Abstract
Validamycin, is classified as an environmentally friendly fungicide. It has high efficacy with little associated pollution risk, and it has been used in China on tobacco for many years especially during leaf spot season. To understand changes in microbial communities and functional aspects of the tobacco phyllosphere after exposure to validamycin, the chemical was sprayed on tobacco leaves during brown spot epidemic periods caused by Alternaria alternata, and asymptomatic and symptomatic leaves of tobacco were sampled at different times (0 day before, 5, 10, and 15 days after application). The fungal and bacterial population diversity and structure were revealed using Illumina NovaSeq PE250 high-throughput sequencing technology, and Biolog-ECO technology which analyzes the metabolic differences between samples by using different carbon sources as the sole energy source. The results showed that the microbial community structure of both asymptomatic and symptomatic tobacco leaves changed after the application of valproate, with the microbial community structure of the asymptomatic tobacco leaves being more strongly affected than that of the symptomatic leaves, and the diversity of bacteria being greater than that of fungi. Phyllosphere fungal diversity in asymptomatic leaves increased significantly after application, and bacterial abundance and diversity in both asymptomatic and symptomatic leaves first increased and then decreased. Validamycin treatment effectively reduced the relative abundance of Alternaria, Cladosporium, Kosakonia, and Sphingomonas in leaves showing symptoms of tobacco brown spot, while the relative abundance of Thanatephorus, Pseudomonas, and Massilia increased significantly after application. Furthermore, the ability to metabolize a variety of carbon sources was significantly reduced in both types of leaves after validamycin application, and both types had a weaker ability to metabolize α-Ketobutyric Acid after application. This study reveals phyllosphere micro-ecological changes in symptomatic and asymptomatic tobacco leaves during different periods after validamycin application and the effects on the metabolic capacity of phyllosphere microorganisms. It can provide some basis for exploring the effect of validamycin on the control of tobacco brown spot.
Keywords: tobacco brown spot, validamycin, high-throughput sequencing, microbial composition, phyllosphere microorganisms
Introduction
Tobacco is an economically important crop in China, which accounts for 39.6% of global tobacco production and approximately 40% of global tobacco consumption (Wang et al., 2013, 2014). Many fungal and bacterial diseases occur on tobacco leaves during production (Xu et al., 2014), even during the curing of leaves (Chen et al., 2020). Common diseases on tobacco leaves include target spots, brown spots, wildfire and powdery mildew (Xiao et al., 2003; Wu et al., 2012; Feng et al., 2022). Once leaf spots are prevalent, they can lead to lower-quality tobacco leaves, significantly reduce yields, and cause huge economic losses. For tobacco leaf spot disease management, validamycin is the only agent in China registered for controlling tobacco target spot, caused by Rhizoctonia solani. This fungicide has also been reported to have high activity against many other diseases, such as Rhizoctonia diseases, rice sheath blight, corn southern leaf blight, and rice false smut (Lueas, 1975; Kazuho, 1985; Asano et al., 1987; Müller et al., 1995). Validamycin is an antimicrobial pesticide isolated from an actinomycete, which interferes with and inhibits the growth and development of fungi after being absorbed by the fungal cells (Masaru et al., 1986; Copping and Menn, 2000). Years of large-scale field application have fully demonstrated its high efficacy, and it is considered harmless, pollution-free, and environmentally friendly for sheath blight disease. However, the effects of validamycin on tobacco brown spot caused by the ascomycete Alternaria alternata is not certain, and its effects on phyllosphere microorganisms has not been well studied.
The types and numbers of phyllosphere microbiota are closely related to plant growth development and metabolic activities (Lindow and Brandl, 2003; Leveau, 2015). Host plants and phyllosphere microorganism are in a dynamic equilibrium process (Blacquière et al., 2012; Carrión et al., 2019). Additionally, the microbial structures showed complex dynamic changes as affected by weather, soil, and other environmental factors. The study of Lindow and Leveau (2002) showed that bacterial community diversity in olive leaves was lowest during the warmest and driest months of the year and highest during cooler and wetter months. Carbon sources are the basis of microbial life activities. Biolog ECO metabolic phenotype technology can reflect the metabolic capacity of all microorganisms in the ecological environment to 31 common carbon sources, then reflect the nutritional requirements characteristics of environmental microorganisms, and understand the changes of microorganisms (Liu et al., 2021). Analysis of substrate utilization profiling in the tobacco brown spot revealed that the microbial community was highly utilized by carbohydrates, followed by carboxylic acids, amino acids, polymers and amines, however the utilization of phenolic compounds was minimum (Dai et al., 2022). Exploring the carbon metabolic ability of tobacco phyllosphere microorganisms after the application of the agent can confirm the influence of the agent on the microorganisms.
The population structure of phyllosphere microorganisms is closely related to the occurrence of plant disease (Mejia et al., 2008; Huang Y. et al., 2021). Xiang et al. (2022) found that microflora structure and diversity are different in symptomatic and asymptomatic tobacco leaves. At the early stage of tobacco brown spot, symptomatic leaves of different age had similar sets of dominant fungi and bacteria than the asymptomatic leaves. The dominant fungi included species of Alternaria, Didymella, Cladosporium, and the dominant bacteria included species of Pseudomonas and Sphingomonas (Xiang et al., 2022). In the middle and late stages of tobacco brown spot development, Pantoea was abundant in the bacterial community (Dai et al., 2022). The dominant phyllosphere fungi associated with leaves bearing tobacco brown spot were Alternaria, Phoma, Fusarium, and Cladosporium (Liu et al., 2009). In addition to the pathogenic fungus, Alternaria, the cause of tobacco brown spot, there are also species of Phoma and Fusarium associated with leaves. Whether these pathogen also cause tobacco leaf disease or increase the incidence of tobacco brown spot still need to be further investigated.
The composition of fungi and bacteria of tobacco brown spot diseased leaves have been widely reported (Dai et al., 2022). The occurrence of diseases changes the structure and diversity of the microbial community, and the composition of the microbial flora also is change by the host leaf maturity, the environment, and the use of control agents (Miller et al., 2018). The study of the changes of phyllosphere microorganisms can confirm the effect of agents on diseases. In tobacco leaves, the application of low fungicide dosages can significantly affect the population structure of beneficial bacteria such as species of Stenotrophomonas, Serratia, and Flavobacterium, which are known to be beneficial for distinct agricultural plants (Chen et al., 2020). When conditions are conducive to tobacco target spot caused by R. solani, the application of azoxystrobin can reduce disease incidence, and also cause significant changes in the diversity, community composition and relative abundance of both target and non-target microbes on asymptomatic and symptomatic tobacco leaves (Sun M. et al., 2023). Dimethachlon, a dicarboximide fungicide, can significantly affect the change of the population structure of beneficial bacteria such as species of Stenotrophomonas, Sphingomonas, Flavobacterium, and Serratia in tobacco leaves (Chen et al., 2021). Validamycin can provide effective control of Alternaria, which was tobacco brown spot diseased pathogen (Aftab et al., 2012). However, the phyllosphere microecological mechanisms of validamycin are poorly understood, especially its effects on microbial community structure and microbial metabolic function groups, whether on symptomatic or asymptomatic tobacco leaves.
Therefore, the objective of this study was to (i) analyze the structure and diversity of microbial communities in the leaf phyllosphere before and after validamycin application, (ii) investigate the changes of carbon metabolic functions in the phyllosphere before and after validamycin application, and (iii) compare changes in the structure, diversity, and metabolic functions of microbial communities in symptomatic and asymptomatic tobacco leaves after validamycin application.
