Abstract
Large full-thickness skin lesions have been one of the most challenging clinical problems in plastic surgery repair and reconstruction. To achieve in situ skin regeneration and perfect clinical outcomes, we must address two significant obstacles: angiogenesis deficiency and inflammatory dysfunction. Recently, black phosphorus has shown great promise in wound healing. However, few studies have explored the bio-effects of BP to promote in situ skin regeneration based on its nanoproperties. Here, to investigate whether black phosphorus nanosheets have positive bio-effects on in situ skin repair, we verified black phosphorus nanosheets’ positive effects on angiogenic and anti-inflammatory abilities in vitro. Next, the in vivo evaluation performed on the rat large full-thickness excisional wound splinting model more comprehensively showed that the positive bio-effects of black phosphorus nanosheets are multilevel in wound healing, which can effectively enhance anti-inflammatory ability, angiogenesis, collagen deposition, and skin re-epithelialization. Then, multiomics analysis was performed to explore further the mechanism of black phosphorus nanosheets’ regulation of endothelial cells in depth. Molecular mechanistically, black phosphorus nanosheets activated the JAK-STAT-OAS signaling pathway to promote cellular function and mitochondrial energy metabolism in endothelial cells. This study can provide a theoretical basis for applying two-dimensional black phosphorus nanosheets as nanomedicine to achieve in situ tissue regeneration in complex human pathological microenvironments, guiding the subsequent optimization of black phosphorus.
Keywords: black phosphorus nanosheets, wound healing, angiogenesis, multiomics analysis, JAK-STAT signaling, oligoadenylate synthetase, mitochondrial function
Large full-thickness skin lesions caused by extensive burns, severe trauma, and resection of giant tumors have been one of the most challenging clinical problems in plastic surgery repair and reconstruction. The standard treatment for sizable full-thickness skin defects is split-thickness skin grafting.1 Still, grafted skin patients suffer from dysfunctional and disfiguring scarring, often causing unbearable pain to the patient’s organism, especially in the pediatric population.2 Besides, split-thickness skin grafts increase the patient’s wound healing burden and are always limited by the donor-site skin source, leading to infection, sepsis, multiple organ dysfunction, and death.3 Although the current skin substitutes can partially accelerate wound healing, they still have many problems, such as slow revascularization after transplantation, poor survival, and lack of anti-infection ability and mechanical properties.4 Therefore, it is urgent to develop biodegradable dressings that can temporarily protect large skin defects while reducing the inflammatory response and promoting angiogenesis, thus stimulating the in situ regeneration of damaged skin to achieve wound closure.
Anatomical integrity with the comparable function of the skin is finally restored through the complex and dynamic process of wound healing. A typical wound healing process is made up of four multilevel and overlapping phases: coagulation, inflammation, proliferation, and remodeling.5 Endothelial cells (ECs) function and homeostasis are critical to all stages of large skin defect wound repair. When receiving injury signals, ECs must respond rapidly to the inflammatory environment, proliferate, and migrate to develop new blood vessels, thus providing oxygen and necessary nutrients for repairing skin tissues.6 Moreover, it acts as a hub to release biosignals (growth factors or cytokines) to communicate with stem cells, immunocytes, fibroblasts, keratinocytes, and other cells in the injured sites to regenerate the tissue.7 Complex cross-talk ensures these cells’ activation, migration, and plasticity during tissue repair. The ECs regulation on skin tissue repair is greatly orchestrated by the Janus kinase (JAK) signal transducer and activator of transcription (STAT) pathway.8 The proliferation, migration, cell survival, and immunity of ECs are stimulated by a variety of JAK/STAT ligands, including multifarious growth factors, cytokines, and chemokines.9 These biological processes are precisely controlled essential cellular mechanisms for tissue regeneration, energy metabolism, and immune response. For instance, the JAK-STAT signaling contributes to the EC activation, which physiologically recruits leukocytes and platelets to the injury site as well as facilitates hemostasis, immune regulation, and angiogenesis.10 The Oligoadenylate synthetase (OAS) family comprises OAS1, OAS2, OAS3, and OASL. OAS, one of the downstream target proteins of JAK-STAT, can also be directly induced by IFN and has been widely studied in the antiviral field. Functionally, OAS can exert antiviral activity via the classical ribonuclease L-dependent pathway.11 OAS family genes have been linked to a variety of immune-related illnesses and pivotal cellular functions like apoptosis, growth, breathing, and energy metabolism, in addition to their antiviral effects. For example, the OAS family has not only been reported in tumor progression12 and autoimmune diseases13 but has also been found to be closely related to the function and homeostasis of ECs.14 However, little is known regarding the role of JAK-STAT signaling and the OAS family of ECs’ function in wound healing.
Nanotechnology is rapidly flourishing, and various nanomaterials are developed in wound healing, such as inorganic nanoparticles, polymeric nanostructures, liposomes, etc.15−17 The tunable physicochemical properties of nanomaterials can be employed not only as multifunctional drug-delivery platforms but also as bioactive reagents to modulate cellular functions through precise design.18−20 Compared with biochemical agents, we could manipulate the intracellular distribution and degradation rate by controlling the physicochemical properties of the nanomaterials. Even through the modification of nanomaterials, it is possible to precisely regulate cellular biological behaviors by targeting specific organelles and genes. Black phosphorus (BP), a inorganic nanomaterial composed of a single phosphorus element, sparks substantial interest among scientists because of its physicochemical properties, such as high surface activity, adjustable bandgap, excellent electrical conduction, and good biocompatibility.15 In particular, the biofriendly property that BP can degrade nontoxically to phosphate and phosphonate significantly reduces concerns about its biosafety, a prerequisite for bioactive reagents. Notably, recent studies focus on developing BP as drug-delivery platforms or composite on bio-scaffolds utilizing photodynamic or photothermal therapy for various types of wounds.21−24 It has been ignored that BP itself could possibly regulate a cell biological process, which hinders the concept and research to develop BP as nanobioactive medicine. Lately, He et al. demonstrated that BP quantum dots could act as immunomodulatory agents that maintain NK cell viability and immunity via the interaction with Toll-like receptors, thus activating downstream NF-κB signaling pathways to regulate cytokine secretion and reprogram cell metabolism.25 However, there are few studies to discuss and explore BP’s interface and druggability on skin regeneration: whether BP can enhance cellular function to promote in situ tissue regenerative capacity by activating specific biochemical signaling pathways during wound repair. Nonetheless, before investigating how to innovate and optimize the design of BP as nanomedicines for wound therapy, the molecular mechanisms and potential targets based on the properties of BP interacting with biological systems need to be clarified first.
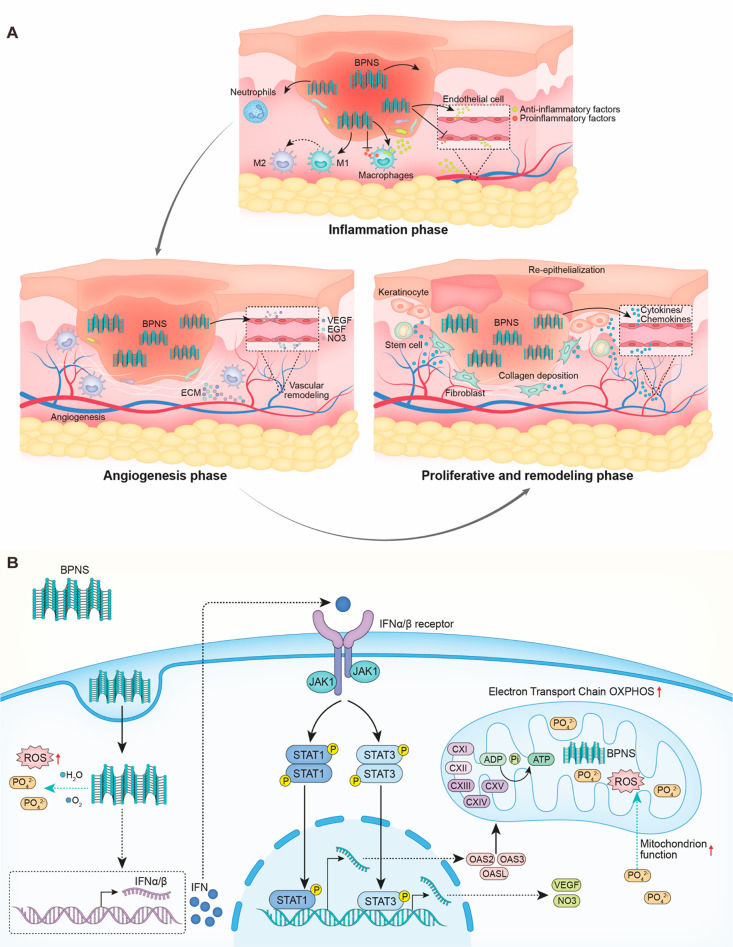
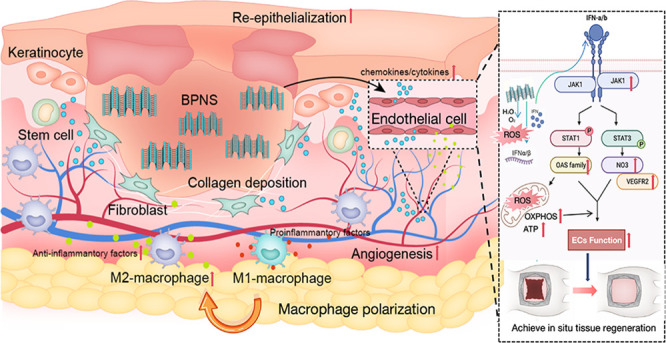
Herein, we explored the molecular mechanism of BP nanosheets (BPNS) regulation of ECs’ function in-depth and provided the theoretical basis of BP’s druggability in wound healing. In this study, we have found that BPNS could enhance angiogenesis and anti-inflammation in vitro. Then, we applied GelMA/BPNS on the rat excisional wound splinting model and observed that it effectively reduced the inflammatory response and promoted angiogenesis, collagen deposition, and skin re-epithelialization via releasing BPNS (Scheme 1A). Multiomics further revealed that BPNS could induce an intracellular virus-likely immune response, thus activating the downstream JAK-STAT-OAS signaling pathways to regulate cellular functions and mitochondrial energy metabolism (Scheme 1B).
Scheme 1. (A) BPNS Exerting Multilevel Bio-effects on Full-Span Wound; (B) BPNS Entering ECs and Inducing the Virus-Likely Immune Response, Thus Activating JAK-STAT-OAS Signaling and Enhancing the Mitochondrion Function.
In healing inflammation phase, BPNS could affect macrophages polarization and ECs activation to rapidly shut off inflammatory response; in angiogenesis phase, BPNS enter the ECs and promote ECs functions, such as angiogenic capacity, CAMs, ECM organization; in proliferative and remodeling phase, BPNS regulate ECs’ cytokines secretion to communicate with stem cells, fibroblasts, and keratinocytes at the injury site, thus promoting the re-epithelialization and collagen deposition. .
Results and Discussion
Characterization of BPNS Nanomaterials
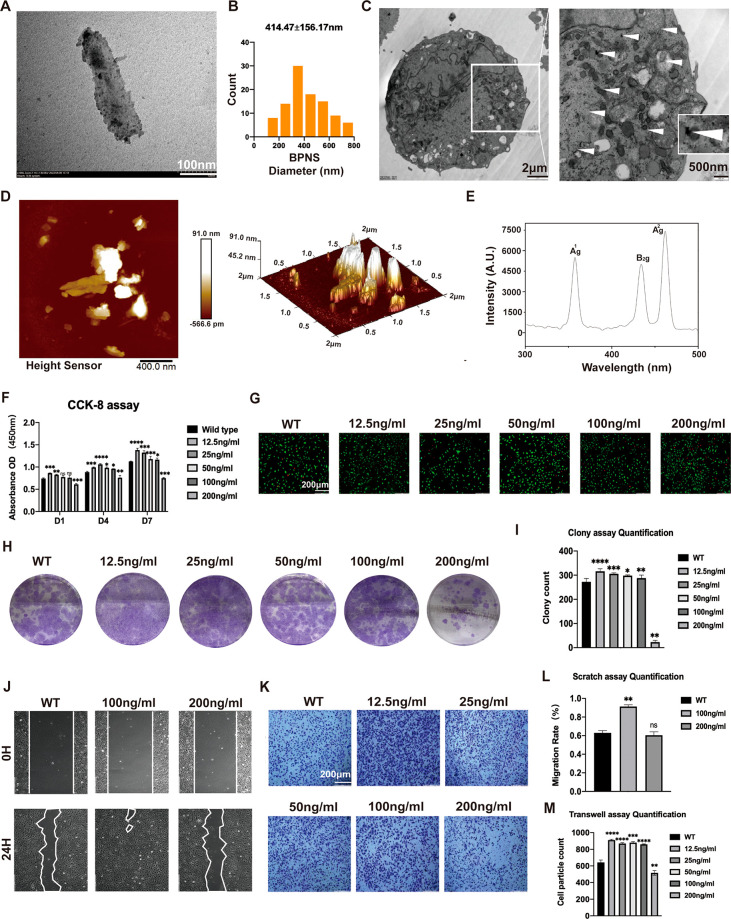
Two-dimensional (2D) BPNS were purchased and used in the research to investigate the bio-effects of 2D BP nanomaterials. Since we wanted to explore in depth the biological mechanisms triggered by the physicochemical properties of BPNS itself, we did not apply additional modifications to the BPNS. The morphologies of BPNS were characterized via transmission electron microscopy (TEM) and atomic force microscope (AFM) (Figure 1A–D). TEM determined the size distribution of BPNS (100 BPNS analyzed), and results showed that the diameter of BPNS is 100–800 nm (mean diameter = 414.47 ± 156.17 nm) (Figure 1B). Then, we employed TEM to verify whether BPNS could enter the cells and intracellular distribution. TEM images showed the BPNS could enter the human umbilical vein endothelial cells (HUVECs) when HUVECs were cultured with EC medium containing BPNS for 24 h. After entering the cell, BPNS can be distributed in different compartments, such as cell membranes, cytoplasm, and organelles (Figure 1C). AFM showed the 2D and 3D morphologies of BPNS (Figure 1D). Similar to what was previously reported, BPNS displayed three distinct peaks based on Raman spectra. Three distinct characteristic peaks of BPNS around 364.2, 438.5, and 467.2 cm–1 are attributed to its Ag1, B2g, and Ag2 modes (Figure 1E). These results confirmed the BPNS we purchased.
Figure 1.
Characterization and biocompatibility of BPNS nanomaterials. (A) TEM image of 2D morphology of BPNS. Scale bar: 100 nm. (B) 100 BPNS particle diameter analysis: diameter of BPNS, 100–800 nm (mean diameter = 414.47 ± 156.17 nm). (C) TEM image of intracellular distribution of BPNS in HUVECs after 24 h treatment. Scale bar in the left column: 2 μm. Scale bar in the right column: 500 nm. (D) AFM showing the 2D and 3D morphologies of BPNS. Scale bar: 400 nm. (E) Raman spectra of BPNS with three distinct characteristic peaks. (F) Effects of BPNS with different concentrations on proliferation of HUVECs by CCK8 assay. (G) Effects of BPNS with different concentrations on proliferation of HUVECs by Live/Dead assay. Scale Bar: 200 μm. (H) Effects of BPNS with different concentrations on proliferation of HUVECs by Colony assay. (I) Colony assay quantification. (J) Effects of BPNS with different concentrations on horizontal migration ability of HUVECs by Scratching assay. (K) Effects of BPNS with different concentrations on vertical migration ability of HUVECs by Transwell migration assay. (L) Scratching assay quantification. (M) Transwell migration assay quantification. Data are shown as the mean ± standard deviation (n = 4). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; ns, not significant.
