Abstract
Facioscapulohumeral dystrophy (FSHD) has a unique genetic aetiology resulting in partial chromatin relaxation of the D4Z4 macrosatellite repeat array on 4qter. This D4Z4 chromatin relaxation facilitates inappropriate expression of the transcription factor DUX4 in skeletal muscle. DUX4 is encoded by a retrogene that is embedded within the distal region of the D4Z4 repeat array. In the European population, the D4Z4 repeat array is usually organized in a single array that ranges between 8 and 100 units. D4Z4 chromatin relaxation and DUX4 derepression in FSHD is most often caused by repeat array contraction to 1–10 units (FSHD1) or by a digenic mechanism requiring pathogenic variants in a D4Z4 chromatin repressor like SMCHD1, combined with a repeat array between 8 and 20 units (FSHD2).
With a prevalence of 1.5% in the European population, in cis duplications of the D4Z4 repeat array, where two adjacent D4Z4 arrays are interrupted by a spacer sequence, are relatively common but their relationship to FSHD is not well understood. In cis duplication alleles were shown to be pathogenic in FSHD2 patients; however, there is inconsistent evidence for the necessity of an SMCHD1 mutation for disease development.
To explore the pathogenic nature of these alleles we compared in cis duplication alleles in FSHD patients with or without pathogenic SMCHD1 variant. For both groups we showed duplication-allele-specific DUX4 expression. We studied these alleles in detail using pulsed-field gel electrophoresis-based Southern blotting and molecular combing, emphasizing the challenges in the characterization of these rearrangements. Nanopore sequencing was instrumental to study the composition and methylation of the duplicated D4Z4 repeat arrays and to identify the breakpoints and the spacer sequence between the arrays. By comparing the composition of the D4Z4 repeat array of in cis duplication alleles in both groups, we found that specific combinations of proximal and distal repeat array sizes determine their pathogenicity. Supported by our algorithm to predict pathogenicity, diagnostic laboratories should now be furnished to accurately interpret these in cis D4Z4 repeat array duplications, alleles that can easily be missed in routine settings.
Keywords: facioscapulohumeral muscular dystrophy, FSHD, D4Z4, duplications, DUX4
Facioscapulohumeral muscular dystrophy (FSHD) is caused by shortening of the D4Z4 repeat array. Occasionally duplications of this array are found with unknown pathogenicity. Lemmers et al. have developed a formula to predict pathogenicity of these duplications and have unravelled the mechanism by which they cause FSHD.
Introduction
Facioscapulohumeral dystrophy (FSHD; MIM 158900) is one of the most prevalent inherited muscular dystrophies in adults. The disease is characterized by progressive and often asymmetric weakness and wasting of facial, shoulder girdle and upper arm muscles, typically starting in the second decade of life. FSHD is, however, highly variable in disease presentation ranging from life-long non-penetrant or asymptomatic pathogenic variant carriers to early onset cases with rapid progression and wheelchair dependency. With progression, other muscle may become affected. Extramuscular symptoms, such as retinal vasculopathy, are rare and often remain undiagnosed.1,2
FSHD is caused by ectopic expression of the DUX4 retrogene in skeletal muscle, which disturbs normal muscle homeostasis eventually resulting in apoptosis.3-5 DUX4 is a cleavage stage and germline transcription factor that under normal conditions is silenced in somatic tissues, such as skeletal muscle.6-8 A copy of the DUX4 open reading frame is embedded within each 3.3 kb large and CpG-rich D4Z4 unit, which is organized in a polymorphic array of 8–100 units on chromosome 4 in non-affected individuals.9-12 In this size range the D4Z4 repeat array shows high CpG methylation and is decorated with repressive chromatin marks to enforce a closed chromatin environment in somatic tissue. The level of CpG methylation correlates linearly with the size of the D4Z4 repeat array.13 In FSHD, this repressive chromatin structure is partially absent, as evidenced by reduced CpG methylation and loss of repressive histone modifications, resulting in sporadic DUX4 expression in skeletal muscle.13-16 The D4Z4 repeat array maps to the subtelomere of the long arm of chromosome 4 of which two major variants exist: 4qA and 4qB.17 Only D4Z4 repeat arrays on 4qA can express DUX4 in skeletal muscle as this haplotype uniquely contains a somatic DUX4 polyadenylation signal immediately distal to the D4Z4 array.18
D4Z4 chromatin relaxation and DUX4 expression in FSHD are caused by either a contraction of the D4Z4 repeat array to sizes between 1 and 10 units on a disease permissive (i.e. DUX4 polyadenylation signal-containing) 4qA haplotype (FSHD1; >95% of cases) or by pathogenic variants in chromatin factors that contribute to a repressive D4Z4 chromatin structure, most often SMCHD1 (FSHD2: <5% of cases) and rarely DNMT3B or LRIF1.19-22 The disease severity in FSHD1 roughly and inversely correlates with D4Z4 repeat array size.23 This also holds true for FSHD2 as SMCHD1 variant carriers with medium-sized D4Z4 arrays between 8 and 20 units on 4qA are symptomatic, while variant carriers with arrays >20 units on 4qA generally remain asymptomatic.24 The most severe and early onset patients carry an array size between 1 and 3 units for FSHD1, and 8 and 10 units for FSHD2, a situation in which FSHD1 and FSHD2 overlap.25-27
In 10–30% of the cases, FSHD1 is caused by a de novo D4Z4 repeat array contraction, and in half of these events this rearrangement occurs during early cell divisions leading to gonosomal mosaicism.28,29 The disease severity in patients with gonosomal mosaicism depends on the proportion of cells that carry the FSHD1 allele in skeletal muscle and the repeat array size. Because of the rearrangement occurring early in development, the detection of somatic mosaicism in peripheral white blood cells is representative for mosaicism in skeletal muscle.30
The D4Z4 repeat array can also be found on chromosomes 10q and 4qB, the latter being almost equally common to 4qA in the European and Asian population.31 However, on 4qB chromosomes, the adjacent region distal to D4Z4 that encodes the 3′UTR of the DUX4 retrogene present on chromosome 4qA, is missing. The DUX4 sequence on chromosome 10q is highly homologous to 4qA, but a single nucleotide polymorphism in the polyadenylation signal prohibits stable transcription from this chromosome.18 Consequently, chromatin derepression of 4qB and 10q by repeat array contraction or by variants in D4Z4 chromatin repressors does not result in stable DUX4 transcription from these repeat arrays in skeletal muscle and does not lead to FSHD (Supplementary Fig. 1).
In addition to D4Z4 repeat array contractions, other rare D4Z4 rearrangements, such as D4Z4 proximally extended deletions (DPED) and 4; 10 translocations can be associated with FSHD.32,33 Recently, in cis duplications of the D4Z4 repeat array have been identified: these are characterized by a D4Z4 repeat array that is followed by a second and sometimes a third D4Z4 repeat array with a spacer sequence in between.24,34 Typically, the proximal D4Z4 repeat array in these duplication alleles is of normal size, while the distal repeat array is <10 units, with both arrays ending with a 4qA sequence. Approximately 1.5% of all 4qA chromosomes have an in cis D4Z4 duplication, but they have not yet been reported in 4qB chromosomes.24
The major 4qA and 4qB variants can be subdivided in several sub-haplotypes based on sequence variations at the proximal and distal end of the D4Z4 repeat array. At the proximal end, this is a simple sequence length polymorphism (SSLP) ranging from 157 to 171 bp. Regarding the distal end of the repeat array, two 4qA variants have been described: the common 4A161S variant and the five times less common and European-specific 4A161L haplotype. Remarkably, the vast majority of duplication alleles identified thus far are of the 4A161L haplotype (Supplementary Fig. 1).
Recently, we showed that in FSHD2 patients, ∼8% of the 4qA alleles have an in cis D4Z4 duplication.24 Segregation analysis in these families showed that a 4qA duplication allele is only pathogenic when combined with a pathogenic SMCHD1 variant. Despite the distal D4Z4 repeat array of these 4qA duplication alleles being mostly <7 units, they seem not to be FSHD-causing in the absence of a variant in a D4Z4 chromatin repressor. In contrast, a French study reported on two FSHD patients in whom the duplication allele was the only permissive allele without pathogenic SMCHD1 variant, suggesting a direct role of 4qA duplication alleles in FSHD.34 More recently, we also identified several FSHD patients with a 4qA duplication allele in the absence of hypomethylation patterns typical for FSHD2 or a pathogenic variant in any of the known FSHD2 genes. This prompted us to study these individuals in more detail.
