Abstract
Background
Human viruses released into the environment can be detected and characterized in wastewater. The study of wastewater virome offers a consolidated perspective on the circulation of viruses within a population. Because the occurrence and severity of viral infections can vary across a person’s lifetime, studying the virome in wastewater samples contributed by various demographic segments can provide valuable insights into the prevalence of viral infections within these segments. In our study, targeted enrichment sequencing was employed to characterize the human virome in wastewater at a building-level scale. This was accomplished through passive sampling of wastewater in schools, university settings, and nursing homes in two cities in Catalonia. Additionally, sewage from a large urban wastewater treatment plant was analysed to serve as a reference for examining the collective excreted human virome.
Results
The virome obtained from influent wastewater treatment plant samples showcased the combined viral presence from individuals of varying ages, with astroviruses and human bocaviruses being the most prevalent, followed by human adenoviruses, polyomaviruses, and papillomaviruses. Significant variations in the viral profiles were observed among the different types of buildings studied. Mamastrovirus 1 was predominant in school samples, salivirus and human polyomaviruses JC and BK in the university settings while nursing homes showed a more balanced distribution of viral families presenting papillomavirus and picornaviruses and, interestingly, some viruses linked to immunosuppression.
Conclusions
This study shows the utility of building-level wastewater-based epidemiology as an effective tool for monitoring the presence of viruses circulating within specific age groups. It provides valuable insights for public health monitoring and epidemiological studies.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40246-024-00580-1.
Keywords: Human age-related virome, Building-level, Passive samplers, Wastewater-based epidemiology, Targeted enrichment sequencing
Background
Human viruses can be released into the environment through faeces, urine, saliva or desquamation of skin cells, [1] and, as other excreted substances, they can be identified and detected in wastewater. Analysing the sewage virome offers a significant understanding into the viruses that are actively circulating at a given population and has also the potential to elucidate the introduction of emerging viruses and their modes of transmission [2]. From the initial polio surveillance efforts in the early 1930s to the present-day COVID-19 pandemic, wastewater-based epidemiology (WBE) has been widely employed to monitor vaccine-derived viruses and evaluate population immunity in areas where inactivated virus vaccines have been administered [3].
Numerous studies have documented the virome of urban wastewater, revealing the coexistence of viral families associated with persistent infections (Anelloviridae, Parvoviridae, Adenoviridae, Papillomaviridae, Polyomaviridae, Herpesviridae) alongside commonly viral families that contribute more directly to viral disease burdens (including Astroviridae, Picornaviridae, Coronaviridae) [4, 5].
It should be noted that the occurrence and severity of viral infections can vary across different stages of an individual's life. School-age children generally experience milder symptoms compared to other age groups, while the severity of viral infections tends to increase well before reaching old age [6].
To the best of our knowledge, various studies have explored the application of WBE at the building level with a focus on SARS-CoV-2 occurrence [7–14]. However, the building scale has been hardly considered for virome studies. Notably, McCall et al. [15] recently reported distinct viral profiles in wastewater collected from different types of buildings. Obtaining representative wastewater samples at a building-scale is challenging, but passive sampling has recently reemerged as an effective solution to address this issue [13, 16].
Next-generation sequencing (NGS) techniques are evolving as valuable complements to traditional molecular methods for analysing environmental samples. NGS provides insights into viral sequences present in these samples without the need for prior information. In recent years, advancements in sample multiplexing, streamlined library preparation protocols and bioinformatic tools have made NGS a more practical and accessible approach for viral detection [17]. Although viral detection by NGS in complex samples such as wastewater is still challenging, targeted enrichment sequencing (TES) strategies have shown to be a promising tool for viral detection and discovery in wastewater [2, 18]
The objective of this work was to characterize the virome present in wastewater at building-level, specifically in schools, university settings and nursing homes hosting residents of different age’s ranges, social habits and levels of vulnerability. The goal was to assess how different age groups contribute with different viral profiles to wastewater. To achieve this, passive samplers were utilized to collect wastewater samples from schools, university settings and nursing homes in two different cities. These samples were then subjected to virome characterization using a TES approach. Obtained results were compared with the virome profile of a large urban wastewater treatment plant (WWTP), which served as a reference.
Results
Abundance and diversity of viral reads in different types of wastewater samples
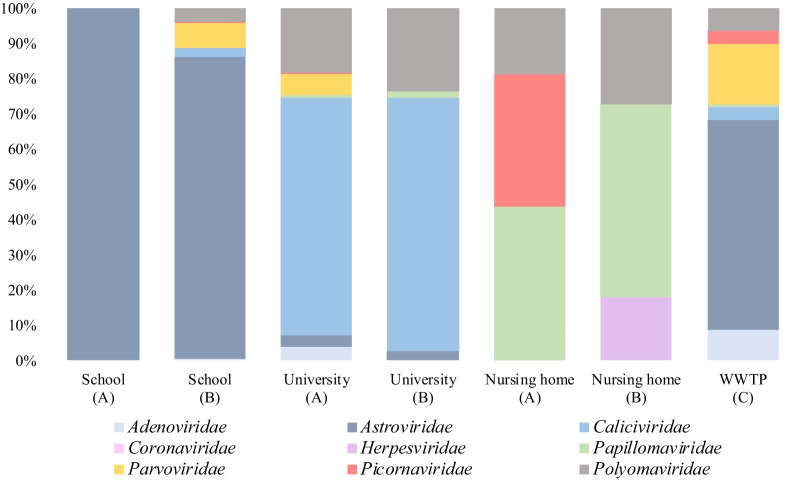
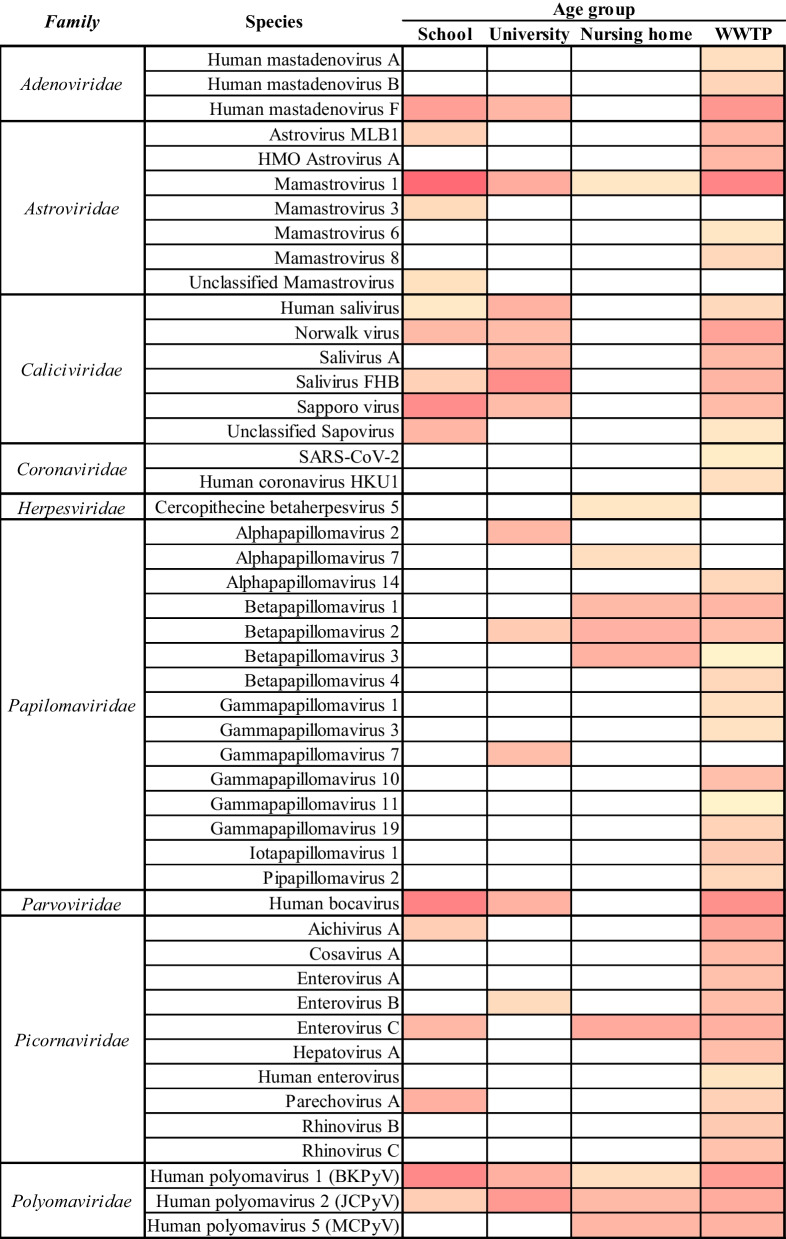
Bioinformatic analysis of the raw data generated from TES applied to the four distinct wastewater types analysed yielded a total of 3,019,032 reads. These reads were taxonomically ascribed to 11 vertebrate viral families, 9 of which included families containing human viruses, encompassing 21 genera. Within the comprehensive analysis of wastewater samples, we identified a total of 48 distinct viral species, and in certain instances, we were able to further classify them into serotypes, serogroups, or genotypes. After manually trimming, a total of 142,643 human viral reads were obtained from the WWTP influent and the wastewater collected from schools, university settings, and nursery homes yielded 1,495,358, 50,520, and 142,643 human viral reads, respectively. Shannon indexes were calculated in the sites presenting a higher number of reads being of 1.77, 1.69, 1.48 and 1.31 in the WWTP, School (B), University (A) and Nursing Home (A), respectively. The relative abundances of different viral families comprising human viruses detected at each sampling site are depicted in Fig. 1, while the number of different viral species identified for each viral family is illustrated in Fig. 2.
