Abstract
The UspA surface antigen of Moraxella catarrhalis was recently shown to be comprised of two different proteins (UspA1 and UspA2) which share an internal region containing 140 amino acids with 93% identity (C. Aebi, I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen, Infect. Immun. 65:4367–4377, 1997). Isogenic uspA1, uspA2, and uspA1 uspA2 mutants were tested in a number of in vitro systems to determine what effect these mutations, either individually or together, might exert on the phenotype of M. catarrhalis 035E. Monoclonal antibodies specific for UspA1 or UspA2 were used in an indirect antibody accessibility assay to prove that both of these proteins were expressed on the surface of M. catarrhalis. All three mutants grew in vitro at the same rate and did not exhibit autoagglutination or hemagglutination properties that were detectably different from those of the wild-type parent strain. When tested for the ability to adhere to human epithelial cells, the wild-type parent strain and the uspA2 mutant readily attached to Chang conjunctival cells. In contrast, the uspA1 mutant and the uspA1 uspA2 double mutant both attached to these epithelial cells at a level nearly 2 orders of magnitude lower than that obtained with the wild-type parent strain, a result which suggested that expression of UspA1 by M. catarrhalis is essential for attachment to these epithelial cells. Both the wild-type parent strain and the uspA1 mutant were resistant to the bactericidal activity of normal human serum, whereas the uspA2 mutant and the uspA1 uspA2 double mutant were readily killed by this serum. This latter result indicated that the presence of UspA2 is essential for expression of serum resistance by M. catarrhalis.
Moraxella catarrhalis is an important pathogen of the respiratory tract of both children and adults. This unencapsulated, gram-negative organism accounts for up to 20% of cases of acute bacterial otitis media (6, 7, 17, 37) and is associated with approximately one-third of infectious exacerbations of chronic obstructive pulmonary disease in adults (14, 24, 40, 44). As a consequence of its emerging medical importance, M. catarrhalis has become the focus of research efforts aimed at elucidating its interaction with the human host and at developing strategies for a vaccine to protect against this pathogen (3, 15–18, 20–22, 36, 39, 47).
Efforts to identify potential vaccine candidates among the surface antigens of M. catarrhalis have focused primarily on the outer membrane proteins of this organism. In M. catarrhalis, outer membrane protein profiles examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) display remarkably little strain-specific variability (5, 41). Some of these outer membrane proteins, especially CopB (outer membrane protein B2 [OMP B2]) (2, 8, 26, 28, 51), OMP CD (32, 42), and the UspA antigen (high-molecular-weight outer membrane protein [HMW-OMP]) (27, 35), which consists of two related proteins, UspA1 and UspA2 (1), have been characterized in some detail. CopB and OMP CD, as well as either or both UspA1 and UspA2, have been shown to induce the synthesis of antibodies that are biologically active against M. catarrhalis (11, 26, 27, 58).
The UspA1 and UspA2 proteins are of particular interest because of their unusual characteristics. In M. catarrhalis 035E, the uspA1 and uspA2 genes encode predicted proteins of 88 and 62 kDa, respectively (1). In SDS-PAGE, the native forms of these two proteins apparently form oligomers or aggregates, each of which migrates in SDS-PAGE with an apparent molecular weight of greater than 250,000. Apparently monomeric forms of these proteins can be detected in Western blot analysis as minor bands of approximately 120 kDa (UspA1) and 85 kDa (UspA2) (1). The amino acid sequences of UspA1 and UspA2 are 43% identical, but an internal region in each protein contains 140 amino acids where the level of identity is 93%. This latter region contains an epitope that is present in both UspA1 and UspA2 and which is defined by its reactivity with the monoclonal antibody (MAb) 17C7 (1). This epitope is present in all disease-associated isolates of M. catarrhalis tested to date and induces the synthesis of antibodies that, when used to passively immunize mice, enhanced the elimination of M. catarrhalis in a pulmonary clearance model (27). Equally important, the very high molecular weight UspA antigen composed of UspA1 and UspA2 has been shown to be a target for antibodies present in convalescent sera of patients recovering from M. catarrhalis infections (13, 25, 27), indicating that one or both of these proteins are expressed in vivo.
To assess and differentiate functional characteristics of the UspA1 and UspA2 proteins, we constructed a set of isogenic mutants of M. catarrhalis 035E that lacked the ability to express UspA1 or UspA2 or both of these proteins. These mutants were compared to the wild-type strain in a number of in vitro systems, including assessment of their abilities to adhere to human epithelial cells and to resist killing by normal human serum.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. M. catarrhalis strains were routinely grown at 37°C on brain heart infusion (BHI) agar plates (Difco Laboratories, Detroit, Mich.) in an atmosphere of 95% air–5% CO2 supplemented, when necessary, with kanamycin (20 μg/ml) (Sigma Chemical Co., St. Louis, Mo.) or chloramphenicol (0.5 μg/ml) (Sigma); in some cases, cells were grown in BHI broth. The BHI broth used to grow M. catarrhalis cells for attachment assays was sterilized by filtration. Escherichia coli strains were cultured on Luria-Bertani agar plates (38) supplemented, when necessary, with ampicillin (100 μg/ml), kanamycin (30 μg/ml), or chloramphenicol (30 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| M. catarrhalis | ||
| 035E | Wild-type isolate from middle ear fluid | 27 |
| 035E.1 | Isogenic mutant of 035E with a kan cartridge in the uspA1 structural gene | 1 |
| 035E.2 | Isogenic mutant of 035E with a kan cartridge in the uspA2 structural gene | 1 |
| 035E.12 | Isogenic mutant of 035E with a kan cartridge in the uspA2 structural gene and a cat cartridge in the uspA1 structural gene | This study |
| P-44 | Wild-type isolate that exhibits rapid hemagglutination | 52 |
| P-48 | Wild-type isolate that exhibits slow hemagglutination | 52 |
| Escherichia coli DH5α | Host for cloning experiments | Stratagene |
| Plasmids | ||
| pBluescript II SK+ | Cloning vector; Ampr | Stratagene |
| pUSPA1 | pBluescript II SK+ with a 2.7-kb insert containing most of the uspA1 gene of M. catarrhalis 035E | 1 |
| pUSPA1CAT | pUSPA1 with a cat cartridge replacing the 0.6-kb BglII fragment of the uspA1 gene | This study |
Characterization of outer membrane proteins.
Outer membrane vesicles of M. catarrhalis strains were prepared as described previously (43, 45). Proteins present in these preparations were resolved by SDS-PAGE and detected by staining with Coomassie blue or by Western blot analysis as described elsewhere (26).
MAbs.