Biocompatibility of BPNS Nanomaterials
We performed biocompatibility tests with different concentrations of BPNS to explore the optimal concentration of BPNS-regulated ECs. The capacity of BPNS to affect cell viability and proliferation was tested by Cell Counting Kit 8 (CCK-8) and Live/Dead assay. The proliferation ability of HUVECs cultured with EC medium containing 0, 12.5, 25, 50, 100, and 200 ng/mL BPNS for 1, 4, and 7 days was quantified by CCK-8 (Figure 1F). Except for 200 ng/mL, the O.D. values increased with time, showing that the cells maintained good vitality and could proliferate. Besides, BPNS exert their bio-effects on cell proliferation rate, which varies with different concentrations. BPNS with a concentration below 200 ng/mL could boost the proliferation rate of HUVECs on days 4 and 7. In the Live/Dead assay, we found the same cell viability tendency as in the CCK-8 assay (Figure 1G). The 200 ng/mL group had less living cell count with a notable dead cell rate, which indicated the importance of the concentration of BPNS on proliferation. In addition, the colony formation assay verified that BPNS with 0, 12.5, 25, 50, and 100 ng/mL groups have positive effects on the cloning ability of HUVECs, and the differences were statistically significant (Figure 1H,I).
ECs’ migration is a hallmark of angiogenesis and vascular remodeling. To investigate whether BPNS could elevate the migration ability of HUVECs, we employed the Transwell migration and the Scratch assay. In the Scratch assay (Figure 1J,L), HUVECs cultured in EC medium with 100 ng/mL BPNS significantly promote the horizontal migration ability of HUVECs. In the Transwell migration assay, HUVECs cocultured with BPNS (coBPNS) concentration lower than 200 ng/mL significantly enhanced the capability of vertical migration ability compared with the WT group (Figure 1K,M). After these assays to screen the BPNS concentration, we applied 100 ng/mL BPNS to the subsequent ECs function study.
BPNS Nanomaterials Accelerate Angiogenesis In Vitro
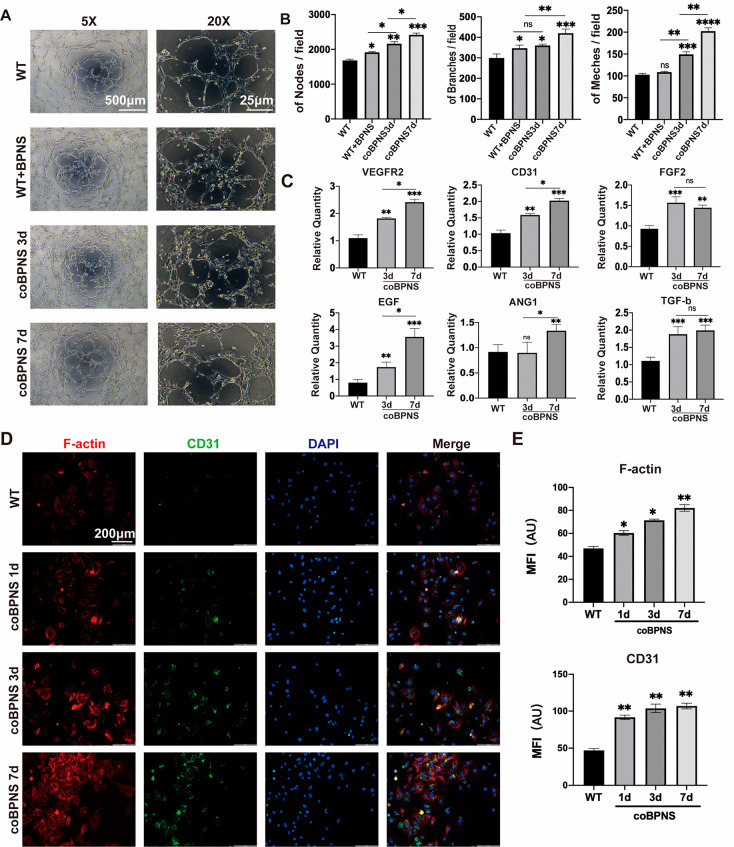
Many nanomaterials have been reported to modulate angiogenesis, such as carbon nanotubes, borate-based glasses, ceria nanozyme, gold nanoparticles, etc.26−29 Their effects in regulating angiogenesis were mainly related to morphology, size, dose, and treatment duration.30 Previous studies have demonstrated that BPNS can promote angiogenesis through photothermal effects. Still, whether BPNS itself has the function of regulating angiogenesis has yet to be explored. To study the bio-effects of BPNS on angiogenesis regulation, we conducted the tube formation assay to test the angiogenic capacity of HUVECs cocultured with BPNS. Before the assay, HUVECs were cocultured with EC medium containing BPNS for 3 or 7 days (coBPNS3d and coBPNS7d). During the assay, we also set a group in which BPNS had been added into the wells at the beginning of the assay (WT+BPNS group) to verify whether BPNS exhibits an instant bio-effect on angiogenesis (Figure 2A). After 6–8 hours of incubation, the HUVECs on the matrigel were able to form tiny tubular structures. Calculating the average number of nodes, meshes, and branches per field revealed the positive bio-effect of the BPNS (Figure 2B). We observed that both the WT+BPNS group and coBPNS groups exhibited significantly promoted angiogenic ability in vitro. Obviously, the stimulatory effect of BPNS intensified with increasing coculture time (Figure 2B). The WT+BPNS group partially accelerated the angiogenesis of HUVECs: adding additional BPNS during the tube formation assay can significantly boost the formation of nodes and branches, which indicated that BPNS exhibited instant bio-effects on ECs. Next, we evaluated the angiogenesis-related gene expression level of HUVECs cultured with BPNS via qRT-PCR (Figure 2C). VEGFR2, CD31, FGF2, ANG1, TGF-b, and EGF were upregulated in the coBPNS3d and coBPNS7d groups. Interestingly, we found that BPNS regulated the angiogenic gene profile of ECs differently. For example, BPNS has a coculture time-dependent trend for VEGF2, CD31, and EGF. However, for FGF2 and TGF-b, coculture for 3 days significantly increased gene expression. For ANG-1, 7 day coculture was sufficient to induce upregulation of its expression.
Figure 2.
Angiogenesis regulation in vitro. (A) Tube formation assay of HUVECs after 6 hours. Scale bar in the left column: 500 μm. Scale bar in the right column: 20 μm. (B) Quantitative results of tube formation assay. BPNS exhibit positive bio-effects on angiogenesis. (C) BPNS’s bio-effects on promoting expression of angiogenesis-related genes (VEGFR-2, CD31, FGF-2, ANG-1, TGF-b, and EGF). (D) IF image of F-actin and CD31 in HUVECs. Scale bar: 200 μm. (E) MFI quantitative analysis of F-actin and CD31. Cocultured with BPNS could significantly enhance F-actin and CD31 signal of ECs, and the effect is culture time-dependent. Data are shown as the mean ± standard deviation (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001; ns, not significant.
Moreover, BPNS could nontoxically degrade to phosphate and phosphonate. As the donor of the phosphate group, BP degradation products as reaction substrates can participate in a series of protein phosphorylation and promote various cellular biological processes. Therefore, we also assessed whether the promotion of angiogenesis of BPNS results from their degradation products. However, cocultured with different concentrations of H3PO4 and Na2HPO4 could not significantly promote the tube formation of HUVECs (Supporting Information Figure S2D,E). These results indicated that the nanomaterial structure might be the primary mechanism of BPNS’ positive bio-effects rather than the metabolites.
CD31 was a classic marker of ECs, and its expression level was related to the angiogenic capacity.31 Also, the constant remodeling of the actin cytoskeleton is essential for ECs migration in angiogenesis.32 Thus, we test the expression of CD31 and F-actin characterized by immunofluorescence (IF) staining (Figure 2D,E). IF staining was performed after treatment of HUVECs with BPNS for 0, 1, 3, and 7 days. The intensive signal of the F-actin and CD31 protein in ECs represented the activation of angiogenesis. Quantitative analysis of the pictures by the mean fluorescence intensity (MFI) showed that the expression of F-actin and CD31 was significantly enhanced with increasing BPNS coculture time of cells. Coculturing with BPNS for 1 day could significantly enhance F-actin and CD31 expression of ECs, and the effect gradually increased with the prolongation of the culture time. These results confirmed that the entering of nanomaterials into cells may provide structural support, cause skeletal remodeling, and enhance the vascular regenerative ability of ECs.
Anti-inflammation Regulation of BPNS Nanomaterials In Vitro
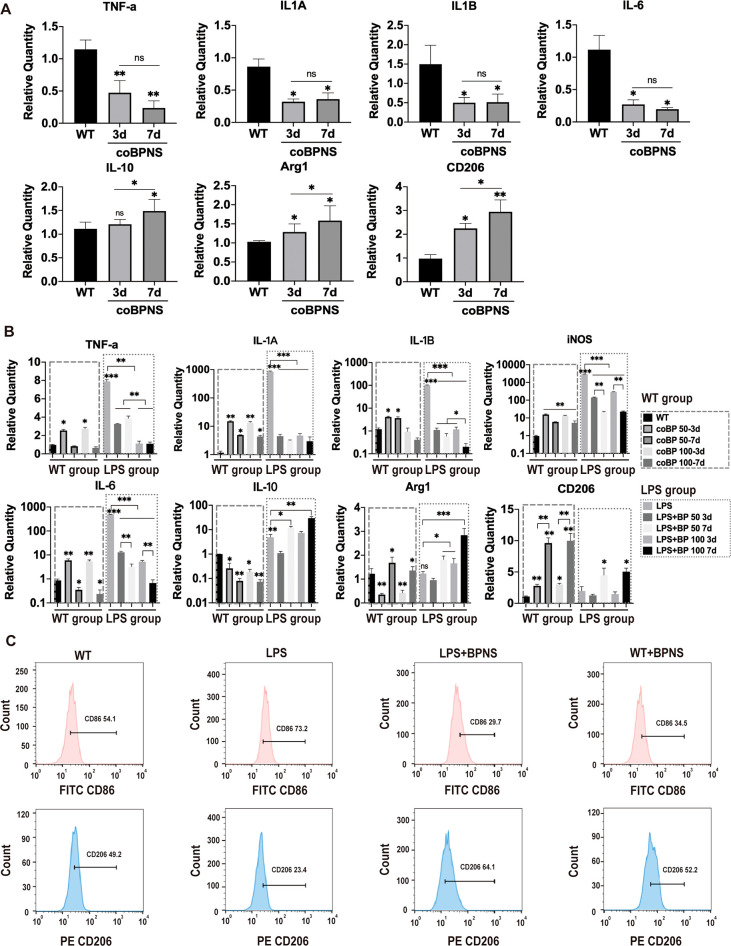
Persistent, hard-to-resolve inflammation caused by large full-thickness skin defects is the main reason for preventing skin regeneration. Vascular ECs could change their phenotypes to support different stages of inflammation even though the identity and functions of immunocytes have historically been the focus of inflammation research.33 ECs could also reactively secrete anti-inflammatory and pro-inflammatory factors when accepted injury signals.34 Therefore, we evaluated the inflammation-related gene expression profile of HUVECs cultured with BPNS via qRT-PCR (Figure 3A). The pro-inflammatory factors (e.g., TNF-a, IL-1A, IL-1B, and IL-6) were significantly downregulated at 3 days of coculture with BPNS and were not significantly different from the those of the coBPNS7d group, indicating that BPNS could suppress pro-inflammatory genes of ECs in a short exposure time. In contrast, the expression of anti-inflammatory factor-related genes (e.g., IL-10, Arg1, and CD206) was significantly higher in the coBPNS7d group than in the coBPNS3d group, showing the anti-inflammatory effects enhanced with increasing coculture time.
Figure 3.
Anti-inflammation regulation in vitro. (A) Downregulated expression pro-inflammatory genes TNF-a, IL-1A, IL-1B, and IL-6 and upregulated anti-inflammatory genes including IL-10, Arg1, and CD206 on day 3/day 7 in ECs. (B) Expression of pro-inflammatory genes TNF, IL-1A, IL-1B, iNOS, and IL-6 and anti-inflammatory genes including IL-10, Arg 1, and CD206 on day 3/day 7 in macrophages. (C) Flow cytometry results showing BPNS’ bio-effects on macrophage polarization. Data are shown as the mean ± standard deviation (n = 3). *p < 0.05, **p < 0.01, and ***p < 0.001; ns, not significant.
Macrophages play a crucial role in the inflammatory process of wound repair. Given its function in halting inflammation, eliminating apoptotic cells from the wound bed, assuring the start of the proliferative phase, and limiting autoreactivity to released self-antigen, the polarization of macrophages from the M1 to the M2 subgroup is a significant event in wound healing.5 Moreover, more angiogenic potential exists in the M2 phenotype than in other subsets.35 Herein, lipopolysaccharide (LPS) was employed in this study to drive RAW264.7 cells to the M1 subgroup to simulate the in vivo late-phase inflammatory conditions of wound healing. After 12 h treatment, a fresh DMEM-LPS medium with BPNS was added to the LPS medium for 3 or 7 days. We also set a parallel WT group cultured with DMEM LPS-free medium with the same coculture concentration and time of BPNS to evaluate the bio-effects of BPNS on resting macrophages. It was found that BPNS could reduce the pro-inflammatory factors (e.g., TNF-a, IL-1A, IL-1B, iNOS, and IL-6) caused by acute inflammation while promoting the secretion of anti-inflammatory factors (e.g., IL-10, Arg1, and CD206), and the effect intensity is related to dose and treatment duration (Figure 3B LPS group). The flow cytometry results further confirmed BPNS’ bio-effects on macrophage polarization (Figure 3C). Cocultured with BPNS could downregulate the expression M1 marker CD86 (LPS group 73.2% vs LPS+BPNS group 29.7%) and upregulate the M2 marker CD206 (LPS group 23.4% vs LPS+BPNS group 64.1%) on LPS-induced cells’ surface. These findings suggest that BPNS could elevate anti-inflammatory factors to inhibit the inflammation development and further promote the macrophage polarization to the M2 type, ensuring the course of regeneration.