Fig. 1.
Relative abundance (% reads matching a viral family/total reads) of the detected viral families at building and WWTP-level representing different age-group wastewater contributors. A–C represents the different cities studied
Fig. 2.
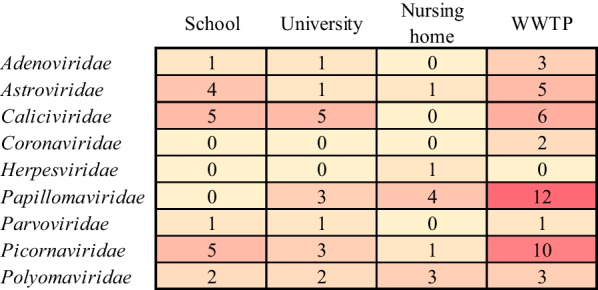
Heatmap profile showing the abundance of viral species detected in building and WWTP-level wastewater. Numbers indicate the quantity of different species that has, at least, one sequence with a positive BLAST hit that passed all the selection criteria. Data spanned from yellow (not detected) to red (high abundance)
Wastewater treatment plant
Urban wastewater encompassed a wide range of viral families and viral species, displaying remarkable diversity (Fig. 3 and Additional file 1: Table S1). The higher number of reads were associated with the family Astroviridae, particularly with Mamastrovirus 1 (AstV-1). Notably, reads matching other Astroviridae species like AstV-6, AstV-8, and HMO-AstV-A were exclusively detected in urban wastewater, distinguishing it from the other wastewater samples analysed from the buildings.
Fig. 3.
Relative viral species abundance (Log10 number of reads) detected at building and WWTP-level
The second-highest number of reads matched Human Bocavirus (Parvoviridae). The third-highest number of reads corresponded to three different subgroups within the Adenoviridae family (AdV). Among these, AdV-40 and -41 (group F) exhibited the highest relative number of classified AdVs reads.
Less abundantly, we also detected sequences matching the persistently excreted viruses JC, BK and MC polyomaviruses (JCPyV, BKPyV and MCPyV) and 12 different species of human papillomaviruses, that together with identified members of the family Picornaviridae displayed the greatest diversity observed in these samples (Fig. 2). The majority of the Picornaviridae reads matched Aichivirus A and Enterovirus-C (EV-C) species. Among the observed Caliciviridae sequences, those corresponding to Norwalk Virus, Salivirus A (Sal-A), Salivirus FHB (Sal-FHB), Human Salivirus (HSalV) and Sapovirus (SapV) were the most prevalent. Finally, reads corresponding to the Coronaviridae family, including SARS-CoV-2 and Human Coronavirus HKU1, were also detected being WWTP the only site where this viral family was detected along this study.
Schools
School wastewater primarily contained viruses associated with gastroenteritis. Among the viral reads detected in school samples, sequences matching members of the Astroviridae family were found to be predominant resulting in over 6 logs of reads. In the site A school, Astroviridae was the sole viral family with only a few reads identified. More in detail, a high number of reads matching AstV-1 were consistently found in schools, especially in samples collected in January. Sequences matching other AstV such as AstV-3 and AstV-MLB1 were also identified.
Caliciviridae and Picornaviridae were the two families presenting the highest number of viral species (Fig. 2). Members of the family Caliciviridae in these buildings included HSalV, Norwalk Virus, Sal-FHB, and Sapporo virus. Picornaviridae species detected were Aichivirus A, Enterovirus C, and Parechovirus A (HPeV-A). AdV-40 and -41 (group F) and Human Bocavirus (HBoV), belonging to the Parvoviridae family, and Human polyomavirus 1 (HPyV-1 or BKPyV) and Human polyomavirus 2 (HPyV-2 or JCPyV) were also observed in school samples.
University settings
Viral signatures identified in wastewater collected at the university settings in both cities showed remarkably similar patterns of viral excretion in their sewer outlets, with 60–70% of viral reads matching members of the family Caliciviridae, which also encompassed the highest diversity of viral species (HSalV, Sal-A, Sal-FHB and SapV). Additionally, reads matching members of the Polyomaviridae, JCPyV and BKPyV (15–20%), and the Astroviridae (AstV-1), (< 10%), were also consistently detected in both sites. Moreover, in site A university setting, there were reads belonging to families Parvoviridae (HBoV) (6%) and Picornaviridae (EV-B) (0,13%). Viral reads assigned to Alphapapillomavirus 2 (α-PV-2), Betapapillomavirus 2 (β-PV-2) and Gammapapillomavirus 7 (γPV-7) were also detected.
Nursing homes
The viromes of the wastewater collected at the nursing homes were less diverse than those obtained from the other wastewater types. In nursing homes from both cities, the relative abundances of the viral families detected ranged between 15 and 30%. Sequences assigned to families Polyomaviridae (JCPyV, BKPyV and MCPyV) and Papillomaviridae (PV-7, β-PV-1, β-PV-2, and β-PV-3) were found in both locations, with Papillomaviridae having the highest diversity of viral species. In site A, viral reads matching the Picornaviridae family were identified (EV-C), while site B also presented few reads matching the Cercopithecine betaherpesvirus 5, also known as human cytomegalovirus (CMV), were unique to this type of setting.
Discussion
In this study, WBE has been conducted at building-scale to study the human virome excreted by different aged populations. This has been achieved by analysing different types of wastewater using torpedo passive samplers previously reported as useful tools for viral detection and characterization in wastewater, especially at sites where composite samples are not always easy to obtain [13, 19]. Moreover, the use of TES allowed the detection of a high number of reads matching viral families that specifically infect humans.
Schools, university settings and nursing homes from two different cities were sampled and analysed for virome determination. The results, in terms of relative viral abundance, demonstrated similar viral excretion patterns for the same types of buildings in two different cities. It should be noted that some differences among buildings from the same type but from different cities arose, as well as variations within the same building sampled at different times. These differences were less pronounced when analysing results from the WWTP at different times (data not shown). Despite being derived from a restricted sample size, these results align with a recent study conducted by McCall and co-workers [15], who described higher variations among viral groups observed when comparing different buildings of the same type, as opposed to the variations observed among WWTPs. This observation appears reasonable when we consider the smaller number of individuals contributing to the wastewater virome at the building level. It also considers the possibility of small outbreaks being limited to very small communities, which are typically observed at the WWTP level, where the excretions of a significant percentage of the population are consolidated.
The wastewater virome obtained from the largest urban WWTP in Catalonia, analysed as the most representative sample of the complete human excreted virome in the studied geographical area, revealed a similar viral profile than reported in previous studies using the same TES approach [2, 19]. Astrovirus were the most prevalent viral family, followed by Human Bocavirus, Adenovirus F and Human Polyomaviruses. Interestingly, Papillomaviridae was the family with the highest number of different viral species. These findings align with the results reported by McCall and co-workers [15] who also reported Astroviridae and Parvoviridae as the predominant viral families detected in WWTP after applying a TES approach. Of the 37 distinct viral species detected in the WWTP, 18 were exclusively found in these WWTP samples and not in the building-scale wastewater. This probably indicates these viruses are excreted at low concentrations or by a small number of people and are detected when analysing samples integrating the shedding of a high number of contributors. On the other side, there were species belonging to the Astroviridae family which were solely identified in school wastewater, while some members of the Papillomaviridae family were exclusively detected in university settings and nursing homes but not in the WWTP. Furthermore, the relative abundances of certain viral species that were present at both building and WWTP-level were different, emphasizing the potential of this type of analysis to provide insights into the viral burden within different demographic groups. Given this finding, the significance of the abundance and observed occurrence trends of the detected viral species in the various samples analysed in this study will be further explored in the subsequent sections.