Materials and methods
Experimental design
A field experiment was conducted in commercial tobacco fields of N. tabacum cv. Yunyan 105 in August 2020 in Heishitou Town, Weining County, Guizhou Province, China. The region is a humid subtropical monsoon climatic zone. The field was planted with tobacco cultivar Yunyan 105 during 2 years. Two-month old seedlings were transplanted into the field in April each year, and management conditions were typical for tobacco plants. The soil was laterite. Prior to the experiment, tobacco brown spot had sporadically appeared on the tobacco plants in the field, and the incidence rate was about 5–10%. And by the end of the full sampling period, the disease severity was moderate, the incidence rate was more than 20%. On 29 August 2020, nine symptomatic and nine asymptomatic tobacco plants at least 3.5 m apart from each other were randomly selected for first sampling, and tobacco leaves at the same leaf position were collected. In the test, a multifunctional sprayer (model: DSF01A-20-100, Guizhou Qianfengyuan Agricultural Science and Technology Development Co., LTD.) was used to evenly spray the surface of the blade until the droplets disappeared. Then at 5 (September 3), 10 (September 8), and 15 (September 13) days, diseased and healthy tobacco leaves were taken for samples. The classification of tobacco brown spot followed the Chinese National Standard (GB/T 23222–2008) from the Standardization Administration of China.
Sampling and processing of leaves
Three symptomatic and three asymptomatic tobacco plants were randomly selected from experiment plot, and the middle and lower mature leaves were sampled, respectively. Three strains of tobacco were mixed as one sample and repeated in triplicate. At the time of 0, 5, 10, and 15 days after application, sampling was performed separately. The letters B and J represent diseased and healthy tobacco leaves, respectively (Table 1). All samples were clipped carefully from the leaves using sterile hand shears, and placed into separate 50 mL plastic centrifuge tubes. The tubes were separately packed in sterile zip bags and placed in a portable Styrofoam box with ice packs. Upon arrival in the laboratory, the samples were immediately placed at 4°C for culturing, or at −80°C for DNA extraction for high-throughput sequencing.
Table 1.
Samples information.
| Sampling location | 0 day before application | 5 days after application | 10 days after application | 15 days after application | |
|---|---|---|---|---|---|
| Asymptomatic tissue | Sample No. | J01 | J11 | J21 | J31 |
| J02 | J12 | J22 | J32 | ||
| J03 | J13 | J23 | J33 | ||
| Group No. | J0 | J1 | B2 | B3 | |
| Symptomatic tissue | Sample No. | B01 | B11 | B21 | B31 |
| B02 | B12 | B22 | B32 | ||
| B03 | B13 | B23 | B33 | ||
| Group No. | B0 | B1 | B2 | B3 | |
Response of microbial communities exposed to validamycin
DNA extraction, PCR amplification, and high-throughput sequencing
The DNA of tobacco leaves was extracted by the EasyPure plant genome DNA kit (TransGen Biotech, China) in CTAB method. The solution was visualized using 2% agarose gel electrophoresis, and concentration and purity were assessed by a NanoDrop ND-2000 (Thermo Fisher Scientific, Waltham, MA, United States).
The hypervariable V4 region of bacterial 16S rRNA was amplified with the primers 515F (5′-CCTACGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′) (Walters et al., 2015). The fungal ITS1 region was amplified using primers ITS5-1F-F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS1-1F-R (5′-GCTGCGTTCTTCATCGATGC-3′) (Wang et al., 2020). All PCR reactions were carried out with 15 μL of Phusion®High-Fidelity PCR Master Mix (New England Biolabs), 2 μM of forward and reverse primers, and about 10 ng template DNA. Thermal cycling consisted of initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, with a final 72°C for 5 min to complete extension. PCR products were purified with the Qiagen Gel Extraction Kit (Qiagen, Germany). Amplification products were used in library construction and sequenced on an Illumina NovaSeq platform targeting 250 bp paired-end reads.
Phyllosphere microorganisms sequencing and analysis
Sequences with at least 97% similarity were assigned to the same operational taxonomic unit (OTU) using Uparse software V7.0.1001 (Edgar, 2013). Based on the Mothur algorithm, the taxonomic information of each representative sequence of each OTU was annotated using the UNITE and Silva database. The OTU abundance was normalized to the sample with the fewest sequences, to analyze the alpha and beta diversity of the microbial communities for each sample. The community composition of each sample was counted at the kingdom, phylum, class, order, family, genus, and species levels using the program Unit (V7.24) (Altschul et al., 1990; Kõljalg et al., 2013).
After obtaining the sequencing results and calculation of the OTU matrix, QIIME (V. 1.9.1) was used for full-sample similarity comparison to analyze the alpha diversity and to enumerate taxa. R software (Version 2.15.3) was used to draw dilution curves, rank abundance curves, and species accumulation boxes (White et al., 2009). Beta diversity on both weighted and unweighted unifrac distances was calculated using QIIME software (V.1.9.1). ITS sequences were aligned using Muscle (V.3.8.31) (Edgar, 2004). Phylogenetic analysis was conducted with FastTree 2 (V. 1.9.1) using Maximum Likelihood (ML). Functional composition of fungal and bacterial OTUs was predicted using PICRUSt (Price et al., 2010). Spearman correlation analysis was used to study the relationship between environmental factors and microbial species richness (alpha diversity), and to study relationships between environmental factors (temperature, relative humidity, and rainfall) and species abundance (Algina and Keselman, 1999). The top 50 most abundant species were selected for calculation of correlation coefficients, and correlation coefficients greater than −0.5 and less than 0.5, and p-values less than 0.05 were selected for symbiotic network interactions mapping (Qin et al., 2012 Jiao et al., 2017). The FUNGuild database was used to estimate phyllosphere microorganisms trophic mode (Nguyen et al., 2016).
Phyllosphere microorganisms carbon metabolism
Fifty Milliliters of 0.8% NaCl (27 mL, w/v) were each mixed with 1 g of tobacco leaves in 100 mL conical flask of different samples (cut into 2 mm x 2 mm pieces), and shaken for 1 h at 28°C. The mixture was then diluted at a ratio of 1:1000 with 0.8% NaCl. Into each of the Biolog Eco - Plates™ (Hayward, California), 150 μL of the dilutions were added and incubated at 28°C in darkness for 7 days in an OmnLog incubator following Deng et al. (2014). The Biolog OmniLog® system (BIOLOG, Hayward, CA, USA) in this study was equipped with a charge-coupled device camera system and an incubator. The color level of each well was measured in OmniLog Units (OU) every 30 min by OmniLog digital camera. The OU values reflected the dynamics of microbial carbon metabolism and represented the utilization of carbon sources. The Biolog ECO plate has 96 microtiter wells, for three replicates per plate. Each of the 32 microtiter wells, contains 1 different organic carbon source and the same amount of tetrazolium violet dye. The organic carbon source in the wells is the only source of energy for the microorganisms, and the tetrazolium violet dye turns purple if the microorganisms can utilize the carbon source after injection into the microtiter wells. The color’s depth reflects the microorganisms’ energy to utilize the carbon source, which indirectly reflects the change in the community composition of microorganisms. Kinetic and Parametric software from Biolog Inc. D5E_OKA_data.exe was used to collect the color intensity data and OL_FM_1.2.exe was used for data analysis” (Cruz et al., 2021), and heat maps were used to analyze the metabolic functions of the samples using HemI software (Baker et al., 2010).
Statistical analysis
Initial data analysis included taxonomic annotation of OTUs sequences, cluster analysis of all valid sequences of samples using Uparse v7.0.1001 software, annotation analysis, Alpha diversity analysis, Beta diversity analysis, and inter group analysis using Qiime software (Version 1.9.1). IBM SPSS Statistics 23 (IBM Corp., New York, NY, USA) was used to compare the differences of Alpha diversity and Beta diversity indexes of fungal and bacterial communities (p < 0.05). The co-occurrence networks of phyllosphere microbial genera and the relationships between environmental factors and microbial genera richness were demonstrated using Spearman’s rank analysis based on significant (p < 0.05) and strong positive correlations (r > 0.5) or strong negative correlations (r < −0.5; Li and Wu, 2018). Statistics were calculated by using Excel 2019, pictures were processed using Adobe Photoshop CS5.
