Abstract
In the human gastrointestinal tract, microorganisms are present in large numbers in the colon but are sparse in the proximal small intestine. In this study, we have shown that acid extracts of fresh human terminal ileal mucosal samples mediate antimicrobial activity. Following cation-exchange chromatography, one of the eluted fractions demonstrated antibacterial activity against bacteria normally resident in the human colonic lumen. This activity was further fractionated by reverse-phase high-performance liquid chromatography and identified as histone H1 and its fragments. We have also shown that in tissue sections, immunoreactive histone H1 is present in the cytoplasm of villus epithelial cells. In vitro culturing of detached (from the basement membrane) villus epithelial cells led to the release of antimicrobial histone H1 proteins, while the cells demonstrated ultrastructural features of programmed cell death. Our studies suggest that cytoplasmic histone H1 may provide protection against penetration by microorganisms into villus epithelial cells. Moreover, intestinal epithelial cells released into the lumen may mediate antimicrobial activity by releasing histone H1 proteins and their fragments.
Most of the exposure of the human body to bacteria occurs in the gastrointestinal tract. In the upper gut, this exposure occurs in association with ingestion of nutrients, whereas in the colon, there is a large resident population of microorganisms (estimated to be 1014 [31]). A variety of host responses are required to mediate protection against invasion by pathogens and to achieve relative sterility in the small intestine, the lumen of which is rich in nutrients. These host responses can be divided into innate and adaptive defense systems (17). The latter entails the development of specific responses embodied in the gastrointestinal immune system and mediated by secretory immunoglobulin A (IgA) antibodies (34). Specific secretory IgA-mediated protection takes time to develop (8); therefore, a preexisting or rapidly responsive antimicrobial defense is required. Surface epithelial cells are of critical importance in mediating innate protection against microbes in the lumen of the gastrointestinal tract. Recent studies with animals (3, 18–20, 29) and humans (13, 24, 25) suggested the existence of a novel form of preexisting innate host protection which is operative in the mucus layer and the lumen and which is mediated by peptides with broad-spectrum antibacterial activities. These peptides, of the defensin family, are expressed by Paneth cells, which are located at the base of small intestinal crypts (3, 13, 18–20, 24, 29). In addition to this preexisting innate protection, intestinal epithelial cells may be capable of mediating antimicrobial activity following injury.
In studies to investigate the expression of antimicrobial activities in extracts of human terminal ileal mucosa, we have isolated and characterized antimicrobial histone H1 proteins and their fragments. In immunohistochemical studies on tissue sections, histone H1 was found to be expressed in the cytoplasm of villus epithelial cells. In addition, detached ileal epithelial cells were shown to release antimicrobial histone H1 while undergoing apoptosis (programmed cell death). Our studies suggest that cytoplasmically expressed histone H1 may provide protection against penetration by microorganisms into villus epithelial cells. In addition, intestinal epithelial cells released into the lumen may be capable of releasing antimicrobial histone H1 protein while undergoing apoptosis.
MATERIALS AND METHODS
Mucosal tissue.
Fresh human terminal ileal mucosal samples were obtained from operation resection specimens (right hemicolectomies; samples obtained >5 cm from tumor; 38 patients) and brain-dead organ donors (approved by the Ethics Committee of Nottingham University Hospitals; 8 donors).
Isolation and purification of histone H1.
Terminal ileal mucosa was dissected from submucosa and extracted with 10% (vol/vol) acetic acid. The mucosal samples were homogenized, sonicated, and stirred overnight. After centrifugation (at 67,000 × g for 60 min at 4°C), the pellets were reextracted with 10% acetic acid. The supernatants from both extraction procedures were pooled and applied to a cation-exchange column (SP Sepharose Fast Flow; Pharmacia Biotech, Uppsala, Sweden), and positively charged molecules were eluted with a 0 to 1 M NaCl gradient. The eluted fractions were dialyzed with a 1-kDa-cutoff dialysis membrane (Sectra Por membrane; Pierce & Warriner, Chester, United Kingdom), lyophilized, and tested for antimicrobial activity. Fractions expressing antimicrobial activity were purified further by reverse-phase high-performance liquid chromatography (RP-HPLC) on an Aquapore C4 column (Brownlee column; Applied Biosystems Ltd., Foster City, Calif.) with a linear water-acetonitrile gradient that contained 0.1% trifluoroacetic acid. The purity of the polypeptides was assessed by RP-HPLC (C18 column) and acid urea-polyacrylamide gel electrophoresis (AU-PAGE).
AU-PAGE.
Extracted mucosal samples were analyzed by AU-PAGE as previously described (15, 21). In brief, acid urea (6.25 M)–15% polyacrylamide minigels were prepared and prerun with 5% acetic acid for 45 to 60 min at 150 V. Samples (in 3 M urea with 5% acetic acid) were electrophoresed with 5% acetic acid at 150 V until the methyl green dye front had migrated near the end of the gel. The gels were subsequently stained with Coomassie blue for 15 min.
For protein sequence analysis, the proteins and peptides were electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Applied Biosystems Ltd.) (33). In brief, samples were electrophoresed as described above. With a Trans Blot cell (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom), the gel and the PVDF membrane were sandwiched between filter papers in the transfer cassette. This was inserted into a tank containing 1% (vol/vol) acetic acid, with the PVDF membrane facing the cathode. A constant current of 70 mA was applied for 24 h.
Amino acid analysis.
The amino acid compositions of purified proteins and peptides were determined by use of an Applied Biosystems 420H automated amino acid analyzer. This method was also used to quantify the amount of histone in a given sample. A known volume was applied to the analyzer, and the height of the RP-HPLC peak for each amino acid was recorded. These values were compared to those of standards; from this comparison, the quantity of each amino acid in the unknown sample was determined. The total protein in the known volume was calculated from the amino acid data and thus revealed the concentration of the sample. Proteins were also quantified by the Bradford assay (1).
Protein sequencing.