Materials and methods
Subjects
This study was focused on individuals with a self-reported European descent and approved by the Medical Ethical Committees from participating hospitals. A detailed clinical description of relevant family members of the families with a dominant in cis duplication allele can be found in the Supplementary material. Clinical evaluation of all FSHD cases was performed by an experienced neurologist after informed consent. For the clinical severity, we used the age corrected severity score (ACSS), based on the 10-scale Ricci score [ACSS = (Ricci score / age at examination) × 1000].35,36
Genetic and methylation analysis of D4Z4 repeat arrays
These studies were carried out as previously described.37 Briefly, genomic DNA from peripheral blood mononuclear cells or fibroblasts cultures (Supplementary Fig. 2) embedded in agarose plugs were digested with EcoRI/HindIII, EcoRI/BlnI and XapI for D4Z4 repeat array sizing and HindIII for determining the distal haplotype and separated by pulsed-field gel electrophoresis (PFGE). After separation, DNA was transferred by Southern blotting to charged nylon membranes (Hybond-XL) and serially hybridized with radioactively labelled probes p13E-11, D4Z4, 4qA and 4qB. Hybridizing fragments were visualized by phosphor imaging on a Typhoon scanner (Amersham). SSLP and 4A161S and 4A161L (S/L) analysis for specifying the haplotype of the D4Z4 alleles was performed as described previously.10,38 Most individuals that carried an in cis duplication allele were also analysed by molecular combing, as previously described.24 Briefly, DNA was combed on a glass slide, which was hybridized with antibody-labelled FSHD-specific probes and scanned by the Fibervision HeliXScan. D4Z4 alleles were selected and counted using the general procedure by Fiberstudio 0.9.12 software. Southern blot-based methylation analysis was done using the methylation-sensitive restriction endonuclease FseI, as described, and delta1 methylation values (i.e. D4Z4 methylation levels corrected for repeat array length) were calculated as before.13
Nanopore sequencing
Targeted nanopore sequencing and CpG methylation analysis for samples Rf2704.102, Rf2704.201 and Rf2988.202 was done as previously described.39 Briefly, amplification-free libraries were prepared using the Cas9 Sequencing Kit protocol (Oxford Nanopore Technologies) with ∼5 μg of genomic DNA and CRISPR/Cas guide RNAs targeting the p13E-11, pLAM and D4Z4 regions. The Cas9 libraries were loaded onto MinION version R9.4.1 flow cells and data collected with a Mk1B sequencing device using MinKNOW software (Oxford Nanopore Technologies). Modified base calling was performed using the guppy_basecaller (v6.3.8) with the dna_r9.4.1_450bps_modbases_5mc_cg_sup.cfg config file. Minimap2 (v2.22) and samtools (v1.12) were used to align nanopore reads to the T2T CHM13v2.0 reference genome that included an accessory 4qB D4Z4 region from HG002 T2T assembly. Reads were scored for mismatches and assigned to haplotypes with custom scripts that analysed Smith–Waterman local alignments to 4qA, 10q and 4qB T2T reference sequences. From the mapped 3.3 kb D4Z4 gRNA-cut reads, base-calling accuracy was estimated from these Smith-Waterman alignments, which showed a median nucleotide identity of: 98.7% (10q; 1495 reads), 98.6% (4qA, 787 reads) and 98.7% (4qB, 691 reads). Reference-based modified base-calling was performed using Megalodon (v2.5.0, https://github.com/nanoporetech/megalodon) using the Remora models. Single-read methylation plots were generated with modbamtools40 and kernel smoothing of averaged methylation levels from the reference anchored base calls was performed in R using the smooth_ksmooth function (smoothr package) and plotted with ggplot2. The methylation levels of D4Z4 cut reads were summarized as % 5mC per read using a probability of >0.8 for modified and <0.2 for unmodified base calls from the ∼320 CpG sites detected per 3.3 kb read.
Gene expression analysis
.For gene expression analysis we generated primary fibroblast cultures from the indicated individuals according to previously described methods (detailed protocols at www.urmc.rochester.edu/fields-center/). Unaffected control and confirmed FSHD fibroblast cell cultures originated from the University of Rochester Medical Center bio-repository. Fibroblasts were transformed into myocytes using MyoD transductions, as described previously.41 Differentiation into myotubes was induced by serum reduction at 80–100% confluency. Expression analysis for DUX4, GUSB, DUX4 target genes (MBD3L2, ZSCAN4 and TRIM43) and myogenic differentiation genes (MYH3 and MYOG) was performed in triplicate by previously described PCR conditions and primer pairs.32
Statistical analysis
Expression results are represented as mean ± standard deviation (SD). To establish the significance of the mean between the groups, one-way ANOVA was used followed by a Bonferroni post-test. P < 0.05 was considered statistically significant.
Results
Genetic analysis of patients that are genetically not FSHD1 or FSHD2
Over the past decades, we have studied the D4Z4 repeat array by PFGE in combination with Southern blotting in patients with a clinical phenotype consistent with FSHD from more than 1000 families. In 920 families we could genetically confirm FSHD, but the remaining patients were—according to the genetic criteria—neither FSHD1 nor FSHD2. For most of these genetically inconclusive cases, the clinical diagnosis of FSHD was unclear and sometimes further genetic analysis confirmed a different diagnosis. For seven of these families, as well as two previously diagnosed FSHD1 families, we identified a 4qA cis D4Z4 duplication allele in the affected individuals, which were followed-up in detail in this study.
FSHD Family Rf2704
The proband presented with asymmetric mild facial weakness and scapular winging at the age of 8 years. At age 10, pronounced facial weakness, obvious scapula dyskinesia (right-side more pronounced) and abdominal muscle weakness were noted. No extra-muscular signs (visual, hearing, respiratory function) were observed. He achieved all psychomotor milestones on time, but endurance was always less than his healthy brother. The child’s mother was referred for evaluation after her son was diagnosed. She reported easy fatigability in childhood and a sore feeling in her legs after walking longer distances. This fatiguability was clearly more pronounced than her peers, in spite of being physically active in daily life and sports. At the age of 48, she reported difficulties in lifting heavy objects. She has no limitations in raising her arms, but carrying heavy objects above her head was very difficult. Furthermore, she reported moderate-to-severe myalgia in neck and shoulder girdle since childhood. A physical examination had not yet been performed due to COVID.
Upon Southern blot-based FSHD genetic analysis by linear gel electrophoresis and hybridization with probe p13E-11, which is the gold standard in diagnostic laboratories, the patient did not appear to have a contraction of D4Z4 seen in FSHD1, and had normal CpG methylation at the D4Z4 repeat array excluding FSHD2. More detailed genetic analysis using PFGE and Southern blotting on agarose embedded DNA and probe D4Z4 revealed an extra fragment in both mother and son, which was shorter in the affected son (Fig. 1A). Interestingly, the shorter allele is also faintly visible in the DNA of the mother, indicating that she is mosaic for this D4Z4 repeat array contraction. The extra bands were confirmed by 4qA hybridization and the subsequent S/L typing and SSLP analysis suggested the presence of an in cis duplication allele of the haplotype 4A161L. The in cis duplicated D4Z4 repeat array was somatically contracted in the mosaic mother resulting in gonosomal mosaicism and transmitted to the affected son (Fig. 1B). Mosaicism for a D4Z4 repeat array rearrangement in blood is indicatory for mosaicism in skeletal muscle, which may explain her mild disease presentation.30 A broad and comparable mosaicism throughout the body was indeed verified by demonstrating similar levels of mosaicism in cultured skin fibroblasts (Supplementary Fig. 2). To confirm this finding, we performed molecular combing in the mother and identified a duplication allele, which consisted of a 17-unit D4Z4 array followed in cis by a 9-unit array. In addition, we identified some molecular combing signals in which the 17-unit D4Z4 array was followed by a 2-unit array, which confirmed the mosaic contraction of the duplication allele in mother. The molecular combing analysis in the son only revealed the shorter duplication allele of two units, in combination with an unremarkable chromosome 4 allele (Fig. 1C).
