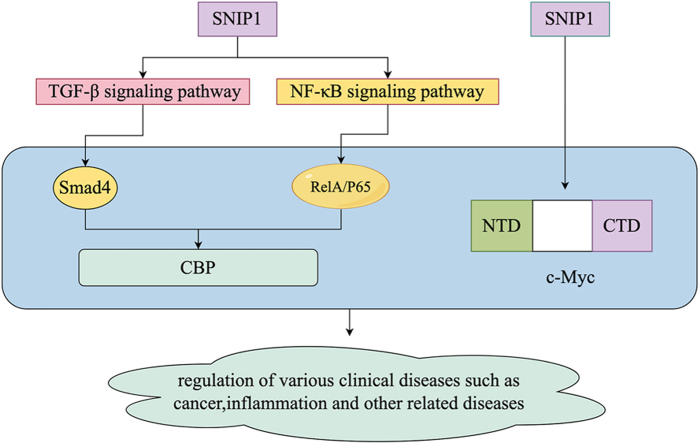
Abstract
Smad intranuclear binding protein 1 (SNIP1), a highly conserved nuclear protein, functions as a transcriptional regulator and exerts a significant influence on disease progression. In addition, the N-terminal domain of SNIP1 facilitates its interaction with Smad4, a signaling protein associated with the TGF-β family, and RelA/p65, a transcription factor connected to NF-κB. This interaction further enhances the transcriptional activation of c-Myc-dependent genes. Presently, the primary emphasis in research is directed towards targeting the catalytic domain of SNIP1, as it holds promise as a potential therapeutic target for various diseases. While the significance of SNIP1 in pathological mechanisms remains uncertain, this review aims to comprehensively examine the existing literature on the association between SNIP1 and proteins implicated in the regulation of diverse clinical conditions, including cancer, inflammation, and related diseases.
Keywords: SNIP1, C-Myc, NF-κB pathway, TGF-β pathway
Graphical abstract
Highlights
-
•
SNIP1 exerts significant influence on disease progression.
-
•
The mechanism of SNIP1 is described.
-
•
This article presents a comprehensive review of SNIP1 and its related regulatory factors in modulating clinical diseases.
1. Introduction
SNIP1 is a protein composed of 396 amino acids, featuring a bipartite nuclear localization signal (NLS) at its N-terminus and an FHA domain at its C-terminus. The FHA domain, which is a phosphothreonine recognition motif found in various nuclear proteins, is postulated to possess a docking function akin to the recognition site of the modular phosphotyrosine domain [1,2]. The FHA domain of SNIP1 is accountable for cellular damage repair, cell cycle regulation, and apoptosis induction. Moreover, the N-terminal region of SNIP1 harbors a nuclear localization signal, which strengthens its association with Smad4 and RelA/p65. Additionally, the C/H1 domain of SNIP1 has the potential to bind CBP/p300 transcriptional coactivators. SNIP1, a member of the Smad transcription factor family, demonstrates expression in various mammalian cell types and plays a crucial role in regulating cell growth, proliferation, and differentiation [2]. Following the inhibition of SNIP1 expression via siRNA interference, a subsequent decrease in the levels of c-Myc and cyclin D1 was observed [3]. This observation implies that the downregulation of SNIP1 gene expression coincided with a concurrent reduction in the expression of c-Myc and cyclin D1, ultimately leading to G1 arrest and apoptosis [3]. Prior investigations have documented the pivotal involvement of c-Myc, an oncogene, in the modulation of various cellular processes, such as cell growth, differentiation, and apoptosis.
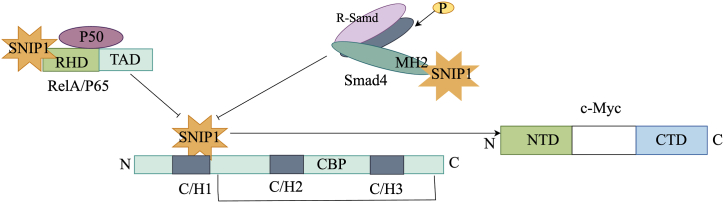
SNIP1 is frequently upregulated in both animal and human tumors, with a 15 % increase in expression observed in human tumors [4]. Its role involves enhancing the transcriptional activity of the proto-oncogene c-Myc [5]. Through its binding to the N-terminal of c-Myc, SNIP1 hinders the ubiquitination process, thereby promoting the stability of the c-Myc protein. This interaction facilitates the interaction of c-Myc with the transcription enhancer P300, and promotes c-Myc transcriptional activity. As a result, elevated transcriptional of c-Myc increases proliferation, migration, and invasiveness of cancer cells [5]. The downregulation of the SNIP1 gene has been shown to result in a decrease in the expression of the cyclin D1 gene and its promoter activity, ultimately causing cell cycle arrest at the G1 phase [6]. Additionally, Fig. 1 illustrates the model depicting the binding sites of SNIP1.
Fig. 1.
Model of SNIP1 binding sites.
The N-terminal region of SNIP1 is capable of direct interaction with various factors through the utilization of the RHD domain to bind to RelA/p65 and the MH2 domain to bind to Smad4. Moreover, it impedes the interaction between the C-terminal activation domain of RelA/p65 and inhibits the connection between Smad4 and the C/H1 region of CBP/p300, ultimately resulting in the inhibition of transcriptional activity. SNIP1 exhibits the ability to significantly enhance the transcriptional activity and stability of c-Myc by attaching to the N-terminus of c-Myc via the FHA domain located on the C-terminus.
2. Structure and function of SMAD proteins
The SMAD group consists of transcription factors that serve as substrates for TGF-β receptors and are present in vertebrates, insects, and nematodes [3].
The SMAD proteins, with molecular weights ranging from 42 to 60 kDa, possess conserved MH1 and MH2 functional domains at their N-terminal and C-terminal ends, respectively. These domains exhibit interactivity and exert inhibitory effects [7]. Furthermore, these regions demonstrate high conservation across various species. Activation of serine/threonine receptors induces the formation of heterotrimeric SMAD complexes, which subsequently translocate from the cytoplasm to the nucleus and initiate the transcription of downstream target genes [8]. Smad proteins play crucial roles in a multitude of intricate biological processes, including cellular differentiation. These proteins exhibit a wide range of biological activities when interacting with various signaling proteins [9]. For instance, they possess the capability to modulate the response of specific genes to TGF-β by forming long-lasting protein-protein interactions with additional transcription factors [10]. The regulation of cellular growth entails the involvement of SMAD proteins, wherein BR-Smad binding to diverse signaling proteins. Co-Smad, a ubiquitous SMAD, forms complexes with R-Smad to govern the transcription and translation processes of nuclear target genes.
