Abstract
Selenocysteine (Sec) is co-translationally inserted into selenoproteins in response to codon UGA with the help of the selenocysteine insertion sequence (SECIS) element. The number of selenoproteins in animals varies, with humans having 25 and mice having 24 selenoproteins. To date, however, only one selenoprotein, thioredoxin reductase, has been detected in Caenorhabditis elegans, and this enzyme contains only one Sec. Here, we characterize the selenoproteomes of C.elegans and Caenorhabditis briggsae with three independent algorithms, one searching for pairs of homologous nematode SECIS elements, another searching for Cys- or Sec-containing homologs of potential nematode selenoprotein genes and the third identifying Sec-containing homologs of annotated nematode proteins. These methods suggest that thioredoxin reductase is the only Sec-containing protein in the C.elegans and C.briggsae genomes. In contrast, we identified additional selenoproteins in other nematodes. Assuming that Sec insertion mechanisms are conserved between nematodes and other eukaryotes, the data suggest that nematode selenoproteomes were reduced during evolution, and that in an extreme reduction case Sec insertion systems probably decode only a single UGA codon in C.elegans and C.briggsae genomes. In addition, all detected genes had a rare form of SECIS element containing a guanosine in place of a conserved adenosine present in most other SECIS structures, suggesting that in organisms with small selenoproteomes SECIS elements may change rapidly.
INTRODUCTION
Selenocysteine (Sec) is a rare amino acid that is co-translationally inserted into protein in response to codon UGA (1–4). Since termination of translation is the common function of UGA, for Sec insertion selenoprotein genes employ a cis-acting stem–loop mRNA structure, the selenocysteine insertion sequence (SECIS) element (5–7). In eukaryotes, SECIS elements are located in 3′-untranslated regions (3′-UTRs) (8) and recruit several trans-acting factors, including SECIS-binding protein 2 (4,9) and Sec-specific elongation factor EFSec/eSelB (10,11).
Sec exhibits scattered evolutionary distribution in eukaryotes (12–14). For most selenoproteins, there are homologs that contain Cys in place of Sec, so there are organisms that prefer Sec, whereas others prefer Cys, in critical sites of certain redox proteins. Humans have 25 selenoprotein genes (15), of which 23 have a single Sec. In addition, one thyroid hormone deiodinase isozyme (type 2 deodinase) encodes two Sec (although the second is not needed for activity and may alternatively function as a stop) (16) and Selenoprotein P has 10 Sec residues (17). A total of 24 selenoproteins have been reported in mice (15), but only three in Drosophila melanogaster (18,19). In addition, multiple selenoproteins have also been reported in Chlamydomonas (12) and zebrafish, including one protein that had 17 Sec residues (20–22).
In contrast to other animals, only one selenoprotein, thioredoxin reductase (TR-Se), has been reported in Caenorhabditis elegans (23,24). In this protein, Sec is located in the C-terminal penultimate position, and an unusual form of SECIS element is present in the 3′-UTR (24). A second thioredoxin reductase gene also occurs in C.elegans, but it has Cys in place of Sec (23). In D.melanogaster, both thioredoxin reductases contain Cys rather than Sec in the C-terminal penultimate position (25,26). The presence of functional Cys-containing enzymes in animals suggests that TR-Se could evolve into a protein in which Sec is dispensable for thioredoxin reductase function. These observations raise a question of whether additional selenoproteins exist in C.elegans, which are responsible for the conservation of the Sec insertion machinery in this organism, including Sec tRNA, SBP2, EFSec/eSelB, selenophosphate synthetase (SelD, SPS) and other trans-acting factors.
In this paper, we employed three independent methods for the characterization of selenoprotein gene sets in C.elegans and Caenorhabditis briggsae genomes, including prediction of SECIS elements and identification of conserved alignments containing Sec–Sec or Sec–Cys pairs. The data suggest that in these nematodes TR-Se is the only selenoprotein and thus the Sec insertion machinery inserts only one residue. In contrast, other nematodes contain additional selenoproteins. It appears that the set of selenoprotein genes in nematode genomes has been reduced during evolution, with the extreme reduction case being C.elegans and C.briggsae genomes.
METHODS
Sequences, programs and databases
The sequences of the C.elegans chromosomes (27) were downloaded from the NCBI site (ftp://ftp.ncbi.nih.gov/genbank/genomes/C_elegans/) and combined into a single database. The C.briggsae genome (28) was downloaded from the Sanger Institute's ftp server (ftp://ftp.sanger.ac.uk/pub/wormbase/cbriggsae/cb25.agp8). C.elegans (22 227 predicted proteins) and C.briggsae (25 111 proteins) proteomes were obtained from ftp://ftp.sanger.ac.uk/pub/databases/wormpep/wormpep and ftp://ftp.wormbase.org/pub/wormbase/briggsae/brigpep1.gz, respectively.
Nematode expressed sequence tags (ESTs) were obtained from wormbase (ftp://genome.wustl.edu/pub/estmgr/est/est_exp.Z) and NCBI ‘est_others’ database (ftp://ftp.ncbi.nih.gov/blast/db/FASTA/est_others.gz). In addition, Dirofilaria immitis EST sequences were obtained from http://www.nematode.net. Other eukaryotic ESTs and cDNAs were downloaded from the Eukaryotic Gene Ortholog database (EGO, release 5 at http://www.tigr.org/tdb/tgi/ego/). For manual sequence analyses and homology searches, various programs from the BLAST suite and other NCBI databases were used. SECIS elements were visualized with an RNAnice program (15).
SECISearch analyses of nematode sequences
Several versions of SECISearch have been previously developed and reported (15,29), and similar approaches were also used in other studies (30). This program searches nucleotide sequences in several steps, including analyses of primary sequence conservation and secondary structures and, finally, evaluation of the free energy for each candidate structure. For two closely related genomes, an additional step is also incorporated that uses BLASTN to identify pairs of conserved sequences between the two sets of SECIS candidates.
C.elegans and C.briggsae genomes were separately searched with SECISearch 2.0 and with a modified version of SECISearch adapted for nematode searches. The major difference between the two versions of the program was that the nematode version allowed up to four mismatched or bulged nucleotides in helix 2 compared with two in the default version of SECISearch 2.0. These relaxed parameters increased the number of hits 10- to 20-fold; however, the searches were manageable owing to subsequent incorporation of evolutionary conservation criteria. To apply these criteria, the sequences selected by SECISearch or the modified version of SECISearch were combined into C.elegans and C.briggsae databases in FASTA format. The two files were analyzed against each other using the following BLASTN parameters: gap penalty = 1, extension = 1 and expectation = 0.00001. Pairs of sequences that matched these criteria were then analyzed manually against annotated genomes and NCBI nr and EST databases.
