Abstract
Background
PTEN is a tumour suppressor gene and well-known for being frequently mutated in several cancer types. Loss of immunogenicity can also be attributed to PTEN loss, because of its role in establishing the tumour microenvironment. Therefore, this study aimed to represent the link between PTEN and cGAS-STING activity, a key mediator of inflammation, in tumour samples of glioblastoma patients.
Methods
Tumour samples of 36 glioblastoma patients were collected. After DNA isolation, all coding regions of PTEN were sequenced and analysed. PTEN expression status was also evaluated by qRT-PCR, western blot, and immunohistochemical methods. Interferon-stimulated gene expressions, cGAMP activity, CD8 infiltration, and Granzyme B expression levels were determined especially for the evaluation of cGAS-STING activity and immunogenicity.
Results
Mutant PTEN patients had significantly lower PTEN expression, both at mRNA and protein levels. Decreased STING, IRF3, NF-KB1, and RELA mRNA expressions were also found in patients with mutant PTEN. Immunohistochemistry staining of PTEN displayed expressional loss in 38.1% of the patients. Besides, patients with PTEN loss had considerably lower amounts of IFNB and IFIT2 mRNA expressions. Furthermore, CD8 infiltration, cGAMP, and Granzyme B levels were reduced in the PTEN loss group.
Conclusion
This study reveals the immunosuppressive effects of PTEN loss in glioblastoma tumours via the cGAS-STING pathway. Therefore, determining the PTEN status in tumours is of great importance, like in situations when considering the treatment of glioblastoma patients with immunotherapeutic agents.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11060-023-04556-4.
Keywords: Glioblastoma, PTEN, cGAS-STING pathway, Interferon response, Immunogenicity
Introduction
Glioblastoma is the most common and aggressive brain tumour that frequently displays the loss of phosphatase and tensin homologue (PTEN) activity, both in primary and recurrent tumour types [1]. PTEN has phosphatase activities on target protein as well as on target lipids, thus regulating a wide range of cellular functions from cell death to motion [2]. Loss of PTEN activity can be caused by gene mutations, epigenetic regulation, or post-transcriptional and -translational regulations. PTEN loss was reported in 55.9% of gastric cancer cases with PTEN mutations [3]. PTEN mutations have a negative impact on survival and are reported around 40% and 33% in all glioblastoma and IDH wild type glioblastoma cases, respectively [4, 5]. Mutations of genes that regulate oncogenic signalling pathways are associated with immunosuppressive tumour microenvironment [6–8]. GBM studies have shown that programmed death ligand-1 (PD-L1) expression increases after PTEN loss [9] and PTEN is significantly mutated in anti-PD-1 immunotherapy non-responsive tumours [10].
cGAS and STING are cytoplasmic DNA sensors and key regulatory proteins triggering innate immune response [11]. After activation and transmission of their signal to the nucleus via the TANK-binding kinase 1 (TBK1) and Interferon regulatory factor 3 (IRF3) proteins, transcription of type I interferon (IFN) and interferon-stimulated genes (ISGs) are activated, which in turn regulate many innate and adaptive immune system cells. The increase of cytosolic chromatin fragments and micronuclei along with replicative stress during malignant transformation increases the possibility of DNA leakage in cancer cells and thus activates the cGAS-STING pathway [12].
It has been shown that PTEN has protein phosphatase activity on IRF3, which is the transcription factor of type I IFN. Therefore, PTEN provides a direct link between tumour suppression and antiviral innate immunity [13]. If PTEN displays antitumor activity, in part through the stimulation of type I IFN response, its loss might disrupt the cGAS-STING pathway and contribute to the immunosuppressive microenvironment of GBM tumours. Based on this hypothesis, this study aimed to explore the effects of PTEN mutations and expressional status on the cGAS-STING activity in glioblastoma tumours.
Methods
Patients and tumour samples
Our study protocol was approved by the Institutional Clinical Research Ethics Committee (2018, 18-12.1T/17). Written informed consent was obtained prior to enrolment. Patients diagnosed with primary or recurrent GBM were included into this study. GBM samples were collected during surgical intervention and processed for immunohistochemistry and molecular analysis. All tissue samples were classified based on the WHO classification [14] and IDH-positive samples were excluded from the study. A tissue aliquot was used for DNA, RNA, and protein purification; whereas, another aliquot was fixed in a 4% paraformaldehyde solution. Since the Neurosurgery Department of Ege University Faculty of Medicine constitutes the reference clinic of West Turkey, patients were preferentially transferred to other health care centres for further adjuvant treatment and follow-ups, after early follow-up CT scans were taken.