The TGF-β superfamily encompasses approximately 30 mammalian proteins, which can be classified into two primary groups: activated AR-Smad (Activin/R-Smad) and BR-Smad (BMP/R-Smad) [11,12]. AR-Smad interacts with various inhibitors or activators to regulate the transcription of nuclear target genes, while BR-Smad binds to additional signaling proteins. A total of 10 SMAD proteins, namely SMAD1-SMAD10, have been identified. These SMAD proteins can be further categorized into three groups: the universal/common type (SMAD4), the receptor-activated type (SMAD1, SMAD2, SMAD3, SMAD5, SMAD8), and the inhibitory type (SMAD6, SMAD7) [13].
Additional research is required to investigate the classification, structure, and function of SMAD9 [14]. Abnormalities in the TGF-β signaling network system often leads to various clinical diseases, such as tumors, bone dysplasia, and vascular disease [15,16]. Previous studies have shown that the Type I and Type II receptors of serine/threonine kinase exhibit an affinity for the TGF-β superfamily. Upon binding to the Type I receptor, the ligands stimulate for activating SMAD proteins. A receptor-modulating SMAD (R-SMAD) acts as the immediate substrate for the type I receptor. The bone morphogenetic protein type I receptor is responsible for the activation of SMAD1, SMAD5, SMAD8, and SMAD2. Specifically, SMAD3 is activated by activin and nodal/DA, as well as the type I receptor [17].
3. SNIP1 and SMAD proteins
SNIP1, a nuclear protein companion of Smad, is present in the Drosha complex [18]. In order for the effective processing of pri-miRNA into pre-miRNA to occur, the Arabidopsis homolog of SNIP1, known as DAWDLE (DDL), plays a crucial role by facilitating the recognition and binding of pri-miRNA to the Arabidopsis homolog of Drosha [19]. The reduction of SNIP1 in mammalian cells leads to a decrease in the expression of multiple miRNAs, including miRNA-21. These findings suggest that SNIP1 may play a role in miRNA production by interacting with the Smad protein to enhance the processing of the Drosha enzyme [19].
4. Possible mechanism of SNIP1
The N-terminal domain of SNIP1 interacts with Smad4, a protein involved in TGF-β family signaling, as well as RelA/p65, a transcription factor associated with NF-κB. Additionally, SNIP1's identical field binds to CBP/P300's C/H1 domain, competing with the attachment of Smad4 and RelA/p65 to CBP/P300 [1]. The overexpression of SNIP1 or its N-terminal region inhibits the transcriptional activation of Smad4 and NF-κB, potentially by interfering with their interaction with CBP/P300. This highlights the significance of SNIP1 as a suppressor of various transcriptional pathways in the C/H1 domain that depend on CBP/P300 [1]. Moreover, SNIP1 exhibits the potential to enhance the transcriptional activation of genes that rely on c-Myc. This is achieved through direct interaction with c-Myc. SNIP1 prevents the degradation of c-Myc by the proteasome, thereby maintaining its stability. Furthermore, SNIP1 aids in the formation of the c-Myc/P300 complex, and augmenting the transcriptional efficacy of c-Myc.
4.1. SNIP1 interacts with TGF-β
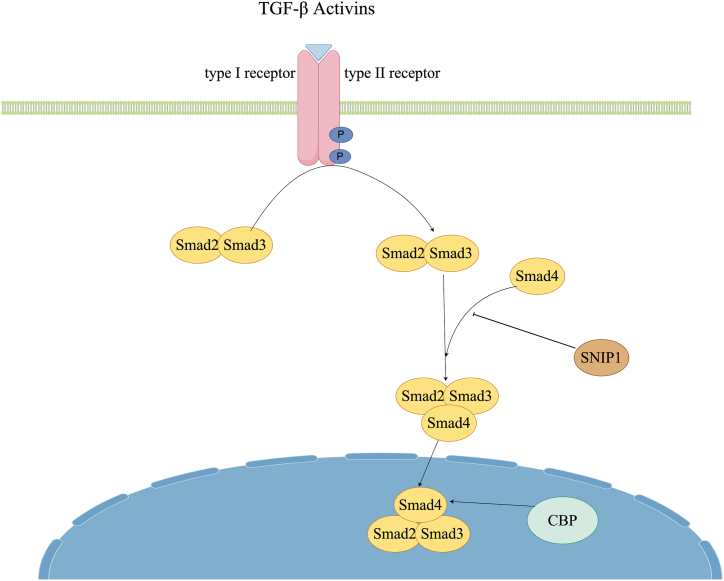
In TGF-β signaling pathway, SNIP1 interacts with Smad4 to inhibit the recruitment of CBP/P300 by Smad4. This interaction hinders the activation of downstream genes in the TGF-β pathway, thereby impeding the signaling pathway [20]. CBP and P300, possessing nearly identical structure and function, form a coactivator family known as CBP/P300. Within the cell, CBP and P300 bind to multiple transcription factors, thereby assisting in the activation of downstream genes [1]. CBP/P300 possesses histone acetyltransferase (HAT) activity that is involved in promoting the initiation of transcription by loosening the chromatin structure. Additionally, it has the ability to interact with Smad4 and act as an intermediary in the transcriptional process of downstream genes facilitated by Smad4 (Fig. 2).
Fig. 2.
SNIP1 can inhibit the recruitment of Smad4's coactivator CBP/P300 by interacting with Smad4 in the TGF-β signaling pathway.
TGF-β engages in interactions with two pairs of transmembrane serine/threonine protein kinases, namely TGFBR1 (type I receptor) and TGFBR2 (type II receptor), to establish receptor complexes. TGFBR2 is responsible for the phosphorylation and subsequent activation of TGFBR1 kinase, which subsequently phosphorylates the transcription factors SMAD2 and SMAD3. Following phosphorylation, these SMADs undergo trimerization with SMAD4 and translocate into the nucleus to facilitate the transcriptional activation of target genes. However, the interaction between SNIP1 and Smad4 can disrupt the binding of Smad4 to CBP/p300, leading to insufficient binding.
