Summary
In early neurodevelopment, the central nervous system is established through the coordination of various neural organizers directing tissue patterning and cell differentiation. Better recapitulation of morphogen gradient production and signaling will be crucial for establishing improved developmental models of the brain in vitro. Here, we developed a method by assembling polydimethylsiloxane devices capable of generating a sustained chemical gradient to produce patterned brain organoids, which we termed morphogen-gradient-induced brain organoids (MIBOs). At 3.5 weeks, MIBOs replicated dorsal-ventral patterning observed in the ganglionic eminences (GE). Analysis of mature MIBOs through single-cell RNA sequencing revealed distinct dorsal GE-derived CALB2+ interneurons, medium spiny neurons, and medial GE-derived cell types. Finally, we demonstrate long-term culturing capabilities with MIBOs maintaining stable neural activity in cultures grown up to 5.5 months. MIBOs demonstrate a versatile approach for generating spatially patterned brain organoids for embryonic development and disease modeling.
Keywords: morphogen gradient, diffusion, brain organoids, patterning, ganglionic eminence, GABAergic projection neurons, interneurons, dorsal-ventral axis
Graphical abstract
Highlights
-
•
We engineer a microdevice for mimicking morphogen gradients in 3D human hPSC tissues
-
•
The non-invasive approach enables long-term culture out of the device
-
•
We produce mature organoids with ganglionic eminence cell types
Motivation
The use of small molecules for the guided differentiation of brain organoids has proven to be a useful tool for modeling various aspects of early neurodevelopment. Still, the embryonic brain is patterned through a combination of morphogen gradients stemming from organizer regions. To address this inconsistency, we designed a device capable of mimicking neural organizers by maintaining a steady morphogen gradient. We used this device to expose forebrain organoids to multiple gradient conditions and cataloged the diversity of cell types produced.
In early fetal development, the central nervous system forms through intricate interactions of patterning factors. Pavon et al. engineer a device capable of reproducing signaling molecule gradients for generating morphogen-gradient-induced brain organoids, spatially patterned brain organoids that better recapitulate embryonic development.
Introduction
Early human neurodevelopment relies on a wide variety of signaling factors to accurately and efficiently pattern the brain. Prior studies in model systems revealed that the diffusion and reaction-diffusion of various morphogens can instruct neural induction and, subsequently, cell fate patterning, leading to the establishment of the dorsal-ventral (D-V) and anterior-posterior (A-P) axes of the central nervous system (CNS). At the most anterior region of the developing neural tube, the primary brain vesicle prosencephalon further develops into telencephalon, which encompasses the cerebrum.1,2 The telencephalon itself produces a wide range of diverse cell types in a D-V-dependent manner.3 The dorsal-most regions of the telencephalon become the cortex, and the ventral regions give rise to the subpallium.
Transiently, during the embryonic stages of neurodevelopment, the subpallium holds the ganglionic eminences (GEs) comprising the medial GE (MGE), lateral GE (LGE), and caudal GE (CGE). The GEs contain ventral ventricular zones (VZs) responsible for the production of GABAergic interneurons (INs) and projection neurons.4,5,6 The GABAergic cell types go on to innervate a wide variety of regions in the telencephalon depending on place of origin. The MGE produces INs that will migrate into the cortex and striatum.6,7,8,9,10,11 The LGE generates medium spiny neurons (MSNs) for the striatum and INs that will migrate into the olfactory bulb (OB).12,13,14,15,16 Finally, the CGE produces INs that will travel to the striatum, cortex, and amygdala.13,17,18 While much of what we know of the GEs has come from animal models, recent characterizations of postmortem human fetal brain tissue have aided in enlightening the unique transcriptomes of the human GEs.19,20,21,22,23 Due to the unique progenitor pools of the GEs, reproducing the anatomical regionalization in GEs is a key component of accurately modeling brain development.
To facilitate further investigation into the complexity of human neurodevelopment, in recent years, 3D human pluripotent stem cell (hPSC)-derived cell cultures, termed neural organoids, have been developed to serve as models for various regions of the CNS depending on the protocol used.24,25,26,27,28,29,30,31,32,33,34,35,36,37,38 Particularly, forebrain organoids have been well established using guided differentiation techniques to generate region-specific organoids modeling single brain regions such as the cortex, striatum, MGE, and optic cup, respectively.27,28,39,40,41,42 To model the connectivity of multiple embryonic brain regions, organoids representing different brain regions have been derived separately, before fusing to form assembloids.39,40,41,43,44,45 While proven useful, current protocols do not fully recapitulate the early neural patterning processes.
At present, protocols that enable the generation of D-V or A-P axes in a single organoid through establishment of morphogenic gradients are extremely limited. To overcome these limitations, unique methods have been developed including the chimeric organoid model, which fused an aggregate of Sonic hedgehog (Shh)-expressing cells to a forebrain organoid, thus mimicking the effect of an SHH organizer (SHH organoid).46 SHH organoids were grown and matured to 70 days, where they confirmed topography established at day 20 remained largely discrete and contained cell types of cortical, MGE, and hypothalamic lineages. Alternatively, microfluidic devices have been designed to generate defined exogenous chemical gradients by a planar dilution network to pattern the A-P axis of an entire neural tube.47,48,49,50 While such a microfluidic gradient generator offers the adaptability of creating chemical gradients for a wide variety of signaling molecules in a controlled manner, it is still challenging to establish a steep gradient within tissues with low aspect ratios such as most organoids. In addition, microfluidic technologies have been applied mostly to adherent tissues, and the limited space in enclosed microfluidic channels prevents long-term culture or transferring of patterned tissues, which is essential for developing mature neural organoids. Moreover, to generate the chemical gradient in a microfluidic system, the use of peristaltic pumps and copious amounts of media are required, hindering its ability to scale up and be adopted by other research labs. Hence, a more accessible alternative method with the capability to reliably produce a gradient and allow for long-term culture is desired.
In the current study, we describe a bioengineering approach to generate morphogen gradient -induced brain organoids (MIBOs). Leveraging the properties of passive diffusion to create a sustained chemical gradient of purmorphamine (Pur), a Shh agonist, we designed and optimized a microdevice for 3D tissue culture to induce D-V patterning within a single forebrain organoid. Using this technology, MIBOs were patterned for 19 days and further cultured out of the device long-term with observable maturation up to 5.5 months. Gradient quantification of 3.5-week-old MIBOs revealed robust gradient magnitudes for both NKX2.1 and PAX6. Single-cell transcriptomic analysis of 4-month-old MIBOs revealed composition of cell types primarily arising from the LGE and CGE with contribution of the MGE to a smaller degree. Finally, MIBOs were functionally active as indicated by live calcium imaging analysis. Thus, MIBOs can be fine-tuned with specific morphogenic gradients to achieve spatially patterned and functionally mature brain organoids.
Results
Fabricating diffusion gradient device for 3D cultures
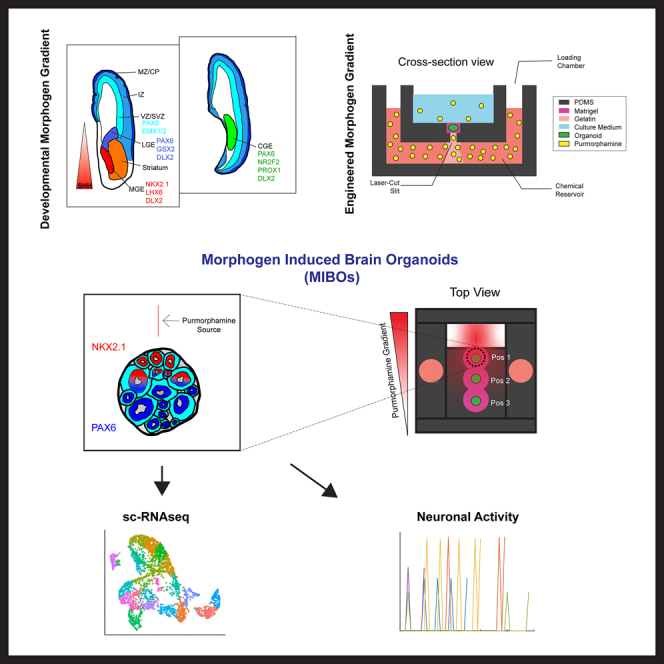
We have previously developed a localized passive diffusion (LPaD) device to induce sustained concentration gradients of Pur to pattern 2D neuroepithelial cell sheets.51 In the current study, we have altered the design of LPaD for 3D organoid culture (Figure S1A). First, we added polydimethylsiloxane (PDMS) sidewalls to self-contain the culture medium (Figure S1B). Doing so allowed us to reduce media usage by 10-fold. Further, to ensure that organoids are continuously exposed to the Pur gradient from the same direction, we embedded organoids in Matrigel to limit the rotation of the organoids (Figures 1A and 1B). To facilitate the Matrigel embedding, we added a structure within the culturing area, allowing for the droplet formation needed to Matrigel embed our organoids at three varying distances from the Pur source (Figures 1B, S1A, and S1B).
Figure 1.
Design and generation of MIBOs
(A) Schematic overview of culturing protocol in gradient device with representative bright-field images. All scale bars are 500 μm.
(B) Cross-section of PDMS device showing the controlled passive diffusion of Pur molecules from the chemical reservoir into the culturing medium. Top view of PDMS device showing the three possible positions within the device and the effect of Pur gradient with distance from the source. (Dotted lines) Phase image of a freshly embedded organoid in a device with gelatin drop (white dotted line) over 200-μm slit. Scale bar, 500 μm.
(C) (Left) Cartoon representation of a patterned organoid in a PDMS device. Regions closest to the Pur source produce NKX2.1+ rosettes, while areas further away produce PAX6+ rosettes.
(D) Cartoon renderings of coronal sections of the human telencephalon at 15 post-conception weeks (pcw) made with references from the Allen Brain Atlas. The colored rostral telencephalon (right) depicts MGE, LGE, and cortical regions of the embryonic brain along with markers expressed by the progenitors of each region. Black and gray rostral telencephalon depicts migratory routes populating the striatum and cortex with GABAergic projection neurons and INs from the LGE, CGE, and MGE along with common markers associated with each. Caudal telencephalon shows CGE and markers expressed by its progenitors and INs. See also Figure S1.
Given the alterations to the original 2D design and the drastic change in media volumes, we reanalyzed the gradient profile established by our new device. The fluorescent dye, DAPI, was chosen for having a molecular weight (277 g/mol) similar to many small molecule morphogens that could be used with our device. DAPI intensity retained within the Matrigel across three conditions helped to quantify and verify the efficacy of our device’s ability to generate gradients using passive diffusion. In the uniform DAPI condition, 1× DPBS containing 10 μg/mL of DAPI was added directly into the culturing medium, completely dousing the Matrigel layer. As such, the uniform DAPI condition shows indiscriminate DAPI fluorescence throughout all three positions (Figure S1C). In contrast, when DAPI-containing media is added only to the chemical reservoir and DPBS alone is added to the culturing medium, the concentration gradient takes effect, and the Matrigel area closest to the source (position 1) displays the highest degree of DAPI intensity (“gradient DAPI,” Figure S1C). The gradient steepness is displayed as intensity greatly decreases near positions 2 and 3 (Figure S1D). Finally, the “no DAPI” condition shows that without the presence of DAPI, our Matrigel layer has little to no background signal as expected (Figures S1C and S1D).
