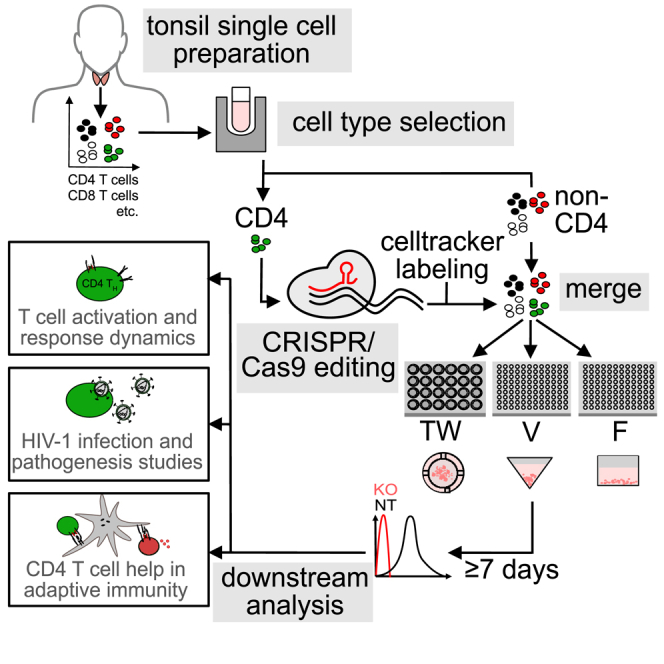
Summary
The molecular and immunological properties of tissue-resident resting CD4 T cells are understudied due to the lack of suitable gene editing methods. Here, we describe the ex vivo culture and gene editing methodology ediTONSIL for CD4 T cells from human tonsils. Optimized CRISPR-Cas9 RNP nucleofection results in knockout efficacies of over 90% without requiring exogenous activation. Editing can be performed on multiple cell types in bulk cultures or on isolated CD4 T cells that can be labeled and reintroduced into their tissue environment. Importantly, CD4 T cells maintain their tissue-specific properties such as viability, activation state, or immunocompetence following reassembly into lymphoid aggregates. This highly efficient and versatile gene editing workflow for tonsillar CD4 T cells enables the dissection of molecular mechanisms in ex vivo cultures of human lymphoid tissue and can be adapted to other tonsil-resident cell types.
Keywords: ex vivo human tonsil histoculture, gene editing, HIV infection, adaptive immunity
Graphical abstract
Highlights
-
•
ediTONSIL is a versatile gene editing workflow for tonsillar CD4 T cells
-
•
Optimized RNP nucleofection enables highly efficient editing
-
•
ediTONSIL preserves tissue-resident properties of the edited cells
-
•
Underlying molecular mechanisms can be dissected by studying reintegrated edited cells
Motivation
CD4 T cells are central players of adaptive immunity that rapidly change activation states in response to exogenous T cell receptor (TCR) stimulation. While molecular processes in activated human CD4 T cells from peripheral blood are well studied, resting CD4 T cells are refractory to gene editing transduction and transfection methods without prior activation. Knowledge on the molecular biology of truly resting CD4 T cells in their tissue environment is therefore lacking. We present here the culture and editing workflow ediTONSIL that allows for gene editing of tissue-resident CD4 T cells without compromising their activation state or immunological function. Reintegration in their natural cell environment enables molecular mechanistic analyses of this cell type in their native/physiological tissue context.
Morath et al. present a workflow for highly efficient gene editing of tonsil CD4 T cells without disturbing their physiological functions and unique properties. Reintegration of edited and labeled cells in their physiological cell environment enables mechanistic dissection of cell-cell interactions, e.g., in adaptive immunity or during HIV infection.
Introduction
Adaptive immune reactions involve fine-tuned, complex, and dynamic interactions between different cell types that define the remarkable potency of our immune system. In the case of CD4 T cells, interactions with antigen-presenting cells (APCs) that display MHC-II-restricted specific antigenic peptides in the context of close cell-cell conjugates (immunological synapse) drive CD4 T cell activation, proliferation, and differentiation to elicit effector function that includes helper functions to B and CD8 T cells toward humoral and cytotoxic immune responses, respectively.1 These APC-effector interactions take place in lymphoid organs and tissue architecture, composition, and density are essential parameters for efficacy and outcome of these reactions. Despite this essential impact of the tissue environment, most studies on CD4 T cell function employ CD4 T cells from readily available peripheral blood, which display activation and differentiation states that are markedly distinct from that of tissue-resident CD4 T cells. An alternative approach is the use of small animal models (mostly mouse), in which complex immune reactions can be studied. While these models provide fundamental insight, they also have important limitations with respect to human pathogenesis as they differ in tissue composition, the spectrum of cell surface markers to identify cell populations, life span and exposure to a complex immune-stimulatory environment.2 These differences are particularly relevant for studies of pathogen-host interactions such as HIV infection as even transgenic or humanized mouse models do not fully mirror the complex pathogenesis induced by the infection in humans.3,4 To overcome this limitation, organotypic ex vivo cultures of lymphoid tissue explants were developed and cultures of human tissue organ proved most useful. Grown as tissue blocks (human lymphoid histoculture [HLH])5,6,7,8,9,10,11,12 and in suspension (human lymphoid aggregate culture [HLAC]),9,10,13,14,15,16 human tonsil tissue is permissive to HIV-1 infection and mirrors hallmarks of HIV pathogenesis such as CD4 T cell depletion. In important contrast to peripheral blood CD4 T cells that require activation for productive HIV infection, tonsil CD4 T cells are fully permissive although they are phenotypically resting and do not proliferate.5 Of note, the original version of HLH retained the ability to elicit adaptive antibody responses to recall antigen7,17,18 and a recent optimization of cell purification and reassembly also improved the immunological features of HLACs, allowing the testing of vaccine candidates.19,20
The key limitation in this organotypic and immunocompetent cell culture system is the lack of methodology for targeted gene editing of specific cell types, which currently precludes detailed molecular dissection of cell function in this tissue-like context. Most available methods for gene editing of primary human lymphocytes require prior cell activation for efficient delivery of viral vectors, expression of transgenes, or sufficiently high transfection rates.21,22,23 These approaches are typically associated with high cytotoxicity and editing rates that require subsequent isolation of edited cells. Moreover, these editing approaches require prior cell activation and analyses are therefore limited to the second stimulation of cells that had returned to a phenotypical resting state after a first round of activation. We recently introduced a CRISPR-Cas9 RNP nucleofection approach for peripheral blood CD4 T cells that resulted in highly efficient gene editing without compromising the resting state of these cells.21 In this study, we adapted this protocol to tissue-resident cells in ex vivo tonsil cultures that are first characterized with respect to different culture setups and define the ediTONSIL workflow that enables the activation-neutral editing of bulk and isolated tonsil cell populations. Reintegration of these cells in their physiological cell context enables the molecular dissection of complex immune cell interactions in these organotypic cultures.
Results
Optimizing culture conditions of ex vivo tonsil suspension cultures
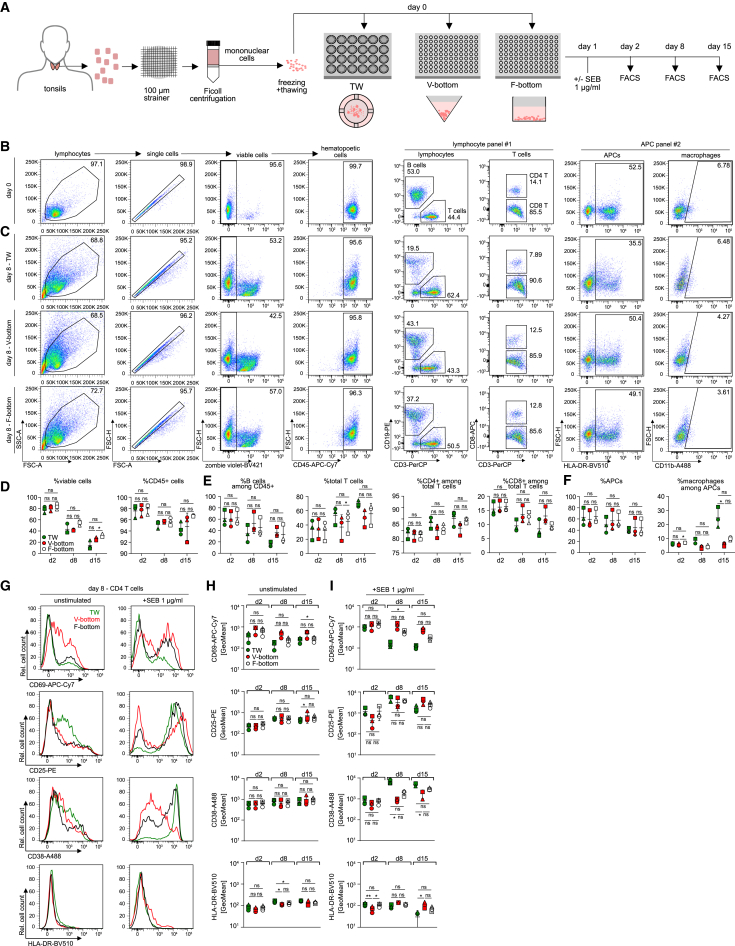
The viability of ex vivo tonsil cultures is currently limited to 2–3 weeks.5,19,24 To establish optimized conditions, we compared the impact of different culture conditions on cell viability, activation/differentiation, and response to stimulation. Tonsil tissue from healthy donors that underwent routine tonsillectomy or tonsillotomy were dispersed through a cell strainer for subsequent isolation of mononuclear cells (Figure 1A). At this step, cells can be either plated for immediate culture or frozen in aliquots for future use. For comparison, we plated cells in transwells (TW) or V- or F-bottom 96-well plates for subsequent phenotypic characterization by flow cytometry (Figure 1A). Cell viability was assessed by staining with the dead-cell dye zombie violet (over 95% viable cells at day 0) and cell type composition was determined by the detection of specific cell surface markers (Figure 1B). Expectedly, cell viability decreased over time, reaching approximately 20% after 15 days of culture. This decline was independent of the culture format (Figure 1D). With respect to specific cell populations, the relative amount of T cells slightly increased with time at constant CD4 vs. CD8 T cell ratios at the expense of APCs. The only notable impact of the culture format was a marked enrichment of macrophages among all APCs in TW cultures (Figure 1F). Activation of CD4 T cells with staphylococcal enterotoxin B (SEB) superantigen triggered potent cell surface exposure of the activation markers CD69, CD25, and CD38 within 7 days, while HLA-DR surface levels remained largely unaltered (Figures 1G–1I; Figures S1A–S1E). While the activation pattern was similar in all three culture conditions, exposure of the early activation marker CD69 was more transient and induction of surface CD38 more pronounced in TW cultures.
Figure 1.
Comparison of cell culture plates for human tonsil cell culture
(A) Schematic of the experimental setup. Cell suspensions were prepared from human tonsils, frozen, and thawed for culture in three different cell culture plate systems, 24-well transwell culture (TW), 96-well V-bottom (V-bottom), or 96-well F-bottom (F-bottom) plates at 10 × 106 cells/mL. Cell composition was analyzed on day 2, day 8, and day 15 post seeding by flow cytometry analysis. Activation response of CD4 T cells in the different setups was measured 1, 7, and 14 days post addition of SEB at 1 μg/mL by flow cytometry.
(B) Representative dot plots and gating strategy for identification of cell populations within tonsil bulk cell cultures at day 0, using two antibody cocktails for characterization of lymphoid cells (panel #1) and myeloid cells (panel #2).
(C) Representative dot plots at day 8 of culture for TW, V-bottom, and F-bottom setups.
(D–F) Quantification of (B) and (C) for three tonsils in TW (green), V-bottom (red), or F-bottom (white) setups.