Wastewater virome associated with persistent infections
Different families of viruses known to persistently infect humans have been found in the different wastewater samples analysed. A discernible trend reveals a higher excretion of these viral families among the elderly, followed by adults, and finally, by children. It is essential to emphasize that persistent infection does not necessarily culminate in clinical manifestations, although viruses that persistently infect humans can sporadically lead to pathological symptomatology.
Human adenoviruses
With more than 110 types described, different organ tropisms and a wide variety of clinical manifestations (revised in Rusiñol and Girones [1]), Human AdV are persistently excreted and make a significant contribution to the viral community present in sewage [2] being proposed as a human viral faecal indicator. The urban wastewater analysed contained reads that matched AdV-F, -A and -B. Among those AdV types identified (Additional file 1: Table S1), AdV-7, -21 and –B3 (Human mastadenovirus B), first isolated from young children, are associated with several respiratory diseases [20], while types A and F have been linked to gastrointestinal infections. Wastewater from the schools and the university settings wastewater only presented AdV-F (Adv-70 and -41)[21].
While both, elderly individuals and young children, are at a heightened risk of AdV infection [22], AdVs reads were not detected in the wastewater from nursing homes. The detection of human AdVs in wastewater have been correlated with the size of the contributing population being sometimes not detected in small size sewer systems [19, 23].
Human polyomaviruses
Human polyomaviruses, with 14 different members, are known to be transmitted among humans during the first years of live through close contact and are excreted by a significant percentage of the population in their urine, typically without recognizable clinical symptoms. Clinical manifestations of these viruses are primarily linked to deep immunosuppression. Among these, Merkel Cell polyomavirus (MCPyV) was discovered to cause Merkel Cell Skin Carcinoma linked to immunosuppression in elderly people and that ageing is a risk factor to develop it.
In this study, sequences matching BK, JC and MCPyV were detected in the WWTP influent. BK and JCPyV were detected in all building samples, whereas MCPyV reads were only observed in the nursing home. An observed seroprevalence increasing with age for JCPyV and decreasing for BKPyV has been previously described [24] and results here obtained are almost in fully agreement with this. The nursing homes studied are contributed to by a small number of people, which would explain the low number of JCPyV reads found. Its presence, along with the absence of human AdV, corroborates previously reported observations describing JCPyV as a faecal viral indicator [25] useful in small sewers [19, 23].
Human papillomaviruses
Within the Papillomaviridae family, β-PV and α-PV infect non-genital mucosa and skin. Their excretion has been linked to epithelial shedding in urine, and the presence of these viruses and their high diversity in the environment could be considered emerging [26, 27]. Members of the genus α-PV primarily infect oral and genital mucosa, as well as external genitals, and are associated with mucosal tumour development in humans [28]. Members of this genus are differentiated by their potential to induce cancer, classified as either low-risk Human Papillomavirus (LR-HPV) or high-risk Human Papillomavirus (HR-HPV), and are known to be causing persistent infections [29]. Their occurrence has been widely documented within the environment [30] reinforcing that investigating these viruses in these matrices is highly valuable for describing their prevalence in specific population groups.
Mucosal HPV infections have traditionally been considered sexually transmitted diseases, although could infect children through horizontal transmission, primarily from mother to child [31]. In this study, HPV reads were observed at WWTPs, university settings and nursing homes but not in school’s wastewater. In fact, this family was the more diverse found in urban wastewater with sequences matching 12 different viral species being HPV-90, a rare emergent LR-HPV, one of the more prevalent types found [32]. Out of the 12 different HPV species detected in urban sewage, 9 were not found at building-level wastewaters. β-PV 1 and 2 species (HPV-22, HPV-100 and HPV-75) and α-PV 7 (HPV-70) were described exclusively in nursing homes wastewater. HPV-70 has been linked to the development and progression of cervical intraepithelial neoplasia and other squamous intraepithelial lesions [33, 34] and to scrotal calcinosis development [34] and showed high prevalence among some unvaccinated male groups [35]. Lastly, α-PV s 7 (HPV-149) and α-PV 2 (HPV-28) have been detected exclusively in university settings, they are known to cause skin warts due to the cutaneous tropism of this species, unlike most α-PV which exhibits mucosal tropism [36, 37].
Herpesviruses
Regarding members of Herpesviridae family, which establish latency and persist for the life of the individual [38], human cytomegalovirus (CMV) is the only member of the family present in one of the nursing homes tested. CMV is the most common opportunistic pathogen in immunocompromised patients [39] but it can also infect immunocompetent individuals causing self-limiting illness resembling mononucleosis [40, 41]. Interestingly, this virus was also detected in a building wastewater by McCall [15].
Wastewater virome associated with acute infections
Additionally, sequences ascribed to viruses belonging to families commonly associated with acute infections have been identified in the different wastewaters analysed. The overall observed trend suggests a higher excretion of these viruses by children and lower excretion by adults and the elderly. It is important to highlight that the presence of these viruses in wastewater indicates viral replication but does not necessarily translate to clinical manifestations, as these viruses can often cause subclinical infections, particularly in children.
Astroviruses
Human Astrovirus (HAstV) is one of the most important causes of viral gastroenteritis worldwide. Although described in faecal samples from adults, they are responsible for the 10% of acute viral gastroenteritis in children [42]. In this study, the only species common to all wastewater types analysed was Mamastrovirus-1, which decreased in reads number in agreement with the age of the contributor individuals (Fig. 3). HAstV sequences were consistently more prevalently detected in January samples (data not shown), aligning with their known seasonal pattern [43]. A deeper analysis of the sequences of Mamastrovirus-1, allowed a further classification into serotypes in some of the pooled samples (Additional file 1). Serotype HAstV-1 was identified in all samples which aligns with reported data indicating its high prevalence within the population [43]. These results agree with a prior study conducted in Spain, specifically targeting children under the age of 5 who suffered from acute gastroenteritis, and that study reported a 11.5% prevalence rate [44]. The abundance of reads corresponding to Astroviridae (AstV), and, in particular to AstV-1, were higher in school samples compared to other studied locations. AstV reads account for more than 80% of reads resulting from school wastewater analysis. This explains why the total numbers of reads from schools was higher than those obtained from the WWTP possibly indicating a higher viral load in the school environment due to elevated AstV excretion. AstV-6, AstV-8 and HMO Astrovirus A were only detected at WWTP-level. HAstV-2 and HAstV-3 reads have also been detected in school and university settings whereas HAstV-4 reads were only found in nursing home wastewater.
Caliciviruses
Norwalk virus is the only species belonging to the Norovirus genus (NoV). NoV infections are the leading cause of gastroenteritis outbreaks [45], are usually self-limiting in healthy patients although in immunocompromised individuals, elderly people and young children can cause severe complications [46, 47]. Norwalk species are divided into genogroups, that are divided into 30 genotypes. In this study, NoV was detected in schools, university settings and at the urban WWTP. A deeper analysis of the sequences obtained allowed a classification into genogroups in some samples (Additional file 1). Despite, genogroups GI, GII and GIV are prevalent human pathogens, only GI and II were found in this study. In particular, the GII.17 genotype, that was identified in the school from city B and at the WWTP, has gained prominence in recent years, overtaking GII.4 as the dominant strain in Asia [48] and has also been described in Europe [49–51].
Sapovirus is an important cause of diarrhoea in children, especially in countries where vaccines for rotavirus are available [52, 53]. They also have been identified as pathogen in adults, especially in immunocompromised individuals or those who live in nursing homes [54–56]. The results obtained showed Sapovirus reads in the wastewater collected from schools and university settings, which is consistent with previous reports indicating that it represents one of the highest burdens of disease in young children [57] as well as its association with sporadic and limited outbreaks [58].
All in all, results obtained suggest children and young adults are the main shedders of members of the Caliciviridae family while these viruses were not detected in nursing home wastewater suggesting no active infections or low excretion levels of these viruses at the time these analyses were conducted.
Coronaviruses
To date, seven human coronaviruses (HCoVs) have been described. In this study, only Human coronaviruses HKU1 (HCOV-HKU1) and SARS-CoV-2 were reported in the urban wastewater collected at the studied WWTP. HCOV-HKU1 has been associated with severe complications in vulnerable populations presenting cold-like symptoms [59] and SARS-CoV-2 with the Covid-19 [60]. These results suggest that if HCoV were excreted at building-level, which is likely, excretion levels were low and under the detection limits of techniques employed. It should be said that SARS-CoV2 was detected in all the buildings by applying qPCR [13] which is a more sensitive technique than NGS [18]. Recently, a specific probe-based capture panel designed for the specific sequencing of CoV has been successfully applied to the detection of a high diversity of human and animal CoV in wastewater (Martinez-Puchol et al., submitted to publication).
