Abstract
The Halopseudomonas species, formerly classified as Pseudomonas pertucinogena lineage, form a unique phylogenetic branch within the Pseudomonads. Most strains have recently been isolated from challenging habitats including oil‐ or metal‐polluted sites, deep sea, and intertidal zones, suggesting innate resilience to physical and chemical stresses. Despite their comparably small genomes, these bacteria synthesise several biomolecules with biotechnological potential and a role in the degradation of anthropogenic pollutants has been suggested for some Halopseudomonads. Until now, these bacteria are not readily amenable to existing cultivation and cloning methods. We addressed these limitations by selecting four Halopseudomonas strains of particular interest, namely H. aestusnigri, H. bauzanensis, H. litoralis, and H. oceani to establish microbiological and molecular genetic methods. We found that C4‐C10 dicarboxylic acids serve as viable carbon sources in both complex and mineral salt cultivation media. We also developed plasmid DNA transfer protocols and assessed vectors with different origins of replication and promoters inducible with isopropyl‐β‐d‐thiogalactopyranoside, l‐arabinose, and salicylate. Furthermore, we have demonstrated the simultaneous genomic integration of expression cassettes into one and two attTn7 integration sites. Our results provide a valuable toolbox for constructing robust chassis strains and highlight the biotechnological potential of Halopseudomonas strains.
This study aimed to access four different Halopseudomonas species isolated from challenging habitats. Therefore, we developed cultivation strategies on the one hand, and provided, on the other hand, a valuable toolbox for constructing robust chassis strains.
INTRODUCTION
Microbes play an important role as production platforms in various sectors of the biotechnological and chemical industries. Their use under industrial process conditions requires tolerance to different stressors, such as toxic reaction intermediates or products, osmotic stress, or pH shifts (Bitzenhofer et al., 2021). Several bacteria are well established as biotechnological workhorses, including Escherichia coli, Pseudomonas putida, Pseudomonas taiwanensis, Corynebacterium glutamicum, and Bacillus subtilis (Blombach et al., 2022; Gießelmann et al., 2019; Loeschcke & Thies, 2015; Reva et al., 2006; Rodrigues et al., 2014; Tabor et al., 2011; van Dijl & Hecker, 2013). These bacteria were optimised towards stress tolerance, e.g. by engineering metabolic fluxes towards bioproduction, reducing the genome, additional stress‐resistance genes such as those enabling compatible solute production, or applying adaptive laboratory evolution (Cárdenas Espinosa et al., 2023; Gießelmann et al., 2019; Kuepper et al., 2020). A different strategy harnesses billion years of natural evolution by establishing microorganisms that have naturally adapted to specific challenges (Blombach et al., 2022; Czajka et al., 2017; Fatma et al., 2020; Riley & Guss, 2021). Here, their natural habitat already serves as an important indicator of the type of natural stress tolerance to be expected (Czajka et al., 2017; Volmer et al., 2015).
Among such potentially stress‐tolerant bacteria, many members of the Halopseudomonas genus were isolated from habitats with toxic contaminations, elevated or constantly changing temperatures, or high osmotic pressure. Most of the currently identified 25 species have been described for the first time in the last 20 years (Bollinger, Thies, Katzke, & Jaeger, 2020). Previously, they were placed as the Pseudomonas pertucinogena lineage within the genus Pseudomonas, but considerable molecular and metabolic differences from other Pseudomonas spp. called for a recent reclassification as Halopseudomonas (syn. Neopseudomonas) within the Pseudomonadaceae (Rudra & Gupta, 2021).
Halopseudomonads appear to have in common a small genome with a size of approximately 4 Mb, which is uncommon among Pseudomonaceae. This implies a limited metabolic flexibility of these bacteria, which, in contrast to the versatile group of Pseudomonas spp., may reflect a specific adaption to ecological niches. Consequently, Halopseudomonads are described to metabolise hardly any sugars but grow on organic acids like acetic acid, lactic acid, and succinic acid (Pascual et al., 2012; Sánchez et al., 2014; Wang & Sun, 2016; Zhang et al., 2011). Further, they metabolise Tween20 and sebacic acid. Genome analyses revealed genes encoding interesting biocatalysts such as esterases, halohydrin dehalogenases, and ω‐transaminases or the biosynthesis potential to produce ectoine and polyhydroxyalkanoates (Bollinger, Thies, Katzke, & Jaeger, 2020). Furthermore, a role of Halopseudomonads in the degradation of anthropogenic pollutants such as oil or plastic waste was suggested (Bollinger, Thies, Katzke, & Jaeger, 2020; Gomila et al., 2017; Villela et al., 2023). Until now, Halopseudomonas spp. have not been studied in detail, presumably associated with a lack of reference genomes, metabolic models, and robust molecular genetic tools. We have investigated the growth of four selected strains under typical laboratory conditions. Furthermore, we characterised different plasmid‐based expression systems and evaluated the genomic integration of expression modules into two attTn7‐sites via Tn7‐transposition using H. litoralis as one of the studied model strains.
EXPERIMENTAL PROCEDURES
Bacterial strains and plasmids
Escherichia coli DH5α λpir (Penfold & Pemberton, 1992) was used for cloning; the strain S17‐1 λpir (Simon et al., 1983) was used for the conjugational transfer of expression plasmids. The E. coli strains were either cultivated on LB agar plates or in liquid LB medium (Luria/Miller, Carl Roth®) at 37°C, supplemented with 10 μg/mL gentamycin (Gm), 100 μg/mL ampicillin (Amp) or 50 μg/mL kanamycin (Km) when needed. H. aestusnigri VGXO14 (Sánchez et al., 2014), H. bauzanensis BZ93 (Zhang et al., 2011), H. litoralis 2SM5 (Pascual et al., 2012), and H. oceani KX20 (Wang & Sun, 2016) were grown on LB‐agar plates supplemented with a total of 3% (w/v) NaCl at 30°C for 48 h. The increased salt concentration prevented the cells from drying out and enabled storage of the plates for up to one month at 4°C. For the cultivation of the R‐strains (VGXO14R, BZ93R, 2SM5R, and KX20R), 25 μg/mL rifampicin (Rif) was supplemented. When other antibiotics were needed, 25 μg/mL Km or 25 μg/mL Gm were added. To identify and distinguish the four different species from one another, hydrolysis of synthetic plastic material was determined by halo formation on Impranil DLN agar plates. Therefore, 4 mL Impranil® DLN‐SD (Covestro AG) were added to 1 L of LB‐agar (containing 3% (w/v) NaCl and antibiotics when needed) and mixed thoroughly (Molitor et al., 2020). The liquid cultivation, unless stated otherwise, was performed in LB medium enriched with 45 mM succinate (Merck KGaA, Cas: 6106‐21‐4) (LBsuc) as a complex medium. The used mineral salt medium (MSM medium) was prepared according to (Hartmans et al., 1989), supplemented with 18 mM sebacic acid (Merck KGaA, Cas: 111‐20‐6) as the sole carbon source. The carbon source was supplemented in C‐equimolar concentrations derived from comparable studies (C‐equimolar to 30 mM adipic acid [Ackermann et al., 2021]). Sebacic acid was prepared as a sterile‐filtered 10‐fold concentrated stock solution in H2O while adjusting to pH 10 with sodium hydroxide and supplemented to the medium before cultivation. The cultivation was either performed in 100 mL Erlenmeyer flasks (10% filling volume) at 130 rpm or in a 48‐well Round Well Plate® (1 mL filling volume) sealed with “Breathable rayon film seals for biological cultures” (VWR®, Radnor) at 1000 rpm in the microbioreactor BioLector I (Beckman Coulter GmbH) for at least 24 h at 30°C unless otherwise stated. Two to three colonies were added to the cultivation medium for pre‐culture inoculation. Bacterial strains harbouring a vector‐based antibiotic resistance gene were cultivated under the required selection pressure (25 μg/mL Gm or 25 μg/mL Km). The growth rate at the logarithmic growth phase was calculated using the following equation (Christian et al., 1982):
r(t), growth rate [h−1]; N 1, mean of a biological triplicate of the biomass at early logarithmic state; N 2, mean of a biological triplicate of the biomass at late logarithmic state; t, time [h].
All bacterial strains and plasmids used in this study are listed in Table S1, along with their construction and genetic properties.
Cultivation of selected Halopseudomonas spp. for target gene expression
Main cultures of H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20 were inoculated to an optical density OD580 nm of 0.1 (determined with a Photometer, cuvette: 1 cm path length) and grown in Round Well Plates® using the microbioreactor BioLector I (Beckman Coulter GmbH). The cell density was monitored online every 20 min via scattered light intensity at 620 nm, and GFP fluorescence intensity was measured using an Ex508 nm/Em532 nm filter, whereas mCherry fluorescence was detected with an Ex580 nm/Em610 nm filter. The induction of heterologous gene expression was achieved during early logarithmic growth phase (approximately 4.5 h after inoculation) by adding the inducer molecule isopropyl‐β‐d‐thiogalactopyranoside [IPTG] [Cas: 367‐93‐1], l‐arabinose [Cas: 5328‐37‐0], or salicylic acid [Cas: 69–72‐7] [Merck KGaA]), respectively. Stock solutions were prepared in 100x concentration in water or 70% ethanol. The dynamic range is defined as the ratio of the fluorescence intensity of the highest signal and the signal obtained with the uninduced control.