Next generation sequencing
DNA was isolated from fresh GBM tissue samples with the QIAamp DNA Isolation Kit (Qiagen). All coding sequences of PTEN were amplified with 10x specific primer sets. The Nextera DNA Library Preparation Kit (Illumina) was used for library preparation from pooled PCR amplicons and sequenced with the NextSeq 550 System (Illumina). Quality control and evaluation of gene sequencing results were performed by use of appropriate programs as previously described [15].
Quantitative RT-PCR
Total RNA was isolated from fresh GBM tissue samples with the RNeasy Kit (Qiagen) and 1 µg was transcribed to cDNA by using the iScript Reverse Transcription Supermix Kit (Bio-Rad). qRT-PCR was performed for the cGAS-STING pathway members and interferon-regulated genes using the primer pairs below (5’→3’) and iTaq Universal SYBR Green Supermix (Bio-Rad) on the LightCycler 480 Instrument (Roche). The relative target gene mRNA expression level was normalized to the GAPDH housekeeping gene by using the ΔΔCt method.
GAPDH (F: CATTGCCCTCAACGACCACTTT; R: GGTGGTCCAGGGGTCTTACTCC)
GZMB (F: CTTCCTGATACGAGACGACTTC; R: CGGCTCCTGTTCTTTGATATTG)
IFIT2 (F: GCGTGAAGAAGGTGAAGAGG; R: GCAGGTAGGCATTGTTTGGT)
IFI44 (F: GATGTGAGCCTGTGAGGTCC; R: CTTTACAGGGTCCAGCTCCC).
IFNB1 (F: CAGCATCTGCTGGTTGAAGA; R: CATTACCTGAAGGCCAAGGA)
IL6 (F: AGACAGCCACTCACCTCTTCAG; R: TTCTGCCAGTGCCTCTTTGCTG)
IRF3 (F: AGAGGCTCGTGATGTGGTCAAG; R: AGGTCCACAGTATTCTCCAGG).
ISG15 (F: CAGCCATGGGCTGGGAC; R: GCCGATCTTCTGGGTGATCT)
NF-KB1 (F: GAAGCACGAATGACAAGAGGC; R: GCTTGGCGGATTAGCTCTTTT)
PTEN (F: CCACAAACAGAACAAGATGCT; R: GCTCTATACTGCAAATGCTATCG)
RELA (F: TCACCCCCACGAGCTTGTA; R: TTGTTGTTGGTCTGGATGCG).
STING (F: GCAGTGTGTGAAAAAGGGAAT; R: AGGTCCACAGTATTCTCCAGG)
Western blot
Fifty mg of fresh GBM tissue sample was lysed in RIPA buffer, before 14 µg of the protein lysate was loaded onto a 12% SDS-PAGE gel for separation. PTEN (ProteinTech, 1:2000), STING (D2P2F; Cell Signaling, 1:1000), IRF3 (D6I4C; Cell Signaling, 1:1000), NF-KB (L8F6; Cell Signaling, 1:1000), Phospho-NF-KB (Ser536; 93H1; Cell Signaling, 1:1000), Beta-Actin (Cell Signaling, 1:1000) primary antibodies; and, Anti-Rabbit IgG (Cell Signaling, 1:1000) and Anti-Mouse IgG (Cell Signaling, 1:1000) secondary antibodies were used. The Clarity Western ECL Substrate (Bio-Rad) and C-DiGit Blot Scanner (Licor) were used for the detection of ECL signals.
cGAMP activation
The 2’3’-cGAMP ELISA Kit was used according to the manufacturer’s instructions (Cayman Chemical). Protein lysates were prepared using RIPA buffer (Thermo Scientific) and Protease inhibitors (Thermo Scientific). In the experiments, 30 µg of protein lysates were used and all samples were run in duplicates.
