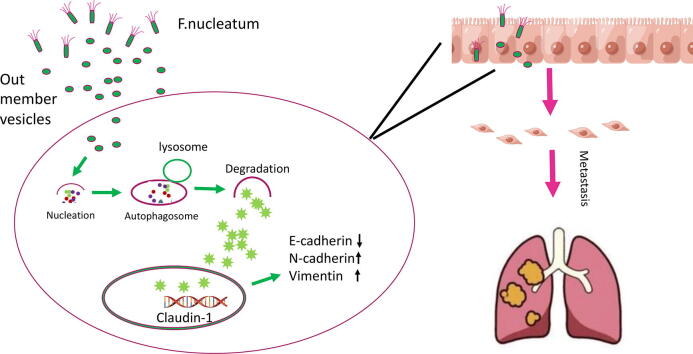
Graphical abstract
Keywords: Fusobacterium nucleatum, Autophagy, Outer membrane vesicles, Epithelial-mesenchymal transition
Highlights
-
•
In vivo, Fn OMVs promoted lung metastasis in tumour-bearing mice, while chloroquine (CHQ; an autophagy inhibitor) reduced the number of pulmonary metastases resulting from intratumoral Fn OMV injection.
-
•
Fn OMVs promoted cancer cell invasion and migration in vivo, leading to changes in the expression of EMT related proteins.
-
•
RNA-seq showed that Fn OMVs activate intracellular autophagy pathways. Blocking autophagic flux with CHQ not only reduced Fn OMV-induced cancer cell migration in vitro and in vivo but also reversed the expression changes in EMT-related proteins.
Abstract
Introduction
Metastasis is an important cause of high mortality and lethality of oral cancer. Fusobacterium nucleatum (Fn) can promote tumour metastasis. Outer membrane vesicles (OMVs) are secreted by Fn. However, the effects of Fn-derived extracellular vesicles on oral cancer metastasis and the underlying mechanisms are unclear.
Objectives
We aimed to determine whether and how Fn OMVs mediate oral cancer metastasis.
Methods
OMVs were isolated from brain heart infusion (BHI) broth supernatant of Fn by ultracentrifugation. Tumour-bearing mice were treated with Fn OMVs to evaluate the effect of OMVs on cancer metastasis. Transwell assays were performed to determine how Fn OMVs affect cancer cell migration and invasion. The differentially expressed genes in Fn OMV-treated/untreated cancer cells were identified by RNA-seq. Transmission electron microscopy, laser confocal microscopy, and lentiviral transduction were used to detect changes in autophagic flux in cancer cells stimulated with Fn OMVs. Western blotting assay was performed to determine changes in EMT-related marker protein levels in cancer cells. Fn OMVs’ effects on migration after blocking autophagic flux by autophagy inhibitors were determined by in vitro and in vivo experiments.
Results
Fn OMVs were structurally similar to vesicles. In the in vivo experiment, Fn OMVs promoted lung metastasis in tumour-bearing mice, while chloroquine (CHQ, an autophagy inhibitor) treatment reduced the number of pulmonary metastases resulting from the intratumoral Fn OMV injection. Fn OMVs promoted the migration and invasion of cancer cells in vivo, leading to altered expression levels of EMT-related proteins (E-cadherin downregulation; Vimentin/N-cadherin upregulation). RNA-seq showed that Fn OMVs activate intracellular autophagy pathways. Blocking autophagic flux with CHQ reduced in vitro and in vivo migration of cancer cells induced by Fn OMVs as well as reversed changes in EMT-related protein expression.
Conclusion
Fn OMVs not only induced cancer metastasis but also activated autophagic flux. Blocking autophagic flux weakened Fn OMV-stimulated cancer metastasis.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a highly malignant cancer of the head and neck region. It ranks seventh among the most commonly occurring cancers worldwide and causes a large financial burden for both families and society. Metastasis remains an important cause of death in patients with HNSCC; however, there are limited effective treatment strategies for preventing metastasis [1], [2]. Recent studies have revealed the role of bacteria in various tumours [3], [4], [5]. In breast cancer, bacteria enhance cancer cell resistance to shear stress of fluids, thereby causing lung metastasis [6]. In oral cancer patients, the structure of the oral flora is remarkably different from that of normal individuals, and transplantation of this abnormal flora was found to promote oral tumour development in an oral cancer mouse model [7], [8], [9]. Thus, researchers are presently investigating specific tumorigenic bacteria and the precise mechanisms by which they influence oral cancer development [10], [11], [12], [13].
Fusobacterium nucleatum (Fn), a gram-negative anaerobe, is an abundant species in the oral cavity [14], [15]. According to several epidemiological investigations, Fn is highly associated with the development, occurrence, prognosis and treatment of colorectal cancer (CRC) [16], [17], [18]. Fn uses various mechanisms to promote CRC development and occurrence [19], [20], [21]. Fn also enhances the development and occurrence of other tumours [22], [23]. Metagenomic sequencing revealed a positive correlation between Fn abundance and oral carcinogenesis [24]. However, the mechanisms by which Fn promotes oral cancer progression remain undetermined.
Bacterial extracellular vesicles (bEVs) bounded by a protein lipid bilayer enclose the contents from parental bacterial cells [25]. Outer membrane vesicles (OMVs) are bEVs derived from gram-negative bacterial species. These vesicles develop from the outer membrane and contain cytoplasmic and periplasmic components [26]. OMVs are released by bacteria into the extracellular space. OMVs elicit intracellular signals through ligand-receptor interactions; furthermore, target cells internalize them through the processes of phagocytosis, endocytosis, membrane fusion, or micropinocytosis [27], [28]. OMVs release their contents into target cells to generate the corresponding biological activity. For example, OMVs from Escherichia coli induce proinflammatory responses in receptor cells [29]. However, it remains unclear whether Fn OMVs are associated with oral cancer metastasis.
Autophagy, a conserved catabolic process, has key functions in every step of metastasis, including local invasion, intravasation, vasodilation, extravasation, and colonization [30], [31], [32], [33]. Crosstalk between autophagy and bacteria can affect cancer progression in many ways [34]. According to recent studies, Fn enhances CRC metastasis by promoting autophagy signalling [35]. However, it remains to be determined whether autophagy is associated with the mechanisms by which Fn OMVs promote oral cancer metastasis.
The present study aimed to address the following three questions:
-
(1)
can Fn OMVs cause metastasis of oral tumours?
-
(2)
can Fn OMVs induce autophagic flux in tumour cells?
-
(3)
is autophagy involved in Fn OMV-mediated changes in biological functions of oral tumours?
We found that Fn OMVs can cause lung metastasis of tumours, an effect that may be related to the phenotype of epithelial-mesenchymal transition (EMT) in cancer cells induced by Fn OMVs. Autophagy inhibition can block the stimulating effect of Fn OMVs on cancer cells, thus indicating that the regulatory effects of Fn OMVs on cancer cells involve autophagy.
Materials and methods
Culture and preparation of Fn OMVs
The Fn ATCC 25586 strain was provided by the microecology laboratory of Zhujiang Hospital. The strain was cultured at 37 °C under anaerobic conditions (10% H2, 80% N2, and 10% CO2) in BHI (brain heart infusion) broth containing hemin (1 μg/mL; Hopebio, China) and menadione (1 μg/mL; Hopebio).
Streptococcus salivarius (134928) and Lactobacillus rhamnosus (134266) were provided by BeNa Culture Collection (BNCC; Beijing, China). S. salivarius was cultured at 37 °C in BHI broth for 24 h under 5% CO2 condition. L. rhamnosus was propagated microaerophilically at 37 °C in MRS (de Man, Rogosa, and Sharpe) broth. The E. coli strain was obtained from the microecology laboratory of Zhujiang Hospital. These bacterial strains were used as controls.