MAb 17C7 is a murine immunoglobulin G (IgG) antibody that reacts with a conserved epitope of both UspA1 and UspA2 from M. catarrhalis 035E (1). To produce MAbs individually specific for UspA1 and UspA2, mice were immunized by intraperitoneal injection with 50 μg of either purified UspA1 or purified UspA2 from strain 035E (40a) suspended in 50% (vol/vol) Freund’s complete adjuvant (Difco). One month later, the mice received an intraperitoneal injection with 25 μg of the appropriate protein suspended in 50% (vol/vol) Freund’s incomplete adjuvant (Difco). Approximately 2 weeks later, the mice were injected intravenously with 25 μg of the purified protein. Three days later, the mice were euthanized and their spleens were removed for use in the hybridoma fusion protocol (48). Hybridoma culture supernatants were screened for the presence of UspA1- or UspA2-specific MAbs, using purified UspA1 or UspA2 as antigen in an enzyme-linked immunosorbent assay. The IgG1 MAb 11A6 was shown to be specific for UspA1, and the IgG1 MAb 17H4 was shown to be specific for UspA2. MAb 3F12, an IgG MAb specific for the major outer membrane protein of Haemophilus ducreyi (34), was used as a negative control in the indirect antibody accessibility assay. These MAbs were used in the form of hybridoma culture supernatant fluid in both the indirect antibody accessibility assay and the colony blot radioimmunoassay (23). MAbs 17C7 and 11A6 were purified with protein G-Sepharose 4 Fast Flow (Amersham Pharmacia Biotech, Piscataway, N.J.) for use in attachment inhibition assays.
Mutant construction method.
The 1.3-kb chloramphenicol resistance (cat) cartridge was prepared by excision (using BamHI) from pUCΔECAT (kindly provided by Bruce A. Green, Wyeth-Lederle Vaccines). The cat cartridge was subsequently ligated into BglII restriction sites located in the mid-portion of the cloned segment from the uspA1 gene in pUSPA1 (1); after transformation of competent E. coli DH5α cells, recombinant clones were identified by selection on solidified media containing chloramphenicol.
Transformation of M. catarrhalis.
The electroporation method used for transformation of M. catarrhalis 035E has been described in detail elsewhere (28). In this study, M. catarrhalis cells were electroporated with 5 μg of linear DNA (a PCR product containing the truncated uspA1 gene with the cat cartridge insertion) in 5 μl of water.
Southern blot analysis.
Chromosomal DNA purified from wild-type and mutant M. catarrhalis strains was digested with either PvuII or HindIII (New England Biolabs), and Southern blot analysis was performed as described previously (50). Double-stranded DNA probes were labeled with 32P by using a Random Primed DNA labeling kit (Boehringer Mannheim, Indianapolis, Ind.).
Indirect antibody accessibility assay.
Overnight BHI broth cultures of M. catarrhalis 035E and its isogenic mutants were diluted in phosphate buffered saline (PBS) containing 10% (vol/vol) fetal bovine serum and 0.025% (wt/vol) sodium azide (PBS-FBS-A) to a density of 110 Klett units (ca. 5 × 108 CFU/ml) as measured with a Klett-Summerson colorimeter (Klett Manufacturing Co., New York, N.Y.). Portions (100 μl) of this suspension were added to 1 ml of hybridoma culture supernatant. After incubation at 4°C for 1 h with gentle agitation, the bacterial cells were washed once and suspended in 1 ml of PBS-FBS-A. Affinity-purified goat anti-mouse immunoglobulin, radiolabeled with 125I to a specific activity of 108 cpm per μg, was added and the mixture was incubated for 1 h at 4°C with gentle agitation. The cells were then washed four times with 1 ml of PBS-FBS-A, suspended in 500 μl of triple detergent (26), and transferred to glass tubes. The radioactivity present in each sample was measured by using a gamma radiation counter.
Autoagglutination and hemagglutination assays.
The ability of M. catarrhalis strains to autoagglutinate was assessed by using bacterial cells grown overnight on a BHI agar plate. These cells were resuspended in PBS to a turbidity of 400 Klett units in a glass tube and subsequently allowed to stand at room temperature for 10 min, at which time the turbidity of this suspension was again determined. Rapid and slow autoagglutination were defined as turbidities of less than and greater than 200 Klett units, respectively, after 10 min. The hemagglutination slide assay using heparinized human blood group O Rh+ erythrocytes was performed as previously described (52).
Serum bactericidal assay.
Complement-sufficient normal adult human serum was prepared by standard methods. Complement inactivation was achieved by heating the serum for 30 min at 56°C. An M. catarrhalis broth culture in early logarithmic phase was diluted in Veronal-buffered saline containing 0.10% (wt/vol) gelatin to a concentration of 1 × 105 CFU/ml, and 20-μl portions were added to 20 μl of native or heat-inactivated normal human serum together with 160 μl of Veronal-buffered saline containing 5 mM MgCl2 and 1.5 mM CaCl2. This mixture was incubated at 37°C in a stationary water bath. At time 0 and at 15 and 30 min, duplicate 10-μl aliquots were removed, suspended in 75 μl of BHI broth, and spread onto prewarmed BHI agar plates.
Adherence assay.
A method used to measure adherence of Haemophilus influenzae to Chang conjunctival cells in vitro (55) was adapted for use with M. catarrhalis. Briefly, 2 × 105 to 3 × 105 HEp-2 cells (ATCC CCL 23) or Chang conjunctival cells (ATCC CCL 20.2) were seeded into each well of a 24-well tissue culture plate (Corning-Costar) and incubated for 24 h before use. A 0.3-ml volume from an antibiotic-free overnight culture of M. catarrhalis was inoculated into 10 ml of fresh BHI medium lacking antibiotics, and this culture was subsequently allowed to grow to a density of approximately 5 × 108 CFU/ml (120 Klett units) with shaking in a gyratory water bath. The culture was harvested by centrifugation at 6,000 × g at 4 to 8°C for 10 min. The supernatant was discarded, and a Pasteur pipette was used to gently resuspend the bacterial cells in 5 ml of pH 7.4 PBS or PBS containing 0.15% (wt/vol) gelatin (PBS-G). The bacterial cells were centrifuged again, and this final pellet was gently resuspended in 6 to 8 ml of PBS or PBS-G.
Portions (25 μl containing 107 CFU) of this suspension were inoculated in duplicate into the wells of a 24-well tissue culture plate containing monolayers of HEp-2 or Chang cells. For attachment inhibition assays, the bacterial cells were incubated with various concentrations of purified MAbs for 30 min at 37°C immediately prior to addition of these bacterial cells to the monolayers. These tissue culture plates were centrifuged for 5 min at 165 × g and then incubated for 30 min at 37°C. Nonadherent bacteria were removed by rinsing the wells gently five times with PBS or PBS-G, and the epithelial cells were then released from the plastic support by adding 200 μl of PBS containing 0.05% trypsin and 0.02% EDTA. This cell suspension was serially diluted in PBS or PBS-G and spread onto BHI plates to determine the number of viable M. catarrhalis present. Adherence was expressed as the percentage of bacteria attached to the human cells relative to the original inoculum added to the well.