Moreover, we were surprised that BPNS might transiently trigger a slight inflammatory response in resting macrophages. Notably, BPNS did not seem to trigger an acute immune response as LPS (Figure 3B). For resting macrophages, cocultured with BPNS for 3 days promoted the secretion of pro-inflammatory factors, which gradually decreased with increasing dose or treatment duration (Figure 3B WT group). The results indicated that short-term coculture of BPNS could activate local resting macrophages to respond to early stage acute inflammation, then gradually inhibit the secretion of pro-inflammatory factors, and produce anti-inflammatory factors (Arg1 and CD206) as the coculture time and dose increase. Also, BPNS was able to promote the expression of the M2-type surface characteristic marker CD206 in macrophages at a resting state, suggesting that BPNS might induce the direct transition of resting macrophages to the M2-type. The data of flow cytometry further provided evidence for this, which showed BPNS reduced the CD86′s expression (WT group 54.1% vs WT+BPNS group 34.5%) and promoted the CD206′s expression (WT group 49.2% vs WT+BPNS group 52.2%) in the WT+BPNS group (Figure 3C). Currently, the regulation of macrophage M1 to M2 polarization by nanomaterials, consequently promoting tissue regeneration, is an emerging area of tissue engineering.36,37 Our results showed that BPNS can affect the phenotypes of macrophages in the resting state, but not so significantly as in the acute state. These results together indicated that BPNS can stimulate the activation of immune cells like biological macromolecules, and this proper activation is beneficial for tissue regeneration. Similarly, several nanomaterials (e.g., nanohydroxyapatite, nano metal–organic frameworks, etc.) could evoke immune responses like biomolecules, either.38,39 Their physicochemical properties can be modified to modulate the cellular triggered immune response, applied as a disease therapeutic strategy.37,40
In addition, inflammation and ROS go hand in hand, so we performed ROS detection to assess whether BPNS caused excessive elevation of intracellular ROS and leakage in normal cells, exacerbating the local inflammatory response in wound. Kong et al. have demonstrated BPs’ ROS-mediated mechanism on different cell lines: BP might specifically increase intracellular ROS levels in cancer cell lines, but normal cell lines would only see a modest increase.41 Thus, we assessed the ROS level in ECs and tumor cells. As shown in Figure S1A,B, compared with the positive control (exposed with H2O2 for 6 hours), coculturing with BPNS moderately increased the intracellular ROS level in HUVECs with a time-dependent effect. While in tumor cells (143B), we observed elevated levels of intracellular ROS after coculturing with the same BPNS concentration for 1 hour in a time-dependent manner. There was no statistically significant difference in ROS levels at 1 day of treatment and oxidative stress (positive control) (results are only shown up to 1 day due to extensive tumor cell death after 1 day of BPNS treatment) (Figure S1C,D). These interesting results confirmed that the ability of BPNS to promote intracellular ROS production varies for different cell lines. Furthermore, ROS is an upstream messenger of numerous pathways involved in cellular biological events.42 Thus, we believe moderate intracellular ROS production in ECs contributes to multiple biological processes during wound healing, such as immune response, angiogenesis, etc.42
Multilevel Positive Effects of BPNS Nanomaterials on Wound Healing Process In Vivo
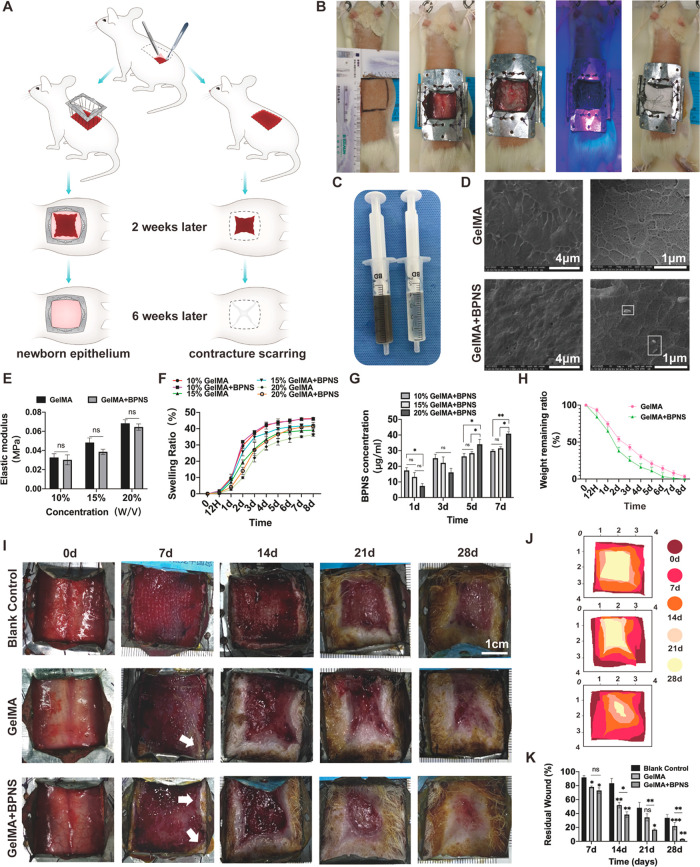
We constructed a large full-thickness skin defect splinting rat model to investigate the BPNS’ bio-effects covering the full-time wound repair span. Because rodents and humans have very different wound-healing characteristics, rats have a contracture-based closure compared to humans, who mend wounds by generating newborn tissue.43 Therefore, we designed anticontracture molds for rats’ skin defects to limit the contracture nature during wound healing so that the healing process is more similar to the physiological function of human wound healing: the healing site is dominated by newborn epithelium, rather than contracture scarring (Figure 4A). We excised 10 cm2 (3.3 cm × 3.3 cm) of full-thickness skin on the backs of the rats, which is about 40–50% of the total skin area on the backs of the rats. The molding process was described in detail in the Experimental Section (Figure 4B).
Figure 4.
BPNS’ positive bio-effects on wound healing in vivo. (A) Animal experimentation pattern diagram and the advantages of rat excisional wound splinting model. Applying anticontracture molds for rats’ skin defects makes the healing site be dominated by newborn epithelium rather than contracture scarring. (B) Rat excisional wound splinting model surgery procedure and application of GelMA and GelMA/BPNS hydrogel in vivo: the incision was marked in the back of the rat as a 3.3 cm × 3.3 cm square. Then, the proposed excision area was incised and removed. Following that, the anticontracture mold was firmly sutured to the rat’s skin. Finally, the hydrogel was injected into the wound surface, cured by 25 s blue light irradiation, and fixed by gauze coverage. (C) Appearance image of GelMA and GelMA/BPNS hydrogel. (D) SEM images of GelMA and GelMA/BPNS hydrogels. Scale bar in the right column: 4 μm. Scale bar in the left column: 1 μm. (E) Quantitative analysis of the elastic modulus of hydrogels. (F) Swelling ratio curves of GelMA and GelMA/BPNS hydrogels soaked in PBS at 37 °C. (G) Quantitative analysis of BPNS release from different concentrations of GelMA/BPNS at 1, 3, 5, and 7 days. (H) Weight remaining ratio curves of 20% GelMA and GelMA/BPNS hydrogels. (I) Micrographs of large full-thickness wounds healing in rats. Scale bar: 1 cm. (J) Schematic illustration of wound morphological changes in the different groups over 28 days. (K) Quantitative results of the residual wound. The GelMA/BPNS group showed the most effective promotion of wound closure. Data are shown as the mean ± standard deviation (for panels E and F, n = 4; for panels G and H, n = 3; for panels I and K, n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001; ns, not significant.
Methacrylate gelatin (GelMA) hydrogel is widely applied to deliver therapeutic molecules in wound healing. As the nanomaterial-delivery GelMA systems have shown significantly enhanced regenerative behaviors in various tissues that are rarely observable with GelMA, GelMA is a suitable platform to study nanomaterial function.44 Thus, we packaged BPNS in GelMA for in vivo experiments. Compared with blank GelMA, GelMA/BPNS appeared deep gray due to the presence of BPNS (Figure 4C). The scanning electron microscope (SEM) could observe the morphology of some larger BPNS (about 400 nm) after loading into the GelMA hydrogel (Figure 4D, white square). Based on the hydrogel manufacturer’s instruction and previous literature, we tested three concentrations of GelMA (10, 15, and 20% (w/v)) by comparing elastic modulus, swelling behavior, and BPNS release.45−47 The elastic modulus of the hydrogel increased with a higher concentration from 10 to 20% (w/v) GelMA. Although incorporating BPNS slightly decreased the elastic modulus, it was not statistically different from the blank group (Figure 4E). In contrast, the swelling behavior decreased with increasing GelMA concentration. Moreover, in the 15 and 20% (w/v) groups, we found that adding BPNS improved the swelling properties of GelMA, which may be related to the fact that the addition of BPNS increased the porous structure of GelMA with higher concentration. 20% (w/v) GelMA/BPNS showed the ability to continuously absorb water within 7 days, indicating that the surface exudate can be continuously absorbed when applied to the wound surface (Figure 4F). In addition, we characterized BPNS release by measuring the concentration of BPNS in phosphate-buffered saline (PBS) (simulated in vivo environment) containing type I and II collagenases (GelMA degradation solutions) at 1, 3, 5, and 7 days using UV–vis.48 The BPNS release in 10 and 15% (w/v) are higher on the first day compared to that of the 20% (w/v) group. Also, a similar trend was observed at 3 days (although not statistically significant). Under the same other preparing conditions, the degradation rate decreased with increasing GelMA concentration.49 Thus, early and rapid degradation in the group with lower-concentration GelMA can release more BPNS. Interestingly, on the fifth and seventh days, the 20% (w/v) group significantly released more BPNS than the other two groups (Figure 4G). Two factors may lead to this phenomenon. First, it is related to the easy degradation of bare BPNS when they are exposed to air and oxygenated water for an extended time.48 Therefore, the early stage rapid release of BPNS in the 10 and 15% (w/v) groups was difficult to detect later by UV–vis. Second, the higher stiffness and smaller pore size of GelMA may enhance its encapsulation performance for BPNS, thereby providing a protective effect. We analyzed the pore diameter of the GelMA/BPNS, which showed the mean size of the pore is 268.25 ± 78.42 nm (range from 100 to 600 nm) (Figure S5). The distribution of pore sizes is appropriately smaller than the diameter of BPNS (Figure 1B), facilitating better encapsulation and minimizing oxygen interaction. Conversely, 10 and 15% (w/v) with larger pore sizes49 may expose the BPNS surface to more air and oxygenated water. Hence, we posit that 20% (w/v) GelMA, characterized by denser pore size and higher stiffness, contributes to slow degradation, protecting BPNS and averting sudden release effects in the early stages, thereby achieving a gradual release of BPNS. The large wound also requires the mechanical property of the hydrogel dressing to temporarily replace the deficient full-thickness skin to protect the deeper tissues. Hence, we finally chose 20% (w/v) GelMA for subsequent animal experiments. Next, we performed a degradation assay by soaking 20% (w/v) GelMA and GelMA/BPNS hydrogels in GelMA degradation solutions. We found that both hydrogels totally degraded in about 8 days, and the BPNS-doped GelMA degraded slightly faster than the blank GelMA (Figure 4H). Based on the above experimental data, we changed the hydrogel dressing every 7 days. Meanwhile, we set up a blank control group and a GelMA group to exclude the influence of other factors as much as possible (n = 6 in each group).
Wounds were photographed with a digital camera at 0, 7, 14, 21, and 28 days postoperatively (Figure 4I): newborn epithelium could be seen at the wound edge in the GelMA/BPNS group on day 7 (white arrow). The rate of wound closure was calculated for four consecutive weeks: GelMA/BPNS group > GelMA group > blank control group (Figure 4J,K). In contrast to Blank control, GelMA, as a breathable, protective dressing, can partially promote wound healing. However, the GelMA/BPNS group showed the most effective promotion of wound closure. On day 28, the residual wound rate of the GelMA/BPNS group was 3.28 ± 0.56%, which was significantly lower than that of the other two groups (Figure 4K). Notably, the tissue at the wound healing site in the GelMA/BPNS group on day 28 was newborn epithelium rather than contracture-acting normal skin or scar.
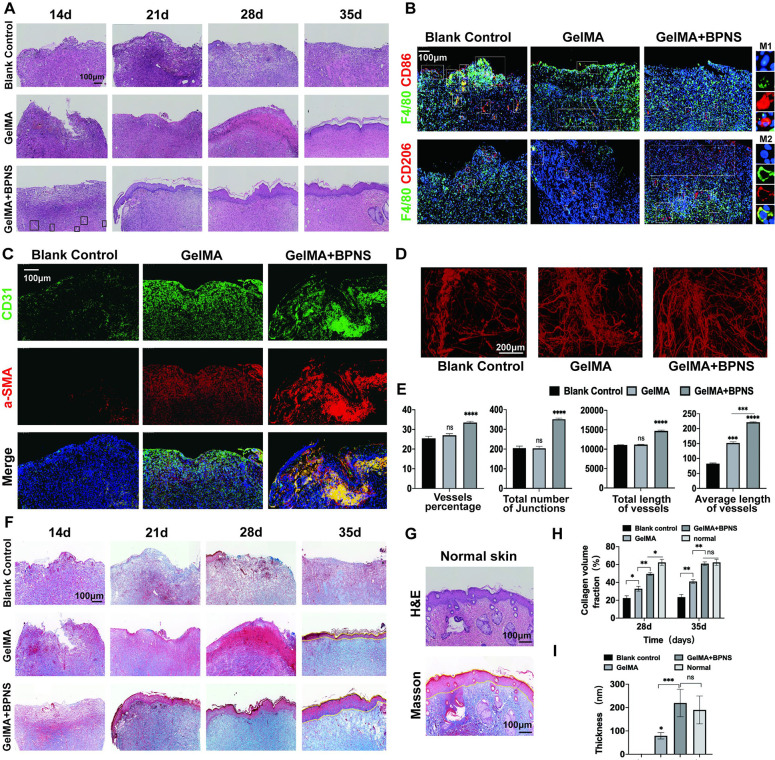
Skin samples were taken weekly, and Histological staining was performed at 14, 21, 28, and 35 days to present an overall picture of the wound healing. Hematoxylin and eosin (H&E) staining showed that wound healing was the fastest in the GelMA/BPNS group, with local inflammatory reactions beginning to subside on day 14. In other groups, on day 14, we could still observe the inflammatory granulation tissue with many immune cells owing to the relatively large proportion of defects we had molded before (Figure 5A). We further used IF staining of F4/80, CD86, and CD206 to characterize the macrophages’ polarization states of the 14 day local wound. On day 14, the prominent phenotype of macrophages is M2 in the GelMA/BPNS group, while in the other two groups were M1 (Figure 5B). The results indicated that when the other two groups were still in an inflammatory state, the wound had already shut off the inflammatory response and begun the proliferative stage of repair in the GelMA/BPNS group.
Figure 5.
In vivo histological evaluation of large full-thickness skin defects wound healing in rats. (A) H&E staining of the wound treated with GelMA or GelMA/BPNS hydrogel. (black squares, typical neovascular sprout structures). Scale bar: 100 μm. (B) IF staining of F4/80, CD206, and CD86 of macrophages on day 14. Scale bar: 100 μm. (C) IF staining of CD31 and a-SMA of vascular signal on day 21. Scale bar: 100 μm. (D) Micro-CT evaluation of newly formed vessels in the wounds on day 28. Scale bar: 200 μm. (E) Quantitative results of Micro-CT newly formed vessels analysis. (F) Masson staining of the wound. Scale bar: 100 μm. (G) H&E and Masson staining of normal skin. (H) Quantitative results of collagen volume fraction on days 28 and 35. (I) Quantitative analysis of epidermis thickness on day 35. Data are shown as the mean ± standard deviation (n = 6). *p < 0.05, **p < 0.01, and ***p < 0.001; ns, not significant.
For the angiogenic stage, we could observe some neovascular sprout structures in the GelMA/BPNS group on day 14 while in the other two groups on day 21 (Figure 5A, black square, long tubular structure). Thus, we further applied IF staining of CD31 and a-SMA to characterize the neovascularization in vivo on day 21 (Figure 5C). a-SMA and CD31 were selected to demonstrate the development of blood vessels at the local injury site. These two plasma proteins denote smooth muscle cells or ECs of mature blood vessels, respectively. IF staining of CD31 and a-SMA coexpression showed that the signal of blood vessels significantly enhanced on day 21 (Figure S2A), indicating that the angiogenesis in the GelMA/BPNS group was more active than the other two groups. The vascular signal at this time is clump-like, which may be clusters of blood vessels that have not yet undergone vascular remodeling in the early stage angiogenesis and smaller capillaries close to the epidermis. Later, micro-CT was applied to evaluate the neovascularization on day 28. We observed that the GelMA/BPNS hydrogel group had more mature vascular networks, significantly higher than those of the other two groups in qualification of the percentage of vessels, the number of junctions, the total vessels length, and the average vessels length (Figure 5D,E). The above validation results of BPNS in vivo are consistent with the cellular-level assays.
In the GelMA/BPNS group, initial epithelial structures appeared on day 21. However, at this time, epithelial structures had not appeared in the Blank control group and GelMA. We did not observe epithelial structures in the Blank control group at the center of the 35 days wound due to the difficulty in subsiding inflammation and angiogenesis disability caused by the large skin defect. Although the GelMA group also formed a complete but thin structure of the epithelium, the GelMA/BPNS group was the thickest and most complete, similar to typical skin epithelial structures on day 35 (Figure 5A,F,G,I). For the Masson staining, wounds treated with GelMA/BPNS at 28 and 35 days revealed substantial collagen deposition and thick, wavy collagen fibers, in contrast to the other two groups (Figure 5F,G,I). These results indicated that GelMA/BPNS effectively promoted collagen generation, epithelial resurfacing, and matrix remodeling in wound healing.
The above in vivo results provide us with a more comprehensive understanding of BPNS’s bio-effects on stimulating skin tissue regeneration in situ: BPNS plays essential roles in anti-inflammatory, angiogenic, re-epithelialization, and matrix remodeling processes, indicating that BPNS’ bio-effect is multilevel. These results also suggest that BPNS are promising to be developed as nanomedicine for resolving the complex pathologic environment of large skin defect wounds, while the molecular mechanisms of its interface with biological systems need to be further clearly explored.
