Graphical abstract
Keywords: Upland cotton, Gossypium purpurascens, Genome, Structural variation, Domestication, Origin
Highlights
-
•
A high-quality genome of G. purpurascens is assembled for the first time.
-
•
The HIC belongs to G. purpurascens and is best classified as a primitive race of G. hirsutum.
-
•
G. purpurascens may have been partly domesticated and were used for YAZHOUBU (Yazhou cloth) in Sanya.
-
•
G. purpurascens seeds possibly disperse from Central America to South China by ocean.
-
•
SVs are found to have important effects on cotton domestication and improvement.
Abstract
Introduction
Allotetraploid upland cotton (Gossypium hirsutum L.) is native to the Mesoamerican and Caribbean regions, had been improved in the southern United States by the mid-eighteenth century, was then dispersed worldwide. However, a Hainan Island Native Cotton (HIC) has long been grown extensively on Hainan Island, China.
Objectives
Explore HIC’s evolutionary relationship and genomic diversity with other tetraploid cottons, its origin and whether it was used for YAZHOUBU (Yazhou cloth, World Intangible Cultural Heritage) weaving, and the role of structural variations (SVs) in upland cotton domestication.
Methods
We assembled a high-quality genome of one HIC plant. We performed phylogenetic analysis, divergence time estimation, principal component analysis and population differentiation estimation using cotton assemblies and/or resequencing data. SVs were detected by whole-genome comparison. A F2 population was used for linkage analysis and to study effects of SVs. Buoyancy and salt water tolerance tests for seeds were conducted.
Results
We found that the HIC belongs to G. purpurascens. G. purpurascens is best classified as a primitive race of G. hirsutum. The potential for long range transoceanic dispersal of G. purpurascens seeds was proved. A set of SVs, selective sweep regions between G. hirsutum races and cultivars, and quantitative trait loci (QTLs) of eleven agronomic traits were obtained. SVs, especially large-scale SVs, were found to have important effects on cotton domestication and improvement. Of them, eight large-scale inversions strongly associated with yield and fiber quality have probably undergone artificial selection in domestication.
Conclusion
G. purpurascens including HIC is a primitive race of G. hirsutum, probably disperse to Hainan from Central America by floating on ocean currents, may have been partly domesticated, planted and was likely used for YAZHOUBU weaving in Hainan much earlier than the Pre-Columbian period. SV plays an important role in cotton domestication and improvement.
Introduction
Cotton has been cultivated and used to make fabrics for at least 7,000 years. Allotetraploid upland cotton (Gossypium hirsutum L.), the largest source of renewable textile fiber in the world and accounting for >90 % of the world’s production, originated from one hybridization event 1–1.5 million years ago (MYA) [1], [2]. It has generally been accepted that the modern American upland cotton crop was firstly established in the southern United States from domesticated early-cropping perennials through extensive human selection, and then subsequently dispersed worldwide. Historically, most of the cotton produced in China was short, thick-fiber Asian cotton (Gossypium arboreum), which had been cultivated for >2,000 years. Upland cotton cultivars from the United States were introduced into China several times from 1865 to 1950, and upland cotton eventually completely replaced Asian diploid cottons [3]. The excellent upland cotton cultivars have made a huge contribution to cotton production in China since 1950. Thus, cotton is a successful example of crop introduction in China.
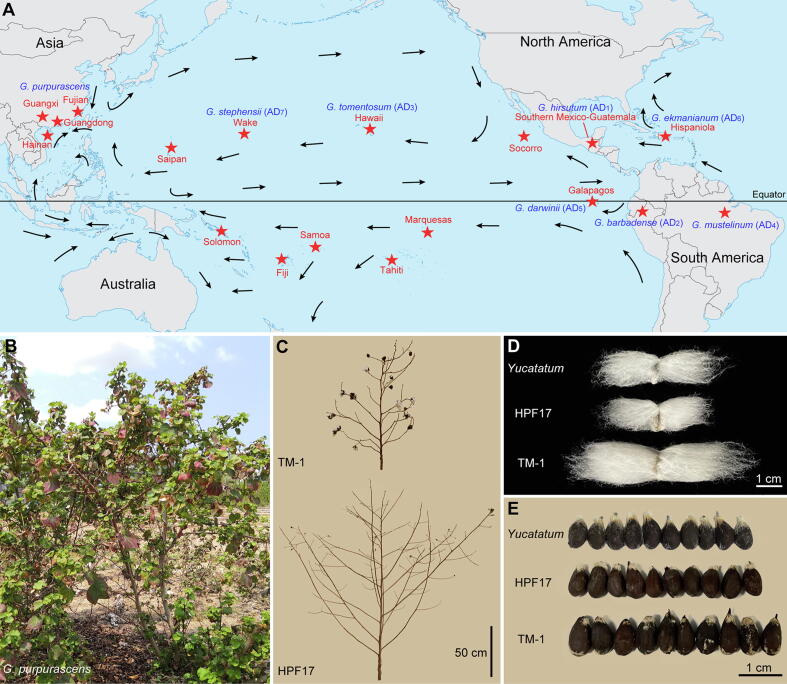
Gossypium wild relatives grow primarily as perennial upright shrubs or small trees and exist in various stages of domestication as feral derivatives. Through intensive study of germplasm collections, Hutchinson [4] identified one wild and six partly domesticated races (not botanical varieties) of G. hirsutum based on their morphologies and distinct geographic distributions, mainly in the Americas. The introduction of the annual race Latifolium into the southern United States led to the establishment of the American upland cotton crop. However, the wild species or landraces of Gossypium are also extensively present on Pacific islands including the Galapagos (G. darwinii), Hawaii (G. tomentosum), Wake Island (G. stephensii), Socorro Island, the Marquesas Islands, Tahiti, Samoa, Fiji, Solomon, Saipan Islands, and the South China Sea Islands (G. purpurascens) [5], [6], [7], [8], [9], [10], [11], [12] (Fig. 1A). G. purpurascens, widely distributed in China, India, Congo, Africa, Egypt, and Brazil during the 17th century [13], [14], was firstly classified as a distinct species (G. purpurascens Poir.) [14], [15], [16], and then as a G. hirsutum race [17]; however, it was not afterward included in the geographical races of G. hirsutum [4]. There also exists a native cotton (Hainan Island cotton, HIC) that has long been grown extensively in South China; HIC plants can be easily found in coastal and insular habitats in Hainan, Guangdong, Guangxi, and Fujian provinces [18]. This cotton is generally thought to be G. purpurascens, and might have been naturally growing on the South China Sea Islands for centuries [19]. Its origin, evolutionary relationship and when and how it was introduced to these regions have not been reported.
Fig. 1.
The distribution of discovered wild tetraploid cottons in the Pacific Ocean and typical phenotypes of G. purpurascens. HPF17 is a G. purpurascens and TM-1 is a cultivated line of G. hirsutum. (A) Distribution of discovered wild tetraploid cottons and the main ocean currents in the Pacific Ocean. The red stars indicate the locations where exist wild tetraploid cottons. The black arrows indicate the directions of ocean currents during Northern Hemisphere winter. The distribution and directions of ocean currents refer to Encyclopedia Britannica (https://www.britannica.com/science/ocean-current). The map is a screenshot of the original map drawn by Ministry of Natural Resources of the P. R. China (Drawing review No. GS (2016)1665). (B) A perennial G. purpurascens plant grew in Yacheng, Hainan Island, China. (C) Plant architectures of HPF17 and TM-1. (D) Phenotypes of fiber length. (E) Phenotypes of seed size. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Structural variations (SVs) are important in plant evolution, domestication and genomic diversity [20], [21], [22]. Here, we assembled the genome of a wild HIC plant that grew naturally in Yacheng, Sanya, Hainan Island, one of the birthplaces of China’s textile industry. High-quality assembly of HIC allows us to identify that SVs, especially large-scale SVs are strongly associate with cotton domestication and improvement, by direct whole-genome comparison of twelve tetraploid cotton assemblies and association analysis between SVs and agronomic traits. Our work illustrates the important effects of SVs on crop domestication and improvement.
Materials and methods
Plant material
HPF17 is one Hainan Island cotton (HIC) belonging to G. purpurascens and was collected from Yacheng, Sanya, Hainan Province, China (Fig. 1B–E, Fig. S1 and Table S1). HPF17 is a perennial that has the strong vegetative growth habit, small seeds with hard seed coats and long dormancy, strong photoperiod response, small boll size and low boll number per plant, low lint yield, and poor fiber quality common to most wild species of Gossypium; it also has few (or no) trichomes on its stems and leaves. The original seeds collected from the wild have been preserved and the HPF17 is one of these original seeds. Self-pollinating for HPF17 and its progenies is being conducted to get self-bred lines.
Sampling and sequencing
Young fresh leaves from HPF17 plant were collected and genomic DNA from leaves was isolated with CTAB method. For whole-genome Illumina sequencing, DNA sequencing libraries were constructed according to the TruSeq DNA Sample Preparation Guide, and sequenced on Illumina NovaSeq 6000 platform with PE150 mode.
For PacBio sequencing, circular consensus sequencing (CCS) libraries were constructed using the Pacific Biosciences SMRTbell Template Prep Kit 1.0, and were size-selected by Sage ELF for 14–17-Kb molecules, followed by primer annealing and the binding of SMRTbell templates to polymerases with the DNA/Polymerase Binding Kit. Sequencing was performed on the PacBio Sequel II System (https://www.pacb.com/products-and-services/pacbio-systems/sequel/) with HiFi mode. A total of five SMRT cells were sequenced.
For Hi-C sequencing, the Hi-C library was prepared according to the standard procedure. Nuclear DNA was cross-linked in situ, extracted, and then digested by Mbo I restriction endonucleases, the sticky ends of the digested fragments were subsequently biotinylated, diluted and ligated randomly. The sequencing library was constructed with the enriched biotinylated DNA fragments, and sequenced on Illumina NovaSeq 6000 platform with PE150 mode, generating 569 Gb data for HPF17.
For RNA-seq, total RNAs were extracted from the roots, stems and leaves of 2-week-old HPF17 plants and subjected to RNA-seq on Illumina NovaSeq 6000 platform with PE150 mode, yielding on average 22 M reads for each biological replicate (three biological replicates for each tissue). PacBio iso-seq data were downloaded from NCBI Nucleotide database [23].
Genome assembly
Consensus reads (HiFi reads) were generated using ccs software (v4.0.0) in SMRTLink (v8.0) (https://github.com/pacificbiosciences/unanimity), yielding 130.66 Gb of CCS reads. Firstly, contigs were assembled using CCS reads by hifiasm (v0.2.0) [24] with default parameters. Illumina paired-end Hi-C reads were mapped to the contig assembly by Bowtie2 (v2.2.3) [25]. Subsequently, HiC-Pro (v2.7.8) (https://github.com/nservant/HiC-Pro) was used to process the mapped Hi-C reads to obtain valid read pairs and generate normalized contact maps. Finally, Pseudochromosomes were organized using LACHESIS [26] with key parameters including “CLUSTER_MIN_RE_SITES = 26, CLUSTER_MAX_LINK_DENSITY = 10, CLUSTER_NONINFORMATIVE_RATIO = 4”, followed by manual correction.