The bacterial culture medium was centrifuged after the cell cultures reached an OD600 (optical density at 600 nm) reading of 0.5. The obtained supernatant was filtered through a 0.22-μm filter (Merck Millipore) to exclude debris of the parental bacterial strain and other contaminants after subjecting the bacterial cultures to pelleting (30 min, 5,000 × g). Ultracentrifugation of the obtained supernatant was performed for 120 min at 100,000 × g at 4 °C. Crude OMVs were obtained by washing the supernatant twice with phosphate-buffered saline (PBS). Subsequently, refiltration of the purified OMVs was performed using a 0.22-μm filter (Merck Millipore). The OMVs were then subjected to density gradient ultracentrifugation under the following conditions: 45 Ti rotor, 100,000 × g, 2 h, and 4 °C. The obtained final pellets were again suspended in PBS and stored at −80 °C. A BCA protein assay kit (Beyotime, China) was used to determine the concentration of the obtained OMVs.
Cell culture
CAL27 and HSC3 human HNSCC cell lines were provided by ATCC. CAL27-luc cells were constructed by Genechem Co., Ltd. These cells were routinely cultured at 37 °C in DMEM (Thermo Fisher Scientific) containing 10% foetal bovine serum (FBS) and 1% antibiotics (100 U/mL penicillin and 100 U/mL streptomycin) under 5% CO2 condition. A Mycoplasma PCR detection kit (Beyotime, China) was used to detect Mycoplasma contamination in the cultured cells.
Migration and invasion assays
Cells were cocultured for 24 h with Fn OMVs (5 µg/mL). Treated and untreated cells were harvested, resuspended and plated in serum-free DMEM at the density of 5 × 104 cells/well. Next, 200 μL of the suspension was placed in the upper compartment of a Transwell insert (Corning, 3422), while 600 μL of complete DMEM with 10% FBS was placed in the lower compartment. Cells that migrated through the insert’s microporous membrane were fixed and stained with a crystal violet solution (0.5%) after incubation for 24 h. Five visual fields for each insert were randomly selected and viewed through a microscope at 200 × magnification. After images were acquired, the number of cells in each visual field was determined.
Serum-free DMEM was used to dilute Matrigel (354234, BD Biosciences). Fifty microlitres of the mixture was then coated on the membrane of a Transwell insert for 2 h at 37 °C. The remaining steps were the same as described above for the migration assay.
Immunoblot analysis
RIPA buffer with a protease and phosphatase inhibitor cocktail (Keygen, Nanjing, China) were used for cell lysis. The protein extracts were separated by SDS-PAGE, and the separated proteins were transferred onto PVDF membranes. After blocking with 5% skimmed milk at room temperature (RT) for 1 h, the membranes were then incubated at 4 °C with the primary antibodies for 24 h. The membranes were washed with TBST and incubated with the secondary antibodies at RT for 1 h. An electrochemiluminescence detection kit (Merck Millipore, USA) was used to visualize the protein signals. Supplementary Table 1 lists the antibodies used in the present study.
Animal experiments
CAL27-Luc cells (5 × 106 cells/100 μL) were subcutaneously injected into the axillae of BALB/c nude mice (nu/nu, weighing approximately 20 g, 4 weeks of age) on day 0. After the average diameter of the xenografts reached 5 mm on day 7, the animals were randomly divided into groups and received the following treatments: (1) PBS; (2) Fn OMVs; (3) chloroquine (CHQ); and (4) Fn OMVs + CHQ.
Fn OMVs were administered intratumourally at the dose of 1 mg/kg once a week. CHQ was administered at 25 mg/kg dose intraperitoneally twice a week. Tumour size and animal weight were monitored once a week and calculated as follows: volume (mm3) = length × width2 × 0.5.
Small animal imaging
Mice were first anaesthetized after 8 weeks. Next, D-luciferin from Promega at the dose of 150 mg/kg was administered intraperitoneally into mice at 10 min before imaging. Bruker molecular imaging software was used to monitor lung metastasis progression. Following the appearance of metastatic foci in the lungs, the mice were killed, and their lung tissues were extracted. Haematoxylin and eosin (H&E) staining was used to evaluate lung metastasis. Supplementary Figure 6 shows the flowchart of the experimental scheme.
H&E staining
Five-micrometre-thick paraffin-embedded mouse lung tissue sections were stained with H&E for visualizing metastatic foci in mouse lungs and oral cancer tissues. An Olympus microscope (Tokyo, Japan) was used to acquire the images.
Transmission electron microscopy
Autophagosomes/autolysosomes were evaluated by transmission electron microscopy (TEM). Cultured cells were fixed overnight at 4 °C with 4% glutaraldehyde; subsequently, after washing with PBS, the cells were fixed with 1% osmium tetroxide (OsO4) buffer for 1.5 h at 4 °C. The cells were washed with PBS, dehydrated using graded ethanol series, and embedded in an Epon812 epoxy resin. Ultrathin sections (80 nm thickness) were mounted on a copper grid and subjected to double staining with 0.2% lead citrate and 1% uranium acetate. An HT7800 transmission electron microscope was used to observe the sections.
Confocal microscopy
To detect autophagic flux in cells stimulated with Fn OMVs, CAL27 cells were transfected with GFP-LC3B and mRFP-GFP-LC3 adenoviruses (purchased from Hanbio, Shanghai, China). The transfection process was performed based on the instructions provided by Hanbio. After transfection with GFP-LC3B or mRFP-GFP-LC3, the cells were first stimulated with Fn OMVs for 12 h and then fixed with 4% paraformaldehyde for 30 min. Nuclei were re-stained with DAPI for 5 min. All procedures were conducted at RT in the dark. Finally, the cells in confocal dishes were imaged by a confocal laser scanning microscope.
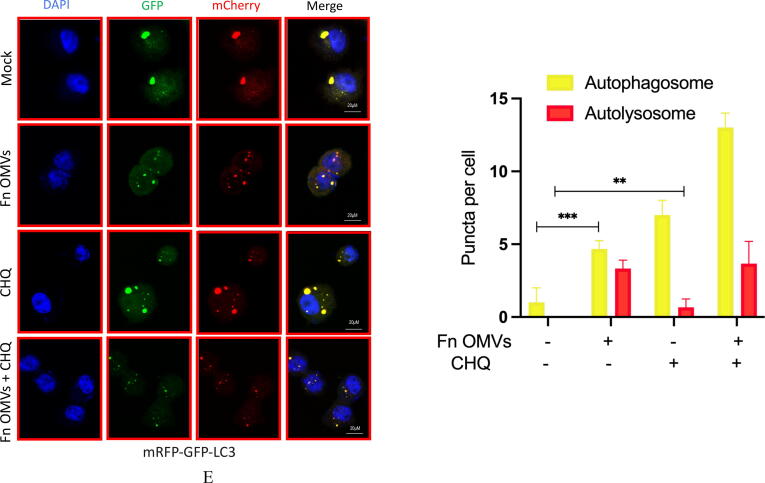
This determination method is based on varying pH stability of green and red fluorescent proteins. Under acidic conditions (pH < 5), GFP fluorescence signals can be quenched, while mRFP fluorescence signals do not change significantly. In the merged green and red images, autophagosomes are visualized as yellow dots (i.e., RFP+GFP+). An increase in the signals of both yellow dots and red dots in the cell indicates an increase in autophagic flux. In contrast, autophagic flux is considered to be blocked when only yellow dot signal increases with no change in red dot signal or when the cell shows a decrease in both yellow dot and red dot signals.
