Abstract
This review investigates the role of aneuploidy and chromosome instability (CIN) in the aging brain. Aneuploidy refers to an abnormal chromosomal count, deviating from the normal diploid set. It can manifest as either a deficiency or excess of chromosomes. CIN encompasses a broader range of chromosomal alterations, including aneuploidy as well as structural modifications in DNA. We provide an overview of the state-of-the-art methodologies utilized for studying aneuploidy and CIN in non-tumor somatic tissues devoid of clonally expanded populations of aneuploid cells.
CIN and aneuploidy, well-established hallmarks of cancer cells, are also associated with the aging process. In non-transformed cells, aneuploidy can contribute to functional impairment and developmental disorders. Despite the importance of understanding the prevalence and specific consequences of aneuploidy and CIN in the aging brain, these aspects remain incompletely understood, emphasizing the need for further scientific investigations.
This comprehensive review consolidates the present understanding, addresses discrepancies in the literature, and provides valuable insights for future research efforts.
Keywords: Aging, cancer, tumor, glioblastoma, brain, neurons, astrocytes, aneuploidy, mutations, genomic instability, chromosome instability, copy number alterations, mutation frequency, disease, neurodegeneration
2. Introduction
Advances in biomedical science and genome analysis technologies have enabled the study of genome-wide DNA sequence content at the single-cell level in humans and model organisms, providing high-resolution information on the variability of genomes between individual cells. These technologies have significantly advanced the study of somatic mosaicism, particularly in disease-free tissues where clonally expanded cells are usually not present. It is now evident that virtually all adult humans are genetic mosaics of postzygotic mutations1–3.
Genomic instability refers to the propensity of genetic material within a cell or organism to undergo changes, frequently leading to mutations, chromosomal aberrations, and other forms of genetic variation4–8. This instability can arise from various factors, such as errors in DNA replication, exposure to environmental mutagens, or defects in DNA repair mechanisms. Genomic instability is a hallmark of most cancer cells and is thought to play a critical role in the progression and heterogeneity of tumors9–12. Furthermore, genomic instability has been associated with aging13–15, as the accumulation of genetic changes over time can contribute to cellular senescence and functional decline in various tissues and organs16–19. This can increase the likelihood of age-related diseases, such as cancer and neurodegeneration4,20–22.
Genomic instability can manifest as modifications sequence, structure, or number of chromosomes, as well as epigenetic changes to DNA methylation or histone patterns. This can lead to modifications in gene expression programs, which can have functional implications for cellular processes and contribute to the development of diseases. Maintaining genome integrity is essential for cell fitness and proper physiological functioning of the organism. The significance of genome maintenance is evidenced by the presence of numerous redundant, evolutionarily conserved pathways that ensure genome integrity is retained. These pathways act as caretakers of DNA replication fidelity and ensure the precise segregation of the genome content into daughter cells. Despite the activities of these numerous molecular mechanisms, errors still occur, and mutations are inevitable. Indeed, in contrast to physicochemical DNA damage, mutations cannot be repaired and are permanent. Mutations in the DNA sequence can be inherited by daughter cells and adversely affect cell fitness23–27.
This review specifically focuses on chromosome instability (CIN), which represents a form of genomic instability that can result in numerous chromosomal alterations, including deletions, insertions, duplications, translocations, inversions, aneuploidy (changes in chromosome number), or polyploidy (changes in the number of the entire set of chromosomes). CIN is a well-established hallmark of tumors, and it plays a critical role in promoting the accumulation of genetic alterations that fuel the progression of tumors28. CIN contributes to the genomic heterogeneity of tumors29–32, and enables the selection of clones with advantageous phenotypes33, such as increased proliferative potential, resistance to therapy34,35, and immune evasion36,37. Additionally, CIN has been shown to contribute to the development of more aggressive tumor subtypes and the acquisition of metastatic potential, which are critical determinants of clinical outcomes.
While in the context of cancer cells CIN is considered an important factor, in non-transformed cells, it can lead to the loss of cellular function and contribute to developmental disorders38–41. This is because aneuploidy can disrupt the balance of gene dosage42–44, impair cellular pathways45,46, and interfere with chromosome segregation during cell division47,48, leading to genomic instability and detrimental cell fates49–53. Thus, while aneuploidy can be a selectable benefit to cancer cells, it can be detrimental to normal cellular function and contribute to disease in non-transformed cells. The phenomenon of aneuploidy being both beneficial to cancer cells and detrimental to normal cellular function is known as the aneuploidy paradox54. Mounting evidence suggests that aneuploidy, a consequence of CIN, is not only a hallmark of cancer but can also manifest in non-transformed cells, resulting in impaired cellular function, decreased viability, and heightened vulnerability to disease53,55–57.
The brain is a highly specialized and complex organ that plays a critical role in maintaining normal physiological functions, and its dysregulation is associated with a range of diseases. Therefore, investigating molecular changes that occur during aging in the brain and how they contribute to either healthy aging or disease is crucial in understanding the underlying mechanisms of neurodegenerative diseases such as Alzheimer's disease, Parkinson's disease, and Huntington's disease. In the context of brain aging, there is evidence for and against the presence of CIN in neurons and glial cells. There are several possible explanations for the discrepancies in the literature regarding the frequency of somatic aneuploidy in the aging brain4,58–60. One factor is the sensitivity of the methodologies used to measure somatic aneuploidy. Unlike in tumor cells where aneuploidies are clonal, in somatic disease-free tissues, aneuploidies are expected to be uniquely present in single cells, at most at sub-clonal levels. Another factor is the variation of aneuploidy levels among different age groups, brain areas and or different cell types, which may explain some of the discrepancies reported in the literature.
Because of the challenges in measuring CIN in complex organs, much remains unknown about the prevalence and consequences of CIN in the aging brain. This review aims to summarize current understanding of aneuploidy, CIN, genome structural variation (SV) and large copy number alterations (CNA) in the mammalian brain, with a particular focus on their associations with aging and the factors that may contribute to the wide range of reported aneuploidy frequencies across studies. Firstly, we will describe the state-of-the-art methodologies used to measure aneuploidy and CIN in somatic tissues. Subsequently, we will provide an overview of the studies reporting the presence of aneuploidy or CIN in the brain, as well as studies that found no evidence of these somatic alterations and discuss the potential reasons for the discrepancies in findings across studies.
3. Methodologies to measure genomic instability and technical challenges.
Mutations, including aneuploidy, cannot be detected in bulk tissues unless they are clonal. While methods are available to measure aneuploidy in single cells, technical challenges remain. Detection of sub-clonal structural variation in single cells poses an even greater challenge. Moreover, the ability to simultaneously analyze multiple types of genomic instability in a single cell has been limited until recently, with emerging technologies allowing for such analyses61–63. The challenges of analyzing different mutation types in the same cell, particularly in the context of aneuploidy, have been a limitation of many of the methodologies used thus far. In addition, CIN can be defined as an increased rate of chromosome gains and losses that manifests as cell-to-cell karyotypic heterogeneity within a tissue, and in general can be measures by tracking chromosome numbers within a single cell and its progeny over time, and by quantitatively assessing cell-to-cell heterogeneity within a given population64.
There are three main molecular cytogenetic methodologies that have been utilized for the quantification of aneuploidy in both brain tissues and individual cells: Spectral Karyotyping (SKY), Fluorescence in situ Hybridization (FISH), and Single-Cell Low-Coverage Whole-Genome Sequencing (scL-WGS). It is worth noting that some studies applied FACS based methods, and they will be also discussed in this section.
3.1. Spectral karyotyping (SKY)
SKY is a type of molecular cytogenetic technique that uses a set of chromosome-specific fluorescent probes that bind to and label each chromosome with a unique color65–67 (Figure 1A). The labeled chromosomes are then visualized under a specialized fluorescent microscope equipped with a filter cube that contains a series of interference filters, dichroic mirrors, and a beam splitter, allowing for the simultaneous capture of multiple fluorochromes. As a result, SKY allows visualization of all the chromosomes from a single cell, where each chromosome is labeled with a unique combination of different fluorochromes that can be distinguished by their emission spectra. This technique allows for the detection of both numerical and structural chromosomal abnormalities, and it is particularly useful in identifying complex rearrangements such as translocations, especially those involving multiple chromosomes that would be difficult to detect using other cytogenetic methods.
Figure 1: Molecular Cytogenetic Techniques commonly used to quantify aneuploidy and large copy number variation.
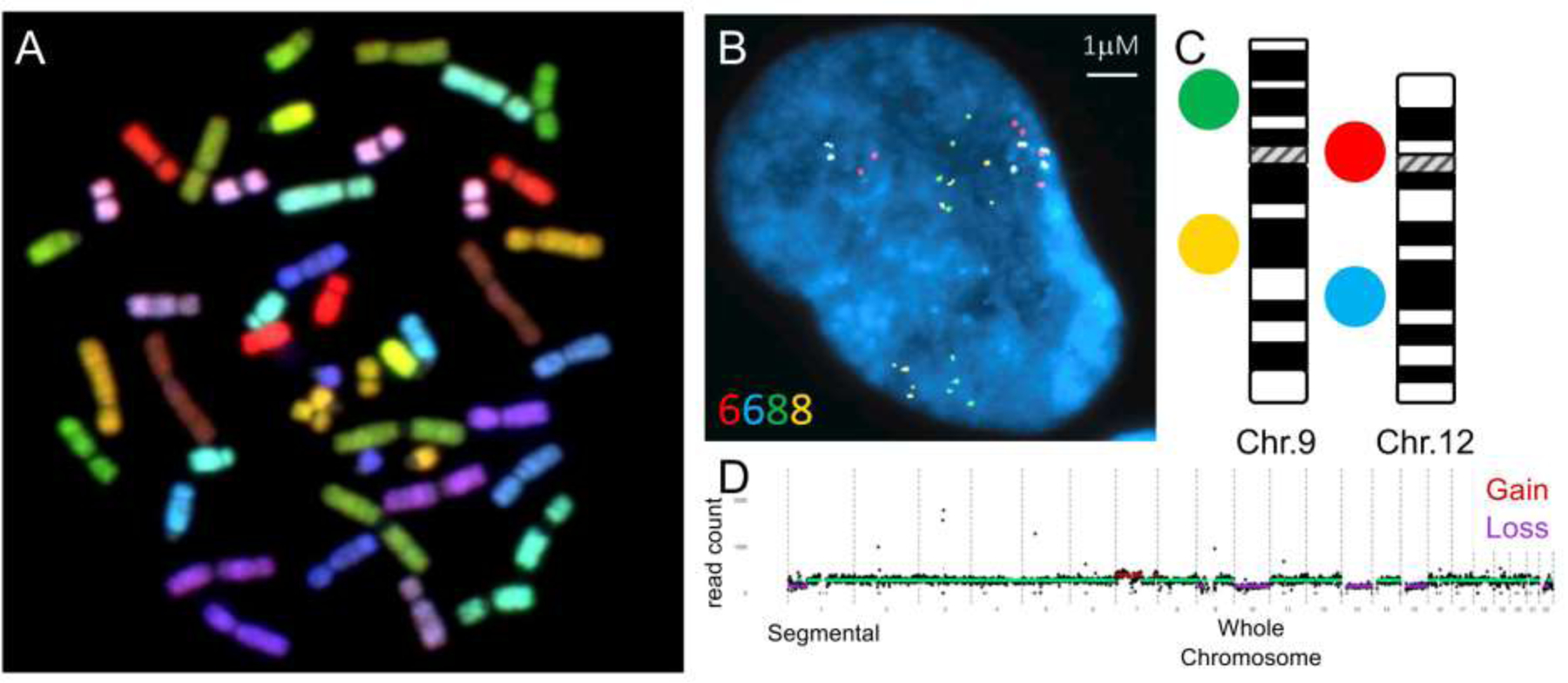
A) Spectral Karyotyping (SKY) image of metaphase chromosomes illustrating human chromosomes labeled with unique fluorophores or combinations. This allows the identification of all human chromosomes in a single image based on the emission spectra assigned to each autosome or sex chromosome1. B-C) Representative interphase nucleus analyzed using interphase FISH (iFISH) to quantify copy number changes to infer chromosome specific aneuploidies. Copy number alterations are determined by enumerating signals at locus-specific probes of interest. A 4-color iFISH approach is employed, utilizing 2-locus specific probes mapping to a single chromosome, which enable the measurement of aneuploidy events (C)2. D) Representative example of copy number estimates across the entire genome quantified by scL-WGS3. After normalizing mappability, GC content, and amplification bias, the results are presented as a copy number variation plot. Each black dot represents a genomic bin, while green horizontal lines indicate regions with 2 copies. Purple regions represent chromosome loss, and red regions represent chromosome gain. The plots were generated using Ginko4.