Immunohistochemistry
Five µm sections were prepared from formalin-fixed and paraffin-embedded GBM tissue samples, mounted on poly-L-lysine-coated slides, and processed according to a standard immunohistochemical (IHC) procedure as previously described [16]. Slides were incubated with PTEN (ProteinTech, 1:200) or CD8 (Clone 1A5, Novocastra, 1:30) primary antibodies for 32 min at RT. All slides were reviewed by two pathologists (TA and YE). Immunohistochemical expression patterns of PTEN were interpreted in accordance with the literature [17]. CD8 density was evaluated in 3 grades according to the distribution density within the tumour. The presence of CD8 + lymphocyte clusters, i.e. ≥ 20 CD8 + lymphocytes in one HPF within the tumour was evaluated as +++. The presence of ≥ 20 CD8 + lymphocytes per mm2 (⁓5 HPF) in the densest area of the tumour was evaluated as ++. If the presence of CD8 + lymphocytes per mm2 (⁓5 HPF) in the densest area of the tumour was < 20, it was evaluated as +.
Bioinformatic analysis
The Cancer Genome Atlas (TCGA) database was used to evaluate PTEN gene expression in a total of 162 samples, all selected from Glioblastoma Multiforme and containing PTEN expression data (TCGA, Firehose Legacy) [18, 19]. Samples were split into two groups based on median PTEN expression (high vs. low). The p-value derived from Student’s t-test and q-value from Benjamini-Hochberg procedure. Tumor IMmune Estimation Resource (TIMER), on the other side, is a comprehensive resource that systematically analyses the infiltration of various immune cells and their clinical impact across a spectrum of cancer types [20]. In this study, it was used for the analyses of PTEN expression in correlation with immune infiltrating cells, including CD8 + and CD4 + T cells, by the purity-corrected partial Spearman method. p-values < 0.05 were considered statistically significant.
Statistical analysis
Statistical analyses were performed using SPSS for Windows version 26.0 (SPSS, Chicago, IL, USA). Normality was evaluated using Kolmogorov-Smirnov and Shapiro-Wilk tests. Non-parametric tests Mann-Whitney, Kruskal-Wallis, and ANOVA were used to compare distributions of variables. A p-value of < 0.05 (two-sided) was considered as statistically significant.
Results
Study group
From February 2019 to July 2022, a total of 36 patients diagnosed with grade IV GBM were recruited for this study. Tissue samples were obtained from 23 primary and 13 recurrent tumours. Altogether, the study group consisted of 13 female and 23 male GBM patients; more patient demographics and baseline characteristics are given in Table 1. Baseline characteristics of patients between the wild-type and mutant PTEN genotypes were similar.
Table 1.
Patient demographics and baseline characteristics
| Characteristic | Total | PTEN WT |
PTEN Mutant |
|
|---|---|---|---|---|
|
Age, years, no (%) |
≤ 50 | 8 (22.2%) | 8 (50.0%) | 0 |
| > 50 | 28 (77.8%) | 8 (50.0%) | 20 (100%) | |
|
Gender, no (%) |
Female | 13 (36.1%) | 6 (37.5%) | 7 (35.0%) |
| Male | 23 (63.9%) | 10 (62.5%) | 13 (65.0%) | |
|
Karnofsky Performance Scale (KPS) |
100 | 11 (30.6%) | 3 (18.75%) | 8 (40.0%) |
| 90 | 9 (25.0%) | 4 (25.0%) | 5 (25.0%) | |
| 80 | 8 (22.2%) | 4 (25.0%) | 4 (20.0%) | |
| 70 | 3 (8.3%) | 2 (12.5%) | 1 (5.0%) | |
| ≤ 60 | 5 (13.9%) | 3 (18.75%) | 2 (10.0%) | |