High-Throughput sequencing
CAL27 cells were incubated with Fn OMVs for 12 h and then lysed with control cells. This experiment was repeated three times. In brief, following the manufacturer’s instructions, TRIzol reagent (Invitrogen, USA) was used to extract total RNA. Subsequently, an ABclonal mRNA-seq Lib prep kit (ABclonal, China) was used to prepare paired-end libraries in accordance with the manufacturer’s instructions. The sequencing of the library preparations was performed on the MGISEQ-T7 platform, followed by the generation of 150 bp paired-end reads. Mapped reads were obtained by separately aligning the clean reads with the reference genome under the orientation mode by using HISAT2 software. The reads mapped to each gene were counted by FeatureCounts. Gene expression levels, represented by fragments per kilobase transcript per million mapped reads (FPKM), were determined by the gene length and read count mapped to this gene. The DESeq2 algorithm was used to identify the differentially expressed genes between the two groups. By using the KEGG pathway database, KEGG pathway analysis was performed to determine the biological functions of different pathways.
Extraction of RNA and qRT-Pcr
As described earlier, TRIzol was used to extract total RNA from cells [36]. The extracted total RNA was reverse transcribed into cDNA by using Reverse Transcriptase Premix. In keeping with the manufacturer’s protocol, qRT-PCR was conducted on an ABI vii7 system by using SYBR Green Master Mix and primers specific for the indicator genes. By using GAPDH as an internal reference, the relative quantitative expression level of each gene was calculated using the 2–ΔΔCt method. The primers used (Shanghai Sangen Biotechnology Co., Ltd., China) are shown in Supplementary Table 2.
Fluorescence in situ hybridization
Five-micrometre-thick sections fixed in formalin and embedded in paraffin were used to localize Fn in HNSCC and adjacent normal tissues by fluorescence in situ hybridization (FISH). The digoxigenin-labelled Fn probe (digoxigenin-5ʹ-CGCAATACAGAGTTGAGCCCTGC-3ʹ-digoxigenin; dilution 1:100) and the universal bacterial probe (digoxigenin-5ʹ-GCTGCCTCCCGTAGGAGT-3ʹ-digoxigenin) synthesized by Sangon Biotech Company (Shanghai, China) were used to detect Fn.
Statistical analysis
Prism version v8.4.0.671 (GraphPad software) was used for statistical analysis. An unpaired t test or analysis of variance (ANOVA) was data comparison, as required. The description is provided in figure legends. All data are expressed as mean ± standard deviation (SD). Statistical significance was indicated by *p < 0.05 and **p < 0.01, and ***p < 0.001,was set as the statistical significance threshold.
Ethics statement
The animal care and study protocols were approved by the Experimental Animal Ethics Committee of Southern Medical University (Approval Number: LAEC-2020-224FS).
Results
Characterization of Fn OMVs
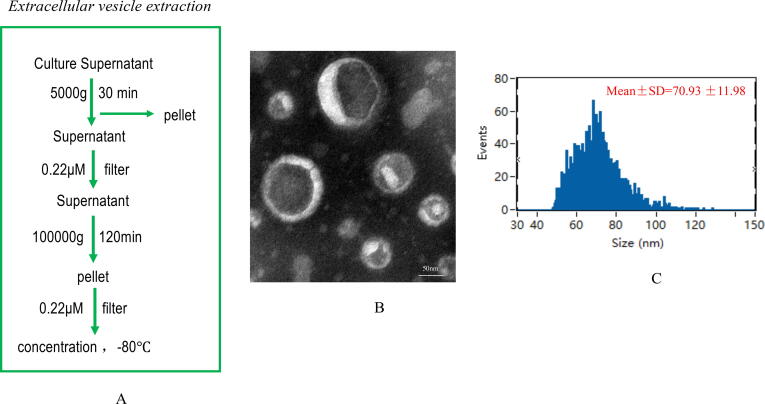
OMVs were isolated from the culture broth supernatant of Fn by gradient centrifugation; the bacterial concentration was 1 × 109 colony-forming units/mL (Fig. 1A). A transmission electron microscope was used to determine the morphology and size of OMVs (Fig. 1B). Based on NTA (nanoparticle tracking analysis), the size range of OMVs was 62–310 nm (peak: 131 ± 25 nm) (Fig. 1C). TEM and NTA revealed that the particle size and morphology of Fn OMVs were identical to those previously described. OMVs derived from S. salivarius, L. rhamnosus, and E. coli (used as controls) were also isolated using a similar method (Supplementary Fig. 1). These purified OMVs were used in the subsequent experiments.
Fig. 1.
Extraction and identification of F. nucleatum OMVs.(A) Extraction of F. nucleatum OMVs by ultracentrifugation followed these steps. (B) F. nucleatum OMVs were observed by TEM and exhibited a similar tray structure. (C) NTA showed that the OMVs ranged in size from 62 to 310 nm, with a peak at 131 ± 25 nm.
Fn OMVs induce EMT phenotype in cancer cells to promote lung metastasis of oral squamous cell carcinoma xenografts
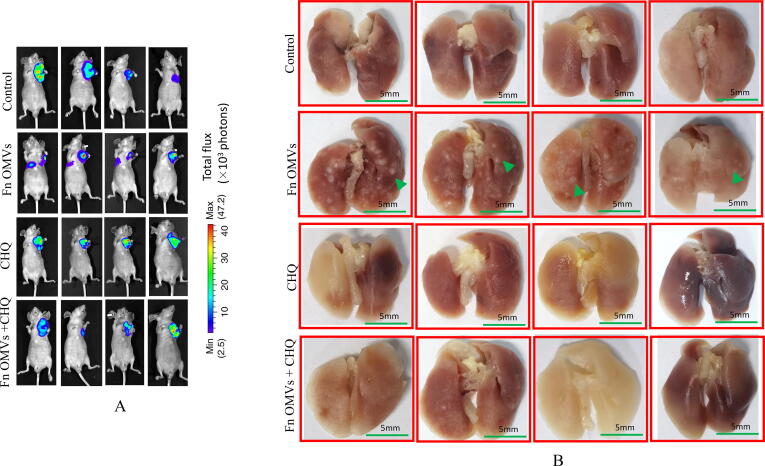
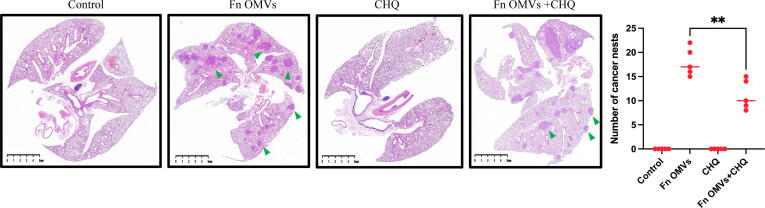
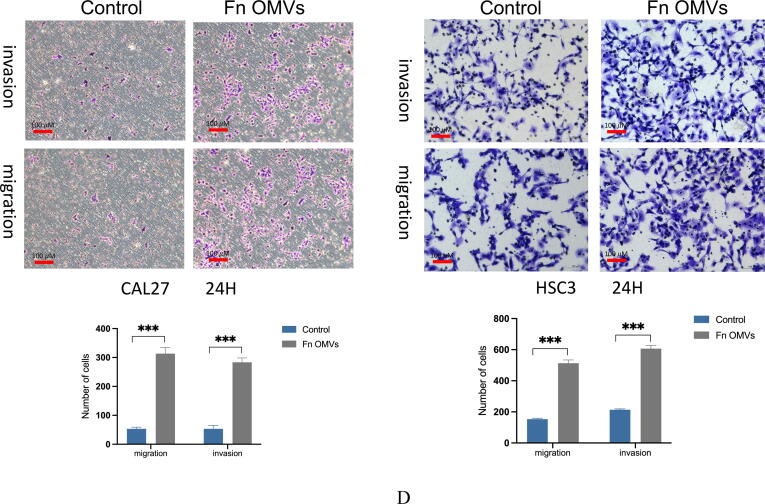
An in vivo assessment of the effect of Fn OMVs on tumour metastasis was performed. The axillae of nude mice were subcutaneously injected with OSCC cells. Following the establishment of the tumour-bearing mouse model, Fn OMVs were injected into the tumour tissue for 7 weeks. The fluorescence imaging results showed that 75% of the mice treated with Fn OMVs exhibited multiple fluorescence signals in their lungs and that the lungs of the control group mice did not show fluorescence signals; this finding indicated that Fn OMVs promoted lung metastasis of oral cancer (Fig. 2A). After mouse lung tissue was removed and fixed with paraformaldehyde, metastatic nodules were found on the lung surfaces of all mice treated with Fn OMVs (Fig. 2B). H&E staining showed that following treatment with Fn OMVs, the lungs of mice showed several cancer nests (Fig. 2C). Transwell assays showed that migration and invasion of CAL27 and HSC3 cells were enhanced after treatment with Fn OMVs for 24 h (Fig. 2D). However, enhancement of invasion and migration was not observed after the treatment of cancer cells with other bacterial OMVs (Supplementary Fig. 2).