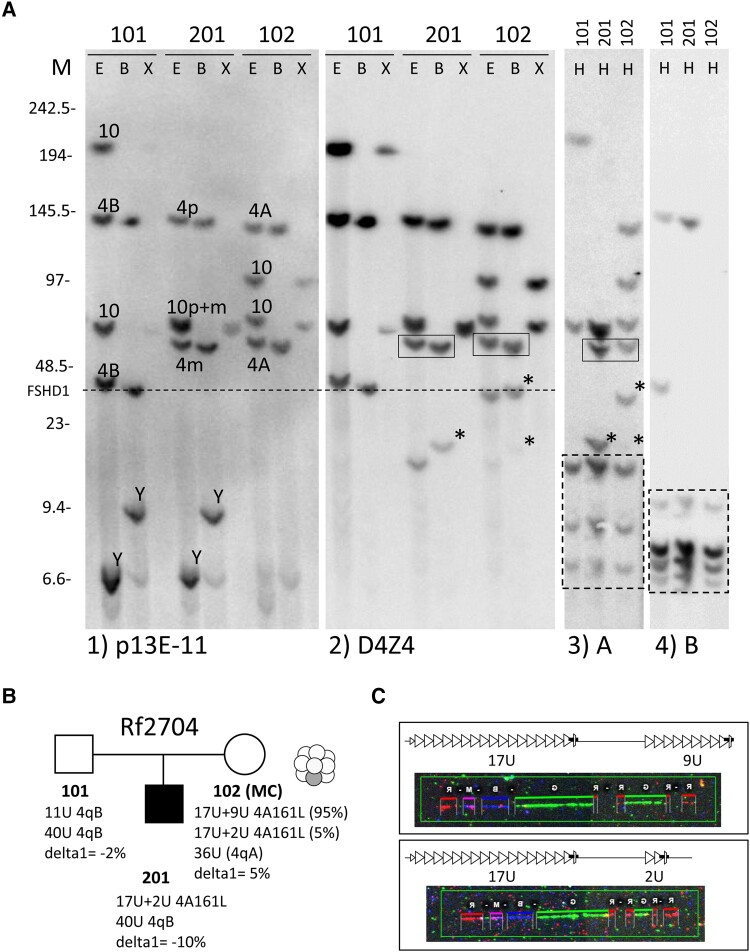
Figure 1.
Genetic analysis of Family Rf2704. (A) Southern blot (SB) analysis of Family Rf2704 showing father (Individual 101), mother (Individual 102) and affected son (Individual 201). Each sample is digested with restriction enzymes EcoRI and HindIII (E), EcoRI and BlnI (B) and XapI (X) and hybridized with probes p13E-11 (Blot 1) and D4Z4 (Blot 2) or digested with HindIII (H) and hybridized with probes A (Blot 3) and B (Blot 4). The identified chromosome 4 and 10 derived D4Z4 fragments and haplotypes are indicated on Blot 1. Initial hybridization with probe p13E-11 did not reveal an FSHD1-allele but subsequent D4Z4 hybridization (Blot 2) of the same blot revealed extra chromosome 4-like (BlnI-sensitive; XapI-insensitive) fragments in Individuals 102 and 201. The ‘A’ hybridization (Blot 3) showed that these extra fragments were of the 4qA haplotype (indicated with an asterisk on Blots 2 and 3) and co-inherited with the boxed 4qA fragment on Blots 2 and 3. This suggests the presence of an in cis duplication allele in Individual 201, which is inherited from Individual 102, in whom the allele is mosaic. The fragment size threshold for FSHD1 is indicated with a dotted line. Size markers (M) in kilobases are indicated on the left. Cross hybridizing chromosome Y fragments on Blot 1 are indicated (Y). Dotted boxed fragments on the bottom of Blots 3 and 4 are cross-hybridizing fragments unrelated to chromosomes 4qter and 10qter. (B) Summary of genotype and methylation analysis in Family Rf2704 (mosaicism in mother is illustrated by graphic). (C) Molecular combing (MC) analysis confirmed an in cis duplication allele with 17U+9U (Individual 102) and 17U+2U (Individual 201 and uncommonly in Individual 102) D4Z4 units.
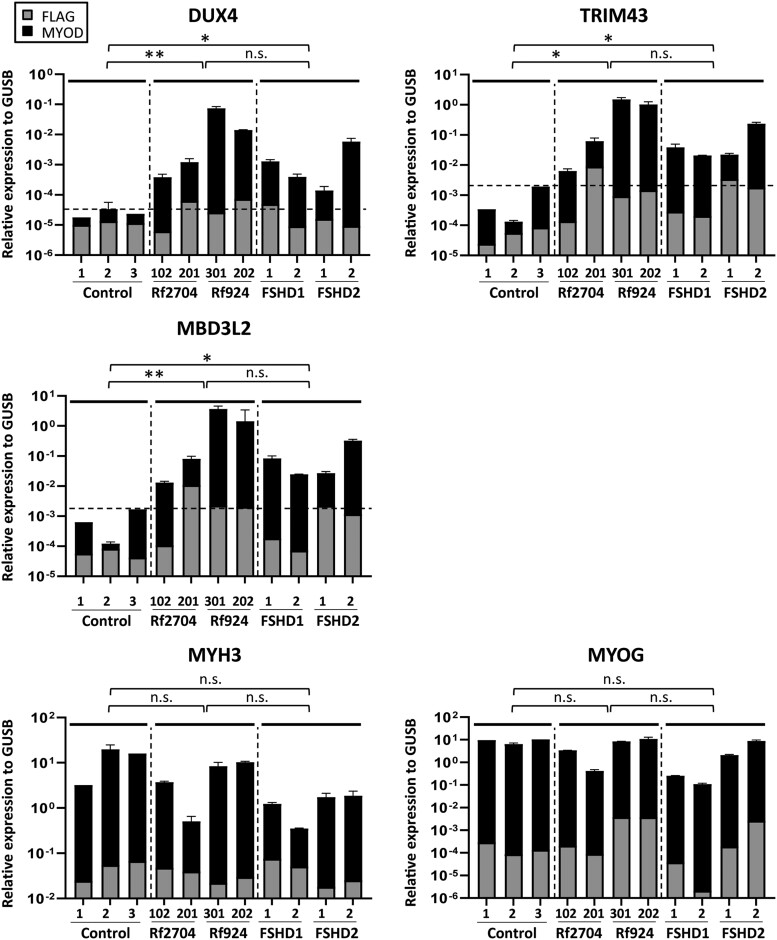
This finding suggests that the 17U+2U duplication allele is responsible for the clinical presentation of FSHD in the son, and that this allele causes a mild phenotype in the mother because of somatic mosaicism. To strengthen this interpretation, we performed DUX4 expression analysis after forced myogenesis by MYOD1 transduction in fibroblasts of mother and proband. In both samples, we found FSHD-level expression of DUX4 and its target genes, which was probably because of the mosaicism 10× lower in the mildly affected mother (Fig. 2). Methylation levels on the D4Z4 array were normal, excluding a modifying role for FSHD2 genes in this family. Collectively, this analysis confirmed FSHD1 by a de novo FSHD1-sized in cis duplication allele in both individuals.
Figure 2.
Expression analysis of Families Rf2704 and Rf924. To study DUX4 and DUX4 target gene expression, fibroblast of in cis duplication allele carriers, unaffected and disease controls (FSHD1 and FSHD2) were transduced with MyoD (black bar) in duplo or control FLAG (grey bar) in singular to enforce myogenic differentiation. MyoD transduced fibroblast of Individuals Rf2704.102 and 201 and of Individuals Rf924.202 and 301 showed FSHD-level expression of DUX4 and the target genes TRIM43 and MDB3L2, which is lower in the mosaic carriers of the duplication allele (Individuals Rf2704.102 and Rf924.202). The dotted line in graphs DUX4, TRIM43 and MDB3L2 shows the expression level in controls. Myogenic markers (MYH3 and MYOG) confirmed the myogenic conversion. Expression levels are in log scale and are normalized to GUSB. Data shown are means ± standard deviation. Groups of unaffected controls, standard FSHD controls (FSHD1 and FSHD2) and duplication allele carriers were compared using the Bonferroni multiple-comparison test. **P ≤ 0.01; *P ≤ 0.05. n.s. = not significant.