Results
Microbial community composition and diversity
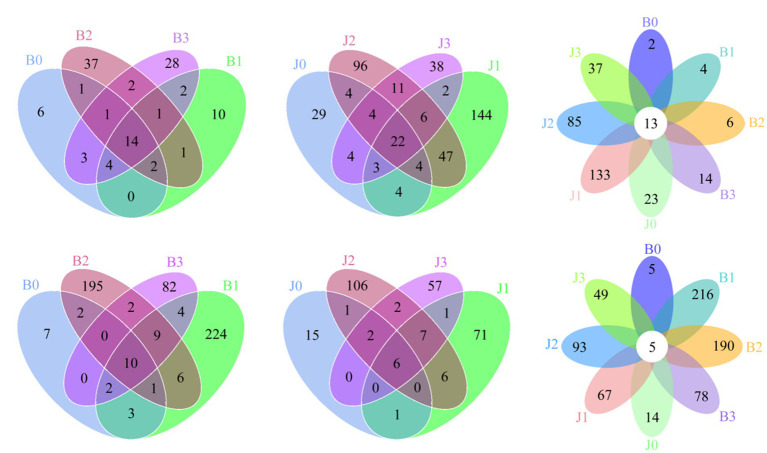
At the OTUs level, the differences between the communities at symptomatic and asymptomatic leaves were depicted with Venn diagrams. A total of 317 fungal OTUs were discovered, and there were 14, 22, and 13 common fungal OTUs in the symptomatic leaves, asymptomatic leaves, and all leaves, respectively (Figures 1A–C). The OTUs in symptomatic leaves gradually increased after validamycin application, and the OTUs in asymptomatic leaves increased first and then decreased. The number of OTUs was higher in asymptomatic leaves than in symptomatic leaves. A total of 712 bacterial OTUs were discovered in these samples, and there were 10, 6, and 5 core bacterial OTUs for bacterial communities in the symptomatic leaves, asymptomatic leaves, and all leaves at four sampling time points, respectively (Figures 1D–F). The number of OTUs in symptomatic leaves was higher than in asymptomatic leaves. After validamycin application, the independent OTUs numbers in symptomatic and asymptomatic leaves increased and then decreased.
Figure 1.
Venn diagram showing the number of microbial OTUs detached in tobacco leaves. (A–C) For fungi, (D–F) for bacteria. Numbers in the non-overlapping region indicate unique OTUs for the single sample; numbers in the overlapping region indicate shared OTUs for multi-samples; numbers in the core region indicate shared OTUs for each sample.
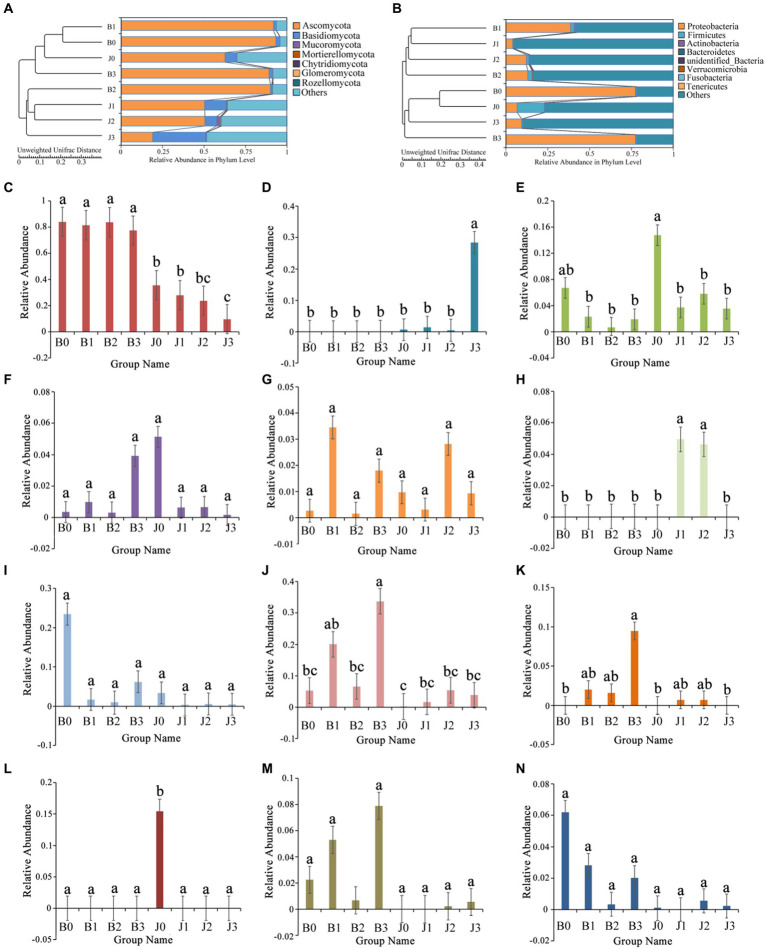
The main fungal phyla were Ascomycota, Basidiomycota, Mucoromycota, Mortierellomycota, and Chytridiomycota (Figure 2A; Table 2). The “Others” fungal group represented all microorganisms that were unidentified or whose relative abundance was less than 0.1% (Figures 2A,B). After validamycin application, the community composition of microorganisms significantly changed, and the relative abundance in symptomatic and asymptomatic leaves decreased for Ascomycota (0.04 and 0.44%) and increased for Basidiomycota (0.01 and 0.25%). At the genus level, the dominant genus in tobacco leaves at the four sampling time points was Alternaria, with relative abundances between 77 and 84%, and the relative abundance in asymptomatic leaves was significantly lower than that of symptomatic leaves (Figure 2C). The six most abundant genera were Alternaria, Thanatephorus, Cladosporium, Boeremia, Epicoccum, and Archaeorhizomyces (Figures 2D–H). After validamycin application, the relative abundance of Thanatephorus (0.27%) increased and Archaeorhizomyces (0.05%) decreased significantly in asymptomatic leaves while Thanatephorus was almost absent in symptomatic leaves.
Figure 2.
Changes in microbial community (A for fungi, B for bacteria) compositions at the genus level in tobacco leaves. (C) Alternaria, (D) Thanatephorus, (E) Cladosporium, (F) Boeremia, (G) Epicoccum, (H) Archaeorhizomyces, (I) Kosakonia, (J) Pseudomonas, (K) Serratia, (L) Weissella, (M) Massilia, (N) Sphingomonas. “a, b, and c” represents a statistical difference between groups (p < 0.05).
Table 2.
List of relative abundance in the fungal and bacterial community of the group.