Analysis of sequence alignments containing a paired TGA codon
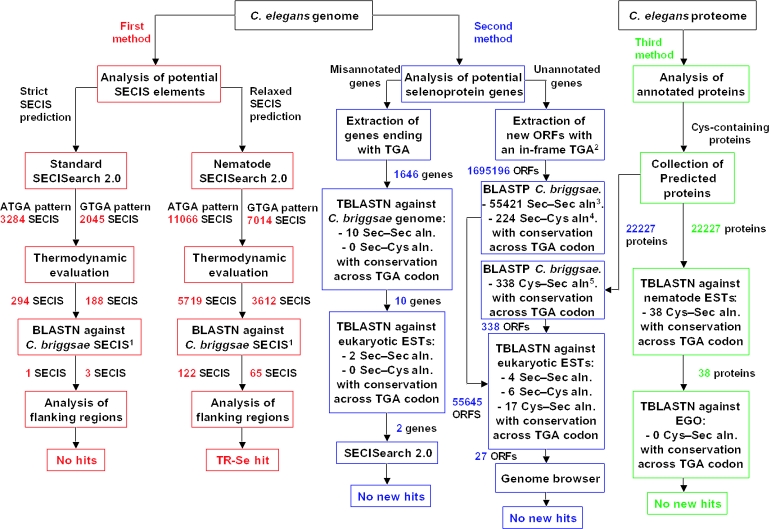
Selenoproteins can be identified by homology procedures that search for alignments where a Sec codon in a known (query) selenoprotein gene is aligned to a TGA codon or a Cys codon in the target. Because the majority of selenoproteins have both Sec- and Cys-containing homologs, in pairwise alignments these residues correspond to the Sec codon in the selenoprotein under study. Therefore, known or predicted coding regions [anonymous open reading frames (ORFs)] containing Sec codons can be compared at the amino acid level against other amino acid or DNA sequences to obtain such alignments. The resulting Sec–Sec (TGA–TGA) or Sec–Cys (TGA–TGT/TGC) pairs may be indicative of Sec coding function. However, selenoproteins and their Cys-containing homologs show a scattered distribution across eukaryotes (12–15), in which some species prefer Sec-containing proteins while others opt for Cys-containing versions. An example of this situation is the SelU selenoprotein, which has been shown to possess Sec in fishes, birds, diatoms, sea urchins and a green algae, but Cys in mammals, frogs, worms, slime molds, yeasts and plants (14). BLAST and other sequence similarity programs are well suited to provide initial alignments involving Sec-containing polypeptides; however, these alignments are false-positive prone and require further assessment as discussed below (Figure 1).
Figure 1.
A general schema of selenoprotein identification in C.elegans, C.briggsae and other nematodes. Three computational approaches, SECIS-dependent and SECIS-independent, are depicted, including (i) SECIS prediction (shown in red); (ii) analysis of missannotated or non-predicted selenoprotein genes in C.elegans (shown in blue); and (iii) search of Sec-containing homologs of C.elegans proteins (shown in green). Other designations are as follows: 1: BLASTN analysis of 382 ATGA and 307 GTGA C.briggsae SECIS elements using a standard SECISearch and of 6998 ATGA and 4119 GTGA C.briggsae SECIS elements using nematode SECISearch. 2: Length at least 60 nt. 3: BLASTP against 2 098 402 candidate C.briggsae ORFs containing TGA in-frame. 4: BLASTP against 25 111 predicted C.briggsae proteins. 5: BLASTP against 2 098 402 C.briggsae ORFs containing TGA in-frame.
Analysis of sequence conservation in regions flanking an aligned TGA codon
The fact that the sequences located downstream of a recoded stop codon (UGA in selenoprotein genes) correspond to actual coding regions implies that these regions must have typical patterns of sequence conservation (31). Therefore, at appropriate phylogenetic distances, alignments containing Sec–Sec or Sec–Cys pairs are suggestive of selenoprotein function provided that conservation involves both flanking regions of Sec. This approach, the analysis of sequence conservation in regions flanking a UGA triplet, has already been used to uncover novel selenoproteins in bacterial, mammalian and fish genomes (14,15,32). We have applied a similar procedure in our search for selenoprotein genes in the C.elegans genome (searches for misannotated and unannotated genes) and in the analysis of the C.elegans proteome (annotated proteins) (Figure 1).
Search for potential misannotated selenoprotein genes
For all C.elegans genes annotated as having a TGA stop signal, 30 nt on either side of the TGA codon were translated in-frame with the TGA and compared using TBLASTN against the C.briggsae genome. Those sequences, in which the annotated TGA was aligned to TGA (Sec) or TGT/TGC (Cys) and which had an E-value <0.1, were retained. These sequences were further extended by extracting 60 nt on either side of the TGA, translated and analyzed using TBLASTN against the NCBI collection of non-human and non-mouse EST sequences. Of the resulting HSPs (high-scoring segment pairs), we kept those with an E-value <0.1 and at least 10 conserved residues on both sides of the aligned TGA codon (Figure 1).
Search for unannotated selenoprotein genes
All C.elegans and C.briggsae TGA-containing ORFs [sequences between two in-frame non-TGA stop signals, which contain an in-frame TGA (e.g. TAA/TAG–TGA–TAA/TAG)] were predicted in regions with no known genes. The TGA-containing ORFs were predicted after masking the genomic sequence for known genes in wormpep and brigpep. ORFs shorter than 60 nt were discarded. Remaining ORFs and the annotated sets of proteins for these nematodes were compared using BLASTP in the three possible combinations of selenoprotein conservation between C.elegans and C.briggsae:
Sec in C.elegans and Sec in C.briggsae. C.elegans translated TGA-containing ORFs were compared against C.briggsae translated TGA-containing ORFs.
Sec in C.elegans and Cys in C.briggsae. C.elegans translated TGA-containing ORFs were compared against the set of annotated C.briggsae proteins.
Cys in C.elegans and Sec in C.briggsae. The set of annotated C.elegans proteins was compared against C.briggsae translated TGA-containing ORFs.