Assessment of the assembly quality
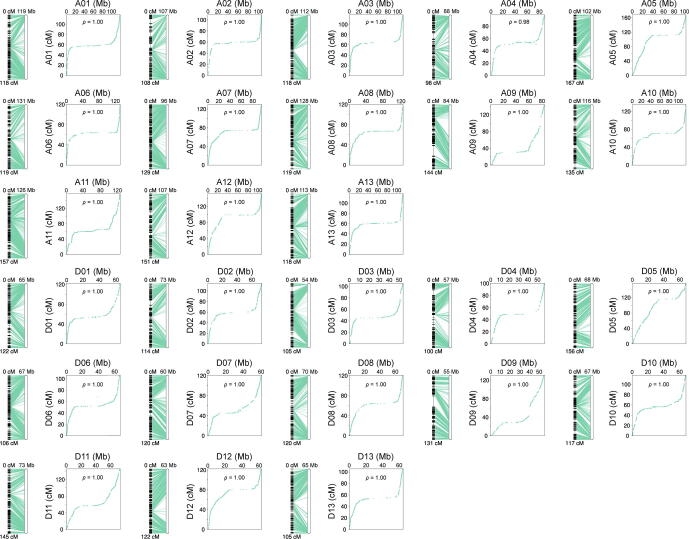
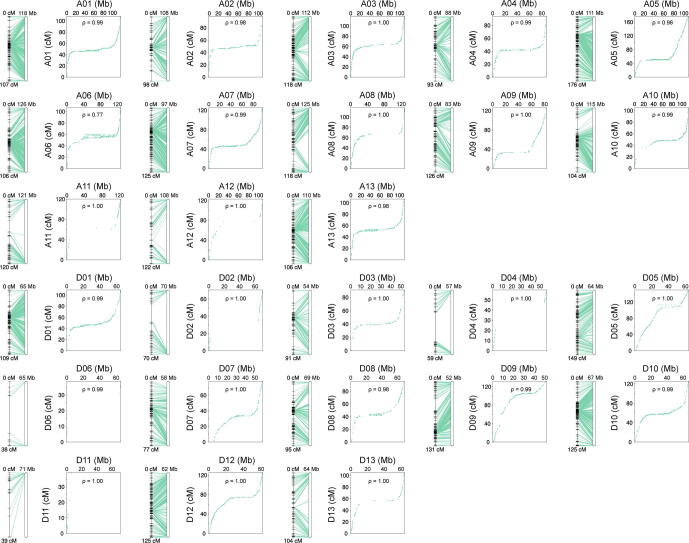
Quality of the genome assembly was evaluated as follow: (1) A genetic map for 177 (TM-1 × HPF17)F2 individuals was constructed. Collinearity analysis and Spearman coefficient calculation of each chromosome were conducted to assess consistency between the assembly and the genetic map. (2) 36 full-length BAC sequences downloaded from NCBI Nucleotide database [23] were mapped to the assembly using MUMmer (v4.0.0rc1) [27], and the coverage and identity were calculated. (3) The whole-genome Illumina paired-end reads were mapped to the assembly using the Burrows-Wheeler Aligner (BWA) software (v0.7.17) [28] for calculating the mapping rate and genome coverage. (4) Benchmarking Universal Single-Copy Orthologs (BUSCO) (v5.3.3, embryophyta_odb10) [29] was applied to assess the integrity of genic regions. (5) Transcripts from Illumina RNA-seq and PacBio iso-seq were aligned to the assembly by blat (v36) [30], and the coverage and mapping ratio of transcripts were calculated to estimate the completeness of the assembly. (6) Assembly quality assessment index based on LTR (LAI) [31] was calculated by LTR_retriever (v2.6) [32]. LTRs were detected by LTRharvest (v1.5.1) [33] with parameters “-similar 85 -vic 10 -seed 20 -seqids yes -minlenltr 100 -maxlenltr 20000 -mintsd 4 -maxtsd 6 -motif TGCA -motifmis 1” and LTR_FINDER (v1.05) [34] with parameters “-D 20000 -d 1000 -L 7000 -l 100 -p 20 -C -M 0.85”, the results from the initial predictions were then integrated according to the LTR_retriever pipeline [32].
Gene prediction and annotation
Gene structures were predicted using three strategies: transcriptome-based, homolog-based, and ab initio prediction. Proteins from seven plant genomes (Arabidopsis thaliana, Hibiscus syriacus, Theobroma cacao, G. herbaceum, G. arboretum, G. raimondii and G. hirsutum) (Table S2) were aligned to the genome assembly by TBLASTN (v2.2.26) [35] (E-value of 1e−5), and then used to predict gene structures by GeMoMa (v1.6) [36]. Two pieces of software, Augustus [37] and GeneMark-ES [38] were employed for ab initio prediction. RNA-seq data from multiple tissues were assembled by Trinity (v2.4.0) [39]. The assembled transcripts and high quality Iso-Seq transcripts were mapped to the genome with GMAP [40] and used in gene structure prediction with PASA [41]. All these evidences were combined by EvidenceModeler (EVM) [42], generating a high-confidence, non-redundant set of gene structures. Alternative splicing and untranslated regions (UTR) were subsequently added by PASA (https://github.com/PASApipeline/PASApipeline/projects).
The functional annotation of the predicted proteins was performed by BLAST (v 2.2.26) [35] searching against various functional databases including NT (https://www.ncbi.nlm.nih.gov/nucleotide/), Swissprot (https://www.uniprot.org/), NR (ftp://ftp.ncbi.nlm.nih.gov/blast/db/FASTA/nr.gz), eggnog (https://eggnogdb.embl.de/), GO (https://geneontology.org/page/go-database) and KEGG (https://www.genome.jp/kegg/), and HMMER (v3.1) [43] searching against PFAM (https://xfam.org/). A program to identify genome-wide plant transcription factors (TFs), iTAK (v1.7) [44] was used to predict TFs in the genome with default parameters.
Identification of short variations and structural variations (SVs)
Eleven tetraploid cotton genomes (Table S3) were aligned against the HPF17 genome by MUMmer (v4.0.0rc1) [27] with key parameters of “-c 100 -l 40”. One-to-one genomic alignment results were extracted with the “delta-filter -1” parameter, and single nucleotide polymorphisms (SNPs) and small insertion and deletions (InDels) were identified by delta2vcf module in MUMmer from the one-to-one alignment blocks. SNPs and InDels were then merged into a non-redundant set by BCFtools [45]. The putative functional effects of SNPs and InDels were annotated by ANNOVAR [46].
For identification of SVs, the eligible alignment blocks from MUMmer were extracted with the “delta-filter -m -i 90 -l 100” parameter, and SVs were then detected using SyRI [47] with default parameters. The detected variations from SyRI comprise two hierarchy structures: genome rearrangements and sequential variations. The rearrangements and sequential variations in syntenic regions were used for further analysis. According to the variation definitions in SyRI outputs, these variations were converted into three types of SVs: Presence/Absence variations (PAVs), inversions (INVs), and translocations (TRANSs) according to the method described previously [21].
For merging SVs from the twelve genomes, the following criterion was used: two SVs were merged as one if the overlapping ratio of these two SVs (the length of the overlapping sequence/the length of non-redundant genome segment covered by the two SVs) exceeds 90 %. The criterion tests were performed with the SURVIVOR (v1.0.7) software (https://github.com/fritzsedlazeck/SURVIVOR).
SV validation
Eight large-scale INVs were validated by Hi-C data, PCR amplification and sequencing read mapping. Hi-C data from HPF17 were mapped to the HPF17, TM-1 and Hai7124 genome, and the normalized contact maps were subsequently generated. Primers were designed for the PCR amplification of inversion borders using appropriate templates. PacBio reads and Illumina paired-end reads (Table S4) were mapped to the TM-1 genome, and subsequently the pairs of split reads and complete reads covering the inversion borders were manually counted.
Re-alignment of the Illumina paired-end reads by BWA (v0.7.17) [28] showed mapped reads aligned to inversion borders, which could be classified into three cases: For one read and one inversion’s two borders, (1) The whole read covered one border; (2) A part of the read mapped to one border’s one side, and the other part of this read mapped to the other border’s same side; (3) The whole read mapped to one border’s one side, and the other read of this pair mapped to the other border’s same side. For PacBio long reads, there were only case (1) and case (2). Case (1) negates the existence of inversion and case (2) and case (3) support. When three cases occur in one accession, it is suggested that this accession is heterozygous on this inversion. Only reads of primary alignments were considered.
Genetic map construction using the TM-1 × HPF17 mapping population
Two genetic maps were constructed using a F2 population from a cross of TM-1 × HPF17. (1) For assembly assessment, DNA libraries of 177 F2 individuals were sequenced (∼5 × genome coverage) on the Illumina NovaSeq 6000 platform with PE150 mode. SNPs between TM-1 and HPF17 were identified using Samtools (v1.10) [48] and Genome Analysis Toolkit (GATK) (v4.1.9.0) [49]. A total of 2,548,643 simple SNPs were retained after filtering by Bcftools (v1.11) [45] with key parameters of “-m2 -M2 -i ‘F_MISSING <= 0.2 & MAC >= 3 & MAF >= 0.05’”. Simple SNPs from the parents and SNPs with the same genotype in the population were classified into a bin using SNPbinner (v1.0.0) [50]. Bins were assigned to linkage groups and ordered by LepMap3 (v0.2) [51] with a logarithm of odds (LOD) score of 9. The final map consisted of 9,762 bins. (2) For QTL mapping, SNPs of 326 F2 individuals and two parents were detected using “ZJU CottonSNP40K” liquid chip [52]. SNPs showing non-polymorphism in parents or progenies, or missing in any parent were deleted. Finally, a genetic map was constructed by LepMap3 (v0.2) [51] with a LOD score of 9 using 6,301 SNPs filter by P-value for segregation distortion > 0.01.
Identification of quantitative trait loci (QTLs) using the TM-1 × HPF17 mapping population
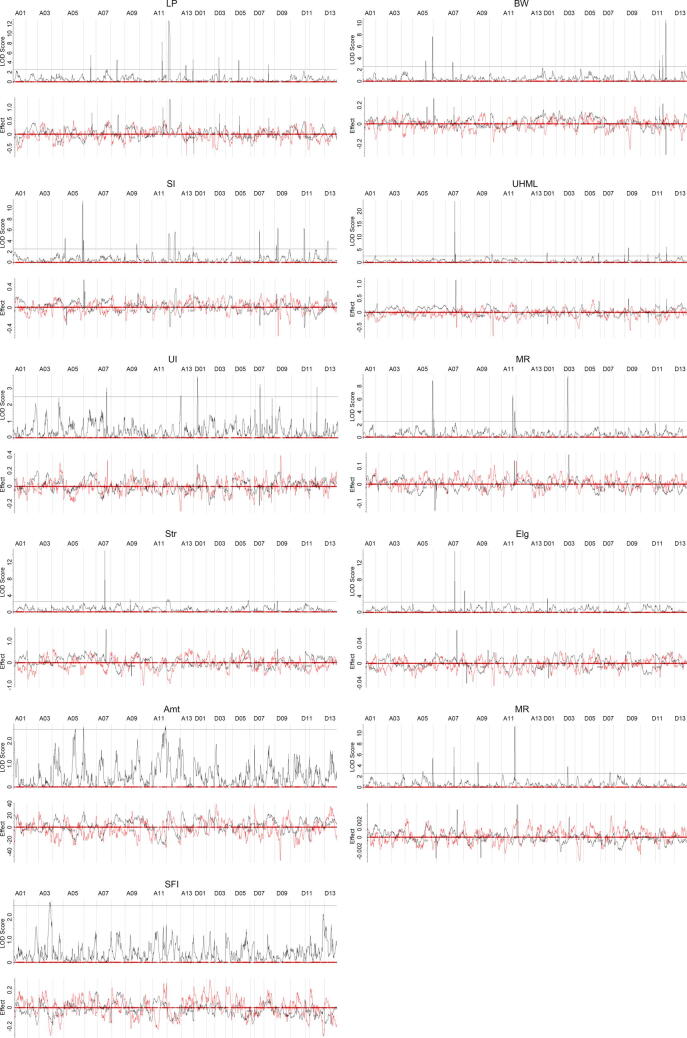
The F2 individuals were planted in the field in Dangtu, Anhui Province, China in 2020 for phenotype collection of yield and fiber quality, including link percentage (LP), boll weight (BW), seed index(SI), upper half mean length (UHML), uniformity index (UI), Micronaire value (Mic), breaking tenacity (Str), breaking elongation (Elg), Amount of fibers (Amt), maturity ratio (MR) and short fiber index (SFI). Standard field management practices were applied, including fertilization, irrigation, weed management and pest management, following the usual local management practices. The mature fiber traits for each F2 plant were measured using a high-volume fiber-test instrument (HVI9000; Uster Technologies AG, Charlotte, NC, USA). The identification of QTLs was performed using QTL IciMapping (v4.2) [53] software with the “ICIM-ADD” mapping method and a LOD threshold of 2.5. The walk step was 1 cM and the PIN value was 0.001.
