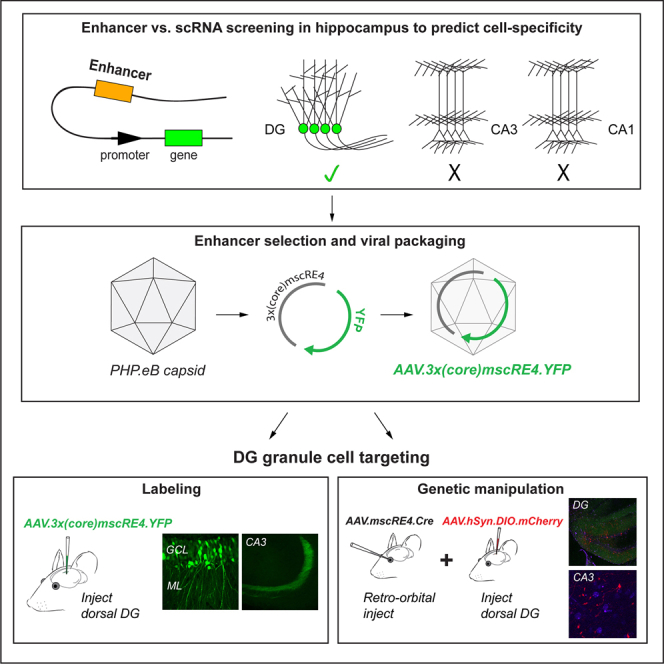
Summary
The mammalian brain contains a diverse array of cell types, including dozens of neuronal subtypes with distinct anatomical and functional characteristics. The brain leverages these neuron-type specializations to perform diverse circuit operations and thus execute different behaviors properly. Through the use of Cre lines, access to specific neuron types has improved over past decades. Despite their extraordinary utility, development and cross-breeding of Cre lines is time consuming and expensive, presenting a significant barrier to entry for investigators. Furthermore, cell-based therapeutics developed in Cre mice are not clinically translatable. Recently, several adeno-associated virus (AAV) vectors utilizing neuron-type-specific regulatory transcriptional sequences (enhancer-AAVs) were developed that overcome these limitations. Using a publicly available RNA sequencing (RNA-seq) dataset, we evaluated the potential of several candidate enhancers for neuron-type-specific targeting in the hippocampus. Here, we demonstrate that a previously identified enhancer-AAV selectively targets dentate granule cells over other excitatory neuron types in the hippocampus of wild-type adult mice.
Keywords: dentate gyrus, granule cells, AAV, enhancer, viral targeting, hippocampus, CA3, gene therapies, perforant path
Graphical abstract
Highlights
-
•
Recently identified enhancer-AAV “mscRE4” shows selectivity for dentate granule cells
-
•
mscRE4-driven fluorophore expression is present in all subcellular compartments
-
•
CaMKII-AAV co-expression reveals negligible overlap with nearby excitatory cell types
-
•
mscRE4-driven Cre expression permits genetic manipulation at mossy fiber-CA3 presynapse
Motivation
Precise subfield-specific operations of the hippocampus work together to enable diverse cognitive and memory-related functions, and thus a methodology that targets distinct hippocampal neuron types for isolated study is necessary. Cre approaches have become the standard for cell-type-specific investigation in the hippocampus, but these approaches are often expensive due to the need for extensive cross-breeding to other (floxed) mouse lines and cannot be directly translated to clinical therapies. Enhancer adeno-associated viruses (enhancer-AAVs), a more versatile and translatable cell-specific vector targeting method, are garnering increasing interest in neuroscience. However, enhancer-AAVs with selectivity for distinct hippocampal principal neurons are currently limited.
Banks et al. demonstrate that a previously identified enhancer-AAV abundantly and selectively targets dentate gyrus granule cells in the dorsal hippocampus, driving transgene expression in all subcellular compartments with no measurable expression in neighboring excitatory cell types. This vector targeting method is suitable for expressing Cre recombinase in dentate granule cells, thus providing a tool for specific presynaptic (mossy fiber boutons and axons) genetic modulation.
Introduction
The hippocampus plays critical roles in pattern recognition, spatial navigation, and episodic memory. To accomplish these diverse tasks, a series of operations are executed in the “trisynaptic circuit” (i.e., dentate gyrus [DG], Cornu Ammonis 3 [CA3], and CA1), where information is functionally transformed by distinct operations at each location.1 This is well illustrated by the highly divergent spatial coding schemes of distinct hippocampal subfields2,3,4 and their contributions to distinct aspects of mnemonic function.5 However, due to the highly sequential manner in which information is processed through the trisynaptic circuit, the hippocampus is particularly susceptible to pathophysiology including epilepsy and various psychiatric disorders.6 Therefore, subfield-specific cellular targeting methods are essential to understand the function of hippocampal areas in health and disease and to develop effective preventative and therapeutic interventions.
Our ability to investigate the function of hippocampal subfields in isolation has steadily improved in past decades. Researchers have long exploited the anatomically well-separated nature of the subfields for lesion studies.7,8 More recently, genetic targeting (i.e., Cre mice) approaches have become the standard for identification and discrimination between functionally heterogeneous cell populations. Cre lines with selectivity for specific excitatory cell types of each hippocampal subfield are available.9,10 Despite their utility, Cre approaches for investigation of specific brain cell populations are nonetheless limited to studies in mice alone, are often expensive due to the need for extensive cross-breeding to other (floxed) mouse lines, and cannot be directly translated to clinical therapies. Thus, more versatile and translatable cell-specific vector targeting methods (enhancer adeno-associated viruses [enhancer-AAVs]) are garnering increasing interest in neuroscience.11,12,13,14,15 Enhancer-AAVs incorporate short transcriptional regulatory elements to target expression of transgenes selectively within distinct cell populations in wild-type animals in vivo and often maintain cell-type specificity across species, including humans.12,13,16 Despite a growing appreciation of their utility, enhancer-AAVs with selectivity for distinct hippocampal principal neurons are currently limited.
A recent study rigorously investigated differences in chromatin accessibility of single-cell regulatory elements (scREs) across distinct neocortical pyramidal neurons, identifying several putative novel cell-type-specific enhancer elements.16 Based on these findings, enhancer-AAVs incorporating these scREs successfully targeted functionally distinct pyramidal cells residing in different cortical layers.16 As an analogous approach in the hippocampus has not been established, we examined whether the scREs identified by Graybuck and colleagues discriminate between hippocampal excitatory neurons. By comparing the expression of these scRE-regulated genes with an established RNA sequencing (RNA-seq) map of the hippocampus,10 we found that a target gene regulated by the scRE4 enhancer14 showed high expression in dorsal DG granule cells over other excitatory neuron types. When incorporated into an enhancer-AAV to drive YFP expression, we observed abundant and highly selective labeling of DG granule cells in the dorsal hippocampus. Expression was present in all subcellular compartments of the granule cells, and importantly, no measurable expression was detected in other neighboring excitatory cell types in the CA3 or hilus regions. Further, we demonstrate that systemic delivery of this enhancer-AAV to drive Cre expression will allow for genetic or activity manipulation of DG granule cells across mouse models. Together, we identified a viral approach to selectively target hippocampal granule cells, which allowed for specific presynaptic (mossy fiber boutons and axons) genetic modulation and thus may provide a tool for targeted clinical interventions at the mossy fiber-CA3 synapse in the future.
Results
Identification of a DG granule-cell-selective AAV targeting approach
Selective viral targeting of distinct excitatory cell classes in the cortex and hippocampus is currently limited. The most widely used excitatory neuron promotor (i.e., CaMKIIα) induces transgene expression across most excitatory neuron classes in these regions. Furthermore, the CaMKIIα promoter also induces off-target transgene expression in inhibitory neurons.17 Thus, identifying novel REs with specificity to functionally distinct excitatory neuron subclasses is essential. A recent study identified distinctions in chromatin accessibility among different cortical excitatory neurons within the same cortical region,16 revealing several REs selective for distinct deep-layer pyramidal neurons. The hippocampus is also comprised of several neighboring excitatory neuron subtypes, which currently are not accessible using analogous single-viral (i.e., enhancer-AAV) approaches.