4.2. SNIP1 interacts with NF-κB
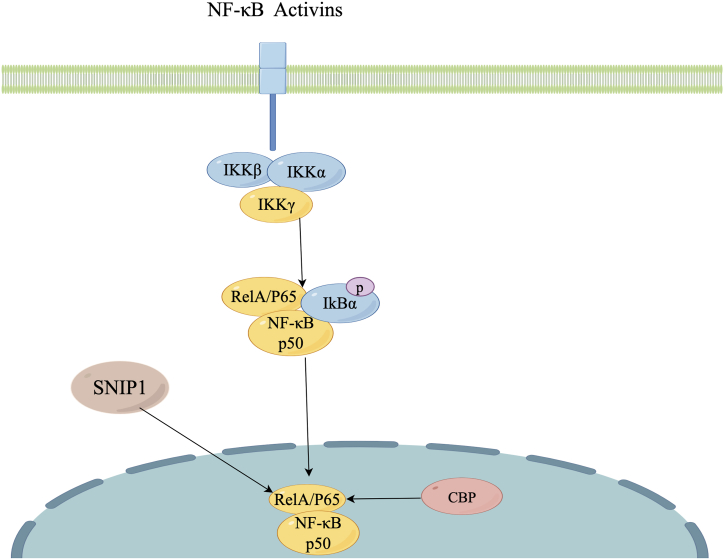
The suppressive effect on NF-κB activity by SNIP1, which interacts with the C/H1 domain of CBP/P300 as depicted in Fig. 3, is noteworthy. Nevertheless, SNIP1 does not impede the activity of transcription factors, such as p53, that bind to other important regions of P300. It also does not affect factors, such as VP16, that do not rely on co-activators [1]. The inhibitory effect of SNIP1's N-terminal domain on NF-κB activity is mediated through its competition with the NF-κB subunit RelA/P65 for P300 binding, similar to its interference with Smad signal transmission [21]. Immunohistochemical analysis demonstrates the precise regulation of SNIP1 expression during development. Moreover, the observation of nuclear staining for RelA/P65 and P300 in specific tissues suggests their potential involvement in the spatially and temporally controlled regulation of NF-κB activity.
Fig. 3.
Within the NF-κB signaling pathway, SNIP1 demonstrates the capacity to interact with RelA/P65.
Inflammatory mediators, including proinflammatory cytokines, lipopolysaccharide (LPS), growth factors, and antigen receptors, initiate the activation of an IKK complex that comprises IKKβ, IKKα, and IKKγ. This complex then phosphorylates IκB proteins, which binding to NF‐κB, preventing their nuclear translocation and binding to DNA. The phosphorylation of IκB causes its own ubiquitination and subsequent degradation through the proteasome, leading to the liberation of the NF-κB/Rel complex. The active NF-κB/Rel complex undergoes further activation through posttranslational modifications such as phosphorylation, acetylation, and glycosylation. Subsequently, it translocates into the nucleus and, in conjunction with CBP or other transcriptional factors, exerts its transcriptional regulatory functions. Furthermore, SNIP1 engages in competition with CBP for the binding to RelA/p65, resulting in the inhibition of NF-κB activity.
4.3. SNIP1 regulates the activity of c-Myc
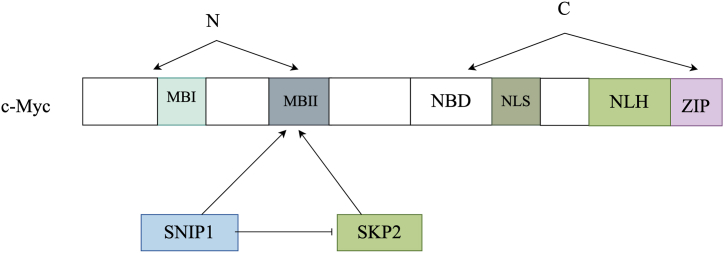
In recent years, there has been a notable surge in the quantity of proteins that show a binding affinity for c-Myc. c-Myc is a vital regulator of cellular growth and development. The domain map analysis demonstrated that the C-terminal FHA domain of SNIP1, along with its adjacent residues, interacts with 147 amino acids located at the N-terminus of c-Myc, resulting in an enhancement of c-Myc-dependent gene transcription (Fig. 4) [22]. This effect is closely related to SNIP1's recruitment with the c-Myc gene,and the association of p300/CBP coactivator with SNIP1. Moreover, the SNIP1/c-Myc complex was detected using the yeast two-hybrid screening technique.
Fig. 4.
The activity of c-Myc is controlled by SNIP1.
c-Myc's N-terminal region contains the Myc boxes (MBI and MBII), which are crucial for its transcriptional activity. Conversely, the C-terminal contains a nonspecific DNA-binding domain (NDB), a nuclear localization signal (NLS), and a helix-loop-helix (HLH) motif. The FHA-containing C terminus of SNIP1 interacts with the N-terminal region of c-Myc, thereby enhancing its transcriptional activity. S phase kinase-associated protein 2 (Skp2) facilitates c-Myc ubiquitylation by binding to the MBII domain, regulating the degradation of c-Myc through the proteasome pathway. Both SNIP1 and Skp2 compete for binding to c-Myc.
SNIP1 has been observed to interact with the C-terminus of c-Myc, thereby enhancing its transcriptional capacity by stabilizing c-Myc and impeding its degradation. Furthermore, SNIP1 acts as a bridge, connecting the c-Myc/P300 complex. In vitro transformation experiments have indicated that the interaction between SNIP1, c-Myc target genes, and the H-Ras oncogene may contribute to a synergistic effect on tumor progression and anchorage-independent growth [23]. The strong correlation observed between SNIP1 and c-Myc staining in a tissue microarray of non-small cell lung cancer provides further evidence supporting SNIP1 and c-Myc interaction. This suggests that SNIP1 may play a significant role as a regulator of c-Myc function in tumor progression.
Additionally, it was found that SNIP1 competes with Skp2 for binding to c-Myc [24]. The contrasting effects of Skp2 and SNIP1 on c-Myc protein levels were also observed. Skp2 functions as a component responsible for substrate recognition in the SCF E3 ubiquitin ligase, thereby facilitating the ubiquitination and degradation of c-Myc. In contrast, it has been observed that SNIP1 exerts a suppressive effect on the ubiquitination process of c-Myc. Previous investigations have revealed a competitive interaction between Skp2 and SIP1 for binding to c-Myc through its MBII domain. This suggests that specific regions within MBII play a crucial role in regulating c-Myc stability through SNIP1. Moreover, recent studies have demonstrated that SNIP1 is capable of recruiting TET2 to the promoter regions of c-Myc target genes. Those targets are involved in DNA damage response and cell survival [25]. Notably, SNIP1-dependent activation of TET2 serves as a protective mechanism against apoptosis induced by DNA damage. The relationship between epigenetic regulation and the maintenance of genomic stability is elucidated by the participation of the TET2-SNIP1-c-Myc pathway in the response to DNA damage [26].