FSHD Family Rf924
The proband (Individual 301) of Family Rf924 was first seen in the hospital at the age of 9 years because of facial asymmetry. From the age of 13, she showed clinical weakness in elevation of both arms above the shoulders, scapula alata and bilateral mild weakness of the pectoralis major muscle. Her serum creatine kinase was slightly elevated (CK 468 U/l; normal range <123). On electromyography of upper and lower limb muscles, no obvious anomalies were noted. Light microscopy examination of a deltoid muscle biopsy revealed no abnormalities and common forms of limb girdle muscular dystrophy were excluded based on western blot analysis of muscle tissue. Re-examination at the age of 14 showed asymmetric weakness of the rhomboid muscle and a mild weakness of left ankle dorsiflexion, particularly of the left tibial anterior muscle. At the age of 16, the proband showed mild weakness of the left orbicularis oculi muscle, weakness of the left orbicularis oris muscle, scapula alata, weakness of abduction, adduction and forward flexion of both upper limbs, weakness of hamstring muscles and ankle dorsiflexion, and hypoactive deep tendon reflexes. At 22 years of age, she presented with a positive Beevor’s sign and at 27 years she reported difficulty in rising from a supine to a sitting position and difficulty in climbing stairs. She showed moderately pronounced Popeye arms, horizontal clavicles and a protruding abdomen with lumbar hyperlordosis. At 50 years of age, the mother (Individual 202) of the proband has no complaints of progressive muscle weakness. When specifically asked, she reports having difficulty blowing up a balloon and she cannot whistle. She has overexertion symptoms and reports unexplained falls. On physical examination however, there were no abnormalities. No signs of facial weakness, in particular she is able to bury her eyelashes, had a normal smile and symmetric pouting of her lips. No scapular winging and no Beevor’s sign were noted. MRC grading of all muscles is normal. The proband’s father and mother showed no signs of myopathy.
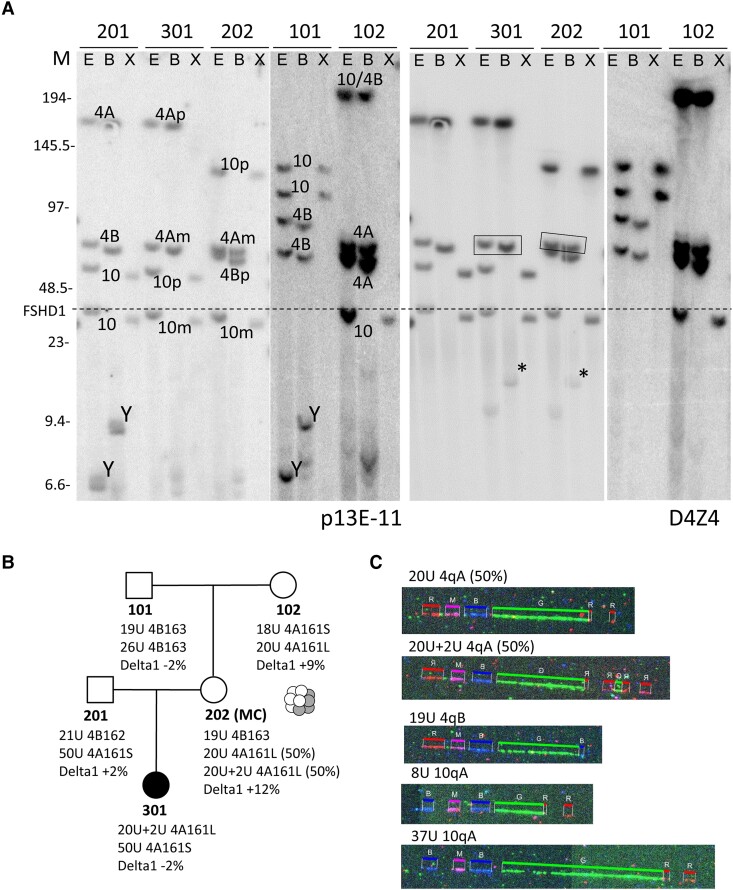
The patient was tested negative for FSHD1 using the regular linear gel electrophoresis in combination with Southern blotting and hybridization with probe p13E-11. More detailed PFGE with Southern blotting analysis identified a 4A161L-type duplication allele (20U+2U) in the proband, Subject 301, which was inherited from her unaffected mother (Fig. 3A and B). The mother (Individual 202) is mosaic for the 20U+2U duplication allele and the parental 20U allele (without duplicated array), both in 50% of her cells. The mosaicism explains why she is not affected and was confirmed by molecular combing in blood from Individual 202 (Fig. 3C). Most probably, the de novo duplication allele derived from the 20-unit 4A161L allele that she inherited from her mother (Individual 102). D4Z4 methylation levels were normal, excluding FSHD2. Testing MYOD1 transduced fibroblasts from Individuals 301 and 202 revealed expression of DUX4 and target genes in both individuals, confirming FSHD1 by an in cis duplication allele (Fig. 2).
Figure 3.
Genetic analysis Family Rf924. (A) Southern blot analysis of Family Rf924 consisting of grandfather (Individual 101), grandmother (Individual 102), father (Individual 201), mosaic mother (Individual 202) and affected daughter (Individual 301). Labelling as in Fig. 1, but now only showing the Southern blot with p13E-11 (left) and D4Z4 (right) hybridizations. The extra fragments (indicated with an asterisk) from the in cis duplication alleles in Individuals 202 and 301 are of the 4qA haplotype (Supplementary Fig. 3) and co-inherited with the boxed 4qA fragment. (B) Summary of genotype and methylation analysis in Family Rf924 (mosaicism in Individual 202 is illustrated by graphic). (C) Molecular combing (MC) analysis in Individual 202 showed 19U 4qB and mosaic alleles 20U 4qAL and 20U+2U 4qAL.
Additional duplication allele families
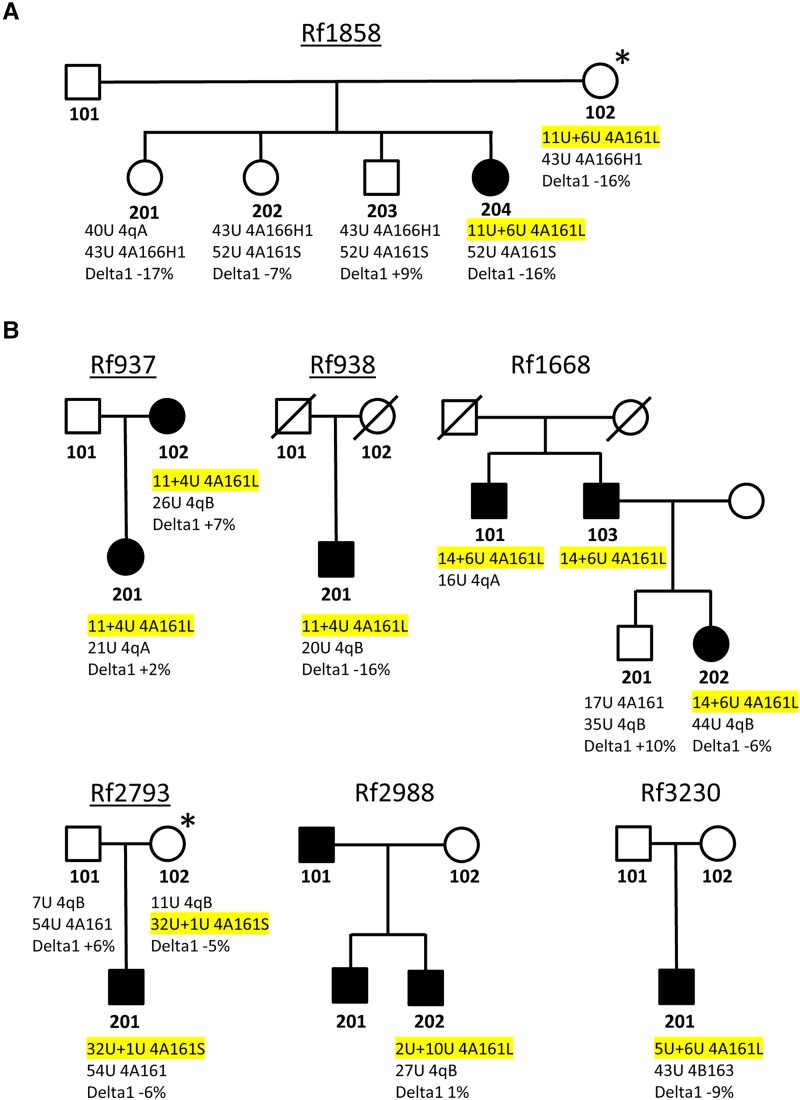
We identified seven other families in which FSHD is associated with an autosomal dominant in cis duplication allele with normal levels of FseI-D4Z4 methylation. The pedigrees and the genetic and methylation details are summarized in Fig. 4 and Supplementary Fig. 3 and the clinical details for the carriers of a duplication allele are described in the Supplementary material, ‘Clinical data’ section. For four of the seven families (Families Rf1858, Rf937, Rf938 and Rf2793), we obtained fibroblasts from the carriers of the in cis duplication allele and for all, we found expression of DUX4 and DUX4 target genes after MyoD transduction (Supplementary Fig. 4). For Family Rf1858, we also obtained fibroblasts from three siblings (Individuals 201, 202 and 203) of the proband, who do not have the duplication allele and did not show DUX4 and target gene expression after forced myogenesis by MyoD. The mothers of the proband in Families Rf1858 and Rf2793 are asymptomatic carrier of the in cis duplication allele. Thus, in total we identified nine families in which we found evidence of an autosomal dominant D4Z4 duplication allele.