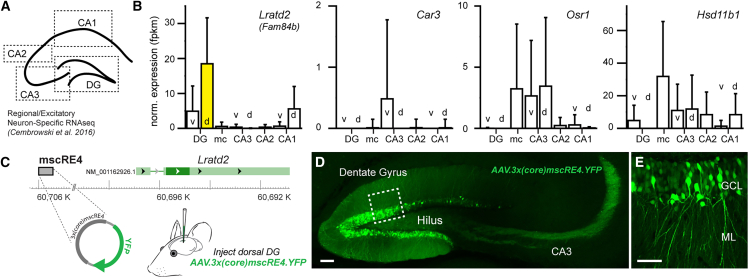
Using a well-curated RNA-seq database from distinct hippocampal subregions,10 we first examined whether four candidate genes from the Graybuck study (Lratd2, Car3, Osr1, Hsd11b1) displayed differential expression in hippocampal excitatory neuron subclasses in the mouse brain (Figure 1A). In cortical neurons, the Lratd2, Car3, Osr1, and Hsd11b1 genes are associated with unique mouse scREs (mscREs) (mscRE4, mscRE10, mscRE13, and mscRE16, respectively).14 When cross-referenced with hippocampal neurons, we noted that three of these four genes were selectively expressed in distinct excitatory types (Figure 1B). The gene Lratd2 (otherwise known as Fam84b), associated with the enhancer region mscRE4 (Figure 1C), was highly expressed in dorsal DG granule cells (Figure 1B, yellow bar), and to a lesser extent in ventral granule cells and dorsal CA1 pyramidal neurons, relative to other hippocampal neurons. The gene Car3 displayed selective expression in ventral CA3 pyramidal neurons. The Osr1 gene displayed relatively high expression in mossy cells and CA3 pyramids. Lastly, Hsd11b1 showed relatively similar expression across all excitatory cells.
Figure 1.
Identification of dentate granule-cell-selective enhancer for AAV approach
(A) A schematic showing the regional sampling strategy for excitatory-neuron-specific single-cell RNA-seq (scRNA-seq) data in the hippocampus.10
(B) Of the target genes from previously identified enhancer-AAVs selectively targeting cortical excitatory neuron types,13Lratd2 showed the highest overall normalized expression in dentate gyrus (DG) granule cells in the dorsal hippocampus compared to other hippocampal excitatory neuron types.
(C) A plasmid containing three copies of a short core sequence of mscRE4 transcript were packaged into an AAV (PHP.eB) capsid for intracerebral injections in the DG to drive YFP expression.
(D and E) AAV.3x(core)mscRE4.YFP expression in DG granule cells after 1–2 weeks. Scale bars: 100 μm (D) and 50 μm (E).
v, ventral; d, dorsal; DG, dentate gyrus; mc, mossy cells; CA3, Cornu Ammonis 3; CA2, Cornu Ammonis 2; CA1, Cornu Ammonis 1; ML, molecular layer; GCL, granule cell layer.
If RNA expression levels (Figure 1B) correlate with chromatin availability of the associated mscREs in these hippocampal cell classes,18 then AAVs that incorporate these enhancer sequences (enhancer-AAVs) might electively drive expression in distinct hippocampal cell classes. Because the mscRE4-associated Lratd2 gene showed both the greatest overall expression and high specificity to a single excitatory cell class, we packaged a plasmid (Addgene# 164458) containing three repeats of a truncated “core” mscRE4 enhancer sequence (155 bp)14 to express a YFP reporter into the PHP.eB capsid19 (referred to as “AAV.3x(core)mscRE4.YFP” hereafter; see STAR Methods for full description of vector components) (Figure 1C). ∼150 nL AAV.3x(core)mscRE4.YFP virus (titer 1.90E+13 vg/mL) was stereotaxically injected into the dorsal DG of adult C57BL/6J mice. In accordance with RNA expression of the mscRE4-associated Lratd2 gene (Figure 1B), DG granule cells and their mossy fiber axons displayed abundant YFP labeling (Figures 1D and 1E) 1–2 weeks postinjection. Remarkably, somatic YFP expression in regions near the granule cell layer (i.e., the hilus and CA3 regions) was absent (Figure 1D). We also evaluated whether AAV.3x(core)mscRE4.YFP selectively targets pyramidal tract (i.e., layer 5B) neurons in the cortex, as previously reported in the primary visual area,16 using stereotaxic injections in the primary motor cortex. As anticipated, we observed highly selective expression in M1 layer 5B pyramidal neurons (Figure S1). Together, our results indicate that mscRE4-based enhancer-AAVs are well suited for granule cell targeting with local stereotaxic injections.
AAV.3x(core)mscRE4.YFP labels DG granule cells but not neighboring hilar mossy cells
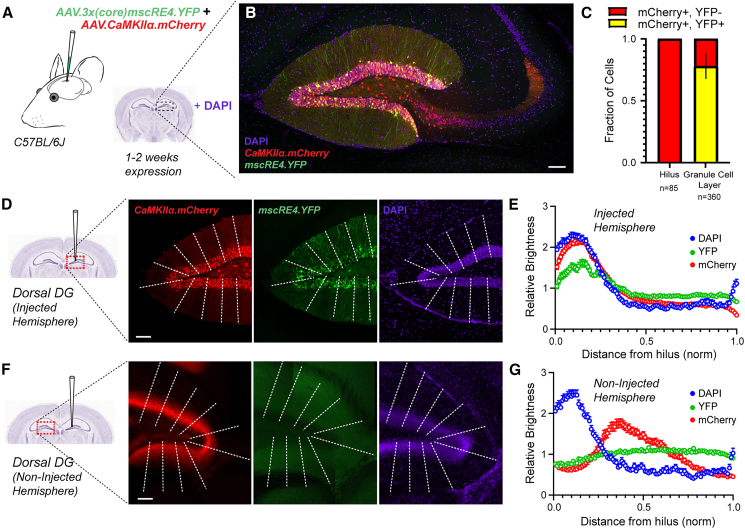
Our transcriptomic and histological surveys suggest that the mscRE4 enhancer preferentially drives expression in granule cells with respect to neighboring excitatory neurons in the dorsal hippocampus. The CaMKIIα promoter has traditionally been incorporated in AAVs to target excitatory neuron types. However, CaMKIIα promotes reporter expression indiscriminately across all major excitatory cell classes in close proximity to the DG, including granule cells, hilar mossy cells, and pyramidal neurons.17,20,21 Thus, we examined the co-expression patterns of AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry (Addgene# 114469) from mice unilaterally co-injected with both AAVs (>113 vg/mL each) in the dorsal hippocampus. To control for potential differences in serotype tropism,22 the AAV.CaMKIIα.mCherry construct was also packaged into the PHP.eB capsid. 1–2 weeks postinjection in dorsal DG, slices were prepared using a derivative of the “magic cut”23 to preserve granule cell mossy fiber axons and were mounted for confocal imaging (Figure 2A). mCherry labeling was consistently observed in granule cells somas and their dendrites in the DG, in pyramidal cells in the CA3 stratum pyramidale, and in putative hilar mossy cells. However, co-labeling of YFP and mCherry was highly selective for granule cells (Figure 2B), with no apparent co-labeling of somas in the hilus. Specifically, hilar cells positive for mCherry were consistently negative for YFP despite their proximity to the injection site (Figure 2C). In contrast, 77.2% mCherry-positive cells in the DG granule cell layer were also YFP positive, indicating relatively strong transgene expression directed by mscRE4 in these cells (Figure 2C). Importantly, AAV.CaMKIIα.mCherry labeling extended to neurons well outside of the DG, indicating that spatial restriction of AAV diffusion from our injection site was unlikely to account for the granule cell selectivity of mscRE4.