Fig. 2.
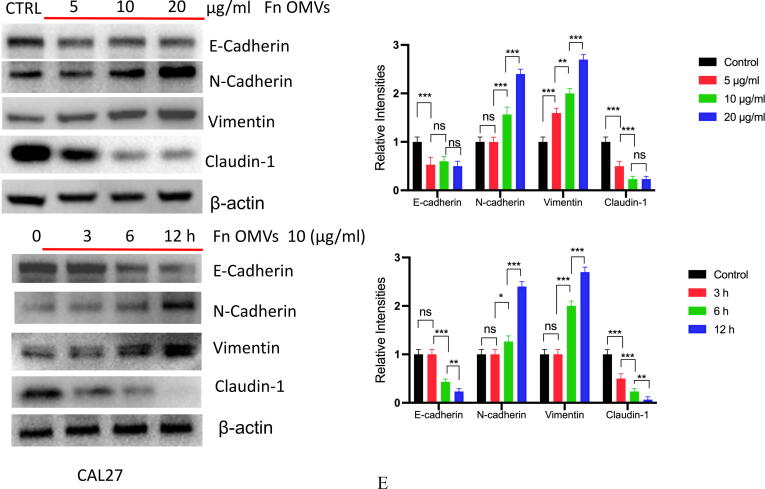
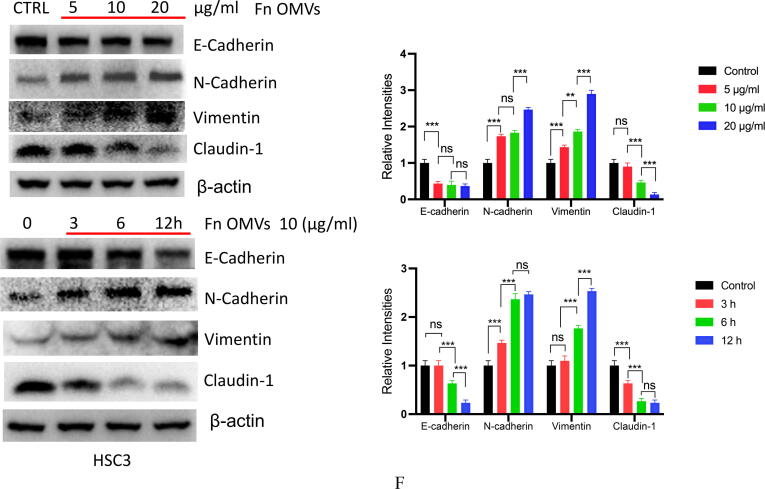
Fn OMVs induce EMT phenotype of cancer cells to promote cancer Metastasis.(A) After the cal27- luc cells expressing luciferase gene were subcutaneously injected into the armpit of mice, the mice lungs could detect fluorescent signals after 7 weeks of Fn OMVs treatment.(B) Lung specimens of mice. After Fn OMVs treatment, white metastatic foci were seen on the lung surface. The arrow shows lots of metastatic nodules on the surface of the lung. (C) H&E staining showed that a large number of cancer nests could be seen in the lung sections of Fn OMVs group mice. Pulmonary metastatic nodules from slices of lung specimens per mice were quantified. Arrows indicated metastatic nodules. Data are presented as mean ± SD (two-way ANOVA). Each dot indicates an individual mouse. The significant difference between Fn OMVs group and Fn OMVs/CHQ group, **p < 0.01. (D) After treatment with Fn OMVs for 24 h, the number of CAL27 and HSC3 cells passing through the transwell membrane increased, indicating that the invasion and migration of cancer cells were enhanced. The migrated and invaded cells were quantified by counting in five fields. The significant difference among the groups, ***p < 0.001. Scale bar, 100 μm. (E) The expression of EMT markers (E-Cadherin, N-Cadherin, Vimentin, Claudin-1) in cal27 was regulated by the stimulation time and concentration of Fn OMVs. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, *p < 0.05, **p < 0.01, ***p < 0.001, ns, no significant difference. (F) The expression of EMT markers (E-Cadherin, N-Cadherin, Vimentin, Claudin-1) in hsc3 was regulated by the stimulation time and concentration of Fn OMVs. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, *p < 0.05, **p < 0.01,***p < 0.001, ns, no significant difference.
EMT is closely associated with metastasis. We further investigated whether Fn OMVs can increase oral cancer cells’ metastatic potential by regulating EMT. For this purpose, CAL27 and HSC3 cells were cocultured with Fn OMVs. As shown in Fig. 2E-2F, the results of western blotting (WB) assay revealed that Fn OMV exposure decreased E-cadherin expression and increased Vimentin expression in CAL27 and HSC3 cells in a time/concentration-dependent manner. Thus, Fn OMVs can promote lung metastasis of OSCC xenografts, probably because they can induce the EMT phenotype in cancer cells.