| Site of target lesion(s) | Temporal | 12 (33.3%) | 3 (18.75%) | 9 (45.0%) |
| Frontal | 11 (30.5%) | 6 (37.5%) | 5 (25.0%) | |
| Parietal | 6 (16.7%) | 4 (25.0%) | 2 (10.0%) | |
| Occipital | 4 (11.1%) | 2 (12.5%) | 2 (10.0%) | |
| Temporo-occipital | 2 (5.6%) | 1 (6.25%) | 1 (5.0%) | |
| Other | 1 (2.8%) | 0 | 1 (5.0%) | |
| Resection | Gross total | 32 (88.9%) | 14 (87.5%) | 18 (90%) |
| Subtotal | 3 (8.3%) | 2 (12.5%) | 1 (5.0%) | |
| Biopsy* | 1 (2.8%) | 0 | 1 (5.0%) | |
| Invasion | No | 14 (38.9%) | 6 (37.5%) | 8 (40.0%) |
| Corpus callosum | 11 (30.5%) | 5 (31.25%) | 6 (30.0%) | |
| Multiple | 1 (2.8%) | 0 | 1 (5.0%) | |
| Multifocal | 4 (11.1%) | 3 (18.75%) | 1 (5.0%) | |
| Multifocal + corpus callosum | 3 (8.3%) | 1 (6.25%) | 2 (10.0%) | |
| Brainstem + corpus callosum | 1 (2.8%) | 0 | 1 (5.0%) | |
| Other | 2 (5.6%) | 1 (6.25%) | 1 (5.0%) | |
*3 cc incisional (via mini-craniotomy) biopsy sample
PTEN mutations and expression profile
Coding regions of the PTEN gene were sequenced in a total of 36 tissue samples. No mutations were detected in 15 (41.67%) tumour samples; whereas, a total of 28x PTEN mutations were identified in the other 21 (58.33%) tumour samples. As shown in Supplementary Table 1; 15x missense, 6x frameshift, 5x start/stop loss, 1x stop gained, and 1x synonymous mutation were found and classified according to the VarSome database.
Mutant PTEN patients had significantly lower PTEN expression, both at mRNA (p = 0.008) and protein (p = 0.006) levels (Fig. 1c-e). For one sample, the tissue lysate did not work for western blot analyses; therefore, protein expression was only evaluated by IHC. IHC analysis showed that whereas PTEN expression in 3 patients (5.6%) was intact; decreased expression could be detected in 26 patients (72.2%) and complete loss in 8 patients (22.2%) (Fig. 1f-g). All patients with PTEN loss carried the mutant PTEN genotype. Besides, PTEN expression did not significantly change between primary and recurrent tumours (Supp. File, Fig. 1).
Fig. 1.
PTEN expression status in wild-type (WT) and mutant (M) genotypes. (A) qRT-PCR results of PTEN mRNA expression normalised to GAPDH (*p = 0.008); (B) Western Blot analysis of PTEN protein expression normalised to B-actin (**p = 0.006); (C) Western Blot images of PTEN in the GBM tissues. PTEN genotypes are indicated as WT and M at the top of the gel; (D) Immunohistochemistry analysis of PTEN showing intact expression; (E) Weaker staining of PTEN with focal loss. Cases were clustered as PTEN low; (F) PTEN immunohistochemistry image demonstrates complete PTEN loss, images at 100x magnification; (G) Comparison of PTEN IHC expression in relation to PTEN genotypes
Effects of PTEN status on the cGAS-STING pathway
The expression patterns of cGAS-STING pathway members in GBM tissue samples were compared in relation to primary and recurrent tumours, and to PTEN genotypes of the patients. Accordingly, mRNA expressions did not significantly differ between primary and recurrent tumour samples (Supp. File, Fig. 1). Interestingly, STING, IRF3, NF-KB1, and RELA mRNA expressions decreased significantly in patients with the mutant PTEN genotype (Fig. 2a). However, protein expressions did not significantly differ between patients with wild-type or mutant genotypes (Fig. 2b-c).
Fig. 2.