Figure 4.
Pedigrees of other families with an autosomal dominant in cis duplication allele. (A) Pedigree of Family Rf1858 showing a summary of genotype and methylation analysis. For each individual both chromosome 4 D4Z4 arrays (size and haplotype) are indicated and the delta1 methylation score. The duplication allele is highlighted in yellow. The duplication allele was identified in the proband and her mother (Individuals 102 and 204), but only proband Individual 204 is affected. The proband and her mother (Individual 102) show a rather low delta1 methylation level, but not below the FSHD2 threshold. (B) Pedigree of Families Rf1668, Rf937, Rf938, Rf2793, Rf2988 and Rf3220 with similar description. For all underlined pedigrees, we analysed DUX4 and target gene expression for one or more family members (Supplementary Fig. 3). Clinically affected individuals are marked with a filled symbol and individuals marked with an asterisk are asymptomatic or non-penetrant cis duplication allele carriers.
Interestingly, in Families Rf2988 and Rf3230, we identified an in cis duplication allele with a composition that is opposite to all other in cis duplication alleles. Here, the most proximal array is shorter than the distal array (2U+10U for Family Rf2988 and 5U+6U for Family Rf3230). Initial standard Southern blot-based genetic testing for both cases with probe p13E-11 revealed a normal FSHD1 allele, but missed the presence of the distal in cis duplicated D4Z4 array. However, the D4Z4 hybridization revealed an extra 4qA fragment, which was explained by an in cis duplication at the FSHD1-sized allele and confirmed by molecular combing (Supplementary Fig. 3).
Composition of duplication alleles predicts pathogenicity
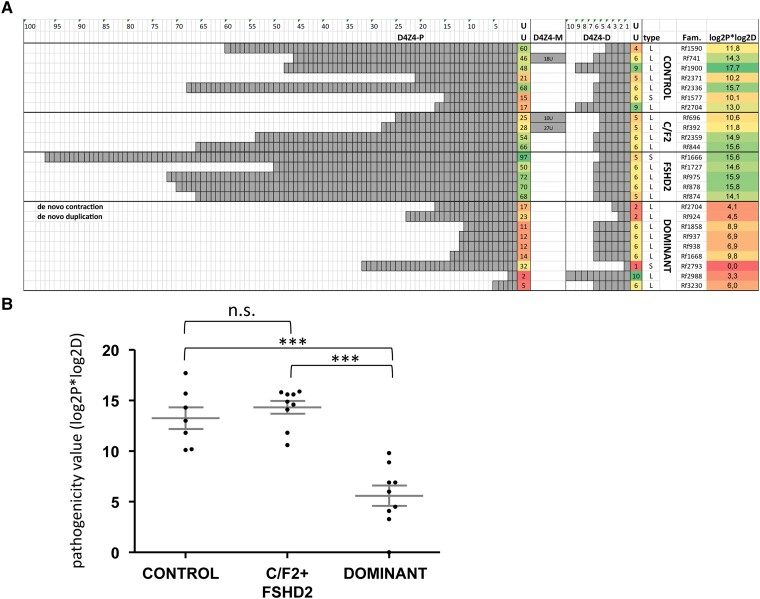
Based on the clinical description and expression of DUX4 and its target genes, all cases described here can be considered FSHD. For the patients in two of the nine families (Families Rf1858 and Rf938), we found reduced D4Z4 methylation suggestive of genetic modifiers that affect D4Z4 methylation. But since none of these delta1 methylation values are below the threshold for FSHD2 and mutation analysis of SMCHD1 was unremarkable, and our data suggest that some duplication alleles can be dominantly pathogenic, as in FSHD1. To explore the possible basis for this pathogenicity, we compared the composition of these dominantly pathogenic alleles (n = 9) with those found in control individuals (n = 7) and in FSHD2 families (i.e. only pathogenic when combined with a SMCHD1 variant; n = 9) (Fig. 5A). Three of these in cis duplication alleles were composed of three D4Z4 arrays, as previously shown by molecular combing.24 To compare, we calculated only with the size of the proximal D4Z4 array (p13E-11-linked) and the most distal D4Z4 array closest to the telomere. A quick inspection shows that the proximal D4Z4 array of the dominant pathogenic group is generally shorter than for the others. And for some autosomal dominant duplication alleles, the most distal array is very short. These observations suggest that specific combinations of proximal and distal repeat sizes might determine dominant pathogenicity. To predict the pathogenicity of duplication alleles, we devised a formula in which we multiplied the log2 values of the proximal and most distal D4Z4 array sizes. A log2 transformation of the D4Z4 array size was used as it was previously successfully applied to identify correlations between D4Z4 methylation and repeat array size and between clinical severity and repeat array sizes in FSHD2.13,38 The product showed to be ≤10 for autosomal dominant alleles and ≥10 or higher for duplication alleles identified in controls and in FSHD2 families (Fig. 5B). We observed a significant difference (P < 0.001) between the product found in autosomal dominant duplication alleles versus the other alleles.
Figure 5.
Composition of in cis duplication alleles predicts pathogenicity. (A) Representation of in cis duplication alleles identified in independent families, showing the repeat array size in units (U) of the most proximal (D4Z4-P) and most distal repeat array (D4Z4-D). In three families we identified an in cis duplication allele that is composed of three D4Z4 arrays. The size of this extra D4Z4 array is indicated in the middle column (D4Z4-M). We identified four groups of carriers: unaffected individuals in the population (‘Controls’), carriers in FSHD2 families (FSHD2 and C/F2, respectively) and carriers of an autosomal dominant in cis duplication allele (‘Dominant’). The in cis duplication alleles that were identified in FSHD2 patients and their unaffected family members are marked C/F2 and sporadic FSHD2 patients for whom we did not know if the duplication allele was non-pathogenic without an SMCHD1 mutation, are marked FSHD2. The haplotype (type) of the alleles is indicated (‘S’ for 4A161 and ‘L’ for 4A161L) as well as the family number (Fam.). The value in the last column is the product of the log2 values of the proximal and most distal D4Z4 arrays. Conditional formatting colours of this value indicate the pathogenicity of the duplication allele ranging from non-pathogenic (green) to pathogenic (red). (B) Comparison of the pathogenicity value (log2P*log2D) of the duplication alleles between the different groups shows a significant lower value (one-way ANOVA) for the autosomal dominant duplication alleles compared to those found in the other groups. ***P < 0.001. n.s. = not significant.
Nanopore sequencing analysis of samples with a duplication allele
Previously, we and others have showed that CpG methylation of the D4Z4 array is an important epigenetic marker in FSHD.13-16 Based on the significant differences in array sizes of in cis duplication alleles in controls, FSHD2 families and the dominant cases described here, we hypothesize that the methylation of the shortest D4Z4 array in the duplication allele is below the threshold of DUX4 repression in skeletal muscle. To explore the methylation status of duplication alleles and to obtain high resolution structural information about these alleles, we analysed two cis duplication families by nanopore sequencing.