SKY offers numerous advantages over other cytogenetic methods, primarily due to its ability to provide comprehensive analysis of chromosomal abnormalities throughout the entire genome at the level of a single cell in one single experiment. However, a limitation of this technique is that it relies on metaphase chromosome spreads, which requires the use of actively proliferating cells, restricting its applicability to studying only proliferating cells and in relatively small numbers of cells, due to the complexity and time required for analysis. Metaphase chromosome-based analyses can be very accurate for establishing whole chromosome gain or loss or large copy number alterations, as it has been the standard of care for pre-natal diagnosis until recently, when array-based methods have largely replaced them. However, outside CLIA Certified standard operating procedure (SOP), accuracy needs to be maintained to ensure that chromosome metaphase spreads preparations follow well validate protocols to limit chromosome loss during the process of metaphase spreading or overlapping metaphases, the inclusion of which will result in false positives.
In addition, SKY is technically challenging and requires specialized expertise and equipment to perform and interpret the results. Another limitation is that SKY is unable to detect small-scale chromosomal abnormalities, such as insertions and deletions smaller than 10Mb and it cannot detect inversions. Finally, SKY is relatively expensive compared to other cytogenetic techniques.
Multiplex-FISH (M-FISH)68 has also been applied to measure aneuploidy in the brain. It is similar to SKY but mainly differs in the imaging methodology. SKY can detect all fluorophores in one single imaging, but M-FISH requires serial imaging acquiring one fluorophore at the time followed by merging single channels, which can increase background. M-FISH can however still allow for the detection of chromosomal abnormalities in a single experiment.
3.2. Fluorescent in situ Hybridization (FISH)
FISH is one of the most commonly used methodologies to detect chromosome abnormalities in clinical settings. It allows for the detection of copy number alterations at regions of interest by identifying those sites with fluorescently labeled probes. FISH can facilitate the analysis of entire chromosome aneuploidies by utilizing probes labeled with two different fluorophores that are mapped to sub-centromeric and sub-telomeric regions of a specific chromosome69,70. However, complex chromosomal alterations outside the locus specific probes may not be detected using this approach.
FISH provides several advantages over other methods for detecting chromosomal abnormalities, including its ability to measure aneuploidy at specific regions of interest in both proliferating and non-proliferating cells (interphase FISH, or iFISH), as well as in tissue sections (Figure 1B). It enables the analysis of individual cells, allowing the detection of aneuploidy and copy number alterations at the single-cell level. Moreover, FISH offers the advantage of analyzing a large number of cells, more than 1000 cells per sample, which allows for a more representative sample analysis, particularly in cases where clonal expansion is not anticipated. This makes FISH a valuable tool for studying somatic aneuploidy in healthy tissues. The technique also enables the combined analysis of aneuploidy and large copy number alterations with morphometric analysis of nuclear size and morphology in single cells, as well as the combined analysis of DNA FISH with immunofluorescence staining for the expression of proteins of interest. This approach is particularly advantageous when locus-specific probes are generated in-house69, as the cost per cell analyzed is greatly reduced, making this technology highly affordable. In addition, the main instrumentation required for this technique includes hybridization ovens and access to fluorescence microscopy, which are widely available at most institutions.
Historically, FISH has been limited to the analysis of a handful of probes, typically four. Recent advances in FISH techniques, including multicolor interphase FISH (mi-FISH), have overcome the limitation of analyzing only a few chromosomes simultaneously. mi-FISH enables the analysis of up to three sets of four probes, allowing for the simultaneous detection of multiple aneuploidies within a single cell71. However, it is worth nothing that aneuploidy analysis for the full chromosome complement in a single cell is still challenging.
The accuracy of using FISH to detect aneuploidy levels in the brain has been questioned due to the potential detection of false positives due to high backgrounds, localized lack of hybridization signals, variation in hybridization efficiency, and probe clustering, which can lead to both false positive and false negative results. In addition, inferring aneuploidy based on the count of a subset of chromosome has the potential to amplify the rate of false positive counts. This issue is particularly prevalent when applying FISH to tissue section analysis using a single probe per chromosome. However, it should be noted that using two probes, labeled with different fluorophores for each chromosome tested, remedies this drawback by greatly reducing the number of false positives60,69,70.
3.3. Single-Cell Low-Coverage Whole-Genome Sequencing (scL-WGS).
scL-WGS for detecting aneuploidy is a technique that uses next-generation sequencing to analyze the DNA content of individual cells72,73. In this approach, single cells are isolated and subjected to whole-genome amplification (WGA), a process that produces multiple copies of the entire genome from a single cell. The amplified DNA is then sequenced using low coverage sequencing, which provides a relatively shallow read depth of the genome but reduces costs, allowing for a higher number of cells to be analyzed. The sequencing data is then analyzed to detect local variation in sequencing read depth of individual cells, such as the loss or gain of entire chromosomes or chromosome segments, which are indicative of aneuploidy (Figure 1C). There are numerous established protocols that comprehensively describe the methodologies of WGS, including limitations of certain WGS protocols and sequencing library construction, as well as dedicated analytical tools designed to facilitate the mapping of reads and the identification of aneuploidies30,74–79. Here we will limit our discussion to the application of this method to the analysis of somatic aneuploidy in brain.
In comparison to other molecular cytogenetic techniques, low coverage single cell sequencing has the advantage of being able to analyze multiple chromosomes or chromosome segments simultaneously, whereas FISH typically only analyzes a limited number of regions of interest. Thus, low coverage single cell sequencing can detect copy number variations (CNVs) and other genomic aberrations that may be missed by traditional molecular cytogenetic methods. However, scL-WGS has its own limitations and can yield both false positive and false negative results80, particularly for the detection of aneuploidy in a polyploid context70. False positive and negative results can occur as an outcome of uneven sequencing coverage or allelic drop out due to amplification bias. In addition, PCR amplification cycles needs to be carefully controlled, as amplification outside the log phase can lower the sensitivity of detecting aneuploid chromosomes over the entire genome content in single cells. These types of artifacts make it difficult to identify genomic variants, however, single-cell variant callers are implementing strategies to try and overcome some of these artifacts leading to false positive and negative results81.
3.4. Other methodologies
Some other molecular techniques to measure aneuploidy or DNA content have been applied more sporadically to the analysis of aneuploidy in the mammalian brain. Slide-based cytometry (SBC)82 can combine the quantification of DNA content with immunolabeling and Chromogenic in situ hybridization.
Fluorescence-activated cell sorting (FACS) is a high-throughput method that separates cells based on their physical and/or chemical properties, such as size, granularity, and fluorescence intensity. The cells are stained with a fluorescent dye that binds to DNA, allowing for the measurement of DNA content in individual cells. While FACS is high throughput, it lacks the sensitivity to detect aneuploidy but is rather more suited to quantify ploidy changes.
Flow cytometry can also be utilized to understand the relative DNA content and composition of chromosomes and their relative frequencies in mitotic cells. A technique for examining the complete set of human chromosomes using flow cytometry involves the staining of chromosomal DNA with two specific dyes such as Hoechst 33342 and chromomycin A3 (CA3), which have distinct interactions with DNA. Hoechst 33342 binds preferentially to adenine and thymine bases, while chromomycin A3 attaches to DNA regions rich in guanine and cytosine bases. Based on their different binding preferences, it is possible to create a detailed map of the chromosomes by generating Bivariate flow karyotypes plot of chromosomes83,84.
Single-nucleotide polymorphism (SNP) arrays can be used to measure aneuploidy by comparing the ratio of alleles in the sample to a reference genome after hybridization of a DNA sample of interest to microarrays containing probes that detect specific SNPs across the genome. By comparing the ratio of the SNP alleles in the sample to the reference genome, it is possible to determine the number of copies of each chromosome in the sample. Arrays find application in a technique known as array painting. This procedure combines two vital elements: the isolation of derivative chromosomes (those affected by translocations) using a MoFlo sorter and the execution of high-resolution microarray analysis. This distinctive combination of techniques enables to accurately determine the exact genomic sites where translocation breakpoints occurred. Essentially, array painting harmonizes the strategic isolation of translocated chromosomes with state-of-the-art microarray technology, facilitating an enhanced mapping of genetic alterations85.
These techniques are based on the analysis of bulk DNA and their sensitivity to quantify aneuploidy is generally around 5% Even though the detectable levels are still variable among different chromosome microarrays platforms86,87, these methods cannot measure stochastic non-clonal aneuploidy changes.
3.5. Considerations for Accurate Analysis of Aneuploidy and Chromosomal Instability in Somatic Genome Variation
Although scL-WGS and FISH are the most commonly applied techniques for measuring somatic aneuploidy in the mammalian brain, they have distinct advantages and limitations for detecting aneuploidy, and none should be considered superior over the other. Instead, a combinatorial approach that incorporates multiple techniques may provide the most accurate analysis of somatic aneuploidy.