Plasmid isolation
Plasmids were isolated from bacterial cells by alkaline lysis using the ‘innuPREP Plasmid Mini Kit’ (Analytik Jena) according to the manufacturer's instructions. For elution, nuclease‐free water warmed to 65°C was used. Isolated plasmid DNA was then stored at −20°C. DNA sequences were determined by Eurofins Genomics.
Transformation of bacteria with plasmid DNA
Chemically competent E. coli cells prepared with the calcium‐chloride/magnesium‐chloride method were transformed using the heat‐shock method (Hanahan, 1983) with slight modifications. 1–2 μL of the plasmid DNA were added to 100 μL chemically competent cells and then incubated on ice for 30 min. Subsequently, the heat shock was performed at 42°C for 90 s and 700 μL of LB medium was added. Afterwards, the cells were incubated in a rotator wheel for 90 min (KmR, or GmR) at 37°C. Then, 100 μL of the cells were plated on LB agar plates and incubated at 37°C overnight under respective selection pressure.
To transform H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20 with plasmid DNA, a slightly modified version of the room temperature protocol for electroporation was used (Datsenko & Wanner, 2000; Tu et al., 2016). An overnight pre‐culture was harvested via centrifugation (1 min, 21,000 g) and then washed thoroughly with 1 mL of sterile water. After another centrifugation step for 1 min, the cell pellet was resuspended in 80 μL of sterile water, and 1 μL (~ 100 ng) of the external DNA was added. Afterwards, the sample was transferred to an electroporation cuvette (electrode spacing 1 mm: Bio‐Budget Technologies GmbH). The electroporation was performed by using the ‘EC1’ program (25 μF, 200 Ω, 4.5–5 ms, 20 kV/cm) of MicroPulser (Bio‐Budget Technologies GmbH). After that, 700 μL of LBsuc medium was added to the cell suspension and then cultivated under agitation for 2 h at 30°C. Afterwards, 100 μL of the cells were plated on LB‐agar containing 3% (w/v) NaCl with glass beads and incubated at 30°C for 48 h under respective selection pressure.
Tn7 transposition
For the Tn7 transposition, the miniTn7 tools pUC18R6KT‐miniTn7T‐Km, which was a gift from Herbert Schweizer (Addgene plasmid # 64969; http://n2t.net/addgene:64969, RRID: Addgene_64969) (Choi et al., 2005) and the pBG‐13 (Zobel et al., 2015) were used. Triparental conjugational gene transfer (Elhai & Wolk, 1988) was applied to transform the Halopseudomonads with these plasmids. To this end, E. coli S17‐1 λpir was transformed with the plasmid of interest. A second transformation of E. coli S17‐1 λpir with the helper plasmid pTNS2 was performed. Subsequently, 250 μL of each overnight pre‐culture of these E. coli strains, as well as 500 μL of a H. litoralis 2SM5R pre‐culture, were combined and mixed gently. After centrifugation (1 min, 21,000 g), the obtained cell pellet was resuspended in 100 μL LBsuc and transferred onto a membrane filter, which was placed on LB‐agar without any additives. After incubation for at least 5 h at 30°C in the dark, the cells were washed off the filter with 1 mL LBsuc. Finally, the cell suspension was centrifuged for 1 min at 21000 g and resuspended in 100 μL of the supernatant. The suspension was plated on LB‐agar (3% (w/v) NaCl, 25 μg/mL Rif) using glass beads, incubated for two days at 30°C under the required selection pressure, and seeded on agar plates again. Further, the iridescent phenotype of Halopseudomonas colonies and their polyester hydrolase activity (Figure 1B) can be utilised to confirm the isolation of transformed Halopseudomonas after the conjugation.
FIGURE 1.
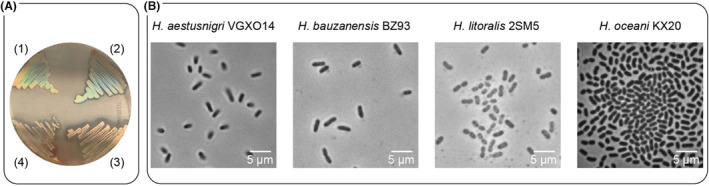
Phenotypes of selected Halopseudomonas spp. (A) Cell lawns of Halopseudomonas strains after cultivation on LB‐agar containing Impranil® DLN‐SD. (1) H. aestusnigri VGXO14, (2) H. bauzanensis BZ93, (3) H. litoralis 2SM5, (4) H. oceani KX20. The agar plate was transilluminated with daylight while the picture was taken. (B) Phase contrast microscopy images, taken of motile bacteria from stationary phase cultures of H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20 grown in LBsuc medium.
A PCR protocol was established to evaluate the successful genomic integration, including primers, each binding specifically at one of the two glmS genes (oligos 12 and 13; Table S1). The other primer binds within the integration cassette (oligo 9 or 10; Table S1). Thus, only a genomic integration led to a PCR product. To illustrate the localisation of glmS1 (locus tag: BLU11_RS05420) and glmS2 (locus tag: BLU11_RS09655) in relation to the origin of replication oriC of the chromosomes, Tubic Ori‐Finder 2022 (Dong et al., 2022) was used to identify oriC of the chromosomes (GenBank: LT629748.1 and CP000010.1). The region 3,507,165‐3,507,739 nt of LT629748.1, and the regions 3,006,500‐3,006,897 nt as well as 3,055,348‐3,055,787 nt of CP000010.1 were identified as the possible origin of replication (the sequences can be found in the Supplementary Material S1).
Phase‐contrast microscopy
Approximately 10 μL of the cells obtained from stationary phase cultures cultivated in LBsuc medium in a BioLector® were placed on a microscope slide and covered by a coverslip. Images were taken on a motorised inverted microscope (Nikon Eclipse Ti, Nikon Europe B.V.) equipped with a high‐resolution camera from Zyla (Andor Neo SCC‐01462, Andor, Belfast) and a CFI Plan Aprochromat λ100x Oil Ph3 DM oil‐Immersion objective (ΝΑ 1.45; W.D.0.13; Nikon). Images were captured using the Nikon NIS‐Elements AR software package. The analysis was performed using Fiji, ImageJ2 software (Schindelin et al., 2012).
RESULTS AND DISCUSSION
We selected four strains from the genus Halopseudomonas as model organisms to establish standard cultivation conditions and probe their genetic accessibility using a set of replicative and integrative vectors. The strains were selected based on expected eurythermia and euryhalinity, presumably resulting from their natural habitats.
H. aestusnigri VGXO14 was isolated from a crude oil‐contaminated sand sample of the intertidal zone (Sánchez et al., 2014). It harbours a remarkable number of lipolytic enzymes, some with astonishing substrate promiscuity and resistance to organic solvents (Bollinger, Molitor, Thies, et al., 2020; Coscolín et al., 2019). The soil bacterium H. bauzanensis BZ93 was isolated from an industrial site (Zhang et al., 2011). Both strains were thus expected to exhibit tolerance to various chemicals and may contribute to the biodegradation of pollutants as indicated by the degradation of poly(ethylene terephthalate) for enzymes from H. aestusnigri VGXO14 and H. bauzanensis BZ93 at ambient temperature (Avilan et al., 2023; Bollinger, Thies, Knieps‐Grünhagen, et al., 2020). Additionally, we chose two marine water isolates, H. litoralis 2SM5 and H. oceani KX20. H. litoralis was also isolated from the tidal zone and is thus expected to cope with constantly changing environmental conditions. It can tolerate NaCl concentrations up to 15%, representing the highest reported osmotolerance among Halopseudomonas spp. (Bollinger, Thies, Katzke, & Jaeger, 2020; Pascual et al., 2012). In contrast, the deep‐sea organism H. oceani was isolated from a habitat with high pressure and low temperature, reflected by the ability to grow at 4°C but tolerating up to 42°C (Wang & Sun, 2016).
Growth of Halopseudomonas spp. under typical laboratory conditions
As a first step, we aimed to establish reliable cultivation conditions for the selected Halopseudomonas species and monitor growth. As previously reported, these strains form small beige, non‐pigmented colonies on LB‐agar plates (Pascual et al., 2012; Sánchez et al., 2014; Wang & Sun, 2016; Zhang et al., 2011). On LB‐Impranil agar, the strains are known to form halos, indicating polyester‐hydrolysing activity (Molitor et al., 2020). When cultivated on LB‐Impranil agar, H. litoralis showed the largest halo, followed by H. oceani, whereas the smallest halo was observed around H. aestusnigri colonies (Figure 1A). Furthermore, bacteria grown in the form of dense lawns showed an iridescent, rainbow‐coloured diffuse glow upon transillumination (Figure 1A). This observation indicated a coordinated movement and aggregation of cells in specific patterns within the lawn or the production of exopolysaccharides leading to structural colours (Kientz et al., 2012; Mizuno et al., 2022; Petruzzi et al., 2017), a phenomenon that, to our knowledge, has not been reported for any Halopseudomonas species before.