5. Application of SNIP1 in clinical diseases
The regulatory function of SNIP1 significantly impacts the development of various diseases. Recent studies suggest that SNIP1 is closely associated with the progression, management, and postoperative outcomes of specific malignancies and inflammatory conditions. The reduction of SNIP1 gene expression in embryos through siRNA has been found to potentially lead to fetal death. Additionally, SNIP1 may also exerts an influence on tumor development and therapy by regulating tyrosine phosphorylation of their target proteins. Furthermore, SNIP1 plays a vital role in regulating the activity of the prominent oncogene c-Myc. Additionally, it has been observed that an increased expression level of SNIP1 leads to the competitive binding of SNIP1 and RB [26].
5.1. The roles of SNIP1 in cancer
SNIP1 serves as a transcriptional regulator in tumor development, involved in various cellular processes such as miRNA generation, cellular growth, DNA damage repair, and multiple signaling pathways [27]. The mechanism of SNIP1involved in tumor development may be a target for cancer therapy.
The suppression of SNIP1 expression in NSCLC cells by miR-138-5p has been found to impede the expression of full-length SNIP1 or its amino terminus [28].What is more, SNIP1 is a non-histone substrate of lysine methyltransferase KMT5A, which undergoes KMT5A-mediated mono-methylation to promote breast cancer cell growth and invasion. KMT5A-mediated K301 methylation of SNIP1 inhibits the hippo protein kinase cascade through transcriptional activation of Mark 4, enhancing the metastasis of triple-negative breast cancer (TNBC) [29]. It enriches our understanding of the role of SNIP1 in cancer dissemination and provides a novel basis for the clinical intervention of breast cancer. N6-methyladenosine mediate the progress of LncRNA BCAN-AS1 binding to SNIP1, and LncRNA BCAN-AS1- SNIP1 complex can enhance the stability of c-Myc [30]. This interaction facilitates the progression of pancreatic cancer [31]. In addition, functional studies showed that overexpression of miR-29a-3p inhibited the migration and proliferation of cervical cancer HeLa cells, and miR-29a-3p down-regulated the mRNA expression of SNIP1 downstream genes, including HSP27, c-Myc, and cyclin D1 [32,33]. Moreover, Over-expression of SNIP1 promoted cell invasion and migration in osteosarcoma cells. Interestingly, elevated levels of SNIP1 have been detected in osteosarcoma tissues and cell lines. The upregulation of miR-335 has been observed in osteosarcoma, and its overexpression has been shown to inhibit the proliferation and metastasis of osteosarcoma both in vitro and vivo models [34]. Recent research findings suggest that MKRN1 is significantly upregulated in CRC and activates the TGF-β signaling pathway through the process of ubiquitination and subsequent degradation of SNIP1.This, in turn, enhances epithelial-mesenchymal transition (EMT) and facilitates the movement of CRC cells [35].
Thus, SNIP1 can directly or indirectly regulates gene transcription associated with the progression of various human cancers and tumorigenesis (Table 1).
Table 1.
SNIP1 linked to Cancers.
| Diseases | Methods | Markers | Conclusion | Reference |
|---|---|---|---|---|
| Non-small cell lung cancers (NSCLCs) | The expression of miR-138-5p and SNIP1 were analyzed in lung cancer serum samples. The effects of miR-138-5p on cell proliferation and metastasis were investigated by CCK-8, colony formation tests, Western blot were used to detect the protein expression of SNIP1 and related genes. | Cyclin D1 E-cadherin GAPDH SNIP1 miR-138-5p c-Myc |
SNIP1 competes with HDAC bind to RB and decrease HDAC activity in vitro. Down-regulation of SNIP1 decreased the colony-forming ability of lung cancer cells. What is more, miR-138-5p suppressed lung cancer cell proliferation and migration by targeting SNIP1. | [28] |
| Triple-negative breast cancer (TNBC) | Mass spectrometry (MS) analysis to identify KMT5A. Coimmunoprecipitation assay and LC-MS/MS analysis SNIP1-interacting proteins from MDA-MB-231 cells. Western blot and qPCR were used to detect the protein expression of SNIP1 |
MARK4 c-MYC KAT2A SNIP K301 KMT5A |
Methylation of SNIP1 by KMT5A enhances key oncogenic pathway-Hippo signaling, thereby promoting TNBC metastasis. KMT5A-mediated SNIP1 methylation could be regarded as an essential step for c-MYC/KAT2A complex formation and c-MYC target activation. | [29] |
| (Supplemental). SNIP1 linked to Cancers | ||||
|---|---|---|---|---|
| Diseases | Methods | Markers | Conclusion | Reference |
| Pancreatic ductal adenocarcinoma (PDAC) | Surgically removed PDAC samples. The c-Myc RIP-seq analysis was performed on PDAC cells,and a panel of c-Myc-associated lncRNAs was identified. | BCAN-AS1 c-Myc SNIP1 SKP2 PAICS MCM7 HADHB MYO18A KIF5B |
SNIP1 binds to BCAN-AS1 by recognizing its m6A modification. This progress block ubiquitination and degradation of c-Myc mediated by S-phase kinase-associated protein 2 (SKP2), then effectively inhibit tumor growth and metastasis. | [31] |
| Cervical cancer | The expression of miR-29a-3p and SNIP1 were analyzed in cervical cancer HeLa cells. The protein expression of SNIP1 and related genes were measured through Western blot and RT-qPCR. | SNIP1 HSP27 c-Myc cyclin D1 MMP9 MAPK1 N-cadherin E-cadherin CDK2 |
miR-29a-3p suppressed the migration and proliferation in HeLa cells by directly targeting SNIP1.And miR-29a-3p/SNIP1 axis could provide new insight into the development of cervical cancer. | [32] |
| Osteosarcoma | Use the qRT-PCR assessed circUSP48, miR-335 and SNIP1 expression in OS cell lines and tissues. Sanger sequencing, RNase R processing and FISH detection were performed for circUSP48 validation. | SNIP1 MMP-2 MMP-7 c-Myc NF-κB p65 UBE2G1 KRTAP10-1 CRTAC1 PEBP4 |
SNIP1 is a direct target of miR-335, and miR-335 inhibit SNIP1exprssion. Inaddition, overexpression of SNIP1 weakens the inhibitory effect of miR-335. SNIP1 may be a potential target for future osteosarcoma treatment. | [34] |
| Colorectal cancer (CRC) | Tumour tissues and matched normal fresh tissues were collected from three CRC patients. Immunohistochemistry, western blotting, and quantitative real-time PCR analysis were performed to test the level of related index. | MKRN1 SNIP1 TRA2A LUC7L HT29 HCT116 HCT15 RKO |
SNIP1 exhibits potential as a significant biomarker in preventing and treating colorectal cancer. Additionally, the MKRN1/SNIP1/TGF-β pathway might be regarded as a novel target for the development of anti-metastatic therapeutics in CRC. | [35] |
5.2. SNIP1 and inflammatory disorders
SNIP1 has primarily been characterized as a facilitator of TGF-β-triggered genetic manifestation, exhibiting properties that effectively mitigate inflammation [35].The cloning and characterization of the SNIP1 protein have revealed its ability to inhibit the TGF-β signal transduction pathway. Moreover, there is evidence indicating a significant decrease in SNIP1 levels in peripheral blood mononuclear cells of IBD patients [36]. Additionally, SNIP1 has been found to impede the recruitment of Smad4 into the coactivator CBP/P300 and TGF-β downstream genes to alleviate acute pancreas injury [37]. Furthermore, overexpression SNIP1 reduced the expression of TNF-α and IL-6, alleviated ECM degradation, reduced the phosphorylation levels of p65 and IκBα, and alleviated osteoarthritis(OA) inflammation [38]. Moreover, recent investigations have revealed that under conditions of excessive stress, the absence of SNIP1 accelerates the development of pathological cardiac hypertrophy by activating the NF-κB pathway [39,40]. In contrast, overexpression of SNIP1 in cardiomyocytes has been shown to effectively exacerbate pathological cardiac remodeling by blocking the NF-κB signaling pathway. (Table 2).