For example, while scL-WGS can provide high-resolution information on copy number changes across the entire genome, it may have limited sensitivity to detect aneuploidy in a polyploid background because the ratio of genome mapping to the gained or lost chromosome is lower when the entire genome is duplicated. It may also have low sensitivity in detecting copy number changes in chromosomal regions that lack mapped reads due to the low coverage nature of scL-WGS. Specialized analytical tools to overcome these limitations are being developed, which should provide enhanced methodologies to increase sensitivity and specificity88. On the other hand, FISH can specifically target and visualize a particular chromosome of interest in a very large number of cells, allowing for more sensitive detection of aneuploidy in a polyploid background, yet limited to the regions of interest.
4. The developing mammalian brain.
Data obtained from mice and rats suggest the presence of aneuploid cells in the developing brain with frequencies as high as 30% of analyzed cells per tissue (Table 1). These levels are considerably higher than that of the adult (4 month) brain which are reported around 1%.
Table 1:
Summary of aneuploidy and CIN results in brain of mice and rats.
| AUTHOR | YEAR | SPECIES | TECHNIQUE | PROBES | CASES (N) | AGE | TISSUE | CELL TYPE | NUMBER OF NUCLEI | DETECTION METRIC | AREA RESULT | CHROMOSOME | CHR AFFECTED % | AVERAGE CHR % | % PER TISSUE | NOTES | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| REHEN 5 | 2001 | Mo (BALB/c) | SKY-FISH | Chr X/Y | unk | E11-14-adult | Cortex | Mitotic neuroblasts and post mitotic bulk | 220 (SKY) 300 (FISH) | Aneuploidy | Embryonic | Y | 1–5.9 | unk | 33 | ||
| Embryonic | X | 2.1 | unk | ||||||||||||||
| KAUSHAL 6 | 2003 | Mo (GFP transgenic) | SKY-FISH | Chr X/Y | 65 | P5-P10 | SVZ and OB | NPCs (SKY) bulk (FISH) | 65 (SKY) unk (FISH) | Aneuploidy | SVZ | X/Y | 5 | unk | 33 | ||
| OB | X/Y | 6 | unk | ||||||||||||||
| YANG 7 | 2003 | Mo (BALB/c) | SKY | n/a | unk | E11-14 | Cortex | Mitotic NPC | 220 | Aneuploidy | NPC | all | unk | unk | 33.2 | ||
| KINGSBURY 8 | 2005 | Mo (BALB/c) | FISH and paint | Chr X/Y | 10 | 10 wks | Whole brain | Neurons | 1000 | Hyperdiploidy | X/Y | 0.2 | unk | unk | |||
| WESTRA 9 | 2008 | Mo (BALB/c) | FISH and M-FISH | Chr 16/X | unk | P0, P7 and adult | Cerebellum | Mitotic NPCs, neurons, and non-neurons | >18,000 | Aneuploidy | X | 1.1 | 1 | 20 | P0 (15.3) | ||
| 16 | 1.6 | 1 | P7 (20.8) | ||||||||||||||
| FAGGIOLI 10 | Mo (C57B6) | FISH-dual | Chr 1/7/14/15/6/18/19/Y | 21 | 4–28 mo. | Cortex and cerebellum | NeuN and NeuN-neg | >10,000 | Aneuploidy | Cerebellum | 1/7/14/15/6/18/19/Y | 0.7–1.6 | 1.2–9.8 | up to 30 | 28 mo significantly increased vs. 4 mo | ||
| Cortex | 1/7/14/15/6/18/19/Y | <1 | <1 | ||||||||||||||
| KNOUSE 11 | 2014 | Mo (Nestin-GFP mice, C57BL/6) | sc-WGS | n/a | unk | Embryonic and adult | Cerebral Cortex | Bulk, neuron/non (NeuN) | 43 nuclei and 19 nuclei | Aneuploidy | Embryonic | 15 | 2.3 | unk | 1 | ||
| Adult | na | 0 | 0 | ||||||||||||||
| ANDRIANI 12 | 2016 | Mo (Ercc1−/Δ7 and Bub1bH/H) | FISH-dual | Chr 1/7/18 | 18 | E13.5-6 mo | Cerebral cortex and cerebellum | Bulk | 1000–1500 | Aneuploidy | Embryonic | 1/18 | 2.3–37.7 | variable | 30 | ||
| Adult | <1 | <1 | |||||||||||||||
| PETERSON 13 | 2012 | Mo (C57B6) | SKY and FISH | Chr 8/16 | unk | E19 | Cerebral Cortex | Mitotic NPS and post mitotic bulk | 100 (SKY) 2500–3500 (FISH) | Aneuploidy | E19 | 16 | 2.1 | unk | 24–29 | ||
| 8 | 1.6 | unk | |||||||||||||||
| NING 14 | 2015 | rat | sc-WGS | n/a | 2 | E18 | Hippocampus | NeuN | 19 | CNV | 8 | unk | unk | unk | large scale CNV detected | ||
Rehen89 reported the occurrence of chromosomal aneuploidy in developing and adult neurons. In this study, embryonic mouse brain (E11-15) was dissected, and intact cortical hemispheres were treated with colcemid to induce metaphase arrest. The resulting cells were dissociated and fixed for SKY which revealed approximately 33% aneuploid cells when examining all chromosomes. Subsequently, the same technical approach was applied for the analysis of the chromosome content of cells in the postnatal (P5-P10) subventricular zone (SVZ) of mice, an area that harbors neuronal and progenitor stem cells90. The results showed that 33% of mitotic SVZ cells had lost or gained chromosomes in vivo, confirming that dividing neuronal stem and progenitor cells show aneuploidy. If each chromosome has an equal probability of being gained or lost, it can be extrapolated that the rate of aneuploidy for a single chromosome is approximately 1.65%. These results support the theory that neuronal precursors undergo chromosomal segregation defects, resulting in the generation of aneuploid neurons, ultimately leading to genetic mosaicism91,92. Using chromosome paint probes for iFISH specific for both the X and Y chromosome, Rehen and colleagues90 demonstrated that among adult neurons, 1.16% carried numerical alterations for either of these chromosomes.
Kaushal and collaborators90 conducted a study in mice that were hemizygous for the enhanced green fluorescent protein (eGFP) inserted at a single locus on Homo Sapiens Autosome 15 (HSA15) and ubiquitously expressed under the control of the chicken β-actin promoter. The researchers utilized the level of GFP fluorescence and the number of HSA 15 copies to identify aneuploid cells and isolate them from their normal counterparts. A loss of one copy of HSA 15 resulted in loss of GFP expression, similar to loss of heterozygosity (LOH). Based on metaphase spreads, an average chromosome loss rate of 5.13% was observed in SVZ cells, while interphase cells exhibited an average loss rate of 4.96%. These estimates are concordant with previous data89.
By profiling gene expression of GFP+ and GFP- cells, they were able to identify 22 differentially expressed genes between the two populations (diploid vs. hypo-aneuploid). While the study did not further investigate the identified genes, the 22 genes mapped to various regions of the mouse genome, suggesting a potentially widespread genome-wide deregulation. Notably, Annexin A1, a gene with potential implications in stroke and neurodegenerative conditions, was among the identified genes.
Kaushal and colleagues assessed the functional consequences of the loss of one copy of HSA 15 by evaluating the proliferation and survival index of aneuploid cells in culture. They stained neuronal cells with markers for proliferation and cell death, demonstrating that aneuploid cells remain viable and competent to divide under these experimental conditions. Additionally, they provided evidence that aneuploidy persists in subventricular zone (SVZ)-born cells that migrate to the olfactory bulb (OB), supporting the notion that low levels of aneuploidy may result in functional cells with distinct biological properties.
It has been postulated that aneuploidy during mammalian brain development may play a crucial role in generating genetic diversity that contributes to the functional complexity of the central nervous system. To test this hypothesis, Kingsbury and colleagues devised an experimental approach to trace hyperdiploid neurons in vivo and demonstrated that aneuploid cells are functionally integrated into the neuronal circuitry and actively participate in brain functions90. These studies suggest that hyperdiploid neurons carrying numerical alterations for both X and Y chromosomes constitute around 0.2% of neurons in the brain, and that they are functionally active and integrated into the neuronal network. Furthermore, aneuploidy has been observed in various areas of the brain. Evidence for aneuploid cells throughout the mammalian neuroaxis has also been demonstrated by studies that found 15.3% and 20.8% of cerebellar cells aneuploid at postnatal day P0 and P7 when all chromosomes were analyzed93. Chromosome segregation defects contribute to the generation of aneuploid cells, as evidenced by immunofluorescent staining for histone H3 and vimentin of cerebellar Neuronal Stem Cells (NPCs). Immunostaining using anti-pericentrin identified a subset of cells with supernumerary centrosomes (up to 3 per cell), suggesting the presence of cells that have the potential to undergo multipolar mitosis. Accordingly, neuronal, and non-neuronal cells in the adult cerebellum were found to harbor aneuploid cells, as shown in the NeuN+ and NeuN- enriched populations by FISH analysis using probes for murine chromosomes X and 16, which revealed a frequency of aneuploidy of 2.7%.
A recent investigation utilized sc-WGS to explore copy number variations (CNVs) in rat neurons at embryonic day 18 (E18) and identified large CNVs94. One notable advantage of this study was its comparative approach involving various whole genome amplification methods (GenomePlex WGA4, MDA, and MALBAC). Collectively, these methods provide substantial evidence for the presence of significant copy number alterations in the mammalian brain. Nonetheless, the frequency of CNVs events could not be precisely determined due to the technical constraints of the study. To note, in a prior investigation employing scWGS to examine aneuploidy in neural progenitor cells of mouse embryos, the analysis of 36 cells revealed the absence of aneuploidy95. Although technical challenges may contribute to discrepancies in reported findings, it is important to recognize the potential influence of biological variations within the tested cell types. These differences could elucidate why certain studies identify aneuploidy in the brain while others fail to detect it. An emerging perspective underscores the divergence in genome maintenance programs between germline and progenitor cells compared to somatic differentiated cells. Notably, progenitor cells exhibit a more adept genome maintenance program, a concept supported by recent research96,97.
Although certain discrepancies remain, numerous studies converge to suggest that aneuploidy is a common occurrence in the development of the murine nervous system. This supports the hypothesis that aneuploidy may contribute at various levels to the genetic variability necessary for the functional and structural mosaicism that characterizes the brain. Furthermore, the percentage of aneuploid cells across all chromosomes in the cerebellum (around 20%) is lower than that of the cerebral cortex or olfactory bulb (around 33%), suggesting that there may be inherent differences in the rate of mosaic aneuploidy between brain regions during development. The fate of aneuploid cells could be directed toward programmed cell death, as demonstrated by the reduction of aneuploidy in the adult brain when compared to development, as it normally occurs in neuroproliferative zones. Alternatively, aneuploid cells could be committed to producing genetic and functional diversity, as demonstrated by the low level of aneuploid cells that remain in the adult brain.