| Community Structure | Relative abundance (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B0 | B1 | B2 | B3 | J0 | J1 | J2 | J3 | |||
| Fungi | Phyla | Ascomycota | 0.93 ± 0.05 a | 0.92 ± 0.03 a | 0.90 ± 0.08 a | 0.89 ± 0.03 a | 0.63 ± 0.14 b | 0.50 ± 0.03 b | 0.51 ± 0.15 b | 0.19 ± 0.04 c |
| Basidiomycota | 0.02 ± 0.02 cde | 0.02 ± 0.02 de | 0.01 ± 0.01 e | 0.02 ± 0.01 cde | 0.07 ± 0.02 cd | 0.13 ± 0.02 b | 0.07 ± 0.04 c | 0.32 ± 0.06 a | ||
| Mucoromycota | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.01 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.02 ± 0.04 a | 0.01 ± 0.01 a | ||
| Genera | Alternaria | 0.84 ± 0.16 a | 0.81 ± 0.12 a | 0.84 ± 0.07 a | 0.77 ± 0.08 a | 0.36 ± 0.11 b | 0.28 ± 0.02 b | 0.24 ± 0.06 bc | 0.10 ± 0.03 c | |
| Thanatephorus | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.01 ± 0.01 b | 0.01 ± 0.00 b | 0.01 ± 0.01 b | 0.28 ± 0.06 a | ||
| Cladosporium | 0.07 ± 0.10 ab | 0.02 ± 0.03 b | 0.01 ± 0.00 b | 0.02 ± 0.01 b | 0.15 ± 0.05 a | 0.04 ± 0.05 b | 0.06 ± 0.03 b | 0.04 ± 0.05 b | ||
| Boeremia | 0.00 ± 0.00 a | 0.01 ± 0.01 a | 0.00 ± 0.00 a | 0.04 ± 0.05 a | 0.05 ± 0.08 a | 0.01 ± 0.00 a | 0.01 ± 0.00 a | 0.00 ± 0.00 a | ||
| Epicoccum | 0.00 ± 0.00 a | 0.03 ± 0.06 a | 0.00 ± 0.00 a | 0.02 ± 0.02 a | 0.01 ± 0.01 a | 0.00 ± 0.00 a | 0.03 ± 0.02 a | 0.01 ± 0.00 a | ||
| Archaeorhizomyces | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.05 ± 0.02 a | 0.05 ± 0.04 a | 0.00 ± 0.00 b | ||
| Bacteria | Phyla | Proteobacteria | 0.38 ± 0.40 b | 0.39 ± 0.13 b | 0.13 ± 0.08 bc | 0.77 ± 0.30 a | 0.07 ± 0.10 bc | 0.04 ± 0.01 c | 0.12 ± 0.06 bc | 0.09 ± 0.03 bc |
| Firmicutes | 0.01 ± 0.01 b | 0.02 ± 0.02 b | 0.02 ± 0.02 b | 0.01 ± 0.01 b | 0.16 ± 0.14 a | 0.01 ± 0.01 b | 0.02 ± 0.01 b | 0.01 ± 0.01 b | ||
| Actinobacteria | 0.00 ± 0.00 ab | 0.01 ± 0.01 a | 0.00 ± 0.00 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 ab | 0.00 ± 0.00 b | 0.00 ± 0.00 ab | 0.00 ± 0.00 b | ||
| Genera | Kosakonia | 0.23 ± 0.40 a | 0.02 ± 0.01 a | 0.01 ± 0.00 a | 0.06 ± 0.04 a | 0.03 ± 0.05 a | 0.00 ± 0.00 a | 0.01 ± 0.01 a | 0.00 ± 0.01 a | |
| Pseudomonas | 0.05 ± 0.05 bc | 0.20 ± 0.13 ab | 0.07 ± 0.09 bc | 0.34 ± 0.27 a | 0.00 ± 0.00 c | 0.02 ± 0.01 bc | 0.05 ± 0.01 bc | 0.04 ± 0.01 bc | ||
| Serratia | 0.00 ± 0.00 b | 0.02 ± 0.02 ab | 0.02 ± 0.00 ab | 0.09 ± 0.15 a | 0.00 ± 0.00 b | 0.01 ± 0.00 ab | 0.01 ± 0.01 ab | 0.00 ± 0.00 b | ||
| Weissella | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.15 ± 0.13 a | 0.00 ± 0.00 b | 0.00 ± 0.00 b | 0.00 ± 0.00 b | ||
| Massilia | 0.02 ± 0.04 a | 0.05 ± 0.04 a | 0.01 ± 0.01 a | 0.08 ± 0.12 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.01 ± 0.00 a | ||
| Sphingomonas | 0.06 ± 0.11 a | 0.03 ± 0.03 a | 0.00 ± 0.00 a | 0.02 ± 0.02 a | 0.00 ± 0.00 a | 0.00 ± 0.00 a | 0.01 ± 0.00 a | 0.00 ± 0.00 a | ||
There was significant difference between different letters for the data in row direction (p < 0.05) according to Duncan’s new multiple range test.
The bacterial OTUs were placed in the Proteobacteria, Firmicutes, Actinobacteria, Verrucomicrobia, and Bacteroidetes, as well as “others” accounting for a large percentage (Figure 2B; Table 2). The relative abundance of Proteobacteria in symptomatic leaves was higher than that in asymptomatic leaves, whereas the relative abundance of Firmicutes on asymptomatic leaves was higher than that on symptomatic leaves. At the genus level, the dominant genera in symptomatic and asymptomatic leaves were Kosakonia, Pseudomonas, Serratia, Weissella, Massilia, Sphingomonas, Pantoea, Erwinia, Methylobacterium and Akkermansia (Figures 2I–N). After validamycin application, the relative abundance of Kosakonia (0.03%) in asymptomatic tissues was greatly reduced, while those of Pseudomonas (0.04%) and Massilia (0.01%) in asymptomatic leaves increased. Weissella was almost absent on symptomatic leaves, while its relative abundance was higher on asymptomatic leaves, but Weissella (0.15%) almost disappeared after the application of validamycin.
Microbial community alpha diversity
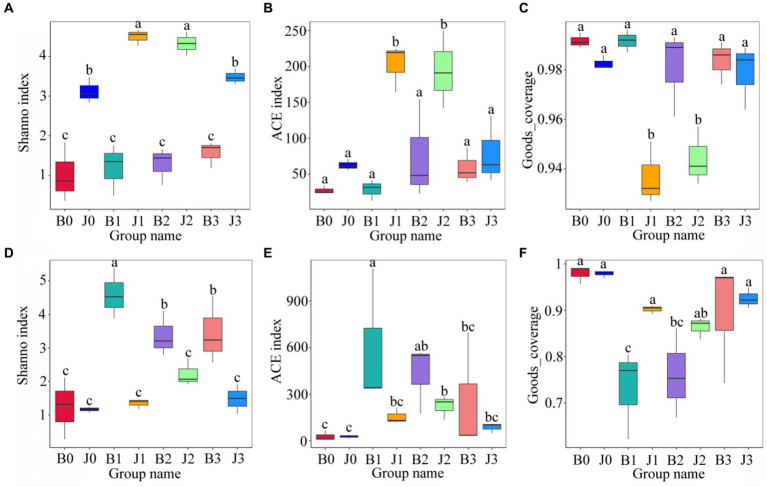
Statistics of Alpha Diversity analysis index (Shannon index, ACE index, goods_coverage) for each sample with 97% threshhold for OTUs were used to assess the differences in species richness and diversity of microbial communities in each sample (Figure 3). Before validamycin application, the Ace index and the Shannon index of fungal communities in asymptomatic leaves were significantly higher than those of symptomatic leaves. At 5 days and 10 days after spraying, these indices of fungal community in asymptomatic leaves increase significantly. There were no significant differences in the Goods_coverage of each sample (Figure 3C).
Figure 3.
Alpha diversity of the microbial (A–C for fungi, D–F for bacteria) communities in tobacco leaves. (A,D) Show the Shanno index; (B,E) show the ACE index; (C,F) Show the Goods_coverage. “a, b, and c” represents a statistical difference between groups (p < 0.05).
Significant differences between the Shannon index and the ACE index of bacterial communities in symptomatic and asymptomatic leaves (Figures 3D,E). At 5 days after validamycin application, the Ace index and the Shannon index of bacterial communities increased significantly both in symptomatic leaves and asymptomatic leaves from pre-treatment, and then they decreased significantly.
Microbial community beta diversity
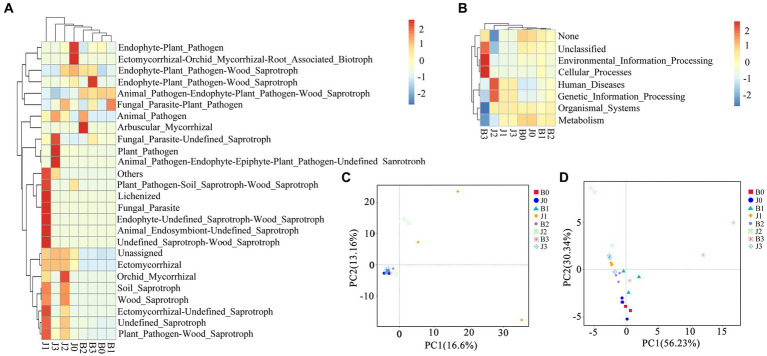
Principal coordinate analysis (PCoA) of the microbial community composition of different samples was based on Unifrac distances. For fungal community composition, nearly all samples were clustered together in the bottom left corner (Figure 4C), and only asymptomatic samples 1 and 2 (out of three replicates) at pretreatment were dispersed on the graph. This indicated that all the symptomatic samples (three each from days 0, 5, 10 and 15) showed little difference in Unifrac distances implying that they share the same OTUs (Figure 4C). The group of J1 and J2 did not cluster with other samples, but J3 had no significant differences from the other samples. The differences between the fungal communities of symptomatic leaves and those of asymptomatic leaves gradually decreased after the application of validamycin.
Figure 4.
Microbial functional categories and principal coordinate analysis of tobacco leaves. (A,C) For fungal communities, (B,D) for bacterial communities.
In the bacterial community composition, most samples were dispersed (Figure 4D). The difference between symptomatic and asymptomatic leaves was more significant, and the difference between after and before application of validamycin was equally significant. Differences between replicate samples of symptomatic and asymptomatic leaves at the same time points after application, and surface validamycin caused a greater effect on the bacterial community composition of symptomatic and asymptomatic leaves.