Table 2.
SNIP1 associated with inflammatory diseases.
| Diseases | Methods | Markers | Conclusion | Reference |
|---|---|---|---|---|
| Inflammatory bowel disease (IBD) | Colonoscopic and ileoscopic biopsies were collected from inflamed and unaffected sites of the colons patients. Intestinal barrier function was measured. Total RNA was extracted from primary IEC, and immunohistochemical staining was used for SNIP1. |
TNF-α IFN-γ IL-17A IL-23 SNIP1 protein and mRNA IκBα |
SNIP1 plays a crucial role in maintaining the integrity of the intestinal epithelium and reducing inflammation in the intestinal mucosa. Moreover, SNIP1 inhibits proinflammatory signaling in colonic epithelial cells through suppressing NF-κB activation. | [36] |
| Acute pancreas (AP) injury | Establish AP model and treated with modafinil. The relevant indexes were detected by histopathology and cell biology, including Hematoxylin and Eosin (H&E) staining, RT-qPCR,CCK8 assay and ELISA. |
TNF-α IL-6 CRP SOD MDA Amylase and Lipase |
The sumoylation-mediated modification of SNIP1 leads to a reduction in downstream TGF-β inhibitory genes. Modafinil treatment in an AP rat model significantly restored the expression of SNIP1, resulting in a reduction in inflammation, oxidative stress, and apoptosis. | [37] |
| Table2 (supplemental). SNIP1 Associated with Inflammatory Diseases | ||||
|---|---|---|---|---|
| Diseases | Methods | Markers | Conclusion | Reference |
| Osteoarthritis (OA) | The expression of SNIP1 was confirmed by clinical trials and animal experiments. Inflammatory cytokines and NF-κB related mediators were observed | SNIP p-IκBα IκBα NF-κB p65 TNF-α IL-6 MMP13 collagen-II |
SNIP1 alleviated OA and repressed inflammation by inhibiting the activation of NF-κB. This study might provide a new insight into OA treatment. | [38] |
| Cardiac Hypertrophy | Gain-of-function and loss-of-function methodologies were utilized to explore the role of SNIP1 in the progression of cardiac hypertrophy. | Atrial natriuretic peptide (ANP) β-myosin heavy chain (β-MHC) IKKβ NF-κB p65IκBα |
SNIP1 contributes to hemodynamic overload and adverse cardiac remodeling in the presence of AngII by modulating the NF-κB pathway And overexpression of SNIP1 diminishes the hypertrophic effect on NRCMs caused by AngII. |
[40] |
5.3. SNIP1 and cell apoptosis
The regulation of stem cell survival and apoptosis is a fundamental aspect of developmental processes. It serves as a critical mechanism for maintaining the integrity of morphogenesis, size, genomic stability, and cellular fate. Despite the prevalence of programmed cell death in development, the underlying mechanisms governing this process are not fully understood. Recent studies have highlighted the significance of SNIP1 in the viability of neural precursor cells and the generation of new neurons, underscoring its essential role in brain maturation [41]. Dysplasia and caspase 9-dependent apoptosis were significantly upregulated in the brain lacking SNIP1. SNIP1 exerts mechanistic control over target genes that promote cell viability and neurogenesis [42]. The functionality of SNIP1 is influenced by the TGFβ and NF-κB signaling pathways. Scholars have observed that SNIP1 facilitates neurogenesis and inhibits cell death in the murine brain development model. Further investigations have also implicated SNIP1 in the prevention of cell death [42,43]. Moreover, the presence of SNIP1 has been found to enhance the interaction between the Polycomb complex PRC2 and the genome, leading to the removal of H3K27me3 from specific target genes [44]. It has been demonstrated that removing PRC2 alone reduces apoptosis and brain dysplasia, and partially restores the genetic programming in SNIP1-deficient animals. The precise control of PRC2 and H3K27 markers at specific sites in the developing brain can modulate cell survival and apoptosis. Both SNIP1 and PRC2 collaboratively occupy chromatin targets to regulate genetic programming in the brain. The interaction between PRC2 and SNIP1 plays a crucial role in regulating various cellular processes, including cell cycle progression, programmed cell death, and brain development [45]. However, the specific mechanism by which SNIP1-PRC2 modulates chromatin and coordinates caspase 9-mediated cell death remains incompletely understood, highlighting the need for further investigation. Additionally, mutations in SNIP1 have been associated with neurodevelopmental and psychiatric disorders. Specifically, psychomotor delay, epilepsy, and craniofacial malformations have been associated with the SNIP1 variant E366G [45]. Furthermore, an examination was conducted on publicly available data from the Human Gene Mutation Database and Mastermind, revealing a significant association between epilepsy and cranial dysplasia with a specific SNIP1 variant, namely R111C [46]. Accordingly, SNIP1 plays a crucial role in the intricate development of the human nervous system.