Figure 2.
AAV.3x(core)mscRE4.YFP labels dentate granule cells in dorsal DG
(A) AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry were unilaterally injected together in the dorsal DG in one hemisphere of young adult (7–11 weeks old) wild-type mice (n = 5; 3 females and 2 males) and expressed for 1–2 weeks prior to acute slice preparation.
(B and C) YFP/mCherry co-expression appeared selective for granule cells in confocal images (40× objective; oil) of “magic cut” slices (B), which was further assessed by (C) cell counting in the DG-hilus region, where mCherry+ hilar cells were consistently YFP−, and 77.2% mCherry+ cells in the granule cell layer were YFP+.
(D–G) Line scan profiles in the DG (D and E) in the injected hemisphere and (F and G) in the non-injected hemisphere depicting the relative brightness of DAPI, YFP, and mCherry signals across a normalized distance from the hilus. Scale bars: 100 μm throughout.
To further examine the neuron-type specificity of AAV.3x(core)mscRE4.YFP within the DG-hilus region, we next quantified the relative brightness of YFP, mCherry, and DAPI signals in the DG of the injected hemisphere as well as the non-injected hemispheres (Figures 2D and 2F) of the same mice. In both injected and non-injected hemispheres, the peak DAPI signal delineated the granule cell layer in the DG (Figures 2E and 2G). In injected hemispheres, mCherry and YFP signals followed the DAPI signal, rapidly tapering by around 50% (Figure 2E), supporting our claim that AAV.3x(core)mscRE4.YFP selectively labels granule cell bodies and their neurites.
Hilar mossy cell axons make strong contralateral projections, terminating in the inner molecular layer of the contralateral DG.24 Thus, we also examined the labeling via mCherry and YFP in the contralateral, non-injected DG (Figures 2F and S2). In contralateral hemispheres, the mCherry signal was well structured in the DG but highly disjointed from DAPI (Figure 2G), signifying AAV.CaMKIIα.mCherry labeling of commissural hilar mossy cell axons. The mscRE4-driven YFP signal was notably flat in contralateral hemispheres (Figures 2F and 2G), showing only background autofluorescence (Figures 2F and S2), which provides further evidence for mscRE4 as a granule-cell-selective enhancer that spares mossy cells.
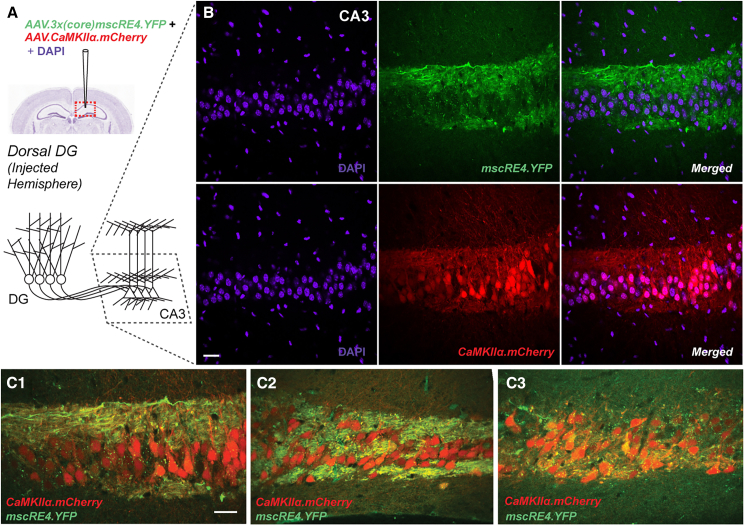
AAV.3x(core)mscRE4.YFP labels mossy fiber axons but not neighboring CA3 pyramidal neurons
We next evaluated expression in the CA3 region following co-injection of AAV.CaMKIIα.mCherry and AAV.3x(core)mscRE4.YFP in the dorsal DG. Granule cells make direct projections to CA3 via mossy fibers (Figure 3A), which were preserved in our slices using the magic cut (see STAR Methods). We observed strong somatic expression of mCherry in both cell somas and neuritic structures of CA3 (Figure 3B), likely due to mCherry expression in both mossy fiber axons and CA3 pyramidal cells. As described above, strong YFP expression was present in the mossy fibers extending into CA3. However, in contrast to the mCherry signal, expression of YFP was absent from cell somas in CA3 (Figure 3B). Furthermore, YFP expression was absent from mCherry-positive CA3 pyramidal neurons surrounded by co-labeled mossy fiber axons projecting from the DG (Figures 3C1–3C3 and S3; examples from 3 different mice). Combined, our findings indicate high selectivity of the mscRE4 enhancer-AAV for granule cells over neighboring excitatory cell types.
Figure 3.
AAV.3x(core)mscRE4.YFP expression in mossy fibers but not CA3 pyramidal neurons
(A) Co-injection of AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry into the dorsal DG in one hemisphere of adult (7–11 weeks old) wild-type mice produced (B) mCherry expression in CA3 somas along with a lack of YFP expression in CA3 somas, as well as (C1–C3) co-expression of mCherry and YFP in mossy fiber axons projecting from the DG (as seen in confocal images; 40× objective, oil). Scale bars: 50 μm.
A recent study found that anterograde AAV serotypes (i.e., PHP.eB) may induce dose-dependent toxicity specifically in adult-born granule cells.25 This problem could be circumvented through use of AAV-retro capsids to direct expression in granule cells. As DG granule cells make direct projections to CA3, we examined whether a retrograde form of AAV.3x(core)mscRE4.YFP would show a similar targeting profile when injected in the CA3 region. Thus, we packaged the same YFP-expressing mscRE4 construct into a retrograde capsid (referred to as “AAV.3x(core)mscRE4.YFP (retrograde)” hereafter) and injected it at the CA3b/CA3a border (Figure S4A). After 1–2 weeks, we observed a YFP expression pattern comparable to that produced by the anterograde (PHP.eB) AAV.3x(core)mscRE4.YFP capsid described earlier. Specifically, in the DG, we observed YFP expression in granular layer cell somas and dendrites in the molecular layer (Figure S4B), while in CA3, punctate YFP expression surrounding, but not overlapping with, pyramidal cell somas was again apparent (Figure S4C). Together, these results indicate that AAV.3x(core)mscRE4.YFP (retrograde) injections in the CA3 region target DG granule cells via their mossy fiber axons. The density of YFP expression in the DG granular layer appeared reduced compared to that produced by anterograde (PHP.eB) AAV.3x(core)mscRE4.YFP expression, likely due to the different serotype and injection site. To confirm the extent and cell-type selectivity of AAV.3x(core)mscRE4.YFP (retrograde) expression using another method, we also performed immunohistochemistry to visualize expression with diaminobenzidine (DAB). We observed abundant dark brown reaction products in DG granular layer cell somas and molecular layer dendrites (Figure S4D). Further, we found punctate expression in the CA3 injection region with no visible reaction product in CA3 cell bodies (Figure S4E), thus complementing our AAV.3x(core)mscRE4.YFP (retrograde) expression seen with fluorescent confocal imaging.