Gene family identification, phylogenetic tree construction, divergence time estimation and synonymous substitution rates estimation based on orthologous genes
Protein sequences from HPF17 and other ten accessions (T. cacao, G. raimondii, G. herbaceum, G. arboreum, and the two subgenomes each from G. hirsutum, two G. barbadense, G. tomentosum, G. mustelinum, G. darwinii) (Table S5) were used for gene family identification. For genes with alternative splicing variations, the longest transcripts were kept as representative. An all-vs-all BLASTP (v2.2.28) (E-value threshold 1e−5) analysis was carried out and OrthoMCL (v1.4) [54] was used to identify orthologous and paralogous genes with default parameters. Multiple sequence alignment for all single-copy orthologous genes was performed using MUSCLE [55]. Finally, the phylogenetic tree was constructed using PhyML (v3.1) [56] by maximum-likelihood (ML) method with 1,000 bootstrap replicates. T. cacao was used as the outgroup. Divergence times between these species were estimated using MCMCTREE software in the PAML package (v4.9d) [57]. Calibration times came from TimeTree database (https://www.timetree.org/). Synonymous substitution rates (Ks) between subgenomes of each species were estimated using the Codeml program in PAML package (v4.9d) [57] with the free-ratio “branch” model based on orthologous gene sets.
Phylogenetic tree construction with high-depth resequencing data
Illumina resequencing data of 4 diploids and 24 allotetraploids at high depth (Table S6) were mapped to the subgenome A of TM-1 [23] (for diploids) or the whole genome of TM-1 (for allotetraploids) using BWA (v0.7.17) [28] with the MEM algorithm. After alignment, only reads mapped to the subgenome A were selected for next analysis. Mapped reads were sorted according to their aligned chromosomal positions, the potential PCR duplicates were removed, and only unique alignments were retained using SAMtools (v1.10) [48]. SNP calling was performed using the HaplotypeCaller and GenotypeGVCFs modules in GATK (v4.1.9.0) [49]. To obtain high-quality SNPs, only variations passed the filtering by BCFtools (v1.11) [45] with key parameters of “-m2 -M2 - 'F_MISSING > 0.15 | MAC < 3 | MAF < 0.05'“ were retained for further analysis. The phylogenetic tree in ML method was constructed using the SNPs on fourfold degenerate sites by PhyML (v3.1) [56] with 1,000 bootstrap replicates. T. cacao was used as the outgroup.
Phylogenetic analysis, principal component analysis (PCA), genetic diversity and population differentiation estimation with resequencing data of 382 cotton accessions
Resequencing data of 382 tetraploid cotton accessions (Table S7) were mapped to the TM-1 genome [23] to conduct SNP calling using the mpileup and call modules of BCFtools (v1.11) [45]. A subset of 1,797,864 SNPs was filtered from the raw SNP set (QUAL > 30, MQ > 30, MAF > 0.05, missing rate < 0.1). Subsequently a phylogenetic tree was constructed by MEGA-X (v10.2.5) [58] with neighbor-joining (NJ) method. PCA of SNP diversity was performed using plink (v1.90) [59].
Nucleotide diversity (π) [60] was calculated using VCFtools (v0.1.12b) [61]. The diversity ratios (ROD, πlandraces/πcultivars) between different groups were calculated as an estimate of selective sweeps. Fixation index (Fst) [62] values were measured using VCFtools. Cross-population composite likelihood ratio (XP-CLR) values were calculated using xplcr software python version (v1.1.2) [63]. All the three values were calculated with 100-Kb windows sliding in 20-Kb steps. Windows in which SNPs<10 were filtered. The average π or Fst of all sliding windows were considered as the values at a whole-genome level in/between different groups. Putative selective sweep regions were identified as the genomic regions with the top 5 % ROD, weighted Fst or normalized XP-CLR values between landraces and cultivars.
Buoyancy and salt water tolerance tests for seeds
Seeds of G. purpurascens (HPF17) and G. hirsutum cultivated line (TM-1) were used in the tests. Artificial seawater consisted of sea salt (Haibao Ltd.) and ultra-pure water (35 g sea salt per litre of water), and was sterilized. All the tests were conducted in the conditions of 28℃ and 14 h Light/10 h Dark. To test buoyancy, seeds with long fibers were placed in buckets to which 1 L of artificial seawater had been added, and the seeds remaining floating were counted after a number of days (three replicates for each sample). To test the salt water tolerance, seeds after acid-delinting and disinfection using 5 % H2O2 were immersed in seawater continuously, and were randomly sampled for germination test at monthly intervals. For germination test, seeds were washed with ultra-pure water several times, the seed coats were removed, and the seeds were placed on germination pads to test viability [8].
Statistical analysis
The two-tailed Student’s t-test, correlation analysis and the descriptive statistics were performed in R (v3.6.3). The significance level was set at P = 0.05 for the two-tailed Student’s t-test.
Results
Assembly and annotation of the high-quality genome
One wild HIC cotton plant named “HPF17” grew naturally in Yacheng was selected for genome assembly (Fig. 1B–E, Fig. S1 and Table S1). The genome size of HPF17 was estimated to be ∼ 2,055 Mb based on K-mer analysis [64] (Tables S8 and S9). We produced 130.65 Gb (∼57 × genome equivalent) (Table S10) of high-quality PacBio circular consensus sequencing (CCS) reads and assembled the genome using hifiasm [24]. The initial assembly generated 3,394 contigs with an N50 value of 13.20 Mb (Table S11). Subsequently, we categorized and ordered the contigs to produce chromosome-scale scaffolds using approximately 50-fold coverage of valid unique interacting paired-end data generated through high-throughput chromosome conformation capture (Hi-C) (Fig. S2 and Table S12). The final assembly included 2,558,691,823 bases and 3,343 scaffolds, resulting in a scaffold N50 value of 88.21 Mb (Table S11). A total of 2,267,310,418 (88.61 %) bases from the contigs were anchored and oriented onto pseudochromosomes (Table S13). A small number of contigs could not be anchored mainly because of the high heterozygosity in corresponding genomic regions (detailed in next section).
To validate our assembly, we randomly sequenced 177 individuals of a F2 population derived from a cross between the G. hirsutum genetic standard line Texas Marker 1 (TM-1) [1] and HPF17, and constructed a genetic map using 2,548,643 high-quality single nucleotide polymorphism (SNP) loci. The map consisted of 9,762 bins (6,490 non-redundant bins) spanning a total distance of 3,260.23 cM with an average marker interval of 0.50 cM in 26 linkage groups (Fig. S3 and Table S14). Assembly quality was evaluated by conducting collinearity analysis of each chromosome with the genetic map (Fig. S4 and Tables S14 and S15). The average Spearman coefficient of chromosomes was 0.998 and the smallest was 0.966, suggesting high consistency between the assembly and the genetic map. In addition, the accuracy and completeness of the assembly were confirmed by perfect matches with 36 completely sequenced bacterial artificial chromosome sequences of G. hirsutum [1] (Table S16), and by the high mapping ratio (99.66 %) and coverage ratio (99.55 %) of the 268.83 Gb of Illumina data generated from HPF17 (Tables S8 and S17). Likewise, the identification of 1,601 (99.19 %) of the 1,614 highly conserved core proteins in the embryophyta Benchmarking Universal Single-Copy Orthologs (BUSCOs) [29] dataset (Table S18) and the high coverage rate (99.70 %) of mRNA sequences from HPF17 (Table S19) evidenced the completeness of the assembly in genic regions. Moreover, the high long terminal repeat (LTR) assembly index (LAI) [31] score (16.16), as well as the great contig N50 size all indicated that we had assembled a high-quality genome (Fig. 2A and Table 1).
Fig. 2.
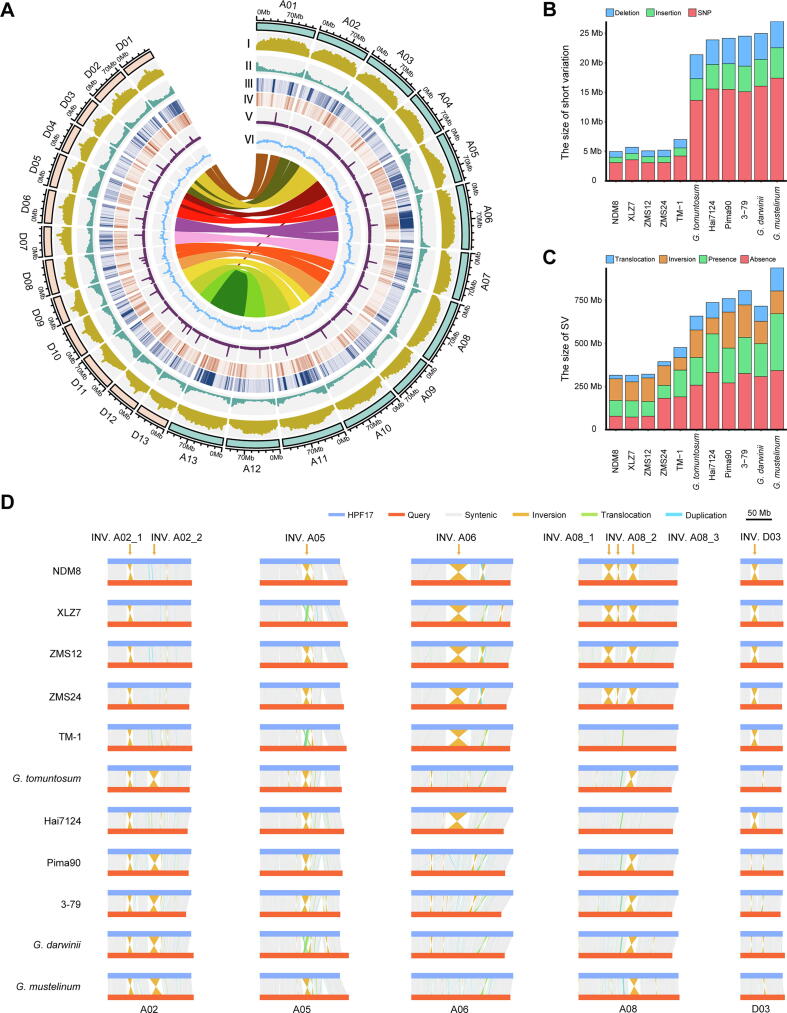
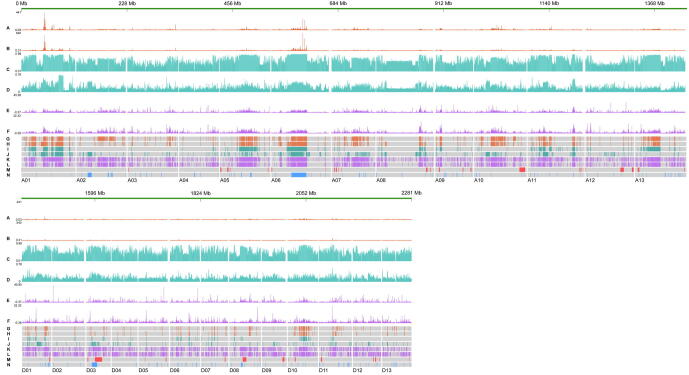
Chromosomal features and genome alignment of the HPF17 genome. (A) Multi-dimensional display of genomic components of HPF17 across chromosomes. I, Transposable element (TE) density. II, Gene density. III, Density of SNPs between HPF17 and TM-1. IV, Density of InDels between HPF17 and TM-1. V, Chip-seq signals for centromeres. VI, GC content. All these data are shown in 1-Mb windows. The inner lines show genic synteny blocks in homoeologous chromosomes between the subgenome A and D. (B, C) The total size of short variations (B) and SVs (C) in each cotton genome compared to HPF17. (D) The genome alignment of five chromosomes between HPF17 and other tetraploid cottons. Eight large-scale inversions were marked by orange arrows. In these synteny plots, the reference genome (HPF17) is represented by blue horizontal lines and the query genome by red horizontal lines. Vertical lines represent syntenic (grey), inverted (orange), translocated (green), and duplicated (light blue) regions, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Table 1.