Fn OMVs promote Cancer-Related autophagy activation
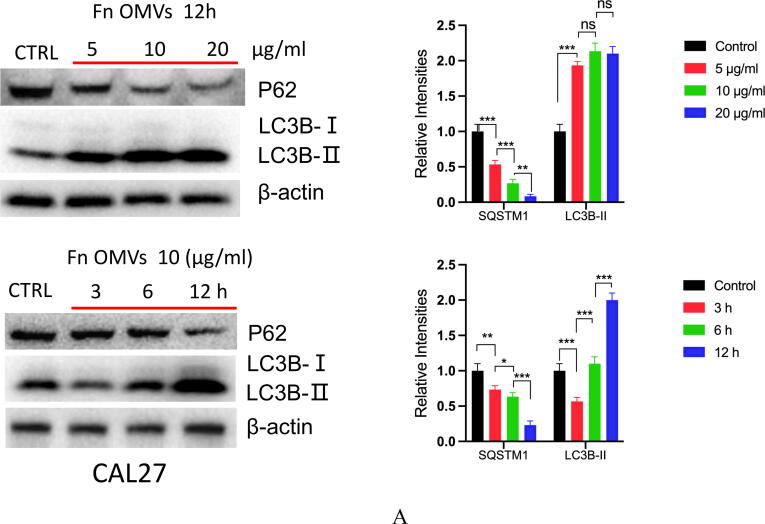
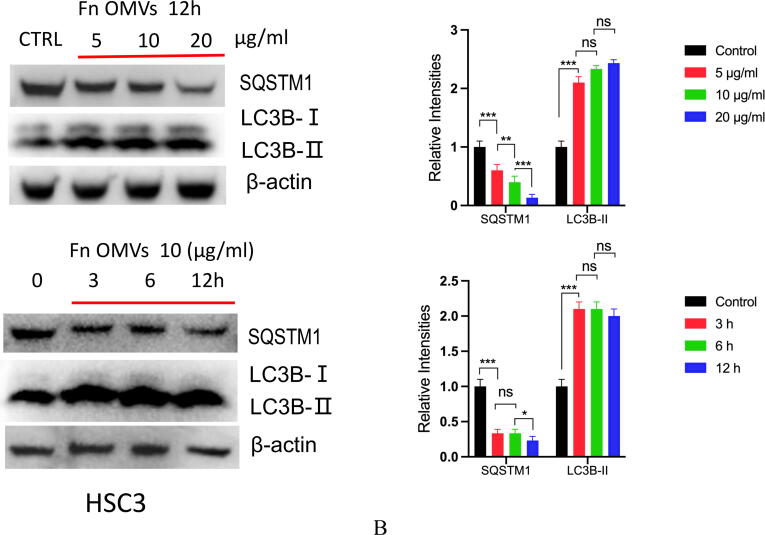
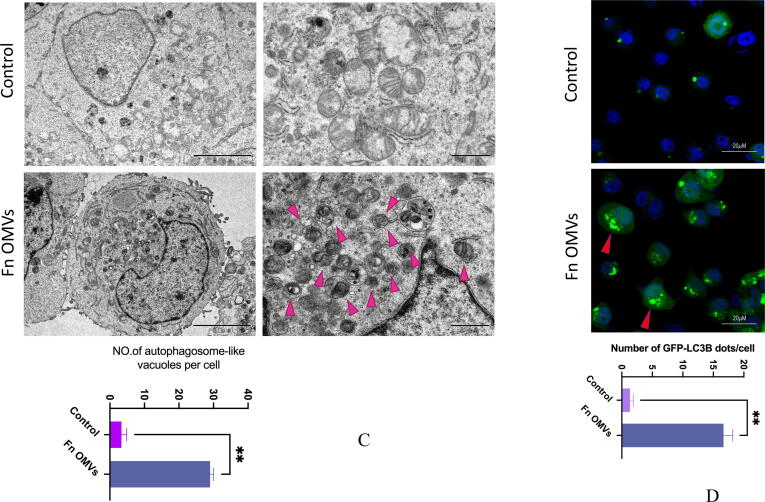
To determine the mechanism of action, we cocultured CAL27 cells with Fn OMVs. The cocultured were then subjected to RNA-seq analysis, and the gene expression profiles of CAL27 cells cocultured with and without Fn OMVs were compared. Coculture with Fn OMVs downregulated 2814 genes and upregulated 1454 genes in CAL27 cells. According to the results of the pathway analysis, Fn OMV infection of CAL27 cells stimulated the autophagy pathway (Supplementary Fig. 3A). Heatmap analysis showed that autophagy-related gene expression was upregulated in cancer cells cocultured with Fn OMVs. qRT-PCR confirmed that Fn OMV exposure affected the mRNA levels of Beclin-1, ATG3, and ATG7 in cancer cells (Supplementary Fig. 3B). WB assay demonstrated that Fn OMVs altered P62 and LC3B expression levels in cancer cells (Fig. 3A-3B). CAL27/HSC3 cells showed a low level of LC3 cleavage under basic conditions. After stimulation with Fn OMVs, LC3B-II expression level increased significantly. TEM showed that higher autophagic vesicle formation in CAL27 cells stimulated with Fn OMVs (Fig. 3C). After coculture of CAL27 cells stably expressing the GFP-LC3B fusion protein with Fn OMVs for 24 h, confocal imaging revealed increased LC3B expression in the cells, thus indicating the production of a large number of autophagosomes (Fig. 3D). Autolysosomes are formed by the fusion of autophagic vesicles and lysosomes. Cancer cells with stable expression of the mRFP-EGFP-LC3 fusion protein were generated and cocultured with Fn OMVs for 24 h. Confocal imaging showed partial disappearance of green fluorescence; this finding indicated that exposure to Fn OMVs enhanced autophagic flux in cancer cells (Fig. 3E). In general, the study results show that Fn OMVs induce activation of autophagy pathways in oral cancer cells.
Fig. 3.
Fn OMVs Promotes Cancer-Related Autophagy Activation (A) When CAL27 cells were cocultured with different concentrations of Fn OMVs or cocultured with the same concentration of Fn OMVs for different times, the expression of LC-3I, LC-3II and p62 in cancer cells was time or concentration dependent. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, *p < 0.05, **p < 0.01,***p < 0.001, ns, no significant difference. (B) When HSC3 cells were cocultured with different concentrations of Fn OMVs or cocultured with the same concentration of Fn OMVs for different times, the expression of LC-3I, LC-3II and p62 in cancer cells was time or concentration dependent. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, *p < 0.05, **p < 0.01,***p < 0.001, ns, no significant difference. (C) Top: Representative transmission electron microscopy (TEM) images showed after treatment of CAL27 with Fn OMVs, lots of membrane structures appeared in cancer cells. Arrowheads indicate autophagosomes. Scale bars: 5 μm or 1 μm. Bottom: Quantitative analysis of the numbers of autophagic vacuoles. Data are presented as mean ± SD. The difference between the groups, **p < 0.01. (D) Top: Representative confocal microscopic images of LC3B puncta in CAL27 transfected with GFP-LC3B cocultured with Fn OMVs. Arrowheads indicate green fluorescent proteins were LC3B. Bottom: Quantitative analysis of green fluorescent proteins per cell. Data are presented as mean ± SD. The difference between the groups, **p < 0.01. (E) Representative confocal microscopic images of LC3B puncta in CAL27 transfected with mRFP-GFP-LC3 cocultured with Fn OMVs. Autophagosomes are indicated by yellow dots with both GFP and RFP, and autolysosomes by free red dots with only RFP in merged images. Scale bar: 20 μm. Right: Quantitative analysis of autophagosomes and autolysosomes per cell. Data are presented as mean ± SD. The significant difference among the groups, **p < 0.01, ***p < 0.001. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fn induces metastasis of cancer cells through the autophagy pathway
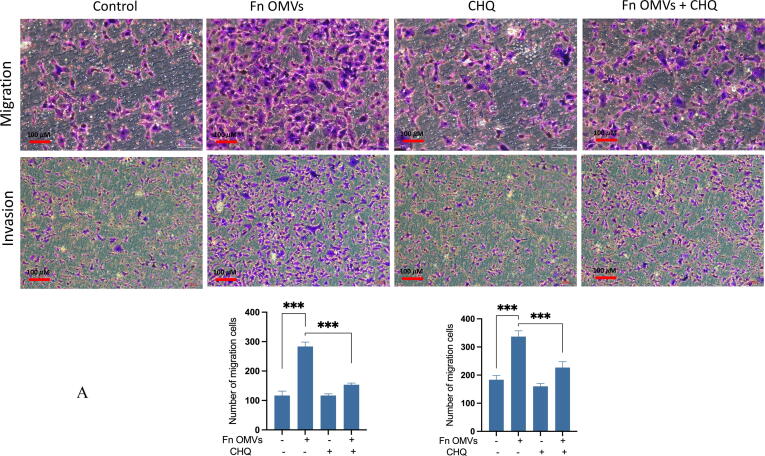
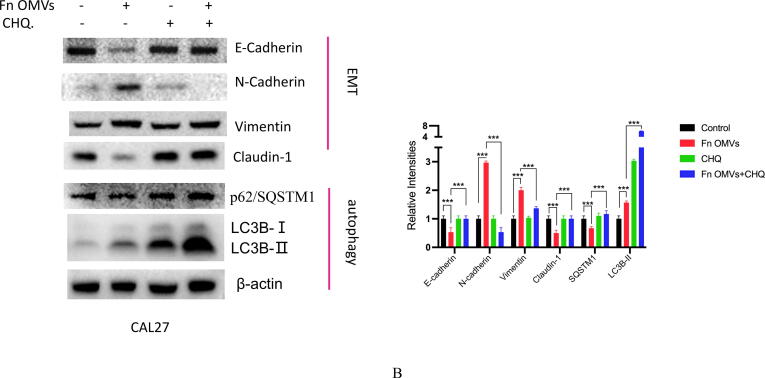
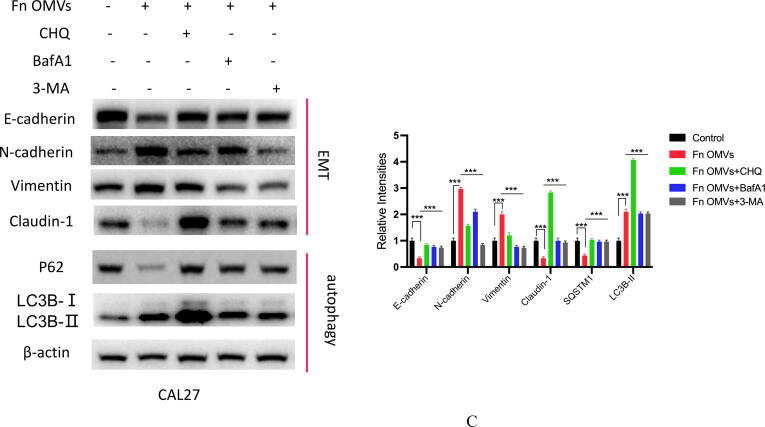
Coculture of Fn OMVs with CAL27 cells changed their biological function. What role does autophagy play in the process by which Fn OMVs promote cancer metastasis? To answer this question, we treated CAL27 cells with Fn OMVs and chloroquine, an autophagic lysosomal inhibitor, for 24 h. Transwell experiments showed that CHQ addition blocked Fn OMVs’ activity in promoting cancer cell migration and invasion (Fig. 4A). EMT has a critical function in cancer metastasis. WB assay revealed that CHQ attenuated the Fn OMV-mediated upregulation of claudin-1, Vimentin, and N-Cadherin expression and downregulated P62 and E-cadherin expression in CAL27 cells; this finding showed that CHQ can inhibit the EMT in cancer cells induced by Fn OMVs (Fig. 4B). After blocking autophagic flux in cells by CHQ, the function of Fn OMVs in promoting metastasis was affected. To eliminate interference, we selected two other autophagy pathway inhibitors, namely bafilomycin A1 (BafA1) and 3-methyladenine (3MA), to verify whether a similar phenomenon occurred. BafA1 and 3MA inhibited the upstream autophagy pathway. WB assay demonstrated that BafA1 and 3MA inhibited EMT caused by Fn OMVs (Fig. 4C). These conclusions were verified in HSC3, another oral cancer cell line (Supplementary Fig. 4A-C).