All protein sequencing was carried out by use of an Applied Biosystems 473A automated sequencer with protocols and chemicals supplied by the manufacturer and based on Edman degradation chemistry (2). Large protein fragments were sequenced directly following their separation by AU-PAGE and subsequent transfer to PVDF membranes (see above). Visualization of transferred fragment bands was achieved by staining the membranes with amido black and destaining with 50% methanol in water.
RP-HPLC-purified proteins, which were assumed to be modified at the N terminus, were digested with endoprotease Lys-C (Promega UK Ltd., Southampton, United Kingdom). Proteins were digested overnight at 37°C in 200 mM ammonium bicarbonate. The generated peptide fragments were separated by RP-HPLC on an Aquapore C4 column (Applied Biosystems Ltd.) with an acetonitrile gradient of 3 to 40% over 35 min. Selected peptide peaks were collected and applied to Briobrene-coated glass fiber discs for subsequent automated sequencing. Generated sequence data were analyzed with Applied Biosystems 610A data analysis software, and protein sequences were identified from the SwissProt database by use of the FASTA algorithm (23).
Antimicrobial assays.
The antimicrobial activities of mucosal extracts and their purified fractions were studied by use of radial diffusion (15) and 96-well microtiter plate (established in our laboratory) antimicrobial assays. These assays were performed with Salmonella typhimurium CS015, a phoP mutant of the wild-type strain (5) (generous gift from S. Miller, Massachusetts General Hospital, Boston); Salmonella typhimurium ATCC 14028 (wild-type strain), Streptococcus faecium ATCC 19434, and Streptococcus faecalis ATCC 19433 (from the National Collection of Type Cultures); Escherichia coli ATCC 25922 (from the National Collection of Industrial and Marine Bacteria); Escherichia coli ATCC 29648 (from the American Type Culture Collection); and Staphylococcus aureus ATCC 9144 and a beta-hemolytic Streptococcus group G clinical isolate (both generous gifts from R. Edwards, Queens Medical Centre, Nottingham, United Kingdom).
Radial diffusion assays were performed as previously described (14). In brief, bacteria (in the mid-logarithmic phase) were added to warm (approximately 37°C) 1% (wt/vol) low-electroendosmosis-type agarose (Sigma)–0.02% (wt/vol) Tween 20 (Sigma)–0.03% (wt/vol) Trypticase soy broth (TSB; Becton Dickinson) in 10 mM sodium phosphate buffer (pH 7.4). This agarose mixture was poured onto petri dishes and allowed to set for 30 to 60 min at 4°C. Wells (4-mm diameter) were made in this underlay, and samples (5 to 20 μl in 0.1% [vol/vol] acetic acid [16 mM]) were applied to each well. Acetic acid (0.1%) alone was used as a control. After incubation for 2 h at 37°C, the lower layer of agarose was covered with a nutrient-rich top layer (1% [wt/vol] agarose–6% [wt/vol] TSB in distilled water). Bacteria in the underlay were allowed to grow by incubation for 18 h at 37°C. Antimicrobial activity in samples was demonstrated by the presence of a bacterium-free zone around the wells. Antimicrobial activity was expressed in square millimeters and was calculated with the formula [(y − x)/2]2, where y is the diameter of the zone of bacterial clearance (millimeters) and x is the diameter of the well (4 mm). For example, a clear zone whose total diameter was 10 mm (including the sample well) would represent 9 mm2 of activity, whereas one whose total diameter was 5 mm (including the sample well) would represent 0.25 mm2 of activity.
We also used a microtiter plate antimicrobial assay to allow bacterial growth and its inhibition to be assessed over 5 h. Bacteria were grown to the mid-logarithmic phase and diluted 1:2 with fresh broth, and 10 μl was applied to wells of a 96-well microtiter plate (polystyrene, flat bottom, sterile; Costar Corporation, Cambridge, Mass.) containing 50 to 80 μl of 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 7.4) and 10 to 40 μl of sample (in 0.1% acetic acid) to achieve a final volume in the well of 100 μl. Based on the fact that an optical density at 620 nm (OD620) of 0.2 was equal to 5 × 107 CFU/ml (15), a range of 6.25 × 107 to 8.75 × 107 CFU/ml was added to each assay well. After incubation of the plate at 37°C for 1 h, 100 μl of prewarmed (to 37°C) double-strength (6% [wt/vol]) TSB was applied to each well (final pH range, 6.8 to 6.9). The OD620 of the wells was determined soon after the application of TSB with a Labsystems iEMS Reader MF. The plate was subsequently incubated at 37°C for 5 h, and optical density readings were taken every hour. The same volume of 0.1% acetic acid as in the test sample was used as a control. The antimicrobial activity of test samples was expressed as the percent reduction in bacterial growth (OD620) after 5 h of incubation at 37°C compared to the bacterial growth (OD620) of the 0.1% acetic acid control with the following equation: antimicrobial activity = [(OD620 of control − OD620 of sample)/OD620 of control] × 100. For example, the antimicrobial activity for a sample with an OD620 of 0.1 and a control OD620 of 0.6 (both after 5 h of incubation at 37°C) would be 83.3%.
Immunohistochemical analysis.
Cryostat sections (6 μm thick) of human small intestine and cytospin preparations of EDTA-isolated terminal ileal epithelial cells (see below) were air dried, fixed in acetone for 10 min, and labelled with anti-human histone H1 monoclonal antibodies (Cambio, Cambridge, United Kingdom). Bound antibodies were detected with biotinylated anti-mouse immunoglobulin G followed by an avidin-biotinylated horseradish peroxidase complex (Vectastain Elite ABC kit; Vecta Laboratories, Burlingame, Calif.). Peroxidase activity was developed with 3′-diaminobenzidine tetrahydrochloride, but nuclear counterstaining was not performed. Control sections and cytospin preparations were incubated with phosphate-buffered saline (PBS) (pH 7.4) instead of the primary anti-human histone antibodies. All other steps were identical.
Dot blot analysis.
Samples (10 μl) of acetic acid extracts of supernatants of detached and cultured terminal ileal epithelial cells (see below) were applied to PVDF membranes by use of a microfiltration apparatus attached to a water-vac system. The PVDF membranes were subsequently incubated with anti-human histone H1 antibodies or control buffer (PBS [pH 7.4]). Bound antibodies were detected with the Vectastain Elite ABC kit as described above.