RESULTS
Construction of an isogenic M. catarrhalis mutant lacking expression of both UspA1 and UspA2.
Construction of M. catarrhalis mutants lacking the ability to express either UspA1 (mutant strain 035E.1) or UspA2 (mutant strain 035E.2) has been described elsewhere (1). For constructing a double mutant that lacked expression of both UspA1 and UspA2, the 0.6-kb BglII fragment within the incomplete uspA1 open reading frame of pUSPA1 (1) was replaced with a cat cartridge, yielding the recombinant plasmid pUSPA1CAT. Oligonucleotide primers (5′-CGGGATCCGTGAAGAAAAATGCCGCAGGT-3′ and 5′-CGGGATCCCGTCGCAAGCCGATTG-3′) were used in PCR to amplify the 3.2-kb insert of pUSPA1CAT; this PCR product was used to electroporate the kanamycin-resistant uspA2 mutant 035E.2. Southern blot analysis was used to prove that a chloramphenicol- and kanamycin-resistant transformant (strain 035E.12) derived from this experiment was a uspA1 uspA2 double mutant (data not shown).
Characterization of selected proteins expressed by the wild-type and mutant M. catarrhalis strains.
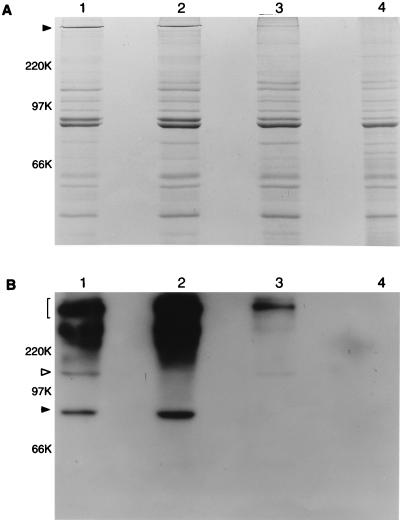
Proteins present in outer membrane vesicles extracted from the wild-type strain and these three mutant strains were resolved by SDS-PAGE and either stained with Coomassie blue (Fig. 1A) or probed with MAb 17C7 in Western blot analysis (Fig. 1B). The wild-type parent strain 035E possessed a very high molecular weight band detectable by Coomassie blue staining (Fig. 1A, lane 1) that was also similarly abundant in the uspA1 mutant 035E.1 (Fig. 1A, lane 2). The uspA2 mutant 035E.2 (Fig. 1A, lane 3) had a much reduced level of expression of a band in this same region of the gel; this band was not visible in the uspA1 uspA2 double mutant 035E.12 (Fig. 1A, lane 4).
FIG. 1.
Detection of the UspA1 and UspA2 proteins in wild-type and mutant strains of M. catarrhalis 035E. Proteins (10 μg) present in EDTA-extracted outer membrane vesicles from the wild-type strain (lane 1), uspA1 mutant 035E.1 (lane 2), uspA2 mutant 035E.2 (lane 3), and isogenic uspA1 uspA2 double mutant 035E.12 (lane 4) were resolved by SDS-PAGE and either stained with Coomassie blue (A) or transferred to nitrocellulose and probed with MAb 17C7 followed by radioiodinated goat anti-mouse immunoglobulin in Western blot analysis (B). In panel A, the closed arrowhead indicates the very high molecular weight form of the UspA antigen, in which UspA2 likely predominates. In panel B, the bracket on the left indicates the region of the autoradiograph containing the very high molecular weight forms of the UspA1 and UspA2 proteins that bind MAb 17C7. The open arrowhead indicates the 120-kDa, putative monomeric form of UspA1. The closed arrowhead indicates the 85-kDa, putative monomeric form of UspA2. This autoradiograph was overexposed to allow detection of the relatively minor, putative monomeric form of UspA1 in lanes 1 and 3; this overexposure resulted in detection of a diffuse region of MAb 17C7 reactivity in the region of the autoradiograph near and immediately above the 220-kDa position marker in lanes 1 and 2. This material is present only in those strains which express UspA2 (lanes 1 and 2) and likely represents different size aggregates of UspA2. Molecular weight position markers (in kilodaltons) are present on the left.
Western blot analysis using the UspA1- and UspA2-reactive MAb 17C7 revealed that the wild-type strain (Fig. 1B, lane 1) expressed abundant amounts of MAb 17C7-reactive antigen, most of which had a very high molecular weight, in excess of 220,000. The wild-type strain also exhibited discrete antigens with apparent molecular weights of approximately 120,000 and 85,000 which bound this MAb (Fig. 1B, lane 1). The uspA1 mutant 035E.1 (Fig. 1B, lane 2) lacked expression of the 120-kDa antigen, which was proposed to be the monomeric form of UspA1 (1), but still expressed the 85-kDa antigen. The amount of very high molecular weight MAb 17C7-reactive antigen expressed by this uspA1 mutant appeared to be equivalent to that expressed by the wild-type strain. The uspA2 mutant 035E.2 (Fig. 1B, lane 3) expressed the 120-kDa antigen but lacked expression of the 85-kDa antigen, which was proposed to be the monomeric form of the UspA2 protein (1). In contrast to the uspA1 mutant, the uspA2 mutant had relatively little very high molecular weight antigen reactive with MAb 17C7. Finally, the uspA1 uspA2 double mutant 035E.12 (Fig. 1B, lane 4) expressed no detectable MAb 17C7-reactive antigens.
Binding of UspA1- and UspA2-specific MAbs to whole cells of the wild-type and mutant strains.