(G) Representative histograms of CD4 T cell surface activation marker expression of CD25, CD69, CD38, and HLA-DR on day 8 of culture with (+SEB 1 μg/mL) or without (unstimulated) exogenous activation.
(H and I) Quantification of CD69, CD25, CD38, and HLA-DR signal on CD4 T cells without (H) or with (I) exogenous activation as shown in (G) for three tonsils in TW (green), V-bottom (red), or F-bottom (white) setups. Shown are means with SD. Each symbol represents one donor. Statistical significance was assessed by repeated measures one-way ANOVA for all datasets except where data were not normally distributed and statistical testing was performed using Friedman test (% macrophages among APCs, F; CD38 expression [unstimulated/+SEB] and CD25 expression in the unstimulated condition, H). ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
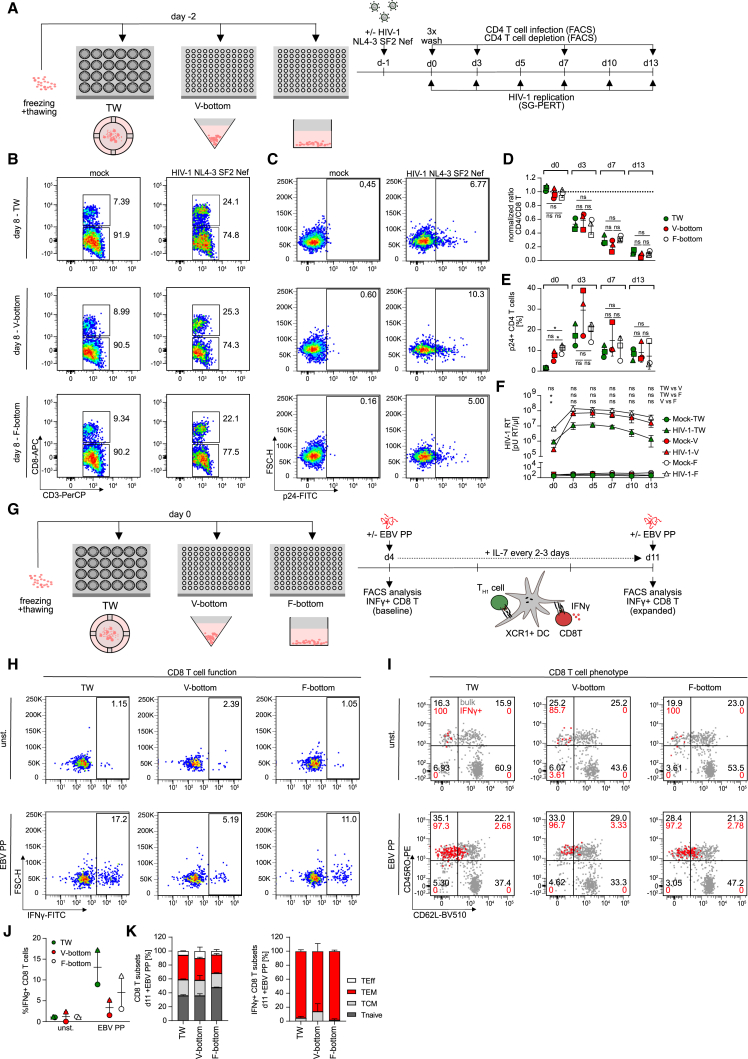
We next conducted a function characterization of ex vivo tonsil cultures under these different conditions. Since HIV-1 typically replicates efficiently in ex vivo tonsil cultures and induces the specific depletion of CD4 T cells that is characteristic for AIDS pathogenesis,5,7,25,26 we first analyzed their permissivity for HIV-1 infection. Following plating, cultures were infected with the full-length, replication-competent HIV-1 variant NL4.3 SF2nef 1 day later and HIV-1 replication and CD4 T cell depletion were quantified over time by assessing the activity of the viral enzyme reverse transcriptase in the cell culture supernatant (SG-PERT assay27,28), determining the number of productively infected cells by intracellular staining for the viral capsid protein p24, and defining the ratio of CD4 vs. CD8 T cells by flow cytometry (Figures 2B–2F). As indicated by the reduction in CD4 T cells paralleled by a relative increase in CD8 T cells, CD4 T cell depletion upon HIV-1 infection was observed with comparable magnitude and kinetics in all three culture conditions. The frequency of productively infected, p24+ cells reached a peak 3 days post infection (dpi) and then declined, but V-bottom cultures tended to allow the detection of higher frequencies of productively infected cells among viable cells (Figure 2E). Finally, virus replication in the culture as determined by the SG-PERT assay also rapidly increased in the first dpi and then slowly declined with time but the virus titers observed in TW cultures were markedly lower compared with those in the other culture conditions (Figure 2F).
Figure 2.
Different culture setups for human tonsil cells support HIV-1 infection to similar extend but show distinct preservation of immunocompetency
(A) Schematic of the experimental setup. Cryopreserved human tonsil cells were thawed and seeded in 24-well transwell (TW), 96-well V-bottom or 96-well F-bottom cell culture plates 1 day prior to infection with 1.5 × 105 BCU per 2 × 106 cells using HIV-1 NL4.3 SF2nef virus. On subsequent days, HIV-1 infection was characterized by analyzing CD4 T cell depletion by a decreased CD4/CD8 T cell ratio and CD4 T cell infection by intracellular flow cytometry staining for HIV-1 capsid protein (p24) and last, detection of newly produced HIV-1 virions in the culture supernatant by SG-PERT.
(B and C) Representative dot plots showing CD4 and CD8 T cells subgated from CD3 T cells as well as p24 signal in CD4 T cells for the three culture setups.
(D and E) Quantification of (B) and (C) for three tonsils for the respective harvesting timepoints cultured in TW (green), V-bottom (red), or F-bottom (white) setups. Shown are means with SD. Each symbol represents one donor.
(F) SG-PERT analysis of HIV-1 RT activity in the supernatant for mock or HIV-1 infected cultures in TW (green), V-bottom (red), or F-bottom (white) setups. Of note, the higher culture volume in TW plates is a confounding factor for lower RT activity/μL. Shown are means with SD of three tonsils per data point.
(G) Schematic of the experimental setup for analysis of EBV-specific memory CD8 T cell reactivation in tonsil cultures. Briefly, cells were thawed and seeded in 24-well transwell (TW) or 96-well V- or F-bottom plates as depicted. On day 4, EBV peptide pool was added together with interleukin (IL)-7 for 7 days. On day 4 and day 11, baseline and expanded EBV-specific CD8 T cells, respectively, were identified by production of IFNγ following stimulation with the EBV peptide pool.
(H) Representative dot plots of CD8 T cells in bulk tonsil cell cultures on day 11 of culture with (EBV PP) or without (unst.) EBV peptide pool.
(I) Characterization of memory subsets based on expression of CD45RO and CD62L among total (gray) or superimposed IFNγ+ (red) CD8 T cells on day 11 following peptide stimulation of tonsil cells cultured with (EBV PP) or without (unst.) EBV peptide pool for 7 days.
(J) Quantification of (H) for two tonsils per condition. Shown are means with range.
(K) Quantification of (I) for total or IFNγ+ CD8 T cells on day 11 in EBV PP-stimulated cultures. TEff = T effector cells, TEM = effector memory cells, TCM = central memory cells, Tnaive = naive T cells. Statistical significance of CD4 T cell infection and SG-PERT data were assessed by repeated measures one-way ANOVA (E) and (F). The CD4 T cell depletion data in (D) were normalized and Friedman test was used for analysis. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To assay the immune competence of these cultures, we probed their ability to mount CD4 T cell help to CD8 T cells in response to presentation of a recall antigen by XCR1+ dendritic cells (DCs)29,30,31,32 (Figure 2G; Figures S1F–S1P). To this end, cultures were treated with a pool of peptides derived from Epstein-Barr virus (EBV), a very common and lifelong infection in the general population that can trigger efficient CD4 and CD8 T cell responses.33,34,35 The presence and expansion of reactive memory cells was then probed 1 week later by the quantification of interferon gamma (INFγ)-producing CD8 T cells upon peptide restimulation. We observed expansion of INFγ+ CD8 T cells in response to EBV peptides in all culture conditions but this effect was clearly more pronounced in TW and F-bottom cultures and V-bottom cultures only allowed for residual expansion of INFγ+ CD8 T cells (1.5%/5.2% IFNγ+ CD8 T cells for two tonsils compared with above 3.0/8.9 and 11.0%/17.2% in F-bottom and TW cultures for the same tonsils, respectively) (Figures 2H and 2J). The subset distribution of total or CD8 T INFγ+ cells, however, was comparable in all culture conditions (mean of 36.0%–47.8% Tnaive, 20.3%–23.1% TCM, 26.5%–35.1% TEM and 5.4%–10.5% TEff cells among total CD8 T cells across the culture setups compared with 1.7%–13.9% TCM and 86.1%–98.4% TEM among IFNγ+ CD8 T cells) (Figures 2I and 2K). Of note, prior depletion of CD4 T cells from these cultures abrogated the induction of INFγ+ CD8 T cells, indicating that CD4 T cell help is critical for this expansion (Figures S1M–S1P). The culturing format thus has important implications for the functional properties of these cultures with all three culture systems supporting HIV-1 replication yet superior preservation of immunocompetence in TW and F-bottom cultures, which are therefore more suitable for the analysis of complex immune reactions.