Parvoviruses
Human bocavirus (HBoV) is associated with respiratory symptoms and gastroenteritis in children [61] although its role as a causative agent has been not described yet [62]. HBoV is usually attributed to infectious in infants although it can affect children older than 5 years and adults with higher incidences in winter and spring [63, 64]. In this study, HBoV was present in school, university, and WWTP samples with the highest number of reads obtained from school samples in agreement with excretion patterns described for this virus and with previous studies regarding its presence in sewage [65–67]
Picornaviruses
Picornavirus sequences were detected in all the wastewater types analysed although WWTP samples presented the highest diversity of viral species suggesting excretion related to small outbreaks within small communities.
Aichivirus (AiV) is associated with childhood infections [68] responsible of a low proportion of gastroenteritis outbreaks [69], it has been detected in the stool of patients with gastroenteritis among different age groups in Spain [70] and in this study has been found in WWTP samples collected from schools and nursing homes.
The genus Cosavirus include five species of Human Cosavirus (HCoSV-A, B, D, E, F). It was first detected in stool samples of healthy children and children with non-polio acute flaccid paralysis [71, 72] and, later, in children with or without diarrhoea [73–76]. HCoSVs have also been found in sewage and rivers [67, 77, 78]. Despite their presence and diversity in sewage has been described [79], only few clinical studies have been published, showing a very low detection rate if any [80–83] that could explain why sequences matching HCoSV-A were only detected at the WWTP level but not at building-level.
The same happened with Hepatitis A virus (HAV) which usually cause self-limited infections being transmitted by faecal-oral route but also be spread by person-to-person transmission causing large epidemics [84]. The prevalence of HAV correlates with the socioeconomic status of the studied area [85]. In developed countries, where vaccination programs against HAV exist, the prevalence of HAV has decreased [86]. In this study, HAV was only detected at the WWTP-level. In Spain, specifically in Catalonia, vaccination against HAV is systemic and included in the vaccination schedule for children at 15 months old. Although it is not technically mandatory, it is widely practiced. We hypothesize that there was no detection at schools and university settings as children and young people are heavily immunised but people travelling to endemic areas, men having sex with men, prisoners, homeless and other susceptible collectives may contribute to the HAV observed at WWTP-level [87].
Both enterovirus (EV) and Human Parechovirus (HPeV) are picornaviruses causing a common infection in children. HPeV causes mainly respiratory and gastrointestinal symptoms [88] although in children have been associated with central nervous system infections [89]. EV have been associated with wide range of illness: from mild febrile illness, respiratory infection, gastroenteritis, meningitis, encephalitis, feet-hand-mouth disease and other pathologies [90]. However, EV is usually seen in older children and adults while HPeV is not frequent at these ages [91]. This aligns with our results since HPeV reads were only present at school and WWTP while EV reads were observed in all samples studied with differences in the distribution of the three species (A, B and C).
Human rhinoviruses (HRV) are the most prevalent human respiratory viruses and responsible for more than a half of cold-like illnesses each year. In this study, Rhinovirus B and C were only found in the WWTP and was no detected in building’s wastewater probably because the sampling was conducted in winter when influenza and respiratory syncytial viruses are predominant over HRV [92–94].
Salivirus (SaV) was first described from stool samples from children with gastroenteritis [95]. Its relationship with gastroenteritis remains still unclear [96] However, SaV has been detected worldwide in clinical samples and urban sewage [97, 98]. In our study, two species of SaV were identified: SaV-A at university settings and at the WWTP and SaV-FHB in schools, universities, and at the WWTP. These results agree with previous publications describing its association with infection in patients always under 20-year-old and its presence in children’s stool [99, 100]. SaV reads were the more abundant reads at the university settings even when comparing to WWTP, suggesting young adults as the SaV target population.
In summary, it can be hypothesized that the virome in human wastewater comprises a mosaic of viruses originating from a variety of sources. Some viruses may originate from individuals of various age groups, while others might be specific to certain populations. Additionally, our findings indicate that viruses associated with acute infections causing limited outbreaks are more frequently excreted by children than by the older population, and these viruses are typically detectable in WWTPs.
These viruses can also be detected at the building level during an ongoing outbreak at the time of sampling, or when asymptomatic individuals contribute to increased viral load in wastewater. On the other hand, certain viral species that are restricted to specific individuals or demographic groups, such as certain HPV or astrovirus species, may be more readily detectable at the building level and could become diluted to undetectable levels in the influent of larger wastewater treatment systems. Furthermore, the observed virome profiles can be valuable for understanding the epidemiology of these viruses, including their excretion patterns and transmission pathways.
Conclusions
Passive sampling, in the form of torpedo devices, was applied as a straightforward and efficient method for the wastewater virome characterization at a building-scale. TES provided a comprehensive list of viruses present at WWTP and building-level scale.
The virome profiles observed in the two cities studied for the same type of building linked to a specific demographic group were similar suggesting different viral excretion profiles for each sewer system analysed.
WWTP virome, analysed as a reference, presented astrovirus, particularly MAstV-1, as the more abundant viruses followed by HBoV, adenoviruses, polyomaviruses, picornaviruses and caliciviruses.
Astrovirus (particularly AstV-1) predominated in school samples, while salivirus and human polyomaviruses JC and BK dominated in university settings. Nursing homes showed a more balanced distribution of viral families presenting papillomavirus and picornaviruses. Notably, viruses associated with immunosuppression, such as MCPyV and CMV.
A discernible trend emerged: viruses associated with acute infections were more frequently detected in schools but less so in elderly residences, while viruses linked to persistent infections exhibit an inverse trend, being more commonly detected in elderly residences and less frequently in schools.
Overall, the study provides insights into the diversity and distribution of human viruses in different types of wastewaters sources. The results highlight the potential of building-scale WBE as a tool for tracking communal infectious diseases. Such information can be useful in gaining insights into these infections and in designing targeted public health intervention strategies for specific populations.
Methods
Sample collection
While numerous WBE studies predominantly utilize composite samples gathered by automatic samplers at WWTP inlets, the advent of the COVID-19 pandemic has spurred the adoption of flexible passive sampling techniques [14, 16]. These approaches facilitate the monitoring and viral profiling of various waterborne pathogens within compact sewer catchments [16, 19].
Our study focussed on three distinct building types—schools, university settings, and nursery homes—in two cities in Catalonia, Northeast Spain, as outlined in Table 1. School buildings mainly accommodate students aged 3 to 12 but also a diverse range of users, including teachers, administrative staff, parents, guardians, maintenance, and support personnel, as well as participants in various after-school programs. Buildings at the university campus primarily cater to individuals aged 18 to 30, but given the dynamic nature of campuses, these spaces also host researchers, administrators, and other faculty members throughout the day.
Table 1.
Sampling sites and building users description
| Age group | City | Sampling sites | Population represented |
|---|---|---|---|
| 3–12 | A | School sewer outlet | 500 |
| 18–30 | A | University campus sewer outlet | 100 |
| > 65 | A | Nursing home sewer outlet | 300 |
| 3–12 | B | School sewer outlet | 500 |
| 18–30 | B | University campus sewer outlet | 400–1000 |
| > 65 | B | Nursing home sewer outlet | 500 |
| All ages | C | Inlet urban wastewater treatment plant | 1.5 M |
The studied nursing homes, accommodating 300 predominantly elderly residents as full-time inhabitants, represent facilities with comparatively lower user capacity. These residents constitute their primary users.
Additionally, wastewater from a third city (C), with an urban population of about 1.5 million, was obtained from the main WWTP inlet. This wastewater served as a reference for a comprehensive analysis of the excreted virome.
Buildings were sampled two/three times during January and March 2022 except for the nursery home at city A where only samples in March were collected (detailed in Additional file 1). The sampling was conducted using 3D printed torpedo-shaped passive sampling units (kindly donated by Prof. McCarthy) fitted with 2 electronegative membranes (EZ-Pak filters 0.45 µm, Merck Millipore). Torpedoes were deployed for 24 h at the outlet of each building sewer and at the inlet of the WWTP. They were retrieved and transported inside a plastic bag to the laboratory in a portable icebox.
Elution of the viral particles, nucleic acid extraction and DNase treatment
Prior to nucleic acids (NA) extraction, the elution of the viruses adsorbed to the electronegative membranes was performed. Electronegative membranes were carefully introduced, using sterile tweezers, inside Power Bead Tubes (glass 0.1 mm Qiagen), 700 µl of glycine (0.25 N, pH = 8) were added and then bead-beating was applied for 30 s at 4 m/s using FastPrep-24™ (MP Bio, USA). Sample tubes were centrifuged for 1 min at 20.000×g and the resulting 60 µl supernatant was treated with Turbo DNase (Invitrogen, Carlsbad, CA, USA) for 1 h at 37 ºC. The QIAamp Viral RNA mini kit and the Qiacube Automatic system (Qiagen) were used for the NA extraction into a final volume of 50 µl.