Microbial functional prediction
Fungal and bacterial ecological functions were predicted using FUNGuild based on community composition (Figure 4A). For fungal communities, the dominant functions in symptomatic leaves were animal pathogen-endophyte-plant pathogen-wood saprotroph, unassigned, plant pathogen, endophyte-plant pathogen, undefined saprotroph. In asymptomatic leaves in the fungal community, the dominant functions were animal pathogen-endophyte-plant pathogen-wood saprotroph, the dominant functions in symptomatic leaves were unassigned, followed as endophyte-plant pathogen, undefined saprotroph. Asymptomatic leaves were richer in fungal functional groups than that of symptomatic leaves. Asymptomatic leaves had more ecologically functional groups than symptomatic leaves for fungal communities.
Bacterial families were estimated using PICRUSt (Figure 4B). The diversity of functional groups of bacteria in asymptomatic and symptomatic leaves was significantly lower than that of fungi. In this study, metabolism, genetic information processing, unassigned, environmental information processing, and cellular processes were the dominant bacterial community functions common to all samples, only differing in their relative abundance from sample to sample. The functional groups of B3 and J2 were relatively more concentrated in distribution.
The relationship between microorganisms and environmental conditions
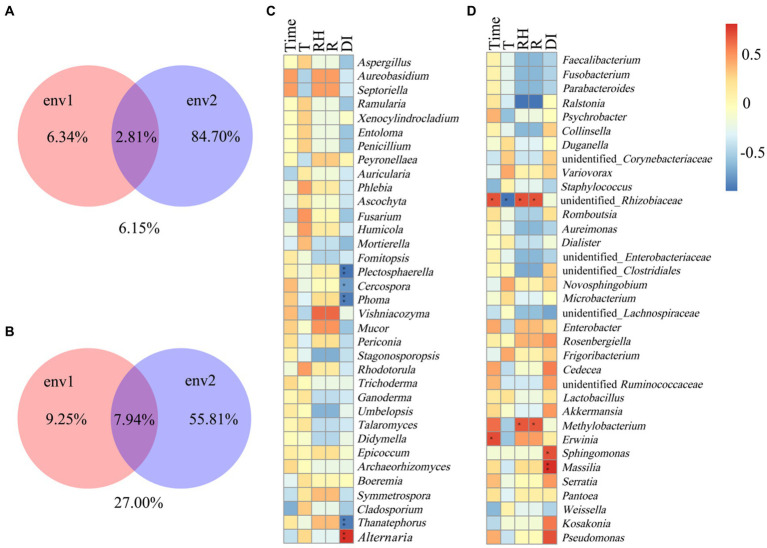
The Spearman correlation coefficient was used as a measure to study the relationship between environmental factors (time, temperature, relative humidity, rainfall) and microorganisms. Variance Partitioning analysis was used to account for the proportion of different environmental factors explaining changes in microbial community occurrence (Figure 5). Env1 included time, temperature, env2 included rainfall and disease index. In the fungal community, env2 occupied 84.70% of the community structure and env1 occupied 6.34% of the fungal community structure (Figure 5A). For fungal communities, the disease index was significantly and positively correlated with Alternaria, while negatively correlated with Thanatephorus, Phoma, Cercospora, and Plectosphaerella (Figure 5C). In the bacteria community, env2 occupied 55.81% of the community structure and env1 occupied 9.25% of the community structure (Figure 5B). Time was significantly and positively correlated with unidentified_Rhizobiaceae and Erwinia, the temperature was negative correlated with unidentified_Rhizobiaceae, relative humidity, and rainfall were positively correlated with unidentified_Rhizobiaceae and Methylobacterium, and disease index was significantly and positively correlated with Sphingomonas and Massilia (Figure 5D).
Figure 5.
Spearman correlation analysis between microbial communities and environmental factors in tobacco leaves. (A,C) For fungal communities, (B,D) for bacterial communities. Temperature (T), relative humidity (RH), rainfall (R), disease index (DI). “*” Represents a statistical difference between groups (p < 0.05), “**“represents a significant statistical difference between groups (p < 0.01) (env1: Time, T, RH; env2: R, DI).
Co-occurrence network analysis between microorganisms
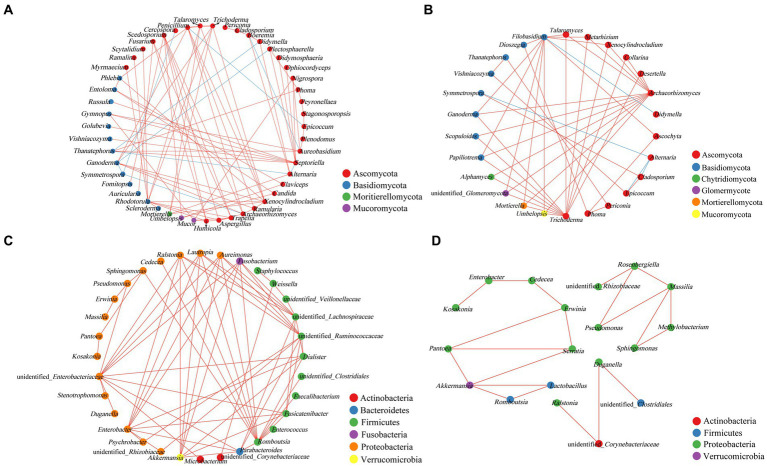
Network analysis of the co-occurrence of the most abundant 50 fungal species revealed that 49 fungal species were showing positive co-occurrence with each other (p < 0.05, r < −0.5). In fungal genera, 8 strong negatives and 4 strong negative correlations were identified from asymptomatic and symptomatic leaves, respectively (Figures 6A,B). As the pathogen of tobacco brown spot, the positively correlation was showed between Alternaria with Mortierella, Boeremia, and Didymella, and negatively correlated with Thanatephorus and Septoriella. In asymptomatic leaves, Thanatephorus showed positive co-occurrence with Didymella, Epicoccum with Penicillium, and Ganoderma with Plectosphaerella and Symmetrospora. Alternaria was positively correlated with Symmetrospora, Epicoccum, and Cladosporium in asymptomatic leaves, and Didymella was positively correlated with Filobasidium. There were only positive correlations between bacterial genera (Figures 6C,D).
Figure 6.
Co-occurrence networks of microbial communities at genus level. (A,B) For fungal communities, (C,D) for bacterial communities (A,C for asymptomatic leaves, B,D for symptomatic leaves). Node color represents the phylum of the genus and the size corresponds to the centrality score. Red lines indicate positive relationships, and blue lines indicate negative relationships.
Microbial community carbon metabolic profiles
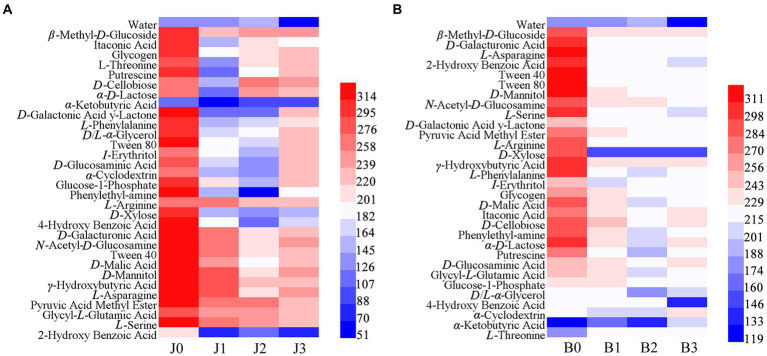
The Biolog ECO microplate was used to explore the metabolism of 31 basal carbon sources in symptomatic and asymptomatic leaves after validamycin application (Figure 7). The results showed that microbial communities were more capable of utilizing carbohydrates before validamycin, the metabolic capacity was reduced after the agent treatment. In asymptomatic tissues, L-arginine, γ-hydroxybutyric acid, and pyruvic acid methyl ester had strong metabolic capacity, α-keto butyric acid, and 2-hydroxy benzoic acid had lower metabolism. The metabolic capacity of asymptomatic tissues for 2-hydroxy benzoic was significantly reduced after validamycin treatment, with most metabolic capacities rising before decreasing. The metabolism of carbon sources from symptomatic was significantly reduced after the drug treatment. Reduced metabolism of D-xylose, 4-hydroxy benzoic acid, and D/L-α-glycerol and increased metabolism of α-cyclodextrin, α-ketobutyric acid and L-threonine in symptomatic leaves after validamycin treatment.