A study98 has been conducted to measure the frequency of aneuploidy by FISH in three autosomes in the cerebral cortex and cerebellum of adult and developing brains of two mutant mouse models: Bub1bH/H mice, which have a faulty mitotic checkpoint99, and Ercc1−/Δ7 mice100, defective in nucleotide excision repair and inter-strand cross-link repair. During embryonic development (E13.5), it was found that Bub1bH/H mice, but not Ercc1−/Δ7 mice, had a significantly higher frequency of aneuploid nuclei relative to wild-type controls in the cerebral cortex, reaching a frequency as high as 40.3% for each chromosome tested. However, aneuploid cells in these mutant mice were likely eliminated early in development through apoptosis and/or immune-mediated clearance mechanisms, which would explain the low levels of aneuploidy during adulthood in the cerebral cortex of Bub1bH/H mice. It is noteworthy that a more recent sc-WGS analysis of 21 nuclei isolated from the same Bub1bH/H adult mouse model detected a frequency of 31% aneuploidy95. However, the specific brain region analyzed, the age of the mouse, and the cell types analyzed were not specified, making direct comparison of findings difficult.
Taken together, these findings suggest that aneuploidy may be tolerated in the brain of mice, even at extremely high levels (30–50%), albeit at the cost of fitness. This underscores a remarkable degree of plasticity in a highly complex organ such as the brain and raises important questions about the potential role of aneuploidy in brain function and disease.
5. Genomic instability in the adult, aging brain
Although limited studies have been conducted on the developing human brain, several studies have investigated aneuploidy in the adult human brain. These studies have generally shown significantly lower levels of aneuploidy in the adult human brain compared to the developing brain, which is consistent with observations in the adult mouse brain. A general observation from these studies is that across all the specific loci and chromosomes examined, the frequency of aneuploidy per chromosome in the healthy human brain has shown considerable variability.
Rehen et al.90 and Thomas and Fenech101 conducted separate studies analyzing hippocampal cells from older patients (>80 y.o.). Rehen's study assessed aneuploidy for HSA 21 alone, while Thomas and Fenech's study evaluated aneuploidy for both HSA 21 and HSA 17. The results showed that aneuploidy for HSA 21 was 5.2%, while aneuploidy for HSA 21 and HSA 17 was 29.8% in both studies combined. Additionally, Thomas and colleagues found that hippocampal cells from older patients had an aneuploidy frequency of 18% for HSA 17 and 11.8% for HSA 21, indicating a wide range of aneuploidy between two chromosomes or an elevated biological variability between individuals. Furthermore, the two studies on HSA 21 revealed a significant discrepancy in the frequency of aneuploidy, with Rehen's study reporting 5.2%, a frequency much lower than the 11.8% reported by Thomas and collaborators.
Iourov and colleagues102 analyzed the cerebral cortex of seven healthy individuals with an age range of 24.6 to 12.9 years using a cocktail of 5 probes (HSA 13, 18, 21, X, Y). The study demonstrated that the average level of aneuploidy was comparable for each chromosome tested, with a frequency of approximately 0.5% for each chromosome, except for the Y chromosome, which showed a lower frequency of 0.1%. The study conducted by the Westra group93 analyzed patients of a comparable age range and observed aneuploidy for HSA 6 (1.2% for NeuN+ and NeuN- combined) and HSA 21 (1.8% for NeuN+ and NeuN- combined), resulting in an overall aneuploidy frequency of about 3% for both autosomes combined. However, this frequency differs significantly from the results reported by the Iourov group, indicating that there is considerable variation in the frequency of aneuploidy between different chromosomes and between different studies.
Standard molecular cytogenetic techniques are laborious and time-consuming. To address this issue, studies have been conducted to increase the throughput of aneuploidy detection using cytometry-based methods103,104 where up to 100,00 nuclei can be analyzed. These results suggest that about 10% of neurons in the healthy human brain are hyperdiploid, but they lack genomic resolution.
Recent studies utilizing sc-WGS technologies have questioned the results from studies based on cytogenetics. Knouse and colleagues95 isolated neuronal progenitor cells and adult neurons from mice and analyzed them using NGS and CNV calling. Their results revealed minimal levels of aneuploidy, at approximately 1%, in both the embryonic and adult brain. Subsequent studies conducted by the same research team suggest low level of aneuploidy in the adult brain (approximately 0.8% for entire-chromosome copy number variations105). This observation is supported by studies involving the AD brain, wherein a slightly elevated occurrence of CNV events was detected in AD cells in comparison to cells from control subjects (4.1% vs. 1.4%, or 0.9% vs. 0.7%, utilizing distinct filtering methods)106. Iťs noteworthy, however, that these differences did not reach statistical significance.
It should be noted that the number of cells analyzed in many scWGS experiments is limited, with as few as nine cells being examined. Van den Bos et al.107 conducted a sc-WGS study to investigate somatic aneuploidy in approximately 1500 brain cells, including a subset of cells from healthy brain tissue. Consistent with other sc-WGS studies, the authors found that only approximately 0.7% (4 out of 589) of cells were determined to be aneuploid. A recent report by McConnell et al., who analyzed over 2000 neurons from a single individual88,108 reported a percentage of aneuploidy in the adult neurons to be less than 3%.
McConnell and group analyzed postmortem frontal cortex neurons for CNAs using single cell approaches and noted that somatic CNAs are common feature of neuronal genomes averaging 13 to 24% in neurons108. These results are supported by additional studies as well as from others88,109,110.
Through these studies utilizing sc-WGS technology, it has emerged that somatic sub chromosomal CNV may be prevalent within the brain. McConnell and colleagues have reported that approximately 41% of neurons analyzed contain at least one large-scale CNA108, while Cai and colleagues110 have reported that approximately 68% of neurons analyzed contain at least one large-scale CNA. The size range of these CNAs has not been consistently defined across these studies, but efforts and new analytical tools are being devoted to overcoming this limitation.
With the cost of sequencing continuing to decline and the availability of more throughput, it is expected that in the near future sc-WGS may become applicable to the analysis of large numbers of cells in multiple individuals, allowing for a better understanding of the extent and consequences of somatic aneuploidy in the human brain.
Future prospects
The form of genomic instability primarily discussed in this review pertains to aneuploidy and large copy number alterations. Genomic instability encompasses a multitude of aberrations that have been observed in the healthy brain, even during its developmental stages. However, elucidating the underlying etiology and resolving the reported inconsistencies in frequencies of aneuploidy and large copy number alterations pose a formidable challenge. The application of FISH techniques tends to overestimate the levels of these genomic aberrations, whereas sc-WGS may underestimate them.
Nonetheless, despite the ongoing debate surrounding the frequency of such genomic changes, substantial evidence indicates that aneuploidy and large copy number alterations significantly impair cellular fitness in healthy cells. This suggests their detrimental effects on brain cell function, which may potentially manifest through cell non-autonomous effects, even when their occurrence across the tissue is infrequent.
It is important to recognize and consider various biological variables that extend beyond technical challenges when studying aneuploidy in tissues. These variables include brain region and cell type, age, disease state, environmental factors, underlying genetic causes, and inter-individual variability. These factors are more closely linked to true biological or physiological functions, and can be challenging to control experimentally, especially in the analysis of human brain tissue.
Most of the research investigating genomic instability in the human brain has primarily focused on post-mitotic neurons. However, non-neuronal cells, which exhibit mitotic activity, retain the capacity to accumulate mutations and transmit them to their progeny.
Given the extraordinary complexity of the brain, much remains to be elucidated regarding genomic instability within this context. Several pivotal questions persist unanswered. For instance, is there a finite threshold for the number of genomic alterations that neurons and non-neuronal cells can tolerate before compromising their health? Furthermore, do these thresholds remain consistent across different cell types or brain regions? How do diverse forms of genomic instability, including aneuploidy, large copy number alterations, copy number variations (CNVs), single nucleotide variants (SNVs), insertions and deletions (indels), and retrotranspositions, impact neuronal and non-neuronal cells? Do they exhibit similar patterns of genomic instability? What mechanisms underlie these genetic changes?
Additionally, it is crucial to investigate whether aneuploidy and other forms of genomic instability increase in the human brain with age, as observed in mice, and explore their association with age-related diseases. Can one type of genomic alteration confer benefits in certain cell types while proving detrimental in others (e.g., SNVs promoting genomic heterogeneity within the neuronal network but causing cell death in non-neuronal cells)? Addressing these critical questions will advance our understanding of genomic instability in the brain and its implications for neurological function and disease.
Table 2:
Summary of aneuploidy and CIN results in brain of human subjects.