Establishment of ediTONSIL gene knockout in bulk tonsil cultures
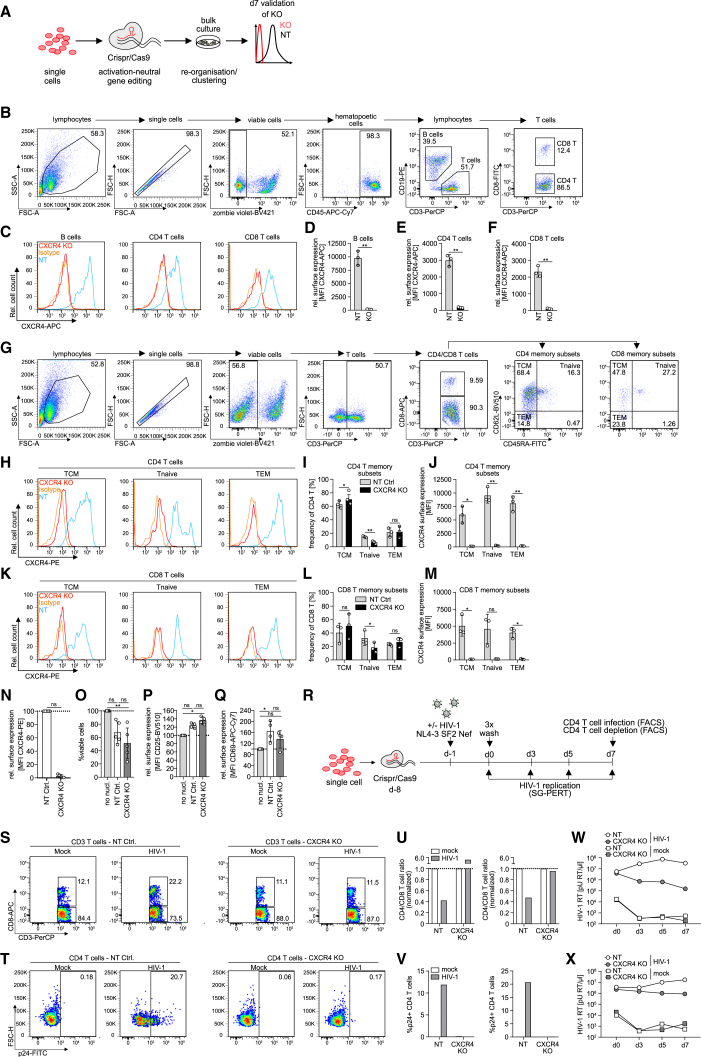
We next attempted to perform gene editing of tonsil cells, here termed ediTONSIL, by example of the chemokine receptor CXCR4 that serves as an essential co-receptor for entry of X4-tropic HIV-1 strains and is abundantly expressed on all major immune cell populations. To this end, thawed bulk tonsil cells were nucleofected with CRISPR-Cas9 RNP complex and cultured for 7 days. Knockout (KO) was assessed on the protein level by flow cytometry, which enabled the analysis of KO efficiency in distinct cell (sub)populations and differentiation states (Figures 3A–3G). Among the lymphocyte populations, B cells and CD4 and CD8 T cells displayed highly efficient KO of the cxcr4 gene compared with a non-targeting (NT) RNP control within 7 days (Figures 3C–3F; mean B cell CXCR4 surface level for NT 9793.0 ± 1338.7 arbitrary units [AU] vs. KO 166.3 ± 41.7, CD4 T cells 2996.3 ± 341.8 vs. 171.3 ± 26.8 and CD8 T cells 2321.0 ± 328.4 vs. 103.6 ± 31.4) irrespective of the memory differentiation state of CD4 or CD8 T cells (Figures 3G–3M; CD4 T TCM CXCR4 surface level mean for NT 5977.0 ± 1556.3 AU vs. KO 114.3 ± 3.2, Tnaive 9543.7 ± 1419.5 vs. 230.3 ± 66.0 and TEM 8065.0 ± 1392.8 vs. 173.0 ± 22.3; CD8 T TCM 5051.3 ± 1527.6 vs. 99.9 ± 8.1, Tnaive 4606.3 ± 2177.5 vs. 137.7 ± 14.4 and TEM 4049.0 ± 765.2 vs. 117.8 ± 70.4) and KO efficiency did not require further exogenous stimulation by SEB (Figures S2A–S2E). Among myeloid cells, macrophages and DCs, although far less abundant in tonsil cultures than lymphocytes, were edited with similar efficiency (macrophage CXCR4 surface levels: NT 3845.7 ± 742.4 AU vs. KO 165.2 ± 68.3; DC CXCR4 surface levels: 8464.0 ± 640.9 vs. 67.4 ± 21.1; Figures S2F–S2J). Such highly efficient KO was consistently obtained with tonsil cultures from different donors; however, the extent by which the procedure impaired cell viability varied between cells from different donors. Reduction in cell viability was accompanied by an upregulation of the T cell activation markers CD25 and CD69, indicating general stress induced by the nucleofection (Figures 3N–3Q). Nevertheless, KO of cxcr4 in bulk tonsil cultures was functional and prevented productive HIV-1 infection, virus-induced CD4 T cell depletion, and virus replication (Figures 3R–3X). Altogether, these results establish that genes can be efficiently knocked out in bulk tonsil cultures; however, the results also reveal that this approach is associated with significant cytotoxicity and unspecific CD4 T cell activation.
Figure 3.
Gene editing of bulk tonsil cells by CRISPR-Cas9 nucleofection is highly efficient in all major tonsil cell populations and across T cell differentiation states
(A) Schematic of experimental setup. Tonsil cells were thawed, nucleofected with non-targeting or cxcr4 RNP complex, and cultured in 96-well F-bottom plates for 1 week until analysis of KO efficiency in distinct cell populations by flow cytometry.
(B) Representative dot plots and gating strategy for identification of lymphocyte subsets among tonsil cell cultures.
(C) Representative histograms showing CXCR4 surface expression on B cells and CD4 and CD8 T cells on day 7 post nucleofection with CXCR4-targeting RNP (cxcr4 KO) with superimposed histograms for the non-targeting RNP (NT) condition and isotype control.
(D)-(F) Quantification of (C) for NT or cxcr4 KO in B cells, CD4 or CD8 T cells. Each dot represents one tonsil. Shown are means with SD.
(G) Representative dot plots and gating strategy for identification of memory subsets based on CD45RA and CD62L expression among CD4 and CD8 T cells in tonsil cell cultures.
(H and K) Representative histograms showing CXCR4 surface expression on central memory (TCM), naive (Tnaive) and effector memory (TEM) CD4 (H) or CD8 (K) T cell subsets on day 7 post nucleofection with cxcr4-targeting RNP (CXCR4 KO) with superimposed histograms for the non-targeting RNP (NT) condition and isotype control.
(I and L) Distribution of memory subsets among CD4 (I) and CD8 (L) T cells for the NT or cxcr4 KO condition.
(J and M) Quantification of (H) and (K) for NT or cxcr4 KO conditions in the respective memory subsets. Each dot represents one tonsil. Shown are means with SD.
(N) Relative surface expression of CXCR4 on cxcr4 KO or NT tonsil cells on day 7 post nucleofection, calculated by normalizing the MFI in cxcr4 KO conditions to the NT control.
(O–Q) Frequency of viable cells (O) or surface expression of CD25 (P) and CD69 (Q) on CD4 T cells among NT or cxcr4 KO cultures on day 7, normalized to non-nucleofected (no nucl.) cells, respectively.
(N)–(Q) Each dot represents one tonsil. Shown are means with SD. Tonsils were cultured in 24-well transwell plates.
(R) Schematic of experimental setup for analysis of HIV-1 infection in cxcr4 KO tonsil cultures. Briefly, tonsil cells were thawed, nucleofected with CRISPR-Cas9 RNP complex as indicated and infected with HIV-1 NL4.3 SF2nef at 1.5 × 105 BCU per 2 × 106 cells 7 days later by addition of the virus to the cultures. On the following day (d0), cells were washed 3x and infection was assessed by analysis of CD4 T cell infection and depletion by flow cytometry analysis upon final harvest and production of HIV-1 virions in the supernatant by SG-PERT throughout the infection course.
(S and T) Representative dot plots showing CD4 and CD8 T cells subgated from CD3 T cells (S) as well as p24 signal in CD4 T cells (T) in NT or cxcr4 KO mock or HIV-1 infected conditions, respectively on day 7 post infection.
(U and V) Quantification of (S) and (T) for two tonsils with the ratio of CD4 vs. CD8 T cells in (U) normalized to the mock condition for NT or cxcr4 KO, respectively.
(W and X) HIV-1 RT activity in the supernatant of mock or HIV-1 infected NT or cxcr4 KO cultures at indicated timepoints. Cells were cultured in 96-well V-bottom plates. Statistical significance was assessed by paired t test for comparison of two groups in (D), (E), (F), (I), (J), (L), (M), or by Wilcoxon test in (N) (normalized data). Data in (O)–(Q) were normalized and Friedman test was used for comparison of more than two groups. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Establishment of ediTONSIL gene KO in tonsil CD4 T cells
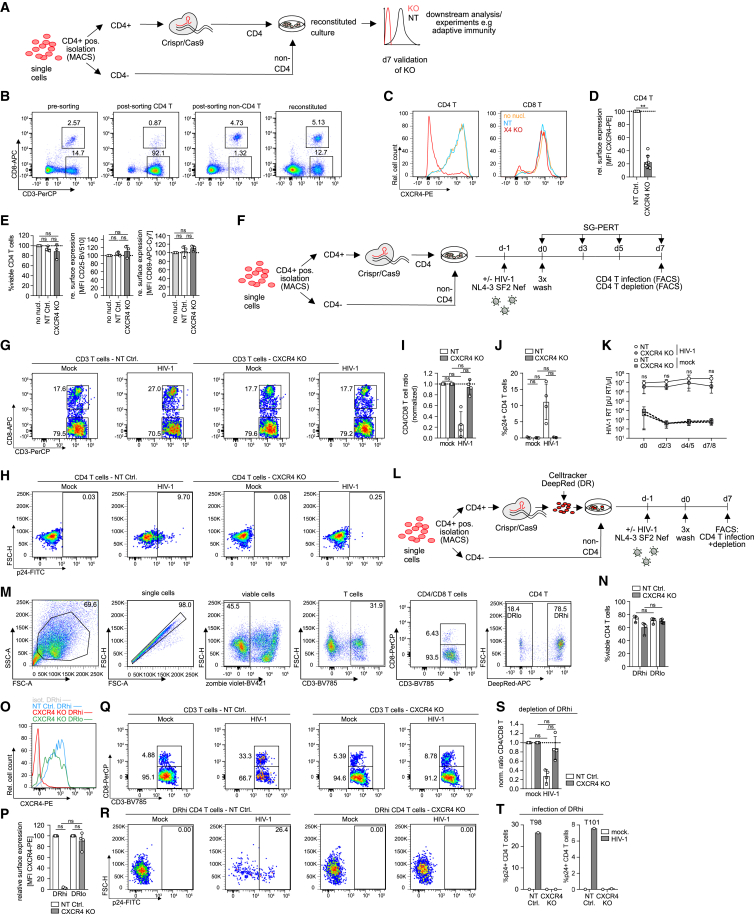
We reasoned that nucleofection conditions would need to be optimized for each cell population in tonsil cultures to limit cytotoxicity. Since there is no approach that renders nucleofection specific for a distinct cell subset in a complex heterogeneous mix of different cells, we attempted to improve cell viability following gene editing by isolating specific cell populations from tonsil cultures for editing. An additional requirement that gene editing of tissue-resident cells should fulfill is that subsequent reuniting these cells with the unedited tonsil cells allows for reconstituting of the original cell composition and function of the culture to support the precise molecular dissection of cell-cell interactions in the natural tissue context (Figure 4A). Focusing on CD4 T cells, we first established positive selection of CD4 T cells by magnetic beads (magnet-assisted cell sorting, MACS) as the most efficient method for cell isolation (CD4 T cell purity 94.5%/55.6% for two donors after negative isolation vs. 91.7%/85.6% for the same tonsils after positive selection, respectively and 16.5%/6.1% vs. 2.5%/1.5% CD4 T cells left in the non-CD4 cell fraction; Figures S3A–S3D; Table S1) which enabled us to faithfully reconstitute tonsil cultures following isolation and add back (Figure 4B). Importantly, efficiency of the positive isolation with respect to CD4 T cell depletion from the non-CD4 fraction is underestimated by negative gating for CD4 T cells based on CD3 and CD8 staining since a large fraction of the residual population does not express CD4, likely representing unconventional T cells and further validating the efficiency of the CD4 positive selection (Figures S3A–S3H). Of note, the positive isolation method did not affect viability, activation or susceptibility to HIV-1 infection (Figures S3I–S3T). Nucleofection of isolated CD4 T cells with cxcr4-specific CRISPR-Cas9 RNP complexes and immediate reconstitution resulted in highly efficient reduction of CXCR4 on CD4 T cells within 7 days post nucleofection (mean of 22.4% ± 10.1% CXCR4 surface expression left for KO normalized to NT CD4 T cells, Figures 4C and 4D). Importantly, this procedure did not affect CD4 T cell viability or activation state and exogenous cytokine supplementation was thus not required to enhance survival, likely explained by the presence of T cell survival factors that are already provided by the tonsil microenvironment36,37 (Figure 4E; Figures S4A–S4F). Similar KO efficiency was obtained in positively vs. negatively isolated CD4 T cells, excluding adverse effects of the positive isolation strategy, e.g., by residual magnetic particles on CD4 T cells during nucleofection. Moreover, independently prepared RNP complexes yielded comparable KO efficiencies, validating the reproducibility of the workflow (Figures S4G–S4I). Infecting tonsil cultures that were reconstituted after cxcr4 gene editing in CD4 T cells with HIV-1 (Figure 4F) revealed that the removal of the entry co-receptor strongly reduced virus-induced depletion of CD4 T cells (∼5% relative CD4 T cell reduction in in cxcr4 KO vs. ∼75% in the NT condition) and fully abrogated virus replication in these cultures to a similar extent as treatment with the HIV-1 entry-inhibitor T20 (Figures 4G–4K; Figures S4J–S4O). Since the tonsillar CD4 T cells were permissive to HIV-1 infection in the absence of exogenous activation, this important functional property of tissue-resident CD4 T cells was not impaired by the editing procedure.