Figure 7.
Cluster heat map carbon sources in Biolog ECO-Plate of microbial communities from different samples (A for asymptomatic leaves, B for symptomatic leaves).
Discussion
Field application of validamycin (0.11 Kg a.i./ ha−1) presented a higher effect on Alternaria on asymptomatic leaves than that of symptomatic leaves. In our study, validamycin had significant effects on the phyllosphere microorganisms of tobacco leaves. After the application of validamycin, the relative abundance of Alternaria decreased on both symptomatic leaves and asymptomatic leaves. The relative abundance of Alternaria was reduced more on asymptomatic leaves. When 50% Kresoxim-methyl was used, the relative abundance of Alternaria was reduced in both healthy and diseased leaves, but the effect of the agent was greater on healthy leaves than on diseased leaves (Xiang et al., 2022). In studies on the phyllosphere microorganisms at different stages after the onset of tobacco brown spot, the relative abundance of Alternaria increased with the degree of disease (Dai et al., 2022). They found that validamycin can control the relative abundance of the pathogen at the onset of the tobacco brown spot. Validamycin had limited effect in the treatment of existing tobacco brown spot, but the relative abundance of Alternaria in asymptomatic leaves can be effectively suppressed. We recommend that validamycin be used preventively and not curatively. However, quantitative measurements of the microorganisms and more field trials on validamycin are still needed to further determine the efficacy of validamycin.
The species composition of phyllosphere microorganisms in tobacco changed after validamycin application. The species richness and community diversity of phyllosphere microorganisms increased at the beginning but decreased with the increase in application time. This implies that validamycin can inhibit the increase in the relative abundance of potential pathogens on asymptomatic leaves quickly, but low persistence and multiple applications can be considered if using validamycin for disease prevention. In a study of phyllosphere microorganisms in tobacco brown spot, the species richness and community diversity of fungi were higher in asymptomatic leaves than in symptomatic leaves, while the species richness and community diversity of bacteria were higher in asymptomatic leaves than in symptomatic leaves (Xiang et al., 2022). The dominant fungal genera were Alternaria, Thanatephorus, Cladosporium, Boeremia, and Epicoccum, and the dominant bacterial genera were Kosakonia, Pseudomonas, and Serratia. In previous studies, tobacco’s dominant fungi and bacteria were Alternaria, Didymella, Cladosporium, Pseudomonas and Sphingomonas, respectively. There were slight differences in the results of the two studies. However, in addition to tobacco target spot pathogens, the dominant genera were equally high in Alternaria, Pseudomonas and Sphingomonas during the onset of tobacco target spot (Sun M. L. et al., 2023). This indicates that the four fungi can exist together and even affect each other’s disease development. However, Pseudomonas are a group of bacteria that are widely found in the environment, plants, and air. Pseudomonas syringan pv. tabaci and P. syringae pv. angula are the causal agents of tobacco wildfire disease and tobacco bacterial angular spot disease, respectively, that can cause bacterial diseases of tobacco (Turner et al., 2020). Sphingomonas, a non-pathogenic bacterial genus, shows a strong antagonistic effect against Diaporthecitri on citrus (Li et al., 2022), but its relative abundance on asymptomatic leaves was significantly reduced by validamycin. Thanatephorus and Archaeorhizomyces were relatively abundant on symptomatic leaves but almost absent on asymptomatic leaves. Cladosporium, Kosakonia, Pseudomonas, and Sphingomonas were present on both symptomatic and asymptomatic leaves. These bacteria were also found in phyllosphere microbial investigations of tobacco leaf spot caused by Didymella and tobacco leaf mildew and rot (Huang Z. et al., 2021), implying that these bacteria may be resident on tobacco leaves. Validamycin application reduced the relative abundance of Cladosporium, Kosakonia and Sphingomonas in addition to the pathogen, Alternaria. Cladosporium as a potential pathogen, can also cause diseases in some crops such as cucumber, tomato, and canola (Batta, 2004; Rivas and Thomas, 2005 Idnurm et al., 2021). Guo et al. (2023) explored the effect of azoxystrobin on phyllosphere microorganisms of tobacco brown spot, and showed a similar decrease in the relative abundance of Cercospora in addition to Thanatephorus, but an increase in the relative abundance of Pseudomonas and Sphingomonas. When the fungicide STROBY (containing azoxystrobin) was used to control tobacco brown spot, the agent caused a decrease in community diversity in symptomatic leaves and an increase in bacterial community diversity, with STROBY having a significantly greater effect on the fungal community than on the bacterial community (Xiang et al., 2022). When agents are applied for disease control, they affect the entire phyllosphere microecology, and act on multiple pathogenic or beneficial bacteria at the same time, resulting in changes in the structure and diversity of phyllosphere microorganisms to achieve control of disease exacerbation (Wang et al., 2016).
The metabolic capacity of asymptomatic leaves remained higher than that of symptomatic leaves after validamycin application, and both had a lower metabolic capacity for 2-hydroxy benzoic. The results are the same with those of the metabolic capacity for 2-hydroxy benzoic in studies on the metabolism of phyllosphere microorganisms of tobacco brown spot at different spatiotemporal environments (Dai et al., 2022). And 2-hydroxy benzoic also named salicylic acid, it was closely related to plant immunity and disease resistance (An and Mou, 2011). However, further experiments are needed to determine whether this metabolic decline was due to changes in phyllosphere microorganisms. After validamycin, the metabolic capacity of symptomatic and asymptomatic leaves was significantly reduced. But 15 days after the application, the metabolic capacity of asymptomatic tobacco leaves started to recover. It is possible that the number of phyllosphere microorganisms decreased and their metabolic capacity for carbon sources was reduced since the agents significantly controlled the relative abundance of the pathogenic Alternaria already present in asymptomatic tobacco leaves. This result was similar to the results of phyllosphere microbial metabolism capacity after Bordeaux mixture for tobacco target spot control (Li et al., 2022). But afterward, the community diversity started to recover, the diversity increased and the metabolic capacity for carbon sources started to recover. In contrast, the asymptomatic tissues were unable to recover their metabolic capacity, because the integrity of the tobacco leaf was damaged from the onset of the disease and the relative abundance of Alternaria was too high. The relative abundance of beneficial bacteria could not be restored to normal values, Phyllosphere microbes unable to restore homeostasis. This is important for plant leaves to stay healthy (Liu et al., 2020).
The relationship between different microorganisms on the tobacco leaves was showed in the co-occurrence network (Deng et al., 2012; Vacher et al., 2016). In this study, there were significantly more interactions between fungi than bacteria, and the interrelationships between asymptomatic leaves were stronger than those of symptomatic leaves. The results were in common with the variation in the relative abundance of species, with negative correlations between Alternaria and Thanatephorus. In Spearman correlation analysis, the disease index was significantly and positively correlated with Alternaria, while negatively correlated with Thanatephorus, Phoma, Cercospora, and Plectosphaerella. The two results suggest a possible competitive relationship between Alternaria and Thanatephorus, which was found to be similarly negatively correlated with Alternaria and Thanatephorus after validamycin application in a study of tobacco target spot disease (Guo et al., 2023). In tobacco leaves, Alternaria and Thanatephorus may have a competitive relationship, the two are different tobacco disease pathogens, when the disease occurs at the same time, the pathogen proliferation generously, the environment, nutrients, the same increase in the demand for habitation, the leaf resources are limited, one of them first increase, will inhibit the growth of the other pathogen, and the relative abundance of the non-pathogenic bacteria is low, the energy required is less, will not be affected from it too much.