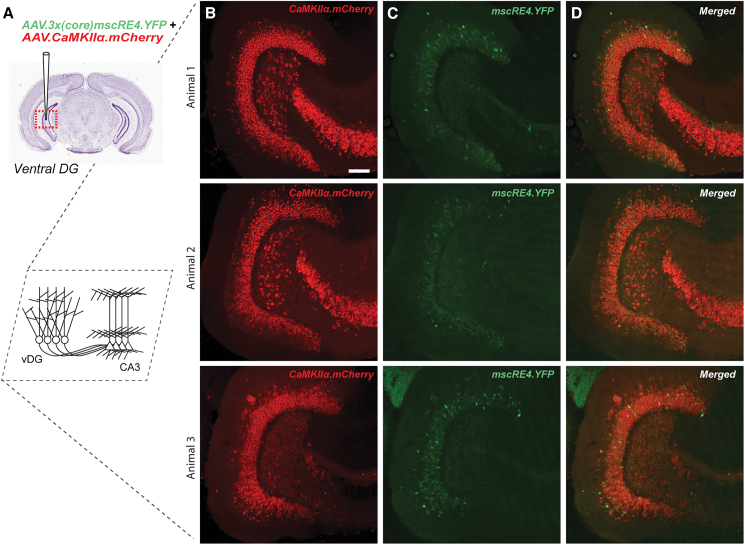
AAV.3x(core)mscRE4.YFP labeling in ventral hippocampus
The mscRE4-associated Lratd2 gene also showed expression in ventral DG granule cells (Figure 1B), albeit far less than that observed in the dorsal DG. Thus, we also examined the expression patterns of AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry from mice co-injected with both AAVs in the ventral DG (Figure 4A). As seen in the dorsal hippocampus, we observed mCherry labeling in DG granule cell somas, as well as putative hilar mossy cells and CA3 pyramidal cells (Figure 4B). We also observed YFP labeling in a subset of DG granule cells somas, with a noticeable absence of somatic labeling in the hilus or CA3 (Figure 4C). The overlap in mCherry and YFP labeling was again restricted to the DG granular layer in the ventral hippocampus (Figure 4D). These findings were again in concordance with predictions from the RNA-seq dataset (Figure 1B), as the density of YFP expression in the ventral DG granular layer appeared reduced compared to that produced in the dorsal hippocampus.
Figure 4.
AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry labeling in ventral hippocampus
(A) Adult wild-type mice were co-injected with AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry in the ventral DG (vDG).
(B) After 1–2 weeks, in confocal images (20× objective; horizontal sections), we observed mCherry labeling in DG granule cell somas, as well as putative hilar mossy cells and CA3 pyramidal cells.
(C and D) We also observed (C) YFP labeling in a subset of DG granule cells somas, with a noticeable absence of somatic labeling in the hilus or CA3, and thus (D) co-expression of mCherry and YFP labeling was restricted to the DG granular layer. Scale bars: 100 μm.
AAV.mscRE4.Cre allows for genetic manipulation of DG granule cells
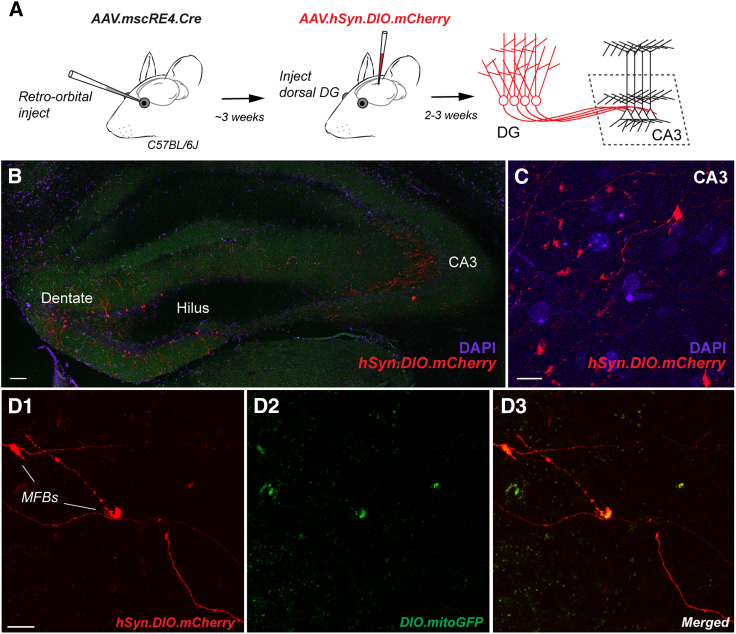
A vast array of floxed mouse lines and AAV vectors are available for immediate “off-the-shelf” use. To assess whether mscRE4 could be used generically for granule cell genetic manipulations, we packaged a Cre-expressing construct (Addgene# 163476)16 driven by the “full” mscRE4 sequence into the PHP.eB capsid (referred to as “AAV.mscRE4.Cre” hereafter). In multiple experiments, we recently noted that local stereotaxic injections of Cre-expressing AAVs viruses often produced off-target expression, regardless of the fidelity of its promoter or enhancer (data not shown). However, we have found that cell-type specificity is apparently well maintained with Cre-expressing viruses when delivered systemically.26 Thus, we next tested the granule cell selectivity of mscRE4-directed Cre expression using retro-orbital (RO) injections.
To examine this question, we first injected ∼1E+11 viral genome copies of AAV.mscRE4.Cre in the RO sinus of wild-type (WT) mice (Figure 5A). ∼3 weeks after RO injections, AAVs expressing floxed constructs were then stereotaxically injected in the dorsal DG (e.g., AAV.hSyn.DIO.mCherry [Addgene# 50459; titer 2.20E+13]; Figures 5A and 5B). 2–3 weeks following stereotaxic surgery, tissue was procured for confocal imaging. Sparse mCherry labeling was evident in DG granular layer somas and in processes within the CA3, with a distinct lack of somatic expression again notable within the hilus and CA3 (Figures 5A and 5B). Upon higher-magnification examination in CA3, sparse red axonal labeling was noted, including many giant mossy fiber boutons characteristic of CA3-granule presynapses (Figure 5C). Multiple floxed constructs could be expressed together in granule cells simultaneously in this way. We co-injected AAV.EF1α.DIO.mitoGFP (Addgene# 174112; titer 4.00E+12) with AAV.hSyn.DIO.mCherry, which allowed for mitochondrial labeling, as recently demonstrated,27 here in granule cells. mCherry and mito.GFP co-expression was often found in DG granule cells, including in their mossy fiber boutons (Figure 5D). Together, these results demonstrate that systemic delivery of AAV.mscRE4.Cre, combined with local stereotaxic injection of floxed transgenes, can be used for selective genetic manipulation of DG granule cells in dorsal hippocampus.
Figure 5.
Genetic manipulation of DG granule cells using AAV.mscRE4.Cre
(A) A schematic of the experimental layout to achieve genetic manipulation of DG granule cells through retro-orbital (RO) injections of AAV.mscRE4.Cre, followed later by stereotaxic injection of the floxed construct AAV.hSyn.DIO.mCherry.
(B) Sparse mCherry labeling in the DG granular layer somas and in CA3 observed 2–3 weeks after stereotaxic injection in confocal images (10× objective).
(C) Higher-magnification images (60× objective) in CA3 revealed sparse red axonal labeling.
(D1–D3) Stereotaxic co-injection of two floxed constructs, AAV.EF1α.DIO.mitoGFP and AAV.hSyn.DIO.mCherry, produced mitochondrial labeling in DG granule cells, including their mossy fiber boutons (MFBs).
Scale bars: 100 μm (B) and 10 μm (C and D1–D3).
Non-granule-cell “off-target” mscRE4 labeling caveats in dorsal hippocampus
While mscRE4-directed expression was primarily restricted to granule cells following local injection in dorsal DG, when injections were improperly targeted, we also noted dense YFP expression in the dorsal subiculum (Figure S5). Furthermore, as anticipated by our RNA-seq analysis earlier, we also saw sparse YFP expression in the putative CA1 pyramidal neurons when injections spread into that region (Figure S5). In sum, these data indicate that mscRE4 is selective but not completely specific for granule cells in the dorsal hippocampal region.
