However, any such difference must be dictated by sequences outside the putative −10 and −35 hexamers, since these sequences are identical in the two strains. An important finding is that the Tox− (strain 86-313) vacA promoter is capable of initiating higher levels of vacA mRNA synthesis in the strain 60190 background than in the strain 86-313 background. Therefore, it seems likely that the vacA transcription level difference between these two strains is not due solely to a difference in vacA promoter strength. One possibility is the expression of a trans-acting repressor factor in strain 86-313, or alternatively, that strain 60190 produces an activator factor which is absent or reduced in quantity or function in strain 86-313.
Another possible explanation for these data is that strains 60190 and 86-313 differ in vacA transcript stability. To investigate this possibility, we attempted to determine the half-lives of vacA transcripts in these two strains by using serial quantitative primer extension analyses of bacterial cells that had been treated with rifampin to inhibit RNA polymerase activity. These experiments repeatedly yielded nonlinear patterns of primer extension signal decay, and therefore, it remains unclear whether strains 60190 and 86-313 differ in vacA transcript stability. Important determinants of mRNA stability in prokaryotic organisms include stem-loop structures located at either the 5′ or the 3′ ends of transcripts (4, 12, 13). In the promoter switching experiments described in this report, we replaced the entire 5′ untranslated region of vacA from strain 86-313 with that from strain 60190 and failed to demonstrate any significant increase in vacA transcription in the chimeric strain (86-313 VXC-1, Fig. 3). This suggests that sequences at the 5′ end of vacA mRNA do not significantly alter vacA mRNA stability. Both Tox+ and Tox− strains that have been analyzed thus far contain prominent stem-loop structures at the 3′ ends of vacA transcripts (2, 10), and thus, there is also no evidence that sequence differences in this region would contribute to different vacA mRNA stability.
Determinants of the vacuolating cytotoxin phenotype.
The two groups of H. pylori strains analyzed in this study (Tox+ and Tox−) clearly differ in the capacity to induce vacuolation of HeLa cells. One explanation for this difference, supported by data in this study, as well as previous studies (6, 8), is that there are higher concentrations of VacA in broth culture supernatant from Tox+ strains than in supernatant from Tox− strains. Heterogeneity among strains in the level of vacA transcription would undoubtedly be a factor that contributes to this phenomenon. In addition, there also may be heterogeneity among strains in the efficiency of vacA secretion, possibly related to differences in vacA signal sequences (2). In support of this hypothesis, in the present study, we detected 10-fold higher concentrations of VacA in supernatant from Tox+ H. pylori 60190 than in supernatant from Tox− H. pylori 86-338 but found that the two strains did not differ substantially in the level of vacA transcription (Fig. 1 and Table 1).
A second explanation for different vacuolating phenotypes is that the Tox+ and Tox− strains analyzed in this study produce vacA products (types s1/m1 and s2/m2, respectively) that have markedly different amino acid sequences. Specifically, type s1/m1 and type s2/m2 VacA proteins are only about 58% identical within a 250-amino-acid midregion segment (2). These substantial differences would be expected to result in considerably different structural and functional properties. Nevertheless, in previous studies, we have demonstrated that a type s2/m2 VacA protein is capable of assembling into a complex oligomeric structure that is almost identical to that of type s1/m1 VacA proteins (9). To determine whether the different amino acid sequences of type s1/m1 and s2/m2 VacA proteins are important determinants of the vacuolating cytotoxin phenotype, we purified VacA oligomers from culture supernatants of strains 60190 (type s1/m1 VacA) and 86-338 (type s2/m2 VacA) and tested equal concentrations of the two acid-activated proteins in a HeLa cell vacuolation assay. This experiment indicated that the s1/m1 VacA protein produced prominent cell vacuolation, as expected, whereas the type s2/m2 VacA protein lacked any detectable activity in this assay. Thus, equal concentrations of VacA from Tox+ and Tox− strains are not equal in toxicity.
In summary, the vacuolating cytotoxin phenotype of an H. pylori strain is dependent on the amino acid sequence of its vacA product but may also be modulated by other strain-specific factors, such as the level of vacA transcription or the efficiency of VacA secretion. The considerable variation in these determinants among H. pylori strains is consistent with the high level of genetic diversity that exists in the H. pylori species (15) and may be relevant to the occurrence of different clinical outcomes in H. pylori-infected humans.
ACKNOWLEDGMENTS
This work was supported by grant AI 39657 from the National Institutes of Health and by the Medical Research Service of the Department of Veterans Affairs. J.A. is the recipient of a Clinician Scientist Fellowship from the Medical Research Council (United Kingdom).
We thank Beverly Hosse for technical assistance and Mikio Karita for his gift of the xylE/km cassette.