| AUTHOR | YEAR | SPECIES | TECHNIQUE | PROBES | CASES (N) | AGE | TISSUE | CELL TYPE | NUMBER OF NUCLEI | DETECTION METRIC | CHROMOSOME | CHR AFFECTED % | AVERAGE CHR % | % PER TISSUE | NOTES |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YANG 15 | 2001 | Hu | FISH | Chr 11/18/21 | Control 4 AD 7 | 69–87 | Hippocampus | Bulk | 400 | Polyploidy | n/a | none | 0 % Polyploidy | 0: polyploid | |
| YUROV 16 | 2001 | Hu | FISH | Chr 1/7/ 8/13/16/18/21/22/X/Y | Control 2 Schizophrenia 6 | 28 and 76 | Prefrontal cortex | Neurons | 1800 | Trisomy | n/a | none | 0 % Trisomy | 0: trisomy | |
| REHEN 17 | 2005 | Hu | FISH-dual | Chr 21 | 6 | 2–96 | Hippocampus, frontal, occipital cortex | NeuN and NeuN-neg | 500–1500 | Aneuploidy and T21 | 21 | 3.2–5.2% aneuploidy, 2.4–3.8% tetrasomy | 4% Aneuploidy | unk | |
| PACK 18 | 2005 | Hu | FISH and LOH | Chr 1/3/6/7/8/9/11 | 8 | 40–75 | Hippocampus, cerebral cortex, cerebellum | Bulk | unk | Aneuploidy | unk | unk | unk | 38–47 | |
| YUROV 19 | 2007 | Hu | Q-FISH | Chr 1/9/15/16/17/18/X/Y | 12 | 8–11 gest wks | Telencephalic region | Bulk | 5000 | Aneuploidy | 1, 9, 15, 16, 17, 18, X, Y | 1.32, 0.89, 1.17, 1.31, 0.93, 1.25, 2.33, 0.78 | 1.25–1.45% Aneuploidy | 30–35 | |
| WESTRA 9 | 2008 | Hu | FISH | Chr 6/21 | 6 | 45–76 | Cerebellum | NeuN and NeuN-neg | 3000 | Aneuploidy | 6, 21 | 1.2, 1.8 | 1% Aneuploidy | unk | |
| THOMAS FENECH 20 | 2008 | Hu | Q-FISH | Chr 17/21 | 9 | 60–98 | Hippocampus | NeuN and NeuN-neg | 1000 | Aneuploidy | 17, 21 | 18, 11.8 | unk | unk | |
| IOUROV 21 | 2009 | Hu | Q-FISH | Chr 1/7/11/13/14/17/18/21/ X/ Y | 7 | 8–47 | Cerebral cortex | NeuN and NeuN-neg | 7000 | Aneuploidy | 13, 18, 21, X, Y | 0.5, 0.6, 0.4, 0.4, 0.1 | 0.4–0.9 Aneuploidy | 10–22 | |
| MOSCH 22 | 2007 | Hu | SBC | n/a | 13 | 71.7 +/- 10.3 | Entorhinal cortex | Neurons | 80,000–120,000 | DCV | n/a | n/a | n/a | 10: hyperdiploid | |
| WESTRA 23 | 2010 | Hu | FACS | n/a | 24 | 35–95 | Frontal cortex and Cerebellum | NeuN | 10,000 | DCV | n/a | n/a | n/a | 4: Cortical neurons | |
| ARENDT 24 | 2010 | Hu | SBC | n/a | 14 | 71.7 +/- 10.3 | Entorhinal cortex | Neurons | 80,000–120,000 | DCV | n/a | n/a | n/a | 10: hyperdiploid | |
| FISCHER 25 | 2012 | Hu | SBC | n/a | 18 | 31–88 | Frontal, Temporal, Parietal, Entorhinal, Occipital cortex | Neurons | 500,000 | DCV | n/a | n/a | n/a | 11.5: hyperdiploid | |
| PAMPHLETT 26 | 2011 | Hu | SNP array | n/a | 28 | Adult | Frontal cortex | Bulk | Bulk DNA | CNV and aneuploidy | unk | unk | unk | 410 CNV observed | |
| GOLE 27 | 2013 | Hu | sc-WGS | n/a | 1 | unk | Brain | NeuN | 2 | CNV and aneuploidy | unk | unk | unk | CNV detected | |
| MCCONNELL 28 | 2013 | Hu | sc-WGS | n/a | 3 | 20–26 | Frontal cortex | NeuN | 36 (110 total) | CNV and aneuploidy | unk | unk | unk | 2.7: aneuploidy 13–41: large CNVs | |
| KNOUSE 11 | 2014 | Hu | sc-WGS | n/a | 4 | 48–70 | Frontal lobe | NeuN | 22 (89 total) | CNV and aneuploidy | 18, 22 | 1.12, 1.12 | unk | 2.2 | |
| CAI 29 | 2014 | Hu | sc-WGS | n/a | 3 | Adult | Cortex | NeuN | 32 (97 total) | CNV and aneuploidy | unk | unk | unk | 0: aneuploid 68: large-scale CNV (≥1) | |
| VAN DEN BOS 30 | 2016 | Hu | sc-WGS | n/a | 6 | 69–95 | Frontal cortex | NeuN and NeuN-neg | 98 (598 total) | CNV and aneuploidy | 4, 6, 13, 16, 18, 21, 22 | 0.2, 0.2, 0.2, 0.34, 0.2, 0.2, 0.2 | 0–0.34 % Aneuploidy | 0.7 | |
| CHRONISTER 31 | 2019 | Hu | sc-WGS | n/a | 5 | 0.36–95 | Prefrontal cortex | NeuN and NeuN-neg | 474 | CNV | unk | unk | unk | 11.9 | |
| SUN 32 | 2023 | Hu | 1 | 49 | Dorsolateral prefrontal cortex | NeuN | 2125 | unk | unk | unk | ≥ 1 226 CNV neurons deletion | Deletions ranged in size from 1Mb to entire chromosomes | |||
| KIM 33 | 2014 | Hu | WGS | n/a | Controls 2 Schizophrenia 3 | Controls (52–59) schizophrenia (32–60) | Prefrontal cortex | Bulk | unk | CNV | unk | unk | unk | unk | 106 somatic deletions |
Acknowledgments
The authors on this review are supported by funds of the National Institute of Health: R01#AG068908 to CM and JC; P01#P01AG017242 to JV, JC, ZZ and YS; U01#U01CA238726 to CM. CM is supported by CCGS #P30CA072720.
Footnotes
Ethical Statement
This work constitutes a comprehensive review of published literature, and no studies involving human or vertebrate animal subjects were conducted. Moreover, no personal data is described or disclosed in this review article.
Conflicts of Interest (CoI)
The authors have no conflict to declare.
References
- 1.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, Cullen M, Epstein CG, Burdett L, Dean MC, Chatterjee N, Sampson J, Chung CC, Kovaks J, Gapstur SM, Stevens VL, Teras LT, Gaudet MM, Albanes D, Weinstein SJ, Virtamo J, Taylor PR, Freedman ND, Abnet CC, Goldstein AM, Hu N, Yu K, Yuan JM, Liao L, Ding T, Qiao YL, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Aldrich MC, Amos C, Blot WJ, Bock CH, Gillanders EM, Harris CC, Haiman CA, Henderson BE, Kolonel LN, Le Marchand L, McNeill LH, Rybicki BA, Schwartz AG, Signorello LB, Spitz MR, Wiencke JK, Wrensch M, Wu X, Zanetti KA, Ziegler RG, Figueroa JD, Garcia-Closas M, Malats N, Marenne G, Prokunina-Olsson L, Baris D, Schwenn M, Johnson A, Landi MT, Goldin L, Consonni D, Bertazzi PA, Rotunno M, Rajaraman P, Andersson U, Beane Freeman LE, Berg CD, Buring JE, Butler MA, Carreon T, Feychting M, Ahlbom A, Gaziano JM, Giles GG, Hallmans G, Hankinson SE, Hartge P, Henriksson R, Inskip PD, Johansen C, Landgren A, McKean-Cowdin R, Michaud DS, Melin BS, Peters U, Ruder AM, Sesso HD, Severi G, Shu XO, Visvanathan K, White E, Wolk A, Zeleniuch-Jacquotte A, Zheng W, Silverman DT, Kogevinas M, Gonzalez JR, Villa O, Li D, Duell EJ, Risch HA, Olson SH, Kooperberg C, Wolpin BM, Jiao L, Hassan M, Wheeler W, Arslan AA, Bueno-de-Mesquita HB, Fuchs CS, Gallinger S, Gross MD, Holly EA, Klein AP, LaCroix A, Mandelson MT, Petersen G, Boutron-Ruault MC, Bracci PM, Canzian F, Chang K, Cotterchio M, Giovannucci EL, Goggins M, Hoffman Bolton JA, Jenab M, Khaw KT, Krogh V, Kurtz RC, McWilliams RR, Mendelsohn JB, Rabe KG, Riboli E, Tjonneland A, Tobias GS, Trichopoulos D, Elena JW, Yu H, Amundadottir L, Stolzenberg-Solomon RZ, Kraft P, Schumacher F, Stram D, Savage SA, Mirabello L, Andrulis IL, Wunder JS, Patino Garcia A, Sierrasesumaga L, Barkauskas DA, Gorlick RG, Purdue M, Chow WH, Moore LE, Schwartz KL, Davis FG, Hsing AW, Berndt SI, Black A, Wentzensen N, Brinton LA, Lissowska J, Peplonska B, McGlynn KA, Cook MB, Graubard BI, Kratz CP, Greene MH, Erickson RL, Hunter DJ, Thomas G, Hoover RN, Real FX, Fraumeni JF Jr., Caporaso NE, Tucker M, Rothman N, Perez-Jurado LA & Chanock SJ Detectable clonal mosaicism and its relationship to aging and cancer. Nature genetics 44, 651–658, doi: 10.1038/ng.2270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costantino I, Nicodemus J & Chun J Genomic Mosaicism Formed by Somatic Variation in the Aging and Diseased Brain. Genes (Basel) 12, doi: 10.3390/genes12071071 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breuss MW, Yang X, Schlachetzki JCM, Antaki D, Lana AJ, Xu X, Chung C, Chai G, Stanley V, Song Q, Newmeyer TF, Nguyen A, O'Brien S, Hoeksema MA, Cao B, Nott A, McEvoy-Venneri J, Pasillas MP, Barton ST, Copeland BR, Nahas S, Van Der Kraan L, Ding Y, Network NBSM, Glass CK & Gleeson JG Somatic mosaicism reveals clonal distributions of neocortical development. Nature 604, 689–696, doi: 10.1038/s41586-022-04602-7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andriani GA, Vijg J & Montagna C Mechanisms and consequences of aneuploidy and chromosome instability in the aging brain. Mech Ageing Dev, doi: 10.1016/j.mad.2016.03.007 (2016). [DOI] [PMC free article] [PubMed]
- 5.Schumacher B, Pothof J, Vijg J & Hoeijmakers JHJ The central role of DNA damage in the ageing process. Nature 592, 695–703, doi: 10.1038/s41586-021-03307-7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tubbs A & Nussenzweig A Endogenous DNA Damage as a Source of Genomic Instability in Cancer. Cell 168, 644–656, doi: 10.1016/j.cell.2017.01.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holland AJ & Cleveland DW Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nature reviews. Molecular cell biology 10, 478–487, doi: 10.1038/nrm2718 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zasadil LM, Britigan EM & Weaver BA 2n or not 2n: Aneuploidy, polyploidy and chromosomal instability in primary and tumor cells. Seminars in cell & developmental biology 24, 370–379, doi: 10.1016/j.semcdb.2013.02.001 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Rawi DH & Bakhoum SF Chromosomal instability as a source of genomic plasticity. Curr Opin Genet Dev 74, 101913, doi: 10.1016/j.gde.2022.101913 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nowell PC The clonal evolution of tumor cell populations. Science 194, 23–28, doi: 10.1126/science.959840 (1976). [DOI] [PubMed] [Google Scholar]
- 11.Lengauer C, Kinzler KW & Vogelstein B Genetic instability in colorectal cancers. Nature 386, 623–627, doi: 10.1038/386623a0 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Negrini S, Gorgoulis VG & Halazonetis TD Genomic instability--an evolving hallmark of cancer. Nature reviews. Molecular cell biology 11, 220–228, doi: 10.1038/nrm2858 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Niedernhofer LJ, Gurkar AU, Wang Y, Vijg J, Hoeijmakers JHJ & Robbins PD Nuclear Genomic Instability and Aging. Annual review of biochemistry 87, 295–322, doi: 10.1146/annurev-biochem-062917-012239 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Vijg J, Dong X, Milholland B & Zhang L Genome instability: a conserved mechanism of ageing? Essays Biochem 61, 305–315, doi: 10.1042/EBC20160082 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeijmakers JH DNA damage, aging, and cancer. The New England journal of medicine 361, 1475–1485, doi: 10.1056/NEJMra0804615 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Busuttil RA, Dolle M, Campisi J & Vijga J Genomic instability, aging, and cellular senescence. Annals of the New York Academy of Sciences 1019, 245–255, doi: 10.1196/annals.1297.041 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Rodier F, Munoz DP, Teachenor R, Chu V, Le O, Bhaumik D, Coppe JP, Campeau E, Beausejour CM, Kim SH, Davalos AR & Campisi J DNA-SCARS: distinct nuclear structures that sustain damage-induced senescence growth arrest and inflammatory cytokine secretion. Journal of cell science 124, 68–81, doi: 10.1242/jcs.071340 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez-Otin C, Blasco MA, Partridge L, Serrano M & Kroemer G The hallmarks of aging. Cell 153, 1194–1217, doi: 10.1016/j.cell.2013.05.039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montagna J. V. a. C. Genome instability and aging: Cause or effect? Translational Medicine of Aging 1, 5–11 (2017). [Google Scholar]