Deriving MIBOs via an exogenous Pur gradient
We next cultured forebrain organoids using a modified version of the protocol developed by Cederquist et al. in 2019 (Figure 1A). Without activation of the SHH pathway, a default dorsal-anterior forebrain fate was anticipated. Once organoids were Matrigel embedded in our device, organoids were immobile without significant hydrogel degradation and were capable of being held in place for the subsequent 19 days in the device. Organoids continued to grow stably and exhibited neuroepithelial organization during this period despite immobilization (Figure 1A). At day 25 (3.5 weeks), organoids were carefully removed from devices and transferred into ultra-low attachment dishes, which were then placed on orbital shakers for the remainder of the culturing scheme. Long-term organoids grew to maturity for further analysis using immunohistochemistry (IHC), single-cell RNA sequencing, and calcium imaging. Using this two-step protocol, MIBOs cultured in our devices are not limited by the space constraints of the device and can be cultured long-term for further maturation and characterization after the initial patterning period.
Successful dorsal-ventral patterning in MIBOs
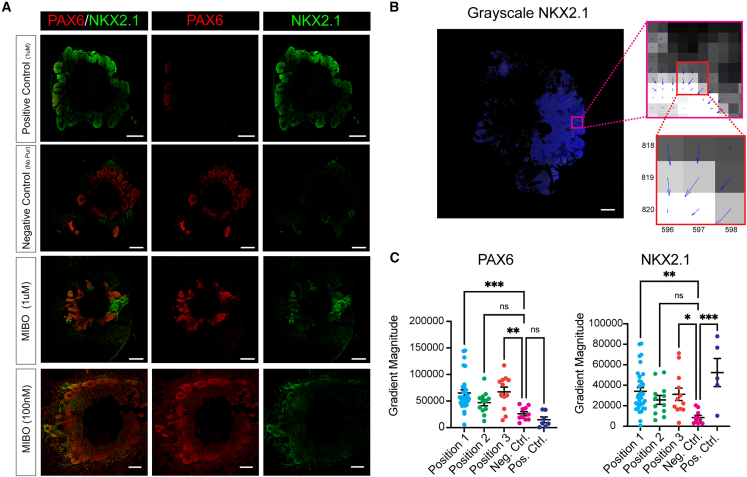
Having established organoid viability using our device, we next sought to characterize MIBOs that had been exposed to a concentration gradient of 1 μM Pur. Here, we observed that MIBOs had simultaneous but mutually exclusive expression of the dorsal (PAX6) and ventral (NKX2.1) forebrain markers (Figures 2A and S2). With 1 μM Pur, we robustly observed NKX2.1-expressing as well as PAX6-expressing neural rosettes within a single organoid (Figures 2A and S2A and S2B). This unique expression pattern further emphasizes the ability for our device to procure ventral gradation even within a single progenitor domain. Next, we developed a MATLAB code (see STAR Methods) that uses a Sobel gradient operator to quantify the gradient magnitude of a grayscale image of PAX6 or NKX2.1 staining (Figures 2B and 2C). The analysis showed a significant increase in the gradient magnitude for MIBOs compared to our positive controls (adding 1 μM Pur directly into the culture medium) and negative controls (no Pur added). As expected, the positive control showed a high degree of NKX2.1 expression in organoids (Figures 2A and 2C). In contrast, the negative control exhibited homogeneous PAX6 expression throughout the organoid (Figures 2A and 2C).
Figure 2.
Characterizing the patterning effects of Pur gradient on forebrain organoids
(A) Comparison of brain organoids grown in PDMS devices and subjected to varying concentrations of Pur with positive (adding 1 μM Pur directly into the culture medium) and negative controls (no Pur added). All scale bars represent 500 μm. Scale bar for 100-nM condition has a 200-μm scale bar.
(B) Breakdown of how gradient magnitude is calculated for each image. (Left) MATLAB analysis of grayscale NKX2.1 gradient magnitude. (Top) Zoomed in image showing blue quivers for each pixel. Size of quiver corresponds to size of gradient magnitude, and quiver angle displays the direction of greatest increase in pixel intensity. (Bottom) 3 × 3 neighborhood of pixels illustrating how the center pixel establishes a gradient direction and gradient magnitude based on its neighbors.
(C) (Left) Gradient magnitude measured in sections stained for PAX6 across all conditions, while MIBO (right) gradient magnitude is measured in sections stained for NKX2.1 across all conditions. Error bars represent +/− SEM. Statistical significance was obtained by one-way ANOVA with Dunnett’s test for multiple comparisons. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S2.
To determine if the cell fate patterning in MIBOs depends on the source concentration of Pur, we tested the effects of adding a 100 μM or 100 nM Pur concentration to the chemical reservoir. These two concentrations had originally been used for varying degrees of patterning effects in the 2D culture system.51 We observed full ventralization of the organoids exposed to a 100 μM Pur gradient (Figure S2C). NKX2.1 was uniformly expressed throughout these organoids. Interestingly, at 100 nM condition, we found that PAX6 was largely expressed by the radially arranged progenitors of the rosette structures. On the other hand, NKX2.1 expression was still detected, albeit minimally scattered across the regions just distal to the rosettes (Figure 2A).
Finally, we examined the effect of distance from Pur source on D-V patterning. We designed the Matrigel embedding area with three equally ascending distances from the Pur source, which we have termed here position 1, position 2, and position 3 (Figure 1B). Position 1 showed the most consistency in producing D-V patterning within the forebrain organoids. Within position 1, we observed consistent generation of regions expressing NKX2.1 within VZ progenitors, indicating emergence of MGE-like regions from the side of the organoid exposed to the highest concentration of Pur (Figure 2A). The rest of the organoid expressed PAX6, indicating a more dorsal fate. While position 2 and 3 organoids occasionally showed similar D-V patterning, the PAX6 and NKX2.1 expression was deemed stochastic (Figure S2A). As a result, hereafter, we focused our efforts on further characterizing the MIBOs produced in position 1 induced by 1 μM Pur source.
Investigating cell fates arising from progenitors of MIBOs through single-cell RNA-seq
Cederquist et al. had previously documented a stark difference in the size of PAX6+- versus NKX2.1+-expressing rosettes in the SHH organoids, where NKX2.1+ rosettes tended to be elongated when compared to the more circular PAX6+ rosettes. Within our patterned organoids, at the border of where markers transition, we noted several rosettes that partially expressed NKX2.1 and PAX6 (Figure S2B). Additionally, many PAX6-expressing rosettes also showed elongated morphologies, prompting us to further investigate the fate of mature cells produced by these patterned organoids in single cells.
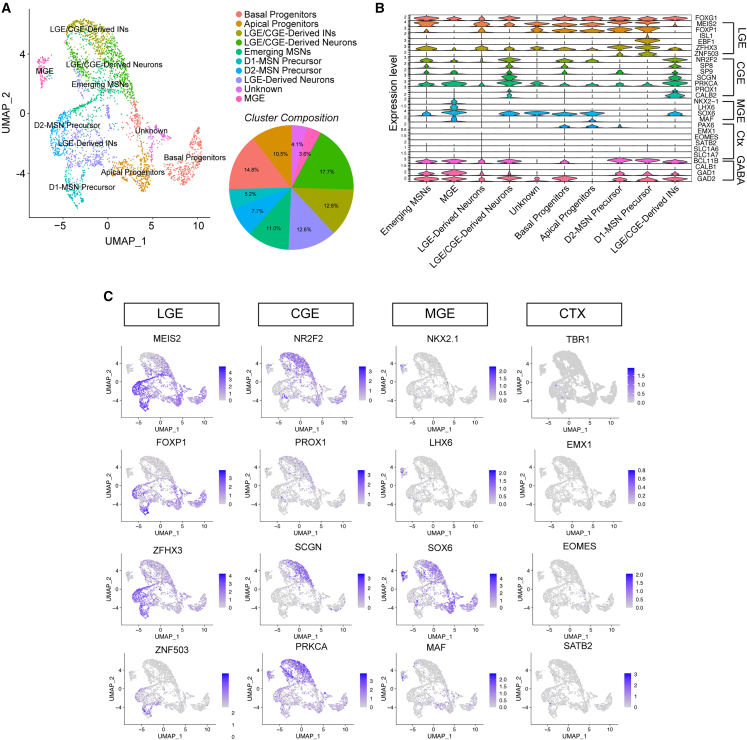
Single-cell transcriptomic analysis of 4-month-old MIBOs revealed minimal cortical or excitatory markers within our sample (Figure 3). Instead, we found that a large majority of our cells were GE derived in nature. Specifically, cluster cell annotation based on canonical markers identified LGE- and CGE-derived GABAergic cells with a small group of MGE-derived cells (Figures 3A–3C and S3C–S3H). From these data, we reasoned that PAX6 staining pattern observed at 3.5 weeks specifically marked dorsal subpallial progenitors of the LGE/CGE and not cortical progenitors of the pallium.11,19 Therefore, the patterning created in MIBOs was more representative of the D-V patterning observed in the GEs.
Figure 3.
Single-cell transcriptomics reveals GE fates in MIBOs
(A) UMAP embedding of 3,366 cells dissociated from position 1 MIBOs at 4 months. Pie chart displays percentage of cells pertaining to each annotated cluster.
(B) Violin plot highlighting representative lineage marker genes across annotated cell clusters.
(C) Feature maps showing expression for the top marker genes in the LGE, CGE, MGE, and cortex. See also Figure S3.
Indeed, cell annotations of our single-cell data found evidence of LGE-derived clusters with robust expression of known LGE markers, including MEIS2, FOXP1, and EBF1 (Figure 3C). Additionally, emerging MSN and MSN precursors for D1 and D2 subtypes were identified within our LGE-derived clusters (Figures 3A–3C).21 The GABAergic projection neurons of the LGE are thought to be primarily produced in the ventral LGE (vLGE), with GABAergic INs more heavily produced by the dorsal (dLGE).11,52,53 Interestingly, the GABAergic IN population in our sample was almost entirely CALB2+ cells. Some of these CALB2+ INs could be LGE derived, while others derive from CGE progenitors due to high co-expression with NR2F2, SP8, PROX1, and PRKCA20,21,54,55 (Figure 3C). The LGE/CGE-derived neuron cluster was identified by co-expression of LGE/CGE markers with the presence of BCL11B+ and SOX6+ (Figures 3C and S3E). BCL11B is found with high fidelity in mature MSNs, as well as early postmitotic MSNs.56 Moreover, while SOX6 is strongly expressed in the MGE and CGE mantle zone (MZ), the MZ of the LGE (FOXP1+) lacks SOX6 expression and is notably the region of MSN maturation.56,57 Finally, MGE-derived neurons were identified in a small cluster that expressed the canonical markers NKX2.1, LHX6, and SOX6 (Figure 3C).