Effects of PTEN genotypes on cGAS-STING pathway. (A) mRNA expression graphs of STING, IRF3, NF-KB1, and RELA genes in wild-type (WT) and mutant (M) patients; (B) Protein expressions of STING, IRF3, NF-KB, and Phospho-NF-KB normalized to B-actin; (C) Western Blot images of protein expressions. PTEN genotypes are indicated as WT and M at the top of the gel
Effects of PTEN expression onto tumour immunogenicity
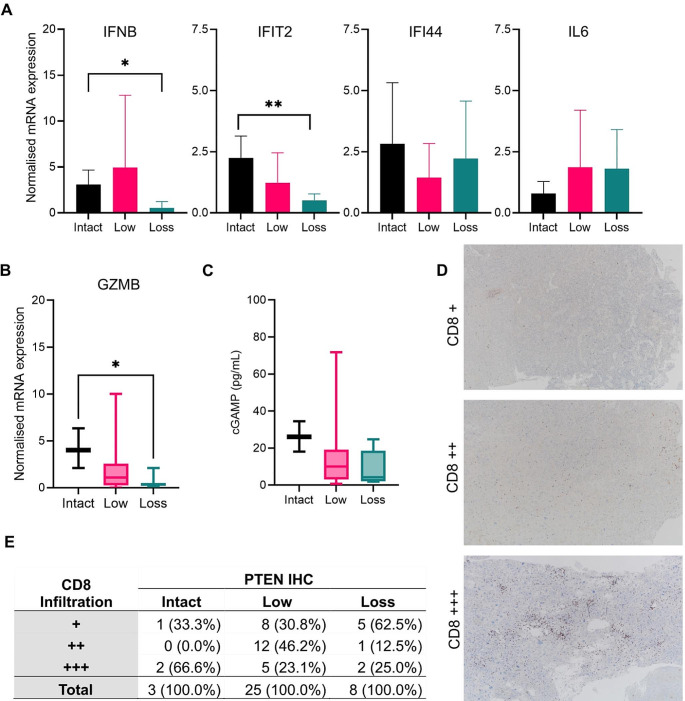
For evaluating the role of PTEN expression on cGAS-STING activity and tumour immunogenicity, mRNA expression levels of interferon-stimulated genes, especially IFNB, IFIT2, IFI44, and IL6 were analysed. The results showed that IFNB (p = 0.033) and IFIT2 (p = 0.015) mRNA levels significantly decreased in patients with PTEN loss (Fig. 3a). Consistently, Granzyme B (GZMB) expression levels also decreased in patients with PTEN loss (p = 0.045) (Fig. 3b). Analyses of cGAMP production in whole tumour protein lysates revealed a wide range of cGAMP expression levels in GBM tumour samples (Fig. 3c). Although not statistically significant, mean cGAMP was lowest in patients with PTEN loss.
Fig. 3.
Effects of PTEN expression to tumour immunogenicity. (A) Transcription levels of interferon-stimulated genes according to immunohistochemically expression status of PTEN (*: p = 0.033; **: p = 0.015); (B) Normalised mRNA expression of GZMB (*: p = 0.045); (C) cGAMP production measured by ELISA in tumour lysates; (D) Immunohistochemistry analysis of CD8 showing 3 grades of infiltration. E: Comparison of CD8 infiltration in relation to PTEN status, images at 40x magnification
To reveal if CD8 infiltration is altered in relation with PTEN expression, tumour samples were immunohistochemically stained with anti-CD8 antibody and evaluated by two pathologists. Considering the staining intensity, three groups could be established (Fig. 3d). Samples of patients with PTEN loss showed significantly decreased CD8 infiltration (p = 0.04) (Fig. 3e).
Bioinformatics analysis was performed on data from 162 IDH negative GBM samples obtained from the TCGA database for validation studies. Different ISG genes, such as MX1 and OAS, and infiltration of additional immune cells were also investigated and similar results obtained; i.e., the low PTEN mRNA expressing group displayed significantly decreased ISGs (IFNB, IFIT2, IFI44, MX1, and OAS1); and, CD4 + and CD8 + cell infiltration (Fig. 4a-c).
Fig. 4.
TCGA database analysis. Median PTEN expression was used to discriminate low (n = 81) and high (n = 81) expression groups. (A) mRNA expression levels of interferon-stimulated genes; (B) CD4+; and (C) CD8 + cell infiltration in relation to PTEN expression
Discussion
Immunosuppressive effects of PTEN have been studied extensively in several tumours. However, the mechanism of these effects has not been fully established to date. This study reports 28x PTEN mutations in 21 (58.33%) GBM tumour samples and its significantly low mRNA and protein expression levels in those PTEN mutant tumours. A GBM prognostic model based on immune-regulated genes revealed that patients with PTEN mutation are in the high-risk group [21]. Increased immune cell infiltration, more aggressive immune activity, higher expression of immune checkpoint genes, and less benefit from immunotherapy are listed among the characteristic features of these high-risk group patients [21]. These results are also in accordance with the PTEN-associated immune prognostic signature in GBM [22, 23].