Isolation of epithelial cells.
Villus epithelial cells of fresh human terminal ileal mucosal samples were detached with EDTA as described previously (16). In brief, strips (approximately 5 by 2 mm) of mucosa were incubated with 1 mM EDTA in a shaking water bath at 37°C for 30 min. The detached epithelial cells were washed three times with PBS (pH 7) by centrifugation at 400 × g for 10 min each time. After the third wash, the cells were resuspended in PBS (pH 7) and incubated at 37°C in 5% CO2 for 3 or 18 h.
Cell culture supernatants were obtained by centrifugation (at 13,400 × g for 30 min at 4°C). These were subsequently extracted with 10% (vol/vol) acetic acid by overnight stirring, followed by centrifugation (at 13,400 × g for 30 min at 4°C). The extracts were lyophilized, resuspended in 0.1% acetic acid, and subsequently used in antimicrobial assays, AU-PAGE, and immunoblot studies (see above). Cultured epithelial cells were studied by transmission electron microscopy (TEM) (see below). Cytospin preparations of detached epithelial cells were also made (approximately 50,000 cells per slide), fixed in acetone, and stored at −20°C before immunohistochemical studies (see above).
Electron microscopy.
Cultured epithelial cells (see above) were obtained for TEM after each incubation period. These cells were fixed in 2.5% glutaraldehyde (in 0.1 M cacodylate buffer [pH 7.4]) for 24 h at 4°C and processed according to standard procedures (28). Sections (80 nm) were mounted on copper grids and stained with uranyl acetate and lead citrate prior to examination on a JEOL 1010 transmission electron microscope.
Immunoelectron microscopy.
Sections (15 μm) of normal human small intestine were incubated with anti-human histone H1 monoclonal antibodies. Labelled antibodies were detected with the Vectastain Elite ABC kit as described above. After development of peroxidase activity, sections were fixed in 2.5% glutaraldehyde (in 0.1 M cacodylate buffer [pH 7.4]), postfixed, dehydrated in ascending grades of alcohol, and infiltrated with Taab resin (28). A resin-filled capsule was inverted over the section at the region of interest and allowed to polymerize at 60°C for 18 h. After removal of the capsules, 0.5-μm sections were cut and stained with 1% toluene blue. Areas containing peroxidase-positive epithelial cells were selected, and ultrathin sections (which were not counterstained with toluidine blue or hematoxylin) were examined by TEM.
RESULTS
Unfractionated terminal ileal mucosal extracts.
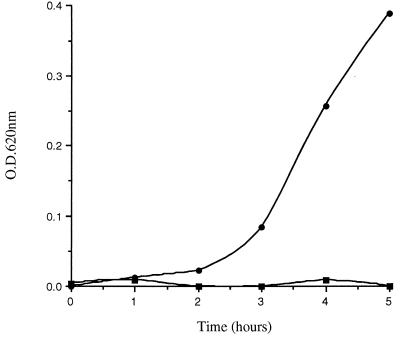
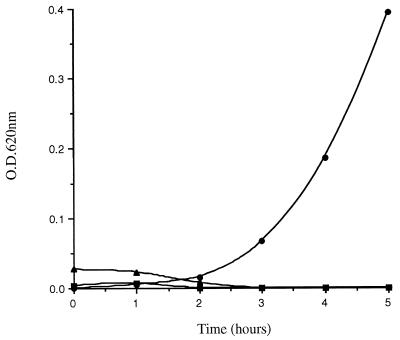
All acetic acid extracts (some of which were pooled) of terminal ileal mucosal samples obtained from a total of 38 resected intestines demonstrated potent activity against S. typhimurium CS015 (mean ± standard deviation [SD] reduction in bacterial OD620 at 5 h, 90.7% ± 6.0% [n = 29]; Fig. 1) and against two strains of E. coli, ATCC 25922 (reduction in bacterial OD620, 89.3% ± 6.1%) and ATCC 29648 (reduction in bacterial OD620, 66.1% ± 3.5%). Bacterial growth was always observed in the control sample (containing the same volume of 0.1% acetic acid), as shown in the representative experiment in Fig. 1.
FIG. 1.
Activity against S. typhimurium CS015 by acid extracts of human terminal ileal mucosa. The mucosal extract (in 0.1% acetic acid) (▪) and the control (0.1% acetic acid only) (•) were applied, together with bacteria, to 96-well microtiter plates, and bacterial growth was assessed hourly. The data represent one of 29 experiments performed.
Activity against S. typhimurium CS015 was also observed in the radial diffusion assay (17.0-mm2 zone of bacterial clearance).
Fractionation of mucosal extracts by cation-exchange chromatography.
Aqueous acetic acid extracts of terminal ileal mucosal samples were applied to a cation-exchange column incorporated into a fast protein liquid chromatography apparatus. Bound, positively charged proteins were eluted with a 0 to 1 M NaCl gradient. After extensive dialysis, four eluted fractions demonstrated activity against S. typhimurium CS015. One of the active fractions (designated fraction 15) was used for further studies as it demonstrated impressive antimicrobial activity and distinct bands on Coomassie blue-stained AU-PAGE. This fraction also demonstrated activity against E. coli ATCC 25922, E. coli ATCC 29648, and S. faecium ATCC 19434 (each tested once) and against S. faecalis ATCC 19433 and S. aureus ATCC 9144 (each tested twice) (Table 1). Although the fraction demonstrated activity against the phoP mutant of S. typhimurium (CS015), no activity was present against wild-type S. typhimurium ATCC 14028 or against a beta-hemolytic Streptococcus group G clinical isolate (each tested twice) (Table 1).
TABLE 1.
