Abstract
Cyanide is a highly toxic compound that is found in wastewaters generated from different industrial activities, such as mining or jewellery. These residues usually contain high concentrations of other toxic pollutants like arsenic and heavy metals that may form different complexes with cyanide. To develop bioremediation strategies, it is necessary to know the metabolic processes involved in the tolerance and detoxification of these pollutants, but most of the current studies are focused on the characterization of the microbial responses to each one of these environmental hazards individually, and the effect of co‐contaminated wastes on microbial metabolism has been hardly addressed. This work summarizes the main strategies developed by bacteria to alleviate the effects of cyanide, arsenic and heavy metals, analysing interactions among these toxic chemicals. Additionally, it is discussed the role of systems biology and synthetic biology as tools for the development of bioremediation strategies of complex industrial wastes and co‐contaminated sites, emphasizing the importance and progress derived from meta‐omic studies.
This work summarizes the main strategies developed by bacteria to alleviate the effects of cyanide, arsenic, and heavy metals, analyzing interactions among these toxic chemicals. Additionally, it is discussed the role of systems biology and synthetic biology as tools for the development of bioremediation strategies of complex industrial wastes and co‐contaminated sites, emphasizing the importance and progresses derived from meta‐omic studies.
INTRODUCTION
Although cyanide is a natural compound produced by cyanogenic plants and some arthropods, fungi and bacteria, with defensive or offensive purposes, it is a powerful environmental hazard generated at large scale in a great variety of industrial activities, such as mining and jewellery, coke production, synthesis of organic chemicals and food processing (Luque‐Almagro et al., 2016). Manufacture of cyano‐derivatives includes, mainly, hydrogen cyanide, sodium cyanide, ferrocyanide and acrylonitrile. Exclusively in mining activities, approximately one million tons per year of cyanide are used in a process called cyanidation, which is widely applied for the extraction of gold and other metals from ores (Dash et al., 2009). Therefore, large amounts of cyanide‐containing wastes are produced, within a range of 10,000–30,000 mg/L cyanide, but some of them reach extremely high concentrations of cyanide, up to 100,000 mg/L (Wild et al., 1994). Additionally, these cyanide‐containing wastes usually contain metals and metalloids like mercury, lead, copper, cadmium, chromium, zinc and arsenic (Table S1, Supplementary Material), which contribute to elevate their toxicity, constituting a severe risk for human health (Alvillo‐Rivera et al., 2021; Ćwieląg‐Drabek et al., 2020; Kumar et al., 2016; Razanamahandry et al., 2017). Heavy metals are present in nature with a natural origin like volcanic activity and weathering of metal‐bearing rocks, but environmental and human health threats derived from these inorganic elements are caused by anthropic activities like mining, industrial processing and agricultural practices. Toxicity of heavy metals and metalloids depends on the element and their different chemical forms, but in general, they cause oxidative stress and damage, inactivation of a wide variety of enzymes by binding to thiol and other reactive groups or replacing their natural metal cofactors, and even tumoral processes in different organs (Ali et al., 2019).
The toxicity of the cyano compounds depends on their ability to release free cyanide (Baxter & Cummings, 2006; Kumar et al., 2016). Hydrogen cyanide gas (cyanhydric acid, HCN) is considered the most toxic form of cyanide when inhaled or absorbed. Another highly toxic forms of cyanide are the inorganic salts, such as NaCN and KCN, which dissociate in aqueous solution to produce the anion cyanide (CN−). Thus, at alkaline pH, the predominant form is the anion cyanide, while at neutral and acidic pH prevails hydrogen cyanide. Less toxic forms of cyanide are metal‐cyanide complexes, both including weak acid dissociable (WAD) complexes (with Ni, Cu or Zn) and strong acid dissociable (SAD) complexes (with Fe or Co); more oxidized forms like cyanate (OCN−) and thiocyanate (SCN−); organic cyanides like nitriles; and cyanolipids and other biological compounds (Kumar et al., 2016; Luque‐Almagro et al., 2016, 2018). Cyanide causes severe negative effects on living beings, provoking relevant metabolic changes. These include inhibition of several key metalloenzymes like the cytochrome c oxidase involved in cellular respiration, which occurs through direct chelation of iron present as metallic centre, leading also to the generation of oxidative stress (Hariharakrishnan et al., 2009; Malmir et al., 2022). Inhibition of relevant enzymes of the TCA cycle also occurs by reaction of cyanide with keto groups present in organic acids like pyruvate, 2‐oxoglutarate and oxaloacetate to produce nitrile derivatives (Luque‐Almagro et al., 2018; Roldán et al., 2021).
The presence of metal and cyanide‐containing wastes in the environment is a major ecological threat, especially considering that they are stored usually in artificial ponds that are prone to leaching, discharges or dam breaks, causing environmental disasters, economic losses and even direct human tragedies (Piciullo et al., 2022). The uncertainties on physicochemical characteristics of tailings and their behaviour, the dam construction method, the management practices and the poor performance and monitoring processes contribute to an increase in the risk of dam failures, and climate change could increase this threat due to the high frequency and intensity of extreme weather episodes (i.e. floods). In this sense, almost 50% of the released tailings volumes have been documented after 2000, with about 640 direct fatalities (Piciullo et al., 2022). Some relevant disasters that released high volumes of toxic contaminants causing severe environmental, economic and human fatalities are listed in Table 1, but a complete and detailed information about tailing dam failures is available online in several databases, and it was recently analysed by Piciullo et al. (2022). Even more, releases of toxic residues are frequent and not documented in artisanal and small‐scale gold mining, where amalgamation of gold with mercury is used together with the cyanidation process, thus combining two highly toxics in these wastewaters that are often discharged directly into the environment (Chen et al., 2017). The pH of cyanide‐containing liquid wastes from mining usually are very acidic, and under these conditions cyanide volatilizes to the atmosphere as hydrogen cyanide, which can undergo oxidation and photolysis, thus reducing cyanide toxicity in the surrounding the cyanide leaching facilities (Brüger et al., 2018).
TABLE 1.
Some relevant mine tailings dam failures.
| Mine (location, year) | Principal ore/material released | Volume of tailings released (m3) | Direct human effects |
|---|---|---|---|
| Kearl oil and sands mine (Canada, 2023) | Bitumen | 5300 | |
| Williamson mine (Tanzania, 2022) | Diamond | 12,800,000 | Household and agricultural damages |
| Jagersfontein (South Africa, 2022) | Diamond | 5,040,000 | Agricultural damage and hundreds of people severely injured |
| Brumadinho dam (Brazil, 2019) | Fe | 10,000,000 | Human fatalities: 270 |
| Kokoya gold mine (Liberia, 2017) | Cyanide | 11,500 | |
| Bento Rodrigues dam (Brazil, 2015) | Fe | 60,000,000 | Human fatalities: 19 |
| Proyecto Magistral Mine (Mexico, 2014) | Cyanide | 1900 | |
| Ajka alumina plant Magyar Aluminium (Hungary, 2010) | Al | 30,000,000 | Human fatalities: 10. More than 7000 people evacuated or directly affected |
| Kingston plant (Tennessee, USA, 2008) | As, Hg | 4,100,000 | Cleanup workers suffered severe health problems |
| Baia Mare/Baia Borsa (Romania, 2000) | Ag, Au, cyanide | 100,000 | Interruption of water supply |
| Aznalcóllar (Spain, 1998) | Sulphide, As | 6,800,000 | Agricultural damage and natural reserve failure |
| Omai gold mine (Guyana, 1995) | Cyanide | 4,200,000 | |
| Harmony, Merriespruit (South Africa, 1994) | Au | 600,000 | Human fatalities: 17 |
| Cerro Negro No. 4 (Chile, 1985) | Cu | 2,000,000 | |
| El Cobre Old Dam (Chile, 1965) | Cu | 1900,000 | Human fatalities: ~300 |
The European Parliament has requested to stop using cyanide in mining activities, considering its elevated toxicity and environmental impact. However, related legislation has not been developed in other countries, and even in the USA and New Zealand, cyanide is used for pest control in agriculture (Roldán et al., 2021; Warburton & Livingstone, 2015). Cyanide‐containing wastes must be treated to reduce cyanide concentrations before they are legally discharged into the environment, although cyanide solutions in tailings undergo natural attenuation reactions leading to a decrease in cyanide concentration. The main mechanism of natural attenuation is volatilization, but it also includes chemical and biological oxidation, hydrolysis, precipitation, complexation and sorption. Cyanide elimination treatments may use physicochemical or biological methods (Alvillo‐Rivera et al., 2021; Dash et al., 2009; Malmir et al., 2022). Most of the chemical treatments are oxidative processes in which cyanide is oxidized with Cl2, SO2 + O2, H2O2 or O3 as oxidants, usually producing cyanate and ammonium as intermediates. There are other physicochemical treatments based on non‐oxidative processes, such as iron‐cyanide precipitation, activated carbon polishing and ion exchange/reverse osmosis. Additionally, cyanide may be recovered by acidification. The main limitations of these decontamination methods are the high stability of some metal complexes, the presence of co‐contaminants, the generation of toxic by‐products, the operating cost and the volume of reagents used (Table 2). Furthermore, these methods require equipment, maintenance and licences for the discharge of the newly generated products, which could contain additional toxic pollutants like chlorine. In recent decades the development of eco‐friendly treatments to degrade cyanide, which are efficient and cheap, is on high demand. This is not an easy task, considering that these complex wastes are heterogenous mixtures of co‐contaminants, and, therefore, the implemented treatment methodology requires to be compatible with an efficient removal of all the different chemicals (Coudert et al., 2020).
TABLE 2.
Physicochemical and biological methods used for cyanide removal.
| Treatment | Removal of SAD metal‐CN complexes | Other pollutants removed | Generation of toxic by‐products | Operating cost/reagent volume |
|---|---|---|---|---|
| Physicochemical methods | ||||
| Oxidative processes | ||||
| Alkaline chlorination | Yes | Thiocyanate | No | Low/high |
| SO2 / Air | Yes | No | Copper | High/high |
| Hydrogen peroxide | Yes | No | No | Low/high |
| Ozonation | Yes | Thiocyanate | Cyanate, ammonium, nitrate | High/high |
| Non‐oxidative processes | ||||
| Iron‐cyanide precipitation | Yes | No | Cyanide waste solids | Low/high |
| Activated carbon polishing | Yes | No | Cyanide waste solids | Low/low |
| Ion exchange/reverse osmosis | No | Thiocyanate, cyanate, nitrate, nitrite | Waste brine | High/low |
| Biological methods | ||||
| Bioremediation | Yes/No a | Yes/No a | No | Low/low |
This capacity depends on the bacterial strain used.
The application of organisms to the removal of environmental pollutants from air, water, soil or industrial effluents, in natural or artificial settings, is called bioremediation. This process has been described to be eco‐friendly, cheap and sustainable for environmental management. Many different organisms like bacteria, microalgae, fungi and plants can be used for bioremediation purposes. However, bacteria display the greatest degradative biotechnological potential. This is related to their advantageous properties, such as small genome size, relative cell simplicity, short replication cycles and rapid evolution/adaptation, which make bacteria the best suitable candidates for the development of bioremediation technologies. In this sense, different bacteria have been described to be able to resist and degrade elevated concentrations of cyanide, metals and metalloids. In the development of a process for bioremediation of these toxic wastewaters it must be considered the possible interactions of microorganisms with metals and cyanide, the formation of different cyano‐derivatives and the catalysed and non‐catalysed reactions initiated by the reactivity of cyanide with other molecules (Biełło, Cabello, et al., 2023, Biełło, Olaya‐Abril, et al., 2023; Luque‐Almagro et al., 2016; Newsome & Falagán, 2021).
The different mechanisms displayed by microorganisms to tolerate and detoxify industrial wastes that contain cyanide, arsenic, mercury, copper and cadmium are presented in this review. Interactions of these toxic chemicals with other cellular molecules, and their metabolic consequences, are also discussed. Omics approaches, including meta‐omics, applied to the study of bacterial degradation of cyanide‐containing wastes to design further bioremediation strategies are analysed.
BACTERIAL RESISTANCE AND DETOXIFICATION OF INDUSTRIAL CYANIDE‐ AND METAL‐CONTAINING WASTEWATERS
Microorganisms use different strategies to tolerate cyanide and heavy metals, but they share some similarities that may have relevance in the development of bioremediation processes contributing to detoxify industrial complex mixtures with cyanide and metals as co‐contaminants. Thus, cyanotrophic organisms use cyanide as the sole nitrogen source for growth, but alkaliphilic microorganisms are needed to avoid hydrogen cyanide volatilization. Also, the presence of a cyanide‐insensitive terminal oxidase like the cytochrome bd oxidase CioAB is required for cyanide resistance, in addition to an enzymatic cyanide degradation pathway (Luque‐Almagro et al., 2005). In the alkaliphilic, cyanotrophic bacterium Pseudomonas pseudoalcaligenes CECT 5344, both cyanide‐insensitive respiration and the cyanide assimilation pathway are connected through a malate: quinone oxidoreductase that generates oxaloacetate, which reacts with cyanide forming a cyanohydrin (2‐hydroxynitrile). This nitrile is hydrolyzed further by the nitrilase NitC, which releases ammonium that is used as a nitrogen source for bacterial growth (Estepa et al., 2012; Luque‐Almagro et al., 2018; Roldán et al., 2021). On the contrary, considering that metals are not degraded easily by microorganisms, resistance mechanisms are mainly based on metal detoxification and/or immobilization. Different types of resistance mechanisms to metals have been described in bacteria (Figure 1), including extracellular metal barriers like cell wall, plasma membrane or capsule, metal extrusion through efflux pumps, intracellular, periplasmic or extracellular sequestration, precipitation, biosorption and metal biotransformation (Bazzi et al., 2020; Choudhury & Srivastava, 2001; Kabiraj et al., 2022). Several bacteria also use some of these mechanisms to tolerate radionuclides (López‐Fernández et al., 2021). All these mechanisms could be targets in the development of biotechnologies. In this sense, metal efflux systems, which are the most prevalent mechanisms that confer bacterial resistance to heavy metals, could be subjected to modification or overexpression to increase the resistance or tolerance to different metals (Bazzi et al., 2020; Delmar et al., 2015). However, it is worth noting that metal resistance mechanisms based only on cell extrusion may not solve the contamination problem because the metal remains in the polluted site, except when the metal was previously biotransformed to a volatile form, as occurs in the reduction of Hg2+ to Hg0.
FIGURE 1.
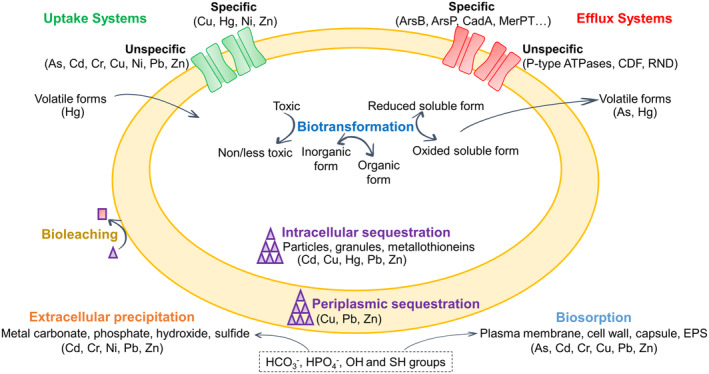
Different general strategies developed by bacteria to tolerate heavy metals.