In each case, ORFs were selected, which contained at least three (out of six analyzed) conserved residues on both sides of the Sec. Each set was then compared using TBLASTN against non_human and non_mouse ESTs to search for additional evidence of Sec–Sec, Sec–Cys or Cys–Sec alignments and sequence conservation in flanking sequences. ORFs were further extracted that were represented by at least 5 unique alignments with an E-value <0.1 and at least 5 (out of 10) conserved residues on both sides of the aligned TGA codon. Finally, to assess whether the frame in which the ORF has been defined is the true coding frame, we used TBLASTX to align the ORFs against their conserved ESTs and only kept those ORFs whose highest-scoring HSP matched the previously predicted ORF with the aligned Sec codon. The resulting sequences for each set were examined manually using the Ensembl C.elegans genome browser (Figure 1).
Analysis of the C.elegans proteome for Sec-containing homologs in eukaryotes
All 22 227 predicted C.elegans ORFs were compared using TBLASTN against nematode EST sequences (a non-redundant set of 751 292 ESTs made by combining the wormbase EST collection and all nematode ESTs from the NCBI ‘est_others’ database). Cys–Sec alignments, which were supported by at least five different cDNAs/ESTs (to avoid false positives caused by sequencing errors) irrespective of identity and E-values, were selected. Predicted proteins so supported were analyzed using TBLASTN against the EGO database of homologous genes to identify selenoprotein homologs in either Sec or Cys form across the eukaryotic domain (Figure 1).
In addition, to search specifically within the nematode lineage, the wormpep-EST alignments (whether supported after the search in the EGO database or not) were further screened. Since any homologs are likely to be highly conserved within nematodes, fairly strict criteria of significance and identity were applied to the BLAST HSPs. Of all Cys–Sec alignments, we only kept those with an E-value of at least 0.001, identity of at least 60% and which were supported by at least 5 unique ESTs.
This procedure restricted the set to 8 peptides supported by 66 ESTs. We discarded any peptides that had a higher-scoring HSP against their target EST in another frame and with no in-frame stop codons. We then translated each EST and kept only those with no non-UGA stop codons in-frame. Finally, the annotations of these predicted proteins were examined in wormbase, which allowed us to discard proteins of known function.
Expression constructs and experiments
The plasmid pTRc for the expression of C.elegans TR-Se in Escherichia coli has been described previously (23). In short, the pET-21b(+) vector (Novagen) carries the ORF of C.elegans TR-Se with an E.coli formate dehydrogenase H SECIS element (23,33,34) that was cloned immediately downstream of the Sec TGA codon. The C.elegans TR-Se cDNA was introduced into the vector via NdeI/BamHI restriction sites. The internal NdeI site of the C.elegans TR-Se was eliminated by site-directed mutagenesis.
Constructs for expression in mammalian cells were based on a pCR3.1-Uni vector (Invitrogen) to allow strong expression under the CMV promoter. pTRc-Uni was created by amplifying the coding region of C.elegans TR-Se with primers cTR_Uni_F 5′-GTACCATGAAATCTCTCACCGAGTTATTC-3′ and cTR_Uni_R 5′-GACCCTTCTTAACCTCAGCAACC-3′ and inserting the resulting PCR product into pCR3.1-Uni vector (Invitrogen) according to the manufacturers' protocol. pTRc3-Uni was created by transferring the full-length cDNA fragment NotI/ApaI from initial pBluescript SK vector (Stratagene) into respective restriction sites of circular pCR3.1Uni vector.
CV-1 cells were grown, transfected and metabolically labeled with 75Se as described previously (20,29). Metabolic labeling of bacteria was also carried out as reported previously (23).
RESULTS AND DISCUSSION
Analysis of C.elegans and C.briggsae genomes with SECISearch 2.0
We previously developed SECISearch to identify selenoprotein genes by searching sequence databases for SECIS elements (15). It has a high true-positive rate and incorporates relatively stringent criteria that describe SECIS elements in selenoprotein genes. The default version of SECISearch recognizes over 95% of animal SECIS elements. Although it was developed to search for animal SECIS elements, most eukaryotic SECIS elements can be identified using this program.
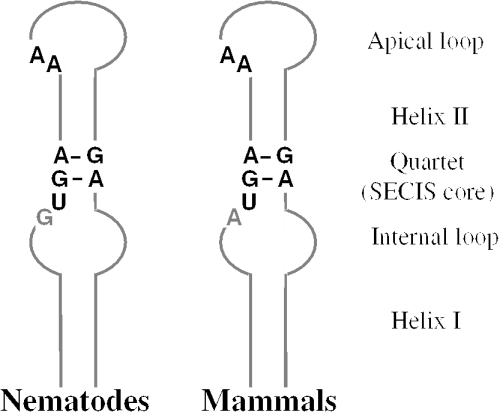
The majority of eukaryotic selenoprotein genes have several conserved nucleotides within their SECIS elements, including a TGAN……NGAN (N is any nucleotide) quartet of non-Watson–Crick base-paired nucleotides (the SECIS core), an unpaired A preceding the quartet and an unpaired AA motif in the apical loop or bulge (Figure 2). In addition, two mammalian selenoprotein genes have CC in place of AA (15), and several thyroid hormone deiodinase SECIS elements (mostly in fish; none in mammals) contain a guanosine in place of the unpaired adenosine (35). The C.elegans thioredoxin reductase (TR-Se) gene also contains the unpaired G rather than the A (24) (Figure 3).
Figure 2.
SECIS element consensus structures in nematodes and mammals. Both structures conserve the UGA_GA pattern within the quartet (SECIS core) and AA in the apical loop (or bulge). In nematode SECIS elements, a G is present immediately upstream of the quartet, whereas the majority of mammalian SECIS elements have an A in this position. The mammalian type of SECIS element in prevalent in most other eukaryotes. The nematode form of SECIS element is also present in some thyroid hormone deiodinase genes in fish, whereas none of the 25 mammalian selenoproteins has this form.
Figure 3.