Gene Ontology analysis of MIBOs
To gain a better understanding of the cell-type specification obtained through the sustained exposure of a Pur concentration gradient, we performed differentially expressed gene (DEG) analysis (false discovery rate adjusted p < 0.01) of each annotated cluster. Using cell-type-specific DEGs, we performed a Gene Ontology (GO, org.Hs.eg.db: genome-wide annotation for human) analysis with an emphasis on understanding the biological processes associated with the top 100 DEGs (Figure 4). As expected, our apical and basal progenitor clusters were enriched for terms relating to proliferation, division, and differentiation (Figure 4B). In addition, the LGE/CGE-derived INs were enriched for synaptic signaling, neuronal migration, and telencephalon development (Figure 4C), which is in line with our initial classification and is indicative of migrating INs. Interestingly, our D1 and D2 MSN precursor clusters were both enriched for genes relating to the establishment of axonal projections and synaptic transmission, supporting their designation as potential GABAergic projection neurons. Likewise, LGE/CGE-derived neurons also showed high expression of axonal development genes but also included genes associated with OB development, possibly hinting that the annotated cluster might contain a portion of dLGE INs fated for the OB as well as vLGE/CGE GABAergic projection neurons. Finally, the cluster of MGE-like cells exceedingly expressed a variety of synaptic transmission and regulation genes.
Figure 4.
Gene Ontology focused on the biological processes associated with top 100 DEGs in annotated clusters (related to Figure 3)
(A) Heatmap emphasizing the top 5 DEGs across annotated clusters.
(B) Enrichment of genes associated with neural progenitor cells.
(C) Representative enriched gene sets mapped to biological processes across annotated clusters. Enrichment scores were defined as –log10 (false discovery rate adjusted p values).
Confirmation of protein expression in 3.5-month-old MIBOs using immunohistochemistry
We next sought to validate our results by immunostaining MIBOs that had been matured and collected at 3.5 months. Again, we saw wide expression of BCL11B+ and even some DARPP32+ cells indicating the presence of GABAergic projection neurons and possibly emerging MSNs (Figure S4A). The most abundant INs in MIBOs were CABL2+ cells that seemed to be undergoing tangential migration with most cells located at the outermost region of the organoid58,59 (Figure S4B). While many cells within our MIBOs appear to be undergoing migration, a significant portion of them were differentiated neurons expressing RBFOX3 (NEUN), in conjunction with mature GABAergic markers, such as GAD1and SLC32A1 (Figure S4C).
In summary, we have shown that MIBOs can produce a wide variety of cells derived from the GEs. At the current concentration of 1 μM Pur, NKX2.1+ regions of MIBOs are exclusive to small sections that continue to produce MGE-derived cells into maturity. The rest of the MIBOs represents the D-V axis of the LGE and CGE, thus producing a range of GABAergic projection neurons and primarily CALB2+ INs. These results support the notion that PAX6 expression and rosette morphology at 3.5 weeks were likely representative of dorsal LGE/CGE apical progenitors.
MIBOs show differential cell composition and gene expression profiles when compared to conventional brain organoid protocols
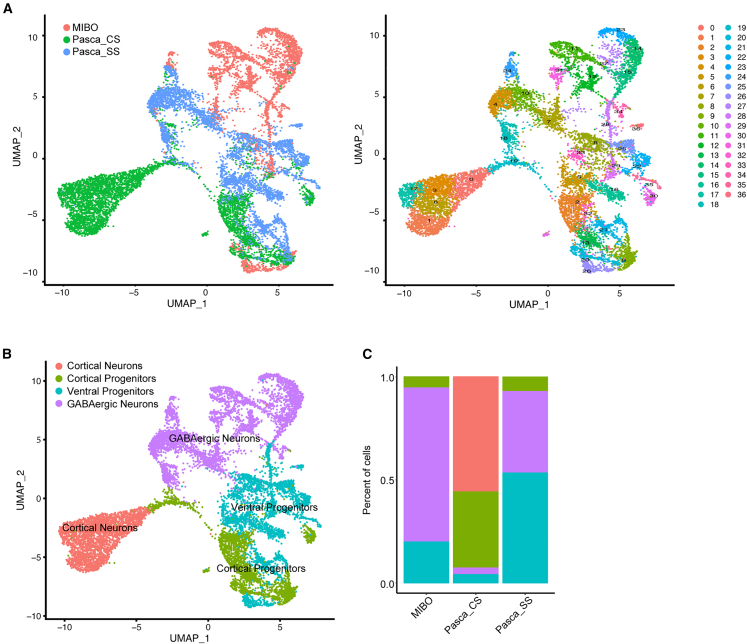
To further probe the unique features of MIBOs compared to other conventionally grown brain organoids cultured without a gradient device, we utilized and integrated the published single-cell transcriptomes of 105-day-old cortical (CS) and subpallial (SS) organoids from Birey et al. (2017)43 with our MIBO single-cell dataset for further analyses. Quality control was applied to filter cells and attained a total of 12,399 cells projected onto a Uniform Manifold Approximation and Projection (UMAP) containing all three datasets (Figure 5A). A total of 37 unique clusters were identified in the integrated dataset with all clusters being annotated into four broad groups for initial screening (Figure 5B). The first group, “cortical neurons,” was chosen by expression of glutamatergic synaptic marker SLC17A7, along with conventional mature neuronal markers, STMN2 and RBFOX3. “Cortical progenitors” were designated by the expression progenitor markers NES and HES1 with dorsal progenitor marker PAX6. Designated “GABAergic neurons” required expression of GAD1, GAD2, and SLC32A1 along with STMN2 and RBFOX3. Lastly, “ventral progenitors” were distinguished by NES and HES1 expression in conjunction with CRABP1 and LIN28A. From this initial subgrouping, the CS sample was clearly differentiated from others, containing a large percentage of cortical neurons and cortical progenitors. However, both SS and MIBO shared a large portion of the GABAergic neurons and ventral progenitors. We next sought to tease apart the unique intricacies of genetic expression that differentiated all three samples (Figure S5A).
Figure 5.
Integrated UMAPs identifying cluster contributions from each sample
(A) Integrated UMAP comparing MIBO dataset with conventional cortical and subpallial organoid protocols.
(B) Initial broad annotations for integrated dataset to allow first-pass interpretation.
(C) Stacked bar plot showing cell population yield based on annotations. See also Figures S5 and S6.
As previously described, MIBOs contain a large portion of BCL11B+ cells that seem to indicate the presence of a projection neuron subtype. The BCL11B+ cell population in MIBOs are mostly GABAergic and typically co-express GAD2. In contrast, the BCL11B+ cells in CS are glutamatergic, highly expressing SLC17A7, and likely representative of deep layer pyramidal neurons (Figure S5B). SLC32A1 was found broadly in our GABAergic neuron cluster but seemed to carry a stronger expression profile in the SS sample.
To further examine the BCL11B+ population of the CS, we sought to characterize the lineage of cortical progenitors that give rise to the mature glutamatergic cells. What we found was that despite MIBOs containing PAX6+ progenitors, it was the presence of EMX2 and to a lesser degree EMX1 that better indicated a cluster’s propensity for giving rise to cortical tissue (Figure S6). These cortical-fated cells also seem to uniquely undergo an EOMES+ intermediate stage that gives rise to a mature TBR1+ excitatory cell type.
Next, because all three samples contained a population of GABAergic neurons, we investigated the IN composition in the integrated dataset (Figures 5B and 5C). The two IN markers that were notably expressed were somatostatin (SST) and CALB2 (Figure S6A). The SST-containing cluster was made up in large part from SS cells. This is likely the case because SS cultures were continually exposed to the SHH pathway agonist SAG, thereby producing more MGE-representative tissue within their organoids.43 However, the presence CALB2+ cells were observed across all three samples (Figure S6A). Interestingly, across the SS and MIBO samples, the CALB2+ populations were quite distinct. In MIBO, CALB2+ cells were also strongly expressing the migration marker ERBB4, supporting our IHC results (Figure S6A). Moreover, CALB2+ INs in MIBO tended to more strongly express GAD2, whereas CALB2+ INs in SS displayed higher GAD1 and SLC32A1 expression, potentially hinting at an observable difference between dorsal GE (dGE)-derived CALB2+ INs and MGE-derived CALB2+ INs (Figure S6B). Curiously, the CS sample exhibited a large population of CALB2+ cells that were grouped with excitatory neurons. This population of CALB2+ cells within CS was puzzling as it had very little gene expression related to GABA production (Figure S6B). Still, this cluster shared the expression profile with the neuronal development gene, NRN1, which is involved in promoting neurite outgrowth and possibly hints at mature neuronal subtype.60,61
Characterizing the functioning neural circuits in MIBOs using calcium imaging analysis
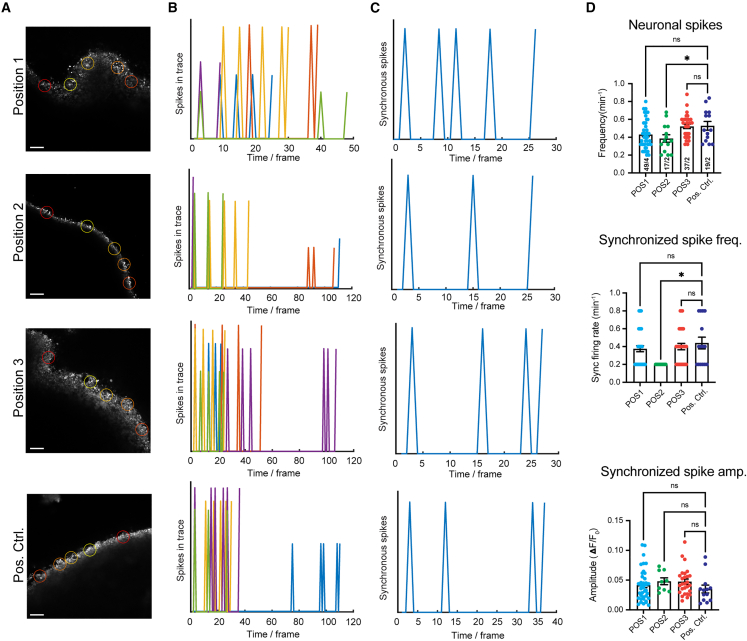
To better characterize the neural activities within MIBOs, we performed live Ca2+ imaging in MIBOs and a subpallial control organoid at 5.5 months (Figure 6). We observed stable and consistent levels of neural activity in their spontaneous Ca2+ spikes, synchronicity of spikes, and their amplitudes (Figures 6B–6D). We investigated if cell fate patterning influences neural maturation by testing Ca2+ activities in MIBOs cultured across three positions on the device. We found that Ca2+ activity in MIBOs across three positions was comparable to that of positive control organoids (Figure 6D). Though we observed a minor position variability in their neural activity, all three positions showed robust synchronous and spontaneous firing with expected metrics as previously published.62 Hence, our results show free-floating MIBOs that were cultured long term out of the device yield active and viable organoids with mature neural network characteristics.