Figure 4.
CD4 T cells can be specifically targeted for gene editing by CRISPR-Cas9 RNP nucleofection in tonsil culture by magnetic cell isolation and reconstitution
(A) Schematic of experimental setup. Briefly, tonsil cells were thawed and subjected to positive magnetic cell isolation of CD4 T cells. Magnetic beads were then removed from CD4 T cells prior to CRISPR-Cas9 RNP nucleofection and CD4 T cells were merged with the negative, non-CD4 T cell fraction, forming the reconstituted culture. Seven days post nucleofection, efficiency of the KO was assessed by flow cytometry.
(B) Representative dot plots showing CD4 and CD8 T cell frequencies among bulk cells prior to CD4 sorting (pre-sorting) and CD4 fraction after sorting (post-sorting CD4 T) and non-CD4 fraction after sorting (post-sorting non-CD4 T) as well as the reconstituted culture post merging of CD4 and non-CD4 fraction (reconstituted).
(C) Representative histograms of CXCR4 expression on CD4 and CD8 T cells on day 7 following CD4 T cell isolation, nucleofection and merging with non-CD4 T cells. Shown is the CXCR4 surface signal on non-nucleofected (no nucl.), NT control (NT), or cxcr4 KO (X4 KO) cultures.
(D) Quantification of CXCR4 on CD4 T cells for eight tonsils. CXCR4 signal was normalized to the NT condition for each tonsil. Tonsil cells were cultured in 24-well transwell or 96-well V-bottom plates.
(E) Frequency of viable CD4 T cells and surface expression of CD25 and CD69 among NT or cxcr4 KO CD4 T cells on day 7 for three tonsils, normalized to non-nucleofected cells, respectively. Tonsil cells were cultured in 24-well transwell plates. (D) and (E) Each dot represents one donor. Shown are means with SD.
(F) Schematic of experimental setup to assess HIV-1 infection in tonsil cultures with edited CD4 T cells. Briefly, tonsil cells were thawed and CD4 T cells were isolated and nucleofected prior to merging with non-CD4 T cells. On day 7 post nucleofection, cultures were infected with HIV-1 NL4.3 SF2nef at 1.5 × 105 BCU per 2 × 106 cells and HIV-1 infection was assessed on subsequent days by analysis of CD4 T cell depletion, CD4 T cell infection by intracellular p24 staining, and production of new virions in the supernatant by SG-PERT.
(G and H) Representative dot plots showing frequencies of CD4 and CD8 T cells among tonsil CD3 T cells (G) or intracellular p24 staining among tonsil CD4 T cells (H) in mock or HIV-1 infected cultures on day 7 post infection for NT or cxcr4 KO cultures, respectively.
(I) Quantification of (G) for four tonsils, normalized to the mock condition, respectively.
(J) Quantification of (H) for four tonsils. Tonsil cells were cultured in 24-well transwell or 96-well V-bottom plates. (I), (J) Each dot represents one donor. Shown are means with SD.
(K) SG-PERT quantification of HIV-1 RT activity in the supernatant at indicated timepoints for mock or HIV-1 infected cultures nucleofected with NT or cxcr4 KO RNP. Shown are means with range of three donors per condition. Tonsil cells were cultured in 24-well transwell or 96-well V-bottom plates.
(L) Schematic of the experimental setup for identification of nucleofected CD4 T cells among total CD4 T cells in tonsil culture by celltracker staining.
(M) Representative dot plots and gating strategy for identification of nucleofected, celltracker DeepRed high intensity (DRhi) CD4 T cells in tonsil bulk cultures.
(N) Quantification of viable CD4 T cells among DRhi and DRlo cells in NT and cxcr4 KO conditions for four tonsils.
(O) Representative histogram of CXCR4 expression on celltracker-stained (DRhi), cxcr4 KO CD4 T cells with superimposed histogram of the DRhi population in the NT condition and isotype control as well as DRlo CD4 T cell population in the CXCR4 KO condition.
(P) Relative surface expression of CXCR4 on the DRhi and DRlo population in cxcr4 KO tonsil CD4 T cells on day 7 post nucleofection, normalized to the NT control for four tonsils. Shown are means with SD.
(Q and R) Representative dot plots showing frequencies of CD4 and CD8 T cells among tonsil CD3 T cells (Q) or intracellular p24 staining among DRhi tonsil CD4 T cells (R) in mock or HIV-1 infected cultures on day 7 post infection for NT or cxcr4 KO cultures, respectively.
(S) Quantification of the CD4 vs. CD8 T cell ratio as calculated for the DRhi population among total CD4 T cells for four tonsils, normalized to the mock condition, respectively. (N), (P), (S) Each dot represents one donor. Shown are means with SD.
(T) Quantification of R in HIV-1 infected cultures for two tonsils as depicted. Tonsil cells for celltracker experiments were cultured in 96-well V-bottom plates. Statistical significance was assessed by Wilcoxon test for comparison of two groups in (D) or by Friedman test for comparison of more than two groups in (E), (I), (K), (N), (P), and (S). For normally distributed data in (J), repeated-measures one-way ANOVA was used. ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
The KO procedure was highly efficient as only ∼10% of CD4 T cells in the reconstituted cultures were precluded from editing. These represent residual CD4 T cells present in the non-CD4 T cell fraction that could not be removed during initial cell separation (Figure 4B; Figures S3E–S3H). While not sufficient to drive appreciable HIV-1 replication in cxcr4 KO cultures, this residual population may exert immune functions that are relevant for other scenarios. We therefore tested if labeling of the isolated and gene-edited CD4 T cell population allows for phenotypically distinguishing edited and unedited cell populations in reconstituted tonsil cultures after embedding of CD4 T cells (Figure 4L). Staining of gene-edited CD4 T cells with DeepRed celltracker prior to culture reconstitution followed by 1 week of culture indeed allowed the identification of gene-edited (DeepRed high [DRhi]) and unedited (DeepRed low [DRlo]) populations with unaffected viability of celltracker-stained cells (74.2% ± 4.4% viable cells in NT DRhi CD4 T vs. 71.1% ± 3.9% in DRlo and 60.2% ± 8.3% in cxcr4 KO DRhi CD4 T vs. 69.7% ± 3.2% in DRlo; Figures 4M and 4N). Of note, CXCR4 cell surface expression was undetectable on DRhi cells (Figures 4O and 4P), revealing that the residual CXCR4+ CD4 T cell population in the experiment described above (Figure 4D) reflects the presence of unedited cells, largely consisting of CD4/CD8-negative T cells and a minor fraction of CD4 T cells that resisted positive selection. Consistently, DRhi cells were fully protected from HIV-1 infection (Figures 4Q–4T). Isolation and modification of CD4 T cells combined with subsequent reconstitution of tonsil cultures thus allows for highly efficient and activation-neutral gene editing where edited cell populations can be further labeled by celltracker staining to unambiguously identify cells of interest while enabling direct comparison with unedited cells in the same condition.
Gene editing to study complex adaptive immune reactions
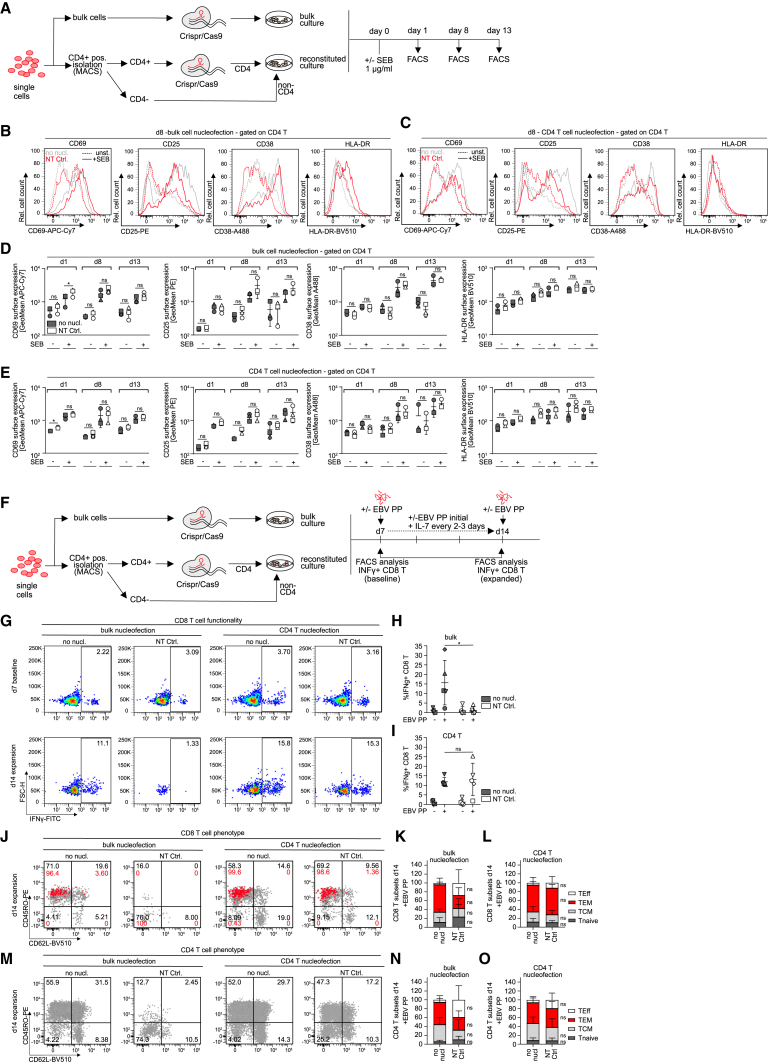
In contrast to HIV-1 replication and virus-induced depletion of CD4 T cells, complex immune reactions such as mounting of recall CD8 T cell responses (Figures S1F–S1P) depend on complex functional interactions of several immune cell types. To address if gene editing affects the ability of cells to conduct such activities following reconstitution, we assessed next the impact of ediTONSIL on adaptive immune reactions. First, we performed RNP nucleofection on bulk tonsil cultures or on isolated CD4 T cells with subsequent reconstitution into a complex organotypic culture and assessed the response of these cultures to T cell activation by SEB (Figure 5A). Under both conditions, the analysis of cell surface activation markers revealed an efficient response of CD4 T cells to stimulation that was not significantly altered in magnitude and kinetic by the nucleofection procedure (Figures 5B–5E). We next applied the analogous experimental setting to the expansion of EBV-specific CD8 T cells in response to the EBV peptide pool (Figures 2G and 5F). While RNP nucleofection of bulk tonsil cells abrogated the ability of these cultures to expand IFNγ+ CD8 T cells, editing of isolated CD4 T cells did not impair their ability to provide help for CD8 T cell expansion in their tonsil environment (Figures 5G–5I). Consistently, the CD8 T cell subset distribution was slightly skewed toward effector cells (TEff) upon bulk nucleofection while isolated editing of CD4 T cells did not significantly affect the memory T cell subset composition of these cultures (Figures 5J–5O). EdiTONSIL of isolated tonsil CD4 T cells with subsequent embedding in the physiological cell environment thus preserves their capacity to undergo complex adaptive immune reactions.
Figure 5.
CRISPR-Cas9-mediated gene editing of tonsil CD4 T cells can be used as a tool to mechanistically dissect T cell activation and CD4 T cell helper function
(A) Schematic of the experimental setup to assess activation response of CD4 T cells after nucleofection of bulk or isolated CD4 T cells. Briefly, bulk cells or positively isolated CD4 T cells after bead removal were either left untreated or nucleofected with NT RNP complex prior to seeding, i.e., merging with non-CD4 T cells for the reconstituted culture. One day after seeding, cultures were stimulated with SEB or left untreated and activation response was assessed on subsequent days by flow cytometry.