In order to access Halopseudomonads for biotechnological applications, suitable cultivation conditions need to be established. We first cultivated the strains in different cultivation vessels filled with LB medium to identify a suitable bioreactor system. The type and filling of cultivation vessels are decisive for oxygen transfer (by surface‐to‐volume ratio and shaking geometry and frequency) as well as shear forces (e.g. in the case of baffles). Growth in a 100 mL Erlenmeyer flask (10% filling volume) was compared with growth in two types of microbioreactors available for the BioLector I® system (Beckman Coulter GmbH), i.e. a Round Well Plate® (RWP) (1 mL filling volume) and a FlowerPlate® (FP) (1 mL filling volume; oxygen transfer rate: 45 mmol/L/h) (Figure S1A). Here, the cultivation in FPs led to strong aggregate formation within all strains' cultures, hampering the growth monitoring using light scattering (see jagged curve in Figure S1B) or optical density. Aggregation of cells indicates stress that may here be caused by shear forces (Fakhruddin & Quilty, 2007; Trunk et al., 2018; Tsagkari et al., 2022). Comparable growth with less aggregate formation was observed when flasks and RWP were used as cultivation vessels. Next, we examined the cell morphology by phase contrast microscopy, revealing that the stationary phase cells of Halopseudomonas spp. maintained their rod‐shaped morphology after 24 h of cultivation under these conditions (Figure 1B). The sampled cells appeared fully motile, but H. litoralis 2SM5 and H. oceani KX20 still tended to aggregate (Figure S3). The number of colony‐forming units (CFU) per mL determined for stationary phase cells was in the same order of magnitude after growing in all three cultivation vessels, as exemplarily assessed for H. bauzanensis (Figure S1C). Thus, we decided to use RWPs for all further cultivation experiments allowing to monitor growth of bacteria at higher throughput.
We observed multiphasic growth curves upon cultivation in LB medium indicating that different carbon sources are used up sequentially (Figure S1D, Figure S2A–D). After 24 h of growth in flasks and RWP, the cells reached a final cell density OD580 nm of approximately 1, which is low when compared with cultures of Pseudomonas sp. or E. coli sp. grown in the same medium, indicating that the nutrients provided by this medium were not depleted. Hence, we tried to identify additives and set up a suitable mineral salt medium (MSM). Previous studies reported that these strains hardly metabolised carbohydrates, but instead used dicarboxylic acids (Figure S4) (Pascual et al., 2012; Sánchez et al., 2014; Wang & Sun, 2016; Zhang et al., 2011). We therefore supplied the complex media with succinic acid (C4) and assessed their use as sole carbon source in MSM (Figure 2).
FIGURE 2.
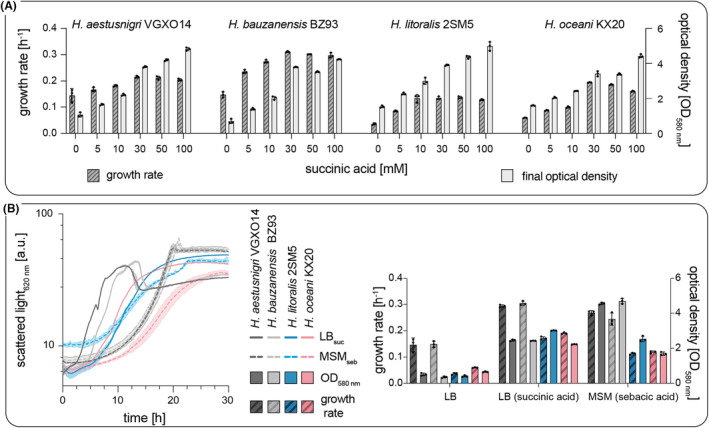
Growth of H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20 in complex and mineral salt media compositions. (A) Growth in LB medium supplemented with increasing concentrations of succinic acid. The growth rate (dark grey, left y‐axis) and the optical density at 580 nm after 24 h (light grey, right y‐axis) are shown. (B) Comparison of the growth in LB medium with 45 mM succinic acid (continuous line) and MSM with 18 mM sebacic acid as sole carbon source (dotted line). Growth curves determined by light scattering are shown on the left, and growth rates (striated bars, left y‐axis) and the optical densities after 24 h of cultivation (filled bars, right y‐axis) are shown on the right. The shown data represent the mean of biological triplicates. The calculated standard deviations are either indicated by shadows or error bars.
Supplementation of LB medium with succinic acid at the beginning of the logarithmic growth phase (4.5 h) improved the growth of all Halopseudomonas strains during batch cultivations (Figure 2A). Here, highest biomass densities (determined as OD580 nm) were observed with concentrations of 50 mM and 100 mM succinic acid. Maximal growth rates were measured upon the addition of 30 mM succinic acid. A shift from pH 7 to pH 10, presumably evoked by the metabolism of the added acid, was ruled out as main reason for the observed growth characteristics, as an elevated pH per se did not improve the growth of Halopseudomonads (Figure S5).
Succinic acid is an intermediate of the citrate cycle and is known as a suitable carbon source for many bacteria including Pseudomonas species (Collier et al., 1996; Dhamale et al., 2022; Mendonca et al., 2020). According to previous reports, Halopseudomonads are also able to grow on less common dicarboxylic acids (de Witt et al., 2023; Pascual et al., 2012; Sánchez et al., 2014; Wang & Sun, 2016; Zhang et al., 2011). Hence, we assessed the growth of H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20 upon supplementation of both LB and MSM medium with short‐chain (C2 and C3) and long chain (C5‐C10) dicarboxylic acids in C‐equimolar concentrations (Table 1; Figure S6). In general, the addition of dicarboxylic acids with shorter chain length, such as oxalic acid (C2‐dicarboxylic acid), malonic acid (C3‐dicarboxylic acid) as well as glutaric acid (C5‐dicarboxylic acid), and pimelic acid (C7‐dicarboxylic acid) led to minor growth improvements compared to LB medium or inhibited growth. C6‐ and C8‐dicarboxylic acids supplemented as additional (LB) and as sole carbon source (MSM) evoked a prolonged lag phase, but the cultures reached comparable final biomasses to those supplemented with succinic acid (C4‐dicarboxylic acid). Notably, supplementation of MSM with long‐chain acids azelaic acid (C9‐dicarboxylic acid) and sebacic acid (C10‐dicarboxylic acid) resulted in comparable or even better growth performance of all species, indicated by a shorter lag phase, higher growth rate, and /or higher optical densities. (Figure S6).
TABLE 1.
Overview of the metabolism of various dicarboxylic acids as an additional or sole carbon source by H. aestusnigri VGXO14, H. bauzanensis BZ93, H. litoralis 2SM5, and H. oceani KX20.
| Dicarboxylic acids | LB medium | MSM medium | ||||||
|---|---|---|---|---|---|---|---|---|
| H. aestusnigri | H. bauzanensis | H. litoralis | H. oceani | H. aestusnigri | H. bauzanensis | H. litoralis | H. oceani | |
| C2 | − | − | − | − | − | − | − | − |
| C3 | − | ++ | ++ | + | − | − | − | − |
| C4 | +++ | +++ | +++ | ++ | ++ | ++ | +++ | +++ |
| C5 | + | − | + | + | − | − | − | − |
| C6 | + | − | ++ | +++ | + | + | +++ | +++ |
| C7 | + | − | + | + | + | − | − | − |
| C8 | +++ | +++ | +++ | +++ | ++ | + | + | ++ |
| C9 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
| C10 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ |
The C2–C10 dicarboxylic acids were tested in concentrations C‐equimolar to 45 mM succinic acid and were used as additional carbon sources in LB medium and as sole carbon sources in MSM medium. The indicators correlate to relative biomass values after 48 h. The values determined for LB medium with additives were related to the biomass achieved for a specific strain with LB medium without additives (100%): − growth inhibiting; + a range up to 150%; ++ a range up to 180%; +++ a range greater than 180%. The highest biomass achieved for a specific strain in MSM with a respective carbon source was set to 100%. − growth inhibiting; + a range up to 50%; ++ a range up to 80%; +++ a range up to 100%. The respective growth curves are shown in Figure S4.
Comparing the results among strains, we observed that cultures of H. aestusnigri VGXO14 and H. bauzanensis BZ93 grown in MSM achieved higher final biomass than complex media cultures, whereas H. litoralis 2SM5 and H. oceani KX20 reached higher final biomasses when using the complex medium (Figure 2B). H. litoralis and H. oceani metabolised a larger spectrum of dicarboxylic acids than H. formosensis FZJ (de Witt et al., 2023), whereas growth of H. aestusnigri and H. bauzanensis was even inhibited by C6 dicarboxylic acids (Table 1).
In summary, succinic acid and sebacic acid were the most beneficial supplements to LB medium, resulting in short lag phases, high growth rates, and biomass for all tested species. Interestingly, sebacic acid appeared superior to succinate when present as sole carbon source. The maximal growth rates achieved for the four species with the optimised media (MSM with sebacic acid) were 0.267 ± 0.08 h−1, 0.243 ± 0.023 h−1, 0.112 ± 0.004 h−1, and 0.118 ± 0.005 h−1 for H. aestusnigri, H. bauzanensis, H. litoralis, and H. oceani, respectively. Reported growth rates of the Proteobacteria Cupravidus necator and Pseudomonas nitroreducens on sebacic acid were within a similar range (Janota‐Bassalik & Bohdanowicz‐Strucinska, 1974; Lang et al., 2007; Strittmatter et al., 2022). From Sullivan et al. (2022), approx. growth rates can be deduced for Acinetobacter baylyi ADP‐1 of 0.27 h−1 growing on sebacic acid and of 0.43 h−1 on adipic acid.