Discussion
Here, we first examined the likelihood that REs with specificity for neocortical excitatory neuron subtypes would also map onto subsets of excitatory neurons in the hippocampus in WT mice. Predictive analysis was performed by comparing hippocampal excitatory cell-specific mRNA expression for genes10 associated with newly identified regulatory regions of interest.16 Several candidates with potential selectivity for different excitatory neurons were identified. A single promising candidate (mscRE4), which displayed high selectivity for dentate granule cells in dorsal hippocampus, was validated via local injection. To our knowledge, this is the first vector targeting method suitable for a single excitatory cell type within the hippocampal trisynaptic circuit. Our results suggest that highly distinct neuron types situated in different brain regions may share a similar chromatin accessibility signature and, thus, effective enhancer elements. Thus, high-resolution comparisons of enhancer viruses between these and other brain regions may advance cell-specific targeting technologies more rapidly.
Cell-type-specific targeting with AAVs in the brain
Cre mouse lines are beneficial for cell-specific studies throughout the brain10,28,29,30; however, they are often limited by off-target31,32,33 or incomplete34 expression. The use of AAV vectors with cell-type-selective cis-active DNA control elements has opened an alternative strategy avenue to advance circuit-, cellular-, and synapse-specific questions in neuroscience. Inclusion of full-size or truncated promotor sequences can direct AAV expression within particular neuron classes.35,36,37 However, the large size of most promotor sequences represents a significant issue due to the relatively small AAV packaging size. Shorter regulatory (i.e., enhancers) elements present an ideal opportunity for inclusion in AAV-targeting experiments, with several recent studies demonstrating exquisite neuron-subclass specificity.12,14,15,38 An additional benefit of many enhancer-AAVs is their cross-species adaptability, with specific enhancer elements often exhibiting neuron-type specificity across rodents, non-human primates, and mature human neurons.12,13,14
mscRE4-driven expression in dorsal hippocampus
Recent work from the Allen Institute identified several short REs with specificity for distinct neocortical pyramidal cells, which were then validated in the V1 region after incorporation into enhancer-AAVs.16 While many of the genes regulated by these newly identified enhancers showed interesting expression patterns in hippocampus,10 here, we focused on one candidate (mscRE4) showing selective and relatively high expression in excitatory granule cells in the DG of the dorsal hippocampus. When injected locally, we found that mscRE4-directed YFP expression in granule cells, their dendrites in the dentate, and broadly throughout their mossy fiber axons. Intriguingly, there was an evident lack of expression in the nearby excitatory mossy cells and CA3 pyramidal neurons, matching our expectations based on the expression pattern of the Lratd2 mRNA expression. Further, as our RNA-seq results suggested, mscRE4-directed YFP expression was present, albeit to a much lower extent, in ventral hippocampal DG granule cells. Importantly, recent work suggests that adult-born granule cells may be susceptible to AAV-induced toxicity in a dose-dependent manner following direct injections into the dentate.25 Thus, viral titer may be a critical variable when studying granule cells with enhancer-AAVs. Alternatively, mscRE4 constructs may be packaged into the AAV-retro capsid and injected into CA3 to avoid potential toxicity in adult-born cells.25
We further demonstrated that AAV.mscRE4.Cre maintains cell-type specificity when delivered systemically (via RO injections) and combined with subsequent stereotaxic co-injection of multiple floxed constructs in the dorsal DG. This approach produced sparse labeling in DG granule cells, allowing for localization of mitochondria in these cells, including the mossy fiber boutons in CA3. By varying the amount of viral particles introduced via RO injections, differential levels of sparsification or dense expression should be possible. This approach may be especially useful for leveraging the Cas9 system to knock out genes of interest in granule cells. In addition, independent pre- and postsynaptic manipulations should be feasible using a combination of mscRE4 and CaMKII-driven AAVs to target DG granule cells and CA3 pyramidal cells, respectively. Finally, co-injection of AAV-retro-packaged mscRE4 constructs with anterograde CaMKII AAVs would, in principle, permit simultaneous pre- and postsynaptic for ex vivo or in vivo evaluation of the input-output relationship at mossy fiber-CA3 synapses in WT or disease model mice.
AAV.3x(core)mscRE4 also appears to provide excellent utility for directed studies of the subiculum, where it strongly labeled putative excitatory neurons in this region. As with studies in the DG described here, accurately positioned, low-volume viral injections will undoubtedly be necessary for subiculum-specific targeting. In addition, YFP expression was occasionally seen in putative CA1 pyramidal neurons. While this off-target effect does lessen the specificity of mscRE4 in granule cell studies, it could provide an avenue for experiments requiring highly sparse expression in CA1 in mice and potentially across other species. Finally, the extent to which mscRE4 targets expression in non-glutamatergic neurons (i.e., inhibitory GABAergic interneurons) requires further evaluation. Our current work has not distinguished whether sparse YFP+ cell bodies in the DG granule cell layer may be GABAergic interneurons. GABAergic neurons also reside in the hilus and CA3; however, the lack of somatic YFP expression in these regions suggests that AAV.3x(core)mscRE4.YFP expression in GABAergic interneurons is low.
Basic science and clinical applications for granule cell targeting in dorsal hippocampus
While our findings indicate mscRE4-directed expression selectively in granule cells, additional studies employing mscRE4-driven expression of neuronal effectors (e.g., ChR2) need to be performed in the same context to validate its specificity more thoroughly. Control of mossy fiber axon firing using ChR2 has been shown using a granule-cell-targeting Cre line.39 mscRE4 should allow for viral deployment of ChR2, genetically encoded Ca2+ indicators, and any of their derivatives in vivo across any mouse line. Thus, this enhancer virus has the potential to rapidly accelerate research into dentate-CA3-related questions, such as mechanisms of memory formation and storage, pattern separation, and exploratory behaviors,5,40,41,42 and into similar questions in primates43 if cross-species expression is maintained.
Beyond basic science applications, mscRE4-granule cell targeting may have important preclinical applications. One of the prime advantages of cell-specific targeting with enhancer-AAVs is their rapid deployment in mouse models of different brain disorders.44 Relevant to our findings, recent work using a multiplexed Cre approach demonstrated online control of epileptic seizures through optogenetic modulation of DG excitability in vivo.45 Thus, temporally well-controlled opto- or chemogenetic modulation of granule cell excitability with mscRE4 targeting represents a promising future direction for translational research in epilepsy and other disorders.
Limitations of the study
In addition to DG granule cells, systemic delivery of mscRE4-directed constructs will result in expression in layer 5B cortical pyramidal neurons, as previously found in the primary visual16 and motor areas (Figure S1). However, region-specific manipulations such as optogenetic stimulation or stereotaxic injection of floxed constructs in the hippocampus should restrict manipulation to DG granule cells. It is also possible that a small number of CA1 pyramidal cells in the dorsal hippocampus may be targeted by mscRE4-directed constructs (Figures 1B and S5). Lastly, though recently identified mscREs have been shown to maintain cell-type specificity across species in previous work,12,13,16 it is still unclear whether this mscRE4-AAV method is suitable for targeting DG granule cells in other species.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-GFP antibody | Life Technologies, Carlsbad, CA | Cat#A111222; RRID: AB_221569 |
| Biotinylated donkey anti-rabbit antibody | Jackson Immunoresearch Laboratories, West Grove, PA | Cat#711-065-152; RRID: AB_2340593 |
| Bacterial and virus strains | ||
| pAAV.3x(core)mscRE4.minCMV.SYFP2 | Graybuck et al., 202116 | Addgene# 164458 |
| pAAVmscRE4.minBGpromoter.iCre | Graybuck et al., 202116 | Addgene# 163476 |
| pAAV.CaMKIIα.mCherry | Karl Deisseroth | Addgene# 114469 |
| PHP.eB capsid | Chan et al., 201719 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 3,30-diaminobenzidine tetrahydrochloride (DAB) | Sigma Aldrich, St. Louis, MO | Cat#D5905 |
| Avidin-biotin complex (ABC Elite) | Vector Laboratories, Burlingame, CA | Cat#PK-6100 |
| Deposited data | ||
| ‘Hipposeq’: RNA-seq database of gene expression in hippocampal principal neurons | Cembrowski et al., 2016 | http://hipposeq.janelia.org |
| Experimental models: Organisms/strains | ||
| Mouse: C57Bl/6J | The Jackson Laboratory | RRID:IMSR_JAX:000664 |
| Software and algorithms | ||
| ImageJ/FIJI | Wayne S. Rasband; NIH | Version: 2.7.0 |
| Line scan normalization custom algorithm | This paper | https://doi.org/10.5281/zenodo.10161946 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Matthew J.M. Rowan (mjrowan@emory.edu).