(B and C) Representative histograms showing CD69, CD25, CD38, and HLA-DR expression on tonsil CD4 T cells on day 7 post nucleofection of bulk cells (B) or isolated CD4 T cells (C). Superimposed histograms show expression profiles of the non-nucleofected culture (gray) and cultures nucleofected with non-targeting control (NT Ctrl) RNP (red). Dashed lines represent unstimulated conditions while solid lines represent SEB-stimulated cultures.
(D and E) Quantification of (B) and (C) for day 1, day 8, and day 13 post SEB stimulation for non-nucleofected or NT conditions with and without stimulation after bulk (D) or isolated CD4 T cell (E) nucleofection. Each symbol represents one donor. Shown are means with SD. Tonsil cells were cultured in 96-well F-bottom plates.
(F) Schematic of the experimental setup for assessing immunocompetency of tonsil bulk or CD4 T cells after nucleofection. Briefly, bulk cells or positive isolated CD4 T cells after bead removal were either left untreated or nucleofected with NT RNP complex prior to seeding, i.e., merging with non-CD4 T cells for the reconstituted culture. Seven days after seeding, EBV peptide pool was added together with interleukin (IL)-7. On day 7 and day 14, baseline and expanded EBV-specific CD8 T cells, respectively, were identified by production of IFNγ following stimulation with the EBV peptide pool.
(G) Representative dot plots showing IFNγ+ CD8 T cells in tonsil cultures for bulk or isolated CD4 T cell nucleofection (NT Ctrl.) in comparison with untreated (no nucl.) conditions on day 7 or day 14 of culture upon restimulation with EBV PP, respectively.
(H and I) Quantification of (G) for five tonsils, showing frequency of IFNγ+ CD8 T cells on day 14 in cultures were either bulk cells or isolated CD4 T cells were left untreated (no nucl) or nucleofected with NT Ctrl. Shown are means with SD. Each symbol represents one donor.
(J and M) Characterization of memory subsets based on expression of CD45RO and CD62L among total CD8 (J) or CD4 (M) T cells (gray) or superimposed IFNγ+ CD8 T cells (J) (red) on day 14 following peptide stimulation of tonsil cells cultured with EBV peptide pool for 7 days (K) and (L). Quantification of (J) for total CD8 T cells in cultures where bulk cells (left) or isolated CD4 T cells (right) were nucleofected (NT Ctrl.) or left untreated (no nucl.).
(N and O) Quantification of (M) as described above. TEff = T effector cells, TEM = effector memory cells, TCM = central memory cells, Tnaive = naive T cells. Shown are means with SD for five tonsils. Tonsil cells were cultured in 96-well F-bottom plates. Statistical significance was assessed by repeated-measures one-way ANOVA for data in (D) and (E), except for CD38 expression after bulk nucleofection and HLA-DR expression after CD4 nucleofection where data were not normally distributed and Friedman test was used instead. For (H) and (I), Friedman test was used for comparison of more than two groups and Wilcoxon test was used for data in (K), (L), (N), and (O) for comparison of two groups, ns, p > 0.05; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Discussion
Increasing efforts are made to study pathobiological principles and their therapeutic targeting in complex organoid and organotypic models to account for the impact of tissue environments and cell type heterogeneity at complexity levels between individual cell types (cell line or isolated primary cells) and small animal models/patient samples. In particular for lymphoid organs that predominately consist of hematopoietic cells, which are difficult to generate by iPS technology,38,39 ex vivo tonsil cultures are a well-established organotypic model that proved invaluable to study complex pathogen replication and complex immune reactions ex vivo.5,7,9,18,19,20,24,40,41,42,43 However, the full potential of this culture model for gaining molecular insight into immune reactions could so far not be exploited due to the lack of gene editing methods that allow for modification of gene expression with high precision and without impairing the activation state and immune competence of tonsil-resident cells. In the case of HLAC, a particular challenge for such an approach lies in the fact that close to 100% KO efficiency is required to conduct meaningful studies, since even minor contamination with unedited subpopulations may be sufficient to exert complex functions, expand in the course of the experiment, and thereby potentially mislead conclusion on the relevance of the gene of interest in the process studied. Furthermore, the gene editing approach per se should not affect the intrinsic properties of the edited cells. The ediTONSIL workflow established herein offers these possibilities as it allows gene editing with over 90% efficacy without requiring exogenous activation. Editing can in principle be done in bulk cultures to effectively target all major cell populations in parallel, although this comes at the cost of a significantly reduced cell viability. However, because HLACs are launched as suspension cultures that subsequently reorganize into immune-competent structures, specific cell populations can be isolated and edited at the beginning of an experiment and embedded in their natural cellular environment immediately after editing. Importantly, several lines of evidence document that ediTONSIL does not alter the tissue-resident characteristics of the edited tonsillar CD4 T cells: upon reintroduction into their natural cell environment they (1) are permissive to productive HIV infection without exogenous stimulation, (2) respond to TCR stimulation, and (3) exert helper function for CD8 T cell responses. ediTONSIL of isolated CD4 T cells with subsequent reconstitution of the culture therefore neither cause cell activation per se nor does it impair the activation response to external stimulation such as antigenic stimulation of CD4 T cells and also preserves the competence for complex functional cell-cell interaction such as CD4 T cell help to a recall CD8 T cell response. As illustrated at the example of HIV-1 infection, kinetics and magnitude of virus replication as well as CD4 T cell depletion are unaffected by the editing approach but targeting essential host cell genes such as the entry co-receptor fully abrogates virus infection and spread. Moreover, dye labeling of edited cells prior to reintegration in the complex organotypic cultures provides a convenient tool to identify and visualize them over extended periods of time in their natural cell environment. Adapting the methodology established here for tonsillar CD4 T cells to, e.g., CD8 T cells, B cells, and myeloid cells will offer a versatile toolbox to study mechanisms in all cell types present in the culture. This allows precise dissection of molecular mechanisms in basic immunology but also complex cell-cell interactions in the tissue context. This may be instrumental for, e.g., dissecting the mechanisms underlying CD4 T cell help to CD8 T cell responses, which is an emerging concept in infectious diseases and cancer44,45,46,47 and insights yielded from ediTONSIL studies in a human lymphoid tissue system will complement in vivo mouse studies. Importantly, this culture system is also suitable to study humoral immunity, e.g., B cell antibody production to recall antigens but also de novo vaccine responses and ediTONSIL opens further possibilities to study B cell responses.7,19,20 Furthermore, studies on viral pathogenesis and cell interactions in ex vivo tonsil cultures, exemplified herein with HIV-1, can be expanded to other pathogen infections including HTLV, human herpesviruses (HHV-6, HHV-7, HCMV, HSV-2, EBV), vaccinia virus, measles virus, and West-Nile virus.17,40,41,42,43,48,49,50,51,52,53 ediTONSIL thus opens avenues toward the mechanistic dissection in organotypic lymphoid cultures and will be widely applied to studies of pathogen-host interaction but also of fundamental principles of adaptive immunity.
Limitations of the study
The workflow developed herein allows for the efficient gene editing of bulk or individual cell populations in ex vivo tonsil cultures. For individual cell populations, however, the isolation efficiency is a critical parameter and the success of ediTONSIL depends on the availability and quality of commercial or in-house isolation procedures for cells of interest. Furthermore, it is important to note that the half-life of each individual target protein is a key determinant. Since the viability of these ex vivo cultures significantly decreases after 2 weeks, proteins with low turnover will not be sufficiently depleted within the lifespan of the culture. The fact that the applicability of this workflow depends on these individual features of each target implies that KOs must be monitored specifically for each target, which currently precludes the use of this method for high-throughput screening approaches. Establishing conditions that expand the lifespan of these model cultures will be a central goal of future developments.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD3-PE (clone SK7) | BD Biosciences | Cat#345765; RRID:AB_2868796 |
| CD3-PerCP (clone SK7) | Biolegend | Cat#344814; RRID:AB_10639948 |
| CD3-BV785 (clone UCHT1) | Biolegend | Cat#300472; RRID:AB_2687178 |
| CD4-BV421 (clone RPA-T4) | BD Biosciences | Cat#562424; RRID:AB_11154417 |
| CD4-A647 (clone OKT4) | Biolegend | Cat#317422; RRID:AB_571941 |
| CD8-FITC (clone SK1) | Biolegend | Cat#344703; RRID:AB_1877178 |
| CD8-APC (clone SK1) | Biolegend | Cat#344722; RRID:AB_2075390 |
| CD8-PerCP (clone SK1) | Biolegend | Cat#344708; RRID:AB_1967122 |
| CD19-PE (clone HIB19) | Biolegend | Cat#302207; RRID:AB_314238 |
| CD45-APC-Cy7 (clone 2D1) | Biolegend | Cat#368515; RRID:AB_2566376 |
| HLA-DR-BV510 (clone L243) | Biolegend | Cat#307646; RRID:AB_2561396 |
| CD38-A488 (clone HIT2) | Biolegend | Cat#303512; RRID:AB_493088 |
| CD25-PE (clone M-A251) | Biolegend | Cat#356103; RRID:AB_2564145 |
| CD69-APC-Cy7 (clone FN50) | Biolegend | Cat#310914; RRID:AB_314849 |
| CD11c-BV605 (clone 3.9) | Biolegend | Cat#301636; RRID:AB_2563796 |
| CD11b-A488 (clone ICRF44) | Biolegend | Cat#301318; RRID:AB_493017 |
| CD45RA-FITC (clone MEM-56) | Immunotools | Cat#21279453 |
| CD45RO-PE (clone UCHL1) | Biolegend | Cat#304205; RRID:AB_2564160 |
| CD62L-BV510 (clone DREG-56) | Biolegend | Cat#304843; RRID:AB_2617003 |
| CXCR4-PE (clone 12G5) | Biolegend | Cat#306506; RRID:AB_314611 |
| CXCR4-APC (clone 12G5) | Biolegend | Cat#306510; RRID:AB_314616 |
| P24-FITC (clone KC57) | Beckman Coulter | Cat#6604665; RRID:AB_1575987 |
| IFNγ-FITC (clone 4S.B3) | Biolegend | Cat#502507; RRID:AB_315230 |
| Bacterial and virus strains | ||
| HIV-1 NL4.3 SF2nef | Fackler et al. (2006)54 | N/A |
| Biological samples | ||
| Human tonsil tissue | Department of Otorhinolaryngology, Head and Neck Surgery, University Hospital Heidelberg, Germany | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| PepTivator® EBV Consensus – premium grade | Miltenyi Biotec | Cat#130-103-462 |
| HIV-1 Gag epitope pool | NIH AIDS Reagent Program | Cat#ARP-12437 |
| HIV-1 Env epitope pool | NIH AIDS Reagent Program | Cat#ARP-12698 |
| ITS-G (Insulin-Transferrin-Selen) Cocktail 100x | Thermo Fisher Scientific | Cat#41400045 |
| FBS, heat-inactivated | Capricorn | Cat#FBS-HI-12A |
| RPMI 1640 | Gibco/Thermo Fisher Scientific | Cat#61870036 |
| Non-essential amino acids 100x | Gibco/Thermo Fisher Scientific | Cat#11140050 |
| Amphotericin B/Fungizone 100x | Gibco/Thermo Fisher Scientific | Cat#15290026 |
| Penicillin/Streptomycin Solution 100x | Capricorn | Cat#PS-B |
| DMEM, high glucose | GlutaMAX™ Supplement | Cat#61965026 |
| Ficoll® Paque Plus | Merck | Cat#GE17144003 |
| T20/Fuzeon/Enfuvirtide | Roche | N/A |
| DeepRed celltracker dye | Thermo Fisher Scientific | Cat#C34565 |
| Zombie violet fixable viability kit | Biolegend | Cat#423113 |
| Brefeldin A | Sigma | Cat#B6542 |
| IL-7 | Biolegend | Cat#581902 |
| IL-15 | Biolegend | Cat#570302 |
| IL-2 | Biomol | Cat#50442 |
| Critical commercial assays | ||
| BD Cytofix/Cytoperm kit | BD Biosciences | Cat#554714 |
| EasySep™ Release Human CD4 Positive Selection Kit |
Stemcell Technologies | Cat#17752 |
| EasySep™ Human CD4+ T cell Isolation Kit |
Stemcell Technologies | Cat#17952 |
| Experimental models: Cell lines | ||
| Human embryonic kidney 293 T cells (HEK293T) | Pear et al. (1993)55 | RRID:CVCL_1926; ATCC Cat#CRL-11268 |
| TZM-bl cells, human carcinoma cell, derived from HeLa | NIH AIDS Reagents Program (Platt et al., 1998)56 | PRID: CVCL_B478; Cat#8129-442 |
| Oligonucleotides | ||
| SG-PERT fwd primer; TCCTGCTCAA CTTCCTGTCGAG |
Vermeire et al. (2012)57 | N/A |
| SG-PERT rev primer; CACAGGTCAA ACCTCCTAGGAATG |
Vermeire et al. (2012)57 | N/A |
| CXCR4 sgRNA1 | Albanese et al. (2021)21 | N/A |
| CXCR4 sgRNA2 | Albanese et al. (2021)21 | N/A |
| Recombinant DNA | ||
| pHIV-1NL4.3 SF2 Nef X4 | Fackler et al. (2006)54 | N/A |
| Software and algorithms | ||
| BD FACSDiva Software | BD Biosciences | www.bdbiosciences.com |
| BD FACSuite Software | BD Biosciences | www.bdbiosciences.com |
| FlowJo software 10.4.2 | BD Biosciences | www.flowjo.com |
| Microsoft Office Excel 2016 | Microsoft Office | www.microsoft.com |
| Prism 8.0.1 | GraphPad | www.graphpad.com |
| Other | ||
| Stemcell EasySep Magnet | Stemcell technologies | Cat#18000 |
| 100 μm PET cell strainer | PluriSelect Life Science | Cat#43-50100-51 |
| 24-Well Insert PET | cellQART | Cat#932 04 02 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Prof. Dr. Oliver T. Fackler (oliver.fackler@med.uni-heidelberg.de).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
(1)
All primary data generated in this study are available from the lead contact upon request. The flow cytometry datasets will be publicly available on FlowRepository when sufficient storage capacity is available.