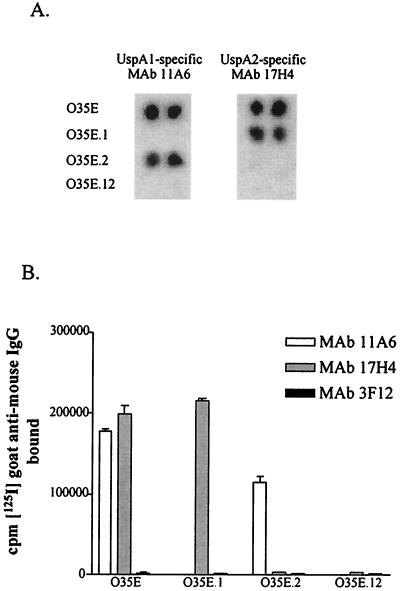
The indirect antibody accessibility assay was used to determine whether both UspA1 and UspA2 are exposed on the surface of M. catarrhalis and accessible to antibody. MAbs 11A6 and 17H4, specific for UspA1 and UspA2, respectively, were produced for use in this assay. The specificity of each of these MAbs was first confirmed in the colony blot radioimmunoassay where MAb 11A6 bound the wild-type strain 035E and the uspA2 mutant but did not bind the uspA1 mutant 035E.1 or the uspA1 uspA2 mutant 035E.12 (Fig. 2A). MAb 17H4 bound the wild-type strain 035E and the uspA1 mutant 035E.1 but did not react with the uspA2 mutant 035E.2 or with the double mutant 035E.12 (Fig. 2A). Both of these MAbs bound to the surface of whole cells of the wild-type strain 035E in the indirect antibody accessibility assay (Fig. 2B), a result which indicated that both UspA1 and UspA2 are exposed on the surface of M. catarrhalis 035E. In this same assay, each MAb bound only to the mutant strain that expressed its homologous antigen (e.g., the UspA1-specific MAb 11A6 bound to whole cells of the uspA2 mutant 035E.2) (Fig. 2B). Neither MAb bound to cells of the uspA1 uspA2 mutant 035E.12 (Fig. 2B).
FIG. 2.
Binding of MAbs to wild-type and mutant strains of M. catarrhalis. Colony paste of wild-type strain 035E, uspA1 mutant 035E.1, uspA2 mutant 035E.2, and uspA1 uspA2 double mutant 035E.12 spotted in duplicate on filter paper was probed with MAbs 11A6 and 17H4 in the colony blot radioimmunoassay (A) to prove the specificity of these MAbs for UspA1 and UspA2, respectively. These two MAbs were then tested for the ability to bind to whole cells of these same strains in the indirect antibody accessibility assay (B). Binding of the UspA1- and UspA2-specific MAbs to whole cells of these four strains is reflected by the amount (in counts per minute) of radioiodinated goat anti-mouse IgG bound to MAbs attached to the surface of the bacterial cells. MAb 3F12, a murine IgG MAb specific for the major outer membrane protein of H. ducreyi (34), was used as a negative control.
Characterization of the growth, autoagglutination, and hemagglutination properties of the wild-type and mutant strains.
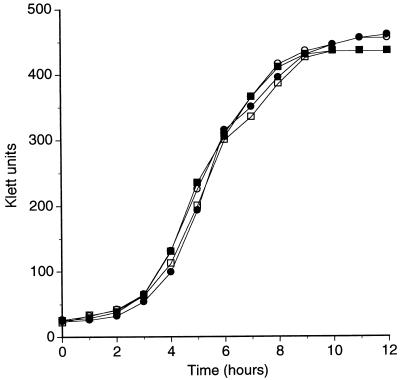
The colony morphology of these three mutant strains grown on BHI agar plates did not differ from that of the wild-type parent strain (data not shown). Similarly, the rates and extents of growth of all four of these strains in BHI broth were very similar if not identical (Fig. 3). In an autoagglutination assay performed as described in Materials and Methods, all four strains exhibited the same, relatively low rate of autoagglutination (data not shown). Finally, there was no detectable difference between the wild-type parent and the three mutants in a hemagglutination assay using human erythrocytes (52). Control hemagglutination experiments were performed with a pair of M. catarrhalis isolates (strains P-44 and P-48) previously characterized as having rapid and slow rates, respectively, of hemagglutination (52).
FIG. 3.
Comparison of the growth of the wild-type and mutant strains of M. catarrhalis in vitro. Wild-type strain 035E (closed squares), uspA1 mutant 035E.1 (open squares), uspA2 mutant 035E.2 (closed circles), and uspA1 uspA2 double mutant 035E.12 (open circles) from overnight broth cultures were diluted to a density of 35 Klett units in BHI broth and subsequently allowed to grow at 37°C with shaking. Growth was followed by means of turbidity measurements.
Effect of the uspA1 and uspA2 mutations on the ability of M. catarrhalis to adhere to human cells.
Preliminary experiments revealed that the wild-type M. catarrhalis strain 035E adhered readily to HeLa cells, HEp-2 cells, and Chang conjunctival cells in vitro (data not shown). To determine whether lack of expression of UspA1 or UspA2 affected this adherence ability, the wild-type strain and the three mutant strains were first used in an attachment assay with HEp-2 cells. In this set of experiments, PBS was used to wash the HEp-2 cell monolayers and as the diluent for serial dilution of the trypsinized HEp-2 cell monolayer at the completion of the assay. The wild-type strain and the uspA2 mutant 035E.2 exhibited similar levels of attachment to HEp-2 monolayers (Table 2). The uspA1 mutant 035E.1, however, was less able to adhere to these HEp-2 cells; lack of expression of UspA1 reduced the level of attachment approximately sixfold (Table 2). The uspA1 uspA2 double mutant 035E.12 exhibited a similarly reduced level of attachment (Table 2).
TABLE 2.
Adherence of wild-type and mutant strains of M. catarrhalis to HEp-2 and Chang conjunctival cells in vitro
| Strain | Adherencea to:
|
|
|---|---|---|
| HEp-2 cellsb | Chang cellsc | |
| 035E (wild type) | 14.7 ± 4.9 | 51.4 ± 30.8 |
| 035E.1 (uspA1 mutant) | 2.4 ± 0.9 (0.006d) | 0.8 ± 0.5 (0.002) |
| 035E.2 (uspA2 mutant) | 19.1 ± 7.0 (0.213) | 55.9 ± 16.7 (0.728) |
| 035E.12 (uspA1 uspA2 double mutant) | 2.3 ± 1.8 (0.011) | 0.6 ± 0.2 (0.002) |
Expressed as percentage of the original inoculum that was adherent to the human epithelial cells at the end of the 30-min incubation period. Each value represents the mean ± standard deviation of two independent experiments.
PBS was used for washing of the monolayers and for serial dilutions of adherent M. catarrhalis.
PBS-G was used for washing of the monolayers and for serial dilutions of adherent M. catarrhalis.
P value compared to wild-type strain 035E, using the two-tailed Student t test.