Materials availability
This study did not generate any new unique reagents.
Data and code availability
-
•
This paper analyzes an existing, publicly available dataset (‘Hipposeq’ from Cembrowski et al., 2016). Access to this dataset is listed in the key resources table. All microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
A custom algorithm was generated to normalize line scan measurements; the DOI to access this code can be found in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (mjrowan@emory.edu) upon request.
Experimental model and study participant details
Young adult (7–11 week old) C57Bl/6J mice of both sexes were used for all experiments, with data collected from ≥3 mice per experimental condition. C57Bl/6J mice were originally purchased from The Jackson Laboratory (RRID:IMSR_JAX:000664) and breeding colonies are maintained in our Division of Animal Resources facility at Emory. All animal procedures were approved by the Emory University Institutional Animal Care and Use Committee (IACUC).
Method details
Intracranial viral injections
One group of mice was initially injected with AAV(PHP.eB).3x(core)mscRE4.minCMV.SYFP2 (AAV.3x(core)mscRE4.YFP; titer 1.90E+13 vg/mL) alone, while another group was injected simultaneously with both AAV.3x(core)mscRE4.YFP and AAV(PHP.eB).CaMKIIα.mCherry (AAV.CaMKIIα.mCherry; titer 6.00E+14 vg/mL) at a 1:1 ratio in the dentate gyrus region of the dorsal hippocampus. A final group of mice were injected with AAV(retro).mscRE4.minCMV.SYFP2 (AAV.3x(core)mscRE4.YFP (retrograde); titer 7.36E+11 vg/mL) in the CA3b/CA3a border region of the dorsal hippocampus. When performing viral microinjections, mice were head-fixed in a stereotaxic platform (David Kopf Instruments) using ear bars while under isoflurane anesthesia (1.8–2.2%). Thermoregulation was provided by a heating plate using a rectal thermocouple for biofeedback, thus maintaining core body temperature near 37°C. A craniotomy was gently made in the skull (<0.5 μm in diameter) to allow access for a glass microinjection pipette. For dorsal dentate gyrus targeting, coordinates (in mm from bregma) for AAV.3x(core)mscRE4.YFP microinjection were X = +/−1.50; Y = −1.91; α = 0°; Z = −2.20. For ventral dentate gyrus targeting, coordinates were X = +/−2.61; Y = −3.52; α = 0°; Z = −3.65, −3.35 (virus delivered at two Z depths). For dorsal CA3b/CA3a border targeting, coordinates for AAV.3x(core)mscRE4.YFP (retrograde) microinjection were X = +/−2.50; Y = −2.00, α = 0°, Z = −2.25. For cortical M1 injection, coordinates were X = −1.75; Y = 1.41; α = 0°; Z = −0.850 and −0.55. Virus was injected slowly (∼0.02 μL min−1) using a Picospritzer (for dentate, 0.15–0.2 μL; for M1, ∼0.25 μL for each Z injection depth). After ejection of virus, micropipettes were held in place (≥5 min) before withdrawal. The scalp was closed with surgical sutures and secured with Vetbond (3M) tissue adhesive. After allowing for onset of expression (hippocampus, 1–2 weeks; cortical, 3 weeks), animals were sacrificed and acute slices harvested.
Retro-orbital injections
Male and female C57Bl6/J adult mice (7–11 week old) were given AAV retro-orbital injections which cross the blood-brain barrier, as previously described in Chan et al., 2017.19 Mice were first anesthetized with 1.8–2% isoflurane. AAV(PHP.eB).mscRE4.minBGpromoter.iCre (‘AAV.mscRE4.Cre’; titer 6.88E+12 vg/mL) was titrated to approximately 1 × 1011 viral genome copies in sterile saline. Injections of the titrated virus were performed in the retro-orbital sinus of the left eye using a 31G × 5/16 TW needle on a 3/10 mL insulin syringe. Mice were kept on a heating pad for the duration of the procedure until recovery, followed by a 5-6-week viral expression period until sample collection.
‘Magic cut’ slice preparation and tissue fixation
Mice were anesthetized with isoflurane and sacrificed by decapitation. The brain was immediately removed by dissection and submerged in ice-cold cutting solution (in mM) 87 NaCl, 25 NaHO3, 2.5 KCl, 1.25 NaH2PO4, 7 MgCl2, 0.5 CaCl2, 10 glucose, and 7 sucrose. Brain slices (100–200 μm) were sectioned using a vibrating blade microtome (VT1200S, Leica Biosystems) in the same solution. The ‘magic cut’23 was employed to maintain the GC-CA3 mossy fibers within the slice. To obtain the magic cut, following dissection from the cranium brains were cut with a scalpel to remove the cerebellum (α = 90°) and the prefrontal cortex (α =90°). A medial cut was then made straight through the midline (α = 90°). Each hemisphere then received a cut angled ∼22° inward to the midline at the anterior side of the brain. Acute slices were then transferred to a well-plate to fix in 4% paraformaldehyde (in PB) for 1–1.5 h at room temperature. After fixation, slices were washed in 1xPBS for 5 min. A subset of slices were then incubated in DAPI in 1xPBS (1:1000) for 5 min. Slices were then mounted onto glass slides with Slowfade Diamond Antifade (Invitrogen), cover-slipped, and stored at 4°C until imaging.
Immunohistochemistry
For a subset of slices obtained from mice injected with AAV.3x(core)mscRE4.YFP (retrograde), immunohistochemistry was performed to further assess viral expression. Free-floating sections from one series were incubated in 0.1% (v/v) Triton X-100 and 3% (w/v) hydrogen peroxide to eliminate endogenous peroxidase, rinsed in PBS, and pre-blocked in 5% (v/v) normal donkey serum for 30 min at room temperature (RT). Sections were incubated in rabbit anti-GFP antibody (1:1000, A111222, Life Technologies, Carlsbad, CA) in PBS at RT for 24 h, then rinsed and incubated for 1 h at RT in biotinylated donkey anti-rabbit antibody (1:250, 711-065-152, Jackson Immunoresearch Laboratories, West Grove, PA) in PBS. After several rinses in PBS, the sections were incubated in avidin–biotin complex (ABC Elite; Vector Laboratories, Burlingame, CA, USA) for 90 min at 4°C. Immunoreactivity was visualized by incubation in 0.05% 3,30-diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO, USA) and 0.01% hydrogen peroxide in PBS, until a dark brown reaction product was evident (∼12 min). Sections were rinsed and mounted on glass slides, air dried, stained with cresyl violet and coverslipped.