Does PTEN mutations effect tumour immunogenicity through the cGAS-STING pathway? It is known that PTEN has an essential phosphatase activity on IRF3, a master transcription factor responsible for IFNB synthesis [13]. PTEN mutations in the catalytic phosphatase domain can impair phosphatase activity, either by gain or loss of function; some mutations can even effect just lipid, but not protein phosphatase activity [24, 25]. Therefore, mutation-dependent gain or loss of function of phosphatase activity might have potential effects on interferon release. This study shows that expression of cGAS-STING pathway members like STING, IRF3, NF-KB1, and RELA indeed decreases in patients carrying the mutant PTEN genotype.
In a recently published article, the presence of STING has particularly been shown in the tumour vasculature in human GBM samples [26]. Several reports also highlighted the positive immunogenic effects of STING agonist treatments in the GBM microenvironment [26–30]. Circulation of the STING protein through the vesicular traffic pathway includes sorting into Rab7-positive endolysosomes before degradation [31]. Rab7 and TBK1, on the other side, are substrates of PTEN, by which way it can contribute to the vesicular traffic of STING [32]. Disruption of STING trafficking, regulated by the Rab7 and TBK1 axis, in breast cancer cells with PTEN loss also supports the role of PTEN on tumour immunogenicity through the cGAS-STING pathway [32].
PTEN deficiency induces galectin-9 secretion, which drives M2 macrophage polarization and therefore is associated with angiogenesis and glioma progression [33]. A recent study showed that macrophage PTEN deficiency regulates nuclear factor (erythroid-derived 2)-like 2 (NRF2) function, and interrelates with NICD before it controls STING-mediated TBK1 function, resulting in enhanced inflammatory response in liver injury [34]. Both studies indicate that PTEN loss may also influence STING regulation in the tumour microenvironment by affecting macrophage polarization. However, further studies are needed to elucidate the mechanism of how tumour-specific regulations occur.
Contributions of PTEN to tumour immunity through the cytosolic cGAS-STING pathway may provide new insights into the treatment of PTEN loss tumours. For instance, PTEN-null triple-negative breast cancer (TNBC) cell lines were hyper-responsive to STING agonists because of diminished STING turnover and increased production of IRF3 targets [32]. Recently, it has been shown that PTEN deficiency in high-grade serous ovarian cancer resulted in decreased sensitivity to carboplatin alone. However, STING activation potentiates response to carboplatin chemotherapy, prolongs overall survival, repolarizes M2-like suppressive macrophages, and rescues T-cell activation in the tumour microenvironment [35]. Due to the immune privilege of the brain, GBM has a “cold tumour” phenotype, and displays low numbers of tumour-infiltrating lymphocytes and other immune effector cell types. This current study shows that PTEN-deficient GBM patients have decreased CD8 infiltration, Granzyme B activity, and interferon expression. Bioinformatics analysis using the TCGA database also revealed decreased ISGs and T cell infiltration in GBM patients, validating our findings. Therefore, STING activation might turn the GBM microenvironment into a “hot tumour” to benefit from improved immunotherapy.
Nevertheless, there are some limitations of this study. Surgical operations and follow-ups of patients have not solely been accomplished by Ege University Faculty of Medicine. Because of this, data was not accessible for all patients and survival analyses could not be performed. However, the main aim of this study was to determine immunogenic effects that might occur by PTEN loss through the cGAS-STING pathway, rather than its survival effects. Another limitation is the low number of cases included into this study, as we hope that soon other studies will support the relationship between PTEN and cGAS-STING in GBM [36].
In conclusion, immunosuppressive effects of PTEN loss through the cGAS-STING pathway in GBM tumours could be shown in this study. The future perspective of this study would be if these results could be applied to translational studies for the clinical treatment of PTEN-null tumours. Finally, we propose that the PTEN protein is required for the proper functioning of the cGAS-STING pathway and that STING agonists may not be effective in GBM tumours with PTEN loss.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the Ege University Scientific Research Projects Coordination (TGA-2019-20329). We would like to thank Timur Kose for his statistical advice.
Author contributions
EO, NA, HB, KEC and TY collected tumour samples; ED, ZY and AES performed the analysis; TA and YE performed the pathological diagnosis and immunohistochemically staining; VB and BK wrote the main manuscript text; ZY prepared the figures; CA performed bioinformatics analysis; VB designed and supervised the study. All authors reviewed the results and approved the final version of the manuscript.