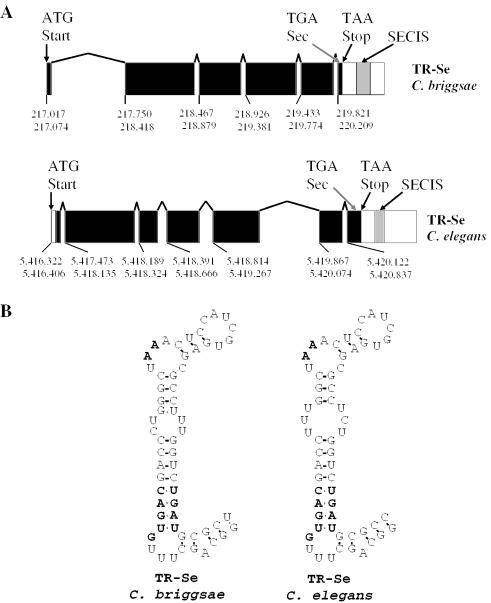
Organization of C.briggsae and C.elegans thioredoxin reductase (TR-Se) genes and structures of their SECIS elements. (A) TR-Se genes are shown with boxes indicating exons and lines connecting boxes representing introns. Filled boxes show coding regions, and open boxes represent untranslated regions within exons. The locations of initiator ATG codons, Sec-encoding TGA codons, and terminator signals in the C.elegans and C.briggsae genes are indicated. Gray boxes indicate the location of SECIS elements in 3′-UTRs. Upper and lower row numbers correspond to the beginning and the end of each exon, respectively. The C.briggsae TR-Se gene sequence is located in a contig cb25.fpc0143 from assembly cb25.agp8 (gi:22417497; CAE61785). The C.elegans TR-Se gene is located on chromosome IV (accession no. AF148217). (B) SECIS elements in C.briggsae and C.elegans. The quartet of non-Watson–Crick interacting nucleotides, an unpaired G preceding the quartet, and an unpaired AA motif in the apical bulge are shown in bold.
We applied the previously developed version of SECISearch 2.0 to analyze C.elegans and C.briggsae genomes for the presence of selenoprotein genes. The searches were performed with both ATGA_AA_GA and GTGA_AA_GA motifs (Figure 2), which were used separately (Table 1). The analysis of primary sequences and secondary structures using the GTGA_AA_GA motif resulted in 2045 hits for C.elegans and 2229 for C.briggsae. After applying free energy criteria, these numbers were reduced to 188 and 307 hits, respectively. These two sets of candidate SECIS elements were then analyzed using BLASTN against each other to identify pairs of conserved sequences, which resulted in three significant alignments. All three corresponded to coding regions and had no candidate Sec UGA codon, and therefore were highly unlikely to function as SECIS elements. A similar analysis using the ATGA_AA_GA motif and secondary structure, free energy and evolutionary conservation criteria described above for the GTGA_AA_GA motif also filtered out all hits and did not produce true positives (Table 1). Thus, the search of the two genomes with the standard version of SECISearch did not identify selenoprotein genes in the two nematode genomes.
Table 1.
Analysis of C.elegans and C.briggsae genomes for the presence of SECIS elements with modified and default patterns of SECISearch
| GTGA_AA_GA motif with the modified pattern (default pattern) | ATGA_AA_GA motif with the modified pattern (default pattern) | |||
|---|---|---|---|---|
| C.elegans | C.briggsae | C.elegans | C.briggsae | |
| Primary sequence and secondary structure criteriaa | 7014 (2045) | 7384 (2229) | 11 066 (3284) | 12 418 (3792) |
| Free energy criteriab | 3612 (188) | 4119 (307) | 5719 (294) | 6998 (382) |
| Evolutionary conservation criteriac | 65 (3) | 65 (3) | 122 (1) | 122 (1) |
| Not annotatedd | 3 (0) | 3 (0) | 4 (0) | 4 (0) |
| SECIS elementse | 1 (0) | 1 (0) | 0 (0) | 0 (0) |
aNumber of candidate sequences after the initial analysis of genomes for primary sequences and secondary structures.
bNumber of candidate sequences after application of free energy criteria.
cNumber of significant alignments of candidate sequences (BLASTN, gap = 1, ext = 1, E = 0.00001).
dNumber of candidate SECIS elements that corresponded to unannotated sequences.
eNumber of true SECIS elements. First number in each cell shows the results of searches with the modified pattern and that in parenthesis with the default pattern.
Analysis of C.elegans and C.briggsae genomes with the modified (nematode) version of SECISearch
The reason as to why the search did not identify the known selenoprotein (i.e. TR-Se) in the two nematode genomes was that the C.elegans TR-Se SECIS element (24) had three consecutive mismatches within helix 2 of the stem–loop structure (Figure 3). Such potentially unstable structure had no analogs in known eukaryotic selenoprotein genes and raised the possibility that nematode SECIS elements may have structural differences compared with other eukaryotic SECIS elements.
We modified the program by relaxing search parameters to allow up to four mismatches in helix 2, which resulted in a modified (‘nematode’) search pattern. The use of this pattern produced a 10- to 20-fold increase in the number of SECISearch hits (Table 1). However, the subsequent application of evolutionary conservation criteria reduced the hits to 65 and 122 for GTGA_AA_GA and ATGA_AA_GA patterns, respectively. Analysis of these hits against annotated nematode genomes and NCBI non-redundant and EST databases filtered out all sequences except 8, one of which corresponded to the true positive, TR-Se.
The remaining seven sequences (three and four for the GTGA_AA_GA and ATGA_AA_GA patterns, respectively) corresponded to regions in the C.elegans genome, which were not annotated. Past research shows that pairs of true SECIS elements are always located in the 3′-UTRs and show less conservation in primary sequences than in the coding regions. The use of these rules disqualified the seven candidates because: (i) no nematode ESTs were homologous to 1.5 kb sequences upstream of the hits; (ii) no predicted C.briggsae mRNAs matched sequences within these 1.5 kb; (iii) when the 1.5 kb sequences from C.elegans were analyzed using BLASTN against the corresponding 1.5 kb sequences in the C.briggsae genome, there were no homologous regions besides those corresponding to candidate SECIS elements. If a gene is present upstream of a candidate SECIS element, some conservation of upstream sequences is expected; and (iv) predicted SECIS structures, while identified by SECISearch, did not resemble those found in selenoprotein genes in terms of overall shape. Thus, this analysis revealed only one selenoprotein gene, TR-Se, in C.elegans and C.briggsae genomes (Figures 3 and 4).
Figure 4.
Alignment of Sec-containing TR-Se from C.elegans and C.briggsae and their Cys-containing homologs (TR-S). The conserved Sec residue (Cys in TR-S sequences) is shown in yellow and other conserved residues are highlighted.