Bacterial cyanide assimilation
Cyanide degradation pathways lead to the formation of less toxic products like ammonia and carbon dioxide. These biodegradative routes can be of different types, including hydrolytic, oxidative, reductive and substitution/transfer reactions (Cabello et al., 2018; Luque‐Almagro et al., 2018; Roldán et al., 2021). These reactions are catalysed by cyanide hydratases, cyanidases (cyanide dihydratases), cyanide mono‐ and dioxygenases, rhodaneses, 3‐cyanoalanine synthases/hydratases, nitrilases or nitrile hydratases (Figure 2). However, there is no experimental evidence for the presence of a cyanide monooxygenase that could generate cyanate as an intermediate, and hence its participation in cyanide degradation has been only hypothesized. While cyanide hydratase is primarily a fungal enzyme, cyanidase has been described in Pseudomonas stutzeri AK61, Bacillus pumilus C1 and Alcaligenes xylosoxodans subsp. denitrificans. Rhodaneses have been identified in a variety of bacteria, including Azotobacter vinelandii, Escherichia coli and different strains of Thiobacillus (Alvillo‐Rivera et al., 2021; Dash et al., 2009; Gupta et al., 2010; Luque‐Almagro et al., 2018). Organic cyanides (nitriles) are metabolized by nitrile hydratases of Corynebacterium sp. C5, Brevibacterium imperalis BS489‐74, Pseudomonas putida NRRL‐18668, Pseudomonas chlororaphis B23, Arthrobacter sp. J1 and Pseudonocardia thermophila JCM3095, while nitrilases are present in Alcaligenes faecalis JM3, Acinetobacter sp. AK226, Pseudomonas thermophila JCM3095, Klebsiella ozaenae and some Rhodococcus rhodochrous strains (Alvillo‐Rivera et al., 2021; Dash et al., 2009; Gupta et al., 2010; Luque‐Almagro et al., 2018). Additionally, as mentioned above, the nitrilase NitC is essential for cyanide assimilation in P. pseudoalcaligenes CECT 5344 (Estepa et al., 2012).
FIGURE 2.
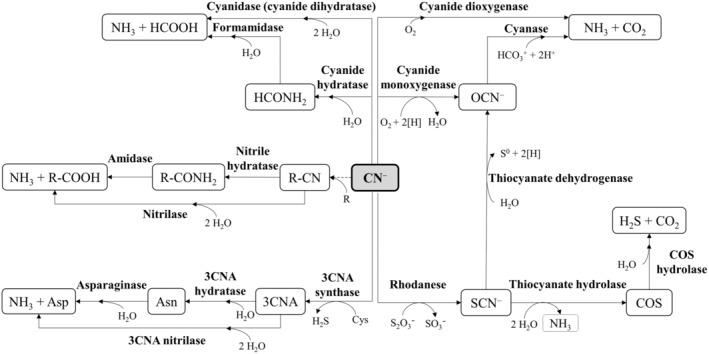
Enzymatic cyanide degradation pathways. 3CNA, 3‐cyanoalanine. Dashed arrows indicate the formation of nitriles (R‐CN) by non‐enzymatic reactions between cyanide and keto acids (R).
Bacterial degradation of free cyanide requires an alkaline pH to avoid volatilization of cyanhydric acid (pKa 9.2), and this may be a critical factor when growing with cyanide as the sole nitrogen source (Luque‐Almagro et al., 2005). However, most cyanotrophic bacteria described so far assimilate cyanide at neutral pH, and only a very limited number of bacteria degrade cyanide under alkaline conditions, such as P. pseudoalcaligenes CECT5344 (Luque‐Almagro et al., 2005), Bacillus marisflavi (Mekuto et al., 2016), Burkholderia cepacia C‐3 (Adjei & Ohta, 2000) and other unidentified isolates (Mirizadeh et al., 2014). Other factors like cyanide concentration or aeration are also important to successfully achieve the bioremediation of cyanide‐containing wastes (Karamba et al., 2017). Therefore, microorganisms used for the bioremediation of complex mixtures of chemicals like cyanide‐containing wastes must display tolerance to elevated concentrations of different metals and metalloids, and to different forms of cyanide and its derivatives, including WAD and SAD metal‐cyanide complexes, cyanate, thiocyanate and nitriles. In this sense, the alkaliphilic bacterium P. pseudoalcaligenes CECT 5344 has been described as a very suitable candidate to be used in the bioremediation of industrial cyanide‐containing wastes (Cabello et al., 2018; Roldán et al., 2021) since it tolerates elevated concentrations of cyanide (up to 30 mM), and it may also grow with cyanide derivatives like cyanate, 3‐cyanoalanine and metal‐cyanide complexes with iron or copper, among others (Luque‐Almagro et al., 2005). In a bioreactor, the strain CECT 5344 was able to detoxify a jewellery waste containing up to 12 mM total cyanide (free and complexed to metals) (Ibáñez et al., 2017). The strain CECT 5344 can also grow in batch cultures with cyanide in the presence of high concentrations of arsenic or mercury (Biełło, Cabello, et al., 2023, Biełło, Olaya‐Abril, et al., 2023). Other bacteria that can degrade metal‐cyanide complexes, including those with zinc, nickel, copper, silver and iron, have been also described (Dimitrova et al., 2020; Patil & Paknikar, 2000; Silva‐Avalos et al., 1990). Although bacteria may degrade cyanide in the presence of metals, in some cases an inhibition of cyanide assimilation was observed. Thus, cyanide degradation is significantly hampered by Cd2+ and Cu2+ in Pseudomonas putida PhCN (Deeb & Altalhi, 2009), and Hg2+ severely affects this process in Serratia marcescens (Karamba et al., 2014). Some studies have described a natural attenuation potential of free cyanide in gold cyanidation tailings based on microbial activity, although under these conditions the cyanide degrading potential of cyanotrophic strains is limited due to the high concentrations of toxic metals as co‐contaminants (Anning et al., 2021; Oudjehani et al., 2001).
Bacterial arsenic detoxification
Arsenic is an abundant natural metalloid that can be found in different oxidation states ranging from −3 (arsine), 0 (arsenic), +3 (arsenite) to +5 (arsenate). All these forms of arsenic display different mobility and toxicity, although it is usually found in the environment as arsenate or arsenite (Kabiraj et al., 2022; Raju, 2022). Inorganic forms of arsenic are present in nature as sulphur‐containing minerals like arsenopyrite (FeAsS), but organic arsenical compounds like methylarsenates are also found. The anthropogenic sources of arsenic are derived from gold mining activities, pesticide production and hydrometallurgical smelter dust treatment to recover mainly copper, gold and lead (Oremland & Stolz, 2003; Sher & Rehman, 2019). Arsenic is a highly toxic element that has been considered a human carcinogen and a major inorganic pollutant that causes an elevated cellular toxicity (Martínez et al., 2011). Arsenic toxicity relies on the capability of arsenite to inactivate a wide variety of enzymes by its binding to sulfhydryl groups and, also on the capability of arsenate to replace phosphate because it displays structural homology to arsenic (Tchounwou et al., 2012). The most toxic forms of arsenic are arsenite and methylarsenates (Petrick et al., 2000). Despite arsenic toxicity, this element has been described to play important roles in the cell, such as in the metabolism of methionine, gene silencing and interaction with the micronutrient selenium (Uthus & Brown‐Borg, 2003).
Arsenic may enter the cells through unspecific mechanisms, such as inorganic phosphate uptake systems as in the case of arsenate because of its structural similarity to phosphate, or through the aquaglyceroporins GlpF of E. coli or AqpS of Sinorhizobium meliloti as for arsenite (Garbinski et al., 2019; Mukhopadhyay et al., 2014). Numerous microorganisms can oxidize arsenite to arsenate, thus providing electrons for oxygen or nitrate respiration in aerobic (AioAB system) or anaerobic (ArxA system) conditions (Mazumder et al., 2020). Bacterial strategies for arsenic resistance include arsenic extrusion from the cytosol (Barral‐Fraga et al., 2018; Biełło, Cabello, et al., 2023; Garbinski et al., 2019), intracellular chelation by metal‐binding peptides (Morelli et al., 2005), bioaccumulation (Diba et al., 2021), immobilization and transformation to potentially less toxic organic forms (Yang & Rosen, 2016) (Figure 3). Arsenite is usually extruded outside the cell through efflux pumps. Additionally, methylated forms of arsenic could be lost by volatilization (Dombrowski et al., 2005). The ars operons are widely distributed among prokaryotes, conferring a mechanism to resist and detoxify arsenic. The arsRB genes encode the transcriptional regulator ArsR that senses arsenite and the energy‐dependent efflux pump ArsB (Rawle et al., 2021; Yang & Rosen, 2016). Additional genes usually present in the ars gene clusters contribute to an increase in the resistance to different forms of arsenic. Thus, accessory genes found in the ars clusters encode the glutaredoxin‐ or thioredoxin‐dependent arsenate reductase ArsC that converts arsenate into arsenite, the ATPase ArsA that improves the function of the ArsB efflux pump, the putative As(III)‐metallochaperone ArsD that transfers arsenite to ArsA, the transmembrane arsenic‐efflux protein Acr3, the S‐adenosylmethionine dependent methyltransferase ArsM, the methylarsenite oxidase/arsenic resistance protein ArsH, the permease ArsP and the putative C‐As lyase ArsI (Andres & Bertin, 2016; Slyemi & Bonnefoy, 2012). The major facilitator superfamily (MFS) permease ArsJ that extrudes 1‐arseno‐3‐phosphoglycerate formed by glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH), and the ArsT and ArsN proteins of unknown function are also encoded by ars gene clusters (Biełło, Cabello, et al., 2023; Chen et al., 2016; Huang et al., 2018) (Figure 3).
FIGURE 3.
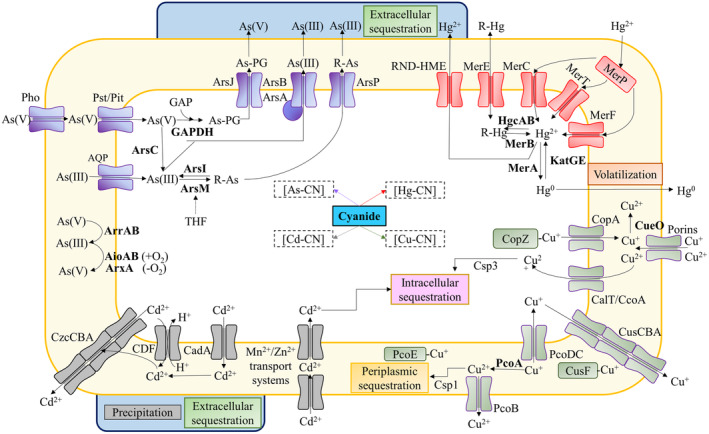
Mechanisms of arsenic, mercury, copper and cadmium resistance and detoxification in bacteria and possible interactions with cyanide in complex wastes. The main transport systems and enzymatic reactions involved in the resistance to As, Hg, Cu and Cd are shown. The enzymes are indicated in bold. In the presence of cyanide, the formation of the different cyanide‐metal complexes (indicated as metal‐CN with squared brackets) may affect the detoxification processes. As‐PG, arseno‐phosphoglycerate; GAP, glyceraldehyde‐3‐phosphate; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; R‐As, R‐Hg: methylated forms of As or Hg; THF, tetrahydrofolate. See the text for further details.
Besides, the cofactor tetrahydrofolate was associated with the mechanism of tolerance to arsenic (Abuawad et al., 2021; Bae et al., 2021). This relationship could be directly linked to the requirement of this cofactor to achieve arsenic methylation via ArsM. Additionally, this process might induce the recycling of S‐adenosylmethione dependent proteins like NitD, which is encoded by a gene of the nit1C cluster of P. pseudoalcaligenes CECT 5344 that is essential for cyanide detoxification/assimilation in this bacterium (Biełło, Cabello, et al., 2023; Estepa et al., 2012). A quantitative proteomic analysis carried out to study the mechanisms of tolerance and resistance to arsenite, in the presence or absence of cyanide, has revealed that in this strain the mechanisms of resistance to arsenic include the extrusion of arsenite through the ArsAB efflux pump and the production of arsenical derivatives, such as arseno‐phosphoglycerate formed by an arsenic inducible glyceraldehyde‐3‐phosphate dehydrogenase, or methylarsenite formed by ArsM‐type methyltransferases, which are further exported through ArsJ, ArsP and Acr3 permeases. Extruded arsenite may be accumulated in the bacterial biofilm (Biełło, Cabello, et al., 2023), as it has been also recently reported in other microorganisms (Barral‐Fraga et al., 2018; Maity et al., 2022; Mathivanan et al., 2021) (Figure 3). In the proteomic analysis of P. pseudoalcaligenes CECT 5344 it was also observed that not only the response to arsenite was different depending on the nitrogen source, ammonium or cyanide, but also that the presence of cyanide or arsenite, independently, induces proteins related to oxidative stress, biofilm formation, cyanide metabolism and arsenic metabolism (Biełło, Cabello, et al., 2023). Therefore, in the detoxification of a complex waste containing cyanide and arsenic, a certain degree of interrelation between these two chemicals may occur. In this sense, the presence of arsenite causes a down‐regulation of the CioAB proteins involved in cyanide‐insensitive respiration (Biełło, Cabello, et al., 2023).
Bacterial mercury detoxification
Mercury is a heavy metal that is considered one of the most toxic elements for human health, it is present in more than 90 minerals, such as cinnabar (HgS), livingstonite (HgSb4S8) and corderoite (Hg3S2Cl2), among others (Beckers & Rinklebe, 2017). Mercury forms strong complexes with inorganic ligands and also interacts either with metals like gold [Au(Hg)2−] and silver [Ag(Hg)2] or with cyanide, producing mercury‐cyanide complexes (Drace et al., 2016; Gworek et al., 2020; Tulasi et al., 2021). Organomercurials, such as methyl‐ and phenyl‐mercury, are also naturally formed (Priyadarshanee et al., 2022). Mercury is released into the environment through natural soil and rock erosion and anthropogenic activities like mining and fuel combustion, but communities that use artisanal small‐scale gold mining are the main producers of large amounts of toxic wastes that contain elevated concentrations of mercury (Aghaei et al., 2019; Bridges & Zalups, 2017; Priyadarshanee et al., 2022). Elemental mercury (Hg0) is volatile, resulting in highly poisonous because it can be inhaled easily (González‐Raymant et al., 2017; Priyadarshanee et al., 2022). Mercury salts, such as mercury chloride, display acute toxicity when they are ingested (Rand & Caito, 2019). Elemental mercury pollution is potentiated by the wind effect for up to 2 years after its releasement and before it is deposited as bivalent mercury (Holzman, 2010). Additionally, organomercurials are considered very potent toxins that threaten human health and ecosystems. Methylmercury causes severe damage at the cellular level, such as mitochondrial dysfunction and oxidative stress because of its affinity to sulfhydryl groups (Zhao et al., 2021). Methylmercury bioaccumulates and biomagnifies within the food chain, displaying a half‐life in the human body of about 44 days and constituting a powerful neurotoxin that crosses the blood–brain barrier, accumulates in brain, kidney and liver, and easily passes through the placenta, affecting the fetus (Bjørklund et al., 2019; Priyadarshanee et al., 2022). Elevated mercury toxicity provokes a broad range of severe diseases, such as Minamata disease, acrodynia, attention deficit disorder and hyperactivity (Kim et al., 2013).
Microorganisms play a major role in the mercury cycle in nature by reduction, oxidation, bioaccumulation and sequestration through precipitation or biosorption processes (Priyadarshanee et al., 2022; Si et al., 2022). The katG and katE genes code for hydroperoxidases that oxidize elemental mercury to bivalent mercury in a wide range of bacterial genera like Escherichia coli, Bacillus and Streptomyces, among others (Colombo et al., 2014). Additionally, mercury methylation is frequent in anaerobic environments by sulphate‐reducing and dissimilatory iron‐reducing bacteria (Gilmour et al., 2011; Podar et al., 2015). Extracellular mercury sequestration requires acidic extracellular pH, but intracellular sequestration is also possible (Gupta & Diwan, 2017; Nurfitriani et al., 2020). Mercury resistance may be driven by efflux pumps of the resistance‐nodulation‐cell division superfamily (RND‐HME). The RND‐based efflux systems are composed of protein complexes that span the whole cell wall of Gram‐negative bacteria, and they include heavy metal extruder (HME) members, among others. However, the best‐characterized mercury resistance mechanism is encoded by the mer operon, which encodes enzymes that transform mercurials into less toxic or volatile forms (Biełło, Olaya‐Abril, et al., 2023; Sone et al., 2013) (Figure 3). Two types of mer gene clusters have been described, showing a narrow spectrum if they only act on inorganic mercury, or a broad spectrum if they are able to use both inorganic and organomercurial forms (Mergeay et al., 2003; Osborn et al., 1997). The Mer system is usually composed of the transcriptional repressor MerR that senses the bivalent form of mercury, the MerD protein that may have a co‐regulatory function helping in the interaction of MerR with mercury, the MerPT mercury transport system, in which MerP binds mercury through two cysteines and, then it transfers directly to four cysteine residues of MerT, facilitating the entrance of mercury at the active side of the cytosolic NADPH‐dependent mercuric reductase MerA that converts Hg2+ into the volatile Hg0 (Champier et al., 2004; Mathema et al., 2011; Sone et al., 2013). MerT may be also responsible for the uptake of organomercurials (Mathema et al., 2011; Zheng et al., 2018). Additionally, other proteins can be present in the Mer systems like the organomercury lyase MerB that cleavages C‐Hg bonds, the methylmercury transporter MerE, the alternative mercury transporter MerH and the specific permease of phenylmercury MerG (Amin et al., 2019; Naguib et al., 2018; Priyadarshanee et al., 2022) (Figure 3).