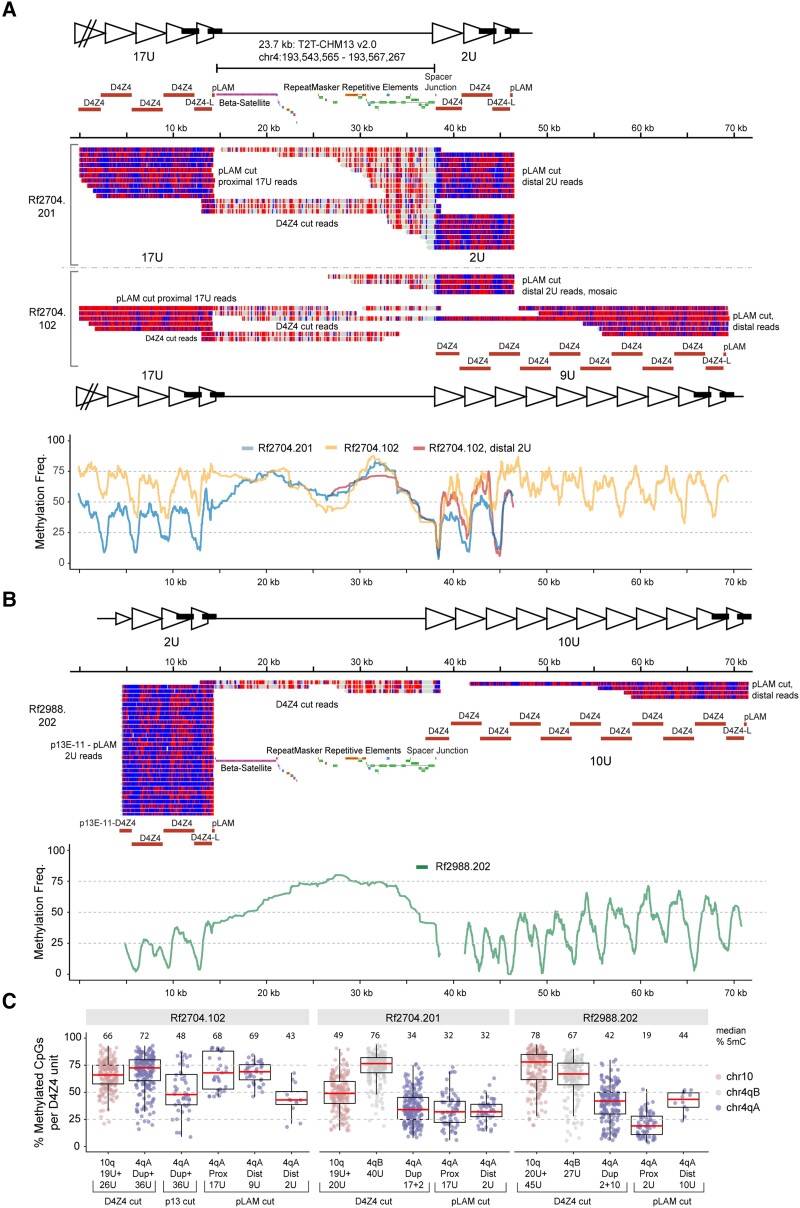
Using a recently developed Cas9 targeted nanopore sequencing technique,39 we determined the sequence and CpG methylation levels of the duplication alleles in Families Rf2704 and Rf2988. These families were selected as they represent the two major classes of D4Z4 repeat array duplications; the majority of cases have a cis duplication consisting of a normal-sized D4Z4 repeat array followed by a contracted D4Z4 repeat array (like in Family Rf2704), but some cases have a cis duplication defined by a contracted D4Z4 repeat array followed by normal-sized D4Z4 repeat array (like in Family Rf2704). We used a combination of guide RNAs to generate nanopore reads anchored to the p13E-11 site at the start of the proximal D4Z4 unit and the pLAM sites flanking the terminal 4qAL D4Z4 unit at both the proximal and distal D4Z4 array (Fig. 6A and B). We found nanopore reads that spanned the 2U arrays from Rf2704.201 (17U+2U) and the 9U distal array from Rf2704.102 (17U+9U). We also detected four distal 2U nanopore reads from Rf2704.102, which were consistent with the inferred maternal mosaicism. The elevated stoichiometry of distal 2U reads versus distal 9U reads in Rf2704.102 was likely due to technical size bias favouring shorter nanopore reads. For the proximal 17U D4Z4 array of the duplication allele in Rf2704 only partial reads were found. For the duplication allele in Rf2988.202 (2U+10U) we found nanopore reads that spanned the 2U arrays but only partial nanopore reads for the distal 10U array were found. The read depth and targeting efficiency from nanopore Cas9 targeted sequencing for all samples from Fig. 6 is summarized in Supplementary Table 1.
Figure 6.
Nanopore sequence analysis in Families Rf2704 and Rf2988. (A) Modbamtools single-read plots of nanopore reads mapped to the proximal D4Z4 arrays, the intervening spacer region and the distal D4Z4 arrays for mother (Individual 102) and son (Individual 201) from Family Rf2704. Reads are coloured by methylation state (5-methyl CpG = red; unmethylated CpG = blue; non-CpG = grey) with locations of the Cas9-guide RNA cut sites indicated. Proximal versus distal reads were mapped by size/sequence differences (Individual 201) or by sequence differences (Individual 102). Bottom: Methylation frequency plot (Savitzky-Golay smoothing filter, order = 2, length = 101) of Individual 102 versus Individual 201 including mosaic pLAM 2U reads from Individual 102. (B) Single-read and methylation frequency plot as in (A) from Individual Rf2988.202. (C) Box plots of D4Z4 methylation levels for all three individuals from A and B, summarized from 3.3 kb D4Z4 guide RNA cut (or single unit) reads (D4Z4 cut, 200 reads per box) or from p13E-11 or pLAM guide RNA cut reads (p13 or pLAM cut, summarized by D4Z4 region).
We also used a D4Z4 guide RNA. This guide RNAs mainly generated 3.3 kb, monomer-size D4Z4 reads, but also some flanking reads that bridged the spacer region between the duplicated arrays. These reads revealed a 23.7 kb spacer region identical to the 4qA sub-telomeric sequence normally found distal to the DUX4 polyadenylation signal. Examination of the distal spacer junction sequence revealed that the duplicated D4Z4 sequence in the second array started ∼295 base pairs distal to the original KpnI restriction recognition site. Interestingly, this spacer junction sequence was identical for both Families Rf2704 and Rf2988, implying that these unrelated families most probably harbour a shared ancestral 4qA in cis duplication allele.
Nanopore sequencing facilitates methylation analysis of the duplicated D4Z4 arrays and comparison of methylation levels within one repeat array and between both repeat arrays of an in cis duplication allele. The methylation plots in Fig. 6A show that the mosaic 17U+9U duplication allele in Individual Rf2704.102 has high methylation levels in both the proximal 17U array and the distal 9U array (yellow), while the 17U+2U duplication allele in Individual Rf2704.201 is highly reduced, both for the distal 2U array and the proximal 17U array (blue). Interestingly, the DNA methylation of the mosaic 17U+2U duplication allele in Individual Rf2704.102 for the distal 2U D4Z4 array (red) is higher compared to this 2U array in Individual Rf2704.201 (blue). In Family Rf2988, for the 2U+10U duplication allele, we detected very low methylation in the proximal 2U array, and also low, but increasing methylation in the distal 10U array (Fig. 6B). We summarized the D4Z4 methylation levels for all three individuals as the percentage of methylated CpGs per 3.3 kb D4Z4 read or repeat unit. It was possible to distinguish 4qB from 4qA repeat units using the 3.3 kb D4Z4 cut reads, which revealed high 4qB methylation of 76% and 67% (median level per D4Z4 unit) for both Rf2704.201 40U 4qB and Rf2988.202 27U 4qB, respectively (Fig. 6C). The aggregate Rf2704.201 17U+2U 4qA methylation was 34% median level, estimated from 4qA D4Z4 cut reads, similar to the 32% median levels seen from D4Z4 regions extracted from both proximal 17U and distal 2U pLAM reads. In contrast, the Rf2988.202 proximal 2U D4Z4 units had a 19% median level versus 44% from distal 10U units (Fig. 6C), agreeing with the aggregate range seen with Rf2988.202 4qA D4Z4 cut reads. The methylation level of the shortest, proximal D4Z4 repeat array in the duplication allele is comparable to the levels found for standard FSHD1 alleles. Thus, we observe a repeat size-dependent methylation level, with the lowest methylation in the 2U arrays for all three duplication alleles.
Discussion
In cis duplications of the D4Z4 repeat array on 4qA chromosomes were previously reported, but the contribution of these alleles to FSHD is unclear. Here we studied the genetic and clinical characteristics of FSHD patients from nine in cis duplication families and showed that duplication alleles with a specific composition can cause FSHD without the requirement of pathogenic variants in D4Z4 chromatin repressors such as SMCHD1. The most striking examples are two families in which we observed a de novo formation of the duplication allele (Family Rf924) or a de novo contraction of the distal D4Z4 array to pathogenic proportions (Family Rf2704), respectively. In six of the nine duplication families we confirmed FSHD by showing DUX4 and DUX4 target expression after myogenic transdifferentiation. We identified several asymptomatic or non-penetrant carriers of the in cis duplication allele, typical for FSHD. Reduced penetrance was also observed for the proband’s mother in Families Rf924, Rf1858 and Rf2793.