Antimicrobial activity of a semipurified acetic acid extract of human terminal ileal mucosaa
| Bacterium | % Antimicrobial activity (mean ± SD) |
|---|---|
| S. typhimurium CS015 | 94.9 ± 5.1 |
| S. typhimurium ATCC 14028 (wild type) | 8.6 ± 8.7 |
| E. coli ATCC 25922 | 93.7 ± 2.8 |
| E. coli ATCC 29648 | 97.1 ± 0.6 |
| S. faecalis ATCC 19433 | 78.5 ± 21.5 |
| S. faecium ATCC 19433 | 95.6 ± 1.6 |
| S. aureus ATCC 9144 | 92.5 ± 6.3 |
| Beta-hemolytic Streptococcus group G | 30.4 ± 0.7 |
The mucosal extract was applied to a cation-exchange column, and an eluted fraction (designated fraction 15) was tested for antimicrobial activity with the 96-well microtiter plate assay. Antimicrobial activity is expressed as percent reduction in bacterial growth compared to bacterial growth in the control (0.1% acetic acid only) after incubation at 37°C for 5 h.
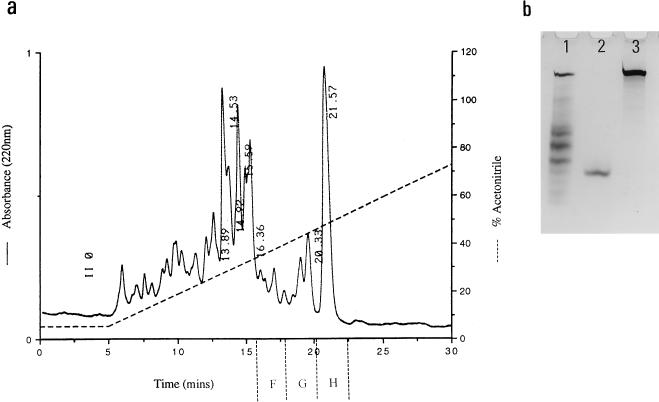
This active fraction (fraction 15) was applied to an Aquapore C4 RP-HPLC column, and of the fractions eluted, three demonstrated activity against S. typhimurium CS015 (Fig. 2a). Analysis of one of these (fraction H) on an Aquapore C18 RP-HPLC column and by AU-PAGE demonstrated the presence of a single entity (Fig. 2b).
FIG. 2.
(a) RP-HPLC fractionation of semipurified ileal mucosal extract. Antimicrobial fraction 15 eluted from the cation-exchange column (1 M NaCl) was applied to an Aquapore C4 column, and of the eluted fractions, F, G, and H demonstrated antimicrobial activity. (b) AU-PAGE of fraction 15 (lane 1), fraction H (lane 3), and synthetic magainin (lane 2). Of the many positively charged peptides and proteins present in fraction 15, one was purified by RP-HPLC (fraction H in panel a).
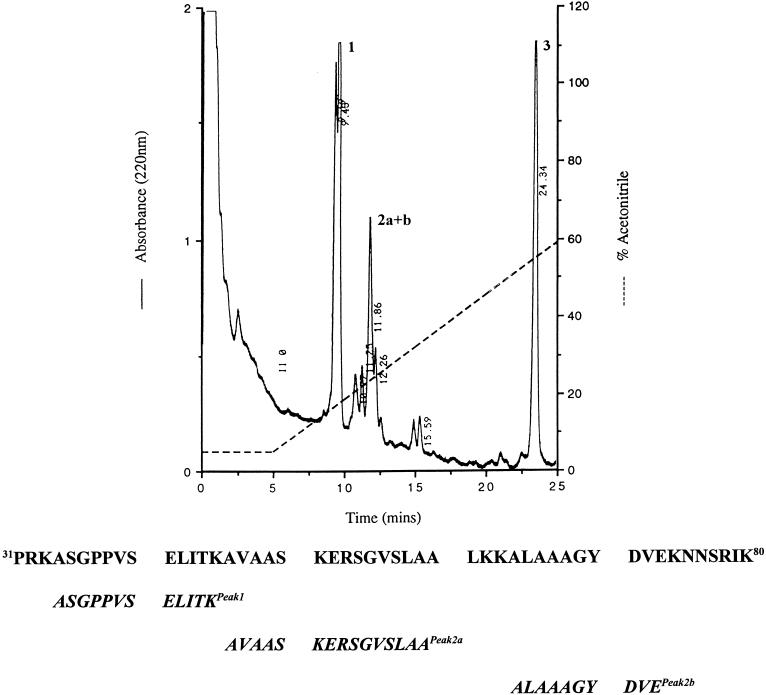
Amino acid analysis of the purified protein showed that it was rich in lysine and alanine (Table 2). N-terminal sequencing failed to provide data, suggesting that it was N terminally blocked. The purified protein was therefore digested with Lys-C, and three peptide fragments were subsequently purified by RP-HPLC. Amino acid sequence analysis of these fragments showed them to be 100% homologous to human histones H1b, H1c, and H1d (Fig. 3).
TABLE 2.
Amino acid composition of an antimicrobial protein purified from acid extract of human terminal ileal mucosaa
| Amino acid | % Composition by molecular weight |
|---|---|
| Aspartic acid | 3.1 |
| Glutamic acid | 4.2 |
| Serine | 8.2 |
| Glycine | 7.6 |
| Histidine | 0.1 |
| Arginine | 2.6 |
| Threonine | 6.3 |
| Alanine | 21.6 |
| Proline | 11.4 |
| Tyrosine | 0.6 |
| Valine | 4.7 |
| Methionine | 0.0 |
| Isoleucine | 1.0 |
| Leucine | 4.8 |
| Phenylalanine | 0.7 |
| Lysine | 22.7 |
Acid extract of ileal mucosa was fractionated by cation-exchange chromatography and RP-HPLC. The amino acid composition of the purified protein present in fraction H (Fig. 2) was determined.
FIG. 3.
C4 RP-HPLC separation of fraction H (Fig. 2) following digestion with Lys-C. Eluted peaks (1, 2, and 3) were collected and sequenced. Peak 1 (retention time, 9.45 min) gave a single unequivocal sequence. Peak 2 (retention time, 11.86 min) gave two signals for some cycles of sequence analysis, indicating a mixture of two peptides. However, these two signals were of sufficiently different intensities to allow the two sequences to be assigned (2a and 2b). Peak 3 gave no sequence, suggesting that it contained the undigested N terminally blocked protein. The amino acid sequence (amino acids 31 to 80) of human histone H1d is shown. Amino acid sequences of peptides 1, 2a, and 2b showed 100% homology to histone H1d. In addition, these peptides also showed 100% sequence homology to histones H1b and H1c.