REFERENCES
- 1.Arico B, Rappuoli R. Bordetella parapertussis and Bordetella bronchiseptica contain transcriptionally silent pertussis toxin genes. J Bacteriol. 1987;169:2847–2853. doi: 10.1128/jb.169.6.2847-2853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atherton J C, Cao P, Peek R M, Jr, Tummuru M K R, Blaser M J, Cover T L. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1994. [Google Scholar]
- 4.Chen C Y, Beatty J T, Cohen S N, Belasco J G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988;52:609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- 5.Cover T L. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 6.Cover T L, Blaser M J. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267:10570–10575. [PubMed] [Google Scholar]
- 7.Cover T L, Blaser M J. Helicobacter pylori infection, a paradigm for chronic mucosal inflammation: pathogenesis and implications for eradication and prevention. Adv Intern Med. 1996;41:85–117. [PubMed] [Google Scholar]
- 8.Cover T L, Cao P, Lind C D, Tham K T, Blaser M J. Correlation between vacuolating cytotoxin production by Helicobacter pylori isolates in vitro and in vivo. Infect Immun. 1993;61:5008–5012. doi: 10.1128/iai.61.12.5008-5012.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cover T L, Hanson P I, Heuser J E. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. J Cell Biol. 1997;138:759–769. doi: 10.1083/jcb.138.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 11.Curcic R, Dhandayuthapani S, Deretic V. Gene expression in mycobacteria: transcriptional fusions based on xylE and analysis of the promoter region of the response regulator mtrA from Mycobacterium tuberculosis. Mol Microbiol. 1994;13:1057–1064. doi: 10.1111/j.1365-2958.1994.tb00496.x. [DOI] [PubMed] [Google Scholar]
- 12.Emory S A, Belasco J G. The ompA 5′ untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emory S A, Bouvet P, Belasco J G. A 5′ terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 14.Garner J A, Cover T L. Analysis of genetic diversity in cytotoxin-producing and non-cytotoxin-producing Helicobacter pylori strains. J Infect Dis. 1995;172:290–293. doi: 10.1093/infdis/172.1.290. [DOI] [PubMed] [Google Scholar]
- 15.Go M F, Kapur V, Graham D Y, Musser J M. Population genetic analysis of Helicobacter pylori by multilocus enzyme electrophoresis: extensive allelic diversity and recombinational population structure. J Bacteriol. 1996;178:3934–3938. doi: 10.1128/jb.178.13.3934-3938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karita M, Tummuru M K R, Wirth H P, Blaser M J. Effect of growth phase and acid shock on Helicobacter pylori cagA expression. Infect Immun. 1996;64:4501–4507. doi: 10.1128/iai.64.11.4501-4507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labigne A, deReuse H. Determinants of Helicobacter pylori pathogenicity. Infect Agents Dis. 1996;5:191–202. [PubMed] [Google Scholar]
- 18.Leunk R D, Johnson P T, David B C, Kraft W G, Morgan D R. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J Med Microbiol. 1988;26:93–99. doi: 10.1099/00222615-26-2-93. [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Turnbough C L., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lupetti P, Heuser J E, Manetti R, Massari P, Lanzaecchia S, Bellon P L, Dallai R, Rappuoli R, Telford J L. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mobley H L T. Defining Helicobacter pylori as a pathogen: strain heterogeneity and virulence. Am J Med. 1996;100:25–115. doi: 10.1016/s0002-9343(96)80223-3. [DOI] [PubMed] [Google Scholar]
- 22.Papini E, de Bernard M, Milia E, Bugnoli M, Zerial M, Rappuoli R, Montecucco C. Cellular vacuoles induced by Helicobacter pylori originate from late endosomal compartments. Proc Natl Acad Sci USA. 1994;91:9720–9724. doi: 10.1073/pnas.91.21.9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phadnis S H, Ilver D, Janzon L, Normark S, Westblom T U. Pathological significance and molecular characterization of the vacuolating cytotoxin gene of Helicobacter pylori. Infect Immun. 1994;62:1557–1565. doi: 10.1128/iai.62.5.1557-1565.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt W, Haas R. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol Microbiol. 1994;12:307–319. doi: 10.1111/j.1365-2958.1994.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 25.Telford J L, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, Tecce M F, Censini S, Covacci A, Xiang Z, Papini E, Montecucco C, Parente L, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:1653–1658. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Taylor D E. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene. 1990;94:23–28. doi: 10.1016/0378-1119(90)90463-2. [DOI] [PubMed] [Google Scholar]
- 27.Zukowski M, M, Gaffney D E, Speck D, Kauffmann M, Findeli A, Wisecup A, Lecocq J P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci USA. 1983;80:1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]