In relation to well‐established model organisms growing under the respective standard conditions, the growth rates obtained here with >1 division/hour were 2–6 times lower but within the range of those reported for batch cultivations of P. putida (0.57 h−1), B. subtilis (1.5 h−1), E. coli (1.1 h−1), or C. glutamicum (0.75 h−1) growing on glucose (Blank et al., 2008; Wittgens et al., 2011). The growth rate of engineered P. putida for growth on adipic acid was determined with 0.35 ± 0.01 h−1 (Ackermann et al., 2021).
Metabolism of long‐chain dicarboxylic acid is not widespread among bacteria. It requires a set of specialised enzymes catalysing the respective analogue of fatty acid, which is best described for A. baylyi ADP1 (Ackermann et al., 2021; Parke et al., 2001). BLASTp homology search uncovered homologues of all the mentioned proteins in H. litoralis and H. bauzanensis (Table S2) and homologues of most of them in H. aestusnigri and H. oceani. The lack of some of the expected proteins may be attributable to the fact that only draft genomes are available for those strains that do not represent closed chromosomes but sets of scaffolds, indicating that parts of the genomes are lacking.
Unlike to A. baylyi, the identified dca‐homologues are distributed among several loci in the genomes of the selected Halopseudomonads, which may indicate functions of these enzymes in other metabolic pathways like terpene/branched amino acid degradation.
Organisms naturally evolved to utilise C6–C10 dicarboxylic acids are of interest in the context of plastic degradation and upcycling of synthetic polymers, as those acids are building blocks for polyesters (like polyethylene adipate terephthalate (PBAT), polyester polyurethane foams and coatings, e.g. Impranil® DLN‐SD, and Nylon 6,6,) as well as part of their hydrolysates and different plasticisers (Ackermann et al., 2021; de Witt et al., 2023; Howard et al., 2012; Sullivan et al., 2022). As it is known that Halopseudomonas spp. secrete enzymes that can depolymerise polyesters (Avilan et al., 2023; Bollinger, Thies, Knieps‐Grünhagen, et al., 2020; de Witt et al., 2023; Haernvall et al., 2017; Molitor et al., 2020), it may be speculated that Halopseudomonads play a role in the breakdown of such polymers present in different habitats. On the other hand, these bacteria might represent interesting targets for establishing biotechnological recycling strategies, which can serve as alternatives to engineered or laboratory‐evolved strains (Ackermann et al., 2021; Sullivan et al., 2022).
Unlocking the genome of Halopseudomonads for engineering purposes
Genetic accessibility requires, among other traits, reliable selection of transformed clones, e.g. after conjugational transfer experiments. Halopseudomonads cannot grow on cetrimide agar as the common Pseudomonas selection medium (Goto & Enomoto, 1970) nor share any antibiotic resistance. In order to enable counterselection, we isolated spontaneously occurring rifampicin (Rif) resistant clones after spreading several colonies of each species on LB‐agar containing 25 μg/mL rifampicin (Rif). The resulting strains were termed H. aestusnigri VGXO14R, H. bauzanensis BZ93R, H. litoralis 2SM5R, and H. oceani KX20R, respectively. To counteract spontaneously occurring Rif‐resistant cells of the E. coli‐donor strain, we exploited the pronounced osmotolerance of Halopseudomonas spp. by using LB agar plates containing Rif and 3% (w/v) NaCl, which impaired the donor's growth (Figure S7). Remarkably, the Rif‐resistant strains formed no aggregates in liquid culture as they were observed for the respective wild‐type strains.
A recent study reported the successful transformation of H. aestusnigri and H. oceani with a pBBR‐derived plasmid by electroporation (Chan et al., 2023). Based on these results, we established an electroporation method using a pJT'Tmcs plasmid harbouring GmR the broad‐host range pRO1600 oriV but no oriT. The washing protocol and the electroporation procedure did not impact the viability of cells obtained from R‐strains (data not shown). For the four selected strains, transformation efficiencies were determined as the number of Gm/Rif‐resistant CFU per mass of DNA: H. aestusnigri VGXO14R – 3*107 CFU/μg; H. bauzanensis BZ93R – 3*104 CFU/μg; H. litoralis 2SM5R – 7*107 CFU/μg; and H. oceani KX20R – 1*108 CFU/μg. Table S3 shows that all strains allowed for the replication of a set of broad‐host‐range plasmids established for Pseudomonas spp. (Martin‐Pascual et al., 2021), harbouring ori's pRO1600, pBBR1, and RSF1010 as well as the respective Rep proteins, except for R6K‐vectors. Interestingly, these strains also appeared to maintain the typical E. coli origins of replication ColE1 / pMB1 / pBR322 that are not applicable in Pseudomonas spp. This observation was confirmed by testing the antibiotic resistance of clones after transformation and the isolation of the plasmids from the Halopseudomonas strains (Figure S8A,B). Nevertheless, curing from such plasmids could be obtained by cultivating the strains in liquid cultures at 37°C for 24 h (Figure S8A).
Next, we evaluated in H. aestusnigri VGXO14R, H. bauzanensis BZ93R, H. litoralis 2SM5R, and H. oceani KX20R for the use of constitutive promoters, namely P em7 and P tac , for the expression of genes encoding the fluorescent proteins GFP and mCherry (Figure S9). Expression of reporter genes from promoter P tac in H. aestusnigri and H. bauzanensis carrying a plasmid with a RO1600 origin resulted in similar fluorescence yields, H. litoralis and H. oceani harbouring the respective plasmid showed only low fluorescence intensities. The highest fluorescence signal was observed with H. litoralis carrying pYT‐P em7 ‐eYFP.
We then tested inducible promoter systems that either lead to an activation (Figure 3A) or de‐repression (Figure 3B) of the gene expression. For all R‐strains evaluated, a gradual induction profile was obtained. Nevertheless, we observed differences in the normalised fluorescence intensities between the different species. On the one hand, H. aestusnigri VGXO14R showed the highest fluorescence intensity compared to the other R‐strains when gene expression was induced with l‐arabinose or salicylic acid. l‐arabinose at a concentration of 20 mM was sufficient to activate expression and reach maximum fluorescence intensity. For H. litoralis 2SM5R and H. oceani KX20R in contrast, 50 mM l‐arabinose was required for maximum intensity, and at least 100 mM for H. bauzanensis BZ93R. Notably, as all strains cannot grow with arabinose as sole carbon source, they probably lack an efficient import mechanism. The highest dynamic range (Figure 3C) of the P araBAD system was achieved in H. bauzanensis BZ93R and H. litoralis 2SM5R, matching the range previously determined for P. putida (Chan et al., 2023). Interestingly, the dynamic ranges achieved here with H. aestusnigri VGXO14R and H. oceani KX20R were higher than previously reported for the respective wild‐type strains (Chan et al., 2023). In E. coli, an arabinose concentration of 13.3 mM was necessary to fully induce gene expression, whereas 133 mM (2% w/v) was required in P. putida (Cook et al., 2018; Guzman et al., 1995). Using the P nagAa/nagR ‐system, a maximum of the normalised fluorescence intensity was reached after adding 1 mM salicylic acid. Notably, the growth of H. aestusnigri was impaired at this concentration, whereas H. bauzanensis, H. litoralis, and H. oceani appeared tolerant to up to at least 10 mM (Figure S10). P. putida KT2440, for comparison, tolerates up to 5 mM salicylic acid; the maximum protein production was achieved when supplementing with 2 mM salicylic acid (Weihmann et al., 2023). However, 10 μM salicylic acid was sufficient to induce expression of the reporter gene mcherry in H. aestusnigri VGXO14R, whereas five times the amount was necessary for H. litoralis 2SM5R and H. oceani KX20R, and ten times the amount for H. bauzanensis BZ93R. These observations may indicate the presence of more efficient mechanisms to keep the inducer out of the cell, matching the observed higher tolerance of those strains. However, fluorescence was also detected when no inducer molecule was added, indicating a leakiness of the promoter system. On the other hand, H. aestusnigri VGXO14R showed the weakest fluorescence intensity when the gene expression was derepressed by the addition of IPTG. Here, H. litoralis 2SM5R and H. oceani KX20R showed the highest sensitivity towards IPTG, since the de‐repression of gfpmut3 expression occurred upon induction with 10 μM IPTG, whereas 50 μM IPTG were needed in H. aestusnigri VGXO14R, and H. bauzanensis BZ93R. An inducer concentration of 80–100 μM IPTG needed for Halopseudomonads represents one‐tenth of concentrations commonly used in Pseudomonads or B. subtilis and is only slightly higher than used for E. coli Tuner (DE3) (Binder et al., 2014, 2016; Hogenkamp et al., 2021; Li et al., 2019). Species‐specific differences may be caused by variances in inductor import, inductor tolerance, but also plasmid copy numbers, as reported for, among others, H. aestusnigri and H. oceani (Chan et al., 2023).