In the presence of cyanide and because of its affinity to mercury, mercury‐cyanide complexes are produced. Interestingly, mercury tolerance in the cyanotrophic bacterium P. pseudoalcaligenes CECT 5344 is higher in the presence of cyanide than when ammonium is used as the sole nitrogen source (Biełło, Olaya‐Abril, et al., 2023). Therefore, the chemical formation of mercury‐cyanide complexes could contribute to decrease the toxicity displayed by both free cyanide and mercury. A quantitative proteomic analysis has been also applied in P. pseudoalcaligenes CECT 5344 to study mercury tolerance in the presence of cyanide (Biełło, Olaya‐Abril, et al., 2023). This study revealed the induction of a glutathione S‐transferase, which is possibly related to the cellular mechanism of mercury detoxification (Bjørklund et al., 2019). Other proteins that could be related to the response to oxidative stress triggered by mercury that was overrepresented in the proteome of the strain CECT5344, grown with mercury and cyanide, included the thiol‐dependent antioxidant protein alkylhydroperoxide reductase AhpC, the thiol‐based redox sensor RegB, the ABC‐type sulfonate transporter and an alkanesulfonate monooxygenase that catalyses the conversion of sulfonates into sulphite and the corresponding aldehyde. In this sense, it has been described that bivalent mercury can react with sulfonates, and in fact, in human intoxication by mercury, a treatment based on oral administration of dimercaptopropane sulfonate is recommended (Bjørklund et al., 2019). Additionally, the dihydropyrimidine dehydrogenase and the dihydropyrimidinase were over‐represented in presence of mercury, as well as the uracil‐xanthine permease and the subunits of the ribonucleoside‐diphosphate reductase, which catalyses the reduction of ribonucleotides into their corresponding deoxyribonucleotides, and its activity relies on glutaredoxins and thioredoxins. This could be related to the well‐described interaction of mercury with pyrimidine nucleosides and nucleotides (Onyido et al., 2004). Mercury also binds to metallic cofactors that are present in enzymes that are essential for cell survival, thus provoking their down‐representation in the proteome of P. pseudoalcaligenes CECT 5344 in the presence of mercury and cyanide. Among these proteins are non‐heme iron dioxygenases, c‐type cytochromes, cytochrome c‐containing proteins, nickel/copper‐dependent superoxide dismutase and heme/Fe‐S bacterioferritin (Biełło, Olaya‐Abril, et al., 2023). The transcriptional regulator MerR2 of P. pseudoalcaligenes CECT 5344 has been proposed as a master regulator that controls the expression of other merR genes like merR5 and merR6, which code for MerR‐type transcriptional regulators, and different structural and regulatory genes (Biełło, Olaya‐Abril, et al., 2023). Curiously, the arsR regulatory gene was also a target of MerR2, and although arsenical compounds were not present in the culture media of the strain CECT 5344 in this proteomic study, several proteins encoded by the ars genes like the thioredoxin‐dependent arsenate reductase ArsC3, the transcriptional regulator ArsR and the arsenical resistance protein ArsH2, were over‐represented in the proteome of the strain CECT 5344 grown with cyanide and mercury. Therefore, a possible interconnection between these two toxic elements, mercury and arsenic, has been postulated, thus highlighting the great biotechnological potential of P. pseudoalcaligenes CECT 5344 for detoxification of mining wastes that are composed of complex mixtures, containing highly toxic compounds like mercury, arsenic and cyanide (Biełło, Olaya‐Abril, et al., 2023).
Microbial remediation of cadmium
Cadmium is a heavy metal widely distributed in nature that is also considered one of the most toxic pollutants persistent in soils. This metal is usually found as a bivalent form, and this ion displays high mobility in aqueous solutions, humic acids and dissolved organic matter (Kubier et al., 2019). In soils used for agricultural purposes, the main sources of cadmium are derived from phosphorus fertilization (Molina‐Roco et al., 2018). Other anthropogenic sources of cadmium are mining, metallurgy, pigment/plastic stabilizers and manufactures of nickel–cadmium batteries (Khan et al., 2022). Cadmium provokes at the cellular level generation of reactive oxygen species, causing DNA damage. In humans, cadmium exposure has been associated with tumoral processes in different organs. Cadmium may enter the cells via manganese, magnesium and zinc transport systems like other heavy metals, which exploit the uptake systems of essential metal ions (Khan et al., 2022).
Many microorganisms have evolved to deal with cadmium toxicity either through efflux transport systems or biosorption. Elevated intracellular cadmium concentrations lead to the induction of efflux systems to achieve its elimination and to maintain homeostasis (Khan et al., 2022). Three different efflux systems could play this function, including P‐type ATPases, cation diffusion facilitators (CDF) and RND systems (Figure 3). The cadmium‐resistance cadAC genes encode the P‐type ATPase CadA, which transports bivalent cadmium from the cytoplasm to the periplasm, and the positive transcriptional regulator CadC (also named CadR), which belongs to the MerR family of transcriptional regulators. In this sense, synthetic biology allowed the development and optimization of a biosensor for the detection of the bivalent cadmium and mercury forms (Guo et al., 2021; Tao et al., 2013). Additional cadB and cadD genes are present in some cad operons, which code for components of the transport system involved in the binding of cadmium to the plasma membrane (Hsieh et al., 2010; Zhang et al., 2015). CDF systems, acting as chemiosmotic antiporters, co‐transport cadmium to the periplasm and protons to the cytoplasm, but like CadA transporters they are unable to extrude metals from the periplasm to the extracellular space. In Pseudomonas, the expression of cadA gene stimulates the rapid induction of the CzcCBA efflux system, which is an RND‐type cation extruder that functions not only for cadmium, but also for bivalent zinc and cobalt (Ducret et al., 2020; Liu et al., 2021). CzcA is the inner membrane component (RND protein) while CzcC is the outer membrane factor (OMF) that spans from the periplasm to the extracellular space. CzcB is the membrane fusion protein (MFP) mainly located in the periplasm, which establishes the attachment between CzcA and CzcC proteins. The CzcCBA system, which is mainly driven by proton motive force, is the only one that may extrude cadmium from the periplasm to the extracellular space (Figure 3). In P. aeruginosa, the expression of czcCBA genes is controlled by the CzcS‐CzcR two‐component system. Thus, the inner membrane‐located sensor CzcS detects elevated levels of periplasmic cadmium or zinc, and activates the response regulator CzcR, allowing expression of the czcCBA genes. Curiously, it has been demonstrated that this regulatory system also regulates the expression of the OprD porin, leading to the resistance to the antibiotic carbapenem (Ducret et al., 2020; Perron et al., 2004).
Other mechanisms of resistance to cadmium are based on either intracellular sequestration, via cadmium binding to metallothioneins or extracellular accumulation in bacterial biofilms (Blindauer, 2011; Naz et al., 2005; White & Gadd, 1998). Also, some microorganisms may induce cadmium precipitation via phosphates, carbonates or sulphides to produce otavite (CdCO3), CdS granules or nanoparticles, which can be accumulated further within biofilms or extracellular polymeric substances (Newsome & Falagán, 2021; Sakpirom et al., 2019).
Bioremediation of cadmium by using different bacteria has been achieved successfully. Thus, cadmium‐resistant strains of E. coli and Klebsiella pneumoniae have been described to detoxify elevated concentrations of this metal. Additionally, it has been reported the biosorption of cadmium by Salmonella enterica and Bacillus cereus (Khan et al., 2022).
In the presence of cyanide, weak cadmium‐cyanide complexes may be formed, and this could contribute to lowering cyanide toxicity, as recently described for mercury and cyanide when they are present simultaneously in the culture media of P. pseudoalcaligenes CECT 5344 (Biełło, Olaya‐Abril, et al., 2023).
Copper bioremediation by bacteria
Copper is an essential transition metal that, at elevated concentrations, results in toxic and highly bioaccumulative (Almeida‐Rodrigues et al., 2022). Copper is found in natural minerals, such as chalcopyrite (CuFeS2) and malachite [Cu2CO3(OH2)]. Although copper is released from volcanic eruptions and forest fires, among others, anthropogenic activities like mining and agricultural practices, including the use of pesticides and fungicides, are the main sources of the large amounts of copper that may threaten the ecosystems and human health. Copper attacks intracellular iron–sulphur centres of various proteins under anoxic conditions, while in aerobiosis catalyses Fenton‐like reactions, thus provoking the generation of reactive oxygen species that cause lipid peroxidation, and DNA and protein damage (Andrei et al., 2020; Fedel‐Moen et al., 2000; Maertens et al., 2021; Scheiber et al., 2013). Thus, in Staphylococcus aureus, enzymes related to oxidative stress response and central carbon metabolism were induced in the presence of copper sulphate (Tarrant et al., 2019). Copper can enter the cells through different types of transporters. Monovalent and bivalent forms of copper can pass to the periplasm through porins, and then, they may be transported to the cytoplasm via PcoDC or CalT/CcoA systems, respectively (Figure 3). The CalT/CcoA system is an MFS‐type Cu(II) transporter that is required for an active cbb3‐type cytochrome oxidase in Rhodobacter capsulatus (Ekici et al., 2014). PcoC and PcoD are membrane proteins located in the inner membrane that participate in the import of monovalent Cu from the periplasm to the cytoplasm (Argüello et al., 2013). Additionally, PcoA and CueO are copper oxidases that convert Cu(I) into Cu(II) in the periplasm, and bivalent copper can be extruded further into the extracellular space via PcoB. In contrast, ABC‐type ATP‐dependent copper transporters have been described to import copper inside the cells. In the soil denitrifier, Paracoccus denitrificans PD1222, an ABC‐type copper transporter has been described to be relevant for the activity of the copper‐dependent nitrous oxide reductase, the last enzyme of the denitrification process, with a key role in emission of the potent greenhouse gas nitrous oxide (Olaya‐Abril et al., 2018).
Hyperaccumulation of copper may affect primary cellular functions. In E. coli and Bacillus subtilis, it has been described that this condition leads to an increase in iron and sulphur acquisition related to the decrease of iron–sulphur cluster stability, both during their biogenesis and when they are bound as cofactors in proteins. Additionally, elevated concentrations of copper are balanced by homeostasis through different resistance mechanisms to this metal (Andrei et al., 2020; Delmar et al., 2015; Dupont et al., 2011). Detoxification of cytoplasmic copper is carried out by P‐type ATPases that pump out monovalent copper from the cytoplasm to the periplasm. In E. coli, the P‐type ATPase CopA, which is controlled by the MerR‐family transcriptional regulator CueR, allows only very low concentrations of copper inside the cell. Periplasmic copper is extruded by the RND‐HME family member CusCBA, which is driven by proton motive force. CusA is the inner‐membrane RND protein that together with the periplasmic CusB and the outer‐membrane CusC transports copper to the extracellular media (Andrei et al., 2020; Delmar et al., 2015). The CusCBA extruder system is regulated by the two‐component regulatory system CusSR. Other bacterial Cus proteins also include an azurin‐like blue copper protein and the periplasmic copper chaperone CusF (Figure 3).
In addition, alternative mechanisms of copper detoxification have been deciphered, such as compartmentalization of bivalent copper mediated by cupric reductases, periplasmic sequestration through copper‐binding proteins like cupreredoxins, or accumulation of copper in the cytoplasm mediated by granules of polyphosphate or peptide‐based chalkophores (Andrei et al., 2020; Li et al., 2023; Remonsellez et al., 2006; Vargas‐Straube et al., 2020). Likewise, it has been demonstrated that PrtA and Csp proteins are involved in the storage of elevated concentrations of copper, in the periplasm or the cytoplasm, in parallel with a reduction of biofilm formation by decreasing the production of extracellular polymeric substances (Vargas‐Straube et al., 2020; Virieux‐Petit et al., 2022).
Bioremediation of copper‐contaminated soils has been performed through mechanisms by which bacteria mobilize or immobilize this metal in soils, and different techniques have been applied, including bioleaching as a process for the ex situ recovery of copper from Cu‐bearing solids, bioimmobilization to limit the in situ leaching of copper into groundwater and phytoextraction assisted by bioaugmentation for in situ enhancement of copper removal from soils (Cornu et al., 2017; González‐Henao & Thaura, 2021). Copper mining activities generate acid–liquid wastes that may also contain elevated concentrations of cyanide, which could hinder the biodegradation of cyanide (Roldán et al., 2021). In this sense, it has been described that Cu2+ inhibits cyanide degradation in Pseudomonas putida PhCN (Deeb & Altalhi, 2009).
Bacterial detoxification of other metals
Other metals usually found in industrial wastewater are chromium, lead, nickel, zinc and cobalt. Microbes may deal with these pollutants through resistance and detoxification mechanisms like those described above (Figure 1). Hexavalent chromium, Cr(VI), enters into the cells via sulphate transport systems (Cervantes et al., 2001), resulting in highly toxic and carcinogenic and provoking oxidative stress responses (Rai et al., 2013). Mechanisms like extracellular accumulation (Flemming et al., 1990) and reduction to the less toxic trivalent Cr(III) form by Cr(VI) reductases, cytochromes or even by the nitroreductases.NfsA/NfsB, have been described for Cr(VI) resistance (Ramírez‐Díaz et al., 2008; Thatoi et al., 2014).
Lead, whose toxicity relies on its binding to sulfhydryl, phosphate and hydroxyl groups, usually occurs as Pb(II), which enters the cells via calcium channels (Kerper & Hinkle, 1997). Once inside the cell, lead can be expelled by ATPase efflux pumps, such as ZntA and CadA (Newsome & Falagán, 2021), or by proteins encoded by the pbr resistance operon (Wei et al., 2014). Intracellular and periplasmic accumulation of Pb through metallothioneins was also observed (Yu et al., 2023). Alternatively, strategies based on the reduction of its mobility by precipitation with phosphate or oxalic acid (Liang et al., 2016; Sharma et al., 2018) or through siderophores (Liu et al., 2023) have been also described.
Nickel uptake occurs through both specific and non‐specific pathways (Macomber & Hausinger, 2011). Systems driven by proton gradient, metal efflux ATPases or proteins like NreB, DmeF and RcnA are responsible for expelling excess of Ni(II) (Macomber & Hausinger, 2011). However, the reduction of Ni(II) and the formation of NiO nanoparticles were also described as resistance mechanisms (Sathyavathi et al., 2014; Zhan et al., 2012).
Zinc is an essential trace element, but in excess, it is harmful to organisms due to its competition with other metals for the active sites of enzymes. Zn uptake may take place through specific mechanisms, controlled by the sensor protein Zur (zinc uptake regulator) or non‐specific mechanisms, which respond to diffusion gradients. The resistance mechanisms to an excess of Zn involve both its extrusion by active ATPases, CDF or RND systems and its intra‐ or extracellular sequestration by metallothioneins (Blindauer, 2015). Additionally, exopolysaccharide (EPS) biosorption and precipitation as ZnS may occur (Newsome & Falagán, 2021).