Fig. 4.
Fn OMVs promote tumor metastasis through autophagy in vivo and in vitro. (A) Transwell experiment showed that after treatment Cal27 with Fn OMVs, the number of cells across the membrane increased, while after treatment with CHQ and Fn OMVs, the number of cells across the membrane decreased. Bottom: The migrated and invaded cells were quantified by counting in five fields. Scale bar, 100 μm. Data are presented as mean ± SD. The significant difference among the groups, ***p < 0.001. (B) Western blot showed that the expression of EMT markers (E-Cadherin, N-Cadherin, Vimentin, Claudin-1) in Cal27 was regulated by the stimulation of Fn OMVs, while after autophagy inhibitor CHQ treated Cal27 cells, the expression of EMT related proteins was reversed. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, ***p < 0.001. (C) Western blot showed that in addition to CHQ, autophagy upstream inhibitors BafA1 and 3MA could also attenuate the changes of EMT markers induced by Fn OMVs in Cal27 cells. Right: Relative band intensities analyzed by ImageJ. Data are presented as mean ± SD. The significant difference among the groups, ***p < 0.001.
To further confirm the role of autophagy in this process, we also evaluated the in vivo effect of CHQ combined with Fn OMVs. After 6 weeks of treatment, mice in the CHQ–Fn OMV combination group had no metastatic lesions in their lung tissues (Fig. 2A-C). E-cadherin expression was lower in the Fn OMVs group than in the CHQ–Fn OMVs combination group. Compared to those of control mice, the tumour tissues of mice treated with Fn OMVs showed higher expression levels of LC3-II, N-Cadherin, and Vimentin and lower expression levels of P62 and E-cadherin. CHQ attenuated the function of Fn OMVs in promoting the EMT in cancer cells. These findings indicate that Fn OMVs promote tumour metastasis through autophagy in vivo and in vitro.
Discussion
Tumour metastasis is a critical factor associated with mortality due to HNSCC, and effective therapeutic strategies for this cancer remain limited. Hence, for developing effective therapeutic agents, it is crucial to understand the underlying mechanisms regulating tumour metastasis [37], [38], [39]. Because Fn was first reported as a cancer-related bacterium in 2002, its related mechanism of cancer promotion has continued to be elucidated [40]. Fn can interfere with cancer cell proliferation, tumour immunity, chemotherapeutic resistance, and other tumour biological characteristics [41], [42], [43], [44], [45]. As confirmed by previous studies, Fn can promote CRC metastasis. Zhuo et al. found that Fn promotes the adhesion of CRC cells to endothelial cells and facilitates cell metastasis and extravasation [19]. Chen et al. showed that CRC metastasis is promoted by Fn through M2 polarization and the miR-1322/CCL20 axis [46]. Fn also promotes EMT by regulating noncoding RNA expression [47]. A correlation between Fn and oral tumour occurrence has also been reported [48], [49], [50]. In previous studies, we also found a similar correlation by using metagenomic sequencing [24]. Additionally, a FISH assay was used to further confirm the existence of Fn in oral tumour tissues (Supplementary Fig. 5). In conclusion, these studies clarified the association between Fn and CRC metastasis, but did not elucidate the role of vesicles secreted by Fn.
OMVs are the extracellular vesicles of gram-negative bacteria and have 50–250 nm diameter. They are released during bacterial growth and participate in various biological processes, including intercellular communication, antibiotic resistance, and bacteriophage infection [51], [52], [53], [54]. OMVs can cross cell membranes and penetrate tissues to affect and change the behaviour of recipient cells. OMVs have higher advantage than their parental bacterial strain because of their lower malignancy risk and higher biocompatibility [55]. Vaccines against Neisseria meningitidis OMVs have been used clinically [56]. However, the influence of Fn OMVs on tumour metastasis is yet to be reported. Hence, the objective of the present study aimed to elucidate the role of Fn OMVs in HNSCC metastasis as well as identify the corresponding mechanism.
Fn OMVs were first isolated and purified as described previously [57], [58], [59]. TEM and particle size analysis showed that Fn OMVs had a vesicular structure. Subsequently, in in vitro tests, Fn OMVs promoted CAL27/HSC3 cell migration and invasion and induced the EMT phenotype in these cells time- and concentration-dependently. EMT plays a key role in HNSCC metastasis. EMT-related changes in the expression levels of E-cadherin, N-cadherin, and Vimentin are linked with increased HNSCC tumour metastasis [60]. In addition, we also isolated OMVs from Escherichia coli, Lactobacillus rhamnosus and Streptococcus salivarius, and did not find that these OMVs can promote the migration and invasion of oral cancer cells. In vivo experiments showed that all mice had metastatic foci in their lungs, consistent with the in vitro data. Metastasis is a multistep process. In addition to EMT, the immune microenvironment, fluid shear force, and tumour-adjacent cells can affect the metastasis of cancer cells. We established a tumour-bearing model and observed that Fn OMVs can also cause lung metastasis of tumours. In conclusion, similar to Fn, Fn OMVs can promote cancer metastasis to the lungs.