- 20.Iourov IY, Yurov YB, Vorsanova SG & Kutsev SI Chromosome Instability, Aging and Brain Diseases. Cells 10, doi: 10.3390/cells10051256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lodato MA & Walsh CA Genome aging: somatic mutation in the brain links age-related decline with disease and nominates pathogenic mechanisms. Human molecular genetics 28, R197–R206, doi: 10.1093/hmg/ddz191 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Gent DC, Hoeijmakers JH & Kanaar R Chromosomal stability and the DNA double-stranded break connection. Nature reviews. Genetics 2, 196–206, doi: 10.1038/35056049 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Campisi J & d'Adda di Fagagna F Cellular senescence: when bad things happen to good cells. Nature reviews. Molecular cell biology 8, 729–740, doi: 10.1038/nrm2233 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Vijg J From DNA damage to mutations: All roads lead to aging. Ageing research reviews 68, 101316, doi: 10.1016/j.arr.2021.101316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L & Vijg J Somatic Mutagenesis in Mammals and Its Implications for Human Disease and Aging. Annu Rev Genet 52, 397–419, doi: 10.1146/annurev-genet-120417-031501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marteijn JA, Lans H, Vermeulen W & Hoeijmakers JH Understanding nucleotide excision repair and its roles in cancer and ageing. Nature reviews. Molecular cell biology 15, 465–481, doi: 10.1038/nrm3822 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Laconi E, Marongiu F & DeGregori J Cancer as a disease of old age: changing mutational and microenvironmental landscapes. British journal of cancer 122, 943–952, doi: 10.1038/s41416-019-0721-1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lengauer C, Kinzler KW & Vogelstein B Genetic instabilities in human cancers. Nature 396, 643–649 (1998). [DOI] [PubMed] [Google Scholar]
- 29.Bakhoum SF & Landau DA Chromosomal Instability as a Driver of Tumor Heterogeneity and Evolution. Cold Spring Harbor perspectives in medicine 7, doi: 10.1101/cshperspect.a029611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DC, de Jong TV, Halsema N, Kazemier HG, Hoekstra-Wakker K, Bradley A, de Bont ES, van den Berg A, Guryev V, Lansdorp PM, Colome-Tatche M & Foijer F Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome biology 17, 115, doi: 10.1186/s13059-016-0971-7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vendramin R, Litchfield K & Swanton C Cancer evolution: Darwin and beyond. The EMBO journal 40, e108389, doi: 10.15252/embj.2021108389 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGranahan N & Swanton C Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell 168, 613–628, doi: 10.1016/j.cell.2017.01.018 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Ben-David U, Beroukhim R & Golub TR Genomic evolution of cancer models: perils and opportunities. Nature reviews. Cancer 19, 97–109, doi: 10.1038/s41568-018-0095-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, Hardcastle T, Lee A, Roy R, East P, Kschischo M, Endesfelder D, Wylie P, Kim SN, Chen JG, Howell M, Ried T, Habermann JK, Auer G, Brenton JD, Szallasi Z & Downward J Chromosomal instability determines taxane response. Proceedings of the National Academy of Sciences of the United States of America 106, 8671–8676, doi: 10.1073/pnas.0811835106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaki BI, Suriawinata AA, Eastman AR, Garner KM & Bakhoum SF Chromosomal instability portends superior response of rectal adenocarcinoma to chemoradiation therapy. Cancer 120, 1733–1742, doi: 10.1002/cncr.28656 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Bakhoum SF & Compton DA Chromosomal instability and cancer: a complex relationship with therapeutic potential. The Journal of clinical investigation 122, 1138–1143, doi: 10.1172/JCI59954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakhoum SF & Cantley LC The Multifaceted Role of Chromosomal Instability in Cancer and Its Microenvironment. Cell 174, 1347–1360, doi: 10.1016/j.cell.2018.08.027 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang S, Williams JF, Kneissig M, Lioudyno M, Rivera I, Helguera P, Busciglio J, Storchova Z, King MC & Torres EM Suppressing Aneuploidy-Associated Phenotypes Improves the Fitness of Trisomy 21 Cells. Cell reports 29, 2473–2488 e2475, doi: 10.1016/j.celrep.2019.10.059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Cheng L, Jia Y, Liu G, Li C, Song S, Bradley A & Huang Y Aneuploid embryonic stem cells exhibit impaired differentiation and increased neoplastic potential. The EMBO journal 35, 2285–2300, doi: 10.15252/embj.201593103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang YC & Amon A Gene copy-number alterations: a cost-benefit analysis. Cell 152, 394–405, doi: 10.1016/j.cell.2012.11.043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krivega M, Stiefel CM & Storchova Z Consequences of chromosome gain: A new view on trisomy syndromes. American journal of human genetics 109, 2126–2140, doi: 10.1016/j.ajhg.2022.10.014 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M & Storchova Z Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Molecular systems biology 8, 608, doi: 10.1038/msb.2012.40 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Borel C, Li L, Muller T, Williams EG, Germain PL, Buljan M, Sajic T, Boersema PJ, Shao W, Faini M, Testa G, Beyer A, Antonarakis SE & Aebersold R Systematic proteome and proteostasis profiling in human Trisomy 21 fibroblast cells. Nature communications 8, 1212, doi: 10.1038/s41467-017-01422-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schukken KM & Sheltzer JM Extensive protein dosage compensation in aneuploid human cancers. Genome research 32, 1254–1270, doi: 10.1101/gr.276378.121 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu J, Tsai HJ, Gordon MR & Li R Cellular Stress Associated with Aneuploidy. Developmental cell 44, 420–431, doi: 10.1016/j.devcel.2018.02.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng P, Zhao X, Katsnelson L, Camacho-Hernandez EM, Mermerian A, Mays JC, Lippman SM, Rosales-Alvarez RE, Moya R, Shwetar J, Grun D, Fenyo D & Davoli T Proteogenomic analysis of cancer aneuploidy and normal tissues reveals divergent modes of gene regulation across cellular pathways. Elife 11, doi: 10.7554/eLife.75227 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Compton DA Mechanisms of aneuploidy. Current opinion in cell biology 23, 109–113, doi: 10.1016/j.ceb.2010.08.007 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nicholson JM, Macedo JC, Mattingly AJ, Wangsa D, Camps J, Lima V, Gomes AM, Doria S, Ried T, Logarinho E & Cimini D Chromosome mis-segregation and cytokinesis failure in trisomic human cells. Elife 4, doi: 10.7554/eLife.05068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker NE & Montagna C Reducing the aneuploid cell burden - cell competition and the ribosome connection. Disease models & mechanisms 15, doi: 10.1242/dmm.049673 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Santaguida S, Richardson A, Iyer DR, M'Saad O, Zasadil L, Knouse KA, Wong YL, Rhind N, Desai A & Amon A Chromosome Mis-segregation Generates Cell-Cycle-Arrested Cells with Complex Karyotypes that Are Eliminated by the Immune System. Developmental cell 41, 638–651 e635, doi: 10.1016/j.devcel.2017.05.022 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang RW, Vigano S, Ben-David U, Amon A & Santaguida S Aneuploid senescent cells activate NF-kappaB to promote their immune clearance by NK cells. EMBO reports 22, e52032, doi: 10.15252/embr.202052032 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D & Pellman D DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58, doi: 10.1038/nature10802 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Andriani GA, Almeida VP, Faggioli F, Mauro M, Tsai WL, Santambrogio L, Maslov A, Gadina M, Campisi J, Vijg J & Montagna C Whole Chromosome Instability induces senescence and promotes SASP. Sci Rep 6, 35218, doi: 10.1038/srep35218 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sheltzer JM & Amon A The aneuploidy paradox: costs and benefits of an incorrect karyotype. Trends in genetics : TIG 27, 446–453, doi: 10.1016/j.tig.2011.07.003 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Macedo JC, Vaz S, Bakker B, Ribeiro R, Bakker PL, Escandell JM, Ferreira MG, Medema R, Foijer F & Logarinho E FoxM1 repression during human aging leads to mitotic decline and aneuploidy-driven full senescence. Nature communications 9, 2834, doi: 10.1038/s41467-018-05258-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barroso-Vilares M & Logarinho E Chromosomal instability and pro-inflammatory response in aging. Mech Ageing Dev 182, 111118, doi: 10.1016/j.mad.2019.111118 (2019). [DOI] [PubMed] [Google Scholar]
- 57.He Q, Au B, Kulkarni M, Shen Y, Lim KJ, Maimaiti J, Wong CK, Luijten MNH, Chong HC, Lim EH, Rancati G, Sinha I, Fu Z, Wang X, Connolly JE & Crasta KC Chromosomal instability-induced senescence potentiates cell non-autonomous tumourigenic effects. Oncogenesis 7, 62, doi: 10.1038/s41389-018-0072-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shepherd CE, Yang Y & Halliday GM Region- and Cell-specific Aneuploidy in Brain Aging and Neurodegeneration. Neuroscience 374, 326–334, doi: 10.1016/j.neuroscience.2018.01.050 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Rosenkrantz JL & Carbone L Investigating somatic aneuploidy in the brain: why we need a new model. Chromosoma 126, 337–350, doi: 10.1007/s00412-016-0615-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faggioli F, Vijg J & Montagna C Chromosomal aneuploidy in the aging brain. Mech Ageing Dev 132, 429–436, doi: 10.1016/j.mad.2011.04.008 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maslov AY, Makhortov S, Sun S, Heid J, Dong X, Lee M & Vijg J Single-molecule, quantitative detection of low-abundance somatic mutations by high-throughput sequencing. Sci Adv 8, eabm3259, doi: 10.1126/sciadv.abm3259 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu L, Wang X, Mu Q, Tam SST, Loi DSC, Chan AKY, Poon WS, Ng HK, Chan DTM, Wang J & Wu AR scONE-seq: A single-cell multi-omics method enables simultaneous dissection of phenotype and genotype heterogeneity from frozen tumors. Sci Adv 9, eabp8901, doi: 10.1126/sciadv.abp8901 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vandereyken K, Sifrim A, Thienpont B & Voet T Methods and applications for single-cell and spatial multi-omics. Nature reviews. Genetics, 1–22, doi: 10.1038/s41576-023-00580-2 (2023). [DOI] [PMC free article] [PubMed]
- 64.Lepage CC, Morden CR, Palmer MCL, Nachtigal MW & McManus KJ Detecting Chromosome Instability in Cancer: Approaches to Resolve Cell-to-Cell Heterogeneity. Cancers (Basel) 11, doi: 10.3390/cancers11020226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y & Ried T Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996). [DOI] [PubMed] [Google Scholar]
- 66.Dorritie K, Montagna C, Difilippantonio MJ & Ried T Advanced molecular cytogenetics in human and mouse. Expert review of molecular diagnostics 4, 663–676 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Montagna C, Lyu MS, Hunter K, Lukes L, Lowther W, Reppert T, Hissong B, Weaver Z & Ried T The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer research 63, 2179–2187 (2003). [PubMed] [Google Scholar]