Control experiments revealed, however, that M. catarrhalis cells did not survive well in the PBS used for washing of the HEp-2 monolayer and serial dilution of the attached M. catarrhalis organisms. When 108 CFU of each of the wild-type and mutant M. catarrhalis strains was suspended in PBS, serially diluted, and allowed to stand for 30 min on ice, the viable number of bacteria decreased to 107 CFU (data not shown). In contrast, when PBS-G was used for this same type of experiment, there was no reduction in the viability of these M. catarrhalis strains over the duration of the experiment. When the HEp-2 cell-based attachment experiments were repeated with PBS-G for washing the HEp-2 cell monolayer and as the diluent, there was only a threefold reduction in adherence of the uspA1 mutant relative to that obtained with the wild-type parent strain (data not shown). This finding suggested that the original sixfold difference in attachment ability observed between the wild-type and uspA1 mutant strain may have been attributable in part to viability problems caused by the use of the PBS wash and diluent.
In a previous study (1), the M. catarrhalis UspA1 protein was found to be most similar to the hsf gene product of H. influenzae, which itself promoted attachment of both H. influenzae type b and recombinant E. coli to Chang conjunctival cells (54). Subsequent experiments using Chang conjunctival cells as the target for bacterial attachment by these M. catarrhalis strains together with a PBS-G wash and diluent revealed a substantial difference in the attachment abilities of the wild-type strain and the uspA1 mutant (Table 2). Whereas the wild-type strain and the uspA2 mutant exhibited similar levels of attachment to the Chang cells, the extent of attachment of the uspA1 mutant was nearly 2 orders of magnitude less than that of the wild-type parent strain. The uspA1 uspA2 double mutant also exhibited a much reduced level of attachment similar to that obtained with the uspA1 mutant (Table 2).
Additional experiments were performed with the M. catarrhalis CopB major outer membrane protein-specific MAb 10F3 (26) and an indirect immunofluorescence technique to detect M. catarrhalis organisms bound to these human epithelial cells. These experiments proved that the reduced attachment levels observed with the uspA1 and uspA1 uspA2 mutants were not the result of increased sensitivity of these two mutants to the trypsin used to release the epithelial cells for determination of viable bacteria (data not shown). Finally, both the UspA-reactive MAb 17C7 and the UspA1-specific MAb 11A6 were tested for the ability to inhibit attachment of strain 035E to Chang cells. Neither MAb inhibited bacterial attachment in vitro (data not shown).
Effect of the uspA1 and uspA2 mutations on serum resistance of M. catarrhalis.
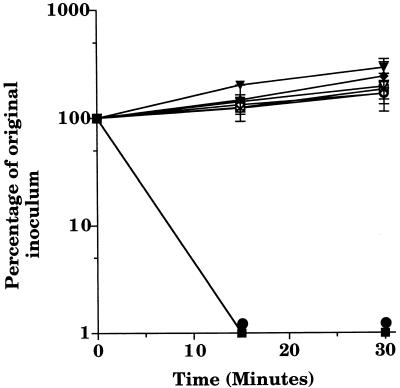
Because the M. catarrhalis UspA2 protein was previously found to resemble most closely the YadA outer membrane protein involved in serum resistance of pathogenic Yersinia species (1), it was appropriate to determine whether UspA2 might play the same functional role in M. catarrhalis. Similar to the majority of disease isolates of M. catarrhalis (29, 30, 57), the wild-type strain 035E was resistant to killing by normal human serum in vitro (28). To examine the effect of the lack of expression of UspA1 or UspA2 on serum resistance, the wild-type strain and the three mutant strains were tested in a serum bactericidal assay. Both the wild-type strain and the uspA1 mutant 035E.1 were able to grow in the presence of normal human serum (Fig. 4), indicating that lack of expression of UspA1 did not adversely affect the ability of strain 035E.1 to resist killing by normal human serum. However, both the uspA2 mutant 035E.2 and the uspA1 uspA2 double mutant 035E.12, having in common the lack of expression of UspA2, were readily killed by normal human serum (Fig. 4). Heat-based inactivation of the complement system present in this normal human serum eliminated the ability of this serum to kill these latter two mutants (Fig. 4).
FIG. 4.
Susceptibility of wild-type and mutant strains of M. catarrhalis to killing by normal human serum. Cells of wild-type strain 035E (diamonds), uspA1 mutant 035E.1 (triangles), uspA2 mutant 035E.2 (circles), and uspA1 uspA2 double mutant 035E.12 (squares) from logarithmic-phase BHI broth cultures were incubated in the presence of 10% (vol/vol) normal human serum (closed symbols) or heat-inactivated normal human serum (open symbols). Data are presented as the percentage of the original inoculum remaining at each time point; error bars are included.
DISCUSSION
Lack of the ability to express UspA1 or UspA2 or both of these proteins had little or no discernible effect on the ability of the respective M. catarrhalis mutants to grow in vitro (Fig. 3). Similarly, it would now appear that UspA1 and UspA2 are not involved in either autoagglutination or hemagglutination in strain 035E because lack of expression of either or both of these macromolecules did not affect the autoagglutination or hemagglutination properties of strain 035E (data not shown). The ability of some M. catarrhalis strains to hemagglutinate may be due to expression of a 200-kDa protein recently described by Fitzgerald et al. (19). In this context, it should be noted that strain 035E apparently does not express this 200-kDa protein and both autoagglutinates and hemagglutinates relatively slowly.
The production of MAbs individually specific for UspA1 and UspA2 permitted unequivocal demonstration of the fact that both of these macromolecules are exposed on the surface of M. catarrhalis 035E (Fig. 2B). Furthermore, the relative level of binding of the UspA1- and UspA2-reactive MAb 17C7 by proteins in outer membrane vesicles of the uspA1 and uspA2 mutants in Western blot analysis (Fig. 1B, lanes 2 and 3) suggested that there is likely more UspA2 than UspA1 present in strain 035E. Subsequent analyses performed in the present study revealed that UspA1 and UspA2 differ by more than just their relative abundance in M. catarrhalis. Specifically, mutations that independently eliminated expression of these two macromolecules had profound effects on at least two different phenotypic traits (adherence ability and serum resistance) of M. catarrhalis 035E.
The similarity between UspA1 and the H. influenzae adhesins encoded by the hsf (54) and hia genes (4) prompted investigation of the possibility that UspA1 is functionally involved in the ability of M. catarrhalis to attach to human epithelial cells. Initial experiments utilizing HeLa, HEp-2, and Chang conjunctival cells revealed that the M. catarrhalis wild-type strain 035E bound readily to these cell lines (data not shown). Additional testing involving the uspA1, uspA2, and uspA1 uspA2 mutants derived from strain 035E revealed that both the uspA1 and uspA1 uspA2 mutants did not exhibit wild-type levels of attachment to HEp-2 cells in vitro (Table 2). Further investigation showed that the uspA1 mutant adhered to Chang conjunctival cells at a level nearly 2 orders of magnitude lower than that obtained with the wild-type parent strain. In contrast, the uspA2 mutant adhered to Chang cells at wild-type levels.