Sequence-independent, single-primer amplification (SISPA) and target enrichment sequencing (TES)
Nucleic acid extractions analysed in this study were pooled according to Additional file 2 and amplified before the sequencing library was constructed as previously described [101]. For this reason, viral sequences identified in each sampling site are the result of the viral sequences present in 2–3 different samples collected in January and March and are presented together to provide a more robust composition of a particular virome.
Briefly, NA were retrotranscribed and tagged using SuperScript IV enzyme (Invitrogen), random nonamer primers and Sequenase 2.0 (Applied Biosystems). The cDNA was then amplified to obtain enough cDNA for the next steps by following 25 PCR cycles using AmpliTaq Gold DNA polymerase (Applied Biosystems). The resulting product of the PCR was purified using Zymo DNA Clean & Concentration kit (Zymo research) and quantified using Qubit 2.0 and the Qubit dsDNA HS Assay Kit (Invitrogen).
Libraries were prepared following manufacture’s instruction using the KAPA HyperPlus Library Preparation Kit (KAPA Biosystems, Roche). Starting with 100 ng of amplified cDNA, enzymatic fragmentation, A-tailing and adapter’s ligation were conducted. Each sample was ligated with KAPA UDI Primer mixes (KAPA Biosystems Roche) and a clean-up was performed with the KAPA HyperPure Beads (KAPA Biosystems, Roche). The libraries were amplified with a LM-PCR of 7 cycles, purified and then quantified using Qubit 2.00 and the Qubit dsDNA HS Assay Kit (Invitrogen).
Capture of viral sequences by VirCapSeq-VERT™ capture panel (Roche)
Libraries were pooled to obtain a final quantity of 1 µg. Then it was hybridised for 20 h with probes of the VirCapSeqVERT™ capture panel (Roche) that contains sequences of viruses infecting vertebrates using the HyperCap Target Enrichment Kit (Roche). The captured DNA was recovered using the HyperCap Bead Kit (Roche), amplified with 14 cycles of LM-PCR, purified (HyperPure Beads, Roche) and quantified with Qubit 2.0. Sequencing of the captured libraries was performed on an Illumina NextSeq2000 platform achieving up to 1000 M 150 × 2 pb pair-end reads.
The generated Pair-end FASTQ files were analysed using ID-seq, an open source and cloud-base bioinformatic tool [102]. Briefly, Illumina adapters, duplicates, low quality and complexity reads were cleaned using Trimmomatic [103] and CD-HITDUP tool v4.6.8 (CD-HIT, PRID:SCR 007105) [104] Reads were paired using the Paired-Reads Interactive Contig Extension (PRICE) computational package (PRICE, RRID:SCR 013063) [105] and the Lempel–Ziv–Wech (LZW) compression score. GSNAPL [106] and RAPsearch2 [107] were used for an assembly-based alignment to the NCBI nucleotide (nt) and non-redundant protein (nr) databases [108]. A minimum of 70% identity and > 100 nt length were considered for analysis. However, for human viruses, all species were assigned with alignments to the NCBI above 90% of identity.
Viral reads of Adenoviridae and Papillomaviridae longer than 100 bp were queried for similarity using BLASTN against the NCBI GenBank nucleotide collection database [108]. For specific characterization, Calicivirus and Enterovirus the Typing Tool developed by RIVM were used (version 2.0 and 0.1, respectively) [109].
Statistical and diversity index calculations
The Shannon diversity indexes were calculated using Excel, as measures of the relative abundances of the species present in a sample. The formula to calculate the Sannon índex (H) is:
where S is the total number of species in the community and pi the proportion of reads belonging to the species.
Supplementary Information
Additional file 1. List of Viral species and serotypes, serogroups or genotypes identified in each building type. Mean ID%: average percentage identity of alignments; Mean CV: the per cent of the reference accession that is covered by at least one contig; Coverage depth: indicates the average read depth across the length of the accession. Metagenomic metrics are mean values obtained at the different samples analysed per each building type and at the WWTP.
Additional file 2. Sample dates and pools of the nucleic acid extractions analysed. Each column corresponds to one pool.
Acknowledgements
The authors express their gratitude to the personnel at both city councils, the staff of the sampled buildings and the wastewater treatment plant involved in this study for their assistance and technical support during sampling.
Abbreviations
- Adv
Adenovirus
- AiV
Aichivirus
- α-PV
Alphapapillomavirus
- β-PV
Betapapillomavirus
- BKPyV
BK polyomavirus
- COVID-19
Coronavirus infectious disease
- DNA
Deoxyribonucleic acid
- EV
Enterovirus
- EV-C
Enterovirus-C
- γ-PV
Gammapapillomavirus
- GI
Genogrup 1
- HAV
Hepatitis A virus
- HR-HPV
High-risk Human Papillomavirus
- HAstV
Human Astrovirus
- HMO-AstV-A
Human astrovirus A
- HBoV
Human Bocavirus
- HCoV
Human coronaviruses
- HCoSV
Human Cosavirus
- CMV
Human cytomegalovirus
- HPV
Human Papillomavirus
- HPyV
Human polyomavirus
- HSalV
Human Salivirus
- JCPyV
JC polyomavirus
- LR-HPV
Low-risk Human Papillomavirus
- AstV
Mamastrovirus
- MCPyV
MC polyomavirus
- NGS
Next-generation sequencing
- NoV
Norovirus
- NA
Nucleic acids
- HPV
Human Papillomavius
- HPeV-A
Paraechovirus A
- qPCR
Quantitative Polymerase Chain Reaction
- SaV
Salivirus
- Sal-A
Salivirus A
- Sal-FHB
Salivirus FHB
- SapV
Sapovirus
- SISPA
Sequence-independent, single-primer amplification
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TES
Target Enrichment approach
- WWTP
Wastewater treatment plant
- WBE
Wastewater-based epidemiology
Author contributions
The investigation, data analysis, and the first original draft were conducted by CMM. APT performed the sampling campaign. HT and NC performed the metagenomic analysis. SMP and MI collaborated in the data analysis. LC, CMB, MR and SBM conceptualized this study. Funding acquisition and project administration were conducted by CMB, NC, LC, and SBM. All the authors commented and revised previous versions of the manuscript. All the authors approved the final manuscript.
Funding
This research was conducted as part of the EPISARS project, which received funding from donations obtained during “La Marató de TV3” dedicated to COVID-19. ICRA authors acknowledge the funding provided by the Generalitat de Catalunya through the Consolidated Research Group grants ICRA-ENV 2017 SGR 1124 and ICRA-TiA 2017 SGR 1318 and the funding from the CERCA program of the Catalan Government.
Availability of data and materials
The datasets generated during the current study are available in zenodo under the DOI numbers: 10.5281/zenodo.8178269, 10.5281/zenodo.10077991, 10.5281/zenodo.10078619, 10.5281/zenodo.10149180, 10.5281/zenodo.10158645.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rusiñol M, Girones R. Summary of excreted and waterborne viruses. Water and Sanitation for the 21st Century: health and microbiological aspects of excreta and wastewater management (Global Water Pathogen Project). Michigan State University; 2019.