Search for potential misannotated selenoprotein genes
The presence of an in-frame TGA codon in selenoprotein genes often leads to misannotation since the TGA is interpreted as a stop signal (18). We conducted a search of the C.elegans annotations for such errors in genes ending with annotated TGA stop codons in the C.elegans genome (Figure 1). For this analysis, we restricted the searches to the 5599 curated genes in the wormpep database. We first selected genes, annotated as having TGA stop signals (1646 genes) and translated 30 nt on either side of the TGA (in frame with the TGA). The regions around the annotated TGA stop codons were examined first against the C.briggsae genome for pairs of aligned Sec–Sec or Sec–Cys residues flanked by conserved sequences. This resulted in only 10 Sec–Sec alignment pairs. We then extended the search to cDNA sequences from a wide range of species, and only 2 of these candidates showed flanking conservation in Sec–Sec alignments. Although ESTs suggested a possible TGA read-through in these two genes, no stable SECIS elements were found in their 3′-UTR regions as well as in the regions 1 kb upstream and 1 kb downstream of TGA, allowing us to conclude that neither was a bona fide selenoprotein.
Search for unannotated genes
Some selenoprotein genes are completely missed by standard computational methods owing to the presence of TGA codons. Therefore, we searched for novel selenoproteins by predicting possible unannotated TGA-containing ORFs in the intergenic regions of C.elegans and C.briggsae genomes (Figure 1). We analyzed three possible situations of selenoprotein conservation between C.elegans and C.briggsae separately (see Methods).
Sec in C.elegans and Sec in C.briggsae. We compared 1 695 196 TGA-containing ORFs predicted in C.elegans with 2 098 402 predicted in C.briggsae and identified 55 421 Sec–Sec alignments with conservation across the TGA codon. However, only four of these ORFs had Sec–Sec alignment pairs surrounded by strong sequence conservation when compared with eukaryotic ESTs. We manually examined each of our four candidate sequences using the Ensembl C.elegans genome browser. One was on the antisense strand of exon 1 of the Ensembl protein K10D6.2b. The second candidate overlapped with emb|X07828.1, a snRNA gene. The remaining two candidates overlapped with ensembl tRNA genes (V:14233953-14234024 and X:9078188-9078259). Thus, no selenoprotein genes were identified.
Sec in C.elegans and Cys in C.briggsae. We compared TGA-containing ORFs predicted in C.elegans with the 25 111 known proteins of C.briggsae and searched for alignments, in which Sec in a C.elegans ORF was paired to Cys in a C.briggsae protein (Sec–Cys pairs). A total of 224 candidates were found, of which only 6 were supported by homology to eukaryotic ESTs. These could be further discarded by manual examination using the C.elegans genome browser because tblastx analysis showed they all had better HSPs against their subject ESTs in another frame and with no in-frame stop codons.
Sec in C.briggsae and Cys in C.elegans. We compared the TGA-containing ORFs predicted in C.briggsae with 22 227 known C.elegans proteins and searched for alignments, in which Sec in a C.briggsae ORF was paired to a Cys in a C.elegans protein (Cys–Sec pairs). A total of 338 candidate ORFs were detected. Eukaryotic ESTs supported 17 candidates, but these again were discarded by manual examination using the C.elegans genome browser because tblastx analysis showed they all had better HSPs against their subject ESTs in another frame and with no in-frame stop codons.
Analysis of the C.elegans proteome for Sec-containing homologs in eukaryotes
To search for possible additional missannotated or unpredicted selenoprotein genes that could have escaped our genome analysis, we screened the annotated set of C.elegans proteins with a third SECIS-independent method (Figure 1), which compared 22 227 proteins in the wormpep database against nematode ESTs. A total of 38 candidate C.elegans proteins, in which a Cys residue was paired to a TGA triplet in otherwise strong alignments involving ESTs, were identified. These 38 sequences were analyzed using TBLASTN against the EGO database in order to identify additional Sec-containing orthologs across the eukaryotic lineage. None of the alignments showed significant conservation across TGA codons (Figure 1).
Because of the limited coverage of the EGO database, and because nematode-specific selenoprotein genes could potentially exist, we focused on the stronger among the 38 candidates. Of eight initially promising C.elegans peptides all but two were discarded either because of in-frame non-TGA stop signals or because there was a far better alignment between a polypeptide and an EST with no in-frame stop signals. We examined the remaining two candidates for the presence of SECIS elements both within supporting ESTs and within the 1 kb flanking sequences of the predicted genes. No stable SECIS elements were found. These tests allowed us to conclude that none of these candidates was likely bona fide selenoproteins.
The analysis of sequence conservation in regions flanking an aligned TGA triplet is a powerful approach to identify novel selenoprotein genes independently of SECIS search. In this work, none of the methods that searched for conserved Sec–Sec, Sec–Cys and Cys–Sec pairs revealed additional selenoprotein genes in the C.elegans or C.briggsae genomes that were missannotated or unpredicted.
Comparative analysis of nematode thioredoxin reductases
Analysis of genomic regions upstream of SECIS elements in C.elegans and C.briggsae characterized genomic structures of TR-Se genes (Figure 3). The C.elegans TR-Se gene was composed of 7 exons, while that in C.briggsae had 6 exons. The difference was due to lack of an intron in C.briggsae that separated exons 3 and 4 in the C.elegans TR-Se gene. In both TR-Se genes, coding regions started in the first exon, whereas the Sec codon and SECIS element were located in the last exon. The 3′-UTR sequences in both enzymes were unusually long compared with other nematode untranslated regions.
The gene for C.elegans TR-Se has been previously cloned and its SECIS could support Sec insertion into mammalian thyroid hormone deiodinase 1 in mammalian cells (24). However, Sec insertion into C.elegans TR-Se using its natural SECIS element had not been attempted. In addition, a question remained in regard to the TR-Se ORF, with one study (23) predicting that the coding region had 669 amino acids and resulted in a 74 kDa protein, whereas the other study (24) proposed that the protein had 525 amino acids and was a 55 kDa polypeptide.