FIGURE 3.
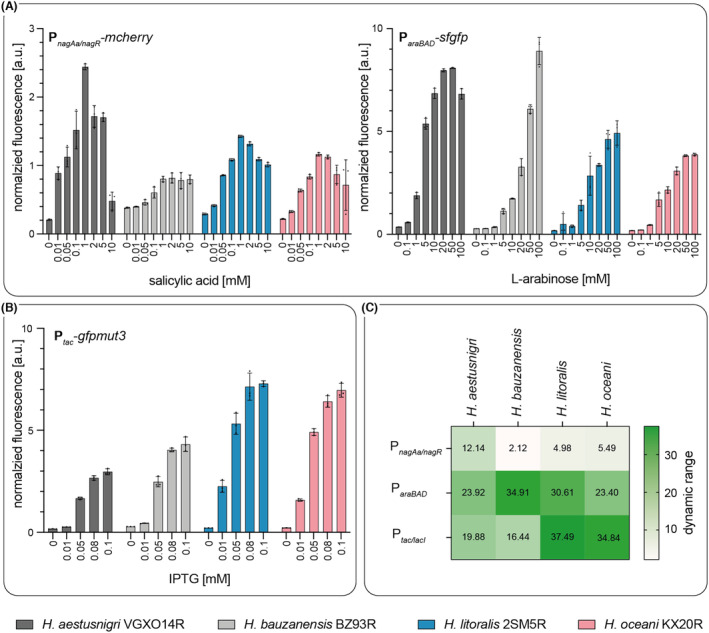
Evaluation of heterologous gene expression in Halopseudomonas strains using different promoters. Induction profiles of different promoters requiring activation (P araBAD , P nagAa/nagR ) (A) or de‐repression (P tac ) (B). Expression of reporter genes was induced by l‐arabinose, salicylic acid, or IPTG at the beginning of the logarithmic growth phase after 4.5 h of cultivation. Cells treated with 70% EtOH served as negative controls. The data represent the means of biological triplicates, and error bars indicate the calculated standard deviations. (C) The dynamic range is shown as a heatmap, increasing from white to dark green.
Chromosomal integration of reporter genes by Tn7 transposition
Expression of a chromosomally integrated gene of interest is often favourable as compared to a plasmid‐based gene since it enables the cultivation without the addition of antibiotics and factors such as plasmid loss and plasmid copy number do not play a role (Jahn et al., 2016). Chromosomal integration can occur either at randomised positions, e.g. via Tn5 transposition, or site‐specific, via homologous recombination or Tn7 transposition into the attTn7‐site. The attTn7‐site is located in a neutral region around 27 bp downstream of the gene glmS encoding a transaminase involved in cell wall biosynthesis (Mitra et al., 2010; Peters & Craig, 2001). Interestingly, the here investigated Halopseudomonads carry two copies of this essential gene instead of one. Therefore, their genomes apparently provide two sites for Tn7 integration: H. aestusnigri VGXO14 (WP_088273794.1, WP_088276615.1); H. bauzanensis BZ93 (WP_074778335.1, WP_036992462.1); H. litoralis 2SM5 (WP_090273138.1, WP_090272410.1); and H. oceani KX20 (WP_104738233.1, WP_104738216.1). Even though the two copies of these genes in each strain are not identical in sequence, the TnsD recognition sites (gene sequences that translate to PRNLAKSVTVE [Mitra et al., 2010]) were found to be highly conserved. H. litoralis 2SM5 is the only strain with a closed genome sequence available that allows for precise localisation of these sites; we, therefore, chose to investigate this strain for the accessibility of both potential sites for Tn7 transposition. To this end, we constructed two mini‐Tn7 vectors differing in selection markers and fluorescence reporter genes, namely pUC18‐R6KT‐miniTn7T‐Km‐P tac/lacI ‐mcherry, and pBG‐13 (Zobel et al., 2015) harbouring sfgfp under the control of the constitutive promoter P em7 .
The Tn7 transposition was performed via triparental conjugational gene transfer with the donor strains E. coli S17‐1/pUC18R6KT‐miniTn7T‐Km‐P tac/lacI ‐mcherry and E. coli S17‐1/pTNS2 and the acceptor strain H. litoralis 2SM5R. The success of Tn7 integration was determined using an analytic PCR with two site‐specific forward primers that bind within the sequence of either glmS1 or glmS2, and a reverse primer that binds within the integration cassette. The obtained PCR products (Figure 4A) were confirmed by sequencing analysis. Of 12 clones analysed after transposition with miniTn7T‐Km‐P tac/lacI ‐mcherry, five had the transposon integrated downstream of glms1, and likewise five carried the transposon downstream of glmS2; for two, the PCR results were inconclusive. Hence, both copies of glmS were indeed addressable for Tn7 transposition. Only a limited number of bacterial genomes are reported to contain more than one copy of glmS accompanied by two attTn7‐sites adjacent to these genes; among them are different Burkholderia spp., β‐proteobacteria with usually two chromosomes (Choi et al., 2006). B. mallei, which harbours two glmS copies on chromosome 1, was studied in more detail. Here, Tn7 transposition was found to occur at both sites; however, in contrast to our observations with H. litoralis, one attTn7‐site was clearly preferred, with 92% of the clones having the transposon integrated downstream of glmS1 only (Choi et al., 2006). In B. mallei, glmS‐1 is located closer to the origin of replication. In H. litoralis, in contrast, both glmS genes are located at similar distances from oriC (Figure 4A). This might point to an explanation for a presumable more randomised mode of integration in the latter.
FIGURE 4.
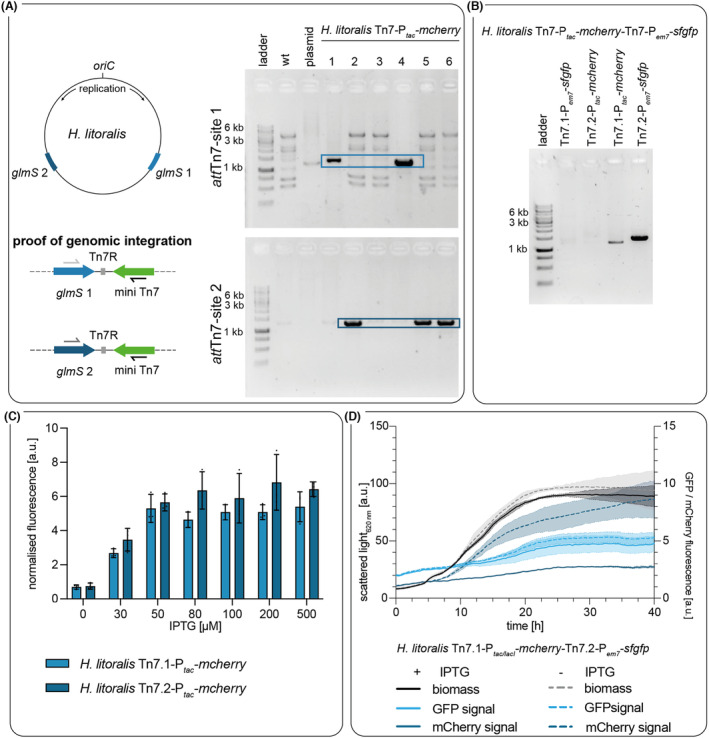
Genomic integration of a reporter gene construct into two attTn7‐sites of H. litoralis 2SM5R. (A) Identification of integration sites. The localisation of genes glmS1 (light blue) and glmS2 (dark blue) within the genome of H. litoralis 2SM5 is shown (illustration not to scale). Transposition events were verified by PCR using the primer pairs shown by convergent arrows with a glmS1 or glmS2 specific forward primer in combination with a primer binding in the transposon either within the antibiotic resistance gene (pUC18R6KT‐miniTn7T‐Km‐P tac ‐mcherry) or within gene sfgfp (pBG‐13). The PCR results after transposition into the different attTn7‐sites are shown on the right‐hand side for six strains with one transposon. Crude cell extract of H. litoralis wild type was applied as control. Blue boxes indicate PCR products of the expected size (1300 bp). (B) One of the identified strains was subsequently transformed with a second and different Tn7 transposon. The colony PCR was performed using a primer combination indicating the integration of P em7 ‐sfgfp either into the first (Lane 1) or the second (Lane 4) integration site and integration of Ptac/lacI‐mcherry (lane 2 (attTn7‐site 1); lane 3 (attTn7‐site 2)) (C) Comparison of the gene expression between two strains with miniTn7T‐Km‐P tac/lacI ‐mcherry in either the first (light blue) or second (dark blue) attTn7‐site. The gene expression was induced by adding IPTG after 4.5 h of cultivation at the beginning of the logarithmic growth phase. (D) Simultaneous expression of target genes integrated in different attTn7 sites. The biomass signal (black) of H. litoralis Tn7.1‐P em7 ‐sfgfp‐Tn7.2‐P tac ‐mcherry as well as the sfGFP fluorescence (light blue, Tn7.1‐site) and the mCherry fluorescence (dark blue, Tn7.2‐site) were measured with a microbioreactor system (BioLector I). The cultures were induced with 50 μM IPTG (dashed line) after 4.5 h of cultivation or treated with 70% EtOH as negative controls (continuous line). The data represent the mean of biological triplicates, and error bars or shadows indicate the calculated standard deviations.