It remains to be determined whether UspA1 itself is an adhesin or whether expression of UspA1 is simply required for proper expression or conformation of another macromolecule which itself is the true adhesin. Bearing in mind the complex situation involving adhesins which are positioned in the tips of the pili of gram-negative bacteria such as Neisseria gonorrhoeae (49), it is premature to conclude that UspA1 binds directly to the human epithelial cells used in the present study. A preliminary report that antiserum directed against purified UspA inhibited attachment of M. catarrhalis to human epithelial cells was published prior to the discovery that there are two UspA proteins expressed by M. catarrhalis (10). Whether this polyclonal antibody bound to UspA1 or UspA2 or both proteins is relevant to correct interpretation of the functional basis for this inhibitory effect. In addition, another laboratory has recently reported that the CD protein of M. catarrhalis will bind highly purified human middle ear mucin (47), a finding which reinforces the likelihood that M. catarrhalis possesses multiple systems for binding host factors. Finally, it must be noted that there clearly are many differences between Chang cells and the epithelium of the upper respiratory tract. Investigation of the attachment ability of the uspA1 mutant in human nasopharyngeal organ culture (53) would provide a more stringent test of the role of UspA1 in the attachment process.
The similarity of the UspA2 protein to the YadA protein expressed by pathogenic Yersinia species also had predictive value regarding the involvement of this M. catarrhalis protein in protecting this organism against killing by normal human serum. YadA has been shown to confer serum resistance on Y. enterocolitica by promoting the fixation of factor H (12), which in turn leads to the degradation of C3b deposited on the bacterial cell surface and prevention of formation of the membrane attack complex (46), in a manner similar to that observed with the M protein of Streptococcus pyogenes (31). When the set of three isogenic M. catarrhalis mutants was incubated in complement-sufficient normal human serum, the uspA2 mutant and the uspA1 uspA2 mutant were both readily killed in this serum (Fig. 4). In contrast, the uspA1 mutant resisted killing by this serum as effectively as did the wild-type parent strain (Fig. 4).
It is known that isolates of M. catarrhalis can be divided into those that are sensitive to the bactericidal activity of normal human serum and those that are resistant to this killing (9, 30, 33, 56, 57). Moreover, one group has suggested that complement resistance is a virulence factor for M. catarrhalis, based on their finding that the majority of disease isolates of M. catarrhalis exhibit some degree of complement resistance (30). A preliminary study by Verduin et al. (56) suggested that the HMW-OMP of M. catarrhalis, which is now known to be identical to either UspA1 or UspA2, is responsible for this serum resistance exhibited by disease isolates of M. catarrhalis. Regardless of whether the HMW-OMP is UspA1 or UspA2, the fact remains that lack of expression of UspA2 in an isogenic uspA2 mutant rendered the serum-resistant wild-type parent strain exquisitely sensitive to killing by this serum. It should also be noted that the available data do not allow determination of whether UspA2 exerts a direct or indirect effect on serum resistance of M. catarrhalis. The existence of such a causal relationship remains to be established and must be pursued carefully, especially because a previous study from our own laboratory has shown that lack of expression of the CopB outer membrane protein, which is likely involved in some transport process, resulted in loss of serum resistance by M. catarrhalis (28).
In conclusion, mutations in the uspA1 and uspA2 genes of M. catarrhalis 035E affected two different phenotypic traits of this pathogen. Southern blot analysis has suggested that disease isolates of M. catarrhalis likely possess both uspA1 and uspA2 genes (1), and future studies will be designed to investigate whether lack of expression of UspA1 or UspA2 has similar effects on other strains of this pathogen. Whether both of these genes are present and expressed in all strains of M. catarrhalis also remains to be determined. This is especially important with regard to isolates of M. catarrhalis obtained from healthy children (i.e., nasopharyngeal carriage isolates). Whether UspA1 or UspA2 could be required for expression of some other capability essential to the ability of M. catarrhalis to colonize the upper respiratory tract or to the production of disease remains to be determined.
ACKNOWLEDGMENTS
This study was supported by Public Health Service grant AI36344 and by Texas Advanced Technology Program award 003660-087 to E.J.H. C.A. was supported by a research grant for young investigators from Novartis AG, Basel, Switzerland.
We thank Steven Berk for providing several isolates of M. catarrhalis. We also acknowledge the generosity of Bruce A. Green and John McMichael in providing the cat cartridge and purified UspA1 and UspA2 from M. catarrhalis 035E, respectively.
REFERENCES
- 1.Aebi C, Maciver I, Latimer J L, Cope L D, Stevens M K, Thomas S E, McCracken G H, Jr, Hansen E J. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect Immun. 1997;65:4367–4377. doi: 10.1128/iai.65.11.4367-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi C, Stone B, Beucher M, Cope L D, Maciver I, Thomas S E, McCracken G H, Jr, Sparling P F, Hansen E J. Expression of the CopB outer membrane protein by Moraxella catarrhalis is regulated by iron and affects iron acquisition from transferrin and lactoferrin. Infect Immun. 1996;64:2024–2030. doi: 10.1128/iai.64.6.2024-2030.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed K, Matsumoto K, Rikitomi N, Nagatake T. Attachment of Moraxella catarrhalis to pharyngeal epithelial cells is mediated by a glycosphingolipid receptor. FEMS Microbiol Lett. 1996;135:305–309. doi: 10.1111/j.1574-6968.1996.tb08005.x. [DOI] [PubMed] [Google Scholar]
- 4.Barenkamp S J, St. Geme J W., III Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol Microbiol. 1996;19:1215–1223. doi: 10.1111/j.1365-2958.1996.tb02467.x. [DOI] [PubMed] [Google Scholar]
- 5.Bartos L C, Murphy T F. Comparison of the outer membrane proteins of 50 strains of Branhamella catarrhalis. J Infect Dis. 1988;158:761–765. doi: 10.1093/infdis/158.4.761. [DOI] [PubMed] [Google Scholar]