5.4. SNIP1 and cell migration and invasion
SNIP1 demonstrates the capacity to regulate signaling pathways facilitated by ATR, a kinase associated with Ataxia-telangiectasia mutated and Rad3-related functions. Complementary post-translational modifications collaborate with the phosphorylation cascade of the ATR protein to govern cellular responses to genotoxic stress. Investigation has substantiated the crucial involvement of SNIP1 in the tumor suppression mechanism of ATR's p14. Furthermore, SNIP1 amplifies p53 expression and selectively regulates ATR-induced phosphorylation of p53 in the context of Ultraviolet (UV) treatment [47]. Additionally, the interaction between SNIP1 and TET2 enhances the trans-activation of the c-Myc target gene by TET2, facilitating the immobilization of the TGF-β-mediated inhibition signal through the SUMOylation of SNIP1 [48]. SUMOylation of SNIP1 weakens its inhibition of the TGF-β signaling pathways and TGF-β targeted genes, such as PAI-1 and MMP2, resulting in a decrease in TGF-β-regulated cell metastasis and invasion [49]. PAI-1 mainly acts as a promoter during tumor growth. It induces tumor vascularization, promotes cell proliferation and facilitates tumor metastasis. MMP2 is known to degrade the extracellular matrix in tumor development. This degradation can mediate tumor angiogenesis, metastasis, and invasion [49]. Consequently, the loss of inhibition of TGF-β-target genes PAI-1 and MMP2 may ultimately amplify TGF-β-regulated cell migration and invasion. Recent investigations into tumor metastasis have demonstrated that the depletion of E-cadherin is associated with the upregulation of genes within the TGF-β signaling pathway at the transcriptional level. Furthermore,TGF-β downregulates the expression of E-cadherin. With an increased concentration of TGF-β1, the expression level of E-cadherin gradually decreases. What is more, the colony formation of E-cadherin-negative cells was rescued by inhibiting TGF-β-induced transcription [50].
6. Prospection
The significance of SNIP1 in the identification and management of various illnesses has been established, making it a promising candidate for therapeutic interventions. Currently, the focus lies on the catalytic SNIP1 domain in design, and the discovery of a specific inhibitory compound for SNIP1 holds potential as a valuable target for disease treatment. Moving forward, it is imperative to conduct comprehensive examinations, investigations, and evaluations to fully understand and explore the medically significant applications of SNIP1. In order to fully comprehend the influence of SNIP1 on cancer and other diseases, it is crucial to utilize all-sided methodologies such as proteomics to uncover all protein substrates acknowledged by SNIP1. This would provide valuable insights into the mechanisms that underlie the gene transcription process mediated by SNIP1.
Funding
This research was funded by the National Natural Science Foundation of China (No. 81871785), the Science and Technology Research Project of Shandong Geriatric Association (No. LKJGG2021W093) and Scientific and Technological Research Plan of Tai'an (2021NS142), Scientific and Technological Research Plan of Tai'an (2022NS220), Scientific and Technological Research Plan of Tai'an (2021NS137)
Data availability statement
This is a review article,no data was used.
CRediT authorship contribution statement
Yinzhong Chen: Writing – review & editing, Writing – original draft, Investigation, Funding acquisition. Wei Guo: Visualization, Investigation, Funding acquisition. Xiucheng Guo: Investigation. Qiao Wanqing: Visualization, Investigation, Funding acquisition. Zongsheng Yin: Writing – review & editing, Visualization, Supervision, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Kim R.H., Wang D., Tsang M., Martin J., Huff C., de Caestecker M.P., Parks W.T., Meng X., Lechleider R.J., Wang T., Roberts A.B. A novel smad nuclear interacting protein, SNIP1, suppresses p300-dependent TGF-beta signal transduction. Genes Dev. 2000;14(13):1605–1616. [PMC free article] [PubMed] [Google Scholar]
- 2.Kim R.H., Flanders K.C., Reffey S.B., Anderson L.A., Duckett C.S., Perkins N.D., Roberts A.B. SNIP1 inhibits NF-kappa B signaling by competing for its binding to the C/H1 domain of CBP/p300 transcriptional co-activators. J. Biol. Chem. 2001;276(49):46297–46304. doi: 10.1074/jbc.M103819200. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh A.K., Yuan W., Mori Y., Varga J. Smad-dependent stimulation of type I collagen gene expression in human skin fibroblasts by TGF-beta involves functional cooperation with p300/CBP transcriptional coactivators. Oncogene. 2000;19(31):3546–3555. doi: 10.1038/sj.onc.1203693. [DOI] [PubMed] [Google Scholar]
- 4.Bracken C.P., Wall S.J., Barré B., Perkins N.D. Regulation of cyclin D1 RNA stability by SNIP1. Cancer. 2008;Res68(18):7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii M., Lyakh L.A., Bracken C.P., Fukuoka J., Hayakawa M., Tsukiyama T., Soll S.J., Harris M., Rocha S., Roche K.C., Tominaga S.I., Jen J., Perkins N.D., Lechleider R.J., Roberts A.B. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24(5):771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 6.Larsson L.G. SNIP1: Myc's new helper in transcriptional activation. Mol Cell. 2006;24(6):811–812. doi: 10.1016/j.molcel.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature425(6958) 2003:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H., Wen J.Y., Yu J.J., Liao J.Y., Liu S., Cai N., Liang H.F., Chen X.P., Ding Z.Y., Zhang B.X. MicroRNA-148a-3p inhibits progression of hepatocelluar carcimoma by repressing SMAD2 expression in an Ago 2 dependent manner. J Exp Clin Cancer. 2020;Res39(1):150. doi: 10.1186/s13046-020-01649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jadhao M., L Chen C., Liu W., Deshmukh D., Liao W.T., Chen J.Y., Urade R., Tsai E.M., Hsu S.K., Wang L.F., Chiu C.C. Endoglin modulates TGFβR2 induced VEGF and proinflammatory cytokine Axis mediated angiogenesis in Prolonged DEHP-exposed breast cancer cells. Biomedicines10. 