Figure 6.
Neuronal activity captured through calcium imaging of MIBOs grown long term
(A) Representative confocal images of intact brain organoids during live Ca2+ imaging using X-Rhod-1 dye. Colored circles represent regions of interest (ROIs) selected for analysis.
(B) Corresponding spike activities captured within ROIs during recording.
(C) Network activity detected in brain organoids by synchronous spiking across multiple ROIs.
(D) Averaged data for synchronous spikes detected in each condition, as well as amplitudes (dF/F0) and frequencies of spontaneous spike activity, are shown in scatterplots. Data points represent averaged data from a single field of view (FOV) consisting of 5 ROIs per FOV. At least 5–7 FOVs are taken from each organoid, and 2–4 organoids were used per experiment. Error bars represent +/− SEM. Statistical significance was obtained by one-way ANOVA with Dunnett’s test for multiple comparisons. ∗p < 0.05.
Discussion
Developmental biology studies strongly support the diffusion and reaction-diffusion of morphogens in inducing tissue patterning. The D-V patterning in the developing neural tube is believed to be induced by Shh produced by the notochord and floor plate, as well as BMP produced in the roof plate. However, the D-V patterning mechanism in the GEs is still poorly defined. Here, we demonstrated that exposing forebrain organoids in a gradient of Pur is sufficient to generate GEs with segregated MGE region and CGE/LGE region. Compared with microfluidics-based approaches, our device takes advantage of passive diffusion of chemicals to generate a sustained chemical gradient without the need for continuous pumping and significant media consumption. Our open-chamber design allows for convenient organoid retrieval and extended culture, which is usually challenging in enclosed microfluidic devices.
Typically, in both 2D and 3D systems, the use of dual SMAD inhibition with WNT inhibition is thought to produce a primarily cortical phenotype.44,63,64 Similarly, in chimeric SHH organoids, the PAX6+ regions also represented cortical fate.46 Surprisingly, PAX6 expression observed in 3.5-week-old MIBOs was primarily identifying the dorsal GE regions of the LGE and CGE. However, given that MIBOs were exposed to a Pur gradient without antiparallel source of Shh antagonism (e.g., BMP4), we speculate that the low levels of persistent Pur throughout the media were sufficient to push a dorsal subpallial fate in the regions furthest from the Pur source. This was supported by the homogeneous expression of GABAergic markers and lack of glutamatergic cells in our MIBO single-cell data. In MIBOs, Pur gradient begins at around week 1, immediately after the establishment of forebrain neural stem cell lineage in the EBs. Namely, the “temporal adaptation” model suggests that, initially, cells will be highly responsive to Shh stimulation.65,66 Furthermore, protocols attempting to produce LGE-derived cell types will typically use lower concentrations of Pur, ranging from 0.5 to 0.65 μM.39,67,68,69,70 Therefore, it is likely that the Pur gradient created by our MIBO device was more conducive for the generation of dGE tissue in general.
It bares discussion that dGE cell types have previously been observed in unguided protocols.33,71,72,73 In fact, many cortical-generating protocols in both 2D and 3D display the spontaneous presence of GABAergic cell types or unintended inhibitory signals within their cultures.74 This might indicate that an SHH source is not necessarily required for the induction of dGE phenotypes or GABAergic cell-type generation. However, in these examples of spontaneous dGE generation, they are typically found in conjunction with a substantial production of glutamatergic excitatory cell types. In the current study, MIBOs were shown to be almost entirely GABAergic and representative of the three GE regions (MGE/LGE/CGE). Further, the key distinction in MIBO is the spatial topography of GE progenitors that go on to, in part, give rise to MGE-derived cells but also MSN precursors and dGE INs. In part, the spatial topography observed in MIBO parallels the D-V patterning of the GEs in vivo, further exhibiting the usefulness of a gradient-generating device and the effects of sustained a Pur gradient.
Excitingly, the device platform is promising for testing and characterizing the manipulation of different Pur concentrations in such a way that different concentration effects could be used to establish new protocols with varying degrees of MGE to LGE/CGE ratios. For example, we predict that an increase in Pur concentration in our chemical reservoir would likely generate increasingly larger MGE regions within our organoid. Potentially, Pur could be increased to such a degree that the D-V patterning of the MGE itself is induced, allowing closer examination of the development of dorsal MGE SST+ and ventral MGE PV+ INs.11,75,76,77
Our single-cell transcriptomic analysis of MIBOs revealed a fascinating diversity of GE-derived cell types. Specifically, we identified a small cluster of MGE-derived cells in addition to vLGE-derived MSNs and dLGE/CGE-derived INs. Cell types were annotated using key marker genes that have been well documented for each region.11,19,20,21,54,57,77,78,79,80,81,82,83,84 While the markers for delineating MGE-derived cells have been established and are unique to the region, the LGE and CGE tend to share many of the same markers (i.e., NR2F2, PAX6, MEIS2) with differences in expression levels serving as the only indicator of regional identity.55,85 Accurately discerning the LGE from the CGE still remains a challenge for the field. Traditionally, the only way to definitively discriminate the regions of the GEs was by anatomical landmarks (bulges). Recently, the field has made great strides that could help improve our understanding of these GEs through the use of single-cell RNA sequencing on rodent and human embryonic tissues to further identify unique markers for each region.20,21,81,84 Due to this ambiguity, it is possible that some clusters in our dataset house cells from both the LGE and CGE jointly. For example, the dLGE is known to produce CALB2+ INs that are fated for the OB, migrating through the rostral migratory stream.4,16,77,86 Likewise, the CGE produces CABL2+ INs that will tangentially migrate toward the cortex and/or striatum.9,87 Due to the similarity of gene expression between both regions, it is possible that the CALB2+ INs of both the LGE and CGE are clustered together in our dataset.
Interestingly, the majority of INs found in our MIBOs were CALB2+ cells. However, the CGE has been well characterized in its ability to produce CALB2+, VIP+, and RELN+ INs.17,88 This begs the question as to why our organoids primarily produced CALB2+ INs. It is plausible that we sequenced MIBOs at an age when diverse IN production was just beginning. Developmentally, the MGE produces cortical INs at an earlier time point than the CGE.17,89 MGE-like subpallial organoids were reported to begin IN production at around day 50 for SST+ and day 200 for PV+.28 Therefore, it is possible that allowing MIBOs to mature beyond 4 months would yield a higher amount of VIP+ or RELN+ INs. Further, CGE-derived INs were reported not to express their subtype markers until they have finished their migration and settled in their final destination, usually within the outermost layers of the cortex.17,90,91,92 It is theorized that, post migration, exogenous signals and local electrophysiological input from the local environment trigger mature marker expression. VIP+ cells often co-express CALB2+ as well. Therefore, there is potential for a portion of the currently identified migrating CALB2+ INs to mature into VIP+ or RELN+ co-expressing INs. Finally, we must consider the temporal effects on IN production in the CGE. Previously, it has been shown that the MGE alters its ratio of SST+ and PV+ IN production temporally.89 In contrast, the IN ratio of VIP+, CALB2+, and RELN+ produced by the CGE remains roughly unchanged throughout embryonic maturation, further hinting at the possibility of even distribution of INs if MIBOs are allowed to mature beyond 4 months.17
Though we have focused on the ability of our devices to D-V pattern the GE, ultimately, our device also has the potential to be a platform that can be further modified to produce more complex neural structures. One option is to make use of temporal adaptation by introducing the Pur gradient at a later time point, once cortical fates have been established and sensitivity to Shh has waned. Doing so might generate pallial and subpallial tissues within a single organoid using a single morphogen. Alternatively, this could also be achieved with an antiparallel gradient of morphogens mimicking BMP and WNT. In the current iteration of our device, we used a single source of morphogen for the ventralization of our tissues. Future modifications could introduce an additional source that releases a dorsalizing small molecule such as cyclopamine (Shh antagonist) or BMP4. We predict that the exposure to the opposing gradients could create MIBOs that express both subpallial and cortical markers in a topographically faithful manner. An organoid model that intrinsically produces subpallial and cortical features could be extremely useful for teasing apart the questions surrounding embryonic neurodevelopment and could potentially provide a useful model for studying the effects of neurodevelopmental disorder risk genes on IN production and migration.
In conclusion, in this work, we report the fabrication of an easy-to-use PDMS device to generate a sustained chemical gradient for organoid patterning. We showed that Pur gradient could induce MIBOs mimicking the D-V patterning observed in the GEs. Single-cell transcriptomic analysis revealed a rich cell diversity of GE-derived GABAergic subtypes, including dorsally derived CALB2+ INs, vLGE MSN precursors, and a small population of MGE-derived cells. Live Ca2+ imaging analysis showed that MIBOs matured into intact organoid structures with functional neurocircuitry. Our system, therefore, could provide a new strategy for generating properly patterned and regionalized neural organoids, a vital foundation for modeling neurodevelopment and disease states.