Summary of the genome assembly and annotation for HPF17.
| Genomic features | HPF17 |
|---|---|
| Total length of scaffolds (Mb) | 2,558.69 |
| Anchored and oriented scaffolds (Mb) | 2,267.31 |
| Genome in chromosomes (%) | 88.61 |
| Estimated genome size (Mb) | 2,055.07 |
| Gap size (Mb) | 0.0383 |
| Number of contigs | 3,394 |
| Contig N50 (Mb) | 13.20 |
| Number of scaffolds | 3,343 |
| Scaffold N50 (Mb) | 88.21 |
| GC content (%) | 34.73 |
| Repeat sequences (%) | 64.67 |
| Predicted protein-coding genes | 79,146 |
| Annotated protein-coding genes | 78,259 |
| Average gene length (bp) | 3,065 |
| Average exon number per gene | 5.79 |
| Complete BUSCOs (%) | 99.19 |
| Illumina read mapping rate (%) | 99.66 |
| LAI | 16.16 |
The current HPF17 assembly consisted of 79,146 predicted protein-coding genes with an average length of 3,065 bp and 5.79 exons per gene (Table 1 and Table S20). Among these predicted genes, 73,539 genes were on pseudochromosomes, and 78,259 (98.88 %) were functionally annotated, including 36,030 in the A subgenome and 37,509 in the D subgenome (Table S21). Within the set of annotated genes, 5,558 putative transcription factors from 69 families (Table S22) were identified, representing 7.02 % of protein-coding genes. In addition, de novo prediction and homology-based annotation of noncoding RNAs identified 48,491 ribosomal RNA (rRNA) genes, 4,854 transfer RNA (tRNA) genes, 823 microRNA (miRNA) genes, and 12,903 small nuclear RNA (snRNA) genes (Table S23). Approximately 1,588.94 Mb of genomic sequences were annotated as repeat sequences, covering 64.67 % of the HPF17 genome (Table S24). Tandem repeats and interspersed repeats (transposons) were found to cover 0.47 % and 58.50 % of the genome, respectively. The main type of transposons was retrotransposons (56.89 %), classified as long interspersed nuclear elements (LINEs), short interspersed nuclear elements (SINEs), long terminal repeats (LTRs) and others, respectively covering 0.35 %, 0.0001 %, 56.40 %, and 0.14 % of the genome. Among LTRs, the Copia family was observed to cover far fewer genomic regions than Gypsy (3.59 % vs 43.65 %).
Next, the centromeric DNA regions were identified by centromere-specific H3 (CenH3) ChIP-Seq [65]. In this process, a total of 53.71 M Illumina reads from G. hirsutum [23] were mapped to the HPF17 assembly. Peak calling was performed, and signals were calculated. This analysis yielded potential centromeric regions for all 26 chromosomes of HPF17 (Fig. 2A).
Discovery of short variations and SVs in allotetraploid cottons
Previous study has compared three SV identification pipelines using Illumina short reads, PacBio long reads and high-quality assemblies, respectively, and concluded that the vast majority of SVs cannot be identified without use of sequence comparisons with high-quality assemblies, especially the large-scale SVs [21]. Thus, the genomes of eleven other tetraploid cottons (Table S3) were aligned with the HPF17 genome, including G. hirsutum cultivars (NDM8, XLZ7, Zhongmiansuo (ZMS) 12, ZMS24 and TM-1), G. barbadense (Hai7124, Pima90 and 3–79), G. tomuntosum, G. mustelinum and G. darwinii. SNPs, small insertion and deletions (InDels) (<= 50 bp) were then identified from the one-to-one alignment blocks. On average, 10,037,323 SNPs (range 3,090,642 in Gh-ZMS12 to 17,413,133 in G. mustelinum) with 4.43 SNPs per kilobase and 1,365,444 small InDels (484,284 in Gh-NDM8 to 2,327,121 in Gb-3–79) covering 3,531,119 bp of genomic sequences (1,168,656 bp in Gh-NDM8 to 6,135,163 bp in G. mustelinum) were identified per genome (Table S25). Short variations across all accessions were subsequently merged into a non-redundant set. In total, 41,365,880 non-redundant SNPs and 6,408,143 non-redundant small InDels affecting 18,710,318 bp of genomic sequences were cataloged (Table S26).
The high-quality assemblies allowed us to identify SVs by means of direct comparative genomic analysis in tetraploid species. Eleven cotton assemblies were compared against the HPF17 genome using the whole-genome comparison and sequence variation annotation tool SyRI [47] (Fig. S5). Three types of SVs (>50 bp) were detected, i.e., presence/absence variations (PAVs), inversions (INVs), and translocations (TRANSs). An average of 171,231 SVs (49,213 in Gh-ZMS12 to 325,544 in G. mustelinum) were identified, affecting an average of 363.8 Mb (212.7 Mb in Gh-ZMS24 to 596.2 Mb in G. mustelinum) of genomic sequences per accession (Table S25). Next, SVs across all accessions were merged into a non-redundant set, yielding a total catalog of 805,397 non-redundant SVs affecting 2.93 Gb of genomic sequences, including 728,987 PAVs, 1,767 INVs and 74,643 TRANSs (Table S26). Of these eleven tetraploid cottons, far fewer short variations and SVs were identified between HPF17 and G. hirsutum, suggesting a closer relationship between HPF17 and G. hirsutum (Fig. 2B, C).
SVs are associated with domestication and differentiation of G. hirsutum
As compared to G. purpurascens and other wild species or races, cultivated upland cottons possess a common set of agronomic features known as “domestication syndrome” [66] traits due to extensive human selection. These traits include a full annual growth habit and photoperiod insensitivity, decreased seed dormancy, large boll size and more bolls per plant, high lint percentage and superior fiber quality. To explore domestication and improvement in G. hirsutum, we developed a F2 population consists of 326 individuals derived from a cross between cultivated line TM-1 and race HPF17 of G. hirsutum. SNPs of this F2 population were detected using “ZJU CottonSNP40K” liquid chip [52] and 24,551 high-confidence SNPs were then retained after filtering. A genetic map of this (TM-1 × HPF17)F2 population was constructed using 6,301 filtered SNPs. The final map consisted of 6,299 SNP loci (2,904 non-redundant SNP loci) spanning a total distance of 2769.88 cM with an average marker interval of 0.96 cM in 26 linkage groups (Figs. S6 and S7, Tables S27 and S28). A total of 69 quantitative trait loci (QTLs) (59 non-redundant QTLs) for eleven yield and fiber quality traits (Tables S29–S31) were identified using the inclusive composite interval mapping (ICIM) method [53] (Figs. S8 and S9, Table S32). These QTLs explained 1.99 %–20.60 % of the phenotypic variation. Notably, the traits belonging to “domestication syndrome” had more QTLs identified, with the most being 12 QTLs for seed size (SI), followed by fiber length (UHML) and lint percentage (LP). We next searched for selection signatures in modern cultivars through identifying regions having reduced diversity and/or increased differentiation between G. hirsutum modern cultivars (GHC) and G. purpurascens (GHP). Totally, 2,489 putative selective sweep regions were identified after integrating (Fig. S10 and Tables S33 and S34). Interestingly, 4.3 % of SV regions between HPF17 and TM-1 (Table S35) were QTL regions (Table S36), 1.1 % bigger than that of non-SV regions (3.2 %). Likewise, 27.1 % of SV regions between HPF17 and TM-1 were selective sweep regions (Table S37), 8.4 % bigger than that of non-SV regions (18.7 %). The mean coverage of QTLs and selective sweep regions on the all chromosomes were 3.4 % and 20.0 %, respectively. Notably, of all types of SVs, selective sweep regions were particularly rich in INVs as 55.5 % of INV regions were selective sweep regions, followed by TRANSs (24.5 %) (Table S38). SVs, particularly INVs and TRANSs, were thus supposed to have probably impacted the domestication and improvement of G. hirsutum.