Transcriptomic Profiling Revealing Multilevel Positive Effects of BPNS Nanomaterials on ECs in Wound Healing
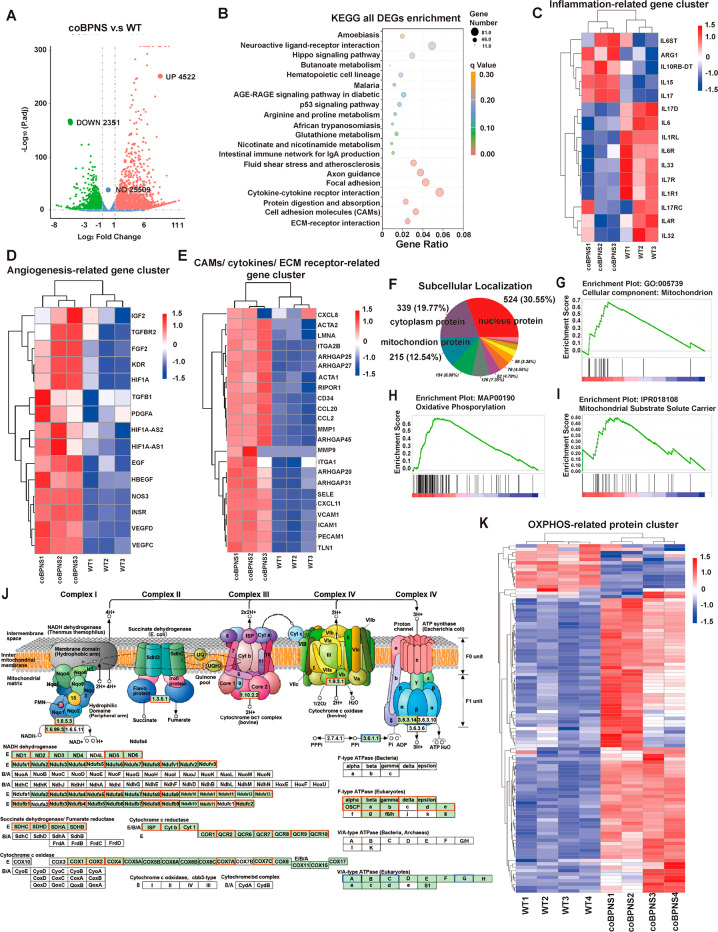
The previous cellular function results revealed that BPNS promoted both the angiogenic and anti-inflammatory capacity of ECs. Furthermore, in vivo experiments showed that BPNS have multilevel bio-effects on promoting wound healing. Therefore, we focused on the bio-effects of BPNS on ECs’ function at the transcriptomic level to uncover the biological interface between BPNS and ECs. We performed high-throughput RNA-seq sequencing of HUVECs WT cells and HUVECs cocultured with BPNS for 7 days. We found that BPNS significantly upregulated the expression of 4522 genes and downregulated the expression of 2351 genes (Figure 6A).
Figure 6.
Transcriptomic and proteomic results. (A) Volcano map showing DEGs in the WT and coBPNS groups. (B) KEGG enrichment of DEGs. (C) Clustering heatmap of inflammation-related genes’ expression. (D) Clustering heatmap of angiogenesis-related genes’ expression. (E) Clustering heatmap of genes’ expression on cytokine–cytokine receptor interaction, CAMs, and ECM–receptor interaction pathway related to wound healing. (F) Subcellular localization of DEPs. 215 mitochondrial proteins (12.54%) were affected. (G) GSEA analysis of GO:cellular component:mitochondrion. (H) GSEA analysis of KEGG:oxidative phosphorylation. (I) GSEA analysis of IPR:mitochondrial substrate solute carrier. (J) KEGG Mapper-Color Pathway of OXPHOS. (K) Clustering heatmap of OXPHOS related to protein.
First, similar to the in vitro results, we observed that BPNS treatment significantly enhanced genes directly related to inflammation and angiogenesis. The heatmap of inflammation-related genes cluster showed that BPNS upregulated the expression of ARG1, IL-10, and IL-7 anti-inflammatory factor genes and downregulated the expression of IL-1RL, IL-6ST, IL-32, IL-17, and other pro-inflammatory factor genes in ECs (Figure 6C). In clustering analysis of crucial genes associated with angiogenesis, BPNS significantly upregulated the expression of the VEGF family, KDR (known alternatively as VEGF-R2), HIF1A, NOS3, HB-EGF, FGF2, PDGFA, EGF, etc. (Figure 6D). In turn, the bio-effects generated by the expression of these genes are not limited to inflammation and angiogenesis alone. They are also involved in other biological processes of wound healing, such as matrix remodeling, epithelialization, and forming a complex network of tissue repair interactions.5,6,50
Furthermore, the enrichment of differential expressed genes (DEGs) reveals positive bio-effects of BPNS on ECs’ function at the molecular level: DEGs were mainly enriched in the Kyoto Encyclopedia of Genes and Genome (KEGG) database for cytokine–cytokine receptor interactions, cell adhesion molecules, and extracellular matrix (ECM) receptor interactions, among other pathways (Figure 6B). Our results suggest that BPNS enhanced the expression of CAMs on ECs’ surface: platelet-endothelial cell adhesion molecule 1 (PECAM-1, known alternatively as CD31), E-selectin (SELE), vascular cell-adhesion molecule 1 (VACAM-1), intercellular adhesion molecule 1 (ICAM-1), and CD34 (Figure 6E). These adhesion molecules likely play essential roles in maintaining the normal structure and function of the vascular, as well as participating in inflammation, hemostasis, angiogenesis, and tissue repair.51 For instance, in an acute inflammatory environment, leukocytes and ECs interact via various adhesive modules, which is a critical event determining the mode of immunocytes transmigration.52,53 What is more, angiogenesis is mediated by EC–EC contacts underlying vascular permeability and integrity, which are also regulated by ECs adhesion molecules.54 Some studies also demonstrated that the expression of CD34 facilitates cell survival and increases the secretion of angiogenic cytokines.55 Thus, BPNS-activated ECs not only promote vascular permeability and recruit leukocytes to the injury site to clear pathogens but also encourage angiogenesis during wound healing.
The ECM of ECs creates an organ-specific vascular niche that combines mechanosignals, growth factors, and autocrine signaling required for tissue homeostasis and regeneration.56 Transcriptomics showed that BPNS enhanced the expression of genes related to dynamic adhesion of ECs to the ECM: TLN1 (talin1), ITGA2B, ITGA1, ACTA1/2, RhoA family, MMP family, etc. (Figure 6E). BPNS dynamically regulated these genes to ensure key signaling events of ECs during skin repair, involving cell migration, invasion, proliferation, and survival.57 For example, talin binding to the integrin cytoplasmic tail is an essential step in the cell signaling cascade that activates integrin. It has been proved that ECs’ talin1 is indispensable for postnatal development angiogenesis58 and regulates wound healing.59 Liu et al. identified the activation of Src and Rho as crucial mechanisms by which collagen I provokes capillary morphogenesis of microvascular ECs.60 It was reported that MMP1 and MMP9 were related to ECs’ tubular morphogenesis.61 Also, MMP1 is unambiguously associated with re-epithelialization, playing a role in initiating the migration of keratinocytes in wound healing.62
BPNS also positively affected the secretion of cytokines/chemokines related to wound healing promotion (Figure 6E). In wound repair, ECs must interact with other cells and affect their cell migration and proliferation, which are orchestrated by various cytokines/chemokines. For instance, we found that BPNS also enhanced the expression of the CXCL8 and CCL20, which are potent chemoattractants for neutrophils and immune cells that produce IL-17A, respectively.63 Emerging studies proved their roles in accelerating epithelial migration during wound healing.50,63 The upregulating expression of chemokine CCL-2 could promote the transition from a neutrophil-rich infiltrative state to a monocyte-rich infiltrative state, contributing to tissue homeostasis and regeneration.64 Re-epithelialization is a prerequisite for complete and effective wound healing. Leading-edge keratinocyte proliferation, migration, and differentiation must be integrated into this process to restore epidermal integrity.65 Satish et al. reported that enhancing expressed CXCL11 could promote motility in undifferentiated keratinocytes and re-epithelialization of the remaining wound.66 The above results indicated that BPNS possibly affected other cells’ behavior via regulating the secretion cytokines/chemokines of ECs.
Transcriptomics has given us a general picture of the bio-effects of BPNS on EC function. The BPNS act synergistically at all stages of wound healing by upregulating inflammatory response genes, angiogenic genes, cytokines/chemokines, cell adhesion molecule expression, and ECM receptor expression, which enable ECs to act as a hub to interact and communicate with neutrophils, macrophages, stem cells, and keratinocytes, as well as ECM at skin injured sites through these biosignals.
Proteomic Profiling Revealing That BPNS Nanomaterials Could Enhance Mitochondrial Function and Oxidative Phosphorylation of ECs
We next performed Tandem Mass Tags (TMT) quantitative proteomic analysis of HUVECs WT and BPNS cocultured cells. The proteomic perspective provides us with independent information on the positive bio-effects of BPNS on ECs function that transcriptomics failed to show. Subcellular localization analysis of differential proteins showed that 215 mitochondrial proteins were affected, accounting for 12.54% of the overall number of DEPs (Figure 6F), 172 of which were positively affected (Figure S2B). Afterward, we performed Gene set enrichment analysis (GSEA) of Gene ontology (GO), KEGG, and InterPro (IPR) database in DEPs (Figure 6G–I): it was seen that GO significantly enriched the cellular components: mitochondrion (GO: 0005739) (adjust P = 0.00030); KEGG was most significantly enriched in the oxidative phosphorylation (OXPHOS) signaling pathway (MAP: 00190) (adjust P = 0); IPR was significantly enriched in the mitochondrial matrix solution carrier (IPR: 018108) (adjust P = 0.033). These proteomic results suggested that BPNS may have intrinsic capability in regulating mitochondrial function and energy metabolism in ECs.
In wound healing, in situ skin regeneration depends on the angiogenesis of ECs to provide the necessary nutrients, macromolecules, and oxygen for injured tissue. At this time, ECs must migrate, proliferate, differentiate, and communicate rapidly to initiate capillary network formation and provide the primitive vascular plexus for further angiogenesis. The demand for biosynthesis and bio-energy is also increasing. Previous studies have considered ECs’ metabolism to be dominated by aerobic glycolysis and ignored mitochondrial function for the long term.67 In contrast, Diebold et al. in 2019 revealed the necessity of ECs’ mitochondrial metabolism during angiogenesis: reducing equivalents NADH and FADH2 generated through the tricarboxylic acid cycle provide electrons to complexes I and II, respectively, to initiate respiration.68 Generally, angiogenesis requires coordinated conversion and coengagement between glycolysis and OXPHOS. In the complex in vivo microenvironment, ECs must respond to the surrounding pathological and physiological signals, orchestrating their metabolism to maintain cellular function and homeostasis.69 However, continuous inflammatory stimulation caused by large skin defects may induce ECs’ metabolic regulation dysfunction. For instance, ECs accompanied by sustaining perivascular inflammatory infiltration would express increased glycolytic activity and decreased mitochondrial OXPHOS activity.70 Therefore, targeted modulation of ECs metabolism is a potential therapeutic strategy for large skin defect wound repair. At the proteomic level, we demonstrated that BPNS upregulated complexes I, II, III, and IV, and F-type adenosinetriphosphatases (ATPase) related protein expression quantity in the electron transport chain, enhancing OXPHOS and promoting ATP synthesis in ECs (Figure 6J,K). This part of the results provides a preliminary theoretical basis for developing BP as a nanomedicine to modulate the mitochondrial metabolic reprogramming of ECs.
Besides, because mitochondria undergo cycles of fusion and fission that preserve their integrity and cross paths with apoptosis pathways, they are highly active organelles.71 Mitochondria fusion is an integral element in angiogenesis that can expedite VEGF responses and eNOS signaling activation.72 Our proteomics results showed that genes-related fusion (especially for OPA1) was upexpressed after BPNS treatment. In contrast, BPNS also downexpressed the fission-related protein DNM1L, which could help ECs cope with mitochondrial overfission under stress conditions, like injury, hypoxia, inflammation, and ischemia caused by large wounds (Figure S2C). In existing studies, nanoparticles always induce mitochondrial fission instead of fusion after endocytosis, for instance, TiO2 nanoparticles, silica nanoparticles, silver nanoparticles, and gold nanoparticles.73−76 The effect of BPNS to cause mitochondrial fusion is seemingly different, and the mechanism of its interaction with mitochondria is the direction of our subsequent exploration. In brief, the proteomic results provided us with the subcellular bio-effects of BPNS on ECs function by regulating mitochondrial dynamic protein expression and respiratory metabolism.
Multiomics Analysis Identified the Molecular Mechanism of BPNS Regulation on ECs’ Function
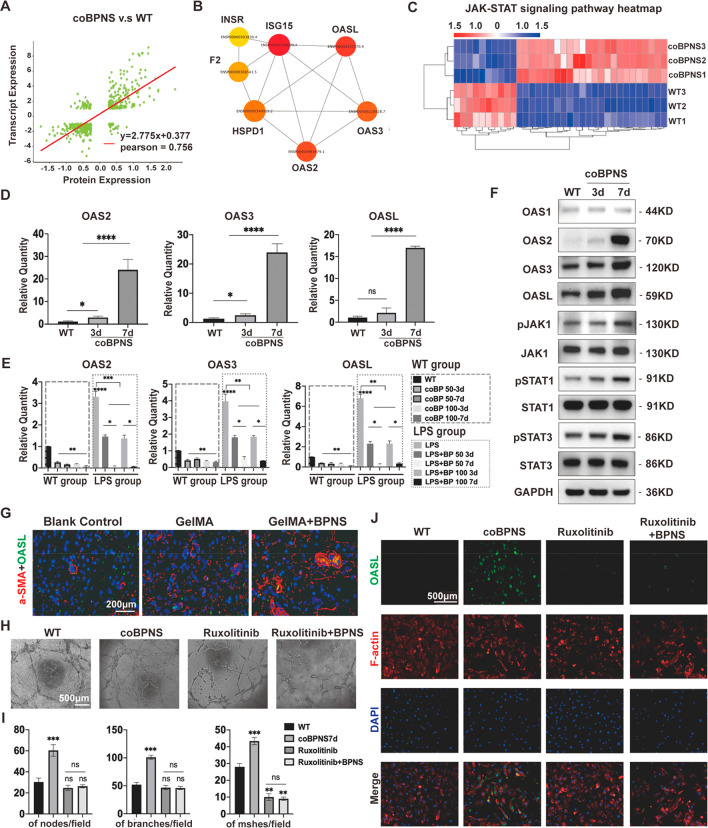
The co-analysis of transcriptomic and proteomic results was performed to deeply explore the molecular mechanism of how BPNS enhances ECs function and to uncover potential wound-healing therapeutic targets. A total of 497 DEGs/DEPs were coregulated by the two omics, and the correlation analysis showed that they were strongly correlated (Pearson index = 0.756) (Figure 7A). The top 10 proteins in TMT quantification results were selected to intersect with the DEGs upregulated in the transcriptome (n = 7). Then, the DEPs interaction network was analyzed to form the pivotal protein interaction (PPI) and identify the hub proteins, which showed that the OAS family members were significantly upregulated (Figure 7B). The multiomics analysis revealed that DEGs and DEPs of KEGG enrichment mainly enriched in interferon-related signaling pathways: e.g., PI3K-Akt, JAK-STAT, MAPK signaling pathway, etc. In the transcriptomics, we observed that BPNS enhanced the elevated levels of IFNA1 (Padj = 0.036), IFNB1 (Padj = 3.63 × 10–11), and IFNAR1 (Padj = 0.0004). Correspondingly, there was also significantly enhanced expression of IFN-induced protein in proteomics.
Figure 7.