Despite a clinical diagnosis of FSHD, the probands of seven of the nine families tested negative for FSHD1 by standard Southern blot-based genetic analysis with probe p13E-11 and they also tested negative for FSHD2 based on D4Z4 methylation analysis. This emphasizes the challenge of genetically confirming a clinical diagnosis of FSHD in D4Z4 duplication carriers. In addition to the routinely used diagnostic Southern blot method, alternative methods were developed including PFGE-based Southern blotting, molecular combing, optical genome mapping (OGM) and long read (nanopore) sequencing.37,39,42,43 Extensive PFGE and Southern blot-based genetic analysis using additional probes hybridizing to the FSHD locus revealed the presence of a duplication allele in each case. Molecular combing can also identify duplication alleles and was used here for confirming the composition of the 4qA duplication allele after Southern blot-based identification, especially for assigning the duplicated 4qA-type fragment to one of the two chromosome 4 repeat arrays if both were of the 4qA type. However, segregation analysis of duplication alleles, or in cases where the chromosome 4 homologue is of the 4qB type, the composition of the duplication alleles can be deduced directly from Southern blot analysis without additional molecular combing analysis.
None of the affected carriers of an in cis duplication allele reported here had D4Z4 methylation levels below the FSHD2 threshold measured on chromosomes 4 and 10, using the methylation sensitive restriction enzyme assay (FseI) corrected for D4Z4 array size (delta1 value).13 This assay is highly discriminative between FSHD2 and controls, or FSHD1, indicating that for all these cases the duplication allele is dominantly causing FSHD, like in FSHD1. However, this methylation assay and bisulphite sequencing-based methylation assays cannot study the methylation of individual D4Z4 arrays of these in cis duplication alleles and therefore we applied nanopore sequencing. Nanopore sequencing revealed a clear reduction of methylation in the shortest, proximal 2U array in the 2U+10U duplication allele and distal 2U array in the 17U+2U duplication allele, comparable to levels found in standard FSHD1 alleles. The distal 10U array in the 2U+10U duplication allele showed intermediate methylation levels consistent with its repeat array number. Unexpectedly, we observed that the proximal 17U array of the 17U+2U duplication allele showed a methylation profile comparable to the distal 2U allele and lower than the 17U array of the 17U+9U duplication allele. This suggests that DUX4 may be expressed from both the proximal and distal 4qAL repeat arrays in the 17U+2U allele. The effect of the distal 9U array on the methylation level of the proximal 17U arrays may suggest a directional spreading of D4Z4 methylation although more duplication alleles need to be sequenced to achieve a full understanding of these methylation profiles.
Our data suggest that the size of both the proximal array and the distal array in a duplication allele plays a role in their pathogenicity. We designed a formula in which we amplified the log2 value of the proximal D4Z4 array with that of the most distal D4Z4 array and the product of the dominant duplication alleles seems significantly lower than that of control and FSHD2 duplication alleles. The formula can also be applied to the reverted duplication allele that we identified. This suggests that the pathogenicity of duplication alleles can be predicted using this formula.
Interestingly, we observed an unexpected preponderance (88%) of 4A161-L type duplication alleles, while 4A161L alleles are only found in 20% of the 4qA alleles and are specific for the European population.38 This observation might indicate that 4A161L alleles are more susceptible to rearrangements. Interestingly, recently two FSHD-causing de novo translocations of chromosome 4qA to chromosome 10 have been described, and also in these cases a 4A161L allele was involved.32 This finding also offers possibilities in genetic diagnosis of duplication alleles in families because the 4A161L-type FSHD allele will be unique in most cases, it can be detected by a simple PCR and is therefore easy to also follow in prenatal diagnosis.
Although most of the duplication alleles were haplotype 4qA-L, we also identified three 4qA haplotype in cis duplication alleles (Fig. 5). Furthermore, the duplication allele identified in patient’s mother Individual Rf924.202 seems to originate from a de novo duplication event of the standard 4A161L allele. On the other hand, by nanopore sequencing of duplication alleles in Families Rf2704 and Rf2988, we identified the same spacer regions and breakpoint sequence. Subsequent analysis of molecular combing data for 16 duplication alleles showed that the spacer region between the cis duplicated D4Z4 arrays ranges from 22 to 28 kb (Supplementary Fig. 5). These findings suggest that although duplication alleles can be formed de novo, some are most probably derived from the same ancient founder.
The clinical description of these individuals combined with the methylation and transcriptional observations and the two de novo rearrangements identified strengthen the interpretation that duplication alleles in the absence of FSHD2 mutations can cause FSHD. Genetic laboratories should be aware of these rare autosomal dominant duplication alleles, which can be easily missed in routine diagnostics.
Supplementary Material
Acknowledgements
We thank all FSHD families for participating in our studies. We are grateful to the platform for immortalization of human cells from the Institut de Myologie, Paris, France, who provided the cells for Family Rf2793. Jan de Bleecker, Corrie Erasmus, Baziel van Engelen, Teresinha Evangelista, Peter van den Bergh, Nicol Voermans, John Vissing and Silvère van der Maarel are members of the European Reference Network for Rare Neuromuscular Diseases (ERN EURO-NMD).
Contributor Information
Richard J L F Lemmers, Department of Human Genetics, Leiden University Medical Center, 2300 RC, Leiden, The Netherlands.
Russell Butterfield, Department of Pediatrics, University of Utah, Salt Lake City, UT 84112, USA.
Patrick J van der Vliet, Department of Human Genetics, Leiden University Medical Center, 2300 RC, Leiden, The Netherlands.
Jan L de Bleecker, Department of Neurology, University Hospital, 9000 Gent, Belgium.
Ludo van der Pol, University Medical Center Utrecht, 3584 EA, Utrecht, The Netherlands.
Diane M Dunn, Department of Human Genetics, University of Utah, Salt Lake City, UT 84112, USA.
Corrie E Erasmus, Neuromuscular Centre Nijmegen, Radboud University Nijmegen Medical Centre, 6525 GA, Nijmegen, The Netherlands.
Marc D'Hooghe, Department of Neurology, Algemeen Ziekenhuis Sint-Jan, 8000, Brugge, Belgium.
Kristof Verhoeven, Department of Neurology, Algemeen Ziekenhuis Sint-Jan, 8000, Brugge, Belgium.
Judit Balog, Department of Human Genetics, Leiden University Medical Center, 2300 RC, Leiden, The Netherlands.
Anne Bigot, Sorbonne Université, Inserm UMRS974, Institut de Myologie, Centre de Recherche en Myologie, F-75013 Paris, France.
Baziel van Engelen, Neuromuscular Centre Nijmegen, Radboud University Nijmegen Medical Centre, 6525 GA, Nijmegen, The Netherlands.
Jeffrey Statland, University of Kansas Medical Center, Kansas City, KS 66103, USA.
Enrico Bugiardini, National Hospital For Neurology and Neurosurgery, UCL Queen Square Institute of Neurology, London, WC1N 3BG, UK.
Nienke van der Stoep, Department of Clinical Genetics, Leiden University Medical Center, 2300 RC, Leiden, The Netherlands.
Teresinha Evangelista, Unité de Morphologie Neuromusculaire, Institut de Myologie, AP-HP, F-75013, Paris, France.
Chiara Marini-Bettolo, The John Walton Muscular Dystrophy Research Centre, Faculty of Medical Sciences, Newcastle upon Tyne, NE1 3BZ, UK.
Peter van den Bergh, Department of Neurology, Saint-Luc UCL, 1200, Brussels, Belgium.
Rabi Tawil, Department of Neurology, University of Rochester Medical Center, NY 14642, Rochester, USA.
Nicol C Voermans, Neuromuscular Centre Nijmegen, Radboud University Nijmegen Medical Centre, 6525 GA, Nijmegen, The Netherlands.
John Vissing, Department of Neurology, University of Copenhagen, DK-2100 Copenhagen, Denmark.
Robert B Weiss, Department of Human Genetics, University of Utah, Salt Lake City, UT 84112, USA.
Silvère M van der Maarel, Department of Human Genetics, Leiden University Medical Center, 2300 RC, Leiden, The Netherlands.
Data availability
Nanopore sequencing data are available through the EGA.
Funding
Funding for the nanopore sequencing was supported by the National Institutes of Health (NICHD, P50 HD060848) and the FSHD Society. This study was in part supported by a Medical Research Council UK strategic award to establish an International Centre for Genomic Medicine in Neuromuscular Diseases (ICGNMD) MR/S005021/1.
Competing interests
S.M.v.d.M. declares that he has acted as consultant and/or is a member of the advisory board for Avidity Biosciences, Dyne Therapeutics and Fulcrum Therapeutics and is a Board member for Renogenyx. R.J.L.F.L and S.M.v.d.M are co-inventors on FSHD patent applications. The other authors declare that they have no conflict of interest.