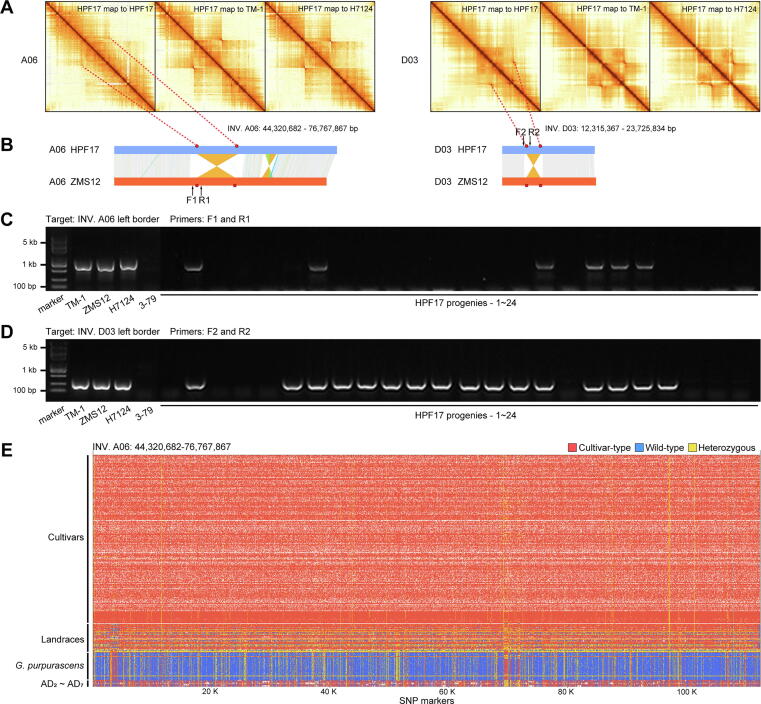
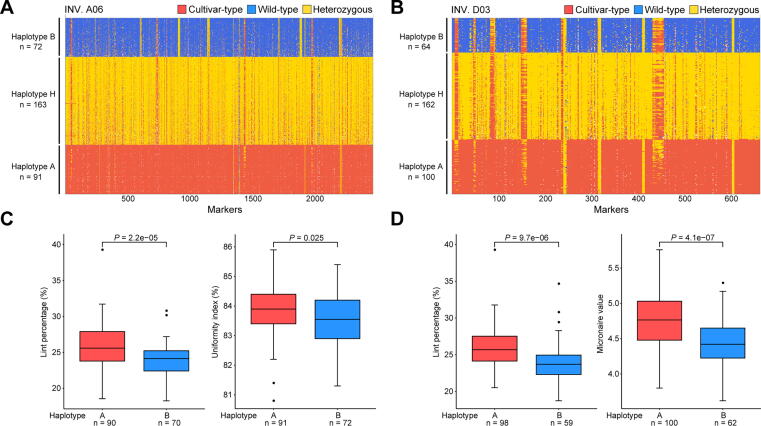
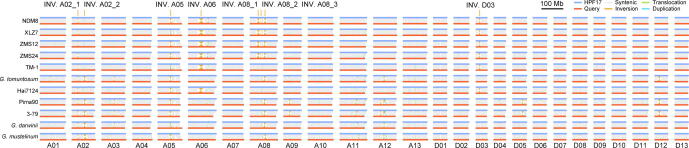
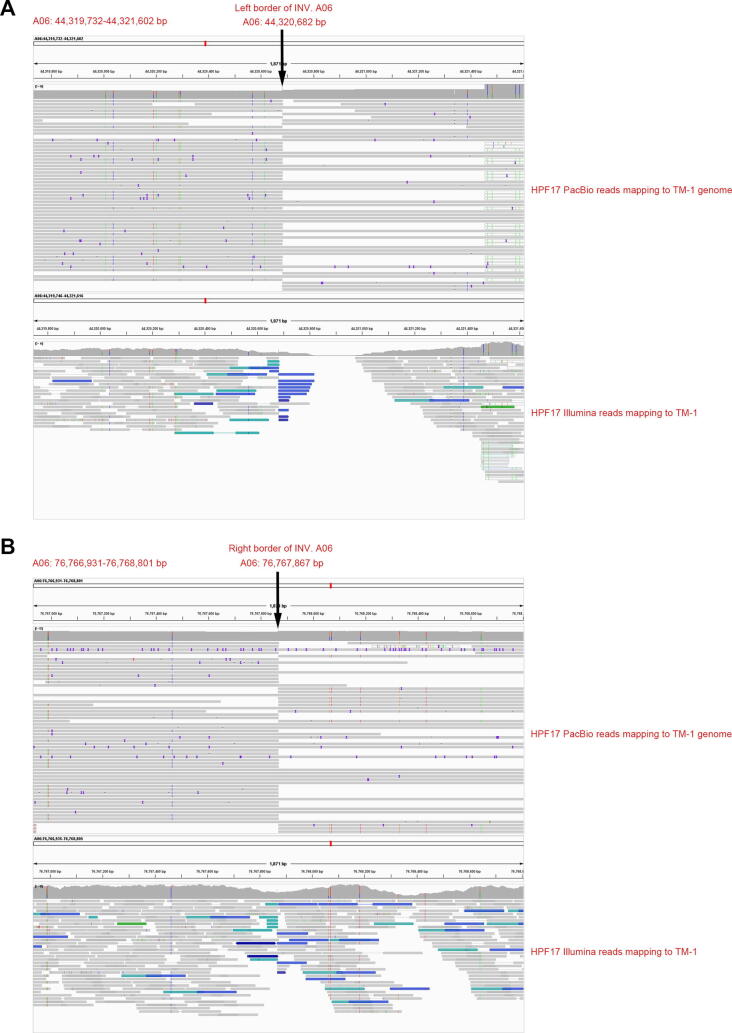
Since the great importance of INVs, eight large-scale INVs diverse in upland cottons were selected for further study. They were in the size range of 4.9–32.4 Mb and on chromosomes A02, A05, A06, A08 and D03 (named as INV. A02_1, INV. A02_2, INV. A05, INV. A06, INV. A08_1, INV. A08_2, INV. A08_3 and INV. D03) (Fig. 2D and Fig. S5). Of those, three (INV. A08_1, INV. A08_2, INV. A08_3) had been reported in previous studies [67]. These INVs were further confirmed by Hi-C data (Fig. 3A) and PCR amplification of borders (Fig. 3B-D and Table S39). Very interestingly, while the results of cultivars used in validations (TM-1, H7124, ZMS12 and 3–79) were consistent with the genome alignment (Fig. 2D), HPF17 was heterozygous on all eight of these INVs. To validate the heterozygous state of these INVs, we performed a manual check, mapping PacBio HiFi reads and Illumina paired-end reads from hundreds of accessions to the TM-1 genome [23] and subsequently counting the pairs of split reads and complete reads covering the INV borders (Fig. S11 and Table S4). The results showed that not only HPF17, but also populations of other G. purpurascens and seven G. hirsutum races from Central America had heterozygous or multiple genotypes (different individuals in the population of one race had different genotypes) on these INVs, suggesting they are still in an unselected state.
Fig. 3.
Example validations of large-scale inversions. (A) Example validations of two large-scale inversions (INV. A06 and INV. D03) based on mapping Hi-C reads to genome assemblies at 500-Kb resolution. The track shows the chromatin interaction matrix with HPF17 Hi-C data mapping against the HPF17, TM-1 and Hai7124 genome. Red dots show higher probability of chromatin contact while yellow dots show lower probability of contact. (B) Diagrams of the two large-scale inversions (INV. A06 and INV. D03) with the red dots representing the inversion borders. (C, D) Example validations of two large-scale inversions (C for INV. A06 and D for INV. D03) based on PCR amplification for inversion borders. (E) Example haplotype classification of large-scale inversion INV. A06 in the natural population consists of 382 accessions. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As most cotton accessions only had short-read sequencing data, we attempted haplotype classification using high-confidence SNPs called with these short-read sequencing data in these large-scale INV regions. Through analyzing the SNP genotypes of each INV (including 2 Mb flanking sequence on each side) from a natural population comprised of 382 cotton accessions (Table S7), three haplotypes were identified for each INV (Fig. 3E and Table S40). The classification results show high consistency with the read mapping results, indicating the accuracy and feasibility of this method. Specifically, the wild tetraploid cottons, G. purpurascens and G. hirsutum races from Central America had heterozygous or multiple genotypes on all eight INVs as expected. However, interestingly, modern cultivars (even the early cultivars such as King, Long star, Mebane and so on) had the same homozygous haplotype (henceforth termed the cultivar-type haplotype) for INV. A02_1, INV. A02_2, INV. A05, INV. A06 and INV. D03 (Table S41), implying that these INVs are strongly associated with G. hirsutum domestication and improvement. However, with regard to INV. A08_1, INV. A08_2, and INV. A08_3, modern cultivars harbored three haplotypes, but the accessions with the wild-type haplotype (another homozygous haplotype opposite to the cultivar-type haplotype) and heterozygous haplotype (Table S41) were much less. These results revealed that these large-scale INVs probably have undergone artificial selection in the early stage of G. hirsutum domestication.
To unveil the relationships between the large-scale INVs and agronomic performance associated with “domestication syndrome”, we further characterized the (TM-1 × HPF17)F2 population. We genotyped three haplotypes from the SNPs on each INV (Fig. 4A, B), then determined the impacts of these INVs on eleven yield and fiber quality traits based on phenotypic differences between individuals with the wild-type and cultivar-type INV haplotypes (Fig. 4C, D and Table S41). Interestingly, INV. A02_1, INV. A02_2, and INV. A06 were significantly associated with lint percentage (LP) and fiber uniformity (UI); INV. A05 with Micronaire value (Mic, a fiber fineness index) and UI; INV. D03 with LP, Mic and maturity ratio (MR); INV. A08_2 with LP, fiber length (UHML), Mic, fiber strength (Str) and fiber elongation (Elg); and INV. A08_3 with LP, Mic, Str, Elg and short fiber ratio (SFI). These results suggested that seed size, lint percentage, fiber qualities and other traits have been significantly improved in modern cotton cultivars probably due to the selection of these INVs, revealing inversions to have played an important role in the domestication and differentiation of G. hirsutum. Notably, the Micronaire value of the cultivar-type haplotype for INV. A05 was significantly lower than that obtained with the wild-type, suggesting that INV.A05 is an important domesticated SV for improving fiber fineness (lower Micronaire value). Furthermore, six INVs were related to increasing LP, the trait of greatest importance in early upland cotton domestication and improvement. However, for three INVs (INV. A08_2, INV. A08_3 and INV. D03), increased LP accompanied with the unduly higher Micronaire value (∼4.5–5.0), i.e. fibers becoming too thick and the maturity of fibers becoming too high. The linkage of high LP and high Mic suggests that the identification of these INVs may contribute to solving the persistent challenge in cotton breeding, namely that plants with higher LP tend to have unduly higher Mic as well. One pleiotropic gene or two (or more) closely-linked genes may exist in the above INVs.
Fig. 4.
Associations between the large-scale inversions and yield and fiber quality traits. (A, B) Example haplotype classification of large-scale inversions (A for INV. A06 and B for INV. D03) in the (TM-1 × HPF17)F2 population. Haplotype A indicates a homozygous haplotype which is the majority in cultivars, haplotype B indicates the opposite homozygous haplotype of haplotype A, and haplotype H indicates the heterozygous haplotype. The F2 population consists of 326 individuals. (C, D) Example box plots for yield and fiber quality traits using F2 individuals with different INV haplotypes (C for INV. A06 and D for INV. D03). The center line denotes the median; box limits indicate the upper and lower quartiles; whiskers mark the 1.5 × interquartile range and points show outliers; n indicates the number of individuals. The significance of phenotypic differences was analyzed with two-tailed t-test.