META‐OMICS APPLIED TO DETOXIFICATION INDUSTRIAL WASTES CONTAINING CYANIDE AND HEAVY METALS
Considering that 99% of microbes present in nature are not cultivable, culture‐independent analysis like meta‐omics, which are based on omics analyses applied to the study of the complex and diverse microbial communities, have been applied in the last decades to investigate biodegradation of environmental hazards (Birrer et al., 2019; Hemmat‐Jou et al., 2021; Kantor et al., 2015; Lloyd et al., 2018; Sonthiphand et al., 2019). However, omic analyses applied to bioremediation of cyanide‐contaminated environments like mining areas, which also contain elevated concentrations of metals, are remarkably scarce. Most of these meta‐omic approaches have been focused on the study of acid mine drainage (AMD) ecosystems, which are generated in mining of gold, copper and nickel ores, but they have not been applied to bioremediation purposes. In addition to their low pH, AMD environments are characterized by the presence of high concentrations of heavy metals, arsenic and sulphates. Furthermore, the cyanidation process used in gold mining for gold extraction from the ores, can also contribute to the presence of cyanide in these acid drainages. In the last decades, culture‐independent approaches have revealed the diversity and metabolic potentials and activities of AMD communities (Bertin et al., 2011; Kantor et al., 2015; López‐Pérez et al., 2021; Munyai et al., 2021; Reis et al., 2016), although these studies did not allow the identification of genes and proteins specifically involved in the resistance and/or the assimilation/detoxification of cyanide. A direct search for cyanide degradation genes/proteins in the current available meta‐omic databases could provide information about the presence and distribution of cyanide‐degrading microorganisms in AMD ecosystems. In addition, the identification of genes involved in siderophore production could also be an indicative of the microbial utilization of metal‐cyanide complexes in these environments.
Metaproteomic studies on sites polluted with cyanide and metals have not been performed up to date, and in the case of metals, proteomic analyses were focused only on sentinel organisms (Abril et al., 2015; Michán et al., 2019). Probably this may be related to the limitations of protein extraction from microorganisms in natural environments, based on: (i) there are no standard and reliable protocols (Bastida et al., 2014; Starke et al., 2019), and those already developed present low extraction yields and low overlap (Leary et al., 2014), (ii) the theoretically high microbial diversity of the samples, although in contaminated environments it is expected to be lower than in non‐polluted areas, (iii) the temporal and spatial variations throughout the different seasons and in the different microenvironments, (iv) the presence of inhibitory substances like humic acids or clays (Bouchez et al., 2016) and (v) the lack of curated and annotated reference databases. Some of these reasons might influence on the experimental design, as temporal/spatial variations that require sampling several times per year using grid systems. Likewise, protocols for the extraction of proteins require optimization, including a reliable post‐alkaline protein extraction protocol to achieve protein isolation from environments contaminated with metals (Herruzo‐Ruiz et al., 2021). Nevertheless, the low overlap derived from the different extraction methods requires samples extracted by using different protocols, making them a pool for each replica. Regarding to the lack of reference databases, proteogenomic approaches are a good alternative (Nesvizhskii, 2014; Ruggles et al., 2017). For this, de novo transcriptomes could be constructed to apply them, after appropriate computational analysis, as databases for proteomics (Amil‐Ruiz et al., 2021). Furthermore, also considering the limitations of proteomic and meta‐proteomic techniques (Kunath et al., 2019), the use of cutting‐edge methodologies as data‐independent acquisition (DIA) mode, the construction of in‐silico libraries (Rosenberger et al., 2017) and the application of deep learning for proteomics (Distler et al., 2014; Wen et al., 2020) could provide essential information for the development of bioremediation strategies based on proteo‐genomics. In any case, there is an urgent need to develop meta‐omic analyses on sites contaminated by cyanide, arsenic and metals to obtain a global and holistic view of the processes triggered by the simultaneous presence of these toxic co‐pollutants that are present in wastewaters generated in mining and other industrial activities. In this sense, a recent study that combines geochemical and meta‐genomic techniques has revealed new insights into natural cyanide biodegradation in a gold tailings environment contaminated with cyanide, identifying indigenous bacteria that induce their genomic machinery for hydrolytic cyanide degradation in the absence of other nitrogen sources, thus representing the first barrier to detoxify cyanide in mining tailings (Welman‐Purchase et al., 2024). Meta‐omics could enrich current knowledge about the mechanisms used by living organisms against environmental contaminants, and therefore, systems and synthetic biology could be applied to develop and optimize bioremediation processes (Dvořák et al., 2017; Roldán et al., 2021).
SYSTEMS BIOLOGY AND SYNTHETIC BIOLOGY‐BASED BIOREMEDIATION
The design of efficient biodegradation processes to detoxify industrial residues containing complex mixtures, such as cyanide‐ and metal‐containing wastes, requires a deep knowledge of the mechanisms displayed by bacteria to tolerate and degrade these pollutants, and their regulatory links, followed by integration on global microbial metabolism. Therefore, it is necessary not only to consider the specific mechanisms conferring resistance and capacity to detoxify cyanide, arsenic and metals, but also those processes required to deal with the metabolic burden, and the changes in metabolic fluxes as a consequence of the presence of these toxic pollutants. In this sense, systems biology may provide the necessary molecular tools and the global knowledge required for the design, construction and optimization of bioremediation strategies, both in vivo and ex vivo, mainly through genetic engineering of model organisms (i.e, E. coli) or new isolated bacterial strains or consortia able to degrade different pollutants (Figure 4). Thus, systems biology may allow the development of specific, sustainable and efficient methods to detect and bioremediate cyanide and heavy metals. Application of synthetic biology to this purpose is still in its infancy, and although significant advances have been addressed on heavy metals (recently reviewed by Thai et al., 2023), some relevant issues, such as ethical concerns, biosafety and legal regulations, must be overcome.
FIGURE 4.
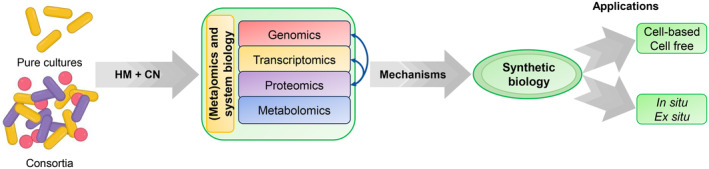
Application of (meta)omics and synthetic biology for cyanide and heavy metal bioremediation.
The first step in the implementation of a bioremediation process is sensing and monitoring the presence of pollutants in the environment. Synthetic biology directed to heavy metal detection has allowed the development of different biosensors, but currently, there are limited examples of the use of synthetic biology for metal bioremediation due to the ethical and legal implications of the utilization and release of genetically modified organisms, and because many of the developed toolkits are relatively new (Capeness & Horsfall, 2020; Somayaji et al., 2022; Thai et al., 2023). Biosensors are multi‐component and modular devices with some biological circuits that include, at least, a sensing element that detects the analyte (i.e. a metal‐binding protein), a signal processing module that also amplifies and tunes the signal, and an output system that allows the measurement and quantitation of the signal, usually based on fluorescence, bioluminescence, colourimetric, electrical or chemotactic detection (Kim et al., 2018; Singh & Kumar, 2021; Thai et al., 2023). Efficient biosensors have been developed for arsenic, copper and other heavy metals (Fan et al., 2022; Roggo & van der Meer, 2017; Thai et al., 2023). Cell‐free biosensors use some biological components like enzymes, proteins, peptides, antibiotics or nucleic acids as elements or modules. However, synthetic biology has been mainly focused to the development of whole‐cell biosensors, based either on engineered transcriptional metalloregulators like ArsR, MerR, CadR, CueR or ZntR (for detection of As, Hg, Cd, Cu or Zn, respectively), or on proteins acting as sensor kinase and response regulator in different two‐component regulatory systems, such as CusSR (for Cu), ZraSR (for Zn) or even the chemotaxis CheAY proteins adapted to detect different metals (Table 3). Synthetic biology approaches applied to metal bioremediation are mainly based on intracellular or extracellular sequestration mechanisms (Table 3). Metals may be trapped, without entering the cells, by binding to the carboxyl, amino, phosphate or hydroxyl groups naturally present on the bacterial cell surface, but synthetic peptides (usually rich in Cys or His) or engineered outer membrane proteins like OmpA, OmpC and LamB (Gram‐negative bacteria), staphylococcal protein SpA, Bacillus surface spore CotB protein (Gram‐positive bacteria) and fimbriae/flagella proteins, are frequently used for metal attachment to the cell envelop. Alternatively, extracellular sequestration may be achieved by metal binding to exopolysaccharides and cell biofilm or extracellular nanofibers (curli) that adsorb metals like mercury. On the contrary, intracellular sequestration requires the transport of the metal into the cells by different uptake systems, and its further accumulation bound to metalloregulatory proteins (AsrR, MerR), or native or engineered Cys‐rich metal‐binding proteins like bacterial metallothioneins or plant phytochelatins. This bioaccumulation usually presents solubility problems, and therefore, co‐expression with glutathione S‐transferase or other tags may increase metal accumulation. Also, some studies have applied other alternative methods like micro‐ or nano‐compartmentation, encapsulation using bacterial encapsulins to form proteinaceous shells, formation of nanoparticles, precipitation with sulphates and phosphates and bioconversion, mainly by reduction mediated by reductases, cytochromes or glutathione (Capeness & Horsfall, 2020; Somayaji et al., 2022; Thai et al., 2023). Synthetic biology can be also used to design dual systems that allow both biosensing and bioremediation of heavy metals (Table 3). Thus, a system for detecting and remediation of cadmium based on the cadR promoter of P. putida, the green fluorescent protein GFP and a fusion protein with the Cd‐binding domain derived from CadR and Lpp‐OmpA protein for membrane attachment, has been recently constructed (Guo et al., 2021).
TABLE 3.
Application of synthetic biology approaches to sensing and removal of heavy metals.
| Applications | Remarks and technical approaches |
|---|---|
| Heavy metal sensing | |
| Cell‐free biosensors | Use of some biocomponents (enzymes, peptides, proteins, antibiotics, nucleic acids) for sensing. |
| Whole‐cell biosensors | Sensing by molecular‐engineered microbial cells. |
| Transcriptional regulators | Based on regulatory proteins like ArsR (As), MerR (Hg), CadR (Cd), CueR (Cu) and ZntR (Zn). |
| Two‐component systems | Based on two‐component regulatory systems like CusSR (Cu), ZraSR (Zn) and CheAY (Cd and other metals) proteins. |
| Heavy metal remediation | |
| Sequestration | Biosorption (chelation/complexation process) and/or bioaccumulation (metabolically active process) as main remediation mechanism. |
| Extracellular | Trapping metal on the cell surface without entering the cells. |
| Functional groups | Metal binding to carboxyl, amino, phosphate or hydroxyl groups in the cell membrane. |
| Peptides and proteins | Metal binding to synthetic peptides rich in Cys or His, engineered outer membrane proteins (OmpA, OmpC, LamB, SpA, CotB) or fimbriae/flagella proteins. |
| Extracellular polymers | Metal binding to exopolysaccharides (biofilm) or extracellular adsorbent nanofibers (curli). |
| Intracellular | Metal attached to native or engineered peptides or proteins after its transport into the cells by uptake systems. |
| Regulatory proteins | Binding to regulatory proteins like ArsR (As), MerR (Hg) or other metalloregulators. |
| Chelating proteins | Binding to Cys‐rich proteins like microbial metallothioneins or plant phytochelatins. Also, co‐expression with a soluble fusion tag like glutathione S‐transferase to increase solubility. |
| Compartmentation/encapsulation | Accumulation in artificial organelles and micro‐ or nano‐compartments, and encapsulation (bacterial encapsulins) to form proteinaceous shells. |
| Precipitation | Formation of nanoparticles and precipitation with sulphates and phosphates. |
| Bioconversion | Metal reduction by reductases (MerA for Hg2+), cytochromes and other proteins or by glutathione. |
| Dual sensing and remediation | Systems designed for both metal biosensing and bioremediation, mainly through extracellular sequestration. |
Conversely, omic techniques have been hardly applied to cyanide biodegradation up to date, and the application of systems and synthetic biology to cyanide bioremediation is at present merely an attractive proposal. Holistic analyses based on transcriptomics (Luque‐Almagro et al., 2015), regulation through small RNAs (Olaya‐Abril et al., 2019), and proteomics (Ibáñez et al., 2017; Olaya‐Abril et al., 2020; Pérez et al., 2021) of the degradation of sodium cyanide and/or cyanide‐containing jewellery residues have been only performed in P. pseudoalcaligenes CECT 5344, the first cyanide‐assimilating bacterium whose genome was completely sequenced (Luque‐Almagro et al., 2013; Wibberg et al., 2016). Systems and synthetic biology approaches could allow the improvement of the biodegradative capacities of this bacterial strain (Roldán et al., 2021).
The implementation of biodegradation processes using natural samples displays some difficulties, and strategies based on natural or artificial microbial consortia could be an alternative more effective than approaches based on the isolation, purification and characterization of individual species (Massot et al., 2022), as recently reported for bacterial degradation of synthetic plastics (Skariyachan et al., 2021). Therefore, the application of bioaugmented samples, composed of natural or artificial consortia, to bioremediate environmental hazards may allow a successful completion of the discovery phase. In fact, it was observed that culture‐dependent or ‐independent methods capture different microbial communities, but enriched in the organisms of interest, as shown in the case of arsenic‐metabolizing microorganisms (Hamood‐Altowayti et al., 2020; Ziegelhöfer & Kujala, 2021). Although this strategy has been applied successfully to achieve thiocyanate biodegradation (Kantor et al., 2015; Watts et al., 2019; Watts & Moreau, 2016), in the case of cyanide biodegradation is unknown, regardless of the presence or absence of heavy metals. Once studied the biodegradation of cyanide in complex mixtures with arsenic and heavy metals (Biełło, Cabello, et al., 2023, Biełło, Olaya‐Abril, et al., 2023), a system that increases the efficiency of biodegradation could be developed by applying synthetic biology. For this, in vivo or ex vivo strategies could be used, with their advantages and inconveniences (Roldán et al., 2021) (Figure 4). Considering the wide chemical structures of the contaminants present in the mining and metal industry wastes, including cyanide, heavy metals, arsenic and metalloids, sulphates, nitrates, among others, a possible biotechnological application may involve the design of mixed reactors, in which cyanide, metals and other pollutants could be detoxified independently. Thus, cyanide could be degraded by using either optimized organisms with greater resistance and detoxification capacities, or the enzymes necessary to carry out synthetic metabolic pathways. Metals, on the contrary, could be bioremediated through the generation of hyperaccumulative microorganisms, which may remove these toxins when growing at a large scale in bioreactors. Another alternative to reduce cyanide concentrations in industrial residues could be a pretreatment based on incubation with organic molecules that chemically react with cyanide like keto acids, among others. This approach would raise the formation of less toxic cyano‐derivatives (nitriles), and hence wastes with high concentrations of cyanide could be detoxified meanwhile renewable raw materials could be produced (Nikodinovic‐Runic et al., 2013; Roldán et al., 2021). However, the efficiency, economic costs and viability of these approaches should be evaluated, particularly for the removal of metal‐cyanide complexes and the treatment of wastes co‐contaminated with different toxic chemicals.
As mentioned above, understanding the mechanisms of resistance and detoxification of cyanide and metals is essential to develop effective bioremediation strategies. However, from complex cyanide mixtures that, in addition to cyanide, contain other very toxic pollutants like arsenic and metals, interactions within all these toxic compounds could have synergistic effects, leading to a global process that is less difficult to achieve than bioremediation of its components individually. In this sense, it has been recently described that P. pseudoalcaligenes CECT 5344 tolerates higher concentrations of mercury when growing with cyanide as a nitrogen source than with ammonium, probably because the mercury‐cyanide complexes formed are less toxic than free cyanide and mercury (Biełło, Olaya‐Abril, et al., 2023). However, more studies on different bacteria and with other metals are necessary to establish possible synergic effects between cyanide and pollutants like arsenic and heavy metals. In fact, cyanide degradation was inhibited by Cu2+ in P. putida and by Hg2+ in Serratia marcescens (Deeb & Altalhi, 2009; Karamba et al., 2014).