What is the underlying mechanism by which Fn OMVs induce the EMT phenotype? To answer this question, we performed RNA-seq and found that the autophagy pathway was activated in cancer cells. qRT-PCR also demonstrated that the expression levels of autophagy-related genes, i.e., ATG3, ATG7, Beclin1, etc., were significantly increased. Some bacterial OMVs can activate autophagy, while others can inhibit autophagy [29], [61]. The findings of TEM and confocal microscopy further supported the conclusion that Fn OMVs can promote autophagy. Treatment with CHQ to block autophagic flux inhibited cancer cell migration and invasion. H&E staining also revealed that CHQ administration in mice reduced the number of lung metastases caused by intratumoral injection of Fn OMVs. According to the results of WB assay, CHQ reversed the EMT phenotype induced by Fn OMVs. To eliminate interference, two other autophagy inhibitors were selected, and the same result was obtained. Overall, Fn OMVs activated autophagy, altered protein homoeostasis in cancer cells, and thus promoted EMT induction in cancer cells. The next question is how Fn OMVs activate autophagy. Because Fn OMVs are a bacterial component, their protein and nucleic acid components are currently unclear, and identifying the components that play an active role in Fn OMVs is challenging.
Limitations of this study
Although our research revealed the signalling flow of Fn OMVs in relation to autophagy and cell migration, the oral microbiome and immune system may work together with Fn OMVs to control cancer metastasis. More specifically, innate immunity induced by the intratumoral injection of Fn OMVs may also participate in some steps of Fn OMV-driven metastasis. Moreover, although the identification of functional components related to the transfer of Fn OMVs is challenging, it is the key to understand the related biological phenomena.
Funding
The present study was funded by the National Natural ScienceFoundation of China, Science Fund for Distinguished Young Scholars (81925026), the National Natural Science Foundation of China Youth Fund (81702979), and Shanghai East Hospital, Tongji University School of Medicine (DFRC2021006).
Ethics statement
The Experimental Animal Ethics Committee of Southern Medical University approved all of the animal care and study protocols (LAEC-2020-224FS).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Yan He, Kun Wen, Muxuan Chen, and Jun Man for their assistance in in vitro assays and their guidance. We thank the members of the Microbiome Medicine Center for useful discussions. We would like to thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.04.002.
Contributor Information
Yubo Xu, Email: 1247845178@qq.com.
Buling Wu, Email: Wubuling623@163.com.
Hongwei Zhou, Email: biodegradation@gmail.com.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Maghami E., Ismaila N., Alvarez A., et al. Diagnosis and Management of Squamous Cell Carcinoma of Unknown Primary in the Head and Neck: ASCO Guideline. J Clin Oncol. 2020;38:2570–2596. doi: 10.1200/JCO.20.00275. [DOI] [PubMed] [Google Scholar]
- 2.Mody M.D., Rocco J.W., Yom S.S., et al. Head and neck cancer. Lancet. 2021;398:2289–2299. doi: 10.1016/S0140-6736(21)01550-6. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12:31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 4.Nejman D., Livyatan I., Fuks G., et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science (New York, NY) 2020;368:973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sepich-Poore GD, Zitvogel L, Straussman R, et al. The microbiome and human cancer. Science (New York, NY) 2021; 371. [DOI] [PMC free article] [PubMed]
- 6.Fu A., Yao B., Dong T., et al. Tumor-resident intracellular microbiota promotes metastatic colonization in breast cancer. Cell. 2022;185(1356–1372):e1326. doi: 10.1016/j.cell.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 7.Weyrich L.S. The evolutionary history of the human oral microbiota and its implications for modern health. Periodontol. 2000;2021(85):90–100. doi: 10.1111/prd.12353. [DOI] [PubMed] [Google Scholar]
- 8.Stasiewicz M., Karpinski T.M. The oral microbiota and its role in carcinogenesis. Semin Cancer Biol. 2021 doi: 10.1016/j.semcancer.2021.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Frank D.N., Qiu Y., Cao Y., et al. A dysbiotic microbiome promotes head and neck squamous cell carcinoma. Oncogene. 2022;41:1269–1280. doi: 10.1038/s41388-021-02137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibanez L., de Mendoza I., Maritxalar Mendia X., Garcia de la Fuente A.M., et al. Role of Porphyromonas gingivalis in oral squamous cell carcinoma development: A systematic review. J Periodontal Res. 2020;55:13–22. doi: 10.1111/jre.12691. [DOI] [PubMed] [Google Scholar]
- 11.Zheng D.W., Deng W.W., Song W.F., et al. Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat Biomed Eng. 2021;6:32–43. doi: 10.1038/s41551-021-00807-9. [DOI] [PubMed] [Google Scholar]
- 12.Mauceri R., Coppini M., Vacca D., et al. Salivary Microbiota Composition in Patients with Oral Squamous Cell Carcinoma: A Systematic Review. Cancers (Basel) 2022;14:5441. doi: 10.3390/cancers14215441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh S., Singh A.K. Porphyromonas gingivalis in oral squamous cell carcinoma: a review. Microbes Infect. 2022;24 doi: 10.1016/j.micinf.2021.104925. [DOI] [PubMed] [Google Scholar]
- 14.Teles F., Wang Y., Hajishengallis G., et al. Impact of systemic factors in shaping the periodontal microbiome. Periodontol. 2000;2021(85):126–160. doi: 10.1111/prd.12356. [DOI] [PubMed] [Google Scholar]
- 15.Meng Q., Gao Q., Mehrazarin S., et al. Fusobacterium nucleatum secretes amyloid-like FadA to enhance pathogenicity. EMBO Rep. 2021;22:e52891. doi: 10.15252/embr.202152891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan C.A., Garrett W.S. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nat Rev Microbiol. 2019;17:156–166. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu T., Guo F., Yu Y., et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell. 2017;170(548–563):e516. doi: 10.1016/j.cell.2017.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo S., Li L., Xu B., et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin Chem. 2018;64:1327–1337. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y., Zhang L., Zheng S., et al. Fusobacterium nucleatum promotes colorectal cancer cells adhesion to endothelial cells and facilitates extravasation and metastasis by inducing ALPK1/NF-kappaB/ICAM1 axis. Gut Microbes. 2022;14:2038852. doi: 10.1080/19490976.2022.2038852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu H., Du J., Chao S., et al. Fusobacterium nucleatum Promotes Colorectal Cancer Cell to Acquire Stem Cell-Like Features by Manipulating Lipid Droplet-Mediated Numb Degradation. Adv Sci (Weinh) 2022;9:e2105222. doi: 10.1002/advs.202105222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong C., Yan X., Zhu Y., et al. Fusobacterium Nucleatum Promotes the Development of Colorectal Cancer by Activating a Cytochrome P450/Epoxyoctadecenoic Acid Axis via TLR4/Keap1/NRF2 Signaling. Cancer Res. 2021;81:4485–4498. doi: 10.1158/0008-5472.CAN-21-0453. [DOI] [PubMed] [Google Scholar]