Multiomics analysis. (A) Correlation analysis of DEGs and DEPs. (B) PPI of hub DEPs. (C) Clustering heatmap of JAK-STAT pathway genes’ expression. (D) qRT-PCR results showing that BPNS upregulated the expression of OAS family genes in HUVECs. (E) qRT-PCR results showing BPNS did not induce the upexpression of OAS family genes in RAW264.7. (F) BPNS upregulated the expression of OAS family, JAK1, p-JAK1, STAT1, p-STAT1, STAT3, and p-STAT3 proteins in HUVECs. (G) Expression of OASL in vivo. Scale bar: 200 μm. (H) Tube formation assay for HUVECs treated with Ruxolitinib. Scale bar: 500 μm. (I) Quantitative results of Tube formation assay. (J) IF images of OASL expression in HUVECs treated with Ruxolitinib. Scale bar: 500 μm. Data are shown as the mean ± standard deviation (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001; ns, not significant.
Nanomaterials’ physicochemical properties are similar to biomolecules such as bacteria and viruses. Studies have shown that BP can enter cells through endocytosis mediated by niche structures when cocultured with cells. This entry mode is similar to the cellular internalization pathway of viruses.77 Tao et al. demonstrated that caveolae-dependent endocytosis and micropinocytosis are primary mechanisms by which BPNS enter cells,78 consistent with our transcriptomic results (the upregulated gene expression of caveolin-1 and cytoskeleton) and IF staining results (the upregulated signals of F-actin). Moreover, as mentioned earlier, BPNS can also mediate a modest elevation of intracellular ROS levels in ECs, triggering an immune response that may serve as one of the chemical cues for regulating cellular function. From the above, we hypothesized that the BPNS virus likely entered the cells, elevated the intracellular ROS level, and activated interferon signaling pathways, further causing the expression of viral defense enzymes, like the OAS family.
We analyzed the upregulating DEGs enrichment pathways to determine the underlying regulatory network of BPNS-mediated OAS family expression. Cocultured with BPNS broadly enhanced the JAK-STAT signaling among those IFN pathways (Figure 7C). Then, we performed qRT-PCR and Western blot to verify whether BPNS activated the JAK-STAT-OAS signaling in ECs. Coculture with BPNS for 7 days significantly upregulated the mRNA expression of OAS2, OAS3, and OASL, and their protein expression level (Figure 7D–F). Western blot further confirmed that cocultured with BPNS for 7 days could promote the phosphorylation of JAK1, STAT1, and STAT3 (Figure 7F). The JAK-STAT pathway is regarded as one of the most essential signaling in cells that transmit messages for hormones, growth factors, and cytokines, contributing to wound healing.8,79 Moreover, different STAT isoform functions vary: STAT1 contributes to the ECs activation, cell growth, and immune response.79−83 while STAT3 target genes are mainly involved in macrophage polarization, angiogenesis, collagen deposition, and tissue remodeling.79,81,84,85 STAT1 and STAT3 act in concert with each other, making the effect of BPNS on wound healing mutilevel (Scheme 1B).
In the context of antiviral and innate immunity, the functions of the OAS family have been thoroughly investigated. Recently, it has been discovered that their biological expressions are intimately linked to the onset of various diseases. Of note, it has been proved that OASL is a vital homeostatic mediator in ECs,14 and also prevents excessive type I IFN signaling.86 Given that OASL has the highest weighting among members of the OAS family in PPI, we further focused on the effect of BPNS on the OASL expression in vivo. IF was employed to verify the coexpression of OASL with vascular endothelium (marked with a-SMA). In the deep vascular network of the epidermis on day 28, not only could we see that the GelMA/BPNS group formed thicker and mature vascular structures but we could also observe that the vascular endothelium expressed and secreted OASL protein (Figure 7G). To further investigate whether BPNS-promoted OASL expression depends on JAK signaling, we used JAK1 inhibitor Ruxolitinib to treat ECs with BPNS. In tube formation assay, we found that Ruxolitinib treatment inhibits the angiogenesis of ECs. The simultaneous addition of BPNS could not rescue the suppression effect of Ruxolitinib (Figure 7H,I). Then, IF results revealed that cocultured with BPNS significantly enhanced the OASL and F-actin signal. When the JAK pathway in ECs is blocked by Ruxolitinib, the OASL signal cannot be elevated by BPNS, while the F-actin signal is still upregulated (Figure 7J). Thus, these results confirmed that intracellular BPNS promote the ECs function via the JAK-STAT-OASL pathway.
Moreover, the OAS can be activated by an inflammatory state.14 We wondered whether BPNS cause the expression of the OAS in immunocytes, thereby triggering a sustained immune response. Thus, we performed qRT-PCR to test whether BPNS could affect the expression level of the OAS family in RAW 264.7. Surprisingly, we found that BPNS suppressed the OAS family’s expression in resting macrophages instead of stimulation. In addition, as a kind of virus defense protein, OAS could be enhanced by LPS and then alleviated after coculturing with BPNS for 3 or 7 days in the LPS medium (Figure 7E). These exciting results indicated that BPNS with proper concentration could not activate macrophages’ immune defense, although its physical nature activated virus defense signaling in ECs. It is worth mentioning that we also focused on the expression of OASL at the overall level of the wound at the inflammation phase (Figure S3). On day 14, the blank control and GelMA groups significantly expressed OASL in the superficial layer, whereas the GelMA/BPNS group presented it in the deep dermis. We hypothesized that OASL expression in the blank control group and GelMA at this time was related to the fact that the inflammatory response had not yet subsided, as this was consistent with the location of expression of local inflammatory cell infiltration in the H&E staining (Figure 5A). The GelMA/BPNS group did not observe an increased expression of OASL in the epidermal, which confirmed that BPNS can inhibit the increase of OASL in immune cells triggered by inflammation in vivo. The above results suggest that the application of BPNS does not exacerbate the inflammatory stress response in the wound site triggered by injury but instead helps the injured tissue to rapidly pass through the inflammatory phase and accelerate angiogenesis and re-epithelialization, which is in line with our previous results.
In addition, the JAK-STAT-OAS axis is also essential for cellular mitochondrial respiration and metabolic reprogramming. McGarry et al. demonstrated that JAK inhibitor tofacitinib treatment significantly affected mitochondrial function by inhibiting the production of ROS, mitochondrial mass, and membrane potential.87 Moreover, some studies have shown that OAS can not only enter the mitochondria to affect mitochondrial membrane potential and promote ROS production,88 but also some of their subunits can be localized to the mitochondrial membrane to affect the mitochondrial function directly.89 Through quantitative proteomic exploration, our study indicated for the positive role of BPNS in promoting mitochondrial OXPHOS in ECs. Nonetheless, a previous study reported that BP is mainly distributed in the endoplasmic reticulum and lysosomes instead of mitochondria after entering the cell.90 Besides, BP requires some specific modifications to achieve colocalization with mitochondria.91 We observed the intracellular distribution and degradation of BPNS using TEM for 7 days and found no significant accumulation of BPNS in mitochondria (Figure S4). Therefore, we suggest that BPNS has a limited direct targeting effect on mitochondria. Referring to the multiomics results and previous reports, we speculate that BPNS would likely affect mitochondrial function through the JAK-STAT-OAS axis. It is worth mentioning that BPNS-mediated elevation of intracellular ROS levels and degradation of the resulting phosphate groups could also contribute to mitochondrial function.92 This part deserves to be explored in more depth subsequently.
Conclusion
Multiomics reveals that BPNS, a 2D biodegradable, safe, and nontoxic nanomaterial, can activate JAK1-STAT1/3-OAS signaling, thus enhancing the ECs’ function. Moreover, through proteomics, we revealed for the possible positive impact of BPNS on ECs’ mitochondrial function and energy metabolism. In addition, we employed GelMA/BPNS hydrogel to gradually release BPNS on the wound surface, exerting a positive and multilevel effect of anti-inflammatory, angiogenesis, collagen deposition, and re-epithelialization in wound healing. Our study provides the theoretical basis for the feasibility of developing two-dimensional BP as nanomedicine-potentiated in situ regenerative therapy for large skin full-thickness defects. It enlightens the fundamental mechanisms of BP’s interface on biological system from an ECs perspective.
Experimental Section
Preparation of Hydrogels
GelMA was purchase from SunP biotech Co., Ltd. (Beijing, China). GelMA freeze-dried powder was dissolved with sterile deionized water at 70 °C. Then after bacteria filtration with 0.22 μm filter, the sterile GelMA hydrogel, BPNS solution (200 μg/mL), and LAP solution were mixed to form a homogeneous prepolymer solution of GelMA hydrogel with gently stirring. The final concentration of GelMA/BPNS hydrogel prepolymer solution is 10, 15, or 20% (w/v) GelMA, 80 μg/mL BPNS, and 0.25% (w/v) LAP solution. The same procedure was followed to prepare the control group hydrogel, which used the same volume of sterile deionized wate instead of BPNS solution. The prepolymer solution were in injection pump stored at 4 °C protecting from light. Rewarming the prepolymer solution at 37 °C for a few minutes before they need to inject on the wound, and then gelatinized under UV light for 25 s.
Characterization of BPNS and Hydrogels
BPNS was purchased from XFNANO Materials Tech. Co., Ltd. (Jiangsu, China). The morphologies of BPNS were observed by TEM(FEI Tecnai Spirit G2 BioTWIN) and AFM (Bruker Dimension Icon, German), respectively. TEM was operated at 200 kV voltage. AFM was operated under the tapping mode in air. Raman spectra were measured using the Raman Microscope equipment (Horiba Scientific LabRAM HR Evolution, Japan) with 632 nm laser excitation at room temperature (RT).
The subcellular distribution and degradation of BPNS in HUVECs were observed by TEM; the cell samples were fixed with 2.5% glutaraldehyde (Sigma-Aldrich, USA) for 2 hours at RT. Following washing, samples were postfixed for 1.5 h with 1% osmium tetroxide (Sigma-Aldrich, USA) and dehydrated with an ethanol series. Samples were infiltrated, embedded in Epon Resin, and polymerized for 12 hours at 60 °C. 60 nm ultrathin sections were produced and dyed with uranyl acetate and lead citrate.
Following freeze-drying, hydrogels were sputtered with a thin layer of gold. A scanning electron microscope (Cyto-SEM, FEI Quanta 450 equipped with Quorum PP3000T Refrigeration transmission system) performing at an accelerating voltage of 10 kV was applied to study the hydrogels’ surface morphology.
Mechanical properties of hydrogels were analyzed under compression conditions. Four cylinder-shaped hydrogels samples (9 mm in diameter and 10 mm in height) were included in each group. The hydrogels were compressed at 5 mm/min on the platform of a universal testing machine (INSTRON 3343, U.S.) until they broke. The elastic modulus was calculated from the data collected during compression.
The hydrogels were soaked in PBS at 37 °C. At certain intervals, the dilative hydrogel specimen was taken out and the extra moisture on its surface was wiped and weighed and the specimen then put back into the PBS. The procedure was repeated until constant weight data remained consistent. The swelling ratio (SR) was determined using following equation: SR = (Wt – W0)/W0 × 100%, where W0 is the initial weight of the dry hydrogel and Wt refers to the weight of the dry hydrogel at time point.
The hydrogel specimens (9 mm in diameter and 10 mm in height) were produced and soaked in 1 mL of PBS containing collagenase types I and II and subsequently shaken at 60 rpm at 37 °C. After 0 and 12 hours and 1, 2, 3, 4, 5, 6, 7, and 8 days, hydrogel specimens were removed. The degradation process was assessed by evaluating dry weight loss between immersions, and the dry weight was measured. The weight remaining ratio (WRR) of the hydrogel was examined as follows: WRR = Wt × W0 × 100%, where Wt and W0 are the dry weights of the remaining and initial hydrogel at time point t, respectively.
The BPNS concentration in GelMA degradation solutions was examined by UV–vis absorption spectroscopy. First, the prepared BPNS suspension was diluted by adding different volumes of GelMA degradation solutions to generate diluted BPNS suspensions of series of BP concentrations (200 ng/mL ∼ 200 μg/mL). Second, following ultrasonic dispersion, UV–vis spectroscopy was operated, and the absorbance values acquired at 460 nm were used to calculate the standard curve of BPNS concentration: A460 = 0.0066 – 0.0027 (R2 = 0.999), where A460 is the absorbance value of hydrogel degradation solution and C is BPNS concentration. Then, the absorption values of the degradation solutions at 460 nm were measured by UV–vis and brought into the standard curve equation to calculate the concentration.
Cell Culture
HUVECs (BNCC, China) were cultured with Endothelial cell medium (EC medium, Sciencell, USA) containing with 1% Endothelial Cell Growth Supplement (Sciencell, USA), 10% fetal bovine serum (FBS), and 1% penicillin–streptomycin. They were grown in a 37 °C incubator with 5% CO2. Every 3 days, culture medium was replaced.
RAW 264.7 were obtained from the Chinese Academy of Science and cultured in the Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, USA) with 15% FBS (Hyclone, USA). They were grown in a 37 °C incubator with 5% CO2. Culture medium was replaced every 1 or 2 days.
For BPNS coculture, cells were seeded on culture plates for 24 hours and then incubated in complete culture medium with BPNS at different concentrations for the coculture period of time.
For Ruxolitinib (Selleck, US) treatment, cells were seeded on culture plates for 24 hours and then incubated in a complete culture medium with 0.4 μM Ruxolitinib. HUVECs were cultured in ECM containing 0.4 μM Ruxolitinib for 4 h, followed by adding 100 ng/mL BPNS in ECM with Ruxolitinib. According to the attenuation of Ruxolitinib efficacy, every 48 h the conditioned medium was changed.
Biocompatibility Assessment
CCK-8 assay: HUVECs were equally seeded in 96-well plates and cocultured with 0, 12.5, 25, 50 100, 200 ng/mL BPNS. After 1, 4, and 7 days of incubation, the culture medium was changed with 100 μL of fresh EC medium with 10% (v/v) CCK-8 reagent (Cell Counting Kit-8, MCE, China) and incubated for 90 min. Then, the cell viability was examined by a 450 nm microplate reader (Thermo Electron Corp., USA).
Living/dead cell staining: The living/dead kit (Solarbio, China) consisted of calcein-AM (green fluorescence), and propidium iodide (red fluorescence) was employed to determine the viability of HUVECs in EC medium with different concentrations of BPNS for 3 days. The culture medium was replaced, and the cells were washed with PBS. Then cells were incubated with 2 μM calcein-AM and 4.5 μM propidium iodide at RT for over 30 min and washed. In addition, the florescent images were observed and photographed via fluorescence microscope.
Colony Formation Assay
An equal number of HUVECs were seeded in 6-well plates and incubated with different concentrations of BPNS in EC medium for 10 consecutive days. The colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 15 min at RT. Then the dye was washed and photographed with a camera. Cell colony images were analyzed by ImageJ software.
Scratch Assay
An equal number of HUVECs were seeded in 6-well plates. When the fusion rate of HUVECs reached 90%, a 1000 μL pipet tip was used to generate a cell scratch, and the shed cells were rinsed. Then the culture media containing different concentrations of BPNS were added to the wells, respectively. The cells were observed and photographed at 24 hours via an inverted light microscope. The images were analyzed by ImageJ software. The cell migration ratio was obtained using the following equation: cell migration ratio (%) = (A0 – At)/A0 × 100%, where A0 and At are scratched areas before and after BP coculture, respectively.
Transwell Assay
We performed Transwell assay to determine the bio-effects of BPNS on HUVECs’ vertical migration ability. The cells were inoculated into each upper chamber of 24-well Transwell plates (aperture, 8 μm; Corning, NY, USA) and serum-starved for 24 hours in serum-free EC medium, containing different concentrations of BPNS. The lower chambers were filled with EC medium containing 20% fetal bovine and cultured at 37 °C at 5% CO2 for 24 hours. After 24 hours, the upper chambers were removed and the cells at the top side were gently wiped by cotton swabs. The cells were fixed with 4% paraformaldehyde at 37 °C for 30 min and then washed. Finally, cells were stained with 5% crystal violet solution. The images of each group were obtained via an inverted microscope and analyzed by ImageJ software.