CONCLUDING REMARKS AND FUTURE DIRECTIONS
To develop a successful bioremediation process to remove complex mixtures of highly toxic contaminants present in cyanide‐containing industrial wastes is crucial to monitor not only the enzymes necessary for the biodegradation of cyanide in the presence of metals through in vivo bioremediation strategies, but also to learn in advance the compensatory mechanisms that allow bacterial survival under these conditions. Additionally, for the reconstruction of artificial metabolic pathways applied to ex vivo strategies, it will be necessary to define all the required components, such as cofactors involved, beyond the enzymes that participate in the different biodegradative processes. Therefore, it is necessary to apply and develop omic/meta‐omic approaches, which provide a global and holistic view of the degradative processes, regardless of the specific strategy in which they are addressed. In the future, further attention will deserve deep in situ analysis of microbial communities like those colonizing acid mine drainage sites. In this sense, the taxonomic data obtained from different acid mine drainage sites have shown an unexpectedly high microbial biodiversity. The comparative meta‐genomic and meta‐transcriptomic analyses from the community‐wide comparative analyses of the active taxa revealed that there are environment‐dependent gene transcriptional profiles (Chen et al., 2015). Another interesting approach derives from microbial community studies and extracellular substances acting as metal chelators that are produced from anaerobic granular sludges, which constitute an ideal carrier for microorganisms used for the treatment of wastewaters containing selenate, cadmium and zinc (Zeng et al., 2023). Furthermore, new omic studies from samples taken from extreme environments have contributed to elucidate linkages between community function and environmental variables, allowing identification and characterization of new lineages, which expand microbial diversity and vary the structure of the tree of life (Shu & Huang, 2022). In addition, natural biodegradation may require not only the action of microbial communities like those described in granular anaerobic treatments (Zeng et al., 2023), but also the cooperation among microbes, plants and fungi, especially to deal with edaphic ecosystem pollutants. Therefore, to provide a holistic approach to environment restauration, it is also necessary to consider the complex relationships between different potential hosts and their associated microbes, as described for the seaweed ‘holobiont’ that facilitates the bioremediation of organic pollutants and heavy metals (Ren et al., 2022).
AUTHOR CONTRIBUTIONS
Alfonso Olaya‐Abril: Conceptualization (equal); software (equal); writing – original draft (equal). Karolina Biełło: Conceptualization (equal); formal analysis (equal); writing – original draft (equal). Gema Rodríguez‐Caballero: Formal analysis (equal); writing – original draft (equal). Purificación Cabello: Formal analysis (equal); writing – original draft (equal). Lara P. Sáez: Formal analysis (equal); writing – original draft (equal). Conrado Moreno‐Vivián: Conceptualization (equal); funding acquisition (equal); writing – original draft (equal); writing – review and editing (equal). Víctor Manuel Luque‐Almagro: Formal analysis (equal); software (equal); writing – original draft (equal). María Dolores Roldán: Conceptualization (equal); funding acquisition (equal); writing – original draft (equal); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
Supporting information
Table S1. Natural or anthropogenic sources of cyanide, arsenic and heavy metals, permissible limits and concentrations in different complex wastes. The numbers in parentheses correspond to the references.
ACKNOWLEDGEMENTS
The authors thank GEMASUR, SAVECO and AVENIR for fruitful collaborations. This work is part of the projects RTI2018‐099573‐B‐100 funded by the Ministerio de Ciencia, Innovación y Universidades (Spain), also supported by FEDER (EU), Junta de Andalucía, Spain (Grant P18‐RT‐3048) and University of Córdoba (PPIT_2022E_025814).
Olaya‐Abril, A. , Biełło, K. , Rodríguez‐Caballero, G. , Cabello, P. , Sáez, L.P. , Moreno‐Vivián, C. et al. (2024) Bacterial tolerance and detoxification of cyanide, arsenic and heavy metals: Holistic approaches applied to bioremediation of industrial complex wastes. Microbial Biotechnology, 17, e14399. Available from: 10.1111/1751-7915.14399
Alfonso Olaya‐Abril and Karolina Biełło have contributed equally to this work.
REFERENCES
- Abril, N. , Chicano‐Gálvez, E. , Michán, C. , Pueyo, C. & López‐Barea, J. (2015) iTRAQ analysis of hepatic proteins in free‐living Mus spretus mice to assess the contamination status of areas surrounding Doñana National Park (SW Spain). Science of the Total Environment, 523, 16–27. Available from: 10.1016/j.scitotenv.2015.03.116 [DOI] [PubMed] [Google Scholar]
- Abuawad, A. , Spratlen, M.J. , Parvez, F. , Slavkovich, V. , Ilievski, V. , Lomax‐Luu, A.M. et al. (2021) Association between body mass index and arsenic methylation in three studies of Bangladeshi adults and adolescents. Environment International, 149, 106401. Available from: 10.1016/j.envint.2021.106401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adjei, M.D. & Ohta, Y. (2000) Factors affecting the biodegradation of cyanide by Burkholderia cepacian strain C‐3. Journal of Bioscience and Bioengineering, 89, 274–277. [DOI] [PubMed] [Google Scholar]
- Aghaei, E. , Diaz‐Alorro, R. , Tadesse, B. & Browner, R. (2019) A review on current practices and emerging technologies for sustainable management, sequestration and stabilization of mercury from gold processing streams. Journal of Environmental Management, 249, 109367. Available from: 10.1016/j.jenvman.2019.109367 [DOI] [PubMed] [Google Scholar]
- Ali, H. , Khan, E. & Hahi, I. (2019) Environmental chemistry and ecotoxicology of hazardous heavy metals: environmental persistence, toxicity, and bioaccumulation. Journal of Chemistry, 2019, 6730305. Available from: 10.1155/201976730305 [DOI] [Google Scholar]
- Almeida‐Rodrigues, P. , Gomes‐Ferrari, R. , Seiko‐Kato, L. , Hauser‐Davis, R.A. & Conte‐Junior, C.A. (2022) A systematic review on metal dynamics and marine toxicity risk assessment using crustaceans as bioindicators. Biological Trace Element Research, 200, 881–903. Available from: 10.1007/s12011-021-02685-3 [DOI] [PubMed] [Google Scholar]
- Alvillo‐Rivera, A. , Garrido‐Hoyos, S. , Buitrón, G. , Thangarasu‐Sarasvathi, P. & Rosano‐Ortega, G. (2021) Biological treatment for the degradation of cyanide: a review. Journal of Materials Research and Technology, 12, 1418–1433. Available from: 10.1016/j.jmrt.2021.03.030 [DOI] [Google Scholar]
- Amil‐Ruiz, F. , Herruzo‐Ruiz, A.M. , Fuentes‐Almagro, C. , Baena‐Angulo, C. , Jiménez‐Pastor, J.M. , Blasco, J. et al. (2021) Constructing a de novo transcriptome and a reference proteome for the bivalve Scrobicularia plana: comparative analysis of different assembly strategies and proteomic analysis. Genomics, 113, 1543–1553. Available from: 10.1016/j.ygeno.2021.03.025 [DOI] [PubMed] [Google Scholar]
- Amin, A. , Sarwar, A. , Saleem, M.A. , Latif, Z. & Opella, S.J. (2019) Expression and purification of transmembrane protein MerE from mercury‐resistant Bacillus cereus . Journal of Microbiology and Biotechnology, 29, 274–282. Available from: 10.4014/jmb.1704.04062 [DOI] [PubMed] [Google Scholar]
- Andrei, A. , Öztürk, Y. , Khalfaoui‐Hassani, B. , Rauch, J. , Marckmann, D. , Trasnea, P.I. et al. (2020) Cu homeostasis in bacteria: the ins and outs. Membranes, 10, 242. Available from: 10.3390/membranes10090242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres, J. & Bertin, P.N. (2016) The microbial genomics of arsenic. FEMS Microbiology Reviews, 40, 299–322. Available from: 10.1093/femsre/fuv050 [DOI] [PubMed] [Google Scholar]
- Anning, C. , Asare, M.O. , Junxiang, W. , Yao, G. & Xianjun, L. (2021) Effects of physicochemical properties of Au cyanidation tailings on cyanide microbial degradation. Journal of Environmental Science and Health, 56, 413–433. Available from: 10.1080/10934529.2021.1885259 [DOI] [PubMed] [Google Scholar]
- Argüello, J.M. , Raimunda, D. & Padilla‐Benavides, T. (2013) Mechanisms of copper homeostasis in bacteria. Frontiers in Cellular and Infection Microbiology, 3, 73. Available from: 10.3389/fcimb.2013.00073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae, S. , Kamynina, E. , Guetterman, H.M. , Farinola, A.F. , Caudill, M.A. , Berry, R.J. et al. (2021) Provision of folic acid for reducing arsenic toxicity in arsenic‐exposed children and adults. Cochrane Database of Systematic Reviews, 10, CD012649. Available from: 10.1002/14651858.CD012649.pub2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral‐Fraga, L. , Martina‐Prieto, D. , Barral, M.T. , Morin, S. & Guasch, H. (2018) Mutual interaction between arsenic and biofilm in a mining impacted river. Science of the Total Environment, 636, 985–998. Available from: 10.1016/j.scitotenv.2018.04.287 [DOI] [PubMed] [Google Scholar]
- Bastida, F. , Hernández, T. & García, C. (2014) Metaproteomics of soils from semiarid environment: functional and phylogenetic information obtained with different protein extraction methods. Journal of Proteomics, 101, 31–42. Available from: 10.1016/j.jprot.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Baxter, J. & Cummings, S. (2006) The current and future applications of microorganism in the bioremediation of cyanide contamination. Antonie Van Leeuwenhoek, 90, 1–17. Available from: 10.1007/s10482-006-9057-y [DOI] [PubMed] [Google Scholar]
- Bazzi, W. , Abou‐Fayad, A.G. , Nasser, A. , Haraoui, L.P. , Dewachi, O. , Abou‐Sitta, G. et al. (2020) Heavy metal toxicity in armed conflicts potentiates AMR in A. baumannii by selecting for antibiotic and heavy metal co‐resistance mechanisms. Frontiers in Microbiology, 11, 68. Available from: 10.3389/fmicb.2020.00068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckers, F. & Rinklebe, J. (2017) Cycling of mercury in the environment. Sources, fate, and human health implications: a review. Critical Reviews in Environmental Science and Technology, 47, 693–794. Available from: 10.1080/10643389.2017.1326277 [DOI] [Google Scholar]
- Bertin, P.N. , Heinrich‐Salmeron, A. , Pelletier, E. , Goulhen‐Chollet, F. , Arsène‐Ploetze, F. , Gallien, S. et al. (2011) Metabolic diversity among main microorganisms inside an arsenic‐rich ecosystem revealed by meta‐ and proteo‐genomics. The ISME Journal, 11, 1735–1747. Available from: 10.1038/ismej.2011.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biełło, K.A. , Cabello, P. , Rodríguez‐Caballero, G. , Sáez, L.P. , Luque‐Almagro, V.M. , Roldán, M.D. et al. (2023) Proteomic analysis of arsenic resistance during cyanide assimilation by Pseudomonas pseudoalcaligenes CECT 5344. International Journal of Molecular Sciences, 24, 7232. Available from: 10.3390/ijms24087232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biełło, K.A. , Olaya‐Abril, A. , Cabello, P. , Rodríguez‐Caballero, G. , Sáez, L.P. , Moreno‐Vivián, C. et al. (2023) Quantitative proteomic analysis of cyanide and mercury detoxification by Pseudomonas pseudoalcaligenes CECT 5344. Microbiology Spectrum, 11, e0055323. Available from: 10.1128/spectrum.00553-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birrer, S.C. , Dafforn, K.A. , Sun, M.Y. , Williams, R.B.H. , Potts, J. , Scanes, P. et al. (2019) Using meta‐omics of contaminated sediments to monitor changes in pathways relevant to climate regulation. Environmental Microbiology, 21, 389–401. Available from: 10.1111/1462-2920.14470 [DOI] [PubMed] [Google Scholar]
- Bjørklund, G. , Crisponi, G. , Nurchi, V.M. , Cappai, R. , Djordjevic, A.B. & Aaseth, J. (2019) A review on coordination properties of thiol‐containing chelating agents towards mercury, cadmium, and lead. Molecules, 24, 3247. Available from: 10.3390/molecules24183247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blindauer, C.A. (2011) Bacterial metallothioneins: past, present, and questions for the future. Journal of Biological Inorganic Chemistry, 16, 1011–1024. Available from: 10.1007/s00775-011-0790-y [DOI] [PubMed] [Google Scholar]
- Blindauer, C.A. (2015) Advances in the molecular understanding of biological zinc transport. Chemical Communications, 51, 4544–4563. Available from: 10.1039/c4cc10174j [DOI] [PubMed] [Google Scholar]
- Bouchez, T. , Blieux, A.L. , Dequiedt, S. , Domaizon, I. , Dufresne, A. , Ferrerira, S. et al. (2016) Molecular microbiology methods for environmental diagnosis. Environmental Chemistry Letters, 14, 423–441. Available from: 10.1007/s10311-016-0581-3 [DOI] [Google Scholar]
- Bridges, C. & Zalups, R. (2017) Mechanism involved in the transport of mercuric ions in target tissue. Archives of Toxicology, 91, 63–81. Available from: 10.1007/s00204-016-1803-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüger, A. , Fafilek, G. , Restrepo, O.J. & Rojas‐Mendoza, L. (2018) On the volatilisation and decomposition of cyanide contaminations from gold mining. Science of the Total Environment, 627, 1167–1173. Available from: 10.1016/j.scitotenv.2018.01.320 [DOI] [PubMed] [Google Scholar]
- Cabello, P. , Luque‐Almagro, V.M. , Olaya‐Abril, A. , Sáez, L.P. , Moreno‐Vivián, C. & Roldán, M.D. (2018) Assimilation of cyanide and cyano‐derivatives by Pseudomonas pseudoalcaligenes CECT5344: from omic approaches to biotechnological applications. FEMS Microbiology Letters, 365, fny032. Available from: 10.1093/femsle/fny032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capeness, M.J. & Horsfall, L.E. (2020) Synthetic biology approaches towards the recycling of metals from the environment. Biochemical Society Transactions, 48, 1367–1378. Available from: 10.1042/BST20190837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes, C. , Campos‐García, J. , Devars, S. , Gutiérrez‐Corona, F. , Loza‐Tavera, H. , Torres‐Guzmán, J.C. et al. (2001) Interactions of chromium with microorganisms and plants. FEMS Microbiology Reviews, 25, 335–347. Available from: 10.1111/j.1574-6976.2001.tb00581.x [DOI] [PubMed] [Google Scholar]
- Champier, L. , Duarte, V. , Michaud‐Soret, I. & Coves, J. (2004) Characterization of the MerD protein from Ralstonia metallidurans CH34: a possible role in bacterial mercury resistance by switching off the induction of the mer operon. Molecular Microbiology, 52, 1475–1485. Available from: 10.1111/j.1365-2958.2004.04071.x [DOI] [PubMed] [Google Scholar]