- 2.Martínez-Puchol S, Rusiñol M, Fernández-Cassi X, Timoneda N, Itarte M, Andrés C, et al. Characterisation of the sewage virome: comparison of NGS tools and occurrence of significant pathogens. Sci Total Environ. 2020;713. [DOI] [PubMed]
- 3.World Health Organization. World Health Organization WHO Guidelines for environmental surveillance of poliovirus circulation Vaccines and Biologicals. 2003. Available from: www.who.int/vaccines-documents/
- 4.McCall C, Wu H, Miyani B, Xagoraraki I. Identification of multiple potential viral diseases in a large urban center using wastewater surveillance. Water Res. 2020;184. [DOI] [PMC free article] [PubMed]
- 5.Schaeffer J, Desdouits M, Besnard A, Le Guyader FS. Looking into sewage: how far can metagenomics help to detect human enteric viruses? Front Microbiol. 2023;14. [DOI] [PMC free article] [PubMed]
- 6.Glynn JR, Moss PAH. Systematic analysis of infectious disease outcomes by age shows lowest severity in school-age children. Sci Data. 2020;7. [DOI] [PMC free article] [PubMed]
- 7.Acer PT, Kelly LM, Lover AA, Butler CS. Quantifying the relationship between SARS-CoV-2 wastewater concentrations and building-level COVID-19 prevalence at an isolation residence: a passive sampling approach. Int J Environ Res Public Health. 2022;19. [DOI] [PMC free article] [PubMed]
- 8.Liu P, Ibaraki M, VanTassell J, Geith K, Cavallo M, Kann R, et al. A sensitive, simple, and low-cost method for COVID-19 wastewater surveillance at an institutional level. Sci Total Environ. 2022;807. [DOI] [PMC free article] [PubMed]
- 9.Corchis-Scott R, Geng Q, Seth R, Ray R, Beg M, Biswas N, et al. Averting an outbreak of SARS-CoV-2 in a University Residence Hall through Wastewater Surveillance. Microbiol Spectr. 2021;9. [DOI] [PMC free article] [PubMed]
- 10.Jain N, Hamilton D, Mital S, Ilias A, Brinkmann M, McPhedran K. Long-term passive wastewater surveillance of SARS-CoV-2 for seven university dormitories in comparison to municipal surveillance. Sci Total Environ. 2022;852. [DOI] [PMC free article] [PubMed]
- 11.Wang Y, Liu P, Zhang H, Ibaraki M, VanTassell J, Geith K, et al. Early warning of a COVID-19 surge on a university campus based on wastewater surveillance for SARS-CoV-2 at residence halls. Sci Total Environ. 2022;821. [DOI] [PMC free article] [PubMed]
- 12.Mangwana N, Archer E, Muller CJF, Preiser W, Wolfaardt G, Kasprzyk-Hordern B, et al. Sewage surveillance of SARS-CoV-2 at student campus residences in the Western Cape, South Africa. Sci Total Environ. 2022;851. [DOI] [PMC free article] [PubMed]
- 13.Pico-Tomàs A, Mejías-Molina C, Zammit I, Rusiñol M, Bofill-Mas S, Borrego CM, et al. Surveillance of SARS-CoV-2 in sewage from buildings housing residents with different vulnerability levels. Sci Total Environ. 2023;872. [DOI] [PMC free article] [PubMed]
- 14.Bivins A, Kaya D, Ahmed W, Brown J, Butler C, Greaves J, et al. Passive sampling to scale wastewater surveillance of infectious disease: lessons learned from COVID-19. Sci Total Environ. 2022;835:155347. 10.1016/j.scitotenv.2022.155347 [DOI] [PMC free article] [PubMed]
- 15.McCall C, Leo Elworth RA, Wylie KM, Wylie TN, Dyson K, Doughty R, et al. Targeted metagenomic sequencing for detection of vertebrate viruses in wastewater for public health surveillance. ACS ES and T Water. 2023.
- 16.Schang C, Crosbie ND, Nolan M, Poon R, Wang M, Jex A, et al. Passive sampling of SARS-CoV-2 for wastewater surveillance. Environ Sci Technol. 2021;55:10432–10441. doi: 10.1021/acs.est.1c01530. [DOI] [PubMed] [Google Scholar]
- 17.Huang B, Jennsion A, Whiley D, McMahon J, Hewitson G, Graham R, et al. Illumina sequencing of clinical samples for virus detection in a public health laboratory. Sci Rep. 2019;9. [DOI] [PMC free article] [PubMed]
- 18.Martínez-Puchol S, Itarte M, Rusiñol M, Forés E, Mejías-Molina C, Andrés C, et al. Exploring the diversity of coronavirus in sewage during COVID-19 pandemic: Don’t miss the forest for the trees. Sci Total Environ. 2021;800. [DOI] [PMC free article] [PubMed]
- 19.Mejías-Molina C, Pico-Tomàs A, Beltran-Rubinat A, Martínez-Puchol S, Corominas L, Rusiñol M, et al. Effectiveness of passive sampling for the detection and genetic characterization of human viruses in wastewater. Environ Sci. (Cambridge). 2023.
- 20.Chuang Y-Y, Chiu C-H, Wong KS, Huang J-G, Huang Y-C, Chang L-Y, et al. Severe adenovirus infection in children. J Microbiol Immunol. 2018;3740. [PubMed]
- 21.Lee B, Damon CF, Platts-Mills JA. Pediatric acute gastroenteritis associated with adenovirus 40/41 in low-income and middle-income countries. Curr Opin Infect Dis. Lippincott Williams and Wilkins; 2020. p. 398–403. [DOI] [PMC free article] [PubMed]
- 22.Saint-Pierre Contreras G, Conei Valencia D, Lizama L, Vargas Zuñiga D, Avendaño Carvajal LF, Ampuero Llanos S. An old acquaintance: could adenoviruses be our next pandemic threat? Viruses. MDPI; 2023. [DOI] [PMC free article] [PubMed]
- 23.Mayer RE, Bofill-Mas S, Egle L, Reischer GH, Schade M, Fernandez-Cassi X, et al. Occurrence of human-associated Bacteroidetes genetic source tracking markers in raw and treated wastewater of municipal and domestic origin and comparison to standard and alternative indicators of faecal pollution. Water Res. 2016;90:265–276. doi: 10.1016/j.watres.2015.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamminga S, Van Der Meijden E, Feltkamp MCW, Zaaijer HL. Seroprevalence of fourteen human polyomaviruses determined in blood donors. PLoS One. 2018;13. [DOI] [PMC free article] [PubMed]
- 25.Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66:238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.la Rosa G, Fratini M, Accardi L, D’Oro G, Della Libera S, Muscillo M, et al. Mucosal and cutaneous human papillomaviruses detected in raw sewages. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed]
- 27.Itarte M, Martínez-Puchol S, Forés E, Hundesa A, Timoneda N, Bofill-Mas S, et al. Ngs techniques reveal a high diversity of rna viral pathogens and papillomaviruses in fresh produce and irrigation water. Foods. 2021;10. [DOI] [PMC free article] [PubMed]
- 28.Zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nat Rev Cancer. European Association for Cardio-Thoracic Surgery; 2002. p. 342–50. [DOI] [PubMed]
- 29.Della Fera AN, Warburton A, Coursey TL, Khurana S, McBride AA. Persistent human papillomavirus infection. Viruses. MDPI AG; 2021. [DOI] [PMC free article] [PubMed]
- 30.Iaconelli M, Petricca S, Libera S Della, Di Bonito P, La Rosa G. First detection of human papillomaviruses and human polyomaviruses in River Waters in Italy. Food Environ Virol. 2015;7:309–15. [DOI] [PubMed]
- 31.Syripinen S, Puronen M. Human papillomavirus infections in children: the potential role of maternal transmission. [DOI] [PubMed]
- 32.Quiroga-Garza G, Zhou H, Mody DR, Schwartz MR, Ge Y. Unexpected high prevalence of HPV 90 infection in an underserved population. Arch Pathol Lab Med. 2013. p. 1569–73. [DOI] [PubMed]
- 33.De Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004. p. 17–27. [DOI] [PubMed]
- 34.Lu CW, Hong HS, Lu CF, Ko YS. Human papillomavirus-70 infection as a possible pathogenesis of eruptive scrotal calcinosis. Dermatol Sin. 2017;35:152–154. [Google Scholar]
- 35.O’Leary MC, Sinka K, Robertson C, Cuschieri K, Lyman R, Lacey M, et al. HPV type-specific prevalence using a urine assay in unvaccinated male and female 11- to 18-year olds in Scotland. Br J Cancer. 2011;104:1221–1226. doi: 10.1038/bjc.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovanda A, Kocjan BJ, Potočnik M, Poljak M. Characterization of a novel cutaneous human papillomavirus genotype HPV-125. PLoS One. 2011;6. [DOI] [PMC free article] [PubMed]
- 37.Favre M, Obalek S, Jablonska S, ORTHl G. Cutaneous warts: clinical, histologic, and virologic correlations. J Virol. 1989. Available from: https://journals.asm.org/journal/jvi [DOI] [PubMed]
- 38.Weidner-Glunde M, Kruminis-Kaszkiel E, Savanagouder M. Herpesviral latency—common themes. Pathogens. 2020;9. [DOI] [PMC free article] [PubMed]
- 39.Fulkerson HL, Nogalski MT, Collins-McMillen D, Yurochko AD. Overview of human cytomegalovirus pathogenesis. Methods in molecular biology. Humana Press Inc.; 2021. p. 1–18. [DOI] [PubMed]