As the localisation of an integron in the chromosome often influences the expression strength (Chaves et al., 2020; Englaender et al., 2017; Sauer et al., 2016), we compared the output of the LacI/P tac expression systems of transposons integrated at each of the attTn7‐sites of H. litoralis based on the mCherry fluorescence 20 h after induction of gene expression. Notably, no significant differences in fluorescence intensities were detected between both integration loci (Figure 4C). This observation matches previous reports regarding the correlation of protein production with the location of the genomic integration locus with regard to oriC (Chaves et al., 2020), independent of the strand polarity (Sauer et al., 2016).
Afterwards, we evaluated the simultaneous occupation of both attTn7‐sites by transforming a strain carrying Tn7‐ LacI/P tac ‐mcherry in attTn7.1 with a second Tn7 transposon harbouring a sfgfp gene behind a constitutively active promoter on plasmid pBG‐13. Another reverse primer was designed to bind within the sfgfp sequence to verify the second integration. Thus, the presence of a PCR product indicated that the transposition occurred into the respective site (Figure 4B); sequencing of the PCR products confirmed subsequent transpositions into both sites. During cultivation and expression, it was observed that the fluorescence reporter genes of both sites were expressed, as confirmed by recording the specific emission spectra of mCherry and sfGFP (Figure S11A). In the majority of the cultures, the mCherry signal appeared immediately after the addition of IPTG, whereas the sfGFP signal unexpectedly did not correlate with the biomass signal despite the presence of a constitutive promoter (Figure 4D). The growth of the cultures appeared not to be affected by the expression of both recombinant genes (Figure S11C).
However, in four out of 45 cultures, the sfGFP signal correlated with the biomass signal (Figure S11B). Notably, the mCherry signal increased in these cultures only in the stationary growth phase despite the time point of IPTG supplementation (Figure S11B), which was not observed when only one locus harboured the transposon. Thus, interference between both loci acting either on the expression level or strain stability may be assumed, leading to repression or loss of the target genes. Studies regarding the simultaneous occupation of both Tn7‐sites in B. mallei or exploitation of three artificial attTn7‐sites in Salmonella enterica reported, in contrast to some extent a correlation of the expression signal with the integron copy number (Bruckbauer et al., 2015; Roos et al., 2015). At present, the underlying mechanism remains elusive; it may involve an unknown regulatory mechanism.
In summary, we have demonstrated that both attTn7‐sites in the genome of H. litoralis are suitable for integrating expression modules. They may be exploited to carry homologous target or reporter genes, for complementation of mutants, or to establish novel biocatalytic functions or reporter genes (Choi et al., 2006; Norris et al., 2010).
CONCLUSION
Halopseudomonas constitutes an only recently established phylum of Proteobacteria. These bacteria, discovered in various challenging habitats over the last two decades, possess only limited metabolic flexibility and apparently produce a variety of polyester hydrolases. Here, we have studied four selected species of Halopseudomonas and could demonstrate the metabolism of several dicarboxylic acids with sebacic acid and succinic acid as preferred carbon sources. We further isolated rifampicin‐resistant strains designated H. aestusnigri VGXO14R, H. bauzanensis BZ93R, H. litoralis 2SM5R, and H. oceani KX20R and provide protocols for electroporation and conjugational gene transfer. Moreover, a genetic toolbox was successfully established, including plasmids with different origins of replication and constitutive and inducible promoter systems. The species‐specific differences in tolerance and induction profiles observed for different Halopseudomonas species suggest a chassis‐a‐la‐carte model (Czajka et al., 2017; Fatma et al., 2020; Riley & Guss, 2021), allowing to select the species which best fits the desired properties rather than aiming for a one‐fits‐all Halopseudomonas model (Czajka et al., 2017; Fatma et al., 2020; Riley & Guss, 2021). We further demonstrated for H. litoralis the presence of two attTn7‐sites suitable for stable chromosomal integration of expression modules. Hence, the results reported here will contribute to further exploring the physiology and biotechnological applications of Halopseudomonas species.
AUTHOR CONTRIBUTIONS
Luzie Kruse: Conceptualization (supporting); formal analysis (lead); investigation (lead); methodology (lead); validation (lead); visualization (lead); writing – original draft (lead). Anita Loeschcke: Conceptualization (supporting); funding acquisition (equal); project administration (supporting); writing – review and editing (supporting). Jan de Witt: Investigation (supporting); methodology (supporting). Nick Wierckx: Conceptualization (supporting); funding acquisition (supporting); project administration (supporting); writing – review and editing (supporting). Karl‐Erich Jaeger: Conceptualization (supporting); funding acquisition (supporting); project administration (lead); writing – review and editing (supporting). Stephan Thies: Conceptualization (lead); funding acquisition (equal); project administration (supporting); writing – review and editing (lead).
CONFLICT OF INTEREST STATEMENT
The authors declare that there are no competing interests associated with the manuscript.
Supporting information
Data S1:
ACKNOWLEDGEMENTS
The authors would like to thank Lennart Ole Witting and Prof. Dietrich Kohlheyer for their support in taking the microscopy pictures. This work was supported by the German Federal Ministry of Education and Research via the Project NO‐STRESS [grant number 031B0852B (to L.K., S.T., A.L. and K.‐E.J.) and 031B085A (to N.W.)]. JdW received funding from the Bio‐based Industries Joint Undertaking (JU) under the European Union’s Horizon 2020 research and innovation program under grant agreement No 887711; we also acknowledge FuturEnzyme funded by the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 101000327. Open Access funding enabled and organized by Projekt DEAL.
Kruse, L. , Loeschcke, A. , de Witt, J. , Wierckx, N. , Jaeger, K.‐E. & Thies, S. (2024) Halopseudomonas species: Cultivation and molecular genetic tools. Microbial Biotechnology, 17, e14369. Available from: 10.1111/1751-7915.14369
Contributor Information
Karl‐Erich Jaeger, Email: k.-e-jaeger@fz-juelich.de.
Stephan Thies, Email: s.thies@fz-juelich.de.
DATA AVAILABILITY STATEMENT
All data are available upon request.
REFERENCES
- Ackermann, Y.S. , Li, W.J. , Op de Hipt, L. , Niehoff, P.J. , Casey, W. , Polen, T. et al. (2021) Engineering adipic acid metabolism in Pseudomonas putida . Metabolic Engineering, 67, 29–40. [DOI] [PubMed] [Google Scholar]
- Avilan, L. , Lichtenstein, B.R. , König, G. , Zahn, M. , Allen, M.D. , Oliveira, L. et al. (2023) Concentration‐dependent inhibition of mesophilic PETases on poly(ethylene terephthalate) can be eliminated by enzyme engineering. ChemSusChem, 16, 1–12. [DOI] [PubMed] [Google Scholar]
- Binder, D. , Grünberger, A. , Loeschcke, A. , Probst, C. , Bier, C. , Pietruszka, J. et al. (2014) Light‐responsive control of bacterial gene expression: precise triggering of the lac promoter activity using photocaged IPTG. Integrative Biology, 6, 755–765. [DOI] [PubMed] [Google Scholar]
- Binder, D. , Probst, C. , Grünberger, A. , Hilgers, F. , Loeschcke, A. , Jaeger, K.E. et al. (2016) Comparative single‐cell analysis of different E. coli expression systems during microfluidic cultivation. PLoS One, 11, e0160711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzenhofer, N.L. , Kruse, L. , Thies, S. , Wynands, B. , Lechtenberg, T. , Rönitz, J. et al. (2021) Towards robust Pseudomonas cell factories to harbour novel biosynthetic pathways. Essays in Biochemistry, 65, 319–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank, L.M. , Ionidis, G. , Ebert, B.E. , Bühler, B. & Schmid, A. (2008) Metabolic response of Pseudomonas putida during redox biocatalysis in the presence of a second octanol phase. FEBS Journal, 275, 5173–5190. [DOI] [PubMed] [Google Scholar]
- Blombach, B. , Grünberger, A. , Centler, F. , Wierckx, N. & Schmid, J. (2022) Exploiting unconventional prokaryotic hosts for industrial biotechnology. Trends in Biotechnology, 40, 385–397. [DOI] [PubMed] [Google Scholar]