After validamycin application, the relative abundance of Alternaria on asymptomatic leaves decreased and the relative abundance of Thanatephorus increased significantly. The proliferation of pathogenic bacteria led to a dramatic increase in the demand for space and nutrients in the leaves, and the proliferation of one of them inhibited the growth of the other. The interactions between the bacteria showed positive correlations, which may be related to the population abundance of each pathogen, or it may be that the stronger effect of validamycin on phyllosphere bacteria directly caused the relative abundance of a large number of bacteria to decrease, but the competition, fitness, and environmental adaptability of these pathogens when coexisting deserve further study. Meanwhile, this experiment provides a limited reference for the control of tobacco target spot disease by validamycin. We still need further field investigations on the specific efficacy of validamycin, and quantitative analysis of phyllosphere microorganisms to gain a deeper understanding of the control effects.
Conclusion
The results of this study showed that the dominant phyllosphere microorganisms of tobacco brown spot were Alternaria, Thanatephorus, Kosakonia, and Pseudomonas, while phytopathogenic fungi such as Boeremia, and Epicoccum were also present in the leaves. The community diversity indices of fungi and bacteria of symptomatic and asymptomatic leaves increased after the application of validamycin, and the microbial community diversity of asymptomatic leaves was higher than that of symptomatic leaves. Microbial community richness increased and then decreased in both symptomatic and asymptomatic leaves. The effect of validamycin on the fungal community on asymptomatic leaves was greater than that on symptomatic leaves, and the structure of the bacterial community on symptomatic leaves was greater than that on asymptomatic leaves. The effect of validamycin on phyllosphere microorganisms gradually decreased with increasing application time.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found a: https://www.ncbi.nlm.nih.gov/, PRJNA982433.
Author contributions
MG: Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Writing – original draft. JH: Conceptualization, Supervision, Writing – review & editing. CJ: Conceptualization, Supervision, Writing–review & editing. YZ: Data curation, Project administration, Writing – review & editing. HW: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. XZ: Data curation, Project administration, Writing – review & editing. TH: Methodology, Supervision, Writing – review & editing. CS: Conceptualization, Writing – review & editing. QW: Data curation, Project administration, Writing – review & editing. FW: Resources, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by China National Tobacco Corporation [110202101048(LS-08) and 110202001035(LS-04)], National Natural Science Foundation of China (31960550 and 32160522), the Hundred Level Innovative Talent Foundation of Guizhou Province [GCC(2022)028–1], the Guizhou Science Technology Foundation (ZK[2021]Key036), Guizhou Province Applied Technology Research and Development Funding Post-subsidy Project, and the Guizhou Tobacco Company (2020XM22, 2020XM03). The authors declare that this study received funding from the China National Tobacco Corporation and Guizhou Tobacco Company. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aftab A. K., Akhtar N., Nafisa S. A., Malik Z. U. (2012). In vitro screening methods using chemical fungicides against canola black spot pathogen. Mycopathologia 10, 63–66. [Google Scholar]
- Algina J., Keselman H. J. (1999). Comparing squared multiple correlation coefficients: Examination of a confidence interval and a test significance. Psychol. Methods. 4, 76–83. doi: 10.1037/1082-989X.4.1.76 [DOI] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- An C., Mou Z. (2011). Salicylic acid and its function in plant immunity. J. Integr. Plant Biol. 53, 412–428. doi: 10.1111/j.1744-7909.2011.01043.x [DOI] [PubMed] [Google Scholar]
- Asano N., Yamaguchi T., Kameda Y., Matsui K. (1987). Effect of validamycins on glycohydrolases of Rhizoctonia solani. J. Antibiot. 40, 526–532. doi: 10.7164/antibiotics.40.526 [DOI] [PubMed] [Google Scholar]
- Baker C. M., Chitrakar R., Obulareddy N., Panchal S., Williams P., Melotto M. (2010). Molecular battles between plant and pathogenic bacteria in the phyllosphere. Braz. J. Med. Biol. Res. 43, 698–704. doi: 10.1590/s0100-879x2010007500060, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batta Y. (2004). Cladosporium tenuissimum Cooke (Deuteromycotina: Hyphomycetes) as a causal organism of new disease on cucumber fruits. Eur. J. Plant Pathol. 110, 1003–1009. doi: 10.1007/s10658-004-1604-7 [DOI] [Google Scholar]
- Blacquière T., Smagghe G., van Gestel C. A., Mommaerts V. (2012). Neonicotinoids in bees: a review on concentrations, side-effects and risk assessment. Ecotoxicology (London, England) 21, 973–992. doi: 10.1007/s10646-012-0863-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrión V. J., Perez-Jaramillo J., Cordovez V., Tracanna V., de Hollander M., Ruiz-Buck D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science (New York, N.Y.) 366, 606–612. doi: 10.1126/science.aaw9285 [DOI] [PubMed] [Google Scholar]
- Chen Q. L., Cai L., Wang H. C., Cai L. T., Goodwin P., Ma J., et al. (2020). Fungal composition and diversity of the tobacco leaf phyllosphere during curing of leaves. Front. Microbiol. 11:554051. doi: 10.3389/fmicb.2020.554051, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Wicaksono W. A., Berg G., Cernava T. (2021). Bacterial communities in the plant phyllosphere harbour distinct responders to a broad-spectrum pesticide. Sci. Total Environ. 751:141799. doi: 10.1016/j.scitotenv.2020.141799, PMID: [DOI] [PubMed] [Google Scholar]
- Copping L. G., Menn J. J. (2000). Biopesticides: a review of their action, applications and efficacy. Pest Manag. Sci. 56, 651–676. doi: 10.1002/1526-4998(200008)56:8 [DOI] [Google Scholar]
- Cruz C. D., Esteve P., Tammela P. (2021). Evaluation and validation of Biolog OmniLog® system for antibacterial activity assays. Lett. Appl. Microbiol. 72, 589–595. doi: 10.1111/lam.13450, PMID: [DOI] [PubMed] [Google Scholar]
- Dai Y. F., Wu X. M., Wang H. C., Li W. H., Cai L. T., Li J. X., et al. (2022). Spatio-temporal variation in the phyllospheric microbial biodiversity of Alternaria alternata-infected tobacco foliage. Front. Microbiol. 13:920109. doi: 10.3389/fmicb.2022.920109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Jiang Y. H., Yang Y., He Z., Luo F., Zhou J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Wang Y., Liu Z., Cheng H., Xue Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604, PMID: [DOI] [PubMed] [Google Scholar]
- Feng Z., Li Y., Ma X., Duan Y., Zhang R., Hsiang T., et al. (2022). Draft genome sequence of Alternaria longipes causing tobacco brown spot. Plant Dis. 106, 734–736. doi: 10.1094/PDIS-06-21-1274-A, PMID: [DOI] [PubMed] [Google Scholar]
- Guo M. Y., Xiong J., Wang H. C., Zhang Y., Cai L. T., Chen X. J., et al. (2023). Inhibitory effect of dimetachlone on tobacco target spot pathogens and the effect on the structure of phyllosphere microbial community in tobacco. Chinese. J. Pestic. Sci. 25, 858–869. doi: 10.16801/j.issn.1008-7303.2023.0061 [DOI] [Google Scholar]
- Idnurm A., Beard C., Smith A., Hills A. L., Chambers K. R. (2021). Emergence of Cladosporium macrocarpum disease in canola (Brassica napus). Australas. Plant Pathol. 50, 687–694. doi: 10.1007/s13313-021-00819-8 [DOI] [Google Scholar]