Immunofluorescence Staining
The cells were fixed with 4% paraformaldehyde at 37 °C for 10 min and then washed with PBS buffer containing 0.1% Triton X-100, and 5% BSA blocked cells for 30 min. Then cells were incubated with indicated primary antibody CD31 (Cell Signaling Technology, CST, USA) at 4 °C overnight. Cells were washed and incubated for 1 hour at RT with appropriate secondary antibody conjugated to Goat anti Mouse Alexa-Fluoro 488 (Invitrogen, USA) and then washed. Phalloidin conjugate stock solution was added from F-actin Stain kit (Abcam, USA) and washed. DNA was stained with DAPI (Solarbio, China) at the final step. The MFI of images were analyzed by ImageJ software.
Tube Formation Assay
First, HUVECs were treated with 100 ng/mL BPNS for 3 or 7days. We also set a group that add 100 ng/mL BPNS into the well at the beginning of assay. Then 80000 HUVECs were reseeded in 96-well plates coated with Matrigel matrix (Corning, USA). After 6 h incubation periods, the newly formed tubular structures were imaged. The node amount, mesh amount, and branch amount were measured and counted by ImageJ software.
Macrophage Regulation In Vitro
To simulate in vivo acute inflammation, LPS (Sigma, USA) was used to induce RAW264.7 cells to M1-phenotype (LPS induced group). After 12 hours, the medium with 200 ng/mL LPS was replaced with a new LPS containing medium 50 or 100 ng/mL BPNS and culture continued for 3 or 7 days.
qRT-PCR
HUVECs were cultured with EC medium containing 100 ng/mL BPNS for 3 or 7 days to evaluate the regulation of BPNS on expression of VEGFR2, CD31, ANG-1, FGF2, EGF, TNF-a, IL-1A, IL-1B, IL-6, IL-10, Arg-1, and CD206. WT group or LPS induced group RAW264.7 was cultured with DMEM medium containing 50 or 100 ng/mL BPNS for 3 or 7 days to evaluate the bio-effects of BP on expression of TNF-a, IL-1A, IL-1B, IL-6, IL-10, Arg-1, and CD206. TRIzol reagent (Invitrogen, USA) was used to extract total RNA from cells. RNA concentration quantified by NanoDrop 8000 spectrophotometer (ThermoFisher Scientific, USA). Total RNA was reverse-transcribed into cDNA using the HiScript III first Strand cDNA Synthesis kit (Vazyme, China). RT-PCR was performed using the Roche LightCycler 480II RT-PCR System (Rohce, Switzerland) and ChamQ Universal SYBR qPCR Master Mix kit (Vazyme, China). The primer sequences of human genome GAPDH, VEGFR2, CD31, ANG-1, FGF2, EGF, TNF-a, IL-1A, IL-1B, IL-6, IL-10, Arg-1, and CD206 are shown in Table 1. And the primer sequences of mouse genome TNF-a, IL-1A, IL-1B, IL-6, iNOS, IL-10, Arg-1, and CD206 are shown in Table 2.
Table 1. HUVECs Primer Sequences.
| Gene | Primer sequences (5′ to 3′) |
|---|---|
| GAPDH | F: ACTTCCTGTGCTCGGTGCT |
| R: GACGGTTATGGTCAAGGTGAA | |
| VEGFR2 | F: CATTGTGTTTGCTCTGGTGGAGGT |
| R: CGTAACGGTCTGGAAGGAACTCTC | |
| ANG-1 | F: CTTCAAGGCTTGGTTACTCGTC |
| R: CTCTTCCTCTCTTTTTCCTCCC | |
| CD31 | F: CTACAGGCACAGAACCGTCTCC |
| R:GCACAGCGTCAGGAACACAGA | |
| FGF2 | F:GCCAGGTCATTGAGATCCATCCA |
| R:GAGACACAGCGGTTCGAGAAGTT | |
| EGF | F:TTGGTGTTCGTGTCGTGAAGGTT |
| R: CCACAGGAGCACAGTCATCTTGAT | |
| TNF-a | F: GGAAAGGACACCATGAGCACTGAA |
| R: GGAAGGAGAAGAGGCTGAGGAACA | |
| IL-1A | F: ACCAACCAGTGCTGCTGAAGG |
| R: GACAAGTGAGACTCCAGACCTACG | |
| IL-1B | F: GCCTTAGGGTAGTGCTAAGAGGA |
| R: GGAGCGAATGACAGAGGGTTT | |
| IL-6 | F: ATGAGGAGACTTGCCTGGTGA |
| R: GTTGGGTCAGGGGTGGTTAT | |
| IL-10 | F: GCCGTGGAGCAGGTGAAGAATG |
| R: GAGTCGCCACCCTGATGTCTCA | |
| Arg-1 | F: CAGTCGTGGGAGGTCTGACATACA |
| R: GCAAGTCCGAAACAAGCCAAGGT | |
| CD206 | F: ACCTCACAAGTATCCACACCATCG |
| R: TGCCTTTCATCACCACACAATCCT |
Table 2. RAW264.7 Primer Sequences.
| Gene | Primer sequences (5′ to 3′) |
|---|---|
| GAPDH | F: TACGCCAACACGGTGCTGTC |
| R: GTACTCCTGCTTGCTGATCCACAT | |
| TNF-a | F: TAACTTAGAAAGGGGATTATGGCT |
| R: TGGAAAGGTCTGAAGGTAGGAA | |
| IL1A | F: ATGCCAGTTGAGTAGGATAAAGGT |
| R: GAGTAAAACCCACTGAGGTAGGAA | |
| IL1B | F: TCACAAGCAGAGCACAAGCC |
| R: CATTAGAAACAGTCCAGCCCATAC | |
| iNOS | F:GACGAGACGGATAGGCAGAGATTG |
| R:GAACTCTTCAAGCACCTCCAGGAA | |
| IL-6 | F: TTGCCTTCTTGGGACTGATG |
| R: ACTCTTTTCTCATTTCCACGATTT | |
| IL-10 | F: GGACAACATACTGCTAACCGACTC |
| R: CCTGGGGCATCACTTCTACC | |
| Arg-1 | F: GGAAGACAGCAGAGGAGGTGAAGA |
| R: GGTAGTCAGTCCCTGGCTTATGGT | |
| CD206 | F: GACCTGGCAAGTATCCACAGCATT |
| R: GCAGTCCTCCTGTCTGTTGTTCTC |
Western Immunoblotting
The HUVECs protein was extracted by RIPA lysis buffer, measured using a BCA protein assay kit (Beyotime, China), separated using SDS electrophoresis, and transferred to a polyvinylidene difluoride membrane (PVDF, Millipore, U.S.). After that, PVDF membranes were soaked in 5% BSA for 1 hour. Then, the membrane was co-incubated with primary antibodies, anti-JAK1 (CST, USA), anti-p-JAK1 (CST, USA), anti-STAT1 (CST, USA), anti-p-STAT1 (CST, USA), anti-STAT3 (CST, USA), anti-p-STAT3 (CST, USA), anti-OAS2 (ThermoFisher, USA), anti-OAS3 (CST, USA), and anti-OASL (Abcam, USA) overnight at 4 °C. The PDVF membranes washed with TBST, and then co-incubated with secondary antibodies (CST, USA) at RT for 1 h. Finally, the blots were washed with TBST and visualized by the enhanced chemiluminescence detection system.
Intracellular ROS Levels of HUVECs and 143B via DCF-DA Staining
HUVECs and 143B were cocultured with BPNS for different culture times and exposed to DCF-DA before evaluating ROS level. Because of the selective killing effect of BPNS at 100 ng/mL, normal cell lines HUVECs were cocultured with BPNS for 1 hour, 6 hours, 1 day, 3 days, 7 days, and 10 days, while tumor cell lines 143B were cocultured with BPNS for 1 hour, 3 hours, 6 hours, 12 hours, and 1 day. ROS was measured by 2′,7′-dichlorofluorescin diacetate (DCF-DA, Sigma, USA). With FBS-free medium, 10 mM DCF-DA stock solution in dimethyl sulfoxide was diluted to 1 mM working solution. The cell samples were incubated with 600 μL of working solution for 30 min at 37 °C in complete darkness. Following that, the cells were washed with FBS-free medium and photographed using a fluorescence microscope. The MFI of images were analyzed by ImageJ software.
Evaluation of the Effectiveness of GelMA/BPNS Hydrogels in Promoting Wound Healing In Vivo
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Beijing Jishuitan Hospital and approved by the Animal Ethics Committee of the Beijing Jishuitan Hospital (202105-02). The study involved 24 male Sprague–Dawley rats weighing 250–280 g at 6–8weeks. Pentobarbital sodium was injected intraperitoneally at a dose of 50 mg/kg to anesthetize the rats. After anesthetizing the rat, skin preparation was performed, and the incision was marked with a marker in the back of the rat as a 3.3 cm × 3.3 cm square. Then, the proposed excision area was incised and removed, after which the anticontracture mold was firmly sutured to the skin of the rat, and the hydrogel was applied to the wound surface, cured by 25 s blue light irradiation, and fixed by local gauze coverage. (Figure 4B). Rats were randomly divided into three treatment group: blank control, GelMA, and BPNS-GelMA. The size of the wound was observed daily. We changed hydrogels and photographed the wound with a digital camera at a fixed focal length on 0, 7, 14, 21, 28, and 35 days. Then the size of wound was quantified by ImageJ software, and the residual wound (%) was calculated by the following formula: residual wound (%) = (At/A0) × 100%, where A0 refers to the original wound area and At refers to the wound area at the appointed time.
Vascular Perfusion
To evaluate the BPNS bio-effects on newly formed vascular in vivo, the rats were perfused with Microfil (Flow Tech, USA) at postoperative 28 days. Rats were anesthetized, and then 50 mL of PBS containing 1% heparin and 36 mL of Microfil were injected into the left ventricle in succession. The wound area was cut off and overnight kept at 4 °C. An in vivo micro-CT scanner (SkyScan1276, Bruker Biospin, Belgium) with a resolution of 18 μm was used to examine each specimen. Following the micro-CT scan, Mimics Research 20.0 was used to reconstruct 3D vascular enhancement images.
Histological Analysis and Immunostaining
To dynamically observe the wound healing, we obtained the samples with skin sampler at 14, 21, and 28 days. On day 35, the rat was sacrificed and the wound along with surrounding skin were extracted. All samples were fixed with 4% paraformaldehyde at 4 °C for 24 hours. Tissue samples were embedded in paraffin wax blocks and sectioned into slices with a thickness of 4 μm. These sections were processed and stained with H&E and Masson stainings. IF staining was performed via applying the following primary antibodies: anti-F4/80 (CST, USA), anti-CD206 (Invitrogen, USA), anti-CD86 (Invitrogen, USA), anti-CD31 (Invitrogen, USA), anti-a-SMA (Invitrogen, USA), anti-OASL (Abcam, USA).
RNA Sequencing and Bioinformatics
HUVECs were cultured in the EC medium containing 100 ng/mL BPNS for 7 days, and total RNA was isolated using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions. Using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, USA), total quantities and integrity of RNA were examined. Novogene Biotechnology Co., Ltd. (China) carried out the transcriptome RNA sequencing. To obtain clean, high-quality data for upcoming analysis, reads including adaptor or ploy-Nb reads and low-quality reads were removed.
Using the DESeq2 R package (1.20.0), differential expression analysis of two groups was carried out. Using a model based on the negative binomial distribution, DESeq2 provides statistical algorithms for identifying differential expression in digital gene expression data. The Benjamini and Hochberg method for reducing the false discovery rate was used to modify the obtained P values. The criteria for significant DEGs were established at Padj = 0.05 and |log 2(fold change)| ≥ 1.
The clusterProfiler R package (3.8.1) was employed to implement GSEA of DEGs and correct gene length bias. DEGs were deemed to significantly enrich GO terms with a corrected P value less than 0.05. KEGG is a database used to decipher high-level biological system functions and utilities from molecular-level data, particularly large-scale molecular data sets produced by genome sequencing and other high-throughput experimental techniques (http://www.genome.jp/kegg/). We tested the statistical enrichment of differentially expressed genes in KEGG pathways using the clusterProfiler R package (3.8.1).
TMT Proteomics and Bioinformatics
HUVECs were cultured in the EC medium containing 100 ng/mL BPNS for 7 days, and total cell samples were isolated using lysis buffer, which consisted if M-PER mammalian protein extraction reagent (Thermofisher, USA) and Roche complete protease inhibitor cocktail (Roche, USA). After ultrasonication on ice for 5 min, the lysate was centrifuged at 12000g for 15 min at 4 °C. Then, the supernatant was added with 1 M dl-dithiothreitol to react for 1 hour at 56 °C and successively alkylated with sufficient iodoacetamide for 1 hour at RT in darkness followed by an ice bath for 2 min. Then samples were quantified by BCA assay (Beyotime, China) and separated by SDS electrophoresis. Coomassie bright blue R-250 was used to stain the gel, and it was then lightened until the bands could be seen clearly.
Each protein sample was obtained and a volume of 100 μL with DB dissolving buffer (8 M urea, 100 mM TEAB, pH 8.5) was added. Trypsin and 100 mM TEAB buffer were incorporated into the sample, which was then combined and digested at 37 °C for 4 hours. After that, formic acid was mixed with digested sample, the pH was adjusted to less than 3, and the sample was centrifuged at 12000g for 5 min at RT. The supernatant was slowly loaded to the C18 desalting column, washed with washing buffer, and then eluted by some elution buffer. Each sample was collected and lyophilized. For the TMT assay, to reconstitute, 100 μL of 0.1 M TEAB buffer was added and followed by 41 μL of acetonitrile-dissolved TMT labeling reagent; the sample was blended with vibrating for 2 hours at RT. The process was ceased through the addition of 8% ammonia. All labeling samples were combined with equal volumes before being desalted and lyophilized. UHPLC-MS/MS analyses were operated in Novogene Co., Ltd. (China), utilizing an EASY-nLCTM 1200 UHPLC system (ThermoFisher Scientific, Germany) in conjunction with a Q Exactive HF-X (ThermoFisher, Germany).
Proteome Discoverer (PD, Thermo, HFX and 480) scanned the resultant spectra from each run separately against the *** database. The retrieval results were further filtered by the software PD to ensure the quality of the analysis results. t test was employed to statistically examine the protein quantitation results. DEPs are proteins whose quantification differs significantly between experimental and control groups ( < 0.05 and |log 2(FC)| > * (FC > * or FC * [fold change, FC]).
The interproscan program was applied to perform GO and IPR analysis against the nonredundant protein database, and the KEGG databases were utilized to examine the protein family function and pathway. DEPs were used for volcanic map analysis, cluster heatmap analysis, and GO, IPR, and KEGG enrichment analysis. The STRING-db server (http://string.embl.de/) was implemented to determine the relevant protein–protein interactions.
Statistical Analysis
All studies were repeated at least three times and yielded similar findings. The representative images exhibited are from at least three different experiments. Unless otherwise specified, data are expressed as mean ± SD from at least three independent repeats. For two-group comparisons, a two-tailed Student’s t test was employed, and for multiple comparisons, a one-way analysis of variance (ANOVA) followed by Tukey post hoc test was utilized. To compare proportions, the two-sided Fisher’s exact test or χ2 test was employed. GraphPad Prism (V.9.0.0) was employed for all statistical analysis. P < 0.05 was regarded as statistically significant.
Acknowledgments
The authors gratefully acknowledge the financial support for this research from the National Key R&D Program of China (Grant Nos. 2022YFB3804400, and 2021YFC2400500), the Beijing Municipal Health Commission (BJRITO-RDP-2023 and BMHC-2021-6), and the Beijing Jishuitan Hospital Nova Program (XKXX202115 and XKXX202114). For the animal experiment mold is improve upon the Chinese utility model patent (ZL202221885123.4). Thanks to the lead inventor of this patent, Dr. Xiaohua Hu, who is also the third author of this article.