2022;(2):417. doi: 10.3390/biomedicines10020417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S.N., Wu J.F. TGF-β/SMAD signaling regulation of mesenchymal stem cells in adipocyte commitment. Stem Cell Res Ther11. 2020;41(1) doi: 10.1186/s13287-020-1552-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Söderström L.Å., Tarnawski L., Olofsson P.S. CD137: a checkpoint regulator involved in atherosclerosis. Atherosclerosis. 2018;272:66–72. doi: 10.1016/j.atherosclerosis.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Hata A., Chen Y.G. TGF-Β signaling from receptors to smads. Cold Spring Harb Perspect Biol. 2016;8(9):a022061. doi: 10.1101/cshperspect.a022061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pangas S.A. Bone morphogenetic protein signaling transcription factor (SMAD) function in granulosa cells. Mol. Cell. Endocrinol. 2012;356(1–2):40–47. doi: 10.1016/j.mce.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai J., Pardali E., Sánchez-Duffhues G., ten Dijke P. BMP signaling in vascular diseases. FEBS Lett. 2012;586(14) doi: 10.1016/j.febslet.2012.04.030. 1993–2002. [DOI] [PubMed] [Google Scholar]
- 15.Santibanez J.F., Quintanilla M., Bernabeu C. TGF-beta/TGF-beta receptor system and its role in physiological and pathological conditions. Clin. Sci. (Lond.) 2011;121:233–251. doi: 10.1042/CS20110086. [DOI] [PubMed] [Google Scholar]
- 16.Henderson N.C., Arnold T.D., Katamura Y., Giacomini M.M., Rodriguez J.D., McCarty J.H., Pellicoro A., Raschperger E., Betsholtz C., Ruminski P.G., Griggs D.W., Prinsen M.J., Maher J.J., Iredale J.P., Lacy-Hulbert A., Adams R.H., Sheppard D. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nature medicine. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murakami M., Kawachi H., Ogawa K., Nishino Y., Funaba M. Receptor expression modulates the specificity of transforming growth factor-beta signaling pathways. Genes. 2009;Cells14(4):469–482. doi: 10.1111/j.1365-2443.2009.01283.x. [DOI] [PubMed] [Google Scholar]
- 18.Batlle E., Massague J. Transforming growth factor-beta signaling in immunity and cancer. Immunity. 2019;50:924–940. doi: 10.1016/j.immuni.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garg V., Agarwal G., Pazhamala L.T., Nayak S.N., Kudapa H., Khan A.W., Doddamani D., Sharma M., Kavi Kishor P.B., Varshney R.K. Genome-wide identification, characterization, and expression analysis of small RNA biogenesis purveyors reveal their role in regulation of biotic stress responses in three legume crops. Front. Plant Sci. 2017;8:488. doi: 10.3389/fpls.2017.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q., An J., Liu X., Zhang M., Ling Y., Wang C., Zhao J., Yu L. SNIP1: a new activator of HSE signaling pathway. Mol. Cell. Biochem. 2012;362(1–2):1–6. doi: 10.1007/s11010-011-1120-y. [DOI] [PubMed] [Google Scholar]
- 21.Ikeda A., Sun X., Li Y., Zhang Y., Eckner R., Doi T.S., Takahashi T., Obata Y., Yoshioka K., Yamamoto K. p300/CBP-dependent and -independent transcriptional interference between NF-kappa B RelA and p53. Biochem. Biophys. Res. Commun. 2000;272(2):375–379. doi: 10.1006/bbrc.2000.2786. [DOI] [PubMed] [Google Scholar]
- 22.Bracken C.P., Wall S.J., Barré B., Panov K.I., Ajuh P.M., Perkins N.D. Regulation of cyclin D1 RNA stability by SNIP1. Cancer Res. 2008;68(18):7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roche K.C., Wiechens N., Owen-Hughes T. The FHA domain protein SNIP1 is a regulator of the cell cycle and cyclin D1 expression. Oncogene. 2004;23(50):8185–8195. doi: 10.1038/sj.onc.1208025. [DOI] [PubMed] [Google Scholar]
- 24.Fujii M., Lyakh L.A., Bracken C.P., Fukuoka J., Hayakawa M., Tsukiyama T., Soll S.J., Harris M., Rocha S., Roche K.C., Tominaga S.I., Jen J., Perkins N.D., Lechleider R.J., Roberts A.B. SNIP1 is a candidate modifier of the transcriptional activity of c-Myc on E box-dependent target genes. Mol Cell. 2006;24(5):771–783. doi: 10.1016/j.molcel.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Chen L.L., Lin H.P., Zhou W.J., He C.X., Zhang Z.Y., Cheng Z.L., Song J.B., Liu P., Chen X.Y., Xia Y.K., Chen X.F., Sun R.Q., Zhang J.Y., Sun Y.P., Song L., Liu B.J., Du R.K., Ding C., Lan F., Huang S.L., Zhou F., Liu S., Xiong Y., Ye D., Guan K.L. SNIP1 recruits TET2 to regulate c-Myc target genes and cellular DNA damage response. Cell Rep. 2018;25(6):1485–1500. doi: 10.1016/j.celrep.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon H.S., Choi Y.Y., Fukuoka J., Fujii M., Lyakh L.A., Song S.H., Travis W.D., Park J.Y., Jen J. High expression of SNIP1 correlates with poor prognosis in non-small cell lung cancer and SNIP1 interferes with the recruitment of HDAC1 to RB in vitro. Lung Cancer. 2013;82(1):24–30. doi: 10.1016/j.lungcan.2013.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Cui H.H., Yi H.Y., Bao H.Y., Tan Y., Tian C., Shi X.Y., Gan D.W., Zhang B., Liang W.Z., Chen R., H Zhu Q., Fang L., Gao X., Huang H.D., Tian R.J., Sperling S.R., Hu Y.H., Chen W. The SWI/SNF chromatin remodeling factor DPF3 regulates metastasis of ccRCC by modulating TGF-β signaling. Nat. Commun. 2022;13(1):4680. doi: 10.1038/s41467-022-32472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J., Han X., Yang X., Li Y., Liang Y., Sun G., Wang R., Wang P., Xie S., Feng J., Sun H. MiR-138-5p suppresses the progression of lung cancer by targeting SNIP1. Thorac Cancer. 2023;14(6):612–623. doi: 10.1111/1759-7714.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu B., Su J., Shi Q., Liu Q., Ma J., Ru G., Zhang L., Zhang J., Hu X., Tang J. KMT5A-methylated SNIP1 promotes triple-negative breast cancer metastasis by activating YAP signaling. Nat. Commun. 2022;13(1):2192. doi: 10.1038/s41467-022-29899-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhong Y., Yang L., Xiong F., He Y., Tang Y., Shi L., Fan S., Li Z., Zhang S., Gong Z., Guo C., Liao Q., Zhou Y., Zhou M., Xiang B., Li X., Li Y., Zeng Z., Li G., Xiong W. Long non-coding RNA AFAP1-AS1 accelerates lung cancer cells migration and invasion by interacting with SNIP1 to upregulate c-Myc. Signal Transduct Target Ther. 2021;6(1):240. doi: 10.1038/s41392-021-00562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu G.D., Su J.C., Zeng L.X., Deng S., Huang X.D., Ye Y., Li R., Bai R.H., Zhuang L.H., Li M., Zhou Q.B., Zheng Y.F., Deng J.G., Zhang S.P., Chen R.F., Lin D.X., Zhang J.L., Zheng J. LncRNA BCAN-AS1 stabilizes c-Myc via N6-methyladenosine-mediated binding with SNIP1 to promote pancreatic cancer. Cell Death Differ. 