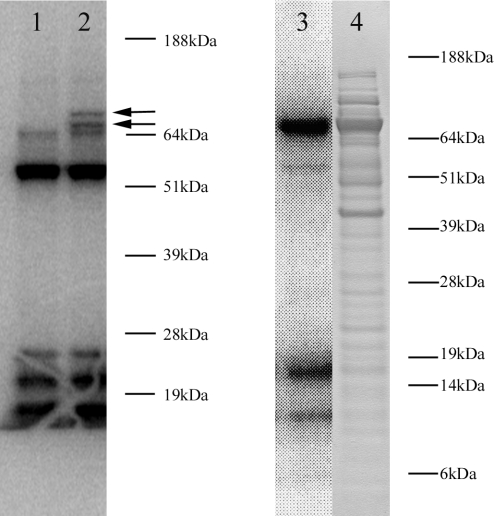
To address these questions, we cloned the entire cDNA for C.elegans TR-Se into a mammalian expression vector and expressed the enzyme in CV-1 cells. When transfected cells were metabolically labeled with 75Se, two additional protein bands could be seen at ∼74 kDa (Figure 5). The reason for the presence of two bands is not clear, but could be due to a differential use of first three in-frame ATG codons to initiate protein synthesis (all three were conserved between C.elegans and C.briggsae TR-Se sequences; see Figure 4). Thus, Sec insertion machinery in mammalian cells supported Sec insertion into C.elegans TR-Se with the help of its natural SECIS element. In addition, it was clear that the full-sized 74 kDa protein was synthesized. Expression of C.elegans TR-Se in E.coli from a construct that contained a bacterial SECIS element also resulted in the synthesis of ∼74 kDa 75Se-labeled polypeptide (Figure 5). Whether the 55 kDa protein could also be synthesized using an alternative initiation codon (Met143; it is not conserved between C.elegans and C.briggsae sequences) is not known. The 55 kDa form would include an entire minimal thioredoxin reductase domain. If expressed, this form could not be seen in our experiment due to a high expression of endogenous 55 kDa thioredoxin reductases in CV-1 cells.
Figure 5.
Expression of C.elegans TR-Se in mammalian cells and E.coli. Lane 1, CV-1 cells were transfected with a TR-Se construct containing only the coding region of the protein. Lane 2, CV-1 cells were transfected with a TR-Se construct containing the full size TR-Se cDNA, which included the SECIS element in the 3′-UTR. Lanes 3 and 4 show E.coli cells expressing TR-Se using a bacterial SECIS element. All samples were prepared from extracts of cells that were metabolically labeled with 75Se. Lanes 1–3 show PhosphorImager detection of 75Se-labeled proteins. Lane 4 shows Coomassie blue protein staining patterns. The location of TR-Se is indicated by arrows. Other major 75Se-labeled bands in lanes 1 and 2 correspond to endogenous selenoproteins. The location of molecular weight standards is shown on the right of the images.
Additional evidence for the presence of the 74 kDa form of TR-Se comes from the comparison of C.elegans and C.briggsae sequences (Figure 4). The C.briggsae TR-Se, similar to the C.elegans enzyme, also has the extended N-terminal region and the two extensions were highly homologous. N-terminal extensions in animal thioredoxin reductases are not uncommon. For example, in thioredoxin glutathione reductases present in mammals (36,37) and parasites (38), the N-terminal extension consists of a glutaredoxin domain, which serves as a substrate for a C-terminal GCUG tetrapeptide. The functional role of the N-terminal region in nematode TR-Se enzymes is not clear as it is not homologous to any known sequences. A Cys residue that is conserved between the two TR-Se sequences is present in this region. Whether it could be a substrate for the C-terminal GCUG tetrapeptide is not known.
Searches for selenoprotein genes in other nematodes
Nematode ESTs from representative organisms were analyzed with a modified nematode version of SECISearch. The analysis of D.immitis ESTs is shown in Table 2. Only one SECIS element (EST with accession no. BQ457177) was identified among 3938 EST sequences. Homology analyses of a region upstream of the predicted SECIS element revealed a coding region homologous to mammalian SelK and also to a Cys-containing homolog in C.elegans. A SECIS was also detected in a SelK EST sequence from Necator americanus, a human parasitic hookworm responsible for debilitating anemia (accession no. AI857092).
Table 2.
Analysis of the D.immitis EST database for the presence of SECIS elements with modified and default patterns of SECISearch
| D.immitis | ||||
|---|---|---|---|---|
| GTGA search with the modified pattern | ATGA search with the modified pattern | GTGA search with the default pattern | ATGA search with the default pattern | |
| Primary sequence and secondary structure criteriaa | 130 | 346 | 40 | 86 |
| Free energy criteriab | 36 | 123 | 5 | 10 |
| Evolutionary conservation criteriac | 25 | 75 | 2 | 5 |
| SECIS elementsd | 1 | 0 | 1 | 0 |
aNumber of candidate sequences after the initial analysis of the entire genomes for primary sequences and secondary structures.
bNumber of candidate sequences after application of free energy criteria.
cNumber of significant alignments of candidate sequences.
dNumber of true SECIS elements.
The alignment of nematode SelK and TR-Se sequences with corresponding SECIS elements in other invertebrate animal species are shown in Figure 6. Interestingly, similar to the SECIS elements in C.elegans and C.briggsae TR-Se, all other nematode SECIS elements had the GTGA_AA_GA pattern. In contrast, all detected SelK and TR-Se sequences in other invertebrates had the ATGA_AA_GA pattern (Figure 6). Thus, it appears that the rare form of SECIS element that was initially seen in C.elegans TR-Se (24) is the predominant SECIS form in nematodes.
Figure 6.
An alignment of predicted SECIS elements in SelK and TR families. The figure illustrates that nematode SECIS elements conserve G upstream of the quartet, whereas SECIS element in homologous genes in other organisms contain A in this position. (A) SelK (G-rich) SECIS elements. Nematode (top two sequences) SelK SECIS elements in D.immitis (accession no. BQ457177) and in N.americanus (AI857092) as compared with insect SECIS elements (two bottom sequences) in A.gambiae (gb|AAAB01008846) and D.melanogaster (AF396454). (B) Thioredoxin reductase SECIS elements. Nematode (top four sequences) TR-Se SECIS elements in C.elegans (AF148217), C.briggsae (gi:22417497), O.volvulus (AI692161) and T.vulpis (CB188966) TR-Se genes as compared with SECIS elements (bottom three sequences) in A.metacerebral, S.mansoni and S.japonicum TR-Se homologs. The location of helices I and II, apical loops (or bulges) and the quartet are indicated above the sequences.
Nematode ESTs were also screened for homologs of known selenoproteins using TBLASTN (Figure 7). This analysis identified several nematode sequences (homologs of the 15 kDa selenoprotein, SelK, selenophosphate synthetase, selenoprotein T, selenoprotein W and glutathione peroxidase) that had a predicted Sec that corresponded to Sec in mammalian selenoproteins, (e.g. Trichinella spiralis glutathione peroxidase homolog; ESTs with accession nos BG521074, BG519951 and BQ738601), but the EST sequences did not extend to SECIS elements.
Figure 7.