Limitations of the study
Our current version of MIBO device fabrication requires multiple PDMS slabs being cut out, carefully assembled, and sealed, which can be time consuming. However, since a majority of the device is made of PDMS, 3D printing molds and consolidating key pieces for a PDMS cast of the device would greatly decrease assembly time. In this study, we have fully relied on NKX2.1 staining to reliably indicate the region of the organoid closest to the Pur source (laser-cut slit). To further validate the direction of the gradient, a polymer microbead could be manually embedded to the side of the organoid closest to the Pur source. Confirmation would require colocalization of NKX2.1 expression on the same side as the microbead. Additionally, while we observed reproducible patterning in a single human embryonic stem cell line (WA-01), reproducibility and robustness of the technology could be tested with additional human embryonic and induced pluripotent stem cell lines.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rabbit anti-NKX2.1, 1:500 | Abcam | Cat# ab76013; RRID:AB_1310784 |
| mouse anti-PAX6, 1:500 | BD Pharmigen | Cat# 561462 |
| rat anti-CTIP2, 1:1000 | Abcam | Cat# ab18465; RRID:AB_2064130 |
| rabbit anti-CALB2, 1:1000 | Swant | Cat# CR7679 |
| chicken anti-MAP2, 1:1000 | Abcam | Cat# ab5392; RRID:AB_2138153 |
| rabbit anti-NeuN, 1:500 | Millipore | Cat# ABN78; RRID:AB_10807945 |
| rabbit anti-DARPP32, 1:200 | Abcam | Cat# ab40801; RRID:AB_731843 |
| mouse anti-GAD67, 1:400 | Millipore | Cat# MAB5406; RRID:AB_2278725 |
| Alexa Fluor 647 goat anti-rabbit Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-21245 (also A21245); RRID:AB_2535813 |
| Alexa Fluor 546 goat anti-mouse Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-11003 (also A11003); RRID:AB_2534071 |
| Alexa Fluor 647 goat anti-rat Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-21247; RRID:AB_141778 |
| Alexa Fluor 488 goat anti-chicken Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-11039; RRID:AB_2534096 |
| Alexa Fluor 546 goat anti-rabbit Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-11010 (also A11010); RRID:AB_2534077 |
| Alexa Fluor 488 goat anti-rabbit Secondary Antibody, 1:1000 | ThermoFisher | Cat# A-11034 (also A11034); RRID:AB_2576217 |
| DAPI, 1:1000 | Sigma | Cat# MBD0015 |
| mTeSR Plus Medium | Stem Cell Technologies | 100–0276 |
| Accutase | Innovative Cell Technologies | AT104-500 |
| E6 | ThermoFisher | A1516401 |
| Matrigel | Corning | 354234 |
| DMEM/F12 | ThermoFisher | 11330032 |
| N2 | ThermoFisher | 17502048 |
| NEAA | ThermoFisher | 11140050 |
| Neurobasal | ThermoFisher | 21103049 |
| B27 without Vitamin A | ThermoFisher | 12587010 |
| GlutaMax | LifeTechnologies | 35050–061 |
| Penecillin-Streptomycin | ThermoFisher | 15140122 |
| Chemicals, peptides, and recombinant proteins | ||
| Y-27632 [10μM] | Axon MedChem | 1683 |
| XAV939 [5 μM] | BioGems | 2848932-25mg |
| LDN193189 [100 nM] | BioGems | 1062443-10MG |
| SB 431542 [10 μM] | BioGems | 3014193-10MG |
| EGF [20 ng/mL] | PeproTech | AF-100-15-500ug |
| FGF [20 ng/mL] | PeproTech | 100-18B-100UG |
| Purmorphamine [1 μM, 100 μM, or 100 nM] | Cayman Chemical | 10009634 |
| BDNF [20 ng/mL] | PeproTech | 450-02-100ug |
| NT3 [20 ng/mL] | PeproTech | 450-03-100 |
| Deposited data | ||
| Single cell transcriptomic data is deposited in GEO | This paper | Accession #: Gene Expression Omniubs: GSE244281 |
| Experimental models: Cell lines | ||
| H1 hESC – WA01 | WiCell | http://hpscreg.eu/cell-line/WAe001-A |
| Software and algorithms | ||
| MATLAB Gradient Analysis | This paper | 10.5281/zenodo.10372539 |
| MATLAB Calcium Imaging | Sun & Sudhof93 | N/A |
| Seurat_4.4.0 | Butler et al.,94 | https://satijalab.org/seurat/ |
| Leica Application Suite X, Version 2.0.0.14332 | LAX Software | N/A |
| Graphpad Prism (9.3.0) | GraphPad software | https://www.graphpad.com/scientific-software/prism/ |
| NIS Elements, Version 5.11.02 | Nikon | N/A |
| ImageJ, Version 4.4 | Schneider et al.,95 | https://imagej.nih.gov/ij/ |
| clusterProfiler (4.0) | Wu et al.,96 | https://bioconductor.org/packages/clusterProfiler |
| AnnotationDbi (1.62.2) | Pages et al.,97 | https://bioconductor.org/packages/AnnotationDbi |
| org.Hs.eg.db (3.17.0) | Carlson98 | https://bioconductor.org/packages/org.Hs.eg.db |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, ChangHui Pak (cpak@umass.edu).
Materials availability
This study did not generate new unique reagents. Unique code was generated, as detailed below.
Data and code availability
-
•
The raw data reported in this publication is archived at NCBI GEO (Gene Expression Omnibus: GSE244281).
-
•
Gradient analysis code is available at https://github.com/Npavo002/MATLAB_SobelGradientAnalysis (10.5281/zenodo.10372539).
-
•
Any additional information required for re-analysis of the data in this paper is available from the lead contact upon request.
Experimental models and subject details
Cell culture
H1 hESC line were grown in a standard 6-well plate with a 9.6 cm^2 culturing surface area coated with Matrigel diluted in DPBS and maintained with mTeSR Plus medium (Stem Cell Technologies) in feeder-free conditions. Y-27632 [10 μM] (Axon MedChem) was added to medium for all cell passaging. Cells were routinely tested for mycoplasma.
MIBOs generation
First, hPSCs were dissociated into single cells using Accutase (Innovative Cell Technologies). We then seeded these cells at a density of 10k cells per well in an Ultra-Low Attachment 96 Well, Round Bottom plate (Costar). Cells are allowed to aggregate overnight in mTeSR Plus with Y-27632 [10 μM] and XAV939 [5 μM] (BioGems). After 24 h, EB cell aggregates will form, and culture medium will switch to E6 (ThermoFisher) containing LDN193189 [100 nM] (BioGems), SB 431542 [10 μM] (BioGems), and XAV939 [5 μM] (BioGems) for 8 days during neural induction. On the eighth day of neural induction, three organoids are transferred into MIBOs device using 200μL wide bore pipette tip (Thermo Scientific), with excess media removed using standard bore pipette tip. 30μL of ice-cold Matrigel is promptly added for each organoid in a MIBOs device, and the pipette tip is used to center each organoid within the device embedding areas. Once embedded, culture medium is switched to a neural differentiation medium consisting of a 1:1 mixture with a base of DMEM/F12 (ThermoFisher) with N2 (ThermoFisher) and NEAA (ThermoFisher) and a base of Neurobasal Medium (ThermoFisher) containing B27 without Vitamin A (ThermoFisher), GlutaMax (LifeTechnologies), and Penicillin-Streptomycin (ThermoFisher). To this 1:1 base mixture we added EGF [20 ng/mL] (PeproTech), and FGF [20 ng/mL] (PeproTech). For the chemical reservoir medium, the same medium was used with the addition of varying concentrations of Purmorphamine [1 μM, 100 μM, or 100 nM] (Cayman Chemical). At Day 12, MIBOs are weaned off N2 by decreasing concentration from 1:100 to 1:200. On Day 25, MIBOs were carefully removed by gently dislodging Matrigel droplets using a standard bore 200μL pipette tip, and promptly transferred into ultra-low attachment 10 cm petri dishes using 1000uL wide bore pipette tips (Genesee Scientific). At Day 25, MIBOs were placed on an orbital shaker and cultured in medium containing only Neurobasal, B27 without vitamin A, Glutamax, Penicillin-Streptomycin, BDNF [20 ng/mL] (Peprotech), NT3 [20 ng/mL] (Peprotech). For long term culture, at day 43, all morphogens were removed and MIBOs culturing continued with Neurobasal, B27 without vitamin A, Glutamax, and Penicillin-Streptomycin alone.
Method details
Cryopreservation and sectioning
Organoid samples were collected at day 25 and day 101. Collected organoids were then washed 3 times with PBS and fixed with 4%Paraformaldahyde for 24 h in 4°C. After fixing is complete, samples are washed with PBS and stored in 30% sucrose solution for 24–48 h in 4°C. Organoids are encased in gelatin solution (10%gelatin with sucrose/PBS) followed by the flash freezing process using a mixture of dry ice and ethanol. Gelatin blocks are then stored in −80°C for long term storage and eventually cryosectioned between 12 and 25 micron section thickness. Sections directly adhere to SuperFrost Plus slides (Fisher) and promptly used for immunohistochemistry. Unused slides are stored in −20°C.
Immunostaining
Sections are washed three times using 0.2% Triton X- in PBS (0.2%PBS/T) and blocked for 1 h at room temperature (RT) using 10% normal goat serum in 0.2%PBS/T (blocking solution). After 1 h, sections receive a quick rinse with 0.2%PBS/T and are incubated overnight at 4°C with blocking solution containing diluted antibodies. The following day slides are washed three times with 0.2%PBS/T and incubated with secondary antibodies and DAPI diluted in blocking solution for 2 h at RT. After the incubation period, slides are washed three times with 0.2%PBS/T and promptly mounted using Fluoromount mounting media (Southern Biotech). Primary antibodies used are as follows: rabbit anti-NKX2.1 [Abcam, AB76013; 1:500], mouse anti-PAX6 [BD Pharmigen, 561462; 1:500], rat anti-BCL11B/CTIP2 [Abcam, AB18465; 1:1000], mouse anti-SATB2 [Abcam, ab51502; 1:400], chicken anti-MAP2 [Abcam, AB5392; 1:1000], rabbit anti-CALB2 [Swant, CR7679; 1:500], rabbit anti-NeuN [Milipore, ABN78; 1:500], rabbit anti-DARPP32 [Abcam, ab40801; 1:200], and mouse anti-GAD67 [Milipore, MAB5406; 1:400]. Secondary antibodies used: Alexa Fluor 647 goat anti-rabbit [ThermoFisher, A21245, 1:1000], Alexa Fluor 546 goat anti-mouse [ThermoFisher, A11003, 1:1000], Alexa Fluor 647 goat anti-rat [ThermoFisher, A21247, 1:1000], Alexa Fluor 488 goat anti-chicken [ThermoFisher, A11039, 1:1000], DAPI [Sigma, MBD0015-1ML; 1:1000], Alexa Fluor 546 goat anti-rabbit [ThermoFisher, A11010; 1:1000], Alexa Fluor 488 goat anti-rabbit [ThermoFisher, A11034; 1:1000].
Image Acquisition
Images of sectioned organoids were imaged using a Leica DMi8 microscope with LAS X Software (Leica Application Suite X, Version 2.0.0.14332). A 10× objective was used to obtain multiple images of a single organoid and then merged to create a large image containing a full organoid section (Figures 2 and S2). 60x, images were captured using Nikon A1R25 confocal microscope.
DAPI z stack image acquisition
MIBO devices were fabricated to characterize gradient profile of small molecule diffusion through Matrigel. To visualize the diffusion of small molecules through the Matrigel, we added 1x DPBS containing 10μg/mL of DAPI into the chemical reservoir. The culture medium was 1x DPBS to induce passive diffusion. After 24 h, all media was aspirated, and the device was taken apart to only retain the culturing area for ease of imaging. The A1R18 confocal microscope was used to capture a large image 6 x 12 scan using a 10× objective across 100μm increments in the z directions. Nikon Elements (Version 5.11.02) was used to stitch captured images together to reconstruct a 3D representation of DAPI uptake by Matrigel. Our positive control received the DAPI solution directly into the culture medium creating a uniform DAPI uptake and the negative control was never exposed to DAPI. Positive control was used to establish and standardize a gain and laser power to be used across all three conditions consistently.
Calcium imaging
Calcium dye mixture is prepared with 1mM X-Rhod-1 a.m. dye (Invitrogen) diluted in modified HEPES buffer (130mM NaCl, 5mM KCL, 2mM CaCl2, 1mM MgCl2, 10mM HEPES, 10mM Glucose, ∼pH 7.4 adjusted with NaOH). Organoids are then incubated in calcium dye mixture for 15 min at RT. After incubation period is over, organoids are quickly washed with the modified HEPES buffer once and imaged using a confocal microscope (Nikon, A1R25). A glass bottom Petri dish (MatTek) was used for all imaging and a stable temperature of 37°C was maintained using an Ibidi stage heater. Time lapse images are captured at 250 ms intervals for a period of 5 min.