- Chen, J. , Yoshinaga, M. , Garbinski, L.D. & Rosen, B.P. (2016) Synergistic interaction of glyceraldehydes‐3‐phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Molecular Microbiology, 100, 945–953. Available from: 10.1111/mmi.13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L.X. , Hu, M. , Huang, L.N. , Hua, Z.S. , Kuang, J.L. , Li, S.J. et al. (2015) Comparative metagenomic and metatranscriptomic analyses of microbial communities in acid mine drainage. The ISME Journal, 9, 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, M. , Lu, W. , Hou, Z. , Zhang, Y. , Jiang, X. & Wu, J. (2017) Heavy metal pollution in soil associated with a large‐scale cyanidation gold mining region in southeast of Jilin, China. Environmental Science and Pollution Research International, 24, 3084–3096. Available from: 10.1007/s11356-016-7968-3 [DOI] [PubMed] [Google Scholar]
- Choudhury, R. & Srivastava, S. (2001) Mutational analysis of zinc resistance in Pseudomonas putida strain S4. Current Microbiology, 43, 316–321. Available from: 10.1007/s002840010309 [DOI] [PubMed] [Google Scholar]
- Colombo, M.J. , Ha, J. , Reinfelder, J.R. , Barkay, T. & Yee, N. (2014) Oxidation of Hg(0) to Hg(II) by diverse anaerobic bacteria. Chemical Geology, 363, 334–340. Available from: 10.1016/j.chemgeo.2013.11.020 [DOI] [Google Scholar]
- Cornu, J.Y. , Huguenot, D. , Jézéquel, K. , Lollier, M. & Lebeau, T. (2017) Bioremediation of copper‐contaminated soils by bacteria. World Journal of Microbiology and Biotechnology, 33, 26. Available from: 10.1007/s11274-016-2191-4 [DOI] [PubMed] [Google Scholar]
- Coudert, L. , Bondu, R. , Rakotonimaro, T.V. , Rosa, E. , Guittonny, M. & Neculita, C.M. (2020) Treatment of As‐rich mine effluents and produced residues stability: current knowledge and research priorities for gold mining. Journal of Hazardous Materials, 386, 121920. Available from: 10.1016/j.jhazmat.2019.121920 [DOI] [PubMed] [Google Scholar]
- Ćwieląg‐Drabek, M. , Piekut, A. , Gut, K. & Grabowski, M. (2020) Risk of cadmium, lead and zinc exposure from consumption of vegetables produced in areas with mining and smelting past. Scientific Reports, 10, 3363. Available from: 10.1038/s41598-020-60386-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, R.R. , Gaur, A. & Balomajumder, C. (2009) Cyanide in industrial wastewaters and its removal: a review on biotreatment. Journal of Hazardous Materials, 163, 1–11. Available from: 10.1016/j.jhazmat.2008.06.051 [DOI] [PubMed] [Google Scholar]
- Deeb, B.E. & Altalhi, A.D. (2009) Degradative plasmid and heavy metal resistance plasmid naturally coexist in phenol and cyanide assimilating bacteria. American Journal of Biochemistry and Biotechnology, 5, 84–93. [Google Scholar]
- Delmar, J.A. , Su, C.C. & Yu, E.W. (2015) Heavy metal transport by the CusCFBA efflux system. Protein Science, 24, 1720–1736. Available from: 10.1002/pro.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diba, F. , Khan, M.Z.H. , Uddin, S.Z. , Istiaq, A. , Shuvo, M.S.R. , Ul‐Alam, A. et al. (2021) Bioaccumulation and detoxification of trivalent arsenic by Achromobacter xylosoxidans BHW‐15 and electrochemical detection of its transformation efficiency. Scientific Reports, 11, 21312. Available from: 10.1038/s41598-021-00745-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrova, T. , Repmann, F. & Freese, D. (2020) Degradation of ferrocyanide by natural isolated bacteria. International Journal of Phytoremediation, 22, 20–28. Available from: 10.1080/15226514.2019.1633996 [DOI] [PubMed] [Google Scholar]
- Distler, U. , Kuharev, J. & Tenzer, S. (2014) Biomedical applications of ion mobility‐enhanced data‐independent acquisition‐based label‐free quantitative proteomics. Expert Review of Proteomics, 11, 675–684. Available from: 10.1586/14789450.2014.971114 [DOI] [PubMed] [Google Scholar]
- Dombrowski, P.M. , Long, W. , Farley, K.J. , Mahony, J.D. , Capitani, J.F. & Di Toro, D.M. (2005) Thermodynamic analysis of arsenic methylation. Environmental Science & Technology, 39, 2169–2176. Available from: 10.1021/es0489691 [DOI] [PubMed] [Google Scholar]
- Drace, K. , Kiefer, A.M. & Veiga, M.M. (2016) Cyanidation of mercury‐contaminated tailings: potential health effects and environmental justice. Current Environmental Health Reports, 3, 443–449. Available from: 10.1007/s40572-016-0113-0 [DOI] [PubMed] [Google Scholar]
- Ducret, V. , Gonzalez, M.R. , Leoni, S. , Valentini, M. & Perron, K. (2020) The CzcCBA efflux system requires the CadA P‐type ATPase for timely expression upon zinc excess in Pseudomonas aeruginosa . Frontiers in Microbiology, 11, 911. Available from: 10.3389/fmicb.2020.00911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont, C.L. , Grassb, G. & Rensing, C. (2011) Copper toxicity and the origin of bacterial resistance–new insights and applications. Metallomics, 3, 1109–1118. Available from: 10.1039/c1mt00107h [DOI] [PubMed] [Google Scholar]
- Dvořák, P. , Nikel, P.I. , Damborský, J. & de Lorenzo, V. (2017) Bioremediation 3.0: engineering pollutant‐removing bacteria in the times of systemic biology. Biotechnology Advances, 35, 845–866. Available from: 10.1016/j.biotechadv.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Ekici, S. , Turkarslan, S. , Pawlik, G. , Dancis, A. , Baliga, N.S. , Koch, H.‐G. et al. (2014) Intracytoplasmic copper homeostasis controls cytochrome c oxidase production. mBio, 5(1), e01055–13. Available from: 10.1128/mbio.01055-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estepa, J. , Luque‐Almagro, V.M. , Manso, I. , Escribano, M.P. , Martínez‐Luque, M. , Castillo, F. et al. (2012) The nit1C gene cluster of Pseudomonas pseudoalcaligenes CECT 5344 involved in assimilation of nitriles is essential for growth on cyanide. Environmental Microbiology Reports, 4, 326–334. Available from: 10.1111/j.1758-2229.2012.00337.x [DOI] [PubMed] [Google Scholar]
- Fan, C. , Zhang, D. , Mo, Q. & Yuan, J. (2022) Engineering Sacharomyces cerevisiae‐based biosensors for copper detection. Microbial Biotechnology, 15, 2854–2860. Available from: 10.1111/1751-7915.14105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedel‐Moen, R. , Ragusa, S.R. , Kimber, R.W.L. & Williams, B.D. (2000) Degradation of metal cyanide complexes by microorganisms. Minerals and Metallurgical Processing, 17, 69–75. [Google Scholar]
- Flemming, C.A. , Ferris, F.G. , Beveridge, T.J. & Bailey, G.W. (1990) Remobilization of toxic heavy metals adsorbed to bacterial wall‐clay composites. Applied and Environmental Microbiology, 56, 3191–3203. Available from: 10.1128/aem.56.10.3191-3203.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbinski, L.D. , Rosen, B.P. & Chen, J. (2019) Pathways of arsenic uptake and efflux. Environment International, 126, 585–597. Available from: 10.1016/j.envint.2019.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, C.C. , Elias, D.A. , Kucken, A.M. , Brown, S.D. , Palumbo, A.V. , Schadt, C.W. et al. (2011) Sulfate‐reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Applied and Environmental Microbiology, 77, 3938–3951. Available from: 10.1128/AEM.02993-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Henao, S. & Thaura, G.H. (2021) Heavy metals in soils and the remediation potential of bacteria associated with the plant microbiome. Frontiers in Environmental Science, 9, fenvs.2021.604216. Available from: 10.3389/fenvs.2021.604216 [DOI] [Google Scholar]
- González‐Raymant, H. , Liu, G. , Liriano, C. , Li, Y. , Yin, Y. & Jiang, G. (2017) Elemental mercury: its unique properties affect its behavior and fate in the environment. Environmental Pollution, 229, 69–86. Available from: 10.1016/j.envpol.2017.04.101 [DOI] [PubMed] [Google Scholar]
- Guo, Y. , Hui, C.‐Y. , Zhang, N.‐X. , Liu, L. , Li, H. & Zheng, H.‐J. (2021) Development of cadmium multiple‐signal biosensing and bioadsorption systems based on artificial cad operons. Frontiers in Bioengineering and Biotechnology, 9, 585617. Available from: 10.3389/fbioe.2021.585617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, N. , Balomajumder, C. & Agarwal, V.K. (2010) Enzymatic mechanisms and biochemistry for cyanide degradation: a review. Journal of Hazardous Materials, 176, 1–13. Available from: 10.1016/j.hazmat.2009.11.038 [DOI] [PubMed] [Google Scholar]
- Gupta, P. & Diwan, B. (2017) Bacterial exopolysaccharide mediated heavy metal removal: a review on biosynthesis, mechanism and remediation strategies. Biotechnology Reports, 13, 58–71. Available from: 10.1016/j.btre.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gworek, B. , Dmuchowski, W. & Baczewska‐Dąbrowska, A.H. (2020) Mercury in the terrestrial environment: a review. Environmental Sciences Europe, 32, 128. Available from: 10.1186/s12302-020-00401-x [DOI] [Google Scholar]
- Hamood‐Altowayti, W.A. , Almoalemi, H. , Shahir, S. & Othman, N. (2020) Comparison of culture‐independent and dependent approaches for identification of native arsenic‐resistant bacteria and their potential use for arsenic bioremediation. Ecotoxicology and Environmental Safety, 205, 111267. Available from: 10.1016/j.ecoenv.2020.111267 [DOI] [PubMed] [Google Scholar]
- Hariharakrishnan, J. , Satpute, R.M. , Prasad, G.B.K.S. & Bhattacharya, R. (2009) Oxidative stress mediated cytotoxicity of cyanide in LLC‐MK2 cells and its attenuation by alpha‐ketoglutarate and N‐acetyl cysteine. Toxicology Letters, 185, 132–1341. Available from: 10.1016/j.toxlet.2008.12.011 [DOI] [PubMed] [Google Scholar]
- Hemmat‐Jou, M.H. , Safari‐Sinegani, A.A. , Che, R. , Mirzaie‐Asl, A. , Tahmourespour, A. & Tahmasbian, I. (2021) Toxic trace element resistance genes and systems identified using the shotgun metagenomics approach in an Iranian mine soil. Environmental Science and Pollution Research International, 28, 4845–4856. Available from: 10.1007/s11356-020-10824-x [DOI] [PubMed] [Google Scholar]
- Herruzo‐Ruiz, A.M. , Fuentes‐Almagro, C.A. , Jiménez‐Pastor, J.M. , Pérez‐Rosa, V.M. , Blasco, J. , Michán, C. et al. (2021) Meta‐omic evaluation of bacterial microbial community structure and activity for the environmental assessment of soils: overcoming protein extraction pitfalls. Environmental Microbiology, 23, 4706–4725. Available from: 10.1111/1462-2920.15673 [DOI] [PubMed] [Google Scholar]
- Holzman, D.C. (2010) Mercury emissions not shrinking as forecast. Environmental Health Perspectives, 118, A198. Available from: 10.1289/ehp.118-a198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh, P.F. , Lin, H.H. , Lin, T.L. & Wang, J.T. (2010) CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae . The Journal of Infectious Diseases, 202, 52–64. Available from: 10.1086/653079 [DOI] [PubMed] [Google Scholar]
- Huang, K. , Xu, Y. , Packianathan, C. , Gao, F. , Chen, C. , Zhang, J. et al. (2018) Arsenic methylation by a novel ArsM As(III) S‐adenosylmethionine methyltransferase that requires only two conserved cysteine residues. Molecular Microbiology, 107, 265–276. Available from: 10.1111/mmi.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez, M.I. , Cabello, P. , Luque‐Almagro, V.M. , Sáez, L.P. , Olaya, A. , Sánchez de Medina, V. et al. (2017) Quantitative proteomic analysis of Pseudomonas pseudoalcaligenes CECT5344 in response to industrial cyanide‐containing wastewaters using liquid chromatography‐mass spectrometry/mass spectrometry (LC‐MS/MS). PLoS One, 12, e0172908. Available from: 10.1371/journal.pone.0172908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabiraj, A. , Biswas, R. , Halder, U. & Bandopadhyay, R. (2022) Bacterial arsenic metabolism and its role in arsenic bioremediation. Current Microbiology, 79, 131. Available from: 10.1007/s00284-022-02810-y [DOI] [PubMed] [Google Scholar]
- Kantor, R.S. , van Zyl, A.W. , van Hille, R.P. , Thomas, B.C. , Harrison, S.T. & Banfield, J.F. (2015) Bioreactor microbial ecosystems for thiocyanate and cyanide degradation unravelled with genome‐resolved metagenomics. Environmental Microbiology, 17, 4929–4941. Available from: 10.1111/1462-2920.12936 [DOI] [PubMed] [Google Scholar]
- Karamba, K.I. , Ahmad, S.A. , Zulkharnain, A. , Yasid, N. , Khalid, A. & Shukor, M.Y. (2017) Biodegradation of cyanide and evaluation of kinetic models by immobilized cells of Serratia marcescens strain AQ07. International Journal of Science & Technology, 14, 1945–1958. Available from: 10.1007/s13762-017-1287-1 [DOI] [Google Scholar]
- Karamba, K.I. , Syed, M.A. , Shukor, M.Y. & Ahmad, S.A. (2014) Effect of heavy metals on cyanide biodegradation by resting cells of Serratia marcescens strain AQ07. Journal of Environmental Microbiology and Toxicology, 2, 17–20. [Google Scholar]
- Kerper, L.E. & Hinkle, P.M. (1997) Cellular uptake of lead is activated by depletion of intracellular calcium stores. The Journal of Biological Chemistry, 272, 8346–8352. Available from: 10.1074/jbc.272.13.8346 [DOI] [PubMed] [Google Scholar]
- Khan, Z. , Elahi, A. , Bukhari, D.A. & Rehman, A. (2022) Cadmium sources, toxicity, resistance and removal by microorganisms—a potential strategy for cadmium eradication. Journal of Saudi Chemical Society, 26, 101569. Available from: 10.1016/j.jscs.2022.101569 [DOI] [Google Scholar]
- Kim, H.J. , Jeong, H. & Lee, S.J. (2018) Synthetic biology for microbial heavy metal biosensors. Analytical and Bioanalytical Chemistry, 410, 1191–1203. Available from: 10.1007/s00216-017-0751-6 [DOI] [PubMed] [Google Scholar]
- Kim, S. , Arora, M. , Fernandez, C. , Landero, J. , Caruso, J. & Chen, A. (2013) Lead, mercury, and cadmium exposure and attention deficit hyperactivity disorder in children. Environmental Research, 126, 105–110. Available from: 10.1016/j.envres.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubier, A. , Wilkin, R.T. & Pichler, T. (2019) Cadmium in soils and groundwater: a review. Applied Geochemistry, 108, 104388. Available from: 10.1016/j.apgeochem.2019.104388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R. , Saha, S. , Dhaka, S. , Kurade, M. , Kang, C. , Baek, S. et al. (2016) Remediation of cyanide‐contaminated environments through microbes and plants: a review of current knowledge and future perspectives. Geosyst Enginer, 20, 28–40. Available from: 10.1080/12269328.2016.1218303 [DOI] [Google Scholar]
- Kunath, B.J. , Minniti, G. , Skaugen, M. , Hagen, L.H. , Vaaje‐Kolstad, G. , Eijsink, V.G.H. et al. (2019) Metaproteomics: sample preparation and methodological considerations. Advances in Experimental Medicine and Biology, 1073, 187–215. Available from: 10.1007/978-3-030-12298-0_8 [DOI] [PubMed] [Google Scholar]
- Leary, D.H. , Hervey, W.J. , Deschamps, J.R. , Kusterbeck, A.W. & Vora, G.J. (2014) Reprint of which metaproteome? The impact of protein extraction bias on metaproteomic analyses. Molecular and Cellular Probes, 28, 51–57. Available from: 10.1016/j.mcp.2014.01.002 [DOI] [PubMed] [Google Scholar]