- 68.Geigl JB, Uhrig S & Speicher MR Multiplex-fluorescence in situ hybridization for chromosome karyotyping. Nat Protoc 1, 1172–1184, doi: 10.1038/nprot.2006.160 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Faggioli F, Vijg J & Montagna C Four-color FISH for the detection of low-level aneuploidy in interphase cells. Methods in molecular biology 1136, 291–305, doi: 10.1007/978-1-4939-0329-0_14 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Andriani GA, Maggi E, Pique D, Zimmerman SE, Lee M, Quispe-Tintaya W, Maslov A, Campisi J, Vijg J, Mar JC & Montagna C A direct comparison of interphase FISH versus low-coverage single cell sequencing to detect aneuploidy reveals respective strengths and weaknesses. Scientific reports 9, 10508, doi: 10.1038/s41598-019-46606-w (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oltmann J, Heselmeyer-Haddad K, Hernandez LS, Meyer R, Torres I, Hu Y, Doberstein N, Killian JK, Petersen D, Zhu YJ, Edelman DC, Meltzer PS, Schwartz R, Gertz EM, Schaffer AA, Auer G, Habermann JK & Ried T Aneuploidy, TP53 mutation, and amplification of MYC correlate with increased intratumor heterogeneity and poor prognosis of breast cancer patients. Genes, chromosomes & cancer 57, 165–175, doi: 10.1002/gcc.22515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bakker B, van den Bos H, Lansdorp PM & Foijer F How to count chromosomes in a cell: An overview of current and novel technologies. BioEssays : news and reviews in molecular, cellular and developmental biology 37, 570–577, doi: 10.1002/bies.201400218 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Nawy T Single-cell sequencing. Nature methods 11, 18, doi: 10.1038/nmeth.2771 (2014). [DOI] [PubMed] [Google Scholar]
- 74.Quispe-Tintaya W, Gorbacheva T, Lee M, Makhortov S, Popov VN, Vijg J & Maslov AY Quantitative detection of low-abundance somatic structural variants in normal cells by high-throughput sequencing. Nature methods 13, 584–586, doi: 10.1038/nmeth.3893 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pique DG, Andriani GA, Maggi E, Zimmerman SE, Greally JM, Montagna C & Mar JC Aneuvis: web-based exploration of numerical chromosomal variation in single cells. BMC Bioinformatics 20, 336, doi: 10.1186/s12859-019-2842-1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garvin T, Aboukhalil R, Kendall J, Baslan T, Atwal GS, Hicks J, Wigler M & Schatz MC Interactive analysis and assessment of single-cell copy-number variations. Nature methods 12, 1058–1060, doi: 10.1038/nmeth.3578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baslan T, Kendall J, Rodgers L, Cox H, Riggs M, Stepansky A, Troge J, Ravi K, Esposito D, Lakshmi B, Wigler M, Navin N & Hicks J Genome-wide copy number analysis of single cells. Nat Protoc 7, 1024–1041, doi: 10.1038/nprot.2012.039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, Liang B, Xu X, Zhou F, Kong L, Shen J, Xia Y, Xuan L, Mao Y, Xue Y, Liu C & Tan J The comparison of the performance of four whole genome amplification kits on ion proton platform in copy number variation detection. Biosci Rep 37, doi: 10.1042/BSR20170252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deleye L, Tilleman L, Vander Plaetsen AS, Cornelis S, Deforce D & Van Nieuwerburgh F Performance of four modern whole genome amplification methods for copy number variant detection in single cells. Sci Rep 7, 3422, doi: 10.1038/s41598-017-03711-y (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rousseau F, Langlois S, Johnson JA, Gekas J, Bujold E, Audibert F, Walker M, Giroux S, Caron A, Clement V, Blais J, MacLeod T, Moore R, Gauthier J, Jouan L, Laporte A, Diallo O, Parker J, Swanson L, Zhao Y, Labelle Y, Giguere Y, Forest JC, Little J, Karsan A & Rouleau G Prospective head-to-head comparison of accuracy of two sequencing platforms for screening for fetal aneuploidy by cell-free DNA: the PEGASUS study. European journal of human genetics : EJHG 27, 1701–1715, doi: 10.1038/s41431-019-0443-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valecha M & Posada D Somatic variant calling from single-cell DNA sequencing data. Comput Struct Biotechnol J 20, 2978–2985, doi: 10.1016/j.csbj.2022.06.013 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mosch B, Mittag A, Lenz D, Arendt T & Tarnok A Laser scanning cytometry in human brain slices. Cytometry A 69, 135–138, doi: 10.1002/cyto.a.20228 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Gray JW, Langlois RG, Carrano AV, Burkhart-Schulte K & Van Dilla MA High resolution chromosome analysis: one and two parameter flow cytometry. Chromosoma 73, 9–27, doi: 10.1007/BF00294840 (1979). [DOI] [Google Scholar]
- 84.Trask B, van den Engh G, Nussbaum R, Schwartz C & Gray J Quantification of the DNA content of structurally abnormal X chromosomes and X chromosome aneuploidy using high resolution bivariate flow karyotyping. Cytometry 11, 184–195, doi: 10.1002/cyto.990110121 (1990). [DOI] [PubMed] [Google Scholar]
- 85.Gribble SM, Ng BL, Prigmore E, Fitzgerald T & Carter NP Array painting: a protocol for the rapid analysis of aberrant chromosomes using DNA microarrays. Nat Protoc 4, 1722–1736, doi: 10.1038/nprot.2009.183 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ballif BC, Rorem EA, Sundin K, Lincicum M, Gaskin S, Coppinger J, Kashork CD, Shaffer LG & Bejjani BA Detection of low-level mosaicism by array CGH in routine diagnostic specimens. Am J Med Genet A 140, 2757–2767, doi: 10.1002/ajmg.a.31539 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Carey L, Scott F, Murphy K, Mansfield N, Barahona P, Leigh D, Robertson R & McLennan A Prenatal diagnosis of chromosomal mosaicism in over 1600 cases using array comparative genomic hybridization as a first line test. Prenat Diagn 34, 478–486, doi: 10.1002/pd.4332 (2014). [DOI] [PubMed] [Google Scholar]
- 88.Sun C, Kathuria K, Emery SB, Kim B, Burbulis IE, Shin JH, Brain Somatic Mosaicism N, Weinberger DR, Moran JV, Kidd JM, Mills RE & McConnell MJ Mapping the Complex Genetic Landscape of Human Neurons. bioRxiv, doi: 10.1101/2023.03.07.531594 (2023). [DOI]
- 89.Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH & Chun J Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences of the United States of America 98, 13361–13366, doi: 10.1073/pnas.231487398 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M & Chun J Constitutional aneuploidy in the normal human brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 2176–2180, doi: 10.1523/JNEUROSCI.4560-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaushal D, Contos JJ, Treuner K, Yang AH, Kingsbury MA, Rehen SK, McConnell MJ, Okabe M, Barlow C & Chun J Alteration of gene expression by chromosome loss in the postnatal mouse brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 5599–5606, doi:23/13/5599 [pii] (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang AH, Kaushal D, Rehen SK, Kriedt K, Kingsbury MA, McConnell MJ & Chun J Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 10454–10462 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Westra JW, Peterson SE, Yung YC, Mutoh T, Barral S & Chun J Aneuploid mosaicism in the developing and adult cerebellar cortex. J Comp Neurol 507, 1944–1951, doi: 10.1002/cne.21648 (2008). [DOI] [PubMed] [Google Scholar]
- 94.Ning L, Li Z, Wang G, Hu W, Hou Q, Tong Y, Zhang M, Chen Y, Qin L, Chen X, Man HY, Liu P & He J Quantitative assessment of single-cell whole genome amplification methods for detecting copy number variation using hippocampal neurons. Sci Rep 5, 11415, doi: 10.1038/srep11415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Knouse KA, Wu J, Whittaker CA & Amon A Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proceedings of the National Academy of Sciences of the United States of America 111, 13409–13414, doi: 10.1073/pnas.1415287111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vermezovic J, Stergiou L, Hengartner MO & d'Adda di Fagagna F Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell death and differentiation 19, 1847–1855, doi: 10.1038/cdd.2012.69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bujarrabal-Dueso A, Sendtner G, Meyer DH, Chatzinikolaou G, Stratigi K, Garinis GA & Schumacher B The DREAM complex functions as conserved master regulator of somatic DNA-repair capacities. Nature structural & molecular biology 30, 475–488, doi: 10.1038/s41594-023-00942-8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andriani GA, Faggioli F, Baker D, Dolle ME, Sellers RS, Hebert JM, Van Steeg H, Hoeijmakers J, Vijg J & Montagna C Whole chromosome aneuploidy in the brain of Bub1bH/H and Ercc1-/Delta7 mice. Human molecular genetics 25, 755–765, doi: 10.1093/hmg/ddv612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P & van Deursen JM BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nature genetics 36, 744–749, doi: 10.1038/ng1382 (2004). [DOI] [PubMed] [Google Scholar]
- 100.Niedernhofer LJ, Garinis GA, Raams A, Lalai AS, Robinson AR, Appeldoorn E, Odijk H, Oostendorp R, Ahmad A, van Leeuwen W, Theil AF, Vermeulen W, van der Horst GT, Meinecke P, Kleijer WJ, Vijg J, Jaspers NG & Hoeijmakers JH A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature 444, 1038–1043, doi: 10.1038/nature05456 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Thomas P & Fenech M Chromosome 17 and 21 aneuploidy in buccal cells is increased with ageing and in Alzheimer's disease. Mutagenesis 23, 57–65, doi:gem044 10.1093/mutage/gem044 (2008). [DOI] [PubMed] [Google Scholar]
- 102.Iourov IY, Vorsanova SG, Liehr T & Yurov YB Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol Dis 34, 212–220, doi: 10.1016/j.nbd.2009.01.003 (2009). [DOI] [PubMed] [Google Scholar]
- 103.Arendt T, Bruckner MK, Mosch B & Losche A Selective cell death of hyperploid neurons in Alzheimer's disease. The American journal of pathology 177, 15–20, doi: 10.2353/ajpath.2010.090955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A & Arendt T Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 6859–6867, doi: 10.1523/JNEUROSCI.0379-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Knouse KA, Wu J & Amon A Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome research 26, 376–384, doi: 10.1101/gr.198937.115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Turan ZG, Richter V, Bochmann J, Parvizi P, Yapar E, Isildak U, Waterholter SK, Leclere-Turbant S, Son CD, Duyckaerts C, Yet I, Arendt T, Somel M & Ueberham U Somatic copy number variant load in neurons of healthy controls and Alzheimer's disease patients. Acta Neuropathol Commun 10, 175, doi: 10.1186/s40478-022-01452-2 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van den Bos H, Spierings DC, Taudt AS, Bakker B, Porubsky D, Falconer E, Novoa C, Halsema N, Kazemier HG, Hoekstra-Wakker K, Guryev V, den Dunnen WF, Foijer F, Tatche MC, Boddeke HW & Lansdorp PM Single-cell whole genome sequencing reveals no evidence for common aneuploidy in normal and Alzheimer's disease neurons. Genome biology 17, 116, doi: 10.1186/s13059-016-0976-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM & Gage FH Mosaic copy number variation in human neurons. Science 342, 632–637, doi: 10.1126/science.1243472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chronister WD, Burbulis IE, Wierman MB, Wolpert MJ, Haakenson MF, Smith ACB, Kleinman JE, Hyde TM, Weinberger DR, Bekiranov S & McConnell MJ Neurons with Complex Karyotypes Are Rare in Aged Human Neocortex. Cell reports 26, 825–835 e827, doi: 10.1016/j.celrep.2018.12.107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A & Walsh CA Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell reports 10, 645, doi: 10.1016/j.celrep.2015.01.028 (2015). [DOI] [PubMed] [Google Scholar]
References (Table1 and 2).