Supplementary material
Supplementary material is available at Brain online.
References
- 1. Padberg GWAM. Facioscapulohumeral disease. PhD thesis. Faculty of Medicine (LUMC), Leiden University; 1982. [Google Scholar]
- 2. Mul K, Lassche S, Voermans NC, Padberg GW, Horlings CG, van Engelen BG. What’s in a name? The clinical features of facioscapulohumeral muscular dystrophy. Pract Neurol. 2016;16:201–207. [DOI] [PubMed] [Google Scholar]
- 3. Kowaljow V, Marcowycz A, Ansseau E, et al. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord. 2007;17:611–623. [DOI] [PubMed] [Google Scholar]
- 4. Geng LN, Yao Z, Snider L, et al. DUX4 Activates germline genes, retroelements, and immune mediators: Implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rickard AM, Petek LM, Miller DG. Endogenous DUX4 expression in FSHD myotubes is sufficient to cause cell death and disrupts RNA splicing and cell migration pathways. Hum Mol Genet. 2015;24:5901–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hendrickson PG, Dorais JA, Grow EJ, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49:925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49:935–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gabriels J, Beckers MC, Ding H, et al. Nucleotide sequence of the partially deleted D4Z4 locus in a patient with FSHD identifies a putative gene within each 3.3 kb element. Gene. 1999;236:25–32. [DOI] [PubMed] [Google Scholar]
- 10. Lemmers RJ, Wohlgemuth M, van der Gaag KJ, et al. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:884–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dixit M, Ansseau E, Tassin A, et al. DUX4, A candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci U S A. 2007;104:18157–18162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scionti I, Fabbri G, Fiorillo C, et al. Facioscapulohumeral muscular dystrophy: New insights from compound heterozygotes and implication for prenatal genetic counselling. J Med Genet. 2012;49:171–178. [DOI] [PubMed] [Google Scholar]
- 13. Lemmers RJ, Goeman JJ, van der Vliet PJ, et al. Inter-individual differences in CpG methylation at D4Z4 correlate with clinical variability in FSHD1 and FSHD2. Hum Mol Genet. 2015;24:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Calandra P, Cascino I, Lemmers RJ, et al. Allele-specific DNA hypomethylation characterises FSHD1 and FSHD2. J Med Genet. 2016;53:348–355. [DOI] [PubMed] [Google Scholar]
- 15. de Greef JC, Lemmers RJ, van Engelen BG, et al. Common epigenetic changes of D4Z4 in contraction-dependent and contraction-independent FSHD. Hum Mutat. 2009;30:1449–1459. [DOI] [PubMed] [Google Scholar]
- 16. Jones TI, King OD, Himeda CL, et al. Individual epigenetic status of the pathogenic D4Z4 macrosatellite correlates with disease in facioscapulohumeral muscular dystrophy. Clin Epigenetics. 2015;7:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lemmers RJ, de Kievit P, Sandkuijl L, et al. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat Genet. 2002;32:235–236. [DOI] [PubMed] [Google Scholar]
- 18. Lemmers RJ, van der Vliet PJ, Klooster R, et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wijmenga C, Hewitt JE, Sandkuijl LA, et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat Genet. 1992;2:26–30. [DOI] [PubMed] [Google Scholar]
- 20. Lemmers RJ, Tawil R, Petek LM, et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet. 2012;44:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van den Boogaard ML, Lemmers RJLF, Balog J, et al. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet. 2016;98:1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamanaka K, Šikrová D, Mitsuhashi S, et al. Homozygous nonsense variant in LRIF1 associated with facioscapulohumeral muscular dystrophy. Neurology. 2020;94:e2441–e2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lunt PW, Jardine PE, Koch MC, et al. Correlation between fragment size at D4F104S1 and age at onset or at wheelchair use, with a possible generational effect, accounts for much phenotypic variation in 4q35-facioscapulohumeral muscular dystrophy (FSHD). Hum Mol Genet. 1995;4:951–958. [DOI] [PubMed] [Google Scholar]
- 24. Lemmers RJLF, van der Vliet PJ, Vreijling JP, et al. Cis D4Z4 repeat duplications associated with facioscapulohumeral muscular dystrophy type 2. Hum Mol Genet. 2018;27:3488–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Goselink RJM, Mul K, van Kernebeek CR, et al. Early onset as a marker for disease severity in facioscapulohumeral muscular dystrophy. Neurology. 2019;92:e378–e385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sacconi S, Briand-Suleau A, Gros M, et al. FSHD1 And FSHD2 form a disease continuum. Neurology. 2019;92:e2273–e2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sacconi S, Lemmers RJ, Balog J, et al. The FSHD2 gene SMCHD1 is a modifier of disease severity in families affected by FSHD1. Am J Hum Genet. 2013;93:744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemmers RJ, Van Overveld PG, Sandkuijl LA, et al. Mechanism and timing of mitotic rearrangements in the subtelomeric D4Z4 repeat involved in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2004;75:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zatz M, Marie SK, Passos-Bueno MR, et al. High proportion of new mutations and possible anticipation in Brazilian facioscapulohumeral muscular dystrophy families. Am J Hum Genet. 1995;56:99–105. [PMC free article] [PubMed] [Google Scholar]
- 30. Tonini MM, Lemmers RJ, Pavanello RC, et al. Equal proportions of affected cells in muscle and blood of a mosaic carrier of facioscapulohumeral muscular dystrophy. Hum Genet. 2006;119(1–2):23–28. [DOI] [PubMed] [Google Scholar]
- 31. Lemmers RJ, van der Vliet PJ, van der Gaag KJ, et al. Worldwide population analysis of the 4q and 10q subtelomeres identifies only four discrete interchromosomal sequence transfers in human evolution. Am J Hum Genet. 2010;86:364–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lemmers RJLF, van der Vliet PJ, Blatnik A, et al. Chromosome 10q-linked FSHD identifies DUX4 as principal disease gene. J Med Genet. 2021;59:180–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemmers RJLF, van der Vliet PJ, Granado DSL, et al. High-resolution breakpoint junction mapping of proximally extended D4Z4 deletions in FSHD1 reveals evidence for a founder effect. Hum Mol Genet. 2022;31:748–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nguyen K, Puppo F, Roche S, et al. Molecular combing reveals complex 4q35 rearrangements in facioscapulohumeral dystrophy. Hum Mutat. 2017;38:1432–1441. [DOI] [PubMed] [Google Scholar]
- 35. Ricci E, Galluzzi G, Deidda G, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann Neurol. 1999;45:751–757. [DOI] [PubMed] [Google Scholar]
- 36. van Overveld PG, Enthoven L, Ricci E, et al. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann Neurol. 2005;58:569–576. [DOI] [PubMed] [Google Scholar]
- 37. Lemmers RJ. Analyzing copy number variation using pulsed-field gel electrophoresis: Providing a genetic diagnosis for FSHD1. Methods Mol Biol. 2017;1492:107–125. [DOI] [PubMed] [Google Scholar]
- 38. Lemmers RJ, van der Vliet PJ, Balog J, et al. Deep characterization of a common D4Z4 variant identifies biallelic DUX4 expression as a modifier for disease penetrance in FSHD2. Eur J Hum Genet. 2018;26:94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Butterfield RJ, Dunn DM, Duvall B, Moldt S, Weiss RB. Deciphering D4Z4 CpG methylation gradients in fascioscapulohumeral muscular dystrophy using nanopore sequencing. bioRxiv [Preprint]. 10.1101/2023.02.17.528868 [DOI] [PMC free article] [PubMed]
- 40.Razaghi R, Hook PW, Ou S, et al. Modbamtools: Analysis of single-molecule epigenetic data for long-range profiling, heterogeneity, and clustering. bioRxiv [Preprint]. [DOI]
- 41. Yao Z, Fong AP, Cao Y, Ruzzo WL, Gentleman RC, Tapscott SJ. Comparison of endogenous and overexpressed MyoD shows enhanced binding of physiologically bound sites. Skelet Muscle. 2013;3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dai Y, Li P, Wang Z, et al. Single-molecule optical mapping enables quantitative measurement of D4Z4 repeats in facioscapulohumeral muscular dystrophy (FSHD). J Med Genet. 2020;57:109–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nguyen K, Walrafen P, Bernard R, et al. Molecular combing reveals allelic combinations in facioscapulohumeral dystrophy. Ann Neurol. 2011;70:627–633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nanopore sequencing data are available through the EGA.