Quantification and statistical analysis
Cell counting quantification
Acute slices containing the dorsal dentate gyrus from the injected hemisphere of male (n = 2) and female (n = 2) C57Bl/6J mice injected with both AAV.3x(core)mscRE4.YFP and AAV.CaMKIIα.mCherry were imaged on a Nikon C2 laser-scanning confocal system with an inverted Nikon ECLIPSE Ti2 microscope (10x objective, dry). Cells were counted in the hilus and the granule cell layer from two acute slices per animal. Cell counting was performed using the freehand selection tool and ROI manager in ImageJ/FIJI. Images were first assessed looking at the TRITC channel alone, so putative cells could first be confirmed as mCherry+ (CaMKIIα-expressing) before assessing the YFP signal. If the fluorescence value of a putative cell in the TRITC channel exceeded that of the mean plus three times the standard deviation of the background signal in that image, then the cell was considered mCherry+. If a putative cell was mCherry+, we then changed the image to the FITC channel to measure the same cell for YFP signal (mscRE4 expression). If the fluorescence value of YFP in that cell exceeded the mean plus three times the standard deviation of the background in the FITC channel, that cell would also be considered YFP+. All putative cells in the hilar region were counted, while in the granule cell layer 45 cells per slice were counted (N = 360). Raw data was analyzed in Excel to determine the percentages of mCherry+/YFP+ and mCherry+/YFP- cells in the hilar region and granule cell layer of each acute slice. These percentages were then inputted into GraphPad Prism for figure generation.
Line scan profiles in the DG
Acute slices containing the dorsal dentate gyrus from both the injected and non-injected hemisphere of co-injected AAV.3x(core)mscRE4.YFP/AAV.CaMKIIα.mCherry male (n = 1) and female (n = 2) C57Bl/6J mice were imaged on a Nikon C2 laser-scanning confocal system with an inverted Nikon Ti2 microscope (10x objective, dry). Three acute slices from each the injected and non-injected hemispheres were analyzed per animal. Line scan profiles were performed using the straight-line tool and ‘Plot Profile’ analysis in ImageJ/FIJI. Ten lines were drawn from the innermost border of the granule cell layer to the edge of the outer molecular layer on the DAPI channel of each image. Lines were drawn approximately equidistant from one another, evenly spanning the upper and lower blades of the dentate gyrus. Line scan values were then obtained on each the DAPI, mCherry, and YFP channels. As the thickness of the hippocampus varies along its axis, fluorescence values from each line scan were fed into a custom written algorithm, which normalized all values to the mean and subsequently interpolated fluorescence values based on a standardized distance (0–1). This data was then inputted into GraphPad Prism for figure generation.
Acknowledgments
We thank Annie M. Goettemoeller (Emory GDDBS) for help in surgical training supporting these studies. The current work was supported by the following grants: 1R56AG072473 (M.J.M.R.), 1RF1AG079269 (M.J.M.R.), and 1R21NS133960 (M.J.M.R.).
Author contributions
E.B., M.J.M.R., and V.J.O. designed experiments and wrote the original manuscript. E.B., C.-A.G., G.A.V., and M.J.M.R. conducted the experiments. E.B., V.J.O., and A.E. completed data analysis. K.A.P., J.Q.Z., and C.J.M. provided guidance on the experiments and manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: January 10, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100684.
Contributor Information
Viktor Janos Oláh, Email: viktor.janos.olah@emory.edu.
Matthew J.M. Rowan, Email: mjrowan@emory.edu.
Supplemental information
References
- 1.Andersen P., Morris R., Amaral D., Bliss T., O’Keefe J., editors. The Hippocampus Book. Oxford University Press; 2006. Copyright Page. [Google Scholar]
- 2.Buzsáki G., Moser E.I. Memory, navigation and theta rhythm in the hippocampal-entorhinal system. Nat. Neurosci. 2013;16:130–138. doi: 10.1038/nn.3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartley T., Lever C., Burgess N., O’Keefe J. Space in the brain: how the hippocampal formation supports spatial cognition. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014;369 doi: 10.1098/rstb.2012.0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oliva A., Fernández-Ruiz A., Buzsáki G., Berényi A. Spatial coding and physiological properties of hippocampal neurons in the Cornu Ammonis subregions. Hippocampus. 2016;26:1593–1607. doi: 10.1002/hipo.22659. [DOI] [PubMed] [Google Scholar]
- 5.Rolls E.T. The mechanisms for pattern completion and pattern separation in the hippocampus. Front. Syst. Neurosci. 2013;7:74. doi: 10.3389/fnsys.2013.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakahara S., Adachi M., Ito H., Matsumoto M., Tajinda K., van Erp T.G.M. Hippocampal Pathophysiology: Commonality Shared by Temporal Lobe Epilepsy and Psychiatric Disorders. Neurosci. J. 2018;2018 doi: 10.1155/2018/4852359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbarosie M., Avoli M. CA3-Driven Hippocampal-Entorhinal Loop Controls Rather than Sustains In Vitro Limbic Seizures. J. Neurosci. 1997;17:9308–9314. doi: 10.1523/JNEUROSCI.17-23-09308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbarosie M., Louvel J., Kurcewicz I., Avoli M. CA3-Released Entorhinal Seizures Disclose Dentate Gyrus Epileptogenicity and Unmask a Temporoammonic Pathway. J. Neurophysiol. 2000;83:1115–1124. doi: 10.1152/jn.2000.83.3.1115. [DOI] [PubMed] [Google Scholar]
- 9.Bazigou E., Lyons O.T.A., Smith A., Venn G.E., Cope C., Brown N.A., Makinen T. Genes regulating lymphangiogenesis control venous valve formation and maintenance in mice. J. Clin. Invest. 2011;121:2984–2992. doi: 10.1172/JCI58050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cembrowski M.S., Wang L., Sugino K., Shields B.C., Spruston N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. Elife. 2016;5 doi: 10.7554/eLife.14997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimidschstein J., Chen Q., Tremblay R., Rogers S.L., Saldi G.-A., Guo L., Xu Q., Liu R., Lu C., Chu J., et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci. 2016;19:1743–1749. doi: 10.1038/nn.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vormstein-Schneider D., Lin J.D., Pelkey K.A., Chittajallu R., Guo B., Arias-Garcia M.A., Allaway K., Sakopoulos S., Schneider G., Stevenson O., et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci. 2020;23:1629–1636. doi: 10.1038/s41593-020-0692-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mich J.K., Graybuck L.T., Hess E.E., Mahoney J.T., Kojima Y., Ding Y., Somasundaram S., Miller J.A., Kalmbach B.E., Radaelli C., et al. Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 2021;34:108754. doi: 10.1016/j.celrep.2021.108754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair R.R., Blankvoort S., Lagartos M.J., Kentros C. Enhancer-Driven Gene Expression (EDGE) Enables the Generation of Viral Vectors Specific to Neuronal Subtypes. iScience. 2020;23 doi: 10.1016/j.isci.2020.100888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blankvoort S., Witter M.P., Noonan J., Cotney J., Kentros C. Marked Diversity of Unique Cortical Enhancers Enables Neuron-Specific Tools by Enhancer-Driven Gene Expression. Curr. Biol. 2018;28:2103–2114.e5. doi: 10.1016/j.cub.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graybuck, L.T., Daigle, T.L., Sedeño-Cortés, A.E., Walker, M., Kalmbach, B., Lenz, G.H., Morin, E., Nguyen, T.N., Garren, E., Bendrick, J.L., et al. (2021). Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron 0. [DOI] [PMC free article] [PubMed]
- 17.Radhiyanti P.T., Konno A., Matsuzaki Y., Hirai H. Comparative study of neuron-specific promoters in mouse brain transduced by intravenously administered AAV-PHP. Neurosci. Lett. 2021;756 doi: 10.1016/j.neulet.2021.135956. [DOI] [PubMed] [Google Scholar]