- 1.Schröck E, du Manoir S, Veldman T, Schoell B, Wienberg J, Ferguson-Smith MA, Ning Y, Ledbetter DH, Bar-Am I, Soenksen D, Garini Y & Ried T Multicolor spectral karyotyping of human chromosomes. Science 273, 494–497 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Faggioli F, Vijg J & Montagna C Four-color FISH for the detection of low-level aneuploidy in interphase cells. Methods in molecular biology 1136, 291–305, doi: 10.1007/978-1-4939-0329-0_14 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Andriani GA, Maggi E, Pique D, Zimmerman SE, Lee M, Quispe-Tintaya W, Maslov A, Campisi J, Vijg J, Mar JC & Montagna C A direct comparison of interphase FISH versus low-coverage single cell sequencing to detect aneuploidy reveals respective strengths and weaknesses. Scientific reports 9, 10508, doi: 10.1038/s41598-019-46606-w (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garvin T, Aboukhalil R, Kendall J, Baslan T, Atwal GS, Hicks J, Wigler M & Schatz MC Interactive analysis and assessment of single-cell copy-number variations. Nature methods 12, 1058–1060, doi: 10.1038/nmeth.3578 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rehen SK, McConnell MJ, Kaushal D, Kingsbury MA, Yang AH & Chun J Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proceedings of the National Academy of Sciences of the United States of America 98, 13361–13366, doi: 10.1073/pnas.231487398 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaushal D, Contos JJ, Treuner K, Yang AH, Kingsbury MA, Rehen SK, McConnell MJ, Okabe M, Barlow C & Chun J Alteration of gene expression by chromosome loss in the postnatal mouse brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 5599–5606, doi:23/13/5599 [pii] (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang AH, Kaushal D, Rehen SK, Kriedt K, Kingsbury MA, McConnell MJ & Chun J Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. The Journal of neuroscience : the official journal of the Society for Neuroscience 23, 10454–10462 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D & Chun J Aneuploid neurons are functionally active and integrated into brain circuitry. Proceedings of the National Academy of Sciences of the United States of America 102, 6143–6147, doi: 10.1073/pnas.0408171102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Westra JW, Peterson SE, Yung YC, Mutoh T, Barral S & Chun J Aneuploid mosaicism in the developing and adult cerebellar cortex. J Comp Neurol 507, 1944–1951, doi: 10.1002/cne.21648 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Faggioli F, Wang T, Vijg J & Montagna C Chromosome-specific accumulation of aneuploidy in the aging mouse brain. Human molecular genetics 21, 5246–5253, doi: 10.1093/hmg/dds375 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knouse KA, Wu J, Whittaker CA & Amon A Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proceedings of the National Academy of Sciences of the United States of America 111, 13409–13414, doi: 10.1073/pnas.1415287111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andriani GA, Faggioli F, Baker D, Dolle ME, Sellers RS, Hebert JM, Van Steeg H, Hoeijmakers J, Vijg J & Montagna C Whole chromosome aneuploidy in the brain of Bub1bH/H and Ercc1-/Delta7 mice. Human molecular genetics 25, 755–765, doi: 10.1093/hmg/ddv612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson SE, Yang AH, Bushman DM, Westra JW, Yung YC, Barral S, Mutoh T, Rehen SK & Chun J Aneuploid cells are differentially susceptible to caspase-mediated death during embryonic cerebral cortical development. The Journal of neuroscience : the official journal of the Society for Neuroscience 32, 16213–16222, doi: 10.1523/JNEUROSCI.3706-12.2012 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ning L, Li Z, Wang G, Hu W, Hou Q, Tong Y, Zhang M, Chen Y, Qin L, Chen X, Man HY, Liu P & He J Quantitative assessment of single-cell whole genome amplification methods for detecting copy number variation using hippocampal neurons. Sci Rep 5, 11415, doi: 10.1038/srep11415 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Y, Geldmacher DS & Herrup K DNA replication precedes neuronal cell death in Alzheimer's disease. The Journal of neuroscience : the official journal of the Society for Neuroscience 21, 2661–2668 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yurov YB, Vostrikov VM, Vorsanova SG, Monakhov VV & Iourov IY Multicolor fluorescent in situ hybridization on post-mortem brain in schizophrenia as an approach for identification of low-level chromosomal aneuploidy in neuropsychiatric diseases. Brain Dev 23 Suppl 1, S186–190 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M & Chun J Constitutional aneuploidy in the normal human brain. The Journal of neuroscience : the official journal of the Society for Neuroscience 25, 2176–2180, doi: 10.1523/JNEUROSCI.4560-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pack SD, Weil RJ, Vortmeyer AO, Zeng W, Li J, Okamoto H, Furuta M, Pak E, Lubensky IA, Oldfield EH & Zhuang Z Individual adult human neurons display aneuploidy: detection by fluorescence in situ hybridization and single neuron PCR. Cell cycle 4, 1758–1760 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, Monakhov VV & Soloviev IV Aneuploidy and confined chromosomal mosaicism in the developing human brain. PloS one 2, e558, doi: 10.1371/journal.pone.0000558 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas P & Fenech M Chromosome 17 and 21 aneuploidy in buccal cells is increased with ageing and in Alzheimer's disease. Mutagenesis 23, 57–65, doi:gem044 [pii] 10.1093/mutage/gem044 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Iourov IY, Vorsanova SG, Liehr T & Yurov YB Aneuploidy in the normal, Alzheimer's disease and ataxia-telangiectasia brain: differential expression and pathological meaning. Neurobiol Dis 34, 212–220, doi: 10.1016/j.nbd.2009.01.003 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Mosch B, Morawski M, Mittag A, Lenz D, Tarnok A & Arendt T Aneuploidy and DNA replication in the normal human brain and Alzheimer's disease. The Journal of neuroscience : the official journal of the Society for Neuroscience 27, 6859–6867, doi: 10.1523/JNEUROSCI.0379-07.2007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra JW, Rivera RR, Bushman DM, Yung YC, Peterson SE, Barral S & Chun J Neuronal DNA content variation (DCV) with regional and individual differences in the human brain. J Comp Neurol 518, 3981–4000, doi: 10.1002/cne.22436 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arendt T, Bruckner MK, Mosch B & Losche A Selective cell death of hyperploid neurons in Alzheimer's disease. The American journal of pathology 177, 15–20, doi: 10.2353/ajpath.2010.090955 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer HG, Morawski M, Bruckner MK, Mittag A, Tarnok A & Arendt T Changes in neuronal DNA content variation in the human brain during aging. Aging cell 11, 628–633, doi: 10.1111/j.1474-9726.2012.00826.x (2012). [DOI] [PubMed] [Google Scholar]
- 26.Pamphlett R, Morahan JM, Luquin N & Yu B Looking for differences in copy number between blood and brain in sporadic amyotrophic lateral sclerosis. Muscle & nerve 44, 492–498, doi: 10.1002/mus.22095 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Gole J, Gore A, Richards A, Chiu YJ, Fung HL, Bushman D, Chiang HI, Chun J, Lo YH & Zhang K Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nature biotechnology 31, 1126–1132, doi: 10.1038/nbt.2720 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McConnell MJ, Lindberg MR, Brennand KJ, Piper JC, Voet T, Cowing-Zitron C, Shumilina S, Lasken RS, Vermeesch JR, Hall IM & Gage FH Mosaic copy number variation in human neurons. Science 342, 632–637, doi: 10.1126/science.1243472 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai X, Evrony GD, Lehmann HS, Elhosary PC, Mehta BK, Poduri A & Walsh CA Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell reports 8, 1280–1289, doi: 10.1016/j.celrep.2014.07.043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Bos H, Spierings DC, Taudt AS, Bakker B, Porubsky D, Falconer E, Novoa C, Halsema N, Kazemier HG, Hoekstra-Wakker K, Guryev V, den Dunnen WF, Foijer F, Tatche MC, Boddeke HW & Lansdorp PM Single-cell whole genome sequencing reveals no evidence for common aneuploidy in normal and Alzheimer's disease neurons. Genome biology 17, 116, doi: 10.1186/s13059-016-0976-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chronister WD, Burbulis IE, Wierman MB, Wolpert MJ, Haakenson MF, Smith ACB, Kleinman JE, Hyde TM, Weinberger DR, Bekiranov S & McConnell MJ Neurons with Complex Karyotypes Are Rare in Aged Human Neocortex. Cell reports 26, 825–835 e827, doi: 10.1016/j.celrep.2018.12.107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C, Kathuria K, Emery SB, Kim B, Burbulis IE, Shin JH, Brain Somatic Mosaicism N, Weinberger DR, Moran JV, Kidd JM, Mills RE & McConnell MJ Mapping the Complex Genetic Landscape of Human Neurons. bioRxiv, doi: 10.1101/2023.03.07.531594 (2023). [DOI]
- 33.Kim J, Shin JY, Kim JI, Seo JS, Webster MJ, Lee D & Kim S Somatic deletions implicated in functional diversity of brain cells of individuals with schizophrenia and unaffected controls. Sci Rep 4, 3807, doi: 10.1038/srep03807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]