Antimicrobial activity of purified histone H1 protein.
The purified histone H1 protein demonstrated potent activity against S. typhimurium CS015 (mean ± SD reduction in bacterial OD620 at 5 h, 93.7% ± 11.9%; tested 10 times; MIC, 3.47 to 6.95 μg/ml) but not against wild-type S. typhimurium ATCC 14028 (mean ± SD reduction in bacterial OD620 at 5 h, 10.4% ± 6.3%; tested 3 times).
Histone H1 fragments mediate antimicrobial activity.
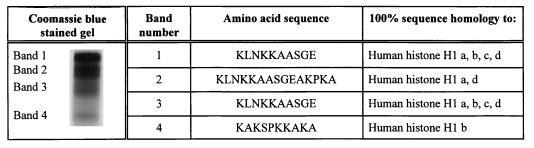
Antimicrobial activity from another human terminal ileal mucosal extract was determined by cation-exchange chromatography and RP-HPLC as described above. One fraction demonstrated potent activity against S. typhimurium CS015 (mean ± SD reduction in bacterial OD620 at 5 h, 90.3% ± 2.0%; tested three times). AU-PAGE showed that this fraction contained four distinct protein entities. Following transfer to a PVDF membrane, amino acid sequence analysis of these four proteins showed them to be 100% homologous to histone H1 proteins (Fig. 4).
FIG. 4.
AU-PAGE of an antimicrobial fraction of human ileal mucosal extract. This fraction was obtained by cation-exchange chromatography, followed by RP-HPLC. AU-PAGE analysis demonstrated the presence of four distinct protein bands (stained by Coomassie blue). Following transfer to a PVDF membrane, amino acid sequence analyses of these four proteins revealed 100% homology to histone H1 proteins.
Expression of histone H1 proteins in human terminal ileal mucosa.
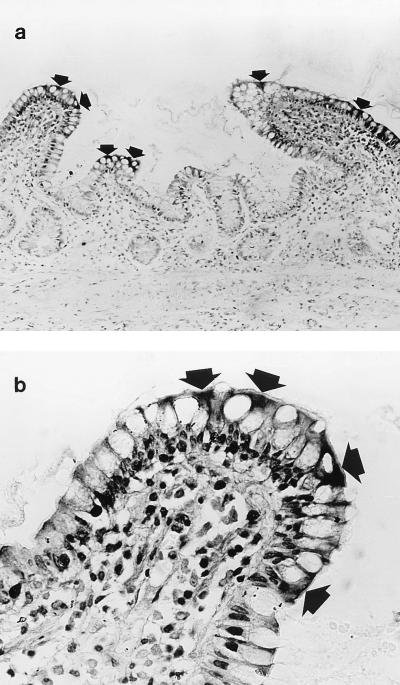
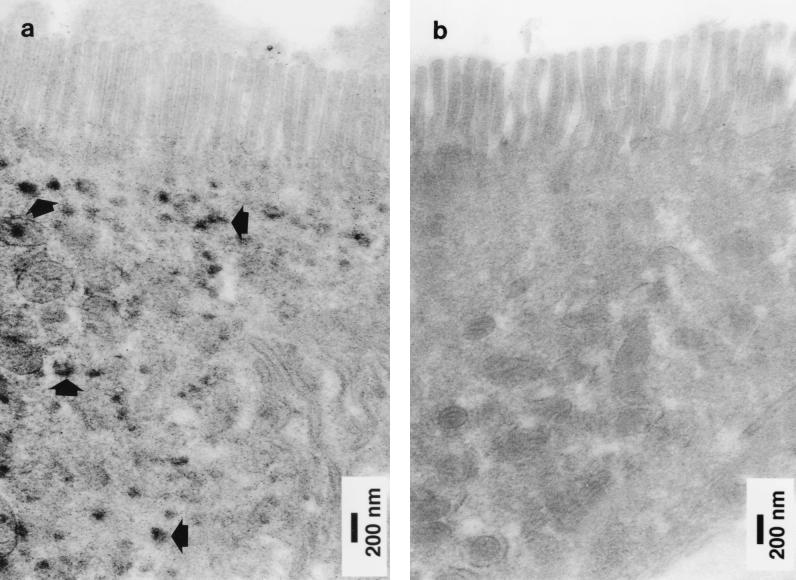
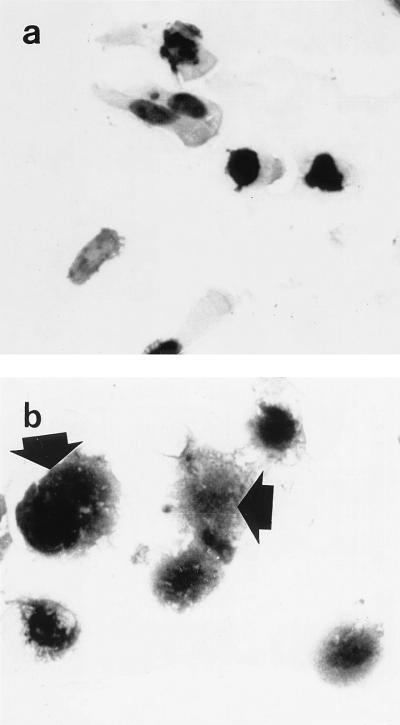
In order to investigate the expression of histone H1 proteins in human terminal ileal mucosa in vivo, immunohistochemical analysis was performed on tissue sections with a monoclonal antibody to histone H1. As expected, these studies (in which no nuclear counterstaining was performed) showed the presence of immunoreactive histone H1 in nuclei of epithelial cells and cells in the lamina propria. In addition, immunoreactivity was also seen in the cytoplasm of some villus epithelial cells. These cells were present at the villus tip and on the lateral aspect of some villi (Fig. 5). Studies by electron microscopy showed that immunoreactive histone H1 was present in the cytoplasm of the epithelial cells (Fig. 6). Immunohistochemical studies on cytospin preparations of detached villus epithelial cells also demonstrated the presence of immunoreactive histone H1 in the cytoplasm of some cells (Fig. 7).