- 40.Bravender T. Epstein-Barr virus, cytomegalovirus, and infectious mononucleosis. Adolesc Med State Art Rev. 2010;21:251–264. [PubMed] [Google Scholar]
- 41.Klemola E, von Essen R, Henle G, Henle W. Article navigation article navigation JOURNAL ARTICLE Infectious-mononucleosis-like Disease with Negative Heterophil Agglutination Test. Clinical Features in Relation to Epstein-Barr Virus and Cytomegalovirus Antibodies. J Infect Dis. 1970;121:608–14. [DOI] [PubMed]
- 42.Lyman WH, Walsh JF, Kotch JB, Weber DJ, Gunn E, Vinjé J. Prospective study of etiologic agents of acute gastroenteritis outbreaks in child care centers. J Pediatr. 2009;154:253–257. doi: 10.1016/j.jpeds.2008.07.057. [DOI] [PubMed] [Google Scholar]
- 43.Guix S, Caballero S, Villena C, Bartolomé R, Latorre C, Rabella N, et al. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J Clin Microbiol. 2002;40:133–139. doi: 10.1128/JCM.40.1.133-139.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vu D-L, Sabrià A, Aregall N, Michl K, Rodríguez Garrido V, Goterris L, et al. Novel Human astroviruses: prevalence and association with common enteric viruses in undiagnosed gastroenteritis cases in Spain. Viruses. 2019;11. [DOI] [PMC free article] [PubMed]
- 45.Atmar RL, Estes MK. The epidemiologic and clinical importance of norovirus infection. Gastroenterol Clin North Am. 2006; p. 275–90. [DOI] [PubMed]
- 46.Harris JP, Edmunds WJ, Pebody R, Brown DW, Lopman BA. Deaths from norovirus among the elderly, England and Wales. Emerg Infect Dis. 2008;14:1546–1552. doi: 10.3201/eid1410.080188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trivedi TK, Desai R, Hall AJ, Patel M, Parashar UD, Lopman BA. Clinical characteristics of norovirus-associated deaths: a systematic literature review. Am J Infect Control. 2013 [cited 2023 Jul 18];41:654–7. [DOI] [PubMed]
- 48.De Graaf M, Van Beek J, Vennema H, Podkolzin AT, Hewitt J, Bucardo F, et al. Emergence of a novel GII.17 norovirus-End of the GII.4 era?. 2005. Available from: http://www.cdc.gov/norovirus/reporting/calicinet/index.html [DOI] [PMC free article] [PubMed]
- 49.Dinu S, Nagy M, Negru DG, Popovici ED, Zota L, Oprişan G. Molecular identification of emergent GII.P17-GII.17 norovirus genotype, Romania, 2015. Eurosurveillance. 2016;21. [DOI] [PubMed]
- 50.Giammanco GM, De Grazia S, Bonura F, Cappa V, Li Muli S, Pepe A, et al. Norovirus GII.17 as major epidemic strain in Italy, winter 2015–16. Emerg Infect Dis. 2017;23:1206–8. [DOI] [PMC free article] [PubMed]
- 51.Leblanc JJ, Pettipas J, Gaston D, Taylor R, Hatchette TF, Booth TF, et al. Outbreak of norovirus GII.P17-GII.17 in the Canadian Province of Nova Scotia. Can J Infect Dis Med Microbiol. 2016;2016. [DOI] [PMC free article] [PubMed]
- 52.Platts-Mills JA, Liu J, Rogawski ET, Kabir F, Lertsethtakarn P, Siguas M, et al. Use of quantitative molecular diagnostic methods to assess the aetiology, burden, and clinical characteristics of diarrhoea in children in low-resource settings: a reanalysis of the MAL-ED cohort study. Lancet Glob Health. 2018;6:e1309–e1318. doi: 10.1016/S2214-109X(18)30349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halasa N, Piya B, Stewart LS, Rahman H, Payne DC, Woron A, et al. The changing landscape of pediatric viral enteropathogens in the post-rotavirus vaccine era. Clin Infect Dis. 2021;72:576–585. doi: 10.1093/cid/ciaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inns T, Wilson D, Manley P, Harris JP, O’Brien SJ, Vivancos R. What proportion of care home outbreaks are caused by norovirus? An analysis of viral causes of gastroenteritis outbreaks in care homes, North East England, 2016–2018. BMC Infect Dis. 2019;20. [DOI] [PMC free article] [PubMed]
- 55.Pietsch C, Liebert UG. Intrahost viral evolution during chronic sapovirus infections. J Clin Virol. 2019;113:1–7. doi: 10.1016/j.jcv.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 56.Daniel-Wayman S, Fahle G, Palmore T, Green KY, Prevots DR. Norovirus, astrovirus, and sapovirus among immunocompromised patients at a tertiary care research hospital. Diagn Microbiol Infect Dis. 2018;92:143–146. doi: 10.1016/j.diagmicrobio.2018.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Becker-Dreps S, Bucardo F, Vinjé J. Sapovirus: an important cause of acute gastroenteritis in children. Lancet Child Adolesc Health. 2019;3:758–759. doi: 10.1016/S2352-4642(19)30270-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker-Dreps S, Gonzalez F, Bucardo F. Sapovirus: an emerging cause of childhood diarrhea. Curr Opin Infect Dis. 2020;33:388–397. doi: 10.1097/QCO.0000000000000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vabret A, Dina J, Phanie Gouarin S, Lle Petitjean J, Corbet S, Ois Freymuth F. Detection of the new human coronavirus HKU1: a report of 6 cases. 2006. [DOI] [PMC free article] [PubMed]
- 60.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, rn Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. 2005. Available from: www.pnas.orgcgi. doi:10.1073pnas.0504666102 [DOI] [PMC free article] [PubMed]
- 62.Guido M, Tumolo MR, Verri T, Romano A, Serio F, De Giorgi M, et al. Human bocavirus: Current knowledge and future challenges. World J Gastroenterol. Baishideng Publishing Group Co; 2016. p. 8684–97. [DOI] [PMC free article] [PubMed]
- 63.Cheng WX, Jin Y, Duan ZJ, Xu ZQ, Qi HM, Zhang Q, et al. Human bocavirus in children hospitalized for acute gastroenteritis: a case-control study. Clin Infect Dis. 2008;47:161–167. doi: 10.1086/589244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bagasi AA, Howson-Wells HC, Clark G, Tarr AW, Soo S, Irving WL, et al. Human Bocavirus infection and respiratory tract disease identified in UK patient cohort. J Clin Virol. 2020;129. [DOI] [PMC free article] [PubMed]
- 65.Salvo M, Lizasoain A, Castells M, Bortagaray V, Castro S, Colina R, et al. Human bocavirus: detection, quantification and molecular characterization in sewage and surface waters in Uruguay. Food Environ Virol. 2018;10:193–200. doi: 10.1007/s12560-017-9334-0. [DOI] [PubMed] [Google Scholar]
- 66.Thongprachum A, Fujimoto T, Takanashi S, Saito H, Okitsu S, Shimizu H, et al. Detection of nineteen enteric viruses in raw sewage in Japan. Infect Genet Evol. 2018;63:17–23. doi: 10.1016/j.meegid.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 67.Blinkova O, Rosario K, Li L, Kapoor A, Slikas B, Bernardin F, et al. Frequent detection of highly diverse variants of cardiovirus, cosavirus, bocavirus, and circovirus in sewage samples collected in the United States. J Clin Microbiol. 2009;47:3507–3513. doi: 10.1128/JCM.01062-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oh DY, Silva PA, Hauroeder B, Diedrich S, Cardoso DDP, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 69.Rivadulla E, Romalde JL. A comprehensive review on human aichi virus. Virol Sin. 2020;35:501–516. doi: 10.1007/s12250-020-00222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rivadulla E, Varela MF, Romalde JL. Epidemiology of Aichi virus in fecal samples from outpatients with acute gastroenteritis in Northwestern Spain. J Clin Virol. 2019;118:14–19. doi: 10.1016/j.jcv.2019.07.011. [DOI] [PubMed] [Google Scholar]
- 71.Kapoor A, Victoria J, Simmonds P, Slikas E, Chieochansin T, Naeem A, et al. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci USA. 2008;105:20482–20487. doi: 10.1073/pnas.0807979105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Victoria JG, Kapoor A, Li L, Blinkova O, Slikas B, Wang C, et al. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol. 2009;83:4642–4651. doi: 10.1128/JVI.02301-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bonanno Ferraro G, Mancini P, Divizia M, Suffredini E, Della Libera S, Iaconelli M, et al. Occurrence and genetic diversity of human cosavirus in sewage in Italy. Food Environ Virol. 2018;10:386–390. doi: 10.1007/s12560-018-9356-2. [DOI] [PubMed] [Google Scholar]
- 74.Nikonov OS, Chernykh ES, Garber MB, Nikonova EY. Enteroviruses: classification, diseases they cause, and approaches to development of antiviral drugs. Biochemistry (Moscow). Maik Nauka Publishing / Springer SBM; 2017. p. 1615–31. [DOI] [PMC free article] [PubMed]