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Data availability
No datasets were generated or analysed during the current study.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Eda Dogan and Zafer Yildirim contributed equally to this work.
References
- 1.Wang J, Cazzato E, Ladewig E, Frattini V, Rosenbloom DI, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. doi: 10.1038/ng.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee YR, Chen M, Pandolfi PP. The functions and regulation of the PTEN tumour suppressor: new modes and prospects. Nat Rev Mol Cell Biol. 2018;19(9):547–562. doi: 10.1038/s41580-018-0015-0. [DOI] [PubMed] [Google Scholar]
- 3.Kim B, Kang SY, Kim D, Heo YJ, Kim KM (2020) PTEN Protein Loss and loss-of-function mutations in gastric cancers: the relationship with microsatellite instability, EBV, HER2, and PD-L1 expression. Cancers (Basel) 12(7). 10.3390/cancers12071724 [DOI] [PMC free article] [PubMed]
- 4.Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferguson SD, Hodges TR, Majd NK, Alfaro-Munoz K, Al-Holou WN, et al. A validated integrated clinical and molecular glioblastoma long-term survival-predictive nomogram. Neurooncol Adv. 2021;3(1):vdaa146. doi: 10.1093/noajnl/vdaa146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peng W, Chen JQ, Liu C, Malu S, Creasy C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2016;6(2):202–216. doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 8.Cetintas VB, Batada NN. Is there a causal link between PTEN deficient tumors and immunosuppressive Tumor microenvironment? J Transl Med. 2020;18(1):45. doi: 10.1186/s12967-020-02219-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsa AT, Waldron JS, Panner A, Crane CA, Parney IF, et al. Loss of Tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 10.Zhao J, Chen AX, Gartrell RD, Silverman AM, Aparicio L, et al. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat Med. 2019;25(3):462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ablasser A, Gulen MF. The role of cGAS in innate immunity and beyond. J Mol Med (Berl) 2016;94(10):1085–1093. doi: 10.1007/s00109-016-1423-2. [DOI] [PubMed] [Google Scholar]
- 12.Li T, Chen ZJ. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215(5):1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li S, Zhu M, Pan R, Fang T, Cao YY, et al. The Tumor suppressor PTEN has a critical role in antiviral innate immunity. Nat Immunol. 2016;17(3):241–249. doi: 10.1038/ni.3311. [DOI] [PubMed] [Google Scholar]
- 14.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 15.Cetintas VB, Duzgun Z, Akalin T, Ozgiray E, Dogan E et al (2023) Molecular dynamic simulation and functional analysis of pathogenic PTEN mutations in glioblastoma. J Biomol Struct Dyn 1–13. 10.1080/07391102.2022.2162582 [DOI] [PubMed]
- 16.Ertan Y, Sezak M, Demirag B, Kantar M, Cetingul N, et al. Medulloblastoma: clinicopathologic evaluation of 42 pediatric cases. Childs Nerv Syst. 2009;25(3):353–356. doi: 10.1007/s00381-008-0784-4. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Piskorz A, Bosse T, Jimenez-Linan M, Rous B, et al. Immunohistochemistry and next-generation sequencing are complementary tests in identifying PTEN Abnormality in Endometrial Carcinoma biopsies. Int J Gynecol Pathol. 2022;41(1):12–19. doi: 10.1097/PGP.0000000000000763. [DOI] [PubMed] [Google Scholar]
- 18.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X, Wang S, Li Q. Comprehensive Analysis of expression and Prognostic Value of sirtuins in Ovarian Cancer. Front Genet. 2019;10:879. doi: 10.3389/fgene.2019.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li T, Fan J, Wang B, Traugh N, Chen Q, et al. TIMER: a web server for Comprehensive Analysis of Tumor-infiltrating Immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang K, Rao C, Li Q, Lu J, Zhu Z, et al. Construction and validation of a glioblastoma prognostic model based on immune-related genes. Front Neurol. 2022;13:902402. doi: 10.3389/fneur.2022.902402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giotta Lucifero A, Luzzi S (2022) Immune Landscape in PTEN-Related Glioma Microenvironment: a bioinformatic analysis. Brain Sci 12(4). 10.3390/brainsci12040501 [DOI] [PMC free article] [PubMed]