G. purpurascens is best classified as a primitive race of G. hirsutum
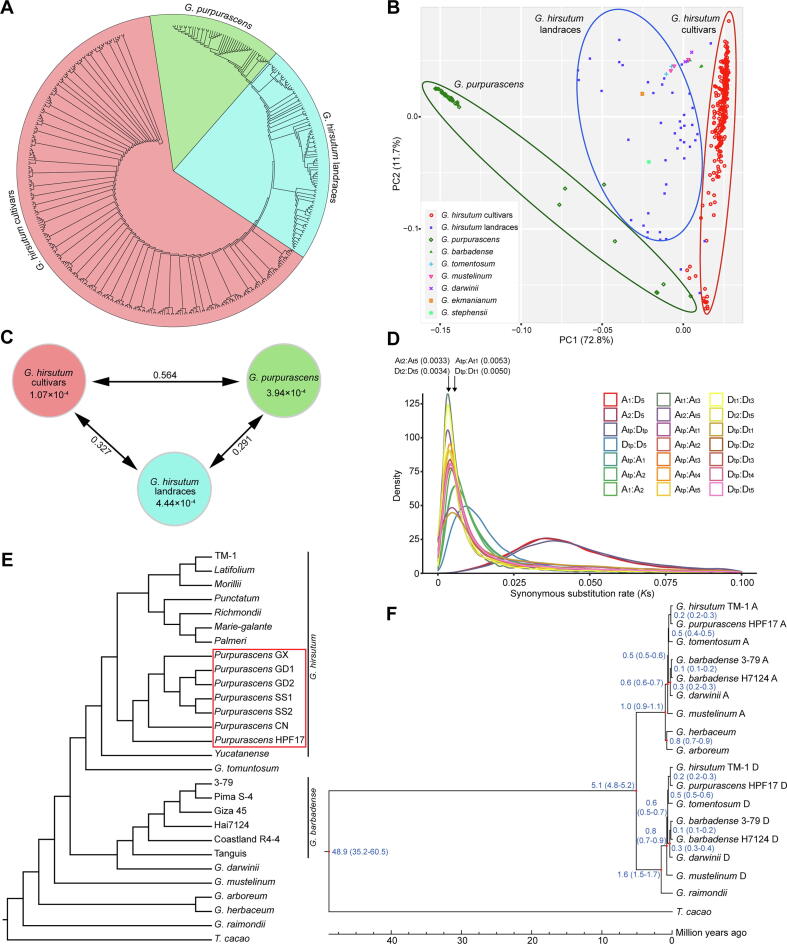
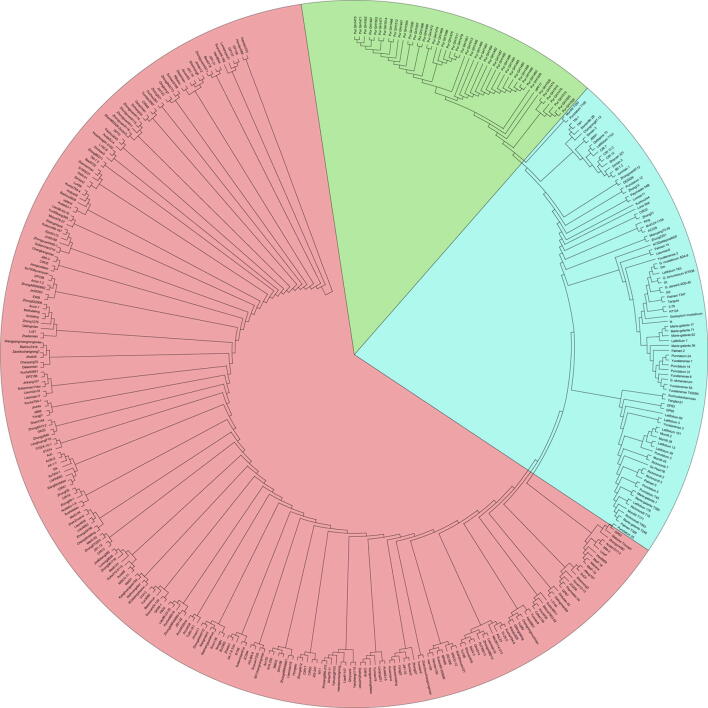
To conduct SNP calling, whole-genome resequencing data of a broad spectrum of G. hirsutum (382 accessions) spanning the full wild to domesticated cotton continuum, including 276 cultivars of G. hirsutum (GHC), 48 G. purpurascens from South China (GHP), 46 landraces or races of G. hirsutum (GHL) from Central America, and 12 other tetraploid cotton species (AD2-AD7) (Table S7) were used. After filtering, 1,797,864 SNPs were identified and used to conduct phylogenetic analysis (neighbor-joining [NJ] method), principal component analysis (PCA), and estimation of the genetic diversity (π) and population differentiation (Fst and XP-CLR). The phylogenetic analysis (Fig. 5A and Fig. S12) and PCA (Fig. 5B) showed that all G. hirsutum accessions clustered into three groups: cultivars (GHC); landraces or races (GHL) including Yucatanense, Richmondi, Morilli, Marie-galante, Palmeri, Punctatum, and Latifolium; and G. purpurascens (GHP). The G. purpurascens accessions including HPF17 formed a tight cluster that was distinct from G. hirsutum cultivars and races, but relatively close to the seven G. hirsutum races.
Fig. 5.
The evolution and domestication of G. purpurascens. (A) A phylogenetic tree constructed using NJ method based on SNPs from 382 tetraploid cotton accessions. Sectors contain the most representatives of each group, respectively. (B) PCA plot of the first two components of SNP diversity. The x-axis shows the first component (PC1) and the y-axis shows the second component (PC2) with contribution values shown in parentheses. Ovals contain the most representatives of each group, respectively. (C) Whole-genome genetic diversity and population differentiation of/among the three groups. The values in the circle represent nucleotide diversity (π) values for each group, and the values on each line indicate weighted population fixation index (Fst) between the two corresponding groups. (D) Ks value density plots for orthologous genes among cotton subgenomes. Peak Ks values are indicated in the parentheses. The Atp and Dtp indicate the subgenome A and D of HPF17, respectively. (E) A phylogenetic tree constructed using ML method based on SNPs on fourfold degenerate sites called with high-depth resequencing data. The red rectangular frame indicates the G. purpurascens accessions. (F) A phylogenetic tree with divergence times constructed using ML method based on single-copy orthologous genes. Divergence times for each node are indicated in blue labels. The red points indicate the nodes of which calibration times were used. Bootstrap values from 1,000 trials were 100 for all the nodes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Further analysis was performed by constructing two other, more accurate phylogenetic trees using the maximum-likelihood (ML) method based on 1) 1,162,978 SNPs from fourfold degenerate sites called with high-depth resequencing data (Fig. 5E and Table S6) and 2) the sequences of 6,191 single-copy orthologous genes (Fig. 5F and Table S5). Both phylogenetic trees demonstrated high consistency in that HPF17 together with other G. purpurascens accessions clustered tightly between the clade of the wild species or race of G. hirsutum, i.e. G. yucatatum, and the clades of the other six geographical races from Central America. This pattern indicated that G. purpurascens is more primitive than the races from Central America except for G. yucatatum, and it is probably also a race of G. hirsutum. The results also indicated that the selected HIC plant belongs to G. purpurascens. The relationships of other accessions were consistent with previously reports [18], [68], [69], [70].
Next, the divergence times for every node in the phylogenetic tree based on single-copy orthologous genes were estimated using MCMCTREE [57] (Fig. 5F). The divergence times between diploid A and diploid D, diploid and tetraploid, G. herbaceum and G. arboreum, and G. hirsutum and G. barbadense were estimated to be 5.1 MYA (4.8–5.2 MYA), 1.6 MYA (1.5–1.7 MYA), 0.8 MYA (0.7–0.9 MYA), and 0.6 MYA (0.5–0.7 MYA), respectively, highly consistent with previous studies [1], [2], [68], [69]. The divergence time between G. purpurascens (HPF17) and G. hirsutum (cultivated line TM-1) was estimated to be 0.2 MYA (0.2–0.3 MYA), similar to 0.3 MYA (0.2–0.4 MYA) between G. darwinii and G. barbadense and 0.5 MYA (0.4–0.6 MYA) between G. tomentosum and G. hirsutum. The peak synonymous substitution rates (Ks) for orthologous genes between G. purpurascens (HPF17) and G. hirsutum (TM-1) were calculated to be 0.0053 and 0.0050 for subgenome A and D, respectively, a little higher than the values between G. darwinii and G. barbadense (0.0033 and 0.0034) and between G. tomentosum and G. hirsutum (0.0038 and 0.0035) (Fig. 5D and Table S42). The peak Ks values of other comparisons were consistent with previous studies [68], [69]. Collectively, the above results suggested a relatively distant relationship between G. purpurascens and cultivated G. hirsutum. Therefore, taking all findings into consideration, G. purpurascens is best classified as a primitive race of G. hirsutum, and may have been grown in Hainan Island much earlier than previously suspected.
Overall nucleotide diversity (π) values in the GHC, GHP, and GHL clusters were estimated to be 1.07 × 10-4, 3.94 × 10-4 and 4.44 × 10-4, respectively (Fig. 5C); the π value in GHC being much lower than those in GHP and GHL indicated a strong genetic bottleneck during upland cotton domestication. Cultivars (GHC) also showed a high degree of differentiation from GHP and GHL with Fst values of 0.564 and 0.327, suggesting that increased selection pressure occurred during the past decades of cultivar development. The similar π values of GHP and GHL, together with the smaller Fst value between GHP and GHL than the value between GHP and GHC also supported G. purpurascens as a race of G. hirsutum. In general, the π and Fst values in this study were higher than those reported several years ago [71], but consistent with more recently reported results [18], [70] due to the increased representation of landraces and the higher sequencing depth.
Salt water tolerance and the origin of G. purpurascens
Perennial tree cottons in G. hirsutum that occupy coastal and insular habitats have hard seed coats and their seeds can remain viable after long periods of immersion in seawater. More broadly, hard seed coats are common to most wild species of Gossypium [8], [72]. Therefore, it is natural to consider the possibility that cotton seeds may be dispersed by ocean currents and arrive in viable condition. In order to explore the potential for long range transoceanic dispersal of G. purpurascens, we tested the buoyancy and salt water tolerance of seeds (Fig. S13 and Table 2). Most seeds with fiber could float on the calm seawater surface for at least six months, whether from HPF17 (64.44 %, n = 3 × 30) or TM-1 (70.00 %, n = 3 × 30), indicating that the seeds of both cultivated and wild cotton may have a certain ability to spread across oceans by floating. However, all the TM-1 seeds absorbed water readily and failed to germinate or lost viability after seven days of seawater immersion. In contrast, after at least 180 days of seawater immersion, 6.25 % (1/16) of HPF17 seeds could germinate normally. Interestingly, we found that all the viable seeds able to germinate normally were not imbibitional before every germination test; in contrast, all the seeds that could not germinate normally were imbibitional. This finding indicates that seed coat is a key tissue for salt water tolerance of wild cotton seeds. Considering that cotton seeds can be attached to floating logs, pumice, tangled mats of vegetation, and the like, seed viability represents the limiting factor for transoceanic dispersal [8]. Our results suggested that modern cultivated cottons germinate promptly as a result of unconscious selection accompanying the breeding of annual cottons.
Table 2.
Summary of buoyancy and salt water tolerance test for G. purpurascens seeds.
| 0 d | 7 d | 14 d | 30 d | 60 d | 90 d | 120 d | 150 d | 180 d | |
|---|---|---|---|---|---|---|---|---|---|
| Remained floating seeds on seawater surface | |||||||||
| HPF17 | 30 | 30 | 30 | 30 | 30 | 29.33 ± 0.47 | 26.33 ± 1.25 | 24.33 ± 0.47 | 19.33 ± 1.25 |
| TM-1 | 30 | 30 | 30 | 30 | 29.67 ± 0.47 | 29.00 ± 0.82 | 27.33 ± 0.47 | 25.00 ± 0.82 | 21.00 ± 0.82 |
| Germinated seeds after seawater immersion | |||||||||
| HPF17 | 30/30 | 23/30 | 21/30 | 19/30 | 14/30 | 10/30 | 6/30 | 1/15 | 1/16 |
| TM-1 | 30/30 | 0/270 | – | – | – | – | – | – | – |
Notes: The results of buoyancy test show the mean number and standard deviation calculated using three replicates of seeds which remained floating on seawater surface after several days. The results of salt water tolerance test show the number of seeds which germinated normally after several days of seawater immersion, and the total number of seeds used in one germination test separated by slash. HPF17 is a G. purpurascens and TM-1 is a cultivated line of G. hirsutum. Lack of test results after >6 months.
Discussion
Origin and evolution of G. purpurascens
G. purpurascens is photoperiod sensitive and needs short-day conditions to complete its reproductive phase; consequently, it is mainly distributed in South China, including the Hainan, Guangdong, Guangxi, and Fujian provinces. Comparative genomic and phylogenetic analyses in this study, together with morphological distinctions and geographic distribution affirmed that G. purpurascens is a primitive race of G. hirsutum. The long-term domestication and improvement of cotton has brought genetic bottlenecks and a loss of genetic diversity to cotton breeding. Compared with domesticated cotton, wild relatives provide a rich resource of genetic diversity and potentially contribute to greater resistance to various abiotic and biotic stresses. Despite the importance of cotton wild relatives, there are few genome assemblies for wild forms of the most widely planted domesticated species, G. hirsutum. In this context, the genome assembly and comparative genomic analysis of a primitive G. hirsutum race, G. purpurascens, will become a valuable resource for cotton improvement and provide insight into the evolution and domestication of Gossypium.