- 6.Bluestone, C. D. 1986. Otitis media and sinusitis in children: role of Branhamella catarrhalis. Drugs 31(Suppl. 3):132–141. [DOI] [PubMed]
- 7.Bluestone C D, Stephenson J S, Martin L M. Ten-year review of otitis media pathogens. Pediatr Infect Dis J. 1992;11:S7–S11. doi: 10.1097/00006454-199208001-00002. [DOI] [PubMed] [Google Scholar]
- 8.Campagnari A A, Shanks K L, Dyer D W. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect Immun. 1994;62:4909–4914. doi: 10.1128/iai.62.11.4909-4914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapman A J, Jr, Musher D M, Jonsson S, Clarridge J E, Wallace R J., Jr Development of bacterial antibody during Branhamella catarrhalis infection. J Infect Dis. 1985;151:878–882. doi: 10.1093/infdis/151.5.878. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, McMichael J, Vandermeid K, Hahn D, Smith R, Eldridge J, Cowell J. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Antibodies to the UspA outer membrane protein of Moraxella catarrhalis block bacterial attachment in vitro and are protective in a murine pulmonary challenge model, abstr. E-53; p. 290. [Google Scholar]
- 11.Chen D, McMichael J C, van der Meid K R, Hahn D, Mininni T, Cowell J, Eldridge J. Evaluation of purified UspA from Moraxella catarrhalis as a vaccine in a murine model after active immunization. Infect Immun. 1996;64:1900–1905. doi: 10.1128/iai.64.6.1900-1905.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.China B, Sory M-P, N’Guyen B T, de Bruyere M, Cornelis G R. Role of YadA protein in prevention of opsonization of Yersinia enterocolitica by C3b molecules. Infect Immun. 1993;61:3129–3136. doi: 10.1128/iai.61.8.3129-3136.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen J J, Renneberg J, Bruun B, Forsgren A. Serum antibody response to proteins of Moraxella (Branhamella) catarrhalis in patients with lower respiratory tract infection. Clin Diagn Lab Immunol. 1995;2:14–17. doi: 10.1128/cdli.2.1.14-17.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies B I, Maesen F P V. The epidemiology of respiratory tract pathogens in Southern Netherlands. Eur Respir J. 1988;1:415–420. [PubMed] [Google Scholar]
- 15.Faden H. Comparison of the local immune response to nontypeable Haemophilus influenzae (nHI) and Moraxella catarrhalis (MC) during otitis media. In: Mestecky J, et al., editors. Advances in mucosal immunology. New York, N.Y: Plenum Press; 1995. pp. 733–736. [PubMed] [Google Scholar]
- 16.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y Tonawanda/Williamsburg Pediatrics. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 17.Faden H, Harabuchi Y, Hong J J Tonawanda/Williamsburg Pediatrics. Epidemiology of Moraxella catarrhalis in children during the first 2 years of life: relationship to otitis media. J Infect Dis. 1994;169:1312–1317. doi: 10.1093/infdis/169.6.1312. [DOI] [PubMed] [Google Scholar]
- 18.Faden H S, Hong J J, Murphy T F. Immune response to outer membrane antigens of Moraxella catarrhalis in children with otitis media. Infect Immun. 1992;60:3824–3829. doi: 10.1128/iai.60.9.3824-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzgerald M, Mulcahy R, Murphy S, Keane C, Coakley D, Scott T. A 200 kDa protein is associated with haemagglutinating isolates of Moraxella (Branhaemella) catarrhalis. FEMS Immunol Med Microbiol. 1997;18:209–216. doi: 10.1111/j.1574-695X.1997.tb01047.x. [DOI] [PubMed] [Google Scholar]
- 20.Goldblatt D, Scadding G K, Lund V J, Wade A M, Turner M W, Pandey J P. Association of Gm allotypes with the antibody response to the outer membrane proteins of a common upper respiratory tract organism, Moraxella catarrhalis. J Immunol. 1994;153:5316–5320. [PubMed] [Google Scholar]
- 21.Goldblatt D, Seymour N D, Levinsky R J, Turner M W. An enzyme-linked immunosorbent assay for the determination of human IgG subclass antibodies directed against Branhamella catarrhalis. J Immunol Methods. 1990;128:219–225. doi: 10.1016/0022-1759(90)90213-f. [DOI] [PubMed] [Google Scholar]
- 22.Goldblatt D, Turner M W, Levinsky R J. Branhamella catarrhalis: antigenic determinants and the development of the IgG subclass response in childhood. J Infect Dis. 1990;162:1128–1135. doi: 10.1093/infdis/162.5.1128. [DOI] [PubMed] [Google Scholar]
- 23.Gulig P A, Patrick C C, Hermanstorfer L, McCracken G H, Jr, Hansen E J. Conservation of epitopes in the oligosaccharide portion of the lipooligosaccharide of Haemophilus influenzae type b. Infect Immun. 1987;55:513–520. doi: 10.1128/iai.55.3.513-520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hager H, Verghese A, Alvarez S, Berk S L. Branhamella catarrhalis respiratory infections. Rev Infect Dis. 1987;9:1140–1149. doi: 10.1093/clinids/9.6.1140. [DOI] [PubMed] [Google Scholar]
- 25.Helminen M E, Beach R, Maciver I, Jarosik G P, Hansen E J, Leinonen M. Human immune response against outer membrane proteins of Moraxella (Branhamella) catarrhalis determined by immunoblotting and enzyme immunoassay. Clin Diagn Lab Immunol. 1995;2:35–39. doi: 10.1128/cdli.2.1.35-39.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Helminen M E, Maciver I, Latimer J L, Cope L D, McCracken G H, Jr, Hansen E J. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect Immun. 1993;61:2003–2010. doi: 10.1128/iai.61.5.2003-2010.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helminen M E, Maciver I, Latimer J L, Klesney-Tait J, Cope L D, Paris M M, McCracken G H, Jr, Hansen E J. A large, antigenically conserved protein on the surface of Moraxella catarrhalis is a target for protective antibodies. J Infect Dis. 1994;170:867–872. doi: 10.1093/infdis/170.4.867. [DOI] [PubMed] [Google Scholar]
- 28.Helminen M E, Maciver I, Latimer J L, Lumbley S R, Cope L D, McCracken G H, Jr, Hansen E J. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival of this organism in vivo. J Infect Dis. 1993;168:1194–1201. doi: 10.1093/infdis/168.5.1194. [DOI] [PubMed] [Google Scholar]
- 29.Hol C, Verduin C M, van Dijke E, Verhoef J, van Dijk H. Complement resistance in Branhamella (Moraxella) catarrhalis. Lancet. 1993;341:1281. doi: 10.1016/0140-6736(93)91185-o. [DOI] [PubMed] [Google Scholar]
- 30.Hol C, Verduin C M, Van Dijke E E A, Verhoef J, Fleer A, van Dijk H. Complement resistance is a virulence factor of Branhamella (Moraxella) catarrhalis. FEMS Immunol Med Microbiol. 1995;11:207–212. doi: 10.1111/j.1574-695X.1995.tb00118.x. [DOI] [PubMed] [Google Scholar]