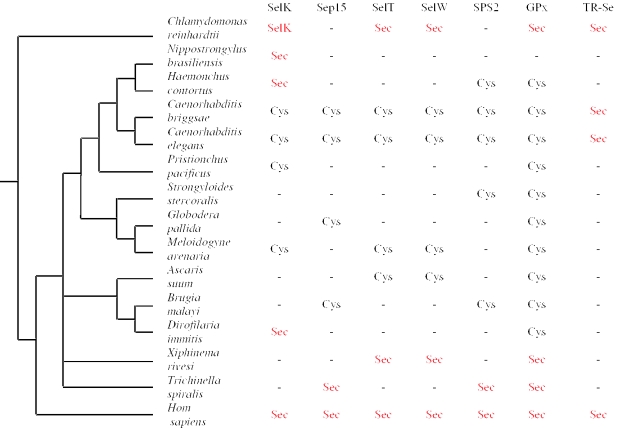
Distribution of selenoprotein genes and their Cys-containing homologs in nematodes. Selenoproteins found in at least one nematode are indicated by Sec and shown in red. Detected Cys homologs are indicated by Cys and shown in black. The nematode tree was obtained from TreeBase (40). The occurrence of human and Chlamydomonas selenoproteins is also indicated. Sequence information for some nematodes was not sufficient to determine whether a particular selenoprotein or its Cys homolog was present.
The completed genome sequences of nematodes other than C.elegans and C.briggsae are not available to determine how many selenoprotein genes are encoded in the genomes of these organisms. Nevertheless, it was clear from our analysis that at least some nematodes had several selenoproteins (Figure 7), which were present in other animals and corresponded to Cys-containing homologs in C.elegans and C.briggsae. In addition, a large number of selenoproteins could be seen in some platyhelminthes, such as Schistosoma (data not shown) and in green algae (12,13). These observations suggest that the presence of only a single selenoprotein in C.elegans and C.briggsae was due to the reduction in the number of selenoproteins in these organisms. In further support of this, reconstruction of ancestral character states of selenoproteins found in the eukaryotic clade (25 widespread taxa under a generalized parsimony model of evolution with unequal character state gain-loss weights which favor Sec to Cys conversion, as derived from currently held biological assumptions of Sec/Cys exchange), consistently suggests a common Sec-containing protein ancestor at the base of eukaryotes for many selenoprotein families, including those shown in Figure 6 (S. Castellano, C. Chapple and R. Guigó, unpublished data).
In conclusion, our study suggests that C.elegans and C.briggsae genomes each contain only one Sec UGA codon and one SECIS element. If so, the entire Sec insertion system is used to insert a single Sec in these animals. However, it should be noted that our searches were based on the assumption that Sec insertion systems and mechanisms are conserved between nematodes and other eukaryotes. While these assumptions agree with all previously characterized systems, these might not necessarily hold true in every case. Analyses of additional nematode and other eukaryotic genomes for selenoprotein genes should further address conservation and evolution of selenoprotein genes and Sec insertion systems.
In our work, searches for other selenoproteins indicated that the occurrence of only one selenoprotein was due to a reduction in the number of selenoproteins in nematodes during evolution. Except for TR-Se, all selenoproteins were lost in C.elegans and C.briggsae, whereas some nematodes still contained additional selenoproteins. A Cys-containing form of thioredoxin reductase that exhibited as high activity as the Sec-containing form was reported in Drosophila (39). Thus, thioredoxin reductase could also be replaced with the Cys homolog, and it would not be surprising if an animal is identified in the future that lacks selenoproteins altogether.
We found that all detected nematode selenoprotein genes contained an unusual form of SECIS element, with G rather than A being in the conserved position preceding the Quartet of non-Watson–Crick base pairs. The low number of SECIS elements in nematode genomes presumably makes it easier for SECIS elements to evolve in coordination with SECIS-binding proteins. Thus, the data suggest that in organisms with small selenoproteomes SECIS elements may rapidly change.
Acknowledgments
This study was supported by NIH grants GM061603 and GM065204 and by grant BIO2000-1358-C02-02 from Plan Nacional de I+D (Spain). Funding to pay the Open Access publication charges for this article was provided by GM061603.
Conflict of interest statement. None declared.
REFERENCES
- 1.Atkins J.F., Gesteland R.F. The twenty-first amino acid. Nature. 2000;407:463. doi: 10.1038/35035189. [DOI] [PubMed] [Google Scholar]
- 2.Bock A. Biosynthesis of selenoproteins—an overview. Biofactors. 2000;11:77–78. doi: 10.1002/biof.5520110122. [DOI] [PubMed] [Google Scholar]
- 3.Hatfield D.L., Gladyshev V.N. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 2002;22:3565–3576. doi: 10.1128/MCB.22.11.3565-3576.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driscoll D.M., Copeland P.R. Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 2003;23:17–40. doi: 10.1146/annurev.nutr.23.011702.073318. [DOI] [PubMed] [Google Scholar]
- 5.Zinoni F., Heider J., Bock A. Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl Acad. Sci. USA. 1990;87:4660–4664. doi: 10.1073/pnas.87.12.4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry M.J., Banu L., Chen Y.Y., Mandel S.J., Kieffer J.D., Harney J.W., Larsen P.R. Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature. 1991;353:273–276. doi: 10.1038/353273a0. [DOI] [PubMed] [Google Scholar]
- 7.Heider J., Baron C., Bock A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO Rep. 1992;11:3759–3766. doi: 10.1002/j.1460-2075.1992.tb05461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low S.C., Berry M.J. Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996;21:203–208. [PubMed] [Google Scholar]
- 9.Copeland P.R., Fletcher J.E., Carlson B.A., Hatfield D.L., Driscoll D.M. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO Rep. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tujebajeva R.M., Copeland P.R., Xu X.M., Carlson B.A., Harney J.W., Driscoll D.M., Hatfield D.L., Berry M.J. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO Rep. 2000a;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novoselov S.V., Rao M., Onoshko N.V., Zhi H., Kryukov G.V., Xiang Y., Weeks D.P., Hatfield D.L., Gladyshev V.N. Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO Rep. 2002;21:3681–3693. doi: 10.1093/emboj/cdf372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L.H., Wang X.F., Eyal Y., She Y.M., Donald L.J., Standing K.G., Ben-Hayyim G. A selenoprotein in the plant kingdom. Mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J. Biol. Chem. 2002;277:25983–25991. doi: 10.1074/jbc.M202912200. [DOI] [PubMed] [Google Scholar]