FIG. 5.
Expression of histone H1 in human terminal ileal mucosa. Immunohistochemical analysis was performed with an anti-histone H1 monoclonal antibody, but a nuclear stain was not used. (a) Histone H1 immunoreactivity was seen in the cytoplasm of epithelial cells, at the tip and on the lateral aspect of villi (arrows), as well as in nuclei. (b) Villus (from the left margin of panel a) at a high magnification. Arrows indicate immunoreactivity. Approximate magnifications: a, ×224; b, ×389.
FIG. 6.
Immunoelectron microscopy of a section of normal human small intestinal mucosa labelled with an anti-human histone H1 monoclonal antibody by the peroxidase technique. Two epithelial cells near the villus tip are shown. (a) In one, immunoreactive histone H1 (arrows) was present in the cytoplasm below the microvilli. (b) The cytoplasm of an adjacent epithelial cell did not contain immunoreactive histone H1.
FIG. 7.
Expression of immunoreactive histone H1 in detached villus epithelial cells of human terminal ileal mucosa. Cytospin preparations of epithelial cells were made following detachment with EDTA (and subsequent washing with PBS). Photomicrographs of the same cytospin preparation were taken. Strong histone H1 immunoreactivity was seen in nuclei. Strong histone H1 immunoreactivity was seen in the cytoplasm (arrows) of some epithelial cells (b). Magnification, ×590.
Extracts of detached villus epithelial cells express antimicrobial activity.
Villus epithelial cells were detached by treatment with EDTA and extracted with acetic acid. These extracts demonstrated activity against S. typhimurium CS105 in the radial diffusion assay (5.9 ± 0.6 mm2).
Detached villus epithelial cells release antimicrobial histone H1 while undergoing apoptosis.
Following treatment with EDTA, detached villus epithelial cells were cultured at 37°C in 5% CO2 for 3 or 18 h. Acetic acid extracts of the supernatants of the cultured epithelial cells demonstrated activity against S. typhimurium CS015 (mean ± SD reduction in bacterial OD620 at 5 h, 98.3% ± 2.4% and 97% ± 4% for 3-h and 18-h supernatants, respectively) (Fig. 8). Immunoblotting studies demonstrated the presence of histone H1 in the supernatants (Fig. 9).
FIG. 8.
Acid extracts (in 0.1% acetic acid) of supernatants of detached and cultured (for 3 h [▪] or for 18 h [▴]) villus epithelial cells mediate antimicrobial activity. Microtiter plate antimicrobial assays were performed with S. typhimurium CS015, and 0.1% acetic acid only (•) was used as a control. The data represent one of three experiments performed.
FIG. 9.
Dot blot analysis demonstrating the presence of immunoreactive histone H1 in supernatants of detached and cultured (for 18 h) terminal ileal villus epithelial cells. Acid extracts were applied to PVDF membranes, which were subsequently incubated with an anti-human histone H1 monoclonal antibody (b) or control buffer (a). Strong histone H1 immunoreactivity was seen with the monoclonal antibody.
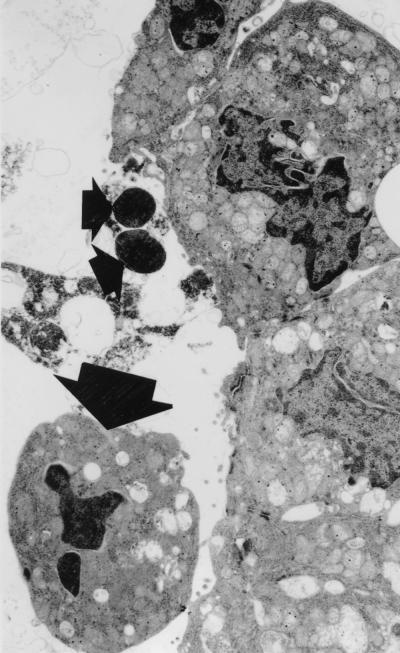
In TEM studies, the detached and cultured villus epithelial cells demonstrated ultrastructural features of cells undergoing apoptosis (Fig. 10).
FIG. 10.
Transmission electron micrograph of detached villus epithelial cells cultured for 3 h. Apoptotic bodies (small arrows) and an apoptotic epithelial cell (large arrow) are shown.
DISCUSSION
There is a very large and complex resident population of bacteria (approximately 1012 per g) in the human colonic lumen (31). In contrast, the lumen of the small intestine is relatively sterile. A marked change in the enteric flora occurs across the ileocecal valve, with up to a 106-fold decrease in the number of microorganisms per g (32). In the lumen of the more proximal small intestine (jejunum), microorganisms are either absent or present in very small numbers (32).
A number of factors are likely to be responsible for the maintenance of this state of relative sterility of the small intestinal lumen. The roles of gastric acid and peristaltic activity have long been recognized, but recent studies have begun to investigate protective defense mechanisms which are operative at the mucosal level and which can be divided into innate immunity and adaptive immunity (17). Innate protection is broad spectrum and provides either preexisting or rapidly responsive antimicrobial defense. Recent studies with animals have suggested that Paneth cells, which are normally restricted to the small intestine, are important in mediating innate antimicrobial defense. In addition to lysozyme (4), these cells have been shown to express antimicrobial phospholipase A2 (PLA2s) (10) and antimicrobial peptides of the defensin family (3, 13, 18–20, 24, 25, 29). Other components of innate antimicrobial defense likely are operative at the mucosal level.