- Li, Y. , Ma, Y. , Xia, Y. , Zang, T. , Sun, S. , Gao, J. et al. (2023) Discovery and biosynthesis of tricyclic copper‐binding ribosomal peptides containing histidine‐to‐butyrine crosslinks. Nature Communications, 14, 2944. Available from: 10.1038/s41467-023-38517-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, X. , Kierans, M. , Ceci, A. , Hillier, S. & Gadd, G.M. (2016) Phosphatase‐mediated bioprecipitation of lead by soil fungi. Environmental Microbiology, 18, 219–231. Available from: 10.1111/1462-2920.13003 [DOI] [PubMed] [Google Scholar]
- Liu, H. , Zhang, Y. , Wang, Y. , Xie, X. & Shi, Q. (2021) The connection between Czc and Cad systems involved in cadmium resistance in Pseudomonas putida . International Journal of Molecular Sciences, 22, 9697. Available from: 10.3390/ijms22189697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Ju, Y. , Mandzhieva, S. , Pinskii, D. , Minkina, T. , Rajput, V.D. et al. (2023) Sporadic Pb accumulation by plants: influence of soil biogeochemistry, microbial community and physiological mechanisms. Journal of Hazardous Materials, 444, 130391. Available from: 10.1016/j.jhazmat.2022.130391 [DOI] [PubMed] [Google Scholar]
- Lloyd, K.G. , Steen, A.D. , Ladau, J. , Yin, J. & Crosby, L. (2018) Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems, 3, e00055‐18. Available from: 10.1128/mSystems.00055-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Fernández, M. , Jroundi, F. , Ruiz‐Fresneda, M.A. & Merroun, M.L. (2021) Microbial interaction with and tolerance of radionuclides: underlying mechanisms and biotechnological applications. Microbial Biotechnology, 14, 810–828. Available from: 10.1111/1751-7915.13718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Pérez, M.E. , Saldaña‐Robles, A. , Zanor, G.A. , Ibarra, J.E. & del Rincón‐Castro, M.C. (2021) Microbiomes in agricultural and mining soils contaminated with arsenic in Guanajuato, Mexico. Archives of Microbiology, 203, 499–511. Available from: 10.1007/s00203-020-01973-1 [DOI] [PubMed] [Google Scholar]
- Luque‐Almagro, V.M. , Acera, F. , Igeño, M.I. , Wibberg, D. , Roldán, M.D. , Sáez, L.P. et al. (2013) Draft whole genome sequence of the cyanide‐degrading bacterium Pseudomonas pseudoalcaligenes CECT5344. Environmental Microbiology, 15, 253–270. Available from: 10.1111/j.1462-2920.2012.02875.x [DOI] [PubMed] [Google Scholar]
- Luque‐Almagro, V.M. , Blasco, R. , Huertas, M.J. , Martínez‐Luque, M. , Moreno‐Vivián, C. , Castillo, F. et al. (2005) Alkaline cyanide biodegradation by Pseudomonas pseudoalcaligenes CECT5344. Biochemical Society Transactions, 33, 168–169. Available from: 10.1042/BST0330168 [DOI] [PubMed] [Google Scholar]
- Luque‐Almagro, V.M. , Cabello, P. , Sáez, L.P. , Olaya‐Abril, A. , Moreno‐Vivián, C. & Roldán, M.D. (2018) Exploring anaerobic environments for cyanide and cyano‐derivatives microbial degradation. Applied Microbiology and Biotechnology, 102, 1067–1074. Available from: 10.1007/s00253-017-8678-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luque‐Almagro, V.M. , Escribano, M.P. , Manso, I. , Sáez, L.P. , Cabello, P. , Moreno‐Vivián, C. et al. (2015) DNA microarray analysis of the cyanotroph Pseudomonas pseudoalcaligenes CECT5344 in response to nitrogen starvation, cyanide and a jewelry wastewater. Journal of Biotechnology, 214, 171–181. Available from: 10.1016/j.jbiotec.2015.09.032 [DOI] [PubMed] [Google Scholar]
- Luque‐Almagro, V.M. , Moreno‐Vivián, C. & Roldán, M.D. (2016) Biodegradation of cyanide wastes from mining and jewellery industries. Current Opinion in Biotechnology, 38, 9–13. Available from: 10.1016/j.copbio.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Macomber, L. & Hausinger, R.P. (2011) Mechanisms of nickel toxicity in microorganisms. Metallomics, 3, 1153–1162. Available from: 10.1039/c1mt00063b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens, L. , Cherry, P. , Tilquin, F. , van Houdt, R. & Matroule, J.Y. (2021) Environmental conditions modulate the transcriptomic response of both Caulobacter crescentus morphotypes to Cu stress. Microorganisms, 9, 1116. Available from: 10.3390/microorganisms9061116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity, S. , Sarkar, D. , Poddar, K. , Patil, P. & Sarkar, A. (2022) Biofilm‐mediated heavy metal removal from aqueous system by multi‐metal‐resistant bacterial strain Bacillus sp. GH‐s29. Applied Biochemistry and Biotechnology, 195, 4832–4850. Available from: 10.1007/s12010-022-04288-7 [DOI] [PubMed] [Google Scholar]
- Malmir, N. , Allahyari‐Fard, N. , Aminzadeh, S. , Moghaddassi‐Jahromi, Z.M. & Mekuto, L. (2022) An overview of emerging cyanide bioremediation methods. Processes, 10, 1724. Available from: 10.3390/pr10091724 [DOI] [Google Scholar]
- Martínez, V.D. , Vucic, E.A. , Becker‐Santos, D.D. , Gil, L. & Lam, W.L. (2011) Arsenic exposure and the induction of human cancers. Journal of Toxicology, 2011, 431287. Available from: 10.1155/2011/431287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massot, F. , Bernard, N. , Alvarez, L.M.M. , Martorell, M.M. , MacCormack, W.P. & Ruberto, L.A.M. (2022) Microbial associations for bioremediation. What does microbial consortia mean? Applied Microbiology and Biotechnology, 106, 2283–2297. Available from: 10.1007/s00253-022-11864-8 [DOI] [PubMed] [Google Scholar]
- Mathema, V.B. , Thakuri, B.C. & Sillanpaa, M. (2011) Bacterial mer operon mediated detoxification of mercurial compounds: a short review. Archives of Microbiology, 193, 837–844. Available from: 10.1007/s00203-011-0751-4 [DOI] [PubMed] [Google Scholar]
- Mathivanan, K. , Uthaya‐Chandirika, J. , Vinothkanna, A. , Yin, H. , Liu, X. & Meng, D. (2021) Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment—a review. Ecotoxicology and Environmental Safety, 226, 112863. Available from: 10.1016/j.ecoenv.2021 [DOI] [PubMed] [Google Scholar]
- Mazumder, P. , Sharma, S.K. , Taki, K. , Kalamdhad, A.S. & Kumar, M. (2020) Microbes involved in arsenic mobilization and respiration: a review on isolation, identification, isolates and implications. Environmental Geochemistry and Health, 42, 3443–3469. Available from: 10.1007/s10653-020-00549-8 [DOI] [PubMed] [Google Scholar]
- Mekuto, L. , Alegbeleye, O.O. , Ntwampe, S.K.O. , Ngongang, M.M. , Mudumbi, J.B. & Akinpelu, E.A. (2016) Co‐metabolism of thiocyanate and free cyanide by Exiguobacterium acetylicum and Bacillus marisflavi under alkaline conditions. 3 Biotech, 6, 173. Available from: 10.1007/s13205-016-0491-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mergeay, M. , Monchy, S. , Vallaeys, T. , Auquier, V. , Benotmane, A. , Bertin, P. et al. (2003) Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal‐responsive genes. FEMS Microbiology Reviews, 27, 358–410. Available from: 10.1016/S0168-6445(03)00045-7 [DOI] [PubMed] [Google Scholar]
- Michán, C. , Chicano‐Gálvez, E. , Fuentes‐Almagro, C.A. & Alhama, J. (2019) Redox and global interconnected proteome changes in mice exposed to complex environmental hazards surrounding Doñana National Park. Environmental Pollution, 252, 427–439. Available from: 10.1016/j.envpol.2019.05.085 [DOI] [PubMed] [Google Scholar]
- Mirizadeh, S. , Yaghmaei, S. & Nejad, Z.G. (2014) Biodegradation of cyanide by a new isolated strain under alkaline conditions and optimization by response surface methodology (RSM). Journal of Environmental Health Science and Engineering, 12, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina‐Roco, M. , Escudey, M. , Antilén, M. , Arancibia‐Miranda, N. & Manquián‐Cerda, K. (2018) Distribution of contaminant trace metals inadvertently provided by phosphorus fertilisers: movement, chemical fractions and mass balances in contrasting acidic soils. Environmental Geochemistry and Health, 40, 2491–2509. Available from: 10.1007/s10653-018-0115-y [DOI] [PubMed] [Google Scholar]
- Morelli, E. , Mascherpa, M.C. & Scarano, G. (2005) Biosynthesis of phytochelatins and arsenic accumulation in the marine microalga Phaeodactylum tricornutum in response to arsenate exposure. Biometals, 18, 587–593. Available from: 10.1007/s10534-005-2998-1 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay, R. , Bhattacharjee, H. & Rosen, B.P. (2014) Aquaglyceroporins: generalized metalloid channels. Biochimica et Biophysica Acta, 1840, 1583–1591. Available from: 10.1016/j.bbagen.2013.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munyai, R. , Ogola, H.J.O. & Modise, D.M. (2021) Microbial community diversity dynamics in acid mine drainage and acid mine drainage‐polluted soils: implication on mining water irrigation agricultural sustainability. Frontiers in Sustainable Food Systems, 5, 701870. Available from: 10.3389/fsufs.2021.701870 [DOI] [Google Scholar]
- Naguib, M. , El‐Gendy, A. & Khairalla, A. (2018) Microbial diversity of mer operon genes and their potential rules in mercury bioremediation and resistance. Open Biotechnology Journal, 12, 56–77. Available from: 10.2174/1874070701812010056 [DOI] [Google Scholar]
- Naz, N. , Young, H.K. , Ahmed, N. & Gadd, G.M. (2005) Cadmium accumulation and DNA homology with metal resistance genes in sulfate‐reducing bacteria. Applied and Environmental Microbiology, 71, 4610–4618. Available from: 10.1128/aem.71.8.4610-4618.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii, A.I. (2014) Proteogenomics: concepts, applications and computational strategies. Nature Methods, 11, 1114–1125. Available from: 10.1038/nmeth.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome, L. & Falagán, C. (2021) The microbiology of metal mine waste: Bioremediation applications and implications for planetary health. Geohealth, 5, e2020GH000380. Available from: 10.1029/2020GH000380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikodinovic‐Runic, J. , Guzik, M. , Kenny, S.T. , Babu, R. , Werker, A. & Connor, K.E.O. (2013) Carbon‐rich wastes as feedstocks for biodegradable polymer (polyhydroxyalkanoate) production using bacteria. Advances in Applied Microbiology, 84, 139–200. Available from: 10.1016/B978-0-12-407673-0.00004-7 [DOI] [PubMed] [Google Scholar]
- Nurfitriani, S. , Arisoesilaningsih, E. , Nuraini, Y. & Handayanto, E. (2020) Bioaccumulation of mercury by bacteria isolated from small scale gold mining tailings in Lombok, Indonesia. Journal of Ecological Engineering, 21, 127–136. Available from: 10.12911/22998993/123247 [DOI] [Google Scholar]
- Olaya‐Abril, A. , Hidalgo‐Carrillo, J. , Luque‐Almagro, V.M. , Fuentes‐Almagro, C. , Urbano, F.J. , Moreno‐Vivián, C. et al. (2018) Exploring the denitrification proteome of Paracoccus denitrificans PD1222. Frontiers in Microbiology, 9, 1137. Available from: 10.3389/fmicb.2018.01137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya‐Abril, A. , Luque‐Almagro, V.M. , Pérez, M.D. , López, C.M. , Amil, F. , Cabello, P. et al. (2019) Putative small RNAs controlling detoxification of industrial cyanide‐containing wastewaters by Pseudomonas pseudoalcaligenes CECT5344. PLoS One, 14, e0212032. Available from: 10.1371/journal.pone.0212032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya‐Abril, A. , Pérez, M.D. , Cabello, P. , Martignetti, D. , Sáez, L.P. , Luque‐Almagro, V.M. et al. (2020) Role of the dihydrodipicolinate synthase DapA1 on iron homeostasis during cyanide assimilation by the alkaliphilic bacterium Pseudomonas pseudoalcaligenes CECT5344. Frontiers in Microbiology, 11, 28. Available from: 10.3389/fmicb.2020.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyido, I. , Norris, A.R. & Buncel, E. (2004) Biomolecule‐mercury interactions: modalities of DNA based‐mercury binding mechanisms. Remediation strategies. Chemical Reviews, 104, 5911–5929. Available from: 10.1021/cr030443w [DOI] [PubMed] [Google Scholar]
- Oremland, R.S. & Stolz, J.F. (2003) The ecology of arsenic. Science, 300, 939–944. Available from: 10.1126/science.1081903 [DOI] [PubMed] [Google Scholar]
- Osborn, M. , Bruce, K.D. , Strike, P. & Ritchie, D.A. (1997) Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiology Reviews, 4, 239–262. Available from: 10.1111/j.1574-6976.1997.tb00300.x [DOI] [PubMed] [Google Scholar]
- Oudjehani, K. , Zagury, G.J. & Deschenes, L. (2001) Natural attenuation potential of cyanide via microbial activity in mine tailings. Applied Microbiology and Biotechnology, 58, 409–415. Available from: 10.1007/s00253-001-0887-2 [DOI] [PubMed] [Google Scholar]
- Patil, Y.B. & Paknikar, K.M. (2000) Biodegradation of silver‐cyanide from electroplating industry wastewater. Letters in Applied Microbiology, 30, 33–37. [DOI] [PubMed] [Google Scholar]
- Pérez, M.D. , Olaya‐Abril, A. , Cabello, P. , Sáez, L.P. , Roldán, M.D. , Moreno‐Vivián, C. et al. (2021) Alternative pathway for 3‐cyanoalanine assimilation in Pseudomonas pseudoalcaligenes CECT5344 under noncyanotrophic conditions. Microbiology Spectrum, 9, e0077721. Available from: 10.1128/Spectrum.00777-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perron, K. , Caille, O. , Rossier, C. , van Delden, C. , Dumas, J.‐L. & Köhler, T. (2004) (2004) CzcR‐CzcS, a two‐component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa . The Journal of Biological Chemistry, 279, 8761–8768. Available from: 10.1074/jbc.M312080200 [DOI] [PubMed] [Google Scholar]
- Petrick, J.S. , Ayala‐Fierro, F. , Cullen, W.R. , Carter, D.E. & Vasken‐Aposhian, H. (2000) Monomethylarsonous acid (MMA(III)) is more toxic than arsenite in Chang human hepatocytes. Toxicology and Applied Pharmacology, 163, 203–207. Available from: 10.1006/taap.1999.8872 [DOI] [PubMed] [Google Scholar]
- Piciullo, L. , Storrøsten, E.B. , Liu, Z. , Nadim, F. & Lacasse, S. (2022) A new look at the statistics of tailing dam failures. Engineering Geology, 303, 106657. Available from: 10.1016/j.enggeo.2022.106657 [DOI] [Google Scholar]
- Podar, M. , Gilmour, C.C. , Brandt, C.C. , Soren, A. , Brown, S.D. , Crable, B.R. et al. (2015) Global prevalence and distribution of genes and microorganisms involved in mercury methylation. Science Advances, 1, e1500675. Available from: 10.1126/sciadv.1500675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshanee, M. , Chatterjee, S. , Rath, S. , Dash, H.R. & Das, S. (2022) Cellular and genetic mechanism of bacterial mercury resistance and their role in biogeochemistry and bioremediation. Journal of Hazardous Materials, 423, 126985. Available from: 10.1016/j.jhazmat.2021.126985 [DOI] [PubMed] [Google Scholar]
- Rai, U.N. , Singh, N.K. , Upadhyay, A.K. & Verma, S. (2013) Chromate tolerance and accumulation in Chlorella vulgaris L.: role of antioxidant enzymes and biochemical changes in detoxification of metals. Bioresource Technology, 136, 604–609. Available from: 10.1016/j.biortech.2013.03.043 [DOI] [PubMed] [Google Scholar]
- Raju, N.J. (2022) Arsenic in the geo‐environment: a review of sources, geochemical processes, toxicity and removal technologies. Environmental Research, 203, 111782. Available from: 10.1016/j.envres.2021.111782 [DOI] [PubMed] [Google Scholar]