2023;10:2213–2230. doi: 10.1038/s41418-023-01225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y., Zhang W., Yan L., Zheng P., Li J. miR-29a-3p directly targets Smad nuclear interacting protein 1 and inhibits the migration and proliferation of cervical cancer HeLa cells. PeerJ. 2020;8:e101. doi: 10.7717/peerj.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gong Y., Wan J.H., Zou W., Lian G.Y., Qin J.L., Wang Q.M. MiR-29a inhibits invasion and metastasis of cervical cancer via modulating methylation of tumor suppressor SOCS1. Future Oncol. 2019;15(15):1729–1744. doi: 10.2217/fon-2018-0497. [DOI] [PubMed] [Google Scholar]
- 34.Xie Y., Deng H., Wei R., Sun W., Qi Y., Yao S., Cai L., Wang Y., Deng Z. Overexpression of miR-335 inhibits the migration and invasion of osteosarcoma by targeting SNIP1. Int. J. Biol. Macromol. 2019;133:137–147. doi: 10.1016/j.ijbiomac.2019.04.016. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Li Q.S., Liu H.L., Tang H.T., Yang H.L., Wu D.Q., Huang Y.Y., Li L.C., Liu L.H., Li M.X. MKRN1 promotes colorectal cancer metastasis by activating the TGF-β signaling pathway through SNIP1 protein degradation. J. Exp. Clin. Cancer Res. 2023;42(1):240. doi: 10.1186/s13046-023-02825-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.He C., Shi Y., Wu R., Sun M., Fang L., Wu W., Liu C., Tang M., Li Z., Wang P., Cong Y., Liu Z. miR-301a promotes intestinal mucosal inflammation through induction of IL-17A and TNF-α in IBD. Gut. 2016;65(12):1938–1950. doi: 10.1136/gutjnl-2015-309389. [DOI] [PubMed] [Google Scholar]
- 37.Wu J.W., Lu Y.J., Qin A.C., Li S., Huang B., Jiang X.W., Qiao Z.M. Modafinil ameliorated pancreatic injury and inflammation through upregulating SNIP1. Gen. Physiol. Biophys. 2020;39(4):383–392. doi: 10.4149/gpb_2020016. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y.Z., Guo W., Lu W.Z., Guo X.C., Gao W.L., Yin Z.S. SNIP1 reduces extracellular matrix degradation and inflammation via inhibiting the NF-κB signaling pathway in osteoarthritis. Arch. Biochem. Biophys. 2023;747 doi: 10.1016/j.abb.2023.109764. [DOI] [PubMed] [Google Scholar]
- 39.Yao L., Chen X., Shen M., Zhao Y., Cao Q. Isosteviol attenuates DSS-induced colitis by maintaining intestinal barrier function through PDK1/AKT/NF-κB signaling pathway. Int Immunopharmacol. 2023;114 doi: 10.1016/j.intimp.2022.109532. [DOI] [PubMed] [Google Scholar]
- 40.Lu Y.Y., Xu D.C., Zhao Y.F., Zhu G.F., Zhu M.Y., Liu W.J., Yu X.J., Chen W., Liu Z., Xu Y.W. Smad nuclear interacting protein 1 acts as a protective regulator of pressure overload-induced pathological cardiac hypertrophy. J. Am. Heart Assoc. 2016;5(11) doi: 10.1161/JAHA.116.003943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ammous Z., Rawlins L.E., Jones H., Leslie J.S., Wenger O., Scott E., Deline J., Herr T., Evans R., Scheid A., Kennedy J., Chioza B.A., Ames R.M., Cross H.E., Puffenberger E.G., Harries L., Baple E.L., Crosby A.H. A biallelic SNIP1 Amish founder variant causes a recognizable neurodevelopmental disorder. PLoS Genet. 2021;17 doi: 10.1371/journal.pgen.1009803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng D., Zheng Q., Zhang X., Piao X., Luo L., Jia Y. A molecular brake that modulates spliceosome pausing at detained introns contributes to neurodegeneration. Protein Cell. 2023;14(5):318–336. doi: 10.1093/procel/pwac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Espín-Palazón R., Traver D. The NF-κB family: key players during embryonic development and HSC emergence. Exp. Hematol. 2016;44(7):519–527. doi: 10.1016/j.exphem.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Matsui Y., Djekidel M.N., Lindsay K., Samir P., Connolly N., Wu G., Yang X., Fan Y., Xu B., Peng J.C. SNIP1 and PRC2 coordinate cell fates of neural progenitors during brain development. Nat. Commun. 2023;14(1):4754. doi: 10.1038/s41467-023-40487-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meng G.D., Zheng Q., Zhang X., Piao X., Luo L., Jia Y. A molecular brake that modulates spliceosome pausing at detained introns contributes to neurodegeneration. Protein Cell. 2023;14(5):318–336. doi: 10.1093/procel/pwac008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Z. Ammous, L.E. Rawlins, H. Jones, J.S. Leslie, O. Wenger, E. Scott, J Deline, T. Herr, R. Evans, A. Scheid, J. Kennedy, B.A. Chioza, R.M. Ames, H.E. Cross, E.G. Puffenberger, L Harries, E.L. Baple, A.H.Crosby, A biallelic SNIP1 Amish founder variant causes a recognizable neurodevelopmental disorder. PLoS Genet. 17(92021):e1009803. [DOI] [PMC free article] [PubMed]
- 47.Roche K.C., Rocha S., Bracken C.P., Perkins N.D. Regulation of ATR-dependent pathways by the FHA domain containing protein SNIP1. Oncogene. 2007 Jul 5;26(31):4523–4530. doi: 10.1038/sj.onc.1210233. [DOI] [PubMed] [Google Scholar]
- 48.Chen L.L., Lin H.P., Zhou W.J., He C.X., Zhang Z.Y., Cheng Z.L., Song J.B., Liu P., Chen X.Y., Xia Y.K., Chen X.F., Sun R.Q., Zhang J.Y., Sun Y.P., Song L., Liu B.J., Du R.K., Ding C., Lan F., Huang S.L., Zhou F., Liu S., Xiong Y., Ye D., Guan K.L. SNIP1 recruits TET2 to regulate c-Myc target genes and cellular DNA damage response. Cell Rep. 2018;25(6):1485–1500. doi: 10.1016/j.celrep.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu S., Long J., Yuan B., Zheng M., Xiao M., Xu J., Lin X., Feng X.H. SUMO modification reverses inhibitory effects of smad nuclear interacting protein-1 in TGF-β responses. J. Biol. Chem. 2016;291(47):24418–24430. doi: 10.1074/jbc.M116.755850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., Ewald A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573:439–444. doi: 10.1038/s41586-019-1526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review article,no data was used.