Calcium imaging analysis
Raw images are extracted using NIS Elements and analyzed using a stimulation-free MATLAB protocol as demonstrated previously.93 Using the MATLAB protocol, we first create a Maximum Intensity Projection (MIP) by ‘stacking’ the time lapse images captured using the confocal microscope. The MIP serves as a guide for selecting 5 active regions of interests (ROIs). Each ROI was contained to a 50 μm diameter. A time interval was determined by taking the recording duration (in seconds) over total frames i.e., for our 5-min recordings we used 300/109 to obtain a time interval of 2.75. The analysis tracks the changes in pixel intensity within ROIs for one field of view (FOV). The recorded changes in intensity for one FOV are used to obtain a frequency of spiking activity and instances where spiking activity occurs across more than one ROI are used to obtain synchronous spikes and a synchronous firing rate. Amplitude is obtained by using the mean value of DF/F0 from individual peaks.
Live single cell dissociation
3.5-month-old organoids were transferred to a Petri dish and rinsed with HBSS (1 bottle HBS salt, Sigma H2387-10X1L; 1 mL 1M HEPES, pH 7.3; 0.35g NaHCO3; 100 mL autoclaved ddH2O). After rinsing, organoids were cut into smaller pieces using a sterile razor blade. A small amount of Digestion Solution (1900μl HBSS (−/−), 50μl papain, 11 μL of 0.5M EDTA, 7 mg L-Cysteine) was added to the Petri dish and a wide bore tip was used to transfer the smaller pieces into the Digestion Solution that was sterile filtered with 0.22 μm. The solution was incubated at 37°C for 15 min after filtering. Once the 15 min of incubation was complete, 50 μL of DNAse Solution (DNAse reconstituted from Worthington Kit with 500μg HBSS) was added to the organoids before gentle trituration using a wide bore tip. Mixture is incubated again for 10 min before being triturated with a p1000 tip and then a p200 tip. After this step, the solution was strained through a 70 μm cell strainer and then a 30 μm cell strainer. Another 1 mL of Neurobasal was used to rinse the cells prior to centrifugation at 300 rcf for 5 min. The supernatant that formed was removed, and the cells were resuspended in 1X PBS and 0.04% BSA. Cells were kept on ice while concentration and viability were assessed with a hemocytometer (Bio-Rad). The volume of cells was transferred to a 1.5 mL microcentrifuge tube for single-cell RNA sequencing (sc-RNA).
10XscRNASeq protocol
Following organoid dissociation and cell count, cell concentration was adjusted to 700–1200 cells/μL for 10X single cell sequencing. For each sample, 7,500 cells were loaded into the 10X Chromium controller to target recovery 3,500 cells and Gel Beads in Emulsion (GEM) was generated. Cell quality was performed using the Cellometer K2 cell counter (Nexcelom Bioscience). The single cell gene expression profiling was performed using the Chromium Next GEM Single Cell 3ʹ v3.1 (Dual Index) kit. Each sample (cell suspensions) were loaded onto a well of Chip G on the 10x Genomics Chromium Controller System following the manufacture’s user manual (10x Genomics). Barcoding and cDNA synthesis were performed according to the manufacturer’s instructions. The qualitative analysis of cDNA was performed using the 2100 Agilent Bioanalyzer High Sensitivity assay. The cDNA libraries were constructed using the 10x Chromium Single cell 3′ Library Kit v3.1 (dual index) according to the manufacturer’s protocol. The quality assessment of final Libraries was done on Qubit fluorometer using DNA High Sensitivity assay (Thermo Scientific), and 2100 Agilent Bioanalyzer High Sensitivity assay (Agilent Technologies). The libraries were sequenced on Illumina NextSeq 500 using NextSeq 500/550 Mid Output Kit v2.5 (150 Cycles) sequencing kit, with following read length: 28 bp Read1 for 10x cell barcode and UMI, 90 bp Read 2 for insert, and 10 bp I7 and I5 for sample index. Phix (Illumina) was spiked in at 1% as per kit manual recommendation (10x Genomics).
Single cell data alignment
10x Could Analysis Cell Ranger Pipeline (Cell Ranger Count v7.0.1) was used for initial data processing. Cell Ranger is a set of analysis pipelines that process Chromium single cell data to align reads, generate feature-barcode matrices, perform clustering and other secondary analysis. Reads were aligned to Human (GRCh38) 2020-A reference genome. The aggr pipeline was used to combine data from multiple samples into an experiment-wide feature-barcode matrix and analysis. The 10x Genomics Loupe Browser was used for visualization, initial quality assessment and filtering of single cell gene expression data. The Single Cell Gene Expression (scRNAseq) was performed at Genomics Resource Laboratory, University of Massachusetts Amherst, MA (RRID:SCR_017907).
Single cell data processing and normalization
Seurat 4 was used analyze raw data from Cell Ranger h5 files. We filtered out low-quality cells by removing cells with less than 1000 or more than 10000 unique genes. To further remove low-quality/dying cells we removed cells with more than 15% mitochondrial transcripts. After quality control, we harvested 26,329 genes and 3366 high-quality cells from MIBOs organoid. Original and processed data can be found in Data Availability. We applied the default normalization approach of Seurat.
Identifying Differentially Expressed Genes
Top 100 Differentially Expressed Genes (DEGs) for each pre-annotated cluster was found using ‘FindAllMarkers.’ The applied function uses Wilcoxon rank-sum test, a non-parametric statistical test that is capable of handling data with non-normal distributions and outliers. The analysis was performed for each annotated cluster individually and the DEGs were determined using user defined thresholds. We only considered genes exhibiting positive fold changes and expressed in a minimum of 25% of cells within the cluster. Genes with a log-fold change greater than or equal to 0.25 were considered DEGs.
Cell annotations
Following quality control, we found a total of 22 clusters generated with a fine resolution (2.0) from dissociated MIBOs. The Top 100 DEGs were used to examine Gene Ontology (GO) within annotated clusters from MIBO sample. We performed a manual annotation for each cluster based on canonical markers from precious studies, such as NES and HES1 for apical progenitors, ASCL1 and HES6 for basal progenitors, and STMN2, MAP2 and DCX for neurons. To assist in cluster annotation, we filtered ribosomal genes post clustering. We then used canonical telencephalic markers to combine initial clusters based on similarity in gene expression profile. A total of 10 cell classes were derived and included various subpopulations of GE derived cells. Cluster 15 was labeled as unknown given that there was no clear association based on expected marker genes.
Gene ontology on R
To complete GO analysis we use Biomanager to install the Bioconductor ‘clusterProfiler’ package, which is designed for the functional enrichment analysis and visualization of gene clusters. Additionally, we use the ‘annotationDbi’ package to provide a unified interface for querying and retrieving information from annotation databases. Finally, we relied on ‘org.Hs.eg.db’ to map gene identifiers and associated annotation information to the human genome database.
Single cell data processing and normalization for integrated dataset
We first downloaded the cortical and subpallial data available from the Gene Expression Omnibus: GSE93811 and used the Seurat integration procedure to reduce the influence of batch effects. First, we loaded the raw data for CS, SS, and MIBO, and created a list of Seurat objects after performing quality control. Next, we normalized each Seurat object and found the top 2000 highly variable genes using the “vst” method in FindVariableFeatures function. The integration features used the ScaleData function for centering and scaling. The Reciprocal PCA (RPCA) procedure was chosen as the method of integration when finding integration anchors for all three datasets. Following the anchor identification, we used the function IntegrateData with our selected anchor set. After integration, the data was scaled using ScaleData and a PCA was performed using RunPCA. Nearest neighbor graph was constructed using 20 k-nearest neighbors with 30 principal components. Finally, we used Louvain clustering on our neighbor graph using FindClusters at a 2.0 resolution. A Uniform Manifold Approximation and Projection (UMAP) based on the top 30 principal components was generated and used for all downstream analysis.
Device fabrication
Individual pieces for MIBOs device were made entirely out of PDMS as previously described by Li et al., 2021. Modifications included a PDMS Matrigel embedding area which used a PDMS film and laser cut (40 W Epilog Mini 18 x 12) three ascending positions at 20% speed, 80% power, 2500 Hz frequency, and 600 DPI resolution with vector job type (Figures 1 and S1). Additionally, two thin PDMS slabs (5 cm × 2 cm) are glued to the front and back the device frame to form a self-contained area for culture medium.
Quantification and statistical analysis
MATLAB gradient quantification
We used MATLAB and its inbuilt operators and functions to develop a program capable of analyzing the local change in pixel intensity. The program was then run on raw images captured by Leica DMi8 microscope. Here we leveraged the ‘Sobel’ gradient operator which uses the weighted sum of pixels in a 3x3 neighborhood to determine the gradient direction and gradient magnitude of the pixel. We also used the ‘quiver’ function to visualize the gradient direction (angle of quiver) and gradient magnitude (size of quiver) of individual pixels within an image. The sum of each pixel’s gradient magnitude was used for our analysis (Figures 2B and 2C). Image intensity was normalized by dividing all analyzed pixels by the value of the brightest pixel in their respective image providing a range of 0–1. An individual binary mask was made to assure that only regions of our organoid sections were measured and that any IHC artifacts were limited. MATLAB code available:https://github.com/Npavo002/MATLAB_SobelGradientAnalysis (10.5281/zenodo.10372539)
Quantification and statistical analysis of calcium imaging data
Data wrangling was done in Microsoft Excel with raw data points transferred to Prism (9.3.0) for basic statistics, outlier detection, significance tests, and graph generation. A ROUT outlier test was performed to identify outliers within each dataset. The data was fitted with nonlinear regression and a false discovery rate of Q = 1%. To test for statistical significance a one-way ANOVA test with multiple comparisons (Dunnett’s test) was used to compare conditions.
Quantification of DAPI gradient in Matrigel
Using ImageJ (4.4) we created an 8-bit grayscale image of the slice acquired through NIS elements. We set our measurements to record ‘Mean gray value’ to measure DAPI intensity within the slice view. The rectangle tool was used to select the entire image as the ROI and ‘Plot profile’ was used to generate a gray value by distance plot. The plot takes the mean gray value of a pixel column (y direction) and plots it corresponding to its distance (in pixels) in the x direction.
Acknowledgments
This work was supported by NIMH (R01 MH122519 to C.P. and R21 MH130843 to Y.S. and C.P.), UMass IALS/BMB Faculty Start up Fund (to C.P.), Tourette Association of America (Young investigator award to C.P.), UMass IONS Seed funding, and the NIGMS T32 BTP training program (T32 GM135096 to N.P.). We would like to thank members of the Sun and Pak labs for critical feedback and acknowledge the support from UMass IALS Light Microscopy Core and Genomics Core.
Author contributions
The study was designed by Y.S., C.P., and N.P. N.P. executed all experiments and analysis with the help of K.D., F.Y., R.S., B.M.-M., and R.R. C.P., Y.S., and N.P. interpreted the data, generated figures, and wrote the manuscript.
Declaration of interests
The authors listed (Y.S., C.P., F.Y., and N.P. ) have filed a provisionary patent related to this work (Docket Number: UMA 23-023, Title: “A chemical gradient inducing device and methods for developing patterned organoids.”).