- Huang Y., Wang H. C., Cai L. T., Li W., Pan D., Xiang L., et al. (2021). Phyllospheric microbial composition and diversity of the tobacco leaves infected by Didymella segeticola. Front. Microbiol. 12:699699. doi: 10.3389/fmicb.2021.699699, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Wang P., Pu Z., Lu L., Chen G., Hu X., et al. (2021). Effects of mancozeb on citrus rhizosphere bacterial community. Microb. Pathog. 154:104845. doi: 10.1016/j.micpath.2021.104845, PMID: [DOI] [PubMed] [Google Scholar]
- Jiao S., Chen W., Wei G. (2017). Biogeography and ecological diversity patterns of rare and abundant bacteria in oil contaminated soils. Mol. Ecol. 26, 5305–5317. doi: 10.1111/mec.14218 [DOI] [PubMed] [Google Scholar]
- Kazuho M. (1985). Characteristics of validamycin a in controlling Rhizoctonia diseases. Natural Products, 301–308. doi: 10.1016/B978-0-08-029223-6.50048-3 [DOI] [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F., Bahram M., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Leveau J. (2015). Life of Microbes on Aerial Plant Parts. Principles of Plant-Microbe Interactions, ed. Lugtenberg B., Springer, Cham. doi: 10.1007/978-3-319-08575-3_4 [DOI] [Google Scholar]
- Li S., Wu F. (2018). Diversity and co-occurrence patterns of soil bacterial and fungal communities in seven intercropping systems. Front. Microbiol. 9:1521. doi: 10.3389/fmicb.2018.01521, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. D., Zhu Z. R., Zhang Y., Xu J., Wang H., Wang Z., et al. (2022). The phyllosphere microbiome shifts toward combating melanose pathogen. Microbiome 10:56. doi: 10.1186/s40168-022-01234-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Brandl M. T. (2003). Microbiology of the phyllosphere. Appl. Environ. Microbiol. 69, 1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindow S. E., Leveau J. H. (2002). Phyllosphere microbiology. Curr. Opin. Biotechnol. 13, 238–243. doi: 10.1016/s0958-1669(02)00313-0 [DOI] [PubMed] [Google Scholar]
- Liu H., Brettell L. E., Singh B. (2020). Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 25, 841–844. doi: 10.1016/j.tplants.2020.06.003, PMID: [DOI] [PubMed] [Google Scholar]
- Liu C., Wang H. C., Xie H. L., Chen Q. L., Yu Z. H., Sun G. J. (2009). Fungal community structure analysis of tobacco leaf infected with brown spot asymptomatic. Guizhou Agric. Sci. 7, 54–59. doi: 10.16135/j.issn1002-0861.2019.0114 [DOI] [Google Scholar]
- Liu T. T., Zeng Y. T., Wang H. C., Cai L. T., Zhang C. Q. (2021). Metabolic and microbial community structure analysis in the phyllosphere of tobacco leaves with different maturity during brown spot occurring season. Chinese Tobacco Sci. 42, 22–29. doi: 10.13496/j.issn.1007-5119.2021.06.004 [DOI] [Google Scholar]
- Lueas G. B. (1975). Disease of tobacco, 3rd Edn. Raleigh, NC: Biological Consulting Associates. 267–296. [Google Scholar]
- Masaru U., Akiko I., Tetsu O., Motoo S. (1986). Effect of validamycin on β-D-Glucan-degrading enzymes from Rhizoctonia solani. Agric. Biol. Chem. 50, 1885–1886. doi: 10.1080/00021369.1986.10867663 [DOI] [Google Scholar]
- Mejia L. C., Rojas E. I., Maynard Z., Zuleyka M., Sunshine V. B., Elizabeth A., et al. (2008). Endophytic fungi as biocontrol agents of theobroma cacao pathogens. Biol. Control 46, 4–14. doi: 10.1016/j.biocontrol.2008.01.012 [DOI] [Google Scholar]
- Miller E. T., Svanbäck R., Bohannan B. J. M. (2018). Microbiomes as metacommunities: understanding host-associated microbes through metacommunity ecology. Trends Ecol. Evol. 33, 926–935. doi: 10.1016/j.tree.2018.09.002 [DOI] [PubMed] [Google Scholar]
- Müller J., Boller T., Wiemken A. (1995). Effects of validamycin a, a potent trehalase inhibitor, and phytohormones on trehalose metabolism in roots and root nodules of soybean and cowpea. Planta 197, 362–368. doi: 10.1007/BF00202658 [DOI] [Google Scholar]
- Nguyen N. H., Song Z., Bates S. T., Branco S., Tedersoo L., Menke J. (2016). Funguild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006 [DOI] [Google Scholar]
- Price M. N., Dehal P. S., Arkin A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 490, 55–60. doi: 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Rivas S., Thomas C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43, 395–436. doi: 10.1146/annurev.phyto.43.040204.140224 [DOI] [PubMed] [Google Scholar]
- Sun M., Shi C., Huang Y., Wang H., Li J., Cai L., et al. (2023). Effect of disease severity on the structure and diversity of the phyllosphere microbial community in tobacco. Front. Microbiol. 13:1081576. doi: 10.3389/fmicb.2022.1081576, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M. L., Wang H. C., Shi C. H., Li J. J., Cai L. T., Xiang L. G., et al. (2023). Effect of azoxystrobin on tobacco leaf microbial composition and diversity. Front. Plant Sci. 13:1101039. doi: 10.3389/fpls.2022.1101039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S. E., Pang Y. Y., O'Malley M. R., Weisberg A. J., Fraser V. N., Yan Q., et al. (2020). A deoR-type transcription regulator is required for sugar-induced expression of type III secretion-encoding genes in Pseudomonas syringae pv. Tomato DC3000. Molecular plant-microbe interactions. MPMI 33, 509–518. doi: 10.1094/MPMI-10-19-0290-R [DOI] [PubMed] [Google Scholar]
- Vacher C., Hampe A., Porté A. J., Sauer U., Compant S., Morris C. E. (2016). The phyllosphere: microbial jungle at the plant-climate interface. Annu. Rev. Ecol. Evol. Syst. 47, 1–24. doi: 10.1146/annurev-ecolsys-121415-032238 [DOI] [Google Scholar]
- Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., et al. (2015). Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1, e00009–e00015. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. C., Huang Y. F., Wang J., Chen X. J., Wei K. S., Wang M. S., et al. (2016). Activities of azoxystrobin and difenoconazole against Alternaria alternata and their control efficacy. Crop Prot. 90, 54–58. doi: 10.1016/j.cropro.2016.08.022 [DOI] [Google Scholar]
- Wang Y., Wang H., Cheng H., Chang F., Wan Y., She X. (2020). Niche differentiation in the rhizosphere and endosphere fungal microbiome of wild Paris polyphylla Sm. PeerJ 8:e8510. doi: 10.7717/peerj.8510, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. C., Wang J., Li W. H., Huang Y. F., Xia H. Q., Wang M. S., et al. (2014). Cladosporium cladosporioides identified in China on tobacco seeds. Plant Dis. 98:1002. doi: 10.1094/PDIS-12-13-1203-PDN, PMID: [DOI] [PubMed] [Google Scholar]
- Wang H. C., Wang M. S., Xia H. Q., Yang S. J., Guo Y. S., Xu D. Q., et al. (2013). First report of fusarium wilt of tobacco caused by fusarium kyushuense in China. Plant Dis. 97:424. doi: 10.1094/PDIS-09-12-0835-PDN [DOI] [PubMed] [Google Scholar]
- White J. R., Nagarajan N., Pop M. (2009). Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 5:e1000352. doi: 10.1371/journal.pcbi.1000352, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y. H., Zhao Y. Q., Fu Y., Zhao X. X., Chen J. G. (2012). First report of target spot of flue-cured tobacco caused by Rhizoctonia solani AG-3 in China. Plant Dis. 96:1824. doi: 10.1094/PDIS-06-12-0551-PDN [DOI] [PubMed] [Google Scholar]
- Xiang L. G., Wang H. C., Cai L. T., Guo T., Luo F., Hsiang T., et al. (2022). Variations in leaf phyllosphere microbial communities and development of tobacco brown spot before and after fungicide application. Front. Microbiol. 13:1068158. doi: 10.3389/fmicb.2022.1068158, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S., Charoenwattana P., Holcombe L., Turner J. G. (2003). The Arabidopsis genes RPW8.1 and RPW8.2 confer induced resistance to powdery mildew diseases in tobacco. Molecular plant-microbe interactions. MPMI 16, 289–294. doi: 10.1094/MPMI.2003.16.4.289, PMID: [DOI] [PubMed] [Google Scholar]
- Xu H., Yang G. H., Zhang M., Dang L., Wang W. Y., Wang M. F., et al. (2014). Analysis of diversity of endophytes and phyllospheric microorgaism from Yunnan tobacco leaves. J. Yunnan Agric. University (Natural Science). doi: 10.3969/j.issn.1004-390X(n).2014.02.001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found a: https://www.ncbi.nlm.nih.gov/, PRJNA982433.