- 23.Yu J, Lai M, Zhou Z, Zhou J, Hu Q, et al. The PTEN-associated immune prognostic signature reveals the landscape of the Tumor microenvironment in glioblastoma. J Neuroimmunol. 2023;376:578034. doi: 10.1016/j.jneuroim.2023.578034. [DOI] [PubMed] [Google Scholar]
- 24.Costa HA, Leitner MG, Sos ML, Mavrantoni A, Rychkova A, et al. Discovery and functional characterization of a neomorphic PTEN mutation. Proc Natl Acad Sci U S A. 2015;112(45):13976–13981. doi: 10.1073/pnas.1422504112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Post KL, Belmadani M, Ganguly P, Meili F, Dingwall R, et al. Multi-model functionalization of disease-associated PTEN missense mutations identifies multiple molecular mechanisms underlying protein dysfunction. Nat Commun. 2020;11(1):2073. doi: 10.1038/s41467-020-15943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger G, Knelson EH, Jimenez-Macias JL, Nowicki MO, Han S, et al. STING activation promotes robust immune response and NK cell-mediated Tumor regression in glioblastoma models. Proc Natl Acad Sci U S A. 2022;119(28):e2111003119. doi: 10.1073/pnas.2111003119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudreau CE, Najem H, Ott M, Horbinski C, Fang D, et al. Intratumoral Delivery of STING Agonist results in clinical responses in Canine Glioblastoma. Clin Cancer Res. 2021;27(20):5528–5535. doi: 10.1158/1078-0432.CCR-21-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bielecki PA, Lorkowski ME, Becicka WM, Atukorale PU, Moon TJ, et al. Immunostimulatory silica nanoparticle boosts innate immunity in brain tumors. Nanoscale Horiz. 2021;6(2):156–167. doi: 10.1039/d0nh00446d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripathi S, Najem H, Mahajan AS, Zhang P, Low JT et al (2022) cGAS-STING pathway targeted therapies and their applications in the treatment of high-grade glioma. F1000Res, 11:1010. 10.12688/f1000research.125163.1 [DOI] [PMC free article] [PubMed]
- 30.Zhou Y, Guo Y, Chen L, Zhang X, Wu W, et al. Co-delivery of phagocytosis checkpoint and STING agonist by a trojan horse nanocapsule for orthotopic glioma immunotherapy. Theranostics. 2022;12(12):5488–5503. doi: 10.7150/thno.73104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonugunta VK, Sakai T, Pokatayev V, Yang K, Wu J, et al. Trafficking-mediated STING degradation requires sorting to Acidified endolysosomes and can be targeted to Enhance Anti-tumor Response. Cell Rep. 2017;21(11):3234–3242. doi: 10.1016/j.celrep.2017.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ritter JL, Zhu Z, Thai TC, Mahadevan NR, Mertins P, et al. Phosphorylation of RAB7 by TBK1/IKKepsilon regulates Innate Immune Signaling in Triple-negative Breast Cancer. Cancer Res. 2020;80(1):44–56. doi: 10.1158/0008-5472.CAN-19-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ni X, Wu W, Sun X, Ma J, Yu Z, et al. Interrogating glioma-M2 macrophage interactions identifies Gal-9/Tim-3 as a viable target against PTEN-null glioblastoma. Sci Adv. 2022;8(27):eabl5165. doi: 10.1126/sciadv.abl5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang T, Qu X, Zhao J, Wang X, Wang Q, et al. Macrophage PTEN controls STING-induced inflammation and necroptosis through NICD/NRF2 signaling in APAP-induced liver injury. Cell Commun Signal. 2023;21(1):160. doi: 10.1186/s12964-023-01175-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shakfa N, Li D, Conseil G, Lightbody ED, Wilson-Sanchez J et al (2023) Cancer cell genotype associated Tumor immune microenvironment exhibits differential response to therapeutic STING pathway activation in high-grade serous Ovarian cancer. J Immunother Cancer 11(4). 10.1136/jitc-2022-006170 [DOI] [PMC free article] [PubMed]
- 36.Shi X, Wang S, Wu Y, Li Q, Zhang T, et al. A bibliometric analysis of the Innate Immune DNA sensing cGAS-STING pathway from 2013 to 2021. Front Immunol. 2022;13:916383. doi: 10.3389/fimmu.2022.916383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.