Published: January 15, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.crmeth.2023.100689.
Contributor Information
Yubing Sun, Email: ybsun@umass.edu.
ChangHui Pak, Email: cpak@umass.edu.
Supplemental information
References
- 1.Rakic P. Specification of Cerebral Cortical Areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 2.Stern C.D. Neural induction: 10 years on since the ‘default model. Curr. Opin. Cell Biol. 2006;18:692–697. doi: 10.1016/j.ceb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Xu Q., Guo L., Moore H., Waclaw R.R., Campbell K., Anderson S.A. Sonic hedgehog signaling confers ventral telencephalic progenitors with distinct cortical interneuron fates. Neuron. 2010;65:328–340. doi: 10.1016/j.neuron.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corbin J.G., Nery S., Fishell G. Telencephalic cells take a tangent: non-radial migration in the mammalian forebrain. Nat. Neurosci. 2001;4:1177–1182. doi: 10.1038/nn749. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima K. Control of tangential/non-radial migration of neurons in the developing cerebral cortex. Neurochem. Int. 2007;51:121–131. doi: 10.1016/j.neuint.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Wonders C.P., Anderson S.A. The origin and specification of cortical interneurons. Nat. Rev. Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- 7.Anderson S.A., Marín O., Horn C., Jennings K., Rubenstein J.L. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- 8.Anderson S.A., Kaznowski C.E., Horn C., Rubenstein J.L.R., McConnell S.K. Distinct Origins of Neocortical Projection Neurons and Interneurons In Vivo. Cerebr. Cortex. 2002;12:702–709. doi: 10.1093/cercor/12.7.702. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q., Cobos I., De La Cruz E., Rubenstein J.L., Anderson S.A. Origins of Cortical Interneuron Subtypes. J. Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Q., Tam M., Anderson S.A. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J. Comp. Neurol. 2008;506:16–29. doi: 10.1002/cne.21529. [DOI] [PubMed] [Google Scholar]
- 11.Flames N., Pla R., Gelman D.M., Rubenstein J.L.R., Puelles L., Marín O. Delineation of Multiple Subpallial Progenitor Domains by the Combinatorial Expression of Transcriptional Codes. J. Neurosci. 2007;27:9682–9695. doi: 10.1523/JNEUROSCI.2750-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandler R.C., Mayer C., Fishell G. Cortical interneuron specification: the juncture of genes, time and geometry. Curr. Opin. Neurobiol. 2017;42:17–24. doi: 10.1016/j.conb.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torigoe M., Yamauchi K., Kimura T., Uemura Y., Murakami F. Evidence That the Laminar Fate of LGE/CGE-Derived Neocortical Interneurons Is Dependent on Their Progenitor Domains. J. Neurosci. 2016;36:2044–2056. doi: 10.1523/JNEUROSCI.3550-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wichterle H., Turnbull D.H., Nery S., Fishell G., Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- 15.Waclaw R.R., Wang B., Pei Z., Ehrman L.A., Campbell K. Distinct Temporal Requirements for the Homeobox Gene Gsx2 in Specifying Striatal and Olfactory Bulb Neuronal Fates. Neuron. 2009;63:451–465. doi: 10.1016/j.neuron.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olsson M., Björklund A., Campbell K. Early specification of striatal projection neurons and interneuronal subtypes in the lateral and medial ganglionic eminence. Neuroscience. 1998;84:867–876. doi: 10.1016/s0306-4522(97)00532-0. [DOI] [PubMed] [Google Scholar]
- 17.Miyoshi G., Hjerling-Leffler J., Karayannis T., Sousa V.H., Butt S.J.B., Battiste J., Johnson J.E., Machold R.P., Fishell G. Genetic Fate Mapping Reveals That the Caudal Ganglionic Eminence Produces a Large and Diverse Population of Superficial Cortical Interneurons. J. Neurosci. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nery S., Fishell G., Corbin J.G. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat. Neurosci. 2002;5:1279–1287. doi: 10.1038/nn971. [DOI] [PubMed] [Google Scholar]
- 19.Alzubi A., Lindsay S., Kerwin J., Looi S.J., Khalil F., Clowry G.J. Distinct cortical and sub-cortical neurogenic domains for GABAergic interneuron precursor transcription factors NKX2.1, OLIG2 and COUP-TFII in early fetal human telencephalon. Brain Struct. Funct. 2017;222:2309–2328. doi: 10.1007/s00429-016-1343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velmeshev D., Chavali M., Nowakowski T.J., Bhade M., Mayer S., Goyal N., Alvarado B., Mancia W., Wang S., Speir M., et al. Molecular diversity and lineage commitment of human interneuron progenitors. bioRxiv. 2021 Preprint at. [Google Scholar]
- 21.Shi Y., Wang M., Mi D., Lu T., Wang B., Dong H., Zhong S., Chen Y., Sun L., Zhou X., et al. Mouse and human share conserved transcriptional programs for interneuron development. Science. 2021;374:eabj6641. doi: 10.1126/science.abj6641. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Z., Zhang D., Yang F., Xu M., Zhao S., Pan T., Liu C., Liu Y., Wu Q., Tu Q., et al. Evolutionarily conservative and non-conservative regulatory networks during primate interneuron development revealed by single-cell RNA and ATAC sequencing. Cell Res. 2022;32:425–436. doi: 10.1038/s41422-022-00635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keefe F., Monzón-Sandoval J., Rosser A.E., Webber C., Li M. Single-Cell Transcriptomics Reveals Conserved Regulatory Networks in Human and Mouse Interneuron Development. Int. J. Mol. Sci. 2023;24:8122. doi: 10.3390/ijms24098122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancaster M.A., Renner M., Martin C.-A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lancaster M.A., Corsini N.S., Wolfinger S., Gustafson E.H., Phillips A.W., Burkard T.R., Otani T., Livesey F.J., Knoblich J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sasai Y. Next-Generation Regenerative Medicine: Organogenesis from Stem Cells in 3D Culture. Cell Stem Cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 27.Paşca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.-Y., O’Rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sloan S.A., Andersen J., Pașca A.M., Birey F., Pașca S.P. Generation and Assembly of Human Brain Region-Specific Three-Dimensional Cultures. Nat. Protoc. 2018;13:2062–2085. doi: 10.1038/s41596-018-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., Ming G.-L. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian X., Su Y., Adam C.D., Deutschmann A.U., Pather S.R., Goldberg E.M., Su K., Li S., Lu L., Jacob F., et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell. 2020;26:766–781.e9. doi: 10.1016/j.stem.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Lullo E., Kriegstein A.R. The use of brain organoids to investigate neural development and disease. Nat. Rev. Neurosci. 2017;18:573–584. doi: 10.1038/nrn.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jo J., Xiao Y., Sun A.X., Cukuroglu E., Tran H.-D., Göke J., Tan Z.Y., Saw T.Y., Tan C.-P., Lokman H., et al. Midbrain-like Organoids from Human Pluripotent Stem Cells Contain Functional Dopaminergic and Neuromelanin-Producing Neurons. Cell Stem Cell. 2016;19:248–257. doi: 10.1016/j.stem.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monzel A.S., Smits L.M., Hemmer K., Hachi S., Moreno E.L., van Wuellen T., Jarazo J., Walter J., Brüggemann I., Boussaad I., et al. Derivation of Human Midbrain-Specific Organoids from Neuroepithelial Stem Cells. Stem Cell Rep. 2017;8:1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muguruma K., Nishiyama A., Kawakami H., Hashimoto K., Sasai Y. Self-Organization of Polarized Cerebellar Tissue in 3D Culture of Human Pluripotent Stem Cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 39.Miura Y., Li M.-Y., Birey F., Ikeda K., Revah O., Thete M.V., Park J.-Y., Puno A., Lee S.H., Porteus M.H., Pașca S.P. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 2020;38:1421–1430. doi: 10.1038/s41587-020-00763-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang Y., Tanaka Y., Patterson B., Kang Y.-J., Govindaiah G., Roselaar N., Cakir B., Kim K.-Y., Lombroso A.P., Hwang S.-M., et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell. 2017;21:383–398.e7. doi: 10.1016/j.stem.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagley J.A., Reumann D., Bian S., Lévi-Strauss J., Knoblich J.A. Fused dorsal-ventral cerebral organoids model complex interactions between diverse brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.-H., Peters A., Park T.S., Zambidis E.T., Meyer J.S., et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N., et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang Y., Tanaka Y., Cakir B., Patterson B., Kim K.-Y., Sun P., Kang Y.-J., Zhong M., Liu X., Patra P., et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24:487–497.e7. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen J., Revah O., Miura Y., Thom N., Amin N.D., Kelley K.W., Singh M., Chen X., Thete M.V., Walczak E.M., et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell. 2020;183:1913–1929.e26. doi: 10.1016/j.cell.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cederquist G.Y., Asciolla J.J., Tchieu J., Walsh R.M., Cornacchia D., Resh M.D., Studer L. SPECIFICATION OF POSITIONAL IDENTITY IN FOREBRAIN ORGANOIDS. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rifes P., Isaksson M., Rathore G.S., Aldrin-Kirk P., Møller O.K., Barzaghi G., Lee J., Egerod K.L., Rausch D.M., Parmar M., et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat. Biotechnol. 2020;38:1265–1273. doi: 10.1038/s41587-020-0525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demers C.J., Soundararajan P., Chennampally P., Cox G.A., Briscoe J., Collins S.D., Smith R.L. Development-on-chip: in vitro neural tube patterning with a microfluidic device. Development. 2016;143:1884–1892. doi: 10.1242/dev.126847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J.H., Sim J., Kim H.-J. Neural Stem Cell Differentiation Using Microfluidic Device-Generated Growth Factor Gradient. Biomol. Ther. 2018;26:380–388. doi: 10.4062/biomolther.2018.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cosson S., Lutolf M.P. Hydrogel microfluidics for the patterning of pluripotent stem cells. Sci. Rep. 2014;4:4462. doi: 10.1038/srep04462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li N., Yang F., Parthasarathy S., Pierre S.S., Hong K., Pavon N., Pak C., Sun Y. Patterning Neuroepithelial Cell Sheet via a Sustained Chemical Gradient Generated by Localized Passive Diffusion Devices. ACS Biomater. Sci. Eng. 2021;7:1713–1721. doi: 10.1021/acsbiomaterials.0c01365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waclaw R.R., Allen Z.J., Bell S.M., Erdélyi F., Szabó G., Potter S.S., Campbell K. The Zinc Finger Transcription Factor Sp8 Regulates the Generation and Diversity of Olfactory Bulb Interneurons. Neuron. 2006;49:503–516. doi: 10.1016/j.neuron.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 53.Chu J., Anderson S.A. Development of Cortical Interneurons. Neuropsychopharmacology. 2015;40:16–23. doi: 10.1038/npp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wei S., Du H., Li Z., Tao G., Xu Z., Song X., Shang Z., Su Z., Chen H., Wen Y., et al. Transcription factors Sp8 and Sp9 regulate the development of caudal ganglionic eminence-derived cortical interneurons. J. Comp. Neurol. 2019;527:2860–2874. doi: 10.1002/cne.24712. [DOI] [PubMed] [Google Scholar]