- Bollinger, A. , Molitor, R. , Thies, S. , Koch, R. , Coscolín, C. , Ferrer, M. et al. (2020) Organic‐solvent‐tolerant carboxylic ester hydrolases for organic synthesis. Applied and Environmental Microbiology, 86, e00106–e00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger, A. , Thies, S. , Katzke, N. & Jaeger, K.E. (2020) The biotechnological potential of marine bacteria in the novel lineage of Pseudomonas pertucinogena . Microbial Biotechnology, 13, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger, A. , Thies, S. , Knieps‐Grünhagen, E. , Gertzen, C. , Kobus, S. , Höppner, A. et al. (2020) A novel polyester hydrolase from the marine bacterium Pseudomonas aestusnigri – structural and functional insights. Frontiers in Microbiology, 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckbauer, S.T. , Kvitko, B.H. , Karkhoff‐Schweizer, R.R. & Schweizer, H.P. (2015) Tn5/7‐lux: a versatile tool for the identification and capture of promoters in Gram‐negative bacteria. BMC Microbiology, 15, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas Espinosa, M.J. , Schmidgall, T. , Pohl, J. , Wagner, G. , Wynands, B. , Wierckx, N. et al. (2023) Assessment of new and genome‐reduced Pseudomonas strains regarding their robustness as chassis in biotechnological applications. Microorganisms, 11, 837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, D.T.C. , Baldwin, G.S. & Bernstein, H.C. (2023) Revealing the host‐dependent nature of an engineered genetic inverter in concordance with physiology. Biodesign Research, 5, 0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves, J.E. , Wilton, R. , Gao, Y. , Munoz, N.M. , Burnet, M.C. , Schmitz, Z. et al. (2020) Evaluation of chromosomal insertion loci in the Pseudomonas putida KT2440 genome for predictable biosystems design. Metabolic Engineering Communications, 11, e00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.H. , DeShazer, D. & Schweizer, H.P. (2006) Mini‐Tn7 insertion in bacteria with multiple glmS‐linked attTn7 sites: example Burkholderia mallei ATCC 23344. Nature Protocols, 1, 162–169. [DOI] [PubMed] [Google Scholar]
- Choi, K.H. , Gaynor, J.B. , White, K.G. , Lopez, C. , Bosio, C.M. , Karkhoff‐Schweizer, R.A.R. et al. (2005) A Tn7‐based broad‐range bacterial cloning and expression system. Nature Methods, 2, 443–448. [DOI] [PubMed] [Google Scholar]
- Christian, R.R. , Hanson, R.B. & Newell, S.Y. (1982) Comparison of methods for measurement of bacterial growth rates in mixed batch cultures. Applied and Environmental Microbiology, 43, 1160–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier, D.N. , Hager, P.W. & Phibbs, P.V., Jr. (1996) Catabolite repression control in the Pseudomonads . Research in Microbiology, 147, 551–561. [DOI] [PubMed] [Google Scholar]
- Cook, T.B. , Rand, J.M. , Nurani, W. , Courtney, D.K. , Liu, S.A. & Pfleger, B.F. (2018) Genetic tools for reliable gene expression and recombineering in Pseudomonas putida . Journal of Industrial Microbiology & Biotechnology, 45, 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscolín, C. , Bargiela, R. , Martínez‐Martínez, M. , Alonso, S. , Bollinger, A. , Thies, S. et al. (2019) Hydrocarbon‐degrading microbes as sources of new biocatalysts. In: Taxonomy, genomics and ecophysiology of hydrocarbon‐degrading microbes. Cham: Springer International Publishing, pp. 353–373. 10.1007/978-3-319-60053-6_13-1 [DOI] [Google Scholar]
- Czajka, J. , Wang, Q. , Wang, Y. & Tang, Y.J. (2017) Synthetic biology for manufacturing chemicals: constraints drive the use of non‐conventional microbial platforms. Applied Microbiology and Biotechnology, 101, 7427–7434. [DOI] [PubMed] [Google Scholar]
- Datsenko, K.A. & Wanner, B.L. (2000) One‐step inactivation of chromosomal genes in Escherichia coli K‐12 using PCR products. PNAS, 97, 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Witt, J. , Molitor, R. , Gätgens, J. , Ortmann de Percin Northumberland, C. , Kruse, L. , Polen, T. et al. (2023) Biodegradation of poly(ester‐urethane) coatings by Halopseudomonas formosensis . Microbiolog and Biotechnology accepted: MICROBIO‐2023‐282‐RA. 10.1111/1751-7915.14362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamale, T. , Saha, B.K. , Papade, S.E. , Singh, S. & Phale, P.S. (2022) A unique global metabolic trait of Pseudomonas bharatica CSV86T: metabolism of aromatics over simple carbon sources and co‐metabolism with organic acids. Microbiology, 168, 001206. [DOI] [PubMed] [Google Scholar]
- Dong, M.J. , Luo, H. & Gao, F. (2022) Ori‐finder 2022: a comprehensive web server for prediction and analysis of bacterial replication origins. Genomics, Proteomics & Bioinformatics, 20, 1207–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhai, J. & Wolk, C.P. (1988) Conjugal transfer of DNA to cyanobacteria. Methods in Enzymology, 167, 747–754. [DOI] [PubMed] [Google Scholar]
- Englaender, J.A. , Jones, J.A. , Cress, B.F. , Kuhlman, T.E. , Linhardt, R.J. & Koffas, M.A.G. (2017) Effect of genomic integration location on heterologous protein expression and metabolic engineering in E. coli . ACS Synthetic Biology, 6, 710–720. [DOI] [PubMed] [Google Scholar]
- Fakhruddin, A.N.M. & Quilty, B. (2007) Measurement of the growth of a floc forming bacterium Pseudomonas putida CP1. Biodegradation, 18, 189–197. [DOI] [PubMed] [Google Scholar]
- Fatma, Z. , Schultz, J.C. & Zhao, H. (2020) Recent advances in domesticating non‐model microorganisms. Biotechnology Progress, 36, e3008. [DOI] [PubMed] [Google Scholar]
- Gießelmann, G. , Dietrich, D. , Jungmann, L. , Kohlstedt, M. , Jeon, E.J. , Yim, S.S. et al. (2019) Metabolic engineering of Corynebacterium glutamicum for high‐level ectoine production: Design, combinatorial assembly, and implementation of a transcriptionally balanced heterologous ectoine pathway. Biotechnology Journal, 14, 1800417. [DOI] [PubMed] [Google Scholar]
- Gomila, M. , Mulet, M. , Lalucat, J. & García‐Valdésa, E. (2017) Draft genome sequence of the marine bacterium Pseudomonas aestusnigri VGXO14T . Genome Announcements, 5, e00765‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, S. & Enomoto, S. (1970) Nalidixic acid cetrimide agar: a new selective plating medium for the selective isolation of Pseudomonas aeruginosa . Japanese Journal of Microbiology, 14, 65–72. [DOI] [PubMed] [Google Scholar]
- Guzman, L.‐M. , Belin, D. , Carson, M.J. & Beckwith, J. (1995) Tight regulation, modulation, and high‐level expression by vectors containing the arabinose PBAD promoter. Journal of Bacteriology, 177, 4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haernvall, K. , Zitzenbacher, S. , Wallig, K. , Yamamoto, M. , Schick, M.B. , Ribitsch, D. et al. (2017) Hydrolysis of ionic phthalic acid based polyesters by wastewater microorganisms and their enzymes. Environmental Science & Technology, 51, 4596–4605. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. (1983) Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology, 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Hartmans, S. , Smits, J.P. , Van Der Werf, M.J. , Volkering, F. & De Bont, J.A.M. (1989) Metabolism of styrene oxide and 2‐phenylethanol in the styrene‐degrading Xanthobacter strain 124X. Applied and Environmental Microbiology, 55, 2850–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenkamp, F. , Hilgers, F. , Knapp, A. , Klaus, O. , Bier, C. , Binder, D. et al. (2021) Effect of photocaged isopropyl‐β‐d‐1‐thiogalactopyranoside solubility on the light responsiveness of LacI‐controlled expression systems in different bacteria. Chembiochem, 22, 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard, G.T. , Norton, W.N. & Burks, T. (2012) Growth of Acinetobacter gerneri P7 on polyurethane and the purification and characterization of a polyurethanase enzyme. Biodegradation, 23, 561–573. [DOI] [PubMed] [Google Scholar]
- Jahn, M. , Vorpahl, C. , Hübschmann, T. , Harms, H. & Müller, S. (2016) Copy number variability of expression plasmids determined by cell sorting and droplet digital PCR. Microbial Cell Factories, 15, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janota‐Bassalik, L. & Bohdanowicz‐Strucinska, B. (1974) Growth of a wild strain and of a pimelic acid‐utilizing mutant of Pseudomonas azelaica on aliphatic dicarboxylic acids. Journal of General Microbiology, 84, 79–84. [DOI] [PubMed] [Google Scholar]
- Kientz, B. , Vukusic, P. , Luke, S. & Rosenfeld, E. (2012) Iridescence of a marine bacterium and classification of prokaryotic structural colors. Applied and Environmental Microbiology, 78, 2092–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuepper, J. , Otto, M. , Dickler, J. , Behnken, S. , Magnus, J. , Jäger, G. et al. (2020) Adaptive laboratory evolution of Pseudomonas putida and Corynebacterium glutamicum to enhance anthranilate tolerance. Microbiology, 166, 1028–1040. [DOI] [PubMed] [Google Scholar]
- Lang, E. , Griese, B. , Spröer, C. , Schumann, P. , Steffen, M. & Verbarg, S. (2007) Characterization of “Pseudomonas azelaica” DSM 9128, leading to emended descriptions of Pseudomonas citronellolis Seubert 1960 (approved lists 1980) and Pseudomonas nitroreducens lizuka and Komagata 1964 (approved lists 1980), including Pseudomonas multiresinivorans as its later heterotypic synonym. International Journal of Systematic and Evolutionary Microbiology, 57, 878–882. [DOI] [PubMed] [Google Scholar]