- 31.Horstmann R D, Sievertsen H J, Knobloch J, Fischetti V A. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–1661. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiao C B, Sethi S, Murphy T F. Outer membrane protein CD of Branhamella catarrhalis—sequence conservation in stains recovered from the human respiratory tract. Microb Pathog. 1995;19:215–225. doi: 10.1016/s0882-4010(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 33.Jordan, K. L., S. H. Berk, and S. L. Berk. 1990. A comparison of serum bactericidal activity and phenotypic characteristics of bacteremic, pneumonia-causing strains, and colonizing strains of Branhamella catarrhalis. Am. J. Med. 88(Suppl. 5A):28S–32S. [DOI] [PubMed]
- 34.Klesney-Tait J, Hiltke T J, Spinola S M, Radolf J D, Hansen E J. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179:1764–1773. doi: 10.1128/jb.179.5.1764-1773.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klingman K L, Murphy T F. Purification and characterization of a high-molecular-weight outer membrane protein of Moraxella (Branhamella) catarrhalis. Infect Immun. 1994;62:1150–1155. doi: 10.1128/iai.62.4.1150-1155.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klingman K L, Pye A, Murphy T F, Hill S L. Dynamics of respiratory tract colonization by Branhamella catarrhalis in bronchiectasis. Am J Respir Crit Care Med. 1995;152:1072–1078. doi: 10.1164/ajrccm.152.3.7663786. [DOI] [PubMed] [Google Scholar]
- 37.Kovatch A L, Wald E R, Michaels R H. Beta-lactamase-producing Branhamella catarrhalis causing otitis media in children. J Pediatr. 1983;102:261–264. doi: 10.1016/s0022-3476(83)80537-x. [DOI] [PubMed] [Google Scholar]
- 38.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 39.Mathers A E, Goldblatt D, Aebi C, Yu R-H, Schryvers A B, Hansen E J. Characterization of an outer membrane protein of Moraxella catarrhalis. FEMS Immunol Med Microbiol. 1997;19:231–236. doi: 10.1111/j.1574-695X.1997.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 40.McLeod D T, Ahmad F, Capewell S, Croughan M J, Calder M A, Seaton A. Increase in bronchopulmonary infection due to Branhamella catarrhalis. Br Med J. 1986;292:1103–1105. doi: 10.1136/bmj.292.6528.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40a.McMichael, J., et al. Submitted for publication.
- 41.Murphy T F. The surface of Branhamella catarrhalis: a systemic approach to the surface antigens of an emerging pathogen. Pediatr Infect Dis J. 1989;8:S75–S77. [PubMed] [Google Scholar]
- 42.Murphy T F, Kirkham C, Lesse A J. The major heat-modifiable outer membrane protein CD is highly conserved among strains of Branhamella catarrhalis. Mol Microbiol. 1993;10:87–97. doi: 10.1111/j.1365-2958.1993.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 43.Murphy T F, Loeb M R. Isolation of the outer membrane of Branhamella catarrhalis. Microb Pathog. 1989;6:159–174. doi: 10.1016/0882-4010(89)90066-1. [DOI] [PubMed] [Google Scholar]
- 44.Nicotra B, Rivera M, Liman J I, Wallace R J. Branhamella catarrhalis as a lower respiratory tract pathogen in patients with chronic lung disease. Arch Intern Med. 1986;146:890–893. [PubMed] [Google Scholar]
- 45.Patrick C C, Kimura A, Jackson M A, Hermanstorfer L, Hood A, McCracken G H, Jr, Hansen E J. Antigenic characterization of the oligosaccharide portion of the lipooligosaccharide of nontypable Haemophilus influenzae. Infect Immun. 1987;55:2902–2911. doi: 10.1128/iai.55.12.2902-2911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pilz D, Vocke T, Heesemann J, Brade V. Mechanism of YadA-mediated serum resistance of Yersinia enterocolitica serotype O3. Infect Immun. 1992;60:189–195. doi: 10.1128/iai.60.1.189-195.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reddy M S, Murphy T F, Faden H S, Bernstein J M. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol Head Neck Surg. 1997;116:175–180. doi: 10.1016/S0194-59989770321-8. [DOI] [PubMed] [Google Scholar]
- 48.Robertson S M, Frisch C F, Gulig P A, Kettman J R, Johnston K H, Hansen E J. Monoclonal antibodies directed against a cell surface-exposed outer membrane protein of Haemophilus influenzae type b. Infect Immun. 1982;36:80–88. doi: 10.1128/iai.36.1.80-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rudel T, Scheurerpflug I, Meyer T F. Neisseria PilC protein identified as type-4 pilus tip-located adhesin. Nature. 1995;373:357–359. doi: 10.1038/373357a0. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sethi S, Surface J M, Murphy T F. Antigenic heterogeneity and molecular analysis of CopB of Moraxella (Branhamella) catarrhalis. Infect Immun. 1997;65:3666–3671. doi: 10.1128/iai.65.9.3666-3671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soto-Hernandez J L, Holtsclaw-Berk S, Harvill L M, Berk S L. Phenotypic characteristics of Branhamella catarrhalis strains. J Clin Microbiol. 1989;27:903–908. doi: 10.1128/jcm.27.5.903-908.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stephens D S, Farley M M. Pathogenic events during infection of the human nasopharynx with Neisseria meningitidis and Haemophilus influenzae. Rev Infect Dis. 1991;13:22–33. doi: 10.1093/clinids/13.1.22. [DOI] [PubMed] [Google Scholar]
- 54.St. Geme J W, III, Cutter D, Barenkamp S J. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J Bacteriol. 1996;178:6281–6287. doi: 10.1128/jb.178.21.6281-6287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.St. Geme J W, III, Falkow S. Haemophilus influenzae adheres to and enters cultured human epithelial cells. Infect Immun. 1990;58:4036–4044. doi: 10.1128/iai.58.12.4036-4044.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verduin C M, Bootsma H J, Hol C, Fleer A, Jansze M, Klingman K L, Murphy T F, van Dijk H. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C: American Society for Microbiology; 1995. Complement resistance in Moraxella (Branhamella) catarrhalis is mediated by a high-molecular-weight outer membrane protein (HMW-OMP), abstr. B137; p. 189. [Google Scholar]
- 57.Verduin C M, Jansze M, Hol C, Mollnes T E, Verhoef J, van Dijk H. Differences in complement activation between complement-resistant and complement-sensitive Moraxella (Branhamella) catarrhalis strains occur at the level of membrane attack complex formation. Infect Immun. 1994;62:589–595. doi: 10.1128/iai.62.2.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y-P, Myers L E, Mcguinness U, Chong P, Kwok Y, Klein M H, Harkness R E. The major outer membrane protein, CD, extracted from Moraxella (Branhamella) catarrhalis is a potential vaccine antigen that induces bactericidal antibodies. FEMS Immunol Med Microbiol. 1997;17:187–199. doi: 10.1111/j.1574-695X.1997.tb01012.x. [DOI] [PubMed] [Google Scholar]