- 55.Kanatani S., Yozu M., Tabata H., Nakajima K. COUP-TFII Is Preferentially Expressed in the Caudal Ganglionic Eminence and Is Involved in the Caudal Migratory Stream. J. Neurosci. 2008;28:13582–13591. doi: 10.1523/JNEUROSCI.2132-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arlotta P., Molyneaux B.J., Jabaudon D., Yoshida Y., Macklis J.D. Ctip2 Controls the Differentiation of Medium Spiny Neurons and the Establishment of the Cellular Architecture of the Striatum. J. Neurosci. 2008;28:622–632. doi: 10.1523/JNEUROSCI.2986-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Azim E., Jabaudon D., Fame R.M., Macklis J.D. SOX6 controls dorsal-ventral progenitor parcellation and interneuron diversity during neocortical development. Nat. Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y., Flandin P., Long J.E., Cuesta M.D., Westphal H., Rubenstein J.L.R. Distinct molecular pathways for development of telencephalic interneuron subtypes revealed through analysis of Lhx6 mutants. J. Comp. Neurol. 2008;510:79–99. doi: 10.1002/cne.21772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gleeson J.G., Lin P.T., Flanagan L.A., Walsh C.A. Doublecortin Is a Microtubule-Associated Protein and Is Expressed Widely by Migrating Neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- 60.Naeve G.S., Ramakrishnan M., Kramer R., Hevroni D., Citri Y., Theill L.E. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nedivi E., Wu G.Y., Cline H.T. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sebastian R., Jin K., Pavon N., Bansal R., Potter A., Song Y., Babu J., Gabriel R., Sun Y., Aronow B., Pak C. Schizophrenia-associated NRXN1 deletions induce developmental-timing- and cell-type-specific vulnerabilities in human brain organoids. Nat. Commun. 2023;14:3770. doi: 10.1038/s41467-023-39420-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosebrock D., Arora S., Mutukula N., Volkman R., Gralinska E., Balaskas A., Aragonés Hernández A., Buschow R., Brändl B., Müller F.J., et al. Enhanced cortical neural stem cell identity through short SMAD and WNT inhibition in human cerebral organoids facilitates emergence of outer radial glial cells. Nat. Cell Biol. 2022;24:981–995. doi: 10.1038/s41556-022-00929-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoon S.-J., Elahi L.S., Pașca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., Fan H.C., et al. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dessaud E., Yang L.L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B.G., Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 66.Dessaud E., McMahon A.P., Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- 67.Krajka V., Naujock M., Pauly M.G., Stengel F., Meier B., Stanslowsky N., Klein C., Seibler P., Wegner F., Capetian P. Ventral Telencephalic Patterning Protocols for Induced Pluripotent Stem Cells. Front. Cell Dev. Biol. 2021;9:716249. doi: 10.3389/fcell.2021.716249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amimoto N., Nishimura K., Shimohama S., Takata K. Generation of striatal neurons from human induced pluripotent stem cells by controlling extrinsic signals with small molecules. Stem Cell Res. 2021;55:102486. doi: 10.1016/j.scr.2021.102486. [DOI] [PubMed] [Google Scholar]
- 69.Chen X., Saiyin H., Liu Y., Wang Y., Li X., Ji R., Ma L. Human striatal organoids derived from pluripotent stem cells recapitulate striatal development and compartments. PLoS Biol. 2022;20:e3001868. doi: 10.1371/journal.pbio.3001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma L., Hu B., Liu Y., Vermilyea S.C., Liu H., Gao L., Sun Y., Zhang X., Zhang S.-C. Human Embryonic Stem Cell-Derived GABA Neurons Correct Locomotion Deficits in Quinolinic Acid-Lesioned Mice. Cell Stem Cell. 2012;10:455–464. doi: 10.1016/j.stem.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weick J.P., Held D.L., Bonadurer G.F., Doers M.E., Liu Y., Maguire C., Clark A., Knackert J.A., Molinarolo K., Musser M., et al. Deficits in human trisomy 21 iPSCs and neurons. Proc. Natl. Acad. Sci. USA. 2013;110:9962–9967. doi: 10.1073/pnas.1216575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Larsen Z.H., Chander P., Joyner J.A., Floruta C.M., Demeter T.L., Weick J.P. Effects of Ethanol on Cellular Composition and Network Excitability of Human Pluripotent Stem Cell-Derived Neurons. Alcohol Clin. Exp. Res. 2016;40:2339–2350. doi: 10.1111/acer.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Floruta C.M., Du R., Kang H., Stein J.L., Weick J.P. Default Patterning Produces Pan-cortical Glutamatergic and CGE/LGE-like GABAergic Neurons from Human Pluripotent Stem Cells. Stem Cell Rep. 2017;9:1463–1476. doi: 10.1016/j.stemcr.2017.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tanaka Y., Cakir B., Xiang Y., Sullivan G.J., Park I.-H. Synthetic Analyses of Single-Cell Transcriptomes from Multiple Brain Organoids and Fetal Brain. Cell Rep. 2020;30:1682–1689.e3. doi: 10.1016/j.celrep.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wonders C.P., Taylor L., Welagen J., Mbata I.C., Xiang J.Z., Anderson S.A. A spatial bias for the origins of interneuron subgroups within the medial ganglionic eminence. Dev. Biol. 2008;314:127–136. doi: 10.1016/j.ydbio.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fogarty M., Grist M., Gelman D., Marín O., Pachnis V., Kessaris N. Spatial Genetic Patterning of the Embryonic Neuroepithelium Generates GABAergic Interneuron Diversity in the Adult Cortex. J. Neurosci. 2007;27:10935–10946. doi: 10.1523/JNEUROSCI.1629-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stenman J.M., Wang B., Campbell K. Tlx Controls Proliferation and Patterning of Lateral Telencephalic Progenitor Domains. J. Neurosci. 2003;23:10568–10576. doi: 10.1523/JNEUROSCI.23-33-10568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim J.Y., Cho B., Moon C. Timely Inhibitory Circuit Formation Controlled by Abl1 Regulates Innate Olfactory Behaviors in Mouse. Cell Rep. 2020;30:187–201.e4. doi: 10.1016/j.celrep.2019.12.004. [DOI] [PubMed] [Google Scholar]
- 79.Long J.E., Swan C., Liang W.S., Cobos I., Potter G.B., Rubenstein J.L.R. Dlx1&2 and Mash1 Transcription Factors Control Striatal Patterning and Differentiation Through Parallel and Overlapping Pathways. J. Comp. Neurol. 2009;512:556–572. doi: 10.1002/cne.21854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ma T., Zhang Q., Cai Y., You Y., Rubenstein J.L.R., Yang Z. A subpopulation of dorsal lateral/caudal ganglionic eminence-derived neocortical interneurons expresses the transcription factor Sp8. Cerebr. Cortex. 2012;22:2120–2130. doi: 10.1093/cercor/bhr296. [DOI] [PubMed] [Google Scholar]
- 81.Su-Feher L., Rubin A.N., Silberberg S.N., Catta-Preta R., Lim K.J., Ypsilanti A.R., Zdilar I., McGinnis C.S., McKinsey G.L., Rubino T.E., et al. Single cell enhancer activity distinguishes GABAergic and cholinergic lineages in embryonic mouse basal ganglia. Proc. Natl. Acad. Sci. USA. 2022;119 doi: 10.1073/pnas.2108760119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kessaris N., Magno L., Rubin A.N., Oliveira M.G. Genetic programs controlling cortical interneuron fate. Curr. Opin. Neurobiol. 2014;26:79–87. doi: 10.1016/j.conb.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Frazer S., Prados J., Niquille M., Cadilhac C., Markopoulos F., Gomez L., Tomasello U., Telley L., Holtmaat A., Jabaudon D., Dayer A. Transcriptomic and anatomic parcellation of 5-HT3AR expressing cortical interneuron subtypes revealed by single-cell RNA sequencing. Nat. Commun. 2017;8:14219. doi: 10.1038/ncomms14219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mi D., Li Z., Lim L., Li M., Moissidis M., Yang Y., Gao T., Hu T.X., Pratt T., Price D.J., et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science. 2018;360:81–85. doi: 10.1126/science.aar6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tripodi M., Filosa A., Armentano M., Studer M. The COUP-TF nuclear receptors regulate cell migration in the mammalian basal forebrain. Development. 2004;131:6119–6129. doi: 10.1242/dev.01530. [DOI] [PubMed] [Google Scholar]
- 86.Lledo P.-M., Merkle F.T., Alvarez-Buylla A. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31:392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Butt S.J.B., Fuccillo M., Nery S., Noctor S., Kriegstein A., Corbin J.G., Fishell G. The Temporal and Spatial Origins of Cortical Interneurons Predict Their Physiological Subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 88.Xu X., Roby K.D., Callaway E.M. Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J. Comp. Neurol. 2006;499:144–160. doi: 10.1002/cne.21101. [DOI] [PubMed] [Google Scholar]
- 89.Miyoshi G., Butt S.J.B., Takebayashi H., Fishell G. Physiologically Distinct Temporal Cohorts of Cortical Interneurons Arise from Telencephalic Olig2-Expressing Precursors. J. Neurosci. 2007;27:7786–7798. doi: 10.1523/JNEUROSCI.1807-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bartolini G., Ciceri G., Marín O. Integration of GABAergic Interneurons into Cortical Cell Assemblies: Lessons from Embryos and Adults. Neuron. 2013;79:849–864. doi: 10.1016/j.neuron.2013.08.014. [DOI] [PubMed] [Google Scholar]
- 91.Marín O., Rubenstein J.L. A long, remarkable journey: Tangential migration in the telencephalon. Nat. Rev. Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- 92.Lim L., Mi D., Llorca A., Marín O. Development and functional diversification of cortical interneurons. Neuron. 2018;100:294–313. doi: 10.1016/j.neuron.2018.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun Z., Südhof T.C. A simple Ca2+-imaging approach to neural network analyses in cultured neurons. J. Neurosci. Methods. 2021;349:109041. doi: 10.1016/j.jneumeth.2020.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2:100141. doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hervé Pagès M.C. 2023. AnnotationDbi. (Bioconductor). 10.18129/B9.BIOC.ANNOTATIONDBI 10.18129/B9.BIOC.ANNOTATIONDBI. [Google Scholar]
- 98.Carlson M. 2023. org.Hs.eg.db. (Bioconductor). 10.18129/B9.BIOC.ORG.HS.EG.DB 10.18129/B9.BIOC.ORG.HS.EG.DB. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw data reported in this publication is archived at NCBI GEO (Gene Expression Omnibus: GSE244281).
-
•
Gradient analysis code is available at https://github.com/Npavo002/MATLAB_SobelGradientAnalysis (10.5281/zenodo.10372539).
-
•
Any additional information required for re-analysis of the data in this paper is available from the lead contact upon request.