Glossary
Abbreviations
- (BP)
black phosphorus
- (BPNS)
BP nanosheets
- (CAMs)
cell adhesion molecules
- (DCF-DA)
2′,7′-dichlorofluorescin diacetate
- (DEGs)
differential expressed genes
- (DEPs)
differentially expressed proteins
- (DMEM)
Dulbecco’s modified Eagle’s medium
- (ECs)
endothelial cells’
- (ECM)
extracellular matrix
- (FBS)
fetal bovine serum
- (GelMA)
gelatin methacrylate
- (GO)
gene ontology
- (GSEA)
gene set enrichment analysis
- (HUVECs)
human umbilical vein endothelial cells
- (ICAM-1)
intercellular adhesion molecule 1
- (IF)
immunofluorescence
- (KEGG)
Kyoto encyclopedia of genes and genome
- (LPS)
lipopolysaccharide
- (MFI)
mean fluorescence intensity
- (OD)
optical density
- (OXPHOS)
oxidative phosphorylation
- (PECAM-1)
platelet-endothelial cell adhesion molecule 1
- (PVDF)
polyvinylidene difluoride membrane
- (qRT-PCR)
quantitative real-time PCR
- (ROS)
reactive oxygen species
- (SELE)
E-selectin
- (TMT)
tandem mass tags
- (VACAM-1)
vascular cell-adhesion molecule 1
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsnano.3c11177.
(Figure S1) BPNS’ ROS-mediated mechanism; (Figure S2) quantitative results of in vivo analyses, clustering heatmap of mitochondrion-related protein expression, mitochondrial dynamic-related protein expression, tube formation assays, and quantitative tube formation assays results; (Figure S3) OASL expression; (Figure S4) BPNS distribution and degradation; (Figure S5) pore diameter analysis (PDF)
Author Contributions
# X.B. and R.W. contributed equally to this work.
The authors declare no competing financial interest.
Supplementary Material
References
- Yin Y.; Zhang R.; Li S.; Guo J.; Hou Z.; Zhang Y. Negative-pressure therapy versus conventional therapy on split-thickness skin graft: A systematic review and meta-analysis. International Journal of Surgery (London, England) 2018, 50, 43–48. 10.1016/j.ijsu.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Berchialla P.; Gangemi E. N.; Foltran F.; Haxhiaj A.; Buja A.; Lazzarato F.; Stella M.; Gregori D. Predicting severity of pathological scarring due to burn injuries: a clinical decision making tool using Bayesian networks. Int. Wound J. 2014, 11, 246–52. 10.1111/j.1742-481X.2012.01080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan M. P.; Cancio L. C.; Elster E. A.; Burmeister D. M.; Rose L. F.; Natesan S.; Chan R. K.; Christy R. J.; Chung K. K. Burn wound healing and treatment: review and advancements. Crit Care 2015, 19, 243. 10.1186/s13054-015-0961-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao S. F.; Ma H.; Wang Y. H.; Wang T. H. Occlusive drainage system for split-thickness skin graft: A prospective randomized controlled trial. Burns 2017, 43, 379–387. 10.1016/j.burns.2016.08.025. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rodriguez N.; Martinez-Jimenez I.; Garcia-Ojalvo A.; Mendoza-Mari Y.; Guillen-Nieto G.; Armstrong D. G.; Berlanga-Acosta J. Wound Chronicity, Impaired Immunity and Infection in Diabetic Patients. MEDICC Rev. 2022, 24, 44–58. 10.37757/MR2021.V23.N3.8. [DOI] [PubMed] [Google Scholar]
- DiPietro L. A. Angiogenesis and wound repair: when enough is enough. J. Leukoc Biol. 2016, 100, 979–984. 10.1189/jlb.4MR0316-102R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich D. B.; David D. T.; Sundararaman A.; Patel J. Endothelial Heterogeneity in Development and Wound Healing. Cells 2021, 10, 2338. 10.3390/cells10092338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jere S. W.; Abrahamse H.; Houreld N. N. The JAK/STAT signaling pathway and photobiomodulation in chronic wound healing. Cytokine Growth Factor Rev. 2017, 38, 73–79. 10.1016/j.cytogfr.2017.10.001. [DOI] [PubMed] [Google Scholar]
- Du H.; Zhou Y.; Suo Y.; Liang X.; Chai B.; Duan R.; Huang X.; Li Q. CCN1 accelerates re-epithelialization by promoting keratinocyte migration and proliferation during cutaneous wound healing. Biochem. Biophys. Res. Commun. 2018, 505, 966–972. 10.1016/j.bbrc.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Beckman J. D.; DaSilva A.; Aronovich E.; Nguyen A.; Nguyen J.; Hargis G.; Reynolds D.; Vercellotti G. M.; Betts B.; Wood D. K. JAK-STAT inhibition reduces endothelial prothrombotic activation and leukocyte-endothelial proadhesive interactions. J. Thromb Haemost 2023, 21, 1366–1380. 10.1016/j.jtha.2023.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. G.; Hur S. Cellular origins of dsRNA, their recognition and consequences. Nat. Rev. Mol. Cell Biol. 2022, 23, 286–301. 10.1038/s41580-021-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho W. J.; Law A. M. K.; Masle-Farquhar E.; Castillo L. E.; Mawson A.; O’Bryan M. K.; Goodnow C. C.; Gallego-Ortega D.; Oakes S. R.; Ormandy C. J. Activation of the viral sensor oligoadenylate synthetase 2 (Oas2) prevents pregnancy-driven mammary cancer metastases. Breast Cancer Res. 2022, 24, 31. 10.1186/s13058-022-01525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. Z.; Zheng Y. X.; Zhou Y.; Xu F.; Cui Y. Z.; Chen X. Y.; Wang Z. Y.; Yan B. X.; Zheng M.; Man X. Y. OAS1, OAS2, and OAS3 Contribute to Epidermal Keratinocyte Proliferation by Regulating Cell Cycle and Augmenting IFN-1–Induced Jak1–Signal Transducer and Activator of Transcription 1 Phosphorylation in Psoriasis. J. Invest Dermatol 2022, 142, 2635–2645 e9. 10.1016/j.jid.2022.02.018. [DOI] [PubMed] [Google Scholar]
- Kim T. K.; Jeon S.; Park S.; Sonn S. K.; Seo S.; Suh J.; Jin J.; Kweon H. Y.; Kim S.; Moon S. H.; Kweon O.; Koo B. H.; Kim N.; Lee H. O.; Kim Y. M.; Kim Y. J.; Park S. H.; Oh G. T. 2′-5′ oligoadenylate synthetase-like 1 (OASL1) protects against atherosclerosis by maintaining endothelial nitric oxide synthase mRNA stability. Nat. Commun. 2022, 13, 6647. 10.1038/s41467-022-34433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Gaihre B.; George M. N.; Li Y.; Tilton M.; Yaszemski M. J.; Lu L. 2D phosphorene nanosheets, quantum dots, nanoribbons: synthesis and biomedical applications. Biomater Sci. 2021, 9, 2768–2803. 10.1039/D0BM01972K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roma-Rodrigues C.; Heuer-Jungemann A.; Fernandes A. R.; Kanaras A. G.; Baptista P. V. Peptide-coated gold nanoparticles for modulation of angiogenesis in vivo. Int. J. Nanomedicine 2016, 11, 2633–9. 10.2147/IJN.S108661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J.; Li X.; Wang C.; Guo H.; Liu J.; Xu H. Carbon nanotubes activate macrophages into a M1/M2 mixed status: recruiting naive macrophages and supporting angiogenesis. ACS Appl. Mater. Interfaces 2015, 7, 3180–8. 10.1021/am507649n. [DOI] [PubMed] [Google Scholar]
- Eid H. M.; Ali A. A.; Ali A. M. A.; Eissa E. M.; Hassan R. M.; Abo El-Ela F. I.; Hassan A. H. Potential Use of Tailored Citicoline Chitosan-Coated Liposomes for Effective Wound Healing in Diabetic Rat Model. Int. J. Nanomedicine 2022, 17, 555–575. 10.2147/IJN.S342504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Wang Y. F.; Li X.; Wang Y.; Huang Q.; Ma X.; Liang X. J. Nanoparticle-Driven Controllable Mitochondrial Regulation through Lysosome-Mitochondria Interactome. ACS Nano 2022, 16, 12553–12568. 10.1021/acsnano.2c04078. [DOI] [PubMed] [Google Scholar]
- Shou X.; Liu Y.; Wu D.; Zhang H.; Zhao Y.; Sun W.; Shen X. Black phosphorus quantum dots doped multifunctional hydrogel particles for cancer immunotherapy. Chem. Eng. J. 2021, 408, 127349 10.1016/j.cej.2020.127349. [DOI] [Google Scholar]
- Ding Q.; Sun T.; Su W.; Jing X.; Ye B.; Su Y.; Zeng L.; Qu Y.; Yang X.; Wu Y.; Luo Z.; Guo X. Bioinspired Multifunctional Black Phosphorus Hydrogel with Antibacterial and Antioxidant Properties: A Stepwise Countermeasure for Diabetic Skin Wound Healing. Adv. Healthc Mater. 2022, 11, 2102791 10.1002/adhm.202102791. [DOI] [PubMed] [Google Scholar]
- Zhang X.; Chen G.; Liu Y.; Sun L.; Sun L.; Zhao Y. Black Phosphorus-Loaded Separable Microneedles as Responsive Oxygen Delivery Carriers for Wound Healing. ACS Nano 2020, 14, 5901–5908. 10.1021/acsnano.0c01059. [DOI] [PubMed] [Google Scholar]
- Xu S.; Chang L.; Hu Y.; Zhao X.; Huang S.; Chen Z.; Ren X.; Mei X. Tea polyphenol modified, photothermal responsive and ROS generative black phosphorus quantum dots as nanoplatforms for promoting MRSA infected wounds healing in diabetic rats. J. Nanobiotechnology 2021, 19, 362. 10.1186/s12951-021-01106-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.; Yu Y.; Fan L.; Li W.; Bian F.; Wang J.; Zhao Y. Sprayable Multifunctional Black Phosphorus Hydrogel with On-Demand Removability for Joint Skin Wound Healing. Adv. Healthcare Mater. 2023, 2302588 10.1002/adhm.202302588. [DOI] [PubMed] [Google Scholar]
- He L.; Zhao J.; Li H.; Xie B.; Xu L.; Huang G.; Liu T.; Gu Z.; Chen T. Metabolic Reprogramming of NK Cells by Black Phosphorus Quantum Dots Potentiates Cancer Immunotherapy. Adv. Sci. (Weinheim, Ger.) 2023, 10, 2202519 10.1002/advs.202202519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri P.; Harfouche R.; Soni S.; Hentschel D. M.; Sengupta S. Shape effect of carbon nanovectors on angiogenesis. ACS Nano 2010, 4, 574–82. 10.1021/nn901465h. [DOI] [PubMed] [Google Scholar]
- Yang Q.; Chen S.; Shi H.; Xiao H.; Ma Y. In vitro study of improved wound-healing effect of bioactive borate-based glass nano-/micro-fibers. Mater. Sci. Eng. C Mater. Biol. Appl. 2015, 55, 105–17. 10.1016/j.msec.2015.05.049. [DOI] [PubMed] [Google Scholar]
- Yuan A.; Xia F.; Bian Q.; Wu H.; Gu Y.; Wang T.; Wang R.; Huang L.; Huang Q.; Rao Y.; Ling D.; Li F.; Gao J. Ceria Nanozyme-Integrated Microneedles Reshape the Perifollicular Microenvironment for Androgenetic Alopecia Treatment. ACS Nano 2021, 15, 13759–13769. 10.1021/acsnano.1c05272. [DOI] [PubMed] [Google Scholar]
- Darweesh R. S.; Ayoub N. M.; Nazzal S. Gold nanoparticles and angiogenesis: molecular mechanisms and biomedical applications. Int. J. Nanomedicine 2019, 14, 7643–7663. 10.2147/IJN.S223941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargozar S.; Baino F.; Hamzehlou S.; Hamblin M. R.; Mozafari M. Nanotechnology for angiogenesis: opportunities and challenges. Chem. Soc. Rev. 2020, 49, 5008–5057. 10.1039/C8CS01021H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Dong L.; Zhong H.; Yang L.; Li Q.; Su C.; Gu W.; Qian Y. Extracellular Vesicles (EVs) from Lung Adenocarcinoma Cells Promote Human Umbilical Vein Endothelial Cell (HUVEC) Angiogenesis through Yes Kinase-associated Protein (YAP) Transport. Int. J. Biol. Sci. 2019, 15, 2110–2118. 10.7150/ijbs.31605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadunandanan Nair N.; Samuel V.; Ramesh L.; Marib A.; David D. T.; Sundararaman A. Actin cytoskeleton in angiogenesis. Biol. Open 2022, 11, bio058899. 10.1242/bio.058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke J. P. Inflammation and Its Role in Regeneration and Repair. Circ. Res. 2019, 124, 1166–1168. 10.1161/CIRCRESAHA.118.314669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pober J. S.; Sessa W. C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol 2007, 7, 803–15. 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- Jetten N.; Verbruggen S.; Gijbels M. J.; Post M. J.; De Winther M. P.; Donners M. M. Anti-inflammatory M2, but not pro-inflammatory M1 macrophages promote angiogenesis in vivo. Angiogenesis 2014, 17, 109–18. 10.1007/s10456-013-9381-6. [DOI] [PubMed] [Google Scholar]
- Zhou J.; Liu W.; Zhao X.; Xian Y.; Wu W.; Zhang X.; Zhao N.; Xu F. J.; Wang C. Natural Melanin/Alginate Hydrogels Achieve Cardiac Repair through ROS Scavenging and Macrophage Polarization. Adv. Sci. (Weinheim, Ger.) 2021, 8, 2100505 10.1002/advs.202100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon O. R.; Browe D. C.; Gonzalez-Fernandez T.; Pitacco P.; Whelan I. T.; Von Euw S.; Hobbs C.; Nicolosi V.; Cunningham K. T.; Mills K. H. G.; Kelly D. J.; Dunne A. Nano-particle mediated M2 macrophage polarization enhances bone formation and MSC osteogenesis in an IL-10 dependent manner. Biomaterials 2020, 239, 119833 10.1016/j.biomaterials.2020.119833. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Deng J.; Cao J.; Wu H.; Feng G.; Zhang R.; Ran B.; Hu K.; Cao H.; Zhu X.; Zhang X. Nano-hydroxyapatite-evoked immune response synchronized with controllable immune adjuvant release for strengthening melanoma-specific growth inhibition. Acta Biomater 2022, 145, 159–171. 10.1016/j.actbio.2022.04.002. [DOI] [PubMed] [Google Scholar]
- Hao F.; Yan Z. Y.; Yan X. P. Intracellular fate and immune response of porphyrin-based nano-sized metal-organic frameworks. Chemosphere 2022, 307, 135680 10.1016/j.chemosphere.2022.135680. [DOI] [PubMed] [Google Scholar]
- Wen T.; Quan G.; Niu B.; Zhou Y.; Zhao Y.; Lu C.; Pan X.; Wu C. Versatile Nanoscale Metal-Organic Frameworks (nMOFs): An Emerging 3D Nanoplatform for Drug Delivery and Therapeutic Applications. Small 2021, 17, 2005064 10.1002/smll.202005064. [DOI] [PubMed] [Google Scholar]
- Kong N.; Ji X.; Wang J.; Sun X.; Chen G.; Fan T.; Liang W.; Zhang H.; Xie A.; Farokhzad O. C.; Tao W. ROS-Mediated Selective Killing Effect of Black Phosphorus: Mechanistic Understanding and Its Guidance for Safe Biomedical Applications. Nano Lett. 2020, 20, 3943–3955. 10.1021/acs.nanolett.0c01098. [DOI] [PubMed] [Google Scholar]
- Dunnill C.; Patton T.; Brennan J.; Barrett J.; Dryden M.; Cooke J.; Leaper D.; Georgopoulos N. T. Reactive oxygen species (ROS) and wound healing: the functional role of ROS and emerging ROS-modulating technologies for augmentation of the healing process. Int. Wound J. 2017, 14, 89–96. 10.1111/iwj.12557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Ge J.; Tredget E. E.; Wu Y. The mouse excisional wound splinting model, including applications for stem cell transplantation. Nat. Protoc 2013, 8, 302–9. 10.1038/nprot.2013.002. [DOI] [PubMed] [Google Scholar]