-
(2)
This Paper does not report original code
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
Human lymphoid aggregate culture (HLAC)
HLACs were generated as previously described.19 Tonsils were removed and received from randomly selected anonymous donors with informed consent in accordance with the ethics vote S-123/2014 from the Heidelberg University Hospital ethics committee. Gender and age of the donors is not known. One-half of each tonsil was collected during standard tonsillectomy while the other half was processed in the department of pathology to assess chronic tonsillitis and exclude any unexpected malignancy. Removed tonsil tissue was immediately transferred to 50mL Falcon tubes containing 1X phosphate-buffered saline (PBS) and processed on the same day as described.19 In brief, dead and burnt tissue was removed and tonsils were sliced into blocks of approximately 2mm size that were passed through a 100 μm mash PET strainer. The suspension was filled up with PBS to 35 mL and erythrocytes and debris was removed from the cell suspension by layering onto 15 mL of Ficoll Separating Solution and centrifugation for 20 min at 2000 rpm (Heraeus MEGAFUGE 40R) without break at room temperature. The mononuclear fraction above the erythrocyte layer was isolated, washed twice with PBS and frozen in 90% FBS with 10% DMSO at −80°C in aliquots of 1x108 cells/ml. For experiments, cells were thawed in tonsil medium (RPMI 1640 GlutaMAX, 10% FCS, 5% Fungizone, 5% non-essential amino acids, 5% Sodium Pyruvate, 5% Penicillin/Streptomycin, 5% ITS-G), washed twice and resuspended in 10 mL medium prior to viability and cell counting in a 1:100 dilution with Trypan blue. Cells were resuspended in tonsil medium at 1x107 cells/ml and 200 μL of the cell suspension was added per well of a V or F-bottom 96-well plate. For transwell culture in 24-well plates, cells were resuspended at 4x107 cells/ml and 50μL cell suspension was added to the upper transwell while the lower well was filled with 500 μL medium. Transwells were equilibrated with medium prior to use. Cells were cultured at 37°C, 5% CO2 under humidified conditions for up to three weeks.
Cell culture
Human embryonic kidney 293T (HEK293T) cells were cultured in Dulbecco’s modified Eagle medium (DMEM; ThermoFisher Scientific) containing 10% FCS, 5% penicillin and 5% streptomycin. The cells are female and have not been authenticated.
TZM-bl cells (JC53BL-13) were obtained from the NIH AIDS Research and Reference Reagent Program (Cat. no. 8129) and are derived from a HeLa cell clone that was engineered to express CD4, CCR5 and CXCR4 to allow entry of X4-and R5-tropic HIV-1 strains56 and further contains integrated reporter genes for firefly Luc and E. coli β-galactosidase under the control of an HIV-1 long terminal repeat. For determination of virus stock infectivity, TZM-bl cells were infected with titrated amounts of the virus and X-β-Gal substrate was added after 2–3 days. The substrate is converted by the β-galactosidase to a blue colored product that is used to assess infection of TZM-bl cells and relative infectivity of the virus stock.
Method details
Virus production, infectivity measurements, infection assays, and CD4 T cell depletion
Virus production from 293T cells, infectivity measurements using SG-PERT for total viral particles and TZM-bl reporter assay were performed as described previously.27,28 2x106 tonsil cells were infected with 1.5x105 BCU/well of the CXCR4 tropic chimera HIV-1NL4.3 SF2nef (X4-tropic HIV-1)54. For F- and V-bottom 96-well plates, virus was diluted in 100 μL/well and added after removal of 100 μL spent medium from each well. For transwell plates, 250μL medium were removed from the lower transwell and virus was added in 250μL fresh medium. Mock wells were treated accordingly without addition of virus to the medium. One day post-infection (dpi) HLACs were washed 3x with 200 μL tonsil medium for V- and F-bottom 96-well plates by centrifugation at 1200 rpm for 5 min at RT and addition of fresh medium after washing. Transwell cultures were washed by 3x renewal of the lower transwell medium (500 μL) with 1 min incubation time respectively to allow full submerging of the upper transwell and transition of input virus in the supernatant to the lower well. HLACs were cultured for 7–15 days post infection. Every two to three days, 5 μL supernatant for determination of viral titers was harvested. The measurement started 1 dpi with the initial supernatant collected after the washing step.
Flow cytometry analysis
Tonsil cells were washed with PBS and stained with zombie violet fixable viability dye (1:1000 in PBS), incubated for 15 min at RT and washed with FACS buffer (1xPBS +0.5% BSA +2mM EDTA) prior to surface antibody staining. Surface staining of CD45, CD3, CD4, CD8, CD19, HLA-DR, CD11b, CD11c, CD38, CD25, CD69, CD45RO, CD45RA, CD62L and CXCR4 was performed at a 1:100 dilution of antibodies in FACS buffer and incubation at 4°C for 20 min, respectively. Cells were then washed and fixed with 3% PFA for 10 min or 90 min for BSL-3 sample inactivation. For intracellular staining of HIV-1 capsid (p24), cells were then resuspended in 0.1% Triton X-100 containing 1:100 diluted anti-p24 antibody (clone KC57, Beckman Coulter) and incubated for 20 min at 4°C and acquired on the same day. For intracellular staining of IFNγ, cells after surface staining and PFA fixation were permeabilized and stained using the BD Cytofix/Cytoperm kit according to the manufacturer’s protocol. Briefly, cells were incubated in Cytofix/Cytoperm solution for 20 min at 4°, washed 2x with 1xPerm buffer and resuspended in 1xPerm buffer containing 1:100 diluted anti-human IFNγ antibody. Samples were incubated for 30 min at 4°C, washed and acquired by flow cytometry on the same day.
Samples were acquired using a BD FACSVerse with BD FACSuite Software or FACS Celesta with BD FACS Diva software. Gating was performed using FlowJo software 10.4.2 and data was processed with Microsoft Office Excel 2016 and GraphPad Prism 8.0.1 software.
Quantification of HIV-1 induced CD4 T cell depletion
CD4 T cell depletion was quantified as described previously.10 In brief, the CD4/CD8 ratio was calculated for each sample and related to the ratio of the corresponding uninfected mock sample that was set to 1.
RNP preparation
RNP complexes were prepared as described.21 Briefly, sgRNAs were obtained from Synthego and reconstituted to 100 μM in TE buffer supplied by the manufacturer. Cas9 was obtained from IDT in a ready-to-use solution of 62 μM. SgRNA and Cas9 were mixed on ice at a ratio of 2.5:1 (sgRNA:Cas9) with PBS to obtain final concentrations of 100 pmol of sgRNA and 40 pmol of Cas9 in 50 μL complex volume, incubated for 15–20 min at RT and frozen at −80°C until usage. Immediately before nucleofection, the RNP complexes for NT Ctrl. or cxcr4 were thawed on ice. Equal amounts of NT Ctrl. RNP and cxcr4 RNP were used per cell in each experiment for proper comparison. The final cxcr4 RNP mix with which cells were nucleofected further consisted of equal amounts of two different sgRNA-Cas9 RNP complexes as described.21
CRISPR/Cas9 RNP nucleofection of tonsil cells
Tonsil bulk cells or isolated and bead-released CD4 T cells were prepared for nucleofection as described.21 Briefly, cells were washed once in 1xPBS and resuspended in P3 primary cell nucleofection buffer (Lonza) as recommended by the manufacturer’s protocol, using 2x106 cells in 20μL nucleofection buffer for 20 μL Nucleocuvette Strip or 1x107 cells in 100μL for single Nucleocuvettes. Cells were mixed briefly with the RNP mix that was prepared shortly before nucleofection on ice and were transferred to the respective nucleofection cuvettes. For 2x106 cells, 6 μL RNP mix in total were used while 24μL were used for 1x107 cells. Lonza 4D-Nucleofector X Unit was used for electroporation and cells were immediately after resuspended in unsupplemented RPMI 1640 to reach a concentration of 4x107 cells/ml and incubated for 15 min prior to dilution in tonsil medium to a final concentration of 1x107 cells/ml prior to merging with non-CD4 T cells. After nucleofection, cells were cultured either in 24-well transwell or 96-well V-/F-bottom plates as indicated. Culture plate setup did not have any influence on KO efficiency.
SEB stimulation of tonsil cell cultures
SEB was added to tonsil cultures at 1μg/ml final concentration directly into the medium of V or F-bottom 96-well plates or into the lower transwell of transwell plates and was left on the cells for the remaining culture period.
Cytokine treatment of tonsil cell cultures for optimization of post-nucleofection conditions
After nucleofection of CD4 T cells, reconstituted cell suspensions were seeded in 24-well transwell plates with IL-7, IL-15 and IL-2 alone or in combination (IL-7+IL-15) at indicated concentrations of 0.5 or 2 ng/ml for IL-7 and IL-15 and 10 or 50 IU/mL for IL-2. Every 2–3 days, medium was refreshed by preparing a 3x concentrated pre-dilution of the concentrated cytokine stocks in tonsil medium which was added to fill up the lower transwells after removal of a third of the spent medium every 2–3 days.
T20 treatment of tonsil cells to inhibit fusion and entry of HIV-1
T20 (Fuzeon, Enfuvirtide) was added to the diluted virus stock for infection of tonsil cultures at 20 μM final concentration on day 7 post nucleofection, was added freshly after washing of the virus input and was renewed on day 4 post infection.