- Li, W.J. , Jayakody, L.N. , Franden, M.A. , Wehrmann, M. , Daun, T. , Hauer, B. et al. (2019) Laboratory evolution reveals the metabolic and regulatory basis of ethylene glycol metabolism by Pseudomonas putida KT2440. Environmental Microbiology, 21, 3669–3682. [DOI] [PubMed] [Google Scholar]
- Loeschcke, A. & Thies, S. (2015) Pseudomonas putida—a versatile host for the production of natural products. Applied Microbiology and Biotechnology, 99, 6197–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin‐Pascual, M. , Batianis, C. , Bruinsma, L. , Asin‐Garcia, E. , Garcia‐Morales, L. , Weusthuis, R.A. et al. (2021) A navigation guide of synthetic biology tools for Pseudomonas putida . Biotechnology Advances, 49, 107732. [DOI] [PubMed] [Google Scholar]
- Mendonca, C.M. , Yoshitake, S. , Wei, H. , Werner, A. , Sasnow, S.S. , Thannhauser, T.W. et al. (2020) Hierarchical routing in carbon metabolism favors iron‐scavenging strategy in iron‐deficient soil Pseudomonas species. Proceedings of the National Academy of Sciences, 117, 32358–32369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra, R. , McKenzie, G.J. , Yi, L. , Lee, C.A. & Craig, N.L. (2010) Characterization of the TnsD‐attTn7 complex that promotes site‐specific insertion of Tn7. Mobile DNA, 1, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K. , Maree, M. , Nagamura, T. , Koga, A. , Hirayama, S. , Furukawa, S. et al. (2022) Novel multicellular prokaryote discovered next to an underground stream. eLife, 11, e71920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor, R. , Bollinger, A. , Kubicki, S. , Loeschcke, A. , Jaeger, K.E. & Thies, S. (2020) Agar plate‐based screening methods for the identification of polyester hydrolysis by Pseudomonas species. Microbial Biotechnology, 13, 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris, M.H. , Kang, Y. , Wilcox, B. & Hoang, T.T. (2010) Stable, site‐specific fluorescent tagging constructs optimized for Burkholderia species. Applied and Environmental Microbiology, 76, 7635–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parke, D. , Garcia, M.A. & Ornston, L.N. (2001) Cloning and genetic characterization of dca genes required for β‐oxidation of straight‐chain dicarboxylic acids in Acinetobacter sp. strain ADP1. Applied and Environmental Microbiology, 67, 4817–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual, J. , Lucena, T. , Ruvira, M.A. , Giordano, A. , Gambacorta, A. , Garay, E. et al. (2012) Pseudomonas litoralis sp. nov., isolated from mediterranean seawater. International Journal of Systematic and Evolutionary Microbiology, 62, 438–444. [DOI] [PubMed] [Google Scholar]
- Penfold, R.J. & Pemberton, J.M. (1992) An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene, 118, 145–146. [DOI] [PubMed] [Google Scholar]
- Peters, J.E. & Craig, N.L. (2001) Tn7: smarter than we thought. Nature Reviews. Molecular Cell Biology, 2, 806–814. [DOI] [PubMed] [Google Scholar]
- Petruzzi, B. , Briggs, R.E. , Swords, W.E. , De Castro, C. , Molinaro, A. & Inzana, T.J. (2017) Capsular polysaccharide interferes with biofilm formation by Pasteurella multocida serogroup A. MBio, 8, e01843‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva, O.N. , Weinel, C. , Weinel, M. , Böhm, K. , Stjepandic, D. , Hoheisel, J.D. et al. (2006) Functional genomics of stress response in Pseudomonas putida KT2440. Journal of Bacteriology, 188, 4079–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley, L.A. & Guss, A.M. (2021) Approaches to genetic tool development for rapid domestication of non‐model microorganisms. Biotechnology for Biofuels, 14, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, A.L. , Becker, J. , de Souza Lima, A.O. , Porto, L.M. & Wittmann, C. (2014) Systems metabolic engineering of Escherichia coli for gram scale production of the antitumor drug deoxyviolacein from glycerol. Biotechnology and Bioengineering, 111, 2280–2289. [DOI] [PubMed] [Google Scholar]
- Roos, K. , Werner, E. & Loessner, H. (2015) Multicopy integration of mini‐Tn7 transposons into selected chromosomal sites of a Salmonella vaccine strain. Microbial Biotechnology, 8, 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudra, B. & Gupta, R.S. (2021) Phylogenomic and comparative genomic analyses of species of the family Pseudomonadaceae: proposals for the genera Halopseudomonas gen. Nov. and Atopomonas gen. Nov., merger of the genus Oblitimonas with the genus Thiopseudomonas . International Journal of Systematic and Evolutionary Microbiology, 71, 005011. [DOI] [PubMed] [Google Scholar]
- Sánchez, D. , Mulet, M. , Rodríguez, A.C. , David, Z. , Lalucat, J. & García‐Valdés, E. (2014) Pseudomonas aestusnigri sp. nov., isolated from crude oil‐contaminated intertidal sand samples after the prestige oil spill. Systematic and Applied Microbiology, 37, 89–94. [DOI] [PubMed] [Google Scholar]
- Sauer, C. , Syvertsson, S. , Bohorquez, L.C. , Cruz, R. , Harwood, C.R. , Van Rij, T. et al. (2016) Effect of genome position on heterologous gene expression in Bacillus subtilis: an unbiased analysis. ACS Synthetic Biology, 5, 942–947. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nature Methods, 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, R. , Priefer, U. & Puhl, A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnology, 1, 784–791. [Google Scholar]
- Strittmatter, C.S. , Eggers, J. , Biesgen, V. , Hengsbach, J.‐N. , Sakatoku, A. , Albrecht, D. et al. (2022) Insights into the degradation of medium‐chain‐length dicarboxylic acids in Cupriavidus necator H16 reveal ß‐oxidation differences between dicarboxylic acids and fatty acids. Applied and Environmental Microbiology, 88, e01873‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan, K.P. , Werner, A.Z. , Ramirez, K.J. , Ellis, L.D. , Bussard, J.R. , Black, B.A. et al. (2022) Mixed plastics waste valorization through tandem chemical oxidation and biological funneling. Science (1979), 378, 207–211. [DOI] [PubMed] [Google Scholar]
- Tabor, J.J. , Levskaya, A. & Voigt, C.A. (2011) Multichromatic control of gene expression in Escherichia coli. Journal of Molecular Biology, 405, 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunk, T. , Khalil, H.S. & Leo, J.C. (2018) Bacterial autoaggregation. AIMS Microbiology, 4, 140–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkari, E. , Connelly, S. , Liu, Z. , McBride, A. & Sloan, W.T. (2022) The role of shear dynamics in biofilm formation. Npj Biofilms and Microbiomes, 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Q. , Yin, J. , Fu, J. , Herrmann, J. , Li, Y. , Yin, Y. et al. (2016) Room temperature electrocompetent bacterial cells improve DNA transformation and recombineering efficiency. Scientific Reports, 6, 24648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijl, J. & Hecker, M. (2013) Bacillus subtilis: from soil bacterium to super‐secreting cell factory. Microbial Cell Factories, 12, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villela, H. , Modolon, F. , Schultz, J. , Delgadillo‐Ordoñez, N. , Carvalho, S. , Soriano, A.U. et al. (2023) Genome analysis of a coral‐associated bacterial consortium highlights complementary hydrocarbon degradation ability and other beneficial mechanisms for the host. Scientific Reports, 13, 12273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volmer, J. , Schmid, A. & Bühler, B. (2015) Guiding bioprocess design by microbial ecology. Current Opinion in Microbiology, 25, 25–32. [DOI] [PubMed] [Google Scholar]
- Wang, M.Q. & Sun, L. (2016) Pseudomonas oceani sp. nov., isolated from deep seawater. International Journal of Systematic and Evolutionary Microbiology, 66, 4250–4255. [DOI] [PubMed] [Google Scholar]
- Weihmann, R. , Kubicki, S. , Bitzenhofer, N.L. , Domröse, A. , Bator, I. , Kirschen, L.‐M. et al. (2023) The modular pYT vector series employed for chromosomal gene integration and expression to produce carbazoles and glycolipids in P. Putida . FEMS Microbes, 4, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittgens, A. , Tiso, T. , Arndt, T.T. , Wenk, P. , Hemmerich, J. , Müller, C. et al. (2011) Growth independent rhamnolipid production from glucose using the non‐pathogenic Pseudomonas putida KT2440. Microbial Cell Factories, 10, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.C. , Liu, H.C. , Zhou, Y.G. , Schinner, F. & Margesin, R. (2011) Pseudomonas bauzanensis sp. nov., isolated from soil. International Journal of Systematic and Evolutionary Microbiology, 61, 2333–2337. [DOI] [PubMed] [Google Scholar]
- Zobel, S. , Benedetti, I. , Eisenbach, L. , De Lorenzo, V. , Wierckx, N. & Blank, L.M. (2015) Tn7‐based device for calibrated heterologous gene expression in Pseudomonas putida . ACS Synthetic Biology, 4, 1341–1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1:
Data Availability Statement
All data are available upon request.