G. hirsutum originated from one hybridization event and then was widely distributed in the Caribbean, Central America, and the Pacific. However, of the many wild cotton plants found on distant islands in the Pacific, some have been identified as new species of Gossypium, including G. darwinii (Galapagos), G. tomentosum (Hawaii), and the recently identified G. stephensii from Wake Atoll, an isolated chain of three islets located in the Western Pacific approximately 3,500 km west of Hawaii and 2,500 km east of Guam [10]. Moreover, we still do not know the identities of cotton plants found on Socorro Island, the Marquesas Islands, Tahiti, Samoa, Fiji, Solomon, Saipan Islands, and elsewhere [5], [6], [7], [8], [9], [10], [11], [12]. More investigations in the West Pacific will likely lead more wild cotton species or races to be identified.
New World cottons were introduced into the Philippines before the end of the sixteenth century largely through the efforts of the early missions [8], [73]. However, Phillips [74] has presented several pieces of evidence which make it unlikely the dispersal in fact occurred so recently. Stephens [8] suggested that Wake Island cotton (G. stephensii) may have originated following dispersal from the western coast of Mexico in his work on oceanic dispersal. The presence of wild forms of G. hirsutum on the western coast of Central America can be explained as human-mediated dispersal. The elongated fibers also aided its dispersal by birds. Meanwhile, much as with Wake Atoll, Hainan Island has a tropical climate and is subject to trade winds and occasional typhoons. The buoyancy and salt water tolerance tests in this study proved the potential for long range transoceanic dispersal of G. purpurascens, thus this species could indeed have reached its present location by floating on ocean currents.
Allotetraploid cotton was likely planted early in Yacheng for YAZHOUBU
Archaeological research has identified the use of cultivated cotton (G. barbadense) in the ancient Andes dating back to at least 7,800 years ago [75]. Upland cottons were developed from the progenitors of the semidomesticated G. hirsutum race latifolium local to the Mexican-Guatemalan border [4]. Upland varieties were established in the seaboard colonies of the southeastern United States by the mid-eighteenth century [76], then distributed worldwide and finally become the world’s largest fiber crop. Notably, Punctatum, the best known of the perennial races of G. hirsutum, became widely distributed in the Old World and was very successful in house yards and small cultivations throughout West Africa and Sudan [4]. Hainan Island, China, has an ancient history of cotton cultivation that stretches back well before the planting of G. arboreum in the mainland of China [3]. Yazhou, now called Sanya, on Hainan Island is especially well known for its cotton textiles. As early as the Neolithic period, textile tools such as spinning wheels and spinning needles were in use in Hainan [77]. As early as 3,000 years ago, the Li ancestors already knew how to plant cotton and spin for weaving [3]. The ancient book named “Book of Documents” dating to the Spring and Autumn Period ∼ 3,000 years ago records, “Foreigners on the island wear (cotton) cloth for covering, can weave baskets and cloth”. According to archaeological work in the Changsha Mawangdui, Hunan Province, samples of cotton-spun Yazhou cloth have been found (193BCE). This shows that since the Western Han Dynasty, people have taken fine cotton cloth out of Yazhou. A book named “Ling Wai Dai Da” from the Southern Song Dynasty (1178) also mentions: “Jibei……, it is rich in Lei, Hua, Lianzhou and Lidong of South China, and can be used instead of silk……Hainan’s people use it to weave many things”. Here, the Bei and Jibei refer to cotton in the Li language. Modern scholars generally believe that the ancient Chinese often confused cotton with kapok, and actually used both cotton and kapok to weave cloth, so the word “cotton” here means cotton or kapok. At the beginning of the Yuan Dynasty (1295–1297), Huang Daopo spread Hainan’s advanced cotton textile techniques, the first of which was a manual cotton ginning instrument, which she had learned in Yazhou from the Li nationality, to the Songjiang River Basin, south of the Yangtze River area; she further created a set of tools for cotton ginning, fluffing, spinning, and cloth weaving, which promoted the development of China’s cotton textile industry. As a result, Hainan’s textile technologies gradually came to be used by the whole country.
So, what cotton was planted and used for cloth weaving in Hainan at that time? G. purpurascens, is most likely to be used as a key raw material for YAZHOUBU (a Yacheng cloth historically well-known in China). Firstly, although Bombax ceiba is still widely planted in Hainan and the ancient Chinese often called it “cotton” as well, its fiber is not the suitable major material for cloth weaving because its fiber is very short and weak, so the Li nationality in Hainan usually blended it with cotton. From historical records, Huang Daopo spread a manual cotton gin learned during her stay in Yacheng, Hainan to Yangtze River Basin, and thus dramatically promote the development of the cotton textile industry and cotton planting industry in China. However, B. ceiba seeds has no fuzzes and therefore the lint fibers can be easily separated from its naked seeds, unlike the adherent lint of cotton which is difficult to gin. Therefore, there should not be such a huge gap in the efficiency of collecting cotton lint fibers by hand and by gin, and there must be another material namely cotton. Secondly, the G. purpurascens plants still naturally grow extensively in South China. Genomic analyses indicated that G. purpurascens is a primitive race of G. hirsutum, different from the other seven races in Central America, so G. purpurascens probably have existed in Hainan Island for a long time. In such a long time, it is possible for the local people to find and use the fibers of G. purpurascens. Thirdly, for other three cultivated cotton species, G. herbaceum is mainly planted in the Western Regions in China. G. arboreum had comprised most of the cotton produced in the Chinese mainland for >2,000 years, but it is not easily found growing naturally in Hainan [3]. G. barbadense was planted on a large scale in Hainan in the 20th century, but because of the adjustment of crop cultivation area, hereafter G. barbadense cultivation in Hainan only stay in the breeding and research level [3]. Therefore, although wild G. barbadense can be found on Hainan Island, no wild native species like G. purpurascens has been found at present, so it is hard to believe that G. barbadense has grown naturally for thousands of years in Hainan.
To sum up, it is reasonable to suppose that G. purpurascens was planted and used as a raw material for YAZHOUBU several thousand years ago. As such, we infer that Sanya may be one of the earliest sites of G. hirsutum domestication and cultivation. G. hirsutum may have been domesticated more than once, in more than one part of its native range, and at different times. An ancestor of G. hirsutum may have dispersed to the South China Sea Islands, adapted to the local environment, and been partly domesticated as G. purpurascens and used for YAZHOUBU by local nationalities.
SVs play an important role in cotton domestication and improvement
SVs are a major source of genetic variation. Decades of studies in human and plants have supported that SVs play an important role in many diseases and various traits such as fruit size, flowering time and stress resistance, and are important in plant evolution [20], [21], [78]. Rapidly increasing high-throughput short-read sequencing data accelerated the discovery of genetic variations among diverse germplasms of crops. However, identifying SVs with short-read sequencing data is difficult and unreliable, leading SVs to be poorly researched [79]. Therefore, SV identification with long-read data is increasingly popular, enabling higher sensitivity and accuracy. However, even with long-read data, identification of large-scale SVs or SVs located near repeat sequences is remaining difficult [80]. Recently, advances in genome assembly have generated many high-quality assemblies of plants such as rice, tomato and cotton [20], [21], allowing us to identify large-scale SVs by means of direct genome comparison.
In the present work, we detected plenty of SVs through direct whole-genome comparison using the HPF17 and other eleven published tetraploid cotton assemblies. More QTL regions and selective sweep regions were identified in SV regions than in non-SV regions, implying the relationship between SVs and quantitative traits. Compared with other types of SVs, selective sweep regions were particularly rich in INVs, followed by TRANSs. Large-scale SVs such as INVs and TRANSs are thus supposed to have greater impacts on the genome, and they may play a more important role in genomic diversity and environment adaptation compared with short variations [78]. These results will enrich the genomic resources for cotton improvement and provide insight into cotton domestication and differentiation.
We provide examples of how effects of large-scale INVs on agronomic traits can be explored with only short-read sequencing data. Eight large-scale INVs diverse in upland cottons are strongly associate with agronomic traits such as lint percentage, Micronaire value and fiber strength. The single genotype in cultivars, and the multiple or heterozygous genotypes in races and wild species of these SVs, indicate that these SVs probably have undergone artificial selection in the early stage of upland cotton domestication. These results illustrate the big effects of SVs, particularly large-scale SVs, on crop domestication and improvement. Furthermore, our findings indicate that great attention should be paid to heterozygous genotypes when sequencing and assembling wild or outcross plant species in which highly heterozygous SVs exist, as different individuals may have different genotypes on a certain SV. SV identification with direct genome alignment is also affected by the heterozygosity, so haplotype-resolved genome assembly may develop rapidly and become popular in the near future [24].
Conclusions
In the present work, we firstly completed a high-quality assembly of the HIC. Multiple analyses indicated that the HIC belongs to G. purpurascens and G. purpurascens is a primitive race of G. hirsutum. The potential for long range transoceanic dispersal of G. purpurascens seeds was proved, thus this species may have reached Hainan Island from the western coast of Mexico by floating on ocean currents. Considering together with historical materials, G. purpurascens may have been partly domesticated, planted successfully in small cultivations in Hainan much earlier than the Pre-Columbian period, and was likely used for YAZHOUBU weaving. Thus, modern upland cotton may stem from diverse origins and different domestication events. SVs, especially large-scale SVs, were found to have important effects on cotton domestication and improvement. Eight large-scale inversions are significantly associated with “domestication syndrome” agronomic traits and have probably undergone artificial selection in upland cotton domestication.
CREDIT author statement
T. Zhang conceived and designed the research. P.W. collected G. purpurascens seeds in Yacheng, Hainan Island. Y.H., L.F., T. Zhao, X.G. and T. Zhang coordinated the project. Y.C., C.H., Z.S., J.C., S.J., F.D. and G.M. managed the field work and collected the phenotypic data. Y.C. prepared DNA and RNA for sequencing and performed the experiments. X.Z., Y.C. and W.Y. performed the genome assembly. Y.C., C.H., S.J. and X.Z. performed the bioinformatics work. Y.C. and T. Zhang analyzed the data and prepared the figures and tables. Y.C. wrote the manuscript. T. Zhang revised the manuscript. All authors read and approved the manuscript.
Compliance with ethics requirement
There was no use of any animals or human patients.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This study was financially supported in part by the 2021 Research Program of Sanya Yazhou Bay Science and Technology City (SKJC-2021-02-001), China, the Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang, China (2019R01002), the Fundamental Research Funds for the Central Universities (226-2022-00100) in China. We thank X. Zhang (Huazhong Agricultural University), Z. Ma (Hebei Agricultural University), Y. Zhu (Wuhan University), F. Li and X. Du (Institute of Cotton Research of the Chinese Academy of Agricultural Sciences) and Z. Chen (The University of Texas at Austin) for sharing sequencing data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2023.03.006.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Supplementary figure 1.
Supplementary figure 2.
Supplementary figure 3.
Supplementary figure 4.
Supplementary figure 5.
Supplementary figure 6.
Supplementary figure 7.
Supplementary figure 8.
Supplementary figure 9.
Supplementary figure 10.
Supplementary figure 11.
Supplementary figure 12.
Supplementary figure 13.
Data availability
The HPF17 assembly and annotation files are available at the website https://cotton.zju.edu.cn/. The raw sequencing data used for de novo genome assembly are available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA884852. The re-sequencing data for the F2 population are deposited in NCBI under accession number PRJNA883829. The Illumina RNA-seq data for HPF17 are available in NCBI under accession number PRJNA884803. Further details on data accessibility are outlined in the methods and supplementary materials.