- Ramírez‐Díaz, M.I. , Díaz‐Pérez, C. , Vargas, E. , Riveros‐Rosas, H. , Campos‐García, J. & Cervantes, C. (2008) Mechanisms of bacterial resistance to chromium compounds. Biometals, 21, 321–332. Available from: 10.1007/s10534-007-9121-8 [DOI] [PubMed] [Google Scholar]
- Rand, M.D. & Caito, S.W. (2019) Variation in the biological half‐life of methylmercury in humans: methods, measurements and meaning. Biochimica et Biophysica Acta, 1863, 1209301. Available from: 10.1016/j.bbagen.2019.02.003 [DOI] [PubMed] [Google Scholar]
- Rawle, R. , Saley, T.C. , Kang, Y.S. , Wang, Q. , Walk, S. , Bothner, B. et al. (2021) Introducing the ArsR‐regulated arsenic stimulon. Frontiers in Microbiology, 12, 630562. Available from: 10.3389/fmicb.2021.630562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razanamahandry, L.C. , Karoui, H. , Andrianisa, H.A. & Yacouba, H. (2017) Bioremediation of soil and water polluted by cyanide: a review. African Journal of Environmental Science and Technology, 6, 272–291. Available from: 10.5897/AJEST2016.2264 [DOI] [Google Scholar]
- Reis, M.P. , Dias, M.F. , Costa, P.S. , Ávila, M.P. , Leite, L.R. , de Araújo, F.M.G. et al. (2016) Metagenomic signatures of a tropical mining‐impacted stream reveal complex microbial and metabolic networks. Chemosphere, 161, 266–273. Available from: 10.1016/j.chemosphere.2016.06.097 [DOI] [PubMed] [Google Scholar]
- Remonsellez, F. , Orell, A. & Jerez, C.A. (2006) Copper tolerance of the thermoacidophilic archaeon Sulfolobus metallicus: possible role of polyphosphate metabolism. Microbiology, 152, 59–66. Available from: 10.1099/mic.0.28241-0 [DOI] [PubMed] [Google Scholar]
- Ren, C.G. , Liu, Z.Y. , Wang, X.L. & Qin, S. (2022) The seaweed holobiont: from microecology to biotechnological applications. Microbial Biotechnology, 15, 738–754. Available from: 10.1111/1751-7915.14014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggo, C. & van der Meer, J.R. (2017) Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Current Opinion in Biotechnology, 45, 24–33. Available from: 10.1016/j.copbio.2016.11.023 [DOI] [PubMed] [Google Scholar]
- Roldán, M.D. , Olaya‐Abril, A. , Sáez, L.P. , Cabello, P. , Luque‐Almagro, V.M. & Moreno‐Vivián, C. (2021) Bioremediation of cyanide‐containing wastes: the potential of systems and synthetic biology for cleaning up the toxic leftovers from mining. EMBO Reports, 22, e53720. Available from: 10.15252/embr.202153720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberger, G. , Bludau, I. , Schmitt, U. , Heusel, M. , Hunter, C.L. , Lui, Y. et al. (2017) Statistical control of peptide and protein error rates in large‐scale targeted data‐independent acquisition analyses. Nature Methods, 14, 921–927. Available from: 10.1038/nmeth.4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggles, K.V. , Krug, K. , Want, X. , Clauser, K.R. , Wang, J. , Payne, S.H. et al. (2017) Methods, tools and current perspectives in proteogenomics. Molecular & Cellular Proteomics, 16, 959–981. Available from: 10.1074/mcp.MR117.000024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakpirom, J. , Kantachote, D. , Siripattanakul‐Ratpukdi, S. , McEvoy, J. & Khan, E. (2019) Simultaneous bioprecipitation of cadmium‐to‐cadmium sulfide nanoparticles and nitrogen fixation by Rhodopseudomonas palustris TN110. Chemosphere, 223, 455–464. Available from: 10.1016/j.chemosphere.2019.02.051 [DOI] [PubMed] [Google Scholar]
- Sathyavathi, S. , Manjula, A. , Rajendhran, J. & Gunasekaran, P. (2014) Extracellular synthesis and characterization of nickel oxide nanoparticles from microbacterium sp. MRS‐1 towards bioremediation of nickel electroplating industrial effluent. Bioresource Technology, 165, 270–273. Available from: 10.1016/j.biortech.2014.03.031 [DOI] [PubMed] [Google Scholar]
- Scheiber, I. , Dringen, R. & Mercer, J.F. (2013) Copper: effects of deficiency and overload. Metal Ions in Life Sciences, 13, 359–387. [DOI] [PubMed] [Google Scholar]
- Sharma, J. , Shamim, K. & Dubey, S.K. (2018) Phosphatase mediated bioprecipitation of lead as pyromorphite by Achromobacter xylosoxidans . Journal of Environmental Management, 217, 754–761. Available from: 10.1016/j.jenvman.2018.04.027 [DOI] [PubMed] [Google Scholar]
- Sher, S. & Rehman, A. (2019) Use of heavy metals resistant bacteria: a strategy for arsenic bioremediation. Applied Microbiology and Biotechnology, 103, 6007–6021. Available from: 10.1007/s00253-019-09933-6 [DOI] [PubMed] [Google Scholar]
- Shu, W.S. & Huang, L.N. (2022) Microbial diversity in extreme environments. Nature Reviews. Microbiology, 20, 219–235. Available from: 10.1038/s41579-021-00648-y [DOI] [PubMed] [Google Scholar]
- Si, L. , Branfireun, B.A. & Fierro, J. (2022) Chemical oxidation and reduction pathways of mercury relevant to natural waters: a review. Water, 14, 1891. Available from: 10.3390/w14121891 [DOI] [Google Scholar]
- Silva‐Avalos, J. , Richmond, M.G. , Nagappan, O. & Kunz, D.A. (1990) Degradation of the metal‐cyano complex tetracyanonickelate(II) by cyanide‐utilizing bacterial isolates. Applied and Environmental Microbiology, 56, 3664–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, A. & Kumar, V. (2021) Recent advances in synthetic biology‐enabled and natural whole‐cell optical biosensing of heavy metals. Analytical and Bioanalytical Chemistry, 413, 73–82. Available from: 10.1007/s00216-020-02953-6 [DOI] [PubMed] [Google Scholar]
- Skariyachan, S. , Taskeen, N. , Kishore, A.P. & Krishna, B.V. (2021) Recent advances in plastic degradation: from microbial consortia‐based methods to data sciences and computational biology driven approaches. Journal of Hazardous Materials, 426, 128086. Available from: 10.1016/j.jhazmat.2021.128086 [DOI] [PubMed] [Google Scholar]
- Slyemi, D. & Bonnefoy, V. (2012) How prokaryotes deal with arsenic. Environmental Microbiology Reports, 4, 571–586. Available from: 10.1111/j.1758-2229.2011.00300.x [DOI] [PubMed] [Google Scholar]
- Somayaji, A. , Sarkar, S. , Balasubramaniam, S. & Raval, R. (2022) Synthetic biology techniques to tackle heavy metal pollution and poisoning. Synthetic and Systems Biotechnology, 7, 841–846. Available from: 10.1016/j.synbio.2022.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone, Y. , Nakamura, R. , Pan‐Hou, H. , Itoh, T. & Kiyono, M. (2013) Role of MerC, MerE, MerF, MerT, and/or MerP in resistance to mercurials and the transport of mercurials in Escherichia coli . Biological & Pharmaceutical Bulletin, 36, 1835–1841. Available from: 10.1248/bpb.b13-00554 [DOI] [PubMed] [Google Scholar]
- Sonthiphand, P. , Ruangroengkulrith, S. , Mhuantong, W. , Charoensawan, V. , Chotpantarat, S. & Boonkaewwan, S. (2019) Metagenomic insights into microbial diversity in a groundwater basin impacted by a variety of anthropogenic activities. Environmental Science and Pollution Research International, 26, 26765–26781. Available from: 10.1007/s11356-019-05905-5 [DOI] [PubMed] [Google Scholar]
- Starke, R. , Jehmlich, N. & Bastida, F. (2019) Using proteins to study how microbes contribute to soil ecosystem services: the current state and future perspectives of soil metaproteomics. Journal of Proteomics, 198, 50–58. Available from: 10.1016/j.jprot.2018.11.011 [DOI] [PubMed] [Google Scholar]
- Tao, H.C. , Peng, Z.W. , Li, P.S. , Yu, T.A. & Su, J. (2013) Optimizing cadmium and mercury specificity of CadR‐based E. coli biosensors by redesign of CadR. Biotechnology Letters, 35, 1253–1258. Available from: 10.1007/s10529-013-1216-4 [DOI] [PubMed] [Google Scholar]
- Tarrant, E. , Riboldi, G. , McIlvin, M.R. , Stevenson, J. , Barwinska‐Sendra, A. , Stewart, L.J. et al. (2019) Copper stress in Staphylococcus aureus leads to adaptive changes in central carbon metabolism. Metallomics, 11, 183–200. Available from: 10.1039/c8mt00239h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchounwou, P.B. , Yedjou, C.G. , Patlolla, A.K. & Sutton, D.J. (2012) Heavy metal toxicity and the environment. Experientia. Supplementum, 101, 133–164. Available from: 10.1007/978-3-7643-8340-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai, T.D. , Lim, W. & Na, D. (2023) Synthetic bacteria for the detection and bioremediation of heavy metals. Frontiers in Bioengineering and Biotechnology, 11, 1178680. Available from: 10.3389/fbioe.2023.1178680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatoi, H. , Das, S. , Mishra, J. , Rath, B.P. & Das, N. (2014) Bacterial chromate reductase, a potential enzyme for bioremediation of hexavalent chromium: a review. Journal of Environmental Management, 146, 383–399. Available from: 10.1016/j.jenvman.2014.07.014 [DOI] [PubMed] [Google Scholar]
- Tulasi, D. , Fajon, V. , Joze, K. , Shlyapnikov, Y. , Adotey, D. , Serfor‐Armah, Y. et al. (2021) Mercury methylation in cyanide influenced river sediments: a comparative study in southwestern Ghana. Environmental Monitoring and Assessment, 193, 180. Available from: 10.1007/s10661-021-08920-7 [DOI] [PubMed] [Google Scholar]
- Uthus, E.O. & Brown‐Borg, H.M. (2003) Altered methionine metabolism in long living Ames dwarf mice. Experimental Gerontology, 38, 491–498. Available from: 10.1016/s0531-5565(03)00008-1 [DOI] [PubMed] [Google Scholar]
- Vargas‐Straube, M.J. , Beard, S. , Norambuena, R. , Paradela, A. , Vera, M. & Jerez, C.A. (2020) High copper concentration reduces biofilm formation in Acidithiobacillus ferrooxidans by decreasing production of extracellular polymeric substances and its adherence to elemental sulfur. Journal of Proteomics, 225, 103874. Available from: 10.1016/j.jprot.2020.103874 [DOI] [PubMed] [Google Scholar]
- Virieux‐Petit, M. , Hammer‐Dedet, F. , Aujoulat, F. , Jumas‐Bilak, E. & Romano‐Bertrand, S. (2022) From copper tolerance to resistance in Pseudomonas aeruginosa towards patho‐adaptation and hospital success. Genes, 13, 301. Available from: 10.3390/genes13020301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton, B. & Livingstone, P. (2015) Managing and eradicating wildlife tuberculosis in New Zealand. New Zealand Veterinary Journal, 63, 77–88. Available from: 10.1080/00480169.2014.981315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts, M.P. & Moreau, J.W. (2016) New insights into the genetic and metabolic diversity of thiocyanate‐degrading microbial consortia. Applied Microbiology and Biotechnology, 100, 1101–1108. Available from: 10.1007/s00253-015-7161-5 [DOI] [PubMed] [Google Scholar]
- Watts, M.P. , Spurr, L.P. , Lê‐Cao, K.A. , Wick, R. , Banfield, J.F. & Moreau, J.W. (2019) Genome‐resolved metagenomics of an autotrophic thiocyanate‐remediating microbial bioreactor consortium. Water Research, 158, 106–117. Available from: 10.1016/j.watres.2019.02.058 [DOI] [PubMed] [Google Scholar]
- Wei, W. , Liu, X. , Sun, P. , Wang, X. , Zhu, H. , Hong, M. et al. (2014) Simple whole‐cell biodetection and bioremediation of heavy metals based on an engineered lead‐specific operon. Environmental Science & Technology, 48(6), 3363–3371. Available from: 10.1021/es4046567 [DOI] [PubMed] [Google Scholar]
- Welman‐Purchase, M.D. , Castillo, J. , Gomez‐Arias, A. , Matu, A. & Hansen, R.N. (2024) First insight into the natural biodegradation of cyanide in a gold tailings environment enriched in cyanide compounds. Science of the Total Environment, 906, 167174. Available from: 10.1016/j.scitotenv.2023.167174 [DOI] [PubMed] [Google Scholar]
- Wen, B. , Zeng, W.F. , Liao, Y. , Shi, Z. , Savage, S.R. , Jiang, W. et al. (2020) Deep learning in proteomics. Proteomics, 20, e1900335. Available from: 10.1002/pmic.201900335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, C. & Gadd, G.M. (1998) Accumulation and effects of cadmium on sulphate‐reducing bacterial biofilms. Microbiology, 144, 1407–1415. Available from: 10.1099/00221287-144-5-1407 [DOI] [PubMed] [Google Scholar]
- Wibberg, D. , Bremges, A. , Dammann‐Kalinowski, T. , Maus, I. , Igeño, M.I. , Vogelsang, R. et al. (2016) Finished genome sequence and methylome of the cyanide‐degrading Pseudomonas pseudoalcaligenes strain CECT5344 as resolved by single‐molecule real‐time sequencing. Journal of Biotechnology, 232, 61–68. Available from: 10.1016/j.jbiotec.2016.04.008 [DOI] [PubMed] [Google Scholar]
- Wild, S.R. , Rudd, T. & Neller, A. (1994) Fate and effects of cyanide during wastewater treatment processes. Science of the Total Environment, 156, 93–107. Available from: 10.1016/0048-9697(94)90346-8 [DOI] [Google Scholar]
- Yang, H.C. & Rosen, B.P. (2016) New mechanisms of bacterial arsenic resistance. Biomedical Journal, 39, 5–13. Available from: 10.1016/j.bj.2015.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Li, Y. , Tian, X. , Zang, X. , Yang, S. , Qiao, H. et al. (2023) Pb exposure causes non‐linear accumulation of Pb in D. melanogaster controlled by metallothionein B and exerts ecological effects. Science of the Total Environment, 900, 165680. Available from: 10.1016/j.scitotenv.2023.165680 [DOI] [PubMed] [Google Scholar]
- Zeng, T. , Hu, Q. , Rene, E.R. & Lens, P.N.L. (2023) Microbial community and extracellular polymeric substances analysis of anaerobic granular sludge exposed to selenate, cadmium and zinc. Microbial Biotechnology, 16, 463–473. Available from: 10.1111/1751-7915.14187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, G. , Li, D. & Zhang, L. (2012) Aerobic bioreduction of nickel(II) to elemental nickel with concomitant biomineralization. Applied Microbiology and Biotechnology, 96, 273–281. Available from: 10.1007/s00253-011-3827-9 [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Zhou, Y. , Bao, H. , Zhang, L. , Wang, R. & Zhou, X. (2015) Plasmid‐borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiological Research, 172, 1–6. Available from: 10.1016/j.micres.2015.01.008 [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Zhou, C. , Guo, X. , Hu, G. , Li, G. , Zhuang, Y. et al. (2021) Exposed to mercury‐induced oxidative stress, changes of intestinal microflora, and association between them in mice. Biological Trace Element Research, 199, 1900–1907. Available from: 10.1007/s12011-020-02300-x [DOI] [PubMed] [Google Scholar]
- Zheng, R. , Wu, S. , Ma, N. & Sun, C. (2018) Genetic and physiological adaptations of marine bacterium Pseudomonas stutzeri 273 to mercury stress. Frontiers in Microbiology, 9, 682. Available from: 10.3389/fmicb.2018.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhöfer, A. & Kujala, K. (2021) Assessing the diversity and metabolic potential of psychrotolerant arsenic‐metabolizing microorganisms from a subarctic peatland used for treatment of mining‐affected waters by culture‐dependent and ‐independent techniques. Frontiers in Microbiology, 12, 648412. Available from: 10.3389/fmicb.2021.648412 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Natural or anthropogenic sources of cyanide, arsenic and heavy metals, permissible limits and concentrations in different complex wastes. The numbers in parentheses correspond to the references.