CD4 positive and negative isolation from tonsil cell suspension
In the experiment directly comparing the sorting and nucleofection efficacy in CD4 negative vs. positive sorted CD4 T cells (Figure S3), tonsil cell suspensions were processed for positive or negative CD4 T cell isolation using stemcell EasySep Human CD4+ T cell Isolation Kit or EasySep Release Human CD4 Positive Selection Kit according to the manufacturer’s recommendations. Briefly, tonsil cells were thawed, counted and concentration adjusted to 5x107 cells/ml for negative isolation, i.e., 1x108 cells/ml for positive isolation in 1xPBS with 2% FBS and 2 mM EDTA. All steps were followed according to the manufacturer’s protocol, yielding an unlabeled CD4 T cell fraction from the negative isolation and a CD4 antibody and magnetic-bead labeled CD4 T cell fraction from positive isolation which underwent bead removal with the supplied release buffer directly after isolation. Purity and efficiency of the isolation procedures were assessed using CD3 and CD4 antibody staining for the negative isolation and CD3 and CD8 staining for the positive isolation as recommended. Unlabeled non-CD4 T cells resulting from the CD4 positive isolation procedure (negative fraction) were used to complement both negative and positive isolated CD4 T cells after nucleofection for the reconstituted culture in Figure S4 G-I to avoid a bias resulting from the presence of magnetic beads and possible induction of activation in cultures with negative isolated CD4 T cells and magnetic bead-labeled non-CD4 T cells. Non-CD4 T cells resulting from the positive CD4 T cell isolation were resuspended in tonsil medium, counted and resuspended at 1x107 cells/ml before merging with CD4 T cells for downstream experiments.
For all experiments except the direct comparison of positive vs. negative CD4 T cell isolation, the non-CD4 T cell fraction underwent a second positive isolation procedure to deplete remaining CD4 T cells from this population or alternatively, double amounts of CD4 isolation cocktail and magnetic particles were used in the first positive isolation process in order to increase purity of the non-CD4 fraction which routinely decreased the frequency of contaminating CD4 T cells in this fraction to 1/5th to 1/6th of the original CD4 T cell population. After isolation, CD4 T cells were resuspended in tonsil medium, counted and processed for downstream applications, e.g., nucleofection before resuspension at 1x107 cells/ml and merging with unlabeled non-CD4 T cells at the same concentration, yielding a typical ratio of 1–4 (20%) CD4 T cells to non-CD4 T cells in the final reconstituted culture.
Celltracker staining of isolated tonsil CD4 T cells
Directly after nucleofection and incubation in unsupplemented RPMI 1640 at 4x107 cells/ml, tonsil CD4 T cells were resuspended at 1x107 cells/ml in celltracker DeepRed (Thermo Fisher) solution by addition of 3x volume of PBS containing 4x concentrated celltracker dye, were incubated for 15 min at 37°C and were washed in tonsil medium 2x before resuspension at 1x107 cells/ml and merging with unlabeled, non-nucleofected non-CD4 T cells for reconstituted cultures.
Reactivation and expansion of EBV-specific memory CD8 T cells in tonsil tissue
Tonsils used for CD8 T cell reactivation experiments were pre-selected in separate screening experiments based on the presence of EBV-specific CD8 T cells measured by EBV peptide-specific IFNγ production compared to background activation induced by unspecific HIV-1 Gag peptides. The EBV peptide pool (EBV Peptivator, Miltenyi Biotec) contains MHC class I and class II-restricted epitopes from EBV lytic and latent cycle proteins for stimulation of CD4 and CD8 T cells, respectively. The peptides contained in the pool are recognized by most common HLA types such as HLA-A∗A02:01 and HLA-B∗07:02, ensuring broad response rates across different tonsil donors. The peptide mix was directly added to the culture medium at 1 μg/mL (0.6 μM for each peptide) at indicated timepoints without disturbing the cell layer and IL-7 was added at a final concentration of 1 ng/mL. Cultures were incubated for 7 days with the peptide pool and IL-7 was refreshed every 2–3 days by removing a third of the spent medium and replacing with fresh medium containing 3x concentrated IL-7. To assess IFNγ-production as a correlate for EBV-spec. CD8 T cell reactivation at designated timepoints, separate wells were exposed to the peptide pool on the start day of culture (baseline frequency of reactive CD8 T cells) and after 7 days for 30 min and subsequently incubated with Brefeldin A at 5 μg/mL final concentration for an additional 2 h to block cytokine export. Cells were then harvested with PBS +2 mM EDTA, washed with PBS and stained with zombie violet fixable viability dye and surface antibodies for human CD3, CD8, CD45RO and CD62L prior to fixation, permeabilization and intracellular cytokine staining. In every experiment, control wells that did not receive the peptide pool but were also stimulated at the end day of culture with the EBV peptide pool served as background controls for unspecific activation and expansion of CD8 T cells over the culture period that was negligible in our experiments. Further, antigen-specificity of the IFNγ production was validated in several experiments by stimulating the EBV-peptide pool cultures with an MHC class I epitope pool derived from HIV-1 Gag peptides in parallel, to which the healthy donors in our experiments did not respond with IFNγ production (example S1 J).
Quantification and statistical analysis
Statistical analysis of datasets was carried out using Prism version 8.0.1 (GraphPad). Normality was tested using Shapiro-Wilk test and statistical significance was calculated using the appropriate tests for paired data. For normally distributed data, statistical significance was calculated using paired t-test for comparison of two conditions or repeated measures (RM) one-way ANOVA with Holm-Sidak’s multiple comparison test for comparison of more than two conditions. For not normally distributed data, Wilcoxon test was used for comparison of two conditions with one being a control condition or Friedman test with Dunn’s multiple comparison test for comparison of more than two conditions as indicated. n.s., not significant; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗∗, p < 0.0001. See figure legends for details.
Acknowledgments
This project is supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation), Projektnummer 240245660 - SFB 1129 (project 8) and Projektnummer 259021520 – FA 378/13-2, OK 742/5-2) and the German Center for Infection Research (DZIF) (TTU 04.820 – HIV reservoir to O.T.F., O.T.K.). O.T.F. is a member of the CellNetworks cluster of excellence (EXC81). We want to especially thank Nadine Tibroni, Ina Ambiel, and the Head and Neck Surgery team for support with the acquisition of tonsil specimens and all anonymous tonsil donors that made this study possible.
Author contributions
Conceptualization, O.T.F.; Methodology, L.S. and O.T.K.; Investigation, K.M.; Visualization: K.M.; Writing – Original Draft, O.T.F. and K.M.; Writing –Review & Editing, O.T.F, K.M., G.D., and O.T.K; Funding Acquisition, O.T.F., O.T.K.; Resources, D.G.; Supervision, O.T.F.; Project administration: O.T.F.
Declaration of interests
The authors declare no competing interests.
Published: January 10, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100685.
Supplemental information
References
- 1.Swain S.L., McKinstry K.K., Strutt T.M. Expanding roles for CD4⁺ T cells in immunity to viruses. Nat. Rev. Immunol. 2012;12:136–148. doi: 10.1038/nri3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medetgul-Ernar K., Davis M.M. Standing on the shoulders of mice. Immunity. 2022;55:1343–1353. doi: 10.1016/j.immuni.2022.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang C., Zaman L.A., Poluektova L.Y., Gorantla S., Gendelman H.E., Dash P.K. Humanized mice for studies of HIV-1 persistence and elimination. Pathogens. 2023;12 doi: 10.3390/pathogens12070879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsden M.D. Benefits and limitations of humanized mice in HIV persistence studies. Retrovirology. 2020;17:7. doi: 10.1186/s12977-020-00516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glushakova S., Baibakov B., Margolis L.B., Zimmerberg J. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nature Medicine. 1995;1:1320–1322. doi: 10.1038/nm1295-1320. [DOI] [PubMed] [Google Scholar]
- 6.Glushakova S., Baibakov B., Zimmerberg J., Margolis L.B. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histocultures inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Hum. Retrovir. 1997;13:461–471. doi: 10.1089/aid.1997.13.461. [DOI] [PubMed] [Google Scholar]
- 7.Glushakova S., Grivel J.C., Fitzgerald W., Sylwester A., Zimmerberg J., Margolis L.B. Evidence for the HIV-1 phenotype switch as a causal factor in acquired immunodeficiency. Nature Medicine. 1998;4:346–349. doi: 10.1038/nm0398-346. [DOI] [PubMed] [Google Scholar]
- 8.Margolis L.B., Glushakova S., Grivel J.C., Murphy P.M. Blockade of CC chemokine receptor 5 (CCR5)-tropic human immunodeficiency virus-1 replication in human lymphoid tissue by CC chemokines. J. Clin. Invest. 1998;101:1876–1880. doi: 10.1172/JCI2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckstein D.A., Penn M.L., Korin Y.D., Scripture-Adams D.D., Zack J.A., Kreisberg J.F., Roederer M., Sherman M.P., Chin P.S., Goldsmith M.A. HIV-1 actively replicates in naive CD4(+) T cells residing within human lymphoid tissues. Immunity. 2001;15:671–682. doi: 10.1016/s1074-7613(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 10.Reif T., Dyckhoff G., Hohenberger R., Kolbe C.C., Gruell H., Klein F., Latz E., Stolp B., Fackler O.T. Contact-dependent inhibition of HIV-1 replication in ex vivo human tonsil cultures by polymorphonuclear neutrophils. Cell Rep. Med. 2021;2:100317. doi: 10.1016/j.xcrm.2021.100317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penn M.L., Grivel J.C., Schramm B., Goldsmith M.A., Margolis L. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc Natl Acad Sci USA. 1999;96:663–668. doi: 10.1073/pnas.96.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jayakumar P., Berger I., Autschbach F., Weinstein M., Funke B., Verdin E., Goldsmith M.A., Keppler O.T. Tissue-resident macrophages are productively infected ex vivo by primary X4 isolates of human immunodeficiency virus type 1. J. Virol. 2005;79:5220–5226. doi: 10.1128/JVI.79.8.5220-5226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homann S., Tibroni N., Baumann I., Sertel S., Keppler O.T., Fackler O.T. Determinants in HIV-1 Nef for enhancement of virus replication and depletion of CD4+ T lymphocytes in human lymphoid tissue ex vivo. Retrovirology. 2009;6:6. doi: 10.1186/1742-4690-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muñoz-Arias I., Doitsh G., Yang Z., Sowinski S., Ruelas D., Greene W.C. Blood-Derived CD4 T Cells Naturally Resist Pyroptosis during Abortive HIV-1 Infection. Cell Host Microbe. 2015;18:463–470. doi: 10.1016/j.chom.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Audigé A., Urosevic M., Schlaepfer E., Walker R., Powell D., Hallenberger S., Joller H., Simon H.U., Dummer R., Speck R.F. Anti-HIV state but not apoptosis depends on IFN signature in CD4+ T cells. JJ. Immunol. 2006;177:6227–6237. doi: 10.4049/jimmunol.177.9.6227. [DOI] [PubMed] [Google Scholar]