In order to isolate and characterize antimicrobial peptides and proteins expressed in the human terminal ileal mucosa, we have investigated extracts of fresh mucosal samples. Acetic acid extracts of terminal ileal mucosa demonstrated potent antimicrobial activity, which appeared to be mediated by a number of antimicrobial peptides and proteins. We have isolated and purified one of these entities and identified it as histone H1. In addition, fragments of histone H1 were also shown to express antimicrobial activity. The activities were demonstrated against S. typhimurium CS015 and bacteria normally resident in the human colonic lumen. Wild-type S. typhimurium appears to be resistant to histone H1 proteins and their fragments. The relative resistance of wild-type S. typhimurium (compared to the phoP mutant strain) has also been demonstrated against murine PLA2s (10). Thus, wild-type S. typhimurium may be capable of invading the host by its ability to overcome different innate host defense mechanisms. We have also recently purified human PLA2s from our terminal ileal mucosal extracts (unpublished observations).
Since histone H1 proteins are normally expressed predominantly in eucaryotic nuclei, we performed further studies to investigate the likely functional significance of our findings. Studies on tissue sections by immunohistochemistry and electron microscopy showed that in addition to being present in nuclei, histone H1 proteins were also present in the cytoplasm of intestinal villus epithelial cells. These cells could therefore mediate host protection in vivo following penetration by microorganisms through the cell membrane. Detached villus epithelial cells also released immunoreactive histone H1 protein during culturing while undergoing apoptosis. Moreover, extracts of supernatants of these cultured epithelial cells demonstrated antimicrobial activity. Since we had previously demonstrated that purified histone H1 protein and its fragments express antimicrobial activity, it is likely that the antimicrobial activity in the supernatants of the cultured detached villus epithelial cells is mediated by the released histone H1 protein. It is possible that other antimicrobial molecules are also present in these supernatants.
In the normal small intestine, there is a rapid turnover of villus epithelial cells, with estimates of 2 × 1011 and 1.9 × 108 cells per day lost into the lumen in humans (27) and mice (26), respectively. Although the mechanism(s) of epithelial cell loss into the lumen is not fully understood, studies with rodents and humans have suggested an important role for apoptosis (7, 9, 30). Epithelial cells exfoliated into the lumen of the human small intestine also demonstrate characteristic features of cells undergoing apoptosis (30). From our studies, we postulate that antimicrobial histone H1 proteins and their fragments are released into the human intestinal lumen in vivo by exfoliated apoptotic epithelial cells.
We previously showed that following exposure to Clostridium difficile toxin A, intestinal epithelial cells detach from the basement membrane and undergo apoptosis (16). Such injured and detached cells are released into the lumen in vivo and may represent an important form of rapidly responsive innate protection against resident luminal microorganisms. There has been recent interest in the bactericidal potential of cells undergoing apoptosis in host defense (14, 36). Our studies suggest a novel mechanism by which intestinal epithelial cells undergoing apoptosis may mediate antimicrobial activity. Such activity would most likely be released into the lumen to provide innate protection against luminal microorganisms.
A potent antimicrobial peptide with amino acid sequence similarity to histone H2A was recently isolated from gastric tissue of the Asian toad Bufo bufo gargarizans (22). Thus, peptide fragments of histone proteins, which may be released by cells undergoing apoptosis and which may also arise following proteolytic digestion in the intestinal lumen, could mediate potent antimicrobial activity. Histone H2B fragments have also been demonstrated in extracts of human wound fluid (6). Histone and histone-like antimicrobial proteins have been extracted from a murine macrophage cell line and shown to be expressed in the cytoplasm of these cells (11). In addition, cytoplasmic expression of histone H1 in mouse liver cells has also been reported (35). Thus, although the antimicrobial activity of calf thymus histone has been known for many years (12), our study and other recent studies suggest potential in vivo sites of activity for this family of proteins, which were previously thought to be present only in nuclei.
In conclusion, we have isolated from extracts of human terminal ileal mucosa antimicrobial proteins which have been identified as histone H1 and its fragments. Immunohistochemical studies suggest that in vivo, histone H1 is expressed in the cytoplasm of villus epithelial cells. Our studies also suggest that intestinal epithelial cells detached into the lumen release antimicrobial histone H1 proteins and their fragments while the cells are undergoing apoptosis. Thus, histone proteins and their fragments derived from epithelial cells lost into the lumen may represent an important innate antimicrobial defense against luminal bacteria in the human intestine.
ACKNOWLEDGMENTS
This study was supported by grants from the Medical Research Council and The Wellcome Trust. The electron microscopy studies used equipment funded by grant 048326 from The Wellcome Trust.
We thank Trevor Gray for assistance in the electron microscopy studies.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Edman P. Method of determination of the amino acid sequence in peptides. Acta Chem Scand. 1950;4:283–293. [Google Scholar]
- 3.Eisenhauer P B, Harwig S S S L, Lehrer R I. Cryptdins: antimicrobial defensins of the murine small intestine. Infect Immun. 1992;60:3556–3565. doi: 10.1128/iai.60.9.3556-3565.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlandsen S L, Parsons J A, Taylor T D. Ultrastructural immunocytochemical localization of lysozyme in the Paneth cells of man. J Histochem Cytochem. 1974;22:401–413. doi: 10.1177/22.6.401. [DOI] [PubMed] [Google Scholar]
- 5.Fields P I, Groisman E A, Heffron F. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science. 1989;243:1059–1062. doi: 10.1126/science.2646710. [DOI] [PubMed] [Google Scholar]