- 22.Parhi L., Alon-Maimon T., Sol A., et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat Commun. 2020;11:3259. doi: 10.1038/s41467-020-16967-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y., Baba Y., Ishimoto T., et al. Fusobacterium nucleatum confers chemoresistance by modulating autophagy in oesophageal squamous cell carcinoma. Br J Cancer. 2020;124:963–974. doi: 10.1038/s41416-020-01198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Z., Chen G., Wang P., et al. Alterations of the Oral Microbiota Profiles in Chinese Patient With Oral Cancer. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.780067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie J., Li Q., Haesebrouck F., et al. The tremendous biomedical potential of bacterial extracellular vesicles. Trends Biotechnol. 2022;40:1173–1194. doi: 10.1016/j.tibtech.2022.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Tiku V., Tan M.W. Host immunity and cellular responses to bacterial outer membrane vesicles. Trends Immunol. 2021;42:1024–1036. doi: 10.1016/j.it.2021.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Sartorio M.G., Pardue E.J., Feldman M.F., et al. Bacterial Outer Membrane Vesicles: From Discovery to Applications. Annu Rev Microbiol. 2021;75:609–630. doi: 10.1146/annurev-micro-052821-031444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chronopoulos A., Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene. 2020;39:6951–6960. doi: 10.1038/s41388-020-01509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.David L., Taieb F., Penary M., et al. Outer membrane vesicles produced by pathogenic strains of Escherichia coli block autophagic flux and exacerbate inflammasome activation. Autophagy. 2022:1–13. doi: 10.1080/15548627.2022.2054040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mowers E.E., Sharifi M.N., Macleod K.F. Autophagy in cancer metastasis. Oncogene. 2017;36:1619–1630. doi: 10.1038/onc.2016.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xing Y., Wei X., Liu Y., et al. Autophagy inhibition mediated by MCOLN1/TRPML1 suppresses cancer metastasis via regulating a ROS-driven TP53/p53 pathway. Autophagy. 2021:1–23. doi: 10.1080/15548627.2021.2008752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rakesh R., PriyaDharshini L.C., Sakthivel K.M., et al. Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis. 2022;1868 doi: 10.1016/j.bbadis.2022.166400. [DOI] [PubMed] [Google Scholar]
- 33.Gundamaraju R., Lu W., Paul M.K., et al. Autophagy and EMT in cancer and metastasis: Who controls whom? Biochim Biophys Acta Mol Basis Dis. 2022;1868 doi: 10.1016/j.bbadis.2022.166431. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y., Du J., Wu X., et al. Crosstalk between autophagy and microbiota in cancer progression. Mol Cancer. 2021;20:163. doi: 10.1186/s12943-021-01461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y., Chen Y., Zhang J., et al. Fusobacterium nucleatum Promotes Metastasis in Colorectal Cancer by Activating Autophagy Signaling via the Upregulation of CARD3 Expression. Theranostics. 2020;10:323–339. doi: 10.7150/thno.38870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen G., Sun Q., Cai Q., et al. Outer Membrane Vesicles From Fusobacterium nucleatum Switch M0-Like Macrophages Toward the M1 Phenotype to Destroy Periodontal Tissues in Mice. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.815638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson D.E., Burtness B., Leemans C.R., et al. Head and neck squamous cell carcinoma. Nat Rev Dis Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cramer J.D., Burtness B., Le Q.T., et al. The changing therapeutic landscape of head and neck cancer. Nat Rev Clin Oncol. 2019;16:669–683. doi: 10.1038/s41571-019-0227-z. [DOI] [PubMed] [Google Scholar]
- 39.Leemans C.R., Snijders P.J.F., Brakenhoff R.H. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18:269–282. doi: 10.1038/nrc.2018.11. [DOI] [PubMed] [Google Scholar]
- 40.Slade D.J. New Roles for Fusobacterium nucleatum in Cancer: Target the Bacteria, Host, or Both? Trends Cancer. 2020;7:185–187. doi: 10.1016/j.trecan.2020.11.006. [DOI] [PubMed] [Google Scholar]
- 41.Rubinstein M.R., Wang X., Liu W., et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinstein M.R., Baik J.E., Lagana S.M., et al. Fusobacterium nucleatum promotes colorectal cancer by inducing Wnt/beta-catenin modulator Annexin A1. EMBO Rep. 2019;20:e47638. doi: 10.15252/embr.201847638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gur C., Maalouf N., Shhadeh A., et al. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology. 2019;8:e1581531. doi: 10.1080/2162402X.2019.1581531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamoto Y., Mima K., Ishimoto T., et al. Relationship between Fusobacterium nucleatum and antitumor immunity in colorectal cancer liver metastasis. Cancer Sci. 2021;112:4470–4477. doi: 10.1111/cas.15126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dahlstrand Rudin A., Khamzeh A., Venkatakrishnan V., et al. Short chain fatty acids released by Fusobacterium nucleatum are neutrophil chemoattractants acting via free fatty acid receptor 2 (FFAR2) Cell Microbiol. 2021;23:e13348. doi: 10.1111/cmi.13348. [DOI] [PubMed] [Google Scholar]
- 46.Xu C., Fan L., Lin Y., et al. Fusobacterium nucleatum promotes colorectal cancer metastasis through miR-1322/CCL20 axis and M2 polarization. Gut Microbes. 2021;13:1980347. doi: 10.1080/19490976.2021.1980347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang S., Li C., Liu J., et al. Fusobacterium nucleatum promotes epithelial-mesenchymal transiton through regulation of the lncRNA MIR4435-2HG/miR-296-5p/Akt2/SNAI1 signaling pathway. FEBS J. 2020;287:4032–4047. doi: 10.1111/febs.15233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bronzato J.D., Bomfim R.A., Edwards D.H., et al. Detection of Fusobacterium in oral and head and neck cancer samples: A systematic review and meta-analysis. Arch Oral Biol. 2020;112 doi: 10.1016/j.archoralbio.2020.104669. [DOI] [PubMed] [Google Scholar]
- 49.Fujiwara N., Kitamura N., Yoshida K., et al. Involvement of Fusobacterium Species in Oral Cancer Progression: A Literature Review Including Other Types of Cancer. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21176207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McIlvanna E., Linden G.J., Craig S.G., et al. Fusobacterium nucleatum and oral cancer: a critical review. BMC Cancer. 2021;21:1212. doi: 10.1186/s12885-021-08903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dell'Annunziata F., Folliero V., Giugliano R., et al. Gene Transfer Potential of Outer Membrane Vesicles of Gram-Negative Bacteria. Int J Mol Sci. 2021;22:5985. doi: 10.3390/ijms22115985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui C., Guo T., Zhang S., et al. Bacteria-derived outer membrane vesicles engineered with over-expressed pre-miRNA as delivery nanocarriers for cancer therapy. Nanomed Nanotechnol Biol Med. 2022;45 doi: 10.1016/j.nano.2022.102585. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Z., Liu S., Zhang S., et al. Porphyromonas gingivalis outer membrane vesicles inhibit the invasion of Fusobacterium nucleatum into oral epithelial cells by downregulating FadA and FomA. J Periodontol. 2022;93:515–525. doi: 10.1002/JPER.21-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C., Zhu L., Wang D., et al. T6SS secretes an LPS-binding effector to recruit OMVs for exploitative competition and horizontal gene transfer. ISME J. 2021 doi: 10.1038/s41396-021-01093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li M., Zhou H., Yang C., et al. Bacterial outer membrane vesicles as a platform for biomedical applications: An update. J Control Release. 2020;323:253–268. doi: 10.1016/j.jconrel.2020.04.031. [DOI] [PubMed] [Google Scholar]
- 56.Micoli F., MacLennan C.A. Outer membrane vesicle vaccines. Semin Immunol. 2020;50 doi: 10.1016/j.smim.2020.101433. [DOI] [PubMed] [Google Scholar]
- 57.Chiu Y.K., Yin T., Lee Y.T., et al. Proteomic Profiling of Outer Membrane Vesicles Released by Escherichia coli LPS Mutants Defective in Heptose Biosynthesis. J Pers Med. 2022;12 doi: 10.3390/jpm12081301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Engevik M.A. Fusobacterium nucleatum Secretes Outer Membrane Vesicles and Promotes Intestinal Inflammation. MBio. 2020;12:e02706–e02720. doi: 10.1128/mBio.02706-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L., Liang L., Yang C., et al. Extracellular vesicles of Fusobacterium nucleatum compromise intestinal barrier through targeting RIPK1-mediated cell death pathway. Gut Microbes. 2021;13:1–20. doi: 10.1080/19490976.2021.1902718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Babaei G., Aziz S.G., Jaghi N.Z.Z. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed pharmacotherapy Biomed pharmacotherapie. 2021;133 doi: 10.1016/j.biopha.2020.110909. [DOI] [PubMed] [Google Scholar]
- 61.Bitto N.J., Cheng L., Johnston E.L., et al. Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J Extracell Vesicles. 2021;10:e12080. doi: 10.1002/jev2.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.