- 14.Castellano S., Novoselov S.V., Kryukov G.V., Lescure A., Blanco E., Krol A., Gladyshev V.N., Guigo R. Reconsidering the evolution of eukaryotic selenoproteins: a novel nonmammalian family with scattered phylogenetic distribution. EMBO Rep. 2004;5:71–77. doi: 10.1038/sj.embor.7400036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryukov G.V., Castellano S., Novoselov S.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 16.Salvatore D., Harney J.W., Larsen P.R. Mutation of the Secys residue 266 in human type 2 selenodeiodinase alters 75Se incorporation without affecting its biochemical properties. Biochimie. 1999;81:535–538. doi: 10.1016/s0300-9084(99)80106-0. [DOI] [PubMed] [Google Scholar]
- 17.Hill K.E., Lloyd R.S., Burk R.F. Conserved nucleotide sequences in the open reading frame and 3′ untranslated region of selenoprotein P mRNA. Proc. Natl Acad. Sci. USA. 1993;90:537–541. doi: 10.1073/pnas.90.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellano S., Morozova N., Morey M., Berry M.J., Serras F., Corominas M., Guigo R. In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep. 2001;2:697–702. doi: 10.1093/embo-reports/kve151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin-Romero F.J., Kryukov G.V., Lobanov A.V., Carlson B.A., Lee B.J., Gladyshev V.N., Hatfield D.L. Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J. Biol. Chem. 2001;276:29798–29804. doi: 10.1074/jbc.M100422200. [DOI] [PubMed] [Google Scholar]
- 20.Kryukov G.V., Gladyshev V.N. Selenium metabolism in zebrafish: multiplicity of selenoprotein genes and expression of a protein containing 17 selenocysteine residues. Genes Cells. 2000;5:1049–1060. doi: 10.1046/j.1365-2443.2000.00392.x. [DOI] [PubMed] [Google Scholar]
- 21.Tujebajeva R.M., Ransom D.G., Harney J.W., Berry M.J. Expression and characterization of nonmammalian selenoprotein P in the zebrafish, Danio rerio. Genes Cells. 2000;5:897–903. doi: 10.1046/j.1365-2443.2000.00375.x. [DOI] [PubMed] [Google Scholar]
- 22.Thisse C., Degrave A., Kryukov G.V., Gladyshev V.N., Obrecht-Pflumio S., Krol A., Thisse B., Lescure A. Spatial and temporal expression patterns of selenoprotein genes during embryogenesis in zebrafish. Gene Expr. Patterns. 2003;3:525–532. doi: 10.1016/s1567-133x(03)00054-1. [DOI] [PubMed] [Google Scholar]
- 23.Gladyshev V.N., Krause M., Xu X.M., Korotkov K.V., Kryukov G.V., Sun Q.A., Lee B.J., Wootton J.C., Hatfield D.L. Selenocysteine-containing thioredoxin reductase in C.elegans. Biochem. Biophys. Res. Commun. 1999;259:244–249. doi: 10.1006/bbrc.1999.0765. [DOI] [PubMed] [Google Scholar]
- 24.Buettner C., Harney J.W., Berry M.J. The Caenorhabditis elegans homologue of thioredoxin reductase contains a selenocysteine insertion sequence (SECIS) element that differs from mammalian SECIS elements but directs selenocysteine incorporation. J. Biol. Chem. 1999;274:21598–21602. doi: 10.1074/jbc.274.31.21598. [DOI] [PubMed] [Google Scholar]
- 25.Kanzok S.M., Fechner A., Bauer H., Ulschmid J.K., Müller H.M., Botella-Munoz J., Schneuwly S., Schirmer R., Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 26.Missirlis F., Ulschmid J.K., Hirosawa-Takamori M., Grönke S., Schäfer U., Becker K., Phillips J.P., Jäckle H. Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J. Biol. Chem. 2002;277:11521–11526. doi: 10.1074/jbc.M111692200. [DOI] [PubMed] [Google Scholar]
- 27.C.elegans Sequencing Consortium. Genome sequence of the nematode C.elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 28.Stein L.D., Bao Z., Blasiar D., Blumenthal T., Brent M.R., Chen N., Chinwalla A., Clarke L., Clee C., Coghlan A., et al. The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 2003;1:E45. doi: 10.1371/journal.pbio.0000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kryukov G.V., Kryukov V.M., Gladyshev V.N. New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J. Biol. Chem. 1999;274:33888–33897. doi: 10.1074/jbc.274.48.33888. [DOI] [PubMed] [Google Scholar]
- 30.Lescure A., Gautheret D., Carbon P., Krol A. Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J. Biol. Chem. 1999;274:38147–38154. doi: 10.1074/jbc.274.53.38147. [DOI] [PubMed] [Google Scholar]
- 31.Parra G., Blanco E., Guigo R. GeneID in Drosophila. Genome Res. 2000;10:511–515. doi: 10.1101/gr.10.4.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kryukov G.V., Gladyshev V.N. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arner E.S., Sarioglu H., Lottspeich F., Holmgren A., Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J. Mol. Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 34.Arner E.S. Recombinant expression of mammalian selenocysteine-containing thioredoxin reductase and other selenoproteins in Escherichia coli. Methods Enzymol. 2002;347:226–235. doi: 10.1016/s0076-6879(02)47022-x. [DOI] [PubMed] [Google Scholar]
- 35.Fagegaltier D., Lescure A., Walczak R., Carbon P., Krol A. Structural analysis of new local features in SECIS RNA hairpins. Nucleic Acids Res. 2000b;28:2679–2689. doi: 10.1093/nar/28.14.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Q.A., Kirnarsky L., Sherman S., Gladyshev V.N. Selenoprotein oxidoreductase with specificity for thioredoxin and glutathione systems. Proc. Natl Acad. Sci. USA. 2001;98:3673–3678. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su D., Gladyshev V.N. Alternative splicing involving the thioredoxin reductase module in mammals: a glutaredoxin-containing thioredoxin reductase 1. Biochemistry. 2004;43:12177–12188. doi: 10.1021/bi048478t. [DOI] [PubMed] [Google Scholar]
- 38.Salinas G., Selkirk M.E., Chalar C., Maizels R.M., Fernandez C. Linked thioredoxin-glutathione systems in platyhelminths. Trends Parasitol. 2004;20:340–346. doi: 10.1016/j.pt.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Gromer S., Johansson L., Bauer H., Arscott L.D., Rauch S., Ballou D.P., Williams C.H., Jr, Schirmer R.H., Arner E.S. Active sites of thioredoxin reductases: why selenoproteins? Proc. Natl Acad. Sci. USA. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morell V. TreeBASE: the roots of phylogeny. Science. 1996;273:569. [Google Scholar]