- 75.Lobo PS, Cardoso JF, Barata RR, Lemos PS, Guerra SFS, Soares LS, et al. Near-complete genome of cosavirus A from a child hospitalized with acute gastroenteritis, Brazil. Infect Genet Evol. 2020;85. [DOI] [PubMed]
- 76.da Costa AC, Luchs A, Milagres FA de P, Komninakis SV, Gill DE, Lobato MCABS, et al. Near full length genome of a recombinant (E/D) cosavirus strain from a rural area in the central region of Brazil. Sci Rep. 2018;8. [DOI] [PMC free article] [PubMed]
- 77.Haramoto E, Otagiri M. Occurrence of human Cosavirus in wastewater and river water in Japan. Food Environ Virol. 2014 [cited 2023 Jul 12];6:62–6. Available from: https://pubmed.ncbi.nlm.nih.gov/23943064/ [DOI] [PubMed]
- 78.Guerrero-Latorre L, Romero B, Bonifaz E, Timoneda N, Rusiñol M, Girones R, et al. Quito’s virome: metagenomic analysis of viral diversity in urban streams of Ecuador’s capital city. Sci Total Environ. 2018;645:1334–1343. doi: 10.1016/j.scitotenv.2018.07.213. [DOI] [PubMed] [Google Scholar]
- 79.Bonanno Ferraro G, Mancini P, Divizia M, Suffredini E, Della Libera S, Iaconelli M, et al. Occurrence and genetic diversity of human Cosavirus in Sewage in Italy. Food Environ Virol. 2018;10:386–90. 10.1007/s12560-018-9356-2 [DOI] [PubMed]
- 80.Campanini G, Rovida F, Meloni F, Cascina A, Ciccocioppo R, Piralla A, et al. Persistent human cosavirus infection in lung transplant recipient, Italy. Emerg Infect Dis. 2013;19:1667–1669. doi: 10.3201/eid1910.130352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rovida F, Campanini G, Piralla A, Adzasehoun KMG, Sarasini A, Baldanti F. Molecular detection of gastrointestinal viral infections in hospitalized patients. Diagn Microbiol Infect Dis. 2013;77:231–235. doi: 10.1016/j.diagmicrobio.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daprà V, Galliano I, Montanari P, Zaniol E, Calvi C, Alliaudi C, et al. Bufavirus, cosavirus, and salivirus in diarrheal Italian infants. Intervirology. 2021;64:165–168. doi: 10.1159/000514384. [DOI] [PubMed] [Google Scholar]
- 83.Nielsen ACY, Gyhrs ML, Nielsen LP, Pedersen C, Böttiger B. Gastroenteritis and the novel picornaviruses aichi virus, cosavirus, saffold virus, and salivirus in young children. J Clin Virol. 2013;57:239–242. doi: 10.1016/j.jcv.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 84.Martin A, Lemon SL. Hepatitis A virus: from discovery to vaccines. Hepatology. 2006;43:164–162. doi: 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- 85.Chung GE, Yim JY, Kim D, Lim SH, Park MJ, Kim YS, et al. Seroprevalence of hepatitis A and associated socioeconomic factors in young healthy Korean adults. Gut Liver. 2011;5:88–92. doi: 10.5009/gnl.2011.5.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Franco E, Meleleo C, Serino L, Sorbara D, Zaratti L. Hepatitis A: epidemiology and prevention in developing countries. World J Hepatol. 2012;4:68–73. doi: 10.4254/wjh.v4.i3.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzog C, Van Herck K, Van Damme P. Hepatitis A vaccination and its immunological and epidemiological long-term effects–a review of the evidence. Hum Vaccin Immunother. 2021;17:1496–1519. doi: 10.1080/21645515.2020.1819742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen H, Yao Y, Liu X, Xiao N, Xiao Y, Huang Y, et al. Molecular detection of human parechovirus in children with acute gastroenteritis in Guangzhou, China. Arch Virol. 2014 [cited 2023 Jul 10];159:971–7. 10.1007/s00705-013-1915-0 [DOI] [PubMed]
- 89.Felsenstein S, Yang S, Eubanks N, Sobrera E, Grimm JP, Aldrovandi G. Human parechovirus central nervous system infections in Southern California Children. Pediatr Infect Dis J. 2014 [cited 2023 Jul 10];33. Available from: https://journals.lww.com/pidj/Fulltext/2014/04000/Human_Parechovirus_Central_Nervous_System.4.aspx [DOI] [PubMed]
- 90.Lo C-W, Wu K-G, Lin M-C, Chen C-J, Min-The Ho D, Tang R-B, et al. Application of a molecular method for the classification of human enteroviruses and its correlation with clinical manifestations molecular classification of human enteroviruses. J Microbiol Immunol Infect. 2010 [cited 2023 Jul 10];43:354–9. Available from: http://www.e-jmii.comhttp://www.e-jmii.com [DOI] [PubMed]
- 91.Esposito S, Rahamat-Langendoen J, Ascolese B, Senatore L, Castellazzi L, Niesters HGM. Pediatric parechovirus infections. J Clin Virol. Elsevier; 2014. p. 84–9. [DOI] [PubMed]
- 92.Gwaltney JM Jr, Hendley JO, Simon G, Jordan WS Jr. Rhinovirus infections in an industrial population. 2009 [cited 2023 Jul 10];275:1261–8. Available from: https://www.nejm.org/doi/10.1056/NEJM196612082752301
- 93.Winther B, Hayden FG, Hendley JO. Picornavirus infections in children diagnosed by RT-PCR during longitudinal surveillance with weekly sampling: association with symptomatic illness and effect of season. J Med Virol. 2006 [cited 2023 Jul 10];78:644–50. 10.1002/jmv.20588 [DOI] [PubMed]
- 94.Miller EK, Lu X, Erdman DD, Poehling KA, Zhu Y, Griffin MR, et al. Rhinovirus-associated hospitalizations in young children. JID. 2007 [cited 2023 Jul 10];773. Available from: https://academic.oup.com/jid/article/195/6/773/876889 [DOI] [PMC free article] [PubMed]
- 95.Greninger AL, Runcker C, Chiu CY, Haggerty T, Parsonnet J, Ganem D, et al. The complete genome of klassevirus: a novel picornavirus in pediatric stool. Virol J. 2009;6. [DOI] [PMC free article] [PubMed]
- 96.Mancini P, Bonanno Ferraro G, Suffredini E, Veneri C, Iaconelli M, Vicenza T, et al. Molecular detection of human salivirus in italy through monitoring of urban sewages. Food Environ Virol. 2020;12:68–74. doi: 10.1007/s12560-019-09409-w. [DOI] [PubMed] [Google Scholar]
- 97.Reuter G, Pankovics P, Boros Á. Saliviruses—the first knowledge about a newly discovered human picornavirus. Rev Med Virol.; 2017. [DOI] [PubMed]
- 98.Mancini P, Bonanno Ferraro G, Suffredini E, Veneri C, Iaconelli M, Vicenza T, et al. Molecular detection of human Salivirus in Italy through monitoring of urban sewages. Food Environ Virol. 2020;12:68–74. 10.1007/s12560-019-09409-w [DOI] [PubMed]
- 99.Yu JM, Ao YY, Liu N, Li LL, Duan ZJ. Salivirus in children and its association with childhood acute gastroenteritis: a paired case-control study. PLoS One. 2015;10. [DOI] [PMC free article] [PubMed]
- 100.Ng TFF, Magaña L, Montmayeur A, Lopez MR, Gregoricus N, Oberste MS, et al. Characterization of a salivirus (Picornaviridae) from a diarrheal child in Guatemala. Genome Announc. 2016;4. [DOI] [PMC free article] [PubMed]
- 101.Fernandez-Cassi X, Rusiñol M, Martínez-Puchol S. Viral concentration and amplification from human serum samples prior to application of next-generation sequencing analysis. Methods Mol Biol. 2018;1838:173–188. doi: 10.1007/978-1-4939-8682-8_13. [DOI] [PubMed] [Google Scholar]
- 102.IDseq Portal. [cited 2022 Sep 20]. Available from: https://czid.org/
- 103.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 105.Ruby JG, Bellare P, DeRisi JL. PRICE: software for the targeted assembly of components of (Meta) genomic sequence data. G3: Genes, Genomes, Genetics. 2013;3:865–80. [DOI] [PMC free article] [PubMed]
- 106.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ye Y, Choi JH, Tang H. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinform. 2011;12. [DOI] [PMC free article] [PubMed]
- 108.Home - Nucleotide - NCBI [cited 2022 Sep 20]. Available from: https://www.ncbi.nlm.nih.gov/nucleotide/
- 109.Kroneman A, Vennema H, Deforche K, Avoort H, Peñaranda S, Oberste MS, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. List of Viral species and serotypes, serogroups or genotypes identified in each building type. Mean ID%: average percentage identity of alignments; Mean CV: the per cent of the reference accession that is covered by at least one contig; Coverage depth: indicates the average read depth across the length of the accession. Metagenomic metrics are mean values obtained at the different samples analysed per each building type and at the WWTP.
Additional file 2. Sample dates and pools of the nucleic acid extractions analysed. Each column corresponds to one pool.
Data Availability Statement
The datasets generated during the current study are available in zenodo under the DOI numbers: 10.5281/zenodo.8178269, 10.5281/zenodo.10077991, 10.5281/zenodo.10078619, 10.5281/zenodo.10149180, 10.5281/zenodo.10158645.