- Kurian A. G.; Singh R. K.; Patel K. D.; Lee J. H.; Kim H. W. Multifunctional GelMA platforms with nanomaterials for advanced tissue therapeutics. Bioact Mater. 2022, 8, 267–295. 10.1016/j.bioactmat.2021.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Sun J.; Yang J.; Yang Y.; Ding H.; Yu B.; Ma K.; Chen M. Gelatin methacryloyl (GelMA) loaded with concentrated hypoxic pretreated adipose-derived mesenchymal stem cells(ADSCs) conditioned medium promotes wound healing and vascular regeneration in aged skin. Biomater Res. 2023, 27, 11. 10.1186/s40824-023-00352-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Cao Z.; Wei Q.; Ma K.; Hu W.; Huang Q.; Su J.; Li H.; Zhang C.; Fu X. VH298-loaded extracellular vesicles released from gelatin methacryloyl hydrogel facilitate diabetic wound healing by HIF-1alpha-mediated enhancement of angiogenesis. Acta Biomater 2022, 147, 342–355. 10.1016/j.actbio.2022.05.018. [DOI] [PubMed] [Google Scholar]
- Haghniaz R.; Montazerian H.; Rabbani A.; Baidya A.; Usui B.; Zhu Y.; Tavafoghi M.; Wahid F.; Kim H. J.; Sheikhi A.; Khademhosseini A. Injectable, Antibacterial, and Hemostatic Tissue Sealant Hydrogels. Adv. Healthcare Mater. 2023, 12, 2301551 10.1002/adhm.202301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Wang H.; Huang H.; Xiao Q.; Xu Y.; Guo Z.; Xie H.; Shao J.; Sun Z.; Han W.; Yu X. F.; Li P.; Chu P. K. Surface Coordination of Black Phosphorus for Robust Air and Water Stability. Angew. Chem., Int. Ed. Engl. 2016, 55, 5003–7. 10.1002/anie.201512038. [DOI] [PubMed] [Google Scholar]
- Shie M. Y.; Lee J. J.; Ho C. C.; Yen S. Y.; Ng H. Y.; Chen Y. W. Effects of Gelatin Methacrylate Bio-ink Concentration on Mechano-Physical Properties and Human Dermal Fibroblast Behavior. Polymers (Basel) 2020, 12, 1930. 10.3390/polym12091930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennekampff H. O.; Hansbrough J. F.; Kiessig V.; Dore C.; Sticherling M.; Schroder J. M. Bioactive interleukin-8 is expressed in wounds and enhances wound healing. Journal of surgical research 2000, 93, 41–54. 10.1006/jsre.2000.5892. [DOI] [PubMed] [Google Scholar]
- Albelda S. M. Endothelial and epithelial cell adhesion molecules. Am. J. Respir. Cell Mol. Biol. 1991, 4, 195–203. 10.1165/ajrcmb/4.3.195. [DOI] [PubMed] [Google Scholar]
- Filippi M. D. Neutrophil transendothelial migration: updates and new perspectives. Blood 2019, 133, 2149–2158. 10.1182/blood-2018-12-844605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elices M. J.; Osborn L.; Takada Y.; Crouse C.; Luhowskyj S.; Hemler M. E.; Lobb R. R. VCAM-1 on activated endothelium interacts with the leukocyte integrin VLA-4 at a site distinct from the VLA-4/fibronectin binding site. Cell 1990, 60, 577–84. 10.1016/0092-8674(90)90661-W. [DOI] [PubMed] [Google Scholar]
- Chrifi I.; Louzao-Martinez L.; Brandt M. M.; van Dijk C. G. M.; Burgisser P. E.; Zhu C.; Kros J. M.; Verhaar M. C.; Duncker D. J.; Cheng C. CMTM4 regulates angiogenesis by promoting cell surface recycling of VE-cadherin to endothelial adherens junctions. Angiogenesis 2019, 22, 75–93. 10.1007/s10456-018-9638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. H.; Lee S. H.; Yoo S. Y.; Asahara T.; Kwon S. M. CD34 hybrid cells promote endothelial colony-forming cell bioactivity and therapeutic potential for ischemic diseases. Arterioscler Thromb Vasc Biol. 2013, 33, 1622–34. 10.1161/ATVBAHA.112.301052. [DOI] [PubMed] [Google Scholar]
- Witjas F. M. R.; van den Berg B. M.; van den Berg C. W.; Engelse M. A.; Rabelink T. J. Concise Review: The Endothelial Cell Extracellular Matrix Regulates Tissue Homeostasis and Repair. Stem Cells Transl Med. 2019, 8, 375–382. 10.1002/sctm.18-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L.; Coller J.; Natu V.; Hastie T. J.; Huang N. F. Combinatorial extracellular matrix microenvironments promote survival and phenotype of human induced pluripotent stem cell-derived endothelial cells in hypoxia. Acta Biomater 2016, 44, 188–99. 10.1016/j.actbio.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulous F. E.; Carnevale J. C.; Al-Yafeai Z.; Pearson B. H.; Hamilton J. A. G.; Henry C. J.; Orr A. W.; Petrich B. G. Talin-dependent integrin activation is required for endothelial proliferation and postnatal angiogenesis. Angiogenesis 2021, 24, 177–190. 10.1007/s10456-020-09756-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haage A.; Goodwin K.; Whitewood A.; Camp D.; Bogutz A.; Turner C. T.; Granville D. J.; Lefebvre L.; Plotnikov S.; Goult B. T.; Tanentzapf G. Talin Autoinhibition Regulates Cell-ECM Adhesion Dynamics and Wound Healing In Vivo. Cell Rep 2018, 25, 2401–2416 e5. 10.1016/j.celrep.2018.10.098. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Senger D. R. Matrix-specific activation of Src and Rho initiates capillary morphogenesis of endothelial cells. FASEB J. 2004, 18, 457–68. 10.1096/fj.03-0948com. [DOI] [PubMed] [Google Scholar]
- Davis G. E.; Senger D. R. Endothelial extracellular matrix: biosynthesis, remodeling, and functions during vascular morphogenesis and neovessel stabilization. Circ. Res. 2005, 97, 1093–107. 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- Agren M. S.; Litman T.; Eriksen J. O.; Schjerling P.; Bzorek M.; Gjerdrum L. M. R. Gene Expression Linked to Reepithelialization of Human Skin Wounds. Int. J. Mol. Sci. 2022, 23, 15746. 10.3390/ijms232415746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vongsa R. A.; Zimmerman N. P.; Dwinell M. B. CCR6 regulation of the actin cytoskeleton orchestrates human beta defensin-2- and CCL20-mediated restitution of colonic epithelial cells. J. Biol. Chem. 2009, 284, 10034–45. 10.1074/jbc.M805289200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro J. M.; Pober J. S.; Cotran R. S. Tumor necrosis factor and interferon-gamma induce distinct patterns of endothelial activation and associated leukocyte accumulation in skin of Papio anubis. Am. J. Pathol. 1989, 135, 121–133. [PMC free article] [PubMed] [Google Scholar]
- Rousselle P.; Braye F.; Dayan G. Re-epithelialization of adult skin wounds: Cellular mechanisms and therapeutic strategies. Adv. Drug Deliv Rev. 2019, 146, 344–365. 10.1016/j.addr.2018.06.019. [DOI] [PubMed] [Google Scholar]
- Satish L.; Yager D.; Wells A. Glu-Leu-Arg-negative CXC chemokine interferon gamma inducible protein-9 as a mediator of epidermal-dermal communication during wound repair. J. Invest Dermatol 2003, 120, 1110–7. 10.1046/j.1523-1747.2003.12230.x. [DOI] [PubMed] [Google Scholar]
- Nolfi-Donegan D.; Braganza A.; Shiva S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674 10.1016/j.redox.2020.101674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold L. P.; Gil H. J.; Gao P.; Martinez C. A.; Weinberg S. E.; Chandel N. S. Mitochondrial complex III is necessary for endothelial cell proliferation during angiogenesis. Nat. Metab 2019, 1, 158–171. 10.1038/s42255-018-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Qiu Z.; Dong W.; Yang Z.; Wang J.; Xu H.; Sun T.; Huang Z.; Jin J. S1PR1 induces metabolic reprogramming of ceramide in vascular endothelial cells, affecting hepatocellular carcinoma angiogenesis and progression. Cell Death Dis 2022, 13, 768. 10.1038/s41419-022-05210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W.; Oldham W. M.; Priolo C.; Pandey A. K.; Loscalzo J. Immunometabolic Endothelial Phenotypes: Integrating Inflammation and Glucose Metabolism. Circ. Res. 2021, 129, 9–29. 10.1161/CIRCRESAHA.120.318805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caja S.; Enriquez J. A. Mitochondria in endothelial cells: Sensors and integrators of environmental cues. Redox Biol. 2017, 12, 821–827. 10.1016/j.redox.2017.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugus J. J.; Ngoh G. A.; Bachschmid M. M.; Walsh K. Mitofusins are required for angiogenic function and modulate different signaling pathways in cultured endothelial cells. J. Mol. Cell Cardiol 2011, 51, 885–93. 10.1016/j.yjmcc.2011.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X.; Wang L.; Chen Y.; Ning X.; Zhang Z.; Xin H.; Lv Q. X.; Hou Y.; Liu F.; Kong L. TiO(2)nanotubes induce early mitochondrial fission in BMMSCs and promote osseointegration. Biomed Mater. 2023, 18, 025008. 10.1088/1748-605X/acb7bc. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Xu H.; Li Y.; Liu Y.; Li X.; Zhou W.; Wang J.; Guo C.; Sun Z.; Li Y. Silica nanoparticles perturbed mitochondrial dynamics and induced myocardial apoptosis via PKA-DRP1-mitochondrial fission signaling. Sci. Total Environ. 2022, 842, 156854 10.1016/j.scitotenv.2022.156854. [DOI] [PubMed] [Google Scholar]
- Li J.; Chang X.; Shang M.; Niu S.; Zhang W.; Li Y.; Sun Z.; Wu T.; Kong L.; Zhang T.; Tang M.; Xue Y. The crosstalk between DRP1-dependent mitochondrial fission and oxidative stress triggers hepatocyte apoptosis induced by silver nanoparticles. Nanoscale 2021, 13, 12356–12369. 10.1039/D1NR02153B. [DOI] [PubMed] [Google Scholar]
- Ke S.; Zhou T.; Yang P.; Wang Y.; Zhang P.; Cheng K.; Ren L.; Ye S. Gold nanoparticles enhance TRAIL sensitivity through Drp1-mediated apoptotic and autophagic mitochondrial fission in NSCLC cells. Int. J. Nanomedicine 2017, 12, 2531–2551. 10.2147/IJN.S129274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola A. V.; Aguilar H. C.; Mercer J.; Ryckman B.; Wiethoff C. M. Virus entry by endocytosis. Adv. Virol 2013, 2013, 469538 10.1155/2013/469538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W.; Zhu X.; Yu X.; Zeng X.; Xiao Q.; Zhang X.; Ji X.; Wang X.; Shi J.; Zhang H.; Mei L. Black Phosphorus Nanosheets as a Robust Delivery Platform for Cancer Theranostics. Adv. Mater. 2017, 29, 1603276. 10.1002/adma.201603276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T.; Comoglio F.; Seimiya M.; Cabuy E.; Paro R. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E2327–E2336. 10.1073/pnas.1423074112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M.; Tchkonia T.; Ding H.; Ogrodnik M.; Lubbers E. R.; Pirtskhalava T.; White T. A.; Johnson K. O.; Stout M. B.; Mezera V.; Giorgadze N.; Jensen M. D.; LeBrasseur N. K.; Kirkland J. L. JAK inhibition alleviates the cellular senescence-associated secretory phenotype and frailty in old age. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E6301–E6310. 10.1073/pnas.1515386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea J. J.; Gadina M.; Schreiber R. D. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell 2002, 109 (Suppl. 1), S121–S131. 10.1016/S0092-8674(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Lerchenmuller C.; Heissenberg J.; Damilano F.; Bezzeridis V. J.; Kramer I.; Bochaton-Piallat M. L.; Hirschberg K.; Busch M.; Katus H. A.; Peppel K.; Rosenzweig A.; Busch H.; Boerries M.; Most P. S100A6 Regulates Endothelial Cell Cycle Progression by Attenuating Antiproliferative Signal Transducers and Activators of Transcription 1 Signaling. Arterioscler Thromb Vasc Biol. 2016, 36, 1854–67. 10.1161/ATVBAHA.115.306415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink A. F.; Ciliberti G.; Popp R.; Sirait-Fischer E.; Frank A. C.; Fleming I.; Sekar D.; Weigert A.; Brune B. IL27Ralpha Deficiency Alters Endothelial Cell Function and Subverts Tumor Angiogenesis in Mammary Carcinoma. Front Oncol 2019, 9, 1022. 10.3389/fonc.2019.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P.; Wang L. L.; Liu J.; Dong F.; Song W.; Liao L.; Wang B.; Zhang W.; Zhou X.; Xie Q.; Sun R.; Liu J. Dihydroartemisinin inhibits endothelial cell tube formation by suppression of the STAT3 signaling pathway. Life Sci. 2020, 242, 117221 10.1016/j.lfs.2019.117221. [DOI] [PubMed] [Google Scholar]
- Huang X.; Wang J.; Guan J.; Zheng Z.; Hao J.; Sheng Z.; Wang M.; Xu T.; Guo G.; Yao L. Exosomal Circsafb2 Reshaping Tumor Environment to Promote Renal Cell Carcinoma Progression by Mediating M2Macrophage Polarization. Front Oncol 2022, 12, 808888 10.3389/fonc.2022.808888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. S.; Kim B.; Oh G. T.; Kim Y. J. OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol 2013, 14, 346–55. 10.1038/ni.2535. [DOI] [PubMed] [Google Scholar]
- McGarry T.; Orr C.; Wade S.; Biniecka M.; Wade S.; Gallagher L.; Low C.; Veale D. J.; Fearon U. JAK/STAT Blockade Alters Synovial Bioenergetics, Mitochondrial Function, and Proinflammatory Mediators in Rheumatoid Arthritis. Arthritis Rheumatol 2018, 70, 1959–1970. 10.1002/art.40569. [DOI] [PubMed] [Google Scholar]
- Skrivergaard S.; Jensen M. S.; Rolander T. B.; Nguyen T. B. N.; Bundgaard A.; Nejsum L. N.; Martensen P. M., The Cellular Localization of the p42 and p46 Oligoadenylate Synthetase 1 Isoforms and Their Impact on Mitochondrial Respiration. Viruses 2019, 11.1122. 10.3390/v11121122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Gil E.; Esteban M. Role of mitochondria in apoptosis induced by the 2–5A system and mechanisms involved. Apoptosis 2006, 11, 725–38. 10.1007/s10495-006-5541-0. [DOI] [PubMed] [Google Scholar]
- Boyd D. A.; Bezares F. J.; Pacardo D. B.; Ukaegbu M.; Hosten C.; Ligler F. S. Small-molecule detection in thiol-yne nanocomposites via surface-enhanced Raman spectroscopy. Anal. Chem. 2014, 86, 12315–20. 10.1021/ac503607b. [DOI] [PubMed] [Google Scholar]
- Yang X.; Wang D.; Zhu J.; Xue L.; Ou C.; Wang W.; Lu M.; Song X.; Dong X. Functional black phosphorus nanosheets for mitochondria-targeting photothermal/photodynamic synergistic cancer therapy. Chem. Sci. 2019, 10, 3779–3785. 10.1039/C8SC04844D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukai T.; Ushio-Fukai M. Cross-Talk between NADPH Oxidase and Mitochondria: Role in ROS Signaling and Angiogenesis. Cells 2020, 9, 1849. 10.3390/cells9081849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.