- 16.Doitsh G., Galloway N.L.K., Geng X., Yang Z., Monroe K.M., Zepeda O., Hunt P.W., Hatano H., Sowinski S., Muñoz-Arias I., Greene W.C. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grivel J.C., Margolis L. Use of human tissue explants to study human infectious agents. Nat. Protoc. 2009;4:256–269. doi: 10.1038/nprot.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaw S., Ananth S., Tsopoulidis N., Morath K., Coban B.M., Hohenberger R., Bulut O.C., Klein F., Stolp B., Fackler O.T. HIV-1 infection of CD4 T cells impairs antigen-specific B cell function. Embo j. 2020;39:e105594. doi: 10.15252/embj.2020105594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagar L.E., Salahudeen A., Constantz C.M., Wendel B.S., Lyons M.M., Mallajosyula V., Jatt L.P., Adamska J.Z., Blum L.K., Gupta N., et al. Modeling human adaptive immune responses with tonsil organoids. Nature medicine. 2021;27:125–135. doi: 10.1038/s41591-020-01145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kastenschmidt J.M., Sureshchandra S., Jain A., Hernandez-Davies J.E., de Assis R., Wagoner Z.W., Sorn A.M., Mitul M.T., Benchorin A.I., Levendosky E., et al. Influenza vaccine format mediates distinct cellular and antibody responses in human immune organoids. Immunity. 2023;56:1910–1926.e7. doi: 10.1016/j.immuni.2023.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albanese M., Ruhle A., Mittermaier J., Mejías-Pérez E., Gapp M., Linder A., Schmacke N.A., Hofmann K., Hennrich A.A., Levy D.N., et al. Rapid, efficient and activation-neutral gene editing of polyclonal primary human resting CD4(+) T cells allows complex functional analyses. Nat. Methods. 2022;19:81–89. doi: 10.1038/s41592-021-01328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Packard T.A., Schwarzer R., Herzig E., Rao D., Luo X., Egedal J.H., Hsiao F., Widera M., Hultquist J.F., Grimmett Z.W., et al. CCL2: a Chemokine Potentially Promoting Early Seeding of the Latent HIV Reservoir. mBio. 2022;13:e0189122. doi: 10.1128/mbio.01891-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hultquist J.F., Schumann K., Woo J.M., Manganaro L., McGregor M.J., Doudna J., Simon V., Krogan N.J., Marson A. A Cas9 Ribonucleoprotein Platform for Functional Genetic Studies of HIV-Host Interactions in Primary Human T Cells. Cell Rep. 2016;17:1438–1452. doi: 10.1016/j.celrep.2016.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt A., Huber J.E., Sercan Alp Ö., Gürkov R., Reichel C.A., Herrmann M., Keppler O.T., Leeuw T., Baumjohann D. Complex human adenoid tissue-based ex vivo culture systems reveal anti-inflammatory drug effects on germinal center T and B cells. EBioMedicine. 2020;53:102684. doi: 10.1016/j.ebiom.2020.102684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masur H., Michelis M.A., Greene J.B., Onorato I., Stouwe R.A., Holzman R.S., Wormser G., Brettman L., Lange M., Murray H.W., Cunningham-Rundles S. An outbreak of community-acquired Pneumocystis carinii pneumonia: initial manifestation of cellular immune dysfunction. N. Engl. J. Med. 1981;305:1431–1438. doi: 10.1056/NEJM198112103052402. [DOI] [PubMed] [Google Scholar]
- 26.Gottlieb M.S., Schroff R., Schanker H.M., Weisman J.D., Fan P.T., Wolf R.A., Saxon A. Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N. Engl. J. Med. 1981;305:1425–1431. doi: 10.1056/NEJM198112103052401. [DOI] [PubMed] [Google Scholar]
- 27.Pizzato M., Erlwein O., Bonsall D., Kaye S., Muir D., McClure M.O. A one-step SYBR Green I-based product-enhanced reverse transcriptase assay for the quantitation of retroviruses in cell culture supernatants. J. Virol. Methods. 2009;156:1–7. doi: 10.1016/j.jviromet.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 28.Ananth S., Morath K., Trautz B., Tibroni N., Shytaj I.L., Obermaier B., Stolp B., Lusic M., Fackler O.T. Multi-functional roles of the N-terminal region of HIV-1SF2Nef are mediated by three independent protein interaction sites. J. Virol. 2019;94 doi: 10.1128/JVI.01398-19. e01398-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janssen E.M., Lemmens E.E., Wolfe T., Christen U., von Herrath M.G., Schoenberger S.P. CD4+ T cells are required for secondary expansion and memory in CD8+ T lymphocytes. Nature. 2003;421:852–856. doi: 10.1038/nature01441. [DOI] [PubMed] [Google Scholar]
- 30.Shedlock D.J., Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003;300:337–339. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 31.Eickhoff S., Brewitz A., Gerner M.Y., Klauschen F., Komander K., Hemmi H., Garbi N., Kaisho T., Germain R.N., Kastenmüller W. Robust Anti-viral Immunity Requires Multiple Distinct T Cell-Dendritic Cell Interactions. Cell. 2015;162:1322–1337. doi: 10.1016/j.cell.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hor J.L., Whitney P.G., Zaid A., Brooks A.G., Heath W.R., Mueller S.N. Spatiotemporally Distinct Interactions with Dendritic Cell Subsets Facilitates CD4+ and CD8+ T Cell Activation to Localized Viral Infection. Immunity. 2015;43:554–565. doi: 10.1016/j.immuni.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 33.Long H.M., Meckiff B.J., Taylor G.S. The T-cell Response to Epstein-Barr Virus-New Tricks From an Old Dog. Front. Immunol. 2019;10:2193. doi: 10.3389/fimmu.2019.02193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hislop A.D., Kuo M., Drake-Lee A.B., Akbar A.N., Bergler W., Hammerschmitt N., Khan N., Palendira U., Leese A.M., Timms J.M., et al. Tonsillar homing of Epstein-Barr virus-specific CD8+ T cells and the virus-host balance. J. Clin. Invest. 2005;115:2546–2555. doi: 10.1172/JCI24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaganti S., Heath E.M., Bergler W., Kuo M., Buettner M., Niedobitek G., Rickinson A.B., Bell A.I. Epstein-Barr virus colonization of tonsillar and peripheral blood B-cell subsets in primary infection and persistence. Blood. 2009;113:6372–6381. doi: 10.1182/blood-2008-08-175828. [DOI] [PubMed] [Google Scholar]
- 36.Moutsopoulos N.M., Vázquez N., Greenwell-Wild T., Ecevit I., Horn J., Orenstein J., Wahl S.M. Regulation of the tonsil cytokine milieu favors HIV susceptibility. J. Leukoc. Biol. 2006;80:1145–1155. doi: 10.1189/jlb.0306142. [DOI] [PubMed] [Google Scholar]
- 37.Kreisberg J.F., Yonemoto W., Greene W.C. Endogenous factors enhance HIV infection of tissue naive CD4 T cells by stimulating high molecular mass APOBEC3G complex formation. JJ. Exp. Med. 2006;203:865–870. doi: 10.1084/jem.20051856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demirci S., Haro-Mora J.J., Leonard A., Drysdale C., Malide D., Keyvanfar K., Essawi K., Vizcardo R., Tamaoki N., Restifo N.P., et al. Definitive hematopoietic stem/progenitor cells from human embryonic stem cells through serum/feeder-free organoid-induced differentiation. Stem Cell Res. Ther. 2020;11:493. doi: 10.1186/s13287-020-02019-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rao I., Crisafulli L., Paulis M., Ficara F. Hematopoietic Cells from Pluripotent Stem Cells: Hope and Promise for the Treatment of Inherited Blood Disorders. Cells. 2022;11 doi: 10.3390/cells11030557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lisco A., Vanpouille C., Tchesnokov E.P., Grivel J.C., Biancotto A., Brichacek B., Elliott J., Fromentin E., Shattock R., Anton P., et al. Acyclovir is activated into a HIV-1 reverse transcriptase inhibitor in herpesvirus-infected human tissues. Cell Host Microbe. 2008;4:260–270. doi: 10.1016/j.chom.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langlois M., Bounou S., Tremblay M.J., Barbeau B. Infection of the Ex Vivo Tonsil Model by HTLV-1 Envelope-Pseudotyped Viruses. Pathogens. 2023;12:182. doi: 10.3390/pathogens12020182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pegtel D.M., Middeldorp J., Thorley-Lawson D.A. Epstein-Barr virus infection in ex vivo tonsil epithelial cell cultures of asymptomatic carriers. J. Virol. 2004;78:12613–12624. doi: 10.1128/JVI.78.22.12613-12624.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grivel J.C., Santoro F., Chen S., Fagá G., Malnati M.S., Ito Y., Margolis L., Lusso P. Pathogenic effects of human herpesvirus 6 in human lymphoid tissue ex vivo. J. Virol. 2003;77:8280–8289. doi: 10.1128/JVI.77.15.8280-8289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu Y.J., Barreira-Silva P., Boyce S., Powers J., Cavallo K., Behar S.M. CD4 T cell help prevents CD8 T cell exhaustion and promotes control of Mycobacterium tuberculosis infection. Cell Rep. 2021;36:109696. doi: 10.1016/j.celrep.2021.109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferris S.T., Durai V., Wu R., Theisen D.J., Ward J.P., Bern M.D., Davidson J.T., 4th, Bagadia P., Liu T., Briseño C.G., et al. cDC1 prime and are licensed by CD4(+) T cells to induce anti-tumour immunity. Nature. 2020;584:624–629. doi: 10.1038/s41586-020-2611-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Borst J., Ahrends T., Bąbała N., Melief C.J.M., Kastenmüller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018;18:635–647. doi: 10.1038/s41577-018-0044-0. [DOI] [PubMed] [Google Scholar]
- 47.Zander R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., Cui W. CD4(+) T Cell Help Is Required for the Formation of a Cytolytic CD8(+) T Cell Subset that Protects against Chronic Infection and Cancer. Immunity. 2019;51:1028–1042.e4. doi: 10.1016/j.immuni.2019.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grivel J.C., Ito Y., Faga G., Santoro F., Shaheen F., Malnati M.S., Fitzgerald W., Lusso P., Margolis L. Suppression of CCR5- but not CXCR4-tropic HIV-1 in lymphoid tissue by human herpesvirus 6. Nature Medicine. 2001;7:1232–1235. doi: 10.1038/nm1101-1232. [DOI] [PubMed] [Google Scholar]
- 49.Lisco A., Grivel J.C., Biancotto A., Vanpouille C., Origgi F., Malnati M.S., Schols D., Lusso P., Margolis L.B. Viral interactions in human lymphoid tissue: Human herpesvirus 7 suppresses the replication of CCR5-tropic human immunodeficiency virus type 1 via CD4 modulation. J. Virol. 2007;81:708–717. doi: 10.1128/JVI.01367-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biancotto A., Iglehart S.J., Lisco A., Vanpouille C., Grivel J.C., Lurain N.S., Reichelderfer P.S., Margolis L.B. Upregulation of human cytomegalovirus by HIV type 1 in human lymphoid tissue ex vivo. AIDS Res. Hum. Retrovir. 2008;24:453–462. doi: 10.1089/aid.2007.0155. [DOI] [PubMed] [Google Scholar]
- 51.Vanpouille C., Biancotto A., Lisco A., Brichacek B. Interactions between human immunodeficiency virus type 1 and vaccinia virus in human lymphoid tissue ex vivo. J. Virol. 2007;81:12458–12464. doi: 10.1128/JVI.00326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Condack C., Grivel J.C., Devaux P., Margolis L., Cattaneo R. Measles virus vaccine attenuation: suboptimal infection of lymphatic tissue and tropism alteration. J. Infect. Dis. 2007;196:541–549. doi: 10.1086/519689. [DOI] [PubMed] [Google Scholar]
- 53.Lim J.K., Lisco A., McDermott D.H., Huynh L., Ward J.M., Johnson B., Johnson H., Pape J., Foster G.A., Krysztof D., et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man. PLoS Pathog. 2009;5:e1000321. doi: 10.1371/journal.ppat.1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fackler O.T., Moris A., Tibroni N., Giese S.I., Glass B., Schwartz O., Krausslich H.G. Functional characterization of HIV-1 Nef mutants in the context of viral infection. Virology. 2006;351:322–339. doi: 10.1016/j.virol.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 55.Pear W.S., Nolan G.P., Scott M.L., Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc. Natl. Acad. Sci. USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Platt E.J., Wehrly K., Kuhmann S.E., Chesebro B., Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vermeire J., Naessens E., Vanderstraeten H., Landi A., Iannucci V., Van Nuffel A., Taghon T., Pizzato M., Verhasselt B. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
(1)
All primary data generated in this study are available from the lead contact upon request. The flow cytometry datasets will be publicly available on FlowRepository when sufficient storage capacity is available.
-
(2)
This Paper does not report original code
-
(3)
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.