- 18.Gray L.T., Yao Z., Nguyen T.N., Kim T.K., Zeng H., Tasic B. Layer-specific chromatin accessibility landscapes reveal regulatory networks in adult mouse visual cortex. Elife. 2017;6 doi: 10.7554/eLife.21883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.-L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasrallah K., Frechou M.A., Yoon Y.J., Persaud S., Gonçalves T., Castillo P.E. Activity-dependent LTP in the dentate gyrus promotes epileptic seizures. bioRxiv. 2021 Preprint at. [Google Scholar]
- 21.Kirschen G.W., Shen J., Tian M., Schroeder B., Wang J., Man G., Wu S., Ge S. Active Dentate Granule Cells Encode Experience to Promote the Addition of Adult-Born Hippocampal Neurons. J. Neurosci. 2017;37:4661–4678. doi: 10.1523/JNEUROSCI.3417-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cearley C.N., Wolfe J.H. Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol. Ther. 2006;13:528–537. doi: 10.1016/j.ymthe.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 23.Bischofberger J., Engel D., Li L., Geiger J.R.P., Jonas P. Patch-clamp recording from mossy fiber terminals in hippocampal slices. Nat. Protoc. 2006;1:2075–2081. doi: 10.1038/nprot.2006.312. [DOI] [PubMed] [Google Scholar]
- 24.Botterill J.J., Gerencer K.J., Vinod K.Y., Alcantara-Gonzalez D., Scharfman H.E. Dorsal and ventral mossy cells differ in their axonal projections throughout the dentate gyrus of the mouse hippocampus. Hippocampus. 2021;31:522–539. doi: 10.1002/hipo.23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnston S., Parylak S.L., Kim S., Mac N., Lim C., Gallina I., Bloyd C., Newberry A., Saavedra C.D., Novak O., et al. AAV ablates neurogenesis in the adult murine hippocampus. Elife. 2021;10 doi: 10.7554/eLife.59291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar P., Goettemoeller A.M., Espinosa-Garcia C., Tobin B.R., Tfaily A., Nelson R.S., Natu A., Dammer E.B., Santiago J.V., Malepati S., et al. Native-state proteomics of Parvalbumin interneurons identifies novel molecular signatures and metabolic vulnerabilities to early Alzheimer’s disease pathology. bioRxiv. 2023 doi: 10.1038/s41467-024-47028-7. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kole K., Voesenek B.J.B., Brinia M.E., Petersen N., Kole M.H.P. Parvalbumin basket cell myelination accumulates axonal mitochondria to internodes. Nat. Commun. 2022;13:7598. doi: 10.1038/s41467-022-35350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong S., Doughty M., Harbaugh C.R., Cummins A., Hatten M.E., Heintz N., Gerfen C.R. Targeting Cre Recombinase to Specific Neuron Populations with Bacterial Artificial Chromosome Constructs. J. Neurosci. 2007;27:9817–9823. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniguchi H., He M., Wu P., Kim S., Paik R., Sugino K., Kvitsiani D., Fu Y., Lu J., Lin Y., et al. A Resource of Cre Driver Lines for Genetic Targeting of GABAergic Neurons in Cerebral Cortex. Neuron. 2011;71:995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amat S.B., Rowan M.J.M., Gaffield M.A., Bonnan A., Kikuchi C., Taniguchi H., Christie J.M. Using c-kit to genetically target cerebellar molecular layer interneurons in adult mice. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song A.J., Palmiter R.D. Detecting and Avoiding Problems When Using the Cre/lox System. Trends Genet. 2018;34:333–340. doi: 10.1016/j.tig.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller-Komorowska D., Opitz T., Elzoheiry S., Schweizer M., Ambrad Giovannetti E., Beck H. Nonspecific Expression in Limited Excitatory Cell Populations in Interneuron-Targeting Cre-driver Lines Can Have Large Functional Effects. Front. Neural Circ. 2020;14:16. doi: 10.3389/fncir.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanahira C., Higo S., Watanabe K., Tomioka R., Ebihara S., Kaneko T., Tamamaki N. Parvalbumin neurons in the forebrain as revealed by parvalbumin-Cre transgenic mice. Neurosci. Res. 2009;63:213–223. doi: 10.1016/j.neures.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Nigro M.J., Kirikae H., Kjelsberg K., Nair R.R., Witter M.P. Not All That Is Gold Glitters: PV-IRES-Cre Mouse Line Shows Low Efficiency of Labeling of Parvalbumin Interneurons in the Perirhinal Cortex. Front. Neural Circ. 2021;15:781928. doi: 10.3389/fncir.2021.781928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delzor A., Dufour N., Petit F., Guillermier M., Houitte D., Auregan G., Brouillet E., Hantraye P., Déglon N. Restricted Transgene Expression in the Brain with Cell-Type Specific Neuronal Promoters. Hum. Gene Ther. Methods. 2012;23:242–254. doi: 10.1089/hgtb.2012.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffin J.M., Fackelmeier B., Fong D.M., Mouravlev A., Young D., O’Carroll S.J. Astrocyte-selective AAV gene therapy through the endogenous GFAP promoter results in robust transduction in the rat spinal cord following injury. Gene Ther. 2019;26:198–210. doi: 10.1038/s41434-019-0075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoshino C., Konno A., Hosoi N., Kaneko R., Mukai R., Nakai J., Hirai H. GABAergic neuron-specific whole-brain transduction by AAV-PHP.B incorporated with a new GAD65 promoter. Mol. Brain. 2021;14:33. doi: 10.1186/s13041-021-00746-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hrvatin S., Tzeng C.P., Nagy M.A., Stroud H., Koutsioumpa C., Wilcox O.F., Assad E.G., Green J., Harvey C.D., Griffith E.C., Greenberg M.E. A scalable platform for the development of cell-type-specific viral drivers. Elife. 2019;8 doi: 10.7554/eLife.48089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandael D., Borges-Merjane C., Zhang X., Jonas P. Short-Term Plasticity at Hippocampal Mossy Fiber Synapses Is Induced by Natural Activity Patterns and Associated with Vesicle Pool Engram Formation. Neuron. 2020;107:509–521.e7. doi: 10.1016/j.neuron.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sasaki T., Piatti V.C., Hwaun E., Ahmadi S., Lisman J.E., Leutgeb S., Leutgeb J.K. Dentate network activity is necessary for spatial working memory by supporting CA3 sharp-wave ripple generation and prospective firing of CA3 neurons. Nat. Neurosci. 2018;21:258–269. doi: 10.1038/s41593-017-0061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cayco-Gajic N.A., Silver R.A. Re-evaluating Circuit Mechanisms Underlying Pattern Separation. Neuron. 2019;101:584–602. doi: 10.1016/j.neuron.2019.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pofahl M., Nikbakht N., Haubrich A.N., Nguyen T., Masala N., Distler F., Braganza O., Macke J.H., Ewell L.A., Golcuk K., Beck H. Synchronous activity patterns in the dentate gyrus during immobility. Elife. 2021;10 doi: 10.7554/eLife.65786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker S., Vieweg P., Gao F., Gilboa A., Wolbers T., Black S.E., Rosenbaum R.S. The Human Dentate Gyrus Plays a Necessary Role in Discriminating New Memories. Curr. Biol. 2016;26:2629–2634. doi: 10.1016/j.cub.2016.07.081. [DOI] [PubMed] [Google Scholar]
- 44.Olah V.J., Goettemoeller A.M., Rayaprolu S., Dammer E.B., Seyfried N.T., Rangaraju S., Dimidschstein J., Rowan M.J.M. Biophysical Kv3 channel alterations dampen excitability of cortical PV interneurons and contribute to network hyperexcitability in early Alzheimer’s. Elife. 2022;11 doi: 10.7554/eLife.75316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bui A.D., Nguyen T.M., Limouse C., Kim H.K., Szabo G.G., Felong S., Maroso M., Soltesz I. Dentate gyrus mossy cells control spontaneous convulsive seizures and spatial memory. Science. 2018;359:787–790. doi: 10.1126/science.aan4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper analyzes an existing, publicly available dataset (‘Hipposeq’ from Cembrowski et al., 2016). Access to this dataset is listed in the key resources table. All microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
A custom algorithm was generated to normalize line scan measurements; the DOI to access this code can be found in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact (mjrowan@emory.edu) upon request.