References
- 1.Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum l. Acc. Tm-1) provides a resource for fiber improvement. Nat Biotechnol. 2015;33(5):531–537. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 2.Wendel J.F. New world tetraploid cottons contain old world cytoplasm. Proc Natl Acad Sci U S A. 1989;86(11):4132–4136. doi: 10.1073/pnas.86.11.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Z. China Agriculture Press; Beijing: 2007. Cotton varieties and their genealogy in China; pp. 13–16. [Google Scholar]
- 4.Hutchinson J.B. Intra-specific differentiation in Gossypium hirsutum. Heredity. 1951;5(2):161–193. [Google Scholar]
- 5.Fosberg F.R. Vegetation and flora of wake island. Atoll Res Bull. 1959;67:1–20. [Google Scholar]
- 6.Fryxell P.A., Moran R. Neglected form of Gossypium hirsutum on socorro island, mexico. Emp Cotton Grow Rev. 1963;40:289–291. [Google Scholar]
- 7.Stephens S.G. Polynesian cottons. Ann Mo Bot Gard. 1963;50(1–4):1–22. [Google Scholar]
- 8.Stephens S.G. The potentiality for long range oceanic dispersal of cotton seeds. Am Nat. 1966;100(912):199–210. [Google Scholar]
- 9.d'Eeckenbrugge GC, Lacape J-M. Distribution and differentiation of wild, feral, and cultivated populations of perennial upland cotton (Gossypium hirsutum l.) in mesoamerica and the caribbean. PLoS ONE 2014;9(9):e107458. [DOI] [PMC free article] [PubMed]
- 10.Gallagher J.P., Grover C.E., Rex K., Moran M., Wendel J.F. A new species of cotton from wake atoll, Gossypium stephensii (malvaceae) Syst Bot. 2017;42(1):115–123. [Google Scholar]
- 11.DeJoode D.R., Wendel J.F. Genetic diversity and origin of the Hawaiian Islands cotton, Gossypium tomentosum. Am J Bot. 1992;79(11):1311–1319. [Google Scholar]
- 12.Stephens S.G. Univ. California Press; Oakland: 1966. Problems on the origin, dispersal, and establishment of the galápagos cottons. [Google Scholar]
- 13.Watt G. Longmans, Green, and Co; London: 1907. The wild and cultivated cotton plants of the world: A revision of the genus gossypium, framed primarily with the object of aiding planters and investigators who may contemplate the systematic improvement of the cotton staple. [Google Scholar]
- 14.Watt G. Gossypium. Bull Misc Inf, R Bot Gard. 1927;1927(8):321–356. [Google Scholar]
- 15.Harland S.C. The genetics of cotton. J Genet. 1937;34(1):153–168. [Google Scholar]
- 16.Harland S.C. New polyploids in cotton by the use of colchicine. Trop Agric (Trinidad) 1940;17:53–54. [Google Scholar]
- 17.Hutchinson J.B., Stephens S.G. Note on the “french” or “ small-seeded” cotton grown in the west indies in the 18th century. Trop Agric. 1944;21(7):123–125. [Google Scholar]
- 18.Nazir M.F., He S., Ahmed H., Sarfraz Z., Jia Y., Li H., et al. Genomic insight into the divergence and adaptive potential of a forgotten landrace G. hirsutum l. Purpurascens. J Genet Genomics. 2021;48(6):473–484. doi: 10.1016/j.jgg.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Yu Q. A brief description of the investigation and research on cotton seed in china. Acta Agric. 1941;6(10–12):715. [Google Scholar]
- 20.Alonge M., Wang X., Benoit M., Soyk S., Pereira L., Zhang L., et al. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell. 2020;182(1):145–161.e23. doi: 10.1016/j.cell.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qin P., Lu H., Du H., Wang H., Chen W., Chen Z., et al. Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations. Cell. 2021;184(13):3542–3558. doi: 10.1016/j.cell.2021.04.046. [DOI] [PubMed] [Google Scholar]
- 22.Stein J.C., Yu Y., Copetti D., Zwickl D.J., Zhang L., Zhang C., et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus oryza. Nat Genet. 2018;50(2):285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y., Chen J., Fang L., Zhang Z., Ma W., Niu Y., et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019;51(4):739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- 24.Cheng H., Concepcion G.T., Feng X., Zhang H., Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18(2):170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B., Salzberg S.L. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burton J.N., Adey A., Patwardhan R.P., Qiu R., Kitzman J.O., Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31(12):1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marçais G., Delcher A.L., Phillippy A.M., Coston R., Salzberg S.L., Zimin A. Mummer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018;14(1):e1005944. doi: 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H., Durbin R. Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. Busco: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 30.Kent W.J. Blat—the blast-like alignment tool. Genome Res. 2002;12(4):656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ou S., Chen J., Jiang N. Assessing genome assembly quality using the ltr assembly index (lai) Nucleic Acids Res. 2018;46(21):e126. doi: 10.1093/nar/gky730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou S., Jiang N. Ltr_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018;176(2):1410–1422. doi: 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ellinghaus D., Kurtz S., Willhoeft U. Ltrharvest, an efficient and flexible software for de novo detection of ltr retrotransposons. BMC Bioinf. 2008;9(1):1–14. doi: 10.1186/1471-2105-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z., Wang H. Ltr_finder: an efficient tool for the prediction of full-length ltr retrotransposons. Nucleic Acids Res. 2007;35(suppl_2):W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGinnis S., Madden T.L. Blast: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(suppl_2):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keilwagen J., Hartung F., Paulini M., Twardziok S.O., Grau J. Combining rna-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinf. 2018;19(1):1–12. doi: 10.1186/s12859-018-2203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanke M., Steinkamp R., Waack S., Morgenstern B. Augustus: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32(suppl_2):W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ter-Hovhannisyan V., Lomsadze A., Chernoff Y.O., Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18(12):1979–1990. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., et al. Full-length transcriptome assembly from rna-seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu T.D., Watanabe C.K. Gmap: a genomic mapping and alignment program for mrna and est sequences. Bioinformatics. 2005;21(9):1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 41.Haas B.J., Delcher A.L., Mount S.M., Wortman J.R., Smith R.K., Jr, Hannick L.I., et al. Improving the arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31(19):5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., et al. Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):1–22. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn R.D., Clements J., Eddy S.R. Hmmer web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011;39(suppl_2):W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng Y., Jiao C., Sun H., Rosli H.G., Pombo M.A., Zhang P., et al. Itak: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016;9(12):1667–1670. doi: 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 45.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., et al. Twelve years of samtools and bcftools. GigaScience. 2021;10(2):giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang H., Wang K. Genomic variant annotation and prioritization with annovar and wannovar. Nat Protoc. 2015;10(10):1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goel M., Sun H., Jiao W.-B., Schneeberger K. Syri: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019;20(1):1–13. doi: 10.1186/s13059-019-1911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and samtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gonda I., Ashrafi H., Lyon D.A., Strickler S.R., Hulse-Kemp A.M., Ma Q., et al. Sequencing-based bin map construction of a tomato mapping population, facilitating high-resolution quantitative trait loci detection. Plant Genome. 2019;12(1) doi: 10.3835/plantgenome2018.02.0010. [DOI] [PubMed] [Google Scholar]
- 51.Rastas P. Lep-map3: robust linkage mapping even for low-coverage whole genome sequencing data. Bioinformatics. 2017;33(23):3726–3732. doi: 10.1093/bioinformatics/btx494. [DOI] [PubMed] [Google Scholar]
- 52.Si Z., Jin S., Li J., Han Z., Li Y., Wu X., et al. The design, validation, and utility of the “zju cottonsnp40k” liquid chip through genotyping by target sequencing. Ind Crops Prod. 2022;188(Part A) [Google Scholar]
- 53.Meng L., Li H., Zhang L., Wang J. Qtl icimapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3(3):269–283. [Google Scholar]
- 54.Li L., Stoeckert C.J., Roos D.S. Orthomcl: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar R.C. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of phyml 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 57.Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 58.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega x: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation plink: rising to the challenge of larger and richer datasets. GigaScience. 2015;4(1):s13742–13015. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nei M., Li W.-H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci U S A. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., et al. The variant call format and vcftools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weir B.S., Cockerham C.C. Estimating f-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen H., Patterson N., Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20(3):393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu B, Shi Y, Yuan J, Hu X, Zhang H, Li N, et al. Estimation of genomic characteristics by analyzing k-mer frequency in de novo genome projects. arXiv: Genomics 2013.
- 65.Solomon M.J., Larsen P.L., Varshavsky A. Mapping proteindna interactions in vivo with formaldehyde: evidence that histone h4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–947. doi: 10.1016/s0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 66.Wilkins A.S. A striking example of developmental bias in an evolutionary process: the “domestication syndrome”. Evol Dev. 2020;22(1–2):143–153. doi: 10.1111/ede.12319. [DOI] [PubMed] [Google Scholar]
- 67.He S., Sun G., Geng X., Gong W., Dai P., Jia Y., et al. The genomic basis of geographic differentiation and fiber improvement in cultivated cotton. Nat Genet. 2021;53(6):916–924. doi: 10.1038/s41588-021-00844-9. [DOI] [PubMed] [Google Scholar]
- 68.Chen Z.J., Sreedasyam A., Ando A., Song Q., De Santiago L.M., Hulse-Kemp A.M., et al. Genomic diversifications of five gossypium allopolyploid species and their impact on cotton improvement. Nat Genet. 2020;52(5):525–533. doi: 10.1038/s41588-020-0614-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang G., Wu Z., Percy R.G., Bai M., Li Y., Frelichowski J.E., et al. Genome sequence of Gossypium herbaceum and genome updates of Gossypium arboreum and Gossypium hirsutum provide insights into cotton a-genome evolution. Nat Genet. 2020;52(5):516–524. doi: 10.1038/s41588-020-0607-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan D., Grover C.E., Hu G., Pan M., Miller E.R., Conover J.L., et al. Parallel and intertwining threads of domestication in allopolyploid cotton. Adv Sci (Weinh) 2021;8(10):2003634. doi: 10.1002/advs.202003634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang L., Gong H., Hu Y., Liu C., Zhou B., Huang T., et al. Genomic insights into divergence and dual domestication of cultivated allotetraploid cottons. Genome Biol. 2017;18(1):1–13. doi: 10.1186/s13059-017-1167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stephens S.G. Salt water tolerance of seeds of Gossypium species as a possible factor in seed dispersal. Am Nat. 1958;92(863):83–92. [Google Scholar]
- 73.Bradley A. Trans-pacific relations of Latin America. Pac Aff. 1942;14(1):97–106. [Google Scholar]
- 74.Phillips L.L. The cytogenetics of Gossypium and the origin of new world cottons. Evolution. 1963;17(4):460–469. [Google Scholar]
- 75.Bird JB, Hyslop J, Skinner MD. The preceramic excavations at the Huaca Prieta, Chicama Valley, Peru. Anthropological papers of the amnh. New York: American Museum of Natural History; 1985.
- 76.Wendel J.F., Brubaker C.L., Seelanan T. Physiology of cotton. Springer; Dordrecht: 2010. The origin and evolution of Gossypium; pp. 1–18. [Google Scholar]
- 77.Rong G. Neolithic tools found in li nationality area of Hainan Island. Archaeology 1956;(2):13+38-41.
- 78.Weischenfeldt J., Symmons O., Spitz F., Korbel J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat Rev Genet. 2013;14(2):125–138. doi: 10.1038/nrg3373. [DOI] [PubMed] [Google Scholar]
- 79.Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., Von Haeseler A., et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ho S.S., Urban A.E., Mills R.E. Structural variation in the sequencing era. Nat Rev Genet. 2020;21(3):171–189. doi: 10.1038/s41576-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HPF17 assembly and annotation files are available at the website https://cotton.zju.edu.cn/. The raw sequencing data used for de novo genome assembly are available in the NCBI Sequence Read Archive (SRA) under accession number PRJNA884852. The re-sequencing data for the F2 population are deposited in NCBI under accession number PRJNA883829. The Illumina RNA-seq data for HPF17 are available in NCBI under accession number PRJNA884803. Further details on data accessibility are outlined in the methods and supplementary materials.