- 6.Frohm M, Gunne H, Bergman A C, Agerberth B, Bergman T, Boman A, Liden S, Jornvall H, Boman H G. Biochemical and antibacterial analysis of human wound and blister fluid. Eur J Biochem. 1996;237:86–92. doi: 10.1111/j.1432-1033.1996.0086n.x. [DOI] [PubMed] [Google Scholar]
- 7.Gavrieli Y, Sherman Y, Ben-Sasson S A. Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall J. Lymphocyte recirculation and the gut: the cellular basis of humoral immunity in the intestine. Blood Cells. 1979;5:479–492. [PubMed] [Google Scholar]
- 9.Hall P A, Coates P J, Ansari B, Hopwood D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J Cell Sci. 1994;107:3569–3577. doi: 10.1242/jcs.107.12.3569. [DOI] [PubMed] [Google Scholar]
- 10.Harwig S S L, Tan L, Qu X, Cho Y, Eisenhauer P B, Lehrer R I. Bactericidal properties of murine intestinal phospholipase A2. J Clin Invest. 1995;95:603–610. doi: 10.1172/JCI117704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiemstra P S, Eisenhauer P B, Harwig S S L, Van Den Barselarr M T, Van Furth R, Lehrer R I. Antimicrobial proteins of murine macrophages. Infect Immun. 1993;61:3038–3046. doi: 10.1128/iai.61.7.3038-3046.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirsch J D. Bactericidal action of histone. J Exp Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones D E, Bevins C L. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–23225. [PubMed] [Google Scholar]
- 14.Laochumroonvorapong P, Paul S, Elkon K, Kaplan G. H2O2 induces monocyte apoptosis and reduces viability of Mycobacterium avium-M. intracellulare within cultured human monocytes. Infect Immun. 1996;64:452–459. doi: 10.1128/iai.64.2.452-459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehrer R I, Rosenman M, Harwig S S S L, Jackson R, Eisenhauer P. Ultrasensitive assays for endogenous antimicrobial polypeptides. J Immunol Methods. 1991;137:167–173. doi: 10.1016/0022-1759(91)90021-7. [DOI] [PubMed] [Google Scholar]
- 16.Mahida Y R, Makh S, Hyde S, Gray T, Borriello S P. Effect of Clostridium difficile toxin A on human intestinal epithelial cells: induction of interleukin 8 production and apoptosis after cell detachment. Gut. 1996;38:337–347. doi: 10.1136/gut.38.3.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahida Y R, Rose F, Chan W C. Antimicrobial peptides in the gastrointestinal tract. Gut. 1997;40:161–163. doi: 10.1136/gut.40.2.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouellette A J, Greco R M, James M, Frederick D, Naftilan J, Fallon J T. Developmental regulation of cryptdin, a corticostatin-defensin precursor mRNA in mouse small intestinal crypt epithelium. J Cell Biol. 1989;108:1687–1695. doi: 10.1083/jcb.108.5.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ouellette A J, Miller S I, Henschen A H, Selsted M E. Purification and primary structure of murine cryptdin-1, a Paneth cell defensin. FEBS Lett. 1992;304:146–148. doi: 10.1016/0014-5793(92)80606-h. [DOI] [PubMed] [Google Scholar]
- 20.Ouellette A J, Hsieh M M, Nosek M T, Cano-Gauci D F, Huttner K M, Buick R N, et al. Mouse Paneth cell defensins: primary structures and antibacterial activities of numerous cryptdin isoforms. Infect Immun. 1994;62:5040–5047. doi: 10.1128/iai.62.11.5040-5047.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panyim S, Chalkley R. High resolution acrylamide gel electrophoresis of histones. Arch Biochem Biophys. 1969;130:337–346. doi: 10.1016/0003-9861(69)90042-3. [DOI] [PubMed] [Google Scholar]
- 22.Park C B, Kim M S, Kim S C. A novel antimicrobial peptide from Bufo bufo gargarizans. Biochem Biophys Res Commun. 1996;128:408–413. doi: 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- 23.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter E M, Liu L, Oren A, Anton P A, Ganz T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997;65:2389–2395. doi: 10.1128/iai.65.6.2389-2395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Porter E M, van Dam E, Valore E V, Ganz T. Broad-spectrum antimicrobial activity of human intestinal defensin 5. Infect Immun. 1997;65:2396–2401. doi: 10.1128/iai.65.6.2396-2401.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Potten C S. Structure, function and proliferative organisation of mammalian gut. In: Potten C S, Hendry J H, editors. Radiation and gut. Amsterdam, The Netherlands: Elsevier Science B.V.; 1995. pp. 1–31. [Google Scholar]
- 27.Potten C S, Hendry J H, Moore J V, Chwalinski S. Cytotoxic effects in gastrointestinal epithelium (as exemplified by small intestine) In: Potten C S, Hendry J H, editors. Cytotoxic insult to tissue: effects on cell lineages. London, England: Churchill Livingstone; 1983. pp. 105–152. [Google Scholar]
- 28.Robinson G, Gray T. Electron microscopy. 2. Tissue preparation, sectioning and staining. In: Bancroft J D, Stevens A, editors. Theory and practice of histological techniques. 3rd ed. London, England: Churchill Livingstone; 1990. pp. 525–562. [Google Scholar]
- 29.Selsted M E, Miller S I, Henschen A H, Ouellette A J. Enteric defensins: antibiotic peptide components of intestinal host defence. J Cell Biol. 1992;118:929–936. doi: 10.1083/jcb.118.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibahara T, Sato N, Waguri S, Iwanaga T, Nakahara A, Fukutomi H, Uchiyama Y. The fate of effete epithelial cells at the villus tips of the human small intestine. Arch Histol Cytol. 1995;58:205–219. doi: 10.1679/aohc.58.205. [DOI] [PubMed] [Google Scholar]
- 31.Simon G L, Sherwood L G. Normal alimentary tract microflora. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press Ltd.; 1995. pp. 53–69. [Google Scholar]
- 32.Toskes P P. Small intestine bacterial overgrowth, including blind loop syndrome. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press Ltd.; 1995. pp. 343–352. [Google Scholar]
- 33.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Underdown B J, Mestecky J. Mucosal immunoglobulins. In: Ogra P L, Lamm M E, McGee J R, Mestecky J, Strober W, Bienenstock J, editors. Handbook of mucosal immunology. New York, N.Y: Academic Press, Inc.; 1994. pp. 79–97. [Google Scholar]
- 35.Zlatanova J S, Srebreva L N, Banchev T B, Tasheva B T, Tsanev R G. Cytoplasmic pool of histone H1 in mammalian cells. J Cell Sci. 1990;96:461–468. doi: 10.1242/jcs.96.3.461. [DOI] [PubMed] [Google Scholar]
- 36.Zychlinsky A, Sansonetti P. Apoptosis in bacterial pathogenesis. J Clin Invest. 1997;100:493–496. doi: 10.1172/JCI119557. [DOI] [PMC free article] [PubMed] [Google Scholar]